- School of Basic Medicine, Heilongjiang University of Chinese Medicine, Harbin, Heilongjiang, China
Serine is crucial for tumor initiation, progression, and adaptive immunity. Metabolic pathways for serine synthesis, acquisition, and utilization in tumors and tumor-associated cells are influenced by various physiological factors and the tumor microenvironment, leading to metabolic reprogramming and amplification. Excessive serine metabolism promotes abnormal macromolecule biosynthesis, mitochondrial dysfunction, and epigenetic modifications, driving malignant transformation, proliferation, metastasis, immune suppression, and drug resistance in tumor cells. Restricting dietary serine intake or reducing the expression of serine synthetic enzymes can effectively slow tumor growth and extend patient survival. Consequently, targeting serine metabolism has emerged as a novel and promising research focus in cancer research. This paper reviews serine metabolic pathways and their roles in tumor development. It summarizes the influencing factors of serine metabolism. The article explores the significance of serine synthesis and metabolizing enzymes, along with related biomarkers, in tumor diagnosis and treatment, providing new insights for developing targeted therapies that modulate serine metabolism in cancer.
1 Introduction
Serine, a non-essential amino acid, is vital for nucleotide and lipid metabolism, serving as a key source of one-carbon (1C) units necessary for biological growth. Within cells, serine has multifunctional roles, participating in DNA and RNA synthesis, regulating cell membrane structure and function, and supporting cell growth (1, 2). The importance of serine in cancer cell proliferation differs from that in normal cells. Compared to normal cells, most cancer cells rely on glycolysis for energy procurement (3). Due to its lower production efficiency, cancer cells increase their uptake and utilization of glucose and amino acids such as glutamine (4). In rapidly proliferating cancer cells, however, glucose and glutamine consumption is often insufficient to support biomacromolecule accumulation. In this process, serine provides most of the carbon and nitrogen units required by cancer cells, which is crucial for their survival (5). Studies have shown that both endogenous serine produced from glucose and functionally acquired exogenous serine are associated with the growth of cancer cells in vitro and in vivo and functionally promote cancer progression (6–10).
2 Serine metabolism
Serine is the primary source for synthesizing one-carbon (1C) units, linking amino acid metabolism to nucleotide metabolism. Thus, serine occupies a central role in cellular metabolic processes.
2.1 Serine as a central node in biosynthesis
Serine serves as a pivotal metabolite in various biosynthetic pathways, providing essential materials for cellular biosynthesis processes (11). Through charging tRNAs, serine participates in protein synthesis and acts as a precursor for amino acids like cysteine and glycine. It also provides head groups for the synthesis of sphingolipids and phospholipids. Notably, in the folate-mediated 1C pathway, the cleavage of serine into glycine and 1C units supports the synthesis of porphyrins, thymidylate, purines, glutathione, and S-adenosylmethionine (SAM) (12).
2.2 Pathways for serine acquisition
Serine, essential for cell growth, is acquired through internal synthesis and external uptake.
2.2.1 Internal synthesis
One-carbon metabolism includes a bicyclic pathway formed by the coupling of the folate cycle, methionine cycle, and trans-sulfuration pathway (13, 14). The folate and methionine cycles are crucially interlinked pathways in 1C metabolism, providing methyl groups for DNA, amino acid, creatine, polyamine, and phospholipid synthesis (15). One-carbon metabolism generates various products by cycling 1C units from different amino acids and integrating multiple cellular nutritional states (13). The primary sources of 1C units are the catabolic pathways of serine, glycine, and histidine. Serine and glycine are interconvertible, with serine serving as the principal donor of 1C units when converted to glycine (16). Serine is derived from the metabolism of nutrients, including proteins and phospholipids. Glycine, synthesized endogenously from serine, acquires a hydroxymethyl group through serine hydroxymethyltransferase (SHMT). Although alternative sources for serine synthesis exist, the Serine Synthesis Pathway (SSP) remains the primary supplier of serine in the body, as other sources are insufficient to meet tumor cell demands (2). The SSP is recognized as a critical growth and stress resistance pathway in cancer cells (17–21).
As illustrated in Figure 1, in the folate cycle, folate is reduced twice by dihydrofolate reductase (DHFR), ultimately converting into tetrahydrofolate (THF). Serine is synthesized from the glycolytic intermediate 3-phosphoglyceric acid (3PG) through three steps. First, 3PG is oxidized to 3-phosphohydroxypyruvic acid (3PHP) by phosphoglycerate dehydrogenase (PHGDH), reducing NAD+ to NADH. First, 3PG is oxidized to 3-phosphohydroxypyruvic acid (3PHP) by phosphoglycerate dehydrogenase (PHGDH), reducing NAD+ to NADH. Secondly, through a transamination reaction, 3PHP receives an amino group from glutamate, producing 3-phosphoserine (3P-Ser) and α-ketoglutarate (α-KG), a step catalyzed by phosphoserine transaminase 1 (PSAT1). Finally, 3P-Ser is dephosphorylated by phosphoserine phosphatase (PSPH) to produce serine (12). Under the catalysis of SHMTs, serine is converted to glycine, and during this process, THF accepts the one-carbon unit from serine, forming 5,10-methylenetetrahydrofolate (5,10-CH2-THF). 5,10-CH2-THF can be converted to 10-formyltetrahydrofolate (F-THF) by methylenetetrahydrofolate dehydrogenase (MTHFD) 1/2/1L, or reduced to 5-methyltetrahydrofolate (mTHF) by methylenetetrahydrofolate reductase (MTHFR). mTHF can be demethylated and converted back to THF, completing the folate cycle and initiating the methionine cycle.
In the methionine cycle, mTHF transfers the one-carbon unit to homocysteine to form methionine, which is then converted by methionine synthase (MTR). Methionine is converted into S-adenosylmethionine (SAM) by methionine adenosyltransferase (MAT). SAM, a substrate for methylation reactions, forms S-adenosylhomocysteine (SAH) upon demethylation, which is converted back to homocysteine by S-adenosylhomocysteine hydrolase (SAHH), completing the methionine cycle (14). In the trans-sulfuration pathway, homocysteine is converted to cystathionine by cystathionine beta-synthase (CBS) and further to cysteine by cystathionine gamma-lyase (CTH), ultimately leading to glutathione (GSH) synthesis.
