- 1Department of Oral Medicine, Academic Centre for Dentistry Amsterdam, University of Amsterdam and VU University, Amsterdam, Netherlands
- 2Department of Oral and Maxillofacial Surgery, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands
- 3City of Hope Comprehensive Cancer Center, Duarte CA and Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical System, Los Angeles, CA, United States
Objectives: The human oral microbiome may play a role in the development of oral squamous cell carcinoma. The aim of this scoping review was to examine microbial diversity and differences in the composition of the oral microbiome between OSCC patients and healthy controls.
Methods: A literature search (in PubMed and Embase.com) was performed on January 9, 2023. The outcome variables used from the included studies of this review were alpha- and beta diversity and oral microbiome composition profiles for each taxonomic level (phylum-, class-, order-, genus- and species level).
Results: Thirteen out of 423 studies were included in this review compromising 1,677 subjects, of which 905 (54.0%) were OSCC patients and 772 (46.0%) were healthy controls. Most studies found a higher alpha diversity in the OSCC patient group and significantly different beta diversities between OSCC patient samples and healthy control samples. Studies reported more abundant Fusobacteria (on phylum level), Fusobacterium (on genus level), Fusobacterium nucleatum, Porphyromonas endodontalis and Prevotella intermedia (on species level) in OSCC patients. The healthy control group had more abundant Actinobacteria (on phylum level), Streptococcus and Veilonella (on genus level) and Veilonella parvula (on species level) according to most studies.
Conclusions: Our findings show differences in oral microbiome diversity and composition in OSCC patients. Clinical implications demand continuing study. Development of internationally accepted standard procedures for oral sample collection and oral microbiota analysis is needed for more conclusive and clinically relevant comparisons in future research.
Introduction
Oral squamous cell carcinoma (OSCC) is the most frequent malignancy in the oral cavity and one of the 10 most common cancers worldwide (1). Identified risk factors of OSCC include tobacco use, alcohol use and areca nut intake (2). Human papillomavirus (HPV) may also have oral malignant potential in OSCC development, specifically in younger patients without exposure to the main risk factors (3), although increasing cases in younger adults without the above potential risk factors remain to be defined.
The human oral microbiome is a complex comprised of more than 700 different species (4). The oral microbiome can be classified in different taxonomic levels: phylum-, class-, order-, family-, genus- and species level. The healthy oral cavity can be broadly categorized into six phyla: Firmicutes, Actinobacteria, Proteobacteria, Fusobacteria, Bacteroidetes and Spirochaetes constituting 96% of total oral bacteria (5). New technologies like next-generation sequencing and metagenomic shotgun sequencing have revealed the complexity of the human oral microbiome (4).
Oral diseases such as caries, oral mucositis, gingivitis and periodontitis are linked to dysbiotic shifts of the oral microbiome (6–8). The ecosystem of the mouth can become dysbiotic due to salivary changes, poor oral hygiene and lifestyle factors like diet, smoking, disease and stress (4). In addition, systemic medications use such as antibiotics, prednisone, cancer chemotherapy, and topicals such as steroids may lead to microbial shifts (9, 10–13). To combat oral diseases, an approach to treatment may be to re-establish symbiosis of the oral microbiome (8, 14).
Evidence that members of the human oral microbiome may be associated oral cancer is growing (15). In OSCC, carcinogenesis is hypothesized to begin with an alteration of the oral microbiome composition due to risk factors of oral cancer, like alcohol intake and tobacco use, leading to chronic inflammation. Bacterial products and metabolic by-products of the altered oral microbiome can induce permanent genetic alterations in epithelial cells. Further, genetically altered epithelial cells proliferate and apoptosis is inhibited leading to dysplasia and neoplastic cell proliferation. Finally, tumour cells infiltrate surrounding tissues and eventually metastasize (16). In this hypothesis OSCC might arise from genetic alterations which can be induced by micro-organisms present in the oral microbiome (17).
Individual microorganisms are thought to contribute to cancer risk through various pathways including promoting cell proliferation and invasion, promoting metastasis, influencing the tumour immune microenvironment, increasing chemoresistance, promote chronic inflammation (18). The role of the oral microbiome in OSCC has been increasingly recognized and studied in individual studies (16, 18). However, it remains unclear whether individual microorganisms or microbial signatures can be linked to OSCC. Therefore, the aim of this scoping review is to compare the microbial diversity and oral microbiome composition differences between OSCC patients and healthy controls.
Methods
A literature search was performed based on the Preferred Reporting Items for systematic Reviews and Meta-Analyses (PRISMA) statement (19). To identify all relevant publications, a systematic search in the bibliographic databases PubMed and EMBASE was conducted from inception to January 9, 2023. In PubMed, the following strings were combined: “Oral microbiome”[tiab] OR “mouth microbiome”[tiab] OR “oral bacteria”[tiab] OR “mouth bacteria”[tiab] OR “Microbiota”[Mesh] AND “Oral cancer”[tiab] OR “mouth cancer”[tiab] OR “cancer of the mouth”[tiab] OR “oral tumor”[tiab] OR “oral tumour”[tiab] OR “mouth tumor”[tiab] OR “mouth tumour”[tiab] OR “Mouth Neoplasms”[Mesh]. In EMBASE, the combined strings were: “Oral microbiome”:ti,ab,kw OR “mouth microbiome”:ti,ab,kw OR “oral bacteria”:ti,ab,kw OR “mouth bacteria”:ti,ab,kw OR “oral microbiome”/exp OR “mouth flora”/exp AND “oral cancer”:ti,ab,kw OR “mouth cancer”:ti,ab,kw OR “cancer of the mouth”:ti,ab,kw OR “oral tumor”:ti,ab,kw OR “oral tumour”:ti,ab,kw OR “mouth tumor”:ti,ab,kw OR “mouth tumour”:ti,ab,kw OR “mouth tumor”/exp OR “mouth cancer”/exp. Specific searches entered in PubMed and EMBASE are added in the Supplementary Appendix File.
