
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Oncol. , 06 February 2025
Sec. Cancer Molecular Targets and Therapeutics
Volume 15 - 2025 | https://doi.org/10.3389/fonc.2025.1534055
This article is part of the Research Topic Renewed Insight into Cancer Mechanism and Therapy View all 24 articles
 Wen-yu Cai1†
Wen-yu Cai1† Xin-xian Cai2†
Xin-xian Cai2† Yi-ran Fei3
Yi-ran Fei3 Rui Ye2
Rui Ye2 Ding-ming Song4
Ding-ming Song4 Dan Hu5
Dan Hu5 Wan-wan Zhang1
Wan-wan Zhang1 Ming-fei Xia1
Ming-fei Xia1 Xiao-xiao Yang1*
Xiao-xiao Yang1*Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer characterized by the lack of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). Chemotherapy remains the primary treatment option, yet TNBC frequently develops resistance, leading to relapse and metastasis. Emerging evidence highlights the potential of combining DNA methylation inhibitors with immune checkpoint inhibitors (ICIs). DNA methylation contributes to immune escape by silencing immune-regulatory genes, thereby reducing the tumor’s visibility to immune cells. Reversing this epigenetic modification can reinvigorate immune surveillance and enhance the efficacy of immunotherapies. This review discusses the role of DNA methylation in TNBC progression and immune evasion, focusing on recent advances in combination therapies involving DNA methylation inhibitors and ICIs. We discuss the underlying mechanisms that enable these therapeutic synergies, preclinical and clinical evidence supporting the approach, and the challenges posed by tumor heterogeneity, drug resistance, and toxicity. Finally, we explore the potential for personalized treatment strategies incorporating multi-omics data to optimize therapeutic outcomes. The integration of epigenetic therapies and immunotherapy offers a promising avenue for improving survival in TNBC patients.
Breast cancer is a heterogeneous disease comprising several subtypes, with Triple-Negative Breast Cancer (TNBC) representing approximately 10-15% of all cases (1). Unlike other subtypes, TNBC lacks expression of ER, PR, and HER2 (2). The absence of these critical receptors makes TNBC non-responsive to hormone-based therapies and correlates with higher metastatic potential and shorter overall survival rates (3–6). TNBC tumors are often more prone to early metastasis, particularly to visceral organs, and show poor differentiation at the cellular level. The tumor microenvironment, including immune cells and stromal interactions, is crucial in driving TNBC’s aggressiveness (7). TNBC is also associated with high genetic and phenotypic heterogeneity, complicating treatment efforts (8).
TNBC comprises multiple subtypes, including basal-like 1 (BL1), basal-like 2 (BL2), mesenchymal (M), mesenchymal stem-like (MSL), and luminal androgen receptor (LAR) subtypes. The BL2 subtype, enriched in growth factor signaling pathways, often exhibits stronger resistance to chemotherapy, whereas the BL1 subtype typically shows greater sensitivity to DNA-damaging agents (9). The LAR subtype is characterized by low immune cell infiltration and M2 macrophage activity, which are associated with poorer responses to immunotherapy and worse prognoses (10). In contrast, the BL1 and BL2 subtypes may benefit more from immunotherapy due to higher immune activation markers.
Chemotherapy remains the mainstay treatment for TNBC, but it is fraught with challenges. Despite being more sensitive to initial chemotherapy compared to other breast cancer subtypes, TNBC frequently develops resistance, leading to relapse and metastasis (8). Various molecular pathways often mediate this resistance, such as the overexpression of ATP-binding cassette (ABC) transporter proteins, which actively efflux chemotherapy drugs from cancer cells (11). The current standard of care for TNBC involves anthracycline and taxane-based chemotherapy regimens. While these therapies have demonstrated efficacy in early-stage disease, their effectiveness diminishes significantly in relapsed and metastatic cases due to the emergence of multi-drug resistance (2). Moreover, newer agents like immune checkpoint and PARP inhibitors have shown promise, but only a subset of patients respond favorably, leaving a substantial proportion with limited options (12). As a result, ongoing research is focused on identifying novel targets and combination therapies to overcome resistance and improve outcomes in TNBC patients.
Cancer immunotherapy has rapidly expanded, revolutionizing treatment strategies across multiple malignancies. The main targets of these therapies are inhibitory receptors such as programmed death-1 (PD-1), programmed death-ligand 1 (PD-L1), and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) (13–15). These molecules play a crucial role in downregulating immune responses, which tumors exploit to escape immune surveillance. Blocking these pathways has significantly improved outcomes for various cancers, including melanoma, non-small cell lung cancer, and renal cell carcinoma (16).
The first immune checkpoint inhibitors were approved for advanced-stage melanoma, showing long-lasting remissions in previously untreatable patients (17). PD-1 inhibitors such as nivolumab and pembrolizumab, as well as CTLA-4 inhibitor ipilimumab, have demonstrated durable responses across multiple cancer types by releasing the “brakes” on the immune system, allowing T cells to attack cancer cells more effectively. However, these treatments are not without challenges, as only a subset of patients exhibit favorable responses due to factors such as tumor mutation burden and the immunosuppressive tumor microenvironment (18).
Despite the success of immune checkpoint inhibitors in other cancer types, their efficacy as monotherapy in TNBC has been limited. Although the introduction of ICIs targeting PD-1 and PD-L1 has shown promise, especially in combination with chemotherapy, the immune landscape of TNBC often presents substantial barriers to effective treatment (19, 20).
