
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oral. Health , 22 March 2021
Sec. Oral Infections and Microbes
Volume 2 - 2021 | https://doi.org/10.3389/froh.2021.640129
This article is part of the Research Topic Oral Biofilms: Microbial Interactions and Host Response View all 7 articles
Dental plaque is the key etiological agent in caries formation and the development of the prevalent chronic oral inflammatory disease, periodontitis. The dental plaque biofilm comprises a diverse range of microbial species encased within a rich extracellular matrix, of which extracellular DNA (eDNA) has been identified as an important component. The molecular mechanisms of eDNA release and the structure of eDNA have yet to be fully characterized. Nonetheless, key functions that have been proposed for eDNA include maintaining biofilm structural integrity, initiating adhesion to dental surfaces, acting as a nutrient source, and facilitating horizontal gene transfer. Thus, eDNA is a potential therapeutic target for the management of oral disease–associated biofilm. This review aims to summarize advances in the understanding of the mechanisms of eDNA release from oral microorganisms and in the methods of eDNA detection and quantification within oral biofilms.
The oral cavity is home to a diverse range of microbes from at least 2,000 taxa, with ~100 species residing in an individual's mouth at any given time [1, 2]. Those microbes that colonize the teeth form the dental plaque biofilm, which is the key etiological agent in oral diseases such as caries and periodontitis. Members of the plaque microbiota are also associated with the onset of systemic diseases, including Alzheimer's disease [3], rheumatoid arthritis [4], atherosclerosis [5], and infective endocarditis [6]. Thus, therapeutic strategies that can modulate plaque biofilm development and composition have potential to promote both oral and systemic health.
Dental plaque biofilm formation is initiated via the complex interactions of primary colonizers such as Streptococcus and Actinomyces species with the salivary pellicle that naturally forms on the tooth surface. In turn, these species promote the acquisition of secondary colonizers, which include bridging organisms such as Fusobacterium nucleatum and Corynebacterium species, together with pathobionts such as Porphyromonas gingivalis [7, 8]. As biofilm formation progresses, bacterial species produce and encase themselves within a rich, extracellular matrix known as the extracellular polymeric substance (EPS). This accounts for >90% of biofilm dry weight [9] and comprises a network of molecules that can include polysaccharides, lipids, proteins, and extracellular DNA (eDNA).
Labeling of double-stranded DNA (dsDNA) by immunofluorescence or using DNA dyes such as YOYO-1 has been used to provide evidence of eDNA in biofilms derived from dental plaque samples [10, 11]. Furthermore, application of DNase has been shown to induce significant reductions in early (<8 h) plaque biofilm formation [11]. Such studies are indicative of the critical role of eDNA in oral biofilms, but also highlight eDNA as an attractive target for the management of plaque biofilm formation. This review aims to summarize current understanding of eDNA within dental plaque biofilms, to highlight the tools that can be exploited to detect and quantify eDNA and to provide a perspective on the knowledge gaps that need to be addressed for therapeutic targeting of eDNA to become a reality.
Initial studies evaluating the role of eDNA in oral biofilm formation revealed that DNase was effective in disrupting biofilm formation of both mixed and single species oral biofilms [12], implying a role for eDNA in maintaining biofilm structural integrity. This was further supported by studies focusing on caries-associated bacterial species Streptococcus mutans [13, 14]. Addition of DNA to biofilms containing S. mutans increased aggregation in a DNA-dependent manner, improved strength of hydrogen bonds between bacteria, and neutralized repulsive van der Waals forces and acid–base interactions [15]. eDNA was also shown to enhance adhesion of S. mutans to hydrophobic surfaces and increased the viscoelasticity of the biofilm [16, 17].
Perhaps linked to a structural role, there is evidence that eDNA may be particularly important in early biofilm formation. For early (<4 h) Enterococcus faecalis biofilms, in conjunction with cell wall–anchored proteins PrgB and PrgA, eDNA was shown to promote binding to abiotic surfaces [18]. The PrgB adhesin domain binds DNA in vitro, which promotes establishment of early biofilms [19, 20]. eDNA levels within S. mutans biofilms increase in a time-dependent manner throughout exponential phase of growth to a maximum 5 h postinoculation [21]. Correlating with this, application of DNase was effective in reducing biofilm formation only up to 8 h postinoculation and had little effect following further incubation [11]. It is hypothesized that as biofilm formation progresses, eDNA may be protected from enzymatic digestion by other macromolecules or potentially replaced by alternative matrix components [22]. Nonetheless, DNase application has been found to weaken the mature biofilm matrix, an effect that may improve the ability of adjunctive antimicrobial agents to penetrate the biofilm [12].
eDNA also acts as a potential reservoir for horizontal gene transfer. Application of eDNA purified from Veillonella dispar was capable of transferring tetracycline resistance to Streptococcus mitis [23]. Similarly, hydrogen peroxide (H2O2)–induced eDNA release from Streptococcus gordonii led to increased competence of the microbial cell population and thus an increased capability for the uptake of antibiotic resistance traits [24]. Alongside promoting spread of antimicrobial resistance, eDNA has potential to provide protection against cationic antimicrobial peptides and can act as a shield against aminoglycoside antibiotics because of its partial negative charge [25].
Because of its abundance in carbon, nitrogen, and phosphorous, eDNA can function as a nutrient source [discussed by Vorkapic et al. [26]]. In phosphate-limiting environments, Pseudomonas aeruginosa (associated with endodontic infection) has been shown to release extracellular DNases, enabling the use of eDNA as a nutrient source [27]. The secretin HofQ, expressed by oral pathogen Aggregatibacter actinomycetemcomitans in nutrient-limiting environments, has also been associated with DNA uptake [28].
