- 1School of Medicine, International Medical University, Kuala Lumpur, Malaysia
- 2Faculty of Medicine and Health, UNSW, Sydney, NSW, Australia
- 3Sungai Rengit Dental Clinic, Johor Health Department, Ministry of Health Malaysia, Kota Tinggi, Malaysia
- 4Clinical Sciences Department, Ajman University, Ajman, United Arab Emirates
- 5Centre of Medical and Bio-Allied Health Sciences Research, Ajman University, Ajman, United Arab Emirates
- 6Basic Medical and Dental Sciences Department, Ajman University, Ajman, United Arab Emirates
Objective: The study aimed to evaluate the impact of tobacco use on the composition and functions of the oral microbiome in healthy adult humans.
Methods: We conducted a systematic search on PubMed, Web of Science, and Cinhal databases for literature published until 15 December 2023, to identify studies that have evaluated the oral microbiome with culture-independent next-generation techniques comparing the oral microbiome of tobacco users and non-users. The search followed the PECO format. The outcomes included changes in microbial diversity and abundance of microbial taxa. The quality assessment was performed using the Newcastle–Ottawa Scale (NOS) (PROSPERO ID CRD42022340151).
Results: Out of 2,435 articles screened, 36 articles satisfied the eligibility criteria and were selected for full-text review. Despite differences in design, quality, and population characteristics, most studies reported an increase in bacterial diversity and richness in tobacco users. The most notable bacterial taxa enriched in users were Fusobacteria and Actinobacteria at the phylum level and Streptococcus, Prevotella, and Veillonella at the genus level. At the functional level, more similarities could be noted; amino acid metabolism and xenobiotic biodegradation pathways were increased in tobacco users compared to non-users. Most of the studies were of good quality on the NOS scale.
Conclusion: Tobacco smoking influences oral microbial community harmony, and it shows a definitive shift towards a proinflammatory milieu. Heterogeneities were detected due to sampling and other methodological differences, emphasizing the need for greater quality research using standardized methods and reporting.
Systematic Review Registration: CRD42022340151.
1 Introduction
The human oral cavity harbors a diverse microbial community comprising over 700 species of bacteria or phylotypes that play a commensal role in protecting oral and systemic health (1). These diverse species have been identified by cultivation or the advancing culture in-dependent molecular approaches (1). These species attach and form biofilms on the mouth's soft and hard tissue surfaces in a structurally organized matrix, inducing a dynamic equilibrium with the immune-inflammatory response of the host (2). The human oral cavity serves as one of the major gateways to the respiratory tract, thus giving microorganisms the substantial prospect of invading these sites (3). Despite the similarities between the core microbial composition within the oral cavities, the type of species may vary depending on diet and nutrition, genetic susceptibility, antibiotic usage, hormonal factors, tobacco and alcohol exposure, and recurrent pathogenic infections of the host (4). This disturbance to the equilibrium results in oral dysbiosis altering oral and systemic health through several pathophysiological processes linked to disease (5). Dysbiosis has reportedly been involved in oral diseases such as periodontitis, gingivitis, and oral cancer (6–8).
The emergence of new genomic technology including next-generation sequencing, has led to the identification of resident bacterial populations in almost all organs and systems of the body, and has sparked an increased interest in the microbiota among researchers. These next generation sequencing helped to reveal the complex nature of the oral microbiome community, which could not be revealed by culture methods and traditional Sanger sequencing methods as less abundant and non-cultivable microbes of the population are often overlooked, which jeopardizes the accuracy of the detailed account of the microbial community (9).
Recent studies show that despite a global decline in tobacco consumption, tobacco use is exponentially rising in parts of the world, leading to a consequential public health concern (10). Tobacco smoke comprises numerous toxicants that come into direct contact with the bacteria in the oral cavity, disrupting the microbial ecology of the mouth. These toxic compounds cause cellular injury and cell death, including N-nitrosamines and polycyclic aromatic hydrocarbons blocking DNA repair and initiating tumorigenesis (11). Smoking has been shown to cause the loss of beneficial oral species, leading to pathogenic alterations by interacting with various host cells and extracellular matrix components, ultimately leading to the risk of disease development (12). This alteration increases the local density of the bacterial pathogens or decreases the prevalence of other bacteria (13, 14). Emerging evidence on the effects of smokeless tobacco on the composition of the oral microbiota in humans suggests it leads to a pro-inflammatory milieu in the oral microenvironment, further leading to diseases (15). To date, the literature on the effects of tobacco use on the oral microbiome in humans has not been systematically evaluated. Therefore, we carried out a systematic review as a first attempt to characterize the impact of tobacco use on the oral microbiome profile in healthy adults and to compare the differences in the oral microbiome profile of tobacco users with non-users. It also aims to highlight the potential effects of smoking on the host's health by analyzing the available data regarding the relationship between the human oral microbiome and tobacco use.
2 Material and methods
2.1 Search strategy
A systematic review was conducted to answer the question: “Is the oral microbiome profile of tobacco users different from non-users?” The present systematic review was registered in the International Prospective Register of Systematic Reviews (PROSPERO) under CRD42022340151) The systematic literature search was performed to identify published studies until Dec 2023 examining the oral microbial community in tobacco users in comparison to controls using broad MeSH terms and other related keywords. The search was performed independently by two investigators (NS and CY). The electronic databases used are PubMed, Web of science and CINHAL. The search was carried out using the specific key keywords with the use of Boolean operators “OR” and “AND.” The search strategy and output for each database is provided as Supplementary Tables S1–S3. Following the elimination of duplicates, the titles and abstracts were evaluated in accordance with the preset eligibility criteria as provided below to determine whether or not they should be included for additional full-text reading. Two independent investigators (NS and CY) scanned the titles and corresponding abstracts. If the abstract clearly indicated what was included or excluded, the record was read in its entirety. In the event that the findings of the two investigators disagreed, DG, the third investigator, was consulted. We manually examined the reference lists of the included publications to find any potentially relevant articles that could be included. The systematic review follows in accordance with the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines (16).
2.2 Eligibility criteria
Inclusion criteria
Cross-sectional or prospective observational studies that compared the oral microbiome analyzed with culture-independent next-generation techniques from tobacco users, including cigarettes, water pipes, smokeless, and other forms of tobacco in comparison to healthy controls were included. The detailed PECO (Population, Exposure, Control, and Outcome) scheme followed is below:
Population: Human adults using tobacco
Exposure: Use of any form of tobacco
Control: Non-users
Outcome: Changes in microbial diversity and abundance of various microbial taxa
Type of studies: Cross-sectional or prospective observational studies that utilized culture-independent next-generation techniques without date limitation.
Exclusion criteria
The studies which did not fit into the inclusion criteria were excluded.
