
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 18 February 2025
Sec. Cancer Epidemiology and Prevention
Volume 15 - 2025 | https://doi.org/10.3389/fonc.2025.1478036
This article is part of the Research Topic Advancing Personalized Cancer Care: Novel Genetic and Statistical Approaches View all 4 articles
Background: Epithelioid hemangioendothelioma (EHE) is a rare malignant vascular tumor. Most EHEs (>90%) cases harbor WWTR1::CAMTA1 fusion gene, and CAMTA1 immunohistochemistry (IHC) is a highly sensitive and specific tool for EHE diagnosis. However, there exist CAMTA1-immunonegative cases, the majority of which harbor YAP1::TFE3 fusion, with a few cases having more rare fusions. Liver is one of the most common sites of EHE, where the CAMTA1 subtype dominates, and the other variants are extremely rare. Hence, we focused on the hepatic CAMTA1-immunonegative EHEs to analyze the clinicopathological and molecular features of these peculiar cases.
Methods: The SNOMED search of the hospital pathology files between January 2016 to November 2023 identified 57 hepatic EHEs and 7 cases were CAMTA1-immunonegative. Fluorescence in situ hybridization (FISH), next generation sequencing (NGS) and Sanger sequencing were performed to identify the genetic change of the 7 cases.
Results: This series included 3 females and 4 males, aged from 33 to 64 years. All the 7 cases were negative for CAMTA1 IHC. Four cases were positive for TFE3 IHC and exhibited YAP1::TFE3 fusion. Another 3 cases were also negative for TFE3, while WWTR1::CAMTA1 fusion were detected by NGS in 1 case and demonstrated by FISH in all the 3 cases. Morphologically, among the 4 TFE3 rearrangement cases, 3 cases showed the TFE3-sutype morphologic appearance, while the histology of 1 case was similar to that of CAMTA1- subtype. In the 3 CAMTA1-rearranged lesions, 2 cases had classic EHE morphology, and 1 case exhibited atypical histology, with higher atypia and well-formed vessels. Surgical resection was performed on five cases and two cases were biopsied and received chemotherapy. Follow-up information was available in 6 patients (median 46 months), including 4 patients were alive without disease and 2 patients were alive with disease.
Conclusion: Our study reported 7 CAMTA1-immunonegative hepatic EHEs and most of them were TFE3-rearranged EHEs with morphology variation. Moreover, there does exist the CAMTA1-immunonegative but CAMTA1-rearranged EHE cases. Therefore, the diagnosis of EHE should be based on morphology, combined with CAMTA1 and TFE3 IHC, and if necessary, supplemented by genetic analysis including FISH and NGS, to establish correct diagnosis.
Epithelioid hemangioendothelioma (EHE) is a rare malignant vascular tumor, whose clinical behavior is between hemangioma and malignant angiosarcoma (1). EHE affects all ages with a peak incidence in the 4-5th decades of life and a slight female preponderance in visceral tumors (2). Histologically, conventional EHE is composed of strands and nests of epithelioid cells in a myxohyaline stroma, with the presence of intracytoplasmic vacuoles which may contain erythrocytes. In 2001, Mendlick et al. found recurrent chromosomal translocation, involving 1p36.3 and 3q25 in EHE, and then Errani et al. demonstrated this recurrent translocation result in the WWTR1::CAMTA1 fusion gene (3, 4). Hence, CAMTA1 immunohistochemistry (IHC) has been developed as a sensitive and specific tool for the diagnosis of EHE cases, and for the challenging cases, fluorescence in-situ hybridization (FISH) for the identification of WWTR1::CAMTA1 fusion could further confirm the diagnosis (5, 6).
While a subset of EHE were found to be CAMTA1-immunonegative and fusion-negative, with distinctive morphology, such as abundant eosinophilic cytoplasm, and well-defined vascular channels. These cases were proved to be TFE3-rearranged, harboring YAP1::TFE3 fusion, and TFE3 IHC serve as a useful screening tool for this subtype (7). Although most CAMTA1-negative cases are proved to be TFE3-subtype. In our clinical practice, we found there existed CAMTA1-immunonegative, but CAMTA1-rearranged EHE cases, and such peculiar cases were also reported in previous studies, with CAMTA1-rearrangemnt or other exceedingly rare fusion variant, including WWTR1::ACTL6A and WWTR1::TFE3 fusions (8–11).
