
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 13 March 2025
Sec. Breast Cancer
Volume 15 - 2025 | https://doi.org/10.3389/fonc.2025.1522262
Background: Major advances have been achieved in the characterization of primary breast cancer genomic profiles. Limited information is available on the genomic profile of tumors originating from different metastatic locations in recurrent/metastatic (R/M) breast cancer, especially in Asian patients. This study aims to decipher the mutational profiles of primary and R/M breast cancer in Chinese patients using next-generation sequencing.
Methods: A total of 563 breast cancer patients were enrolled, and 590 tumor tissues and matched peripheral blood samples were collected and subjected to targeted sequencing with a panel of 1,021 cancer-related genes. The mutation spectrum, DNA damage response (DDR) genes, commonly altered signal pathways, and immunotherapy-related markers were compared between primary and R/M breast cancer. The molecular differences between our cohort and the Memorial Sloan Kettering Cancer Center (MSKCC) dataset were also explored.
Results: A total of 361 samples from primary and 229 samples from R/M breast cancer were analyzed. BRCA2, ATRX, and ATM were more frequently observed in R/M lesions among the 36 DDR genes. An ESR1 mutation and PD-L1 and PD-L2 amplification were enriched in R/M breast cancer (all p<0.05). Compared with the MSKCC dataset, we recruited more patients diagnosed at age 50 or younger and more patients with triple-negative breast cancer (TNBC) subtypes. The TNBC patients in our dataset had a higher percentage of PD-L1 amplification in metastasis tumors (p<0.05).
Conclusions: This study revealed the distinctive mutational features of primary and R/M tumors in Chinese breast cancer patients, which are different from those from Western countries. The enrichment of PD-L1 amplification in metastatic TNBC indicates the necessity to re-biopsy metastatic tumors for immunotherapy.
Breast cancer (BC) is the most common cancer and the fifth leading cause of cancer deaths in women in China (1). Breast cancer is divided into molecularly distinct subtypes based on hormone receptor (HR) and human epidermal growth factor-positive (HER2) status and these lead to different clinical outcomes and choice of therapies (2). Large-scale genomic characterization of breast cancer has established the genomic landscape and has contributed to the current understanding of complex mutational features among different breast cancer molecular subtypes (3–6). The genomic information of primary breast cancer and that of recurrent/metastatic (R/M) tumors have also been reported in several studies using patients from Western countries and provide pivotal insights in identifying genomic drivers associated with metastasis and targetable mutations in metastatic tumors. However, breast cancer patients in China have a much younger age at onset of breast cancer and a distinct molecular subtype distribution compared with patients in Western countries (7). It remains unclear whether the genetic changes in metastatic breast cancer in China are different from those in Western countries. However, the genetic landscape between primary and R/M tumors in Chinese breast cancer patients has not yet been characterized.
Currently, immune checkpoint inhibitor (ICI)-based therapies have shown remarkable promise for early-stage or advanced triple-negative breast cancer (TNBC). Compared with hormone receptor-positive (HR+) breast tumors, both TNBC and HER2+ tumors have a higher degree of stromal and intratumoral tumor-infiltrating lymphocytes (TILs) (20% and 16% of cases, respectively) (8, 9) and high immune-related gene expression (10). Moreover, TNBC has a relatively higher tumor mutation burden (TMB) than other breast cancer subtypes and higher rates of cell surface PD-L1 expression. Thus, currently, PD-1/PD-L1 targeted immunotherapy is mostly used in this subset of patients (11–13).
Indeed, checkpoint inhibition with the anti-PD1 antibody pembrolizumab has been approved for advanced-stage PD-L1-positive TNBC due to improved outcomes when combined with frontline chemotherapy (14). However, the IMpassion131 study, with a combination of paclitaxel and the PD-L1 inhibitor atezolizumab, failed to improve progression-free survival (PFS) or overall survival (OS) in advanced TNBC patients (15). Landmark phase III trials tested immunotherapy in the early-stage neoadjuvant setting KEYNOTE-522 trial indicated that addition of pembrolizumab to chemotherapy improved the pathological complete response (pCR) rate (16). Similarly, IMpassion031, in which atezolizumab was combined with chemotherapy, resulted in improved pCR rates compared to chemotherapy alone, regardless of PDL1 status (17). In spite of the great progress of immune checkpoint inhibitors in TNBC, not all patients respond to immunotherapy and there is a strong need to identify prognostic and predictive biomarkers.
Several studies have demonstrated that the PD-L1 status is insufficient for identifying responder patients (18). In the neoadjuvant setting of TNBC, immunotherapy seems to provide benefits regardless of the PD-L1 status (19). Moreover, a recent analysis of the Tumor Cancer Genome Atlas (TCGA) revealed that PD-L1 positivity is only weakly associated with immunotherapy efficacy (20). In breast cancer, especially TNBC, there were discrepancies in its expression between primary tumors and metastatic sites. Primary tumors tend to have higher rates of PD-L1 expression compared to metastatic disease, especially in the liver, skin, and bones whilst PD-L1 expression in lung and lymph node metastases was comparable to that of the primary site (21) (22).
The KEYNOTE-158 trial showed that patients with previously treated advanced-stage solid tumors responded better to pembrolizumab monotherapy if their tumors had a high TMB in a myriad of primary tumor types including thyroid, anal, cervical, biliary, and endometrial (23). Data from GeparNuevo, a phase II neoadjuvant trial (24), and the TAPUR study (25) showed that TMB and immune cell infiltration could serve as independent predictors of response to immune checkpoint inhibition in early or metastatic TNBC. Indeed, a high TMB is more likely to be associated with a deficiency of the MMR pathway or homologous recombination repair system, genetic alterations in DNA polymerase genes (POLE/POLD1), and the APOBEC mutation signature (26). Of note, compared to early BC, more advanced tumors generally display a higher TMB and less abundant TIL levels (27, 28). However, ICIs are more effective in treating TNBC when given early in the course of the disease, which was generally attributed to immune escape mechanisms emerging during the progression of the disease (29), and less was known about the contribution of the evolving mutation profile.
TNBC often involves high lymphocyte infiltration and the measurement of tumor infiltrated lymphocytes (TILs) has been proposed as a surrogate marker of the adaptive immune response against neoplastic cells (30) (31). Data from KEYNOTE-086, a phase II study, revealed that TIL levels can predict response to immunotherapy, particularly in the first-line setting (32, 33). In the metastatic setting, a higher density of CD8+ TILs was associated with increased PFS and OS in the IMpassion130 trial (34). Interestingly, the phase II FUTURE-C-Plus trial showed that patients with CD8- and/or PD-L1- positive tumors benefited more from immunotherapy while a PKD1 somatic mutation indicated worse progression-free and overall survival (35). This indicates that mutation profiles may also contribute to the diverse response to immunotherapies.
Several studies have also demonstrated that BRCA1/2 mutation was associated with an immune activation signature and might predict immunogenicity in BRCA1/2-deficient BC, including TNBC (36, 37). Recently, the phase II I-SPY2 trial showed that the combination of PD-L1 inhibitor, PARP inhibitor, and standard paclitaxel neoadjuvant chemotherapy has superior efficacy over standard neoadjuvant chemotherapy in HER2-negative breast cancer and in a subset of high-risk HR-positive/HER2-negative patients (38). Besides BRCA1/2, approximately 20% of TNBC have a loss of PTEN, leading to a more immunogenic drive, however, its role in immunotherapy is still controversial (11, 39).
To better understand the mutation spectrum between primary and R/M tumors in Chinese breast cancers, we retrospectively analyzed the genomic data of 600 samples from 570 Chinese patients with various molecular and histological subtypes of early-stage and R/M breast cancer to elucidate their mutational landscape. Furthermore, we also performed a subgroup analysis by metastatic lesion to discover significantly altered genes and compared the genomic data of samples originating from the same metastasis lesions between our cohort and the Memorial Sloan Kettering Cancer Center (MSKCC) breast cancer dataset.
Between May 2017 and December 2021, a total of 563 patients diagnosed with malignant breast cancer were retrospectively enrolled in this study. All patients provided written consent for genetic analysis and the protocol was approved by the Ethics Committee of Sun Yat-sen Memorial Hospital (2019-KY-073). ER/PR positivity in the immunohistochemistry testing was indicated with a cut-off of equal or higher than 1% positively stained cells (40). HER2 status was defined by an immunohistochemistry score of 0 or 1 or a lack of HER2 amplification (ratio <2.2) demonstrated by FISH analysis.
