
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 08 April 2025
Sec. Cancer Molecular Targets and Therapeutics
Volume 15 - 2025 | https://doi.org/10.3389/fonc.2025.1560943
This article is part of the Research TopicRenewed Insight into Cancer Mechanism and TherapyView all 24 articles
Tumor metastasis is a significant contributor to increased cancer mortality. Transfer RNA-derived small RNAs (tsRNAs), a class of endogenous non-coding RNA molecules, play crucial functional roles in various physiological processes, including the regulation of transcription and reverse transcription, the modulation of translation processes, the modification of epigenetic inheritance, the regulation of the cell cycle, etc. Dysregulated tsRNAs are closely related to the occurrence and progression of human malignancies. Accumulating evidence indicates that the abnormal expression of tsRNAs is associated with tumor metastasis through a variety of mechanisms. Hence, we summarize the fundamental structure and biological functions of tsRNAs, with a focus on how tsRNAs influence the tumor metastasis process through downstream targets or the regulation of interactions between upstream and downstream molecules, thereby providing a novel perspective for targeted therapy for tumor metastasis.
Tumor metastasis occurs as a result of a series of biological events that arise from interactions between tumor cells and nontumor cells. This process is commonly referred to as the invasion–metastasis cascade (1). After primary tumor cells grow and proliferate to a certain extent, they detach from the primary site and modulate the activity of adhesion factors and matrix metalloproteinases (MMPs) to penetrate the extracellular matrix (ECM) and basement membrane, subsequently entering the vasculature. When highly aggressive tumor cells reach specific secondary organs, they adhere to the endothelial cortex within blood vessels and proliferate and grow under the influence of various growth factors, ultimately leading to metastatic formation. Additionally, the tumor vascular system can provide oxygen and nutrients to metastatic tumor foci (2).
Emerging research has revealed the critical involvement of non-coding RNAs (ncRNAs) in regulating tumor metastasis, with tsRNAs recently identified as novel multifunctional regulators in this process. tsRNAs are generated through the cleavage of transfer RNAs (tRNAs) by specific nucleases. tRNAs can be categorized into two types, tRNA fragments (tRFs) and tRNA-derived stress-induced small RNAs (tiRNAs), depending on the position at which cleavage occurs (3). tsRNAs are not merely degradation products of tRNAs; they play significant roles in various biological processes, including the regulation of transcription and reverse transcription, the modulation of translation processes, and participation in epigenetic modifications, among others (4). The role of tsRNAs in tumor metastasis has emerged as a significant research focus in recent years. Mechanistically, tsRNAs influence numerous processes by interfacing with epigenetic modifiers, RNA-binding proteins, and signaling cascade proteins involved in tumor metastasis. This is achieved through the regulation of downstream target genes or the modulation of interactions between upstream and downstream molecules (5–8). Advances in high-throughput sequencing technologies have enabled systematic identification of tsRNAs, with individual members exhibiting marked heterogeneity in both molecular characteristics and biological functions. These molecules may serve as novel biomarkers for the prediction of tumor metastasis.
A large number of studies have found that abnormal expression of tsRNA can regulate the process of tumor metastasis through multiple pathways, such as gene expression regulation, epigenetic modification, metabolic regulation, tumor angiogenesis, tumor immune escape, epithelial–mesenchymal transition (EMT), and the tumor microenvironment. This review discusses the various mechanisms through which tsRNAs influence tumor metastasis at the epigenetic, transcription and posttranscription levels. We offer new insights into the molecular mechanisms underlying tumor metastasis and the role of tsRNAs in tumor diagnosis, prognosis, and targeted therapies, and fresh perspectives on research on treatment options.
As ncRNAs involved in protein synthesis, tRNAs facilitate the translation of mRNA nucleotide sequences by mediating the interaction between codons and anticodons during polypeptide chain synthesis. Once the corresponding amino acid sequence is released into the ribosome, tRNAs fulfills its role. In eukaryotes, tRNAs interact with nucleases (such as Dicer) to generate tsRNAs with distinct biological functions during specific maturation processes (3). Under different stress conditions, tsRNAs with diverse functionalities can be generated through cleavage and rearrangement. Under hypoxic stimulation, the expression of tDR-0009 and tDR-7336 in breast cancer cells significantly increases, and regulating downstream signaling pathways can increase the resistance of triple-negative breast cancer to doxorubicin (9). tRNAs cleavage through the induction of angiopoietin (ANG) or knockdown of ribonuclease inhibitor 1 (RNH1) increases under arsenite-induced oxidative stress, resulting in an overall increase in tsRNA expression levels (10). Under the protection of DNA methyltransferase (Dnmt2), ANG has difficulty cleaving tRNA. Heat shock stress inactivates Dnmt2, causing tRNA to lack methylation protection and thus increasing cleavage (11). With increasing numbers of related studies, researchers have discovered more types of tsRNAs. To name them uniformly, tsRNAs are usually named according to their biological origin and breakage site. Therefore, tsRNAs can be divided into two major categories: tRNA-derived stress-induced small RNAs (tiRNAs) and tRNA-derived fragments (tRFs).
tRFs generally originate from the ends of precursor or mature tRNAs and can be divided into tRF-1s, tRF-2s, tRF-3s, tRF-5s and i-tRFs according to different cutting sites (Figure 1). tRF-1s are derived from the 3’ end of pretRNA produced after ELAC2 cleavage and contain an RNA polymerase III transcription termination sequence (12). tRF-2s are produced by cleaving the anticodon loop under hypoxic stress; they contain only the anticodon stem and loop and do not have a conventional 5’ end or 3’ end (13). tRF-3s start from the 3’ end of tRNA and are cleaved by ANG and Dicer at the TΨC loop; they are approximately 18–22 bases in length and can be subdivided into tRF-3a and tRF-3b (13, 14). tRF-5s are derived from tRNAs starting from the 5’ end and cut by the Dicer enzyme in the D loop or the area between the D loop and the anticodon loop; they have an approximate length of 14–30 bases and can be categorized into tRF-5a, tRF-5b, and tRF-5c, depending on the cleavage site (14). i-tRFs originate from any fragment within tRNA, excluding the 5’ and 3’ ends. The naming of i-tRFs differ according to the position of the 5’ end (15).
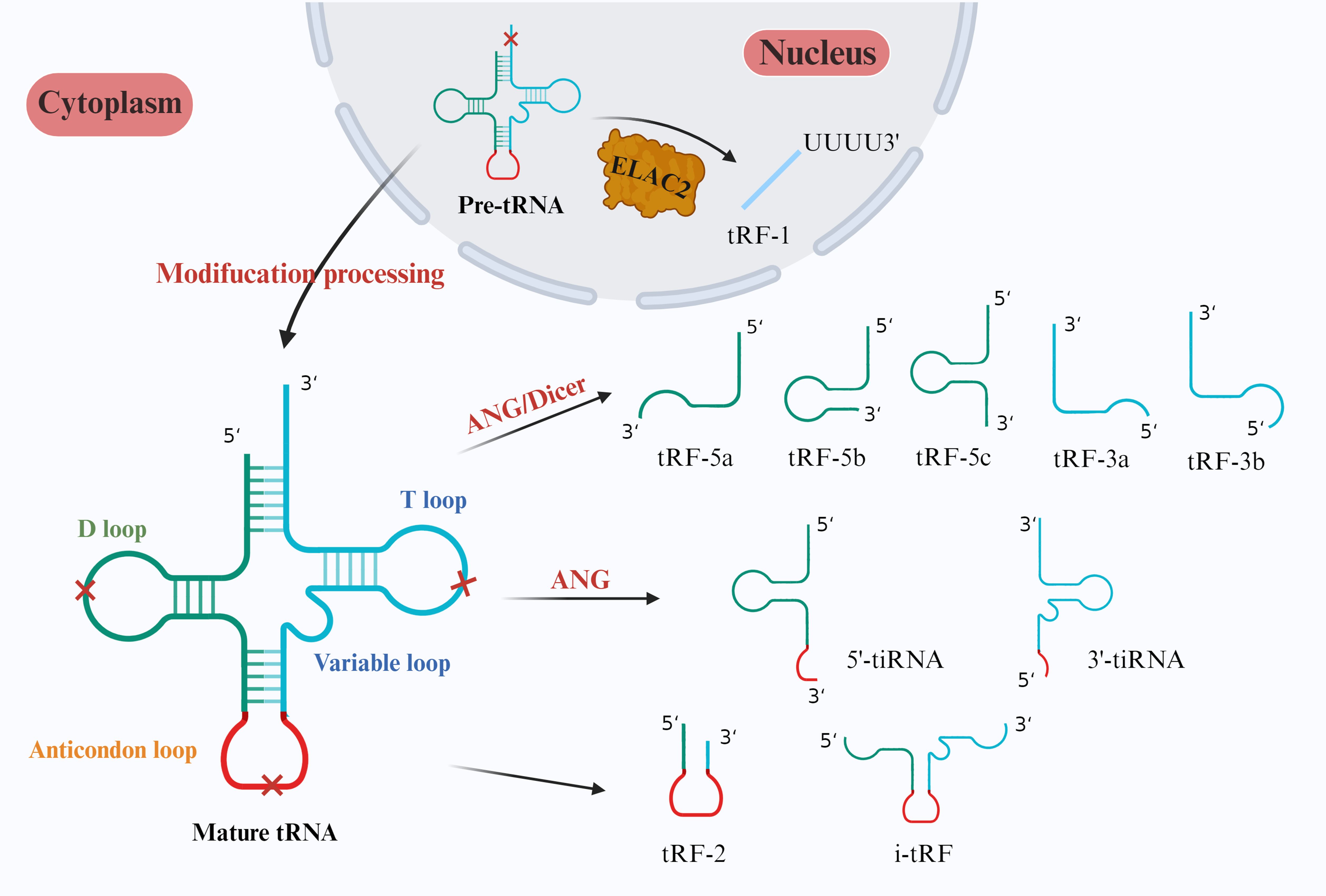
Figure 1. Classification of tsRNAs. In the nucleus, ELAC2 specifically cleaves pretRNAs to form tRF-1s. In mature tRNAs in the cytoplasm, Dicer cleaves the D loop to form tRF-5a, tRF-5b, and tRF-5c. ANG and Dicer then cleave the T loop to form tRF-3a and tRF-3b. Finally, tiRNAs are cleaved by ANG to form anticodon loops, which can be categorized into 5’-tiRNAs and 3’-tiRNAs. Both 2-tRFs and i-tRFs are derived from two cleavage sites in tsRNAs and typically contain anticodon loops.