Liver is one of the most common sites of EHE, in which the CAMTA1-subtype dominates (>90%), and the TFE3-subtype is extremely rare. Hence, we performed this study focusing CAMTA1-immunonegative EHEs involving the liver, to further elucidate the clinicopathological and molecular features of CAMTA1-immunonegative EHE lesions at our institution. To the best of our knowledge, this is the first study of these rare, peculiar hepatic CAMTA1-immunonegative EHE cases.
This study was approved by the Institutional Review Board of West China Hospital. A SNOMED search of hospital surgical pathology documents from January 2016 to November 2023 identified 145 EHEs, including 57 (39.3%) hepatic EHEs. Seven EHE cases with negative CAMTA1 expression were finally identified. All cases were independently reviewed by two pathologists with soft tissue tumor pathology expertise (H.Z. and X.H.). Clinical and follow-up information was collected from the clinical records and pathology reports.
Immunohistochemical staining was performed on 4-μm-thick formalin-fixed, paraffin-embedded (FFPE) tissue sections using the EnVision Plus detection system (Dako, Carpinteria, CA). IHC were performed using the following antibodies: CAMTA1 (clone NBP1-93620, 1:200; Novus Biologicals, Littleton, CO, USA), TFE3 (clone MRQ-37, 1:50; Cell Marque, CA, USA), CD31 (clone JC70A, 1:200; Dako, CA,USA), CD34 (clone QBEnd10, 1:200; Dako, CA,USA), ERG (clone UMAB78, 1:200; ZSGB-Bio, Beijing, China), cytokeratin (clone AE1/AE3, 1:100; Dako, CA,USA), Epithelial Membrane Antigen (EMA, clone GP1.4, 1:150; ZSGB-Bio, Beijing, China), Ki-67 (clone MIB-1, 1:100; Dako, CA, USA). In the immunohistochemical evaluation of CAMTA1 or TFE3, cases showing significant nuclear staining in >5% of tumor cells were considered positive (8). The staining intensity of each case was divided into weak, moderate, or strong, and the extent of nuclear immunoreactivity was scored according to the percentage of positive tumor cells: 1+ (5% to 35%), 2+ (36% to 65%), and 3+ (66% to 100%) (5).
FISH analysis was performed on seven tumors with available material, using the GSP WWTR1:: CAMTA1 fusion gene probe (Anbiping, Guangzhou, China) and GSP TFE3 break apart probe (Anbiping, Guangzhou, China) for the detection of WWTR1::CAMTA1 and TFE3 rearrangement, respectively. The FISH assays were performed on 4-μm-thick sections according to an established laboratory protocol (12). At least 100 nuclei were counted in each case and the tumor was interpreted as positive for WWTR1:: CAMTA1 fusion when at least 10 out of 100 (10%) tumor cells showed a (yellow) fusion signal. The case was interpreted as TFE3-rearranged when at least 10 out of 100 (10%) tumor cells exhibited a split signal pattern which showed that the distance between the green and red signals was greater than the diameter of two signals.
Four cases with available material were analyzed by RT-PCR. Total RNA was isolated from 4-μm sections of FFPE tissue material using the High Pure FFPE RNA Micro Kit (Qiagen, CA, USA) according to the manufacturer’s instructions. CDNA was synthesized using the PrimeScript RT reagent kit (Takara, Tokyo, Japan). The PCR was performed according to standard procedures using the primers (YAP1-exon1-forward: 5’-CTCCGGAAGCTGCCCGACTCC-3’, and TFE3-exon4-reverse: 5’-ACAGGTACTGTTTCACCTGCT-3’). The PCR products were sequenced by the Sanger sequencing (Tsingke Biological Technology, Chengdu, China).
Genomic DNA of case 3 was extracted from the formalin-fixed paraffin- embedded (FFEP) tissue using a QIAamp DNA Mini Kit (Qiagen, CA, USA) and then quantified by Nanodrop spectrophotometer (Thermo Fisher Scientific, DE, USA). Subsequently, the DNA was sheared, purified, ligated with adapters and used for the library construction, and then hybridized to a panel of 1021 genes containing whole exons of 312 genes, selected introns of 38 genes, and selected regions of 709 genes. Sequencing was performed with the Illumina gene+Seq 2000 system (Illumina, San Diego, CA, USA). DNA and RNA of case 7 were extracted from FFPE samples using a QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA) and RNeasy FFPE Kit (Qiagen, Valencia, CA, USA). The NGS were performed using a cancer-related DNA+RNA panel (YuansuS, OrigiMed). DNA and RNA library construction were performed according to the manufacturers’ instructions. The genes were captured and sequenced with a mean coverage of 700× on the Illumina gene+Seq 2000 system (Illumina, San Diego, CA, USA).
