- Department of Child Health, School of Medicine, Cardiff University, Cardiff, UK
The respiratory tract, once believed to be sterile, harbors diverse bacterial communities. The role of microorganisms within health and disease is slowly being unraveled. Evidence points to the neonatal period as a critical time for establishing stable bacterial communities and influencing immune responses important for long-term respiratory health. This review summarizes the evidence of early airway and lung bacterial colonization and the role the microbiome has on respiratory health in the short and long term. The challenges of neonatal respiratory microbiome studies and future research directions are also discussed.
Introduction
Respiratory insufficiency is the major issue limiting viability at the extremes of prematurity. Chronic lung disease of prematurity (CLD), also known as bronchopulmonary dysplasia, is the most common long-term complication of prematurity (1) with reduced lung function persisting into adulthood (2). Many risk factors have been identified in the development of CLD, including prematurity, supplemental oxygen therapy, mechanical ventilation, and patent ductus arteriosus (3). Systemic and pulmonary infections may also contribute to the development of CLD (4, 5). Despite the routine use of antenatal corticosteroids and exogenous surfactant, improved nutrition and ventilatory strategies, CLD remains a significant burden for surviving preterm infants, and an on-going challenge for neonatologists (6).
The microbiome is defined as the whole habitat of microorganisms (or of their genes) and the surrounding environment, with the term microbiota more specifically referring to the community of microorganisms living within a particular environment (7). The diversity of bacterial communities has been linked to important health outcomes (8). Diversity is a measure of how much variety is present in a community of microorganisms, regardless of the identities of the organisms. Diversity is made up of richness, the number of different bacterial species present, and evenness, defined as the relative abundance of the various species within the bacterial community (9). The composition of bacterial communities changes over time. A stable microbiota is resistant to such changes. The concept of dysbiosis refers to a pattern of bacterial colonization predisposing to disease (10).
The human host has evolved in symbiosis with microorganisms, which colonize multiple body sites. The interactions between host and the microbiota are only now being understood with microbiome science demonstrating that bacterial communities living within a human host have an influence over a diverse range of diseases (11). Within neonatal medicine, most of this work has focused on the role of the gut microbiome in the pathogenesis of necrotizing enterocolitis (NEC) (12–14), with widespread use of probiotics, a clinical result of this work (15). Attention is now moving beyond the gut to other body sites to identify the role of microbes in other anatomical locations including the respiratory tract.
The traditional notion of the lungs of healthy individuals being free from bacterial colonization is now defunct (16, 17). The lungs were initially not included in the human microbiome project (18) due to perceived sterility. The sterile lung hypothesis was driven by culture-based studies, designed to identify specific pathogens, failing to detect the low levels of difficult to culture commensal organisms within the lower airways and lungs. Culture-independent techniques utilizing sequencing technology to identify bacterial DNA have led to the discovery of communities of microorganisms within the airways and lungs of healthy individuals (19, 20). The alterations of microbial communities within the airways and lungs in pulmonary disease, and therefore the potential for therapeutic intervention, are only beginning to be grasped. This review summarizes the current evidence surrounding the early colonization of the airways and lungs of infants, and the impact this may have on short- and long-term health consequences.
Methodological Challenges
In addition to the ethical and practical challenges that complicate neonatal research, respiratory microbiome work has to overcome challenges of difficult sample collection, contamination, and the low bacterial load in the lungs.
Bacteria are able to colonize the full length of the airways, from the nostril to the alveoli. The changes in anatomy and environmental exposure along this length lead to the creation of many varied niches for bacterial colonization. Sampling of the proximal airways at any age is straight forward using nasal swabs or nasopharyngeal aspirates. The inaccessibility of the lungs poses significant challenges for researchers. Sampling from the lungs and distal airways in adults has involved using sputum or endoscopic bronchoalveolar lavage (19, 21, 22). In the neonatal population, the lower airways of intubated infants can be accessed using tracheal aspiration to sample the endotracheal tube and trachea (23, 24) or non-bronchoscopic bronchoalveolar lavage (NB-BAL) to sample more distal airways (25). All these techniques are easily contaminated by upper respiratory tract or nasopharyngeal organisms. Avoiding such contamination using diseased explanted lungs and lungs from organ donors for whom no recipient could be found have demonstrated bacterial communities within lung tissue of adults (26, 27). The presence of bacterial DNA within the lungs of even healthy adults is no longer disputed (19, 20).
The particular difficulties of sampling the lung microbiome in neonates results in published studies concentrating on the upper airway or on ventilated infants (23, 28, 29). While the upper airway bacterial communities are related to those in the distal airways (30), it seems that the nasopharynx cannot be used as proxy for lung colonization (26, 30). Within the neonatal population, no studies have thus far reported on the lung microbiome of healthy term-born infants. In neonatal research, the vaginally delivered term-born, exclusively breast fed infant is considered most likely to develop the optimal microbiome (31). Other groups are generally compared to this assumed optimal microbial pattern. The lack of data from the airways and lungs of such infants complicates the interpretation of many neonatal studies. Studies of the gut microbiome reveal that the composition of gut flora is unique to an individual and shows marked variety between individuals (32). A similar variability between individuals has also been observed in the upper airways of children (33, 34). The concept of a “normal” microbiome or “normal” colonization pattern may not exist and may be difficult to define. Patterns of colonization or presence of predominant organisms are, however, distinguishable within populations and maybe associated with favorable or unfavorable outcomes.
Culture-independent techniques utilize sequencing technologies to identify organisms by sequencing target genes. The most commonly used gene for bacterial species identification is the 16S ribosomal RNA gene. The difference in nucleotide sequence within the nine hypervariable regions of the gene are used to identify the bacterial species present within a sample using next generation sequencing (35). All bacterial DNA is identified, not only from resident bacteria but also from dead microbes unable to survive in the sampled niche; and DNA from contaminant organisms acquired at any stage during sample collection or analysis (26). DNA-based techniques may therefore overestimate the community of bacteria residing in a habitat. A transcriptomic approach, using RNA sequences rather than DNA to identify bacteria (36), overcomes the problem of non-viable bacteria, but not contamination. A further alternative to 16S sequencing is a metagenomic approach when all nucleotide sequences are identified rather than an individual gene (37). This provides a more rigorous bacterial identification.
Many respiratory samples, particularly those from neonates, generate a low biomass (38) with low bacterial loads close to the limit of detection of the assays designed to detect such organisms. Careful use of negative controls at each stage of the analysis process is important to ensure validity of the results (39). Contamination during sample collection can be assessed by comparing samples from the lower respiratory tract with samples taken concurrently from the upper respiratory tract. Identical patterns of colonization would imply contamination (38). DNA extraction kits and PCR reagents are also known to be contaminated with bacterial DNA (40). Studies using low biomass samples need to demonstrate thorough record keeping of the lot numbers of reagents used and analyze for contamination from laboratory sources.
