- 1Department of Gastroenterology, Shengjing Hospital of China Medical University, Shenyang, Liaoning, China
- 2Engineering Research Center of Ministry of Education for Minimally Invasive Gastrointestinal Endoscopic Techniques, Shenyang, Liaoning, China
The diagnostic performance of artificial intelligence (AI)-assisted endoscopy for digestive tumors remains controversial. The objective of this umbrella review was to summarize the comprehensive evidence for the AI-assisted endoscopic diagnosis of digestive system tumors. We grouped the evidence according to the location of each digestive system tumor and performed separate subgroup analyses on the basis of the method of data collection and form of the data. We also compared the diagnostic performance of AI with that of experts and nonexperts. For early digestive system cancer and precancerous lesions, AI showed a high diagnostic performance in capsule endoscopy and esophageal squamous cell carcinoma. Additionally, AI-assisted endoscopic ultrasonography (EUS) had good diagnostic accuracy for pancreatic cancer. In the subgroup analysis, AI had a better diagnostic performance than experts for most digestive system tumors. However, the diagnostic performance of AI using video data requires improvement.
1 Introduction
The incidence and mortality rates of gastrointestinal (GI) tumors remain high. The health economic burden of these tumors is of great concern (1, 2). Early diagnosis of GI tumors is critical to achieve the best possible outcome for these patients. Endoscopy is an important method for GI tumor diagnosis, and reducing the rate of missed diagnosis is essential (3–6). Diagnosis of pancreatic tumors and mesenchymal tumors relies heavily on endoscopic ultrasonography (EUS), but the performance among EUS endoscopists varies greatly. Possible blind spots during surgery can lead to compromised patient health (7).
In recent years, the application of artificial intelligence (AI) technology (computer vision) in reducing missed diagnosis of GI tumors and improving the accuracy of EUS has received widespread attention (8, 9). However, whether the ability of AI in diagnosing all types of digestive system tumors is superior to that of experts or nonexperts is unclear (10–13). Although several meta-analyses have measured the ability of AI-assisted endoscopy to diagnose digestive system tumors, there are flaws in their study designs and the results are inconsistent. According to the largest recent surveys of endoscopists’ perceptions of AI, while most endoscopists view it positively, doubts about its diagnostic capabilities persist (14). There are fewer studies on the diagnostic capabilities of diagnostic AI in real-world clinical settings, and most are in the preclinical research stage. To avoid potential risks, a systematic evaluation of the ability of AI to aid in the diagnosis of early digestive system tumors is needed before it can be widely used in clinical practice. Therefore, we conducted a comprehensive umbrella review of this topic in the hope of contributing to the advancement of the literature in this regard.
2 Methods
2.1 Search strategy
This study was prospectively registered in PROSPERO (CRD42023445537). We strictly followed the PRIMA checklist. Institution Review Board approval and written consent are not applicable to this study. The PubMed, Web of Science, Embase, and Cochrane databases were searched to identify all (published and unpublished) meta-analyses and diagnostic studies on AI-assisted endoscopy for the diagnosis of digestive system tumors. The search was completed in July 2023. We searched the databases using a combination of Medical Subject Heading terms and keywords related to digestive system tumors, endoscopy, and AI (see Supplementary Tables S3, S4 for specific search terms). Two authors (C.W.H. and Y.S.) performed separate searches to include relevant studies in the review, and any discrepancies were resolved by consultation with a third author (J.Z.D.). Additionally, meta-analyses and individual diagnostic studies were manually searched using the reference lists of all included articles.
2.2 Selection criteria
Meta-analyses and single diagnostic studies were eligible for inclusion if they included indicators of diagnostic performance, e.g., sensitivity and specificity. Studies were included if the outcome was CRC, pancreatic cancer, esophageal cancer, gastric cancer (GC), mesenchymal tumors, or capsule endoscopy. We extracted data on individual outcomes separately if two or more diagnostic outcomes of the disease were reported in a study. If there was more than one eligible meta-analysis of AI-assisted endoscopy for the same disease diagnostic outcome, we included the most recent study for data extraction, which was generally the study with the largest sample size (15).
The exclusion criteria for this umbrella review were articles with incorrect exposure or design (errors in data count or meta-search design and data organization), studies that did not provide any information regarding the number of patients or images, and studies published in non-English languages.
2.3 Data extraction
The listed authors independently extracted the following information from each eligible study: first author’s name, nationality, year of publication, tumor site, exposure factors, study design (retrospective or prospective), number of patients, and number of images (video or image data). We counted true positives (TPs), false positives (FPs), true negatives (TNs), and false negatives (FNs) for each study of AI. For articles without available TP, FP, FN, and TN data of AI, we emailed the corresponding authors of the studies to request the raw data. Studies wherein the authors did not agree to provide raw data were excluded. Additionally, we performed a grouping analysis according to the location of each tumor (capsule endoscopy, as a specific endoscopic technique, was treated as a separate group). We further sub grouped the studies according to whether the data were collected by image or video, and whether the study design was retrospective or prospective (the number of original studies included in each subgroup was more than three). We also compared the diagnoses of digestive system tumors between experts and nonexperts (experts and nonexperts were both gastroenterologists; experts were defined as having more than 5 years of experience with white light endoscopy or more than 3 years of experience with magnifying endoscopy with narrow-band imaging). It is worth noting that the images and videos involved in most of the AI models were confirmed by histopathology. For studies involving experts and nonexperts, we again extracted TPs, FPs, FNs, and TNs for experts and nonexperts. The third author (J.Z.D.) randomly extracted the data to verify the accuracy.
The AI neural network model used in most of the literature included in this paper was a convolutional neural network, which is a specific class of deep neural network that consists of convolutional and pooling layers in a pattern that resembles the organization of the visual cortex and is hence well suited for image recognition and video analysis (16). Most of the endoscopic images included were white light endoscopy and no magnified narrow-band images.
