
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 25 February 2025
Sec. Cancer Molecular Targets and Therapeutics
Volume 15 - 2025 | https://doi.org/10.3389/fonc.2025.1506577
This article is part of the Research Topic Renewed Insight into Cancer Mechanism and Therapy View all 24 articles
Microorganisms, including bacteria, viruses, and fungi, have been found to play critical roles in tumor microenvironments. Due to their low biomass and other obstacles, the presence of intratumor microbes has been challenging to definitively establish. However, advances in biotechnology have enabled researchers to reveal the association between intratumor microbiota and cancer. Recent studies have shown that tumor tissues, once thought to be sterile, actually contain various microorganisms. Disrupted mucosal barriers and adjacent normal tissues are important sources of intratumor microbiota. Additionally, microbes can invade tumors by traveling through the bloodstream to the tumor site and infiltrating through damaged blood vessels. These intratumor microbiota may promote the initiation and progression of cancers by inducing genomic instability and mutations, affecting epigenetic modifications, activating oncogenic pathways, and promoting inflammatory responses. This review summarizes the latest advancements in this field, including techniques and methods for identifying and culturing intratumor microbiota, their potential sources, functions, and roles in the efficacy of immunotherapy. It explores the relationship between gut microbiota and intratumor microbiota in cancer patients, and whether altering gut microbiota might influence the characteristics of intratumor microbiota and the host immune microenvironment. Additionally, the review discusses the prospects and limitations of utilizing intratumor microbiota in antitumor immunotherapy.
Cancer is a major societal, public health, and economic challenge in the 21st century (1). According to the World Cancer Report 2022 published by the World Health Organization, there were approximately 20 million new cancer cases and 9.7 million cancer-related deaths in 2022 (1). The human body hosts a diverse community of microorganisms, including viruses, bacteria, fungi, archaea, and unicellular eukaryotes. Remarkably, the number of microbial cells in the human body is significantly greater than the number of human cells (2). These microorganisms have previously been found in various parts of the human body, including the skin, mouth, gastrointestinal tract, respiratory tract, urogenital tract, and other mucosal surfaces (3). With breakthroughs in techniques for detecting low microbial biomass and in-depth research on host-microbe interactions, tissues and organs once thought to be sterile, including the liver, pancreas, lungs, breasts, and kidneys, have also shown the presence of low biomass microbial communities (4). With the deepening research on host-microbe interactions, the concept of intratumor microbiota has been proposed.
Notably, recent studies have comprehensively described the cancer mycobiome in 17,401 patient tissue, blood, and plasma samples across 35 cancer types from four independent cohorts, revealing the presence of intratumor mycobiome in these 35 cancer types (5) (Figure 1). The gut microbiome is known to play a significant role in tumor development, resistance, and clinical efficacy (6–8). Similarly, the intratumor microbiome has garnered increasing attention for its involvement in tumor diagnosis, prognosis, and its interactions with the immune system (9). Studies indicate that the diversity and composition of the microbiome within tumor tissues influence immune infiltration, ultimately affecting the survival rate of cancer patients (10). Furthermore, the characteristics of the microbiome are associated with cancer type, cancer risk, pathological type, cancer prognosis, and treatment response (10–13). Recent research has revealed that the distribution of the intratumor microbiome is highly organized, featuring immune and epithelial cell functions that promote cancer progression (14). However, the relationship between the gut microbiome and the intratumor microbiome in cancer patients, as well as whether and how bacteria within tumor cells participate in tumor development and influence treatment responses through immune pathways, still requires further in-depth research. In this review, we summarize the relationship between intratumor microbes and cancer development and progression, and highlight the role of intratumor microbiota in tumor immunity and therapeutic responses. We explore the relationship between the gut microbiome and intratumor microbiome in cancer patients, and propose the possibility of applying intratumor microbiota as novel biomarkers and therapeutic targets for cancer.
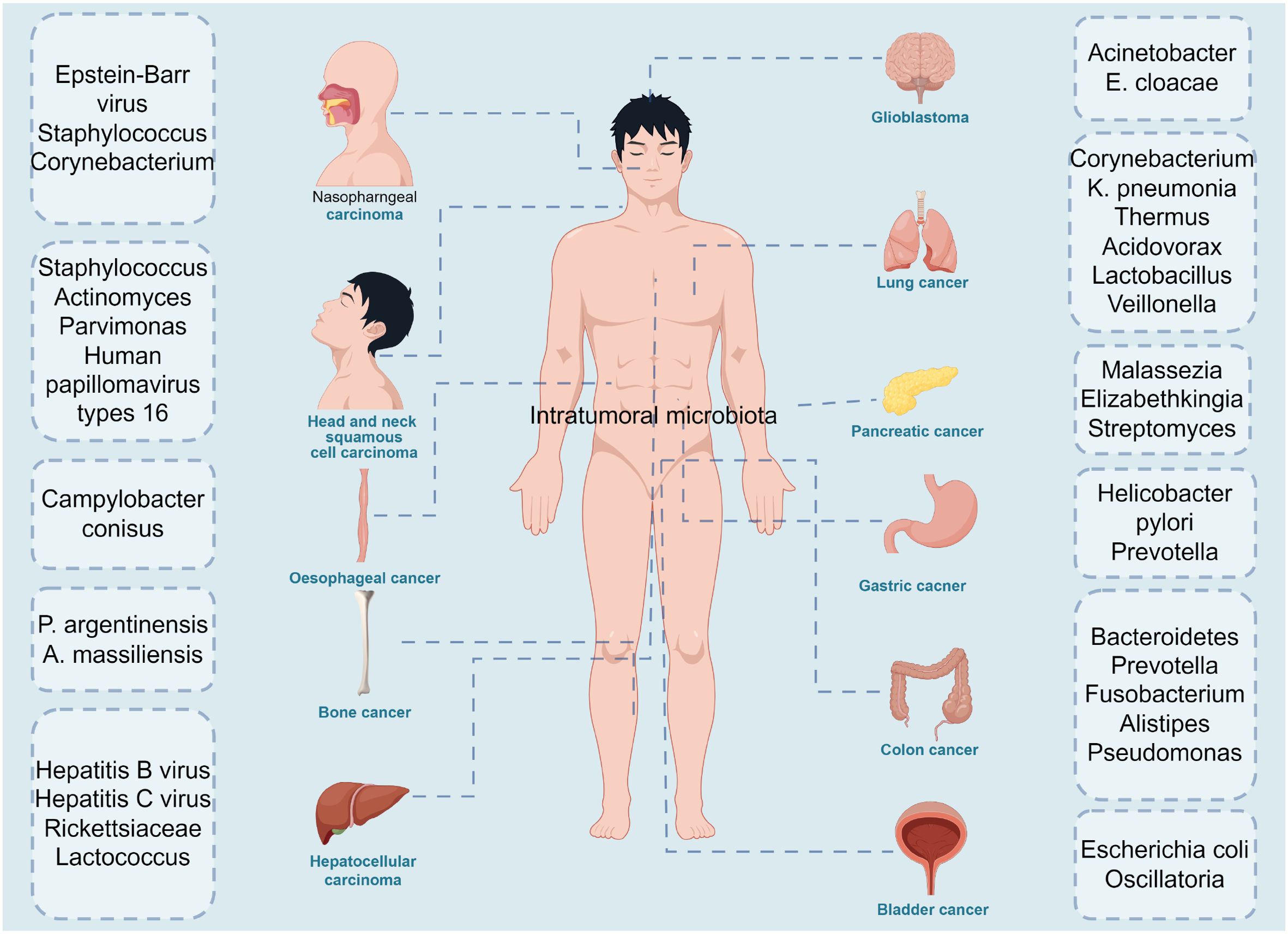
Figure 1. The diversity of intratumoral microbiota in cancers. Microbiota are detected in multiple tumors, including nasopharngeal carcinoma: epstein-Barrvirus, staphylosoccus, corynebacterium, head and neck squamous cell carcinoma: staphylococcus, actinomyces, parvimonas, human papillomavirus type 16, oesophageal cancer: campylobacter conisus, bone cancer: p.argentinensis, a.massiliensis, hepatocellular carcinoma: hepatitis B virus, Hepatitis C virus, Rickettsiaceae, lactococcus, glioblastoma: Acinetobacter, E. cloacae, lung cancer: corynebacterium, K.pneumonia, thermus, acidovorax, lactobacillus, pancreatic cancer: Malassezia, elizabethkingia, streptomyces, gastric cancer: helicobacter pylori, prevotella, colon cancer: Bacteroidetes, prevotella, fusobacterium, alistipes, pseudomonas and bladder cancer: escherichina coli, oscillatoria. The figure was Created by Figdraw.
