- 1School of Public Health, Kalinga Institute of Industrial Technology (KIIT) Deemed to be University, Bhubaneswar, India
- 2Health Technology Assessment in India (HTAIn), ICMR-Regional Medical Research Centre, Bhubaneswar, India
- 3Health Technology Assessment in India (HTAIn), Department of Health Research, Ministry of Health & Family Welfare, Govt. of India, New Delhi, India
- 4All India Institute of Medical Sciences, Jodhpur, India
- 5Kalinga Institute of Dental Sciences, KIIT Deemed to be University, Bhubaneswar, India
- 6School of Computer Engineering, KIIT Deemed to be Uuniversity, Bhubaneswar, India
Objective: Oral cancer is a widespread global health problem characterised by high mortality rates, wherein early detection is critical for better survival outcomes and quality of life. While visual examination is the primary method for detecting oral cancer, it may not be practical in remote areas. AI algorithms have shown some promise in detecting cancer from medical images, but their effectiveness in oral cancer detection remains Naïve. This systematic review aims to provide an extensive assessment of the existing evidence about the diagnostic accuracy of AI-driven approaches for detecting oral potentially malignant disorders (OPMDs) and oral cancer using medical diagnostic imaging.
Methods: Adhering to PRISMA guidelines, the review scrutinised literature from PubMed, Scopus, and IEEE databases, with a specific focus on evaluating the performance of AI architectures across diverse imaging modalities for the detection of these conditions.
Results: The performance of AI models, measured by sensitivity and specificity, was assessed using a hierarchical summary receiver operating characteristic (SROC) curve, with heterogeneity quantified through I2 statistic. To account for inter-study variability, a random effects model was utilized. We screened 296 articles, included 55 studies for qualitative synthesis, and selected 18 studies for meta-analysis. Studies evaluating the diagnostic efficacy of AI-based methods reveal a high sensitivity of 0.87 and specificity of 0.81. The diagnostic odds ratio (DOR) of 131.63 indicates a high likelihood of accurate diagnosis of oral cancer and OPMDs. The SROC curve (AUC) of 0.9758 indicates the exceptional diagnostic performance of such models. The research showed that deep learning (DL) architectures, especially CNNs (convolutional neural networks), were the best at finding OPMDs and oral cancer. Histopathological images exhibited the greatest sensitivity and specificity in these detections.
Conclusion: These findings suggest that AI algorithms have the potential to function as reliable tools for the early diagnosis of OPMDs and oral cancer, offering significant advantages, particularly in resource-constrained settings.
Systematic Review Registration: https://www.crd.york.ac.uk/, PROSPERO (CRD42023476706).
1 Introduction
Cancer is a predominant cause of mortality and a major obstacle to enhancing global survival outcomes. Oral cancer, a critical global health issue, shows significant prevalence, with approximately 377,713 new cases and 177,757 deaths reported annually worldwide (1–3). The projections from the World Health Organisation (WHO) indicate that the rates of incidence and mortality of oral cancer in Asia are expected to rise to 374,000 and 208,000, respectively, by 2040 (4). OSCC (oral squamous cell carcinoma) is the most prevalent form of malignant neoplasm affecting the oral cavity, with low survival rates that vary among ethnicities and age groups. Despite advancements in cancer therapy, mortality rates for oral cancer remain elevated, with an overall 5-year survival rate of approximately 50% (5). Survival rates can reach 65% in high-income countries but drop to as low as 15% in some rural areas, depending on the affected part of the oral cavity (6). Early identification of oral cancer is vital for minimising both morbidity and mortality while optimising patient health and well-being. The diagnosis of pre-malignant and malignant oral cancer generally relies on a comprehensive patient history, thorough clinical examination, and histopathological verification of epithelial changes (7). The World Health Organisation (WHO) classification system stratifies epithelial dysplasia into mild, moderate, or severe categories, determined by the severity of cytological atypia and architectural disruption within the epithelial layer. Clinicians can evaluate the patient's prognosis and devise an appropriate treatment plan by correlating clinical observations with histological findings. Histopathological analysis remains the definitive standard for diagnosing oral potentially malignant disorders (OPMDs) (8). Currently, visual examination by a trained clinician is the primary detection method, but it is subject to variability due to lighting conditions and clinician expertise, which can reduce accuracy (9). In resource-limited environments, the scarcity of trained specialists and healthcare services impedes timely diagnosis and diminishes survival rates. Conventional oral examinations and biopsies, while gold standards, are not appropriate for screening in these areas (4). There is growing interest in using artificial intelligence (AI) models for the early screening of oral cancer in under-resourced and remote areas to address existing limitations.
AI is a rapidly evolving technology that helps with big data analysis, decision-making, and simulation of human thought processes (10). Deep learning, a subfield of AI, is concerned with convolutional neural networks (CNNs) that learn from large datasets and make accurate predictions, particularly in image classification and medical image analysis tasks (11, 12). Recent improvements in deep learning (DL) algorithms have shown that they are very good at finding cancerous lesions in medical imaging methods, such as CT scans for finding lung cancer and mammograms for checking for breast cancer (13). However, we have yet to fully investigate the potential for automatic detection of oral cancer in images. Disease detection through photographic and histopathological medical images is a crucial aspect of contemporary diagnostic medicine. Photographic imaging techniques, such as MRI (magnetic resonance imaging), CT (computed tomography), and x-rays, mobile captured lesions images enable non-invasive visualisation of internal structures, aiding in the detection and characterisation of various conditions (14).
Advancements in artificial intelligence applications further contribute to the analysis of these images, aiding in faster and more accurate disease detection. This multidimensional approach improves diagnostic precision, which leads to better treatment planning and patient outcomes. At present, there is an absence of a thorough quantitative assessment of the evidence regarding AI-based techniques for detecting oral cancer and OPMDs. This research intends to conduct a comprehensive review and meta-analysis of existing studies evaluating the effectiveness of AI algorithms for identifying both oral cancer and OPMDs.
2 Materials and methods
The systematic review was registered with the International Prospective Register of Systematic Reviews (PROSPERO), under Registration Number: CRD42023476706 (https://www.crd.york.ac.uk/prospero/display_record.php?RecordID=476706). The review adhered to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines.
2.1 Databases & search strategy
We conducted an extensive search of the literature to identify all relevant studies by systematically querying the electronic databases PubMed, Scopus, and IEEE. We included articles published in English up to December 31, 2023. The detailed search strategy related to the keywords and concepts “Machine Learning (ML),” “Deep Learning (DL),” “Artificial Intelligence (AI),” “Oral Cancer,” “Oral Pre-cancer,” “Oral Lesions,” and “Diagnostic Medical Images.” We combined each concept's MeSH terms and keywords with “OR” and then joined the concepts with the “AND” Boolean operator. Specific search strategies were tailored for each database (Supplementary File S1).
Two separate reviewers conducted the study screening based on established eligibility criteria, with the literature being organised using EndNote X9.3.3 (Clarivate Analytics, London, UK). Repeated or non-relevant studies were excluded from consideration. In the initial screening phase, the reviewers evaluated the titles and abstracts of articles, classifying them as relevant, irrelevant, or uncertain. Articles considered irrelevant by both reviewers were removed, while those classified as uncertain underwent further review by a third reviewer. During the secondary screening, potentially eligible articles identified from the initial review were assessed by two separate reviewers based on the eligibility criteria. Any disagreements during the full-text review were resolved by involving a third additional reviewer.
2.2 Eligibility criteria
This study includes original research articles focused on the use of AI technologies for diagnosing OPMDs and oral cancer through medical imaging. The included studies provide performance metrics such as sensitivity, specificity, and accuracy, or provide detailed data from the 2 × 2 confusion matrix, covering TP (true positives), TN (true negatives), FP (false positives), and FN (false negatives). Research articles were excluded based on the following criteria: repetition, irrelevant types (including preclinical studies, individual case reports, review articles, or conference proceedings), insufficient data, or lack of reporting on the specified outcomes. These standards were implemented to ensure the rigour and validity of the selected research while minimising potential biases and inaccuracies.