2.2.2 External uptake
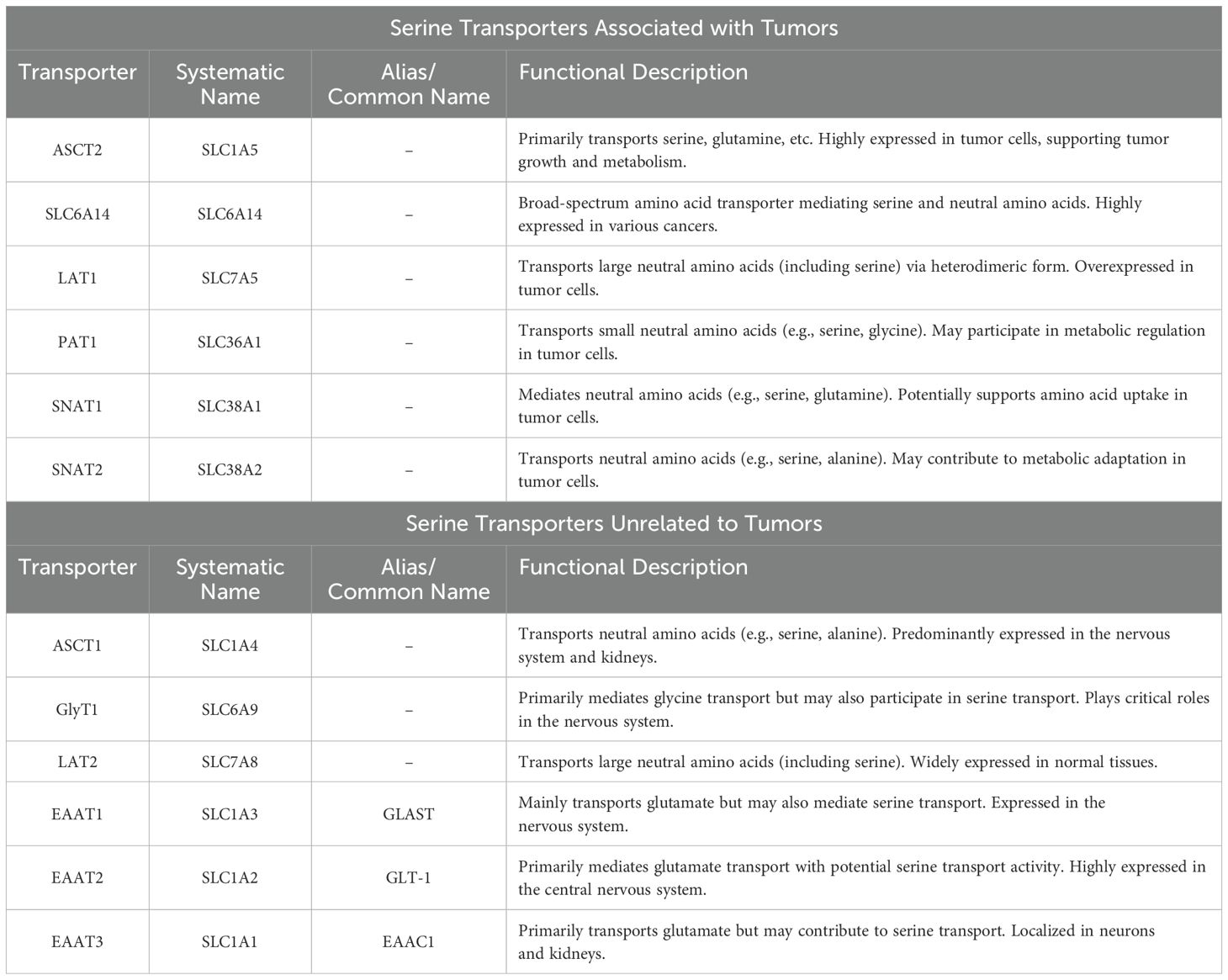
While some cancer cells meet their growth requirements through de novo serine synthesis, others depend on exogenous serine (8, 9, 22). In normal cells, serine is synthesized via the de novo pathway, involving reactions catalyzed by phosphoglycerate dehydrogenase (PHGDH). However, certain cancer cells may have mutations or abnormal expressions in key enzymes of these metabolic pathways, which restrict their ability to synthesize serine de novo. Consequently, these cancer cells often rely on exogenous serine to support their growth and metabolic needs, as observed in Ewing sarcoma and liposarcoma cells (23). The supply of exogenous serine can be achieved through the intake of a serine-rich diet or the availability of serine in the extracellular environment, where serine transport proteins play a critical role. The serine transporters encompass members of the SLC1A, SLC6A, SLC7, SLC36, and SLC38 families, with their specific classifications and functional characteristics detailed in the Table 1. The Solute Carrier Family 1A (SLC1A) includes two major transport systems: Alanine-Serine-Cysteine Transporters 1-2 (ASCT1-2) and human glutamate transporters (Excitatory Amino Acid Transporters 1-5, EAAT1-5). ASCTs are widely expressed and are one of the four primary neutral amino acid transport systems in the human body (24, 25). In particular, ASCT2(SLC1A5) functions as a neutral amino acid exchanger, transporting serine and other amino acids (26). Under serine-limited conditions, ASCT2 is essential for purine nucleotide biosynthesis. Combined depletion of ASCT2 with a serine-free diet induces tumor regression (27). Similarly, in activated T cells, most intracellular serine is obtained from the extracellular environment (28). The main transporter for serine uptake in T cells has not yet been identified, but SLC1A5 is hypothesized to be involved (29). Beyond ASCT2, emerging evidence has revealed ASCT1 (SLC1A4) as a functional serine transporter in astrocytes (30). In parallel, Maddocks and colleagues have identified SLC6A14 and SLC12A4 as contributors to serine transport in cancer cells (31).
This dependence on exogenous serine has significant clinical implications, providing a potential therapeutic target within the serine metabolic pathway. Interfering with cancer cells’ dependency on exogenous serine, such as by inhibiting serine transport proteins or disrupting serine uptake, may hinder cancer cell growth and proliferation. It is important to acknowledge that serine metabolism may differ among various types of cancer, and the specific dependencies and mechanisms of serine metabolism are still under investigation. A deeper understanding of cancer cell dependency on serine is crucial for developing effective treatment strategies for tumors.
2.2.3 Metabolic interplay and plasticity between de novo synthesis and exogenous uptake
Intracellular synthesis and extracellular uptake constitute two primary pathways for serine acquisition in cancer cells, operating independently yet synergistically to sustain tumor metabolism. The de novo synthesis pathway (e.g., serine synthesis pathway, SSP) confers metabolic autonomy by enabling endogenous serine production, particularly under nutrient-replete conditions where SSP suffices to meet proliferative demands (21, 32). For instance, breast cancer cells with PHGDH amplification exhibit marked dependency on SSP (33). Conversely, cancers harboring mutations or dysregulated expression of SSP enzymes (e.g., PHGDH) display compromised biosynthetic capacity, leading to heightened reliance on exogenous serine uptake. This dependency is prominently observed in Ewing sarcoma and liposarcoma (23).
Metabolic plasticity serves as a hallmark of cancer cell adaptation to microenvironmental heterogeneity. Studies demonstrate that cancer cells dynamically rewire serine acquisition strategies based on extracellular serine availability: downregulating SSP when exogenous serine is abundant, while upregulating SSP under serine-depleted conditions (10, 34–36). This bidirectional regulation not only enhances survival under nutritional fluctuations but also underpins adaptive resistance to therapeutic stress.
2.3 Serine catabolism
Serine catabolism serves as a central node linking cellular energy supply and biosynthetic processes. In mammals, serine degradation is predominantly catalyzed by serine hydroxymethyltransferase (SHMT), which transfers the β-carbon of serine to tetrahydrofolate (THF), generating glycine and 5,10-methylenetetrahydrofolate (5,10-CH2-THF). This latter metabolite functions as a one-carbon unit donor, directly participating in the synthesis of purines, thymidylate, and glutathione (11).
Serine catabolism plays pivotal roles in various physiological and pathological contexts. During respiratory impairment, it contributes to NADH regeneration (37), while in hepatic tissues, it generates NADPH to support lipogenesis (38). Under hypoxic conditions, serine catabolism regulates mitochondrial redox homeostasis. Notably, the mitochondrial isoform SHMT2 is highly expressed in proliferating cancer cells, where it enhances one-carbon metabolism to fuel nucleic acid biosynthesis and tumor progression (39). Accumulating evidence positions SHMT2 and methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) — the first and second enzymes in mitochondrial serine catabolism — as independent prognostic markers and potential therapeutic targets across multiple cancer types (40–45).
These multifaceted functions underscore the broad physiological significance of serine catabolism in energy provision, redox equilibrium, and lipid biosynthesis, with its dysregulation being critically implicated in various metabolic disorders and malignancies.
3 Serine metabolism and tumors
Cellular metabolic reprogramming is a common feature of human tumors. In conditions of nutrient limitation and stress, tumor cells readjust metabolic pathways to produce sufficient metabolites for their rapid proliferation (46, 47). As one of the most fundamental components of cell structure, amino acids largely support protein synthesis needed for cell proliferation (48). During tumor development, changes in the rate of amino acid uptake, metabolic pathways, metabolites, or key enzymes are referred to as amino acid metabolic reprogramming (49). Serine metabolism also alters during tumor progression, mainly through changes in metabolites and key enzymes (50).
3.1 Genes related to serine metabolism
Key enzymes for serine synthesis, such as phosphoglycerate dehydrogenase (PHGDH) and phosphoserine aminotransferase (PSAT), are often abnormally expressed in tumor cells. Oncogenes and tumor suppressor genes also regulate the genes encoding enzymes involved in serine synthesis. The following section primarily introduces the related PHGDH and p53 genes.
3.1.1 PHGDH
PHGDH plays a crucial role in the de novo synthesis pathway of serine, acting as both the first enzyme and the rate-limiting enzyme in the reaction process. It is one of the few known metabolic enzymes that are dysregulated in cancer (51–53). Recent studies indicate that activating the serine synthesis pathway (SSP) and upregulating PHGDH expression can promote the growth of various cancer subtypes. Cancer cells use PHGDH and NAD+ to oxidize 10% of the 3-phosphoglycerate (3-PG) produced by glycolysis into the serine precursor 3-phosphohydroxypyruvate (3-PPyr) (54, 55). In addition to catalyzing the first step in serine synthesis, PHGDH accelerates the NADH-dependent reduction of alpha-ketoglutarate (α-KG) to the oncometabolite D-2-hydroxyglutarate (D-2HG), playing an important role in tumorigenesis (56). Amplification of the PHGDH gene on chromosome 1p12 occurs in 6% and 40% of breast cancers and melanomas, respectively (57, 58). In estrogen receptor-negative breast tumors, PHGDH protein levels can be elevated by as much as 70% (7). Increased PHGDH expression levels are also observed in other types of cancers. These studies highlight the important role of PHGDH in tumor biology, offering new directions for further research and cancer treatment.