In a stable state, commensal microbes have a symbiotic relationship with the host, but microbiome dysbiosis can promote the development of enteritis, pneumonia, and cancer (15). Since the 20th century, people have gradually recognized microorganisms that can induce cancer, such as Epstein-Barr virus, hepatitis B virus, human papilloma virus, and Helicobacter pylori (11). The role of gut microbiota in tumors has been extensively studied, and its influence extends beyond just pathogenesis and cancer risk. Microbial signaling also impacts the clinical course of tumors, including the efficacy, bioavailability, and toxicity of chemotherapeutic and immunotherapy drugs (16). At the same time, advances in detection techniques have provided unprecedented opportunities to study the diversity and functional characteristics of intratumor microbiota (17). Direct detection of intratumor microbes provides the most compelling evidence of their presence within tumor tissues (18). Various advanced detection techniques, including correlative light and electron microscopy (CLEM), fluorescence in situ hybridization (FISH) and immunofluorescence (IF), play crucial roles in identifying and studying intratumor microbes. Correlative light and electron microscopy (CLEM) combines the advantages of optical and electron microscopy (19), providing high resolution and environmental context information, making it excellent for detecting intratumor microbiota, but it is costly, time-consuming, and technically complex. Fluorescence in situ hybridization (FISH) has strong specificity and can quickly identify and detect various intratumor microorganisms (20), but it has limited resolution and quantification capabilities, and requires specific sample preparation. Immunofluorescence (IF) offers high specificity and sensitivity, allowing real-time observation and multiple staining, making it very effective for detecting intratumor microbiota (21), though it may suffer from fluorescence spectral interference, and is also costly and technically demanding.
In contrast, high-throughput sequencing technologies, such as 16S rRNA gene sequencing and whole metagenome shotgun sequencing (WMS), can provide more comprehensive microbial data. These techniques not only accurately identify and classify microorganisms but also analyze their taxonomic composition and functional characteristics, despite being more complex and expensive (22, 23). By combining these methods, the intratumor microbiome can be studied more precisely and comprehensively. While metagenomics and other methods provide a wealth of data on intratumor microbiota, pure culturing of these microorganisms remains crucial as it reveals their functions and related mechanisms. Significant progress has been made in recent years in culturing previously unculturable microbes. For instance, using methods that simulate natural environments has successfully cultured Candidatus Pelagibacter ubique and other microbes (24). Additionally, a microfluidic intestine-on-a-chip was designed, successfully culturing bacteria from 11 genera, and reverse genomics was used to isolate and culture specific microbes (25). However, the drawbacks of pure culturing include its complexity, high cost, and the difficulty in isolating and successfully culturing low-biomass intratumor microbes. Traditional in vivo and in vitro experiments, such as flow cytometry, western blot, enzyme-linked immunosorbent assay (ELISA), and mouse models, remain crucial in tumor microbiology research, providing key mechanistic insights by revealing the impact of intratumor microbiota on immune cells and cancer development (18). Omics technologies such as genomics, transcriptomics, and metabolomics are used to study the molecular mechanisms between intratumor microbiota and tumors. Additionally, single-cell RNA sequencing (scRNA-seq) and spatial meta-transcriptomics can elucidate the roles of intratumor microbiota in cancer initiation, progression, metastasis, and treatment responses (26, 27). Other spatial multi-omics techniques, such as spatial genomics, spatial proteomics, and spatial metabolomics, remain indispensable for studying intratumor microbiota and cancer (28). Due to the relatively low biomass of the tumor microbiome, contamination of tumor samples with bacteria or bacterial DNA during sample collection, preparation, and processing can pose significant issues (18). Next-generation sequencing technology acts as a double-edged sword in microbiome research, as its sensitivity can effectively detect both microbial DNA and contaminants, including cross-contamination. Therefore, it is essential to reduce experimental bias and contamination during sampling and processing, include control groups throughout the entire process from sampling to sequencing, and critically assess and report the impact of contamination during analysis (23).
Despite the considerable attention given to research on intratumor microorganisms, their origins remain to be explored. Tumors have characteristics that make them prone to bacterial colonization: imperfect angiogenesis leads to leaky blood vessels, allowing circulating bacteria to embed; tumors are immune-privileged areas, enabling bacterial proliferation; low-oxygen environments favor anaerobic bacteria; and necrotic regions are nutrient-rich, promoting bacterial growth (29). It is currently believed that intratumor microorganisms mainly originate from the following sources: I) disrupted mucosal barrier sources. Intratumor microorganisms may invade through mucosal barriers, including in gastric cancer, colorectal cancer, pancreatic cancer, lung cancer, and cervical cancer (30). The external cavities of these organs are exposed, and during tumorigenesis, the disruption of the mucosal barrier allows microorganisms colonizing the mucosa to infiltrate the tumor.II) adjacent normal tissues. Research has found a high similarity between the microbiome of tumors and that of their adjacent normal tissues (NAT), leading researchers to propose that intratumor bacteria may originate from NAT (19). However, the source of microbes in NAT is not fully understood and may also originate from the tumor microenvironment. Therefore, more evidence is needed to determine whether NAT is a source of intratumor microbiota. III) Through the blood to the tumor site and infiltrate the tumor through damaged blood. Animal experiments have revealed that certain lymph node metastases (LNMs) resurface at distant metastatic sites through blood vessels in the lymph nodes, rather than through the traditionally assumed lymphatic route (31). In colorectal cancer (CRC), bacteria disrupt the gut vascular barrier (GVB) and disseminate to the liver, inducing the formation of a premetastatic niche and promoting the recruitment of metastatic cells (32). Additionally, studies have shown that different cancer cell clones from the primary tumor can colonize distant organs via the systemic blood circulation, indicating that distant metastases may occur independently of lymph node metastasis (32).
