- Centre for Host-Microbiome Interactions, Faculty of Dentistry, Oral & Craniofacial Sciences, King’s College London, London, United Kingdom
Noma is a rapidly progressing periodontal disease with up to 90% mortality in developing countries. Poor, immunocompromised and severely malnourished children (2 to 6 years old) are mostly affected by Noma. Prevention and effective management of Noma is hindered by the lack of sufficient cohesive studies on the microbial etiology of the disease. Research efforts have not provided a comprehensive unified story of the disease. Bridging the gap between existing studies gives an insight on the disease pathogenesis. This current systematic review of etiological studies focuses on the key players of dysbiosis in Noma disease. This review was performed in accordance with the Preferred Reporting Items for Systemic review and Meta-Analyses (PRISMA) statement. Web of Science, MEDLINE via PubMed, Cochrane Library, Scopus, and Science Direct were searched electronically for clinical trials which applied culture dependent or molecular techniques to identify oral microbiota from Noma patients. Trials which involved periodontal diseases except Noma were excluded. After screening 275 articles, 153 full-texts articles were assessed for eligibility of which eight full text articles were selected for data extraction and analysis. The results show that 308 samples from 169 Noma participants (6 months to 15 years old) have been used in clinical trials. There was some variance in the microbiome identified due to the use of 3 different types of samples (crevicular fluid, subgingival plaque, and swabbed pus) and the ambiguity of the stage or advancement of Noma in the studies. Other limitations of the studies included in this review were: the absence of age-matched controls in some studies; the constraints of colony morphology as a tool in distinguishing between virulent fusobacterium genus at the species level; the difficulty in culturing spirochaetes in the laboratory; the choice of primers in DNA amplification; and the selection of probe sets in gene sequencing. This systematic review highlights spirochaetes and P. intermedia as putative trigger organisms in Noma dysbiosis, shows that F. nucleatum promotes biofilms formation in late stages of the disease and suggests that future studies should be longitudinal, with high throughput genome sequencing techniques used with gingival plaque samples from early stages of Noma.
1. Introduction
Noma is a ravaging orofacial gangrenous stomatitis which is characterized by acute necrotizing ulcerative lesions (1, 2). Noma is prevalent in developing countries where most of the victims are children (3–5). Epidemiological case study reports have established risk factors for the disease such as poor hygiene and nutritional status, measles and other eruptive fevers and immunocompromising diseases (6, 7). Although there are studies on Noma dated more than a century ago, the etiological organisms as well as the trigger agents are yet to be sufficiently detailed (8).
For decades, periodontal diseases were reported as infectious diseases caused by singular organisms, but more recent studies have established the host’s response to microbial dysbiosis as pivotal in the pathogenesis (9, 10). Dysbiosis is a microbial community shift or loss of homeostasis which is detrimental to human health (11, 12). Such detrimental effect is influenced by an alteration in ecological diversity, decrease in beneficial species, and an expansion of pathobionts (13, 14). Periodontal diseases are driven by complex dysbiosis of the oral microbiota (15, Deng et al. 2017b). Key players in the microbial community perform a transitory role from healthy state to dysbiosis (16, 17, Wang et al. 2012). Prevalence of these key players inflame the periodontal conditions such that commensal microorganisms are unable to thrive (18, 19). Although some research findings show that the products of metabolic activity in commensal bacteria play a role in periodontal diseases, the initiation of dysbiosis in Noma has not been thoroughly explored (20, 21).
The application of microbial and molecular methods in microbial ecology has elucidated the potential causative agents or trigger organisms in the pathogenesis of several diseases (22, 23). These breakthroughs have resulted in the prevention, diagnosis, and treatment interventions of diseases (24–27). This review of all primary etiological studies on Noma was carried out to establish the extent of consistency in the determination of the oral microbiome and key players of dysbiosis related to Noma disease, to highlight the constraints of the research studies, and to recommend improved strategies for future etiological studies.
2. Methods
This review was performed observing the Preferred Reporting Items for Systemic review and Meta-Analyses (PRISMA) statement (28). A search was conducted electronically in the following database for related papers: Web of Science; MEDLINE via PubMed; Cochrane Library; Scopus; Science Direct. This search used “Cancrum Oris” or “Noma” as context as well the following terms and their combinations: “Isolation” or “bacterial” or “clinical” or “microbiota”. Clinical trials which investigated the microbiome or characterized microorganisms from Noma patients either by culture dependent or molecular techniques were included. Clinical trials which characterized microorganisms from the oral cavity of patients with periodontal or gingival diseases which were not explicitly reported as Noma were excluded. Clinical trials with a generalized deficient description of microorganisms were excluded. Publications which were not available online were sourced directly from corresponding authors. The final search was performed on 29 September 2022.
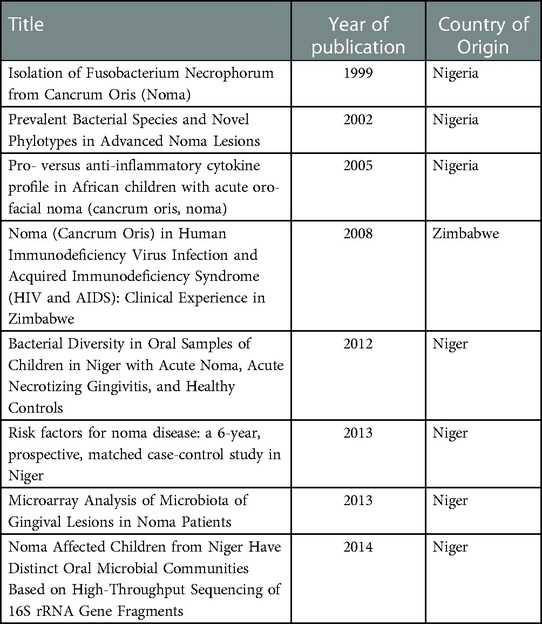
Screening of the title and abstract was performed by the first and last author. The eligibility criteria used were: Primary study, clinical trials, Noma disease, and human patients. The following data items were extracted from searching the full text: article title, first author, year of publication, country of origin of sample population (Table 1). The analysis was conducted manually by reporting individual parameters of the included article in depth. These areas of interest were explored included the subject description, sampling details, methodical approach, method of identification, organisms identified & their relative prevalence, unique features of each clinical trial, bias & limitations of each experimental design.