tiRNAs are produced by cutting the anticodon loop in mature tRNA. They can be divided into 5’-tiRNAs and 3’-tiRNAs according to whether the 5’ end or 3’ end contains an anticodon loop structure. 5’-tiRNAs are derived from the 5’ end of mature tRNA to the end of the anticodon loop, and 3’-tiRNAs are derived from the anticodon loop to the 3’ end of tRNA (16). The cleavage process usually occurs under stressful conditions such as hypoxia, heat shock, oxidative stress, amino acid deficiency, viral infection, and radiation, but tiRNAs can also be generated under specific nonstressful conditions (17). tRNA modifications exert regulatory control over the biogenesis of tiRNAs (18). Besides, and the stress-induced nuclease Rny1 cleaves tRNAs in yeast to produce tiRNAs (19). In summary, the production of tiRNAs is influenced by various factors, such as the type of stress, tRNA modification, and ANG activity (15).
tsRNAs play distinct roles in posttranscriptional regulation and viral reverse transcription. Dicer and the GW182 protein in the RISC complex can have the same effect when bound to tRF-3s (20). Studies in viruses have demonstrated that tRFs may have stem cell functions. In a study of respiratory syncytial virus (RSV), Deng et al. reported that tRF5-GluCTC promoted RSV proliferation by decreasing the expression of apolipoprotein E receptor-2 (ApoER2) (21). Ruggero et al. discovered a tRNA fragment detectable in leukemia virus particles, tRF-3019, which acts as a primer for human T cell lymphotropic virus type 1 (HTLV-1), enabling its reverse transcription (22). Schorn et al. reported that tRF expression was increased in mouse stem cells; the 18 nt 3’CCA tRF inhibited endogenous viral retrotransposition by blocking the reverse transcription process, whereas Dicer-dependent 22 nt tRF-3b inhibited retrotransposition by silencing the retrotransposon gene RPA1 (Figure 2A) (23). In early embryos and germ cells, spermatozoa have high levels of tRFs, where tRNA-GlyGCC controls the Cajal vesicle to regulate the level of histone production to ensure the complete repression of retrotransposable elements in the genome, affecting embryo growth and development processes (24).
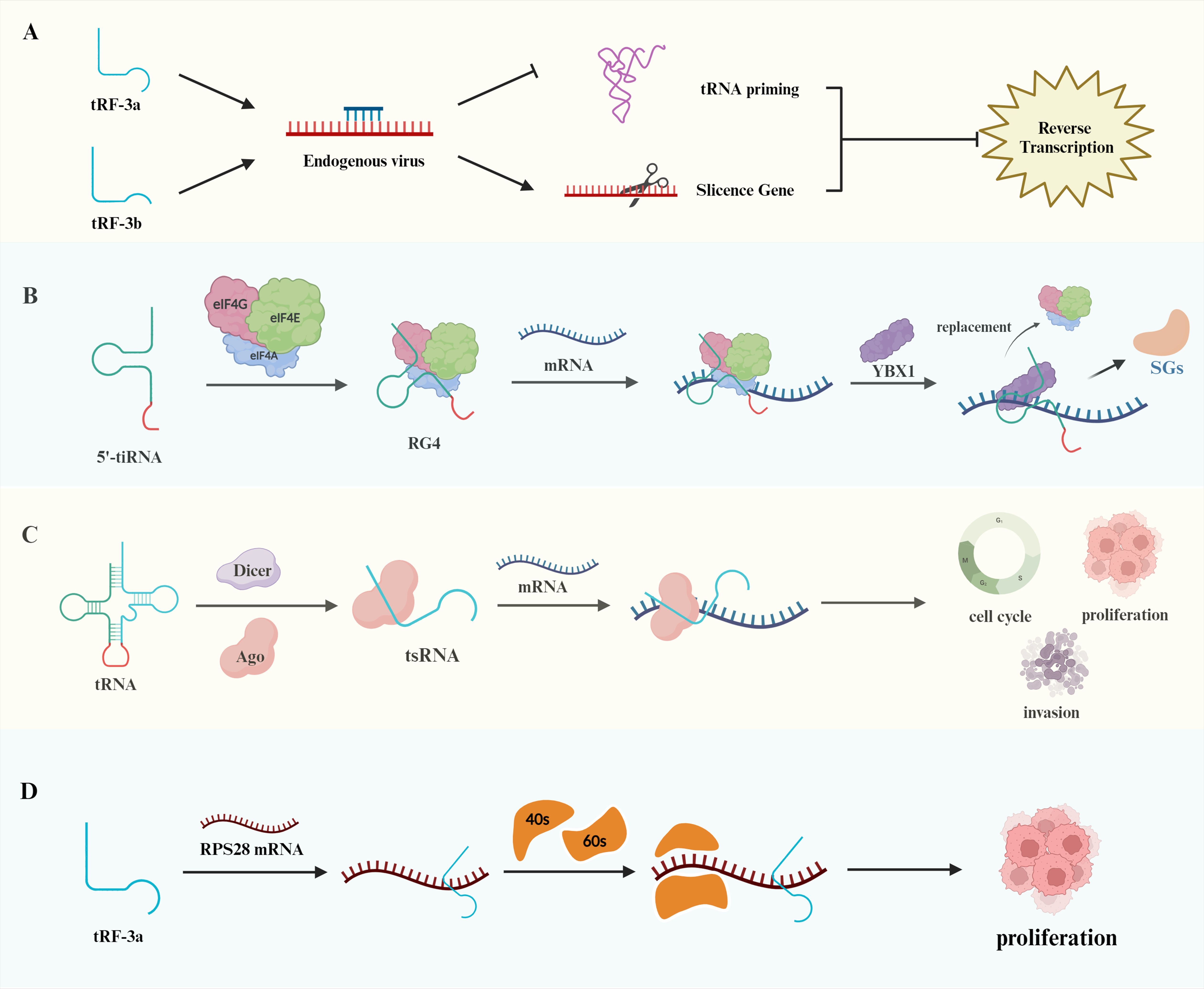
Figure 2. Biological functions of tRNAs. (A) The distinct effects of tRF-3a and tRF-3b on endogenous viruses. tRF-3a exerts its action through tRNA priming, whereas tRF-3b inhibits gene expression through silencing. Both of these mechanisms impede reverse transcription transactivation. (B) Binding of the terminal oligoguanine (TOG) motif at the end of the 5’-tiRNA to eIF, forming an RNA G tetramer that inhibits mRNA translation. Furthermore, YBX1 can displace eIF to further repress translation and release stress granules (SGs). (C) Illustration of the regulation of mRNA structure by tsRNAs, which affects the cell cycle, cell proliferation, and apoptosis. (D) Regulation of ribosomal subunit composition, including 40s and 60s, by tRF-3a binding to RPS28 mRNA, contributing to tumor cell proliferation.
tsRNAs can regulate the translation process of proteins in a unique way. During translation initiation, tsRNAs act by affecting translation initiation factors or translation initiation complexes. Under stress conditions, 5’-tiRNAs produced by ANG cleavage of tRNAs can induce the phosphorylation of the translation initiation factor eIF2 to stall translation (25). A subsequent study revealed that eIF4G is an important scaffolding protein of the translation initiation complex and that the terminal oligoguanine (TOG) motif at the end of tsRNAs can promote the formation of the RNA G tetrameric structure; thus, this combination can effectively inhibit the translation of mRNAs and affect protein synthesis. That study also revealed that the combination of YBX1 and 5’-tiRNA displaces eIF4G in the m7G cap of mRNA and prevents translation initiation more effectively (Figure 2B) (26).
In addition to affecting the process of translation initiation, tsRNAs can affect the process of translation elongation by regulating the secondary structure of mRNAs. This process can be divided into AGO-dependent and AGO-independent regulation. In the AGO-dependent regulation of translational repression, short-stranded tsRNAs bind to AGO1 or AGO2, and long-stranded tsRNAs bind to AGO3 (Figure 2C) (27). Antisense matching of 7-mer motifs in tsRNAs to conserved sites on mRNAs reduces mRNA translation activity (28). Such as, tRF-3s and tRF-5s repress the expression of genes such as miRNAs through interactions with AGO1, AGO3 and AGO4 (29). 3′tsRNA-LeuCAG does not bind to any AGO proteins and opens the secondary structure of mRNA by binding to the corresponding site of the ribosomal protein RPS28 mRNA, which promotes the process of ribosome formation and protein translation of the mRNA. When the expression of 3′tsRNA-LeuCAG is inhibited, the reduction in RPS28 protein synthesis leads to a decrease in the number of 40S and 60S small subunits and impaired ribosome assembly, resulting in the apoptosis of cancer cells such as HeLa cells and HCT-116 cells (Figure 2D) (30). Furthermore, Ying et al. discovered that tRF-Gln-CTG-026 diminishes the binding of pre-40s ribosomes to TSR1, thereby inhibiting protein formation and mitigating liver damage. These findings offer a novel approach for the treatment of liver injury (31).