2.4 Statistical analysis
Pooled sensitivities and specificities with corresponding 95% confidence interval (95% CI) were calculated using a random-effects model, and forest plots (Supplementary Figures S1–S7) were constructed on the basis of these models. The Cochran Q test and I2 statistic were used to assess heterogeneity between the studies (17). I2 >50% or a p-value ≤0.1 was considered to indicate significant heterogeneity. The Egger test was used to detect potential publication bias. Statistical significance was set at a p-value <0.1 (18). Heterogeneity and publication bias were also calculated in the subgroup analysis. All statistical analyses were performed using Stata 16 (StataCorp LLC, College Station, TX, USA) and R version 4.3.1 (R Foundation for Statistical Computing, Vienna, Austria).
2.5 Quality assessment of the methods and evidence
The methodological quality of the meta-analyses was assessed using the AMSTAR2.0 instrument, a 16-item methodological assessment tool (19). Additionally, Grading of Recommendations, Assessment, Development, and Evaluations (GRADE) was used to assess the quality of evidence for each outcome included in the review (20). The GRADE approach categorizes evidence as “high,” “moderate,” “low,” or “very low” quality. The level of evidence can be downgraded by the risk of bias, inconsistency, indirectness, imprecision, and publication bias. The methodological quality of the studies and the quality of the evidence were independently assessed by two authors (C.W.H. and Y.S.).
3 Results
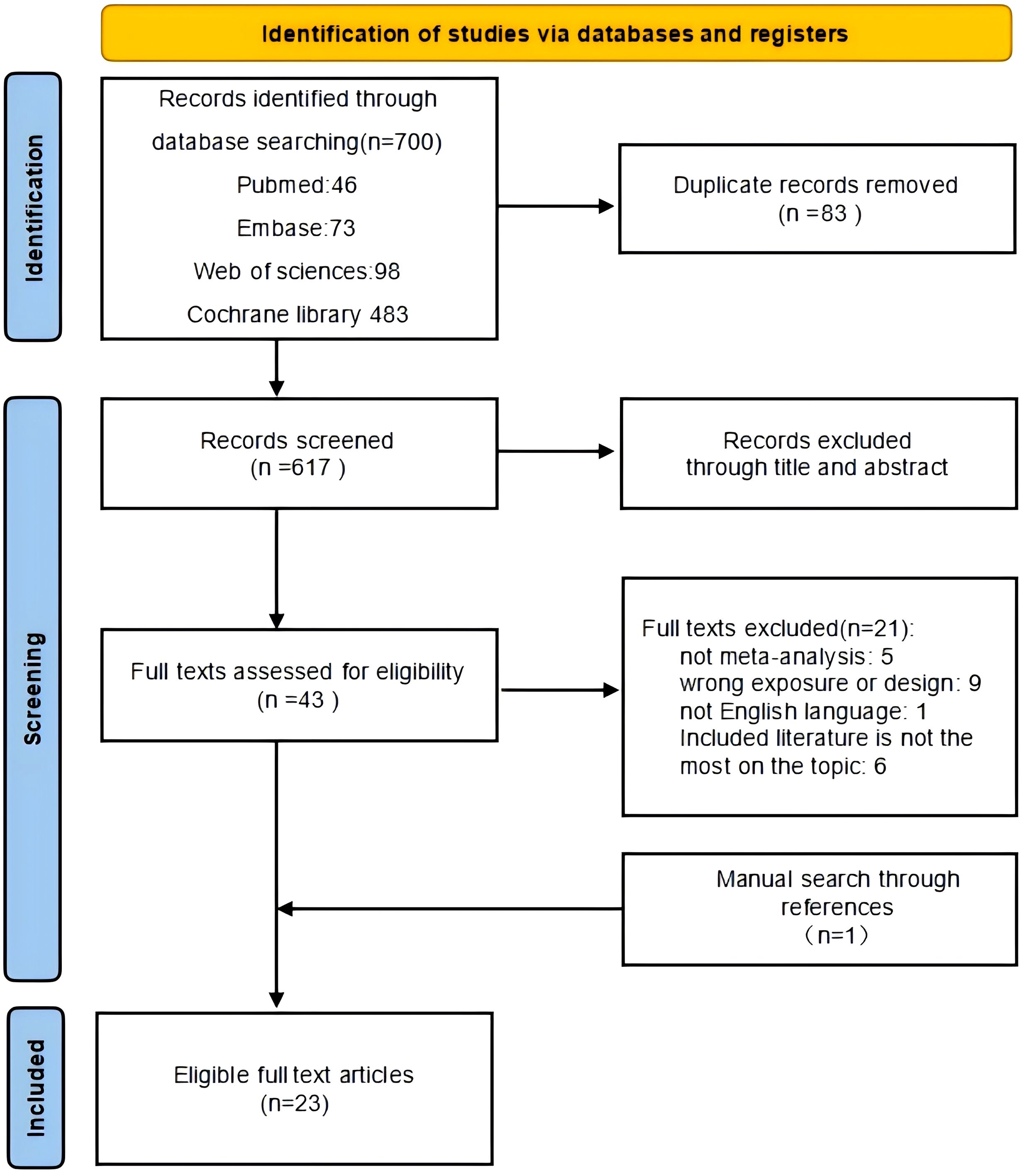
3.1 Characteristics of the meta-analysis (search, deduplication, exclusion, screening, and synthesis)
Figure 1 shows the flowchart of the literature search and screening. After a systematic literature search, 700 articles were identified. After screening titles and abstracts and removing duplicates, 43 articles were included. Then, 22 articles were retrieved for full-text review, of which 21 were discarded for the following reasons: one was not a meta-analysis, two studies had inappropriate designs, and three were published in languages other than English. One meta-analysis was performed by manually searching the reference lists of the included meta-analyses. Finally, 23 meta-analyses were included in this review (Table 1).
