
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 09 April 2025
Sec. Breast Cancer
Volume 15 - 2025 | https://doi.org/10.3389/fonc.2025.1552937
Objective: The purpose of the current study was to determine the relationship between ribonucleotide reductase M1 (RRM1), topoisomerase II alpha (TOP2A), Thymidylate synthase (TYMS), class III beta-tubulin (TUBB3) and phosphatase and tensin homolog (PTEN) expressions and relapse-free survival (RFS) in early-stage invasive breast cancer (IBC) patients with hormone receptor positive (HR+), as well as to develop a nomogram model for forecasting RFS.
Methods: Early-stage IBC patients with HR+ who were diagnosed and treated at the First Affiliated Hospital of Xi’an Jiaotong University from June 2017 to December 2020 were enrolled in this study. The survival analysis was performed by utilizing the Kaplan-Meier method, and the risk factors linked to patient RFS were determined by performing Cox regression analysis. The nomogram for predicting RFS in early-stage IBC patients with HR+ was stablished and validated based on the results of the Cox regression analysis.
Results: In total, 126 early-stage IBC patients with HR+ were included in the current study. Among these patients, 23 cases experienced relapse after surgery, with a median RFS of 29 months. Significant relationships were observed between TYMS, RRM1, TUBB3, TOP2 and PTEN, Ki67 and human epidermal growth factor receptor 2 (HER2) and patient RFS. Cox regression analysis revealed that chemotherapy and higher expression levels of TOP2A, HER2 and Ki67 were independent predictors of RFS in early-stage IBC patients with HR+. The nomogram we constructed using the above independent risk factors exhibited good ability for predicting RFS in early-stage IBC patients with HR+.
Conclusion: Chemotherapy, TOP2A, HER2, and Ki67 expression were independent predictors of RFS in early-stage IBC patients with HR+. The nomogram we developed using these predictors is a reliable tool for predicting RFS in this patient population.
Globally, breast cancer (BC) has become the most common malignancy and a significant public health burden (1). The incidence of BC accounts for approximately 12.5% of new cancer cases, and more than 6 million women worldwide die from BC each year (https://www.wcrf.org/cancer-trends/worldwide-cancer-data/). The incidence of BC has gradually increased due to an aging population and some screening methods, but the survival rate of BC has also improved significantly in the past few decades. A recent study has shown that patients with early BC recurrence have a worse prognosis than those with late recurrence (2). Hormone receptor (HR)-negative (HR-) BC patients have a higher risk of recurrence in the first two years after initial diagnosis, but their risk of recurrence has declined rapidly thereafter, lower than HR-positive (HR+) patients (3, 4). Patients with HR+ BC have an annual risk of recurrence that stays unchanged for a long time, and recurrence may occur within ten years after the initial diagnosis (3). The prognosis of early recurrence is worse than that of late recurrence (5).
It has been reported that the detection of multiple gene expressions, including ribonucleotide reductase M1 (RRM1), topoisomerase II alpha (TOP2A), class III beta-tubulin (TUBB3), Thymidylate synthase (TYMS), can guide individualized chemotherapy to improve therapeutic efficacy and reduce side effects in BC patients (6).Additionally, gene expressions of RRM1, TOP2A, TYMS and TUBB3 have been linked to objective treatment response in BC patients (7). TYMS is a key target gene of 5-fluorouracil in BC patients (8), and increased TYMS expression is associated with a poor prognosis (9). RRM1 expression significantly correlates with sensitivity to gemcitabine and is related to estrogen receptor (ER) status, lymph node metastasis and pathological classification (10). TUBB3 serves as a biomarker for resistance of docetaxel and paclitaxel, while TOP2A expression level obviously is related to anthracyclines efficacy (11). TUBB3 and TOP2A also serve as critical prognostic factors for predicting overall survival (12). It is reported that high TUBB3 expression is associated with low response for taxanes chemotherapy and tumor grade, whereas low TUBB3 and TOP2A expression is associated with improved clinical outcomes (12–14). Individuals with both high human epidermal growth factor receptor 2 (HER2) and TOP2A protein expression present a poor prognosis in T1N0 BC patients and can benefit from anthracycline-based regimens (14). Loss of phosphatase and tensin homolog (PTEN) activity is associated with multiple primary and metastatic malignancies, including BC (15). PTEN serves as a crucial indicator for detecting the occurrence, development, invasion and metastasis of BC, and the loss of PTEN expression may result in lymph node metastasis of BC (16). Therefore, it can serve as an objective indicator toassess the biological behavior of BC.