One major limitation is immune evasion. These include the recruitment of immunosuppressive cells such as regulatory T cells (Tregs) and myeloid-derived suppressor cells, as well as upregulation of inhibitory receptors like TIM-3 and LAG-3, which reduce the efficacy of PD-1/PD-L1 blockade (19). TNBC often exhibits the characteristics of cold tumors, including a lack of tumor antigens, defects in antigen presentation, and insufficient T-cell infiltration in tumor tissues due to the failure of T cells to go home successfully. These mechanisms limit the effectiveness of ICIs (21). Moreover, MYC gene amplification is associated with the absence of immune cell infiltration, while mutations in the PI3K-AKT pathway may suppress the activation of innate immunity (22). To overcome these limitations, ongoing research explores combination therapies that include ICIs with chemotherapy, PARP inhibitors, and other immune-modulatory agents. These combinations aim to turn “cold” tumors into “hot” tumors, enhancing the immunogenicity of TNBC and improving patient outcomes (23, 24).
Epigenetic modifications, including DNA methylation, histone modifications, and non-coding RNA (ncRNA) regulation, are pivotal in cancer development and progression. DNA methylation typically occurs at CpG islands in the promoter regions of genes, silencing tumor suppressor genes in many cancers (25). Histone modifications, such as acetylation and methylation, regulate chromatin structure, influencing gene expression. The enzymes involved in these modifications, such as histone deacetylases (HDACs) and histone methyltransferases, are often dysregulated in cancer, leading to aberrant transcriptional activation or repression (26). Non-coding RNAs, particularly microRNAs and long non-coding RNAs (lncRNAs), further modulate gene expression by affecting mRNA stability and translation (27). These epigenetic mechanisms are reversible, making them attractive targets for therapeutic intervention. Drugs targeting DNA methylation and histone modifications, such as DNA methyltransferase inhibitors (DNMTis) and HDAC inhibitors, are actively explored for cancer therapy, demonstrating promising results in hematologic and solid tumors (28).
Epigenetic modifications, particularly DNA methylation, are intimately involved in TNBC’s immune evasion strategies. DNA methylation is a critical epigenetic mechanism that adds a methyl group to the cytosine ring within CpG islands, primarily in gene promoters. This process is catalyzed by DNA methyltransferases (DNMTs), such as DNMT1, DNMT3A, and DNMT3B, which play a crucial role in maintaining gene silencing (29, 30). Aberrant DNA methylation is prevalent in tumors, with variations in methylation levels across different regions exerting distinct impacts on gene transcription (Figure 1). Aberrant DNA methylation, particularly promoter hypermethylation, leads to the transcriptional repression of tumor suppressor genes, contributing to cancer initiation and progression (31). On the other hand, increased CpG site methylation within gene bodies can enhance gene expression, potentially by stabilizing the transcript (32). Blagitko-Dorfs (33) et al. discovered that the combined use of DNMT and HDAC inhibitors can downregulate oncogenes such as MYC in acute myeloid leukemia cells through the demethylation of gene bodies. This demonstrates that targeting gene body demethylation may represent a viable epigenetic therapeutic strategy. DNA methylation can silence the expression of immune-related genes, including those involved in antigen presentation and interferon signaling pathways, which are critical for an effective immune response (34).
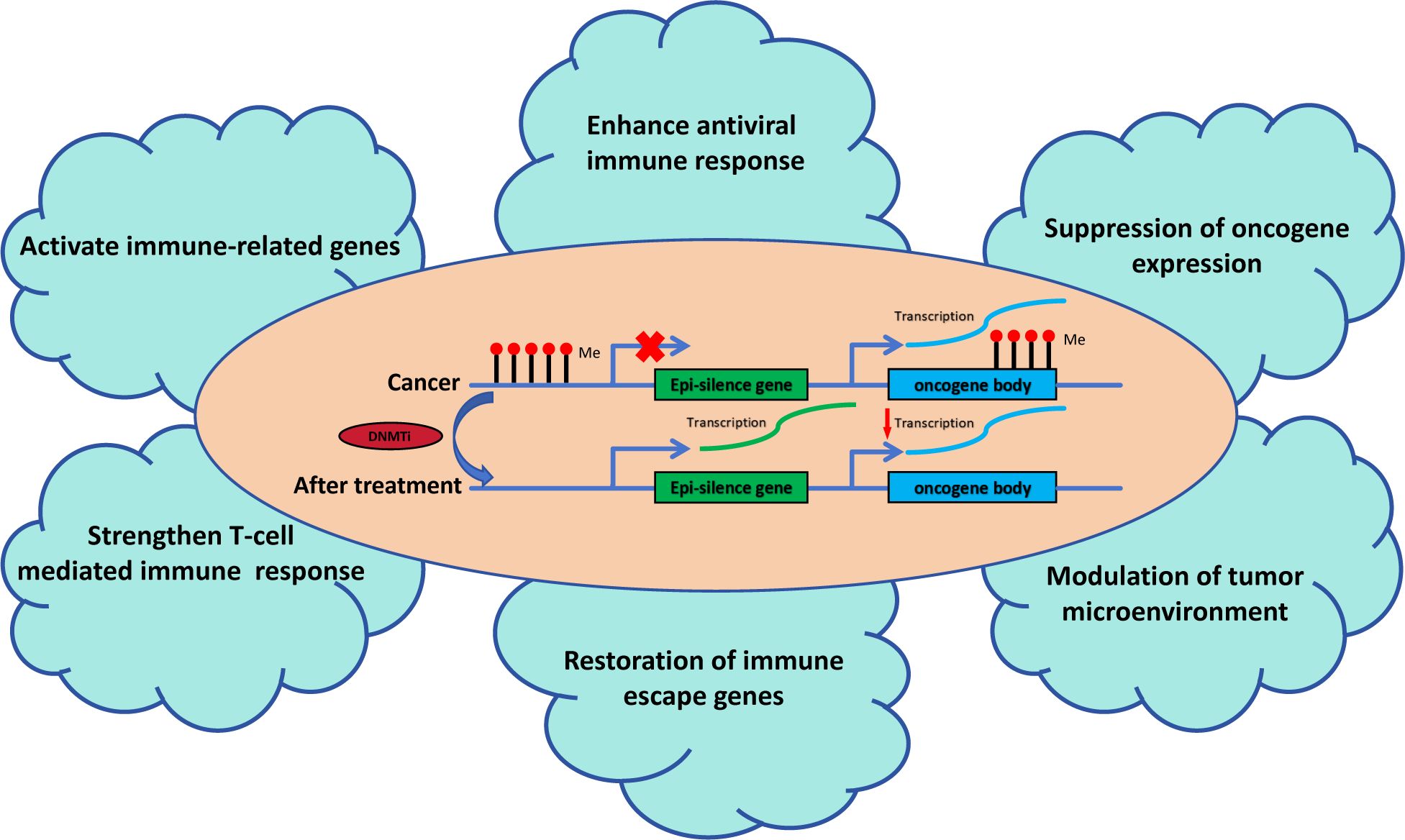
Figure 1. The role of DNA methyltransferase inhibitors in tumor immunity. DNMTis reactivate epi-silenced genes and suppress oncogene expression by reducing DNA methylation. This process enhances antiviral immune responses, activates immune-related genes, and strengthens T-cell-mediated immunity. It modulates the tumor microenvironment, restores immune escape genes, and suppresses oncogene-driven pathways, collectively improving tumor immune surveillance and therapeutic outcomes.