2.3 Data extraction and quality assessment
Two separate reviewers carried out the data extraction process, and any discrepancies were resolved by consulting a third additional reviewer. Data were retrieved using a predefined, pre-tested data extraction sheet designed for this study. The sheet included detailed information on author details, year of publication, image types, machine learning and deep learning models, country, TP, TN, FP, FN, sensitivity, accuracy, specificity, continent, World Bank income groups, WHO region, source of collected dataset, and dataset link. Any discrepancies in data retrieval were addressed through consensus among the entire research team.
In instances of missing or incomplete data, the lead authors of the included studies were reached out to via email. The quality of the included studies was assessed using the Quality Assessment of Diagnostic Accuracy Studies-AI (QUADAS-AI) criteria (15), with evaluations conducted by two independent reviewers. This guideline addresses the risk of bias through four domains: patient selection, index test, reference standard, flow and timing, and applicability concerns through three domains: patient selection, index test, and reference standard. The quality of the methodology employed in the included studies was evaluated using the QUADAS-AI tool in Microsoft Excel (Student—version 365, USA).
2.4 Statistical analysis
The performance of the AI models was assessed through a hierarchical summary receiver-operating characteristic (SROC) curve, which generated combined curves with 95% confidence intervals focused on average sensitivity (SE), specificity (SP), diagnostic odds ratio (DOR), and area under the curve (AUC) estimates. When several AI architectures were evaluated within a single study, the system demonstrating the greatest accuracy or the most comprehensive 2 × 2 confusion matrix was incorporated into the overall meta-analysis.
To enhance the robustness of the results, both positive and negative likelihood ratios (LR + and LR-) were calculated, providing valuable insights into the test's capacity to confirm or exclude a diagnosis across different clinical scenarios and translating its diagnostic performance into practical clinical decision-making. Heterogeneity among the studies was evaluated with the I2 statistic, followed by subgroup analyses to pinpoint the sources of variability. The subgroup analyses included five categories: (1) different AI models (e.g., CNN, VGG, FCN, ResNet, proposed hybrid models, and others); (2) various image types (e.g., histopathological images, photographic images, and optical coherence tomography); (3) diagnostic categories (oral cancer, OPMD, and both); (4) country income levels (high-income wise, upper-middle income and lower-middle income wise); and 5) WHO regions (Americas, Eastern Mediterranean, South-East Asia). All statistical meta-analyses were conducted using MetaDisc (version 1.4, Spain) with a two-tailed significance level of 0.05 (α = 0.05). A cross-hairs plot was constructed using Python (V.3.8.18, Netherlands) to present the discrepancies between sensitivity and specificity estimates (16).
3 Results
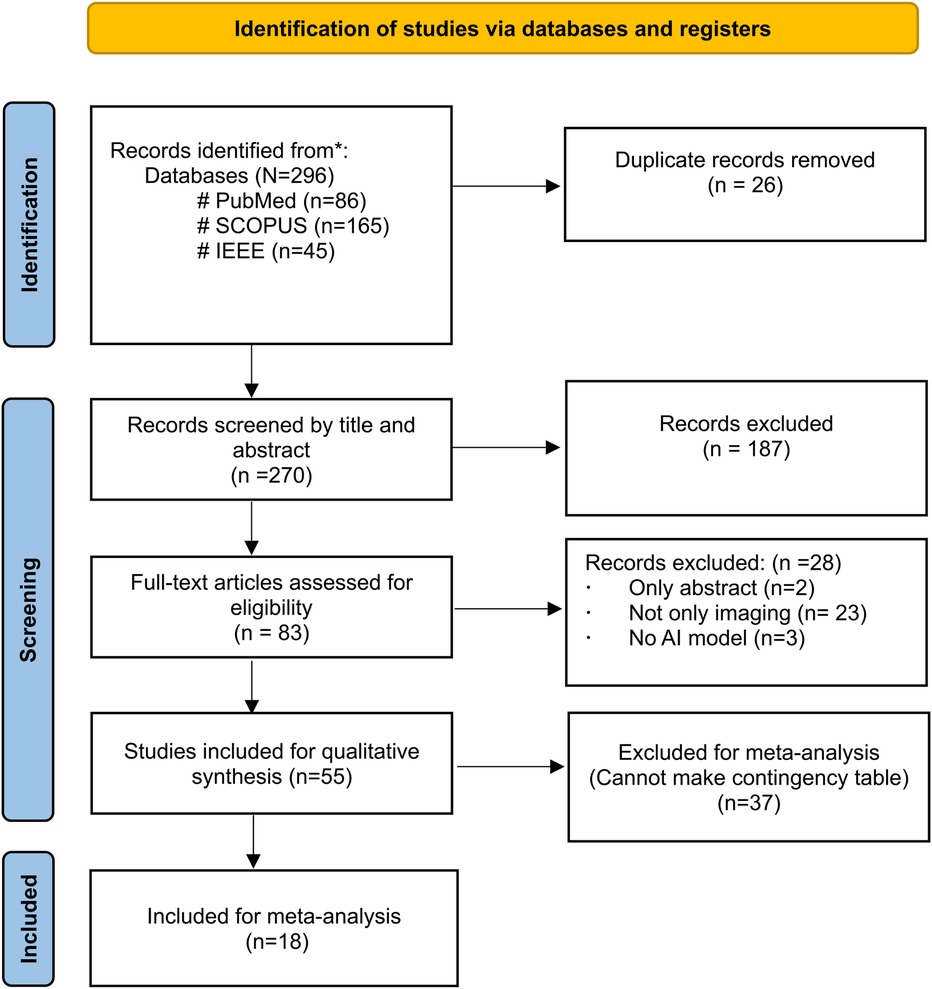
Figure 1 depicts the PRISMA flow diagram for a detailed search and selection of relevant studies. The initial search identified 296 articles. After removing duplicates, 270 were chosen for primary screening. Out of these, 83 were suitable for full-text assessment. The final review included 55 studies (4, 6, 17–69), with only 18 studies considered for the meta-analysis (6, 19–22, 26–31, 36, 44–48, 52–56, 65, 66).
3.1 Characteristics of the included studies
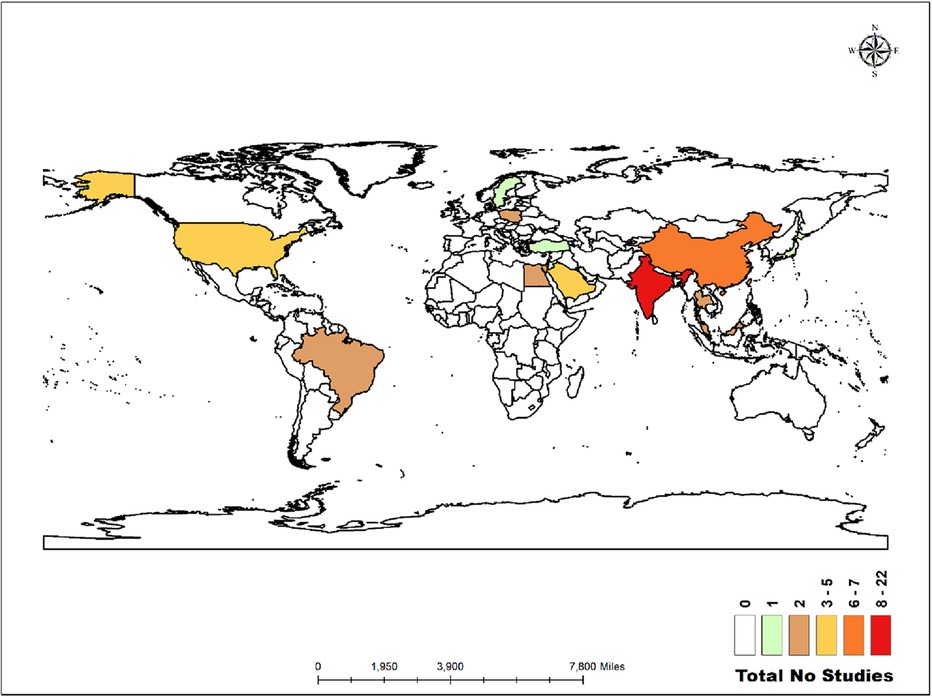
The world map in Figure 2 illustrates the distribution of the studies analysed in this review. Twenty-two studies were conducted in India (4, 17, 22, 23, 25, 28, 29, 31, 32, 39, 48, 51, 53, 55–59, 61–63, 67). Similarly, there were seven studies in China (6, 30, 42–44, 50, 52), five in the United States (24, 40, 47, 65, 69), five in Saudi Arabia (19, 20, 27, 36, 37), and two each in Malaysia (21, 54), Thailand (41, 46), Taiwan (60, 66), Egypt (18, 26), Brazil (45, 68), and Poland (34, 35). Additionally, there was one study each in Japan (38), Jordan (33), Türkiye (49), and Sweden (64).