In relation to oral disease, it has been proposed that eDNA may exacerbate oral inflammation. eDNA isolated from bacterial species associated with endodontic infection (E. faecalis and P. aeruginosa) induced a low-grade inflammatory response from macrophages [29]. However, further work is required to fully evaluate the role of eDNA directly in modulating oral inflammation vs. other molecules that copurify with eDNA as part of the biofilm matrix.
Studies elucidating the molecular mechanisms of eDNA release within oral biofilms have predominantly used monospecies, in vitro biofilm cultures. The main focus to date has been on primary colonizers, in particular members of the Streptococcus genus, but there is some evidence regarding the mechanism of eDNA release for pathobionts, including A. actinomycetemcomitans.
Studies to date suggest that eDNA release from S. mutans can be mediated by extrusion of membrane vesicles or through (auto)lysis [15]. For the latter, murein hydrolase autolysin A (AtlA) has been implicated, the activity of which is calcium dependent. Calcium modulates the two-component system (TCS) VicRK, which in turn alters the N-terminus of AtlA, enabling AtlA to induce cell lysis and thus increased eDNA release [30]. A second TCS, LytST, is also implicated in lysis-dependent eDNA release. LytST regulates expression of lrgAB and thus transcription of the LrgA/B holin-like proteins, inducing degradation of the host cell wall and eDNA release [21, 31]. LytST is also governed by catabolite control protein A (CcpA), which detects fluctuations in the availability of carbohydrates, including sucrose and glucose [32–34]. These mechanisms are summarized in Figure 1. Additional lysis-dependent eDNA release mechanisms identified in S. mutans involve competence-stimulating peptide (CSP) and competence-inducing peptide. The actions of both peptides culminate in the activation of effector ComX, leading to upregulation of autolysis effectors CipB and mutacin V, leading to eDNA release [35–37].
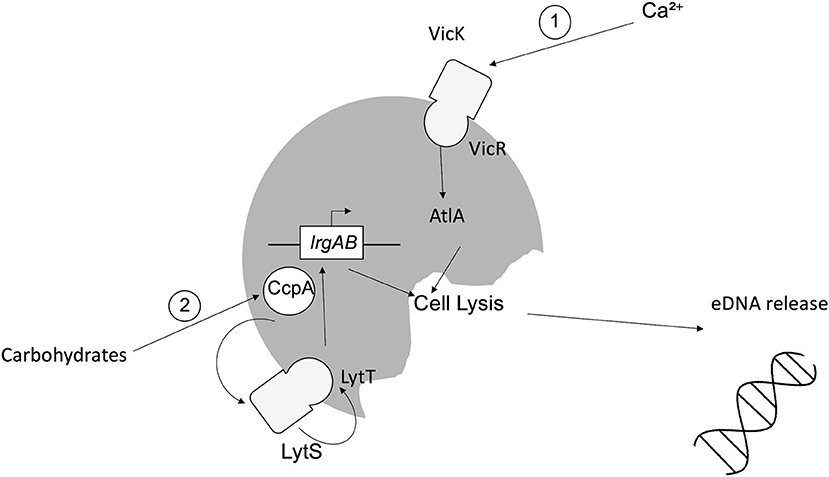
Figure 1. Mechanisms of lysis-dependent eDNA release from Streptococcus mutans. (1) Fluctuations in the availability of calcium induce the activity of the sensor kinase VicK, which in turn induces the activity of intracellular response regulator VicR via phosphorylation, leading to the induction of AtlA, autolysis and eDNA release. (2) Carbohydrates (e.g., sucrose/glucose) induce the activity of CcpA, which modulates TCS LytST, ultimately causing the upregulation of lrgAB and subsequent expression of the LrgA/B holin-like proteins, inducing cell lysis and eDNA release.
Release of eDNA via membrane vesicles is thought to be lysis independent. Membrane vesicle production has been associated with transpeptidase sortase A (SrtA) as, in the absence of SrtA, S. mutans biofilms exhibit reduced protein content within membrane vesicles [21]. However, while eDNA-containing membrane vesicles have been observed within S. mutans biofilms [21], the mechanism of active vesicle release into the extracellular environment has yet to be elucidated.
Streptococcus gordonii is a pioneer colonizer of the oral cavity and ubiquitous constituent of dental plaque. As such, S. gordonii plays an important role in initiating the accretion of dental plaque on the salivary pellicle and can influence whether the microbial community that develops is predisposed to oral health or disease [38]. S. gordonii eDNA is hypothesized to be of chromosomal origin, and its release is associated with H2O2 [24, 39]. Initial studies revealed that, under aerobic conditions, pyruvate oxidase (SpxB) catalyzes the conversion of oxygen to H2O2, which activates TCS VicRK. This, in turn, modulates a second TCS, LytST, which regulates the activity of autolysin AtlS [40]. LytT binds the promoter of AtlS to induce eDNA release [40]. AtlS has been shown to have ~30% homology with AtlA, the autolysin associated with eDNA release by S. mutans [41]. However, it was subsequently inferred that AtlS is not directly involved in eDNA release within S. gordonii biofilms. Rather, following induction, AtlS feeds back to modulate the activity of SpxB and thus intracellular levels of H2O2. H2O2 then induces the activity of murein hydrolase LytF, which is absolutely required for eDNA release [41]. LytF is regulated by the comABCDE competence operon, which controls DNA uptake by S. gordonii [42]. The competence system is inducible by H2O2. As for S. mutans, CSP induces eDNA release in S. gordonii biofilms [43]. Specifically, LytF is proposed to induce eDNA release either via lysis of a subpopulation of the microbial cell population or through incomplete bacterial cell lysis, in which the cell envelope remains largely intact [41]. These proposed mechanisms are illustrated in Figure 2.