Despite the challenges and limitations of studying the airway and lung microbiome, a significant body of work is available relevant to adults, with several comprehensive reviews published (16, 41). The extra difficulties of research in this area in the neonatal population result in fewer published studies, with varying methodology. The evidence relevant to the neonatal population is the focus of this review.
Acquisition of Microbiome
Timing of Colonization
The in utero environment is traditionally considered to be physiologically sterile. This assumption has been challenged with the finding of bacteria in the placenta (42), fetal membranes (43), and amniotic fluid (44) of healthy pregnancies. It remains unclear if there is a low level of bacterial colonization of the in utero fetus, but bacterial presence has been detected in the first passage of meconium from healthy term-born infants (45), previously thought to be sterile, and in cord blood samples (46). Airway colonization may therefore begin in utero in some cases, and this is more likely in those exposed to chorioamnionitis, known to be a significant risk factor for preterm delivery. Bacterial DNA was detected from a larger proportion of preterm lungs and/or gastric fluid within 24 h of birth if delivered due to prelabor premature rupture of membranes or spontaneous preterm labor compared to those delivered by cesarean section for maternal or fetal reasons (47).
Within the first 5 min following birth, microbiological communities can be detected within the oral cavity and nasopharynx of term newborns (48), suggesting that colonization of the upper airways has already commenced. Initial studies searching for the presence of bacterial DNA in the lower respiratory tract of intubated preterm babies showed the presence of a diverse array of bacteria within the first week of life (24). In intubated preterm infants, one study noted that only 2 of 10 tracheal aspirate samples taken at <72 h of age contained detectable bacterial DNA. At 7 days of age, all 10 tracheal aspirates from the same babies contained detectable bacterial DNA (49). In contrast, Lohmann et al. detected bacterial DNA in all tracheal aspirate samples taken immediately after intubation on day 1 of life from 25 preterm neonates ≤32 weeks gestation (23). It appears that the colonization of the airways begins very early in life, at or possibly even before delivery.
Ureaplasma species, implicated in preterm neonatal respiratory infection, have been detected by molecular methods in tracheal aspirates samples at 24 h of age (24). Using RNA-based methodology, another study demonstrated that these organisms are transcriptionally active within the lungs for at least 3 weeks after delivery in some preterm neonates (50).
Studies vary in estimating the time for a stable respiratory microbiota to be established. One study suggests that bacterial density in the nasopharynx of healthy infants increases throughout the first year of life. Reducing diversity over time resulted in stable bacterial communities being established by 1 year of age (51). However, another study did not report a change in bacterial load in the nasopharynx after 1 month of age, but a continual evolution of organisms throughout the first 2 years of life (28).
The timing of bacterial colonization of the neonate remains controversial but a sterile in utero environment can no longer be assumed. Stable airway communities are not established during the neonatal period.
Airway Colonizing Organisms
The adult lung microbiome has been more widely studied using molecular-based techniques. In healthy adult lungs, the phyla Bacteroidetes and Firmicutes predominate (around 80%) with Proteobacteria making up around 10% of the lung microbiome (19, 20, 52). At a genus level, Streptococcus and Veillonella species are the predominant Firmicutes organisms, and Prevotella species make up the majority of Bacteroidetes. Within Proteobacteria; Pseudomonas, Haemophilus, and Neisseria species are most common (41). The presence of known lung pathogens within this list demonstrates that tight immunological control over the microbiome occurs, and challenges the traditional view that respiratory infections are environmentally acquired.
Only limited information is available regarding the initial colonizers of the airways in the early days of life. One study of 10 intubated preterm infants using culture-free methodology noted a dominant organism was present (>50% of total sequences) in 31 of 32 tracheal aspirate samples taken in the first month of life. The most common dominant genus was Staphylococcus, in 19 samples, 17 of these being coagulase negative staphylococci. Ureaplasma species were dominant in nine samples from six subjects. Other species that predominated in a single sample, all between 14 and 21 days of life were Pseudomonas aeruginosa, Enterococcus faecalis, and Escherichia coli (49). A similar finding was reported in a separate study using tracheal aspirates collected during the first week of life. The two most common organisms identified using the 16S RNA gene were Staphylococcus haemolyticus and Staphylococcus epidermidis, both coagulase negative staphylococci (24). A subsequent study also demonstrated the presence of a dominant organism in many tracheal aspirate samples but noted that species within the Proteobacteria phylum, mainly Acinetobacter species, were dominant most often in the first day of life (23). Taken together, this evidence suggests that in the first days of life a pioneering colonizer becomes established but the identity of the organism depends on initial exposure of the infant.
Factors Affecting Airway Colonization
The constituents of the infant gut microbiota have been shown to have been strongly influenced by delivery mode, feeding choices, and the perinatal use of antibiotics (53). The use of drugs such as H2 blockers affecting gastric acid secretion also affect early colonization patterns within the gut (54) and may indirectly affect the lungs. Such factors can have sustained effects on the gut microbiota beyond the neonatal period, with differences between vaginally and cesarean-born infants detectable until 12 months of age (55).
Evidence from the airways is less substantial, but the nasopharynx of term-born infants share the same initial colonizing organisms as various skin sites and the mouth. All sites demonstrated a large influence of the delivery method (48). Infants born by normal vaginal delivery had bacterial communities resembling maternal vaginal flora, while infants born by cesarean section were colonized by maternal skin organisms.
Mechanically ventilated preterm infants exposed to chorioamnionitis appear to have decreased species diversity in their tracheal aspirates although the trend did not reach statistical significance compared to unexposed infants (23). This may reflect overgrowth of pathogenic species in infants exposed to chorioamnionitis. Bacterial load estimation was not made in this study. It would have been interesting to compare the number of organisms present between chorioamnionitis exposed and unexposed infants.
Antibiotic use is widespread in neonatal patients, particularly in those born preterm. Exposure to antibiotics has been shown to reduce diversity and modify colonization patterns of the neonatal gut (56). Antibiotics also induce a significant change in the microbiota of sputum of cystic fibrosis patients (57). The impact on the lungs of neonates is unknown; however, in the upper airways of infants under 12 months of age, those who had received antibiotics in the preceding 4 weeks displayed a reduced proportion of Alloiococcus and Corynebacterium, with an increased proportion of potential pathogens, including Haemophilus, Streptococcus, and Moraxella (58).
Over 200 species of bacteria have been isolated from human breast milk including the beneficial Bifidobacterium and Lactobacillus species (59). Breast milk has the ability not only to pass on desirable colonizing flora from mother to baby but also to provide oligosaccharides to promote development of healthy microbiota (60).
Evidence is divided on the factors affecting the bacterial colonization of the nasopharynx of infants under 12 months of age. One study reported that at 2 months of age, mode of delivery and feeding methods did not affect the composition of the nasopharyngeal microbiome; however, the presence of siblings, recent respiratory tract infections, and attending day care all made a significant difference (58). In contrast, a well-designed study comparing exclusively breast fed and exclusively formula fed infants showed detectable changes in the nasopharyngeal microbiota at 6 weeks of age (61). A similar study showed a significant seasonal effect with a bloom of Corynebacteriaceae in the summer months, but Pasteurellaceae predominated in the winter months (51). These differences may also be explained by geographical differences, with the studies conducted in the Netherlands, Switzerland, and Australia.