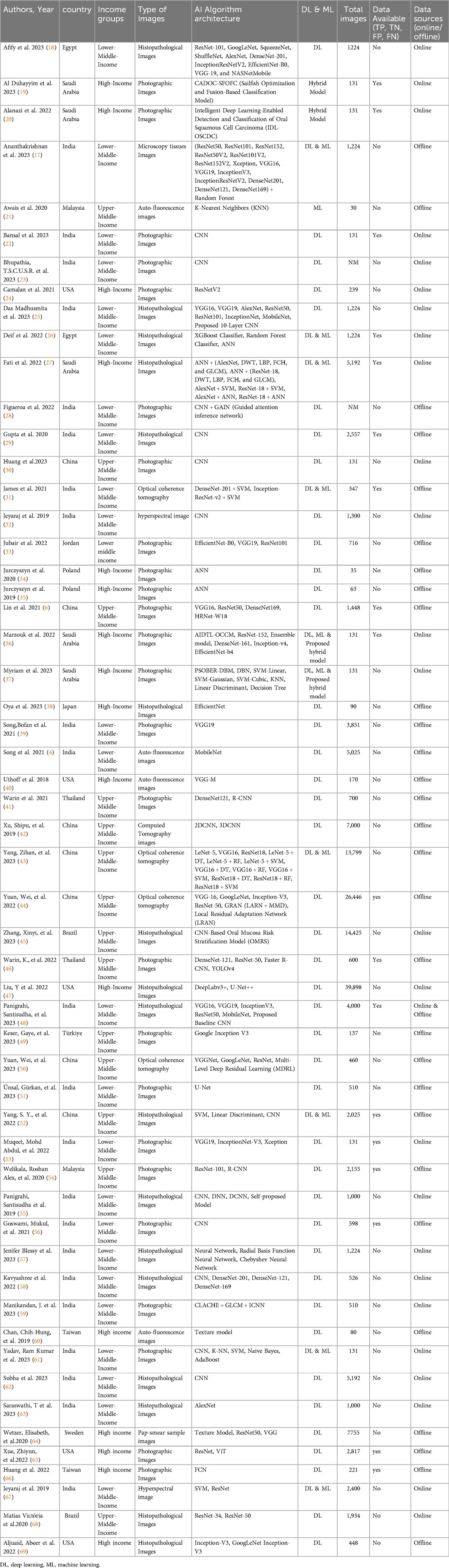
Out of 55 studies, 29 utilised offline patient data from outpatient clinics and inpatient settings across various hospital databases, while 25 relied on online databases. One study incorporated data from both online and offline sources. The studies employed various diagnostic imaging modalities, including photographic images (n = 25), histopathological images (n = 17), optical coherence tomography (OCT) images (n = 4), autofluorescence images (n = 4), hyperspectral images (n = 2), and one study each used pap smear images, microscopy tissue images, and computed tomography. A total of 42 studies utilised deep learning (DL) models, while one study employed a machine learning (ML) model. Eight studies integrated both DL and ML hybrid techniques for feature extraction and classification. Additionally, two studies developed and proposed their hybrid models, and another two studies also proposed their own hybrid models, comparing their performance with pre-trained DL models for classification. The proposed hybrid models include CADOC-SFOFC, IDL-OSCDC, AIDTL-OCCM, and PSOBER-DBM which blend machine learning and deep learning techniques to enhance predictive model, pattern recognition, and classification. Meanwhile, the studies also explored various DL architectures for image classification, such as CNNs and specialized models like ANN, VGG, ResNet, Fully Convolutional Networks (FCNs), etc. Detailed characteristics of these studies are provided in Table 1.
The studies represented a diverse range of settings, with 25 originating from low- and middle-income countries, 14 from upper-middle-income countries, and 16 from high-income countries. In all included studies, retrospective and online data sources provided pre-annotated datasets, whereas datasets collected prospectively were annotated by specialist dentists. Included studies validated their AI models using internal datasets, with one study additionally performing external validation with experts. The studies focused on validating AI algorithms across various imaging modalities, using metrics such as TP, TN, FP, FN, sensitivity, specificity, and AUC.
3.2 Quality assessment
The quality assessment of the studies was assessed using the QUADAS-AI tool (Supplementary File S2). The comprehensive assessment results are depicted in a diagram in the Supplementary Figure. A total of 14 studies showed a low risk of bias in patient selection, while 15 studies demonstrated proper flow and timing management. However, eight studies were at high risk of bias in the index test due to insufficient blinding and inconsistencies. For the reference standard, 10 studies were classified as having a low risk of bias, whereas eight studies exhibited varying levels of risk. Applicability concerns were low during in-patient selection (n = 16), but higher for the index test (n = 7). Many studies demonstrated robustness in multiple domains; however, significant issues were identified in the index test and reference standard, highlighting areas for improvement in future research designs.
3.3 Meta-analysis: pooled performance of AI algorithms
The study evaluates diagnostic accuracy across 18 studies, revealing a high sensitivity of 0.87, identifying 87% of true positive cases, and a specificity of 0.81 recognising 81% of true negative cases. The DOR of 131.63 reflects a strong likelihood of accurate diagnosis, while the SROC curve with an AUC of 0.9758 indicates exceptional diagnostic performance, highlighting the nearly perfect accuracy of the models (Supplementary File S3). These results confirm the reliability and robustness of AI algorithms for precise diagnostic applications. The detailed comparative analysis of pooled sensitivity, specificity, and diagnostic odds ratio (DOR), and the likelihood ratio of various AI Models for detecting oral cancer categorised by image type, oral conditions, and WHO regions are detailed in Table 2.