Studies utilizing culture techniques, studies on diseased populations like periodontitis or caries, which can have an impact on the oral microbiome, animal studies, and studies on e-cigarettes were excluded. Further narrative reviews, systematic reviews, conference reports, and letters to the editor were excluded. The literature search was limited to the English Language.
2.3 Data Extraction
Data was extracted from the selected articles through a separate full-text review by two reviewers. The following study characteristics were extracted from each article: author name, year of publication, study design, sample size, age and gender distribution, type of tobacco, exposure assessment, and significant changes in oral microbial diversity and abundances of taxa.
2.4 Quality assessment
The quality assessment for the included studies was performed independently by two reviewers (NS and CY) using the Newcastle–Ottawa Quality Assessment Scale (17). If there is any discrepancy, then the third author was consulted (DG) and the discrepancy was resolved. This instrument incorporates three separate domains: selection, comparability, and outcomes. The selection domain involves the assessment of four items; comparability has one item, and outcomes include three items. The selected article will receive one star in each item if acceptable, thus obtaining a maximum of four in the selection domain, one in the comparability domain, and three in the outcome domain.
3 Results
3.1 General study characteristics
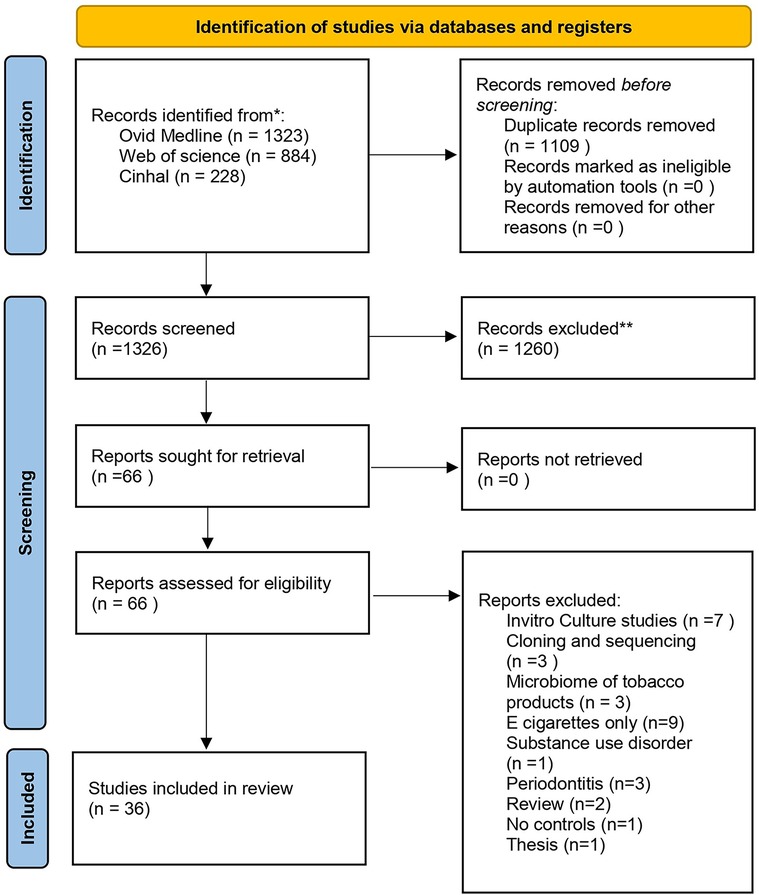
The search yielded 2,435 records from the three databases, of which 1,109 were excluded altogether due to duplicates. Screening of articles by title and abstract and reviewing of full text resulted in 36 eligible articles for full text (Figure 1). Of the 36 articles, nine were from the United States of America (18–26), six were from India (15, 27–31), five were from the United Arab Emirates (32–36),three were from China (37–39), two from Japan (40, 41) and others including Brazil (42), Jordan (43), Hungary (44), Croatia (45), Iran (46), Germany (47), Denmark (48), Italy (49), Sudan (50), Ireland (51) and Korea (52).
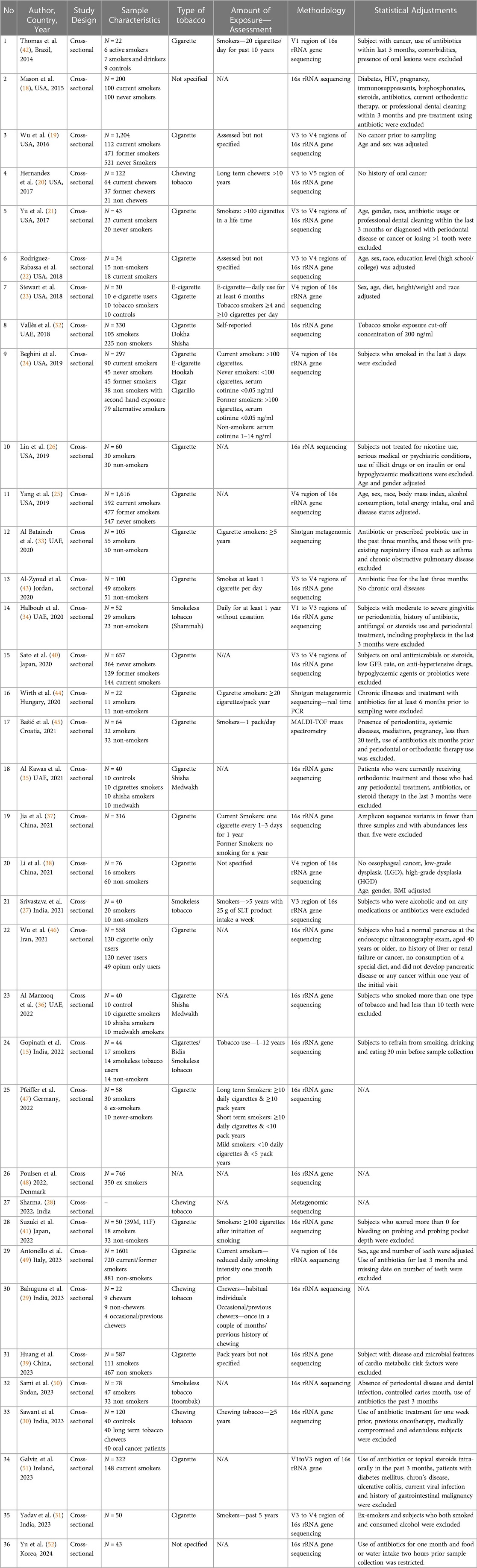
The study design followed cross-sectional studies with a sample size ranging from 22 to 1,616. Most studies were conducted on cigarette users, except seven studies that focused on smokeless tobacco products including chewing tobacco (20, 27–30, 34, 50). The 16s rRNA gene sequencing was the most commonly used methodology except for three studies that used shotgun metagenomic gene techniques (28, 33, 44). A common trait seen in most studies was screening for antibiotic usage before sampling and for the presence of chronic or oral illnesses. Further, some studies also included decisive factors that can influence the microbiome, including alcohol consumption, BMI, and diet, into consideration for profiling of the subjects (25, 27, 31, 38). Sample collection types include saliva, oral and buccal swabs, oral rinses, supragingival, subgingival and tongue scrapes, and mouthwashes. The detailed characteristics are provided in Table 1 (42–52). All controls were deemed healthy except for one study that acquired control subjects from cancer cohorts (19).