The potentially relevant titles and abstracts yielded from the search were screened for eligibility using review manager Rayyan (20). Studies were included that met the following criteria: (a) studies that compare the oral microbiome of OSCC diagnosed patients with healthy controls; (b) observational studies (case-control, cohort, and cross-sectional studies); (c) written in English or Dutch. Studies were excluded if they were: (a) published before 2003; (b) in vitro studies, animal studies, letters or comments on articles, study protocols, preliminary studies, pilot studies, case series (<10 patients) or case reports.
The diversity of the oral microbiome composition in individual studies was reported as alpha- and/or beta diversity. Alpha diversity describes the species diversity/richness within a sample. For alpha diversity, different indices were used, including the Shannon-, Simpson-, InvSimpson-, Chao 1- and Faith's PD index. Beta diversity describes diversity between samples, in this case between OSCC patient samples and healthy control samples. To define beta diversity, Bray-Curtis dissimilarity was calculated. Data on oral microbiome composition profiles was categorized per taxonomic level, presented in relative abundance (%). For each taxonomic level (phylum-, class-, order-, genus- and species level), relative bacterial abundance differences between cases and controls were statistically tested in the individual studies. Only statistically significant differences (p < 0.05) were considered in this review.
Results
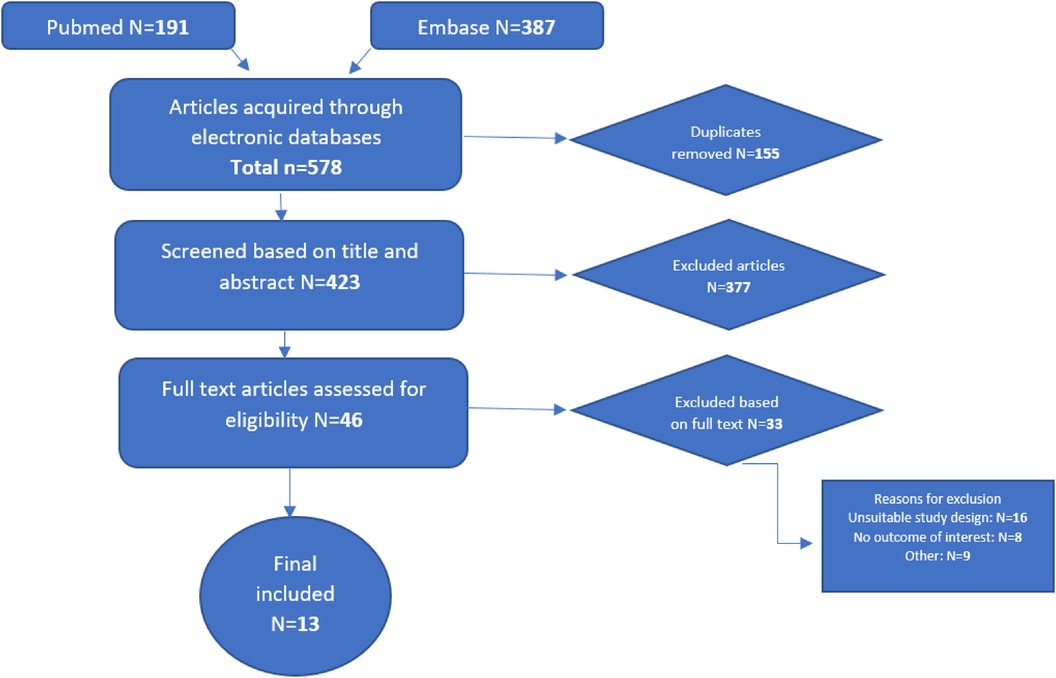
A total of 578 studies were retrieved, of which 155 duplicates were removed. After reading 423 abstracts, 46 full text studies were assessed for eligibility resulting in 13 studies included in this review (Figure 1).
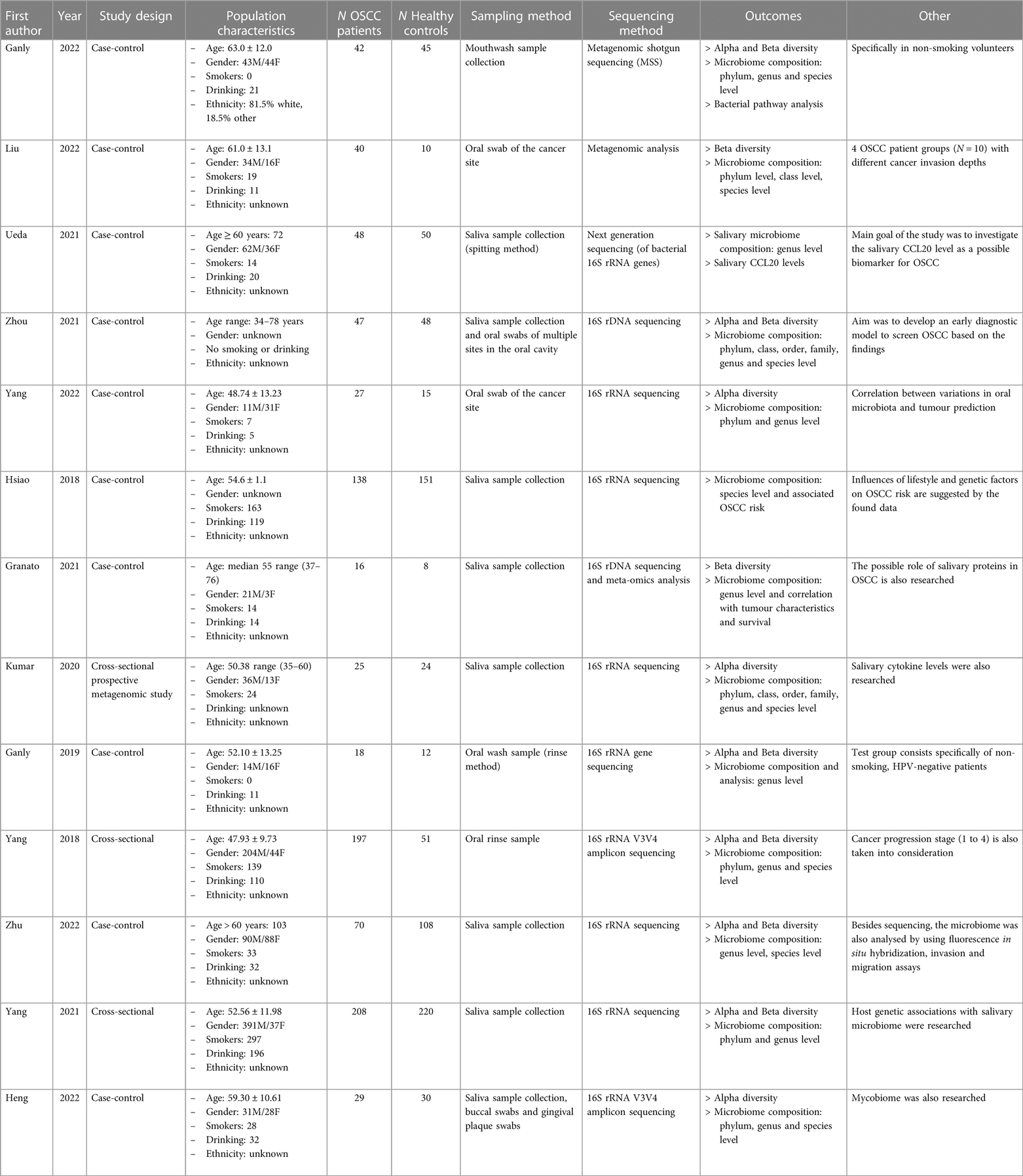
A total of 1,677 subjects from 13 studies were included, of which 905 (54.0%) were OSCC patients and 772 (46.0%) were healthy controls. Of all subjects, 937 (55.9%) were male, 356 (21.2%) were female, and of 384 (22.9%) subjects the gender was unknown. From all subjects, 738 reported smoking (44.0%) and 571 (34.0%) reported regular alcohol consumption. The sampling methods were performed by saliva sampling or via oral swabs. The sequencing method for 9 included studies was performed via 16S rRNA sequencing. Of the remaining studies, two studies performed sequencing via 16S rDNA sequencing, the other two studies used metagenomic shotgun sequencing. An overview of the study characteristics is presented in Table 1.