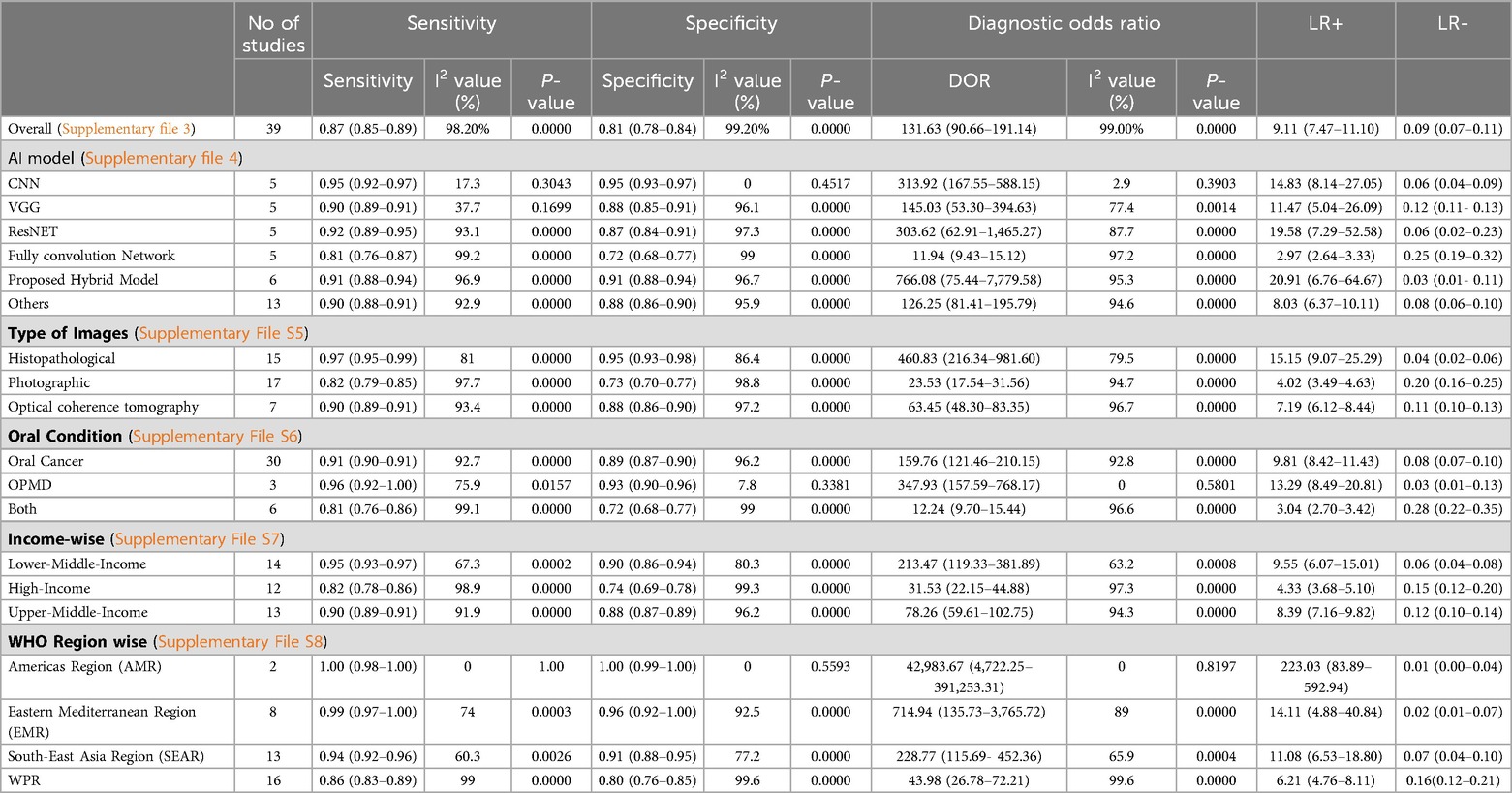
Table 2. Comparative analysis of pooled sensitivity, specificity, diagnostic odds ratio (DOR), and likelihood ratio of Various AI models for detecting OPMDs & oral cancer, categorized by image types, oral conditions, income-wise and wHO regions.
Histopathological images, evaluated in 15 studies, demonstrated the highest sensitivity and specificity, with values of 97% (95% CI: 95%–99%) and 95% (95% CI: 93%–98%), respectively. These images also had a significantly high diagnostic odds ratio (DOR) of 460.83 (95% CI: 216.34–981.60) and an area under the summary receiver operating characteristic (SROC) curve (AUC) of 0.9886. Photographic images, assessed in 17 studies, showed lower sensitivity and specificity at 82% (95% CI: 79%–85%) and 73% (95% CI: 70%–77%), respectively, with a DOR of 23.53 (95% CI: 17.54–31.56) and an AUC of 0.9715 (Figure 3). Optical coherence tomography, evaluated in 7 studies, had a sensitivity of 90% (95% CI: 89%–91%) and specificity of 88% (95% CI: 86%–90%), with a DOR of 63.45 (95% CI: 48.30–83.35) and an AUC of 0.9527 (Supplementary File S5).
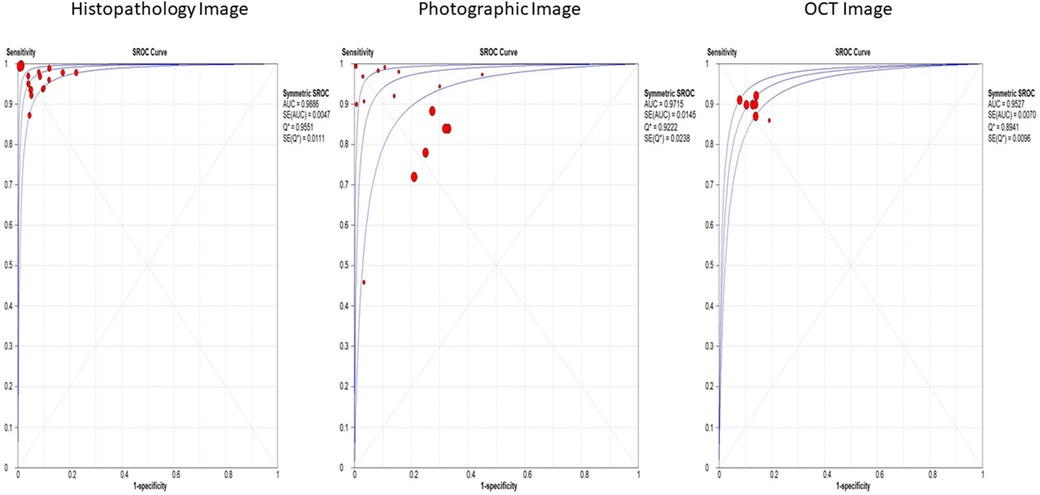
Figure 3. Overall SROC plot for various image-based diagnostic performance of artificial intelligence algorithms for detecting OPMDs & oral cancer.
The crosshair plot below depicts the relationship between the FPR (x-axis) and sensitivity (y-axis) for various data points represented by different colors. It was observed that the majority of the data points were clustered in the top left corner of the plot, indicating high sensitivity (above 0.8) and low FPR (less than 0.3). This suggests that the tested models perform well in terms of accurately identifying TP while generating a low number of FP. A few outliers with lower sensitivity and higher FPR exist, indicating poorer performance in those cases. However, the error bars on each data point show the variability or uncertainty in the measurements (Figure 4).
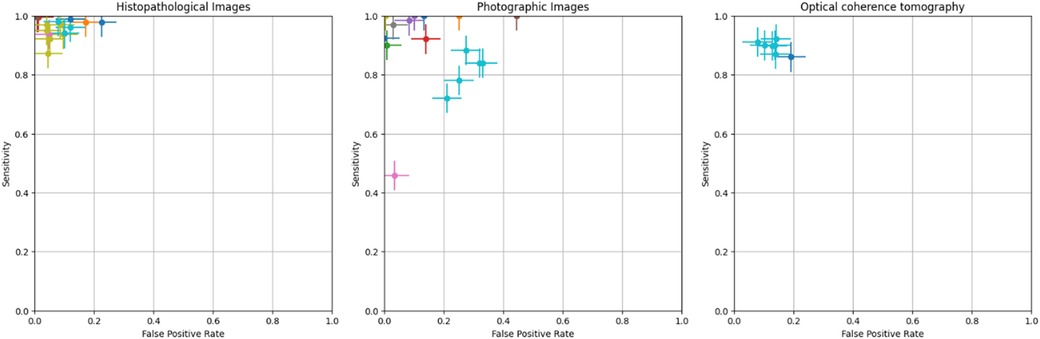
Figure 4. Crosshair plot for various image-based diagnostic performance of artificial intelligence algorithms for detecting OPMDs & oral cancer.
This study categorised and analysed various AI models used for medical image classification, focusing on their performance. The CNN showed high sensitivity and specificity, with a DOR of 313.92 and an AUC of 0.9846. VGG models exhibited slightly reduced sensitivity and specificity, with a DOR of 145.03 and an AUC of 0.9539. ResNet models demonstrated impressive performance, achieving a sensitivity of 92% and specificity of 87%. Fully convolutional networks had lower performance with a sensitivity of 81% and specificity of 72%. The hybrid AI model for enhanced accuracy showed impressive results, with a sensitivity of 91% and specificity of 91%. Other models, integrating various machine learning techniques and deep learning architectures, demonstrated comparable results. (Supplementary File S4).