3.2 Diversity and richness analysis
As displayed in Table 2 (42–52), all included studies except five assessed microbial diversity and richness (23, 28, 31, 38, 45). Five studies reported no difference in diversity difference between the smokers and control groups (34, 36, 41, 49, 52). Four studies (21, 26, 40, 42) reported lower diversity and richness in smokers. The rest of the studies concluded that the richness and phylogenetic biodiversity of smokers or tobacco users were significantly different or higher than non-users or former users.
3.3 Differences in the abundance of various taxa between smokers and non-smokers
Firmicutes were identified as the most abundant phylum across most studies compared to other types of phyla, including Proteobacteria, Bacteroidetes, and Fusobacteria, which varied in abundance among smokers and non-smokers. Four studies reported Fusobacteria being depleted in non-smokers and higher in smokers (18, 22, 27, 49). In contrast, Fusobacteria, in particular, was lower in smokers and drinkers and more abundant in the control group (42). Studies conducted using an oral rinse and saliva of cigarette and tobacco smokers were enriched with phylum Actinobacteria in current users (25, 28, 46, 49). Similarly, an abundance of Actinobacteria in water pipe smokers was reported in another study (36). Bacteroidetes dominated smoker and chewer samples in two studies (22, 29) compared to another, which reported a lower relative abundance in cigarette smokers compared to e-cigarette users and controls (23). In terms of genera, most studies reported different types of genera in various types of samples from smokers and healthy controls. Streptococcus was relatively reported higher in abundance in smokers in several studies. Prevotella and Veillonella, mostly independently, were also found as predominant genus in tobacco users (21, 24, 31, 34, 37, 38, 42–44, 49, 51), while another data reported a significant depletion in the saliva and supragingival plaques of smokeless tobacco users (50). Neisseria was also observed to be higher among other genera in smokers in two studies conducted in China and Denmark (38, 48). The detailed findings are presented in Table 2.
3.4 Differences in metabolic pathways between smokers and non-smokers
Among the 36 studies included, only 9 of them explored the differences in metabolic pathways (15, 19, 26, 27, 30, 37, 39, 40, 49). Wu et al. reported that xenobiotic biodegradation, amino acid metabolism pathways, glycan biosynthesis, and metabolism were enriched in smokers. Further pathways related to aerobic metabolism [tricarboxylic acid (TCA) cycle, oxidative phosphorylation and nitrate reduction] were depleted in current smokers (19, 49). Similarly, Sato et al. also reported significant differences in pathways related to the TCA cycle, glyoxylate cycle, and several compound biosynthesis and degradation between smokers and non-smokers (40). Jia et al. reported that acid production, amino acid-related enzymes and amino sugar, and nucleotide sugar metabolism were all enriched in smokers (37). A recent study on cigarette smokers reported depletion of pathways related to membrane transport and lipid metabolism in smokers as well as xenobiotics biodegradation and enrichment of pathways related to the metabolism of amino acids, nucleotides, vitamins, terpenoids, polyketides, and glycans (26). In the case of smokeless tobacco users, Srivastava et al. reported an increase in amino acid metabolism, xenobiotic biodegradation, and cellular process and signaling (27). Another study also reported an increase in pathways related to amino acid metabolism, synthesis, and degradation (15). Moreover, Sawant et al, observed an increase in pathways related to reductive TCA cycle and pyrimidine biosynthesis in chewing tobacco users (30).
3.5 Methodological quality of the studies
The quality of the studies can be found in Supplementary Table S1. Five studies were graded as very good; twenty-six articles were of good quality, whereas the rest of them were of satisfactory quality.
4 Discussion
This review aimed to evaluate the available evidence on the impact of the use of tobacco in various forms on healthy humans' oral microbiomes. To our knowledge, this is the first systematic and comprehensive review that summarizes the impact of tobacco use on the oral microbiome. Although there were variations in design, quality of the studies, and characteristics, our results highlight that smoking, regardless of the form, altered the normal equilibrium of the oral microbiome. This evidence is in accordance with previous results obtained analyzing oral microbiomes in culture methods and animal models (53, 54). Despite the limited number of studies, other less-known forms of smoking also seemed to be associated with changes in the oral microbiome.
The current review of data from clinical studies emphasizes that cigarette smoking is found to cause alteration in the oral bacterial profiles. Streptococcus was notably a predominant genus in most studies. In healthy populations, streptococci are common members of the subgingival and supragingival habitats and are early commensal invaders of these environments. However, these commensals have been shown to inhibit the proinflammatory response, which is how they predominantly modulate the immune system and aid in biofilm development (55). Notably, the majority of the other bacteria that were significantly increased in smokers were anaerobes, including Prevotella and Veillonella. This could be related to the deprivation of oral oxygen due to cigarette smoking. Smoking may create a depletion of an oxygen environment in the mouth. It would reflect on the oxygen availability of microbes in the oral cavity, leading to the oral microbial ecology alteration. These were also reported to increase smokers' gut microbiome (47, 56). Veillonella and Actinomyces were the anaerobic bacteria found to be higher in smokers, and these could promote the development of biofilms in the oral cavity (37). Interestingly, Actinomyces have also been enriched in several cancers, including liver, esophagus, colorectal cancer, etc. (57–60). Actinomyces has been shown to the production of various immunological and microbial-related genes, such as TLR2, TLR4, and NF-B, which support the growth of colorectal cancer by controlling inflammation by activating the downstream TLR4/NF-B pathway (60). Actinomyces also has been shown to modulate the presence of several other gram-negative bacteria (60). It also reduces antitumor immunity by preventing CD8+ T cell invasion in colorectal cancer (60). Furthermore, nitrate in vegetables is often converted to oral nitrate, which has the potential to make the oral cavity more acidic, and anaerobic bacteria, especially Actinomyces and Veillonella, promote this conversion (61, 62). This acidic environment has been shown to encourage the growth of biofilms and is linked to oral cavity diseases (63). Decreased local oxygen tension and acidic environment are also likely to promote periodontal anaerobes Fusobacterium, Treponema, and P. gingivalis, which are implicated in the development of periodontitis (64).