However, it is still unclear whether RRM1, TOP2A, TYMS, TUBB3 and PTEN can be utilized to prognosticate relapse-free survival (RFS) in early-stage invasive BC (IBC) patients with HR+. Therefore, the aim of this research was to examine the relationship between the aforementioned genes and RFS in IBC patients. Ultimately, a nomogram model was created to predict RFS in early-stage IBC patients with HR+.
In total, 126 IBC patients with HR+ who were diagnosed and treated in the First Affiliated Hospital of Xi’an Jiaotong University between June 2017 and December 2020 were enrolled in this research. The inclusion criteria are as follows: 1) received surgery; 2) operative pathological diagnosis indicated early-stage IBC with HR+ (stage I/II); 3) the availability of gene expression data for Ki67, TYMS, RRM1, TUBB3, TOP2A, HER2 and PTEN; 4) complete clinicopathological data. The exclusion criteria are as follows: 1) surgery was not performed; 2) inability to obtain gene expression data; 3) incomplete clinicopathological data. This study adhered to the Declaration of Helsinki, and was approved and supervised by the Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University (No. XJTU1AF2022LSK-335).
The expression levels of the genes (Ki67, TYMS, RRM1, TUBB3, TOP2A, HER2 and PTEN) in IBC patients were tested by Surexam Co., Ltd (Guangzhou, China). The clinicopathological data, treatment-related data, along with the gene expression data of the patients, were collected for further analysis. Data processing was carried out undoing Microsoft Excel and SPSS26.0 software. The receiver operating characteristic (ROC) curve was used to determine the optimal cut-off value of the continuous data. Following this, the continuous variables were transformed into binary variables based on the cut-off values.
Cox regression analysis was utilized to calculate Hazard ratios (HRs) and 95% confident intervals (CIs) for the variables. According to the results of univariate Cox regression analysis, factors with P<0.05 in the were selected for further multivariate Cox regression analysis to identify independent factors related to the RFS of IBC patients with HR+. Concordance-index (C-index), area under the curve (AUC) of the ROC and Bootstrap calibration curve were conducted to assess the discrimination and calibration of the nomogram.
SPSS 26.0 software was used for data analysis. The difference between the two groups was analyzed by Chi-square test. Survival analysis was conducted by using Kaplan-Meier method, and the Log-rank test was used to analyze the survival difference between two groups. The factors related to the RFS of the patients were determined by Cox regression analysis. Development and validation of the nomogram for forecasting RFS were conducted by RStudio software. P<0.05 was considered to be statistically significant.
126 patients were enrolled in this research according to the inclusion and exclusion criteria. The medium follow-up time was 27 months. Among these patients, 23 cases relapsed after surgery, with a median RFS of 29 months. From the Table 1, we observed that patient age (P=0.009), tumor grade (P=0.032), tumor diameter (P=0.020), sentinel lymph node (P=0.014), Ki67 expression (P<0.001), chemotherapy (P<0.001), and the expression levels of TYMS (P=0.020), RRM1 (P=0.010), TOP2A (P<0.001), HER2 (P=0.004) and PTEN (P<0.001) were associated with the relapse of early-stage IBC patients with HR+.
To determine the factor that might affect patient relapse after surgery, we conducted univariate Cox regression analysis. The results showed that patient age (HR: 0.354, 95%CI: 0.137-0.914, P=0.032), Ki67 expression (HR: 4.684, 95%CI: 1.989-11.033, P<0.001), chemotherapy (HR:3.023, 95%CI: 1.299-7.033, P=0.010), the expressions of TYMS (HR: 2.507, 95%CI: 1.049-5.991, P=0.039), RRM1 (HR: 3.858, 95%CI: 1.658-8.979, P=0.002), TUBB3 (HR: 2.819, 95%CI: 1.041-7.629, P=0.041), TOP2A (HR:6.021, 95%CI: 2.342-15.480, P<0.001), HER2 (HR: 0.200, 95%CI: 0.080-0.501, P=0.001) and PTEN (HR: 0.270, 95%CI: 0.109-0.666, P=0.004) were significantly related to the RFS of the patients (Table 2). Moreover, survival analyses used by Kaplan-Meier method also showed that the above factors were correlated with the RFS of early-stage IBC patients with HR+ (Figure 1). Thus, these factors were enrolled in further multivariate Cox regression analysis. We observed that chemotherapy (HR: 3.601, 95%CI: 1.395-9.298, P=0.008) and the expression levels of Ki67 (HR: 3.143, 95%CI: 1.143-8.647, P=0.027), TOP2A (HR: 3.331, 95%CI: 1.053-10.531, P=0.041) and HER2 (HR: 7.501, 95%CI: 2.674-21.036, P<0.001) were the independent factors affecting the RFS of the patients (Table 3).