Additionally, the study found that oral cancer conditions have a sensitivity of 91% and specificity of 89%, with a diagnostic odds ratio of 159.76 and an AUC of 0.9850. OPMD has a sensitivity of 96% and specificity of 93%, with a DOR of 347.93 and AUC of 0.9849 (Supplementary File S6). In terms of income groups, as classified by the World Bank, diagnostic performance varies: lower-middle-income countries have a sensitivity of 95% and specificity of 90%, while high-income countries exhibit a sensitivity of 82% and specificity of 74%. Upper-middle-income countries show a sensitivity of 90% and a specificity of 88% (Supplementary File S7). Regionally, the Americas Region demonstrated the highest sensitivity and specificity, followed by the Eastern Mediterranean Region, Southeast Asia Region, and Western Pacific Region (Supplementary File S8).
3.4 Heterogeneity analysis
A meta-analysis of 18 studies demonstrated that AI models are effective in diagnosing OPMDs and oral cancer using medical diagnostic images, as indicated by a random-effects model analysis. Nevertheless, substantial heterogeneity was observed among the studies, with sensitivity exhibiting an I2 of 98.2% and specificity showing an I2 of 99.2% (p < 0.01). Detailed results from subgroup analyses, which address the potential sources of inter-study variability, are presented in Table 2.
4 Discussion
This review presents a comprehensive meta-analysis of AI algorithms in medical imaging, specifically focusing on screening for OPMDs and oral cancer. Majority of studies utilised patient data that was collected offline and employed advanced deep learning architectures, such as CNNs, VGG, ResNET, etc. to analyse visual data. The findings indicate that AI algorithms exhibit a high level of diagnostic accuracy in detecting both oral cancer and OPMDs through medical imaging. The pooled sensitivity and specificity were 87% and 81%, respectively, indicating high diagnostic accuracy. Deep learning algorithms, a subfield of AI, have achieved remarkable success in disease classification through the analysis of various medical images. AI-driven medical diagnostic images have proven to be highly accurate and reliable in detecting tuberculosis, as well as cervical, and breast cancer. In tuberculosis detection, deep learning systems analysing chest x-rays have achieved a sensitivity of more than 95%, significantly reducing radiologists’ workload and enabling timely diagnosis (70). In breast cancer detection, AI models interpreting mammograms have outperformed human experts by reducing both false positives and false negatives (71, 72). Similarly, in cervical cancer, AI-based histopathological image analysis has demonstrated a sensitivity of 91%, highlighting its robustness in disease classification (72). The CNN model demonstrated the highest performance, achieving a sensitivity and specificity of 95% in this review. This was particularly notable when compared to other models such as VGG, ResNet, Inception, etc. which, despite being trained with a large number of parameters and being computationally efficient, did not perform as well. Many studies combine machine learning and deep learning techniques to create hybrid models, which also achieved impressive results, with a sensitivity of more than 95% (17).
AI algorithms demonstrated a sensitivity of 95% and specificity of 90% LIMCs compared to all income groups. This suggests that AI models can be effectively trained and utilised in diverse economic settings, potentially offering higher diagnostic accuracy in LMICs where traditional diagnostic resources are scarce. The implementation of AI-enabled portable devices for screening pre-malignant oral lesions may reduce the disease burden and improve the survival rate of oral cancer patients in LMICs (73). A scoping review by Adeoye, John, et al. highlighted the growing application of machine learning to model cancer outcomes in lower-middle-income regions (74). It revealed significant gaps in model development and recommended retraining models with the help of larger datasets; it also emphasised the need to enhance external validation techniques and conduct more impact assessments through randomised controlled trials.
Data is crucial for training AI systems (75). Advanced processing technologies applied to radiology report databases can enhance search and retrieval, aiding diagnostic efforts (76). In this study, we observed that research frequently utilises data from various online sources; however, the datasets are often limited in size and predominantly derived from common databases. Out of 55 studies, 26 used data from different online databases, with many sourcing data from the Kaggle repository, and others from personal medical databases, GitHub, and online libraries. Advocating for globally interconnected networks that aggregate diverse patient data is essential to optimise AI's capabilities, particularly for diseases like OPMDs and Oral Cancer, which require varied image databases. Effective curation of well-annotated medical data into large-scale databases is vital (77). However, inadequate curation remains a significant barrier to AI development (78). Proper curation—encompassing patient selection and image segmentation—ensures high-quality, error-free data and mitigates inconsistencies from varied data collection methods and imaging protocols (78, 79). Global collaborative initiatives, such as The Cancer Imaging Archive which creates extensive labelled datasets, are key to addressing this issue.
In our systematic review, 18 of the 55 studies meeting inclusion criteria provided relevant data for developing contingency tables. Metrics such as precision, F1 score, and recall, while standard in computer science, are insufficient alone for this purpose (80). Additionally, heatmaps from AI models highlight important image features for classification; they also help in the reduction of bias. However, only one-third of studies provide this information (81). Therefore, future AI-based research should prioritise establishing clear and well-defined metrics that bridge the disciplines of healthcare and computer science. In this review, we observed that the same terms are often defined inconsistently across different studies. For instance, the term “validation” is sometimes used to refer to the dataset used for evaluating model performance (82). Most research indicated that training an AI model typically involved dividing the dataset into training and testing subsets. Altman et al. recommended the use of internal validation sets for in-sample assessments and external validation sets for out-of-sample evaluations to enhance the quality of the study (83).
Histopathological imagery demonstrated superior sensitivity and specificity, while photographic images exhibited reduced accuracy. Given that the photos utilised for training deep learning models may not encompass the complete spectrum of oral disease presentations, the algorithm might encounter difficulties in consistently identifying various forms of oral lesions (84). Sub-group analysis revealed histopathological images had the highest DOR (460.83; and sensitivity 0.97), followed by OCT images (DOR 63.45; sensitivity 0.90) and photographic images (DOR 23.53; sensitivity 0.82), differing from previous reviews (85). In AI with deep learning, images are analysed to screen and detect diseases with exceptional accuracy (86) However, medical diagnostic images often reveal significant intra-class homogeneity, which complicates the extraction of nuanced features essential for precise predictions. Additionally, the relatively small size of these datasets compared to natural image datasets restricts the direct application of advanced modelling techniques. Utilising specialised knowledge and contextually relevant features can support the refinement of feature representations and alleviate model complexity, thereby advancing performance in the realm of medical diagnostic imaging (87). Most studies lacked guidelines for image data preparation before training models, Notably, Lin et al. offered a comprehensive procedure for capturing images, using a phone camera grid to ensure the lesion is centred, thus minimising focal length issues in oral photographic images (6).
Despite AI's potential in radiology, challenges persist, such as improving interpretability, reliability, and generalizability. AI's opaque decision-making limits clinical acceptance, requiring further validation through large-scale multicentre studies (88). Effective AI implementation on the one hand can reduce the unnecessary time being invested in conducting procedures, and facilitate early detection as well as improve patient outcomes on the other hand.
This study's constraints may influence both the understanding and broader application of the findings. First, the meta-analysis relies on published literature, which, despite thorough searches, may be subject to publication and language biases, especially because we included studies published only in English. Second, differences in study scale, methodological approaches, and evaluation metrics across studies have introduced inconsistencies that might influence the findings. Despite the execution of sensitivity analyses, the impact of this heterogeneity cannot be entirely discounted. Moreover, variations in imaging tools, equipment standards, and methodologies among studies could affect diagnostic accuracy.
