
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Oncol. , 17 March 2025
Sec. Gynecological Oncology
Volume 15 - 2025 | https://doi.org/10.3389/fonc.2025.1499163
This article is part of the Research Topic Non-coding RNAs as Potential Therapeutics and Biomarkers for Human Diseases View all 11 articles
Background: The aim of this study was to evaluate the association between miR-34 family expression and overall survival (OS) and progression-free survival (PFS) in women with ovarian cancer.
Methods: Literature searches were conducted using databases such as PubMed, Cochrane Library, EMBASE, Web of Science, Wanfang, and CNKI to identify studies reporting pooled hazard ratios (HRs) and 95% confidence intervals (CIs) examining the relationship between miR-34 family expression and overall survival (OS) or progression-free survival (PFS) in female patients with ovarian cancer. All potentially relevant studies were assessed and then pooled.
Results: There were a total of seven literatures included in this systematic review and meta-analysis, which included 672 women. There was a significant improvement in survival for women with ovarian cancer when miR-34s expression was higher (OS, HR = 0.70, 95% CI:0.57–0.86; PFS, HR = 0.48, 95% CI:0.31–0.75). A subgroup analysis of miR-34 family members showed that differences between groups greatly affected PFS (HR = 0.50, 95% CI: 0.40–0.63).
Conclusion: Based on the results of this review, it appears that ovarian cancer women with high expression of miR-34s may have a better chance of surviving.
Systematic Review Registration: PROSPERO (CRD42024499203).
Systematic review registration: https://www.crd.york.ac.uk/PROSPERO/, identifier CRD42024499203.
Among female reproductive system tumors, ovarian cancer is the most common, and it is one of the most aggressive (1). Ovarian cancer is usually diagnosed at an advanced stage with a poor prognosis due to the absence of clinical symptoms in the early stages and the high failure rate of chemotherapy (2). It is estimated that in 2020, there will be around 3,13,959 new cases of ovarian cancer globally, with an incidence of 6.6 and a mortality rate of 4.4, according to the data survey (3). In spite of improvements in survival rates for women with ovarian cancer in the past few decades, the 5-year survival rate for women with stage I epithelial ovarian cancer remains below 30%. It is well known that the miR-34 family (miR-34s) is essential for the pathogenesis of epithelial ovarian cancer, and its inactivation might have significant implications for its progression (2). Therefore, in order to improve the prognosis of ovarian cancer patients, biomarkers relating to prognosis must be identified and new therapeutic targets explored.
There are approximately 22 nucleotides in the length of microRNA (miRNA; miR) that are endogenous non-coding RNA. The miRNA negatively regulates gene expression at the post-transcriptional and translational levels, and plays a significant role in cancer progression (4). miR-34s is a family of three miRNAs that is encoded by two different genes in the human body, miR-34a, miR-34b, and miR-34c. There is an individual transcript for miR-34a on chromosome 1p36.22; however, miR-34b and miR-34c share the same chromosome 11q23.1 for transcription and expression (5). Several processes in tumor development are regulated by the tumor suppressor miR-34s, which is low expressed in most tumors, including proliferation, migration, invasion, metabolism, apoptosis, and cancer stem-cell formation, cell cycle progression, epithelial mesenchymal transformation, and so on, the result is an inhibition of tumor growth and metastasis (2, 6). Direct transcriptional targets of p53 have been shown to be miR-34a and miR-34b/c (7). According to Hannah Welponer et al. (8)show that The levels of miR-34a/b/c in ovarian cancer were found to be significantly decreased compared to control tissues (p=0.002, p<0.001, p<0.001, respectively). Furthermore, the expression of each member of the miR-34 family was determined to have independent prognostic significance in relation to progression-free survival (PFS) (miR-34a: HR=0.6, p=0.033; miR-34b: HR=0.2, p=0.001; miR-34c: HR=0.3, p=0.002, respectively). It was determined that miR-34b (HR=0.4, p=0.016) and miR-34c (HR=0.6, p=0.049) have a prognostic value for overall survival (OS). Research conducted by Jia et al. (9) found no correlation between the presence of miR-34a and overall survival (HR=1.407, p=0.929) or progression-free survival (HR=0.855, p=0.727) in individuals with epithelial ovarian cancer. However, a study by Song et al. (10) found significant effects of tumor size, FIGO stage, histological type, and miR-34c expression on overall survival in ovarian cancer patients. An analysis of multivariate data found a significant association between miR-34c expression and shorter OS time (p=0.038). Consequently, it is necessary to evaluate miR-34s expression and ovarian cancer survival through meta-analysis in order to determine the diagnosis and prognosis assessment of the miR-34s.