Alpha diversity from OSCC patients were compared to healthy controls in several studies. Alpha diversity was reported in nine of 13 included studies. In three studies, no significant difference in alpha diversity between groups was found (21–23). In five studies, alpha diversity was significantly higher in the OSCC patient group, compared to the healthy control group (24–28). On the other hand, in one study, the alpha diversity was significantly higher in the healthy control group, compared to the OSCC patient group (29).
Beta diversity was reported in eight studies. In six studies, beta diversity was significantly different between the OSCC patient group and controls (21, 23, 26–28, 30). One more study concluded a significant difference in beta diversity between groups, however, because of described population bias in this particular study, this result was omitted (24). In one study, no significant difference was found for the beta diversity between groups (26).
Seven of the 13 included studies reported differences in the relative abundance of the oral microbiome between OSCC patient samples and control patients on phylum level (21, 24–28, 30) (Figure 2). Only phyla that were significantly different between patients and controls in two or more studies were included in this graph. Raw results for each taxonomic level can be found in the Supplementary Appendix.
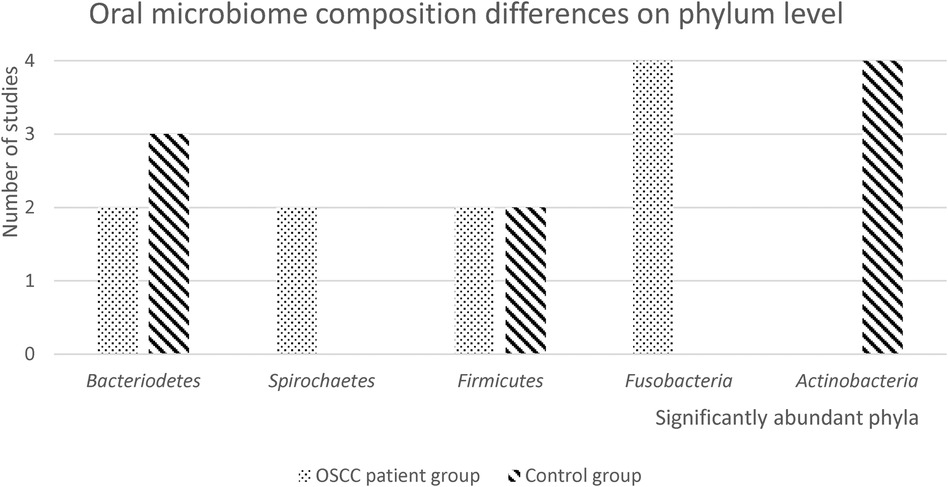
Figure 2. Oral microbiome composition differences on phylum level. In this figure, the number of studies with significantly abundant phyla in either the OSCC patient group or the healthy control group, is presented. Only significantly abundant phyla (p < 0.05), that were reported in 2 or more studies, are depicted.
Four studies found that Fusobacteria were significantly more abundant in OSCC patient samples, and four studies found that Actinobacteria were significantly more abundant in healthy control samples. Firmicutes were found to be significantly more abundant in both OSCC patient and control groups. Two studies reported that Spirochaetes were significantly more abundant in OSCC patient samples. Three studies found that Bacteriodetes were significantly more abundant in OSCC patient samples and two studies found they were more abundant in healthy control samples.
Differences between relative abundances of OSCC patient samples and healthy control samples on class level were reported in two out of the 13 included studies (24, 30). In one study, Flavobacteria and Spirochaetia were significantly more abundant in the OSCC patient group; Bacilli, Betaproteobacteria, Actinobacteria and Negativicutes were more abundant in the control group (30). In the other study, Negativicutes were significantly more abundant in the OSCC patient group; Bacilli, Bacterioidea and Betaproteobacteria were more abundant in the control group (24). On both family and order level, only one study reported data (24). Because of this limitation, results on family and order level were not included in this review.