Tumor DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tumor tissue specimens using the ReliaPrep FFPE gDNA Miniprep System (Promega). Matched germline genomic DNA was isolated from peripheral blood lymphocytes (PBL) using the QIAamp DNA Blood Mini Kit (Qiagen). The concentration and fragment length of DNA were determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.).
Germline genomic DNA and tumor DNA were sheared into fragments at a 200 to 250 bp peak with a Covaris S2 ultrasonicator (Covaris, Inc), and indexed next-generation sequence (NGS) libraries were prepared using an NEBNext Ultra DNA Library Prep Kit for Illumina (NEB). For tumor genomic and matched germline DNA libraries, a previously reported custom-designed panel (Integrated DNA Technologies, Inc.) covering ∼1.5 Mbp of the genome and 1,021 cancer-related genes was used for hybridization enrichment (41). The indexed libraries were sequenced on a Gene+Seq-2000 sequencing system (GenePlus-Suzhou).
The sequenced reads were mapped to the reference human genome (GRCh37) using the default parameters in BWA version 0.7.12 after removing adaptor and low-quality reads. Local realignment around single nucleotide variants (SNVs) and small insertions and deletions (Indels), and quality control assessment, were performed using GATK (version 3.4.46; Broad Institute). Genomic alterations, including SNVs, Indels, copy number alterations (CNA), and gene fusions/rearrangements, were detected with GATK, MuTect (version 1.1.4) and BreakDancer, respectively. PBL DNA was used as a control to identify somatic variations from germline variations.
An MSK dataset (MSKCC, Cancer Cell 2018) was downloaded from the cBioPortal website (https://www.cbioportal.org/study/summary?id=breast_msk_2018, accessed on 11 May 2022). The chi-square test or Fisher’s exact test was used to analyze frequencies of genetic alterations in different groups. As for gene mutation rates among the three subgroups, a chi-square test was performed at first. Then, if p<0.05, Fisher’s exact test and the Bonferroni correction were conducted to analyze the difference between two subtypes. Statistical processing was performed with SPSS version 23 (SPSS Inc., Chicago, IL, USA), and p < 0.05 (two-sided) was considered significant.
In this study, 590 tumor tissues and matched peripheral blood samples were collected from 563 breast cancer patients (Table 1), with 25 patients having two or three tumor samples. Briefly, all the patients were female, and 68.6% were diagnosed at age 50 or younger. Among these 563 patients, 7 patients (1.2%) were diagnosed at stage 0, 84 patients (14.9%) at stage I, 234 patients (41.6%) at stage II, 176 patients (31.3%) at stage III, and 56 patients (9.9%) at stage IV. A total of 361 (61.2%) samples originated from primary breast cancer and 69 (11.7%) samples originated from recurrent breast cancer. The remaining 160 (27.1%) samples represented metastases obtained from lymph nodes (n = 51), liver (n = 38), chest (n = 21), lung (n = 20), brain (n = 12), and distant metastases from other sites (n = 18). The molecular subtypes of tumors include HR+/HER2- (36.8%), HR+/HER2+ (7.1%), HR-/HER2+ (14%), and TNBC (42.1%), based on pathological immunohistochemical staining of sequenced samples.
All 590 breast cancer samples from 563 patients were tested using the 1,021-gene NGS panel. A total of 5,883 somatic alterations were detected in the tumor samples of these patients, including 4,020 SNVs/Indels, 1,814 CNVs, and 49 SVs. The number of alterations detected in each sample ranged from 0 to 124 with a median of 8. No somatic alteration was observed in four tumor samples (4/590, 0.7%). The most common mutational type was a missense mutation (51.1%), followed by CN gain (29.4%), frameshift mutation (6.7%), and nonsense mutation (5.4%). TP53 was the most frequent somatic alteration observed in breast cancer patients and the prevalence of this mutation was 69% (Figure 1). Other genomic variations composed of SNVs/Indels and CNVs with an incidence higher than 10% in all the samples include PIK3CA (35.4%), MYC (26.1%), ERBB2 (24.6%), CDK12 (13.5%), CCND1 (11.6%), KMT2C (11.0%), MCL1(10.6%), and GATA3(10.5%).
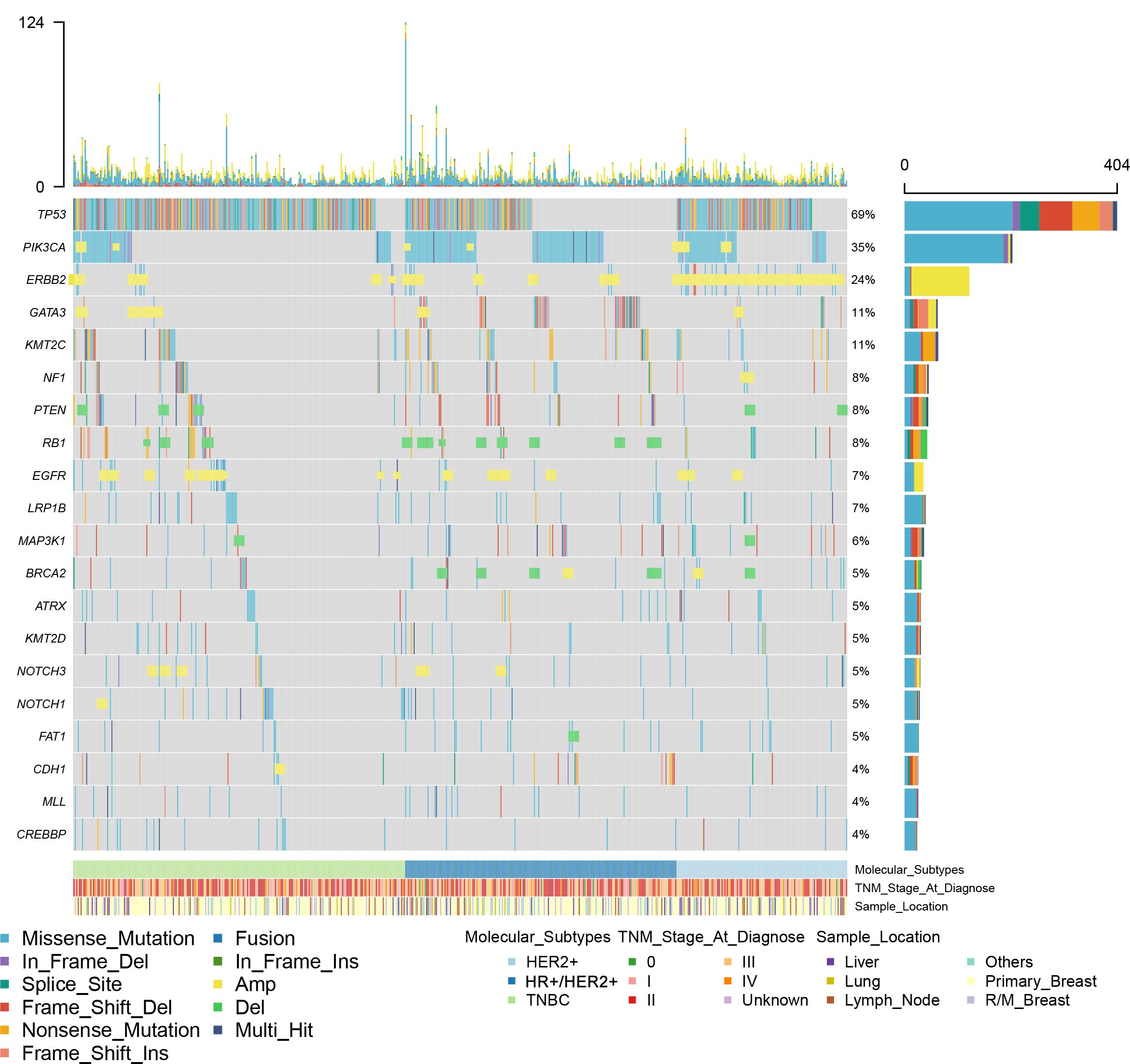
Figure 1. A summary of the genomic characteristics of 590 samples from Chinese patients with breast cancer. Oncoprint showed genetic changes with an incidence of more than 4%. According to HR and HER2 status, the tumor samples were categorized into HR+/HER2- (n = 215), HER2+ (n = 128), or triple-negative (n = 247). Clinicopathological features are annotated at the bottom. Amp, copy number amplification; Del, copy number deletion; HR, hormone receptor; HER2, human epidermal growth factor.