The tumor microbiomes of different cancer types exhibit high heterogeneity (Table 1). A large-scale study by Ravid Straussman’s team demonstrated that the microbial composition of each type of tumor (including pancreatic cancer, breast cancer, lung cancer, ovarian cancer, melanoma, brain cancer, and bone cancer) is distinct, with intratumor bacteria primarily found in cancer cells and immune cells (19). Although bacteria belonging to the Firmicutes and Proteobacteria dominate the microbiota of all cancer types, there is a high degree of heterogeneity in the composition and abundance of bacteria in different cancer types, and the composition of microbial species varies in different subtypes of the same tumor type (17). For example, at the species level, Fusobacterium nucleatum was enriched in breast and pancreatic tumors. Saccharomycetes were more abundant in colon cancer, whereas the relative abundance of Malasseziomycetes was higher in melanoma (17). Additionally, the metabolic functions encoded by intratumor bacteria are associated with the clinical characteristics of certain tumor subtypes. For example, enzymes related to anaerobic respiration are more abundant in breast cancer bacteria (19). Bacteria dominate the tumor microbiome, while fungi are less prevalent (5). The study by Galeano Nino et al. further revealed the spatial and population heterogeneity of the intratumor microbiome (14). The microbial distribution also differs between tumor tissues and peritumoral tissues. For instance, the abundance of certain oral microbes is significantly higher in esophageal and gastric cancer tissues than in adjacent normal tissues (65, 66), whereas Fusobacterium nucleatum, which is enriched in colorectal cancer tissues, is not found in adjacent normal tissues (67). An in-depth investigation of the composition and function of the tumor microbiome will provide new opportunities for cancer treatment.
Gut microbiota and intratumor microbiota are closely linked in terms of their origins and their impact on immunity. First, intratumor microbiota can originate from gut microbiota because microorganisms from the intestines may be transported through the blood to the tumor site (9). For example, the liver is connected to the gut via the hepatic portal vein. Although the gut vascular barrier restricts the dissemination of intestinal bacteria, when the gut vascular barrier is impaired, intestinal bacteria can promote the recruitment of immune cells in the liver, forming a pre-metastatic niche that supports the metastasis of cancer cells to the liver (17). Studies have found that the gut microbiome can colonize pancreatic tumors in patients with pancreatic cancer, and this colonization can alter the overall microbiome of the tumor (10). Most importantly, both intratumor microbiota and gut microbiota have regulatory effects on the tumor microenvironment. Intratumor microbiota may regulate the host immune response similarly to gut microbiota. Gut microbiota has extensive effects on primary lymphoid organs and adaptive immunity. Studies have shown that gut microbiota significantly impacts immune reconstitution, treatment outcomes, and side effects such as infections following hematopoietic stem cell transplantation (HSCT) (68). Immune reconstitution after allogeneic HSCT is closely related to the diversity of gut microbiota, with higher diversity being significantly associated with lower patient mortality. Moreover, gut microbiota influences the host immune status during homeostasis and tumorigenesis. Cancer therapies have demonstrated strong links between different commensal bacteria and protective antitumor T cell responses. Additionally, probiotics, particularly Lactobacillus and Bifidobacterium, are considered safe (69) and have been used to prevent and treat various diseases (70, 71). In cancer, strains of Lactobacillus and Bifidobacterium can alleviate dysbiosis, enhance antitumor immunity, and improve the efficacy of immune checkpoint inhibitors (72–76). These studies indicate that gut microbiota plays an important role in regulating immune responses and antitumor therapies. Due to the complex interactions between the microbiota and the immune system, numerous studies have focused on how the microbiota influences local and systemic anti-tumor immune responses (72). For example, short-chain fatty acids, such as acetate, butyrate, and propionate, are important energy sources for gut microbiota and play a crucial role in regulating host physiology and immune responses (72).
With further research into intratumor microbiota, it has been found that the tumor microbiome plays a crucial role in reshaping the tumor immune microenvironment (17). When intratumor microbiota is recognized by the innate immune system, the adaptive immune system is activated, becoming a pillar of the antitumor response (77). Tumor microecology helps recruit and activate tumor-supportive immune cells. For example, intratumor microbiota induces IL-17 production, supporting the infiltration of B cells into tumor tissues, thereby promoting colon cancer progression (78). Some intratumor microbiota also counteract pro-tumoral immune responses. For instance, patients with F. nucleatum-positive oral squamous cell carcinoma (OSCC) have lower recurrence rates, less frequent lymph node invasion and metastatic relapse, and longer overall survival (OS), relapse-free survival (RFS), and metastasis-free survival (MFS) compared to those with F.nucleatum-negative tumors (79).Intratumor microbiota can slow cancer progression by enhancing antitumor immunity. For example, gram-negative bacteria were detected in the cytoplasm of osteosarcoma cells and tumor-associated macrophages (TAMs). Patients with localized osteosarcoma had an increased number of antitumor M1 macrophages, which may be related to the abundance of gram-negative bacteria (80). The tumor microbiome plays an essential role in tumor immune regulation, with significant differences in the roles of different microbes in various cancers. Future research should focus on elucidating the interaction mechanisms between intratumor microbiota and the host immune system to develop new cancer treatment strategies.
Increasing evidence has shown that intratumor microbiota can have both positive and negative effects on tumor initiation and progression through mechanisms such as DNA mutations, immune evasion, promoting chronic inflammation, and inducing metastasis (81–84) (Figure 2). Importantly, the process of cancer metastasis includes stages such as invasion, dissemination, intravasation, extravasation, and colonization (85). One of the main characteristics of metastasis is its extreme inefficiency; Before successfully reaching and settling in the target organ, cancer cells need to cope with many physical, chemical, and biological challenges (46, 47). Metastasis initiation occurs at an early stage of tumor progression where cancer cells can remotely prepare the pre-metastatic microenvironment (PMN) by secreting components (86). When metastatic cells begin to migrate, they usually invade neighboring tissues en masse to enhance their ability to colonize the new microenvironment (87, 88). Cancer cells usually alter their intrinsic programs to overcome various challenges during metastasis. These programs include the stem cell program/plasticity (for tumor initiation at new sites), the EMT program (for cancer invasion and dissemination), the adhesion program (to prevent apoptosis-induced cell death), and the mechanical stress response program (to resist damage induced by mechanical forces) (89). In addition to directly regulating cancer cells, intra-tumoral bacteria are important inflammatory mediators that shape the specific microenvironment around cancer cells, thus indirectly promoting cancer metastasis (89). Although the number of intratumor microorganisms is low and their biological functions are not yet clear, studies have shown that they are new key players in influencing tumor metastasis and play an important role. For example, studies in breast cancer have found that intratumor bacteria promote host cell survival by reorganizing the actin cytoskeleton, enhancing the resistance of circulating tumor cells to fluid shear stress. The removal of intratumor bacteria can significantly reduce lung metastasis without affecting the growth of the primary tumor (90). Similarly, viable Fusobacterium and its associated microbiota were retained during serial passage in mouse xenografts of human primary colorectal adenocarcinoma (91). Increasing evidence confirms that intratumor bacteria can modulate the intrinsic properties of cancer cells and their external environment, thereby enhancing cancer cell survival and paving the way for cancer metastasis (89) (Table 2). Moreover, studies on tumor-associated host-microbiota have primarily relied on bulk tissue analysis, which obscures the spatial distribution and localized effects of microbiota within tumors (14). Research has found that the distribution of intratumor microbiota is highly organized within tumors, clustering in microniches that are less vascularized, highly immunosuppressive, and associated with malignant cells with lower levels of the cell proliferation antigen Ki-67. Bacteria-infected cancer cells invade the surrounding environment as single cells and recruit myeloid cells to bacterial niches. These findings highlight the significant role of intratumor microbiota in promoting cancer progression (14).