In this study, in order to obtain more accurate information about the prognosis of ovarian cancer, miR-34s expression needs to be further studied. Reviewing relevant literature and using systematic reviews and meta-analyses to assess miR-34s prognostic value in ovarian cancer was the goal of this study.
We searched six electronic databases was conducted for studies published from 2007 until April 2023. and their creation (PubMed, Cochrane Library, EMBASE, Web of Science, Wanfang, and CNKI) for relevant literature. Based on the PICOS tool, a search strategy was formulated: (P) Population: Ovarian cancer patients; (I) Intervention: high expression of miR-34s; (C) Comparator: low expression of miR-34s; (O) Outcomes: OS or PFS; (S) Study Design: retrospective or prospective studies. An overview of the search strategy can be found in Table 1 and Supplementary Materials (The example used here is PubMed).
The results of studies were included if they evaluated the following: (1) OS/PFS of ovarian cancer patients and miR-34s expression; (2) A sufficient amount of information was provided to calculate hazard ratios (HRs) and 95% confidence intervals (CIs); and (3) Retrospective or prospective studies.
(1) Data that is incomplete or unreported in studies; (2) Study results from non-observational studies[including reviews, cell experiments, animal experiments, correspondence, protocols, conference abstracts, or case reports.
Utilizing the literature management software NoteExpress 3.8, we screened and excluded the literature. Research was conducted by two researchers to identify duplication, review papers, conference papers, protocols, and correspondence in the literature. It was then determined which literature should be included and which should be excluded by two researchers after reading the abstracts. Finally, literature not included in the study was read in full by both researchers and was identified for inclusion. This process involved both researchers screening the literature independently, then comparing the remaining literature. Those who remained the same were included, and those who differed were brought in to discuss and resolve the differences.
Data were extracted independently by two reviewers. Data were collected using nine standardized and preselected data extraction forms under the following headings for inclusion in the study: (1) author, (2) date of publication, (3) nation,(4) Study design,(5) miRNA, (6)miRNA cut-off value,(7) disease type, (8) mean age, (9) sample size, (10) assay, and (11) outcome. A 95% CI and p value are provided for the HRs of miR-34s expression for OS and PFS. In the absence of direct reporting of HR and 95% CI, we calculated them based on the number of reported patients and events observed in each group. The HRs were estimated from graphical survival plots if only Kaplan-Meier curves were available (11). In cases where univariate and multivariate analyses were reported simultaneously, only the multivariate analysis was analyzed. Due to the fact that multivariate analysis accounted for confounding factors, the results were more valuable. It was resolved through discussion among the first three authors that differences over data extraction could be resolved.
To evaluate included studies, independent researchers used the Newcastle Ottawa scale (NOS), a method for assessing the quality of observational studies and non-randomized trials. Study groups are compared based on three factors: participant selection, comparability of results, and outcome assessment. In total, nine stars were awarded, one for each item, with the exception of the item “Comparability of cohorts based on design or analysis,” which was awarded two stars. Studies with quality scores less than 6 are considered low-quality studies. A study with a quality score of at least 6 is considered to be included in the meta-analysis. This meta-analysis follows the PRISMA guidelines. Under registration number CRD42024499203, it is registered with PROSPERO (International Register of Prospective Systems Evaluation).
A comprehensive statistical analysis was conducted using RevMan 5.4 software. OS and PFS were defined as the primary outcome indices. The hazard ratio (HR) and its corresponding 95% confidence interval (CI) were utilized as effect indices, calculated using the inverse variance (IV) method.
To assess the heterogeneity among included studies, we applied Cochran’s Q test and calculated the I² statistic (12). If the I² value was less than or equal to 50% and the p-value was greater than or equal to 0.1, it indicated no significant heterogeneity among studies, and a fixed effects model was employed for the meta-analysis. Conversely, if the I² value exceeded 50% and the p-value was less than 0.1, indicating significant heterogeneity, we opted for a random effects model. In the event of high heterogeneity, we conducted subgroup analyses stratified by individual members of the miR-34 family (miR-34a, miR-34b, and miR-34c) to explore potential sources of variability. Additionally, we performed sensitivity analyses to further investigate the origin of the observed heterogeneity. To address the possibility of publication bias, we generated funnel plots for visual assessment.