We further assessed the distribution of mutational signatures between primary breast cancer and R/M breast cancer in order to better understand which mutational processes drive tumor recurrence/metastasis. The observed common SNVs between primary and R/M breast cancer are shown in Figure 2A. The top three most commonly mutated genes were TP53, PIK3CA, and KMT2D in both groups. ESR1 was the only gene among the top 14 genes that had different mutation rates between primary (0.8%) and R/M tumors (7.4%) (p<0.05). In primary breast cancer, an analysis of molecular subtypes demonstrated that TP53 mutations were more frequent in HER2+ and TNBC breast cancer than in other subtypes (p<0.05, Figure 2B). Moreover, PIK3CA mutations were enriched in HR+ patients, GATA3 mutations were more common in HR+/HER2- patients, and ERBB2 was frequently mutated in HER2+ patients (all p<0.05). PTEN was exclusively mutated in HR+/HER2- and TNBC tumors. In R/M breast cancer, the distribution of mutations in TP53, PIK3CA, GATA3, and PTEN were similar to those in primary breast cancer (Figure 2C), indicating these gene mutations have similar roles in distinctive molecular subtypes of both primary and R/M tumors.
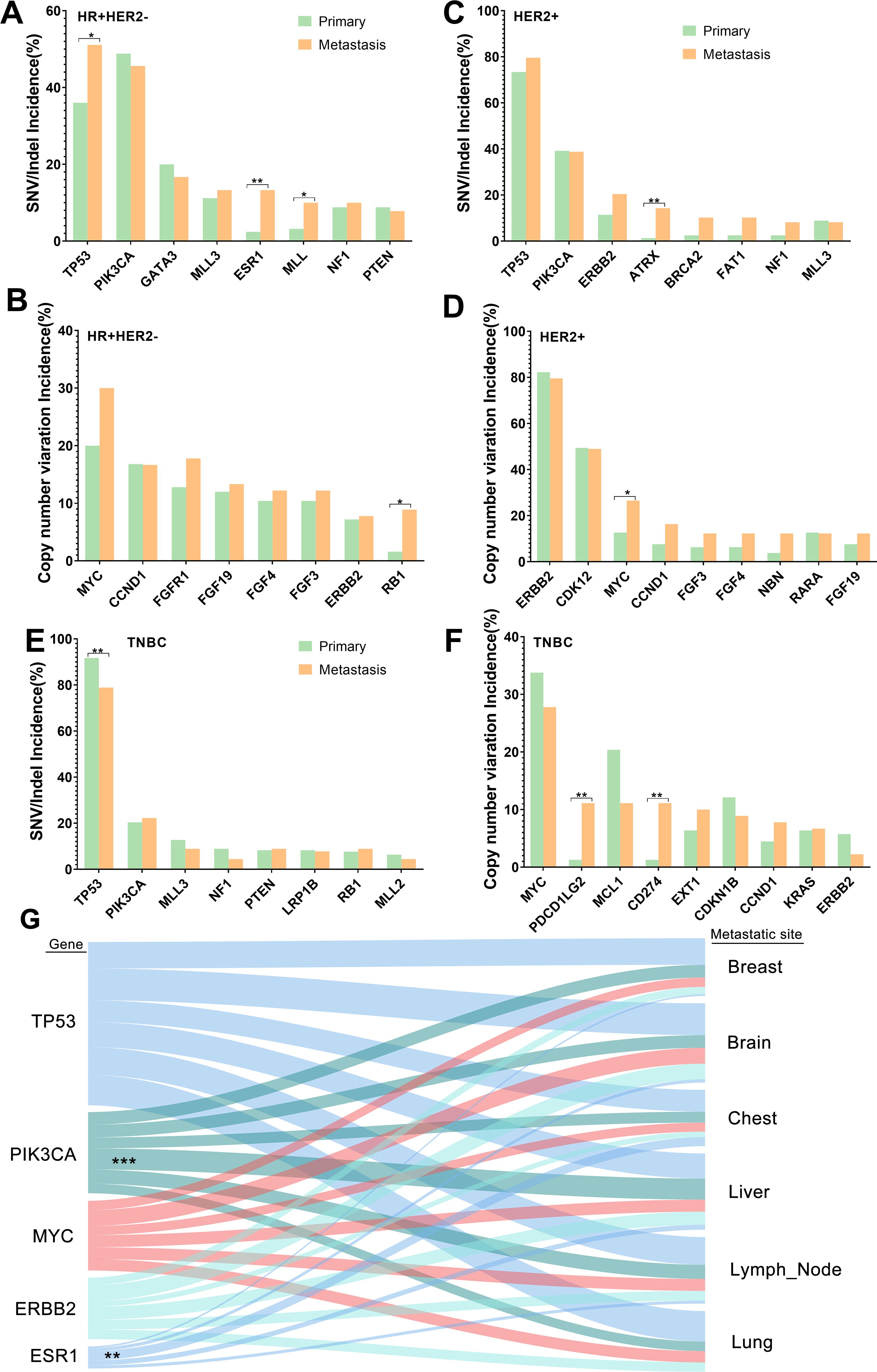
Figure 2. Commonly mutated genes in primary and R/M breast samples. (A, B) Top altered SNVs/Indels and CNVs in HR+/HER2- primary and R/M breast samples. (C, D) Top altered SNVs/Indels and CNVs in HER2+ primary and R/M breast samples. (E, F) Top altered SNVs/Indels and CNVs in TNBC primary and R/M breast samples. (G) Genomic alterations (left) and their association with different organ sites of metastases (right). Line thickness corresponds to the frequency of mutations arising in the indicated metastatic site. Shading identifies the relationships between genes and metastatic sites. Statistically significant associations are shown as asterisks. R/M, recurrent/metastasis; SNV, single-nucleotide variants; Indel, insertion and deletion, CNV, copy number variation; HR, hormone receptor; HER2, human epidermal growth factor; TNBC, triple-negative breast cancer. * denotes p<0.05, ** denotes p<0.01, *** means p<0.001..
The distribution of common CNVs between primary breast cancer and R/M breast cancer is shown in Figure 2D. The top three CNVs observed were MYC, ERBB2, and CDK12 in both groups. The distribution of the top 14 CNVs was similar between the two groups. In primary breast cancer, an analysis of molecular subtypes demonstrated that MYC was more enriched in TNBC than HER2+ (33.7% vs 12.7%, p<0.01) (Figure 2E). Furthermore, ERBB2 and CDK12 amplifications were more likely to be observed in HER2+ patients. Furthermore, CCND1 and FGF3/4/19 were more frequently altered in HR+/HER2- tumors and MCL1 and CDKN1B were more enriched in TNBC (all p<0.05). In the R/M breast cancer cohort, the distribution of mutations in ERBB2, CDK12, and FGFR1 were similar to those in primary breast cancer. However, compared to primary tumors, the frequency of FGF3/4/19 variations in R/M breast cancer was elevated in TNBC and HER2+ (Figure 2F).
Furthermore, we decided to identify mutations that reflected organotropism in their pattern of metastasis (Figure 2G). For example, ESR1 mutations were associated with metastasis to the chest, while PIK3CA mutations were associated with liver metastasis.
We further analyzed the key signaling pathways commonly mutated in all samples. The most common genomic alteration was the P53 pathway (72%), followed by the RTK/Ras/MAPK signaling pathway (66.4%), the PI3K-AKT pathway (54.4%), cell cycle control (28.8%), and the FGF superfamily (9.3%) (Figure 3). Among the genes belonging to the P53 pathway, ATM was the only gene that exhibited statistically different mutation frequencies between primary and R/M breast cancer. In the RTK/Ras/MAPK signaling pathway, the most commonly mutated genes were ERBB2 (24.6%), FGFR1 (9.5%), and NF1 (8.0%). Compared to primary cancer, ABL1(0.3% vs 3.0%, p=0.007) and FGFR1 (7.1% vs 13.1%, p=0.016) were more likely to be observed in R/M sites. The most frequently altered genes in the PI3K-AKT pathway were PIK3CA (35.4%), PTEN (7.5%), and AKT1 (3.7%). No significant differences in the genes of the PI3K-AKT pathway were observed between primary and R/M breast cancer. Among the genes in the cell cycle pathway, CCND1 (11.6%), RB1 (7.3%), and CDKN1B (5.7%) were the top 3 most mutated genes. CCND1 (15.3% vs 9.3%, p=0.027) and CDKN2A (7.6% vs 3.8%, p=0.043) exhibited higher mutation rates in R/M tumors than in primary tumors. Among the genes in the FGF superfamily, FGF19, FGF3, and FGF4 were the most frequently mutated with an incidence of 8.3%, 8.2%, and 7.5%, respectively. R/M tumors more frequently harbored alterations in FGF19 (11.4% vs 6.3%, p=0.026), FGF3 (11.0% vs 6.3%, p=0.039), and FGF4 (10.2% vs 5.8%, p=0.045) than primary tumors.