Epigenetics refers to gene expression that affects gene transcription and translation but does not alter the DNA sequence. Studies have shown that tsRNAs can affect various epigenetic processes to regulate gene expression. PIWI proteins in germ cells can bind to piRNAs to participate in the epigenetic regulation of gene expression (32). tsRNAs are similar in length to piRNAs, and in Tetrahymena thermophila, tsRNAs are bound to the PIWI protein Twi12 to control intranuclear RNA processing (33). Zhang and colleagues reported that human monocytes differentiate into dendritic cells in response to IL-4 stimulation and that 5′tsRNA-Glu interacts with PIWIL-4 to promote histone H3K9 methylation and repress CD1A transcription (34). Transposons are DNA sequences that can potentially damage the genome; however, their activity can be effectively inhibited by epigenetic processes, such as DNA methylation or histone modification (35). 3’CCA tRF is highly expressed in Setdb1-deficient stem cells, and Setdb1 has been shown to mediate H3K9 methylation silencing of retrotransposons (24). Therefore, tsRNAs may act as epigenetic regulators of retrotransposon expression.
The uncontrolled proliferation and apoptosis of cells represent key initiating events in tumor development. Specific fragments of tsRNAs affect the proliferation and apoptosis of cancer cells, which ultimately affects the prognosis of patients with cancer. In ER(+) breast cancer and AR(+) prostate cancer, the expression of SHOT-RNA was shown to be markedly elevated; conversely, the knockdown of SHOT-RNA expression by siRNA significantly reduced the proliferation of cancer cells (36). These findings suggest that SHOT-RNAs can stimulate the corresponding hormone receptors to promote the proliferation of breast and prostate cancer cells. In a study by Fang et al., tsRNA-5001a was markedly elevated in lung adenocarcinoma; furthermore, the overexpression of tsRNA-5001a was shown to suppress the expression of growth arrest and DNA damage-inducible 45 (GADD45), thereby facilitating the proliferation of lung cancer cells (37).
In the process of tumor development, apoptosis also plays a crucial role. The expression of GADD45, a tumor suppressor in prostate cancer, is inhibited by tRF-315, which regulates the cell cycle and prevents apoptosis in prostate cancer cells (38). Similarly, Tao et al. identified 5’tiRNA-His-GTG as a highly expressed molecule in colorectal tumors and demonstrated that reducing 5’tiRNA-His-GTG expression induces apoptosis in cancer cells; through bioinformatics analysis and rescue experiments, they revealed that 5’tiRNA-His-GTG targets the LATS2 protein, thereby upregulating the expression of antiapoptotic genes (39). Furthermore, the aberrant expression of frizzled 3 receptor (FZD3) is typically associated with the development and metastasis of malignant tumors (40). The targeting of the FZD3 protein in breast cancer cells by 5’-tiRNA-Val inhibits the Wnt/β-catenin signaling pathway, which in turn promotes cancer cell apoptosis and slows cancer progression (41).
Tumor metastasis is a complex process involving multiple steps and factors, including cancer cell growth and proliferation, tumor angiogenesis, the epithelial–mesenchymal transition (EMT) process, immune escape, and the formation of the tumor microenvironment. tsRNAs can affect various tumor metastasis processes through different pathways (Figure 3). The following section outlines the molecular mechanisms of tsRNAs in tumor metastasis in detail, and we summarized this information in a Table 1.
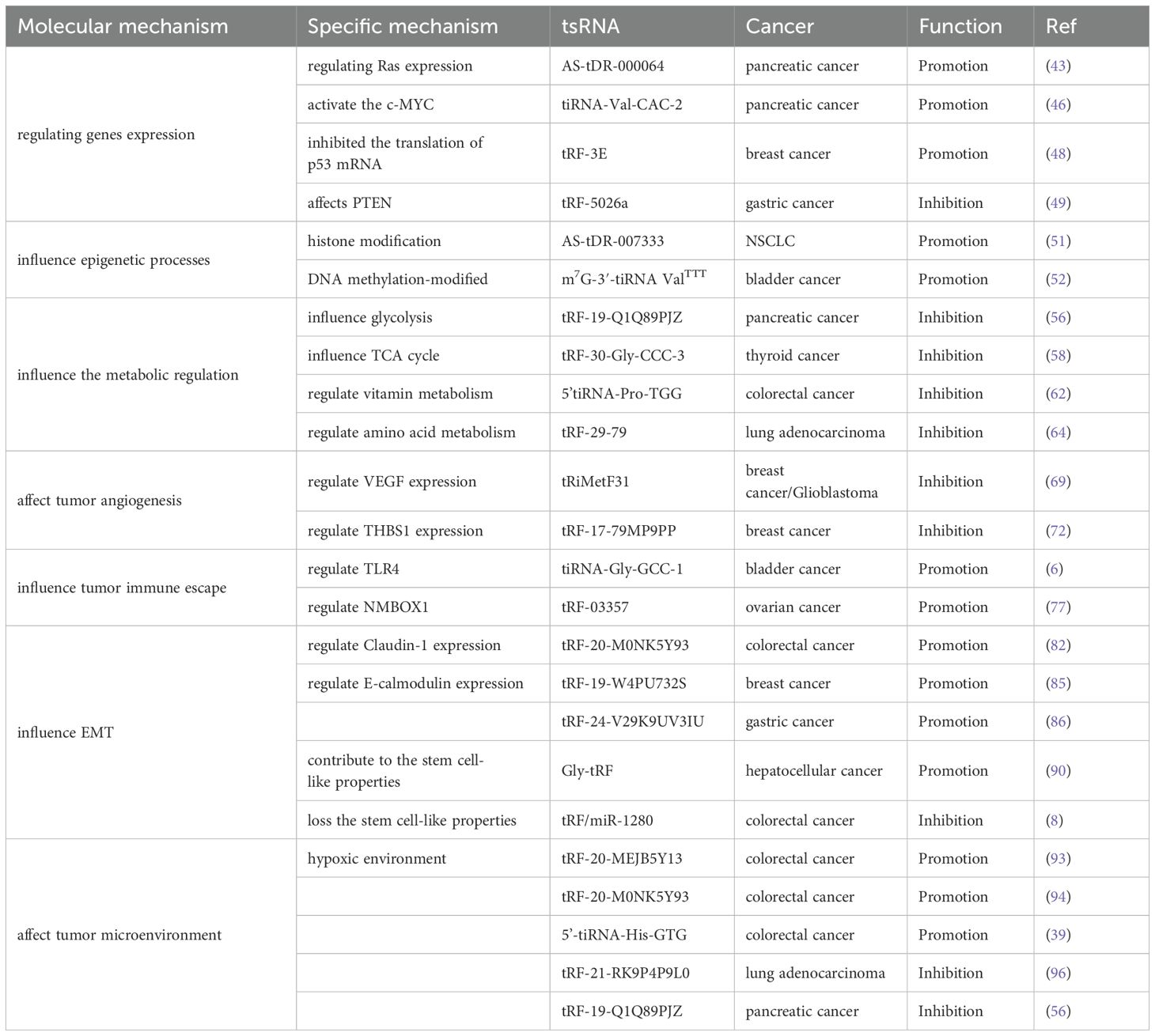
Table 1. The effects of tsRNAs on specific tumor processes and their molecular mechanisms present in metastasis.
At present, there are numerous comprehensive investigations into the mechanisms through which the aberrant expression of tumor-related genes influences the process of tumor invasion and metastasis. It has been demonstrated that the overexpression of oncogenes and the inactivation of antioncogenes can induce the emergence of tumor metastatic phenotypes and contribute to tumor metastasis. Certain specific tsRNAs have the capacity to regulate the expression levels of genes, thereby influencing tumor invasion and metastasis. The Ras gene has been identified as an early oncogene with a demonstrated link to tumor metastasis (42). The downstream target gene of AS-tDR-000064 is enriched in the Ras signaling pathway and has been shown to promote pancreatic cancer (PC) invasion and metastasis by regulating the expression level of the Ras gene (43). The FUBP protein exerts regulatory control over EMT during the metastasis of PC cells via the transforming growth factor beta (TGF-β)/Smad signaling pathway (44); additionally, it has been shown to activate the c-MYC upstream element (FUSE) to regulate c-MYC transcription in lung and breast cancers (45). Xiong et al. reported that the binding of tiRNA-Val-CAC-2 to FUBP1 promotes the metastasis of PC cells (46). To gain further insight into its regulatory mechanism, researchers employed a ChIP assay to determine whether FUBP1 is enriched in the c-MYC promoter region; they reported that FUBP1 accumulated at its upstream element (FUSE). To validate this finding, the researchers conducted a Transwell assay, which demonstrated that reducing FUBP1 expression restored the protein level of c-MYC and attenuated the ability of tiRNA-Val-CAC-2 to promote cancer cell metastasis. In conclusion, tiRNA-Val-CAC-2 has been shown to promote PC metastasis by inhibiting the degradation of FUBP1 and activating c-MYC transcription.
The tumor suppressor gene p53 has been shown to influence the processes of EMT and ECM formation during tumor metastasis (47). The reduced expression of tRF-3E, a tRNA-Glu-derived fragment, leads to diminished interaction with the RNA-binding protein nucleolin, which subsequently inhibits the translational process of p53 mRNA, thereby promoting the proliferation and metastasis of breast cancer cells (48). In gastric cancer, tRF-5026a directly binds to and stabilizes PTEN mRNA through complementary base pairing, resulting in upregulation of the tumor suppressor PTEN, which in turn inhibits the PI3K/Akt pathway, thereby impeding cancer cell proliferation and migration (49). In conclusion, tsRNAs can affect the process of tumor metastasis by modulating the expression of oncogenes or tumor suppressor genes.