An additional two literatures were manually searched in addition to 85 literatures retrieved from six electronic databases. Following the elimination of duplicates, to review the remaining 78 literatures, we read their titles and abstracts, and 34 papers were then excluded again. In total, 34 literatures were read in full, but 27 literatures were excluded (due to incomplete data, review, cell experiment, animal experiment, and interventions included in the review were not met). A total of 7 literatures remain for inclusion in this study. During the period 2009 to 2020, there were 7 literatures published including 672 ovarian cancer patients. Different ethnic groups were involved: 3 from China, 3 from Austria and 1 from Canada. miRs were mainly expressed in tissue samples. The detection method was qRT-PCR. In all 7 studies, 6 studies had survival indicators of OS and 5 studies had survival indicators of PFS. As shown in Figure 1, the included studies have a variety of characteristics.
7 literatures were eligible for inclusion, of which one was of low quality and eight were of medium quality, mainly because none of the papers described the non-response rate (Table 2).
Since there was no heterogeneity in the included literatures (I2 = 35%<50%, p>0.1), the analysis was conducted using a fixed-effects model. There was an association between higher miR-34s expression and improved OS (HR = 0.70,95% CI 0.57-0.86, Figure 2A) in women with ovarian cancer.
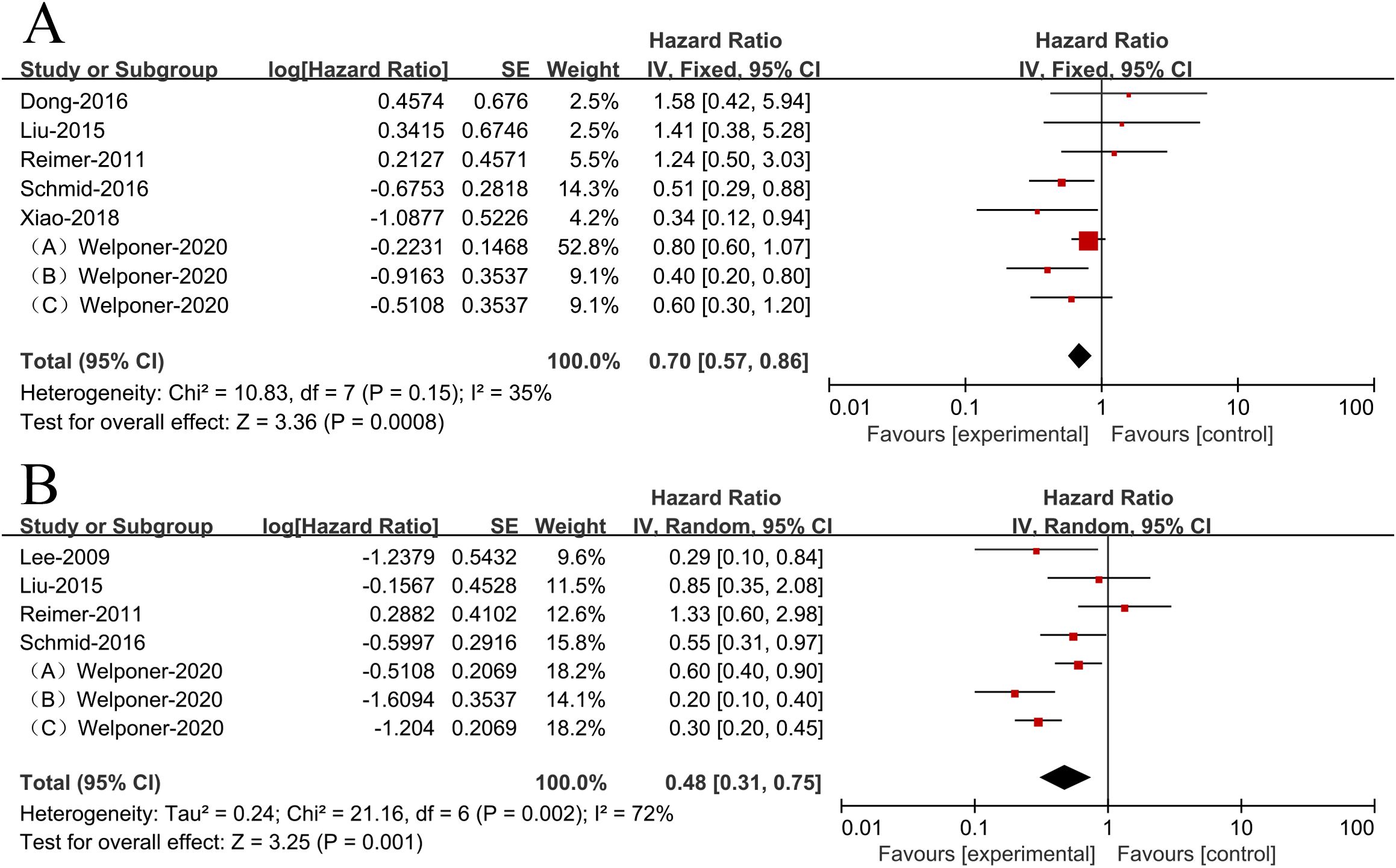
Figure 2. Patient survival plots for miR-34s expression in relation to overall survival (A) and progression-free survival (B).