3.2 Characteristics of the data
The studies reported on AI-assisted endoscopic diagnosis of pancreatic (n=3), esophageal (n=8), gastric (n=6), and colorectal (n=6) cancers; mesenchymal tumors (n=1); and AI-assisted capsule endoscopy for the diagnosis of GI tumors (n=2). Our literature search for individual diagnostic studies not included in the published meta-analyses identified 25 additional studies (the original studies had the same inclusion and exclusion criteria as the meta-analysis): three studies on the diagnosis of pancreatic cancer, two studies on the diagnosis of esophageal cancer, 17 studies on the diagnosis of gastric cancer (GC), and three studies on the diagnosis of mesenchymal tumors. After removing duplicates, 193 original articles were included. We compared AI’s performance with the gold standard pathological diagnosis as follows: TP, correctly diagnosed patients with tumors; TN, correctly diagnosed healthy individuals; FP, incorrectly diagnosed healthy individuals as having tumors; and FN, incorrectly diagnosed individuals with tumors as healthy. Sensitivity (TP/TP+FN) reflects the ability of AI to detect patients, with higher sensitivity indicating fewer missed diagnoses. Meanwhile, specificity (TN/TN+FP) reflects the ability to correctly identify patients without the condition; the higher the specificity, the lower the misdiagnosis rate (10).
3.3 Quality assessment of the meta-analyses
The quality of the included meta-analyses was assessed using AMSTAR (version 2). Supplementary Table S1 presents details of the quality assessment of the 23 included meta-analyses. There was no “high” or “moderate” quality evidence. Eleven of the studies were of “low” quality, and 12 of the studies were of “very low” quality.
3.4 Heterogeneity
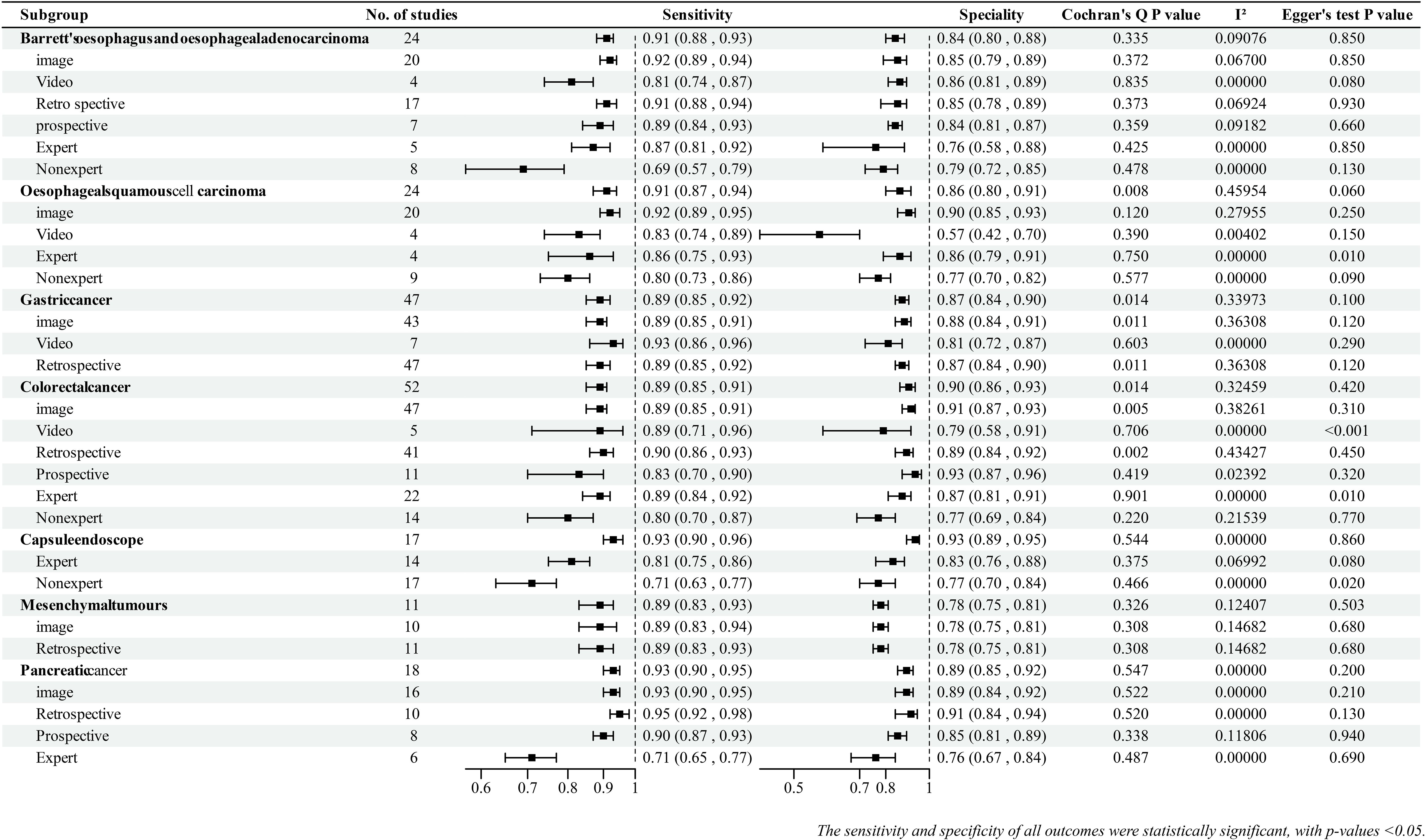
The I2 statistic and Cochran Q test were used to detect possible heterogeneity between the studies. Seven (22%) outcomes had significant heterogeneity (I2 >50% or p-value ≤0.1), and the rest (78%) had no significant heterogeneity (Figure 2). The main reasons for heterogeneity were differences in AI methods and endoscopic imaging techniques, differences in quality and quantity of endoscopic images and videos, and differences in study design. Additionally, the reasons for greater heterogeneity in the image subgroup compared with the video subgroup and in the retrospective subgroup compared with the prospective subgroup were the large number of included studies and differences in AI algorithms and imaging techniques.
3.5 Group and subgroup
3.5.1 GI tumors
The abilities of AI-assisted endoscopy to diagnose esophageal tumors (Barrett esophagus and esophageal adenocarcinoma, and esophageal squamous cell carcinoma), GC, and colorectal cancers (CRCs), and AI-assisted capsule endoscopy to diagnose GI tumors are summarized as follows. Details of the summary effect sizes are shown in Figure 2.