3.1.2 p53
The tumor suppressor gene TP53 (aka, p53) encodes the protein p53, a crucial regulator of cellular metabolism. p53 is pivotal in the stress response to challenges such as DNA damage, hypoxia, and oncogene activation (59). The complexity and diversity of these cellular processes suggest that, beyond its traditional role as a tumor suppressor, p53 also maintains homeostasis in both normal and cancer cells (60). p53’s capacity to respond to nutritional deficiencies arises from its role as a mediator of cellular stress responses. It is closely related to the ability of cancer cells to cope with serine starvation and oxidative stress. Indeed, p53 is crucial in enabling cancer cells to overcome serine starvation and sustain their antioxidant capacity. Cells lacking p53 exhibit reduced survival and severely impaired proliferative capacity due to a failure to effectively cope with serine starvation and oxidative stress. During serine starvation, the p53-p21 signaling pathway is activated, leading to cell cycle arrest. This arrest reallocates depleted serine for glutathione synthesis, thereby promoting cell survival (9).
3.1.3 PSAT1
PSAT1 (phosphoserine aminotransferase 1), a member of the class V pyridoxal 5’-phosphate-dependent aminotransferase family, is a pivotal enzyme in the three-step serine synthesis pathway (SSP). It catalyzes the transamination of 3-phosphohydroxypyruvate (3-PHP) to phosphoserine using glutamate-derived nitrogen, concomitantly generating α-ketoglutarate (α-KG) (61). PSAT1 overexpression is documented across multiple malignancies — including non-small cell lung cancer (NSCLC), breast cancer, gastric cancer, colon cancer, and ovarian cancer — where it drives tumorigenesis and malignant progression (62–67). Elevated PSAT1 levels correlate with aggressive phenotypes such as proliferation, migration, invasion, and therapy resistance (62, 68–74). Recent evidence demonstrates that PSAT1 inhibition attenuates the tumorigenic potential and pulmonary metastasis of clear cell renal cell carcinoma (ccRCC) (75), while its prognostic utility is increasingly recognized (76). Mechanistically, PSAT1 mediates the reversible conversion of 3-PHP to L-phosphoserine, with glutamate serving as a substrate. Structural analyses reveal tight interactions between L-phosphoserine’s phosphate group and conserved arginine (Arg42, Arg328) and histidine (His41, His327) residues, suggesting that analogs targeting these residues could yield potent inhibitors (77). These findings underscore the therapeutic imperative to develop PSAT1 antagonists.
3.1.4 PSPH
PSPH (phosphoserine phosphatase), the terminal enzyme in L-serine biosynthesis, executes the irreversible dephosphorylation of phosphoserine to serine downstream of PHGDH and PSAT1 in the SSP (34). In melanoma, PSPH upregulation promotes tumor growth in vitro and in vivo, whereas its knockdown suppresses proliferation (78). Similarly, elevated PSPH expression in breast (79), liver (80), colorectal (81), NSCLC (82), and cutaneous squamous cell carcinomas (83) correlates with enhanced invasiveness and metastasis. Genomic analyses identify frequent PSPH amplifications and rare mutations in 13% of glioblastomas (84). These oncogenic roles position PSPH as a critical node requiring systematic investigation.
3.1.5 Transcriptional regulator ATF4
Activating transcription factor 4 (ATF4), a basic leucine zipper (bZIP) family member, orchestrates metabolic reprogramming by integrating stress signals (e.g., nutrient deprivation, oxidative/ER stress) (85, 86). ATF4 directly activates SSP enzymes (PHGDH, PSAT1, PSPH) to bolster de novo serine synthesis (87). Its pro-tumorigenic effects manifest via serine pathway activation in colorectal and breast cancers, where ATF4 knockdown suppresses PHGDH expression and chemoresistance (88, 89). Paradoxically, chronic ER stress-induced ATF4 hyperactivation may upregulate pro-apoptotic factors like CHOP, suggesting context-dependent tumor-suppressive potential (90). This functional duality — shaped by tumor type, microenvironmental stress, and genetic background — positions ATF4 as an emerging therapeutic target. Preclinical studies confirm that ATF4 inhibition synergizes with conventional therapies (91), though clinical translation awaits further validation.
3.2 Serine metabolism and the immunosuppressive microenvironment
The immunosuppressive tumor microenvironment (TME) enables immune evasion, with serine metabolism emerging as a key modulator via metabolite allocation, immune cell regulation, and redox control. Serine fuels CD8+ T cell expansion through one-carbon metabolism — a process decoupled from glucose utilization (28). However, tumor cells hijack serine via SSP activation (PHGDH, PSAT1 (68, 92)) or transporters (SLC1A4 (93)), depleting TME serine. This scarcity impairs CD8+ T cell purine synthesis, blunting clonal expansion and IFN-γ production (28). Concurrently, SSP-derived α-KG stabilizes Treg immunosuppression by enhancing KDM5A-mediated Foxp3 demethylation (94). Thus, tumors exploit serine metabolism to simultaneously cripple cytotoxic immunity and bolster Treg activity.
Tumors co-opt serine metabolism (via SSP activation and uptake) to deplete TME serine, directly impairing CD8+ T cells while epigenetically amplifying Treg suppression through α-KG. Targeting this axis — via PHGDH inhibitors or dietary modulation — combined with immune checkpoint blockade may reverse metabolic immunosuppression, offering novel combinatorial therapeutic avenues.
3.3 Tumor treatment strategies targeting serine metabolism
3.3.1 Low-serine diet
Serine starvation can induce cellular stress responses and metabolic reprogramming, thereby inhibiting cancer progression (8–10). Studies show that restricting dietary serine and glycine reduces tumor growth in xenograft and allograft models (9, 95). In genetically engineered mouse models of colorectal cancer (driven by inactivation of the tumor suppressor gene Apc) or lymphoma (driven by activation of the oncogene Myc), restricting dietary serine and glycine intake further improved survival by antagonizing the antioxidant response. Disruption of mitochondrial oxidative phosphorylation (using biguanides) results in complex responses that can either enhance or hinder the antitumor effects of serine and glycine starvation. Mouse models of pancreatic and colorectal cancers driven by the Kras oncogene exhibit a weaker response to serine and glycine depletion, reflecting Kras activation, which upregulates enzymes in the serine synthesis pathway and promotes de novo serine synthesis (10). Initial analyses of cancer cell lines found a close correlation between glycine consumption from the biosynthetic pathway and cell proliferation rates (22). However, subsequent studies revealed that serine, not glycine, is the amino acid consumed most rapidly for proliferation, with some cells resorting to glycine only when serine is depleted (8). Recent studies also show that inhibiting PHGDH enhances the therapeutic effects of a serine-deficient diet (96). Recent in vivo studies have demonstrated that combined dietary intervention and pharmacological inhibition exhibit significant therapeutic efficacy against tumors resistant to single-modality treatments, with evidence of reduced one-carbon metabolic flux (17). Concurrently, our findings reveal that serine deprivation in cancer cells induces mitochondrial dysfunction, leading to cytoplasmic accumulation of mitochondrial DNA (mtDNA) and activation of the cGAS-STING signaling pathway, thereby stimulating tumor-targeted immune responses. This effect is synergistically enhanced when combined with PD-1-targeted therapy (97).
Notably, the role of serine in cancer therapy exhibits duality: studies indicate that serine critically regulates estrogen receptor α (ESR1) gene expression by maintaining histone acetylation levels, and ESR1 expression directly determines the sensitivity of hormone receptor-positive breast cancers to endocrine therapies such as tamoxifen (98). Paradoxically, downstream metabolites of serine catabolism, including formate, exert beneficial effects on antitumor T cells. Formate supplementation synergizes with PD-1 blockade (99). Importantly, serine-restricted diets alone may inadvertently stimulate resistance mechanisms such as PD-L1 upregulation, underscoring the necessity to combine dietary interventions with PD-L1-targeted strategies (100).