Due to heterogeneity among the included literatures (I2 = 72%>50%, p <0.1), analyzing the data was done using a random effects model. According to the results, ovarian cancer patients with higher miR-34s expression had significantly better PFS (HR = 0.48,95% CI 0.31-0.75, Figure 2B).
Since the outcome indicator PFS had significant heterogeneity, we conducted subgroup analysis to find the causes of heterogeneity. According to a stratified analysis by miR-34s member type, studies in the miR-34a and miR-34c groups showed no heterogeneity, while the heterogeneity between the three groups reached a high degree (I2 = 67%, p<0.1, Figure 3), indicating that differences in the groups would greatly affect the results of meta-analysis.
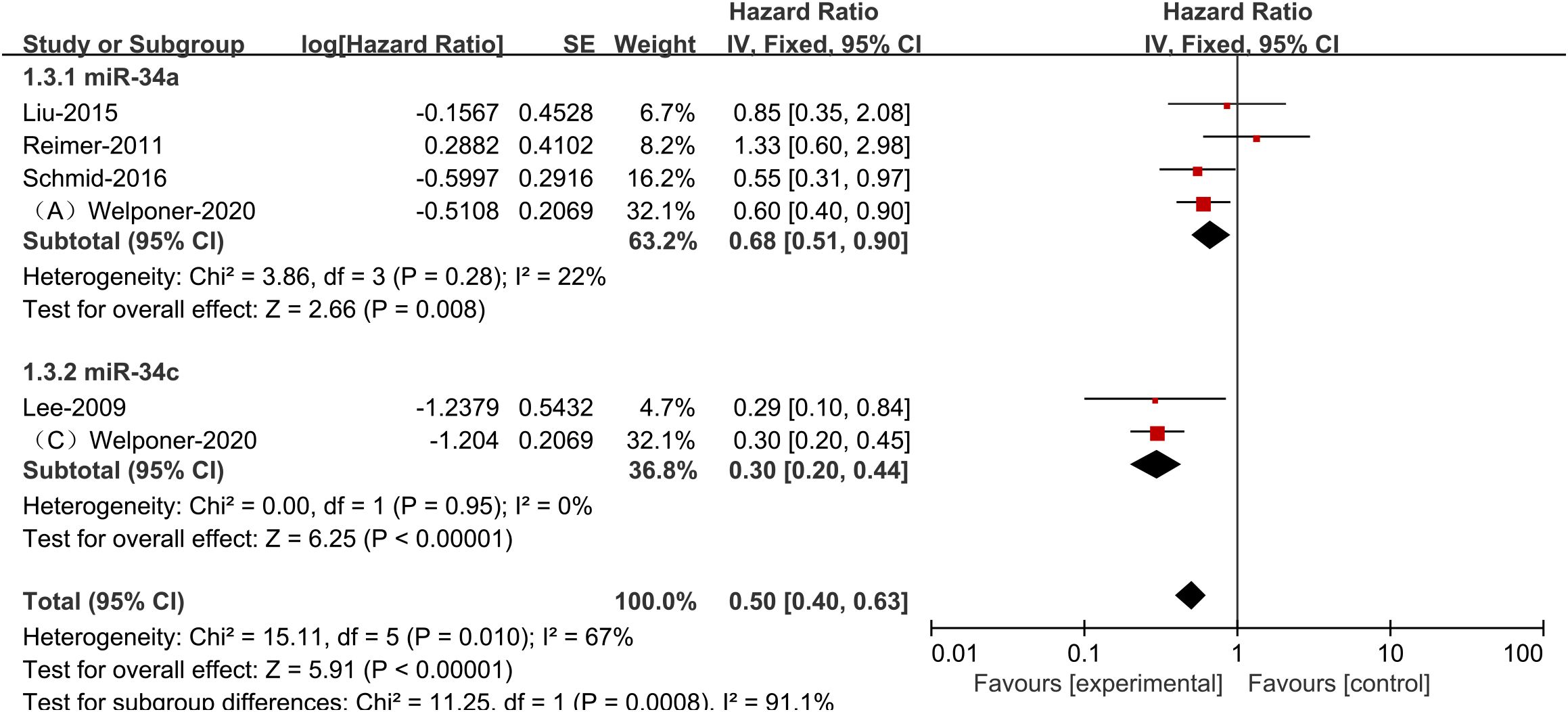
Figure 3. Plots showing specific miR-34s member expression and progression-free survival in ovarian cancer subgroups.