In terms of tumor location, AI-assisted capsule endoscopy showed excellent performance in diagnosing GI tumors with a pooled sensitivity of 0.93 (95% CI: 0.90–0.96) and pooled specificity of 0.93 (95% CI: 0.89–0.95).In gastroenteroscopy, AI-assisted endoscopy exhibited the best diagnostic performance for esophageal squamous cell carcinoma, followed by Barrett esophagus, esophageal adenocarcinoma, colorectal cancer, and GC.
Most groups showed better diagnostic performance with image data than with video data. This was observed in the groups of esophageal adenocarcinoma [EAC], pooled specificity: 0.85 [95% CI: 0.79–0.89]), esophageal squamous cell carcinoma [ESCC], and CRC. The GC group showed similar performance in the picture and video subgroups.
For prospective studies, no data existed on ESCC, and the available prospective studies for GC were limited. Except for the CRC group, the diagnostic performance of AI in retrospective and prospective studies was not significantly different; however, AI performed better in retrospective studies than in prospective studies.
We compared the diagnostic performance of AI with that of experts and nonexperts (expert and nonexpert diagnostic capabilities were meta-analyzed on the basis of the extracted data).he combined results showed that most AI models had better diagnostic capabilities than experts (Summary effect sizes are shown in Figure 2). Interestingly, three studies (40–42) analyzed the ability of nonexperts in diagnosing GC under endoscopy; with the help of AI, the diagnostic performance of nonexperts was found to reach the level of experts. Two studies (43, 44) reported that AI-assisted nonexperts achieved diagnostic performance comparable to that of experts in CRC.
3.5.2 Pancreatic tumors and mesenchymal tumors
Studies on AI-assisted EUS for the diagnosis of digestive system tumors are fewer, focusing on pancreatic tumors and mesenchymal tumors. Overall, the diagnostic performance of AI for pancreatic tumors was superior to that of mesenchymal tumors. Similar to GI tumors, the diagnostic performance of AI-assisted EUS was better than that of experts. There were no significant differences between retrospective and prospective studies.
3.6 Assessment of the risk of bias
Publication bias was found for ESCC (expert and nonexpert subgroups), Barrett esophagus and EAC (video subgroup), CRC (video and expert subgroups), and GC (expert and nonexpert subgroups). The remaining outcomes did not exhibit significant publication bias.
3.7 Grade
We downgraded the evidence according to five factors (risk of bias, inconsistency, indirectness, imprecision, and publication bias). The evidence for the following outcomes was downgraded to “moderate” quality: GC group, GC (image subgroup, retrospective subgroup, expert subgroup, and nonexpert subgroup), CRC group, CRC (image subgroup, video subgroup, retrospective subgroup, and expert subgroup), Barrett esophagus and EAC (video subgroup), and ESCC (expert subgroup and nonexpert subgroup). Only one piece of evidence was downgraded to “low” quality (ESCC group). The primary reasons for this downgrading were inconsistencies and publication bias. The evidence for the remaining outcomes was of “high” quality (Supplementary Table S2).
4 Discussion
Our results showed that the use of AI improved the detection and diagnostic accuracy of early digestive system tumors. Furthermore, AI-assisted endoscopic ultrasonography (EUS) had good diagnostic accuracy for pancreatic cancer.; meanwhile, AI showed high diagnostic performance in capsule endoscopy and esophageal squamous cell carcinoma for early digestive system tumors and precancerous lesions. Additionally, we compared the diagnostic capabilities of AI with those of experts and found that the diagnostic capability of AI was superior to that of experts.
During endoscopy, endoscopists must obtain diagnostic information while performing the endoscopy; therefore, most real-life scenarios are similar to video data formats. Therefore, we subdivided the AI into image subgroups and video subgroups. In this study, most subgroups using video data had lower diagnostic capability than those using image data. Therefore, we concluded that, although AI has a high diagnostic accuracy using image data, its diagnostic capability may be limited during clinical endoscopic procedures.
Prospective studies have a higher level of evidence compared to other studies. We performed meta-analysis on prospective studies of esophageal adenocarcinoma, colorectal cancer, and pancreatic cancer. The results showed that AI’s diagnostic performance in the prospective study group surpassed that of the experts and was higher than video-based diagnostic performance. This supports our conclusion that AI’s diagnostic capability surpasses that of experts, though AI’s performance with video data still requires improvement. Notably, although fewer prospective studies were available for gastric cancer in our review, the results from these studies also align with the conclusion mentioned above. Regarding the outcome of CRC, we found that the retrospective subgroup had higher sensitivity, whereas the prospective subgroup had higher specificity. However, this was not the case in other retrospective and prospective subgroups. We believe that a possible reason for this result is that there may have been selection bias when retrospectively collecting CRC data, which may lead researchers to select images with better bowel preparation scores, resulting in higher sensitivity. Overall, AI’s diagnostic capabilities in prospective studies require further improvement when compared to retrospective studies. This indicates a need for additional prospective studies to validate AI’s diagnostic abilities.
Overall, there was less heterogeneity in the outcomes, except for esophageal squamous carcinoma, GC, and CRC. Regarding the risk of bias, there was greater publication bias associated with esophageal cancer and CRC and almost no publication bias in the remaining outcomes. Selection bias in retrospective studies is inevitable. Many retrospective studies were included in this study; therefore, the selection bias was high. Regarding the evidence rating, the performance of AI in diagnosing ESCC was low-level evidence, whereas those of the remaining studies were medium- and high-level evidences. Although a small number of randomized controlled trials have evaluated the ability of AI to assist in the early diagnosis of cancer in clinical settings, most applications of AI for the endoscopic diagnosis of digestive system tumors are still in the preliminary stage (45–47).