DNA demethylating agents have demonstrated therapeutic or diagnostic potential in hematological disorders (35), and in clinical trials involving patients with recurrent glioblastoma (36) and chemotherapy-resistant hepatocellular carcinoma (37). Furthermore, as reported by Linnekamp et al. (38), partial clinical responses have been observed in several solid tumors, including breast cancer, lung cancer, and colorectal cancer. These responses are characterized by gene demethylation and re-expression in certain patients. DNA methylation frequently silences immune-regulatory genes, impacting immune surveillance and evasion (39, 40). Hypermethylation leads to silencing tumor suppressor genes and immune-related genes, including those involved in antigen presentation and interferon signaling pathways (31). This silencing directly impacts immune escape mechanisms, as it reduces the expression of major histocompatibility complex (MHC) molecules and other immune-related markers, which are essential for effective immune surveillance and response (41). Hypermethylation of immune-regulatory genes like PD-L1 reduces the tumor’s ability to be recognized by cytotoxic T lymphocytes, promoting immune evasion and resistance to therapies (26). These findings suggest that targeting DNA methylation pathways could restore immune function and improve therapeutic outcomes in TNBC.
DNA methylation also plays a role in downregulating immune checkpoints, further enhancing immune evasion. The silencing of STING (stimulator of interferon genes), a crucial component of the innate immune response through DNA methylation, has been observed in TNBC. This silencing reduces the recruitment of immune cells to the tumor site, diminishing the anti-tumor immune response (42). Additionally, epigenetic modifications of immune-related genes contribute to the resistance in immunotherapy treatments, highlighting the potential of combining DNA methylation inhibitors with immunotherapy to overcome immune evasion (43).
Epigenetic modifications also influence the tumor microenvironment, enhancing tumor survival and resistance to therapy. TNBC’s aggressive nature is partly due to the epigenetic remodeling of the extracellular matrix (ECM), which facilitates metastasis and therapeutic resistance (44). TNBC is notorious for its ability to evade the immune system by creating an immunosuppressive tumor microenvironment (TME) (45). One of the primary mechanisms involves the upregulation of immune checkpoint molecules such as PD-L1, which binds to PD-1 on T cells, effectively “turning off” the immune response and allowing the tumor to grow unchecked (46). This immune checkpoint blockade dampens cytotoxic T cell activity and enables TNBC cells to evade immune surveillance. TNBC tumors are also proficient at recruiting immunosuppressive cell populations, including regulatory T cells (Tregs) and myeloid-derived suppressor cells (MDSCs), further suppressing the anti-tumor immune response (47, 48).
Tregs (CD4+Foxp3+) cells, play a crucial role in immune evasion by secreting immunosuppressive cytokines such as IL-10 and TGF-β. These cytokines inhibit the activity of cytotoxic T cells and natural killer cells, thereby protecting the tumor from immune-mediated destruction (46). Similarly, MDSCs are known to suppress T cell proliferation and cytokine production, facilitating the tumor’s ability to escape immune detection (49). Together, these mechanisms create an immune-tolerant environment that allows TNBC to progress.
DNA methylation biomarkers hold significant prognostic value in TNBC. Studies show that aberrant DNA methylation patterns are associated with poor prognosis and resistance to conventional therapies, such as chemotherapy and immunotherapy (50, 51). DNA methylation-induced silencing of tumor suppressor genes and immune-related genes contributes to chemotherapy resistance by preventing the reactivation of apoptotic pathways in cancer cells (52).