5 Conclusion
This review highlights the high accuracy of AI algorithms in diagnosing oral cancer and OPMDs through medical imaging. The findings demonstrate that AI is a reliable approach for early detection, particularly in resource-limited settings. The successful integration of AI-based diagnostics, utilising various imaging modalities, highlights its potential. The widespread use of mobile devices has further expanded the accessibility of this technology, providing crucial healthcare support where specialised medical care is limited. Achieving precise image-based diagnosis with AI requires standardised methodologies and large-scale, multicentric studies. Such measures are significant for ensuring the accuracy and efficiency of screening processes and enhancing overall healthcare outcomes.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
RS: Writing – original draft. KS: Methodology, Writing – review & editing. GD: Formal Analysis, Methodology, Writing – review & editing. GK: Resources, Writing – review & editing. SB: Resources, Writing – review & editing. BP: Supervision, Writing – review & editing. SP: Writing – review & editing, Supervision.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/froh.2024.1494867/full#supplementary-material
References
1. Chi AC, Day TA, Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma–an update. CA Cancer J Clin. (2015) 65(5):401–21. doi: 10.3322/caac.21293
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71(3):209–49. doi: 10.3322/caac.21660
3. Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. (2009) 45(4–5):309–16. doi: 10.1016/j.oraloncology.2008.06.002
4. Song B, Sunny S, Li S, Gurushanth K, Mendonca P, Mukhia N, et al. Mobile-based oral cancer classification for point-of-care screening. J Biomed Opt. (2021) 26(6):065003. doi: 10.1117/1.JBO.26.6.065003
5. Jemal A, Ward EM, Johnson CJ, Cronin KA, Ma J, Ryerson B, et al. Annual report to the nation on the Status of cancer, 1975–2014, featuring survival. J Natl Cancer Inst. (2017) 109(9). doi: 10.1093/jnci/djx030
6. Lin H, Chen H, Weng L, Shao J, Lin J. Automatic detection of oral cancer in smartphone-based images using deep learning for early diagnosis. J Biomed Opt. (2021) 26(8):086007. doi: 10.1117/1.JBO.26.8.086007
7. Kumari P, Debta P, Dixit A. Oral potentially malignant disorders: etiology, pathogenesis, and transformation into oral cancer. Front Pharmacol. (2022) 13:825266. doi: 10.3389/fphar.2022.825266
8. Mello FW, Miguel AFP, Dutra KL, Porporatti AL, Warnakulasuriya S, Guerra ENS, et al. Prevalence of oral potentially malignant disorders: a systematic review and meta-analysis. J Oral Pathol Med. (2018) 47(7):633–40. doi: 10.1111/jop.12726
9. Tanriver G, Soluk Tekkesin M, Ergen O. Automated detection and classification of oral lesions using deep learning to detect oral potentially malignant disorders. Cancers (Basel). (2021) 13(11). doi: 10.3390/cancers13112766
10. Chen YW, Stanley K, Att W. Artificial intelligence in dentistry: current applications and future perspectives. Quintessence Int. (2020) 51(3):248–57. doi: 10.3290/j.qi.a43952
11. Hinton G. Deep learning-A technology with the potential to transform health care. JAMA. (2018) 320(11):1101–2. doi: 10.1001/jama.2018.11100
12. Miotto R, Wang F, Wang S, Jiang X, Dudley JT. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform. (2018) 19(6):1236–46. doi: 10.1093/bib/bbx044
13. Shen L, Margolies LR, Rothstein JH, Fluder E, McBride R, Sieh W. Deep learning to improve breast cancer detection on screening mammography. Sci Rep. (2019) 9(1):12495. doi: 10.1038/s41598-019-48995-4
14. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. (2017) 42:60–88. doi: 10.1016/j.media.2017.07.005
15. Sounderajah V, Ashrafian H, Rose S, Shah NH, Ghassemi M, Golub R, et al. A quality assessment tool for artificial intelligence-centered diagnostic test accuracy studies: QUADAS-AI. Nat Med. (2021) 27(10):1663–5. doi: 10.1038/s41591-021-01517-0
16. Phillips B, Stewart LA, Sutton AJ. ‘Cross hairs’ plots for diagnostic meta-analysis. Res Synth Methods. (2010) 1(3-4):308–15. doi: 10.1002/jrsm.26
17. Ananthakrishnan B, Shaik A, Kumar S, Narendran SO, Mattu K, Kavitha MS. Automated detection and classification of oral squamous cell carcinoma using deep neural networks. Diagnostics (Basel). (2023) 13(5). doi: 10.3390/diagnostics13050918
18. Afify HM, Mohammed KK, Ella Hassanien A. Novel prediction model on OSCC histopathological images via deep transfer learning combined with grad-CAM interpretation. Biomed Signal Process Control. (2023) 83. doi: 10.1016/j.bspc.2023.104704
19. Al Duhayyim M, Malibari AA, Dhahbi S, Nour MK, Al-Turaiki I, Obayya M, et al. Sailfish optimization with deep learning based oral cancer classification model. Comput Syst Sci Eng. (2023) 45(1):753–67. doi: 10.32604/csse.2023.030556
20. Alanazi AA, Khayyat MM, Khayyat MM, Elamin Elnaim BM, Abdel-Khalek S. Intelligent deep learning enabled oral squamous cell carcinoma detection and classification using biomedical images. Comput Intell Neurosci. (2022) 2022:7643967. doi: 10.1155/2022/7643967
21. Awais M, Ghayvat H, Krishnan Pandarathodiyil A, Nabillah Ghani WM, Ramanathan A, Pandya S, et al. Healthcare professional in the loop (HPIL): classification of standard and oral cancer-causing anomalous regions of oral cavity using textural analysis technique in autofluorescence imaging. Sensors (Basel). (2020) 20(20). doi: 10.3390/s20205780
22. Bansal S, Jadon RS, Gupta SK. Lips and tongue cancer classification using deep learning neural network. 2023 6th International Conference on Information Systems and Computer Networks (ISCON) (2023). p. 1–3
23. Bhupathia TSCUSR, Jagadesh SVVD. A cloud based approach for the classification of oral cancer using CNN and RCNN. In: Recent Developments in Electronics and Communication Systems. Advances in Transdisciplinary Engineering. Amsterdam: IOS Press (2023). p. 359–66.
24. Camalan S, Mahmood H, Binol H, Araujo ALD, Santos-Silva AR, Vargas PA, et al. Convolutional neural network-based clinical predictors of oral dysplasia: class activation map analysis of deep learning results. Cancers (Basel). (2021) 13(6). doi: 10.3390/cancers13061291
25. Das M, Dash R, Mishra SK. Automatic detection of oral squamous cell carcinoma from histopathological images of oral Mucosa using deep convolutional neural network. Int J Environ Res Public Health. (2023) 20(3). doi: 10.3390/ijerph20032131
26. Deif MA, Attar H, Amer A, Elhaty IA, Khosravi MR, Solyman AAA. Diagnosis of oral squamous cell carcinoma using deep neural networks and binary particle swarm optimization on histopathological images: an AIoMT approach. Comput Intell Neurosci. (2022) 2022:6364102. doi: 10.1155/2022/6364102
27. Fati SM, Senan EM, Javed Y. Early diagnosis of oral squamous cell carcinoma based on histopathological images using deep and hybrid learning approaches. Diagnostics (Basel). (2022) 12(8). doi: 10.3390/diagnostics12081899
28. Figueroa KC, Song B, Sunny S, Li S, Gurushanth K, Mendonca P, et al. Interpretable deep learning approach for oral cancer classification using guided attention inference network. J Biomed Opt. (2022) 27(1):015001. doi: 10.1117/1.JBO.27.1.015001
29. Gupta RK, Kaur M, Manhas J. Cellular level based deep learning framework for early detection of dysplasia in oral squamous epithelium. Proceedings of ICRIC 2019. Lecture Notes in Electrical Engineering (2020). p. 137–49
30. Huang Q, Ding H, Razmjooy N. Optimal deep learning neural network using ISSA for diagnosing the oral cancer. Biomed Signal Process Control. (2023) 84. doi: 10.1016/j.bspc.2023.104749
31. James BL, Sunny SP, Heidari AE, Ramanjinappa RD, Lam T, Tran AV, et al. Validation of a point-of-care optical coherence tomography device with machine learning algorithm for detection of oral potentially malignant and malignant lesions. Cancers (Basel). (2021) 13(14). doi: 10.3390/cancers13143583
32. Jeyaraj PR, Samuel Nadar ER. Computer-assisted medical image classification for early diagnosis of oral cancer employing deep learning algorithm. J Cancer Res Clin Oncol. (2019) 145(4):829–37. doi: 10.1007/s00432-018-02834-7
33. Jubair F, Al-Karadsheh O, Malamos D, Al Mahdi S, Saad Y, Hassona Y. A novel lightweight deep convolutional neural network for early detection of oral cancer. Oral Dis. (2022) 28(4):1123–30. doi: 10.1111/odi.13825