3. Results
3.1. Included studies
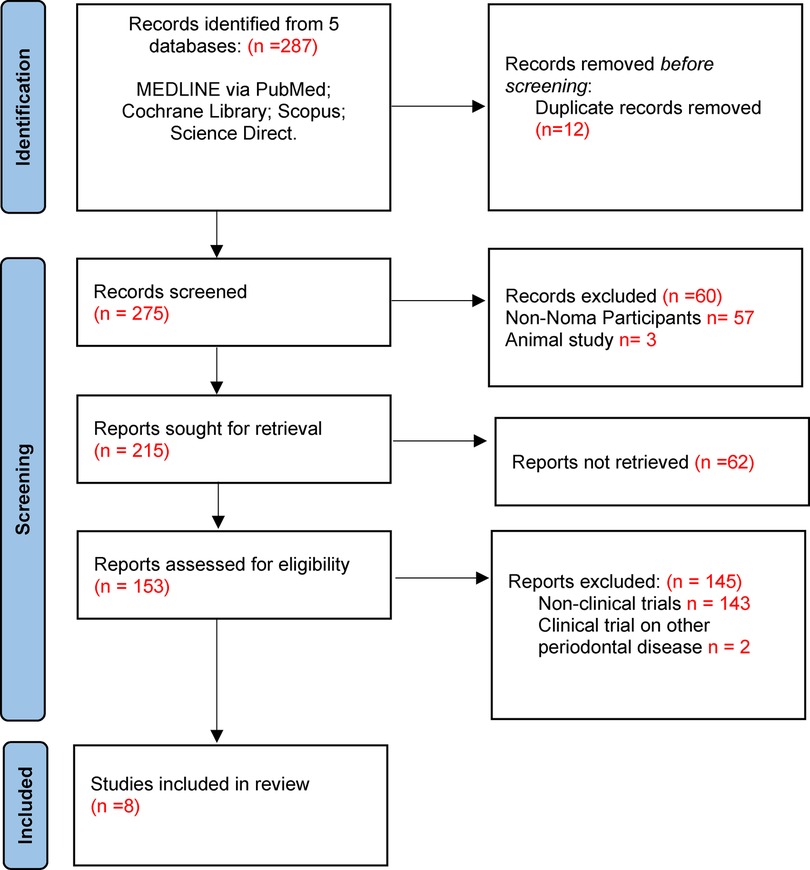
The search was completed on 29 September 2022. A total of 287 article titles were retrieved from five databases. Upon removal of duplicates, 275 articles were then screened, and 153 full-texts articles were further assessed for eligibility. A total of 145 articles were excluded and 8 full text articles were included for data extraction and analysis (Figure 1). Considering the limited research publications about Noma disease, the studies included were of sufficient quality for this review.
The dates of the eight full text articles included in the data extraction spanned the period from 2009 to 2014. Although authors of the publications were globally distributed, the etiological studies were carried out on participants from the Western and Southern geographical regions of Africa namely, Nigeria, Niger, and Zimbabwe (Table 1). Further, all publications were identified as cross-sectional studies with case-control matching based on the Noma disease factor.
3.2. Sampling details
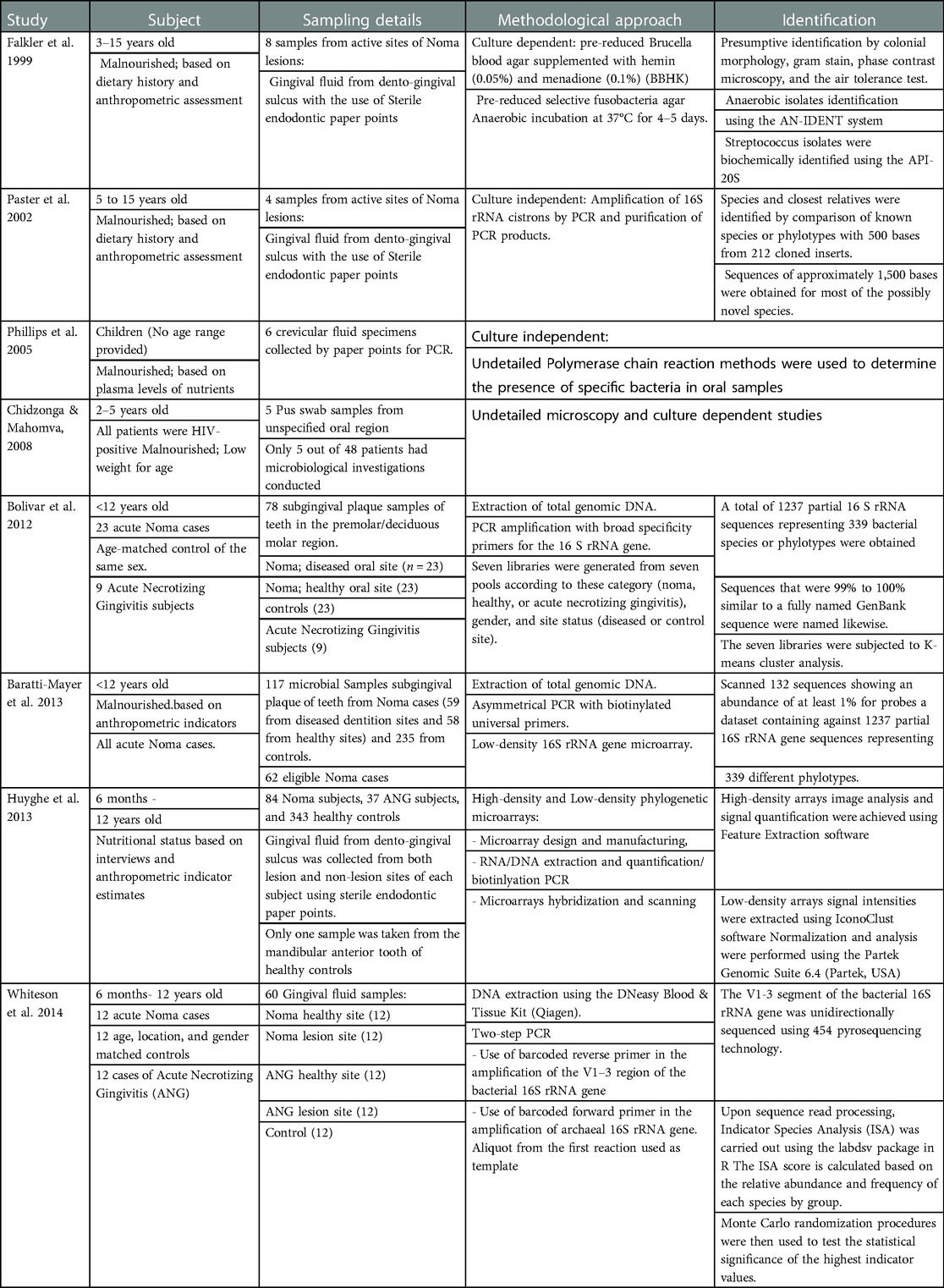
A total of 308 samples from 169 Noma participants and at least 747 case control participants have been used in etiological studies of Noma (Table 2). The age range of the participants span from 6 months to 15 years old. All Noma patients were assessed as malnourished based on standardized protocols of dietary history, anthropometric assessment, and plasma levels of nutrients. There was