- 1Section of Gastroenterology, Department of Immunology and Internal Medicine, University of Manitoba, Winnipeg, MB, Canada
- 2Department of Animal Science, University of Manitoba, Winnipeg, MB, Canada
- 3Department of Medical Microbiology and Infectious Diseases, University of Manitoba, Winnipeg, MB, Canada
- 4Inflammatory Bowel Disease Clinical and Research Centre, University of Manitoba, Winnipeg, MB, Canada
Postnatal maturation of immune regulation is largely driven by exposure to microbes. The gastrointestinal tract is the largest source of microbial exposure, as the human gut microbiome contains up to 1014 bacteria, which is 10 times the number of cells in the human body. Several studies in recent years have shown differences in the composition of the gut microbiota in children who are exposed to different conditions before, during, and early after birth. A number of maternal factors are responsible for the establishment and colonization of gut microbiota in infants, such as the conditions surrounding the prenatal period, time and mode of delivery, diet, mother’s age, BMI, smoking status, household milieu, socioeconomic status, breastfeeding and antibiotic use, as well as other environmental factors that have profound effects on the microbiota and on immunoregulation during early life. Early exposures impacting the intestinal microbiota are associated with the development of childhood diseases that may persist to adulthood such as asthma, allergic disorders (atopic dermatitis, rhinitis), chronic immune-mediated inflammatory diseases, type 1 diabetes, obesity, and eczema. This overview highlights some of the exposures during the pre- and postnatal time periods that are key in the colonization and development of the gastrointestinal microbiota of infants as well as some of the diseases or disorders that occur due to the pattern of initial gut colonization.
Introduction
The human body houses trillions of microbes that are found in different parts of the body. The largest microbial population of the human microbiome is found in the gastrointestinal tract, and the greatest prevalence is in the colon, which is estimated to harbor 1014 bacterial cells and more than 100 times the number of genes in the human genome (1–3). The gut bacteria play an important role in human health by promoting intestinal homeostasis, stimulating the development and maturation of the immune system, protecting against pathogens, digesting fibrous food materials through fermentation, and harvesting nutrients (4–6). An alteration of the gut microbiota has been associated with an increasing number of diseases including inflammatory bowel disease (IBD), obesity, diabetes, asthma, and allergies (6, 7).
The current widespread use of high-throughput molecular microbiology techniques has enhanced our knowledge about the development of the intestinal microbiota to greater levels than were possible with classical culture techniques (8, 9). As a result of comprehensive microbiota analyses over time, insights into fundamental questions about the human microbiome dynamics under different microbial and host conditions are increasing (10). Longitudinal studies including the Human Microbiome Project have investigated the diversity of the bacterial population associated with the human body, its variability within and between individuals, the impact of internal and external factors, as well as characterizing its key components (8, 11, 12), and there is still much to learn. The human gut microbiota of a healthy adult is thought to be highly resilient and stable over time, a condition that may differ from one individual to another (13, 14). However, before reaching maturity, the gut microbiota needs to develop and establish a mutual beneficial co-existence with the host.
It has been suggested that the first contacts with pioneer bacteria could be deterministic for subsequent gut maturation, metabolic and immunologic programing, and consequently for short- and long-term health status (15). In addition, although there are discrepancies, metadata analysis over time has suggested an association between the nature of the initial gut microbiota colonization or microbial dysbiosis and a number of disease conditions in infancy and later in life (16, 17). This review will summarize some of the key factors (pre and postnatal) underlying the establishment of the neonatal gut microbiota and identify its potential impact on some of the immune-, intestinal-, and metabolic-related diseases in childhood or adulthood.
Impact of the Prenatal Period on Gut Microbiota
The intrauterine environment and the unborn infant are generally thought to be sterile until delivery. However, some studies have reported the presence of bacteria in the intrauterine environment, which suggest that these bacteria may influence the microbiota of the infant before birth (18–22). For example, Lactobacillus and Bifidobacterium DNA were detected in the placenta of vaginally and cesarean section-delivered infants (23), and Enterococcus faecium strains that were orally inoculated to pregnant mice were later detected in the amniotic fluid and meconium of the pups following delivery (20). In addition, a unique placental microbiome niche similar to the human oral microbiome composed of non-pathogenic commensal microbiota from different phyla including Firmicutes, Tenericutes, Proteobacteria, Bacteroidetes, and Fusobacteria were characterized from a population-based cohort of placental specimens (24). This suggests that the placenta is not actually sterile and that oral microbiota may play a major role in the colonization of the placenta although the mechanism through which oral microbes find their way into the placenta remains to be elucidated. It is therefore possible that bacteria in the intrauterine environment could result in prenatal colonization of the meconium (6, 25). Jimenez and colleagues showed that the presence of bacterial species in the meconium, such as Escherichia coli, E. faecium, and Staphylococcus epidermidis, could result from the translocation of the mother’s gut bacteria via the bloodstream (19). Despite these findings, it is not clear whether colonization of the infant’s gut microbiota starts before birth, because the presence of bacteria in the amniotic fluid could also be an indication of undetected infection, which may increase the risk of miscarriages or preterm delivery (20). In this case, an association of the placental microbiome with a remote history of antenatal infections such as urinary tract infection in the first trimester and preterm birth has been reported (24). However, as reviewed by Li et al. (6), Bifidobacterium has been reported in meconium, amniotic fluid, fetal membranes, umbilical cord blood, and placenta of healthy mothers and infants with no detectable or known clinical infections or inflammation.
External factors during pregnancy may also influence the future development and behavior of the infant. For example, infant monkeys born from mothers stressed during pregnancy had significantly lower counts of Bifidobacterium and Lactobacillus (26). Probiotic administration (Lactobacillus rhamnosus) to mothers during late pregnancy also resulted in increased fecal Bifidobacterium longum counts in their infants (27), although, it is not clear whether these microbes were acquired from the mother during pregnancy, during birth, or after birth. Several other variables during pregnancy, including the use of antibiotics in the perinatal period, have been associated with delayed colonization by some microbes especially Bifidobacteria and Lactobacillus species (28, 29). This may have long-term impacts since these species are considered to have beneficial properties; for example, allergies, irritable bowel syndrome (IBS), and IBD, have all been frequently reported in antibiotic-exposed children (30–36). Roberts et al. (37) showed that children born to mothers who smoked have a higher risk of IBD, which may be due to disturbed microbial colonization since the cessation of smoking was correlated with increased Firmicutes and Actinobacteria, and a lower proportion of Bacteroidetes and Proteobacteria (38).