The oral cavity is often the first contact with smoke and hence may play an essential role in the degradation of toxic compounds. The depletion of several biodegradation pathways in current smokers suggests potential downstream consequences. A key observation in smokers was the enriched degradation of polycyclic aromatic hydrocarbons and other constituents in cigarette smokers (19). Amino acid-related enzymes and amino sugar and nucleotide sugar metabolism were notably abundant in smokers compared to non-smokers (37). Alternatively, these toxic compounds may saturate the enzymes responsible for their degradation, thus killing the bacteria possessing these enzymes (19). The toxic components in cigarette smoke have been shown to alter the oral immune response, and it has been implicated in the pathogenesis of several oral diseases, including periodontitis and oral cancer (8, 64).
Oral epithelial cells actively participate in oral immune response by expressing specific receptors, including toll-like receptors (TLRs). TLRs are receptors in immune response expressed by cell surfaces and internal vesicles and their stimulation lead to activation multiple intracellular signaling cascades (65) One of the main downstream signaling cascades is the NF-KB, a critical transcription factor that encourages the expression of chemokines, cytokines, and co-stimulatory and adhesion molecules (66). Cigarette smoke has been shown to increase the expression of and alter the functional activation of these receptors, including TLR-2, TLR-4, and others (67, 68). Interestingly, the taxa reported to be enriched in smokers including Fusobacteria, Veillonella, Prevotella, and Actinomyces, as well as other microorganisms, also bind to TLR-2 and TLR-4 using their peptidoglycan and lipopolysaccharide cell walls, and these TLR-2 or TLR-4 mediated signaling leads to up-regulation of several proinflammatory pathways (69–74). TLRs and their signaling machinery have been subsequently implicated in a wide range of human diseases, including several cancers, especially oral cancers (75–77).
Tobacco components have also been shown to increase the virulence of specific periodontal pathogens, particularly for P. gingivalis, which has multiple virulence factors (64, 78, 79). Oxidative stress-related proteins in P. gingivalis are up-regulated in the presence of nicotine and other products, which helps in adaptability and survival ability in a low-oxygen environment and biofilms (78, 80). P. gingivalis biofilms have reduced proinflammatory properties, which can help enhance sustainability (80, 81). However, it was interesting to note that the upregulation of P. gingivalis was reported by two published studies only. P. gingivalis is also known to facilitate many microbial colonizers, including S. oralis, Streptococcus gordonii, Actinomyces viscosus, Fusobacterium spp & Prevotella intermedia (79, 82–84), which has been reported to be upregulated by multitude of studies included in the review.
Interestingly, one of the studies reported that the overall oral microbiome composition of former smokers did not differ in comparison to never smokers; this indicates that changes in the oral microbiome influenced by smoking are permanent (19). Such findings are encouraging and can lay the foundation for microbiome-targeted approaches for smoking cessation and disease prevention.
In our review, we noticed that only very few studies have explored the impact of use of shisha or waterpipe on the oral microbiome. It is now known that waterpipe smoke constitutes many of the same toxicants and is associated with the risk of disease (36). Relative to water pipe smoking, out of the four studies included, Streptococcus sanguinis was found to be higher in smokers (35). Overall, phyla Firmicutes was the most abundant phylum in those combined with other forms of tobacco smoking such as medwakh and cigarettes (35, 36). Few of these bacterial species are known to be a common cause of human respiratory diseases and infections, notably where tobacco consumption is a significant risk factor (85, 86). It is pretty unclear as to what specific bacteria taxa are associated with water pipes due to the scarcity of resources available; however, this could be mainly influenced by the habits of the subjects and other exposures as well.
Smokeless tobacco can also impact oral microbiota, increasing the risk for oral disease pathologies. Due to the nicotine concentration in smokeless tobacco, the growth of S.mutans places the user at an increased risk for dental caries (87). Hung et al. suggested that these tobacco products can increase caries development by fostering S.mutans formation on tooth surfaces (88). Further, streptococci species are known to produce acetaldehyde. Acetaldehyde, a carcinogenic compound, production has been proposed as a mechanism by which bacteria can contribute to oral carcinogenesis (34). This is supported by abundant levels of Streptococcus genera that indicated alterations in smokeless tobacco users compared to controls (15, 27). Furthermore, Fusobacteria abundant in smokeless tobacco users is an opportunistic pathogen and has been known to be capable of growth in acidic conditions (15). Fusobacteria has reportedly been noted in human colorectal carcinoma, suggesting it may have originated from the oral cavity. They promote tumor development by inducing inflammation and the immune response of the host to produce inflammatory factors (89). In addition, these species have reportedly been found to be abundant in head and neck cancer samples (90).
This review noted that sample collection sites in the oral cavity subsequently differed within the studies. This site variation could produce significant bias as the sites may vary in microbial composition. For instance, salivary samples may reflect the bacteria shed from the total oral cavity, whereas tissue sampling would be a deeper representation of the microbiome concerning the host (91). Hence, it wouldn't be rational to assume the impacts of smoking caused by components of tobacco smoke are similar across all microenvironments (44). Further studies are recommended to elucidate the different ecology of these environments, as interpreting the data of a mixture of sample types may obscure meaningful associations and patterns.
The current review highlights that the studies reported until now relied on genetic characterization of the microbiome using 16S sequencing methodology without adequate examination of this functionality. Only three studies employed shotgun sequencing (28, 33, 44). Given that shotgun metagenomic sequencing provides better strain level resolution and functional insights, the field should focus more on this sophisticated methodology, in combination with metabolomics and metaproteomic, in decoding host-microbiome interactions. Microbiome architecture can be highly varied among humans, with inter-individual variation presenting a substantial challenge, necessitating the development of sophisticated machine learning processes that predict the impact of microbiome and metabolites on physiological and pathological situations. Despite these constraints, understanding the ubiquitous activities of microbially regulated metabolites can open up a new avenue for enhancing oral health. One of the potential clinical implication of deciphering host-microbial interactions would be management strategies for tobacco-related illnesses, including smoking cessation strategies by altering the microbiota with probiotics, prebiotics, and other related methods. There is currently insufficient data despite the possibility that several preventive and therapeutic applications might be effective in theory. These are primarily related to the possibility of eubiosis being restored upon smoking cessation. As a matter of fact, we have uncovered a dearth of research on this aspect considering the abundance of studies on tobacco use and oral microbiota and needs to be explored further.
One of the limitations of the current review is the heterogeneity in the methods and the outcome reporting in the included studies, which hindered comparability and quantitative analysis. However, this is a common limitation reported by most of the reviews on microbiome, because of the inherent heterogeneity in the methodology. Further, we have included articles published only in the English language.