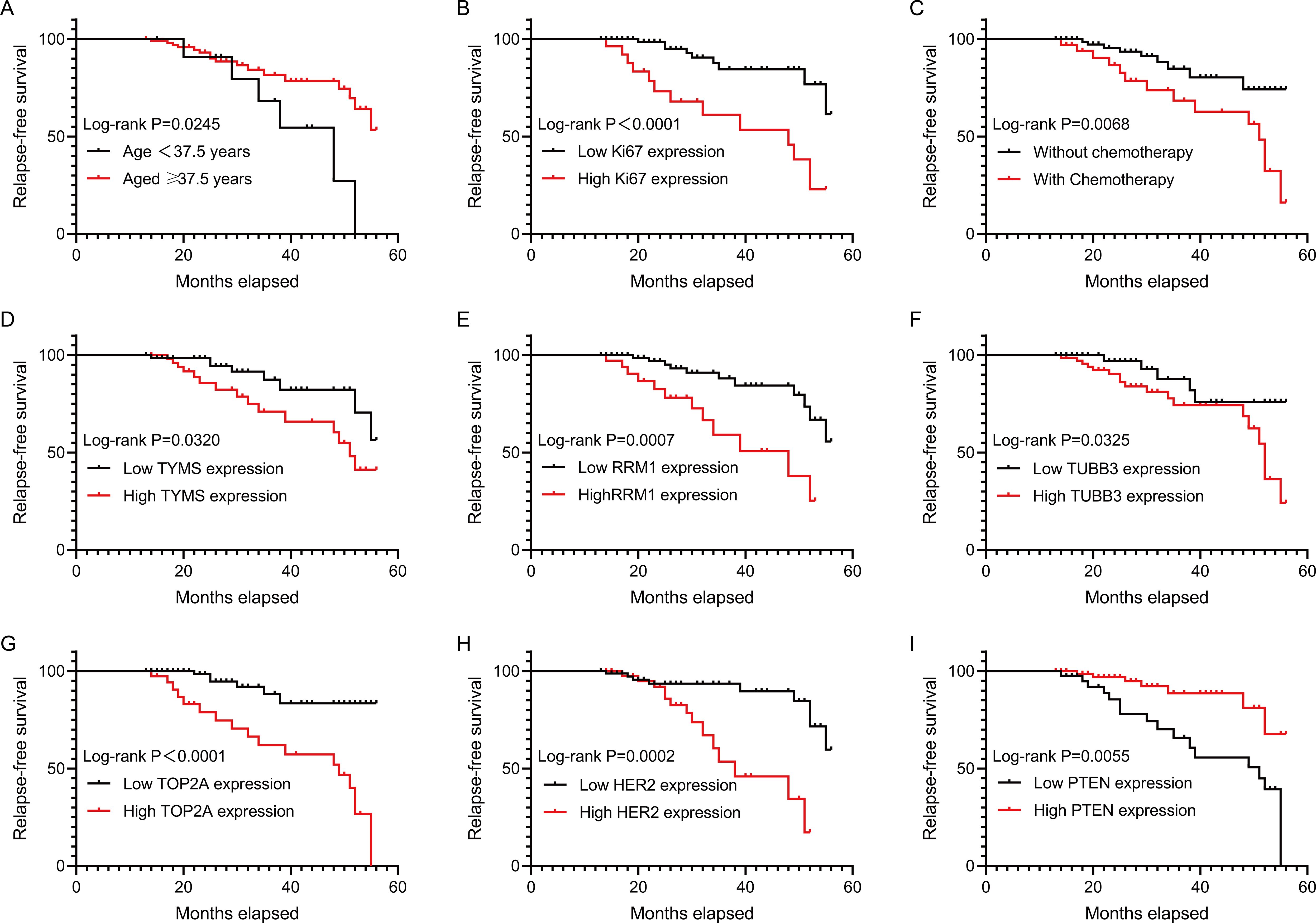
Figure 1. RFS analysis of early-stage IBC patients with HR+. The correlations of RFS of HR+ early-stage IBC patients with age, Ki67 expression, chemotherapy, TYMS expression, RRM1 expression, TUBB3 expression, TOP2A expression, HER2 expression and PTEN expression were analyzed by using Kaplan-Meier method. RFS, relapse-free survival; IBC, invasive breast cancer; HR, hormonal receptor; TYMS, thymidylate synthase; RRM1, ribonucleotide reductase M1; TUBB3, class III beta-tubulin; TOP2A, topoisomerase II alpha; PTEN, phosphatase and tensin homolog; HER2, human epidermal growth factor receptor 2.