Differences in the relative abundance at the genus level, were reported in 10 of the 13 included studies (21, 23–31).
The oral microbiome composition differences on genus level are presented in Figure 3. The genus Fusobacterium was highly abundant in the OSCC patient group in most studies and the genus Streptococcus is highly abundant in the control group. Furthermore, genus Veilonella was mentioned in four studies as the significantly abundant genus in the healthy control group, while one study found that Veilonella was more abundant in the OSCC patient group. Eight out of the 13 included studies reported differences in relative abundance between OSCC patients and controls, on the species level (21, 22, 24, 26, 28–30, 32).
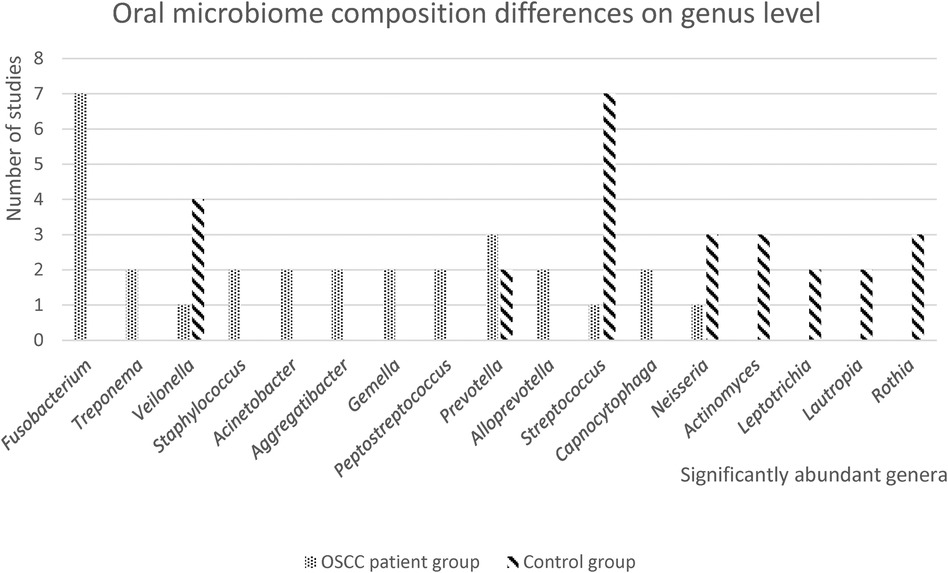
Figure 3. Oral microbiome differences on genus level. In this figure, the number of studies with significantly abundant genera in either the OSCC patient group or the healthy control group, is presented. Only significantly abundant genera (p < 0.05), that were reported in 2 or more studies, are depicted.
The oral microbiome differences are presented in Figure 4. Only significantly abundant species that were mentioned in two or more studies were included. Fusobacterium nucleatum was significantly abundant in two studies, Porphyromonas endodontalis was significantly abundant in three studies and Prevotella intermedia was significantly abundant in two studies for the OSCC patient group. Veilonella parvula was significantly abundant in the OSCC patient group in one study, two studies reported Veilonella parvula was significantly more abundant in the control samples.
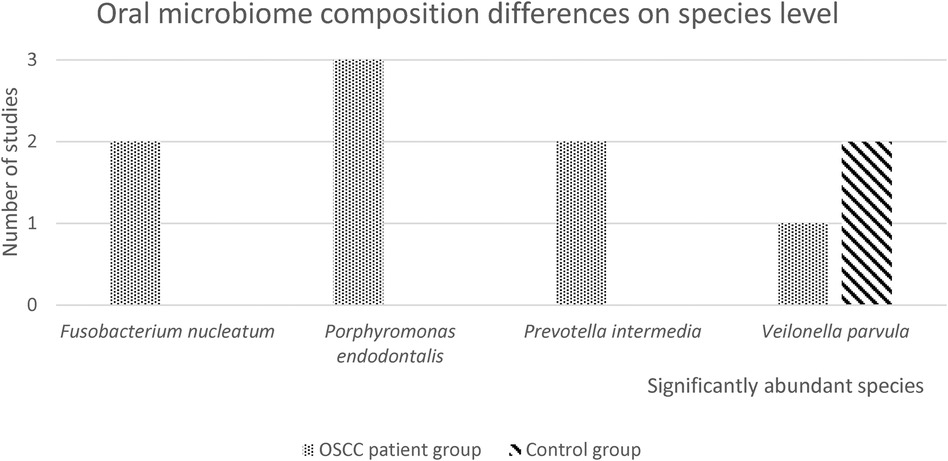
Figure 4. Oral microbiome differences on species level. In this figure, the number of studies with significantly abundant species in either the OSCC patient group or the healthy control group, is presented. Only significantly abundant species (p < 0.05), that were reported in 2 or more studies, are depicted.
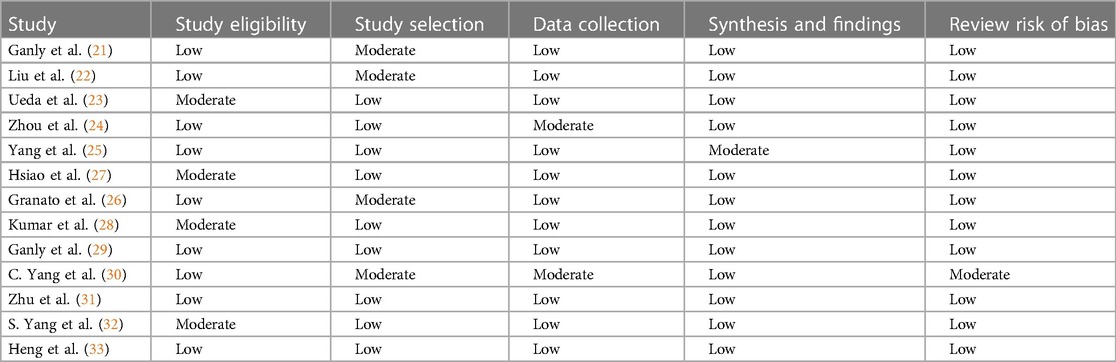
The results of the quality appraisal are presented in Table 2. A guideline for scoring each category was used and is described in more detail in the Supplementary Appendix. The majority of the studies had a case-control study design and almost all studies scored well on the critical appraisal. For one study, the risk of bias was reviewed as moderate (26). The main reason for the higher risk of bias, was the fact that in this study, the samples derived from the healthy controls were not matched to the OSCC patient group with respect to age, gender and oral health status.