5 Conclusion
In this review, it is majorly observed that smoking and smokeless tobacco influence the oral microbial community composition, and there is a definitive shift in the abundance of oral taxa favoring an anaerobic environment, thus promoting a proinflammatory milieu. It is suggested that smoking may perturb the balance of the oral microbiome by affecting the relationships between bacteria and altering their metabolic pathways. However, smokeless and smoking tobacco are a mixture of multiple toxicants, and their direct impact on the oral microbiome is yet unclear. The effect of tobacco on microbial metabolism needs to be elucidated and is critical to our understanding of the etiology of oral and systemic diseases, as oral microbial dysbiosis are associated with several systemic conditions.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
NSe: Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft. NSh: Data curation, Methodology, Validation, Writing – review & editing. DG: Conceptualization, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. CY: Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft.
Funding
The author(s) declare no financial support was received for the research or authorship of this article.
The APC is funded by Ajman University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/froh.2024.1310334/full#supplementary-material
References
1. Deo PN, Deshmukh R. Oral microbiome: unveiling the fundamentals. J. Oral Maxillofac Pathol. (2019) 23(1):122–8. doi: 10.4103/jomfp.JOMFP_304_18
2. Lamont RJ, Koo H, Hajishengallis G. The oral Microbiota: dynamic communities and host interactions. Nat Rev Microbiol. (2018) 16(12):12. doi: 10.1038/s41579-018-0089-x
3. Sharma N, Bhatia S, Sodhi AS, Batra N. Oral microbiome and health. AIMS Microbiol. (2018) 4(1):42–66. doi: 10.3934/microbiol.2018.1.42
4. Li X, Liu Y, Yang X, Li C, Song Z. The oral microbiota: community composition, influencing factors, pathogenesis, and interventions. Front Microbiol. (2022) 13:895537. doi: 10.3389/fmicb.2022.895537
5. Kushwaha SS, Multani RK, Kushwaha NS, Gautam S, Jindal DG, Arora KS, et al. Saliva is a potential diagnostic tool to evaluate the relationship between oral microbiome and potentially malignant disorders for prevention of malignant transformation. Asian Pac J Cancer Prev. (2021) 22(1):125–9. Scopus. doi: 10.31557/APJCP.2021.22.1.125
6. Gopinath D, Kunnath Menon R, Chun Wie C, Banerjee M, Panda S, Mandal D, et al. Salivary bacterial shifts in oral leukoplakia resemble the dysbiotic oral cancer bacteriome. J Oral Microbiol. (2020) 13(1):1857998. doi: 10.1080/20002297.2020.1857998
7. Van Dyke TE, Bartold PM, Reynolds EC. The nexus between periodontal inflammation and dysbiosis. Front Immunol. (2020) 11:511. doi: 10.3389/fimmu.2020.00511
8. Gopinath D, Menon RK, Banerjee M, Su Yuxiong R, Botelho MG, Johnson NW. Culture-independent studies on bacterial dysbiosis in oral and oropharyngeal squamous cell carcinoma: a systematic review. Crit Rev Oncol Hematol. (2019) 139:31–40. doi: 10.1016/j.critrevonc.2019.04.018
9. Cao Y, Fanning S, Proos S, Jordan K, Srikumar S. A review on the applications of next generation sequencing technologies as applied to food-related microbiome studies. Front Microbiol. (2017) 8:1829. doi: 10.3389/fmicb.2017.01829
10. Perez-Warnisher MT, de Miguel MPC, Seijo LM. Tobacco use worldwide: legislative efforts to curb consumption. Ann Glob Health. (n.d.) 84(4):571–9. doi: 10.29024/aogh.2362
11. Chatterjee N, Walker GC. Mechanisms of DNA damage, repair and mutagenesis. Environ Mol Mutagen. (2017) 58(5):235–63. doi: 10.1002/em.22087
12. Jiang Y, Zhou X, Cheng L, Li M. The impact of smoking on subgingival microflora: from periodontal health to disease. Front Microbiol. (2020) 11:66. doi: 10.3389/fmicb.2020.00066
13. Khan I, Bai Y, Zha L, Ullah N, Ullah H, Shah SRH, et al. Mechanism of the gut microbiota colonization resistance and enteric pathogen infection. Front Cell Infect Microbiol. (2021) 11:716299. doi: 10.3389/fcimb.2021.716299
14. Impacts of cigarette smoking on immune responsiveness: Up and down or upside down?—PMC. (n.d.). Available online at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5352117/ (retrieved January 7, 2023)
15. Gopinath D, Wie CC, Banerjee M, Thangavelu L, Kumar RP, Nallaswamy D, et al. Compositional profile of mucosal bacteriome of smokers and smokeless tobacco users. Clin Oral Investig. (2022) 26(2):1647–56. doi: 10.1007/s00784-021-04137-7
16. Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. (2015) 4:1. doi: 10.1186/2046-4053-4-1
17. Wells G, Shea B, O'Connell D, Robertson J, Peterson J, Welch V, et al. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomized Studies in Meta-Analysis. (2014). Available online at: https://www.semanticscholar.org/paper/The-Newcastle-Ottawa-Scale-(NOS)-for-Assessing-the-Wells-Wells/c293fb316b6176154c3fdbb8340a107d9c8c82bf (accessed December 06, 2023).