3.3.2 PHGDH inhibitors
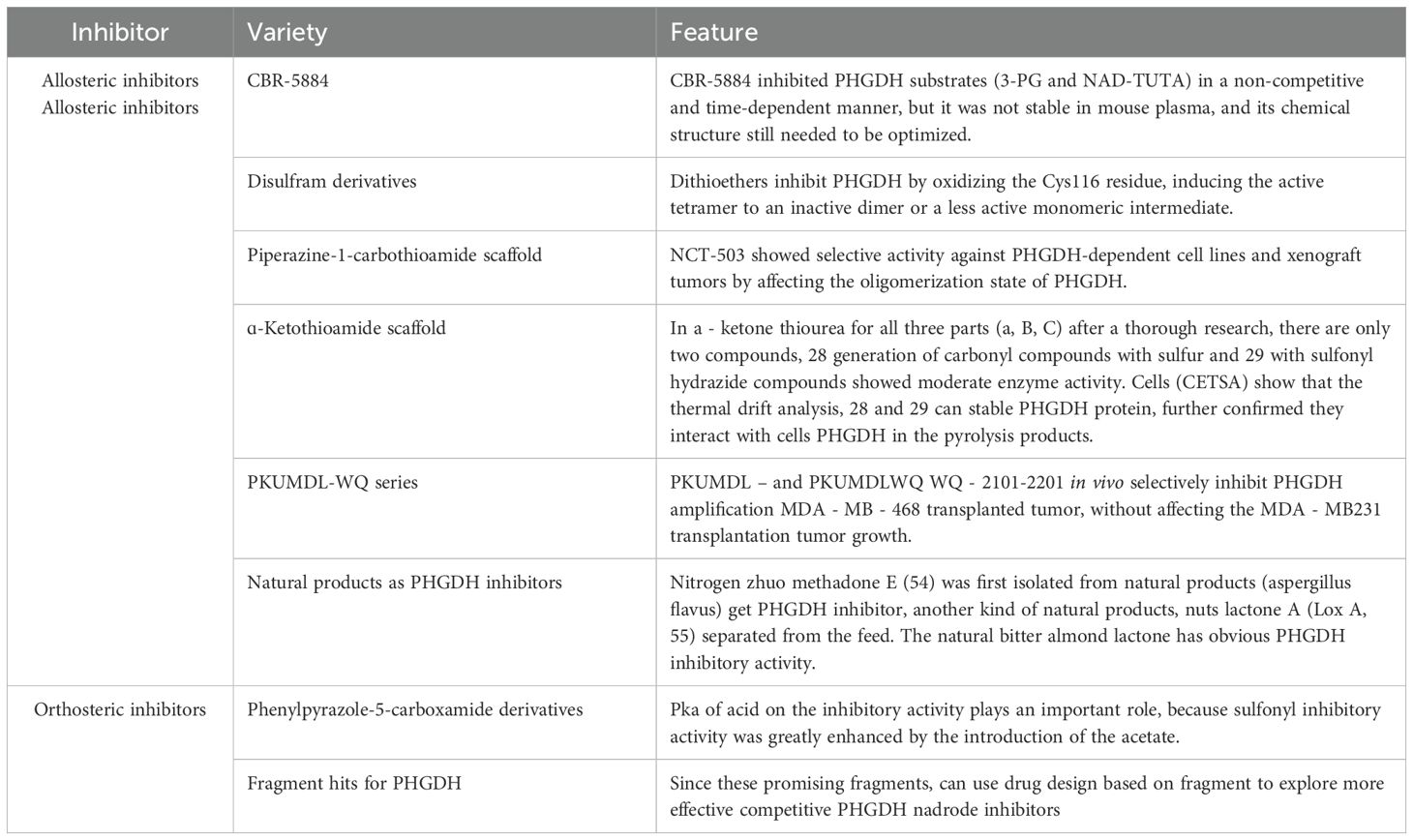
Research into PHGDH’s role in regulating cell growth has deepened, establishing it as a promising direction for targeted drug discovery (92). In 2018, Ishida et al. demonstrated that interfering with the SOG (serine synthesis pathway) using PHGDH inhibitors can induce synergistic cell death in vitro and in vivo (101). Studies show that CBR-5884, a PHGDH inhibitor, has dual effects: it inhibits cell proliferation and increases sensitivity to hypoxia-induced cell death (102). Recent studies indicate that PHGDH expression is reduced by 40-50% in 1p/19q-codeleted gliomas compared to non-codeleted gliomas, suggesting these gliomas have selective vulnerability to serine and glutathione depletion (103).
Numerous PHGDH inhibitors have been reported and can be classified into two categories based on their binding sites: allosteric inhibitors and orthosteric inhibitors. Allosteric inhibitors include the CBR-5884 series, bis-sulfonamide derivatives, piperazine-1-thiocarboxamide scaffolds, α-thio-oxamidamide scaffolds, and pyrazole-5-thiocarboxamide scaffolds, among others. Orthosteric inhibitors include indoleamide derivatives, phenyl-pyrazole-5-carboxamide derivatives, and certain fragments. The characteristics of these inhibitors are summarized in Table 2.
3.3.3 Other inhibitors
Serine hydroxymethyltransferase (SHMT), a pivotal enzyme in one-carbon metabolism, catalyzes the conversion of serine and tetrahydrofolate (THF) to glycine and 5,10-methylenetetrahydrofolate (5,10-CH2-THF), providing one-carbon units for nucleotide synthesis. SHMT is overexpressed in various cancers and strongly associated with tumor proliferation and chemoresistance (22). Recent advances in targeting SHMT have positioned it as a hotspot for metabolic therapy. Pemetrexed, an antifolate chemotherapeutic agent used in non-small cell lung cancer and breast cancer, competitively inhibits cytosolic SHMT1 activity by forming hydrogen bonds at the active site, mimicking THF binding. However, pemetrexed exhibits polypharmacology by targeting multiple one-carbon metabolic enzymes (104). SHIN1, a dual small-molecule inhibitor of SHMT1/2, achieves suppression via competitive THF binding and hydrogen bond interactions between its –NH group and SHMT1/2 (105). Notably, the antidepressant sertraline has recently been identified as a competitive dual SHMT1/2 inhibitor through direct binding to both isoforms (106).
MTHFD2, whose expression is upregulated in tumors such as breast and colorectal cancers, correlates with poor survival (44, 107). DS18561882, a potent and orally bioavailable MTHFD2 inhibitor with demonstrated in vivo antitumor efficacy, has emerged as a promising candidate for breast cancer treatment. However, its biological roles in other malignancies remain to be elucidated (108).
3.4 Traditional Chinese medicine affects serine metabolism
The biosynthesis of serine and glycine is crucial in tumorigenesis (54). Research indicates that a deficiency in serine and glycine reduces glutathione synthesis and increases reactive oxygen species (ROS) levels in cells (32). Moreover, downregulating the rate-limiting enzymes of serine/glycine metabolism, such as phosphoserine phosphatase (PSPH) and phosphoserine aminotransferase 1 (PSAT1), may disrupt the pro-tumor effects mediated by serine/glycine metabolism (7, 75). Targeting amino acid metabolism shows promise for discovering selective inhibitors and new strategies for cancer treatment (109). Recent studies suggest that certain traditional Chinese medicine (TCM) herbs and compounds may exhibit antitumor effects by targeting amino acid metabolic pathways, highlighting TCM’s potential in cancer therapy. Licorice is a typical TCM herb commonly used to treat inflammation and allergies. Recent research suggests that licorice root extract exhibits anticancer effects in nasopharyngeal carcinoma cells. Metabolomic analysis revealed that these anticancer effects may relate to the downregulation of fatty acid biosynthesis metabolites and reductions in glutamic acid, serine, and threonine metabolism (110). Tanshinones, bioactive constituents derived from Salvia miltiorrhiza, have recently been demonstrated to upregulate PHGDH mRNA expression levels (111). Unfortunately, research on how TCM compounds or decoctions regulate serine metabolism and their effects on cancer cell invasion and migration remains scarce. The mechanisms remain unclear and warrant further in-depth studies.