Epigenetic processes are intimately associated with tumor proliferation and metastasis. Recently, it has been demonstrated that tsRNAs can be employed as epigenetic regulators to influence tumor metastasis, as diagnostic targets and as potential avenues for therapeutic intervention in tumors (50). Epigenetic modifications include DNA methylation, histone modification and noncoding RNA regulation. A single epigenetic modification can influence tumor proliferation and metastasis; moreover, different epigenetic modifications can interact to enhance tumor development. Upregulated in non-small cell lung cancer tissues, AS-tDR-007333 exerts dual oncogenic mechanisms. Firstly, this tRF physically interacts with heat shock protein HSPB1 to epigenetically activate MED29 transcription through elevating histone H3K4 monomethylation (H3K4me1) and H3K27 acetylation (H3K27ac) levels at the MED29 promoter; Concurrently, it stimulates ELK4 transcription factor expression, facilitating ELK4 binding to the MED29 promoter for transcriptional reinforcement. These coordinated actions drive MED29-mediated oncogenic proliferation and metastatic dissemination of cancer cells (51).
In addition to the histone modification mechanism, Ying et al. reported a correlation between high expression of the methylation-modified tsRNA m7G-3’-tiRNA ValTTT (mtiRL) and the proliferation and metastasis of bladder cancer cells (52). Subsequent studies revealed that mtiRL promotes bladder cancer metastasis by binding to the ANXA2 protein. mtiRL enhances the binding of ANXA2 to Yes1 and facilitates the phosphorylation of ANXA2 at Tyr24, thereby stimulating the proliferation and metastasis of bladder cancer cells.
Cancer cells develop a distinctive metabolic pattern, which differs from that of normal cells, to meet the energy demands of their aberrant proliferation (53). Glycolysis represents the most prevalent metabolic reprogramming pathway in cancer cells (54), which enables cancer cells to obtain sufficient ATP to facilitate growth, proliferation and metastasis and effectively resist apoptosis. tRF-19-Q1Q89PJZ significantly inhibits the invasive and metastatic ability of PC cells. Furthermore, the glycolysis/gluconeogenesis signaling pathway has been identified as a key factor influencing metastasis in PC cells through bioinformatics analysis. Hexokinase 1 (HK1) is a pivotal enzyme in glycolysis and is linked to prognosis in patients with PC (55). Therefore, the anti-metastatic effect of tRF-19-Q1Q89PJZ overexpression in pancreatic cancer cells is likely mediated through suppression of HK1-dependent glycolysis, which disrupts metabolic reprogramming and thereby attenuates oncogenic progression (Figure 4A) (56). Papillary thyroid carcinoma (PTC) is characterized by an active tricarboxylic acid (TCA) cycle (57), and tRF-30-Gly-CCC-3 is a potential oncogene in PTC that binds to the biotin-dependent enzyme pyruvate carboxylase to inhibit the production of intermediates in the TCA cycle in PTC cells and regulates the metabolic reprogramming process; this ultimately inhibits the proliferation, invasion and metastasis of PTC cells (Figure 4B) (58). Furthermore, it offers a novel approach to the management of PTC.
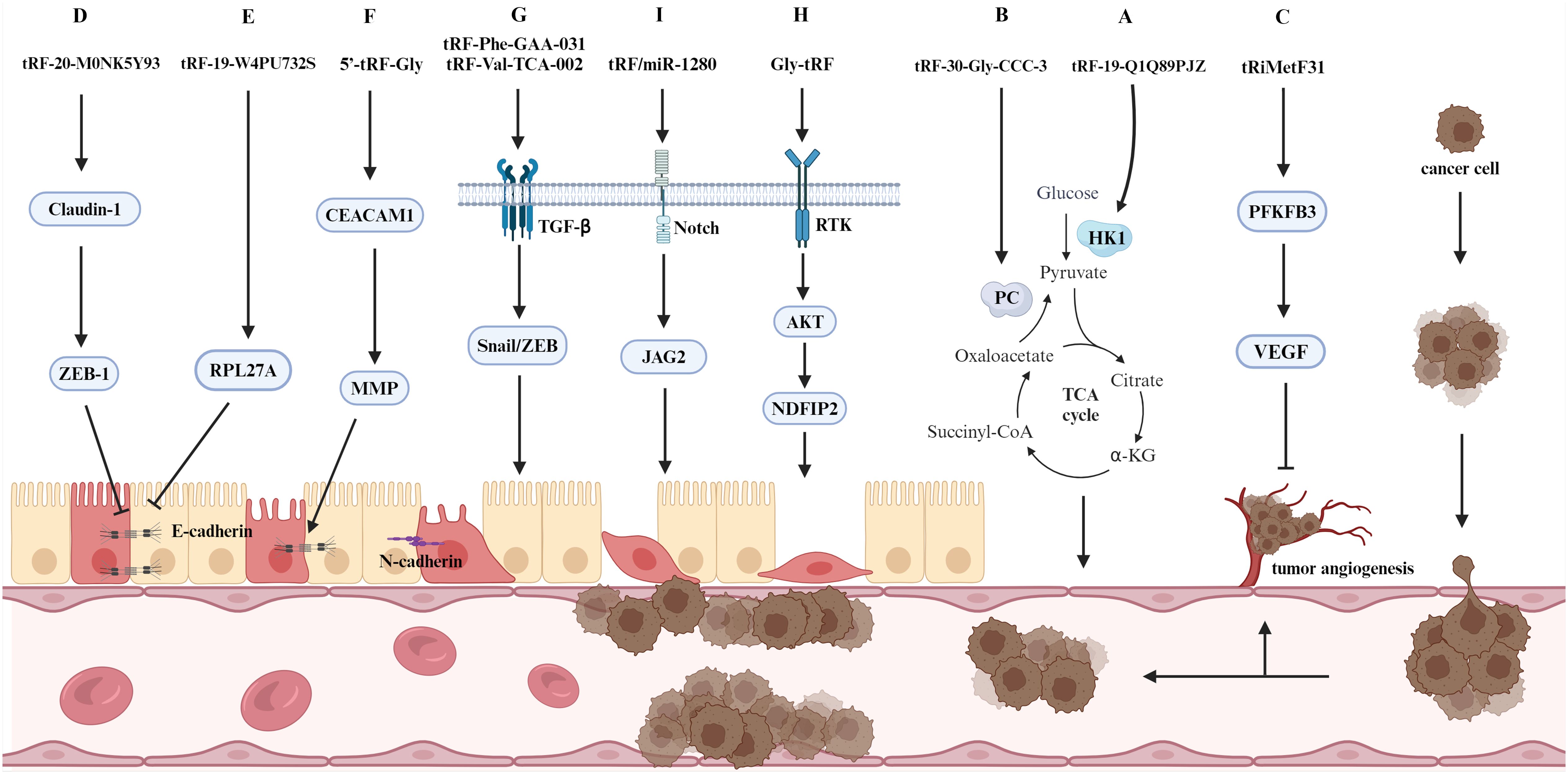
Figure 4. A selection of molecular mechanisms influencing tumor metastasis. The figure delineates the role of tsRNAs in tumor metastasis, specifically their involvement in regulating EMT, signaling pathways, metabolic pathways, and tumor angiogenesis.
There has been a progressive expansion in the field of research investigating the correlation between metabolic pathways associated with amino acid nutrients and vitamins and the process of tumor metastasis (59–61). Wang et al. reported elevated expression of 5’tiRNA-Pro-TGG in colorectal cancer (CRC) specimens, and 5’tiRNA-Pro-TGG targeted HPSE2 to regulate the proliferation and metastasis of cancer cells; metabolic analysis indicated that HPSE2 inhibits multiple metabolic pathways, including riboflavin and retinol, which can effectively reduce CRC development and metastasis (62). CRC consistently demonstrates profound dysregulation of amino acid metabolic networks. Existing literature demonstrates that i-tRF-Glu modulates the expression of glutamine synthetase (GLUL) and related enzymes governing Glu-to-Gln conversion, mechanistically linking this tRNA-derived fragment to the pro-metastatic metabolic reprogramming characteristic of colorectal cancer progression (63). SLC1A5 is a core transporter protein for glutamine metabolism. The binding of tRF-29-79 to PTBP1 affects the stability of SLC1A5 mRNA and mediates glutamine metabolism in lung adenocarcinoma, thereby inhibiting cancer cell proliferation and metastasis (64).
The use of drugs that affect glycolytic and mitochondrial metabolic pathways in cancer cells represents an effective anticancer strategy compared with more general forms of therapy (65). Subsequent research should focus on the identification of tumor markers that influence metabolic pathways.
