
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oral. Health , 21 September 2022
Sec. Oral Health Promotion
Volume 3 - 2022 | https://doi.org/10.3389/froh.2022.1004930
This article is part of the Research Topic Dentistry and Oral Health in Africa View all 5 articles
 Dunstan Kalanzi1*
Dunstan Kalanzi1* Harriet Mayanja-Kizza2
Harriet Mayanja-Kizza2 Damalie Nakanjako2
Damalie Nakanjako2 Fred Semitala2
Fred Semitala2 Gerald Mboowa3
Gerald Mboowa3 Muhammad Mbabali1
Muhammad Mbabali1 Edgar Kigozi3
Edgar Kigozi3 Fred Ashaba Katabazi3
Fred Ashaba Katabazi3 Ivan Sserwadda3
Ivan Sserwadda3 David P. Kateete3
David P. Kateete3 Beatrice Achan4
Beatrice Achan4 Nelson K. Sewankambo2
Nelson K. Sewankambo2 Adrian Muwonge5
Adrian Muwonge5
Background: Dental caries is a multifactorial disease that affects many people. Even though microorganisms play a crucial role in causing dental caries, diagnosis is routinely macroscopic. In order to improve early detection especially in HIV patients who are disproportionately affected, there is need to reconcile the macroscopic and microscopic characteristics of dental caries. Therefore, the aim of this study was to characterize the oral microbiota profile along the decayed, missing, filled teeth (DMFT) index using amplicon sequencing data.
Methods: Amplicon sequencing of the V6-V8 region of the 16S rRNA gene was done on DNA recovered from whole unstimulated saliva of 59 HIV positive and 29 HIV negative individuals. The microbial structure, composition and co-occurrence networks were characterized using QIIME-2, Phyloseq, Microbiome-1.9.2 and Metacoder in R.
Results: We characterized the oral microbiota into 2,093 operational taxonomic units (OTUs), 21 phyla and 239 genera from 2.6 million high quality sequence reads. While oral microbiota did not cluster participants into distinct groups that track with the DMFT index, we observed the following: (a) The proportion of accessory microbiota was highest in the high DMFT category while the core size (∼50% of richness) remained relatively stable across all categories. (b) The abundance of core genera such as Stomatobaculum, Peptostreptococcus and Campylobacter was high at onset of dental caries, (c) A general difference in oral microbial biomass. (d) The onset of dental caries (low DMFT) was associated with significantly lower oral microbial entropy.
Conclusions: Although oral microbial shifts along the DMFT index were not distinct, we demonstrated the potential utility of microbiota dynamics to characterize oral disease. Therefore, we propose a microbial framework using the DMFT index to better understand dental caries among HIV positive people in resource limited settings.
Dental caries is a highly prevalent infectious disease that affects a third of the world's population (1). Dental caries is caused by a number of factors, such as, consumption of refined sugars, inadequate dental hygiene, susceptible tooth surfaces, microorganisms, and time (2). Dental caries is normally diagnosed by visual inspection; and in oral health surveys, it is recorded using the decayed, missing (due to caries) and filled teeth (DMFT) index (3). The DMFT index is a simple and common macroscopic tool (4).
Although dental caries diagnosis is normally based on visual inspection, microorganisms play a significant role in causing caries (5). With the advent of next-generation sequencing (NGS) technology, it is now possible to study the intricate relationships that exist in microbial communities (6), including those found in the oral cavity. Furthermore, the application of this technology in medicine, clinical metagenomics NGS (mNGS), is one of the fastest growing areas of medicine in the developed world (7).
While characterizing oral diseases such as dental caries through oral microbiota is common in the developed world, it remains largely unexplored in developing countries. This is of particular concern because many developing countries have a high burden of disease, which includes HIV. Moreover, it has been reported that people living with HIV/AIDS are at a higher risk of developing dental caries compared to the general population (8, 9). For example, in Uganda, 1.3 million people are infected with HIV (10), and the prevalence of dental caries in this patient population was found to be 83.7% with a mean DMFT of 5.9 (11). This is higher than the prevalence of dental caries in the general adult population reported to be 66.7% with a mean DMFT of 4.71 (12). However, the robust tracking of microbial changes based on the severity of dental caries in people living with HIV has paucity of data.
To address this paucity of research on tracking microbial characteristics in people living with HIV, we characterized oral microbiota differences along the DMFT index in HIV positive and negative individuals using 16S rRNA gene sequencing in a resource limited setting, as a stepping stone towards the application of clinical metagenomics in oral health.
The School of Medicine Research and Ethics committee of Makerere University (#REC REF 2017-053) approved this study, which was conducted in accordance with the ethical standards outlined in the 1964 Declaration of Helsinki and its later amendments. The research team obtained written informed consent from all participants recruited for the study.
This was a cross sectional study carried out at the Mulago Immune Suppression Syndrome (ISS) clinic, which is an HIV care clinic under the Makerere University Joint AIDS Program (MJAP). The Mulago ISS clinic has been in operation for over 15 years and provides HIV related services to approximately 16,000 patients annually, 80% of whom are on anti-retroviral treatment (ART). The clinicians treat on average 300 people every day, which makes it one of few places where HIV related co-morbidities like dental disease can be studied. For this study, we recruited and sampled HIV positive and negative adults at this clinic.
We conveniently sampled 88 persons of the 168 who participated in the study between January and May 2018. This is because only a few studies have reported on oral clinical metagenomics in low and middle income countries (LMICs); therefore, information on powering such studies is limited (13). Studies conducted elsewhere have used 50–65 samples to detect up to 80% of genera (14, 15).
We collected saliva samples for oral microbiome analysis between 9:00 AM and 12:00 noon, in order to minimize the effect of the circadian rhythm, according to a published protocol (16). For measurement of flow rate, saliva was collected for 5 min without any stimulation. Participants were asked not to swallow, but to expel the accumulated saliva into a calibrated plastic centrifuge tube at intervals over a period of 5 min. Saliva samples were collected on ice, transported on ice, and stored at −80°C prior to component analysis. Trained clinicians performed the oral examinations using dental probes and mirrors under suitable artificial light after saliva collection at the same visit.
We assessed caries status using the World Health Organization (WHO) caries classification criteria and reported these findings using the decayed (D), missing (M), filled (F), teeth (DMFT) index (17). A DMFT score of >0 was considered as dental caries and thereafter, we categorized the participants into healthy (DMFT = 0), low (DMFT = 1–3), medium (DMFT = 4–6), high (DMFT = 7–13) and extremely high (DMFT > 14) (Figure 1).