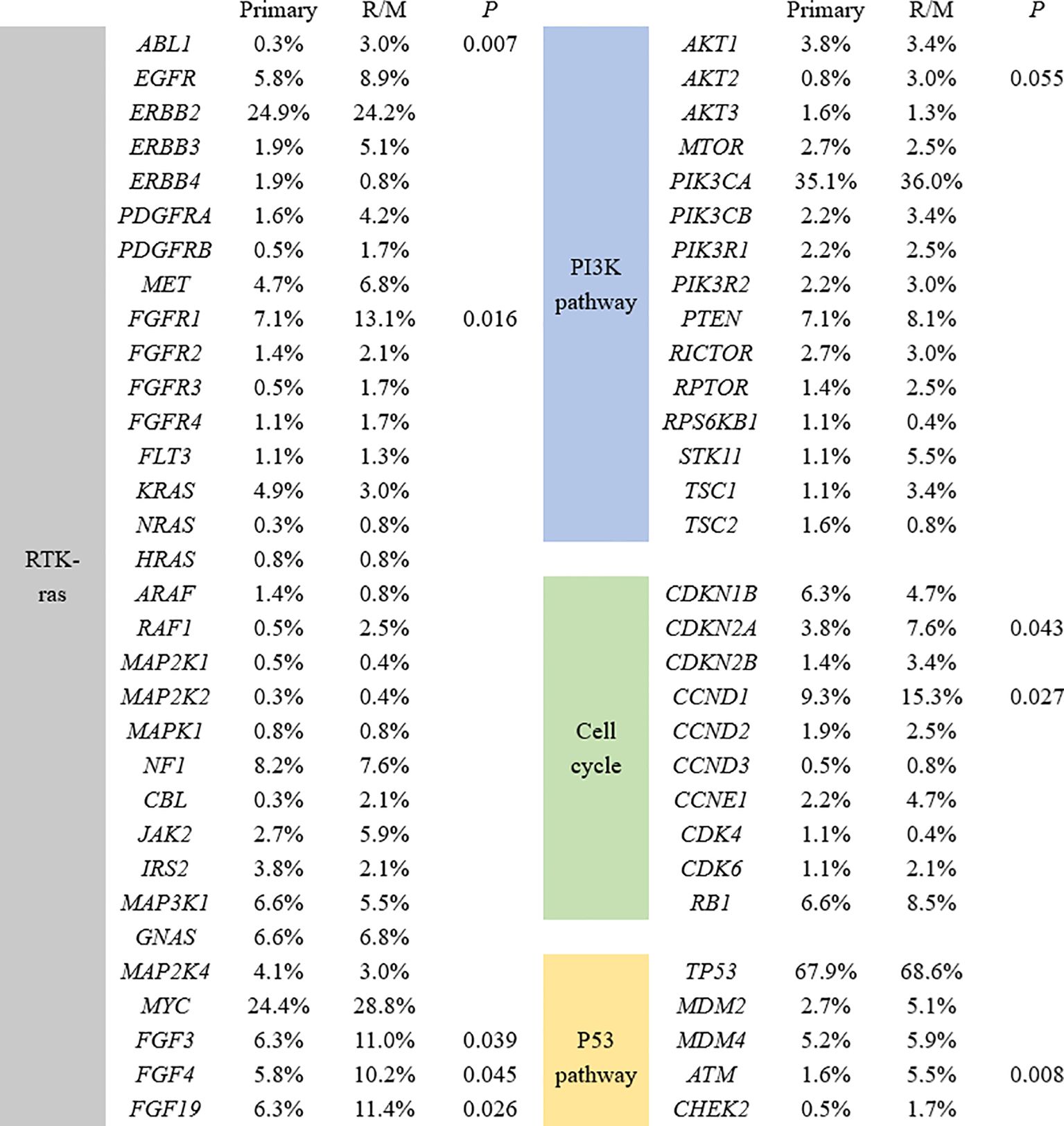
Figure 3. Significant differences in mutant genes between primary and R/M breast samples in four signaling pathways. R/M, recurrent/metastasis samples.
We next evaluated somatic alterations in 36 DDR genes covered by our NGS panels, including ATM, ATR, ATRX BAP1, BLM, BARD1, BRCA1/2, BRIP1, CHEK1/2, CDK12, EMSY, ERCC1, FAM175A FANCA/C/D2/E/F/G/L/M, MRE11A, NBN, RAD50/51/52, RAD51B/C/D, RAD54L, PALB2, RECQL, RECQL4, and WRN. We found 286 (47.6%) breast cancer patients had DDR gene mutations. There were 12 genes detected with more than 2.5% incidence (SNVs and CNVs) across all samples. (Figure 4A). The most frequently altered DDR gene in patients was CDK12 (n =81, 13.5%). The mutation frequencies of CDK12 in primary and R/M breast cancer were 14.0% and 12.7%. R/M breast cancer more frequently harbored alterations in BRCA2, ATRX, and ATM (p<0.05, Figure 4B). There were no significant differences in other DDR genes between primary and metastatic tumors.
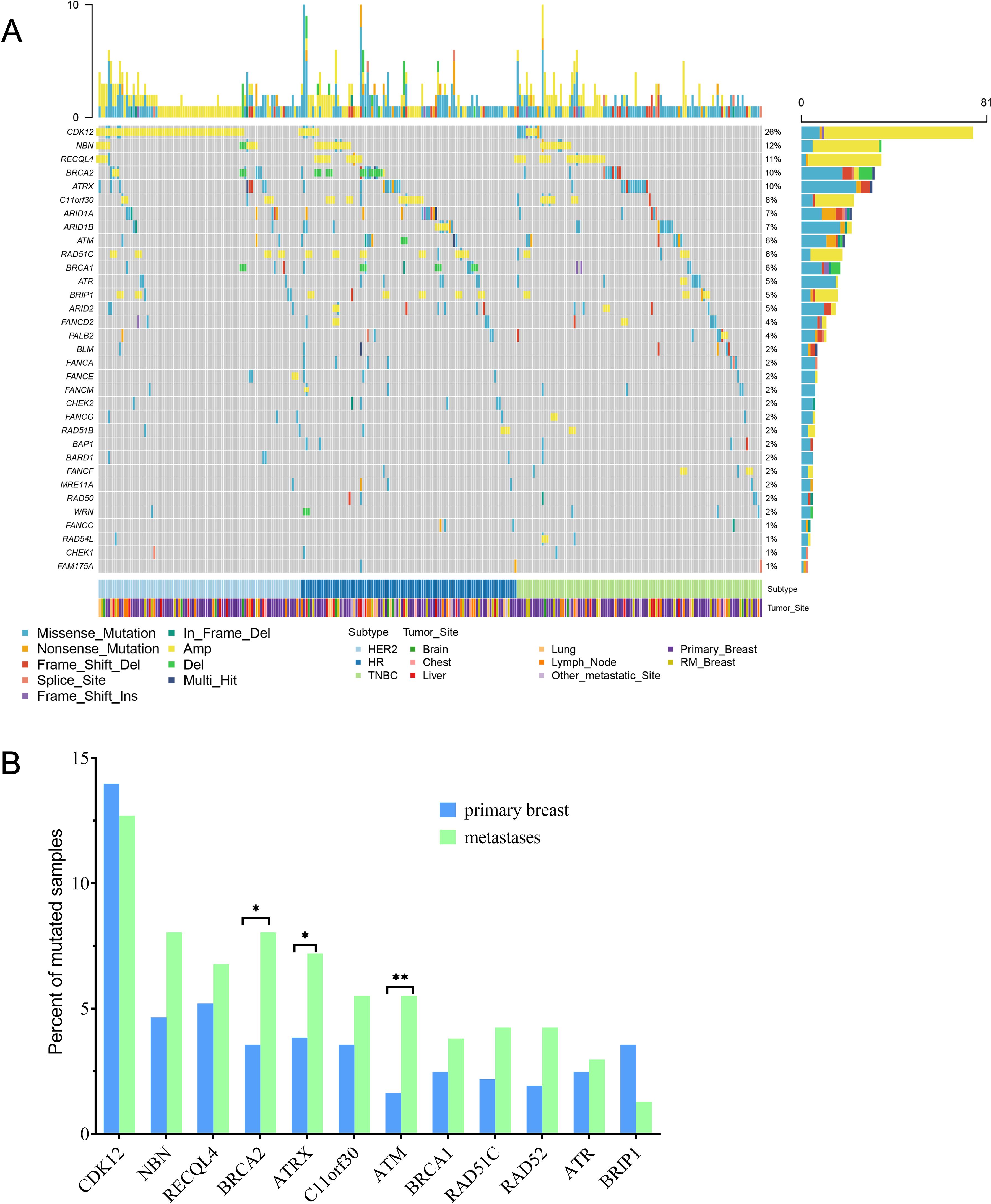
Figure 4. Comparison of 36 DDR genes between primary and R/M breast cancer. (A) Oncoprint showed genetic changes with an incidence of more than 1%. (B) Distribution of the top 12 mutated DDR genes between primary and R/M breast cancer. DDR, DNA damage repair; R/M, recurrent/metastasis; * denotes p<0.05, ** denotes p<0.01.