Based on Ki67 expression, chemotherapy, TOP2A expression and HER2 expression, we developed a nomogram for predicting RFS (Figure 2). The c-index of the nomogram was 0.833, indicating the nomogram had good discrimination. Besides, the Bootstrap calibration curve also indicated that the trend between true values and predicted values was consistent, suggesting that the nomogram had the accuracy for forecasting RFS of early-stage IBC patients with HR+ (Figure 3).
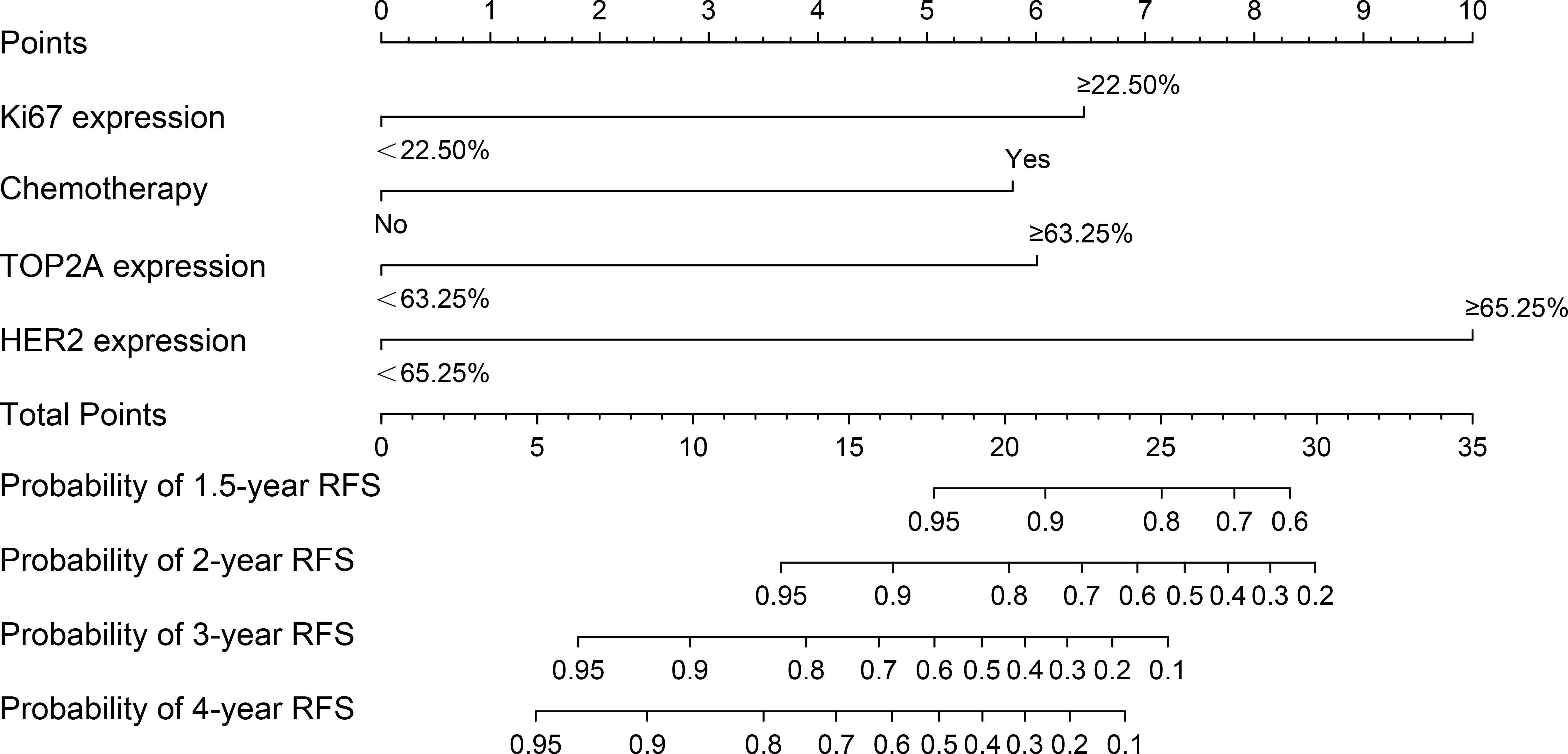
Figure 2. The nomogram for predicting RFS of early-stage IBC patients with HR+. RFS, relapse-free survival; IBC, invasive breast cancer; HR, hormonal receptor; TOP2A, topoisomerase II alpha; HER2, human epidermal growth factor receptor 2.