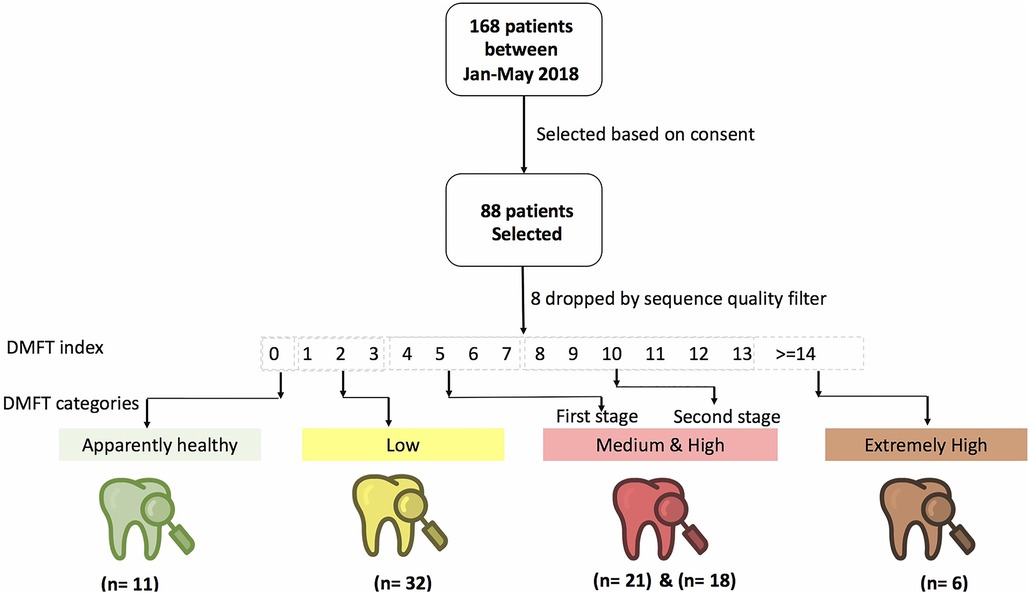
Figure 1. Shows the study design, patient and DMFT categorization of participants recruited from the Mulago Immune Suppression Syndrome (ISS) clinic at Makerere University Joint AIDS Program.
Periodontal health was assessed using a minimum diagnostic criteria of bleeding on probing and periodontal pocket depth measurements. Participants with only bleeding on probing were considered to have gingivitis, while those with periodontal pockets of 4 mm and greater were considered to have some form of periodontitis (18).
Saliva samples were processed as follows prior to DNA extraction using GenoLyse method. While in a biosafety cabinet, 3 milliliters of saliva were transferred to 15 ml sterile centrifuge tubes, and an equal volume of 1% w/v NaCl was added. The samples were vortexed vigorously for 1 min until the specimen was fully liquefied. This was done in order to digest saliva to release the bacteria. Phosphate buffered saline (PBS) pH 6.8 was then added up to the 15 ml mark. The specimens were then centrifuged at 3,000 × g for 15 min at 4 °C, and the supernatant discarded. The resulting pellet was suspended into 1 ml of 1XPBS and then transferred to a 1.5 ml nuclease free centrifuge tube. These were then centrifuged at 10,000 × g for 15 min prior to extraction of DNA using the GenoLyse method, following manufacturer's recommendations (Bruker, USA). Briefly, the supernatant was discarded and the pellet resuspended in 100 mls of GenoLyse lysis buffer (A-LYS) prior to gentle vortexing to lyse the cells of the microorganisms including bacteria. Additional cell lysis was achieved by incubating the tubes at 95 °C for 5 min prior to adding 100 mls of GenoLyse neutralizing buffer (A-NB) to stop the action of A-LYS. The mixture was vortexed again for about 2 s prior to centrifugation at 13,000 rpm for 5 min. DNA was collected from the supernatant and stored prior to use in subsequent analyses.
Aliquots of 30 µl were then shipped under controlled ambient condition to Dalhousie University Integrated Microbiome Resource (IMR, Canada) (19). Following purification of the amplicon pools using AMPure beads, sequencing of the V6-V8 16S rRNA variable loops was performed on the Illumina MiSeq platform (San Diego, CA, USA) using the 400 paired-end MiSeq run according to an established protocol (20). Following sequencing, demultiplexed samples were returned.
We received a total of 88 paired end sequences in fastq format via an html file transfer link from the Dalhousie University Integrated Microbiome Resource (IMR) (19). Demultiplexed reads from the sequencing facility were then imported into the QIIME 2 pipeline (21) for analysis. As part of quality control, the paired-ended reads were trimmed and merged into single ended reads. A QIIME artifact was then generated from these sequences and the metadata file. After dereplication, chimera-removal and denoising using DADA2 (21), an OTUs dataset was generated. We filtered OTUs with a sequence depth of at least 3,000, and retained 90% of the samples and the corresponding metadata (22).
We used QIIME 2 and R-based packages; Phyloseq and Microbiome analyzer for microbial diversity analysis. The OTU-data output was used to estimate the Alpha and Beta diversity indices. We considered observed Shannon for the Alpha diversity analysis (23), whereas Beta diversity was estimated with Bray-Curtis, Constrained analysis of principal coordinates (CAP), Jensen-Shannon divergence (JSD), weighted and unweighted Unifrac distances (21). To examine the association of clinical variables on the oral microbiota structure, we used a permutational multivariate analysis of variances (PERMANOVA) with the Adonis function (9,999 permutations) in phyloseq (23) using the estimated Beta diversity indices as the outcome variable. The results were converted into a bar plot ranking the effect size R2 of each clinical variable and its statistical significance. To examine if participants can be clustered using beta diversity indices, we run CAP, JSD and Bray-Curtis indices. The analysis was constrained by DMFT and periodontal health status (gingivitis and or periodontitis). We also run a principal coordinate analysis (PCoA) of weighted unifrac distance, which was used to estimate the total variance explained by the first five components, henceforth referred to as TVE (24) in Phyloseq. The impact of genera on TVE was used to evaluate the most influential microbes in each DMFT category.
In order to map and track microbial differences along the DMFT index, we characterized changes in the alpha and beta diversity indices, abundance and prevalence, and co-occurrence dynamics.
To investigate differences in abundance, we used Metacoder as described by Foster (25) which combines phylogenetics and abundance. This allowed us to track the different abundant genera along the DMFT index. Briefly, this analysis was done using a Metacoder object generated from QIIME 2 taxonomic classification of OTUs using a naïve Bayes classifier trained on the most recent SILVA database at 97% similarity (26). First, a training dataset was extracted using the primers used for sequencing our samples. The resultant database subset was then used to train the classifier for taxonomically assigning the OTUs. The heat tree highlights branches based on abundance. To determine the influential genera in DMFT categories, we ranked the fifty most abundant genera, and then examined their impact on TVE by sequentially removing them from the data set representing each DMFT category.
To understand core and accessory differences as a proportion of the total richness along the DMFT index, first, we defined the core as the OTUs present in 85% of the samples at each DMFT category. Secondly, the pan-microbiota was taken as the total number of unique OTUs in each category, and the accessory computed as the difference between the pan and core microbiota in each category. We then analyzed the data to detect these microbial components in our samples using microbiome package 1.9.2 (27), the output was summarized and plotted using Tidyrverse 1.2.1 and ggplot2 2.3.1 in R, respectively. To track the core as a proportion of microbial richness, we selected genera shared by all participants in all the DMFT categories then divided that by the richness at each stage. To examine the influence of genera, we first ranked nodes (genera) in the co-occurrence network by their centrality degree after which, we selected the top 50 genera (see co-occurrence network) and then examined their impact on TVE. This was done by sequentially dropping one genus from the selected genera and computing the change in TVE. The change in variance was visualized using bar plots in ggplot colored by the oxygen utilization capacity of each genus. We used the term invaders interchangeably with accessory microbiota, as they represented transient genera. We also used published literature elsewhere to create two other categories i.e., oral-disease associated genera and normal flora (28, 29) (see detailed lists in the Supplementary).
We used the mean abundance correlation matrix of the genera in each DMFT category to map the differences in the co-occurrence network (30, 31). The “associate” function within the microbiome package version 1.9.2 in R (v3.5.1) was used to generate a genus-level spearman correlation matrix, here we set the FDR adjusted p-value at 0.05 (32) and pruned the matrix using a correlation coefficient of >0.5 and <–0.5. The resultant matrix was then converted into a directed-network object from which communities were extracted and visualized in igraph package version 1.2.2. The edges were colored based on statistical significance (Supplementary Figure. S6).