Discussion
It has been hypothesized that OSCC might arise from genetic alterations induced by micro-organisms present in the oral microbiome (17). To gain more insight into which microbial signatures are linked to OSCC, the diversity and oral microbiome composition differences between OSCC patients and healthy controls were compared in this scoping review. A total of 13 studies were included in which oral microbial diversity (alpha- and beta diversity) and oral microbiome compositions (at different taxonomic levels) were compared between OSCC patients and healthy controls.
Most studies found a higher alpha diversity in the OSCC patient group and significantly different beta diversities in OSCC patient samples. Most studies concluded more abundant Fusobacteria (on phylum level), Fusobacterium (on genus level), Fusobacterium nucleatum, Porphyromonas endodontalis and Prevotella intermedia (on species level) in the OSCC patient group. The healthy control group had more abundant Actinobacteria (on phylum level), Streptococcus and Veilonella (on genus level) and Veilonella parvula (on species level) according to most studies.
Alpha diversity describes the species diversity/richness within a sample (33). Most studies found higher alpha diversity in the OSCC patient group. This was not expected because the general consensus is that less diversity in a microbiome indicates dysbiosis which is associated with pathological conditions such as caries, oral mucositis, periodontitis and even oral cancer (8, 34, 35). A possible explanation for this incoherent finding is the variety of sampling methods used in individual studies (35). In this review, some studies collected saliva samples, others took oral swabs of the cancer site or other sites in the oral cavity. This advocates for development of internationally accepted standard procedures for oral sample and metadata collection to promote more conclusive comparisons in future research (36). But even in homogeneous populations, high heterogeneity in microbiome diversity and composition is found (37). This complicates dividing populations into healthy and diseased based on their oral microbial diversity or composition differences.
Beta diversity describes diversity between samples, in this case between OSCC patient samples and healthy control samples (33). Beta diversity was significantly different between cases and controls in most studies, which may indicate a change in the microbiome composition in OSCC patients. This supports the potential contribution of the oral microbiome to OSCC development (16). Yet again, it is important to be cautious when using oral microbiome data to pinpoint differences between groups, because the oral microbiome is highly complex and the diversity measures may over-simplify this complexity (36). Also, different theories about the possible connection between microbiome changes and development of OSCC exist. Either bacteria may be the direct causative factor in the pathogenesis of OSCC and restructure the microbiome to an environment that damages healthy epithelial cells (bacteria before tumor theory), or bacterial presence in the OSCC tumor environment is opportunistic and is established after tumor development (bacteria after tumor theory) (38).
Most studies found Fusobacteria (on phylum level), Fusobacterium (on genus level), Fusobacterium nucleatum, Porphyromonas endodontalis and Prevotella intermedia (on species level) to be more abundant in the OSCC patient group. F. nucleatum is a Gram-negative, anaerobic oral bacterium that is a common inhabitant of the oral microbiome (39). In general, 5.2% of the healthy oral cavity constitutes of Fusobacteria (5). F. nucleatum is one of the main bacteria related to periodontitis and is recently associated to colorectal cancer and breast cancer (40, 41).
A comprehensive review of recent studies by McIlvanna showed F. nucleatum promotes several cancer development related mechanisms including activation of cell proliferation, promotion of cellular invasion, induction of chronic inflammation, and immune evasion (42). Li et al., described that F. nucleatum can promote oral squamous epithelial proliferation, metastasis, and immunomodulation via a large array of pathways (18). For instance, cells infected with F. nucleatum show a rise in gH2AX, a DNA double-strand break marker and a reduced expression of Ku70 and p53, both associated with cell repair (43). F. nucleatum promotes metastasis by activating EMT and the expression of MMPs (44). Moreover, it increases inflammation by influencing the AIM2 and POP1 pathway leading to the increased expression of IL-1B (18). Saikia et al., described the role of specific members of the oral microbiome, such as by influencing the defence mechanisms of oral mucosal stem cells and cancer stem cells (45). So, F. nucleatum has various ways of contributing to OSCC development.
In most studies, more abundant Actinobacteria (on phylum level), Streptococcus and Veilonella (on genus level) and Veilonella parvula (on species level) were found in the healthy control group compared to the OSCC patient group. Actinobacteria are ubiquitous gram-positive bacteria with high guanine and cytosine contents in DNA. Actinobacteria have a characteristic filamentous morphology (42). In general, 11.6% of the healthy oral cavity constitutes of Actinobacteria. Among Firmicutes (36.7% of oral bacteria), Streptococcus (19.2%) is the most abundant genus followed by Veillonella (8.6%) (5). Streptococcus species have been shown to impair F. nucleatum-induced inflammation in oral epithelial cells. A loss of Streptococcus species could therefore promote inflammation that is associated with F. nucleatum (42). What this example shows, is that loss of health associated species, can lead to growth of disease-associated species resulting in inflammation. After this dysbiotic shift in the microbiome and promoted inflammation, OSCC might arise via the hypothesized oral carcinogenesis pathway (16).
Besides studying microbiome makers for OSCC, previous studies used a wide range of salivary markers to detect OSCC including salivary lactate dehydrogenase (46), SLPI (47), salivary microRNA (48), salivary MMP-9 (49) and salivary sialic acid (50). Most studies included relatively small groups of OSCC patients, premalignant patients and controls. And none compared different promising biomarkers to determine relative predictive value of every biomarker. Other directions in research show promising results in predicting tumour progression including the intra-tumour microbiome, premalignant pathological characteristics, and premalignant genetic markers (51–53). In the future, large studies including already described salivary biomarkers, oral microbiome biomarkers and other biomarkers should be conducted to determine the most appropriate (combination) of biomarkers to predict OSCC. This could lead to the development of prediction models based on all risk factors, which might accurately estimate the risk of cancer development. Such prediction models should be AI aided, as the analysis of several biological markers and the determination of the relative risk is very complex.