Evidence from patients with cystic fibrosis followed up to 21 months of age, suggests that species colonizing the upper-airway (oropharyngeal samples) are first present in the stool (62). This suggests that the airway microbiota may be established from that of the gut. In healthy adult subjects who underwent microbiome analysis from the oral cavity, nasal cavity, lungs and stomach, the lung microbiome closely resembled the microbiome of the oral cavity more than other sites (30). Micro-aspiration of saliva into the airway is known to occur during sleep in healthy individuals and may result in aspiration of physiologically significant numbers of bacteria (63).
Accurate determination of the source of the microbiota would require deep sequencing of the genome of organisms from multiple sites including maternal skin, breast milk, birth canal, and from the skin of midwifery or neonatal staff caring for the infant, as well as range of environmental sites – a major undertaking.
Evidence suggests that the airway microbiome is affected by multiple environmental factors. Interpreting the studies is difficult given the different sampling techniques and anatomical sites tested; however, evidence is consistent with studies of bowel colonization showing an effect of delivery mode, antibiotics, and feeding methods.
Chronic Lung Disease of Prematurity
Dysbiosis is defined as an imbalance in the microbes present in a particular niche that contributes or predisposes to disease (10, 16). Adult-based studies have suggested that airway dysbiosis can predict poor outcomes in cystic fibrosis (64) and in asthma (65). It is still unclear if dysbiosis reflects a change in environmental conditions within the airways in those with more severe disease or if dysbiosis is driving the poor prognosis.
Preventing CLD is a significant challenge in neonatology. Pulmonary infection has been suspected as an important contributing factor to CLD (4, 5) with organisms such as Ureaplasma species repeatedly implicated (66). The role of the commensal bacteria is less well studied. Early work using culture independent techniques comparing infants with and without CLD showed that detectable bacterial DNA (16S gene) in NB-BAL fluid was associated with developing CLD. The risk was even higher if the bacteria were present within the first 3 days of life (4). Organisms associated with CLD in this study were E. coli, Haemophilus influenzae, Enterobacter species, and P. aeruginosa. Further work showed the presence bacterial DNA in NB-BAL fluid was associated with higher proteinase activity within the lung, and the chance of developing CLD. Predominant organisms identified with high proteinase activity in the CLD group were Staphylococcus aureus and E. coli (67). The proteinases analyzed in this study were neutrophil derived. A greater immune response due to the presence of bacteria could explain the increased proteinase activity and the mechanism of the bacteria influencing the pathogenesis of CLD. Bacterial proteinases have been implicated in the pathogenesis of gut pathologies (68). The much lower bacterial load in the lungs compared to the gut make the role of colonizing microorganism derived proteinases less likely to be significant in pulmonary diseases, but this remains uncertain.
Another study investigated the difference between airway microbiota in tracheal aspirates from preterm infants with and without CLD using culture-independent techniques (23). Day one colonization patterns were similar in both groups with the predominance of Acinetobacter. Those infants who developed CLD had reduced species diversity and higher proportions of the pathogens Staphylococcus and Klebsiella. Those infants who did not develop CLD showed more stable bacterial communities with greater diversity. This suggests a role for the microbiome in the development of CLD but correlation between pro-inflammatory cytokines and microbial colonization patterns was not witnessed. The microbiome is likely to influence the development of CLD by stimulating pro- or anti-inflammatory responses. The study, however, was not able to provide such a link. Without a plausible mechanism of effect, differences in the microbiome between those with and without CLD may reflect different environmental conditions within the airways of those predisposed to more severe disease due to an independent factor, rather than a causal relationship. Further work is clearly needed to corroborate this study, but the concept of diverse bacterial communities being advantageous follows the pattern of evidence with reports in other diseases including respiratory and gut pathologies (8, 69).
The influence of the microbiome over CLD has been the target of studies due to the chronic nature of CLD and the known impact of bacterial infection. No studies have looked at the impact of the microbiome on other respiratory conditions affecting the neonate.
Microbiological Programing
The early programing theory suggests that early life exposure to microbes is important for long-term health. Epidemiological data suggest a period in infancy that can determine future respiratory health (70, 71). Early colonization patterns are thought to affect immune development and potentially prime the host for later disease. This process was demonstrated in the gut, where early microbial colonization can significantly impact the long-term risk of asthma and allergy (72, 73). The respiratory microbiota also has a role in programing. Infants who were shown to be colonized (culture positive from hypopharyngeal aspirate) at 4 weeks of age with one of Streptococcus pneumoniae, H. influenzae, and/or Moraxella catarrhalis (but not S. aureus) were at significantly higher risk of pneumonia and bronchiolitis by the age of 3 years (74). The same cohort was followed up again at 5 years of age and the same organisms were associated with an increased risk of asthma (75). Interestingly, microbial colonization status at 12 months of age was not correlated with outcomes. The early colonization was important suggesting not only that a critical phase in immune programing exists but also that there may be a window of opportunity early in life to cultivate a healthy or protective microbiome. Cytokine data from these patients showed that the T helper cell type induced by colonization was significantly different. M. catarrhalis and H. influenzae induced a mixed T helper cell response. S. pneumoniae colonization was not associated with a significant change in mucosal cytokines (76). These results demonstrate a possible mechanism of action for the influence of the bacteria over the immune system. By inducing a mixed T helper cell response, the Th1 responses needed for intracellular bacteria destruction may be impaired, potentially leading to chronic inflammation (76). These pathways provide potential therapeutic targets for intervention to influence this process pharmacologically.
An Australian study using culture independent techniques to identify bacterial and viral DNA within infants’ nasopharynx also showed that early (around 2 months of age) asymptomatic carriage of streptococcus species significantly increases the risk of chronic wheeze at 5 years of age (58). In a similar culture-independent study, sampling the nasopharyngeal microbiota repeatedly over the first 2 years of life, the early (6 weeks of age) microbiota composition determined the future stability of the microbiota. Stable profiles were characterized by the presence of Moraxella and Corynebacterium or Dolosigranulum and were associated with breast feeding. Less stable profiles were marked by high abundance of Haemophilus or Streptococcus. Parent-reported respiratory infections were reduced in the stable microbiota group (28).
The combined evidence suggests early colonization of the gut and the respiratory tract may have a role in determining future respiratory health. The factors previously discussed affecting the early colonization patterns may therefore have a lasting impact beyond the neonatal period. Further work to understand the mechanisms of microbiological programing may assist in developing novel treatments or reveal possible ways to influence the microbiota to promote long term health.