4 Summary and outlook
Serine metabolism is crucial in tumor development, and tumor-related biomarkers of this metabolism show promise for clinical applications. In the serine synthesis pathway, key enzymes like PHGDH and PSPH are upregulated in specific tumor types (51, 52, 112). Abnormal expression of PHGDH and PSPH may enhance serine synthesis in tumor cells, positioning them as potential diagnostic biomarkers. Serine metabolic enzymes have also gained attention in tumor research. SHMT2, one of the key enzymes in serine metabolism, is upregulated in certain tumors (113–117). Serine aminotransferase 1 (SAT1) is closely linked to serine metabolism, with abnormal expression associated with tumor development (118, 119). The abnormal expression of these enzymes may affect the metabolic pathways of tumor cells, further influencing tumor proliferation and survival. Thus, they may serve as potential markers for tumor diagnosis and treatment. Research on serine and its metabolites has significant application potential. Alterations in glycine and cysteine concentrations may be linked to tumor development and metabolic disorders (120, 121). In various cancers, serine metabolism, one-carbon (1C) metabolism, and mTOR signaling pathways show abnormal hyperactivation (32, 122, 123). Serine and one-carbon metabolism may serve as key links between mTOR signaling and DNA methylation, promoting tumor growth (124). It is noteworthy that although PHGDH is often amplified in various types of cancer, therapeutic interventions targeting PHGDH are far more complex than initially imagined. This complexity stems from the fact that PHGDH has both tumor-promoting and potential anti-metastatic effects (125, 126). Additionally, exogenous serine can sometimes compensate for the deficiencies caused by the loss of serine biosynthesis (127–129). These findings imply that simply inhibiting PHGDH may not suffice for effective tumor treatment due to its complex dual role in tumor development. Thus, developing effective therapeutic strategies targeting PHGDH necessitates a deeper understanding of its specific functions and regulatory mechanisms in tumors, along with its effects on normal cells. Furthermore, supplementing exogenous serine may pose challenges to therapeutic interventions. Although serine supply can be externally supplemented, achieving a balance in the serine metabolic pathway remains complex for tumor cells. Therefore, relying solely on exogenous serine as a treatment strategy may be infeasible, necessitating comprehensive consideration of the overall regulation of the serine metabolic pathway. Combining these biomarkers with other clinical indicators could enhance the efficacy of tumor diagnosis and analysis. Investigating the link between serine metabolism and tumor development, identifying additional biomarkers, and enhancing research on serine metabolism and related pathways will deepen our understanding of tumor mechanisms and potentially yield new targets and strategies for diagnosis and treatment.
Author contributions
HL: Writing – original draft. SB: Writing – review & editing. LC: Writing – review & editing. MW: Writing – review & editing. YL: Writing – review & editing. YS: Writing – review & editing, Supervision. XH: Writing – review & editing, Supervision, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the National Nature Science Foundation of China (Grant No. 81202638, 81704054).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wu Q, Chen X, Li J, Sun S. Serine and metabolism regulation: A novel mechanism in antitumor immunity and senescence. Aging Dis. (2020) 11:1640–53. doi: 10.14336/AD.2020.0314
2. Holeček M. Serine metabolism in health and disease and as a conditionally essential amino acid. Nutrients. (2022) 14(9):1987. doi: 10.3390/nu14091987
3. Icard P, Shulman S, Farhat D, Steyaert JM, Alifano M, Lincet H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist Update. (2018) 38:1–11. doi: 10.1016/j.drup.2018.03.001
4. Anderson NM, Mucka P, Kern JG, Feng H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell. (2018) 9:216–37. doi: 10.1007/s13238-017-0451-1
5. Hosios AM, Hecht VC, Danai LV, Johnson MO, Rathmell JC, Steinhauser ML, et al. Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Dev Cell. (2016) 36:540–9. doi: 10.1016/j.devcel.2016.02.012
6. Snell K. Enzymes of serine metabolism in normal, developing and neoplastic rat tissues. Adv Enzyme Regul. (1984) 22:325–400. doi: 10.1016/0065-2571(84)90021-9
7. Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. (2011) 476:346–50. doi: 10.1038/nature10350
8. Labuschagne CF, Van Den Broek NJ, Mackay GM, Vousden KH, Maddocks ODK. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. (2014) 7:1248–58. doi: 10.1016/j.celrep.2014.04.045
9. Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. (2013) 493:542–6. doi: 10.1038/nature11743
10. Maddocks ODK, Athineos D, Cheung EC, Lee P, Zhang T, van den Broek NJF, et al. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature. (2017) 544:372–6. doi: 10.1038/nature22056
11. Muthusamy T, Cordes T, Handzlik MK, You L, Lim EW, Gengatharan J, et al. Serine restriction alters sphingolipid diversity to constrain tumour growth. Nature. (2020) 586:790–5. doi: 10.1038/s41586-020-2609-x
12. Li AM, Ye J. The PHGDH enigma: Do cancer cells only need serine or also a redox modulator? Cancer Lett. (2020) 476:97–105. doi: 10.1016/j.canlet.2020.01.036
13. Newman AC, Maddocks ODK. One-carbon metabolism in cancer. Br J Cancer. (2017) 116:1499–504. doi: 10.1038/bjc.2017.118
14. Lan X, Field MS, Stover PJ. Cell cycle regulation of folate-mediated one-carbon metabolism. Wiley Interdiscip Rev Syst Biol Med. (2018) 10:e1426. doi: 10.1002/wsbm.1426
15. Clare CE, Brassington AH, Kwong WY, Sinclair KD. One-carbon metabolism: linking nutritional biochemistry to epigenetic programming of long-term development. Annu Rev Anim Biosci. (2019) 7:263–87. doi: 10.1146/annurev-animal-020518-115206
16. Pan S, Fan M, Liu Z, Li X, Wang H. Serine, glycine and one−carbon metabolism in cancer (Review). Int J Oncol. (2021) 58:158–70. doi: 10.3892/ijo.2020.5158
17. Tajan M, Hennequart M, Cheung EC, Zani F, Hock AK, Legrave N, et al. Serine synthesis pathway inhibition cooperates with dietary serine and glycine limitation for cancer therapy. Nat Commun. (2021) 12:366. doi: 10.1038/s41467-020-20223-y
18. Li X, Gracilla D, Cai L, Zhang M, Yu X, Chen X, et al. ATF3 promotes the serine synthesis pathway and tumor growth under dietary serine restriction. Cell Rep. (2021) 36:109706. doi: 10.1016/j.celrep.2021.10970
19. Jeong S, Savino AM, Chirayil R, Barin E, Cheng Y, Park SM, et al. High fructose drives the serine synthesis pathway in acute myeloid leukemic cells. Cell Metab. (2021) 33:145–59.e6. doi: 10.1016/j.cmet.2020.12.005
20. Sánchez-Castillo A, Heylen E, Hounjet J, Savelkouls KG, Lieuwes NG, Biemans R, et al. Targeting serine/glycine metabolism improves radiotherapy response in non-small cell lung cancer. Br J Cancer. (2024) 130:568–84. doi: 10.1038/s41416-023-02553-y
21. Li L, Qin Y, Chen Y. The enzymes of serine synthesis pathway in cancer metastasis. Biochim Biophys Acta Mol Cell Res. (2024) 1871:119697. doi: 10.1016/j.bbamcr.2024.119697
22. Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. (2012) 336:1040–4. doi: 10.1126/science.1218595
23. Cissé MY, Pyrdziak S, Firmin N, Gayte L, Heuillet M, Bellvert F, et al. Targeting MDM2-dependent serine metabolism as a therapeutic strategy for liposarcoma. Sci Transl Med. (2020) 12(547):eaay2163. doi: 10.1126/scitranslmed.aay2163