Early diagnosis of OSCC plays a critical role in the treatment and prognosis of OSCC as early detection leads to less invasive treatment plans, better survival rates and better quality of life for patients (17, 54). Analyzing salivary samples to assess microbiome and salivary markers for the early diagnosis of OSCC is a potential option due to its ease of collection, non-invasiveness, and cost-efficiency when compared to traditional methods (24). The analysis of the oral microbiome in saliva samples may serve as a potential screening tool for detecting OSCC in the future, particularly in high-risk patients or those with premalignant lesions in the oral cavity. To accomplish this, larger longitudinal studies are required to track the oral microbiome composition of these patients over time, identifying those who develop OSCC and those who do not.
Specific outcomes of different studies are difficult to compare as there are many (small) differences in sampling and analysis leading to (slightly) different outcomes. This includes, but is not limited to, differences in the specific niches on the oral cavity, differences in methods of collection (rinse vs. swabs), differences in rRNA analysis protocols, different sequencing methods, differences in primers, differences in pipelines and differences in statistical packages. For future microbiome studies, it is important that differences in at least sampling methods are avoided and that standards are set.
To conclude, there are data on differences in the human oral microbiome between OSCC patients and healthy individuals. Higher microbial diversity was found in the healthy controls. Some species, e.g., Streptococcus spp, were associated with oral health, while F. nucleatum was associated with OSCC. Translating this data into clinically meaningful insights remains challenging. Further research is necessary to discover practical applications for this acquired knowledge in clinical settings (45, 55).
Author contributions
MD: Visualization, Project administration, Data curation, Writing – original draft, Methodology, Investigation, Formal Analysis, Conceptualization. JP: Writing – review & editing, Supervision. JR-D: Writing – review & editing, Supervision, Conceptualization. JE: Writing – review & editing, Supervision, Conceptualization. AL: Methodology, Formal Analysis, Writing – review & editing, Supervision, Conceptualization.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/froh.2024.1366153/full#supplementary-material
References
1. Reyes L. Porphyromonas gingivalis. Trends Microbiol. (2021) 29(4):376–7. doi: 10.1016/j.tim.2021.01.010
2. Katz J, Onate MD, Pauley KM, Bhattacharyya I, Cha S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int J Oral Sci. (2011) 3(4):209–15. doi: 10.4248/ijos11075
3. Olsen I, Yilmaz Ö. Possible role of Porphyromonas gingivalis in orodigestive cancers. J Oral Microbiol. (2019) 11(1):1563410. doi: 10.1080/20002297.2018.1563410
4. Kilian M, Chapple ILC, Hannig M, Marsh PD, Meuric V, Pedersen AML, et al. The oral microbiome—an update for oral healthcare professionals. Br Dent J. (2016) 221:657–66. doi: 10.1038/sj.bdj.2016.865
5. Panarese I, Aquino G, Ronchi A, Longo F, Montella M, Cozzolino I, et al. Oral and oropharyngeal squamous cell carcinoma: prognostic and predictive parameters in the etiopathogenetic route. Expert Rev Anticancer Ther. (2019) 19(2):105–19. doi: 10.1080/14737140.2019.1561288
6. D’souza S, Addepalli V. Preventive measures in oral cancer: an overview. Biomed Pharmacother. (2018) 107:72–80. doi: 10.1016/j.biopha.2018.07.114
7. Irfan M, Delgado RZR, Frias-Lopez J. The oral microbiome and cancer. Front Immunol. (2020) 11:591088. doi: 10.3389/fimmu.2020.591088
8. Chamoli A, Gosavi AS, Shirwadkar UP, Wangdale KV, Behera SK, Kurrey NK, et al. Overview of oral cavity squamous cell carcinoma: risk factors, mechanisms, and diagnostics. Oral Oncol. (2021) 121:105451. doi: 10.1016/j.oraloncology.2021.105451
9. Vesty A, Gear K, Biswas K, Mackenzie BW, Taylor MW, Douglas RG. Oral microbial influences on oral mucositis duringradiotherapy treatment of head and neck cancer. Support Care Cancer. (2020) 28:2683–91. doi: 10.1007/s00520-019-05084-6
10. Laheij AMGA, Raber-Durlacher JE, Koppelmans RGA, Huysmans M-CDNJM, Potting C, van Leeuwen SJM, et al. Microbial changes in relation to oral mucositis in autologous hematopoietic stem cell transplantation recipients. Sci Rep. (2019) 9:16929. doi: 10.1038/s41598-019-53073-w
11. Bruno JS, Heidrich V, Knebel FH, de Molla VC, Parahyba CJ, Miranda-Silva W, et al. Commensal oral microbiota impacts ulcerative oral mucositis clinical course in allogeneic stem cell transplant recipients. Sci Rep. (2022) 12:17527. doi: 10.1038/s41598-022-21775-3
12. Zaura E, Brandt BW, De Mattos MJT, Buijs MJ, Caspers M, Rashid MDH, et al. Same exposure but two radically different responses to antibiotics: resilience of the salivary microbiome versus long-term microbial shifts in feces. MBio. (2015) 6(6):e01693-15. doi: 10.1128/mbio.01693-15
13. Montero PH, Patel SG. Cancer of the oral cavity. Surg Oncol Clin N Am. (2015) 24(3):491–508. doi: 10.1016/j.soc.2015.03.006
14. Al-Jamaei AAH, Van Dijk BAC, Helder MN, Forouzanfar T, Leemans CR, De Visscher JGAM. A population-based study of the epidemiology of oral squamous cell carcinoma in The Netherlands 1989–2018, with emphasis on young adults. Int J Oral Maxillofac Surg. (2022) 51(1):18–26. doi: 10.1016/j.ijom.2021.03.006
15. Schiff B. Oral Squamous Cell Carcinoma. MSD Manual Professional Edition. (2022). Available online at: https://www.msdmanuals.com/professional/ear,-nose,-and-throat-disorders/tumors-of-the-head-and-neck/oral-squamous-cell-carcinoma (cited October 25, 2022).