no categorization of the stage of Noma in the participants and only one study has reported other pre-existing immunocompromising conditions (HIV/AIDS). The samplings were performed at the dento-gingival sulcus at the active sites of lesions, damaged tissue, and the teeth at diseased dentition sites. Only 3 types of samples were used in the studies (highlighted in Table 2): crevicular/gingival fluid, subgingival tooth plaque, and pus swab. Clinical reports which described spirochetes and Fusobacteriales prior to these studies were excluded from the results presentation due to incomplete or inexplicit classification of isolates.
3.3. Culture-dependent microbial studies
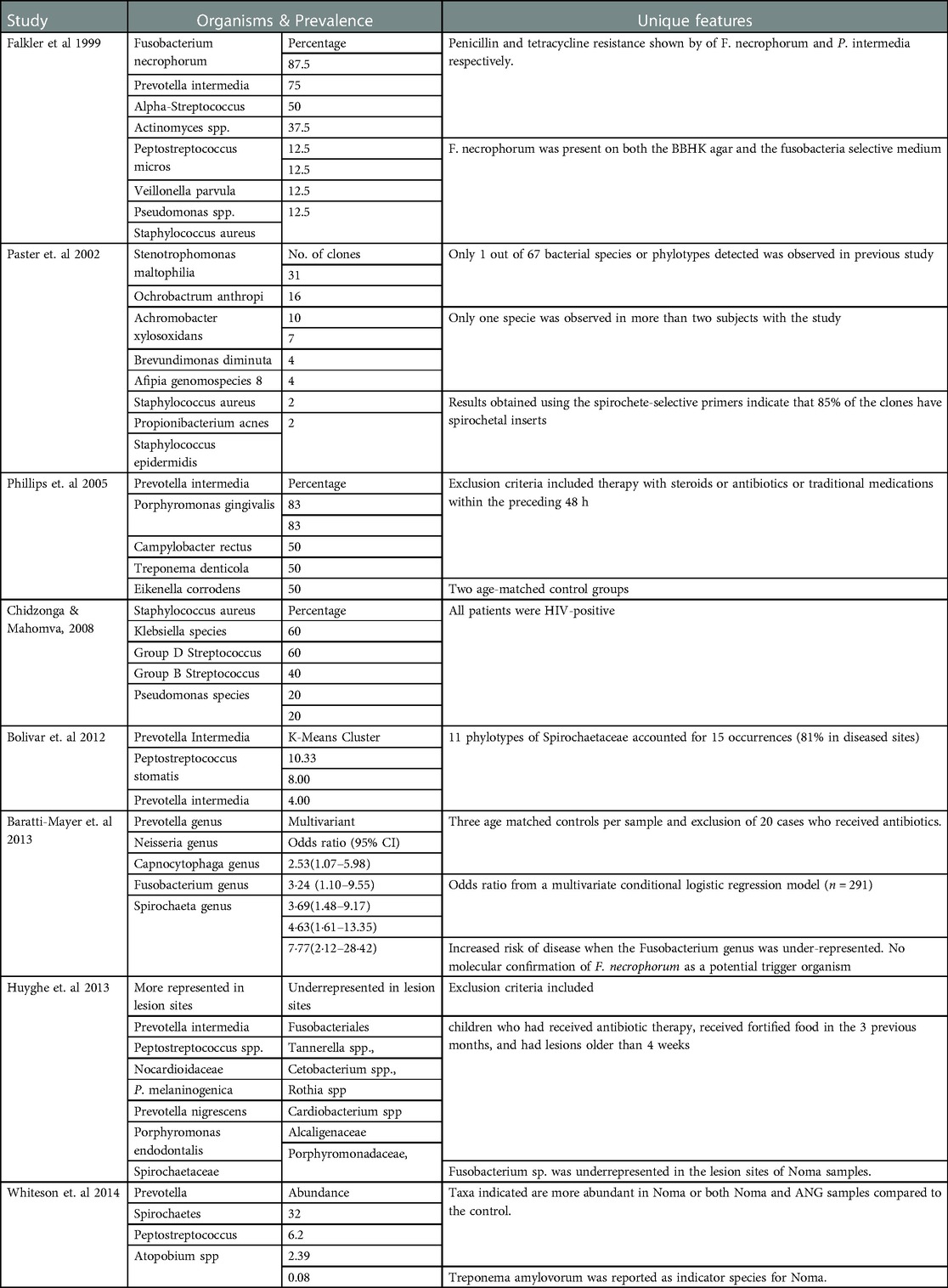
The methodical approaches used in investigating the etiology of Noma are in two categories: the general culture-based method and the more species defining culture-independent methods (Table 2). The use of only pre-reduced Brucella blood agar and pre-reduced selective Fusobacteria agar represented 2.6% of the samples assayed. Beta-hemolytic Fusobacterium necrophorum was reported in 7 out of 8 diseased dento-gingival sulci (Table 3) and is present on both BHI agar and the Fusobacteria selective medium. Prevotella intermedia was identified in 6 out of 8 samples. Moreover, another microbial study identified Staphylococcus aureus, Klebsiella species, group D Streptococcus, and group B hemolytic Streptococcus as the predominant organisms in pus swabs from 5 Noma patients.