Moreover, DNA methylation has emerged as a mechanism of resistance to immunotherapies. Tumors with hypermethylated immune-regulatory genes often show reduced responsiveness to immune checkpoint inhibitors, which aim to enhance the body’s immune response against cancer (53). Combining DNMT inhibitors with immunotherapies has demonstrated the potential to reverse resistance and improve outcomes in TNBC patients.
DNA methylation inhibitors, such as azacitidine and decitabine, are designed to reverse the epigenetic silencing of genes involved in immune regulation and tumor suppression. In the context of TNBC, these inhibitors can restore the expression of immune-related genes, enhancing the effectiveness of immunotherapies like ICIs (54). The hypermethylation of immune-related genes, particularly those involved in antigen presentation, limits immune cell infiltration and recognition, thus contributing to immune evasion in glioma (55). By reversing these epigenetic modifications, DNA methylation inhibitors promote immune activation, increasing tumor antigen visibility to cytotoxic T cells (56, 57). Preclinical studies have demonstrated the synergistic effects of combining DNA methylation inhibitors with ICIs in various cancers, including TNBC. For example, the combination of decitabine with anti-PD-1 therapy in mouse models of pancreatic cancer showed significantly improved survival rates, mediated by increased tumor-infiltrating lymphocytes and a reduction in immunosuppressive cell populations (58). Similar results have been observed in TNBC, where decitabine treatment increases the expression of antigen-presenting genes, such as MHC class I, leading to enhanced T cell-mediated immune responses (59). The underlying mechanisms of synergy between DNA methylation inhibitors and ICIs include enhanced immune cell infiltration, increased antigen presentation, and decreased levels of immunosuppressive factors within the TME. These effects are critical for converting immunologically “cold” tumors into “hot” tumors, which are more responsive to immune checkpoint blockade (53). Preclinical evidence suggests combining these therapies can induce durable anti-tumor responses, even in patients with advanced or resistant cancers.
One of the most significant challenges in treating TNBC with combination therapies, is the intrinsic heterogeneity of TNBC tumors. This diversity leads to varying responses to treatments, as different subclones within a tumor may respond differently to the same therapy (60). Recent studies suggest that identifying TNBC subtypes with specific DNA methylation or immune-related gene expression patterns may help personalize treatment strategies, improving the likelihood of therapeutic success (61). By classifying tumors into more homogeneous subgroups, therapies can be tailored to target the unique molecular features of each subtype, potentially overcoming the challenges posed by heterogeneity. Integrating large-scale multi-omics data requires addressing challenges such as computational demands, data standardization, and robust bioinformatics pipelines. From an ethical perspective, greater attention must be given to patient privacy, data security, and equitable access to personalized treatments derived from multi-omics data.
DNA methylation inhibitors face limitations in clinical application due to the broad spectrum of their target effects. Currently, these inhibitors are primarily used for refractory myelodysplastic syndromes and leukemia, where their therapeutic benefits have been clinically validated. However, the therapeutic efficacy remains unclear in solid tumors, as multiple genes may be epigenetically activated, complicating the treatment outcomes. DNA methylation inhibitors may also trigger compensatory or demethylation events that allow cancer cells to maintain their malignant phenotype despite treatment. Biomarkers such as PD-L1 expression, tumor mutational burden, and specific methylation patterns are being investigated to predict which patients will most likely benefit from these treatments (62).
The combination of DNA methylation inhibitors with immunotherapy introduces potential toxicity concerns, including immune-related adverse events (irAEs) and off-target effects. Immune checkpoint inhibitors can lead to autoimmune reactions, affecting organs such as the liver, lungs, and thyroid (38, 63). Similarly, DNA methylation inhibitors can cause off-target gene demethylation, potentially leading to unwanted gene expression changes that exacerbate toxicity. Studies have reported toxicities such as neutropenia, anemia, and elevated liver enzymes in patients receiving combination therapy.
Biomarkers such as DNA methylation signatures, immune cell infiltration profiles, and circulating tumor DNA (ctDNA) are emerging as potential tools for predicting response to combination therapies in TNBC. Recent advances in single-cell sequencing have enabled more precise characterization of tumor heterogeneity and immune cell interactions, leading to the discovery of novel biomarkers that may guide treatment decisions (64). These biomarkers can also help monitor treatment response and detect early signs of resistance, allowing for timely adjustments in therapy (65). Developing predictive biomarkers such as tumor-infiltrating lymphocyte (TIL) profiles and PD-L1 expression could improve patient selection for immunotherapy-based treatments, increasing the likelihood of therapeutic success (66).
There is growing interest in exploring other epigenetic therapies, such as histone deacetylase (HDAC) inhibitors. HDAC inhibitors can modulate gene expression, enhancing immune recognition of tumors and sensitizing cancer cells to immune checkpoint inhibitors (67). Dual targeting of epigenetic mechanisms, such as combining HDAC inhibitors with DNA methylation or immune checkpoint inhibitors, may lead to more robust anti-tumor responses in TNBC (13). Preclinical studies have shown that HDAC inhibitors can enhance the infiltration of immune cells into tumors and promote the expression of tumor antigens, making them attractive candidates for combination therapies in TNBC (68).