In solid tumors, an increase in the number of tumor vessels is associated with an increased probability of metastasis and a poorer prognosis (66). Numerous different biomolecules have been identified as influencing tumor angiogenesis. These include growth factors, adhesion factors, proteases, angiopoietins and transcription factors, among others (67). tRiMetF31 is a miR-34a-guided tRNA-derived fragment, and its downstream target 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3) reduces cancer cell invasion and metastasis by normalizing the tumor vasculature (Figure 4C) (68). In breast cancer, tRiMetF31 has been mechanistically demonstrated to suppress tumor angiogenesis and metastatic dissemination through direct targeting of PFKFB3. This anti-angiogenic activity is primarily mediated by transcriptional downregulation of vascular endothelial growth factor (VEGF), consequently impairing endothelial cell sprouting and neovascularization (Figure 4I) (69). Similarly, the tRiMetF31/PFKFB3 axis has been shown to exert an inhibitory effect on glioblastoma metastasis and angiogenesis (70). THBS1 is a protein released by activated platelets and has been shown to play an inhibitory role in tumor metastasis through its angiogenic properties (71). In their study, Mo et al. demonstrated that tRF-17-79MP9PP inhibits the THBS1-mediated TGF-β1/Smad3 pathway, thereby suppressing the proliferation and metastasis of breast cancer cells (72). In CRC, ANG expression is typically linked to the metastasis of cancer cells. 5’-tiRNA-Val expression is elevated in CRC tissues and regulates tumor metastasis via the tiRNA-ANG axis (5). This finding offers a novel target for the diagnosis and treatment of metastatic CRC.
In recent years, highly selective targeted drugs have demonstrated some efficacy in antitumor angiogenesis therapy. For example, tRF-1001 activates the downstream targets RBPJ and MAML1 by silencing METLL3, which in turn inhibits vascular endothelial cells and reduces pathological angiogenesis. tRF-1001 can therefore be employed as a targeted therapeutic agent to limit ocular pathological neovascularization (73).
During the process of tumor metastasis, the body’s immune system is capable of recognizing immunogenic cancer cells and limiting their growth and metastasis. However, cancer cells with high metastatic potential can evade immune system recognition and attack through the utilization of specialized molecular mechanisms. This process is referred to as tumor immune escape.
In a recent study, Qin and colleagues reported a potential correlation between the metastasis of uroepithelial bladder cancer (UBC) and tumor immune escape involving tiRNA-Gly-GCC-1 (6). Toll-like receptor 4 (TLR4) has been identified as a potential target gene for UBC and has been shown to increase B7-H1 expression in bladder cancer cells through the ERK signaling pathway, thereby promoting the metastasis of cancer cells (74). tiRNA-Gly-GCC-1 binds to its target, TLR4, and acts as a ‘sponge’, activating the immune escape of cancer cells and promoting the proliferation and metastasis of UBC. High-grade plasmacytoid ovarian cancer (HGSOC) is typically diagnosed at an advanced stage because of its insidious onset and high metastatic potential (75). In a previous study, the expression of the transcription factor HMBOX1 was shown to be markedly lower in HGSOC cells than in normal ovarian epithelial cells, and HMBOX1 was shown to impede tumor progression by curbing immune evasion (76). Zhang et al. demonstrated that tRF-03357 can facilitate the proliferative migration of ovarian cancer cells by downregulating the expression of the target gene NMBOX1 via the MAPK and Wnt signaling pathways (77), suggesting a novel approach for the early diagnosis and targeted treatment of ovarian cancer.
The tumor microenvironment is characterized by the presence of immunosuppressive immune cells that promote cancer cell proliferation and metastasis. T cells are the primary effector cells of antitumor immunity; they recognize tumor-specific antigens on the surface of cancer cells and activate cytotoxic T lymphocytes (CTLs) or T helper (Th) cells to inhibit cancer cell immune escape (78). Gao et al. identified a correlation between tRDFs in lung cancer and the tumor immune microenvironment (TIME). They reported that the increased expression of 5’-tRDF accelerated the activation of memory CD4+ T cells and memory CD8+ T cells in the TIME and that the enrichment of tRNA-Ala-TGC-3-1 in the T cell receptor signaling pathway indicates that tRDFs may play a role in tumor immunomodulation (79). These findings suggest new avenues for influencing the process of tumor metastasis and provide insights into potential immunotherapy strategies.
EMT represents a pivotal step in cancer cell metastasis; it not only affects alterations in adhesion molecules and related signaling pathways but also endows cancer cells with stem cell-like properties, thereby facilitating the rapid proliferation of cancer cells within metastatic foci (80). Calreticulin is primarily responsible for the transformation of micromere cells into tightly bonded cells. The different types of calreticulin and their expression levels significantly influence the EMT process (81). Luan et al. reported that the reduced expression of tRF-20-M0NK5Y93 upregulated Claudin-1 expression and thus promoted CRC metastasis (Figure 4D) (82). Subsequent studies have demonstrated that the transmembrane protein Claudin-1 can regulate EMT marker genes, thereby influencing the EMT process. The specific effects observed depend on the cellular environment (83). In CRC cells, Claudin-1 upregulates ZEB-1, which in turn inhibits the expression of E-calmodulin and thus promotes the EMT process (84). Furthermore, the expression of tRF-19-W4PU732S is elevated in breast cancer and is associated with the proliferation and metastatic process of cancer cells. During the metastatic process, tRF-19-W4PU732S also inhibited the activity of downstream RPL27A, thereby inhibiting the expression of E-calmodulin, which in turn inhibited apoptosis and promoted metastasis (Figure 4E) (85). Similarly, the knockdown of tRF-24-V29K9UV3IU in gastric cancer cells resulted in a reduction in the expression of E-calmodulin, an increase in the expression of N-calmodulin and waveform protein, and the acceleration of the EMT process in cancer cells, which in turn led to an increase in the invasiveness of gastric cancer (86). Furthermore, the expression of calmodulin is also linked to matrix metalloproteinase (MMP) activity. Liu et al. demonstrated that 5’-tRF-Gly facilitates the metastasis of hepatocellular carcinoma (HCC) cells by silencing CEACAM1 (87). The detection of EMT markers revealed that CEACAM1 affects downstream proteins, including MMP2, the cell cycle protein D1 and E-calmodulin. TGF-β, which acts as an inducer, also affects the expression of calmodulin, waveform protein and fibronectin during EMT (Figure 4F). The elevated expression of tRF-Phe-GAA-031 and tRF-Val-TCA-002 in CRC has been found to be associated with a poor prognosis; furthermore, this tRF has been shown to promote the EMT process in tumor metastasis by affecting TGF-β expression (Figure 4G) (88).
Cancer cells that have undergone EMT exhibit properties analogous to those of tumor stem cells, including the promotion of metastasis, invasion and drug resistance (89). The expression of Gly-tRF is increased in HCC and facilitates the metastasis of HCC cells (90). NDFIP2 is a direct target of Gly-tRF, and the overexpression of NDFIP2 modulates the AKT signaling pathway, thereby inhibiting the promoting effect of Gly-tRF on tumor metastasis. In particular, Gly-tRF was shown to increase the expression of stem cell-like phenotype markers, thereby contributing to the stem cell-like properties of metastatic HCC cells. Additionally, NDFIP2 was shown to influence the expression of calmodulin, as determined via EMT core marker assays (Figure 4H). The metastasis of CRC is typically associated with EMT. tRF/miR-1280 binds to and inhibits the expression of the downstream target gene of the Notch pathway, JAG2, which results in the loss of stemness in cancer cells and a reduction in the number of cancer cells with EMT phenotypes, thus inhibiting the growth and metastasis of CRC (Figure 4I) (8).
The tumor microenvironment is a complex system comprising the metabolic environment surrounding tissues and the cellular environment, which includes multiple stromal cells (91). The metabolic environment is characterized by tissue hypoxia and acidosis. This section focuses on the effects of tsRNAs on tumor metastasis within a hypoxic environment.
Hypoxia-inducible factor 1 alpha (HIF-1α) plays a pivotal role in regulating tissue production in hypoxic environments (92). Under hypoxic conditions, the overexpression of Dicer1 has been shown to promote the metastasis of CRC. The mechanism by which this occurs is thought to involve the induction of the expression of EMT-associated factors. Luan et al. reported that the knockdown of tRF-20-MEJB5Y13 inhibited the overexpression of Dicer1 and thus the proliferation and metastasis of CRC (93). In addition, subsequent studies revealed that tRF-20-M0NK5Y93 influences tumor metastasis not only through its impact on the EMT process but also through the metastasis of CRC cells in a hypoxic environment. In a hypoxic environment, tRFs can bind to specific sequences in long noncoding RNAs (lncRNAs), thereby negatively regulating the expression of MATLA1, inducing the SRSF2-activated selective shearing of SMC1A mRNA and promoting the metastasis of CRC (94). Similarly, in CRC, HIF-1α has been shown to accelerate ANG transcription and promote tRNA cleavage, which has been shown to increase the expression level of 5’-tiRNA-His-GTG; this, in turn, activates its downstream target, i.e., the LATS2/Hippo axis, thereby promoting the proliferation and metastasis of CRC (39). The interaction between HIF-1α and the Notch pathway has been shown to influence the process of lung tumor metastasis (95). In the tsRNA–mRNA network established by Wang et al., tRF-21-RK9P4P9L0 was found to be associated with lung adenocarcinoma (LUAD) invasion, metastasis and prognosis. Furthermore, the inhibition of tRF-21-RK9P4P9L0 expression promoted the expression of Notch1, thereby reducing the invasiveness and metastasis of lung adenocarcinoma cells. Conversely, the expression of Notch1 was shown to increase further under hypoxic conditions (96). A reduction in tRF-19-Q1Q89PJZ levels in hypoxic environments has been shown to contribute to increased HK1 transcript levels, thereby alleviating the inhibitory effect of tRF-19-Q1Q89PJZ on PC metastasis (56). These findings indicate that the HIF-1α-mediated hypoxic environment influences both the regulatory function of tsRNAs and the proliferative capacity of tumor metastasis.