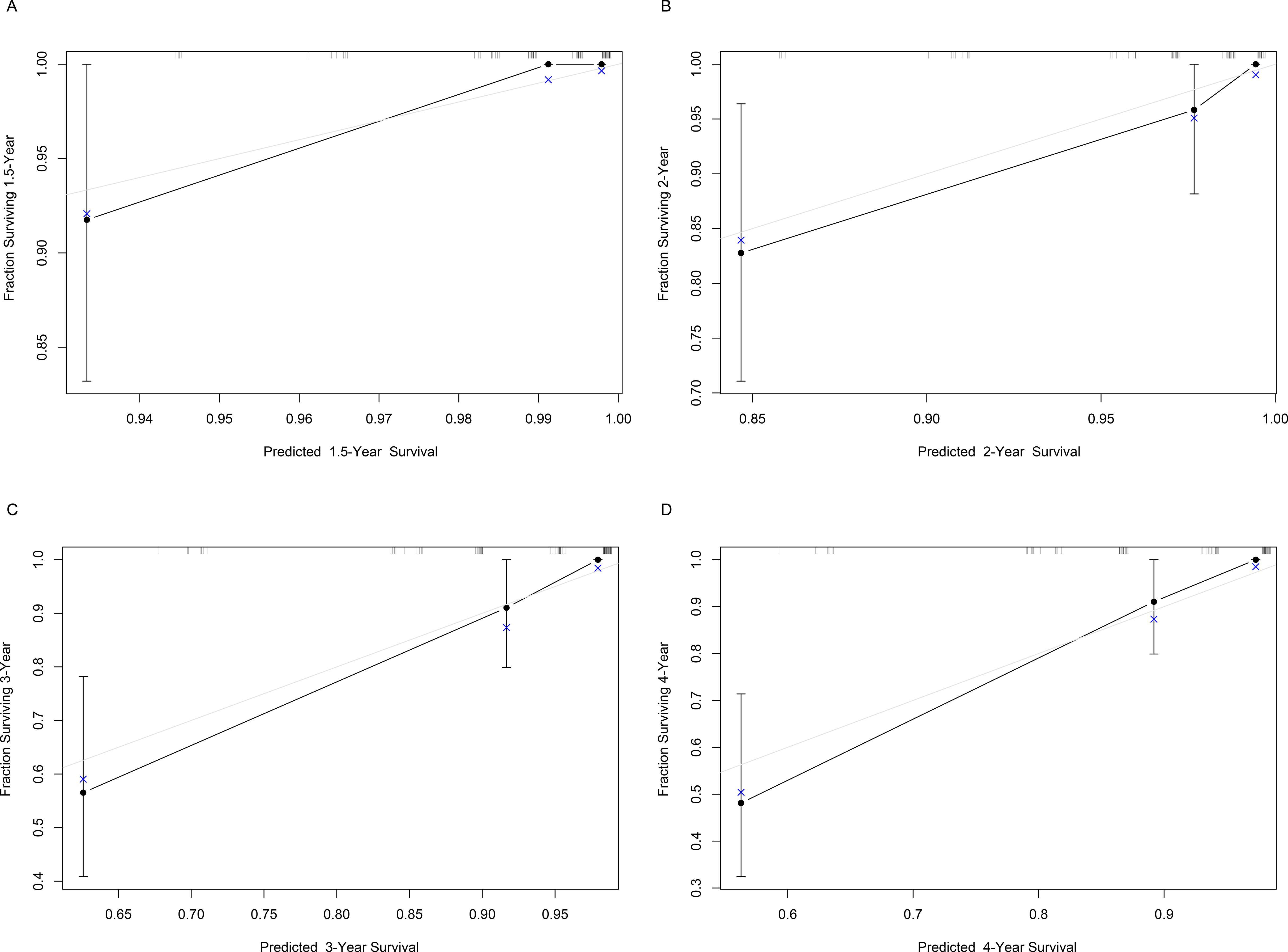
Figure 3. Bootstrap calibration curve of the nomogram for predicting RFS of early-stage IBC patients with HR+. The X-axis represents the predicted probability, and the Y-axis represents the actual probability. The gray line represents the ideal value, and the black line represents the predicted value. It is showed that the trend of the predicted value is consistent with the true value, indicating the good calibration effect of the nomogram. RFS, relapse-free survival; IBC, invasive breast cancer; HR, hormonal receptor.
Although the risk of recurrence and death was decreased after adjuvant endocrine therapy, up to 20% of early BC patients will experience relapse with 10 years after surgery, including local and distant metastasis. Hence, it is imperative to investigate the risk factor for early-stage IBC patients and establish a relevant prediction model.