It is known that primary and R/M breast cancer have distinct immune microenvironments that impact their response to therapy. Thus, we investigated gene variations that are associated with the immunotherapy response. PTEN-inactivating mutations, amplification of MDM2/4, and amplification of the 11q13 regions (including CCND1, FGF3/4/19) were reported to be negative or hyperprogressive biomarkers of immunotherapy. The distribution of PTEN-inactivating mutations and MDM2/4 amplification were similar between primary and R/M breast cancer among three subtypes (Figures 5A, C). Amplification of the 11q13 regions was frequently observed in patients with HR+HER2- and HER2+ breast cancer (Figure 5B). In addition, compared to samples with HR+HER2- and HER2+, PD-L1 amplification was only observed in TNBC (Figures 5D, H). CN gain in PD-L1 was detected in 1.3% (2/157) of primary TNBC and 11.1% (10/90) of metastasis TNBC samples. Similar to PD-L1, PD-L2 CNVs were more often observed in metastasis TNBC (Figure 5E).
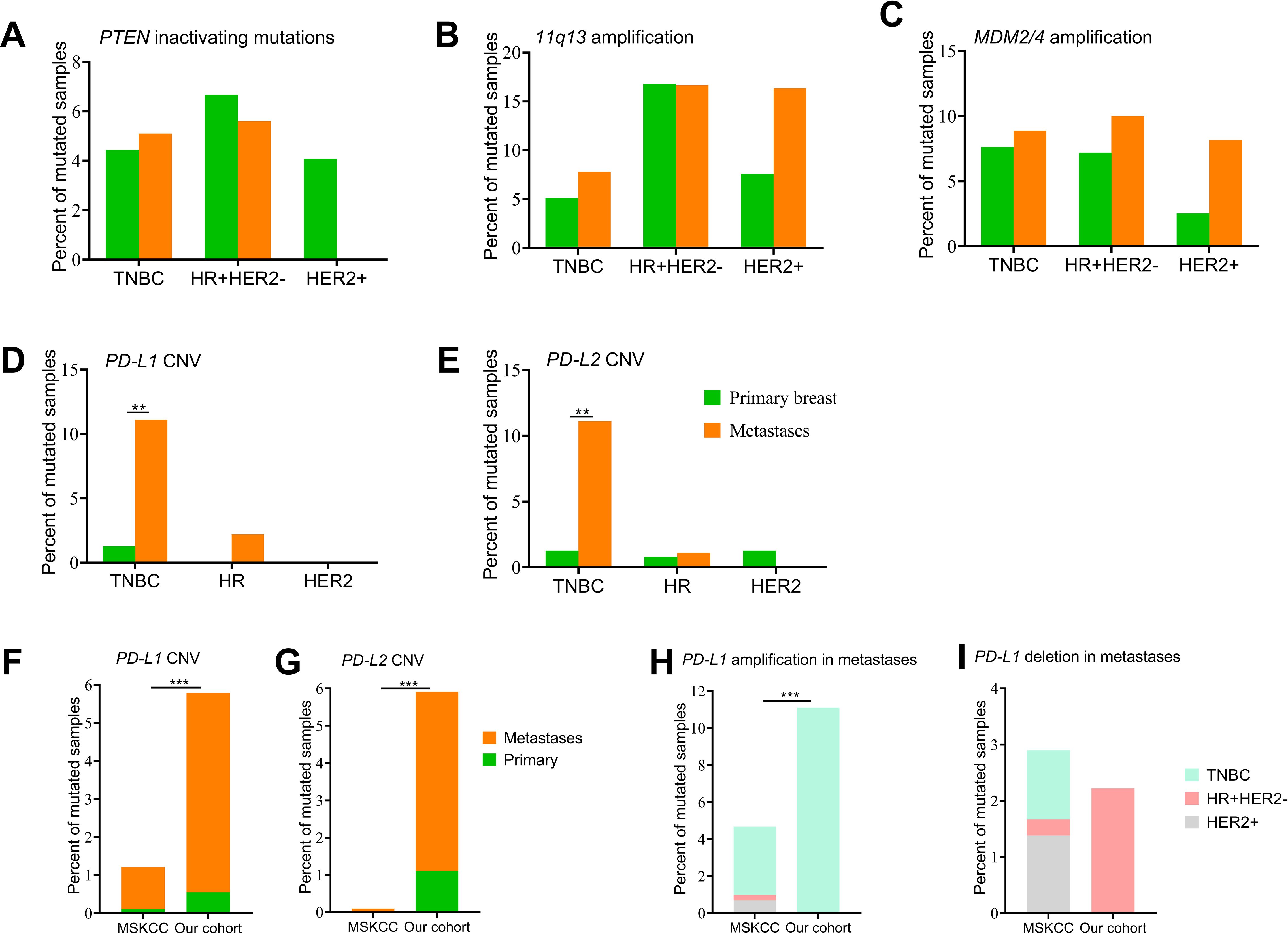
Figure 5. Genomic alterations of immunotherapy biomarkers. (A) PTEN-inactivating mutations between primary breast cancers and metastases. (B-E) Copy number variations of genes in the 11q13 region, MDM2/4, CD274/PD-L1, and PDCD1LG2/PD-L2 between primary breast cancers and metastases. (F, G) Distribution of CD274/PD-L1 and PDCD1LG2/PD-L2 copy number variations between our cohort and the MSKCC dataset. (H, I) Number of PD-L1 amplifications and deletions among the three subgroups between our cohort and the MSKCC dataset. CNV, copy number variation; HR, hormone receptor; HER2, human epidermal growth factor; TNBC, triple-negative breast cancer Del, deletion; Amp, amplification. ** denotes p<0.01, *** means p<0.001.
To better understand molecular differences among primary tumors and metastases between patients from China and Western countries, we obtained the clinical information and results of NGS survival in 1,918 samples from 1,756 breast cancer cases (MSKCC, Cancer Cell 2018), which was available on the cBioPortal website. Compared with the MSKCC dataset, we recruited more breast cancer patients diagnosed aged 50 years or younger and more patients with the TNBC subtype (Supplementary Table 1). The MSKCC dataset included 905 samples of primary breast cancer and 1,000 samples of metastasis disease (Supplementary Table 2). A total of seven CD274/PD-L1 CN gains (0.4%, 7/1918) and five PD-L1 CN deletions (0.3%, 5/1918) were identified, and only one sample originated from primary breast cancer with the HER2 subtype and the other samples were from metastatic sites. Among the 11 metastatic samples, four samples were of the TNBC subtype, four samples were the HR+/HER2- subtype, and three samples were the HER2+ subtype. Only one PDCD1LG2/PD-L2 CNV was identified in a sample of the HR+/HER2- subtype (Figures 5F, G). Higher percentages of PD-L1 and PD-L2 CNVs were identified in metastatic samples in our cohort. We then explored PD-L1 amplification and deletion among the three subtypes between the MSKCC dataset and our cohort and no significant difference was observed (Figures 5H, I).