34. Jurczyszyn K, Gedrange T, Kozakiewicz M. Theoretical background to automated diagnosing of oral leukoplakia: a preliminary report. J Healthc Eng. (2020) 2020:8831161. doi: 10.1155/2020/8831161
35. Jurczyszyn K, Kozakiewicz M. Differential diagnosis of leukoplakia versus lichen planus of the oral mucosa based on digital texture analysis in intraoral photography. Adv Clin Exp Med. (2019) 28(11):1469–76. doi: 10.17219/acem/104524
36. Marzouk R, Alabdulkreem E, Dhahbi S, Nour MK, Al Duhayyim M, Othman M, et al. Deep transfer learning driven oral cancer detection and classification model. Comput Mater Continua. (2022) 73(2):3905–20. doi: 10.32604/cmc.2022.029326
37. Myriam H, Abdelhamid AA, El-Kenawy E-SM, Ibrahim A, Eid MM, Jamjoom MM, et al. Advanced meta-heuristic algorithm based on particle swarm and al-biruni earth radius optimization methods for oral cancer detection. IEEE Access. (2023) 11:23681–700. doi: 10.1109/ACCESS.2023.3253430
38. Oya K, Kokomoto K, Nozaki K, Toyosawa S. Oral squamous cell carcinoma diagnosis in digitized histological images using convolutional neural network. J Dent Sci. (2023) 18(1):322–9. doi: 10.1016/j.jds.2022.08.017
39. Song B, Li S, Sunny S, Gurushanth K, Mendonca P, Mukhia N, et al. Classification of imbalanced oral cancer image data from high-risk population. J Biomed Opt. (2021) 26(10):105001. doi: 10.1117/1.JBO.26.10.105001
40. Uthoff RD, Song B, Sunny S, Patrick S, Suresh A, Kolur T, et al. Point-of-care, smartphone-based, dual-modality, dual-view, oral cancer screening device with neural network classification for low-resource communities. PLoS One. (2018) 13(12):e0207493. doi: 10.1371/journal.pone.0207493
41. Warin K, Limprasert W, Suebnukarn S, Jinaporntham S, Jantana P. Automatic classification and detection of oral cancer in photographic images using deep learning algorithms. J Oral Pathol Med. (2021) 50(9):911–8. doi: 10.1111/jop.13227
42. Xu S, Liu Y, Hu W, Zhang C, Liu C, Zong Y, et al. An early diagnosis of oral cancer based on three-dimensional convolutional neural networks. IEEE Access. (2019) 7:158603–11. doi: 10.1109/ACCESS.2019.2950286
43. Yang Z, Pan H, Shang J, Zhang J, Liang Y. Deep-learning-based automated identification and visualization of oral cancer in optical coherence tomography images. Biomedicines. (2023) 11(3). doi: 10.3390/biomedicines11030802
44. Yuan W, Cheng L, Yang J, Yin B, Fan X, Yang J, et al. Noninvasive oral cancer screening based on local residual adaptation network using optical coherence tomography. Med Biol Eng Comput. (2022) 60(5):1363–75. doi: 10.1007/s11517-022-02535-x
45. Zhang X, Gleber-Netto FO, Wang S, Martins-Chaves RR, Gomez RS, Vigneswaran N, et al. Deep learning-based pathology image analysis predicts cancer progression risk in patients with oral leukoplakia. Cancer Med. (2023) 12(6):7508–18. doi: 10.1002/cam4.5478
46. Warin K, Limprasert W, Suebnukarn S, Jinaporntham S, Jantana P. Performance of deep convolutional neural network for classification and detection of oral potentially malignant disorders in photographic images. Int J Oral Maxillofac Surg. (2022) 51(5):699–704. doi: 10.1016/j.ijom.2021.09.001
47. Liu Y, Bilodeau E, Pollack B, Batmanghelich K. Automated detection of premalignant oral lesions on whole slide images using convolutional neural networks. Oral Oncol. (2022) 134:106109. doi: 10.1016/j.oraloncology.2022.106109
48. Panigrahi S, Nanda BS, Bhuyan R, Kumar K, Ghosh S, Swarnkar T. Classifying histopathological images of oral squamous cell carcinoma using deep transfer learning. Heliyon. (2023) 9(3):e13444. doi: 10.1016/j.heliyon.2023.e13444
49. Keser G, Bayrakdar IS, Pekiner FN, Celik O, Orhan K. A deep learning algorithm for classification of oral lichen planus lesions from photographic images: a retrospective study. J Stomatol Oral Maxillofac Surg. (2023) 124(1):101264. doi: 10.1016/j.jormas.2022.08.007
50. Yuan W, Yang J, Yin B, Fan X, Yang J, Sun H, et al. Noninvasive diagnosis of oral squamous cell carcinoma by multi-level deep residual learning on optical coherence tomography images. Oral Dis. (2023) 29(8):3223–31. doi: 10.1111/odi.14318
51. Unsal G, Chaurasia A, Akkaya N, Chen N, Abdalla-Aslan R, Koca RB, et al. Deep convolutional neural network algorithm for the automatic segmentation of oral potentially malignant disorders and oral cancers. Proc Inst Mech Eng H. (2023) 237(6):719–26. doi: 10.1177/09544119231176116
52. Yang SY, Li SH, Liu JL, Sun XQ, Cen YY, Ren RY, et al. Histopathology-based diagnosis of oral squamous cell carcinoma using deep learning. J Dent Res. (2022) 101(11):1321–7. doi: 10.1177/00220345221089858
53. Muqeet MA, Mohammad AB, Krishna PG, Begum SH, Qadeer S, Begum N. Automated oral cancer detection using deep learning-based technique. 2022 8th International Conference on Signal Processing and Communication (ICSC) (2022). p. 294–7
54. Welikala RA, Remagnino P, Lim JH, Chan CS, Rajendran S, Kallarakkal TG, et al. Automated detection and classification of oral lesions using deep learning for early detection of oral cancer. IEEE Access. (2020) 8:132677–93. doi: 10.1109/ACCESS.2020.3010180
55. Panigrahi S, Swarnkar T. Automated classification of oral cancer histopathology images using convolutional neural network. 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2019 18-21 Nov (2019).
56. Goswami M, Maheshwari M, Baruah PD, Singh A, Gupta R. Automated detection of oral cancer and dental caries using convolutional neural network. 2021 9th International Conference on Reliability, Infocom Technologies and Optimization (Trends and Future Directions) (ICRITO) (2021). p. 1–5
57. Blessy J, Sornam M, Venkateswaran V. Consignment of epidermoid carcinoma using BPN, RBFN and chebyshev neural network. 2023 8th International Conference on Business and Industrial Research (ICBIR) (2023). p. 954–8
58. Kavyashree C, Vimala HS, Shreyas J. Improving oral cancer detection using pretrained model. 2022 IEEE 6th Conference on Information and Communication Technology (CICT) (2022). p. 1–5
59. Manikandan J, Krishna BV, Varun N, Vishal V, Yugant S. Automated framework for effective identification of oral cancer using improved convolutional neural network. 2023 Eighth International Conference on Science Technology Engineering and Mathematics (ICONSTEM) (2023). p. 1–7
60. Chan CH, Huang TT, Chen CY, Lee CC, Chan MY, Chung PC. Texture-map-based branch-collaborative network for oral cancer detection. IEEE Trans Biomed Circuits Syst. (2019) 13(4):766–80. doi: 10.1109/TBCAS.2019.2918244
61. Kumar Yadav R, Ujjainkar P, Moriwal R. Oral cancer detection using deep learning approach. 2023 IEEE International Students’ Conference on Electrical, Electronics and Computer Science (SCEECS) (2023). p. 1–7
62. Subha KJ, Bennet MA, Pranay G, Bharadwaj K, Reddy PV. Analysis of deep learning based optimization techniques for oral cancer detection. 2023 4th International Conference on Electronics and Sustainable Communication Systems (ICESC) (2023). p. 1550–5
63. Saraswathi T, Bhaskaran VM. Classification of oral squamous carcinoma histopathological images using alex net. 2023 International Conference on Intelligent Systems for Communication, IoT and Security (ICISCoIS) (2023). p. 637–43
64. Wetzer E, Gay J, Harlin H, Lindblad J, Sladoje N, editors. When texture matters: texture-focused cnns outperform general data augmentation and pretraining in oral cancer detection. In: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI); 2020 Apr 3–7; Iowa City, IA. Piscataway, NJ: IEEE (2020) p. 517–21. doi: 10.1109/ISBI45749.2020.9098424
65. Xue Z, Yu K, Pearlman PC, Pal A, Chen TC, Hua CH, et al. Automatic detection of oral lesion measurement ruler toward computer-aided image-based oral cancer screening. Annu Int Conf IEEE Eng Med Biol Soc. (2022) 2022:3218–21. doi: 10.1109/EMBC48229.2022.9871610
66. Huang S-Y, Chiou C-Y, Tan Y-S, Chen C-Y, Chung P-C. Deep oral cancer lesion segmentation with heterogeneous features. 2022 IEEE International Conference on Recent Advances in Systems Science and Engineering (RASSE) (2022). p. 1–8
67. Jeyaraj PR, Nadar ERS, Panigrahi BK. Resnet convolution neural network based hyperspectral imagery classification for accurate cancerous region detection. 2019 IEEE Conference on Information and Communication Technology; 2019 6-8 Dec (2019).