Gut–Lung Axis
The GI tract microbiome has the ability to influence diseases anatomically distant from the gut via immunological modulation of mucosal immune responses and the plethora of small metabolite molecules produced by bacteria (77). Figure 1 demonstrates a conceptual model of the mechanisms underlying the gut–lung axis (78). As previously mentioned, the additional influence of the gut over airway and lung bacterial colonization may also be extended to direct transfer of bacteria through micro-aspiration (63). H2 blockers or other drugs affecting the gastric acid secretion may affect lung colonization by affecting the composition of bacteria transferred by micro-aspiration (54). H2 blockers have been shown to be associated with the risk of late onset sepsis and NEC (79, 80) and in one study pneumonia (81) in preterm newborn infants. Thus, use of drugs, which may alter the microbiome, may have wider ranging consequences.
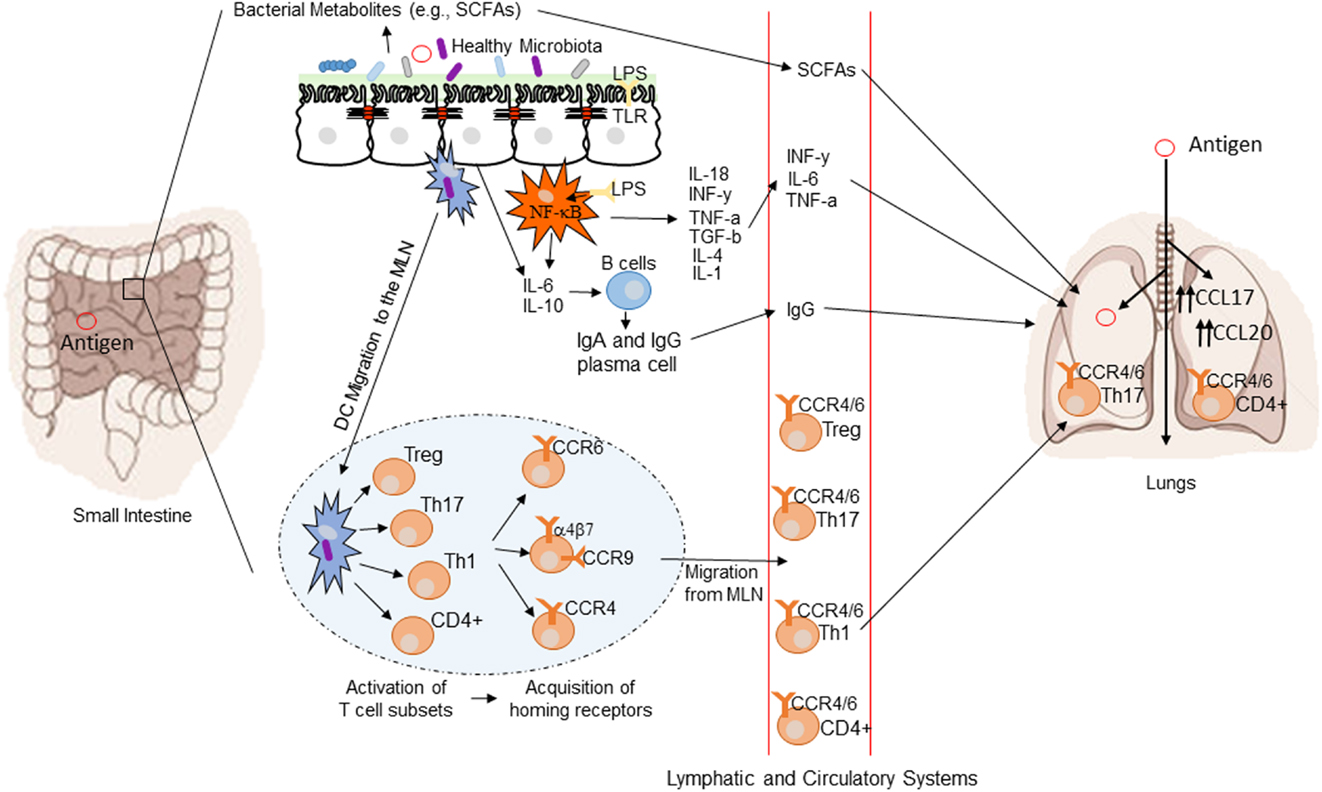
Figure 1. Conceptual figure of the gut–lung axis. Proposed model for the regulatory influence of the gastrointestinal microbiota on the immunology of the lung. Microbes in the intestine are sampled by dendritic cells (DCs) either directly from the lumen or following translocation through M cells to the gut-associated lymphoid tissue (GALT). A combination of signals from the microbes results in phenotypic changes in the DCs and migration to the draining lymph node. DCs promote the activation of various T cell subsets within the mesenteric lymph nodes (MLN) and the production of various regulatory cytokinesm, such as IL-10, TGF-b, INFg, and IL-6. T cell subsets then acquire immune homing molecules (i.e., CCR6, CCR4, and CCR9). Following immune challenge in the airway, cells activated in the GALT and MLN traffic to the respiratory mucosa via CCR4 or CCR6 where they promote protective and anti-inflammatory responses. Production of various bacterial metabolites (e.g., SCFAs) also affects the gut–lung axis, as these products are transported to the lung, where they can alter the levels of inflammation. Modified from Samuelson et al. (78).
The gut microbiome may have a greater effect on respiratory health than previously appreciated (82). Clinical evidence corroborates the presence of the gut–lung axis. Oral probiotics have been shown to be beneficial in preventing ventilator-assisted pneumonia in adults (83) and upper respiratory tract infections (84) but their benefits for respiratory health in neonates remain to be seen.
Bacterial metabolites such as short chain fatty acids (SCFA) may also be a mechanism for gut microbes influencing respiratory health (85). SCFA act directly on both epithelial cells and immune cells significantly affecting the immune response (86). Changes to the fiber content in the diet of mice affect the bacterial composition within the gut and levels of serum SCFA. Mice fed a high fiber diet had more Bacteroides in the gut and raised serum SCFA compared to mice fed a low fiber diet who had more Firmicutes and decreased SCFA. Raised SCFA was associated with reduced allergic airway inflammation (87).
The common mucosal response theory suggests that antigens presented from one mucosal surface can affect lymphoid cell migration to other mucosal sites, influencing immune responses at the distal site (88). The immunological mechanisms explaining this effect are summarized in Figure 1 (78). Bacterial derived antigens presented by dendritic cells (DCs) lead to T-cell activation within mucosa-associated lymphoid tissue. Tissue-specific T-cells expressing chemokine receptor 4 drawn to the lung along with the more generic chemokine receptor 6 (78). Differences in the gut microbiota will result in altered antigen presentation and T-cell activation. Oral probiotics could also influence respiratory health via this mechanism. DCs are critical to antigen presentation. Probiotics lead to DC secretion of the regulatory cytokines IL-10 and IL-12 leading to a shift in the T-helper cell population toward Th1 dominance (89). The improved pathogen clearance and reduction in the Th2 response provides a potential mechanism for beneficial effect of probiotics in the lung.