To determine the factors associated with differences in biomass along the DMFT index, we developed a Poisson regression model in GLM where the outcome variable was mean taxonomic abundance (microbial biomass) using the lme4 package in R version (v3.5.1). We split the data into five subsets each representing a DMFT category, and then run five separate models with the same explanatory variables. It is these models we compared to examine the difference in estimates; the differences here represented the changes at each stage accounting for gender, HIV status and microbes at family level. Model comparison was done using sjtools in in R (v3.5.1).
This study involved 38 and 50 male and female participants (N = 88) with an average age of 39.5 years. The proportion of HIV positive participants was 67% (59/88) with a median CD4 T cell count of 402 cells/mm3, and a viral load of ≤50 copies/ml of blood. All HIV positive participants were on ART. The mean salivary flow rate was 0.9 ml/min with no discernable difference between HIV positive and HIV negative participants. We observed a difference in the mean DMFT among HIV negative and positive participants of 5.9 and 4.9, respectively (Table 1).
A total of 2,601,254 high quality 16S rRNA sequences were recovered from the 88 participants. Samples with the highest and lowest sequence count came from HIV negative and HIV positive participants, respectively (Supplementary Figure S1). When the sequences were filtered to a depth of 3,000, we retained 80 participants (Supplementary Figure S1) from whom 2,093 OTUs were generated with a median frequency of 14,351. There was no statistically significant difference in DMFT categories with regard to alpha diversity indices (Figure 2A, panel A–C) but a statistically significant association (p < 0.005) was observed between DMFT categories and beta diversity indices such as the unweighted Unifrac and Bray-Curtis distances (Figure 2A, panel D–F & Supplementary Figure S2). There was some clustering along the DMFT index specifically separating low and medium categories (Figure 2A, panel D).
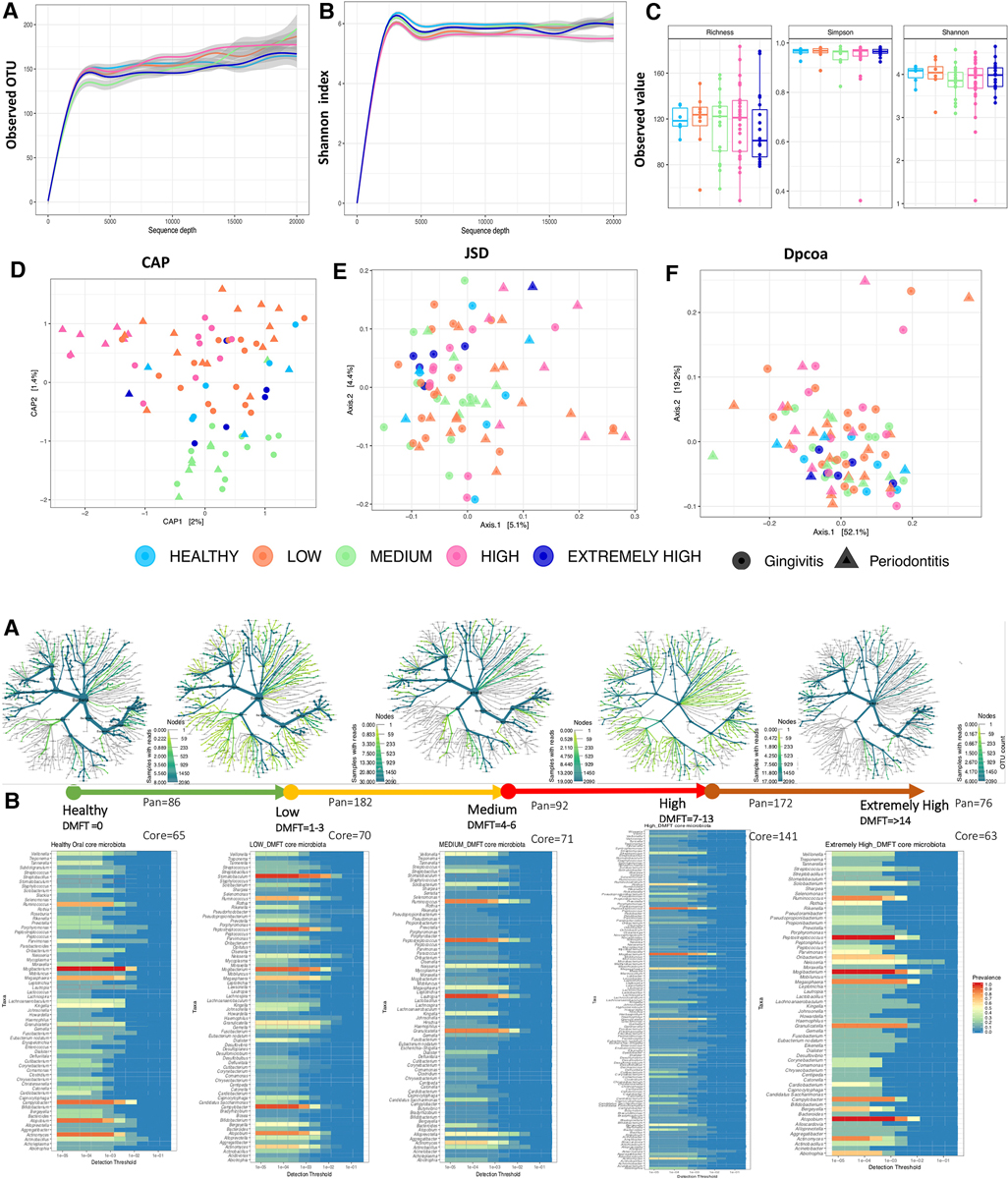
Figure 2. (A) shows the microbial community structure across the DMFT categories. Panel (A–C) show alpha diversity characteristics i.e., the rarefaction curves of observed OTU and Shannon index colored by DMFT category. Panel (C) show a boxplot of alpha diversity indices at a minimum sequence depth set at 3000. Panel (D–F) shows Beta diversity indices. The shapes represent dental caries and periodontal health status (gingivitis/periodontitis). (B) Shows the pan(A) and core(B) oral microbiota along the DMFT index highlighted in arrows colored in order of severity. Panel (A,B) represent genera abundance and prevalence respectively.
The above association was further explored at a taxonomic level i.e., core and accessory microbiota prevalence and abundance (Figure 2B, panel A,B). The 2,093 OTUs clustered into 21 phyla and 239 genera. Firmicutes (33.3%), Bacteroidetes (32.3%) and Proteobacteria (16%) were the dominant phyla, however, at genus level Prevotella (12%), Porphyromonas (3.7%), Leptotrichia (3.4%), Selenomonas (3.4%) were the most predominant (Supplementary Figure S3). Interestingly we detected the genus Mycobacterium DNA from three HIV positive patients and a potential linear relation with CD4 T cell count (Supplementary Figure S4). We also noted that clusters 2 & 5 (Figures 3A, B), were exclusively composed of HIV positive participants; cluster 2 was characterized by a low abundance of genera associated with oral disease while cluster 5 by those that are part of the core.