24. Utsunomiya-Tate N, Endou H, Kanai Y. Cloning and functional characterization of a system ASC-like Na+-dependent neutral amino acid transporter. J Biol Chem. (1996) 271:14883–90. doi: 10.1074/jbc.271.25.14883
25. Arriza JL, Kavanaugh MP, Fairman WA, Wu YN, Murdoch GH, North RA, et al. Cloning and expression of a human neutral amino acid transporter with structural similarity to the glutamate transporter gene family. J Biol Chem. (1993) 268:15329–32. doi: 10.1016/S0021-9258(18)82257-8
26. Christensen HN, Liang M, Archer EG. A distinct Na+-requiring transport system for alanine, serine, cysteine, and similar amino acids. J Biol Chem. (1967) 242:5237–46. doi: 10.1016/S0021-9258(18)99417-2
27. Conger KO, Chidley C, Ozgurses ME, Zhao H, Kim Y, Semina SE, et al. ASCT2 is a major contributor to serine uptake in cancer cells. Cell Rep. (2024) 43:114552. doi: 10.1016/j.celrep.2024.114552
28. Ma EH, Bantug G, Griss T, Condotta S, Johnson RM, Samborska B, et al. Serine is an essential metabolite for effector T cell expansion. Cell Metab. (2017) 25:345–57. doi: 10.1016/j.cmet.2016.12.011
29. Wang W, Zou W. Amino acids and their transporters in T cell immunity and cancer therapy. Mol Cell. (2020) 80:384–95. doi: 10.1016/j.molcel.2020.09.006
30. Kaplan E, Zubedat S, Radzishevsky I, Valenta AC, Rechnitz O, Sason H, et al. ASCT1 (Slc1a4) transporter is a physiologic regulator of brain d-serine and neurodevelopment. Proc Natl Acad Sci U S A. (2018) 115:9628–33. doi: 10.1073/pnas.1722677115
31. Papalazarou V, Newman AC, Huerta-Uribe A, Legrave NM, Falcone M, Zhang T, et al. Phenotypic profiling of solute carriers characterizes serine transport in cancer. Nat Metab. (2023) 5:2148–68. doi: 10.1038/s42255-023-00936-2
32. Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. (2013) 13:572–83. doi: 10.1038/nrc3557
33. Li M, Wu C, Yang Y, Zheng M, Yu S, Wang J, et al. 3-Phosphoglycerate dehydrogenase: a potential target for cancer treatment. Cell Oncol (Dordr). (2021) 44:541–56. doi: 10.1007/s13402-021-00599-9
34. Mattaini KR, Sullivan MR, Vander Heiden MG. The importance of serine metabolism in cancer. J Cell Biol. (2016) 214:249–57. doi: 10.1083/jcb.201604085
35. Pacold ME, Brimacombe KR, Chan SH, Rohde JM, Lewis CA, Swier LJYM, et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol. (2016) 12:452–8. doi: 10.1038/nchembio.2070
36. Geeraerts SL, Heylen E, De Keersmaecker K, Kampen KR. The ins and outs of serine and glycine metabolism in cancer. Nat Metab. (2021) 3:131–41. doi: 10.1038/s42255-020-00329-9
37. Yang L, Garcia Canaveras JC, Chen Z, Wang L, Liang L, Jang C, et al. Serine catabolism feeds NADH when respiration is impaired. Cell Metab. (2020) 31:809–21.e6. doi: 10.1016/j.cmet.2020.02.017
38. Zhang Z, Teslaa T, Xu X, Zeng X, Yang L, Xing G, et al. Serine catabolism generates liver NADPH and supports hepatic lipogenesis. Nat Metab. (2021) 3:1608–20. doi: 10.1038/s42255-021-00487-4
39. Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discovery. (2014) 4:1406–17. doi: 10.1158/2159-8290.CD-14-0250
40. Liu Y, Yin C, Deng MM, Wang Q, He XQ, Li MT. High expression of SHMT2 is correlated with tumor progression and predicts poor prognosis in gastrointestinal tumors. Eur Rev Med Pharmacol Sci. (2019) 23:9379–92. doi: 10.26355/eurrev_201911_19431
41. Ning S, Ma S, Saleh AQ, Guo L, Zhao Z, Chen Y. SHMT2 overexpression predicts poor prognosis in intrahepatic cholangiocarcinoma. Gastroenterol Res Pract. (2018) 2018:4369253. doi: 10.1155/2018/4369253
42. Noguchi K, Konno M, Koseki J, Nishida N, Kawamoto K, Yamada D, et al. The mitochondrial one-carbon metabolic pathway is associated with patient survival in pancreatic cancer. Oncol Lett. (2018) 16:1827–34. doi: 10.3892/ol.2018.8795
43. Yin K. Positive correlation between expression level of mitochondrial serine hydroxymethyltransferase and breast cancer grade. Onco Targets Ther. (2015) 8:1069–74. doi: 10.2147/OTT.S82433
44. Liu F, Liu Y, He C, Tao L, He X, Song H, et al. Increased MTHFD2 expression is associated with poor prognosis in breast cancer. Tumour Biol. (2014) 35:8685–90. doi: 10.1007/s13277-014-2111-x
45. Kim D, Fiske BP, Birsoy K, Freinkman E, Kami K, Possemato RL, et al. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. (2015) 520:363–7. doi: 10.1038/nature14363
46. Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. (2016) 351:728–33. doi: 10.1126/science.aad0489
47. Chen L, Zhang Z, Hoshino A, Zheng HD, Morley M, Arany Z, et al. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat Metab. (2019) 1:404–15. doi: 10.1038/s42255-019-0043-x
48. Peng H, Wang Y, Luo W. Multifaceted role of branched-chain amino acid metabolism in cancer. Oncogene. (2020) 39:6747–56. doi: 10.1038/s41388-020-01480-z
49. Najumudeen AK, Ceteci F, Fey SK, Hamm G, Steven RT, Hall H, et al. The amino acid transporter SLC7A5 is required for efficient growth of KRAS-mutant colorectal cancer. Nat Genet. (2021) 53:16–26. doi: 10.1038/s41588-020-00753-3
50. Li X, Zhang HS. Amino acid metabolism, redox balance and epigenetic regulation in cancer. FEBS J. (2024) 291:412–29. doi: 10.1111/febs.v291.3
51. Mullen AR, Deberardinis RJ. Genetically-defined metabolic reprogramming in cancer. Trends Endocrinol Metab. (2012) 23:552–9. doi: 10.1016/j.tem.2012.06.009
52. Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. (2005) 5:857–66. doi: 10.1038/nrc1737
53. Luo W, Zou Z, Nie Y, Luo J, Ming Z, Hu X, et al. ASS1 inhibits triple-negative breast cancer by regulating PHGDH stability and de novo serine synthesis. Cell Death Dis. (2024) 15:319. doi: 10.1038/s41419-024-06672-z
54. Deberardinis RJ. Serine metabolism: some tumors take the road less traveled. Cell Metab. (2011) 14:285–6. doi: 10.1016/j.cmet.2011.08.004
55. Locasale JW, Cantley LC. Genetic selection for enhanced serine metabolism in cancer development. Cell Cycle. (2011) 10:3812–3. doi: 10.4161/cc.10.22.18224
56. Fan J, Teng X, Liu L, Mattaini KR, Looper RE, Vander Heiden MG, et al. Human phosphoglycerate dehydrogenase produces the oncometabolite D-2-hydroxyglutarate. ACS Chem Biol. (2015) 10(2):510–6. doi: 10.1021/cb500683c
57. Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. (2011) 43:869–74. doi: 10.1038/ng.890
58. Mullarky E, Mattaini KR, Vander Heiden MG, Cantley LC, Locasale JW. PHGDH amplification and altered glucose metabolism in human melanoma. Pigment Cell Melanoma Res. (2011) 24:1112–5. doi: 10.1111/j.1755-148X.2011.00919.x
59. Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. (2009) 137:413–31. doi: 10.1016/j.cell.2009.04.037
61. Yang M, Vousden KH. Serine and one-carbon metabolism in cancer. Nat Rev Cancer. (2016) 16:650–62. doi: 10.1038/nrc.2016.81
62. Yang Y, Wu J, Cai J, He Z, Yuan J, Zhu X, et al. PSAT1 regulates cyclin D1 degradation and sustains proliferation of non-small cell lung cancer cells. Int J Cancer. (2015) 136:E39–50. doi: 10.1002/ijc.v136.4
63. Zhu S, Wang X, Liu L, Ren G. Stabilization of Notch1 and β-catenin in response to ER- breast cancer-specific up-regulation of PSAT1 mediates distant metastasis. Transl Oncol. (2022) 20:101399. doi: 10.1016/j.tranon.2022.101399
64. Ma J, Zhu M, Ye X, Wu B, Wang T, Ma M, et al. Prognostic microRNAs associated with phosphoserine aminotransferase 1 in gastric cancer as markers of bone metastasis. Front Genet. (2022) 13:959684. doi: 10.3389/fgene.2022.959684