The combination of DNA methylation and immune checkpoint inhibitors holds promise for enhancing the therapeutic efficacy of TNBC. By reversing the epigenetic silencing of immune-regulatory genes, tumor visibility to immune cells can be improved, potentially overcoming the immune evasion characteristic of TNBC. While preclinical and early clinical trials have provided encouraging results, challenges such as tumor heterogeneity, resistance, and toxicity must be addressed. Future research should focus on utilizing multi-omics integration techniques within combination therapy groups to explore the mechanisms of personalized treatment, thereby defining specific therapeutic strategies to improve patient outcomes.
W-YC: Conceptualization, Data curation, Formal analysis, Funding acquisition, Validation, Writing – original draft. X-XC: Conceptualization, Data curation, Formal analysis, Validation, Writing – original draft. Y-RF: Data curation, Project administration, Validation, Writing – review & editing. RY: Data curation, Formal analysis, Project administration, Resources, Supervision, Writing – original draft. D-MS: Data curation, Project administration, Visualization, Writing – review & editing. DH: Formal analysis, Methodology, Supervision, Writing – review & editing. W-WZ: Data curation, Project administration, Visualization, Writing – review & editing. M-FX: Data curation, Project administration, Visualization, Writing – review & editing. X-XY: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study design, data collection, data analysis, manuscript preparation, and publication decisions of this work were supported by Zhejiang Province Traditional Chinese Medicine Science and Technology Project (No. 2023ZL409 by X-XY), the Foundation Project of Zhejiang Chinese Medical University (No. 2023FSYYZY03 by W-YC).
Figure 1 in our manuscript was created using Figdraw (https://www.figdraw.com/) online tools. We would like to express our gratitude for these invaluable resources.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that Generative AI was used in the creation of this manuscript. During the preparation of this work, the authors employed ChatGPT-4.0 to enhance the language. The authors will subsequently conduct a comprehensive review and make necessary modifications, assuming full responsibility for the final published version.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Nedeljković M, Damjanović A. Mechanisms of chemotherapy resistance in triple-negative breast cancer—How we can rise to the challenge. Cells. (2019) 8:957. doi: 10.3390/cells8090957
2. Gupta GK, Collier AL, Lee D, Hoefer RA, Zheleva V, Siewertsz Van Reesema LL, et al. Perspectives on triple-negative breast cancer: current treatment strategies, unmet needs, and potential targets for future therapies. Cancers. (2020) 12:2392. doi: 10.3390/cancers12092392
3. Patel R, Kuwar U, Dhote N, Alexander A, Nakhate K, Jain P, et al. Natural polymers as a carrier for the effective delivery of antineoplastic drugs. Curr Drug Delivery. (2023) 21:193–210. doi: 10.2174/1567201820666230112170035
4. Dhote NS, Patel RD, Kuwar U, Agrawal M, Alexander A, Jain P, et al. Application of Thermoresponsive Smart Polymers based in situ Gel as a Novel Carrier for Tumor Targeting. Curr Cancer Drug Targets. (2023) 24:375–96. doi: 10.2174/1568009623666230803111718
5. Raza MA, Sharma MK, Nagori K, Jain P, Ghosh V, Gupta U, et al. Recent trends on polycaprolactone as sustainable polymer-based drug delivery system in the treatment of cancer: Biomedical applications and nanomedicine. Int J Pharmaceutics. (2024) 666:124734. doi: 10.1016/j.ijpharm.2024.124734
6. Sahu N, Jain P, Nagori K, Ajazuddin. Recent advancement and novel treatment strategies for breast fibroadenoma: clinical approach and prospects. Curr Cancer Ther Rev. (2024) 20:1–11. doi: 10.2174/0115733947318171240802100419
7. Gruosso T, Gigoux M, Manem VSK, Bertos N, Zuo D, Perlitch I, et al. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J Clin Invest. (2019) 129:1785–800. doi: 10.1172/JCI96313
8. Medina MA, Oza G, Sharma A, Arriaga LG, Hernández Hernández JM, Rotello VM, et al. Triple-negative breast cancer: A review of conventional and advanced therapeutic strategies. Int J Environ Res Public Health. (2020) 17:2078. doi: 10.3390/ijerph17062078
9. Shaffer C, Robles A, Risinger A, Mooberry S. Abstract P2-13-01: Differential efficacies of DNA damaging agents in basal-like TNBC subtypes. Cancer Res. (2018) 78:P2–13–01. doi: 10.1158/1538-7445.SABCS17-P2-13-01