3.4. Cloning and 16s rRNA gene sequencing
The first culture independent study on Noma etiology involved a subset of the sample population previously used in the microbial method. In 4 samples of gingival fluid (diseased) only Staphylococcus aureus was recurrent in both methods of identification (Table 3) and Stenotrophomonas maltophilia though undetected through microbial culture, was identified in more than two of the four samples by sequencing. The second 16S rRNA sequencing of crevicular fluid from Noma patients showed divergent results. Prevotella intermedia and Tannerella forsythia were identified in 5 out of 6 samples.
3.5. 16s rRNA gene-based oligonucleotide microarray analysis
Low-density and high-density phylogenetic microarrays probed 92.5% of all samples in these reported Noma etiological studies. Although low-density 16S rDNA microarray analysis showed considerable independent associations between microbiota and Noma, it did not report a specific organism as the causative pathogen. While P intermedia was associated with Noma, F necrophorum showed no triggering association. Fusobacterium nucleatum complex was the main reported Fusobacterium species.
High-density phylogenetic microarrays also observed that the Fusobacterium genus was prevalent or more abundant in healthy controls than Noma lesions (Table 3). In fact, fusobacteriales such as Streptobacillus moniliformis, Cetobacterium, and Leptotrichia had higher abundance in healthy donors than in Noma lesions. Prevotella intermedia was the main reported Prevotellaceae genus associated with Noma samples.
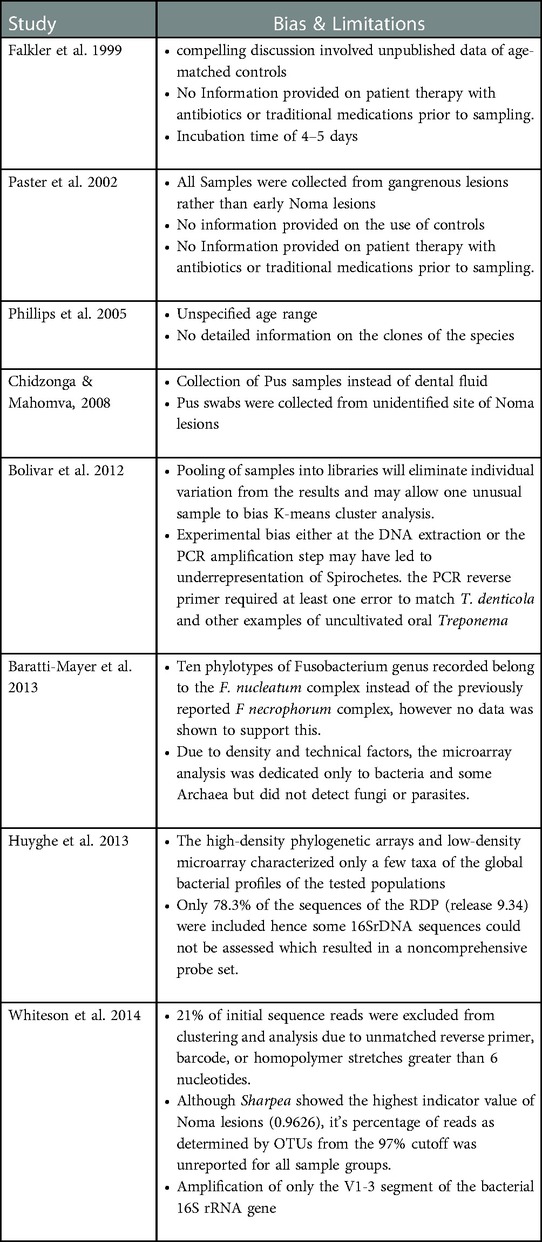
3.6. Limitations and bias in study designs
The limitations and bias associated with the study designs in etiological Noma research are in three categories: The lack of conformity in the type and site for sample collection; the lack of a valid control population to increase statistical reliability of results; the lack of a molecular assessment and analysis that is cognizant of the extensive biodiversity of the oral microbiota (Table 4). Due to the difference in presentation of results, the study heterogeneity could not be quantitatively analyzed by Meta-analysis.
4. Discussion
Distinct ecological habitats of various parts of the oral cavity represent different microbiota and disease dynamics (29, 30). The area of the oral cavity where the sample is collected plays some roles in the microbiome identified (31, 32). Although gingival crevicular fluid (GCF) has been extensively studied for antibodies and immunological inflammatory biomarkers of periodontal diseases (33, 34), saliva, supragingival and subgingival plaque are unequivocally preferred in the microbial characterization of periodontal diseases. Most studies (excluding 35) in this review carried out sampling from GCF and pus which raises the question of whether the organisms characterized are trigger microbes or merely a generic proliferation of the gingival microbiota in response to Noma disease. The analysis of GCF in chronic periodontitis showed that the distinguishing microorganisms and metabolites in these samples between periodontitis patients and healthy individuals are biomarkers (36). Saliva and gingival plaque may provide more information on the microbiota of periodontal dysbiosis than gingival crevicular fluid (37, 38).