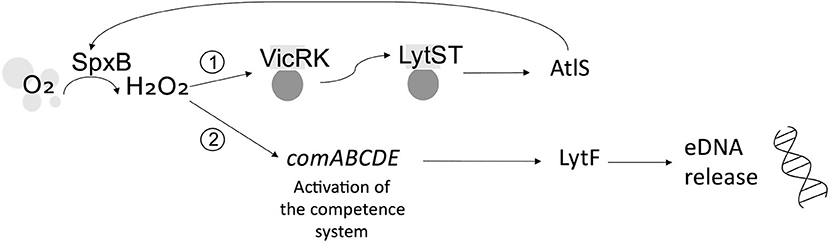
Figure 2. Possible mechanisms of eDNA release from Streptococcus gordonii. Under aerobic conditions, pyruvate oxidase (SpxB) converts oxygen (O2) to hydrogen peroxide (H2O2). (1) Indirect: H2O2 induces TCS VicRK, which, in turn, induces TCS LytST and activation of autolysin AtlS, which feeds back to modulate SpxB and the H2O2-dependent activity of a second pathway. (2) Direct: H2O2 induces the competence system (comABCDE), mediating activation of LytF and resultant eDNA release.
Streptococcus sanguinis is another primary colonizer of the oral cavity, and alike to S. gordonii, the mechanism of eDNA release has been shown to be H2O2 dependent. Nonetheless, there are conflicting reports as to whether or not the H2O2-dependent release of eDNA is observed with or without observable cell lysis [39, 44]. Regardless, as for S. gordonii, pyruvate oxidase (Spx) is pivotal in modulating the release of eDNA in S. sanguinis biofilms via the murein hydrolase LytF [45] and also the catabolite control protein CcpA [46]. Where lysis has been associated with eDNA release, it has been shown that application of either endogenous or exogenous H2O2 induces apoptosis, accompanied by DNA fragmentation and membrane potential depolarization. A link among H2O2, the competence system, and eDNA release was also implicated [44].
Enterococcus faecalis is commonly found in endodontic infections [47]. E. faecalis biofilm formation has been shown to be DNA-dependent, and both lysis-dependent and -independent mechanisms have been described. The first studies revealed that eDNA release from late (>8 h) E. faecalis biofilms can be mediated by a fratricidal mechanism of autolysis, in which a subpopulation of cells within the biofilm is targeted for autolysis and thus eDNA release [48]. Gelatinase (GelE) and serine protease (SprE) were implicated in this fratricidal release of eDNA [49]. GelE induces eDNA release, whereas SprE prevents lysis, with both acting by regulating the activity of autolysin Atn. By contrast, for early (<4 h) biofilms, there is no observable cell lysis but rather an elevated membrane potential indicative of a possible energy requirement for active DNA secretion [48–50]. This is proposed to involve the conjugation apparatus, specifically the matrix channel encoded on plasmid pCF10 [50].
eDNA has also been isolated from biofilms of A. actinomycetemcomitans, a facultative anaerobe commonly associated with the progression of periodontitis. One protein that has been associated with eDNA release from A. actinomycetemcomitans is the secretin HofQ, deletion of which results in decreased eDNA release in the presence of cytokines [28].
Attempts have been made to observe the structure of eDNA in plaque biofilm using high-resolution scanning electron microscopy (SEM). Using an anti-dsDNA primary antibody followed by a 10-nm gold conjugate secondary antibody, eDNA was visualized as a “lattice-” or “net-like” structure comprising strands of eDNA [10, 51]. Similar lattice-like networks have been reported for biofilms of individual oral bacteria, such as S. gordonii [43]. This conforms to the “electrostatic net” model for eDNA, which proposes that in low pH environments, negatively charged eDNA forms electrostatic interactions with positively charged cell membrane–anchored lipoproteins and/or DNA-binding proteins within the biofilm EPS, acting as a net that interconnects the cells [52–54].
For many oral bacteria, histone-like proteins have been associated with the eDNA network within biofilms. For non-typeable Haemophilus influenzae (NTHI), it was found that the strands of eDNA displayed a Holliday junction–like configuration in combination with DNA-binding proteins such as integration host factor and histone-like protein (HU), which served to stabilize these junctions [55] (Figure 3). Likewise, HU was reported within S. gordonii biofilms, and antiserum directed against this protein was shown to disrupt biofilm stability. This antiserum also disrupted S. mitis and Streptococcus oralis biofilms, reflecting the high levels of homology seen for HU across these bacteria [57]. By contrast, the P. gingivalis eDNA matrix is stabilized by histone-like protein-β (HUβ), which is antigenically distinct from HU [58]. Exploiting this, Rocco et al. demonstrated that targeting HUβ could specifically prevent the acquisition of P. gingivalis into preformed biofilms of Streptococcus species, including S. oralis, S. mitis, and S. gordonii [58].
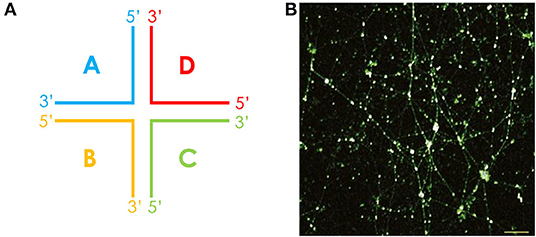
Figure 3. The eDNA lattice within biofilms is structurally related to a Holliday junction. (A) The cruciform-like structure of a Holliday junction, consisting of four double-stranded DNA arms that are joined. The Holliday junction may also adopt alternative conformations. (B) These structures were first observed in a chinchilla middle ear infected with NTHI, where cruciform DNA structures were stained in white and dsDNA in green. Scale bar, 10 μm. Reproduced from [55].
For E. faecalis, the structure of eDNA has been shown to vary in a time-dependent manner. In early (4 h) biofilms, eDNA structures were described as “yarn-like,” appearing as interwoven threads across the biofilm. By contrast, in late (8 h) biofilms, eDNA appeared as a “sweater-like” structure, appearing more as a mat of eDNA. In both instances, however, lattice-like and cruciform structures could still be observed, particularly in early biofilms, providing further support for the notion that eDNA forms Holliday junction–like structures with protein within oral biofilms [50].