AI is not yet widely used in clinical practice; thus, our study systematically summarizes the current research on the diagnostic capability of AI-assisted diagnosis of digestive system tumors and presents a pooled analysis of all data containing comparisons with experts and nonexperts. This study provides the latest evidence of AI-assisted endoscopy for the diagnosis of early digestive system tumors and precancerous lesions. The results of this study have practical implications in guiding the development of real-world applications.
In the past few years, the application of AI to endoscopic clinical practice has received increasing attention. AI can recognize subtle changes that cannot be identified by traditional methods and help identify subtle lesions. During endoscopy, AI can match multiple endoscopic imaging modalities, such as white light endoscopy (WEL) and narrowband imaging (NBI). AI can also help to label suspicious lesions in real time (12, 14).Thus, AI has been shown to improve the detection rate of digestive tumors, especially for less experienced endoscopists. AI-assisted novice endoscopists have lower rates of missed diagnoses, with results not inferior to the expert level (48). Interestingly, AI can also assign monitoring intervals to patients after polypectomy (49), and it can even improve bowel preparation (50). In recent years, automated endoscopic reporting systems have received increasing attention. One study (51) showed that the use of an AI-based endoscopy automatic reporting system significantly improved the accuracy and completeness of esophagogastroduodenoscopy (EGD) reporting, reduced the work burden of endoscopists, and promises to be an enhanced tool for EGD recording services. However, before focusing on the diagnostic capabilities of automated endoscopic reporting systems, it is essential to summarize the diagnostic capabilities of AI. Other prospective studies assessing AI’s performance in diagnosing digestive tumors have yielded important results. Most of these studies reported results aligning with our findings that the application of AI can improve the diagnosis rate of digestive system tumors (52–56). However, some prospective studies have indicated that AI may not enhance the diagnosis rate of these tumors (57, 58). We speculate that this discrepancy may stem from the fact that studies reporting no improvement were primarily single-center studies. One study involved a small sample size, and the other included a high-risk oncology hospital population, potentially influencing the results.
Based on previous meta-analysis and diagnostic tests (36, 41), we defined physicians with 5 years of experience as endoscopists. Although our study provided a uniform definition of “expert,” some diagnostic experiments use more detailed definitions, such as the number of operational cases or recognition by an academic association. This leads to heterogeneity in the definition of “experts.” The AI models or algorithms also differ between studies. These differences will have an unpredictable influence on the research results. Additionally, the prospective and video subgroups included a smaller number of studies, which also affected the results. Therefore, further prospective studies on the diagnostic capabilities of AI models based on video data are required to provide the latest evidence.
Still, before AI can be used in large-scale clinical applications, several problems must be resolved. First, the establishment of AI models requires large amounts of patient data. Owing to the lack of policies regarding the use of training data, there is a substantial risk of patient information leakage, and formulating relevant regulations is recommended to avoid potential risks (59). Additionally, to gain the trust of doctors and patients in the clinical stage of AI application, AI is required to achieve better diagnostic capability and interpretability. Our current research provides the latest evidence for the diagnostic capability of AI in the endoscopic diagnosis of digestive system tumors, indicating that AI has an excellent diagnostic performance; yet, convincing patients of the diagnosis of AI requires more popular science (60, 61).
Although there have been some active attempts to address the “black box” problem in neural networks, we cannot adequately explain the results produced by current AI (62). Most importantly, whether the responsibility for the errors that occur when AI is used in clinical applications lies with the endoscopic technologist, AI developer, or regulator cannot be answered (63–65).
Despite the good diagnostic performance achieved by AI, there are still some problems to be solved before its large-scale clinical application. Sound policies and regulations need to be developed to address the ethical issues associated with AI applications.
5 Conclusion
This is an umbrella review to evaluate the diagnostic performance of artificial intelligence (AI)-assisted endoscopy for digestive tumors. The results indicate that, for early digestive system cancer and precancerous lesions, AI showed a high diagnostic performance in capsule endoscopy and esophageal squamous cell carcinoma. Additionally, AI-assisted endoscopic ultrasonography (EUS) had good diagnostic accuracy for pancreatic cancer. In the subgroup analysis, AI had a better diagnostic performance than experts for most digestive system tumors. However, the diagnostic performance of AI using video data requires improvement.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author/s.
Author contributions