The length of the gestational period may also play an important role in initial infant gut microbial colonization. Colonization in preterm infants has been shown to take place slowly, have a low diversity, and several interindividual differences as opposed to that of full-term infants (6, 39). It is also mostly dominated by potential pathogens including Clostridium species, E. coli, Enterococcus, Streptococcus, Klebsiella, and Staphylococcus (6, 29, 39, 40). Healthy full-term infants are usually colonized by beneficial microbes, such as Bifidobacterium and Lactobacillus, which are not present or are detected in low levels in preterm infants (41). The delayed rate of colonization could result from the events surrounding the delivery, because most preterm infants are delivered through emergency or elective cesarean delivery. Infants born by cesarean section, notably electively, have been shown to have low bacterial richness and diversity (42), which could be a result of less exposure to the mothers’ delivery fluids, delayed oral feeding, and high hygienic care of the preterm infants and use of antibiotics that may consequently lead to colonization by few resistant/notorious microbes that are potentially pathogenic (6).
External Factors Affecting the Development of Gut Microbiota in Infants during and after Birth
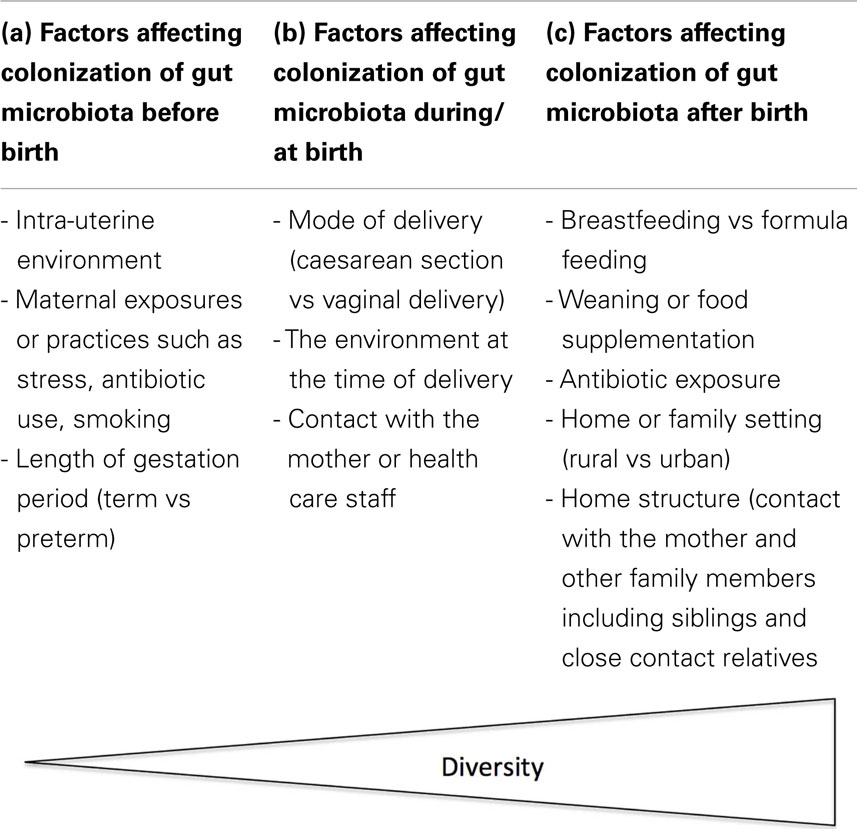
A number of factors (both intrinsic and extrinsic) may influence the process of microbial colonization in infants, which may consequently affect the infant’s health. The following section highlights some of the external factors that affect the initial colonization and establishment of gut microbiota in infants (Table 1).
Delivery Mode
During the birth process and immediately after birth, microbes from the mother and surrounding environment colonize the gastrointestinal tract of the infant leading to the development of a dense complex microbiota (22). The mode of delivery (vaginally or by cesarean section) has been demonstrated to have a strong influence on early gut colonization (43). A review by Mackie and colleagues (22) showed that, in cases of vaginal delivery, a longer birth process has been associated with the presence of viable microbes in the stomach and mouth of the infant, and the same E. coli serotypes were found in both the mouths of babies and in their mothers’ feces immediately after birth. This implies that the proximity of the birth canal and the anus play an important role in the transmission of microbes from the mother to the infant. In addition, bacteria present in the mother’s vagina immediately before birth were reported in the nasopharynxes of over 50% of babies born vaginally (22).
Children born by cesarean section are also exposed to their mothers’ microbiota, but initial exposure is most likely to non-maternally derived environmental isolates from equipment, air, and other infants, with the nursing staff serving as vectors for transfer (44). A number of studies have described altered fecal or intestinal microbiota profiles in cesarean section-delivered infants beginning at 1 day after birth and persisting to 6 weeks (7), 6 months, and even 7 years of age (45). Infants delivered by cesarean section have a lower total microbial diversity within the first 2 years of life associated with a less abundance and diversity of phylum Bacteroidetes (46). They are also less often colonized by Bifidobacteria, Bacteroides, and E. coli, but are more frequently colonized by both Clostridium cluster I and Clostridium difficile (46, 47). It has also been shown that skin microbes including Staphylococcus, Corynebacterium, and Propionibacterium dominate the gut microbiota of cesarean-delivered infants, while vaginally delivered infants have a higher prevalence of vaginal-related microbes such as Lactobacillus, Prevotella, and Sneathia (48–50). Generally, children born by cesarean section have an altered intestinal microbial colonization and studies have highlighted that this may be associated with a subsequent increased risk of developing various diseases including asthma and/or type 1 diabetes (T1D). This could be due to poor development of the immune system since infants born through cesarean section have also been reported to have remarkably lower levels of the Th1-related chemokines CXL10 and CXL11 in their blood, which may translate to less protection (46). A meta-analysis of the association between cesarean section and childhood asthma involving studies with different designs, conducted in different countries and using different measures of asthma reported an increased risk of asthma after cesarean section (17), whereas a 20% increase in the risk of childhood onset of T1D was reported in another meta-analysis of children born through cesarean section (51). An additional meta-analysis investigating the use of antibiotics in infants reported that the use of antibiotics in childhood was associated with asthma and wheezing (16).