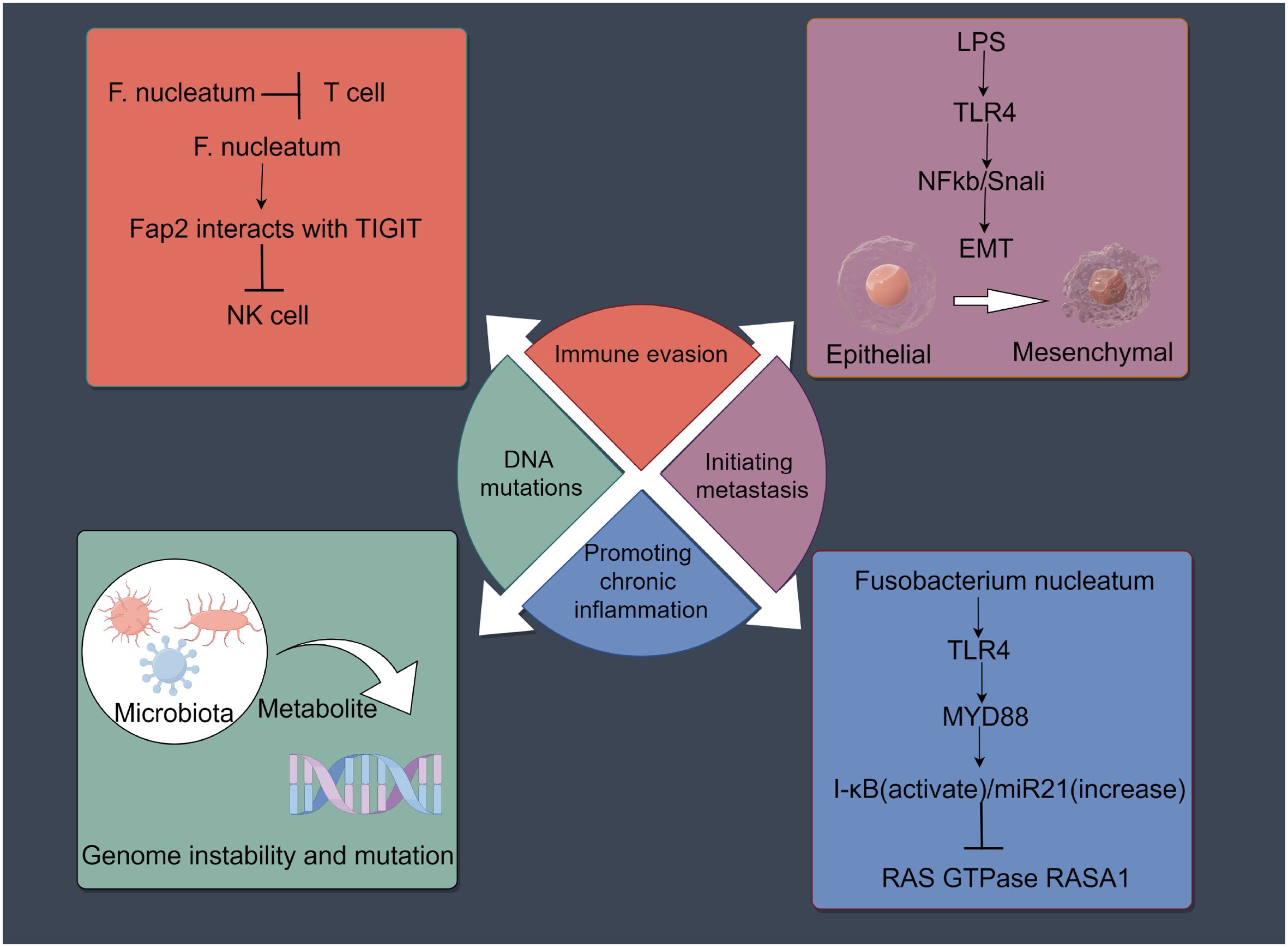
Figure 2. Effects of the intratumoral microbiota on cancers development. Intratumoral microbiota may influence the initiation and progression of cancer through mechanisms such as DNA mutations, immune evasion, promotion of chronic inflammation, and induction of metastasis. The figure was Created by Figdraw.
Given that local microbial infections can cause tumor regression, the engineering of microbiota to improve and enhance their antitumor effects is being explored (9). The microbiota can serve as bacterial vectors expressing cytotoxic drugs, either alone or in combination with other antitumor agents. Tumor-targeting bacteria demonstrate significant advantages as delivery vectors, including enhanced penetration into tumor tissue, maximized efficacy of chemotherapy agents, and reduced systemic toxicity (8). The microbiota can serve as bacterial vectors expressing cytotoxic drugs, either alone or in combination with other antitumor agents. For example, by coupling the radioisotope 188 Rhenium with attenuated Listeria monocytogenes, a unique radioactive Listeria (RL) was created (97). In normal tissues, Listeria is cleared by the immune system, but in the immunosuppressive tumor microenvironment, it is not. In a highly metastatic pancreatic cancer mouse model, RL effectively delivered radioactivity to metastases without harming normal cells. Multiple low-dose RL treatments significantly reduced the number of metastases (by approximately 90%) (97). Additionally, microbiota can induce both innate and adaptive immune responses against tumor cells. Bacterial therapy can activate the host immune system via multiple mechanisms, including the delivery of cytokines, short hairpin RNA (shRNA), and tumor-associated antigens (98). Engineered bacteria can precisely deliver cytokines and short hairpin RNA (shRNA) to tumor tissues, thereby inducing local inflammatory responses and triggering cytotoxic cell death. Apoptotic or necrotic cancer cells release tumor-associated antigens (TAAs) and various stimulatory factors, promoting the maturation of dendritic cells and triggering epitope spreading effects (98). Additionally, microbiota can induce both innate and adaptive immune responses against tumor cells. The systemic use of potent anticancer drugs such as tumor necrosis factor alpha (TNFα) causes high levels of toxicity and severe side effects. Non-pathogenic Escherichia coli strain MG1655, used as a tumor-targeting system to specifically produce TNFα within tumors in mice, demonstrated the potential of non-pathogenic bacteria as platforms for limiting the activity of potent anticancer agents to tumors (99). At the same time, live microbiota can be used as vectors expressing tumor antigens for tumor vaccination (100). The safety and efficacy of bacterial immunotherapy has always been a priority for clinical applications. Bacterial immunotherapy triggers many of the same potential adverse effects as conventional immunotherapy, including potential systemic immune responses, hypertension and fatigue (98, 101).