There may also be a lung–gut axis with the reverse processes in action. Exposing the lungs of mice to lipopolysaccharide results in changes to the bowel flora (90). The impact of acute lung injury and infection may not be restricted to the lungs. Further work is needed to verify these finding and explore any clinically relevant impact.
The gut–lung axis is an example of the potential influence of the gut microbiome over many body systems (91–93). A more detailed understanding of this process may influence feeding and probiotic strategies in neonatology.
Figure 2 summarizes the factors currently shown to affect neonatal bacterial respiratory colonization and the effects colonization patterns may have.
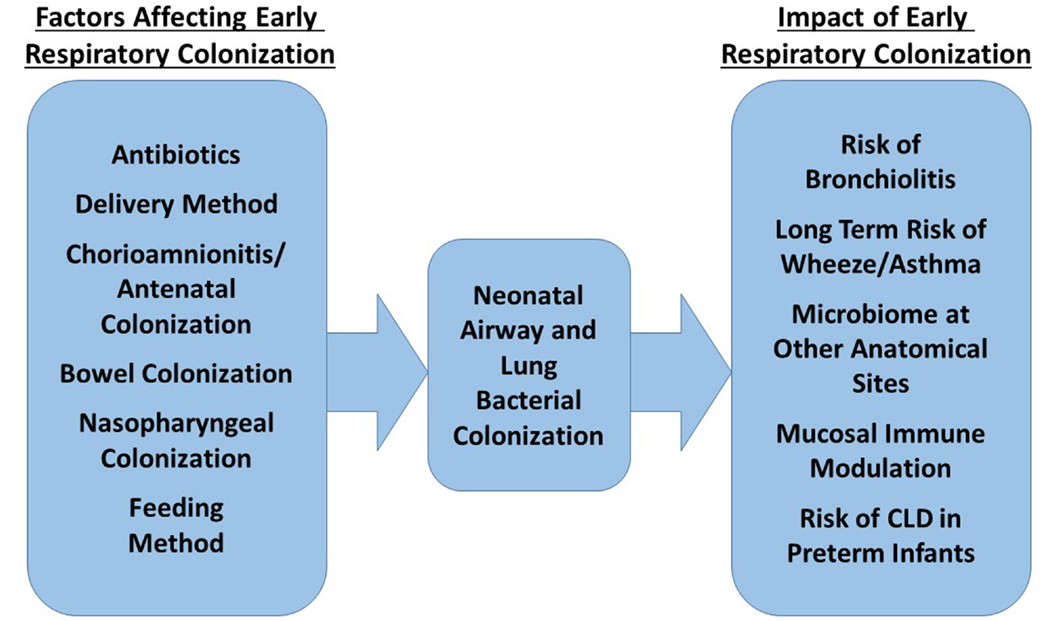
Figure 2. Factors influencing the composition of the neonatal respiratory microbiome and possible impact of this microbiome on future health.
The Future
Identifying the Microbiota and Dysbiosis
Overcoming technical challenges in obtaining samples from the lower airways and lungs of preterm infants can potentially lead to the development of accurate biomarkers for accurately identifying lung microbial colonization. Certainly, molecular methods provide improved sensitivity to traditional culture-based techniques, but modern methods will need to separate infection from colonization, viable from non-viable and commensals from pathogenic bacteria. In clinical work, the transition from culture-based pathogen identification to molecular-based techniques is already underway with PCR tests for, among others, Cytomegalovirus and Neisseria gonorrhoeae used routinely (94). These techniques have already been applied to respiratory bacteria in samples from patients with cystic fibrosis for research purposes (95). It is likely that most routine clinical microbiology will be performed using molecular-based techniques in the future.
Influencing the Microbiota
Probiotics and prebiotics are attempts to manipulate the microbiome of the gut to promote a healthy microbiome. This approach has received widespread support in the neonatal community and a favorable Cochrane review showing efficacy of probiotics in preventing NEC in preterm infants treated with Lactobacillus and/or Bifidobacterium species (96). Alternative strategies for gut microbiome alteration have been tried, with variable success, in adults using fecal transplantation in Clostridium difficile infection (97). Managing the human microbiome with antibiotics has been occurring for decades. The liberal use of antibiotics on neonatal units alters neonatal gut (56), and likely respiratory, microbiota significantly, with unknown long term effects. Routine use of azithromycin to treat possible Ureaplasma infection in pre-terms has been suggested as a way to reduce rates of CLD (98, 99). The impact this would have on other organisms at this important stage of early colonization is unknown and will be important to determine as part of any future trial.
Rising antibiotics resistance and further understanding of host–microbe interactions may lead to a change in emphasis from eradication of pathogens by antibiotics to out competing pathogenic organisms by restoring the healthy, premorbid microbiome. Dysbiosis in the lungs may perhaps be amenable to treatment with nebulized probiotics. Early mouse work has demonstrated nasal administration of probiotic bacteria can protect against respiratory infection (89). Furthermore, as we understand the source of lung colonizing bacteria and factors influencing the early colonizers, manipulation of bacterial acquisition may be possible.
Studying the gut–lung axis may provide significant insights into cultivating the optimal respiratory microbiome. Dietary changes are known to affect the gut microbiome (100). Breast milk is recommended for all infants for many good reasons including reduced upper respiratory tract infections (101). Part of the benefits of breast milk may be through manipulation of the gut and/or respiratory microbiota, but this needs further exploration.
Early microbial programing demonstrates the importance of a long-term view of all areas of neonatal care. Future studies focusing only on the neonatal period will not detect the effects of microbiological programing.
As the understanding of the role of the microbiome increases from observational studies, it is likely that interventional studies will seek to manipulate the respiratory microbiome using established and novel techniques. Detailed work to grasp the complex symbiosis of host and colonizers is needed to minimize the risks of these treatments.
Conclusion
The proponents of microbiome research suggest understanding the microbiome could lead to highly individualized care based on a patients colonization patterns, preventing acute infections, and long-term disease risks by microbiome manipulation (102, 103). The respiratory tract lags behind the gut in terms of the understanding of the role of commensal bacteria in health and disease. In neonatal practice, further work is needed to understand the early colonization and how this can be optimized. The important window of opportunity for influencing long-term health through microbiome-mediated effects is likely to be in the neonatal period.