10. Meisel J, Douglass E, Kalinsky K, Griffiths LM, Li Z, Li X. Abstract P6-01-20: Luminal androgen receptor subtype and M2 macrophage signatures strongly associate with low pathological complete response rates and poor outcomes in patients with triple negative breast cancer receiving neoadjuvant chemotherapy. Cancer Res. (2023) 83:P6–01–20. doi: 10.1158/1538-7445.SABCS22-P6-01-20
11. Abd El-Aziz YS, Spillane AJ, Jansson PJ, Sahni S. Role of ABCB1 in mediating chemo-resistance of triple negative breast cancers. Bioscience Rep. (2021) 41(2):20204092. doi: 10.1042/BSR20204092
12. Ribeiro R, Carvalho MJ, Goncalves J, Moreira JN. Immunotherapy in triple-negative breast cancer: Insights into tumor immune landscape and therapeutic opportunities. Front Mol Biosci. (2022) 9:903065. doi: 10.3389/fmolb.2022.903065
13. Lu Y, Song Y, Xu Y, Ou N, Liang Z, Hu R, et al. The prevalence and prognostic and clinicopathological value of PD-L1 and PD-L2 in renal cell carcinoma patients: a systematic review and meta-analysis involving 3,389 patients. Transl Androl Urol. (2020) 9:367–81. doi: 10.21037/tau.2020.01.21
14. Zhang H, Dai Z, Wu W, Wang Z, Zhang N, Zhang L, et al. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J Exp Clin Cancer Res. (2021) 40:367–81. doi: 10.1186/s13046-021-01987-7
15. Lu Y, Kang J, Luo Z, Song Y, Tian J, Li Z, et al. The prevalence and prognostic role of PD-L1 in upper tract urothelial carcinoma patients underwent radical nephroureterectomy: A systematic review and meta-analysis. Front Oncol. (2020) 10:1400. doi: 10.3389/fonc.2020.01400
16. Shiravand Y, Khodadadi F, Kashani SMA, Hosseini-Fard SR, Hosseini S, Sadeghirad H, et al. Immune checkpoint inhibitors in cancer therapy. Curr Oncol. (2022) 29:3044–60. doi: 10.3390/curroncol29050247
17. Eggermont AMM, Maio M, Robert C. Immune checkpoint inhibitors in melanoma provide the cornerstones for curative therapies. Semin Oncol. (2015) 42:429–35. doi: 10.1053/j.seminoncol.2015.02.010
18. Willsmore ZN, Coumbe BGT, Crescioli S, Reci S, Gupta A, Harris RJ, et al. Combined anti-PD-1 and anti-CTLA-4 checkpoint blockade: Treatment of melanoma and immune mechanisms of action. Eur J Immunol. (2021) 51:544–56. doi: 10.1002/eji.202048747
19. Zhang Y, Chen H, Mo H, Hu X, Gao R, Zhao Y, et al. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell. (2021) 39:1578–93.e8. doi: 10.1016/j.ccell.2021.09.010
20. Saleh R, Toor SM, Khalaf S, Elkord E. Breast cancer cells and PD-1/PD-L1 blockade upregulate the expression of PD-1, CTLA-4, TIM-3 and LAG-3 immune checkpoints in CD4+ T cells. Vaccines. (2019) 7. doi: 10.3390/vaccines7040149
21. Bonaventura P, Shekarian T, Alcazer V, Valladeau-Guilemond J, Valsesia-Wittmann S, Amigorena S, et al. Cold tumors: A therapeutic challenge for immunotherapy. Front Immunol. (2019) 10:168. doi: 10.3389/fimmu.2019.00168
22. Xiao Y, Ma D, Zhao S, Suo C, Shi J, Xue zhu M, et al. Multi-omics profiling reveals distinct microenvironment characterization and suggests immune escape mechanisms of triple-negative breast cancer. Clin Cancer Res. (2019) 25:5002–14. doi: 10.1158/1078-0432.CCR-18-3524
23. Telli ML, Stover DG, Loi S, Aparicio S, Carey LA, Domchek SM, et al. Homologous recombination deficiency and host anti-tumor immunity in triple-negative breast cancer. Breast Cancer Res Treat. (2018) 171:21–31. doi: 10.1007/s10549-018-4807-x
24. Keenan TE, Tolaney SM. Role of immunotherapy in triple-negative breast cancer. J Natl Compr Cancer Network. (2020) 18:479–89. doi: 10.6004/jnccn.2020.7554
25. Jin N, George TL, Otterson GA, Verschraegen C, Wen H, Carbone D, et al. Advances in epigenetic therapeutics with focus on solid tumors. Clin Epigenet. (2021) 13:83. doi: 10.1186/s13148-021-01069-7
26. Li Y, Li Z, Zhu WG. Molecular mechanisms of epigenetic regulators as activatable targets in cancer theranostics. Curr Medicinal Chem. (2019) 26:1328–50. doi: 10.2174/0929867324666170921101947
27. Gray JS, Wani SA, Campbell MJ. Epigenomic alterations in cancer: mechanisms and therapeutic potential. Clin Sci. (2022) 136:473–92. doi: 10.1042/CS20210449
28. Patnaik S, Anupriya. Drugs targeting epigenetic modifications and plausible therapeutic strategies against colorectal cancer. Front Pharmacol. (2019) 10:588. doi: 10.3389/fphar.2019.00588
29. Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer. (2008) 123:8–13. doi: 10.1002/ijc.v123:1
30. Chen S, Jiang Y, Wang C, Tong S, He Y, Lu W, et al. Epigenetic clocks and gliomas: unveiling the molecular interactions between aging and tumor development. Front Mol Biosci. (2024) 11:1446428. doi: 10.3389/fmolb.2024.1446428
31. Yu J, Zayas J, Qin B, Wang L. Targeting DNA methylation for treating triple-negative breast cancer. Pharmacogenomics. (2019) 20:1151–7. doi: 10.2217/pgs-2019-0078