65. Montrose DC, Saha S, Foronda M, McNally EM, Chen J, Zhou XK, et al. Exogenous and endogenous sources of serine contribute to colon cancer metabolism, growth, and resistance to 5-fluorouracil. Cancer Res. (2021) 81:2275–88. doi: 10.1158/0008-5472.CAN-20-1541
66. Zhang Y, Li J, Dong X, Meng D, Zhi X, Yuan L, et al. PSAT1 regulated oxidation-reduction balance affects the growth and prognosis of epithelial ovarian cancer. Onco Targets Ther. (2020) 13:5443–53. doi: 10.2147/OTT.S250066
67. Luo MY, Zhou Y, Gu WM, Wang C, Shen NX, Dong JK. Metabolic and nonmetabolic functions of PSAT1 coordinate signaling cascades to confer EGFR inhibitor resistance and drive progression in lung adenocarcinoma. Cancer Res. (2022) 82:3516–31. doi: 10.1158/0008-5472.CAN-21-4074
68. Metcalf S, Dougherty S, Kruer T, Hasan N, Biyik-Sit R, Reynolds L, et al. Selective loss of phosphoserine aminotransferase 1 (PSAT1) suppresses migration, invasion, and experimental metastasis in triple negative breast cancer. Clin Exp Metastasis. (2020) 37:187–97. doi: 10.1007/s10585-019-10000-7
69. Vié N, Copois V, Bascoul-Mollevi C, Denis V, Bec N, Robert B, et al. Overexpression of phosphoserine aminotransferase PSAT1 stimulates cell growth and increases chemoresistance of colon cancer cells. Mol Cancer. (2008) 7:14. doi: 10.1186/1476-4598-7-14
70. Kottakis F, Nicolay BN, Roumane A, Karnik R, Gu H, Nagle JM, et al. LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature. (2016) 539:390–5. doi: 10.1038/nature20132
71. Liu B, Jia Y, Cao Y, Wu S, Jiang H, Sun X, et al. Overexpression of phosphoserine aminotransferase 1 (PSAT1) predicts poor prognosis and associates with tumor progression in human esophageal squamous cell carcinoma. Cell Physiol Biochem. (2016) 39:395–406. doi: 10.1159/000445633
72. De Marchi T, Timmermans MA, Sieuwerts AM, Smid M, Look MP, Grebenchtchikov N, et al. Phosphoserine aminotransferase 1 is associated to poor outcome on tamoxifen therapy in recurrent breast cancer. Sci Rep. (2017) 7:2099. doi: 10.1038/s41598-017-02296-w
73. Gao S, Ge A, Xu S, You Z, Ning S, Zhao Y, et al. PSAT1 is regulated by ATF4 and enhances cell proliferation via the GSK3β/β-catenin/cyclin D1 signaling pathway in ER-negative breast cancer. J Exp Clin Cancer Res. (2017) 36:179. doi: 10.1186/s13046-017-0648-4
74. Petri BJ, Piell KM, Wilt AE, Howser AD, Winkler L, Whitworth MR, et al. MicroRNA regulation of the serine synthesis pathway in endocrine-resistant breast cancer cells. Endocr Relat Cancer. (2023) 30(11):e230148. doi: 10.1530/ERC-23-0148
75. Ye J, Huang X, Tian S, Wang J, Wang H, Feng H, et al. Upregulation of serine metabolism enzyme PSAT1 predicts poor prognosis and promotes proliferation, metastasis and drug resistance of clear cell renal cell carcinoma. Exp Cell Res. (2024) 437:113977. doi: 10.1016/j.yexcr.2024.113977
76. Zheng MJ, Li X, Hu YX, Dong H, Gou R, Nie X, et al. Identification of molecular marker associated with ovarian cancer prognosis using bioinformatics analysis and experiments. J Cell Physiol. (2019) 234:11023–36. doi: 10.1002/jcp.v234.7
77. Battula P, Dubnovitsky AP, Papageorgiou AC. Structural basis of L-phosphoserine binding to Bacillus alcalophilus phosphoserine aminotransferase. Acta Crystallogr D Biol Crystallogr. (2013) 69:804–11. doi: 10.1107/S0907444913002096
78. Rawat V, Malvi P, Della Manna D, Yang ES, Bugide S, Zhang X, et al. PSPH promotes melanoma growth and metastasis by metabolic deregulation-mediated transcriptional activation of NR4A1. Oncogene. (2021) 40:2448–62. doi: 10.1038/s41388-021-01683-y
79. Wang CY, Chiao CC, Phan NN, Li CY, Sun ZD, Jiang JZ, et al. Gene signatures and potential therapeutic targets of amino acid metabolism in estrogen receptor-positive breast cancer. Am J Cancer Res. (2020) 10:95–113.
80. Zhang J, Wang E, Zhang L, Zhou B. PSPH induces cell autophagy and promotes cell proliferation and invasion in the hepatocellular carcinoma cell line Huh7 via the AMPK/mTOR/ULK1 signaling pathway. Cell Biol Int. (2021) 45:305–19. doi: 10.1002/cbin.11489
81. Sato K, Masuda T, Hu Q, Tobo T, Kidogami S, Ogawa Y, et al. Phosphoserine phosphatase is a novel prognostic biomarker on chromosome 7 in colorectal cancer. Anticancer Res. (2017) 37:2365–71. doi: 10.21873/anticanres.11574
82. Liao L, Ge M, Zhan Q, Huang R, Ji X, Liang X, et al. PSPH mediates the metastasis and proliferation of non-small cell lung cancer through MAPK signaling pathways. Int J Biol Sci. (2019) 15:183–94. doi: 10.7150/ijbs.29203
83. Bachelor MA, Lu Y, Owens DM. L-3-Phosphoserine phosphatase (PSPH) regulates cutaneous squamous cell carcinoma proliferation independent of L-serine biosynthesis. J Dermatol Sci. (2011) 63:164–72. doi: 10.1016/j.jdermsci.2011.06.001
84. Sánchez-Castillo A, Savelkouls KG, Baldini A, Hounjet J, Sonveaux P, Verstraete P, et al. Sertraline/chloroquine combination therapy to target hypoxic and immunosuppressive serine/glycine synthesis-dependent glioblastomas. Oncogenesis. (2024) 13:39. doi: 10.1038/s41389-024-00540-3
85. Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. (2008) 40:14–21. doi: 10.1016/j.biocel.2007.01.020
86. Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. (2003) 11:619–33. doi: 10.1016/S1097-2765(03)00105-9
87. Denicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. N at Genet. (2015) 47:1475–81. doi: 10.1038/ng.3421
88. Yoon BK, Kim H, Oh TG, Oh SK, Jo S, Kim M, et al. PHGDH preserves one-carbon cycle to confer metabolic plasticity in chemoresistant gastric cancer during nutrient stress. Proc Natl Acad Sci U S A. (2023) 120:e2217826120. doi: 10.1073/pnas.2217826120
89. Dai Z, Chen L, Pan K, Zhao X, Xu W, Du J, et al. Multi-omics analysis of the role of PHGDH in colon cancer. Technol Cancer Res Treat. (2023) 22:15330338221145994. doi: 10.1177/15330338221145994
90. Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. (2016) 17:1374–95. doi: 10.15252/embr.201642195
91. Han S, Zhu L, Zhu Y, Meng Y, Li J, Song P, et al. Targeting ATF4-dependent pro-survival autophagy to synergize glutaminolysis inhibition. Theranostics. (2021) 11:8464–79. doi: 10.7150/thno.60028
92. Lee CM, Hwang Y, Kim M, Park YC, Kim H, Fang S. PHGDH: a novel therapeutic target in cancer. Exp Mol Med. (2024) 56:1513–22. doi: 10.1038/s12276-024-01268-1
93. Peng X, Chen R, Cai S, Lu S, Zhang Y. SLC1A4: A powerful prognostic marker and promising therapeutic target for HCC. Front Oncol. (2021) 11:650355. doi: 10.3389/fonc.2021.650355
94. Klysz D, Tai X, Robert PA, Craveiro M, Cretenet G, Oburoglu L, et al. Glutamine-dependent α-ketoglutarate production regulates the balance between T helper 1 cell and regulatory T cell generation. Sci Signal. (2015) 8:ra97. doi: 10.1126/scisignal.aab2610
95. Gravel SP, Hulea L, Toban N, Birman E, Blouin MJ, Zakikhani M, et al. Serine deprivation enhances antineoplastic activity of biguanides. Cancer Res. (2014) 74:7521–33. doi: 10.1158/0008-5472.CAN-14-2643-T
96. Reina-Campos M, Diaz-Meco MT, Moscat J. The complexity of the serine glycine one-carbon pathway in cancer. J Cell Biol. (2020) 219(1):e201907022. doi: 10.1083/jcb.201907022
97. Saha S, Ghosh M, Li J, Birman E, Blouin MJ, Zakikhani M, et al. Serine depletion promotes antitumor immunity by activating mitochondrial DNA-mediated cGAS-STING signaling. Cancer Res. (2024) 84:2645–59. doi: 10.1158/0008-5472.CAN-23-1788
98. Li AM, He B, Karagiannis D, Li Y, Jiang H, Srinivasan P, et al. Serine starvation silences estrogen receptor signaling through histone hypoacetylation. Proc Natl Acad Sci U S A. (2023) 120:e2302489120. doi: 10.1073/pnas.2302489120