Due to the specificity of intratumor microbiota across different tumor types and subtypes, they hold potential as diagnostic tools (Table 3). Studies have found that the presence of oral pathogens, Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans, is associated with an increased risk of pancreatic cancer, while the phylum Fusobacteria and its genus Leptotrichia are associated with a reduced risk of pancreatic cancer (109). Clinical evaluations indicate that tissue and plasma mycobiomes have prognostic and diagnostic capabilities, even in stage I cancers, and exhibit synergistic predictive performance with bacterial communities (5). Some studies have also found that the intratumor microbiota is closely associated with patient prognosis. For example, the prognosis of papillary thyroid carcinoma (PTC) differs between genders and cancer subtypes (13). Further research has found that PTC tissues significantly lack the microorganisms present in adjacent normal tissues, suggesting that these microorganisms are critical in controlling immune cell expression and regulating immune and cancer pathways to slow cancer growth (13). The intratumor load of F. nucleatum may be a potential prognostic factor in stage II/III non-MSI-high/non-sigmoid/non-rectal cancer subset CRC patients receiving oxaliplatin-based adjuvant chemotherapy (110). Therefore, the intratumor microbiota can serve as biomarkers for diagnosis and prognosis.
In conclusion, intratumor microbiota have diverse sources and complex distribution within different tissues, and they are intricately linked with gut microbiota. Intratumor microbiota have potential in the early detection of cancer and in determining patient prognosis. Targeting intratumor microbiota represents a novel clinical approach to cancer treatment, which may improve the efficacy of chemotherapy and enhance the effectiveness of immunotherapy. It is worth noting that although some studies have revealed the mechanisms by which intratumor microbiota influence cancer development, metastasis, and regulation of the tumor microenvironment, the therapeutic effects of microbiota-based treatments may vary depending on tumor type and the immune status of cancer patients. More clinical research is needed to determine their actual value in aiding cancer intervention. Although current experimental technologies have provided us with a deeper understanding of the intratumoral microbiota, we still have limited knowledge of its role in tumorigenesis and progression. This is primarily due to the lack of effective experimental methods to study the occurrence, development, and metastasis of these microorganisms, as well as their spatial distribution and dynamic changes within the tumor.
Although significant progress has been made in recent years in the study of intratumor microbiota, current research still has limitations. For example, the interactions between gut microbiota and intratumor microbiota, and how changes in gut microbiota affect intratumor microbiota and the immune microenvironment, are not well understood. The therapeutic effects and impact of probiotics on cancer and immunotherapy also remain unclear. Further exploration is required to understand the mechanisms through which intratumor microbiota influence antitumor immunity and treatment efficacy, as well as how specific antibiotics can be used to remove immunosuppressive microbes. Future researchers could explore the role of probiotics, antibiotics, and FMT in regulating the intratumoral microbiota in cancer, while dietary patterns also serve as an important modulator of the microbiota.
PF: Conceptualization, Investigation, Project administration, Validation, Writing – review & editing. JY: Conceptualization, Investigation, Validation, Writing – original draft. HZ: Conceptualization, Formal Analysis, Validation, Visualization, Writing – original draft. DS: Formal Analysis, Resources, Supervision, Writing – review & editing. ML: Investigation, Methodology, Validation, Visualization, Writing – review & editing. LC: Conceptualization, Methodology, Resources, Writing – review & editing. LL: Conceptualization, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Foundation of The First Hospital of Lanzhou University, China (ldyyyn2021-91).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin. (2024) 74:229–63. doi: 10.3322/caac.21834
2. Altveş S, Yildiz HK, Vural HC. Interaction of the microbiota with the human body in health and diseases. Bioscience microbiota Food Health. (2020) 39:23–32. doi: 10.12938/bmfh.19-023
3. Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. (2012) 13:260–70. doi: 10.1038/nrg3182
4. Wong-Rolle A, Wei HK, Zhao C, Jin C. Unexpected guests in the tumor microenvironment: microbiome in cancer. Protein Cell. (2021) 12:426–35. doi: 10.1007/s13238-020-00813-8
5. Narunsky-Haziza L, Sepich-Poore GD, Livyatan I, Asraf O, Martino C, Nejman D, et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell. (2022) 185:3789–3806.e3717. doi: 10.1016/j.cell.2022.09.005
6. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. (2018) 359:104–8. doi: 10.1126/science.aao3290
7. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discovery. (2018) 8:403–16. doi: 10.1158/2159-8290.CD-17-1134
8. Liu J, Zhang Y. Intratumor microbiome in cancer progression: current developments, challenges and future trends. biomark Res. (2022) 10:37. doi: 10.1186/s40364-022-00381-5
9. Yang L, Li A, Wang Y, Zhang Y. Intratumoral microbiota: roles in cancer initiation, development and therapeutic efficacy. Signal transduction targeted Ther. (2023) 8:35. doi: 10.1038/s41392-022-01304-4
10. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. (2019) 178:795–806.e712. doi: 10.1016/j.cell.2019.07.008
11. Chai X, Wang J, Li H, Gao C, Li S, Wei C, et al. Intratumor microbiome features reveal antitumor potentials of intrahepatic cholangiocarcinoma. Gut Microbes. (2023) 15:2156255. doi: 10.1080/19490976.2022.2156255
12. Liu Z, Zhang X, Zhang H, Zhang H, Yi Z, Zhang Q, et al. Multi-omics analysis reveals intratumor microbes as immunomodulators in colorectal cancer. Microbiol Spectr. (2023) 11:e0503822. doi: 10.1128/spectrum.05038-22
13. Gnanasekar A, Castaneda G, Iyangar A, Magesh S, Perez D, Chakladar J, et al. The intratumor microbiome predicts prognosis across gender and subtypes in papillary thyroid carcinoma. Comput Struct Biotechnol J. (2021) 19:1986–97. doi: 10.1016/j.csbj.2021.03.032
14. Galeano Niño JL, Wu H, LaCourse KD, Kempchinsky AG, Baryiames A, Barber B, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. (2022) 611:810–7. doi: 10.1038/s41586-022-05435-0
15. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. (2009) 9:313–23. doi: 10.1038/nri2515
16. Murphy CL, O’Toole PW, Shanahan F. The gut microbiota in causation, detection, and treatment of cancer. Am J Gastroenterol. (2019) 114:1036–42. doi: 10.14309/ajg.0000000000000075
17. Wang M, Yu F, Li P. Intratumor microbiota in cancer pathogenesis and immunity: from mechanisms of action to therapeutic opportunities. Front Immunol. (2023) 14:1269054. doi: 10.3389/fimmu.2023.1269054
18. Liu W, Xu J, Pi Z, Chen Y, Jiang G, Wan Y, et al. Untangling the web of intratumor microbiota in lung cancer. Biochim Biophys Acta Rev Cancer. (2023) 1878:189025. doi: 10.1016/j.bbcan.2023.189025
19. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. (2020) 368:973–80. doi: 10.1126/science.aay9189
20. Frickmann H, Zautner AE, Moter A, Kikhney J, Hagen RM, Stender H, et al. Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: a review. Crit Rev Microbiol. (2017) 43:263–93. doi: 10.3109/1040841X.2016.1169990
21. Peng H, Wu X, Zhong R, Yu T, Cai X, Liu J, et al. Profiling tumor immune microenvironment of non-small cell lung cancer using multiplex immunofluorescence. Front Immunol. (2021) 12:750046. doi: 10.3389/fimmu.2021.750046
22. Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, et al. Best practices for analysing microbiomes. Nat Rev Microbiol. (2018) 16:410–22. doi: 10.1038/s41579-018-0029-9
23. Eisenhofer R, Minich JJ, Marotz C, Cooper A, Knight R, Weyrich LS. Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. (2019) 27:105–17. doi: 10.1016/j.tim.2018.11.003
24. Lagier JC, Dubourg G, Million M, Cadoret F, Bilen M, Fenollar F, et al. Culturing the human microbiota and culturomics. Nat Rev Microbiol. (2018) 16:16540–550. doi: 10.1038/s41579-018-0041-0
25. Lewis WH, Tahon G, Geesink P, Sousa DZ, Ettema TJG. Innovations to culturing the uncultured microbial majority. Nat Rev Microbiol. (2021) 19:225–40. doi: 10.1038/s41579-020-00458-8
26. Seferbekova Z, Lomakin A, Yates LR, Gerstung M. Spatial biology of cancer evolution. Nat Rev Genet. (2023) 24:295–313. doi: 10.1038/s41576-022-00553-x
27. Ahmed R, Zaman T, Chowdhury F, Mraiche F, Tariq M, Ahmad IS, et al. Single-cell RNA sequencing with spatial transcriptomics of cancer tissues. Int J Mol Sci. (2022) 23:3042. doi: 10.3390/ijms23063042
28. Vandereyken K, Sifrim A, Thienpont B, Voet T. Methods and applications for single-cell and spatial multi-omics. Nat Rev Genet. (2023) 24:494–515. doi: 10.1038/s41576-023-00580-2
29. Walker SP, Tangney M, Claesson MJ. Sequence-based characterization of intratumoral bacteria-A guide to best practice. Front Oncol. (2020) 10:179. doi: 10.3389/fonc.2020.00179
30. Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. (2020) 30:492–506. doi: 10.1038/s41422-020-0332-7
31. Pereira ER, Kedrin D, Seano G, Gautier O, Meijer EFJ, Jones D, et al. Lymph node metastases can invade local blood vessels, exit the node, and colonize distant organs in mice. Science. (2018) 359:1403–7. doi: 10.1126/science.aal3622
32. Bertocchi A, Carloni S, Ravenda PS, Bertalot G, Spadoni I, Lo Cascio A, et al. Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer Cell. (2021) 39:708–24.e711. doi: 10.1016/j.ccell.2021.03.004
33. Su SC, Chang LC, Huang HD, Peng CY, Chuang CY, Chen YT, et al. Oral microbial dysbiosis and its performance in predicting oral cancer. Carcinogenesis. (2021) 42:127–35. doi: 10.1093/carcin/bgaa062
34. Hooper SJ, Crean SJ, Fardy MJ, Lewis MAO, Spratt DA, Wade WG, et al. A molecular analysis of the bacteria present within oral squamous cell carcinoma. J Med Microbiol. (2007) 56:1651–9. doi: 10.1099/jmm.0.46918-0
35. Li Z, Fu R, Wen X, Wang Q, Huang X, Zhang L. The significant clinical correlation of the intratumor oral microbiome in oral squamous cell carcinoma based on tissue-derived sequencing. Front Physiol. (2023) 13:1089539. doi: 10.3389/fphys
36. Al-Hebshi NN, Nasher AT, Maryoud MY, Homeida HE, Chen T, Idris AM, et al. Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Sci Rep. (2017) 7:1834. doi: 10.1038/s41598-017-02079-3
37. Guidry JT, Birdwell CE, Scott RS. Epstein-Barr virus in the pathogenesis of oral cancers. Oral Dis. (2018) 24:497–508. doi: 10.1111/odi.2018.24.issue-4
38. Kaakoush NO, Deshpande NP, Man SM, Burgos-Portugal JA, Khattak FA, Raftery MJ, et al. Transcriptomic and proteomic analyses reveal key innate immune signatures in the host response to the gastrointestinal pathogen Campylobacter concisus. Infection Immun. (2015) 83:2–45. doi: 10.1128/IAI.03012-14
39. Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, et al. Human microbiome fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res. (2016) 22:5574–81. doi: 10.1158/1078-0432.CCR-16-1786
40. Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. (2018) 67:1024–32. doi: 10.1136/gutjnl-2017-314281
41. Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. (2018) 67:226–36. doi: 10.1136/gutjnl-2017-314205
42. Liu D, Liu Y, Zhu W, Lu Y, Zhu J, Ma X, et al. Helicobacter pylori-induced aberrant demethylation and expression of GNB4 promotes gastric carcinogenesis via the Hippo-YAP1 pathway. BMC Med. (2023) 21:134. doi: 10.1186/s12916-023-02842-6
43. Shao D, Vogtmann E, Liu A, Qin J, Chen W, Abnet CC, et al. Microbial characterization of esophageal squamous cell carcinoma and gastric cardia adenocarcinoma from a high-risk region of China. Cancer. (2019) 125:3993–4002. doi: 10.1002/cncr.v125.22
44. Liu W, Zhang X, Xu H, Li S, Lau HC, Chen Q, et al. Microbial community heterogeneity within colorectal neoplasia and its correlation with colorectal carcinogenesis. Gastroenterology. (2021) 160:2395–408. doi: 10.1053/j.gastro.2021.02.020
45. Wang Y, Yuan Y, Gu D. Hepatitis B and C virus infections and the risk of biliary tract cancers: a meta-analysis of observational studies. Infect Agents Cancer. (2022) 17:45. doi: 10.1186/s13027-022-00457-9
46. Testerman TL, Morris J. Beyond the stomach: an updated view of Helicobacter pylori pathogenesis, diagnosis, and treatment. World J Gastroenterol. (2014) 20:12781–808. doi: 10.3748/wjg.v20.i36.12781
47. Huang JH, Wang J, Chai XQ, Li ZC, Jiang YH, Li J, et al. The intratumoral bacterial metataxonomic signature of hepatocellular carcinoma. Microbiol Spectr. (2022) 10:e0098322. doi: 10.1128/spectrum.00983-22
48. Qu D, Wang Y, Xia Q, Chang J, Jiang X, Zhang H. Intratumoral microbiome of human primary liver cancer. Hepatol Commun. (2022) 6:1741–52. doi: 10.1002/hep4.1908
49. Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. (2019) 574:264–7. doi: 10.1038/s41586-019-1608-2