Author Contributions
DG performed the literature search and wrote the manuscript. SK reviewed the manuscript and authorized the final version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Greenough A. Long term respiratory outcomes of very premature birth (<32 weeks). Semin Fetal Neonatal Med (2012) 17(2):73–6. doi:10.1016/j.siny.2012.01.009
2. Northway WH, Moss RB, Carlisle KB, Parker BR, Popp RL, Pitlick PT, et al. Late pulmonary sequelae of bronchopulmonary dysplasia. N Engl J Med (1990) 323:1793–9. doi:10.1056/NEJM199012273232603
3. Chakraborty M, McGreal EP, Kotecha S. Acute lung injury in preterm newborn infants: mechanisms and management. Paediatr Respir Rev (2010) 11(3):162–70. doi:10.1016/j.prrv.2010.03.002 quiz 170,
4. Beeton ML, Maxwell NC, Davies PL, Nuttall D, McGreal E, Chakraborty M, et al. Role of pulmonary infection in the development of chronic lung disease of prematurity. Eur Respir J (2011) 37(6):1424–30. doi:10.1183/09031936.00037810
5. Davies PL, Maxwell NC, Kotecha S. The role of inflammation and infection in the development of chronic lung disease of prematurity. Adv Exp Med Biol (2006) 582:101–10. doi:10.1007/0-387-33026-7_9
6. Jensen EA, Schmidt B. Epidemiology of bronchopulmonary dysplasia. Birth Defects Res A Clin Mol Teratol (2014) 100:145–57. doi:10.1002/bdra.23235
7. Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome (2015) 3(1):31. doi:10.1186/s40168-015-0094-5
8. Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut (2006) 55(2):205–11. doi:10.1136/gut.2005.073817
9. Robinson CJ, Bohannan BJM, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev (2010) 74(3):453–76. doi:10.1128/MMBR.00014-10
10. Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol (2014) 16(7):1024–33. doi:10.1111/cmi.12308
11. Moran CP, Shanahan F. Gut microbiota and obesity: role in aetiology and potential therapeutic target. Best Pract Res Clin Gastroenterol (2014) 28(4):585–97. doi:10.1016/j.bpg.2014.07.005
12. Morrow AL, Lagomarcino AJ, Schibler KR, Taft DH, Yu Z, Wang B, et al. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome (2013) 1(1):13. doi:10.1186/2049-2618-1-13
13. Mai V, Young CM, Ukhanova M, Wang X, Sun Y, Casella G, et al. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLoS One (2011) 6(6):e20647. doi:10.1371/journal.pone.0020647
14. Unger S, Stintzi A, Shah P, Mack D, O’Connor DL. Gut microbiota of the very low birth weight infant. Pediatr Res (2014) 77(1–2):205–13. doi:10.1038/pr.2014.162
15. AlFaleh K, Anabrees J. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Evid Based Child Health (2014) 9(3):584–671. doi:10.1002/ebch.1976
16. Rogers GB, Shaw D, Marsh RL, Carroll MP, Serisier DJ, Bruce KD. Respiratory microbiota: addressing clinical questions, informing clinical practice. Thorax (2015) 70(1):74–81. doi:10.1136/thoraxjnl-2014-205826
17. Dickson RP, Huffnagle GB. The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog (2015) 11(7):e1004923. doi:10.1371/journal.ppat.1004923
18. Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature (2012) 486(7402):207–14. doi:10.1038/nature11234
19. Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med (2013) 187(10):1067–75. doi:10.1164/rccm.201210-1913OC
20. Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One (2011) 6(2):e16384. doi:10.1371/journal.pone.0016384
21. Stressmann FA, Rogers GB, Klem ER, Lilley AK, Donaldson SH, Daniels TW, et al. Analysis of the bacterial communities present in lungs of patients with cystic fibrosis from American and British centers. J Clin Microbiol (2011) 49(1):281–91. doi:10.1128/JCM.01650-10
22. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One (2010) 5(1):e8578. doi:10.1371/journal.pone.0008578
23. Lohmann P, Luna RA, Hollister EB, Devaraj S, Mistretta T-A, Welty SE, et al. The airway microbiome of intubated premature infants: characteristics and changes that predict the development of bronchopulmonary dysplasia. Pediatr Res (2014) 76(3):294–301. doi:10.1038/pr.2014.85
24. Payne MS, Goss KCW, Connett GJ, Kollamparambil T, Legg JP, Thwaites R, et al. Molecular microbiological characterization of preterm neonates at risk of bronchopulmonary dysplasia. Pediatr Res (2010) 67(4):412–8. doi:10.1203/PDR.0b013e3181d026c3
25. de Blic J, Midulla F, Barbato A, Clement A, Dab I, Eber E, et al. Bronchoalveolar lavage in children. ERS task force on bronchoalveolar lavage in children. European Respiratory Society. Eur Respir J (2000) 15(1):217–31.
26. Goddard AF, Staudinger BJ, Dowd SE, Joshi-Datar A, Wolcott RD, Aitken ML, et al. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci U S A (2012) 109(34):13769–74. doi:10.1073/pnas.1107435109
27. Sze MA, Dimitriu PA, Hayashi S, Elliott WM, McDonough JE, Gosselink JV, et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med (2012) 185(10):1073–80. doi:10.1164/rccm.201111-2075OC
28. Biesbroek G, Tsivtsivadze E, Sanders EAM, Montijn R, Veenhoven RH, Keijser BJF, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med (2014) 190(11):1283–92. doi:10.1164/rccm.201407-1240OC
29. Costello EK, Carlisle EM, Bik EM, Morowitz MJ, Relman DA. Microbiome assembly across multiple body sites in low-birthweight infants. MBio (2013) 4(6):e782–713. doi:10.1128/mBio.00782-13
30. Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio (2015) 6(2):e00037. doi:10.1128/mBio.00037-15
31. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics (2006) 118(2):511–21. doi:10.1542/peds.2005-2824
32. Marchesi J, Shanahan F. The normal intestinal microbiota. Curr Opin Infect Dis (2007) 20(5):508–13. doi:10.1097/QCO.0b013e3282a56a99
33. Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One (2011) 6(2):e17035. doi:10.1371/journal.pone.0017035
34. Sakwinska O, Schmid VB, Berger B, Bruttin A, Keitel K, Lepage M, et al. Nasopharyngeal microbiota in healthy children and pneumonia patients. J Clin Microbiol (2014) 52(5):1590–4. doi:10.1128/JCM.03280-13
35. Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods (2007) 69(2):330–9. doi:10.1016/j.mimet.2007.02.005
36. Castro-Nallar E, Shen Y, Freishtat RJ, Pérez-Losada M, Manimaran S, Liu G, et al. Integrating microbial and host transcriptomics to characterize asthma-associated microbial communities. BMC Med Genomics (2015) 8:50. doi:10.1186/s12920-015-0121-1
37. Morgan XC, Huttenhower C. Meta’omic analytic techniques for studying the intestinal microbiome. Gastroenterology (2014) 146(6):1437.e–48.e. doi:10.1053/j.gastro.2014.01.049