Diet
The succession of microbial colonization in the intestinal tract most occurs during the early development stages especially the first year of life. During this period, the feeding mode shifts from breastfeeding to formula feeding and/or to the introduction of solid food; however, individual instances of gut colonization may vary in terms of microbiota richness and diversity (7, 22, 52). Dynamic balances exist between the gastrointestinal microbiota, host physiology, and diet that directly influence the initial acquisition, developmental succession, and eventual stability of the gut ecosystem (53). Breastfeeding modulates the gut microbiota (54), and this might confer some protective effects to the infant against various forms of diseases or disorders (55), because evidence exists for an entero-mammary pathway that transfers diverse microbes from the mother’s gut to the baby through breast milk (56–59). However, human milk is known to contain complex polysaccharides that act as selective prebiotics and therefore promote the colonization of the infant gut with beneficial microbiota (60, 61), as opposed to children fed with formula. Formula feeding has been associated with an increased microbial richness of species in infants at four months of age with overrepresentation of C. difficile, a known gastrointestinal pathogen (42, 62). In addition, formula feeding induces intestinal hypertrophy and accelerates maturation of hydrolysis capacities; it increases intestinal permeability and bacterial translocation. Therefore, the microbiota may not be the principal actor. However, a recent publication observed more than two times increased numbers of bacteria cells in breast-fed infants, compared to formula-fed ones (63). It is therefore clear that breastfeeding may encourage proliferation or colonization by bacteria that may have protective effects on the growing infant, while formula feeding may predispose children to potential pathogens.
The gut microbiota of children is also influenced by the nature of food (other than formula) they receive, which could also be stratified by income status, mode of upbringing, or geographical location. The microbiota of children in Burkina Faso was found to be dominated by Bacteroidetes, compared with that of Italians, which was dominated by Firmicutes (64). Similarly, the biodiversity of microbiota from USA was lower than that from Malawians or Venezuelan Amerindians (65). Moreover, the effect of the natural environment, such as housing conditions, has also been investigated in animals. In this case, genetically related piglets were housed in either indoor or outdoor environments and sequencing of the 16S rRNA revealed that Lactobacillus strains were dominant in the gut of pigs raised outdoors, compared with hygienic indoor pigs, which had reduced Lactobacillus and more potentially pathogenic phylotypes (66).
Mother and Immediate Family Members
The influence of the mother on the child’s microbiota is evident during the first year after birth (43). This effect is reported to be stronger within the first month of life during which the infant’s intestinal microbiota is both functionally and phylogenetically close to that of the mother. However, at 1 year of age, phylogenetic differences appear while the similarities persist at the functional level (67). With respect to fecal microbiota, close similarities were found between the mother and the infant during the first six months after birth of the infant, which was mainly due to the presence of Bifidobacterium bifidum, Bifidobacterium breve, and Staphylococcus aureus (68).
After birth, environmental, oral, and skin microbes from the mother are mechanically transferred to the newborn in different ways, which may influence the diversity of intestinal microbiota in the neonate. For example, biodiversity in the homes, in the surrounding environment and in family members who have a close or constant contact with the baby have a direct impact on the diversity of microbes that are transferred to the infant (69–71). In addition, hygienic practices (e.g., cleaning of baby’s pacifier through sucking or by other methods) may influence the microbial diversity. Differences in microbial diversity have been associated with the development of allergy and/or asthma later in life (72, 73). Numerous population studies have also confirmed an inverse relationship between allergy prevalence and various measures of “hygiene,” such as growing up on a farm, early day care, and low socioeconomic standards (74, 75). There are also observed differences in the composition of the gut microbiota between infants living in countries with a high and a low prevalence of allergy and between healthy and allergic infants, even very early in life before they have developed any clinical symptoms of disease (76). In addition, an increasing number of older siblings are associated with the colonization of Lactobacilli and Bacteroides at 5 weeks of age, all of which are associated with beneficial effects (47). The mother and family at large, therefore, play a major role in the initial microbial colonization in infants who may have a health impact on the child. Table 1 shows a summary of the factors discussed in this review.
Long-Term Effects of Microbial Colonization
Gut microbiota are generally associated with the development and maturation of the immune system (77). The immune system acquires most of its data from exposure to certain subsets of micro- and macro-organisms (78). For example, early colonization with Escherichia coli and Bifidobacteria is associated with higher numbers of CD20 + B cells that express the memory marker CD27 at 4 and 18 months of age (79). Disruption of exposure to these organisms is at least partly responsible for the immunoregulatory deficits that underlie the increased prevalence of conditions, such as chronic immune inflammatory diseases (i.e., IBD), asthma, and atopic dermatitis (7, 47, 80). Molecular microbiology techniques suggest that a high diversity of the gut microbiota in childhood could be more important as opposed to low diversity, which is associated with increased risk of subsequent allergic diseases, since repeated exposure to different bacterial antigens would enhance the development of immune regulation through inhibition of responses to inappropriate targets, such as gut contents and allergens (81, 82). The microbial diversity and composition of 47 infants as analyzed using barcoded 16S rRNA 454 pyrosequencing in stool samples at 1 week, 1 month, and 12 months of age, revealed that low total diversity of the gut microbiota during the first month of life was associated with asthma in the children at 7 years of age (83). A low gut microbial diversity during the first month of life was also associated with subsequent sensitization and atopic eczema at 2 years of age (82). In addition, reduced bacterial diversity of the infant’s intestinal microbiota was associated with increased risk of allergic sensitization, allergic rhinitis, and peripheral blood eosinophilia, in the first 6 years of life (81). Early-life exposures, including those known to impact gastrointestinal microbiome composition, such as antibiotic administration have also been associated with increased risk for childhood asthma due to altered microbiota profiles or long-term reduction in microbial diversity (7, 84, 85). Alterations of the intestinal microbiota in preterm infants characterized by low microbial diversity and abundance of potentially pathogenic bacteria have been highlighted in the development of necrotizing enterocolitis (NEC), although there are discrepancies among different studies (86, 87). Hällström and colleagues (88) reported a link between cesarean delivery and disturbed intestinal colonization, with an increased frequency of Enterococcus species and Candida albicans, and the probable occurrence of NEC in preterm infants. These findings may not be exclusively due to a disturbed intestinal microbiota because other confounding factors may also be responsible for NEC; however, further investigation will provide useful information on this topic and further clarify the existing discrepancies.