An analysis of OS and PFS sensitivity was conducted by eliminating individual studies, and it was found that no study had a significant impact on the overall results, and the results were relatively stable. According to both OS and PFS publication bias tests, it was found that the funnel plot was roughly symmetric and that there were no significant publication biases (Figure 4).
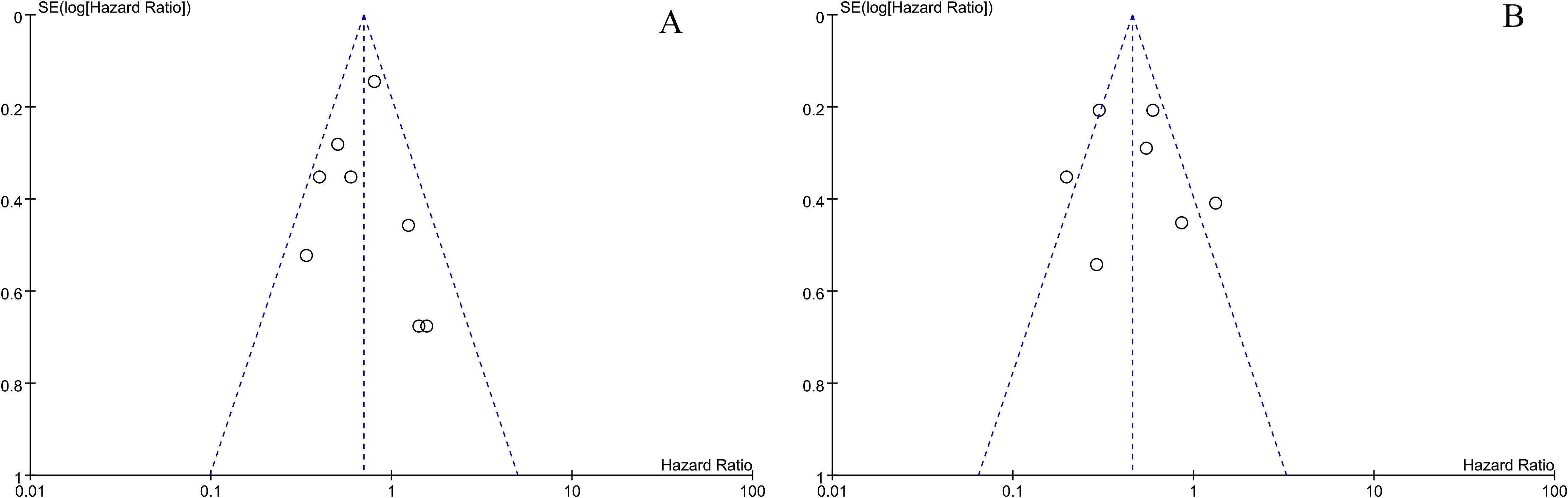
Figure 4. The overall survival and progression-free survival of miR-34s are depicted in a funnel plot (A, B).
Researchers have examined the relationship between miR-34s expression and survival in ovarian cancer patients. A total of 87 literatures were collected through six database and other ways, and 7 literatures were finally included. Firstly, a NOS quality assessment was conducted on the seven included studies, which were generally deemed to be of high quality. A pooled analysis of nine studies with 672 women, higher expression of miR-34s significantly increased OS and PFS for patients with ovarian cancer. Based on the significant heterogeneity of the outcome indicator PFS, stratified analysis by miR-34s member types identified miR-34a and miR-34c expression levels are higher as significantly associated with increased PFS. Lastly, sensitivity analysis found that no single study had a significant impact on overall results, and that the results were relatively stable and reliable. Using a publication bias test, the funnel plot was found to be roughly symmetrical, without evidence of publication bias.