32. Wang Q, Xiong F, Wu G, Liu W, Chen J, Wang B, et al. Gene body methylation in cancer: molecular mechanisms and clinical applications. Clin Epigenet. (2022) 14:154. doi: 10.1186/s13148-022-01382-9
33. Blagitko-Dorfs N, Schlosser P, Greve G, Pfeifer D, Meier R, Baude A, et al. Combination treatment of acute myeloid leukemia cells with DNMT and HDAC inhibitors: predominant synergistic gene downregulation associated with gene body demethylation. Leukemia. (2019) 33:945–56. doi: 10.1038/s41375-018-0293-8
34. Shadbad MA, Safaei S, Brunetti O, Derakhshani A, Lotfinejad P, Mokhtarzadeh A, et al. A systematic review on the therapeutic potentiality of PD-L1-inhibiting microRNAs for triple-negative breast cancer: toward single-cell sequencing-guided biomimetic delivery. Genes. (2021) 12:1206. doi: 10.3390/genes12081206
35. Agrawal K, Das V, Vyas P, Hajdúch M. Nucleosidic DNA demethylating epigenetic drugs – A comprehensive review from discovery to clinic. Pharmacol Ther. (2018) 188:45–79. doi: 10.1016/j.pharmthera.2018.02.006
36. Kirkin AF, Dzhandzhugazyan KN, Guldberg P, Fang JJ, Andersen RS, Dahl C, et al. Adoptive cancer immunotherapy using DNA-demethylated T helper cells as antigen-presenting cells. Nat Commun. (2018) 9:785. doi: 10.1038/s41467-018-03217-9
37. Galle E, Thienpont B, Cappuyns S, Venken T, Busschaert P, Van Haele M, et al. DNA methylation-driven EMT is a common mechanism of resistance to various therapeutic agents in cancer. Clin Epigenet. (2020) 12:27. doi: 10.1186/s13148-020-0821-z
38. Linnekamp JF, Butter R, Spijker R, Medema JP, Van Laarhoven HWM. Clinical and biological effects of demethylating agents on solid tumours – A systematic review. Cancer Treat Rev. (2017) 54:10–23. doi: 10.1016/j.ctrv.2017.01.004
39. Casalino L, Verde P. Multifaceted roles of DNA methylation in neoplastic transformation, from tumor suppressors to EMT and metastasis. Genes. (2020) 11:922. doi: 10.3390/genes11080922
40. Chen C, Wang Z, Ding Y, Wang L, Wang S, Wang H, et al. DNA methylation: from cancer biology to clinical perspectives. Front Bioscience. (2022) 27:326. doi: 10.31083/j.fbl2712326
41. Liu P, Yang F, Zhang L, Hu Y, Chen B, Wang J, et al. Emerging role of different DNA methyltransferases in the pathogenesis of cancer. Front Pharmacol. (2022) 13:958146. doi: 10.3389/fphar.2022.958146
42. Lee K, Lin CC, Servetto A, Bae J, Kandagatla V, Ye D, et al. Epigenetic repression of STING by MYC promotes immune evasion and resistance to immune checkpoint inhibitors in triple negative breast cancer. Cancer Immunol Res. (2022) 10:829–43. doi: 10.1158/2326-6066.CIR-21-0826
43. Gomez S, Tabernacki T, Kobyra J, Roberts PA, Chiappinelli KB. Combining epigenetic and immune therapy to overcome cancer resistance. Semin Cancer Biol. (2019) 65:99–113. doi: 10.1016/j.semcancer.2019.12.019
44. Zolota V, Tzelepi V, Piperigkou Z, Kourea H, Papakonstantinou E, Argentou MI, et al. Epigenetic alterations in triple-negative breast cancer—The critical role of extracellular matrix. Cancers. (2021) 13:713. doi: 10.3390/cancers13040713
45. Feng D, Zhang F, Li D, Shi X, Xiong Q, Wei Q, et al. Developing an immune-related gene prognostic index associated with progression and providing new insights into the tumor immune microenvironment of prostate cancer. Immunology. (2022) 166:197–209. doi: 10.1111/imm.v166.2
46. Zhulai G, Oleinik E. Targeting regulatory T cells in anti-PD-1/PD-L1 cancer immunotherapy. Scandinavian J Immunol. (2021) 95:e13129. doi: 10.1111/sji.13129
47. Kajihara N, Kobayashi T, Otsuka R, Nio-Kobayashi J, Oshino T, Takahashi M, et al. Tumor-derived interleukin-34 creates an immunosuppressive and chemoresistant tumor microenvironment by modulating myeloid-derived suppressor cells in triple-negative breast cancer. Cancer Immunol Immunother. (2023) 72:851–64. doi: 10.1007/s00262-022-03293-3
48. Tao L, Cheng G, Lv F, Wang R, Yang N, Xing Z, et al. Synergistic strategy based on mild phototherapy and deep tumor hypoxia reversal comprehensively remodels the tumor microenvironment for improved immunotherapy. Chem Eng J. (2023) 472:145092. doi: 10.1016/j.cej.2023.145092
49. Takeyama Y, Kato M, Tamada S, Azuma Y, Shimizu Y, Iguchi T, et al. Myeloid-derived suppressor cells are essential partners for immune checkpoint inhibitors in the treatment of cisplatin-resistant bladder cancer. Cancer Lett. (2020) 479:89–99. doi: 10.1016/j.canlet.2020.03.013
50. Lin F, Huang J, Zhu W, Jiang T, Guo J, Xia W, et al. Prognostic value and immune landscapes of TERT promoter methylation in triple negative breast cancer. Front Immunol. (2023) 14:1218987. doi: 10.3389/fimmu.2023.1218987
51. Gómez-Miragaya J, Morán S, Calleja-Cervantes ME, Collado-Sole A, Paré L, Gómez A, et al. The altered transcriptome and DNA methylation profiles of docetaxel resistance in breast cancer PDX models. Mol Cancer Res. (2019) 17:2063–76. doi: 10.1158/1541-7786.MCR-19-0040