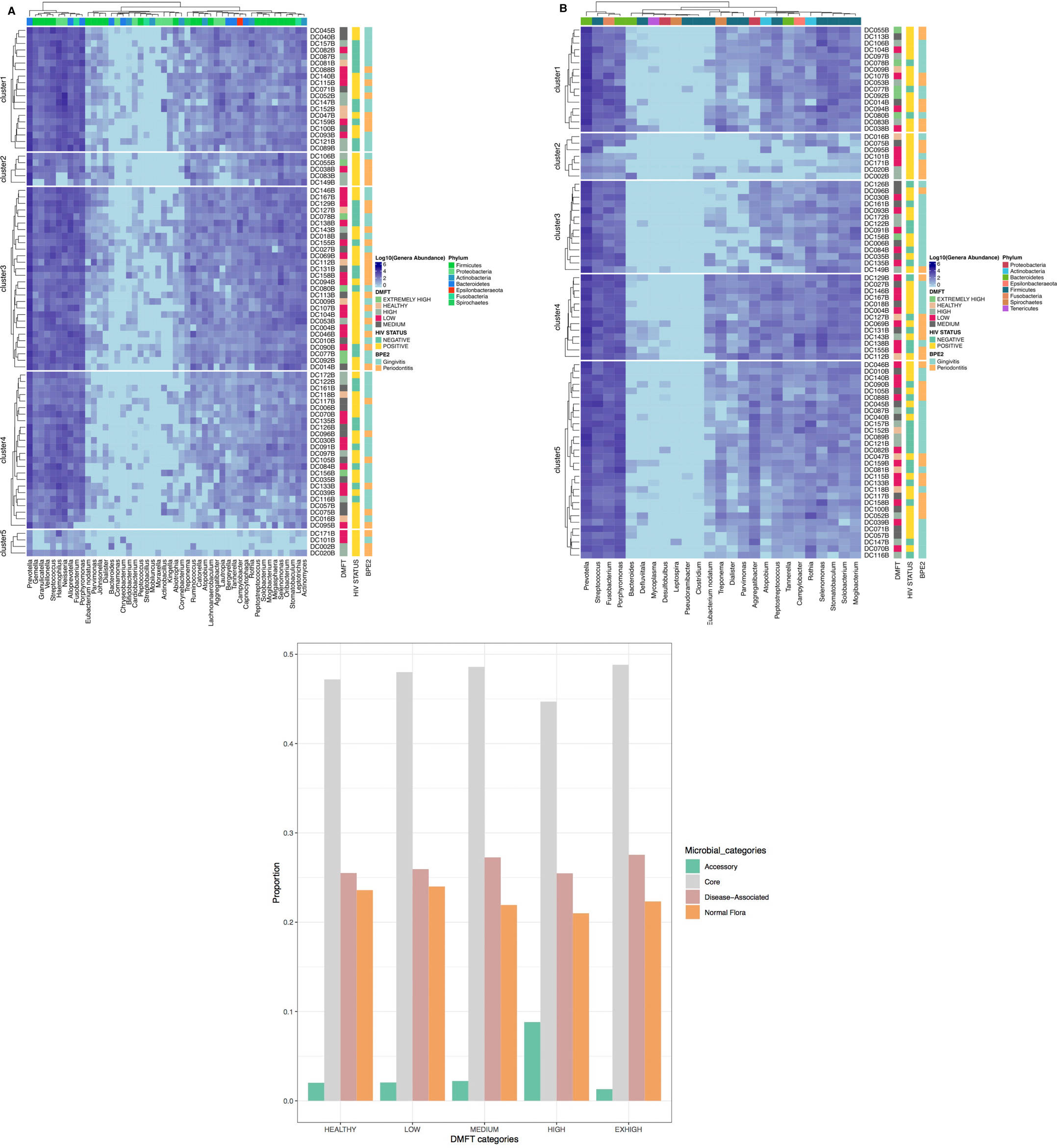
Figure 3. Shows the different microbial components tracked along the DMFT. Panel (A) is the DMFT core, which is the genera present at all stages of the DMFT, Panel (B) shows the genera previously associated with oral disease characterized by patient ID, HIV status and type of oral disease. Panel (C) shows the different proportions of microbial groups along the DMFT categories.
Among the eleven apparently healthy individuals the core and pan microbiota size was 65 and 86 genera respectively, i.e., an accessory microbiota of 21 genera. The most abundant core genera included; Ruminococcus, Mogibacterium, Megasphaera, Campylobacter, Atopobium and Actinomyces (Figure 2B, panel B). The core, normal flora and genera associated with dental caries accounted for 49%, 23% and 27% respectively while the accessory microbiota accounted for 1% (Figure 3C). Of the 65 genera observed among apparently healthy individuals, 49 were shared across the different DMFT categories, hence forth referred to as the DMFT core.
The transition from apparently healthy to low DMFT category was characterized by a slightly higher proportion of the core but the proportion of the accessory microbiota at this stage remained relatively unchanged. This was associated with a greater abundance of Stomatobaculum, Peptostreptococcus and a lower abundance of Atopobium and Actinomyces (Figure 2B, panel B). Here the families with significant differences in abundance included Weeksellacae, Veillonellaceae, Streptococaccae, and Ruminoccocaea (Figure 4).
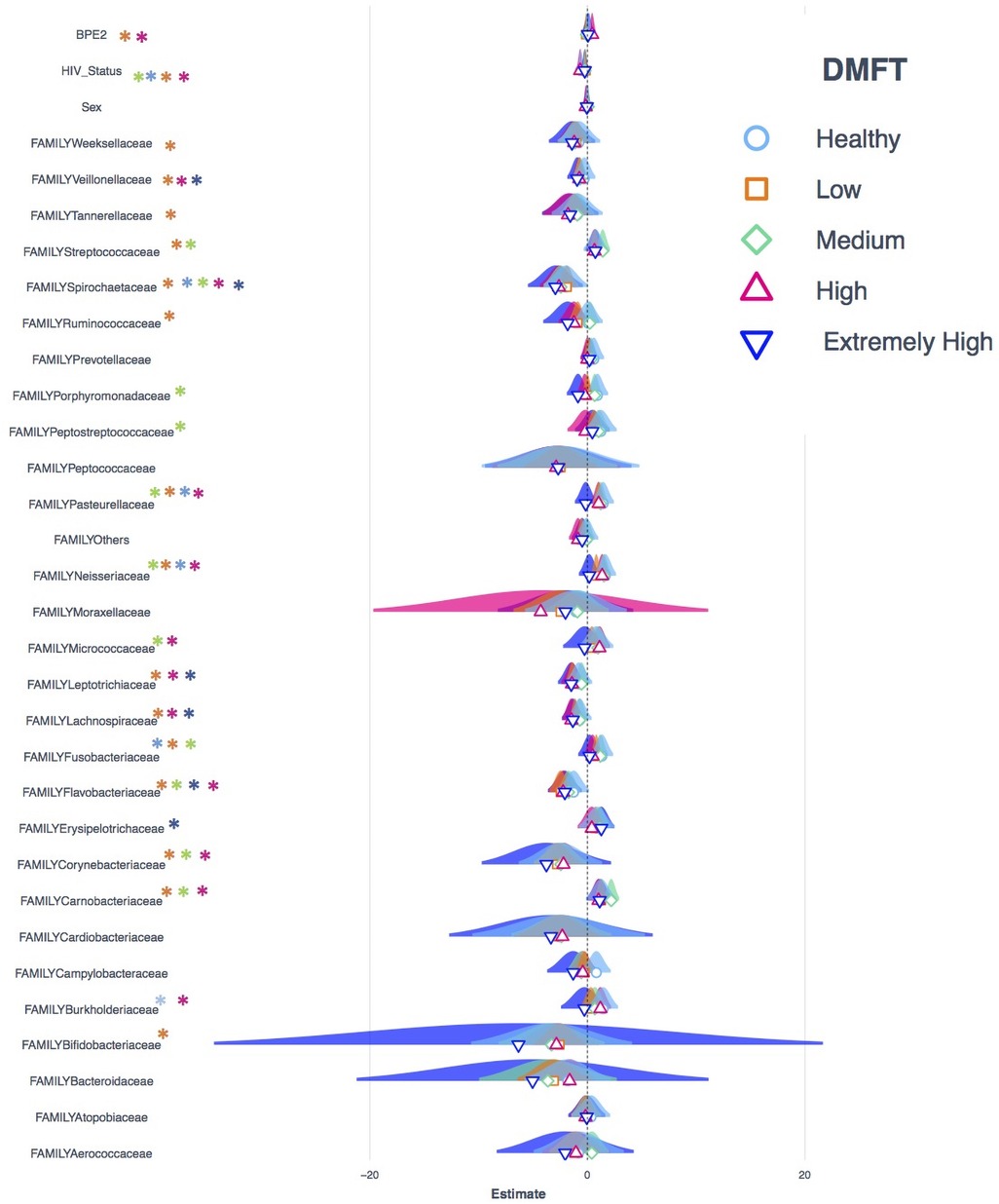
Figure 4. Shows the Poisson regression model of taxonomic abundance and clinical attributes, a comparison of models generated from datasets that represent each DMFT category. The Asterix colors shows in which dataset a variable is statistically significant.