The meta-analysis of research studies revealed that miR-34s expression was associated with a significant improvement in ovarian cancer patients’ OS and PFS. These findings align with the results reported by Schmid et al. (15), indicating that ovarian cancer cases with reduced miR-34a expression exhibited a decrease in PFS (p = 0.039) and OS (p = 0.016). Researchers have found that miR-34s expression levels are lower in cancerous tissues than non-tumorous tissues in ovarian cancer tissues (8). Ovarian cancer exhibiting elevated miR-34a and miR-34c levels may demonstrate reduced malignant potential, decreased invasiveness, and limited dissemination (8). Kwan et al. (17) observed there is a notable decrease in miR-34s expression in a number of malignant tumors, including breast cancer and colon cancer. They further demonstrated that this downregulation facilitated tumor suppression by modulating key molecular pathways involved in tumorigenesis. Inhibition of lung squamous cell carcinoma proliferation, migration, and invasion was associated with high miR-34s expression, according to Sun et al. (18) By targeting miR-34c, Wang et al. (19) demonstrated in vivo and in vitro that miR-34c inhibited malignant behaviors such as invasion, migration, proliferation, and epithelial-mesenchymal transformation of nasopharyngeal carcinoma. According to Elena et al. (20), it is possible that miR-34a can serve as an important indicator of recurrence in patients with non-small cell lung cancer. The diminished expression of miR-34s in osteosarcoma and hepatoblastoma has been correlated with an unfavorable prognosis (21, 22). Hence, it is believed that reduced miR-34s expression is a crucial factor in the initiation and progression of tumor growth.
We acknowledge that the heterogeneity for PFS in our study was high (I² = 72%), indicating significant differences among the included studies. To explore potential sources of this heterogeneity in more detail, we conducted a subgroup analysis to evaluate the impact of different RNA families on the results. Subsequently, we found that after performing the subgroup analysis, the I² value decreased to below 50%, suggesting that these subgroupings explain a substantial part of the heterogeneity. miR-34s represent the initial miRNA identified as being directly regulated by p53, with miR-34a demonstrating the most pronounced level of regulation by p53 (23). Furthermore, p53 and miR-34a expression is influenced by various indirect regulatory factors; for instance, miR-34a can enhance p53 activity by downregulating the expression of SIRT1, a NAD-dependent deacetylase involved in information regulation. Following the inhibition of p53 protein transcription, there is a subsequent down-regulation of p53 protein activity during the process of deacetylation (5, 24). In the context of epithelial-mesenchymal transition (EMT), miR-34 plays a crucial role by regulating the transcription factor Snail, which is pivotal in promoting EMT and invasion (16, 25). Additionally, miR-34a has been shown to negatively regulate E2F3a, a key transcription factor that promotes cell proliferation and facilitates the G1/S transition. In ovarian cancer cells, knockdown of miR-34a resulted in a significant increase in E2F3 expression, highlighting its role in cell cycle regulation (14). Moreover, miR-34a’s role extends to apoptosis, where its expression can influence apoptotic pathways, promoting cell death in cancer cells. This suggests that miR-34a not only acts as a tumor suppressor through cell cycle arrest and EMT regulation but also enhances apoptosis, thereby contributing to the suppression of malignant characteristics in ovarian cancer. For example, upregulation of miR-34a-5p has been shown to suppress the malignant characteristics of ovarian cancer cells by targeting TRIM44, impeding the advancement of the disease (26).Additionally, the miR-34 family members exhibit co-targeting of MET, a receptor protein tyrosine kinase, which influences the motility and invasion of epithelial ovarian cancer (27).Research has identified that MET is a target of miR-34c, enhancing the anti-tumor efficacy of cisplatin in ovarian cancer cells (28).In a study by Lu et al. (29), reverse transcriptional quantitative PCR demonstrated a significant reduction in exosome miR-34b expression in ovarian cancer cells, suggesting that exosome-derived miR-34b can diminish cell proliferation and hinder EMT in the SKOV3 ovarian cancer cell line. Based on these mechanistic studies, the findings collectively emphasize the importance of miR-34s in the cellular biology of ovarian cancer, particularly regarding their suppressive impact on proliferation, invasion, and promotion of apoptosis.
According to Kumar et al. (3), a MeDIP-NGS analysis revealed significant decreases in the relative expression levels of microRNA-34a in both tissue and serum samples of early epithelial ovarian cancer (p<0.0001). Furthermore, the functional analysis of microRNA-34a demonstrated that patients with stage III-IV and I-II epithelial ovarian cancer (EOC) had areas under curves of 92.0 (p<0.0001) and 82.7 (p<0.0001), respectively, suggesting that serum samples from these patients may be useful for monitoring cancer progression. In summary, miR-34s may serve as potential biomarkers in the diagnosis and prognosis of ovarian cancer. However, limitations such as varying sample sources, a paucity of literature, and inadequate sample sizes within studies may compromise the statistical robustness of miR-34s’ prognostic value. By controlling for certain objective conditions, it is feasible to accurately forecast the prognosis of miR-34s expression in ovarian cancer.
To our knowledge, this is the first systematic review and meta-analysis of miR-34s’ prognostic value in ovarian cancer. From the methodological aspect, we conducted a two-person assessment, searched 6 databases, and further collected the literature comprehensively. We formulated inclusion and exclusion criteria according to PICOS principle, and the literature quality was relatively high.