16. Chattopadhyay I, Verma M, Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol Cancer Res Treat. (2019) 18:153303381986735. doi: 10.1177/1533033819867354
17. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. (2009) 6(7):e1000097. doi: 10.1371/journal.pmed.1000097
18. Li R, Li X, Gong T, Liu J, Li Y, Zhou X, et al. Role of oral microbiome in oral oncogenesis, tumor progression, and metastasis. Mol Oral Microbiol. (2022) 38(1):9–22. doi: 10.1111/omi.12403
19. Sami A, Elimairi I, Stanton C, Ross RP, Ryan CA. The role of the microbiome in oral squamous cell carcinoma with insight into the microbiome–treatment axis. Int J Mol Sci. (2020) 21(21):8061. doi: 10.3390/ijms21218061
20. Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan—a web and mobile app for systematic reviews. Syst Rev. (2016) 5:210. doi: 10.1186/s13643-016-0384-4
21. Ganly I, Hao Y, Rosenthal M, Wang H, Migliacci J, Huang B, et al. Oral microbiome in nonsmoker patients with oral cavity squamous cell carcinoma, defined by metagenomic shotgun sequencing. Cancers (Basel). (2022) 14(24):6096. doi: 10.3390/cancers14246096
22. Liu Y, Li ZR, Qi Y, Wen XT, Zhang L. Metagenomic analysis reveals a changing microbiome associated with the depth of invasion of oral squamous cell carcinoma. Front Microbiol. (2022) 13:795777. doi: 10.3389/fmicb.2022.795777/pdf
23. Ueda S, Goto M, Hashimoto K, Hasegawa S, Imazawa M, Takahashi M, et al. Salivary CCL20 level as a biomarker for oral squamous cell carcinoma. Cancer Genomics Proteomics. (2021) 18(2):103–12. doi: 10.21873/cgp.20245
24. Zhou X, Hao Y, Peng X, Li B, Han Q, Ren B, et al. The clinical potential of oral Microbiota as a screening tool for oral squamous cell carcinomas. Front Cell Infect Microbiol. (2021) 11:728933. doi: 10.3389/fcimb.2021.728933/pdf
25. Yang J, He P, Zhou M, Li S, Zhang J, Tao X, et al. Variations in oral microbiome and its predictive functions between tumorous and healthy individuals. J Med Microbiol. (2022) 71(8). doi: 10.1099/jmm.0.001568
26. Granato DC, Neves LA, Trino LD, Carnielli CM, Lopes A, Yokoo S, et al. Meta-omics analysis indicates the saliva microbiome and its proteins associated with the prognosis of oral cancer patients. Biochim Biophys Acta Proteins Proteom. (2021) 1869(8):140659. doi: 10.1016/j.bbapap.2021.140659
27. Hsiao JR, Ip WH, Lee W, Huang CZ, Ou CY, Tsai ST, et al. The interplay between oral microbiome, lifestyle factors and genetic polymorphisms in the risk of oral squamous cell carcinoma. Carcinogenesis. (2018) 39(6):778–87. doi: 10.1093/carcin/bgy053
28. Kumar A, Panda M, Das A, Rahman T, Das R, Das KK, et al. Dysbiosis of salivary microbiome and cytokines influence oral squamous cell carcinoma through inflammation. Arch Microbiol. (2021) 203(1):137–52. doi: 10.1007/s00203-020-02011-w
29. Ganly I, Yang L, Giese R, Hao Y, Nossa CW, Morris LGT, et al. Periodontal pathogens are a risk factor of oral cavity squamous cell carcinoma, independent of tobacco and alcohol and human papillomavirus. Int J Cancer. (2019) 145(3):775–84. doi: 10.1002/ijc.32152
30. Yang CY, Yeh YM, Yu H, Chin CY, Hsu CW, Liu HL, et al. Oral Microbiota community dynamics associated with oral squamous cell carcinoma staging. Front Microbiol. (2018) 9:862. Available online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00862/pdf
31. Zhu W, Shen W, Wang JJ, Xu Y, Zhai RD, Zhang J, et al. Capnocytophaga gingivalis is a potential tumor promotor in oral cancer. Oral Dis. (2024) 30(2):353–62. doi: 10.1111/odi.14376
32. Yang S, Lin C, Chuang CY, Lee YJ, Chung WH, Lai HC, et al. Host genetic associations with salivary microbiome in oral cancer. J Dent Res. (2021) 101(5):590–8. doi: 10.1177/00220345211051967
33. Heng W, Wang W, Dai T, Jiang P, Lu Y, Li R, et al. Oral bacteriome and mycobiome across stages of oral carcinogenesis. Microbiol Spectr. (2022) 10(6):e0273722. doi: 10.1128/spectrum.02737-22
34. Gupta S, Gupta S. Role of human papillomavirus in oral squamous cell carcinoma and oral potentially malignant disorders: a review of the literature. Indian J Dent. (2015) 6(2):91. doi: 10.4103/0975-962X.155877
35. Michálek J, Brychtova S, Pink R, Dvorak Z. Prognostic and predictive markers for perineural and bone invasion of oral squamous cell carcinoma. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. (2019) 163(4):302–8. doi: 10.5507/bp.2019.032
36. Deo PN, Deshmukh R. Oral microbiome: unveiling the fundamentals. J Oral Maxillofac Pathol. (2019) 23(1):122. doi: 10.4103/jomfp.jomfp_304_18
37. Verma D, Garg PK, Dubey AK. Insights into the human oral microbiome. Arch Microbiol. (2018) 200(4):525–40. doi: 10.1007/s00203-018-1505-3
38. Marsh P. In sickness and in health—what does the oral microbiome mean to US? An ecological perspective. Adv Dent Res. (2018) 29(1):60–5. doi: 10.1177/0022034517735295