Previous studies have showed that RRM1, TOP2A, TYMS, TUBBS, and PTEN are prognostic markers and can reflect patients’ response to treatment in BC (6, 7, 16). In the current study, the predictive role of RRM1, TOP2A, TYMS, TUBB3 and PTEN in RFS of early-stage IBC with HR+, and other possible factors associated with RFS were investigated. Our findings indicated that chemotherapy, TOP2A, HER2 and Ki67 were independent predictors of RFS. The nomogram is a convenient tool for forecasting the outcome, which is widely used to quantify risk in various disease (17). Thus, based on the above independent risk factors (chemotherapy, TOP2A, HER2 and Ki67), we established a nomogram for predicting RFS. The c-index of the nomogram indicated a good discrimination, while the Bootstrap calibration curve indicated that the nomogram had accuracy for predicting RFS.
TOP2A is a gene that encodes enzyme topoisomerase IIα, which can break down DNA supercoils through reversible double-stranded DNA breaks during DNA replication and transcription. The enzyme topoisomerase IIα is considered to be the target of anthracyclines. Anthracycline drugs stabilize double-stranded DNA breakage by binding to enzyme topoisomerase IIα, ultimately leading to apoptosis. It has been reported that TOP2A amplification occurs in 2-9% of BC cases (18, 19). Moreover, in patients receiving anthracycline-based adjuvant therapy, TOP2A amplification has a significant impact on RFS and overall survival (18, 20). BC patients with TOP2A amplification showed a better response to adjuvant chemotherapy and a better prognosis (21), while patients with TOP2A deletion showed resistance to anthracyclines (19). There is also a significant association between TOP2A amplification and recurrence in BC patients, including ER+ BC (22, 23). In this research, we observed that TOP2A was an independent predictor for RFS, and the increased expression of TOP2A was related to the increased risk of recurrence in early-stage IBC patients with HR+, which was consistent with the findings of previous studies (12, 14, 22–24). Thus, it is indicated that TOP2A expression can provide prognostic information in HR+ early-stage IBC and may be used to identify patients with high risk of recurrence.
One of the characteristics of malignant tumors is a high proliferation rate (25). Therefore, a high proliferation rate is also an critical variable of tumor prognosis. As a classic proliferation marker, Ki67 has been found to be significantly related to the prognosis, including RFS, of BC patients (24, 26). High Ki67 level is associated with poor prognosis and chemotherapy response (24, 27–29). Similarly, our results showed that higher Ki67 expression was related to worse RFS and was an independent predictor for relapse of HR+ early-stage IBC patients. Besides, we found that patients with higher HER2 level had significantly improved RFS, and HER2 expression was independently relevant to the recurrence of the patients, which is consistent with the previous findings (30). These findings indicated that combined detection of these independent risk factors might provide the clinicians with a simple and convenient tool for predicting the probability of recurrence in early-stage IBC patients with HR+.
However, the limitations in this study cannot be ignored. First, as a single-center and retrospective research, the sample size was relatively small, and only internal validation of the nomogram was performed. Second, only the expression levels of the target genes and some common clinicopathological indexes were analyzed in this study, the laboratory indicators and other potential predictors of the patients were not included for analyzing. Third, although the population of this study was included by inclusion and exclusion criteria, the impact of individual factors on RFS cannot be avoided. Therefore, further studies should be conducted using a prospective, multi-center approach with a larger sample size to verify the findings of the current study. Additionally, external validation of the nomogram should be performed based on a multi-center study.
Our study revealed that chemotherapy, TOP2A, HER2 and Ki67 were independent predictors for RFS in HR+ early-stage IBC patients. The nomogram constructed in this research showed good discrimination and accuracy in predicting RFS of the patients. This provides a convenient tool to predict the recurrence probability in early-stage IBC patients with HR+.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
The studies involving humans were approved by The First Affiliated Hospital of Xi’an Jiaotong University (No. XJTU1AF2022LSK-335). The studies were conducted in accordance with the local legislation and institutional requirements. As a retrospective study, written informed consent was not required in accordance with national legislation and institutional requirements, and the waiver of informed consent was approved by the Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University.