Alongside histone-like proteins, for S. mutans, there is evidence that glucans may support eDNA within the biofilm matrix. Glucans and eDNA have been shown to colocalize within biofilms [59], and in the presence of sucrose, levels of eDNA are elevated [60]. However, even in the absence of glucosyltransferases, eDNA remains abundant within S. mutans biofilms, indicating that the eDNA network is not glucan dependent [59, 61]. In addition, isolation of eDNA from P. aeruginosa biofilm using solid-state analysis revealed eDNA can adopt unusual structures including non-canonical Hoogsteen base pairs, triads, and tetrads [62]. It remains to be seen whether these are present or important in oral biofilms.
To fully elucidate the role of eDNA within oral biofilms, reliable, and reproducible methods of eDNA detection and quantification are required. To date, the majority of studies have utilized extraction of soluble DNA from either monospecies or multispecies biofilms followed by DNA quantification methods including PicoGreen assay, UV-Vis spectrometry or semiquantitative polymerase chain reaction (PCR) (Table 1). These methods allow direct assessment of eDNA within biofilms, and using real-time PCR with species-specific (often 16S rRNA) primers, it can be possible to determine the relative levels of eDNA released from different bacterial strains within mixed species biofilms (Table 1). Nonetheless, a limitation of these quantification approaches is that they provide no information regarding the structural composition or architecture of the eDNA lattice-like networks. Likewise, studies evaluating the capacity for DNase application to impair oral biofilm formation infer the presence of eDNA but provide no direct information regarding the nature of the eDNA network [66, 70, 71]. Rather, to assess these parameters, microscopy approaches are required (Table 1). Two studies have found evidence of the “lattice-” or “net-like” structures of eDNA in dental plaque and dual species (S. gordonii and Candida albicans) biofilms by confocal microscopy using dsDNA-specific antibodies conjugated to a fluorescent secondary antibody [10, 43]. However, using nucleic acid stain SYTO9, Rainey et al. [59] were not able to visualize networks of eDNA in 16 h S. mutans biofilm. This may have been due to the time point at which eDNA release was evaluated, as eDNA release is proposed to be at its maximum in S. mutans biofilm at 5 h postinoculation [21]. Alternatively, it may be due to a limitation of SYTO9, which is commonly used to stain intact bacterial cells rather than extrusions of DNA. Thus, it seems that anti-dsDNA conjugate antibodies or TOTO-1 are most efficacious for the reliable detection of eDNA using fluorescence microscopy. Nonetheless, further work is required to optimize these imaging tools for not only the reliable visualization of eDNA within oral biofilms but also its quantification. SEM can also be used to assess eDNA structures at high resolution [66]. However, without the application of a DNA-specific antibody prior to imaging, it is difficult to ascertain whether structures observed are indeed eDNA or alternative ECM components [50]. Alternatively, extraction with ionic liquids also preserves the structure of eDNA in P. aeruginosa biofilm, providing a method for elucidating the intricacies of eDNA structure and its interactions with nucleic acids including RNA [62]. Going forward, an important consideration will be how to ascertain the relative contributions of different species within an oral biofilm consortium to total eDNA release. It may be possible to combine the fluorescent labeling of dsDNA with techniques such as fluorescence in situ hybridization (FISH), which enables differential labeling of microbes by use of fluorescent probes that bind to genus- or species-specific nucleic acid sequences. For example, CLASI-FISH has been successfully used to investigate the biogeography of dental plaque samples [8]. A combination of these microscopy approaches could help to identify the origin and dynamic of eDNA release within polymicrobial communities.
With growing evidence for the role of eDNA in maintaining oral biofilm structural integrity, eDNA is an attractive potential target for modulating dental plaque formation. One way in which this might be achieved is via application of DNase using toothpaste or mouthwash as a delivery system [discussed by Jakubovics and Burgess [12]]. Bacterial-specific enzymes such as NucB isolated from Bacillus licheniformis have shown efficacy in inhibiting the formation of biofilms produced by oral streptococci, including Streptococcus intermedius and Streptococcus salivarius [72]. Application of NucB to plaque biofilm derived from saliva samples significantly reduced microbial load (evaluated by next-generation sequencing) and modulated biofilm composition. The relative proportion of health-related genera, including Streptococcus and Veillonella species increased following NucB treatment, whereas Porphyromonas and Prevotella species were reduced [10]. These studies indicated a species-specific response to DNase enzymes. This characteristic may prove particularly useful in modulating oral biofilm formation, for which the end goal is to promote a microbial community in homeostasis with the host rather than biofilm eradication. Coating of surfaces could also prove a novel delivery system for DNase. Coating of polymethylmethacrylate, a material commonly used in dental restorations, with DNase I, has been shown to prevent colonization by Staphylococcus aureus and P. aeruginosa [73]. Nonetheless, further work is required to understand the wider spectrum of species-specific responses of oral bacteria to DNases and to characterize DNase enzymes that can exert a desirable modulatory effect on biofilm communities to promote oral health when delivered individually or in combination.
DNase enzymes might also be used as an adjunct to enhance the penetration and thus efficacy of other antibiofilm treatments, such as antibiotics [74]. This approach has also shown promise for antimicrobial photodynamic therapy (aPDT). Treatment of biofilms with DNase I prior to use of aPDT increased biofilm dispersal relative to aPDT alone due to improved ability of the photosensitizer to penetrate the biofilm [75]. Of note, blue light has proven efficacious in inhibiting the formation of F. nucleatum and P. gingivalis biofilm [56]. As blue light (400–450 nm) is close to the UV section of the electromagnetic spectrum, it is possible that one explanation for its antibiofilm effects is the induction of eDNA damage.