50. Guo W, Zhang Y, Guo S, Mei Z, Liao H, Dong H, et al. Tumor microbiome contributes to an aggressive phenotype in the basal-like subtype of pancreatic cancer. Commun Biol. (2021) 4:1019. doi: 10.1038/s42003-021-02557-5
51. Tzeng A, Sangwan N, Jia M, Liu CC, Keslar KS, Downs-Kelly E, et al. Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Med. (2021) 13:60. doi: 10.1186/s13073-021-00874-2
52. Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, et al. The microbiome of aseptically collected human breast tissue in benign and Malignant disease. Sci Rep. (2016) 6:30751. doi: 10.1038/srep30751
53. Parida S, Wu S, Siddharth S, Wang G, Muniraj N, Nagalingam A, et al. A procarcinogenic colon microbe promotes breast tumorigenesis and metastatic progression and concomitantly activates notch and β-catenin axes. Cancer Discovery. (2021) 11:38–57. doi: 10.1158/2159-8290.CD-20-0537
54. Thyagarajan S, Zhang Y, Thapa S, Allen MS, Phillips N, Chaudhary P, et al. Comparative analysis of racial differences in breast tumor microbiome. Sci Rep. (2020) 10:14116. doi: 10.1038/s41598-020-71102-x
55. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell. (2019) 176:998–1013.e1016. doi: 10.1016/j.cell.2018.12.040
56. Yu G, Gail MH, Consonni D, Carugno M, Humphrys M, Pesatori AC, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. (2016) 17:163. doi: 10.1186/s13059-016-1021-1
57. Jungnickel C, Schmidt LH, Bittigkoffer L, Wolf L, Wolf A, Ritzmann F, et al. IL-17C mediates the recruitment of tumor-associated neutrophils and lung tumor growth. Oncogene. (2017) 36:4182–90. doi: 10.1038/onc.2017.28
58. Lee SH, Sung JY, Yong D, Chun J, Kim SY, Song JH, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer (Amsterdam Netherlands). (2016) 102:89–95. doi: 10.1016/j.lungcan.2016.10.016
59. Greathouse KL, White JR, Vargas AJ, Bliskovsky VV, Beck JA, von Muhlinen N, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. (2018) 19:123. doi: 10.1186/s13059-018-1501-6
60. Fassi Fehri L, Mak TN, Laube B, Brinkmann V, Ogilvie LA, Mollenkopf H, et al. Prevalence of Propionibacterium acnes in diseased prostates and its inflammatory and transforming activity on prostate epithelial cells. Int J Med microbiology: IJMM. (2011) 301:69–78. doi: 10.1016/j.ijmm.2010.08.014
61. Samanta M, Harkins L, Klemm K, Britt WJ, Cobbs CS. High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J Urol. (2003) 170:998–1002. doi: 10.1097/01.ju.0000080263.46164.97
62. Ma J, Gnanasekar A, Lee A, Li WT, Haas M, Wang-Rodriguez J, et al. Influence of intratumor microbiome on clinical outcome and immune processes in prostate cancer. Cancers. (2020) 12:2524. doi: 10.3390/cancers12092524
63. Kustrimovic N, Bombelli R, Baci D, Mortara L. Microbiome and prostate cancer: A novel target for prevention and treatment. Int J Mol Sci. (2023) 24:1511. doi: 10.3390/ijms24021511
64. Salachan PV, Rasmussen M, Fredsøe J, Ulhøi B, Borre M, Sørensen KD. Microbiota of the prostate tumor environment investigated by whole-transcriptome profiling. Genome Med. (2022) 14:9. doi: 10.1186/s13073-022-01011-3
65. Yuan X, Liu Y, Kong J, Gu B, Qi Y, Wang X, et al. Different frequencies of Porphyromonas gingivalis infection in cancers of the upper digestive tract. Cancer Lett. (2017) 404:41–7. doi: 10.1016/j.canlet.2017.07.003
66. Chen XH, Wang A, Chu AN, Gong YH, Yuan Y. Mucosa-associated microbiota in gastric cancer tissues compared with non-cancer tissues. Front Microbiol. (2019) 10:1261. doi: 10.3389/fmicb.2019.01261
67. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. (2013) 14:207–15. doi: 10.1016/j.chom.2013.07.007
68. Fredricks DN. The gut microbiota and graft-versus-host disease. J Clin Invest. (2019) 129:1808–17. doi: 10.1172/JCI125797
69. Singh A, Alexander SG, Martin S. Gut microbiome homeostasis and the future of probiotics in cancer immunotherapy. Front Immunol. (2023) 14:1114499. doi: 10.3389/fimmu.2023.1114499
70. Wastyk HC, Fragiadakis GK, Perelman D, Dahan D, Merrill BD, Yu FB, et al. Gut-microbiota-targeted diets modulate human immune status. Cell. (2021) 184:4137–53.e4114. doi: 10.1016/j.cell.2021.06.019
71. Fang Z, Li L, Zhang H, Zhao J, Lu W, Chen W. Gut microbiota, probiotics, and their interactions in prevention and treatment of atopic dermatitis: A review. Front Immunol. (2021) 12:720393. doi: 10.3389/fimmu.2021.720393
72. Li Z, Xiong W, Liang Z, Wang J, Zeng Z, Kołat D, et al. Critical role of the gut microbiota in immune responses and cancer immunotherapy. J Hematol Oncol. (2024) 17:33. doi: 10.1186/s13045-024-01541-w
73. Gamallat Y, Meyiah A, Kuugbee ED, Hago AM, Chiwala G, Awadasseid A, et al. Lactobacillus rhamnosus induced epithelial cell apoptosis, ameliorates inflammation and prevents colon cancer development in an animal model. Biomedicine pharmacotherapy = Biomedecine pharmacotherapie. (2016) 83:536–541. doi: 10.1016/j.biopha.2016.07.001
74. Jacouton E, Chain F, Sokol H, Langella P, Bermúdez-Humarán LG. Probiotic strain lactobacillus casei BL23 prevents colitis-associated colorectal cancer. Front Immunol. (2017) 8:1553. doi: 10.3389/fimmu.2017.01553
75. Kaźmierczak-Siedlecka K, Roviello G, Catalano M, Polom K. Gut Microbiota Modulation in the Context of Immune-Related Aspects of Lactobacillus spp. and Bifidobacterium spp. in Gastrointestinal Cancers. Nutrients. (2021) 13:2674. doi: 10.3390/nu13082674
76. Wan L, Wu C, Wu Q, Luo S, Liu J, Xie X. Impact of probiotics use on clinical outcomes of immune checkpoint inhibitors therapy in cancer patients. Cancer Med. (2023) 12:1841–9. doi: 10.1002/cam4.v12.2
77. Zhang Z, Liao Y, Tang D. Intratumoral microbiota: new frontiers in tumor immunity. Carcinogenesis. (2022) 43:719–27. doi: 10.1093/carcin/bgac063
78. Triner D, Devenport SN, Ramakrishnan SK, Ma X, Frieler RA, Greenson JK, et al. Neutrophils restrict tumor-associated microbiota to reduce growth and invasion of colon tumors in mice. Gastroenterology. (2019) 156:1467–82. doi: 10.1053/j.gastro.2018.12.003
79. Neuzillet C, Marchais M, Vacher S, Hilmi M, Schnitzler A, Meseure D, et al. Prognostic value of intratumoral Fusobacterium nucleatum and association with immune-related gene expression in oral squamous cell carcinoma patients. Sci Rep. (2021) 11:7870. doi: 10.1038/s41598-021-86816-9
80. Heymann CJF, Bobin-Dubigeon C, Muñoz-Garcia J, Cochonneau D, Ollivier E, Heymann MF, et al. Lipopolysaccharide-binding protein expression is associated to the metastatic status of osteosarcoma patients. J Bone Oncol. (2022) 36:100451. doi: 10.1016/j.jbo.2022.100451
81. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of microRNA-21. Gastroenterology. (2017) 152:851–66.e824. doi: 10.1053/j.gastro.2016.11.018
82. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. (2015) 42:344–55. doi: 10.1016/j.immuni.2015.01.010
83. Park SR, Kim DJ, Han SH, Kang MJ, Lee JY, Jeong YJ, et al. Diverse Toll-like receptors mediate cytokine production by Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans in macrophages. Infection Immun. (2014) 82:1914–20. doi: 10.1128/IAI.01226-13
84. Giam CZ, Semmes OJ. HTLV-1 infection and adult T-cell leukemia/lymphoma-A tale of two proteins: tax and HBZ. Viruses. (2016) 8:161. doi: 10.3390/v8060161
85. Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. (2017) 168:670–91. doi: 10.1016/j.cell.2016.11.037
86. Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer. (2017) 17:302–17. doi: 10.1038/nrc.2017.6
87. Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. (2013) 155:1639–51. doi: 10.1016/j.cell.2013.11.029
88. Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. (2014) 158:1110–22. doi: 10.1016/j.cell.2014.07.013
89. Fu A, Yao B, Dong T, Cai S. Emerging roles of intratumor microbiota in cancer metastasis. Trends Cell Biol. (2023) 33:583–93. doi: 10.1016/j.tcb.2022.11.007
90. Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. (2022) 185:1356–1372.e1326. doi: 10.1016/j.cell.2022.02.027
91. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. (2017) 358:1443–8. doi: 10.1126/science.aal5240
92. Domenis R, Cifù A, Marinò D, Fabris M, Niazi KR, Soon-Shiong P, et al. Toll-like receptor-4 activation boosts the immunosuppressive properties of tumor cells-derived exosomes. Sci Rep. (2019) 9:8457. doi: 10.1038/s41598-019-44949-y
93. Heymann CJF, Bard JM, Heymann MF, Heymann D, Bobin-Dubigeon C. The intratumoral microbiome: Characterization methods and functional impact. Cancer Lett. (2021) 522:63–79. doi: 10.1016/j.canlet.2021.09.009
94. Li WT, Iyangar AS, Reddy R, Chakladar J, Bhargava V, Sakamoto K, et al. The bladder microbiome is associated with epithelial-mesenchymal transition in muscle invasive urothelial bladder carcinoma. Cancers. (2021) 13:3649. doi: 10.3390/cancers13153649
95. Yan X, Liu L, Li H, Qin H, Sun Z. Clinical significance of Fusobacterium nucleatum, epithelial-mesenchymal transition, and cancer stem cell markers in stage III/IV colorectal cancer patients. OncoTargets Ther. (2017) 10:5031–46. doi: 10.2147/OTT.S145949
96. Zhang S, Li C, Liu J, Geng F, Shi X, Li Q, et al. Fusobacterium nucleatum promotes epithelial-mesenchymal transiton through regulation of the lncRNA MIR4435-2HG/miR-296-5p/Akt2/SNAI1 signaling pathway. FEBS J. (2020) 287:4032–47. doi: 10.1111/febs.v287.18
97. Quispe-Tintaya W, Chandra D, Jahangir A, Harris M, Casadevall A, Dadachova E, et al. Nontoxic radioactive Listeria (at) is a highly effective therapy against metastatic pancreatic cancer. Proc Natl Acad Sci USA. (2013) 110:8668–73. doi: 10.1073/pnas.1211287110
98. Howell LM, Forbes NS. Bacteria-based immune therapies for cancer treatment. Semin Cancer Biol. (2022) 86:1163–78. doi: 10.1016/j.semcancer.2021.09.006
99. Murphy C, Rettedal E, Lehouritis P, Devoy C, Tangney M. Intratumoural production of TNFα by bacteria mediates cancer therapy. PloS One. (2017) 12:e0180034. doi: 10.1371/journal.pone.0180034
100. Wood LM, Paterson Y. Attenuated Listeria monocytogenes: a powerful and versatile vector for the future of tumor immunotherapy. Front Cell infection Microbiol. (2014) 4:51. doi: 10.3389/fcimb.2014.00051
101. Chacón MR, Enrico DH, Burton J, Waisberg FD, Videla VM. Incidence of placebo adverse events in randomized clinical trials of targeted and immunotherapy cancer drugs in the adjuvant setting: A systematic review and meta-analysis. JAMA network Open. (2018) 1:e185617. doi: 10.1001/jamanetworkopen.2018.5617
102. Pratap Singh R, Kumari N, Gupta S, Jaiswal R, Mehrotra D, Singh S, et al. Intratumoral microbiota changes with tumor stage and influences the immune signature of oral squamous cell carcinoma. Microbiol Spectr. (2023) 11:e0459622. doi: 10.1128/spectrum.04596-22
103. Tan Q, Ma X, Yang B, Liu Y, Xie Y, Wang X, et al. Periodontitis pathogen Porphyromonas gingivalis promotes pancreatic tumorigenesis via neutrophil elastase from tumor-associated neutrophils. Gut Microbes. (2022) 14:2073785. doi: 10.1080/19490976.2022.2073785
104. Cavarretta I, Ferrarese R, Cazzaniga W, Saita D, Lucianò R, Ceresola ER, et al. The microbiome of the prostate tumor microenvironment. Eur Urol. (2017) 72:625–31. doi: 10.1016/j.eururo.2017.03.029
105. Hamada M, Inaba H, Nishiyama K, Yoshida S, Yura Y, Matsumoto-Nakano M, et al. Potential role of the intratumoral microbiota in prognosis of head and neck cancer. Int J Mol Sci. (2023) 24:15456. doi: 10.3390/ijms242015456
106. Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. (2016) 65:1973–80. doi: 10.1136/gutjnl-2015-310101
107. Peters BA, Pass HI, Burk RD, Xue X, Goparaju C, Sollecito CC, et al. The lung microbiome, peripheral gene expression, and recurrence-free survival after resection of stage II non-small cell lung cancer. Genome Med. (2022) 14:121. doi: 10.1186/s13073-022-01126-7
108. Che H, Xiong Q, Ma J, Chen S, Wu H, Xu H, et al. Association of Helicobacter pylori infection with survival outcomes in advanced gastric cancer patients treated with immune checkpoint inhibitors. BMC Cancer. (2022) 22:904. doi: 10.1186/s12885-022-10004-9
109. Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. (2018) 67:120–7. doi: 10.1136/gutjnl-2016-312580
110. Oh HJ, Kim JH, Bae JM, Kim HJ, Cho NY, Kang GH. Prognostic impact of fusobacterium nucleatum depends on combined tumor location and microsatellite instability status in stage II/III colorectal cancers treated with adjuvant chemotherapy. J Pathol Trans Med. (2019) 53:40–9. doi: 10.4132/jptm.2018.11.29
Keywords: intratumor microbiome, cancer, immune system, gut microbiota, therapy
Citation: Fang P, Yang J, Zhang H, Shuai D, Li M, Chen L and Liu L (2025) Emerging roles of intratumoral microbiota: a key to novel cancer therapies. Front. Oncol. 15:1506577. doi: 10.3389/fonc.2025.1506577
Received: 05 October 2024; Accepted: 04 February 2025;
Published: 25 February 2025.
Edited by:
Ahmed Lasfar, Rutgers, The State University of New Jersey, United StatesReviewed by:
Emma Berta Gutiérrez Cirlos, National Autonomous University of Mexico, MexicoCopyright © 2025 Fang, Yang, Zhang, Shuai, Li, Chen and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liping Liu, bGl1bGlwaW5nbGR5eUAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.