DY: Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft. RM: Investigation, Methodology, Supervision, Validation, Visualization, Writing – review & editing. HY: Investigation, Methodology, Validation, Visualization, Writing – review & editing. WL: Conceptualization, Investigation, Methodology, Project administration, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Shaanxi Province Key Research and Development Plan (No. 2024SF-YBXM-114).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Pedersen RN, Mellemkjær L, Ejlertsen B, Nørgaard M, Cronin-Fenton DP. Mortality after late breast cancer recurrence in Denmark. J Clin Oncol. (2022) 40:1450–63. doi: 10.1200/JCO.21.02062
3. Al-Batsh T, Abdel-Razeq N, Al-Masri Y, El-Khatib O, Sharaf B, Tamimi F, et al. Escalation and de-escalation strategies for endocrine therapy in early-stage breast cancer. Biologics. (2025) 19:97–111. doi: 10.2147/BTT.S508634
4. Nguyen Van Long F, Poirier B, Desbiens C, Perron M, Paquet C, Ouellet C, et al. First versus second-generation molecular profiling tests: How both can guide decision-making in early-stage hormone-receptor positive breast cancers? Cancer Treat Rev. (2025) 135:102909. doi: 10.1016/j.ctrv.2025.102909
5. Brenner DR, Brockton NT, Kotsopoulos J, Cotterchio M, Boucher BA, Courneya KS, et al. Breast cancer survival among young women: a review of the role of modifiable lifestyle factors. Cancer Causes Control. (2016) 27:459–72. doi: 10.1007/s10552-016-0726-5
6. Li J, Sun P, Huang T, He S, Li L, Xue G. Individualized chemotherapy guided by the expression of ERCC1, RRM1, TUBB3, TYMS and TOP2A genes versus classic chemotherapy in the treatment of breast cancer: A comparative effectiveness study. Oncol Lett. (2021) 21:21. doi: 10.3892/ol.2020.12282
7. Tsyganov MM, Ibragimova MK, Garbukov EY, Tsydenova IA, Gaptulbarova KA, Dolgasheva DS, et al. Predictive and prognostic significance of mRNA expression and DNA copies aberrations of ERCC1, RRM1, TOP1, TOP2A, TUBB3, TYMS, and GSTP1 genes in patients with breast cancer. Diagnostics (Basel). (2022) 12:405. doi: 10.3390/diagnostics12020405
8. Lone MN, Gul S, Mehraj U, Sofi S, Dar AH, Ganie SA, et al. Synthesis and biological evaluation of novel uracil derivatives as thymidylate synthase inhibitors. Appl Biochem Biotechnol. (2023) 195:6212–31. doi: 10.1007/s12010-023-04367-3
9. Song S, Tian B, Zhang M, Gao X, Jie L, Liu P, et al. Diagnostic and prognostic value of thymidylate synthase expression in breast cancer. Clin Exp Pharmacol Physiol. (2021) 48:279–87. doi: 10.1111/1440-1681.13415
10. Dorman SN, Baranova K, Knoll JH, Urquhart BL, Mariani G, Carcangiu ML, et al. Genomic signatures for paclitaxel and gemcitabine resistance in breast cancer derived by machine learning. Mol Oncol. (2016) 10:85–100.
11. O’malley FP, Chia S, Tu D, Shepherd LE, Levine MN, Huntsman D, et al. Topoisomerase II alpha protein and responsiveness of breast cancer to adjuvant chemotherapy with CEF compared to CMF in the NCIC CTG randomized MA.5 adjuvant trial. Breast Cancer Res Treat. (2011) 128:401–9. doi: 10.1007/s10549-011-1511-5
12. Xu YC, Zhang FC, Li JJ, Dai JQ, Liu Q, Tang L, et al. RRM1, TUBB3, TOP2A, CYP19A1, CYP2D6: Difference between mRNA and protein expression in predicting prognosis of breast cancer patients. Oncol Rep. (2015) 34:1883–94. doi: 10.3892/or.2015.4183
13. Narvi E, Jaakkola K, Winsel S, Oetken-Lindholm C, Halonen P, Kallio L, et al. Altered TUBB3 expression contributes to the epothilone response of mitotic cells. Br J Cancer. (2013) 108:82–90. doi: 10.1038/bjc.2012.553
14. Zhong W, Yang Y, Zhang A, Lin W, Liang G, Ling Y, et al. Prognostic and predictive value of the combination of TOP2A and HER2 in node-negative tumors 2 cm or smaller (T1N0) breast cancer. Breast Cancer. (2020) 27:1147–57. doi: 10.1007/s12282-020-01142-8
15. Kechagioglou P, Papi RM, Provatopoulou X, Kalogera E, Papadimitriou E, Grigoropoulos P, et al. Tumor suppressor PTEN in breast cancer: heterozygosity, mutations and protein expression. Anticancer Res. (2014) 34:1387–400.