52. Zorzan E, Elgendy R, Guerra G, Da Ros S, Gelain ME, Bonsembiante F, et al. Hypermethylation-mediated silencing of CIDEA, MAL and PCDH17 tumour suppressor genes in canine DLBCL: from multi-omics analyses to mechanistic studies. Int J Mol Sci. (2022) 23:4021. doi: 10.3390/ijms23074021
53. Hu C, Liu X, Zeng Y, Liu J, Wu F. DNA methyltransferase inhibitors combination therapy for the treatment of solid tumor: mechanism and clinical application. Clin Epigenet. (2021) 13:166. doi: 10.1186/s13148-021-01154-x
54. Wong KF. DNMT1: A key drug target in triple-negative breast cancer. Seminars in Cancer Biology. (2021) 72:198–213. doi: 10.1016/j.semcancer.2020.05.010
55. McClellan BL, Haase S, Nunez FJ, Alghamri MS, Dabaja AA, Lowenstein PR, et al. Impact of epigenetic reprogramming on antitumor immune responses in glioma. J Clin Invest. (2023) 133(2).e163450. doi: 10.1172/JCI163450
56. Giri A, Aittokallio T. DNMT inhibitors increase methylation in the cancer genome. Front Pharmacol. (2019) 10:385. doi: 10.3389/fphar.2019.00385
57. Katarzyna R, Lucyna B. Epigenetic therapies in patients with solid tumors: Focus on monotherapy with deoxyribonucleic acid methyltransferase inhibitors and histone deacetylase inhibitors. J Can Res Ther. (2019) 15(5):961. doi: 10.4103/jcrt.JCRT_403_17
58. Gonda T, Fang J, Salas M, Do C, Hsu E, Zhukovskaya A, et al. A DNA hypomethylating drug alters the tumor microenvironment and improves the effectiveness of immune checkpoint inhibitors in a mouse model of pancreatic cancer. Cancer Res. (2020) 80:4754–67. doi: 10.1158/0008-5472.CAN-20-0285
59. Alamoudi M, Chipman M, Deleso-Frechette F, Liu E, Zhang R, Wang Z, et al. A therapeutic strategy to inhibit Wnt signaling also reprograms breast tumor-immune cell interactions: Perspectives for conferring immune checkpoint inhibitor susceptibility. Cancer Immunol Res. (2020) 8:A39–9. doi: 10.1158/2326-6074.TUMIMM19-A39
60. Ge JY, Shu S, Kwon M, Jovanović B, Murphy K, Gulvady A, et al. Acquired resistance to combined BET and CDK4/6 inhibition in triple-negative breast cancer. Nat Commun. (2020) 11:2350. doi: 10.1038/s41467-020-16170-3
61. Pack C, Bommireddy R, Munoz LE, Patel JM, Bozeman E, Dey P, et al. Tumor membrane-based vaccine immunotherapy in combination with anti-CTLA-4 antibody confers protection against immune checkpoint resistant murine triple-negative breast cancer. Hum Vaccines Immunotherapeutics. (2020) 16:3184–93. doi: 10.1080/21645515.2020.1754691
62. Raz D. Targeting epigenetic regulators in cancer to overcome targeted therapy resistance. Targeted Therapies Lung Cancer. (2019), 217–32. doi: 10.1007/978-3-030-17832-1_11
63. Damodaran S, Liu D, Schwartz J, Valero V, Ramirez D, Saleem S, et al. A phase Ib trial of bintrafusp alfa and eribulin in patients with metastatic triple-negative breast cancer (TNBC). Cancer Res. (2023) 83:P3–02–03. doi: 10.1158/1538-7445.sabcs22-p3-02-03
64. Yu JX, Guo Z, Wang L. Progress and challenges of immunotherapy predictive biomarkers for triple negative breast cancer in the era of single-cell multi-omics. Life. (2023) 13:1189. doi: 10.3390/life13051189
65. Llinàs-Arias P, Íñiguez-Muñoz S, McCann K, Voorwerk L, Orozco JIJ, Ensenyat-Mendez M, et al. Epigenetic regulation of immunotherapy response in triple-negative breast cancer. Cancers. (2021) 13:4139. doi: 10.3390/cancers13164139
66. Shen H, Yang ESH, Conry M, Fiveash J, Contreras C, Bonner JA, et al. Predictive biomarkers for immune checkpoint blockade and opportunities for combination therapies. Genes Dis. (2019) 6:232–46. doi: 10.1016/j.gendis.2019.06.006
67. Borcoman E, Kamal M, Marret G, Dupain C, Castel-Ajgal Z, Le Tourneau C. HDAC inhibition to prime immune checkpoint inhibitors. Cancers. (2021) 14:66. doi: 10.3390/cancers14010066
Keywords: triple-negative breast cancer, DNA methylation inhibitors, immune checkpoint inhibitors, immune evasion, epigenetic therapy, personalized medicine
Citation: Cai W-y, Cai X-x, Fei Y-r, Ye R, Song D-m, Hu D, Zhang W-w, Xia M-f and Yang X-x (2025) DNA methylation and immune evasion in triple-negative breast cancer: challenges and therapeutic opportunities. Front. Oncol. 15:1534055. doi: 10.3389/fonc.2025.1534055
Received: 25 November 2024; Accepted: 16 January 2025;
Published: 06 February 2025.
Edited by:
Dan Liu, Sichuan University, ChinaReviewed by:
Hongpan Zhang, Affiliated Hospital of North Sichuan Medical College, ChinaCopyright © 2025 Cai, Cai, Fei, Ye, Song, Hu, Zhang, Xia and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-xiao Yang, eWFuZ3hpYW94aWFvMjA1N0AxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.