All studies in this review were performed before the stage categorization of Noma disease was standardized (WHO Regional Office for Africa, 2016). As a result, there is evident disparity in the stage(s) of Noma disease reported in the sampled patients and the organisms identified. The sample population in many of the reviewed studies were patients with advanced lesions. These advanced lesions can be classified as stage 3 (Gangrenous), stage 4 (scarring), and stage 5 (sequelae) of Noma disease. At these stages, Noma is comparable to acute necrotizing ulcerative gingivitis (ANUG) (39, 40, Huyghe et al. 2013b). The identification of a trigger organism in post-virulent stages of Noma is therefore challenging and complex.
Although colony morphology is useful in distinguishing potential pathogens from normal flora, additional molecular techniques are required for discrimination of similar virulent species of the same genus (41). Fusobacterium necrophorum which causes pharyngotonsillitis and peritonsillar abscess but has not previously been linked with periodontal disease was identified by the first morphological study in this review (42). However, the absence of age-matched control groups and exclusion criteria for patients in this study poses a risk of bias in its design.
On the other hand, molecular studies in this review identified F. nucleatum, a periodontal pathogen which is ubiquitous in the oral cavity, as the main species in the Fusobacterium genus associated with Noma (43). The severity of periodontal diseases increases with the prevalence of F. nucleatum since it facilitates the formation of dental plaque (44, Han, 2015b). F. nucleatum is not considered a trigger organism in periodontal dysbiosis, although it connects the initial and later bacterial colonizers such that when F. nucleatum is absent, the prevalence of late colonizers is reduced (45, 46). F. nucleatum and F. necrophorum are the most frequent species implicated in Fusobacterium species bacteremia but morphological classification in the absence of molecular techniques such as 16s rRNA gene sequencing may lead to the misidentification of one for the other (47, 48).
Except for Fusobacterium species, the microbiota observed in ANUG is analogous to that described by molecular studies in this review (49, 50). Although this supports the theory of Noma as a dysbiosis, it does not support P. intermedia as the etiological trigger pathogen due to the advanced stage of the disease in which this species was characterized in (Darveau, Tanner, and Page, 1997 51). Previous studies have established the role of P. intermedia in the initiation and development of periodontitis by promoting periodontal connective tissue and bone matrix destruction through upregulated matrix metalloprotease production (52, Socransky et al. 1998b). Therefore, longitudinal molecular studies on the progression of simple gingivitis and acute necrotizing gingivitis to Noma sequelae will give a clearer picture of both the dysbiosis and trigger microorganism of Noma.
Like P. intermedia, oral spirochetes have been linked to necrotizing ulcerative gingivitis and other periodontal diseases. Particularly, clinical research has shown a correlation between the prevalence of Treponema denticola in periodontal pockets and the progression of periodontal disease in patients (53). Likewise, the higher representation of spirochaetes in Noma lesions is suggestive of its possible role in the dysbiosis of Noma. However, this was only reported in the molecular studies (35, Huyghe et al. 2013c). Due to the difficult nature of culturing Spirochaetaceae in the laboratory, specific culture conditions and specialized media are essential to optimize their growth and isolation (54, 55). Studies have demonstrated that Spirochaetaceae growth is only noticeable 4 weeks after culture and reaches its maximum within 8–12 weeks (56, 57). This contrasts with the longest incubation time of 5 days reported in this review. Also, Spirochaetaceae showed optional growth in Barbour-Stonner-Kelly (BSK) medium with rabbit serum, BSK swine serum + 5 fluorouracil, Cystine Tellurite Blood (CTB) medium, and brain heart infusion broth (56, 57). This potentially accounts for the absence of spirochaetes in the Noma samples since the culture dependent studies used Brucella blood agar supplemented with hemin (0.05%) and menadione (0.1%) (BBHK) (58, Chidzonga and Mahomva, 2008b).
The choice of primers in DNA Amplification (PCR) influences the microbiota detected and quantified using molecular techniques in the studies examined here, especially at the phylum level (59). 16S rRNA gene sequencing which used universally conserved primers in PCR read did not show a high representation of Spirochaetaceae. Whereas using the same sample population, PCR which used spirochete selective primers indicated that 85% of the clones have spirochetal inserts. Spirochaete species have a guanine (G): cytosine (C) ratio ranging from 51% to 65% in DNA (60, 61). This phenomenon not only segments DNA into several linear pieces, but it also makes it a difficult target for universal primers in DNA-DNA Hybridization (62, 63). This further suggests that the amplification achieved by Real-Time qPCR might not be a true reflection of the triggering possibilities of Spirochaete species in the dysbiosis of Noma disease.
The last four studies in this review demonstrate the influence of different molecular techniques on the etiological findings of Noma studies. Although conducted by the Geneva Study Group on Noma (GESNOMA) using the same sample population, the individual study designs and the limitations of each identification approach were discriminating factors in the results. The first 16S rRNA gene sequencing (Bolivar et al. 2012) attributed the underrepresentation (K-mean) of Spirochetes in Noma lesions to possible experimental bias of PCR reverse primer. Nonetheless, 11 phylotypes of Spirochaetaceae accounted for 15 occurrences in diseased sites. The study also emphasized that it’s library cloning yielded ratios of species not absolute numbers. Therefore, an unexpected increase in the representation of non-related Bacteroidetes (83%), might have resulted in a matched decrease for some other related bacteria such as Fusobacteriaceae (44%).