The birth order or family size and the presence or absence of pets have also been implicated in the initial microbial colonization in infants. Over two decades ago, Strachan observed that children who had older siblings were less likely to manifest hay fever as adults, as compared to firstborn children, which could be due to a protective effect of infections brought home by the older siblings (89). Similarly, exposure to livestock or pets, particularly dogs, early in life significantly decreases the risk for asthma and/or allergic reactions since dog ownership is associated with a distinct house dust microbial exposure (74, 90). In addition, mice exposed to dog-associated house dust were found to be protected against airway allergen challenge, because they exhibited less Th2 cytokine production, fewer activated T cells, and a distinct gut microbiome composition that was highly enriched for Lactobacillus johnsonii, which itself can confer airway protection when orally supplemented as a single species (91).
Selective microbial targets have been associated with infants developing eczema (92). Yap and colleagues (93) evaluated the composition of fecal microbiota of infants who developed eczema in the first 5 years of life compared with healthy controls and reported that longitudinal analysis of fecal microbiota composition at 3 days, 1 and 3 months, and 1 year of life showed a higher abundance of Enterobacteriaceae and Clostridium perfringens in children who developed eczema in the first 2 years of life, whereas a lower abundance of Bifidobacterium was observed in those who developed eczema at 5 years of age. The authors suggested that relative abundance of selective microbial targets might contribute to the subsequent development of eczema in childhood. Studies have also associated gut microbiota with the development of T1D (77). For example, metagenomic analysis revealed that the proportion of Bacteroidetes was increased in children with T1D while the proportion of Firmicutes was increased in normal healthy children (78). Conversely, the relative proportion of Bacteroidetes is decreased in obese people compared with lean people, while the proportion of Firmicutes is increased in obese people (78). In addition, colonization with clostridia, at the age of 5 and 13 weeks is associated with an increased risk of developing atopic dermatitis in the subsequent 6 months of life (47). Breastfeeding and formula feeding play major roles in defining the initial microbial colonization, and a previous meta-analysis showed that breast-fed children have a lower risk of being overweight compared to formula-fed children and that the duration of breastfeeding is inversely and linearly associated with the risk of overweight (94).
The use of antibiotics is generally known to change the gut microbiota. A meta-analysis on antibiotic use in infants reported that wheezing and asthma were related to antibiotic use (16). In addition, high-throughput sequencing revealed an incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin (95). The use of antibiotics in this context has been implicated in the development of IBD and current research has shown that children with IBD are more likely to have received antibiotics in their first year of life as compared to healthy controls (35), suggesting that microbial dysbiosis associated with early antibiotic exposure in neonates may be a predisposing factor to IBD, including other disease conditions, such as wheezing and asthma. Table 2 shows a summary of the factors affecting gut microbiota and their effects in neonates, infants or children, as discussed in this review.
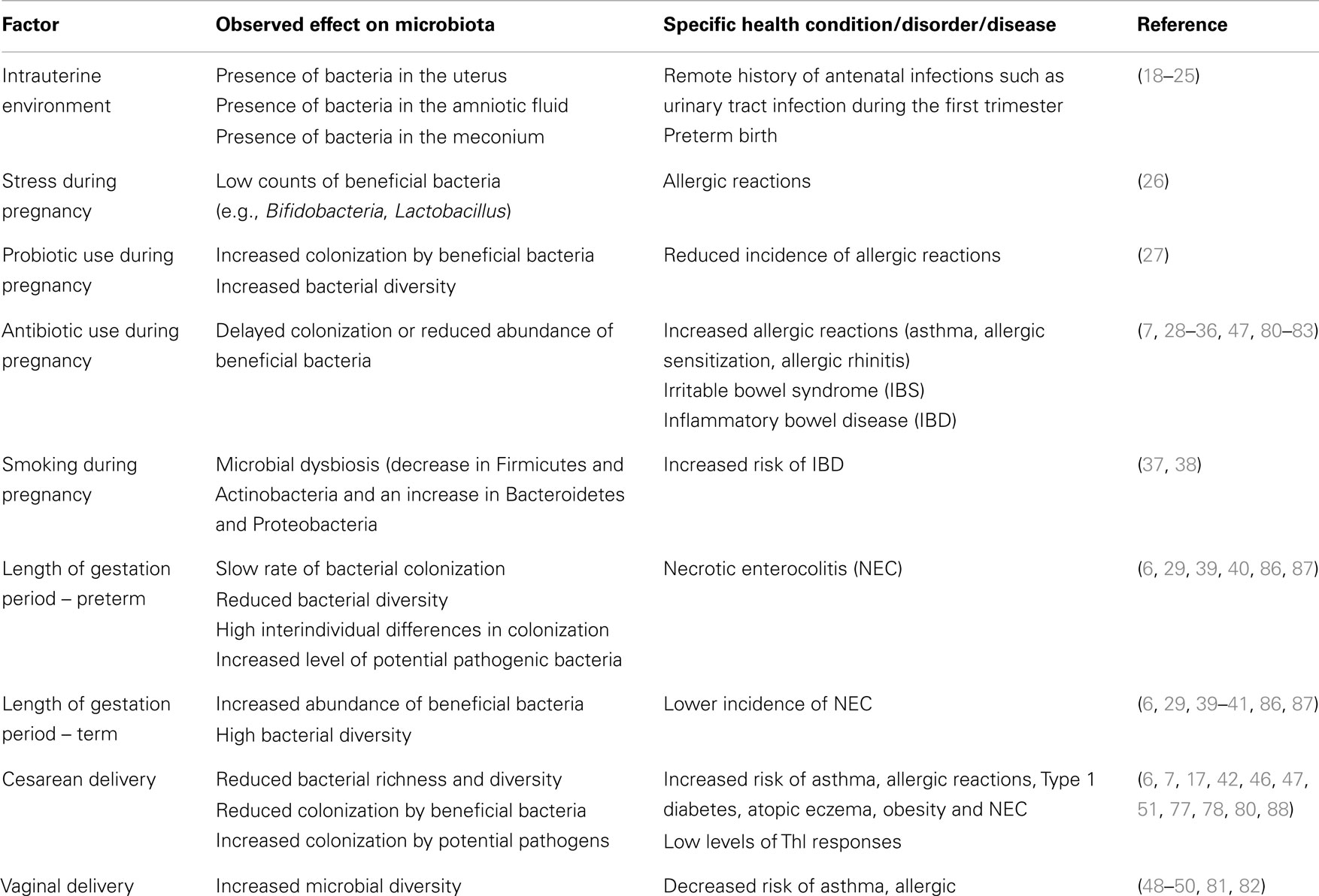
Table 2. Factors affecting colonization of gut microbiota in neonates and infants or children, specific microbial effect, and the resultant health conditions.