Alongside exploitation of DNase enzymes, another strategy could be to target the DNA-binding proteins that associate with eDNA [76]. As discussed, antiserum against P. gingivalis HUβ prevented acquisition of this pathobiont into a preformed oral biofilm [58], whereas antiserum against S. gordonii HU disrupted biofilm formation by not only S. gordonii but also S. mitis and S. oralis [57]. Removal of HU proteins has also been found to increase the porosity of biofilms, indicating that an adjunctive role to make biofilms more accessible to antimicrobial agents may be possible [54]. Further work will be required to determine, for example, the cost efficiency of generating HU antisera and their compatibility with being incorporated into products such as mouthwash or toothpaste.
Natural products have been shown to inhibit eDNA-dependent biofilm formation. Derived from the adzuki bean, 7S globulin 3 and theaflavin-3,3′-digallate inhibited biofilm formation by S. mutans via modulation of eDNA production [67, 77]. The synthesized helical peptide, G3, has also shown efficacy in modulating S. mutans eDNA-dependent biofilm formation. G3 binds and thus destabilizes the eDNA matrix, inducing biofilm dispersal [78]. Sodium hypochlorite, an antiseptic used in endodontic treatments, can impair eDNA-dependent biofilm formation by E. faecalis [66]. However, it is not yet known how such compounds would function in dispersal of the complex biofilm communities found within the oral cavity.
There is strong evidence to suggest that eDNA is an integral component of dental plaque–associated biofilm, with potential to be targeted for the therapeutic manipulation of plaque development to promote oral health. Nonetheless, for such strategies to become a reality and translated into commercial products, a much greater understanding of eDNA architecture and release is required. For some oral bacteria, there is evidence that the mechanism of eDNA release can involve autolysins and the competence system. However, further work is required to fully elucidate not only the molecular basis of eDNA release from these monospecies oral biofilms, but also how interactions between microbes within the complex plaque communities in vivo may further modulate eDNA release. Effective targeting of eDNA will also require improved insight into the molecules with which eDNA complexes to form its characteristic lattice-like networks. A combination of advanced imaging and quantification techniques, used in conjunction with appropriate oral biofilm models, will be needed to achieve such objectives.
HS: contributed to conception, design, drafted and critically revised the manuscript. MJ: critically revised the manuscript. NJ and NR: contributed to conception and critically revised the manuscript. AN: contributed to conception, design and critically revised the manuscript. All authors gave their final approval and agree to be accountable for all aspects of the work.
This work was supported by the Dunhill Medical Trust (RPGF1810101).
The authors declare that this work was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Do T, Devine D, Marsh PD. Oral biofilms: molecular analysis, challenges, and future prospects in dental diagnostics. Clin Cosmet Invest Dentistry. (2013) 5:11–9. doi: 10.2147/CCIDE.S31005
2. Nobbs AH, Jenkinson HF. Interkingdom networking within the oral microbiome. Microbes Infect. (2015) 17:484–92. doi: 10.1016/j.micinf.2015.03.008
3. Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A, et al. Porphyromonas gingivalis in Alzheimer's disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv. (2019) 5:eaau3333. doi: 10.1126/sciadv.aau3333
4. Bingham COIII, Moni M. Periodontal disease and rheumatoid arthritis: the evidence accumulates for complex pathobiologic interactions. Curr Opin Rheumatol. (2013) 25:345–53. doi: 10.1097/BOR.0b013e32835fb8ec
5. Chhibber-Goel J, Singhal V, Bhowmik D, Vivek R, Parakh N, Bhargava B, et al. Linkages between oral commensal bacteria and atherosclerotic plaques in coronary artery disease patients. NPJ Biofilms Microbiomes. (2016) 2:7. doi: 10.1038/s41522-016-0009-7
6. Dadon Z, Cohen A, Szterenlicht YM, Assous MV, Barzilay Y, Raveh-Brawer D, et al. Spondylodiskitis and endocarditis due to Streptococcus gordonii. Annals Clin Microbiol Antimicrobials. (2017) 16:68. doi: 10.1186/s12941-017-0243-8
7. Kolenbrander PE, Palmer RJ, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell–cell distance. Nature Rev Microbiol. (2010) 8:471–80. doi: 10.1038/nrmicro2381
8. Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA. (2016) 113:E791–800. doi: 10.1073/pnas.1522149113
9. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. (2010) 8:623–33. doi: 10.1038/nrmicro2415
10. Rostami N, Shields RC, Yassin SA, Hawkins AR, Bowen L, Luo TL, et al. A critical role for extracellular DNA in dental plaque formation. J Dental Res. (2017) 96:208–16. doi: 10.1177/0022034516675849
11. Schlafer S, Meyer RL, Dige I, Regina VR. Extracellular DNA contributes to dental biofilm stability. Caries Res. (2017) 51:436–42. doi: 10.1159/000477447
12. Jakubovics NS, Burgess JG. Extracellular DNA in oral microbial biofilms. Microbes Infect. (2015) 17:531–7. doi: 10.1016/j.micinf.2015.03.015