CH: Conceptualization, Data curation, Methodology, Software, Writing – original draft. YS: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft. JD: Data curation, Methodology, Writing – original draft. FY: Supervision, Writing – review & editing. JG: Conceptualization, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. SS: Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Liaoning Province Applied Basic Research Program Joint Program Project (2022JH2/101500076), Shenyang Young and Middle-aged Science and Technology Innovation Talent Support Program (grant number: RC200438), and Tree Planting Program of Shengjing Hospital (M1595).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1519144/full#supplementary-material
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. (2022) 72:7–33. doi: 10.3322/caac.21708
2. Miller KD, Nogueira L, Devasia T, Mariotto AB, Yabroff KR, Jemal A. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. (2022) 72:409–36. doi: 10.3322/caac.21731
3. Huang RJ, Laszkowska M, In H, Hwang JH, Epplein M. Controlling gastric cancer in a world of heterogeneous risk. Gastroenterology. (2023) 164:736–51. doi: 10.1053/j.gastro.2023.01.018
4. Wang J, Zhao G, Zhao Y, Zhao Z, Yang S, Zhou A, et al. N6-methylation in the development, diagnosis, and treatment of gastric cancer. J Transl Int Med. (2024) 12:5–21. doi: 10.2478/jtim-2023-0103
5. Xin L, Gao Y, Wang TJ, Meng QQ, Jin ZD, Fu ZJ, et al. EUS development in China: Results from national surveys in 2013 and 2020. Endosc Ultrasound. (2023) 12:90–5. doi: 10.4103/EUS-D-22-00003
6. Jiao Y, Cheng Z, Gao Y, Wang T, Xin L, Lin H, et al. Development and status quo of digestive endoscopy in China: An analysis based on the national census in 2013 and 2020. J Transl Int Med. (2024) 12:177–87. doi: 10.2478/jtim-2023-0115
7. Wu HL, Yao LW, Shi HY, Wu LL, Li X, Zhang CX, et al. Validation of a real-time biliopancreatic endoscopic ultrasonography analytical device in China: a prospective, single-centre, randomised, controlled trial. Lancet Digit Health. (2023) 5:e812–20. doi: 10.1016/S2589-7500(23)00160-7
8. Liu F, Gao A, Zhang M, Li Y, Zhang F, Herman JG, et al. Methylation of FAM110C is a synthetic lethal marker for ATR/CHK1 inhibitors in pancreatic cancer. J Transl Int Med. (2024) 12:274–87. doi: 10.2478/jtim-2023-0128
9. Chahal D, Byrne MFA. primer on artificial intelligence and its application to endoscopy. Gastrointest Endosc. (2020) 92:813–20. doi: 10.1016/j.gie.2020.04.074
10. Luo D, Kuang F, Du J, Zhou M, Liu X, Luo X, et al. Artificial intelligence-assisted endoscopic diagnosis of early upper gastrointestinal cancer: A systematic review and meta-analysis. Front Oncol. (2022) 12:855175. doi: 10.3389/fonc.2022.855175
11. Xu Y, Ding W, Wang Y, Tan Y, Xi C, Ye N, et al. Comparison of diagnostic performance between convolutional neural networks and human endoscopists for diagnosis of colorectal polyp: A systematic review and meta-analysis. PLoS One. (2021) 16(2):e0246892. doi: 10.1371/journal.pone.0246892
12. Mohan BP, Facciorusso A, Khan SR, Madhu D, Kassab LL, Ponnada S, et al. Pooled diagnostic parameters of artificial intelligence in EUS image analysis of the pancreas: A descriptive quantitative review. Endosc Ultrasound. (2022) 11:156–69. doi: 10.4103/EUS-D-21-00063
13. Islam MM, Poly TN, Walther BA, Yeh CY, Seyed-Abdul S, Li YJ, et al. Deep learning for the diagnosis of esophageal cancer in endoscopic images: A systematic review and meta-analysis. Cancers (Basel). (2022) 14:5996. doi: 10.3390/cancers14235996
14. ASGE AI Task Force, Leggett CL, Parasa S, Repici A, Berzin TM, Gross SA, et al. Physician perceptions on the current and future impact of artificial intelligence to the field of gastroenterology. Gastrointest Endosc. (2024) 99:483–9. doi: 10.1016/j.gie.2023.11.053
15. Huang Y, Chen Z, Chen B, Li J, Yuan X, Li J, et al. Dietary sugar consumption and health: umbrella review. BMJ. (2023) 381:e071609. doi: 10.1136/bmj-2022-071609
16. Okagawa Y, Abe S, Yamada M, Oda I, Saito Y. Artificial intelligence in endoscopy. Dig Dis Sci. (2022) 67:1553–72. doi: 10.1007/s10620-021-07086-z
17. Ioannidis JP, Patsopoulos NA, Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. BMJ. (2007) 335:914–6. doi: 10.1136/bmj.39343.408449.80
18. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. (1997) 315:629–34. doi: 10.1136/bmj.315.7109.629
19. Shea BJ, Reeves BC, Wells G, Thuku M, Hamel C, Moran J, et al. AMSTAR 2: a critical appraisal tool for systematic reviews that include randomised or non-randomised studies of healthcare interventions, or both. BMJ. (2017) 358:j4008. doi: 10.1136/bmj.j4008
20. Guyatt GH, Oxman AD, Vist GE, Kunz R, Falck-Ytter Y, Alonso-Coello P, et al. GRADE: an emerging consensus on rating quality of evidence and strength of recommendations. BMJ. (2008) 336:924–6. doi: 10.1136/bmj.39489.470347.AD
21. Dumitrescu EA, Ungureanu BS, Cazacu IM, Florescu LM, Streba L, Croitoru VM, et al. Diagnostic value of artificial intelligence-assisted endoscopic ultrasound for pancreatic cancer: A systematic review and meta-analysis. Diagnostics (Basel). (2022) 12:309. doi: 10.3390/diagnostics12020309
22. Prasoppokakorn T, Tiyarattanachai T, Chaiteerakij R, Decharatanachart P, Mekaroonkamol P, Ridtitid W, et al. Application of artificial intelligence for diagnosis of pancreatic ductal adenocarcinoma by EUS: A systematic review and meta-analysis. Endosc Ultrasound. (2022) 11:17–26. doi: 10.4103/EUS-D-20-00219
23. Guidozzi N, Menon N, Chidambaram S, Markar SR. The role of artificial intelligence in the endoscopic diagnosis of esophageal cancer: a systematic review and meta-analysis. Dis Esophagus. (2023) 36(12):doad048. doi: 10.1093/dote/doad048