In the present study, we explored the genomic characteristics of primary and R/M breast cancer in a large cohort of Chinese patients. Both tumor samples and matched blood specimens from each clinical participant were collected and tested for NGS profiles containing 1,021 cancer-related genes. Our study included patients with TNBC and HR+ disease. In our cohort, 99.3% (597/601) of the samples exhibited at least one mutation. TP53 (68.2%), PIK3CA (35.4%), MYC (26.1%), and ERBB2 (24.6%) were mutated in more than 20% of the cohort. In this subgroup alone, three genes were more commonly mutated in R/M tumors versus primary tumors. We compared the mutational spectrum, DDR genes, common signal pathways, and immunotherapy-related markers between primary and R/M tumors. R/M breast cancer more frequently harbored alterations in BRCA2 and ATM. PD-L1 and PD-L2 amplifications were more likely to be observed in R/M sites in TNBC. Among the multiple genes involved in common altered signal pathways, ABL1, FGFR1, CCND1, CDKN2A, ATM, and FGF3/4/19 were detected more frequently in R/M sites than in primary tumors. Subgroup analysis by metastatic location demonstrated chest wall metastases more frequently harbored alterations in FGFR1, while liver metastases more frequently had mutations in CCND1 and FGF3/4/19.
In the present study, TP53 was the most frequently occurring somatic mutation in all samples and the prevalence of this mutation was 68.2%. The mutation rate of TP53 (53%, n=1314) was significantly lower in a previous study launched by the Fudan University Shanghai Cancer Center (FUSCC) (4), which also included Chinese patients with both primary and metastasis cancer. Compared with FUSCC (4), the MSKCC (42) and TCGA (43) datasets, which enrolled a larger number of HR+ patients, HR+ and TNBC patients were found in similar proportions (43.1% and 41.6%, respectively) to our cohort. The mutation rate of TP53 was nearly 90% in TNBC in our cohort, which may have increased the overall TP53 mutation frequency. In addition, the NGS profiles of 1,021 genes encompassed all introns and exons of TP53 (44), while the FUSCC-BC panel may have omitted the introns of TP53 (4). The PIK3CA frequency was approximately 48% in HR+ tumors, 31.9% in HR-/HER2+ tumors, and 20.4% in TNBC tumors, which were consistent with the mutation frequencies reported in a previous study (6).
We further systematically explored the differences between primary and R/M breast cancer through multiple aspects, including mutation enrichment, DDR genes, oncogenic pathway alterations, and immunotherapy-related markers. ATM was among the DDR genes and involved in the p53 pathway. A higher ATM mutation rate was observed in R/M tumors. This result was consistent with a previous study that showed that patients with ATM mutations develop intermediate- or high-grade disease and have a higher rate of lymph node metastasis (45). ATM has been widely identified as a promising drug target. Up to now, several ATM inhibitors have been developed by different companies and entered clinical trials (46). Due to the development of synthetic lethal targets, higher rates of DDR defects in R/M tumors may expand the subset of patients that derive benefit from PARP inhibitors and other DDR-targeting drugs in the clinic (46). The mutation rate of TP53 in primary and R/M tumors in HR+/HER2- patients was 58.5% and 35.5%, respectively. ESR1 was also enriched in HR+ tumors from metastasis sites. Two recent published studies demonstrated that TP53, ESR1, KMT2C, AKT1, PTEN, and NF1 were more frequently altered in metastatic HR+/HER2− breast cancer compared with the early ones, in accordance with a previous study, indicating their driving role in breast cancer metastasis and relapse (3, 47). Activating mutations of ESR1 are common mechanisms of endocrine therapy resistance in HR +/HER2- advanced patients who exhibited responsiveness to selective ER degraders (48). Therefore, the detection of ESR1 mutations is vital in tailoring effective therapeutic strategies for HR+/HER2- advanced patients.
Consistent with a previous study, breast cancer was associated with a high frequency of mutations in the p53 (72%), RTK-RAS (66.4%), PI3K (54.4%), and cell cycle (28.8%) signaling pathways. Among the genes involved these signaling pathways, ABL1, FGFR1, CCND1, CDKN2A, ATM, and FGF3/4/19 were detected more frequently in R/M sites than primary tumors. The CCND1 and FGF3/4/19 genes are located in adjacent regions in the chromosome 11q13 region. The amplification of the chromosome 11q13 region is often observed in HR+ breast cancers and associated with poor prognosis and treatment failure (49–51). Genomic aberrations of FGFR1 were composed of gene amplification, activating mutations, and gene fusions. Similar to a previous study (43), FGFR1 gene amplification was identified in approximately 15% of patients with ER+ breast cancer. An in vitro study showed that high nuclear FGFR1 expression promotes antiestrogen resistance in ER+ primary tumors (52). More clinical studies are needed to explore the role of FGFR1 overexpression/amplification in estrogen sensitivity in ER+ breast cancer. The comparison of genes involved in common pathways between primary and R/M tumors may help to find the driving mutations accounting for treatment failure, metastasis, and relapse in breast cancer.
A copy number change at 9p24.1, which covered the loci for both PD-L1 and CD274, was a potential biomarker of ICI response (53). Amplification of PD-L1 has recently been evaluated in a pan-cancer analysis of 48,782 tumors, exhibiting a prevalence of 0.7% across tumors (54). In the present study, we found that PD-L1 amplifications were more likely to be observed in R/M sites in TNBC breast cancer. A similar finding was also obtained in a previous study involving 5,399 cases from MSK-IMPACT and TCGA, in which the incidence of 9p24.1 amplifications was 1.0% and showed a significantly higher incidence in TNBC (55). Another study showed that PD-L1 amplification may lead to increased PD-L1 expression in vitro (56). The predictive value of PD-L1 amplifications in immunotherapy has been evaluated in samples of patients with metastatic breast cancer included in the randomized phase II SAFIR02-IMMUNO study, where patients with TNBC had a higher proportion of PD-L1 amplifications and showed an improvement in OS with durvalumab in PD-L1-amplified tumors (hazard ratio = 0.17, 95% CI 0.05–0.55) (57). Recent research on 1,050 urothelial carcinoma cases showed nine tumors with PD-L1 CN gain and nine tumors with PD-L1 CN loss. Patients whose tumors harbored PD-L1 amplification benefited from immunotherapy, while patients whose tumors had PD-L1 CN loss experienced disease progression (58). A case report showed a urothelial carcinoma patient with PD-L2 amplification experienced durable stable disease on pembrolizumab (59). A recent study conducted a longitudinal analysis of PD-L1 expression in surgical samples and recurrent biopsy in non-small cell lung cancer, revealing that PD-L1 expression exhibited dynamic changes during the course of the disease (60). Similar variations of PD-L1 were observed in breast cancer, with approximately one-third of patients showing discrepant PD-L1 expression between primary tumors and matched distant metastases (61). Therefore, it is necessary to re-evaluate the PD-L1 status of recurrent lesions to optimize immunotherapy strategies.
A comparison of the mutation spectrum by metastatic location demonstrated chest wall metastases more frequently harbored alterations in FGFR1, while liver metastases more frequently had mutations in CCND1 and FGF3/4/19. In the MSKCC dataset, the CCND1 and FGF3/4/19 genes were the top 5 CNVs in liver, bone, lymph node, chest wall, and lung metastasis sites (42). The alteration rate of FGFR1 amplification in tumors from the chest wall was lower than in tumors from other metastases, and no significant differences existed among metastases. The ATRX mutation rate was found to be lower in the whole MSKCC cohort. The disparity of the mutation frequencies of the above genes between our cohort and the MSKCC dataset may result from the small number of metastatic tumors involved in our study and the different mutation spectrums due to ethnicity. Further clinical exploration of genomic characteristics by metastatic location is needed in Chinese breast cancer patients.
There are also several limitations in this study. First, this study is a retrospective study that may suffer from selection bias (e.g., different molecular subtypes, metastatic locations). Moreover, a relatively small number of tumors originated from different metastatic locations; thus, some potentially valuable alterations may have been overlooked. Combined and comprehensive NGS and an examination of certain proteins, treatment regimens, and survival analyses may need to be undertaken in the future.
In conclusion, this study revealed the mutational features of primary and R/M tumors in Chinese patients with breast cancer. Our study identified mutational features and genomic signatures of primary and R/M tumors in three subtypes of breast cancers, which may be explored as potential therapeutic targets in our population. Moreover, the enrichment of PD-L1 gene amplification in metastatic TNBC indicates the necessity to re-biopsy metastatic tumors for immunotherapy.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving humans were approved by the Ethics Committee of Sun Yat-sen Memorial Hospital (2019-KY-073). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
LJ: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. ZY: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. WT: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. PY: Formal analysis, Writing – review & editing. RC: Writing – review & editing. YX: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. JZ: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by Natural Science Foundation of China grants (82061148016, 81630074, 81872141, 81702630, 81672622), Guangzhou Science and Technology plan key projects (201804020076), and Natural Science Foundation of Guangdong (2019A1515010146), and Beijing Medical Award Foundation (YXJL-2020-0941-0760), and China Postdoctoral Science Foundation (2021TQ0384, 2021M703731).