The last study by GESNOMA amplified the V1-3 segment of the bacterial 16S rRNA gene prior to 454 pyrosequencing (44). V2-3 hypervariable regions are not suitable for distinguishing all bacteria at genus level. This is the only study that reported Treponema amylovorum as the Spirochaetaceae species for Noma instead of Treponema Denticola. High-throughput sequencing of the entire 16S gene more accurately discriminates bacterial species and copy variants (64, 65). The pyrosequencing analysis ranked fusobacterium as low Noma indicators while placing Sharpea as the highest indicator species of Noma. On the contrary, Sharpea are commonly associated with rumen samples from low-methane-emitting sheep (66). While the abundance of Prevotella was well described in the results, the exclusion of a significant 21% of initial sequence read might have biased the computational analysis.
5. Conclusion
Although this systematic review demonstrated that Noma is a periodontal dysbiosis with Spirochaetes and P. intermedia as putative trigger organisms, F. nucleatum also enhances the late colonization of biofilms in subgingival plaque of advancing lesions. However, the limitations and therefore the bias of the study designs in this review demands stronger evidence from further studies. Longitudinal studies on the acute necrotizing gingivitis and oedema stages of Noma in endemic communities with age-matched controls is needed in the future. Such studies should employ high throughput full genome sequencing techniques on gingival plaque and saliva samples with primers and probe sets that are comprehensive to all possible oral microbiota or the use of PCR-independent molecular deep sequencing techniques.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
IU and MI were involved in the concept and design. IU performed the searches. IU and MI conducted the title and abstract screening. IU, GP, and MI conducted the full text screening, performed the data extraction. IU, DM, GP, and MI Interpreted the results, and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the PGR International Studentship grant EN11089 awarded by Kings College London. The funding institution provided access to articles in restricted databases. The funding body was not involved in the design of the study, analysis, and interpretation of data or in writing the manuscript.
Acknowledgments
Special acknowledgement goes to Yuka Makino, World Health Organization Oral Health Technical (Africa Regional Office), for Inspiring this study on Noma disease, and to Elise Farley for providing valuable literature.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Feller L, Khammissa RAG, Altini M, Lemmer J. Noma (cancrum oris): an unresolved global challenge. Periodontol 2000. (2019) 80(1):189–99. doi: 10.1111/prd.12275
2. Khammissa RAG, Lemmer J, Feller L. ‘Noma staging: a review’, tropical medicine and health. BioMed Central Ltd. (2022) 50(1):40. doi: 10.1186/s41182-022-00431-6
3. Farley E, Mehta U, Srour ML, Lenglet A. Noma (cancrum oris): a scoping literature review of a neglected disease (1843 to 2021). PLoS Negl Trop Dis. (2021) 15(12):e0009844. doi: 10.1371/journal.pntd.0009844
4. Isah S, Amirtharajah M, Farley E, Semiyu Adetunji A, Samuel J, Oluyide B, et al. Model of care, noma children's hospital, northwest Nigeria. Trop Med Int Health. (2021) 26(9):1088–97. doi: 10.1111/tmi.13630
5. Dominic C, Farley E, Elkheir N. More than 100 years of neglect: a bibliometric analysis of global research on noma (cancrum oris). Trans R Soc Trop Med Hyg. (2022) 116(5):479–86. doi: 10.1093/trstmh/trab161
6. Farley E, Lenglet A, Ariti C, Jiya NM, Adetunji AS, van der Kam S, et al. Risk factors for diagnosed noma in northwest Nigeria: a case-control study, 2017. PLoS Negl Trop Dis. (2018) 12(8):e0006631. doi: 10.1371/journal.pntd.0006631
7. Non communicable Diseases Cluster (NCD) regional Programme for noma control (2022). Available at: www.panacee.fr.
8. Huyghe A, François P, Mombelli A, Tangomo M, Girard M, Baratti-Mayer D, et al. Microarray analysis of microbiota of gingival lesions in noma patients. PLoS Negl Trop Dis. (2013) 7(9):e2453. doi: 10.1371/journal.pntd.0002453
9. Hajishengallis G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. (2014) 35(1):3–11. doi: 10.1016/j.it.2013.09.001
10. Deng ZL, Szafrański SP, Jarek M, Bhuju S, Wagner-Döbler I. Dysbiosis in chronic periodontitis: key microbial players and interactions with the human host. Sci Rep. (2017) 7(1):3703. doi: 10.1038/s41598-017-03804-8
11. Kumar PS. Microbial dysbiosis: the root cause of periodontal disease. J Periodontol. (2021) 92(8):1079–87. doi: 10.1002/JPER.21-0245
12. Sedghi L, DiMassa V, Harrington A, Lynch SV, Kapila YL. The oral microbiome: role of key organisms and complex networks in oral health and disease. Periodontol 2000. (2021) 87(1):107–31. doi: 10.1111/prd.12393
13. Payne MA, Hashim A, Alsam A, Joseph S, Aduse-Opoku J, Wade WG, et al. Horizontal and vertical transfer of oral microbial dysbiosis and periodontal disease. J Dent Res. (2019) 98(13):1503–10. doi: 10.1177/0022034519877150
14. Curtis MA, Diaz PI, van Dyke TE. The role of the microbiota in periodontal disease, periodontology 2000. Blackwell Munksgaard. (2020) 83(1):14–25. doi: 10.1111/prd.12296
15. Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. (2012) 13(9):R79. doi: 10.1186/gb-2012-13-9-r79
16. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. (2006) 444(7122):1027–31. doi: 10.1038/nature05414
17. Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol. (2011) 7(10):569–78. doi: 10.1038/nrrheum.2011.121
18. Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. (2010) 8(7):481–90. doi: 10.1038/nrmicro2337
19. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. (2014) 157(1):21–141. doi: 10.1016/j.cell.2014.03.011