18. Mason MR, Preshaw PM, Nagaraja HN, Dabdoub SM, Rahman A, Kumar PS. The subgingival microbiome of clinically healthy current and never smokers. ISME J. (2015) 9(1):268–72. doi: 10.1038/ismej.2014.114
19. Wu J, Peters BA, Dominianni C, Zhang Y, Pei Z, Yang L, et al. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. (2016) 10(10):2435–46. doi: 10.1038/ismej.2016.37
20. Hernandez BY, Zhu X, Goodman MT, Gatewood R, Mendiola P, Quinata K, et al. Betel nut chewing, oral premalignant lesions, and the oral microbiome. PLoS One. (2017) 12(2):e0172196. doi: 10.1371/journal.pone.0172196
21. Yu G, Phillips S, Gail MH, Goedert JJ, Humphrys MS, Ravel J, et al. The effect of cigarette smoking on the oral and nasal microbiota. Microbiome. (2017) 5(1):3. doi: 10.1186/s40168-016-0226-6
22. Rodríguez-Rabassa M, López P, Rodríguez-Santiago RE, Cases A, Felici M, Sánchez R, et al. Cigarette smoking modulation of Saliva microbial composition and cytokine levels. Int J Environ Res Public Health. (2018) 15(11):E2479. doi: 10.3390/ijerph15112479
23. Stewart CJ, Auchtung TA, Ajami NJ, Velasquez K, Smith DP, De La Garza R, et al. Effects of tobacco smoke and electronic cigarette vapor exposure on the oral and gut microbiota in humans: a pilot study. PeerJ. (2018) 6:e4693. doi: 10.7717/peerj.4693
24. Beghini F, Renson A, Zolnik CP, Geistlinger L, Usyk M, Moody TU, et al. Tobacco exposure associated with oral microbiota oxygen utilization in the New York city health and nutrition examination study. Ann Epidemiol. (2019) 34:18–25.e3. doi: 10.1016/j.annepidem.2019.03.005
25. Yang Y, Zheng W, Cai Q-Y, Shrubsole MJ, Pei Z, Brucker R, et al. Cigarette smoking and oral microbiota in low-income and African-American populations. J Epidemiol Community Health. (2019) 73(12):1108–15. doi: 10.1136/jech-2019-212474
26. Lin D, Hutchison KE, Portillo S, Vegara V, Ellingson JM, Liu J, et al. Association between the oral microbiome and brain resting state connectivity in smokers. NeuroImage. (2019) 200:121–31. doi: 10.1016/j.neuroimage.2019.06.023
27. Srivastava A, Mishra S, Verma D. Characterization of oral bacterial composition of adult smokeless tobacco users from healthy Indians using 16S rDNA analysis. Microb Ecol. (2021) 82(4):1061–73. doi: 10.1007/s00248-021-01711-0
28. Department of Biotechnology, Maharaj Vinayak Global University, Jaipur Rajasthan, India, & Sharma MK. Metagenomic analysis of oral microbiota among oral cancer patients and tobacco chewers in Rajasthan, India. Bioinformation. (2022) 18(9):757–63. doi: 10.6026/97320630018757
29. Bahuguna M, Hooda S, Mohan L, Gupta RK, Diwan P. Identifying oral microbiome alterations in adult betel quid chewing population of Delhi, India. PLoS One. (2023) 18(1):e0278221. doi: 10.1371/journal.pone.0278221
30. Sawant S, Dugad J, Parikh D, Srinivasan S, Singh H. Oral microbial signatures of tobacco chewers and oral cancer patients in India. Pathogens. (2023) 12(1):78. doi: 10.3390/pathogens12010078
31. Yadav S, Tripathi V, Saran V. Identification of habit specific bacteria in human saliva through next-generation sequencing. Forensic Sci Int. (2023) 353:111871. doi: 10.1016/j.forsciint.2023.111871
32. Vallès Y, Inman CK, Peters BA, Ali R, Wareth LA, Abdulle A, et al. Types of tobacco consumption and the oral microbiome in the United Arab Emirates healthy future (UAEHFS) pilot study. Sci Rep. (2018) 8(1):11327. doi: 10.1038/s41598-018-29730-x
33. Al Bataineh MT, Dash NR, Elkhazendar M, Alnusairat DMH, Darwish IMI, Al-Hajjaj MS, et al. Revealing oral microbiota composition and functionality associated with heavy cigarette smoking. J Transl Med. (2020) 18(1):421. doi: 10.1186/s12967-020-02579-3
34. Halboub E, Al-Ak’hali MS, Alamir AH, Homeida HE, Baraniya D, Chen T, et al. Tongue microbiome of smokeless tobacco users. BMC Microbiol. (2020) 20(1):201. doi: 10.1186/s12866-020-01883-8
35. Al Kawas S, Al-Marzooq F, Rahman B, Shearston JA, Saad H, Benzina D, et al. The impact of smoking different tobacco types on the subgingival microbiome and periodontal health: a pilot study. Sci Rep. (2021) 11(1):1113. doi: 10.1038/s41598-020-80937-3
36. Al-Marzooq F, Al Kawas S, Rahman B, Shearston JA, Saad H, Benzina D, et al. Supragingival microbiome alternations as a consequence of smoking different tobacco types and its relation to dental caries. Sci Rep. (2022) 12(1):2861. doi: 10.1038/s41598-022-06907-z
37. Jia Y-J, Liao Y, He Y-Q, Zheng M-Q, Tong X-T, Xue W-Q, et al. Association between oral microbiota and cigarette smoking in the Chinese population. Front Cell Infect Microbiol. (2021) 11:658203. doi: 10.3389/fcimb.2021.658203
38. Li Z, Liu Y, Dou L, Zhang Y, He S, Zhao D, et al. The effects of smoking and drinking on the oral and esophageal microbiota of healthy people. Ann Transl Med. (2021) 9(15):1244. doi: 10.21037/atm-21-3264
39. Huang Q, Wu X, Zhou X, Sun Z, Shen J, Kong M, et al. Association of cigarette smoking with oral bacterial microbiota and cardiometabolic health in Chinese adults. BMC Microbiol. (2023) 23(1):346. doi: 10.1186/s12866-023-03061-y
40. Sato N, Kakuta M, Uchino E, Hasegawa T, Kojima R, Kobayashi W, et al. The relationship between cigarette smoking and the tongue microbiome in an east Asian population. J Oral Microbiol. (2020) 12(1):1742527. doi: 10.1080/20002297.2020.1742527
41. Suzuki N, Nakano Y, Yoneda M, Hirofuji T, Hanioka T. The effects of cigarette smoking on the salivary and tongue microbiome. Clin Exp Dent Res. (2022) 8(1):449–56. doi: 10.1002/cre2.489
42. Thomas AM, Gleber-Netto FO, Fernandes GR, Amorim M, Barbosa LF, Francisco ALN, et al. Alcohol and tobacco consumption affects bacterial richness in oral cavity mucosa biofilms. BMC Microbiol. (2014) 14(1):250. doi: 10.1186/s12866-014-0250-2