Authors RC and PY were the employees of Geneplus-Beijing.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1522262/full#supplementary-material
1. Zheng Rs ZS, Zeng HM, Wang SM, Sun KX, Chen R, Li L, et al. Cancer incidence and mortality in China, 2016. J Natl Cancer Center. (2022) 2:1–9. doi: 10.1016/j.jncc.2022.02.002
2. Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U.S.A. (2001) 98:10869–74. doi: 10.1073/pnas.191367098
3. Bertucci F, Ng CKY, Patsouris A, Droin N, Piscuoglio S, Carbuccia N, et al. Genomic characterization of metastatic breast cancers. Nature. (2019) 569:560–4. doi: 10.1038/s41586-019-1056-z
4. Lang GT, Jiang YZ, Shi JX, Yang F, Li XG, Pei YC, et al. Characterization of the genomic landscape and actionable mutations in Chinese breast cancers by clinical sequencing. Nat Commun. (2020) 11:5679. doi: 10.1038/s41467-020-19342-3
5. Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. (2016) 7:11479. doi: 10.1038/ncomms11479
6. Xiao W, Zhang G, Chen B, Chen X, Wen L, Lai J, et al. Characterization of frequently mutated cancer genes and tumor mutation burden in chinese breast cancer. Front Oncol. (2021) 11:618767. doi: 10.3389/fonc.2021.618767
7. Yap YS, Lu YS, Tamura K, Lee JE, Ko EY, Park YH, et al. Insights into breast cancer in the east vs the west: A review. JAMA Oncol. (2019) 5:1489–96. doi: 10.1001/jamaoncol.2019.0620
8. Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol. (2014) 25:1544–50. doi: 10.1093/annonc/mdu112
9. Stanton SE, AdamsML Disis S. Variation in the incidence and magnitude of tumor-infiltrating lymphocytes in breast cancer subtypes: A systematic review. JAMA Oncol. (2016) 2:1354–60. doi: 10.1001/jamaoncol.2016.1061
10. Bianchini G, Qi Y, Alvarez RH, Iwamoto T, Coutant C, Ibrahim NK, et al. Molecular anatomy of breast cancer stroma and its prognostic value in estrogen receptor-positive and -negative cancers. J Clin Oncol. (2010) 28:4316–23. doi: 10.1200/JCO.2009.27.2419
11. Mittendorf EA, Philips AV, Meric-Bernstam F, Qiao N, Wu Y, Harrington S, et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol Res. (2014) 2:361–70. doi: 10.1158/2326-6066.CIR-13-0127
12. Budczies J, Bockmayr M, Denkert C, Klauschen F, Lennerz JK, Gyorffy B, et al. Classical pathology and mutational load of breast cancer - integration of two worlds. J Pathol Clin Res. (2015) 1:225–38. doi: 10.1002/cjp2.25
13. Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. (2012) 486:405–9. doi: 10.1038/nature11154
14. Cortes J, Cescon DW, Rugo HS, Nowecki Z, Im SA, Yusof MM, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet. (2020) 396:1817–28. doi: 10.1016/S0140-6736(20)32531-9
15. Miles D, Gligorov J, Andre F, Cameron D, Schneeweiss A, Barrios C, et al. Primary results from IMpassion131, a double-blind, placebo-controlled, randomised phase III trial of first-line paclitaxel with or without atezolizumab for unresectable locally advanced/metastatic triple-negative breast cancer. Ann Oncol. (2021) 32:994–1004. doi: 10.1016/j.annonc.2021.05.801
16. Schmid P, Cortes J, Pusztai L, McArthur H, Kummel S, Bergh J, et al. Pembrolizumab for early triple-negative breast cancer. N Engl J Med. (2020) 382:810–21. doi: 10.1056/NEJMoa1910549
17. Mittendorf EA, Zhang H, Barrios CH, Saji S, Jung KH, Hegg R, et al. Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): a randomised, double-blind, phase 3 trial. Lancet. (2020) 396:1090–100. doi: 10.1016/S0140-6736(20)31953-X
18. Gibney GT, WeinerMB Atkins LM. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. (2016) 17:e542–51. doi: 10.1016/S1470-2045(16)30406-5
19. Fountzila E, Ignatiadis M. Neoadjuvant immunotherapy in breast cancer: a paradigm shift? Ecancermedicalscience. (2020) 14:1147. doi: 10.3332/ecancer.2020.1147
20. Lee JS, Ruppin E. Multiomics prediction of response rates to therapies to inhibit programmed cell death 1 and programmed cell death 1 ligand 1. JAMA Oncol. (2019) 5:1614–8. doi: 10.1001/jamaoncol.2019.2311
21. Szekely B, Bossuyt V, Li X, Wali VB, Patwardhan GA, Frederick C, et al. Immunological differences between primary and metastatic breast cancer. Ann Oncol. (2018) 29:2232–9. doi: 10.1093/annonc/mdy399
22. Rozenblit M, Huang R, Danziger N, Hegde P, Alexander B, Ramkissoon S, et al. Comparison of PD-L1 protein expression between primary tumors and metastatic lesions in triple negative breast cancers. J Immunother Cancer. (2020) 8:e001558. doi: 10.1136/jitc-2020-001558
23. Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. (2020) 21:1353–65. doi: 10.1016/S1470-2045(20)30445-9
24. Karn T, Denkert C, Weber KE, Holtrich U, Hanusch C, Sinn BV, et al. Tumor mutational burden and immune infiltration as independent predictors of response to neoadjuvant immune checkpoint inhibition in early TNBC in GeparNuevo. Ann Oncol. (2020) 31:1216–22. doi: 10.1016/j.annonc.2020.05.015
25. Alva AS, Mangat PK, Garrett-Mayer E, Halabi S, Hansra D, Calfa CJ, et al. Pembrolizumab in patients with metastatic breast cancer with high tumor mutational burden: results from the targeted agent and profiling utilization registry (TAPUR) study. J Clin Oncol. (2021) 39:2443–51. doi: 10.1200/JCO.20.02923
26. Criscitiello C, Guerini-Rocco E, Viale G, Fumagalli C, Sajjadi E, Venetis K, et al. Immunotherapy in breast cancer patients: A focus on the use of the currently available biomarkers in oncology. Anticancer Agents Med Chem. (2022) 22:787–800. doi: 10.2174/1871520621666210706144112
27. El Bairi K, Haynes HR, Blackley E, Fineberg S, Shear J, Turner S, et al. The tale of TILs in breast cancer: A report from The International Immuno-Oncology Biomarker Working Group. NPJ Breast Cancer. (2021) 7:150. doi: 10.1038/s41523-021-00346-1
28. Barroso-Sousa R, Jain E, Cohen O, Kim D, Buendia-Buendia J, Winer E, et al. Prevalence and mutational determinants of high tumor mutation burden in breast cancer. Ann Oncol. (2020) 31:387–94. doi: 10.1016/j.annonc.2019.11.010
29. Hutchinson KE, Yost SE, Chang CW, Johnson RM, Carr AR, McAdam PR, et al. Comprehensive profiling of poor-risk paired primary and recurrent triple-negative breast cancers reveals immune phenotype shifts. Clin Cancer Res. (2020) 26:657–68. doi: 10.1158/1078-0432.CCR-19-1773
30. Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. (2015) 26:259–71. doi: 10.1093/annonc/mdu450
31. Stanton SE, Disis ML. Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J Immunother Cancer. (2016) 4:59. doi: 10.1186/s40425-016-0165-6
32. Adams S, Schmid P, Rugo HS, Winer EP, Loirat D, Awada A, et al. Pembrolizumab monotherapy for previously treated metastatic triple-negative breast cancer: cohort A of the phase II KEYNOTE-086 study. Ann Oncol. (2019) 30:397–404. doi: 10.1093/annonc/mdy517
33. Adams S, Loi S, Toppmeyer D, Cescon DW, De Laurentiis M, Nanda R, et al. Pembrolizumab monotherapy for previously untreated, PD-L1-positive, metastatic triple-negative breast cancer: cohort B of the phase II KEYNOTE-086 study. Ann Oncol. (2019) 30:405–11. doi: 10.1093/annonc/mdy518