20. Yost S, Duran-Pinedo AE, Teles R, Krishnan K, Frias-Lopez J. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. (2015) 7(1):27. doi: 10.1186/s13073-015-0153-3
21. Herrero ER, Slomka V, Bernaerts K, Boon N, Hernandez-Sanabria E, Passoni BB, et al. Antimicrobial effects of commensal oral species are regulated by environmental factors. J Dent. (2016) 47:23–33. doi: 10.1016/j.jdent.2016.02.007
22. Hamilton-Miller J. Probiotics and prebiotics: scientific aspects G. W. Tannock, Ed. Caister Academic Press, Wymondham, UK, 2005. ISBN 1-904455-01-8. $99, 230 pp. JAC. (2006) 58(1):232–3. doi: 10.1093/jac/dkl171
23. Ippolito MM, Denny JE, Langelier C, Sears CL, Schmidt NW. Malaria and the microbiome: a systematic review. Clin Infect Dis. (2018) 67(12):1831–9. doi: 10.1093/cid/ciy374
24. Ku LC, Boggess KA, Cohen-Wolkowiez M. Bacterial meningitis in infants. Clin Perinatol. (2015) 42(1):29–45. doi: 10.1016/j.clp.2014.10.004
25. Alonso B, Pérez-Granda MJ, Latorre MC, Sánchez-Carrillo C, Bouza E, Muñoz P, et al. Production of biofilm by Staphylococcus aureus: association with infective endocarditis? Enferm Infecc Microbiol Clin. (2021) 40(8):418–22. doi: 10.1016/j.eimc.2021.03.012
26. Liu L, Huang Y, Fu Y, Rao J, Zeng F, Ji M, et al. Hepatitis B virus promotes hepatocellular carcinoma development by activating GP73 to repress the innate immune response. Infect Agents Cancer. (2022) 17(1):52. doi: 10.1186/s13027-022-00462-y
27. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. (2012) 490(7418):55–60. doi: 10.1038/nature11450
28. Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ (Clin Res ed). (2021) 372:n71. doi: 10.1136/bmj.n71
29. Liljemark WF, Bloomquist C. Human oral microbial ecology and dental caries and periodontal diseases. Crit Rev Oral Biol Med. (1996) 7(2):180–98. doi: 10.1177/10454411960070020601
30. Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. (2005) 43(11):5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005
31. Theilade E, Wright WH, Jensen SB, Löe H. Experimental gingivitis in man. II. A longitudinal clinical and bacteriological investigation. J Periodontal Res. (1966) 1:1–13. doi: 10.1111/j.1600-0765.1966.tb01842.x
32. Al-Kamel A, Baraniya D, Al-Hajj WA, Halboub E, Abdulrab S, Chen T, et al. Subgingival microbiome of experimental gingivitis: shifts associated with the use of chlorhexidine and N-acetyl cysteine mouthwashes. J Oral Microbiol. (2019) 11(1):1608141. doi: 10.1080/20002297.2019.1608141
33. Barros SP, Williams R, Offenbacher S, Morelli T. Gingival crevicular fluid as a source of biomarkers for periodontitis. Periodontol 2000. (2016) 70(1):53–64. doi: 10.1111/prd.12107
34. de Aguiar MCS, Perinetti G, Capelli J. The gingival crevicular fluid as a source of biomarkers to enhance efficiency of orthodontic and functional treatment of growing patients. BioMed Res Int. (2017) 2017(3):4–5. doi: 10.1155/2017/3257235
35. Baratti-Mayer D, Gayet-Ageron A, Hugonnet S, François P, Pittet-Cuenod B, Huyghe A, et al. Risk factors for noma disease: a 6-year, prospective, matched case-control study in Niger. The lancet. Global Health. (2013) 1(2):e87–96. doi: 10.1016/S2214-109X(13)70015-9
36. Pei J, Li F, Xie Y, Liu J, Yu T, Feng X. Microbial and metabolomic analysis of gingival crevicular fluid in general chronic periodontitis patients: lessons for a predictive, preventive, and personalized medical approach. EPMA J. (2020) 11(2):197–215. doi: 10.1007/s13167-020-00202-5
37. Yu FY, Wang QQ, Li M, Cheng YH, Cheng YL, Zhou Y, et al. Dysbiosis of saliva microbiome in patients with oral lichen planus. BMC Microbiol. (2020) 20(1):75. doi: 10.1186/s12866-020-01733-7
38. Diao J, Yuan C, Tong P, Ma Z, Sun X, Zheng S. Potential roles of the free salivary microbiome dysbiosis in periodontal diseases. Front Cell Infect Microbiol. (2021) 11:711282. doi: 10.3389/fcimb.2021.711282
39. Paster BJ, Falkler WA Jr, Enwonwu CO, Idigbe EO, Savage KO, Levanos VA, et al. Prevalent bacterial species and novel phylotypes in advanced noma lesions. J Clin Microbiol. (2002) 40(6):2187–91. doi: 10.1128/JCM.40.6.2187-2191.2002
40. Chidzonga MM, Mahomva L. Noma (cancrum Oris) in human immunodeficiency virus infection and acquired immunodeficiency syndrome (HIV and AIDS): clinical experience in Zimbabwe. J Oral Maxillofac Surg. (2008a) 66(3):475–85. doi: 10.1016/j.joms.2007.09.024
41. Workentine ML, Harrison JJ, Weljie AM, Tran VA, Stenroos PU, Tremaroli V, et al. Phenotypic and metabolic profiling of colony morphology variants evolved from Pseudomonas fluorescens biofilms. Environ Microbiol. (2010) 12(6):1565–77. doi: 10.1111/j.1462-2920.2010.02185.x
42. Centor RM, Atkinson TP, Xiao L. Fusobacterium necrophorum oral infections – A need for guidance. Anaerobe. (2022) 75:3. doi: 10.1016/j.anaerobe.2022.102532
43. Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. (2015a) 23:141–7. doi: 10.1016/j.mib.2014.11.013
44. Whitmore SE, Lamont RJ. Oral bacteria and cancer. PLoS Pathog. (2014) 10(3):2. doi: 10.1371/journal.ppat.1003933
45. de Andrade KQ, Almeida-Da-Silva CLC, Coutinho-Silva R. Immunological pathways triggered by porphyromonas gingivalis and fusobacterium nucleatum: therapeutic possibilities? Mediat Inflamm. (2019) 2019:5–9. doi: 10.1155/2019/7241312
46. Binder Gallimidi A, Fischman S, Revach B, Bulvik R, Maliutina A, Rubinstein AM, et al. Periodontal pathogens porphyromonas gingivalis and fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget. (2015) 6(26):22613–23. doi: 10.18632/oncotarget.4209
47. Afra K, Laupland K, Leal J, Lloyd T, Gregson D. Incidence, risk factors, and outcomes of fusobacterium species bacteremia. BMC Infect Dis. (2013) 13:264. doi: 10.1186/1471-2334-13-264
48. Sousa AM, Machado I, Nicolau A, Pereira MO. Improvements on colony morphology identification towards bacterial profiling. J Microbiol Methods. (2013) 95(3):327–35. doi: 10.1016/j.mimet.2013.09.020