The clinical features of hepatic EHE are summarized in Table 1. This series included 4 males and 3 females (ratio 1.33:1), aged from 33 to 64 years (median 44 years; mean 47.8 years). The tumor size ranged from 1.6 to 9 cm (median was 2.7 cm; mean 3.6 cm). Four patients had clinical symptoms, including abdominal pain, cough, fever, and jaundice. The other 3 patients were asymptomatic, and the tumors were found incidentally. Six tumors occurred within the liver, and in 1 case the tumors involved the liver and lung simultaneously. Among the 6 cases, three patients had multicentric lesions involving the whole liver, and another three patients had solitary lesions, with 2 tumors occurring in the right lobe and 1 in the left lobe of the liver.
Immunohistochemically, nuclear CAMTA1 was negative in all the 7 EHE tumors, but the positive control showed positivity. Among the 7 cases, 4 cases exhibited TFE3 positivity and 3 cases were also TFE3-immunonegative. In the 4 TFE3-positive cases, 2 cases showed diffuse nuclear staining (3+) with moderate to strong intensity for TFE3 (case 1 and 2), another 2 cases showed focal nuclear staining (2+) with moderate to strong (case 4) or moderate intensity (case 3). The vascular markers CD31, ERG, and CD34 were generally and strongly expressed in all the 7 cases, and the epithelial markers EMA, cytokeratin was negative in all tested cases. The Ki-67 index was about 5% of the neoplastic cells in 6 cases but was up to 15% in case1.
In the 4 CAMTA1-immunonegative but TFE3-immunopositve cases, all cases were positive for TFE3 rearrangement by FISH, and further demonstrated by RT-PCR. In one case, YAP1::TFE3 fusion was identified by NGS (case 3). All the 4 cases harbored gene fusions between YAP1 exon 1 (NM_001130145.2) and TFE3 exon 4(NM_006521.4).
Among the 3 cases who were immunonegative for both CAMTA1 and TFE3, 3 cases were subjected to FISH analysis, and WWTR1::CAMTA1 fusion gene were identified in each of the 3 cases. In case 7, WWTR1::CAMTA1 fusion was also identified by NGS at both the DNA and RNA levels with the fusion between WWTR1 exon 2 and CAMTA1 exon 9 (WWTR1:NM_015472; CAMTA1: NM_015215). Moreover, the NGS identified a novel breakpoint at nucleotide positions 1663 of CAMTA1 gene.
In the 4 TFE3-rearranged EHE cases, two cases (case 1 and 2) were composed of solid sheets and nests of large epithelioid cells in an inflammatory background (Figure 1A). The tumor cells had prominent nucleoli, with abundant, eosinophilic to vacuolated cytoplasm (Figure 1B), and nuclear pleomorphism can be seen (Figure 1C). The dominant feature was the well-formed, numerous vascular channels, and some areas formed pseudoalveolar-like architecture (Figure 1D). Both of the cases exhibited TFE3 immunopositivity and YAP1::TFE3 fusion gene (Figures 1E–F). In case 3, the tumor intermingled with liver parenchyma in a solid growth pattern with medium-sized epithelioid cells, exhibiting slightly rich cytoplasm, round nuclei and conspicuous nucleoli (Figure 2A). Mature vessel lumen formation which contained prominent erythrocytes, could be identified in some area (Figure 2B). This case harbored focal nuclear staining (2+) for TFE3 and YAP1::TFE3 fusion gene (Figures 2C, D).

Figure 1. Morphological features of TFE3 rearranged EHE case and corresponding immunohistochemical and genetic results. (A) Nest or sheets of epithelioid cells against the background of inflammatory cell (H&E, ×200). (B) The tumor cells had abundant, eosinophilic to vacuolated cytoplasm with prominent nucleoli (H&E, ×400). (C) A few cells with nuclear pleomorphism(arrow) were seen around (H&E, ×400). (D) Some areas formed pseudoalveolar architecture (H&E, ×200). (E) TFE3 positivity was identified in the case (×200), and Sanger sequencing demonstrated the presence of the YAP1::TFE3 fusion gene (F).