Due to the limited number of studies included in our analysis, consisting of three from China, three from Austria, and one from Canada, the data available for certain analyses and subgroup analyses was deemed insufficient. The findings of this study necessitate additional validation through large-scale, multicenter, multi-factorial, and high-quality clinical studies. Secondly, the potential for bias in the study may be influenced by variations in geographic regions and racial demographics. Thirdly, factors such as age, family history, and weight may also introduce bias.
Based on the results of this review, miR-34s could potentially serve as a prognostic indicator for an improved survival outcome for female ovarian cancer patients. However, due to the constraints of the present analysis, it is advisable to exercise caution when interpreting the conclusions. Additional clinical trials with rigorous methodology, a substantial sample size, and an extended follow-up period are necessary to further elucidate the prognostic significance of miR-34s expression in ovarian cancer.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
XLu: Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. XLi: Conceptualization, Methodology, Writing – review & editing. CC: Data curation, Formal analysis, Writing – review & editing. JY: Writing – review & editing, Data curation, Methodology, Supervision, Funding acquisition, Resources. HZ: Conceptualization, Formal analysis, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Guizhou Provincial Health Commission Science and Technology Fund (Project No. gzwkj2023-206).
We are grateful to all the authors for their individual contributions to the present study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1499163/full#supplementary-material
1. Shu C, Liu L, Chen X, Xue J, Fei J, Wang J, et al. ncRNA-mediated low expression of P2RY14 correlates with poor prognosis and tumor immune infiltration in ovarian carcinoma. Ann Transl Med. (2023) 11:10. doi: 10.21037/atm-22-6120
2. Tao F, Tian X, Lu M, Zhang Z. A novel lncrna, lnc-oc1, promotes ovarian cancer cell proliferation and migration by sponging mir-34a and mir-34c. J Genet Genomics. (2018) 45:137–45. doi: 10.1016/j.jgg.2018.03.001
3. Kumar V, Gupta S, Varma K, Chaurasia A, Sachan M. Diagnostic performance of microrna-34a, let-7f and microrna-31 in epithelial ovarian cancer prediction. J Gynecol Oncol. (2022) 33:e49. doi: 10.3802/jgo.2022.33.e49
4. Inoue J, Inazawa J. Cancer-associated mirnas and their therapeutic potential. J Hum Genet. (2021) 66:937–45. doi: 10.1038/s10038-021-00938-6
5. Pan W, Chai B, Li L, Lu Z, Ma Z. P53/microrna-34 axis in cancer and beyond. Heliyon. (2023) 9:e15155. doi: 10.1016/j.heliyon.2023.e15155
6. Rokavec M, Li H, Jiang L, Hermeking H. The P53/mir-34 axis in development and disease. J Mol Cell Biol. (2014) 6:214–30. doi: 10.1093/jmcb/mju003
7. He L, He X, Lim LP, de StanChina E, Xuan Z, Liang Y, et al. A microrna component of the P53 tumour suppressor network. Nature. (2007) 447:1130–4. doi: 10.1038/nature05939
8. Welponer H, Tsibulak I, Wieser V, Degasper C, Shivalingaiah G, Wenzel S, et al. The mir-34 family and its clinical significance in ovarian cancer. J Cancer. (2020) 11:1446–56. doi: 10.7150/jca.33831
9. Liu J ZY, Zhang L, Yan Y, Zheng H. Expression and clinical significance of four mirnas in epithelial ovarian cancer. Tianjin Med J. (2015) 43:996–1000. doi: 10.11958/i.issn.0253-9896.2015.09.0010
10. Xiao S, Li Y, Pan Q, Ye M, He S, Tian Q, et al. Mir-34c/sox9 axis regulates the chemoresistance of ovarian cancer cell to cisplatin-based chemotherapy. J Cell Biochem. (2019) 120:2940–53. doi: 10.1002/jcb.26865
11. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. (2007) 8:16. doi: 10.1186/1745-6215-8-16
12. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. (2003) 327:557–60. doi: 10.1136/bmj.327.7414.557