The medium DMFT category was distinctly separated by microbial clustering (Figure 2A, panel D), and was also characterized by a slightly higher proportion of the core microbiota. The core, normal flora and oral disease associated genera accounted for oral microbial richness of 48%, 22% and 27% respectively (Figure 3C). This difference was associated with an enrichment of Ruminococcus, Peptospteptococcus and Lautropia (Figure 2B, panel B). Participants in the high DMFT category and with periodontitis carried significantly more microbial biomass (Figure 4 and Supplementary Figure S6). Indeed, at this stage the accessory microbes accounted for 9% of the microbiota in the oral cavity (Figure 3C). However, families whose abundance was significantly different primarily belonged to the core; Carnobacteriaceae, Neisseriaceae, and Micrococcaceae.
In comparison with the apparently healthy and low DMFT categories, this stage was characterized by a slightly higher abundance of Solobacterium, Oribacterium, Neisseria, Granulicatella, Atopobium, Abiotrophia and a lower abundance of Campylobacter (Figure 2B, panel B). On the other hand, the proportion of the accessory microbiota fell at this stage (Figure 3C).
There was a characteristic difference in entropy i.e., the proportion of genus pairwise association that are statistically significant (Figure 5A). The low DMFT category was associated with a much lower entropy i.e., ∼99% of pairwise correlations were statistically significant i.e., a change from random to non-random (Figure 5A and Supplementary Figure S7). As the severity of dental caries progressed, entropy returned i.e., co-occurrence clusters gradually changed from non-random to random.
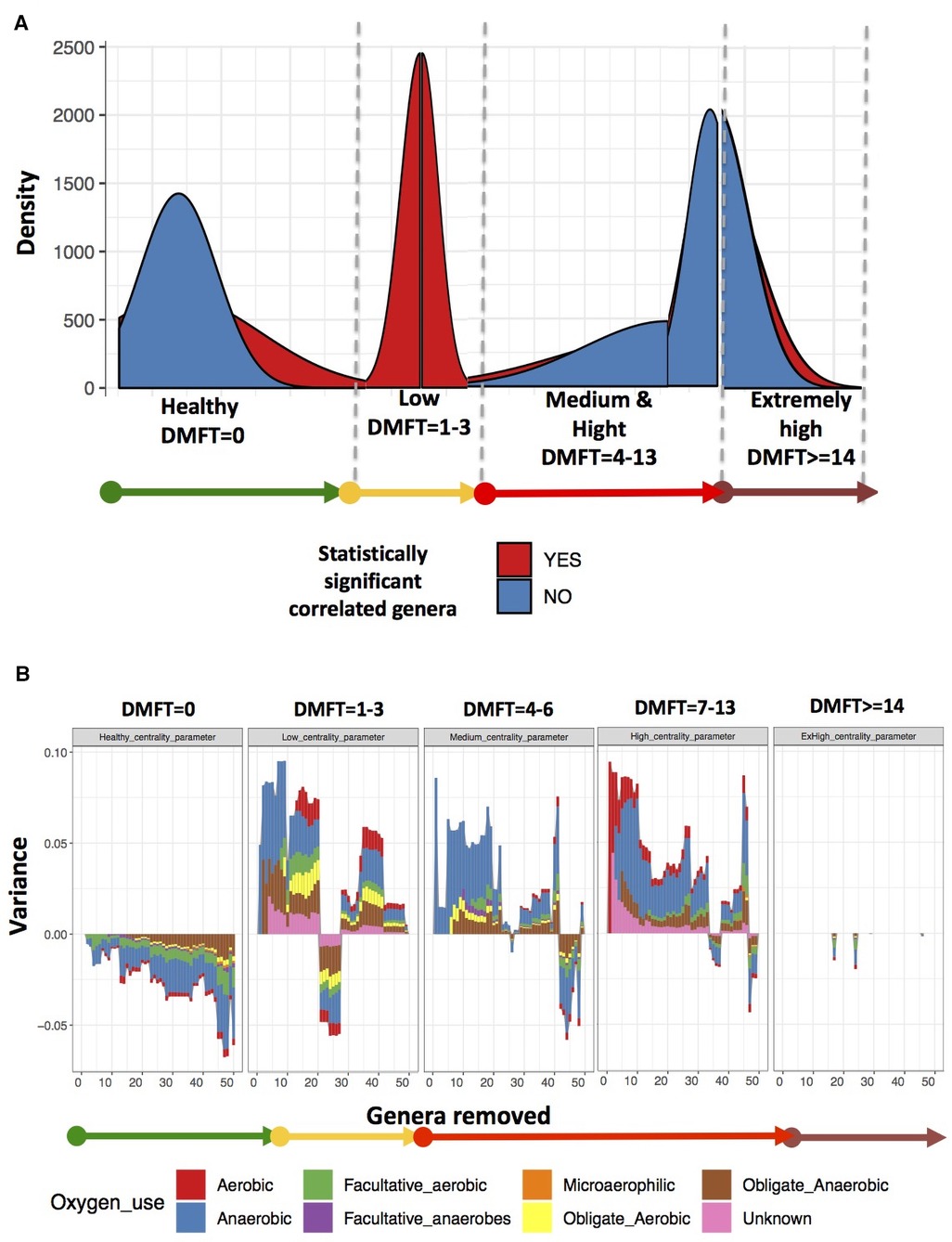
Figure 5. Shows the oral microbial co-occurrence network characteristics. Panel (A) shows the changes in microbial community entropy, Panel (B) shows variance and the genera to which the variance is attributable.
We noted among the apparently healthy participants a difference in variance of up to 5%, most of which was attributable to core genera (Figure 5B). This implied that the most influential genera at this stage belonged to core (see full list of genera identified as influential in Supplementary data). In the low DMFT category, the difference in variance was up to 10%, attributable to 20 genera, most of which belonged to the accessory oral microbiota. These included; Treponema, Desulfoplanes, Desulfosporosinus, Sphaerochaeta, Arenimonas, and Microbacterium among others. Furthermore, these belonged to the families with statistically significant differences in abundance in this category. The medium and high DMFT categories were also characterized by differences in variance of as high as 10%, and we observed that the composition of influential genera changed from aerobic to anaerobic. Unlike the low DMFT category, here 98% of the influential genera belonged to the core (see full list of genera identified as influential in Supplementary data). The extremely high DMFT category was characterized by lower difference in variance, and here too almost all the influential genera were members of the core.