In addition to investigating the effects of hypoxic environments on tumor metastasis, hypoxic conditions have also been found to be responsible for the increased resistance of tumors to chemotherapy. The metastatic tRNA-derived fragments tDR-0009 and tDR-7336, which are formed under hypoxia-specific induction, have been shown to be significantly increased in triple-negative breast cancer (TNBC) cells (9). Bioinformatics analysis revealed that STAT3 is the most frequently interacting downstream target of tDR-0009 and tDR-7336. STAT3 has been shown to activate the transcriptional function of HIF-1α, thereby conferring hypoxia-induced chemoresistance in cancer cells (97). This finding offers a novel avenue for research aimed at enhancing the efficacy of targeted therapy with chemotherapeutic agents and improving the prognosis of individuals with tumors.
Previous research has indicated that metastases are responsible for more than 90% of mortality from malignant tumors and cause varying degrees of damage to other organs (1). Slowing tumor metastasis and improving cancer patient prognosis have become key research areas within the contemporary medical community. tsRNAs play a role in the proliferation and metastasis of malignant tumors due to their structural stability and specificity. Specific tsRNAs associated with metastasis can be screened out by high-throughput RNA sequencing, such as the highly expressed tRF-phe-GAA-031 and tRF-VAL-TCA-002, which promote the dysregulation of EMT process and affect the metastasis of CRC (88). In addition, relevant cell function and mechanism analysis found that tsRNA is expected to be a biomarker for detecting the possibility of tumor metastasis and predicting prognosis. Nevertheless, the field of research remains constrained by several limitations.
First, there is no uniformity in the nomenclature used. The cleavage site for tRNA varies, resulting in a diverse variety of tsRNAs. The conventional nomenclature is based on the source and type of tsRNA without a comprehensive classification system, which has led to the inadvertent exclusion of numerous research-valuable tsRNAs. It is therefore necessary to establish a relatively unified naming system for tsRNAs. Second, although the tsRNA database contains a variety of tRFs and tiRNAs that meet the criteria for research, the data related to tsRNAs in specific cancers are still limited and require further updates to meet the needs of future research. Third, the structure and function of tsRNAs are determined by different chemical modifications. It is yet to be determined whether the effects of these modifications on tsRNAs are correlated with the processes of tumor proliferation and metastasis. Finally, studies on the mechanism of the role of tsRNAs in tumor metastasis are not yet fully comprehensive; for example, it is not known whether tsRNAs regulate DNA methylation to affect tumor metastasis. Moreover, most of the studies are at the theoretical level, and there is a paucity of clinical translational studies. Therefore, further research into the molecular mechanism by which tsRNAs affect tumor metastasis is warranted. The last, emerging evidence underscores the multifaceted regulatory circuits of tsRNAs in tumor metastasis, where context-dependent expression patterns dictate dichotomous biological outcomes. For instance, tRF-3E has been shown to promote breast cancer cell metastasis through NCL binding (48), whereas tRF-17-79MP9PP demonstrates pro-metastatic suppress in the same malignancy (72). Furthermore, through modulation of EMT processes in colorectal cancer metastasis, three identified tsRNAs - tRF-Phe-GAA-031, tRF-Val-TCA-002, and tRF/miR-1280 - demonstrated diametrically opposed regulatory outcomes (8, 88).
In conclusion, recent studies have shown that tsRNAs can influence tumorigenesis and metastasis through the regulation of gene expression, epigenetic processes, the tumor microenvironment, the EMT process and other mechanisms. It is anticipated that tsRNAs will emerge as biomarkers for the clinical screening, prognostic assessment and targeted therapy of tumors. With the advent of high-throughput sequencing technology, the detection of tsRNAs has increased, facilitating further investigation into their potential to modulate tumor metastasis.
HD: Investigation, Methodology, Writing – original draft. CY: Investigation, Writing – original draft. XY: Software, Visualization, Writing – review & editing. JY: Methodology, Visualization, Writing – review & editing. GY: Conceptualization, Funding acquisition, Writing – review & editing. YS: Conceptualization, Funding acquisition, Project administration, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by grants from the Key Scientific and Technological Projects of Ningbo (No. 2021Z133, No.2022Z130), Ningbo Top Medical and Health Research Program (No. 2023020612), and the Medical and Health Research Project of Zhejiang Province (No. 2024KY319).
Thanks for the technical support by the Core Facilities, Health Science Center, Ningbo University. We thank AJE for the language editing work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. (2011) 147:275–92. doi: 10.1016/j.cell.2011.09.024
2. Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. (2003) 3:453–8. doi: 10.1038/nrc1098
3. Zong T, Yang Y, Zhao H, Li L, Liu M, Fu X, et al. tsRNAs: Novel small molecules from cell function and regulatory mechanism to therapeutic targets. Cell Prolif. (2021) 54:e12977. doi: 10.1111/cpr.12977
4. Oberbauer V, Schaefer MR. tRNA-derived small RNAs: biogenesis, modification, function and potential impact on human disease development. Genes (Basel). (2018) 9:607–42. doi: 10.3390/genes9120607
5. Li S, Shi X, Chen M, Xu N, Sun D, Bai R, et al. Angiogenin promotes colorectal cancer metastasis via tiRNA production. Int J Cancer. (2019) 145:1395–407. doi: 10.1002/ijc.32245
6. Qin C, Chen ZH, Cao R, Shi MJ, Tian Y. A novel tiRNA-gly-GCC-1 promotes progression of urothelial bladder carcinoma and directly targets TLR4. Cancers (Basel). (2022) 14:4555–68. doi: 10.3390/cancers14194555
7. Jawad SF, Altalbawy FMA, Hussein RM, Fadhil AA, Jawad MA, Zabibah RS, et al. The strict regulation of HIF-1α by non-coding RNAs: new insight towards proliferation, metastasis, and therapeutic resistance strategies. Cancer Metastasis Rev. (2024) 43:5–27. doi: 10.1007/s10555-023-10129-8
8. Huang B, Yang H, Cheng X, Wang D, Fu S, Shen W, et al. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res. (2017) 77:3194–206. doi: 10.1158/0008-5472.Can-16-3146
9. Cui Y, Huang Y, Wu X, Zheng M, Xia Y, Fu Z, et al. Hypoxia-induced tRNA-derived fragments, novel regulatory factor for doxorubicin resistance in triple-negative breast cancer. J Cell Physiol. (2019) 234:8740–51. doi: 10.1002/jcp.27533
10. Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, et al. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem. (2012) 287:42708–25. doi: 10.1074/jbc.M112.371799
11. Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. (2010) 24:1590–5. doi: 10.1101/gad.586710
12. Zhang L, Liu J, Hou Y. Classification, function, and advances in tsRNA in non-neoplastic diseases. Cell Death Dis. (2023) 14:748. doi: 10.1038/s41419-023-06250-9
13. Kumar P, Kuscu C, Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem Sci. (2016) 41:679–89. doi: 10.1016/j.tibs.2016.05.004
14. Yu X, Xie Y, Zhang S, Song X, Xiao B, Yan Z. tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics. (2021) 11:461–9. doi: 10.7150/thno.51963
15. Yang N, Li R, Liu R, Yang S, Zhao Y, Xiong W, et al. The emerging function and promise of tRNA-derived small RNAs in cancer. J Cancer. (2024) 15:1642–56. doi: 10.7150/jca.89219
16. Li S, Hu GF. Emerging role of angiogenin in stress response and cell survival under adverse conditions. J Cell Physiol. (2012) 227:2822–6. doi: 10.1002/jcp.23051
17. Li S, Xu Z, Sheng J. tRNA-derived small RNA: A novel regulatory small non-coding RNA. Genes (Basel). (2018) 9:246. doi: 10.3390/genes9050246
18. Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. (2018) 361:1346–9. doi: 10.1126/science.aau1646
19. Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol. (2009) 185:43–50. doi: 10.1083/jcb.200811119
20. Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. Rna. (2018) 24:1093–105. doi: 10.1261/rna.066126.118
21. Deng J, Ptashkin RN, Chen Y, Cheng Z, Liu G, Phan T, et al. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol Ther. (2015) 23:1622–9. doi: 10.1038/mt.2015.124
22. Ruggero K, Guffanti A, Corradin A, Sharma VK, De Bellis G, Corti G, et al. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J Virol. (2014) 88:3612–22. doi: 10.1128/jvi.02823-13
23. Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. LTR-retrotransposon control by tRNA-derived small RNAs. Cell. (2017) 170:61–71.e11. doi: 10.1016/j.cell.2017.06.013
24. Boskovic A, Bing XY, Kaymak E, Rando OJ. Control of noncoding RNA production and histone levels by a 5’ tRNA fragment. Genes Dev. (2020) 34:118–31. doi: 10.1101/gad.332783.119
25. Yamasaki S, Ivanov P, Hu GF, Anderson. Angiogenin cleaves tRNA P. and promotes stress-induced translational repression. J Cell Biol. (2009) 185:35–42. doi: 10.1083/jcb.200811106
26. Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. (2011) 43:613–23. doi: 10.1016/j.molcel.2011.06.022
27. Luo S, He F, Luo J, Dou S, Wang Y, Guo A, et al. Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic Acids Res. (2018) 46:5250–68. doi: 10.1093/nar/gky189
28. Shi J, Zhang Y, Zhou T, Chen Q. tsRNAs: the swiss army knife for translational regulation. Trends Biochem Sci. (2019) 44:185–9. doi: 10.1016/j.tibs.2018.09.007
29. Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. (2014) 12:78. doi: 10.1186/s12915-014-0078-0
30. Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. (2017) 552:57–62. doi: 10.1038/nature25005
31. Ying S, Li P, Wang J, Chen K, Zou Y, Dai M, et al. tRF-Gln-CTG-026 ameliorates liver injury by alleviating global protein synthesis. Signal Transduct Target Ther. (2023) 8:144. doi: 10.1038/s41392-023-01351-5
32. Iwasaki YW, Siomi MC, Siomi H. PIWI-interacting RNA: its biogenesis and functions. Annu Rev Biochem. (2015) 84:405–33. doi: 10.1146/annurev-biochem-060614-034258
33. Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K. A Tetrahymena Piwi bound to mature tRNA 3’ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol Cell. (2012) 48:509–20. doi: 10.1016/j.molcel.2012.09.010
34. Zhang X, He X, Liu C, Liu J, Hu Q, Pan T, et al. IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA to upregulate CD1a molecules on monocytes/dendritic cells. J Immunol. (2016) 196:1591–603. doi: 10.4049/jimmunol.1500805
35. Saze H. Epigenetic regulation of intragenic transposable elements: a two-edged sword. J Biochem. (2018) 164:323–8. doi: 10.1093/jb/mvy060
36. Honda S, Loher P, Shigematsu M, Palazzo JP, Suzuki R, Imoto I, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A. (2015) 112:E3816–25. doi: 10.1073/pnas.1510077112
37. Hu F, Niu Y, Mao X, Cui J, Wu X, Simone CB 2nd, et al. tsRNA-5001a promotes proliferation of lung adenocarcinoma cells and is associated with postoperative recurrence in lung adenocarcinoma patients. Transl Lung Cancer Res. (2021) 10:3957–72. doi: 10.21037/tlcr-21-829
38. Yang C, Lee M, Song G, Lim W. tRNA(Lys)-derived fragment alleviates cisplatin-induced apoptosis in prostate cancer cells. Pharmaceutics. (2021) 13(1):55. doi: 10.3390/pharmaceutics13010055
39. Tao EW, Wang HL, Cheng WY, Liu QQ, Chen YX, Gao QY. A specific tRNA half, 5’tiRNA-His-GTG, responds to hypoxia via the HIF1α/ANG axis and promotes colorectal cancer progression by regulating LATS2. J Exp Clin Cancer Res. (2021) 40:67. doi: 10.1186/s13046-021-01836-7
40. Li C, Nguyen V, Clark KN, Zahed T, Sharkas S, Filipp FV, et al. Down-regulation of FZD3 receptor suppresses growth and metastasis of human melanoma independently of canonical WNT signaling. Proc Natl Acad Sci U S A. (2019) 116:4548–57. doi: 10.1073/pnas.1813802116
41. Mo D, Jiang P, Yang Y, Mao X, Tan X, Tang X, et al. A tRNA fragment, 5’-tiRNA(Val), suppresses the Wnt/β-catenin signaling pathway by targeting FZD3 in breast cancer. Cancer Lett. (2019) 457:60–73. doi: 10.1016/j.canlet.2019.05.007
42. Campbell PM, Der. Oncogenic Ras CJ. and its role in tumor cell invasion and metastasis. Semin Cancer Biol. (2004) 14:105–14. doi: 10.1016/j.semcancer.2003.09.015
43. Jin L, Zhu C, Qin X. Expression profile of tRNA-derived fragments in pancreatic cancer. Oncol Lett. (2019) 18:3104–14. doi: 10.3892/ol.2019.10601
44. Zhang Y, Chen J, Zhou N, Lu Y, Lu J, Xing X, et al. FUBP1 mediates the growth and metastasis through TGFβ/Smad signaling in pancreatic adenocarcinoma. Int J Mol Med. (2021) 47:66. doi: 10.3892/ijmm.2021.4899
45. Qian X, Yang J, Qiu Q, Li X, Jiang C, Li J, et al. LCAT3, a novel m6A-regulated long non-coding RNA, plays an oncogenic role in lung cancer via binding with FUBP1 to activate c-MYC. J Hematol Oncol. (2021) 14:112. doi: 10.1186/s13045-021-01123-0
46. Xiong Q, Zhang Y, Xu Y, Yang Y, Zhang Z, Zhou Y, et al. tiRNA-Val-CAC-2 interacts with FUBP1 to promote pancreatic cancer metastasis by activating c–MYC transcription. Oncogene. (2024) 43:1274–87. doi: 10.1038/s41388-024-02991-9
47. Powell E, Piwnica-Worms D, Piwnica-Worms H. Contribution of p53 to metastasis. Cancer Discovery. (2014) 4:405–14. doi: 10.1158/2159-8290.Cd-13-0136
48. Falconi M, Giangrossi M, Zabaleta ME, Wang J, Gambini V, Tilio M, et al. A novel 3’-tRNA(Glu)-derived fragment acts as a tumor suppressor in breast cancer by targeting nucleolin. FASEB J. (2019) 33:13228–40. doi: 10.1096/fj.201900382RR
49. Zhu L, Li Z, Yu X, Ruan Y, Shen Y, Shao Y, et al. The tRNA-derived fragment 5026a inhibits the proliferation of gastric cancer cells by regulating the PTEN/PI3K/AKT signaling pathway. Stem Cell Res Ther. (2021) 12:418. doi: 10.1186/s13287-021-02497-1
50. Han TS, Ban HS, Hur K. H.S. Cho. The epigenetic regulation of HCC metastasis. Int J Mol Sci. (2018) 19:3978. doi: 10.3390/ijms19123978
51. Yang W, Gao K, Qian Y, Huang Y, Xiang Q, Chen C, et al. A novel tRNA-derived fragment AS-tDR-007333 promotes the Malignancy of NSCLC via the HSPB1/MED29 and ELK4/MED29 axes. J Hematol Oncol. (2022) 15:53. doi: 10.1186/s13045-022-01270-y
52. Ying X, Hu W, Huang Y, Lv Y, Ji D, Chen C, et al. A Novel tsRNA, m(7)G-3’ tiRNA Lys(TTT), Promotes Bladder Cancer Malignancy Via Regulating ANXA2 Phosphorylation. Adv Sci (Weinh). (2024) 11:e2400115. doi: 10.1002/advs.202400115
53. DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. (2016) 2:e1600200. doi: 10.1126/sciadv.1600200
54. Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. (2011) 27:441–64. doi: 10.1146/annurev-cellbio-092910-154237
55. Hou Y, Zhang Q, Pang W, Hou L, Liang Y, Han X, et al. YTHDC1-mediated augmentation of miR-30d in repressing pancreatic tumorigenesis via attenuation of RUNX1-induced transcriptional activation of Warburg effect. Cell Death Differ. (2021) 28:3105–24. doi: 10.1038/s41418-021-00804-0
56. Cao W, Zeng Z, Lei S. 5’-tRF-19-Q1Q89PJZ suppresses the proliferation and metastasis of pancreatic cancer cells via regulating hexokinase 1-mediated glycolysis. Biomolecules. (2023) 13:1513. doi: 10.3390/biom13101513
57. Strickaert A, Corbet C, Spinette SA, Craciun L, Dom G, Andry G, et al. Reprogramming of energy metabolism: increased expression and roles of pyruvate carboxylase in papillary thyroid cancer. Thyroid. (2019) 29:845–57. doi: 10.1089/thy.2018.0435
58. Fu B, Lou Y, Lu X, Wu Z, Ni J, Jin C, et al. tRF-1:30-Gly-CCC-3 inhibits thyroid cancer via binding to PC and modulating metabolic reprogramming. Life Sci Alliance. (2024) 7:e202302285. doi: 10.26508/lsa.202302285
59. Gan L, Camarena V, Mustafi S, Wang G. Vitamin C inhibits triple-negative breast cancer metastasis by affecting the expression of YAP1 and synaptopodin 2. Nutrients. (2019) 11:2997. doi: 10.3390/nu11122997
60. Morré DM, Kloppel TM, Rosenthal AL, Fink PC. Chemoprevention of tumor development and metastasis of transplantable hepatocellular carcinomas in rats by vitamin A. J Nutr. (1980) 110:1629–34. doi: 10.1093/jn/110.8.1629
61. Xu J, Li W, Ma J, Liu J, Sha H, Zhou S, et al. Vitamin D - pivotal nutraceutical in the regulation of cancer metastasis and angiogenesis. Curr Med Chem. (2013) 20:4109–20. doi: 10.2174/09298673113209990194
62. Wang XY, Zhou YJ, Chen HY, Chen JN, Chen SS, Chen HM, et al. 5’tiRNA-Pro-TGG, a novel tRNA halve, promotes oncogenesis in sessile serrated lesions and serrated pathway of colorectal cancer. World J Gastrointest Oncol. (2023) 15:1005–18. doi: 10.4251/wjgo.v15.i6.1005
63. Ye C, Cheng F, Huang L, Wang K, Zhong L, Lu Y, et al. New plasma diagnostic markers for colorectal cancer: transporter fragments of glutamate tRNA origin. J Cancer. (2024) 15:1299–313. doi: 10.7150/jca.92102
64. Shi Y, Pan Z, Feng Y, Zhou Q, Wang Q, Wang H, et al. tRF-29-79 regulates lung adenocarcinoma progression through mediating glutamine transporter SLC1A5. Carcinogenesis. (2024) 45:409–23. doi: 10.1093/carcin/bgae010
65. Kubik J, Humeniuk E, Adamczuk G, Madej-Czerwonka B, Korga-Plewko A. Targeting energy metabolism in cancer treatment. Int J Mol Sci. (2022) 23:5572. doi: 10.3390/ijms23105572
66. Ribatti D, Nico B, Floris C, Mangieri D, Piras F, Ennas MG, et al. Microvascular density, vascular endothelial growth factor immunoreactivity in tumor cells, vessel diameter and intussusceptive microvascular growth in primary melanoma. Oncol Rep. (2005) 14:81–4.