34. Adams S, Diamond JR, Hamilton E, Pohlmann PR, Tolaney SM, Chang CW, et al. Atezolizumab plus nab-paclitaxel in the treatment of metastatic triple-negative breast cancer with 2-year survival follow-up: A phase 1b clinical trial. JAMA Oncol. (2019) 5:334–42. doi: 10.1001/jamaoncol.2018.5152
35. Wu SY, Xu Y, Chen L, Fan L, Ma XY, Zhao S, et al. Combined angiogenesis and PD-1 inhibition for immunomodulatory TNBC: concept exploration and biomarker analysis in the FUTURE-C-Plus trial. Mol Cancer. (2022) 21:84. doi: 10.1186/s12943-022-01536-6
36. Kraya AA, Maxwell KN, Wubbenhorst B, Wenz BM, Pluta J, Rech AJ, et al. Genomic signatures predict the immunogenicity of BRCA-deficient breast cancer. Clin Cancer Res. (2019) 25:4363–74. doi: 10.1158/1078-0432.CCR-18-0468
37. Nolan E, Savas P, Policheni AN, Darcy PK, Vaillant F, Mintoff CP, et al. Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci Transl Med. (2017) 9:eaal4922. doi: 10.1126/scitranslmed.aal4922
38. Pusztai L, Yau C, Wolf DM, Han HS, Du L, Wallace AM, et al. Durvalumab with olaparib and paclitaxel for high-risk HER2-negative stage II/III breast cancer: Results from the adaptively randomized I-SPY2 trial. Cancer Cell. (2021) 39:989–998 e5. doi: 10.1016/j.ccell.2021.05.009
39. Thomas A, Routh ED, Pullikuth A, Jin G, Su J, Chou JW, et al. Tumor mutational burden is a determinant of immune-mediated survival in breast cancer. Oncoimmunology. (2018) 7:e1490854. doi: 10.1080/2162402X.2018.1490854
40. Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. (2010) 28:2784–95. doi: 10.1200/JCO.2009.25.6529
41. Zhang Y, Yao Y, Xu Y, Li L, Gong Y, Zhang K, et al. Pan-cancer circulating tumor DNA detection in over 10,000 Chinese patients. Nat Commun. (2021) 12:11. doi: 10.1038/s41467-020-20162-8
42. Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. (2018) 34:427–438 e6. doi: 10.1016/j.ccell.2018.08.008
43. Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. (2012) 490:61–70. doi: 10.1038/nature11412
44. Yi Z, Ma F, Rong G, Guan Y, Li C, Xu B. Clinical spectrum and prognostic value of TP53 mutations in circulating tumor DNA from breast cancer patients in China. Cancer Commun (Lond). (2020) 40:260–9. doi: 10.1002/cac2.12032
45. Stucci LS, Interno V, Tucci M, Perrone M, Mannavola F, Palmirotta R, et al. The ATM gene in breast cancer: its relevance in clinical practice. Genes (Basel). (2021) 12:727. doi: 10.3390/genes12050727
46. Previtali V, Bagnolini G, Ciamarone A, Ferrandi G, Rinaldi F, Myers SH, et al. New horizons of synthetic lethality in cancer: current development and future perspectives. J Med Chem. (2024) 67:11488–521. doi: 10.1021/acs.jmedchem.4c00113
47. Angus L, Smid M, Wilting SM, van Riet J, Van Hoeck A, Nguyen L, et al. The genomic landscape of metastatic breast cancer highlights changes in mutation and signature frequencies. Nat Genet. (2019) 51:1450–8. doi: 10.1038/s41588-019-0507-7
48. Venetis K, Pepe F, Pescia C, Cursano G, Criscitiello C, Frascarelli C, et al. ESR1 mutations in HR+/HER2-metastatic breast cancer: Enhancing the accuracy of ctDNA testing. Cancer Treat Rev. (2023) 121:102642. doi: 10.1016/j.ctrv.2023.102642
49. Arao T, Ueshima K, Matsumoto K, Nagai T, Kimura H, Hagiwara S, et al. FGF3/FGF4 amplification and multiple lung metastases in responders to sorafenib in hepatocellular carcinoma. Hepatology. (2013) 57:1407–15. doi: 10.1002/hep.25956
50. Wang F, Ren C, Zhao Q, Xu N, Shen L, Dai G, et al. Association of frequent amplification of chromosome 11q13 in esophageal squamous cell cancer with clinical benefit to immune check point blockade. J Clin Oncol. (2019) 37:4036–6. doi: 10.1200/JCO.2019.37.15_suppl.4036
51. Karlsson E, Waltersson MA, Bostner J, Perez-Tenorio G, Olsson B, Hallbeck AL, et al. High-resolution genomic analysis of the 11q13 amplicon in breast cancers identifies synergy with 8p12 amplification, involving the mTOR targets S6K2 and 4EBP1. Genes Chromosomes Cancer. (2011) 50:775–87. doi: 10.1002/gcc.20900
52. Servetto A, Kollipara R, Formisano L, Lin CC, Lee KM, Sudhan DR, et al. Nuclear FGFR1 regulates gene transcription and promotes antiestrogen resistance in ER(+) breast cancer. Clin Cancer Res. (2021) 27:4379–96. doi: 10.1158/1078-0432.CCR-20-3905
53. Chic N, Braso-MaristanyA-Prat F. Biomarkers of immunotherapy response in breast cancer beyond PD-L1. Breast Cancer Res Treat. (2022) 191:39–49. doi: 10.1007/s10549-021-06421-2
54. Huang RSP, Haberberger J, Severson E, Duncan DL, Hemmerich A, Edgerly C, et al. A pan-cancer analysis of PD-L1 immunohistochemistry and gene amplification, tumor mutation burden and microsatellite instability in 48,782 cases. Mod Pathol. (2021) 34:252–63. doi: 10.1038/s41379-020-00664-y
55. Gupta S, Vanderbilt CM, Cotzia P, Arias-Stella JA, Chang JC, Zehir A, et al. Next-generation sequencing-based assessment of JAK2, PD-L1, and PD-L2 copy number alterations at 9p24.1 in breast cancer: potential implications for clinical management. J Mol Diagn. (2019) 21:307–17. doi: 10.1016/j.jmoldx.2018.10.006
56. Chen M, Pockaj B, Andreozzi M, Barrett MT, Krishna S, Eaton S, et al. JAK2 and PD-L1 amplification enhance the dynamic expression of PD-L1 in triple-negative breast cancer. Clin Breast Cancer. (2018) 18:e1205–15. doi: 10.1016/j.clbc.2018.05.006
57. Bachelot T FT, Dalenc F. 128O PDL1/CD274 gain/amplification as a predictive marker of checkpoint blockade inhibitor efficacy in metastatic breast cancer: Exploratory analysis of the SAFIR02-IMMUNO randomized phase II trial. Ann Oncol. (2020) 31:S58–9.
58. Gupta S, Vanderbilt CM, Zhang Y, Tickoo SK, Fine SW, Gopalan A, et al. CD274 (PD-L1) copy number changes (Gain) & Response to immune checkpoint blockade therapy in carcinomas of the urinary tract. Bladder Cancer. (2021) 7:395–400. doi: 10.3233/blc-201532
59. George S, Papanicolau-Sengos A, Lenzo FL, Conroy JM, Nesline M, Pabla S, et al. PD-L2 amplification and durable disease stabilization in patient with urothelial carcinoma receiving pembrolizumab. Oncoimmunology. (2018) 7:e1460298. doi: 10.1080/2162402X.2018.1460298
60. John N, Schlintl V, Sassmann T, Lindenmann J, Fediuk M, Wurm R, et al. Longitudinal analysis of PD-L1 expression in patients with relapsed NSCLC. J Immunother Cancer. (2024) 12:e008592. doi: 10.1136/jitc-2023-008592
Keywords: breast cancer, genomic profile, primary tumor, immunotherapy, PD-L1 amplification, TNBC
Citation: Jin L, Yang Z, Tang W, Yu P, Chen R, Xu Y and Zhang J (2025) The evolving landscape of genetic biomarkers for immunotherapy in primary and metastatic breast cancer. Front. Oncol. 15:1522262. doi: 10.3389/fonc.2025.1522262
Received: 04 November 2024; Accepted: 30 January 2025;
Published: 13 March 2025.
Edited by:
Dirk Geerts, Amsterdam University Medical Center, NetherlandsReviewed by:
Guochun Zhang, Guangdong Provincial People’s Hospital, ChinaCopyright © 2025 Jin, Yang, Tang, Yu, Chen, Xu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Zhang, emhhbmdqdW4yMDIxc3pAMTI2LmNvbQ==; Yan Xu, eHk5MzFAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.