68. Victoria Matias A, Cerentini A, Buschetto Macarini LA, Atkinson Amorim JG, Perozzo Daltoe F, von Wangenheim A. Segmentation, detection and classification of cell nuclei on oral cytology samples stained with papanicolaou. 2020 IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS) (2020). p. 53–8
69. Aljuaid MA A, Anwar M. An early detection of oral epithelial dysplasia based on GoogLeNet inception-v3. 2022 IEEE/ACM Conference on Connected Health: Applications, Systems and Engineering Technologies (CHASE) (2022).
70. Qin ZZ, Sander MS, Rai B, Titahong CN, Sudrungrot S, Laah SN, et al. Using artificial intelligence to read chest radiographs for tuberculosis detection: a multi-site evaluation of the diagnostic accuracy of three deep learning systems. Sci Rep. (2019) 9(1):15000. doi: 10.1038/s41598-019-51503-3
71. McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, et al. International evaluation of an AI system for breast cancer screening. Nature. (2020) 577(7788):89–94. doi: 10.1038/s41586-019-1799-6
72. Song J, Im S, Lee SH, Jang HJ. Deep learning-based classification of uterine cervical and endometrial cancer subtypes from whole-slide histopathology images. Diagnostics (Basel). (2022) 12(11). doi: 10.3390/diagnostics12112623
73. Shrestha AD, Vedsted P, Kallestrup P, Neupane D. Prevalence and incidence of oral cancer in low- and middle-income countries: a scoping review. Eur J Cancer Care (Engl). (2020) 29(2):e13207. doi: 10.1111/ecc.13207
74. Adeoye J, Akinshipo A, Koohi-Moghadam M, Thomson P, Su YX. Construction of machine learning-based models for cancer outcomes in low and lower-middle income countries: a scoping review. Front Oncol. (2022) 12:976168. doi: 10.3389/fonc.2022.976168
75. Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts H. Artificial intelligence in radiology. Nat Rev Cancer. (2018) 18(8):500–10. doi: 10.1038/s41568-018-0016-5
76. Zhou LQ, Wang JY, Yu SY, Wu GG, Wei Q, Deng YB, et al. Artificial intelligence in medical imaging of the liver. World J Gastroenterol. (2019) 25(6):672–82. doi: 10.3748/wjg.v25.i6.672
77. Shimizu H, Nakayama KI. Artificial intelligence in oncology. Cancer Sci. (2020) 111(5):1452–60. doi: 10.1111/cas.14377
78. Geras KJ, Mann RM, Moy L. Artificial intelligence for mammography and digital breast tomosynthesis: current concepts and future perspectives. Radiology. (2019) 293(2):246–59. doi: 10.1148/radiol.2019182627
79. Ursprung S, Beer L, Bruining A, Woitek R, Stewart GD, Gallagher FA, et al. Radiomics of computed tomography and magnetic resonance imaging in renal cell carcinoma-a systematic review and meta-analysis. Eur Radiol. (2020) 30(6):3558–66. doi: 10.1007/s00330-020-06666-3
80. Jian J, Xia W, Zhang R, Zhao X, Zhang J, Wu X, et al. Multiple instance convolutional neural network with modality-based attention and contextual multi-instance learning pooling layer for effective differentiation between borderline and malignant epithelial ovarian tumors. Artif Intell Med. (2021) 121:102194. doi: 10.1016/j.artmed.2021.102194
81. Guimaraes P, Batista A, Zieger M, Kaatz M, Koenig K. Artificial intelligence in multiphoton tomography: atopic dermatitis diagnosis. Sci Rep. (2020) 10(1):7968. doi: 10.1038/s41598-020-64937-x
82. Gao Y, Zeng S, Xu X, Li H, Yao S, Song K, et al. Deep learning-enabled pelvic ultrasound images for accurate diagnosis of ovarian cancer in China: a retrospective, multicentre, diagnostic study. Lancet Digit Health. (2022) 4(3):e179–e87. doi: 10.1016/S2589-7500(21)00278-8
83. Altman DG, Royston P. What do we mean by validating a prognostic model? Stat Med. (2000) 19(4):453–73. doi: 10.1002/(SICI)1097-0258(20000229)19:4%3C453::AID-SIM350%3E3.0.CO;2-5
84. Fu Q, Chen Y, Li Z, Jing Q, Hu C, Liu H, et al. A deep learning algorithm for detection of oral cavity squamous cell carcinoma from photographic images: a retrospective study. EClinicalMedicine. (2020) 27:100558. doi: 10.1016/j.eclinm.2020.100558
85. Li J, Kot WY, McGrath CP, Chan BWA, Ho JWK, Zheng LW. Diagnostic accuracy of artificial intelligence assisted clinical imaging in the detection of oral potentially malignant disorders and oral cancer: a systematic review and meta-analysis. Int J Surg. (2024) 110(8):5034–46. doi: 10.1097/JS9.0000000000001469
86. Valente J, Antonio J, Mora C, Jardim S. Developments in image processing using deep learning and reinforcement learning. J Imaging. (2023) 9(10). doi: 10.3390/jimaging9100207
87. Chen X, Wang X, Zhang K, Fung KM, Thai TC, Moore K, et al. Recent advances and clinical applications of deep learning in medical image analysis. Med Image Anal. (2022) 79:102444. doi: 10.1016/j.media.2022.102444
Keywords: oral cancer, AI algorithms, diagnostic performance, deep learning, early detection, medical diagnostic imaging, OPMDs
Citation: Sahoo RK, Sahoo KC, Dash GC, Kumar G, Baliarsingh SK, Panda B and Pati S (2024) Diagnostic performance of artificial intelligence in detecting oral potentially malignant disorders and oral cancer using medical diagnostic imaging: a systematic review and meta-analysis. Front. Oral. Health 5:1494867. doi: 10.3389/froh.2024.1494867
Received: 11 September 2024; Accepted: 22 October 2024;
Published: 6 November 2024.
Edited by:
Ruwan Jayasinghe, University of Peradeniya, Sri LankaReviewed by:
Maria Georgaki, National and Kapodistrian University of Athens, GreeceJessica Maldonado-Mendoza, Metropolitan Autonomous University, Mexico
Copyright: © 2024 Sahoo, Sahoo, Dash, Kumar, Baliarsingh, Panda and Pati. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bhuputra Panda, Ymh1cHV0cmEucGFuZGFAZ21haWwuY29t
 Rakesh Kumar Sahoo
Rakesh Kumar Sahoo Krushna Chandra Sahoo
Krushna Chandra Sahoo Girish Chandra Dash
Girish Chandra Dash Gunjan Kumar
Gunjan Kumar Santos Kumar Baliarsingh6
Santos Kumar Baliarsingh6 Sanghamitra Pati
Sanghamitra Pati