67. Liu ZL, Chen HH, Zheng LL, Sun LP, Shi. Angiogenic signaling pathways L. and anti-angiogenic therapy for cancer. Signal Transduct Target Ther. (2023) 8:198. doi: 10.1038/s41392-023-01460-1
68. O’Neal J, Clem A, Reynolds L, Dougherty S, Imbert-Fernandez Y, Telang S, et al. Inhibition of 6-phosphofructo-2-kinase (PFKFB3) suppresses glucose metabolism and the growth of HER2+ breast cancer. Breast Cancer Res Treat. (2016) 160:29–40. doi: 10.1007/s10549-016-3968-8
69. Wang B, Li D, Ilnytskyy Y, Kovalchuk I, Kovalchuk O. A miR-34a-guided, tRNA(i)(Met)-derived, piR_019752-like fragment (tRiMetF31) suppresses migration and angiogenesis of breast cancer cells via targeting PFKFB3. Cell Death Discovery. (2022) 8:355. doi: 10.1038/s41420-022-01054-w
70. Wang B, Li D, Cherkasova V, Gerasymchuk M, Narendran A, Kovalchuk I, et al. Cannabinol Inhibits Cellular Proliferation, Invasion, and Angiogenesis of Neuroblastoma via Novel miR-34a/tRiMetF31/PFKFB3 Axis. Cancers (Basel). (2022) 14:1908. doi: 10.3390/cancers14081908
71. Isenberg JS, Roberts DD. THBS1 (thrombospondin-1). Atlas Genet Cytogenet Oncol Haematol. (2020) 24:291–9. doi: 10.4267/2042/70774
72. Mo D, He F, Zheng J, Chen H, Tang L, Yan F. tRNA-Derived Fragment tRF-17-79MP9PP Attenuates Cell Invasion and Migration via THBS1/TGF-β1/Smad3 Axis in Breast Cancer. Front Oncol. (2021) 11:656078. doi: 10.3389/fonc.2021.656078
73. Jiang Q, Ma Y, Zhao Y, Yao MD, Zhu Y, Zhang QY, et al. tRNA-derived fragment tRF-1001: A novel anti-angiogenic factor in pathological ocular angiogenesis. Mol Ther Nucleic Acids. (2022) 30:407–20. doi: 10.1016/j.omtn.2022.10.016
74. Wang YH, Cao YW, Yang XC, Niu HT, Sun LJ, Wang XS, et al. Effect of TLR4 and B7-H1 on immune escape of urothelial bladder cancer and its clinical significance. Asian Pac J Cancer Prev. (2014) 15:1321–6. doi: 10.7314/apjcp.2014.15.3.1321
75. Ben-Baruch G, Sivan E, Moran O, Rizel S, Menczer J. D.S. Seidman. Primary peritoneal serous papillary carcinoma: a study of 25 cases and comparison with stage III-IV ovarian papillary serous carcinoma. Gynecol Oncol. (1996) 60:393–6. doi: 10.1006/gyno.1996.0060
76. Zhao H, Jia H, Han Q, Zhang J. Homeobox containing 1 inhibits liver cancer progression by promoting autophagy as well as inhibiting stemness and immune escape. Oncol Rep. (2018) 40:1657–65. doi: 10.3892/or.2018.6551
77. Zhang M, Li F, Wang J, He W, Li Y, Li H, et al. tRNA-derived fragment tRF-03357 promotes cell proliferation, migration and invasion in high-grade serous ovarian cancer. Onco Targets Ther. (2019) 12:6371–83. doi: 10.2147/ott.S206861
78. Kitamura T, Qian BZ, Pollard JW. Immune cell promotion of metastasis. Nat Rev Immunol. (2015) 15:73–86. doi: 10.1038/nri3789
79. Gao Z, Jijiwa M, Nasu M, Borgard H, Gong T, Xu J, et al. Comprehensive landscape of tRNA-derived fragments in lung cancer. Mol Ther Oncolytics. (2022) 26:207–25. doi: 10.1016/j.omto.2022.07.002
80. Huang Y, Hong W, Wei X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J Hematol Oncol. (2022) 15:129. doi: 10.1186/s13045-022-01347-8
81. Wu Y, Xu X, Ma L, Yi Q, Sun W, Tang L. Calreticulin regulates TGF-β1-induced epithelial mesenchymal transition through modulating Smad signaling and calcium signaling. Int J Biochem Cell Biol. (2017) 90:103–13. doi: 10.1016/j.biocel.2017.07.023
82. Luan N, Chen Y, Li Q, Mu Y, Zhou Q, Ye X, et al. TRF-20-M0NK5Y93 suppresses the metastasis of colon cancer cells by impairing the epithelial-to-mesenchymal transition through targeting Claudin-1. Am J Transl Res. (2021) 13:124–42.
83. Zhao X, Zou Y, Gu Q, Zhao G, Gray H, Pfeffer LM, et al. Lentiviral vector mediated claudin1 silencing inhibits epithelial to mesenchymal transition in breast cancer cells. Viruses. (2015) 7:2965–79. doi: 10.3390/v7062755
84. Singh AB, Sharma A, Smith JJ, Krishnan M, Chen X, Eschrich S, et al. Claudin-1 up-regulates the repressor ZEB-1 to inhibit E-cadherin expression in colon cancer cells. Gastroenterology. (2011) 141:2140–53. doi: 10.1053/j.gastro.2011.08.038
85. Zhang Z, Liu Z, Zhao W, Zhao X, Tao Y. tRF-19-W4PU732S promotes breast cancer cell Malignant activity by targeting inhibition of RPL27A (ribosomal protein-L27A). Bioengineered. (2022) 13:2087–98. doi: 10.1080/21655979.2021.2023796
86. Wang H, Huang W, Fan X, He X, Chen S, Yu S, et al. The tRNA-Derived Fragment tRF-24-V29K9UV3IU Functions as a miRNA-like RNA to Prevent Gastric Cancer Progression by Inhibiting GPR78 Expression. J Oncol. (2022) 2022:8777697. doi: 10.1155/2022/8777697
87. Liu D, Wu C, Wang J, Zhang L, Sun Z, Chen S, et al. Transfer RNA-derived fragment 5’tRF-Gly promotes the development of hepatocellular carcinoma by direct targeting of carcinoembryonic antigen-related cell adhesion molecule 1. Cancer Sci. (2022) 113:3476–88. doi: 10.1111/cas.15505
88. Chen H, Xu Z, Cai H, Peng Y, Yang L, Wang Z. Identifying differentially expressed tRNA-derived small fragments as a biomarker for the progression and metastasis of colorectal cancer. Dis Markers. (2022) 2022:2646173. doi: 10.1155/2022/2646173
89. Babaei G, Aziz SG, Jaghi. EMT NZZ. cancer stem cells and autophagy; The three main axes of metastasis. BioMed Pharmacother. (2021) 133:110909. doi: 10.1016/j.biopha.2020.110909
90. Zhou Y, Hu J, Liu L, Yan M, Zhang Q, Song X, et al. Gly-tRF enhances LCSC-like properties and promotes HCC cells migration by targeting NDFIP2. Cancer Cell Int. (2021) 21:502. doi: 10.1186/s12935-021-02102-8
91. Arneth B. Tumor microenvironment. Medicina (Kaunas). (2019) 56:15. doi: 10.3390/medicina56010015
92. Infantino V, Santarsiero A, Convertini P, Todisco S, Iacobazzi V. Cancer cell metabolism in hypoxia: role of HIF-1 as key regulator and therapeutic target. Int J Mol Sci. (2021) 22:5703. doi: 10.3390/ijms22115703
93. Luan N, Mu Y, Mu J, Chen Y, Ye X, Zhou Q, et al. Dicer1 promotes colon cancer cell invasion and migration through modulation of tRF-20-MEJB5Y13 expression under hypoxia. Front Genet. (2021) 12:638244. doi: 10.3389/fgene.2021.638244
94. Luan N, Wang J, Sheng B, Zhou Q, Ye X, Zhu X, et al. tRF-20-M0NK5Y93-induced MALAT1 promotes colon cancer metastasis through alternative splicing of SMC1A. Am J Cancer Res. (2023) 13:852–71.
95. Zong D, Ouyang R, Li J, Chen Y, Chen P. Notch signaling in lung diseases: focus on Notch1 and Notch3. Ther Adv Respir Dis. (2016) 10:468–84. doi: 10.1177/1753465816654873
96. Wang J, Liu X, Cui W, Xie Q, Peng W, Zhang H, et al. Plasma tRNA-derived small RNAs signature as a predictive and prognostic biomarker in lung adenocarcinoma. Cancer Cell Int. (2022) 22:59. doi: 10.1186/s12935-022-02481-6
Keywords: tsRNA, tumor metastasis, signaling pathway, epithelial-mesenchymal transition, tumor microenvironment
Citation: Dong H, Ye C, Ye X, Yan J, Ye G and Shao Y (2025) The biological role and molecular mechanism of transfer RNA-derived small RNAs in tumor metastasis. Front. Oncol. 15:1560943. doi: 10.3389/fonc.2025.1560943
Received: 15 January 2025; Accepted: 24 March 2025;
Published: 08 April 2025.
Edited by:
Dan Liu, Sichuan University, ChinaReviewed by:
Yuan Zhou, Peking University, ChinaCopyright © 2025 Dong, Ye, Ye, Yan, Ye and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongfu Shao, ZnlzaGFveW9uZ2Z1QG5idS5lZHUuY24=; c2hhb3lvbmdmdTExNzNAMTYzLmNvbQ==; Guoliang Ye, eWVndW9saWFuZ0BuYnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.