43. Al-Zyoud W, Hajjo R, Abu-Siniyeh A, Hajjaj S. Salivary microbiome and cigarette smoking: a first of its kind investigation in Jordan. Int J Environ Res Public Health. (2020) 17(1):256. doi: 10.3390/ijerph17010256
44. Wirth R, Maróti G, Mihók R, Simon-Fiala D, Antal M, Pap B, et al. A case study of salivary microbiome in smokers and non-smokers in Hungary: analysis by shotgun metagenome sequencing. J Oral Microbiol. (2020) 12(1):1773067. doi: 10.1080/20002297.2020.1773067
45. Bašić K, Peroš K, Bošnjak Z, Šutej I. Subgingival microbiota profile in association with cigarette smoking in young adults: a cross-sectional study. Dent J. (2021) 9(12):150. doi: 10.3390/dj9120150
46. Wu Z, Han Y, Caporaso JG, Bokulich N, Mohamadkhani A, Moayyedkazemi A, et al. Cigarette smoking and opium use in relation to the oral microbiota in Iran. Microbiol Spectr. (2021) 9(2):e00138–21. doi: 10.1128/Spectrum.00138-21
47. Pfeiffer S, Herzmann C, Gaede KI, Kovacevic D, Krauss-Etschmann S, Schloter M. Different responses of the oral, nasal and lung microbiomes to cigarette smoke. Thorax. (2022) 77(2):191–5. doi: 10.1136/thoraxjnl-2020-216153
48. Poulsen CS, Nygaard N, Constancias F, Stankevic E, Kern T, Witte DR, et al. Association of general health and lifestyle factors with the salivary microbiota—lessons learned from the ADDITION-PRO cohort. Front Cell Infect Microbiol. (2022) 12:1055117. doi: 10.3389/fcimb.2022.1055117
49. Antonello G, Blostein F, Bhaumik D, Davis E, Gögele M, Melotti R, et al. Smoking and salivary microbiota: a cross-sectional analysis of an Italian alpine population. Sci Rep. (2023) 13(1):18904. doi: 10.1038/s41598-023-42474-7
50. Sami A, Elimairi I, Ryan CA, Stanton C, Patangia D, Ross RP. Altered oral microbiome in Sudanese toombak smokeless tobacco users carries a newly emerging risk of squamous cell carcinoma development and progression. Sci Rep. (2023) 13(1):6645. doi: 10.1038/s41598-023-32892-y
51. Galvin S, Anishchuk S, Healy CM, Moran GP. Smoking, tooth loss and oral hygiene practices have significant and site-specific impacts on the microbiome of oral mucosal surfaces: a cross-sectional study. J Oral Microbiol. (2023) 15(1):2263971. doi: 10.1080/20002297.2023.2263971
52. Yu K-M, Cho H-S, Lee A-M, Lee J-W, Lim S-K. Analysis of the influence of host lifestyle (coffee consumption, drinking, and smoking) on Korean oral microbiome. Forensic Sci Int Genet. (2024) 68:102942. doi: 10.1016/j.fsigen.2023.102942
53. Shakhatreh MAK, Khabour OF, Alzoubi KH, Masadeh MM, Hussein EI, Bshara GN. Alterations in oral microbial flora induced by waterpipe tobacco smoking. Int J Gen Med. (2018) 1:47–54. doi: 10.2147/IJGM.S150553
54. Jin J, Guo L, VonTungeln L, Vanlandingham M, Cerniglia CE, Chen H. Smokeless tobacco impacts oral microbiota in a Syrian golden hamster cheek pouch carcinogenesis model. Anaerobe. (2018) 52:29–42. doi: 10.1016/j.anaerobe.2018.05.010
55. Cosseau C, Devine DA, Dullaghan E, Gardy JL, Chikatamarla A, Gellatly S, et al. The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host-microbe homeostasis. Infect Immun. (2008) 76(9):4163–75. doi: 10.1128/IAI.00188-08
56. Antinozzi M, Giffi M, Sini N, Gallè F, Valeriani F, De Vito C, et al. Cigarette smoking and human gut microbiota in healthy adults: a systematic review. Biomedicines. (2022) 10(2):510. doi: 10.3390/biomedicines10020510
57. Wang Q, Rao Y, Guo X, Liu N, Liu S, Wen P, et al. Oral microbiome in patients with oesophageal squamous cell carcinoma. Sci Rep. (2019) 9(1):19055. doi: 10.1038/s41598-019-55667-w
58. Li D, Xi W, Zhang Z, Ren L, Deng C, Chen J, et al. Oral microbial community analysis of the patients in the progression of liver cancer. Microb Pathog. (2020) 149:104479. doi: 10.1016/j.micpath.2020.104479
59. Lu H, Ren Z, Li A, Li J, Xu S, Zhang H, et al. Tongue coating microbiome data distinguish patients with pancreatic head cancer from healthy controls. J Oral Microbiol. (2019) 11(1):1563409. doi: 10.1080/20002297.2018.1563409
60. Xu Z, Lv Z, Chen F, Zhang Y, Xu Z, Huo J, et al. Dysbiosis of human tumor microbiome and aberrant residence of Actinomyces in tumor-associated fibroblasts in young-onset colorectal cancer. Front Immunol. (2022 Sep 2) 13:1008975. doi: 10.3389/fimmu.2022.1008975
61. Li H, Thompson I, Carter P, Whiteley A, Bailey M, Leifert C, et al. Salivary nitrate—an ecological factor in reducing oral acidity. Oral Microbiol Immunol. (2007) 22(1):67–71. doi: 10.1111/j.1399-302X.2007.00313.x
62. Hyde ER, Andrade F, Vaksman Z, Parthasarathy K, Jiang H, Parthasarathy DK, et al. Metagenomic analysis of nitrate-reducing bacteria in the oral cavity: implications for nitric oxide homeostasis. PloS One. (2014) 9(3):e88645. doi: 10.1371/journal.pone.0088645
63. Nolan-Kenney R, Wu F, Hu J, Yang L, Kelly D, Li H, et al. The association between smoking and gut microbiome in Bangladesh. Nicotine Tob Res. (2020) 22(8):1339–46. doi: 10.1093/ntr/ntz220
64. Zhang J, Yu J, Dou J, Hu P, Guo Q. The impact of smoking on subgingival plaque and the development of periodontitis: a literature review. Front Oral Health. (2021) 2:751099. doi: 10.3389/froh.2021.751099
65. Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. (2005) 17(1):1–14. doi: 10.1093/intimm/dxh186
66. Kawai T, Akira S. Signaling to NF-kappaB by toll-like receptors. Trends Mol Med. (2007) 13(11):460–9. doi: 10.1016/j.molmed.2007.09.002
67. Semlali A, Witoled C, Alanazi M, Rouabhia M. Whole cigarette smoke increased the expression of TLRs, HBDs, and proinflammory cytokines by human gingival epithelial cells through different signaling pathways. PLoS One. (2012) 7(12):e52614. doi: 10.1371/journal.pone.0052614
68. Pace E, Ferraro M, Siena L, Melis M, Montalbano AM, Johnson M, et al. Cigarette smoke increases toll-like receptor 4 and modifies lipopolysaccharide-mediated responses in airway epithelial cells. Immunology. (2008) 124(3):401–11. doi: 10.1111/j.1365-2567.2007.02788.x