Modifications of Early Childhood Microbiota
Several methods can be used to attempt to modify disturbed gut microbiota and many of them have been implicated in the improvement of gut microbiota in early childhood. However, an extensive discussion of the methods used to modify gut microbiota is beyond the scope of this review and only brief highlights on probiotics are included. As defined by the Food and Agriculture Organization/World Health Organization, probiotics are live microorganisms that, when administered in adequate amounts, confer health benefits on the host. The mechanisms by which probiotics exert beneficial effects on the host are currently a main area of focus although this may vary depending on the species or the strain involved. In this case, infants fed with a blend of Lactobacillus acidophilus and Bifidobacterium infantis as well as other probiotics had less cases of NEC and a reduced mortality rate compared to the controls (96), supporting the use of probiotics in preterm infants to prevent NEC. Although limited research has been done on the use of probiotics in NEC, probiotics appear promising for use as a prevention strategy for NEC; Abrahamsson and colleagues (97) argue that the time for confirmative NEC probiotic prevention trial in the extremely low birth weight infants in North America is now. On the other hand, the “hygiene hypothesis” suggests that a lack of exposure to microbial stimulus early in childhood is a major factor involved in the development of allergic reactions and immune-related disorders (89, 98), and therefore provides a rationale for using probiotics to modify the gut microbiota to shape the immune response of the host, especially in infancy. In this case, studies have provided evidence for a beneficial effect of different probiotics in the primary prevention and management of allergic diseases (atopic eczema, allergic rhinitis) (99, 100). For further reading on the use of probiotics in allergic diseases, the reader is referred to other detailed review articles (99–102).
Summary and Conclusion
The characteristics of the inherited microbiome are likely very important in understanding offspring health as recent research suggests that aberrant metabolic phenotypes can be directly attributed to intestinal bacterial communities. Here, we have summarized some of the current interactions and associations on early-life exposures that may influence the development and colonization of the gut microbiota in infants and disease conditions that may result due to the nature of colonization. Some studies have already demonstrated the predictive power of the microbiota in enteric diseases while others have actually conducted research and consistently reported similar findings, although discrepancies do exist. In general, a reduction in overall bacterial diversity, a reduced abundance of commensal bacteria, and an increase in abundance of potentially pathogenic bacteria have been associated with immune-related disorders, and in some situations specific bacteria have being involved in the development of allergic reactions in both animal models and human studies. On the other hand, the importance of the gastrointestinal microbiome in defining the immune environment has been demonstrated in a number of studies, indicating that strategies to manipulate gut microbiome membership and function may have far-reaching implications for better health. This, therefore, shows that colonization by a certain subset of microbiota may be a threat or a benefit to the health of the infant during childhood and/or in adulthood. Consequently, future studies should focus on ways of promoting and/or maintaining colonization by the beneficial bacteria in infants who are exposed to compromising situations that encourage colonization by potentially pathogenic phylotypes. Prebiotics and/or probiotics have been suggested to have a promising impact in the prevention and treatment of some immune-related diseases by modulating gut microbiota and regulating host mucosal immune function; however, their efficacy is inconsistent and further studies and the exploration of other treatments are required.
Whether the altered microbiome causes the disease or is the disease affecting the microbiome remains an issue of debate. However, for neonates, it is possible to argue that the first colonizers play a major role in the development of the disease because if this is not the case, then why are some disorders/disease conditions more common in infants born via caesarian delivery compared with vaginally delivered infants? Or, why do formula-fed infants more frequently have metabolic disorders as opposed to breast-fed infants? Other questions include why is it that the majority of potentially known pathogenic microbes are found in children exposed to antibiotics, born by cesarean section, or raised under high hygienic conditions, whereas there are more known beneficial microbes in their counterparts? In support of our argument, differences in the composition of the gut microbiota between infants living in countries with a high and a low prevalence of allergy and between healthy and allergic infants, have been reported even very early in life before development of any clinical symptoms of disease (76), suggesting the role of the gut microbiota in the development of the disease conditions. This topic still remains open for discussion and the answers to the above questions will shed more light on this debate; future research should incorporate extended microbiota analyses, detailed nutrition assessments, and longitudinal measures of disease conditions throughout childhood.
Meta-analyses have substantiated the association between some disease conditions with cesarean section, formula feeding, and use of antibiotics in infants. However, these conditions may result from the impact of differences or changes already observed in the gut microbiota, or they could result from other confounding factors. Advanced molecular microbiology approaches including functional metagenomic analysis may shed more light on factors that affect infant microbial colonization and extend our knowledge on bacterial species that are potentially undesirable or actually beneficial, and that can be considered as potential probiotics for novel formula development in order to closely emulate nature’s models. Future studies should, therefore, focus on functionally characterizing the gut microbiota of children with the aim of identifying some of the metabolic pathways and processes that may be targeted for modulation with beneficial microbiota, and consequently aid in management of the disease conditions that are associated with altered gut microbiota or colonization by potential pathogens.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature (2010) 464:59–65. doi:10.1038/nature08821
2. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell (2006) 124:837–48. doi:10.1016/j.cell.2006.02.017
3. Bjorksten B. Diverse microbial exposure-consequences for vaccine development. Vaccine (2012) 30:4336–40. doi:10.1016/j.vaccine.2011.10.074
4. Young VB. The intestinal microbiota in health and disease. Curr Opin Gastroenterol (2012) 28:63–9. doi:10.1097/MOG.0b013e32834d61e9
5. Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature (2012) 489:231–41. doi:10.1038/nature11551
6. Li M, Wang M, Donovan SM. Early development of the gut microbiome and immune-mediated child disorders. Semin Reprod Med (2014) 32:74–86. doi:10.1055/s-0033-1361825
7. Kozyrskyj AL, Bahreinian S, Azad MB. Early life exposures: impact on asthma and allergic disease. Curr Opin Allergy Clin Immunol (2011) 11:400–6. doi:10.1097/ACI.0b013e328349b166
8. Human Microbiome Project C. A framework for human microbiome research. Nature (2012) 486:215–21. doi:10.1038/nature11209
9. Suau A, Bonnet R, Sutren M, Gordon JJ, Gibson GR, Collins MD, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol (1999) 65:4799–807.
11. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett C, Knight R, Gordon JI. The human microbiome project. Nature (2007) 449:804–10. doi:10.1038/nature06244
12. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende D, et al. Enterotypes of the human gut microbiome. Nature (2011) 473:174–80. doi:10.1038/nature09944
13. Vanhoutte T, Huys G, Brandt E, Swings J. Temporal stability analysis of the microbiota in human feces by denaturing gradient gel electrophoresis using universal and group-specific 16S rRNA gene primers. FEMS Microbiol Ecol (2004) 48:437–46. doi:10.1016/j.femsec.2004.03.001
14. De La Cochetiere MF, Durand T, Lepage P, Bourreille A, Galmiche JP, Dore J. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J Clin Microbiol (2005) 43:5588–92. doi:10.1128/JCM.43.11.5588-5592.2005
15. Rook GA, Lowry CA, Raison CL. Hygiene and other early childhood influences on the subsequent function of the immune system. Brain Res (2014). doi:10.1016/j.brainres.2014.04.004
16. Penders J, Kummeling I, Thijs C. Infant antibiotic use and wheeze and asthma risk: a systematic review and meta-analysis. Eur Respir J (2011) 38:295–302. doi:10.1183/09031936.00105010
17. Thavagnaman S, Fleming J, Bromley A, Shields MD, Cardwell CR. A meta-analysis of the association between caesarean section and childhood asthma. Clin Exp Allergy (2008) 38:629–33. doi:10.1111/j.1365-2222.2007.02780.x
18. Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, Xaus J, et al. Is meconium from healthy newborns actually sterile? Res Microbiol (2008) 159:187–93. doi:10.1016/j.resmic.2007.12.007
19. Jimenez E, Fernandez L, Marin M, Martin R, Odriozola JM, Nueno-Palop C, et al. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol (2005) 51:270–4. doi:10.1007/s00284-005-0020-3
20. DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One (2008) 3:e3056. doi:10.1371/journal.pone.0003056
21. Penders J, Thijs C, Vink C, Stelma FF, Kummeling IS, Van den Brandt PA, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics (2006) 118:511–21. doi:10.1542/peds.2005-2824
22. Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr (1999) 69:S1035–45.
23. Satokari R, Gronroos T, Laitinen K, Salminen S, Isolauri E. Bifidobacterium and Lactobacillus DNA in the human placenta. Lett Appl Microbiol (2009) 48:8–12. doi:10.1111/j.1472-765X.2008.02475.x
24. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med (2014) 6:237ra65. doi:10.1126/scitranslmed.3008599
25. Madan JC, Salari RC, Saxena D, Davidson L, O’Toole GA, Moore JH, et al. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch Dis Child Fetal Neonatal Ed (2012) 97:F456–62. doi:10.1136/fetalneonatal-2011-301373
26. Bailey MT, Lubach GR, Coe CL. Prenatal stress alters bacterial colonization of the gut in infant monkeys. J Pediatr Gastroenterol Nutr (2004) 38:414–21. doi:10.1097/00005176-200404000-00009
27. Lahtinen SJ, Boyle RJ, Kivivuori S, Oppedisano F, Smith KR, Robins-Browne R, et al. Prenatal probiotic administration can influence Bifidobacterium microbiota development in infants at high risk of allergy. J Allergy Clin Immunol (2009) 123:499–501. doi:10.1016/j.jaci.2008.11.034
28. Faa G, Gerosa C, Fanni D, Nemolato S, van Eyken P, Fanos V. Factors influencing the development of a personal tailored microbiota in the neonate, with particular emphasis on antibiotic therapy. J Matern Fetal Neonatal Med (2013) 26(S2):35–43. doi:10.3109/14767058.2013.829700
29. Westerbeek EA, van den Berg A, Lafeber HN, Knol J, Fetter WP, van Elburg RM. The intestinal bacterial colonization in preterm infants: a review of the literature. Clin Nutr (2006) 25:361–8. doi:10.1016/j.clnu.2006.03.002
30. Droste JH, Wieringa MH, Weyler JJ, Nelen VJ, Vermeire PA, Van Bever HP. Does the use of antibiotics in early childhood increase the risk of asthma and allergic disease? Clin Exp Allergy (2000) 30:1548–53. doi:10.1046/j.1365-2222.2000.00939.x
31. Metsala J, Lundqvist A, Virta LJ, Kaila M, Gissler M, Virtanen SM. Mother’s and offspring’s use of antibiotics and infant allergy to cow’s milk. Epidemiology (2013) 24:303–9. doi:10.1097/EDE.0b013e31827f520f
32. Russell SL, Gold MJ, Hartmann M, Willing BP, Thoson L, Wlodarska M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep (2012) 13:440–7. doi:10.1038/embor.2012.32
33. Stensballe LG, Simonsen J, Jensen SM, Bonnelykke K, Bisgaard H. Use of antibiotics during pregnancy increases the risk of asthma in early childhood. J Pediatr (2013) 162:832–8. doi:10.1016/j.jpeds.2012.09.049
34. Villarreal AA, Aberger FJ, Benrud R, Gundrum JD. Use of broad-spectrum antibiotics and the development of irritable bowel syndrome. WMJ (2012) 111:17–20.
35. Shaw SY, Blanchard JF, Bernstein CN. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am J Gastroenterol (2010) 105:2687–92. doi:10.1038/ajg.2010.398
36. Roberts SE, Wotton CJ, Williams JG, Griffith M, Goldacre MJ. Perinatal and early life risk factors for inflammatory bowel disease. World J Gastroenterol (2011) 17:743–9. doi:10.3748/wjg.v17.i6.743
37. Hviid A, Svanstrom H, Frisch M. Antibiotic use and inflammatory bowel diseases in childhood. Gut (2011) 60:49–54. doi:10.1136/gut.2010.219683
38. Biedermann L, Zeitz J, Mwinyi J, Sutter-Minder E, Rehman A, Ott SJ, et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One (2013) 8:e59260. doi:10.1371/journal.pone.0059260
39. Sakata H, Yoshioka H, Fujita K. Development of the intestinal flora in very low birth weight infants compared to normal full-term newborns. Eur J Pediatr (1985) 144:186–90. doi:10.1007/BF00451911
40. Gewolb IH, Schwalbe RS, Taciak VL, Harrison TS, Panigrahi P. Stool microflora in extremely low birthweight infants. Arch Dis Child Fetal Neonatal Ed (1999) 80:F167–73. doi:10.1136/fn.80.3.F167
41. Butel MJ, Suau A, Campeotto F, Magne F, Aires J, Ferraris L, et al. Conditions of Bifidobacterial colonization in preterm infants: a prospective analysis. J Pediatr Gastroenterol Nutr (2007) 44:577–82. doi:10.1097/MPG.0b013e3180406b20
42. Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ (2013) 185:385–94. doi:10.1503/cmaj.121189
43. Matamoro S, Gras-Leguen C, Le Vacon F, Potel G, de La Cochetiere MF. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol (2013) 21:167–73. doi:10.1016/j.tim.2012.12.001
44. Biasucci G, Benenati B, Morelli L, Bessi E, Boehm G. Cesarean delivery may affect the early biodiversity of intestinal bacteria. J Nutr (2008) 138:1796S–800S.