24. Visaggi P, Barberio B, Gregori D, Azzolina D, Martinato M, Hassan C, et al. Systematic review with meta-analysis: artificial intelligence in the diagnosis of oesophageal diseases. Aliment Pharmacol Ther. (2022) 55:528–40. doi: 10.1111/apt.16778
25. Arribas J, Antonelli G, Frazzoni L, Fuccio L, Ebigbo A, van der Sommen F, et al. Standalone performance of artificial intelligence for upper GI neoplasia: a meta-analysis. Gut. (2020) –gutjnl–2020–321922. doi: 10.1136/gutjnl-2020-321922
26. Lui TKL, Tsui VWM, Leung WK. Accuracy of artificial intelligence-assisted detection of upper GI lesions: a systematic review and meta-analysis. Gastrointest Endosc. (2020) 92:821–30. doi: 10.1016/j.gie.2020.06.034
27. Tan JL, Chinnaratha MA, Woodman R, Martin R, Chen HT, Carneiro G, et al. Diagnostic accuracy of artificial intelligence (AI) to detect early neoplasia in barrett’s esophagus: A non-comparative systematic review and meta-analysis. Front Med (Lausanne). (2022) 9:890720. doi: 10.3389/fmed.2022.890720
28. Bang CS, Lee JJ, Baik GH. Computer-aided diagnosis of esophageal cancer and neoplasms in endoscopic images: a systematic review and meta-analysis of diagnostic test accuracy. Gastrointest Endosc. (2021) 93:1006–15. doi: 10.1016/j.gie.2020.11.025
29. Chen PC, Lu YR, Kang YN, Chang CC. The accuracy of artificial intelligence in the endoscopic diagnosis of early gastric cancer: pooled analysis study. J Med Internet Res. (2022) 24(5):e27694. doi: 10.2196/27694
30. Jiang K, Jiang X, Pan J, Wen Y, Huang Y, Weng S, et al. Current evidence and future perspective of accuracy of artificial intelligence application for early gastric cancer diagnosis with endoscopy: A systematic and meta-analysis. Front Med (Lausanne). (2021) 8:629080. doi: 10.3389/fmed.2021.629080
31. Islam MM, Poly TN, Walther BA, Lin MC, Li YJ. Artificial intelligence in gastric cancer: identifying gastric cancer using endoscopic images with convolutional neural network. Cancers (Basel). (2021) 13:5253. doi: 10.3390/cancers13215253
32. Liu XY, Song W, Mao T, Zhang Q, Zhang C, Li XY, et al. Application of artificial intelligence in the diagnosis of subepithelial lesions using endoscopic ultrasonography: a systematic review and meta-analysis. Front Oncol. (2022) 12:915481. doi: 10.3389/fonc.2022.915481
33. Bai J, Liu K, Gao L, Zhao X, Zhu S, Han Y, et al. Computer-aided diagnosis in predicting the invasion depth of early colorectal cancer: a systematic review and meta-analysis of diagnostic test accuracy. Surg Endosc. (2023) 37:6627–39. doi: 10.1007/s00464-023-10223-6
34. Wang A, Mo J, Zhong C, Wu S, Wei S, Tu B, et al. Artificial intelligence-assisted detection and classification of colorectal polyps under colonoscopy: a systematic review and meta-analysis. Ann Transl Med. (2021) 9:1662. doi: 10.21037/atm-21-5081
35. Lui TKL, Guo CG, Leung WK. Accuracy of artificial intelligence on histology prediction and detection of colorectal polyps: a systematic review and meta-analysis. Gastrointest Endosc. (2020) 92:11–22. doi: 10.1016/j.gie.2020.02.033
36. Li MD, Huang ZR, Shan QY, Chen SL, Zhang N, Hu HT, et al. Performance and comparison of artificial intelligence and human experts in the detection and classification of colonic polyps. BMC Gastroenterol. (2022) 22:517. doi: 10.1186/s12876-022-02605-2
37. Bang CS, Lee JJ, Baik GH. Computer-aided diagnosis of diminutive colorectal polyps in endoscopic images: systematic review and meta-analysis of diagnostic test accuracy. J Med Internet Res. (2021) 23(8):e29682. doi: 10.2196/29682
38. Mi J, Han X, Wang R, Ma R, Zhao D. Diagnostic accuracy of wireless capsule endoscopy in polyp recognition using deep learning: A meta-analysis. Int J Clin Pract. (2022) 2022:9338139. doi: 10.1155/2022/9338139
39. Kim HJ, Gong EJ, Bang CS, Lee JJ, Suk KT, Baik GH, et al. Computer-aided diagnosis of gastrointestinal protruded lesions using wireless capsule endoscopy: A systematic review and diagnostic test accuracy meta-analysis. J Pers Med. (2022) 12:644. doi: 10.3390/jpm12040644
40. Liu L, Dong Z, Cheng J, Bu X, Qiu K, Yang C, et al. Diagnosis and segmentation effect of the ME-NBI-based deep learning model on gastric neoplasms in patients with suspected superficial lesions - a multicenter study. Front Oncol. (2023) 12:1075578. doi: 10.3389/fonc.2022.1075578
41. Goto A, Kubota N, Nishikawa J, Ogawa R, Hamabe K, Hashimoto S, et al. Cooperation between artificial intelligence and endoscopists for diagnosing invasion depth of early gastric cancer. Gastric Cancer. (2023) 26:116–22. doi: 10.1007/s10120-022-01330-9
42. Hu H, Gong L, Dong D, Zhu L, Wang M, He J, et al. Identifying early gastric cancer under magnifying narrow-band images with deep learning: a multicenter study. Gastrointest Endosc. (2021) 93:1333–41. doi: 10.1016/j.gie.2020.11.014
43. Chen PJ, Lin MC, Lai MJ, Lin JC, Lu HH, Tseng VS, et al. Accurate classification of diminutive colorectal polyps using computer-aided analysis. Gastroenterology. (2018) 154:568–75. doi: 10.1053/j.gastro.2017.10.010
44. Mori Y, Kudo SE, Wakamura K, Misawa M, Ogawa Y, Kutsukawa M, et al. Novel computer-aided diagnostic system for colorectal lesions by using endocytoscopy (with videos). Gastrointest Endosc. (2015) 81:621–9. doi: 10.1016/j.gie.2014.09.008
45. Nazarian S, Glover B, Ashrafian H, Darzi A, Teare J. Diagnostic accuracy of artificial intelligence and computer-aided diagnosis for the detection and characterization of colorectal polyps: systematic review and meta-analysis. J Med Internet Res. (2021) 23(7):e27370. doi: 10.2196/27370