16. Li K, Li GD, Sun LY, Li XQ. PTEN and SHIP: Impact on lymphatic metastasis in breast cancer. J Cancer Res Ther. (2018) 14:S937–s941. doi: 10.4103/0973-1482.193894
17. Mo S, Zhou Z, Li Y, Hu X, Ma X, Zhang L, et al. Establishment and validation of a novel nomogram incorporating clinicopathological parameters into the TNM staging system to predict prognosis for stage II colorectal cancer. Cancer Cell Int. (2020) 20:285. doi: 10.1186/s12935-020-01382-w
18. Nielsen KV, Ejlertsen B, Møller S, Jørgensen JT, Knoop A, Knudsen H, et al. The value of TOP2A gene copy number variation as a biomarker in breast cancer: Update of DBCG trial 89D. Acta Oncol. (2008) 47:725–34. doi: 10.1080/02841860801995396
19. Moelans CB, De Weger RA, Van Blokland MT, van der Wall E, Van Diest PJ. Simultaneous detection of TOP2A and HER2 gene amplification by multiplex ligation-dependent probe amplification in breast cancer. Mod Pathol. (2010) 23:62–70. doi: 10.1038/modpathol.2009.136
20. Knoop AS, Knudsen H, Balslev E, Rasmussen BB, Overgaard J, Nielsen KV, et al. Retrospective analysis of topoisomerase IIa amplifications and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin, and fluorouracil: Danish Breast Cancer Cooperative Group. J Clin Oncol. (2005) 23:7483–90. doi: 10.1200/JCO.2005.11.007
21. Arriola E, Rodriguez-Pinilla SM, Lambros MB, Jones RL, James M, Savage K, et al. Topoisomerase II alpha amplification may predict benefit from adjuvant anthracyclines in HER2 positive early breast cancer. Breast Cancer Res Treat. (2007) 106:181–9. doi: 10.1007/s10549-006-9492-5
22. Sparano JA, Goldstein LJ, Davidson NE, Sledge GW Jr., Gray R. TOP2A RNA expression and recurrence in estrogen receptor-positive breast cancer. Breast Cancer Res Treat. (2012) 134:751–7. doi: 10.1007/s10549-012-2112-7
23. Tabarestani S, Ghaderian SM, Rezvani H, Mirfakhraie R, Ebrahimi A, Attarian H, et al. Prognostic and predictive value of copy number alterations in invasive breast cancer as determined by multiplex ligation-dependent probe amplification. Cell Oncol (Dordr). (2014) 37:107–18. doi: 10.1007/s13402-013-0165-1
24. Milde-Langosch K, Karn T, Müller V, Witzel I, Rody A, Schmidt M, et al. Validity of the proliferation markers Ki67, TOP2A, and RacGAP1 in molecular subgroups of breast cancer. Breast Cancer Res Treat. (2013) 137:57–67. doi: 10.1007/s10549-012-2296-x
25. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. (2000) 100:57–70. doi: 10.1016/S0092-8674(00)81683-9
26. Colozza M, Azambuja E, Cardoso F, Sotiriou C, Larsimont D, Piccart MJ. Proliferative markers as prognostic and predictive tools in early breast cancer: where are we now? Ann Oncol. (2005) 16:1723–39. doi: 10.1093/annonc/mdi352
27. Urruticoechea A, Smith IE, Dowsett M. Proliferation marker Ki-67 in early breast cancer. J Clin Oncol. (2005) 23:7212–20. doi: 10.1200/JCO.2005.07.501
28. Stuart-Harris R, Caldas C, Pinder SE, Pharoah P. Proliferation markers and survival in early breast cancer: a systematic review and meta-analysis of 85 studies in 32,825 patients. Breast. (2008) 17:323–34. doi: 10.1016/j.breast.2008.02.002
29. Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. (2010) 11:174–83. doi: 10.1016/S1470-2045(09)70262-1
Keywords: invasive breast cancer, hormone receptor positive, relapse-free survival, TOP2A, nomogram
Citation: Yuan D, Ma R, Ye H and Liu W (2025) A prediction model for predicting relapsed-free survival of early-stage invasive breast cancer patients with hormone receptor positive based on Ki67, HER2 and TOP2A. Front. Oncol. 15:1552937. doi: 10.3389/fonc.2025.1552937
Received: 29 December 2024; Accepted: 24 March 2025;
Published: 09 April 2025.
Edited by:
Su Yon Jung, University of California, Los Angeles, United StatesReviewed by:
Sungchan Gwark, Ewha Womans University Seoul Hospital, Republic of KoreaCopyright © 2025 Yuan, Ma, Ye and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenbo Liu, MTczMDE0MzQyNkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.