99. Xu X, Chen Z, Bartman CR, Xing X, Olszewski K, Rabinowitz JD. One-carbon unit supplementation fuels purine synthesis in tumor-infiltrating T cells and augments checkpoint blockade. Cell Chem Biol. (2024) 31:932–43.e8. doi: 10.1016/j.chembiol.2024.04.007
100. Tong H, Jiang Z, Song L, Tan K, Yin X, He C, et al. Dual impacts of serine/glycine-free diet in enhancing antitumor immunity and promoting evasion via PD-L1 lactylation. Cell Metab. (2024) 36:2493–510.e9. doi: 10.1016/j.cmet.2024.10.019
101. Ishida CT, Zhang Y, Bianchetti E, Shu C, Nguyen TTT, Kleiner G, et al. Metabolic reprogramming by dual AKT/ERK inhibition through imipridones elicits unique vulnerabilities in glioblastoma. Clin Cancer Res. (2018) 24:5392–406. doi: 10.1158/1078-0432.CCR-18-1040
102. Engel AL, Lorenz NI, Klann K, Münch C, Depner C, Steinbach JP, et al. Serine-dependent redox homeostasis regulates glioblastoma cell survival. Br J Cancer. (2020) 122:1391–8. doi: 10.1038/s41416-020-0794-x
103. Branzoli F, Pontoizeau C, Tchara L, Stefano AL, Kamoun A, Deelchand DK, et al. Cystathionine as a marker for 1p/19q codeleted gliomas by in vivo magnetic resonance spectroscopy. Neuro Oncol. (2019) 21:765–74. doi: 10.1093/neuonc/noz031
104. Daidone F, Florio R, Rinaldo S, Contestabile R, di Salvo ML, Cutruzzolà F, et al. In silico and in vitro validation of serine hydroxymethyltransferase as a chemotherapeutic target of the antifolate drug pemetrexed. Eur J Med Chem. (2011) 46:1616–21. doi: 10.1016/j.ejmech.2011.02.009
105. Han T, Wang Y, Cheng M, Hu Q, Wan X, Huang M, et al. Phosphorylated SHMT2 regulates oncogenesis through m(6)A modification in lung adenocarcinoma. Adv Sci (Weinh). (2024) 11:e2307834. doi: 10.1002/advs.202307834
106. Geeraerts SL, Kampen KR, Rinaldi G, Gupta P, Planque M, Louros N, et al. Repurposing the antidepressant sertraline as SHMT inhibitor to suppress serine/glycine synthesis-addicted breast tumor growth. Mol Cancer Ther. (2021) 20:50–63. doi: 10.1158/1535-7163.MCT-20-0480
107. Nilsson R, Jain M, Madhusudhan N, Gustafsson Sheppard N, Strittmatter L, Kampf, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. (2014) 5:3128. doi: 10.1038/ncomms4128
108. Kawai J, Toki T, Ota M, Inoue H, Takata Y, Asahi T, et al. Discovery of a potent, selective, and orally available MTHFD2 inhibitor (DS18561882) with in vivo antitumor activity. J Med Chem. (2019) 62:10204–20. doi: 10.1021/acs.jmedchem.9b01113
109. Lukey MJ, Katt WP, Cerione RA. Targeting amino acid metabolism for cancer therapy. Drug Discovery Today. (2017) 22:796–804. doi: 10.1016/j.drudis.2016.12.003
110. Zheng C, Han L, Wu S. A metabolic investigation of anticancer effect of G. glabra root extract on nasopharyngeal carcinoma cell line, C666-1. Mol Biol Rep. (2019) 46:3857–64. doi: 10.1007/s11033-019-04828-1
111. Wang L, Cheng L, Zhang B, Wang N, Wang F. Tanshinone prevents alveolar bone loss in ovariectomized osteoporosis rats by up-regulating phosphoglycerate dehydrogenase. Toxicol Appl Pharmacol. (2019) 376:9–16. doi: 10.1016/j.taap.2019.05.014
112. Peng ZP, Liu XC, Ruan YH, Jiang D, Huang AQ, Ning WR. Downregulation of phosphoserine phosphatase potentiates tumor immune environments to enhance immune checkpoint blockade therapy. J Immunother Cancer. (2023) 11(2):e005986. doi: 10.1136/jitc-2022-005986
113. Liu C, Wang L, Liu X, Tan Y, Tao L, Xiao Y. Cytoplasmic SHMT2 drives the progression and metastasis of colorectal cancer by inhibiting β-catenin degradation. Theranostics. (2021) 11:2966–86. doi: 10.7150/thno.48699
114. Wang W, Wang M, Du T, Hou Z, You S, Zhang S, et al. SHMT2 promotes gastric cancer development through regulation of HIF1α/VEGF/STAT3 signaling. Int J Mol Sci. (2023) 24(8):7150. doi: 10.3390/ijms24087150
115. Qiao Z, Li Y, Cheng Y, Liu S. SHMT2 regulates esophageal cancer cell progression and immune Escape by mediating m6A modification of c-myc. Cell Biosci. (2023) 13:203. doi: 10.1186/s13578-023-01148-7
116. Shan Y, Liu D, Li Y, Wu C, Ye Y. The expression and clinical significance of serine hydroxymethyltransferase2 in gastric cancer. PeerJ. (2024) 12:e16594. doi: 10.7717/peerj.16594
117. Su SW, Chen X, Wang G, Li P, Yang TX, Fang KW, et al. A study on the significance of serine hydroxymethyl transferase expression and its role in bladder cancer. Sci Rep. (2024) 14:8324. doi: 10.1038/s41598-024-58618-2
118. Mou Y, Zhang L, Liu Z, Song X. Abundant expression of ferroptosis-related SAT1 is related to unfavorable outcome and immune cell infiltration in low-grade glioma. BMC Cancer. (2022) 22:215. doi: 10.1186/s12885-022-09313-w
119. Sui X, Hu N, Zhang Z, Wang Y, Wang P, Xiu G. ASMTL-AS1 impedes the Malignant progression of lung adenocarcinoma by regulating SAT1 to promote ferroptosis. Pathol Int. (2021) 71:741–51. doi: 10.1111/pin.v71.11
120. Li AM, Ye J. Reprogramming of serine, glycine and one-carbon metabolism in cancer. Biochim Biophys Acta Mol Basis Dis. (2020) 1866:165841. doi: 10.1016/j.bbadis.2020.165841
121. Wang D, Wan X. Progress in research on the role of amino acid metabolic reprogramming in tumour therapy: A review. BioMed Pharmacother. (2022) 156:113923. doi: 10.1016/j.biopha.2022.113923
122. Menon S, Manning BD. Common corruption of the mTOR signaling network in human tumors. Oncogene. (2008) 27 Suppl 2:S43–51. doi: 10.1038/onc.2009.352
123. Zhang Y, Kwok-Shing-Ng P, Kucherlapati M, Chen F, Liu Y, Tsang YH. A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell. (2017) 31:820–32.e3. doi: 10.1016/j.ccell.2017.04.013
124. Zeng JD, Wu WKK, Wang HY, Li XX. Serine and one-carbon metabolism, a bridge that links mTOR signaling and DNA methylation in cancer. Pharmacol Res. (2019) 149:104352. doi: 10.1016/j.phrs.2019.104352
125. Kim HM, Jung WH, Koo JS. Site-specific metabolic phenotypes in metastatic breast cancer. J Transl Med. (2014) 12:354. doi: 10.1186/s12967-014-0354-3
126. Rossi M, Altea-Manzano P, Demicco M, Doglioni G, Bornes L, Fukano M. PHGDH heterogeneity potentiates cancer cell dissemination and metastasis. Nature. (2022) 605:747–53. doi: 10.1038/s41586-022-04758-2
127. Di Marcantonio D, Martinez E, Kanefsky JS, Huhn JM, Gabbasov R, Gupta A, et al. ATF3 coordinates serine and nucleotide metabolism to drive cell cycle progression in acute myeloid leukemia. Mol Cell. (2021) 81:2752–64.e6. doi: 10.1016/j.molcel.2021.05.008
128. Diehl FF, Lewis CA, Fiske BP, Vander Heiden MG. Cellular redox state constrains serine synthesis and nucleotide production to impact cell proliferation. Nat Metab. (2019) 1:861–7. doi: 10.1038/s42255-019-0108-x
Keywords: serine metabolism, cancer, one-carbon metabolism, serine catabolism, the immunosuppressive microenvironment
Citation: Lyu H, Bao S, Cai L, Wang M, Liu Y, Sun Y and Hu X (2025) The role and research progress of serine metabolism in tumor cells. Front. Oncol. 15:1509662. doi: 10.3389/fonc.2025.1509662
Received: 11 October 2024; Accepted: 21 March 2025;
Published: 08 April 2025.
Edited by:
Trygve Tollefsbol, University of Alabama at Birmingham, United StatesReviewed by:
Xia Ge, Washington University in St. Louis, United StatesErhu Zhao, Southwest University, China
Albert Li, Stanford University, United States
Copyright © 2025 Lyu, Bao, Cai, Wang, Liu, Sun and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Sun, c3VueWFuZ0BobGp1Y20uZWR1LmNu; Xiaoyang Hu, cHNhMTk4MUAxMjYuY29t
 Hanning Lyu
Hanning Lyu Shuchang Bao
Shuchang Bao Lingyun Cai
Lingyun Cai