13. Clarke JK. On the bacterial factor in the aetiology of dental caries. Bri J Exp Pathol. (1924) 5:141–7.
14. Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. (1986) 50:353–80. doi: 10.1128/MR.50.4.353-380.1986
15. Das T, Krom BP, Van Der Mei HC, Busscher HJ, Sharma PK. DNA-mediated bacterial aggregation is dictated by acid-base interactions. Soft Matter. (2011) 7:2927–35. doi: 10.1039/c0sm01142h
16. Das T, Sharma PK, Krom BP, Van Der Mei HC, Busscher HJ. Role of eDNA on the adhesion forces between Streptococcus mutans and substratum surfaces: influence of ionic strength and substratum hydrophobicity. Langmuir. (2011) 27:10113–8. doi: 10.1021/la202013m
17. Peterson BW, van der Mei HC, Sjollema J, Busscher HJ, Sharma PK. A distinguishable role of eDNA in the viscoelastic relaxation of biofilms. mBio. (2013) 4:e00497–13. doi: 10.1128/mBio.00497-13
18. Bhatty M, Cruz MR, Frank KL, Laverde Gomez JA, Andrade F, Garsin DA, et al. Enterococcus faecalis pCF10-encoded surface proteins PrgA, PrgB (aggregation substance) and PrgC contribute to plasmid transfer, biofilm formation and virulence. Mol Microbiol. (2015) 95:660–77. doi: 10.1111/mmi.12893
19. Kohler V, Keller W, Grohmann E. Enterococcus adhesin PrgB facilitates type IV secretion by condensation of extracellular DNA. Mol Microbiol. (2018) 109:263–7. doi: 10.1111/mmi.13994
20. Schmitt A, Jiang K, Camacho MI, Jonna VR, Hofer A, Westerlund F, et al. PrgB promotes aggregation, biofilm formation, and conjugation through DNA binding and compaction. Mol Microbiol. (2018) 109:291–305. doi: 10.1111/mmi.13980
21. Liao S, Klein MI, Heim KP, Fan Y, Bitoun JP, Ahn SJ, et al. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol. (2014) 196:2355–66. doi: 10.1128/JB.01493-14
22. Okshevsky M, Meyer RL. Evaluation of fluorescent stains for visualizing extracellular DNA in biofilms. J Microbiol Methods. (2014) 105:102–4. doi: 10.1016/j.mimet.2014.07.010
23. Hannan S, Ready D, Jasni AS, Rogers M, Pratten J, Roberts AP. Transfer of antibiotic resistance by transformation with eDNA within oral biofilms. FEMS Immunol Med Microbiol. (2010) 59:345–9. doi: 10.1111/j.1574-695X.2010.00661.x
24. Itzek A, Zheng L, Chen Z, Merritt J, Kreth J. Hydrogen peroxide-dependent DNA release and transfer of antibiotic resistance genes in Streptococcus gordonii. J Bacteriol. (2011) 193:6912–22. doi: 10.1128/JB.05791-11
25. Chiang WC, Nilsson M, Jensen PØ, Høiby N, Nielsen TE, Givskov M, et al. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrobial Agents Chemother. (2013) 57:2352–61. doi: 10.1128/AAC.00001-13
26. Vorkapic D, Pressler K, Schild S. Multifaceted roles of extracellular DNA in bacterial physiology. Curr Genet. (2016) 62:71–9. doi: 10.1007/s00294-015-0514-x
27. Mulcahy H, Charron-Mazenod L, Lewenza S. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ Microbiol. (2010) 12:1621–9. doi: 10.1111/j.1462-2920.2010.02208.x
28. Ahlstrand T, Torittu A, Elovaara H, Välimaa H, Pöllänen MT, Kasvandik S, et al. Interactions between the Aggregatibacter actinomycetemcomitans secretin HofQ and host cytokines indicate a link between natural competence and interleukin-8 uptake. Virulence. (2018) 9:1205–23. doi: 10.1080/21505594.2018.1499378
29. Ramirez T, Shrestha A, Kishen A. Inflammatory potential of monospecies biofilm matrix components. Int Endodontic J. (2019) 52:1020–7. doi: 10.1111/iej.13093
30. Jung CJ, Hsu RB, Shun CT, Hsu CC, Chia JS. AtlA mediates extracellular DNA release, which contributes to Streptococcus mutans biofilm formation in an experimental rat model of infective endocarditis. Infect Immunity. (2017) 85:e00252–17. doi: 10.1128/IAI.00252-17
31. Ahn SJ, Qu M-D, Roberts E, Burne RA, Rice KC. Identification of the Streptococcus mutans LytST two-component regulon reveals its contribution to oxidative stress tolerance. BMC Microbiol. (2012) 12:187–187. doi: 10.1186/1471-2180-12-187
32. Klein MI, DeBaz L, Agidi S, Lee H, Xie G, Lin AHM, et al. Dynamics of Streptococcus mutans transcriptome in response to starch and sucrose during biofilm development. PLoS ONE. (2010) 5:e13478. doi: 10.1371/journal.pone.0013478
33. Castillo Pedraza MC, Novais TF, Faustoferri RC, Quivey RG, Terekhov A, Hamaker BR, et al. Extracellular DNA and lipoteichoic acids interact with exopolysaccharides in the extracellular matrix of Streptococcus mutans biofilms. Biofouling. (2017) 33:722–40. doi: 10.1080/08927014.2017.1361412
34. Castillo Pedraza MC, Rosalen PL, Castilho ARFD, Freires IDA, de Sales Leite L, Faustoferri RC, et al. Inactivation of Streptococcus mutans genes lytST and dltAD impairs its pathogenicity in vivo. J Oral Microbiol. (2019) 11:1607505. doi: 10.1080/20002297.2019.1607505
35. Perry JA, Cvitkovitch DG, Lévesque CM. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett. (2009) 299:261–6. doi: 10.1111/j.1574-6968.2009.01758.x
36. Dufour D, Cordova M, Cvitkovitch DG, Lévesque CM. Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J Bacteriol. (2011) 193:6552–9. doi: 10.1128/JB.05968-11