45. Neu J, Rushing J. Caesarean versus vaginal delivery: long term infant outcomes and the hygiene hypothesis. Clin Perinatol (2011) 38:321–31. doi:10.1016/j.clp.2011.03.008
46. Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonization and reduced Th1 responses in infants delivered by caesarean section. Gut (2014) 63:559–66. doi:10.1136/gutjnl-2012-303249
47. Penders J, Gerhold K, Stobberingh EE, Thijs C, Zimmermann K, Lau S, et al. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J Allergy Clin Immunol (2013) 132:601–7. doi:10.1016/j.jaci.2013.05.043
48. Khafipour E, Ghia JE. Mode of delivery and inflammatory disorders. J Immunol Clin Res (2013) 1:1004.
49. Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. Development of the human gastrointestinal microbiota and insights from high- throughput sequencing. Gastroenterology (2011) 140:1713–9. doi:10.1053/j.gastro.2011.02.011
50. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A (2010) 107:11971–5. doi:10.1073/pnas.1002601107
51. Cardwell CR, Stene LC, Joner G, Cinek O, Syenson J, Goldacre MJ, et al. Caesarean section is associated with an increased risk of childhood-onset type 1 diabetes mellitus: a meta-analysis of observational studies. Diabetologia (2008) 51:726–35. doi:10.1007/s00125-008-0941-z
52. Valles Y, Artacho A, Pascual-Garcia A, Ferrus ML, Gosalbes MJ, Abellan JJ. Microbial succession in the gut: directional trends of taxonomic and functional change in a birth cohort of Spanish infants. PLoS Genet (2014) 10:e1004406. doi:10.1371/journal.pgen.1004406
53. Koening JE, Spor A, Scalfone NA, Stombaugh J, Knight R, Angenent LT, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A (2011) 108:4578–85. doi:10.1073/pnas.1000081107
54. Stark PL, Lee A. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year. J Med Microbiol (1982) 15:189–203. doi:10.1099/00222615-15-2-189
55. Kramer MS. Breastfeeding and allergy: the evidence. Ann Nutr Metab (2011) 59(Suppl 1):20–6. doi:10.1159/000334148
56. Donnet-Hughes A, Perez PF, Dore J, Leclerc M, Levenez F, Benyacoub J, et al. Potential role of the intestinal microbiota of the mother in neonatal immune education. Proc Nutr Soc (2010) 69:407–15. doi:10.1017/S0029665110001898
57. Hunt KM, Foster JA, Formey LJ, Schutte UM, Beck DL, Abdo Z, et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One (2011) 6:e21313. doi:10.1371/journal.pone.0021313
58. Jost T, Lacroix C, Braegger C, Chassard C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr (2013) 110:1253–62. doi:10.1017/S0007114513000597
59. Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol (2013) 3(3):203–20. doi:10.1111/1462-2920.12238
60. Garrido D, Barile D, Mills DA. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv Nutr (2012) 3:415S–21S. doi:10.3945/an.111.001586
61. Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Nat Acad Sci U S A (2011) 108:4653–8. doi:10.1073/pnas.1000083107
62. Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut (2007) 56:661–7. doi:10.1136/gut.2006.100164
63. Bezirtzoglou E, Tsiotslas A, Weilling GW. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe (2011) 17(6):478–82. doi:10.1016/j.anaerobe.2011.03.009
64. De Filippo C, Cavalieri D, Di Paola M, Ramazzoti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Nat Acad Sci U S A (2010) 107:14691–6. doi:10.1073/pnas.1005963107
65. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature (2012) 486:222–7. doi:10.1038/nature11053
66. Mulder IE, Schmidt B, Stokes CR, Lewis M, Bailey M, Aminoy RI, et al. Environmentally-acquired bacteria influence microbial diversity and natural innate immune responses at gut surfaces. BMC Biol (2009) 7:79. doi:10.1186/1741-7007-7-79
67. Vaishampayan PA, Kuehl JV, Froula JL, Morgan JL, Ochman H, Francino MP. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol Evol (2010) 2:53–66. doi:10.1093/gbe/evp057
68. Gronlund MM, Grzeskowiak L, Isolauri E, Salminen S. Influence of mother’s intestinal microbiota on gut colonization in the infant. Gut Microbes (2011) 2:227–33. doi:10.4161/gmic.2.4.16799
69. Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrlander C, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med (2011) 364:701–9. doi:10.1056/NEJMoa1007302
70. Sjögren YM, Jenmalm MC, Böttcher MF, Björkstén B, Sverremark-Ekström E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin Exp Allergy (2009) 39:518–26. doi:10.1111/j.1365-2222.2008.03156.x
71. Hanski I, von Hertzen L, Fyhrquist N, Koskinen K, Torppa K, Laatikainen T, et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc Natl Acad Sci U S A (2012) 109:8334–9. doi:10.1073/pnas.1205624109
72. Sherriff AJ. Hygiene levels in a contemporary population cohort are associated with wheezing and atopic eczema in preschool infants. Arch Dis Child (2002) 87:26–9. doi:10.1136/adc.87.1.26
73. Hesselmar B, Sjöberg F, Saalman R, Åberg N, Adlerberth I, Wold AE. Pacifier cleaning practices and risk of allergy development. Pediatrics (2013) 131:e1829–37. doi:10.1542/peds.2012-3345
74. Von Mutius E. Allergies, infections and the hygiene hypothesis – the epidemiological evidence. Immunobiology (2007) 212:433–9. doi:10.1016/j.imbio.2007.03.002
75. Benn CS, Melbye M, Wohlfahrt J, Björkstén B, Aaby P. Cohort study of sibling effect, infectious diseases, and risk of atopic dermatitis during first 18 months of life. Br Med J (2004) 328:1223–30. doi:10.1136/bmj.38069.512245.FE
76. Björkstén B. The hygiene hypothesis: do we still believe in it? Nestle Nutr Workshop Ser Pediatr Program (2009) 64:11–22; discussion 18–22. doi:10.1159/000235780