46. He X, Wu L, Dong Z, Gong D, Jiang X, Zhang H, et al. Real-time use of artificial intelligence for diagnosing early gastric cancer by magnifying image-enhanced endoscopy: a multicenter diagnostic study (with videos). Gastrointest Endosc. (2022) 95:671–8. doi: 10.1016/j.gie.2021.11.040
47. Desai M, Ausk K, Brannan D, Chhabra R, Chan W, Chiorean M, et al. Use of a novel artificial intelligence system leads to the detection of significantly higher number of adenomas during screening and surveillance colonoscopy: results from a large, prospective, US multicenter, randomized clinical trial. Am J Gastroenterol. (2024) 119:1383–91. doi: 10.14309/ajg.0000000000002664
48. Yao L, Li X, Wu Z, Wang J, Luo C, Chen B, et al. Effect of artificial intelligence on novice-performed colonoscopy: a multicenter randomized controlled tandem study. Gastrointest Endosc. (2024) 99:91–9. doi: 10.1016/j.gie.2023.07.044
49. Wu L, Shi C, Li J, Dong Z, Zhou W, Yin A, et al. Development and evaluation of a surveillance system for follow-up after colorectal polypectomy. JAMA Netw Open. (2023) 6(9):e2334822. doi: 10.1001/jamanetworkopen.2023.34822
50. Zhu Y, Zhang DF, Wu HL, Fu PY, Feng L, Zhuang K, et al. Improving bowel preparation for colonoscopy with a smartphone application driven by artificial intelligence. NPJ Digit Med. (2023) 6:41. doi: 10.1038/s41746-023-00786-y
51. Zhang L, Lu Z, Yao L, Dong Z, Zhou W, He C, et al. Effect of a deep learning-based automatic upper GI endoscopic reporting system: a randomized crossover study (with video). Gastrointest Endosc. (2023) 98:181–90. doi: 10.1016/j.gie.2023.02.025
52. Xu H, Tang RSY, Lam TYT, Zhao G, Lau JYW, Liu Y, et al. Artificial intelligence-assisted colonoscopy for colorectal cancer screening: A multicenter randomized controlled trial. Clin Gastroenterol Hepatol. (2023) 21:337–346.e3. doi: 10.1016/j.cgh.2022.07.006
53. Yuan XL, Liu W, Lin YX, Deng QY, Gao YP, Wan L, et al. Effect of an artificial intelligence-assisted system on endoscopic diagnosis of superficial oesophageal squamous cell carcinoma and precancerous lesions: a multicentre, tandem, double-blind, randomised controlled trial. Lancet Gastroenterol Hepatol. (2024) 9:34–44. doi: 10.1016/S2468-1253(23)00276-5
54. Cui H, Zhao Y, Xiong S, Feng Y, Li P, Lv Y, et al. Diagnosing solid lesions in the pancreas with multimodal artificial intelligence: A randomized crossover trial. JAMA Netw Open. (2024) 7:e2422454. doi: 10.1001/jamanetworkopen.2024.22454
55. Dittman LE, Munaretto N, Hinchcliff K, Dutton L, Kakar S. Volar wrist arthroscopy portals using the nanoScope are safer than traditional arthroscopy. Handb (N Y). (2024) 18:15589447231221168. doi: 10.1177/15589447231221168
56. Dong Z, Zhao X, Zheng H, Zheng H, Chen D, Cao J, et al. Efficacy of real-time artificial intelligence-aid endoscopic ultrasonography diagnostic system in discriminating gastrointestinal stromal tumors and leiomyomas: a multicenter diagnostic study. EClinicalMedicine. (2024) 73:102656. doi: 10.1016/j.eclinm.2024.102656
57. Nakao E, Yoshio T, Kato Y, Namikawa K, Tokai Y, Yoshimizu S, et al. Randomized controlled trial of an artificial intelligence diagnostic system for the detection of esophageal squamous cell carcinoma in clinical practice. Endoscopy. (2024) 57(3):210–7. doi: 10.1055/a-2421-3194
58. Schöler J, Alavanja M, de Lange T, Yamamoto S, Hedenström P, Varkey J. Impact of AI-aided colonoscopy in clinical practice: a prospective randomised controlled trial. BMJ Open Gastroenterol. (2024) 11:e001247. doi: 10.1136/bmjgast-2023-001247
59. Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, King D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. (2019) 17:195. doi: 10.1186/s12916-019-1426-2
60. Kerasidou CX, Kerasidou A, Buscher M, Wilkinson S. Before and beyond trust: reliance in medical AI. J Med Ethics. (2022) 48:852–6. doi: 10.1136/medethics-2020-107095
61. Quinn TP, Senadeera M, Jacobs S, Coghlan S, Le V. Trust and medical AI: the challenges we face and the expertise needed to overcome them. J Am Med Inform Assoc. (2021) 28:890–4. doi: 10.1093/jamia/ocaa268
62. de Souza LA Jr., Mendel R, Strasser S, Ebigbo A, Probst A, Messmann H, et al. Convolutional Neural Networks for the evaluation of cancer in Barrett’s esophagus: Explainable AI to lighten up the black-box. Comput Biol Med. (2021) 135. doi: 10.1016/j.compbiomed.2021.104578
63. DeCamp M, Lindvall C. Latent bias and the implementation of artificial intelligence in medicine. J Am Med Inform Assoc. (2020) 27:2020–3. doi: 10.1093/jamia/ocaa094
64. Zhang D, Wu C, Yang Z, Yin H, Liu Y, Li W, et al. The application of artificial intelligence in EUS. Endosc Ultrasound. (2024) 13:65–75. doi: 10.1097/eus.0000000000000053
Keywords: artificial intelligence, endoscopy, endoscopic ultrasound, precancerous lesion, digestive system tumors
Citation: Huang C, Song Y, Dong J, Yang F, Guo J and Sun S (2025) Diagnostic performance of AI-assisted endoscopy diagnosis of digestive system tumors: an umbrella review. Front. Oncol. 15:1519144. doi: 10.3389/fonc.2025.1519144
Received: 29 October 2024; Accepted: 18 March 2025;
Published: 03 April 2025.
Edited by:
Fu Shen, Naval Medical University, ChinaReviewed by:
Calin Cainap, University of Medicine and Pharmacy Iuliu Hatieganu, RomaniaZhen Li, Qilu Hospital of Shandong University, China
Xu Fang, Second Military Medical University, China
Copyright © 2025 Huang, Song, Dong, Yang, Guo and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jintao Guo, eXVlZmFuXzAyMjVAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Changwei Huang1†
Changwei Huang1† Fan Yang
Fan Yang Jintao Guo
Jintao Guo Siyu Sun
Siyu Sun