Even though the pathogenesis of dental caries has for long been linked to certain microorganisms (3), its routine diagnosis is almost exclusively macroscopic. This is mainly because microbial culture-based testing is laborious and complicated (33), which delays comprehensive and timely delivery of clinical results. However, with the advent of culture-independent NGS approaches like 16S rRNA amplicon sequencing (34), we can narrow this gap in the developing countries where HIV may be disproportionately driving the occurrence of oral disease. In this study, we used amplicon sequencing data to characterize the oral microbiota along the routinely used diagnostic DMFT index in HIV positive and negative participants.
Microbiota is integral to the oral cavity and plays a critical role in maintaining its integrity (3). In apparently healthy individuals, this manifests as stability in structure and composition (33, 35). In this study, we observed a relatively stable core size of 49 genera across all DMFT categories. The notion of a microbiome core is commonly investigated as a proxy for resident microbes of any given microbiome (36). Among apparently healthy individuals, we observed genera such as Streptococcus, Staphylococcus, Corynebacterium, Veillonella, Granulicatella and Gemella, most of which have been reported as normal flora of the oral cavity (37). The proportion of accessory microbiota, referred to as invaders, was at its lowest among the apparently healthy individuals. This lends support to the notion that the normal flora inhibit invaders through the production of substances such as fatty acids, peroxides and bacteriocins in order to maintain its integrity in healthy individuals (38). This concerted activity has also been characterised among functional communities (39), which in this study are analogous to co-occurrence communities. Indeed, among healthy individuals we observed that the co-occurrence communities were dominated by members of the core such as Johnsonella, Rikenella, Porphyromonas, Alloprevotella, Tannerella, Fusobacterium and others. These organisms are predominantly anaerobic (28) probably due to microbial succession that occurs during the formation of dental plaque (40).
The onset of pathology is widely associated with oral microbiome dysbiosis and the forces that modulate this, are similar to selection forces i.e., changes in fitness, growth and reproduction of microbes manifested as fluxes in prevalence and abundance (3). In the midst of such forces, the core size remained stable with a slightly higher proportion of accessory microbes, particularly anaerobic lactate fermenters like Sharpea, Lawsonella and Olsenella. These microorganisms have been implicated in endodontic infections and acute apical abscesses (41, 42). There was considerably higher abundance of genera such as Stomatobaculum, Peptostreptococus, Campylobacter and Mogibacterium which have been associated with subgingival pathology (43). In this category, we noted that individuals with gingivitis carried significantly more microbes, and families such as Weeksellacae, Veillonellaceae, Streptococcaceae and Ruminococcacaea accounted for the major difference in abundance. These organisms have been associated with dental caries (28, 44) and labial abscesses (45) and it is most probable that changes in the local environment and microbial interactions favor their growth. We noted the absence of genera such as Roseburia, which is associated with health elsewhere (46), and its absence here probably supports the onset of pathology. The development of dental caries can also be viewed as onset of a constraint to the normal microbial community interaction. In this study, this constraint was detectable as a difference in entropy, that is, 99% of the pairwise correlations were statistically significant. In other words, the probability that the associations observed are occurring randomly is very low, which suggests an overwhelming constraint. This difference in entropy is also associated with up to a 10% variation, attributable to genera such as Lachnospira, Treponema, Aggregatibacter, Corynebacteria and Bifidobacteria whose roles in pathogenesis of oral diseases are well documented (47–50). However, some of the influential genera such as Ferruginibacter and Sphaerochaeta are part of the accessory microbiota.
The proportion of the accessory microbiota was highest in the high DMFT category compared to all the other DMFT categories. It is well established that caries development is associated with the interaction of different microorganisms within a cariogenic biofilm, which change as caries progresses (51). Here, there was a greater abundance of core members, Rothia and Mogibacterium, which are associated with dental caries (52) and periodontal disease (52, 53), and Lautropia, which has been isolated from oral cavities of HIV infected children (54) but is not associated with oral disease (54, 55). However, as severity of dental caries changed, the resident microbes seemed overwhelmed, which was seen as a surge in the accessory microbiota. Microbiota in the high DMFT category appeared to have gone through significant remodeling because most of the invading population in the medium category became established as members of the core. Moreover, it is in these two categories and at low DMFT that we observed the highest proportion of the least biologically characterized organisms.
Although this category was characterized based on only six individuals, nonetheless, we observed a greater abundance of microorganisms although the accessory size was smaller. The core was dominated by genera such as Peptostreptoccus, Mogibacterium and Atopobium which have been implicated in chronic oral pathology (56). We also observed a difference in variation and genera such as Solobacterium, Ruminococcus, Neisseria, Campylobacter, Atopobium and Abitrophia have the largest difference in abundance.
From these findings, we propose a microbial model of the DMFT index (Figure 6) as a foundation for oral clinical metagenomics in resource limited settings. (1) Microbial structure; alpha diversity index does not discriminate along the DMFT index but beta diversity does. (2) Taxonomic composition; the DMFT core genera are approximately twice the normal flora in most categories of the DMFT index. In addition, at dental caries onset/low DMFT, the number of genera whose abundance is significantly different belong to the core, this gradually changes to the accessory in the medium and high DMFT categories. (3) Entropy; dental caries onset creates a microbiome wide constraint detectable as a difference in entropy. Almost all genera-pairwise correlations are statistically significant at this stage interpreted as a reduction in microbial entropy. (4) Periodontal health status; we noted a significant difference in microbial abundance i.e., the low DMFT category best represents gingivitis while the high DMFT category represents periodontitis. With this information, a testable hypothesis about pathobiology, probiotic supplementation and treatment can be generated and tested to improve dental caries management in people living with HIV.
In this study, we did not have equal numbers of patients in each group i.e., 11, 32, 21, 18 and 6 for healthy, low, medium, high and extremely high DMFT categories. This has the potential of increasing the level of uncertainty for the estimates made for groups with fewer individuals. However, we observed a considerable level of consistency in microbes in the different categories, therefore, the impact of sample size per group is likely to be limited.
In conclusion, we characterized the oral microbiota dynamics along the DMFT index in HIV positive and negative individuals. Although the microbial characteristics did not offer a categorical output of the DMFT index, they provided the following insights; (a) the low DMFT category showed significant differences in core genera abundance, (b) the high DMFT category was associated with a higher proportion of accessory microbiota, (c) the influential genera in apparently healthy patients were predominantly core members whereas it was the accessory in the medium and high DMFT categories, (d) low DMFT category was associated with a massive reduction in oral microbial entropy. Therefore, using this information, we have proposed a microbial framework for characterizing the DMFT index to better understand dental caries, and serve as a foundation for improving dental caries diagnosis and management in people living with HIV in resource limited settings.
Dental caries affects a third of the world's population and people living with HIV/AIDS (PLWHIV) have a higher risk of getting dental caries. The diagnosis of dental caries is mostly based on naked eye visualization although the cause includes microorganisms. Conventional laboratory culture methods have not isolated all the organisms associated with oral diseases due to their fastidious growth requirements. However, with the advent of next-generation sequencing (NGS) technology, it is possible to unpack the most intricate relationships in microbial communities. We used NGS to characterize oral microbiota shifts along the decayed missing filled teeth (DMFT) index in a high HIV burden setting as a foundation for improving the diagnosis of dental caries in PLHIV. Although the microbial characteristics did not offer a categorical output of the DMFT index, they provided the following insights; (a) low DMFT category showed significant differences in core and accessory genera abundance and was associated with much lower oral microbial entropy, (b) medium and high DMFT categories were associated with higher accessory microbiota, (c) the influential genera in apparently healthy participants were predominantly core members. Therefore, using this information we proposed a microbial framework for characterizing the DMFT index.