38. Aho VTE, Pereira PAB, Haahtela T, Pawankar R, Auvinen P, Koskinen K. The microbiome of the human lower airways: a next generation sequencing perspective. World Allergy Organ J (2015) 8(1):23. doi:10.1186/s40413-015-0074-z
39. Beck JM. ABCs of the lung microbiome. Ann Am Thorac Soc (2014) 11(Suppl 1):S3–6. doi:10.1513/AnnalsATS.201306-188MG
40. Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol (2014) 12(1):87. doi:10.1186/s12915-014-0087-z
41. Beck JM, Young VB, Huffnagle GB. The microbiome of the lung. Transl Res (2012) 160(4):258–66. doi:10.1016/j.trsl.2012.02.005
42. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med (2014) 6(237):237ra65. doi:10.1126/scitranslmed.3008599
43. Jones HE, Harris KA, Azizia M, Bank L, Carpenter B, Hartley JC, et al. Differing prevalence and diversity of bacterial species in fetal membranes from very preterm and term labor. PLoS One (2009) 4(12):e8205. doi:10.1371/journal.pone.0008205
44. DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One (2008) 3(8):e3056. doi:10.1371/journal.pone.0003056
45. Hansen R, Scott KP, Khan S, Martin JC, Berry SH, Stevenson M, et al. First-pass meconium samples from healthy term vaginally-delivered neonates: an analysis of the microbiota. PLoS One (2015) 10(7):e0133320. doi:10.1371/journal.pone.0133320
46. Jiménez E, Fernández L, Marín ML, Martín R, Odriozola JM, Nueno-Palop C, et al. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol (2005) 51(4):270–4. doi:10.1007/s00284-005-0020-3
47. Miralles R, Hodge R, Mcparland PC, Field DJ, Bell SC, Taylor DJ, et al. Relationship between antenatal inflammation and antenatal infection identified by detection of microbial genes by polymerase chain reaction. Pediatr Res (2005) 57(4):570–7. doi:10.1203/01.PDR.0000155944.48195.97
48. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A (2010) 107(26):11971–5. doi:10.1073/pnas.1002601107
49. Mourani PM, Harris JK, Sontag MK, Robertson CE, Abman SH. Molecular identification of bacteria in tracheal aspirate fluid from mechanically ventilated preterm infants. PLoS One (2011) 6(10):e25959. doi:10.1371/journal.pone.0025959
50. Kotecha S, Hodge R, Schaber JA, Miralles R, Silverman M, Grant WD. Pulmonary Ureaplasma urealyticum is associated with the development of acute lung inflammation and chronic lung disease in preterm infants. Pediatr Res (2004) 55(1):61–8. doi:10.1203/01.PDR.0000100757.38675.50
51. Mika M, Mack I, Korten I, Qi W, Aebi S, Frey U, et al. Dynamics of the nasal microbiota in infancy: a prospective cohort study. J Allergy Clin Immunol (2015) 135(4):905.e–12.e. doi:10.1016/j.jaci.2014.12.1909
52. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med (2011) 184(8):957–63. doi:10.1164/rccm.201104-0655OC
53. Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr (2009) 98:229–38. doi:10.1111/j.1651-2227.2008.01060.x
54. Gupta RW, Tran L, Norori J, Ferris MJ, Eren AM, Taylor CM, et al. Histamine-2 receptor blockers alter the fecal microbiota in premature infants. J Pediatr Gastroenterol Nutr (2013) 56(4):397–400. doi:10.1097/MPG.0b013e318282a8c2
55. Adlerberth I, Lindberg E, Åberg N, Hesselmar B, Saalman R, Strannegård IL, et al. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res (2006) 59(1):96–101. doi:10.1203/01.pdr.0000191137.12774.b2
56. Tanaka S, Kobayashi T, Songjinda P, Tateyama A, Tsubouchi M, Kiyohara C, et al. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol Med Microbiol (2009) 56:80–7. doi:10.1111/j.1574-695X.2009.00553.x
57. Smith DJ, Badrick AC, Zakrzewski M, Krause L, Bell SC, Anderson GJ, et al. Pyrosequencing reveals transient cystic fibrosis lung microbiome changes with intravenous antibiotics. Eur Respir J (2014) 44(4):922–30. doi:10.1183/09031936.00203013
58. Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe (2015) 17(5):704–15. doi:10.1016/j.chom.2015.03.008
59. Jeurink PV, van Bergenhenegouwen J, Jiménez E, Knippels LMJ, Fernández L, Garssen J, et al. Human milk: a source of more life than we imagine. Benef Microbes (2013) 4(1):17–30. doi:10.3920/BM2012.0040
60. Jost T, Lacroix C, Braegger C, Chassard C. Impact of human milk bacteria and oligosaccharides on neonatal gut microbiota establishment and gut health. Nutr Rev (2015) 73(7):426–37. doi:10.1093/nutrit/nuu016
61. Biesbroek G, Bosch AA, Wang X, Keijser BJ, Veenhoven RH, Sanders EA, et al. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med (2014) 190(3):298–308. doi:10.1164/rccm.201401-0073OC
62. Madan JC, Koestler DC, Stanton BA, Davidson L, Moulton LA, Housman ML, et al. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. MBio (2012) 3(4):e00251–12. doi:10.1128/mBio.00251-12
63. Gleeson K, Eggli DF, Maxwell SL. Quantitative aspiration during sleep in normal subjects. Chest (1997) 111(5):1266–72. doi:10.1378/chest.111.5.1266
64. Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A (2012) 109(15):5809–14. doi:10.1073/pnas.1120577109
65. Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol (2011) 127(2):372–3. doi:10.1016/j.jaci.2010.10.048
66. Lowe J, Watkins WJ, Edwards MO, Spiller OB, Jacqz-Aigrain E, Kotecha SJ, et al. Association between pulmonary Ureaplasma colonization and bronchopulmonary dysplasia in preterm infants: updated systematic review and meta-analysis. Pediatr Infect Dis J (2014) 33(7):697–702. doi:10.1097/INF.0000000000000239
67. Davies PL, Spiller OB, Beeton ML, Maxwell NC, Remold-O’Donnell E, Kotecha S. Relationship of proteinases and proteinase inhibitors with microbial presence in chronic lung disease of prematurity. Thorax (2010) 65(3):246–51. doi:10.1136/thx.2009.116061
68. Carroll IM, Maharshak N. Enteric bacterial proteases in inflammatory bowel disease- pathophysiology and clinical implications. World J Gastroenterol (2013) 19(43):7531–43. doi:10.3748/wjg.v19.i43.7531
69. Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One (2010) 5(6):e11044. doi:10.1371/journal.pone.0011044
70. Burke H, Leonardi-Bee J, Hashim A, Pine-Abata H, Chen Y, Cook DG, et al. Prenatal and passive smoke exposure and incidence of asthma and wheeze: systematic review and meta-analysis. Pediatrics (2012) 129(4):735–44. doi:10.1542/peds.2011-2196
71. von Mutius E, Radon K. Living on a farm: impact on asthma induction and clinical course. Immunol Allergy Clin North Am (2008) 28(3):631–47, ix–x. doi:10.1016/j.iac.2008.03.010