39. Xia Y, Sun J, Chen DG. Statistical Analysis of Microbiome Data with R. Singapore: Springer (2018).
40. Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, et al. The microbiome of aseptically collected human breast tissue in benign and malignant disease. Sci Rep. (2016) 6:30751. doi: 10.1038/srep30751
41. Su SC, Chang LC, Da Huang H, Peng CY, Chuang CY, Chen YT, et al. Oral microbial dysbiosis and its performance in predicting oral cancer. Carcinogenesis. (2021) 42(1):127–35. doi: 10.1093/carcin/bgaa062
42. McIlvanna E, Linden GJ, Craig SG, Lundy F, James J. Fusobacterium nucleatum and oral cancer: a critical review. BMC Cancer. (2021) 21(1):1212. doi: 10.1186/s12885-021-08903-4
43. Geng F, Zhang Y, Lu Z, Zhang S, Pan Y. Caused DNA damage and promoted cell proliferation by the/pathway in oral cancer cells. DNA Cell Biol. (2020) 39(1):144–51. doi: 10.1089/dna.2019.5064
44. Shao W, Fujiwara N, Mouri Y, Kisoda S, Yoshida K, Yoshida K, et al. Conversion from epithelial to partial-EMT phenotype by Fusobacterium nucleatum infection promotes invasion of oral cancer cells. Sci Rep. (2021) 11(1):14943. doi: 10.1038/s41598-021-94384-1
45. Saikia PJ, Pathak L, Mitra S, Das B. The emerging role of oral microbiota in oral cancer initiation, progression and stemness. Front Immunol. (2023) 14:1198269. doi: 10.3389/fimmu.2023.1198269
46. Shetty SR, Chadha R, Babu S, Kumari S, Bhat S, Achalli S. Salivary lactate dehydrogenase levels in oral leukoplakia and oral squamous cell carcinoma: a biochemical and clinicopathological study. J Cancer Res Ther. (2012) 8(Suppl 1):S123–5. doi: 10.4103/0973-1482.92226
47. Campbell CMP, Giuliano AR, Torres BN, O'Keefe MT, Ingles DJ, Anderson RL, et al. Salivary secretory leukocyte protease inhibitor (SLPI) and head and neck cancer: the cancer prevention study II nutrition cohort. Oral Oncol. (2016) 55:1–5. doi: 10.1016/j.oraloncology.2016.02.004
48. Fadhil RS, Wei MQ, Nikolarakos D, Good D, Nair RG. Salivary microRNA miR-let-7a-5p and miR-3928 could be used as potential diagnostic bio-markers for head and neck squamous cell carcinoma. PLoS One. (2020) 15(3):e0221779. doi: 10.1371/journal.pone.0221779
49. Smriti K, Ray M, Chatterjee T, Shenoy RP, Gadicherla S, Pentapati KC, et al. Salivary MMP-9 as a biomarker for the diagnosis of oral potentially malignant disorders and oral squamous cell carcinoma. Asian Pac J Cancer Prev. (2020) 21(1):233–8. doi: 10.31557/APJCP.2020.21.1.233
50. Daniel D, Jose J, Harish Kumar A. Is salivary sialic acid a reliable biomarker in the detection of oral potentially malignant disorder and oral squamous cell carcinoma. J Maxillofac Oral Surg. (2021) 20(1):83–9. doi: 10.1007/s12663-019-01309-7
51. Li Z, Fu R, Wen X, Wang Q, Huang X, Zhang L. The significant clinical correlation of the intratumor oral microbiome in oral squamous cell carcinoma based on tissue-derived sequencing. Front Physiol. (2023) 13:1089539. doi: 10.3389/fphys.2022.1089539
52. Wils LJ, Poell JB, Peferoen L, Evren I, Brouns ER, De Visscher JGAM, et al. The role of differentiated dysplasia in the prediction of malignant transformation of oral leukoplakia. J Oral Pathol Med. (2023) 52(10):930–8. doi: 10.1111/jop.13483
53. Wils LJ, Poell JB, Brink A, Evren I, Brouns ER, De Visscher JGAM, et al. Elucidating the genetic landscape of oral leukoplakia to predict malignant transformation. Clin Cancer Res. (2022) 29(3):602–13. doi: 10.1158/1078-0432.ccr-22-2210
54. Sklenicka S, Gardiner S, Dierks EJ, Potter BE, Bell RB. Survival analysis and risk factors for recurrence in oral squamous cell carcinoma: does surgical salvage affect outcome? J Oral Maxillofac Surg. (2010) 68(6):1270–5. doi: 10.1016/j.joms.2009.11.016
55. Booth A, Papaioannou D, Sutton A. Systematic approaches to a successful literature review. (2012). Available online at: http://lib.ugent.be/en/catalog/rug01:002101878/items/000010929920/
Keywords: oral cancer, oral microbiome, oral squamous cell carcinoma (OSCC), microbial diversity, taxonomic level, Fusobacterium nucleatum, Streptococcus
Citation: van Dijk MC, Petersen JF, Raber-Durlacher JE, Epstein JB and Laheij AMGA (2024) Diversity and compositional differences in the oral microbiome of oral squamous cell carcinoma patients and healthy controls: a scoping review. Front. Oral. Health 5:1366153. doi: 10.3389/froh.2024.1366153
Received: 5 January 2024; Accepted: 16 May 2024;
Published: 11 June 2024.
Edited by:
Daniel Lambert, The University of Sheffield, United KingdomReviewed by:
Hope M. Amm, University of Alabama at Birmingham, United StatesZhengrui Li, Shanghai Jiao Tong University, China
Divyashri Baraniya, Temple University, United States
© 2024 van Dijk, Petersen, Raber-Durlacher, Epstein and Laheij. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: A. M. G. A. Laheij, YS5sYWhlaWpAYWN0YS5ubA==
†ORCID:
A. M. G. A. Laheij
orcid.org/0000-0003-1291-2798
 M. C. van Dijk
M. C. van Dijk J. F. Petersen
J. F. Petersen J. E. Raber-Durlacher
J. E. Raber-Durlacher J. B. Epstein
J. B. Epstein A. M. G. A. Laheij
A. M. G. A. Laheij