The sequences generated for this study have been deposited in the Sequence Read Archive of NCBI (Project Number: PRJNA627249). All other data is contained in the main manuscript and supplemental files.
The studies involving human participants were reviewed and approved by the School of Medicine Research and Ethics committee of Makerere University (#REC REF 2017-053). The patients/participants provided their written informed consent to participate in this study.
DK, HMK, DN, FS, DPK, EK and FA generated the study concept and design. DK and MM acquired and organized the data. EK and FA performed the experiments. AM analyzed the data, AM, GM, IS, DK and DPK interpreted the data. DK, AM and GM wrote the first draft of the manuscript. HMK, DN, DPK, EK, FA, BA, AM, MM, NS contributed critical revisions and important intellectual content to the manuscript. All authors contributed to the article and approved the submitted version.
This work is part of a research fellowship supported by Grant Number D43TW010132 supported by Office of The Director, National Institutes of Health (OD), National Institute of Dental & Craniofacial Research (NIDCR), National Institute of Neurological Disorders and Stroke (NINDS), National Heart, Lung, And Blood Institute (NHLBI), Fogarty International Center (FIC), National Institute on Minority Health and Health Disparities (NIMHD). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the supporting office. AM is a Future leader fellow funded by BBSRC (BB/P007767/1) and Wellcome Trust ISSF3 (IS3-R1.09 19/20).
This manuscript has been released as a preprint (https://www.researchsquare.com/articles/rs-25139/v1), ([Dunstan Kalanzi, Harriet Mayanja-Kizza, Damalie Nakanjako, Gerald Mboowa, Muhammad Mbabali, Edgar Kigozi et al.]) (57).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/froh.2022.1004930/full#supplementary-material.
1. Frencken JE, Sharma P, Stenhouse L, Green D, Laverty D, Dietrich T. Global epidemiology of dental caries and severe periodontitis—a comprehensive review. J Clin Periodontol. (2017) 44(Suppl 18):S94–S105. doi: 10.1111/jcpe.12677
2. Hajishengallis E, Parsaei Y, Klein MI, Koo H. Advances in the microbial etiology and pathogenesis of early childhood caries. Mol Oral Microbiol. (2017) 32(1):24–34 doi: 10.1111/omi.12152
3. Proctor D, Shelef K, Gonzalez A, Long C, Dethlefsen L, Burns A, et al. Microbial biogeography and ecology of the mouth and implications for periodontal diseases. Periodontology 2000. (2020) 82(1):26–41. doi: 10.1111/prd.12268
4. Anaise JZ. Measurement of dental caries experience–modification of the DMFT index. Community Dent Oral Epidemiol. (1984) 12(1):43–6. doi: 10.1111/j.1600-0528.1984.tb01408.x
5. Fejerskov O. Changing paradigms in concepts on dental caries: consequences for oral health care. Caries Res. (2004) 38(3):182–91. doi: 10.1159/000077753
6. Morgan XC, Huttenhower C. Chapter 12: human microbiome analysis. PLoS Comput Biol. (2012) 8(12):e1002808. doi: 10.1371/journal.pcbi.1002808
7. Prifti E, Zucker J-D. The new science of metagenomics and the challenges of its use in both developed and developing countries. In: Morand S, Dujardin JP, Lefait-Robin R, Apiwathnasorn C, editors. Socio-ecological dimensions of infectious diseases in Southeast Asia. Singapore: Springer (2015) pp 191–216.
8. Cavasin Filho JC, Giovani EM. Xerostomy, dental caries and periodontal disease in HIV+ patients. Braz J Infect Dis. (2009) 13(1):13–7. doi: 10.1590/S1413-86702009000100005
9. Malhotra A, Ahlawat J, Hegde M, Mahajan A. Dental caries status in human immunodeficiency virus-positive and acquired immunodeficiency syndrome patients [Original Article]. Indian J Oral Sci. (2016) 7(2):103–6. doi: 10.4103/0976-6944.194235
10. Uganda AIDS Commission. The Uganda HIV and AIDS Country Progress Report July 2016-June 2017. (2017).
11. Kalanzi D, Mayanja-Kizza H, Nakanjako D, Mwesigwa CL, Ssenyonga R, Amaechi BT. Prevalence and factors associated with dental caries in patients attending an HIV care clinic in Uganda: a cross sectional study. BMC Oral Health. (2019) 19(1):159. doi: 10.1186/s12903-019-0847-9
12. Kutesa A, Kasangaki A, Nkamba M, Muwazi L, Okullo I, Rwenyonyi CM. Prevalence and factors associated with dental caries among children and adults in selected districts in Uganda. Afr Health Sci. (2015) 15(4):1302–7. doi: 10.4314/ahs.v15i4.33
13. Xian P, Xuedong Z, Xin X, Yuqing L, Yan L, Jiyao L, et al. The oral microbiome bank of China. Int J Oral Sci. (2018) 10(2):16. doi: 10.1038/s41368-018-0018-x
14. Charlson ES, Chen J, Custers-Allen R, Bittinger K, Li H, Sinha R, et al. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One. (2010) 5(12):e15216. doi: 10.1371/journal.pone.0015216
15. Kirst ME, Li EC, Alfant B, Chi YY, Walker C, Magnusson I, et al. Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Appl Environ Microbiol. (2015) 81(2):783–93. doi: 10.1128/aem.02712-14
16. Navazesh M, Kumar SK. Measuring salivary flow: challenges and opportunities. J Am Dent Assoc (1939). (2008) 139(5 suppl):35s–40s. doi: 10.14219/jada.archive.2008.0353
17. Petersen Poul Erik BRJ, World Health Organization. Oral Health Surveys Basic Methods 5th Edition. (2013).
18. Salih Y, Nasr AM, Ahmed ABA, Sharif ME, Adam I. Prevalence of and risk factors for periodontal disease among pregnant women in an antenatal care clinic in Khartoum, Sudan. BMC Res Notes. (2020) 13(1):147. doi: 10.1186/s13104-020-04998-3
19. Centre for Comparative Genomics and Evolutionary Bioinformatics (CGEB) • Dalhousie University. Integrated Microbiome Resource (IMR). (2014).
20. Comeau AM, Douglas GM, Langille MG. Microbiome helper: a custom and streamlined workflow for microbiome research. mSystems. (2017) 2(1):e00127-16. doi: 10.1128/mSystems.00127-16
21. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet C, Al-Ghalith GA, et al. QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Preprints. (2018) 6:e27295v2. doi: 10.7287/peerj.preprints.27295v2
22. Durbin R, Eddy S, Krogh A, Mitchison G. Biological sequence analysis: probabilistic models for proteins and nucleic acids. Cambridge, UK: Cambridge University Press (1998).
23. McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS one. (2013) 8(4):e61217. doi: 10.1371/journal.pone.0061217
24. Gower JC. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. (1966) 53:325–38. doi: 10.1093/biomet/53.3-4.325
25. Foster ZSL, Sharpton TJ, Grünwald NJ. Metacoder: an R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput Biol. (2017) 13(2):e1005404. doi: 10.1371/journal.pcbi.1005404
26. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. (2013) 41(Database issue):D590–6. doi: 10.1093/nar/gks1219
27. Leo L, Sudarshan S, Turaga N, Leung E, Gilmore R, Salojärvi J, et al. Introduction to the microbiome R package. (2019). Available from: https://microbiome.github.io
28. Tanner AC, Kressirer CA, Faller LL. Understanding caries from the oral microbiome perspective. J Calif Dent Assoc. (2016) 44(7):437–46. PMID: 27514155
29. Diaz PI, Hoare A, Hong BY. Subgingival microbiome shifts and community dynamics in periodontal diseases. J Calif Dent Assoc. (2016) 44(7):421–35. PMID: 27514154
30. Mandakovic D, Rojas C, Maldonado J, Latorre M, Travisany D, Delage E, et al. Structure and co-occurrence patterns in microbial communities under acute environmental stress reveal ecological factors fostering resilience. Sci Rep. (2018) 8(1):5875. doi: 10.1038/s41598-018-23931-0
31. Fernandez M, Riveros JD, Campos M, Mathee K, Narasimhan G. Microbial “social networks”. BMC Genomics. (2015) 16(11):S6. doi: 10.1186/1471-2164-16-S11-S6
32. Bioconductor. Microbiome. (2019). Available from: https://bioconductor.org/packages/devel/bioc/html/microbiome.html
33. Kilian M, Chapple IL, Hannig M, Marsh PD, Meuric V, Pedersen AM, et al. The oral microbiome—an update for oral healthcare professionals. Br Dent J. (2016) 221(10):657–66. doi: 10.1038/sj.bdj.2016.865
34. Zarco MF, Vess TJ, Ginsburg GS. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. (2012) 18(2):109–20. doi: 10.1111/j.1601-0825.2011.01851.x
35. Marsh PD, Zaura E. Dental biofilm: ecological interactions in health and disease. J Clin Periodontol. (2017) 44(Suppl 18):S12–S22. doi: 10.1111/jcpe.12679
36. Aguirre de Carcer D. The human gut pan-microbiome presents a compositional core formed by discrete phylogenetic units. Sci Rep. (2018) 8(1):14069. doi: 10.1038/s41598-018-32221-8
37. Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. (2005) 43(11):5721–32. doi: 10.1128/jcm.43.11.5721-5732.2005
38. Devine DA, Marsh PD, Meade J. Modulation of host responses by oral commensal bacteria. J Oral Microbiol. (2015) 7:26941. doi: 10.3402/jom.v7.26941
39. Duran-Pinedo AE, Frias-Lopez J. Beyond microbial community composition: functional activities of the oral microbiome in health and disease. Microbes Infect. (2015) 17(7):505–16. doi: 10.1016/j.micinf.2015.03.014
40. Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology (Reading, England). (2003) 149(Pt 2):279–94. doi: 10.1099/mic.0.26082-0
41. Siqueira JF, Rôças IN. Microbiology and treatment of acute apical abscesses. Clin Microbiol Rev. (2013) 26(2):255–73. doi: 10.1128/cmr.00082-12
42. Siqueira JF Jr, Rocas IN. Uncultivated phylotypes and newly named species associated with primary and persistent endodontic infections. J Clin Microbiol. (2005) 43(7):3314–9. doi: 10.1128/jcm.43.7.3314-3319.2005
43. Shi M, Wei Y, Hu W, Nie Y, Wu X, Lu R. The subgingival microbiome of periodontal pockets with different probing depths in chronic and aggressive periodontitis: a pilot study. Front Cell Infect Microbiol. (2018) 8:124. doi: 10.3389/fcimb.2018.00124
44. Jiang Q, Liu J, Chen L, Gan N, Yang D. The oral microbiome in the elderly with dental caries and health. Front Cell Infect Microbiol. (2018) 8:442. doi: 10.3389/fcimb.2018.00442
45. Slenker AK, Hess BD, Jungkind DL, DeSimone JA. Fatal case of Weeksella virosa sepsis. J Clin Microbiol. (2012) 50(12):4166–7. doi: 10.1128/jcm.01761-12
46. Tamanai-Shacoori Z, Smida I, Bousarghin L, Loreal O, Meuric V, Fong SB, et al. Roseburia spp.: a marker of health? Future Microbiol. (2017) 12:157–70. doi: 10.2217/fmb-2016-0130
47. Liu F, Ma R, Wang Y, Zhang L. The clinical importance of Campylobacter concisus and other human hosted campylobacter species. Front Cell Infect Microbiol. (2018) 8:243. doi: 10.3389/fcimb.2018.00243
48. Visser MB, Ellen RP. New insights into the emerging role of oral spirochaetes in periodontal disease. Clin Microbiol Infect. (2011) 17(4):502–12. doi: 10.1111/j.1469-0691.2011.03460.x
49. Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. (2012) 10(10):717–25. doi: 10.1038/nrmicro2873
50. Mira A, Simon-Soro A, Curtis MA. Role of microbial communities in the pathogenesis of periodontal diseases and caries. J Clin Periodontol. (2017) 44(Suppl 18):S23–S38. doi: 10.1111/jcpe.12671
51. Bowen WH, Burne RA, Wu H, Koo H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. (2018) 26(3):229–42. doi: 10.1016/j.tim.2017.09.008
52. Fridman D, Chaudhry A, Makaryus J, Black K, Makaryus AN. Rothia dentocariosa endocarditis: an especially rare case in a previously healthy man. Tex Heart Inst J. (2016) 43(3):255–7. doi: 10.14503/thij-15-5068
53. Casarin RC, Saito D, Santos VR, Pimentel SP, Duarte PM, Casati MZ, et al. Detection of Mogibacterium timidum in subgingival biofilm of aggressive and non-diabetic and diabetic chronic periodontitis patients. Braz J Microbiol. (2012) 43(3):931–7. doi: 10.1590/s1517-838220120003000012
54. Rossmann SN, Wilson PH, Hicks J, Carter B, Cron SG, Simon C, et al. Isolation of Lautropia mirabilis from oral cavities of human immunodeficiency virus-infected children. J Clin Microbiol. (1998) 36(6):1756–60. doi: 10.1128/JCM.36.6.1756-1760.1998
55. Peterson SN, Snesrud E, Liu J, Ong AC, Kilian M, Schork NJ, et al. The dental plaque microbiome in health and disease. PLoS One. (2013) 8(3):e58487. doi: 10.1371/journal.pone.0058487
56. Xu L, Chen X, Wang Y, Jiang W, Wang S, Ling Z, et al. Dynamic alterations in salivary Microbiota related to dental caries and age in preschool children with deciduous dentition: a 2-year follow-up study [original research]. Front Physiol. (2018) 9:342. doi: 10.3389/fphys.2018.00342. English.29670544
Keywords: human immunodeficiency virus, microbial co-occurrence networks, dental caries, oral microbiota, DMFT index
Citation: Kalanzi D, Mayanja-Kizza H, Nakanjako D, Semitala F, Mboowa G, Mbabali M, Kigozi E, Katabazi FA, Sserwadda I, Kateete DP, Achan B, Sewankambo NK and Muwonge A (2022) Microbial characteristics of dental caries in HIV positive individuals. Front. Oral. Health 3:1004930. doi: 10.3389/froh.2022.1004930
Received: 27 July 2022; Accepted: 31 August 2022;
Published: 21 September 2022.
Edited by:
Maha El Tantawi, Alexandria University, EgyptReviewed by:
Xiongbin Kang, Bielefeld University, Germany© 2022 Kalanzi, Mayanja-Kizza, Nakanjako, Semitala, Mboowa, Mbabali, Kigozi, Katabazi, Sserwadda, Kateete, Achan, Sewankambo and Muwonge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dunstan Kalanzi ZGthbGFuemlAeWFob28uY29t
Specialty Section: This article was submitted to Oral Health Promotion, a section of the journal Frontiers in Oral Health
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.