77. Kondrashova A, Hyoty H. Role of viruses and other microbes in the pathogenesis of type 1 diabetes. Int Rev Immunol (2014) 33(4):284–95. doi:10.3109/08830185.2014.889130
78. Hwang JS, Im CR, Im SH. Immune disorders and its correlation with gut microbiome. Immune Netw (2012) 12:129–38. doi:10.4110/in.2012.12.4.129
79. Lundell AC, Björnsson V, Ljung A, Ceder M, Johansen S, Lindhagen G, et al. Infant B cell memory differentiation and early gut bacterial colonization. J Immunol (2012) 188:4315–22. doi:10.4049/jimmunol.1103223
80. Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med (2002) 347:911–20. doi:10.1056/NEJMra020100
81. Bisgaard H, Li N, Bonnelykke K, Chawes BL, Skov T, Paludan-Muller G, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol (2011) 128:646–52. doi:10.1016/j.jaci.2011.04.060
82. Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol (2012) 129:434–40. doi:10.1016/j.jaci.2011.10.025
83. Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy (2014) 44:842–50. doi:10.1111/cea.12253
84. Joffe TH, Simpson NA. Cesarean section and risk of asthma. The role of intra-partum antibiotics: a missing piece? J Pediatr (2009) 154:154. doi:10.1016/j.jpeds.2008.08.039
85. Johnson CC, Ownby DR, Alford SH, Havstad SL, Williams L, Zoratti EM, et al. Antibiotic exposure in early infancy and risk for childhood atopy. J Allergy Clin Immunol (2005) 115:1218–24. doi:10.1016/j.jaci.2005.04.020
86. Carlisle EM, Morowitz MJ. The intestinal microbiome and necrotizing enterocolitis. Curr Opin Pediatr (2013) 25:382–7. doi:10.1097/MOP.0b013e3283600e91
87. Torraza RM, Neu J. The altered gut microbiome and necrotizing enterocolitis. Clin Perinatol (2013) 40:93–108. doi:10.1016/j.clp.2012.12.009
88. Hällström M, Eerola E, Vuento R, Janas M, Tammela O. Effects of mode of delivery and necrotising enterocolitis on the intestinal microflora in preterm infants. Eur J Clin Microbiol Infect Dis (2004) 23:463–70. doi:10.1007/s10096-004-1146-0
89. Strachan D. Hay fever, hygiene and household size. Br Med J (1989) 299:1259–66. doi:10.1136/bmj.299.6710.1259
90. Ownby DR, Johnson CC, Peterson EL. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. JAMA (2002) 288:963–72. doi:10.1001/jama.288.8.963
91. Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC, et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci U S A (2014) 111:805–10. doi:10.1073/pnas.1310750111
92. Forno E, Onderdonk AB, McCracken J, Litonjua AA, Laskey D, Delaney ML, et al. Diversity of the gut microbiota and eczema in early life. Clin Mol Allergy (2008) 6:11. doi:10.1186/1476-7961-6-11
93. Yap GC, Loo EX, Aw M, Lu Q, Shek LP, Lee BW. Molecular analysis of infant fecal microbiota in an Asian at-risk cohort-correlates with infant and childhood eczema. BMC Res Notes (2014) 7:166. doi:10.1186/1756-0500-7-166
94. Harder T, Bergmann R, Kallischnigg G, Plagemann A. Duration of breastfeeding and risk of overweight. A meta-analysis. Am J Epidemiol (2005) 162:397–403. doi:10.1093/aje/kwi222
95. Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother (2012) 56:5811–20. doi:10.1128/AAC.00789-12
96. Fouhy F, Ross RP, Fitzgerald G, Stanton C, Cotter PD. Composition of the early intestinal microbiota: knowledge, knowledge gaps and the use of high-throughput sequencing to address these gaps. Gut Microbes (2012) 3(3):203–20. doi:10.4161/gmic.20169
97. Abrahamsson TR, Rautava S, Moore AM, Neu J, Sherman PM. The time for a confirmative necrotizing enterocolitis probiotics prevention trial in extremely low birth weight infant in North America is now. J Paediatr (2014) 165:389–94. doi:10.1016/j.jpeds.2014.05.012
98. Daley D. The evolution of the hygiene hypothesis: the role of early life exposures to viruses and microbes and their relationship to asthma and allergic diseases. Curr Opin Allergy Clin Immunol (2014) 14(5):390–6. doi:10.1097/ACI.0000000000000101
99. Kalliomäki M, Antoine J-M, Herz U, Rijkers TG, Wells JM, Mercenier A. Guidance for substantiating the evidence for beneficial effects of probiotics: prevention and management of allergic diseases by probiotics. J Nutr (2010) 140:713S–21S. doi:10.3945/jn.109.113761
100. Isolauri E, Rautava S, Salminen S. Probiotics in the development and treatment of allergic disease. Gastroenterol Clin North Am (2012) 41:747–62. doi:10.1016/j.gtc.2012.08.007
101. Toh ZQ, Anzela A, Tang ML, Licciardi PV. Probiotic therapy as a novel approach for allergic disease. Front Pharmacol (2012) 3:171. doi:10.3389/fphar.2012.00171
Keywords: antibiotics, cesarean section, diet, gut microbiota, immunity, inflammatory diseases
Citation: Munyaka PM, Khafipour E and Ghia JE (2014) External influence of early childhood establishment of gut microbiota and subsequent health implications. Front. Pediatr. 2:109. doi: 10.3389/fped.2014.00109
Received: 27 June 2014; Accepted: 23 September 2014;
Published online: 09 October 2014.
Edited by:
Martin Gerbert Frasch, Université de Montréal, CanadaReviewed by:
Alain Stintzi, Ottawa Institute of Systems Biology, CanadaJoern-Hendrik Weitkamp, Vanderbilt University, USA
Copyright: © 2014 Munyaka, Khafipour and Ghia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean-Eric Ghia, University of Manitoba, 750 McDermot Avenue, Winnipeg, MB R3N 0T5, Canada e-mail:amVnaGlhQG1lZC51bWFuaXRvYmEuY2EsamVnaGlhQHlhaG9vLmZy
 Peris Mumbi Munyaka
Peris Mumbi Munyaka Ehsan Khafipour
Ehsan Khafipour Jean-Eric Ghia
Jean-Eric Ghia