69. Varma AS, Suragimath G, Pisal A, Disale PR, Zope S. Toll-like receptors: molecular microbe sensors in periodontium. World J Dent. (2019) 10(5):396–401. doi: 10.5005/jp-journals-10015-1666
70. Sun Y, Shu R, Zhang M-Z, Wu A-P. Toll-like receptor 4 signaling plays a role in triggering periodontal infection. FEMS Immunol Med Microbiol. (2008) 52(3):362–9. doi: 10.1111/j.1574-695X.2008.00386.x
71. Décanis N, Savignac K, Rouabhia M. Farnesol promotes epithelial cell defense against Candida albicans through toll-like receptor 2 expression, interleukin-6 and human beta-defensin 2 production. Cytokine. (2009) 45(2):132–40. doi: 10.1016/j.cyto.2008.11.011
72. Stathopoulou PG, Benakanakere MR, Galicia JC, Kinane DF. Epithelial cell proinflammatory cytokine response differs across dental plaque bacterial species. J Clin Periodontol. (2010) 37(1):24–9. doi: 10.1111/j.1600-051X.2009.01505.x
73. Mori Y, Yoshimura A, Ukai T, Lien E, Espevik T, Hara Y. Immunohistochemical localization of toll-like receptors 2 and 4 in gingival tissue from patients with periodontitis. Oral Microbiol Immunol. (2003) 18(1):54–8. doi: 10.1034/j.1399-302x.2003.180109.x
74. Vu AT, Baba T, Chen X, Le TA, Kinoshita H, Xie Y, et al. Staphylococcus aureus membrane and diacylated lipopeptide induce thymic stromal lymphopoietin in keratinocytes through the toll-like receptor 2-toll-like receptor 6 pathway. J Allergy Clin Immunol. (2010) 126(5):985–93, 993.e1–3. doi: 10.1016/j.jaci.2010.09.002
75. Pisani LP, Estadella D, Ribeiro DA. The role of toll like receptors (TLRs) in oral carcinogenesis. Anticancer Res. (2017) 37(10):5389–94. doi: 10.21873/anticanres.11965
76. Fukata M, Chen A, Vamadevan AS, Cohen J, Breglio K, Krishnareddy S, et al. Toll-like receptor-4 promotes the development of colitis-associated colorectal tumors. Gastroenterology. (2007) 133(6):1869–81. doi: 10.1053/j.gastro.2007.09.008
77. Lee C-H, Wu C-L, Shiau A-L. Toll-like receptor 4 signaling promotes tumor growth. Journal of Immunotherapy (Hagerstown, Md.: 1997). (2010) 33(1):73–82. doi: 10.1097/CJI.0b013e3181b7a0a4
78. Cogo K, de Andrade A, Labate CA, Bergamaschi CC, Berto LA, Franco GCN, et al. Proteomic analysis of Porphyromonas gingivalis exposed to nicotine and cotinine. J Periodontal Res. (2012) 47(6):766–75. doi: 10.1111/j.1600-0765.2012.01494.x
79. Bagaitkar J, Daep CA, Patel CK, Renaud DE, Demuth DR, Scott DA. Tobacco smoke augments Porphyromonas gingivalis-Streptococcus gordonii biofilm formation. PLoS One. (2011) 6(11):e27386. doi: 10.1371/journal.pone.0027386
80. How KY, Song KP, Chan KG. Porphyromonas gingivalis: an overview of periodontopathic pathogen below the gum line. Front Microbiol. (2016) 7:53. doi: 10.3389/fmicb.2016.00053
81. Olsen I, Lambris JD, Hajishengallis G. Porphyromonas gingivalis disturbs host-commensal homeostasis by changing complement function. J Oral Microbiol. (2017) 9(1):1340085. doi: 10.1080/20002297.2017.1340085
82. Hiratsuka K, Abiko Y, Hayakawa M, Ito T, Sasahara H, Takiguchi H. Role of Porphyromonas gingivalis 40-kDa outer membrane protein in the aggregation of P. gingivalis vesicles and Actinomyces viscosus. Arch Oral Biol. (1992) 37(9):717–24. doi: 10.1016/0003-9969(92)90078-m
83. Nagata H, Iwasaki M, Maeda K, Kuboniwa M, Hashino E, Toe M, et al. Identification of the binding domain of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase for Porphyromonas gingivalis major fimbriae. Infect Immun. (2009) 77(11):5130–8. doi: 10.1128/IAI.00439-09
84. Sufaru I-G, Martu M-A, Solomon SM. Advances in periodontal pathogens. Microorganisms. (2022) 10(7):1439. doi: 10.3390/microorganisms10071439
85. Tomar SL, Hecht SS, Jaspers I, Gregory RL, Stepanov I. Oral health effects of combusted and smokeless tobacco products. Adv Dent Res. (2019) 30(1):4–10. doi: 10.1177/0022034519872480
86. Hani J, Abdel Nour G, Matta J, Jazzar B, Pfaffl MW, Hanna-Wakim L, et al. Shisha microbiota: the good, the bad and the not so ugly. BMC Res Notes. (2018) 11(1):446. doi: 10.1186/s13104-018-3553-9
87. Liu M, Jin J, Pan H, Feng J, Cerniglia CE, Yang M, et al. Effect of smokeless tobacco products on human oral bacteria growth and viability. Anaerobe. (2016) 42:152–61. doi: 10.1016/j.anaerobe.2016.10.006
88. Huang R, Li M, Gregory RL. Effect of nicotine on growth and metabolism of Streptococcus mutans. Eur J Oral Sci. (2012) 120(4):319–25. doi: 10.1111/j.1600-0722.2012.00971.x
89. Wu J, Li Q, Fu X. Fusobacterium nucleatum contributes to the carcinogenesis of colorectal cancer by inducing inflammation and suppressing host immunity. Transl Oncol. (2019) 12(6):846–51. doi: 10.1016/j.tranon.2019.03.003
90. McIlvanna E, Linden GJ, Craig SG, Lundy FT, James JA. Fusobacterium nucleatum and oral cancer: a critical review. BMC Cancer. (2021) 21(1):1212. doi: 10.1186/s12885-021-08903-4
Keywords: microbiome, oral, tobacco, smokers, microbiota, chewers
Citation: Senaratne NLM, Yung on C, Shetty NY and Gopinath D (2024) Effect of different forms of tobacco on the oral microbiome in healthy adults: a systematic review. Front. Oral. Health 5:1310334. doi: 10.3389/froh.2024.1310334
Received: 9 October 2023; Accepted: 11 January 2024;
Published: 20 February 2024.
Edited by:
Praveen S. Jodalli, Manipal College of Dental Sciences, IndiaReviewed by:
Atrey Pai Khot, King George's Medical University, IndiaRamya Iyer, KM Shah Dental College and Hospital, India
© 2024 Senaratne, Yung on, Shetty and Gopinath. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Divya Gopinath ZC5nb3BpbmF0aEBham1hbi5hYy5hZQ==
†ORCID Divya Gopinath orcid.org/0000-0002-4279-7420
 Nikitha Lalindri Mareena Senaratne
Nikitha Lalindri Mareena Senaratne Cheng Yung on3
Cheng Yung on3 Divya Gopinath
Divya Gopinath