13. Lee CH, Subramanian S, Beck AH, Espinosa I, Senz J, Zhu SX, et al. Microrna profiling of brca1/2 mutation-carrying and non-mutation-carrying high-grade serous carcinomas of ovary. PloS One. (2009) 4:e7314. doi: 10.1371/journal.pone.0007314
14. Reimer D, Hubalek M, Kiefel H, Riedle S, Skvortsov S, Erdel M, et al. Regulation of transcription factor E2f3a and its clinical relevance in ovarian cancer. Oncogene. (2011) 30:4038–49. doi: 10.1038/onc.2011.119
15. Schmid G, Notaro S, Reimer D, Abdel-Azim S, Duggan-Peer M, Holly J, et al. Expression and promotor hypermethylation of mir-34a in the various histological subtypes of ovarian cancer. BMC Cancer. (2016) 16:102. doi: 10.1186/s12885-016-2135-2
16. Dong P, Xiong Y, Watari H, Hanley SJ, Konno Y, Ihira K, et al. Mir-137 and mir-34a directly target snail and inhibit emt, invasion and sphere-forming ability of ovarian cancer cells. J Exp Clin Cancer Res. (2016) 35:132. doi: 10.1186/s13046-016-0415-y
17. Wong KY, Yu L, Chim CS. DNA methylation of tumor suppressor mirna genes: A lesson from the mir-34 family. Epigenomics. (2011) 3:83–92. doi: 10.2217/epi.10.74
18. Sun D, Wu Y, Zhang S, Han Y, Shen J, Zheng W, et al. Distinct roles of miR-34 family members on suppression of lung squamous cell carcinoma. BioMed Pharmacother. (2021) 142:111967. doi: 10.1016/j.biopha.2021.111967
19. Wan FZ, Chen KH, Sun YC, Chen XC, Liang RB, Chen L, et al. Exosomes overexpressing mir-34c inhibit Malignant behavior and reverse the radioresistance of nasopharyngeal carcinoma. J Transl Med. (2020) 18:12. doi: 10.1186/s12967-019-02203-z
20. Gallardo E, Navarro A, Viñolas N, Marrades RM, Diaz T, Gel B, et al. Mir-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. (2009) 30:1903–9. doi: 10.1093/carcin/bgp219
21. Wang Y, Jia LS, Yuan W, Wu Z, Wang HB, Xu T, et al. Low mir-34a and mir-192 are associated with unfavorable prognosis in patients suffering from osteosarcoma. Am J Transl Res. (2015) 7:111–9.
22. Jiao C, Zhu A, Jiao X, Ge J, Xu X. Combined low mir-34s are associated with unfavorable prognosis in children with hepatoblastoma: A chinese population-based study. J Pediatr Surg. (2016) 51:1355–61. doi: 10.1016/j.jpedsurg.2016.02.091
23. Hermeking H. Micrornas in the P53 network: micromanagement of tumour suppression. Nat Rev Cancer. (2012) 12:613–26. doi: 10.1038/nrc3318
24. Zhang DG, Zheng JN, Pei DS. P53/microrna-34-induced metabolic regulation: new opportunities in anticancer therapy. Mol Cancer. (2014) 13:115. doi: 10.1186/1476-4598-13-115
25. Jiang X, Ye Z, Jiang Y, Yu W, Fang Q. Lncrna oip5-as1 upregulates snail expression by sponging mir-34a to promote ovarian carcinoma cell invasion and migration. Biol Res. (2020) 53:49. doi: 10.1186/s40659-020-00315-1
26. Li HL, Duan YA, Zhao N. Mir-34a-5p directly targeting trim44 affects the biological behavior of ovarian cancer cells. Eur Rev Med Pharmacol Sci. (2021) 25:1250–60. doi: 10.26355/eurrev_202102_24829
27. Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, et al. Frequent downregulation of mir-34 family in human ovarian cancers. Clin Cancer Res. (2010) 16:1119–28. doi: 10.1158/1078-0432.Ccr-09-2642
28. Yang S, Li Z, Luo R. miR-34c targets MET to improve the anti-tumor effect of cisplatin on ovarian cancer. Onco Targets Ther. (2020) 13:2887–97. doi: 10.2147/ott.S239425
Keywords: meta-analysis, microRNA, miR-34s, ovarian cancer, prognosis
Citation: Luo X, Li X, Chen C, Yang J and Zheng H (2025) The prognostic value of miR-34 family in ovarian cancer: a systematic review and meta-analysis. Front. Oncol. 15:1499163. doi: 10.3389/fonc.2025.1499163
Received: 25 September 2024; Accepted: 28 February 2025;
Published: 17 March 2025.
Edited by:
Alice Conigliaro, University of Palermo, ItalyReviewed by:
Amudha Ganapathy, University of Illinois Chicago, United StatesCopyright © 2025 Luo, Li, Chen, Yang and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Zheng, emhlbmdob25naHFAaG90bWFpbC5jb20=; Jing Yang, ODQ0MjQzMTIzQHFxLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.