72. Huang YJ, Boushey HA. The microbiome and asthma. Ann Am Thorac Soc (2014) 11(Suppl 1):S48–51. doi:10.1513/AnnalsATS.201306-187MG
73. Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol (2001) 107(1):129–34. doi:10.1067/mai.2001.111237
74. Vissing NH, Chawes BLK, Bisgaard H. Increased risk of pneumonia and bronchiolitis after bacterial colonization of the airways as neonates. Am J Respir Crit Care Med (2013) 188(10):1246–52. doi:10.1164/rccm.201302-0215OC
75. Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bønnelykke K, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med (2007) 357(15):1487–95. doi:10.1056/NEJMoa052632
76. Følsgaard NV, Schjørring S, Chawes BL, Rasmussen MA, Krogfelt KA, Brix S, et al. Pathogenic bacteria colonizing the airways in asymptomatic neonates stimulates topical inflammatory mediator release. Am J Respir Crit Care Med (2013) 187(6):589–95. doi:10.1164/rccm.201207-1297OC
77. Russell SL, Finlay BB. The impact of gut microbes in allergic diseases. Curr Opin Gastroenterol (2012) 28(6):563–9. doi:10.1097/MOG.0b013e3283573017
78. Samuelson DR, Welsh DA, Shellito JE. Regulation of lung immunity and host defense by the intestinal microbiota. Front Microbiol (2015) 6:1085. doi:10.3389/fmicb.2015.01085
79. Bianconi S, Gudavalli M, Sutija VG, Lopez AL, Barillas-Arias L, Ron N. Ranitidine and late-onset sepsis in the neonatal intensive care unit. J Perinat Med (2007) 35(2):147–50. doi:10.1515/JPM.2007.017
80. Guillet R, Stoll BJ, Cotten CM, Gantz M, McDonald S, Poole WK, et al. Association of H2-blocker therapy and higher incidence of necrotizing enterocolitis in very low birth weight infants. Pediatrics (2006) 117(2):e137–42. doi:10.1542/peds.2005-1543
81. Terrin G, Passariello A, De Curtis M, Manguso F, Salvia G, Lega L, et al. Ranitidine is associated with infections, necrotizing enterocolitis, and fatal outcome in newborns. Pediatrics (2012) 129(1):e40–5. doi:10.1542/peds.2011-0796
82. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science (2012) 336(6086):1268–73. doi:10.1126/science.1223490
83. Morrow LE, Kollef MH, Casale TB. Probiotic prophylaxis of ventilator-associated pneumonia. Am J Respir Crit Care Med (2010) 182(8):1058–64. doi:10.1164/rccm.200912-1853OC
84. Popova M, Molimard P, Courau S, Crociani J, Dufour C, Le Vacon F, et al. Beneficial effects of probiotics in upper respiratory tract infections and their mechanical actions to antagonize pathogens. J Appl Microbiol (2012) 113(6):1305–18. doi:10.1111/j.1365-2672.2012.05394.x
85. Warner BB, Hamvas A. Lungs, microbes and the developing neonate. Neonatology (2015) 107(4):337–43. doi:10.1159/000381124
86. Geuking MB, Köller Y, Rupp S, McCoy KD. The interplay between the gut microbiota and the immune system. Gut Microbes (2014) 5(3):411–8. doi:10.4161/gmic.29330
87. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med (2014) 20(2):159–66. doi:10.1038/nm.3444
88. Mestecky J. The common mucosal immune system and current strategies for induction of immune responses in external secretions. J Clin Immunol (1987) 7(4):265–76. doi:10.1007/BF00915547
90. Sze MA, Tsuruta M, Yang S-WJ, Oh Y, Man SFP, Hogg JC, et al. Changes in the bacterial microbiota in gut, blood, and lungs following acute LPS instillation into mice lungs. PLoS One (2014) 9(10):e111228. doi:10.1371/journal.pone.0111228
91. Sherwin E, Rea K, Dinan TG, Cryan JF. A gut (microbiome) feeling about the brain. Curr Opin Gastroenterol (2016) 32(2):96–102. doi:10.1097/MOG.0000000000000244
92. Yang I, Corwin EJ, Brennan PA, Jordan S, Murphy JR, Dunlop A. The infant microbiome: implications for infant health and neurocognitive development. Nurs Res (2016) 65(1):76–88. doi:10.1097/NNR.0000000000000133
93. Rossi M, Johnson DW, Campbell KL. The kidney-gut axis: implications for nutrition care. J Ren Nutr (2015) 25(5):399–403. doi:10.1053/j.jrn.2015.01.017
94. Yang S, Rothman RE. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis (2004) 4(6):337–48. doi:10.1016/S1473-3099(04)01044-8
95. Flight WG, Smith A, Paisey C, Marchesi JR, Bull MJ, Norville PJ, et al. Rapid detection of emerging pathogens and the loss of microbial diversity associated with severe lung disease in cystic fibrosis. J Clin Microbiol (2015) 53(7):2022–9. doi:10.1128/JCM.00432-15
96. AlFaleh K, Anabrees J. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst Rev (2014) 4:CD005496. doi:10.1002/14651858.CD005496.pub4
97. van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med (2013) 368(5):407–15. doi:10.1056/NEJMoa1205037
98. Turner MA, Jacqz-Aigrain E, Kotecha S. Azithromycin, Ureaplasma and chronic lung disease of prematurity: a case study for neonatal drug development. Arch Dis Child (2012) 97(6):573–7. doi:10.1136/adc.2010.195180
99. Maxwell NC, Nuttall D, Kotecha S. Does Ureaplasma spp. cause chronic lung disease of prematurity: ask the audience? Early Hum Dev (2009) 85(5):291–6. doi:10.1016/j.earlhumdev.2008.12.002
100. Albenberg LG, Wu GD. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology (2014) 146(6):1564–72. doi:10.1053/j.gastro.2014.01.058
101. Section on Breastfeeding. Breastfeeding and the use of human milk. Pediatrics (2012) 129(3):e827–41. doi:10.1542/peds.2011-3552
102. Kinross JM, von Roon AC, Holmes E, Darzi A, Nicholson JK. The human gut microbiome: implications for future health care. Curr Gastroenterol Rep (2008) 10(4):396–403. doi:10.1007/s11894-008-0075-y
Keywords: bronchopulmonary dysplasia, chronic lung disease, prematurity, microbiome, lung, colonization, 16S RNA gene, neonate
Citation: Gallacher DJ and Kotecha S (2016) Respiratory Microbiome of New-Born Infants. Front. Pediatr. 4:10. doi: 10.3389/fped.2016.00010
Received: 06 November 2015; Accepted: 08 February 2016;
Published: 23 February 2016
Edited by:
Henry J. Rozycki, Children’s Hospital of Richmond at Virginia Commonwealth University, USAReviewed by:
Sule Cataltepe, Brigham and Women’s Hospital and Harvard Medical School, USAGiovanni Vento, Fondazione Policlinico Universitario A. Gemelli Università Cattolica Sacro Cuore, Italy
Stephen Welty, Baylor College of Medicine, USA
Copyright: © 2016 Gallacher and Kotecha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sailesh Kotecha, a290ZWNoYXNAY2FyZGlmZi5hYy51aw==
 David J. Gallacher
David J. Gallacher Sailesh Kotecha
Sailesh Kotecha