37. Wenderska IB, Lukenda N, Cordova M, Magarvey N, Cvitkovitch DG, Senadheera DB. A novel function for the competence inducing peptide, XIP, as a cell death effector of Streptococcus mutans. FEMS Microbiol Lett. (2012) 336:104–12. doi: 10.1111/j.1574-6968.2012.02660.x
38. Kreth J, Merritt J, Qi F. Bacterial and host interactions of oral streptococci. DNA Cell Biol. (2009) 28:397–403. doi: 10.1089/dna.2009.0868
39. Kreth J, Vu H, Zhang Y, Herzberg MC. Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii. J Bacteriol. (2009) 191:6281–91. doi: 10.1128/JB.00906-09
40. Liu Y, Burne RA. The major autolysin of Streptococcus gordonii is subject to complex regulation and modulates stress tolerance, biofilm formation, and extracellular-DNA release. J Bacteriol. (2011) 193:2826–37. doi: 10.1128/JB.00056-11
41. Xu Y, Kreth J. Role of LytF and AtlS in eDNA release by Streptococcus gordonii. PLoS ONE. (2013) 8:62339. doi: 10.1371/journal.pone.0062339
42. Heng NCK, Tagg JR, Tompkins GR. Identification and characterization of the loci encoding the competence-associated alternative σ factor of Streptococcus gordonii. FEMS Microbiol Lett. (2006) 259:27–34. doi: 10.1111/j.1574-6968.2006.00238.x
43. Jack AA, Daniels DE, Jepson MA, Margaret Vickerman M, Lamont RJ, Jenkinson HF, et al. Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology. (2015) 161:411–21. doi: 10.1099/mic.0.000010
44. Li T, Zhai S, Xu M, Shang M, Gao Y, Liu G, et al. SpxB-mediated H2O2 induces programmed cell death in Streptococcus sanguinis. J Basic Microbiol. (2016) 56:741–52. doi: 10.1002/jobm.201500617
45. Cullin N, Redanz S, Lampi KJ, Merritt J, Kreth J. Murein hydrolase LytF of Streptococcus sanguinis and the ecological consequences of competence development. Appl Environ Microbiol. (2017) 83:3–5. doi: 10.1128/AEM.01709-17
46. Zheng L, Chen Z, Itzek A, Ashby M, Kreth J. Catabolite control protein a controls hydrogen peroxide production and cell death in Streptococcus sanguinis. J Bacteriol. (2011) 193:516–26. doi: 10.1128/JB.01131-10
47. Stuart CH, Schwartz SA, Beeson TJ, Owatz CB. Enterococcus faecalis: its role in root canal treatment failure and current concepts in retreatment. J Endodont. (2006) 32:93–8. doi: 10.1016/j.joen.2005.10.049
48. Thomas VC, Hiromasa Y, Harms N, Thurlow L, Tomich J, Hancock LE. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol Microbiol. (2009) 72:1022–36. doi: 10.1111/j.1365-2958.2009.06703.x
49. Thomas VC, Thurlow LR, Boyle D, Hancock LE. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J Bacteriol. (2008) 190:5690–8. doi: 10.1128/JB.00314-08
50. Barnes AMT, Ballering KS, Leibman RS, Wells CL, Dunnya GM. Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. mBio. (2012) 3:e00193–12. doi: 10.1128/mBio.00193-12
51. Holliday R, Preshaw PM, Bowen L, Jakubovics NS. The ultrastructure of subgingival dental plaque, revealed by high-resolution field emission scanning electron microscopy. BDJ Open. (2015) 1:15003. doi: 10.1038/bdjopen.2015.3
52. Dengler V, Foulston L, DeFrancesco AS, Losick R. An electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus. J Bacteriol. (2015) 197:3779–87. doi: 10.1128/JB.00726-15
53. Graf AC, Leonard A, Schäuble M, Rieckmann LM, Hoyer J, Maass S, et al. Virulence factors produced by Staphylococcus aureus biofilms have a moonlighting function contributing to biofilm integrity. Mol Cell Proteom. (2019) 18:1036. doi: 10.1074/mcp.RA118.001120
54. Kavanaugh JS, Flack CE, Lister J, Ricker EB, Ibberson CB, Jenul C, et al. Identification of extracellular DNA-binding proteins in the biofilm matrix. mBio. (2019) 10:19. doi: 10.1128/mBio.01137-19
55. Devaraj A, Buzzo JR, Mashburn-Warren L, Gloag ES, Novotny LA, Stoodley P, et al. The extracellular DNA lattice of bacterial biofilms is structurally related to Holliday junction recombination intermediates. Proc Natl Acad Sci USA. (2019) 116:25068. doi: 10.1073/pnas.1909017116
56. Song HH, Lee JK, Um HS, Chang BS, Lee SY, Lee MK. Phototoxic effect of blue light on the planktonic and biofilm state of anaerobic periodontal pathogens. J Periodontal Implant Sci. (2013) 43:72–8. doi: 10.5051/jpis.2013.43.2.72
57. Rocco CJ, Davey ME, Bakaletz LO, Goodman SD. Natural antigenic differences in the functionally equivalent extracellular DNABII proteins of bacterial biofilms provide a means for targeted biofilm therapeutics. Mol Oral Microbiol. (2017) 32:118–30. doi: 10.1111/omi.12157
58. Rocco CJ, Bakaletz LO, Goodman SD. Targeting the HUβ protein prevents Porphyromonas gingivalis from entering into preexisting biofilms. J Bacteriol. (2018) 200:e00790–17. doi: 10.1128/JB.00790-17
59. Rainey K, Michalek SM, Wen ZT, Wu H. Glycosyltransferase-mediated biofilm matrix dynamics and virulence of Streptococcus mutans. Appl Environ Microbiol. (2018) 85:e02247–18. doi: 10.1128/AEM.02247-18