49. Kaplan D. Acute Necrotizing Ulcerative Tonsillitis and Gingivitis (Vincent’s Infections) (1981).
50. Malek R, Gharibi A, Khlil N, Kissa J. Necrotizing ulcerative gingivitis. Contemp Clin Dent. (2017) 8(3):496–500. doi: 10.4103/ccd.ccd_1181_16
51. Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL, Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. (1998) 25(2):134–44. doi: 10.1111/j.1600-051X.1998.tb02419.x
52. Guan SM, Shu L, Fu SM, Liu B, Xu XL, Wu JZ. Prevotella intermedia upregulates MMP-1 and MMP-8 expression in human periodontal ligament cells. FEMS Microbiol Lett. (2009) 299(2):214–22. doi: 10.1111/j.1574-6968.2009.01748.x
53. Sela MN. Role of Treponema denticola in periodontal diseases. Crit Rev Oral Biol Med. (2001) 12(5):399–413. doi: 10.1177/10454411010120050301
54. Hardham JM, Rosey EL. Antibiotic selective markers and spirochete genetics. J Mol Microbiol Biotechnol. (2000) 2(4):425–32. Available at: http://europepmc.org/abstract/MED/11075914.11075914
55. Paster BJ, Dewhirst FE. Phylogenetic foundation of spirochetes. J Mol Microbiol Biotechnol. (2000) 2(4):341–4. Available at: http://europepmc.org/abstract/MED/11075904.11075904
56. Oliveira A, Fonseca A, Ishikawa M, Yoshinari N. Cinetic growth of Borrelia burgdorferi (Spirochaetaceae) in different culture media. Pesquisa Veterinária Brasileira. (2004) 24:61–4. doi: 10.1590/S0100-736X2004000200002
57. Oliveira A, Fonseca A, Costa C, Mantovani E, Yoshinari N. Growth, cysts and kinetics of Borrelia garinii (Spirochaetales: spirochaetacea) in different culture media. Memórias do Instituto Oswaldo Cruz. (2010) 105:717–9. doi: 10.1590/S0074-02762010000500020
58. Falkler WA Jr, Enwonwu CO, Idigbe EO. Isolation of Fusobacterium necrophorum from cancrum oris (noma). Am J Trop Med Hyg. (1999) 60(1):150–6. doi: 10.4269/ajtmh.1999.60.150
59. Palkova L, Tomova A, Repiska G, Babinska K, Bokor B, Mikula I, et al. Evaluation of 16S rRNA primer sets for characterisation of microbiota in paediatric patients with autism spectrum disorder. Sci Rep. (2021) 11(1):6781. doi: 10.1038/s41598-021-86378-w
60. Karami A, Hosseyni S, Kiarudi M. Molecular characterization of Borrelia burgdorferi linear plasmids by DNA hybridization, PCR, two-dimensional gel electrophoresis, and electron microscopy. Turk J Biol. (2007) 31:73–80.
61. Karami A, Sarshar M, Ranjbar R, Zanjani R. The Phylum Spirochaetaceae. 10.1007/978-3-642-38954-2_156 (2014).
62. Barcellos DE, de Uzeda M, Ikuta N, Lunge VR, Fonseca AS, Kader II, et al. Identification of porcine intestinal spirochetes by PCR-restriction fragment length polymorphism analysis of ribosomal DNA encoding 23S rRNA. Vet Microbiol. (2000) 75(2):189–98. doi: 10.1016/s0378-1135(00)00212-1
63. Lu JJ, Perng CL, Lee SY, Wan CC. Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J Clin Microbiol. (2000) 38(6):2076–80. doi: 10.1128/JCM.38.6.2076-2080.2000
64. Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. (2007) 69(2):330–9. doi: 10.1016/j.mimet.2007.02.005
65. Johnson JS, Spakowicz DJ, Hong BY, Petersen LM, Demkowicz P, Chen L, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. (2019) 10(1):5029. doi: 10.1038/s41467-019-13036-1
Keywords: oral health, Noma, etiology, microbiome, microbiology, molecular biology, dysbiosis
Citation: Uzochukwu I, Moyes D, Proctor G and Ide M (2023) The key players of dysbiosis in Noma disease; A systematic review of etiological studies. Front. Oral. Health 4:1095858. doi: 10.3389/froh.2023.1095858
Received: 11 November 2022; Accepted: 7 February 2023;
Published: 3 March 2023.
Edited by:
Dominic Augustine, M S Ramaiah University of Applied Sciences, IndiaReviewed by:
Miguel Carda Diéguez, Fundación para el Fomento de la Investigación Sanitaria y Biomédica de la Comunitat Valenciana (FISABIO), SpainMahin Ghorbani, Karolinska Institutet (KI), Sweden
© 2023 Uzochukwu, Moyes, Proctor and Ide. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark Ide bWFyay5pZGVAa2NsLmFjLnVr
Specialty Section: This article was submitted to Oral Infections and Microbes, a section of the journal Frontiers in Oral Health
 Ifeanyi Uzochukwu
Ifeanyi Uzochukwu David Moyes
David Moyes Gordon Proctor
Gordon Proctor Mark Ide
Mark Ide