60. Li Y, Du Y, Ye J, Wang B, Liu Y. Effect of extracellular DNA on the formation of Streptococcus mutans biofilm under sucrose environment. Chinese J Stomatol. (2016) 51:81–6. doi: 10.3760/cma.j.issn.1002-0098.2016.02.004
61. Kawarai T, Narisawa N, Suzuki Y, Nagasawa R, Senpuku H. Streptococcus mutans biofilm formation is dependent on extracellular DNA in primary low pH conditions. J Oral Biosci. (2016) 58:55–61. doi: 10.1016/j.job.2015.12.004
62. Seviour T, Winnerdy FR, Wong LL, Shi X, Mugunthan S, Castaing R, et al. The biofilm matrix scaffold of Pseudomonas species contains non-canonically base paired extracellular DNA and RNA. bioRxiv. (2020) 2020:527267. doi: 10.1101/527267
63. Nagasawa R, Sato T, Senpuku H. Raffinose induces biofilm formation by Streptococcus mutans in low concentrations of sucrose by increasing production of extracellular DNA and fructan. Appl Environ Microbiol. (2017) 83:508–18. doi: 10.1128/AEM.00869-17
64. Veerapandian R, Vediyappan G. Gymnemic acids inhibit adhesive nanofibrillar mediated Streptococcus gordonii-Candida albicans mono-species and dual-species biofilms. Front Microbiol. (2019) 10:2328. doi: 10.3389/fmicb.2019.02328
65. Senpuku H, Nakamura T, Iwabuchi Y, Hirayama S, Nakao R, Ohnishi M. Effects of complex DNA and MVs with GTF extracted from Streptococcus mutans on the oral biofilm. Molecules. (2019) 24:E3131. doi: 10.3390/molecules24173131
66. Yu MK, Kim MA, Rosa V, Hwang YC, Del Fabbro M, Sohn WJ, et al. Role of extracellular DNA in Enterococcus faecalis biofilm formation and its susceptibility to sodium hypochlorite. J Appl Oral Sci. (2019) 27:e20180699. doi: 10.1590/1678-7757-2018-0699
67. Wang S, Wang Y, Wang Y, Duan Z, Ling Z, Wu W, et al. Theaflavin-3,3′-digallate suppresses biofilm formation, acid production, and acid tolerance in Streptococcus mutans by targeting virulence factors. Front Microbiol. (2019) 10:1705. doi: 10.3389/fmicb.2019.01705
68. Liu C, Niu Y, Zhou X, Zheng X, Wang S, Guo Q, et al. Streptococcus mutans copes with heat stress by multiple transcriptional regulons modulating virulence and energy metabolism. Sci Rep. (2015) 5:12929. doi: 10.1038/srep12929
69. Kim M, Jeon J, Kim J. Streptococcus mutans extracellular DNA levels depend on the number of bacteria in a biofilm. Sci Rep. (2018) 8:13313. doi: 10.1038/s41598-018-31275-y
70. Mohammed MMA, Nerland AH, Al-Haroni M, Bakken V. Characterization of extracellular polymeric matrix, and treatment of Fusobacterium nucleatum and Porphyromonas gingivalis biofilms with DNase I and proteinase K. J Oral Microbiol. (2013) 5. doi: 10.3402/jom.v5i0.20015
71. Shen M, Yang Y, Shen W, Cen L, McLean JS, Shi W, et al. A linear plasmid-like prophage of Actinomyces odontolyticus promotes biofilm assembly. Appl Environ Microbiol. (2018) 84:e01263–18. doi: 10.1128/AEM.01263-18
72. Shields RC, Mokhtar N, Ford M, Hall MJ, Burgess JG, ElBadawey MR, et al. Efficacy of a marine bacterial nuclease against biofilm forming microorganisms isolated from chronic rhinosinusitis. PLoS ONE. (2013) 8:e55339. doi: 10.1371/journal.pone.0055339
73. Swartjes JJTM, Das T, Sharifi S, Subbiahdoss G, Sharma PK, Krom BP, et al. A functional DNase I coating to prevent adhesion of bacteria and the formation of biofilm. Adv Funct Mater. (2013) 23:2843–9. doi: 10.1002/adfm.201202927
74. Tetz GV, Artemenko NK, Tetz VV. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob Agents Chemother. (2009) 53:1204. doi: 10.1128/AAC.00471-08
75. Panariello BHD, Klein MI, Alves F, Pavarina AC. DNase increases the efficacy of antimicrobial photodynamic therapy on Candida albicans biofilms. Photodiagn Photodyn Ther. (2019) 27:124–31. doi: 10.1016/j.pdpdt.2019.05.038
76. Devaraj A, Buzzo J, Rocco CJ, Bakaletz LO, Goodman SD. The DNABII family of proteins is comprised of the only nucleoid associated proteins required for nontypeable Haemophilus influenzae biofilm structure. Microbiol Open. (2018) 7:e00563. doi: 10.1002/mbo3.563
77. Senpuku H, Mohri S, Mihara M, Arai T, Suzuki Y, Saeki Y. Effects of 7S globulin 3 derived from the adzuki bean [Vigna angularis] on the CSP- and eDNA- dependent biofilm formation of Streptococcus mutans. Arch Oral Biol. (2019) 102:256–65. doi: 10.1016/j.archoralbio.2019.04.010
Keywords: eDNA, plaque, biofilm, periodontitis, caries, extracellular matrices
Citation: Serrage HJ, Jepson MA, Rostami N, Jakubovics NS and Nobbs AH (2021) Understanding the Matrix: The Role of Extracellular DNA in Oral Biofilms. Front. Oral. Health 2:640129. doi: 10.3389/froh.2021.640129
Received: 10 December 2020; Accepted: 09 February 2021;
Published: 22 March 2021.
Edited by:
Jorge Frias-Lopez, University of Florida, United StatesReviewed by:
Bastiaan P. Krom, VU University Amsterdam, NetherlandsCopyright © 2021 Serrage, Jepson, Rostami, Jakubovics and Nobbs. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hannah J. Serrage, aGFubmFoLnNlcnJhZ2VAYnJpc3RvbC5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.