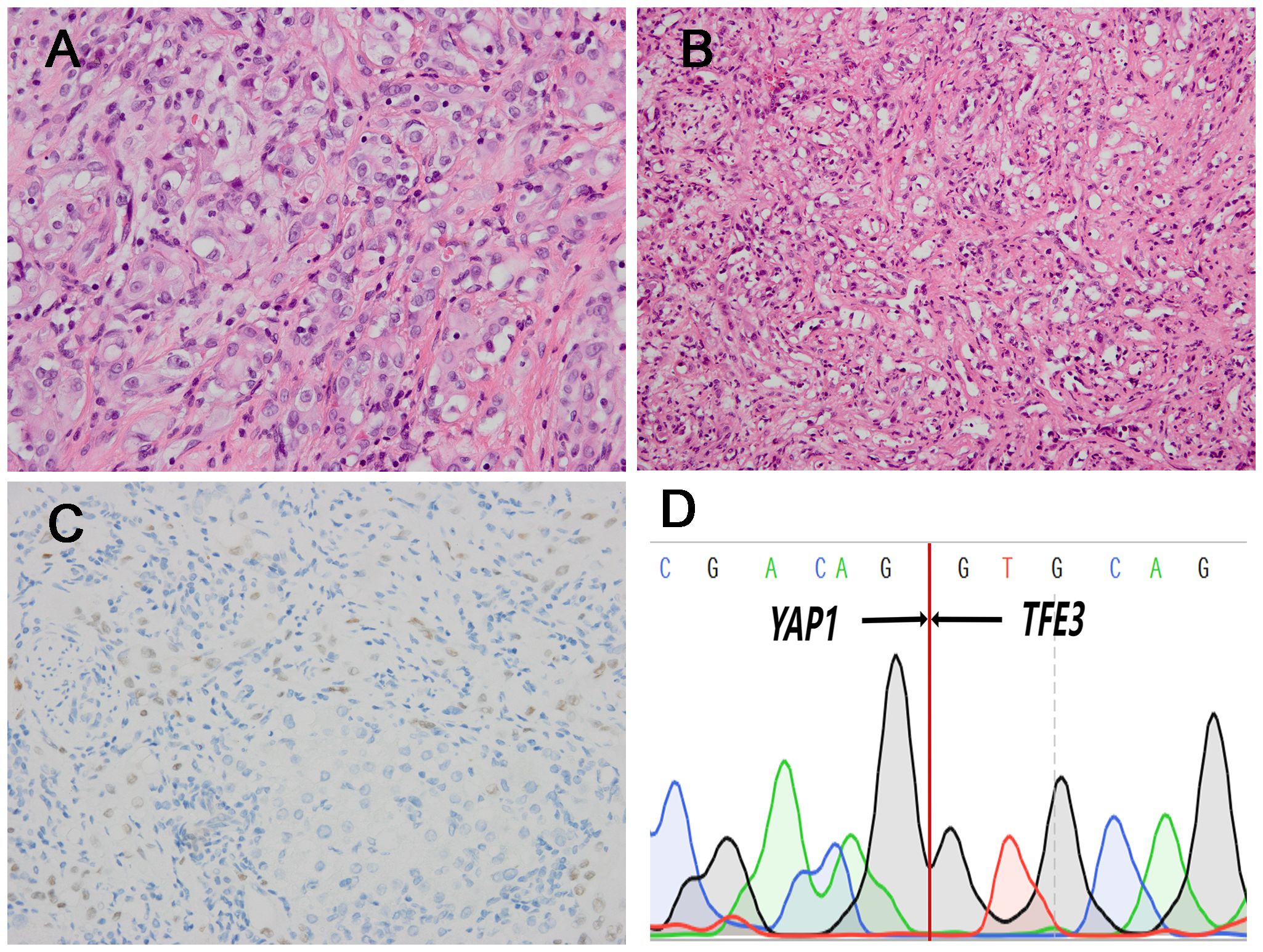
Figure 2. Histopathological features of one TFE3 rearranged (case3) which had lower atypia and corresponding immunohistochemical and genetic results. (A) Medium-sized epithelioid cells exhibited slightly rich cytoplasm, round nuclei and conspicuous nucleoli (H&E, ×400). (B) There were the formation of vascular channels containing erythrocytes (H&E, ×200). (C) The case was positive for TFE3 IHC (×200). (D) Sanger sequencing confirmed the presence of YAP1::TFE3 fusion gene.
In case 4, the tumor cells arranged in discohesive strands or single cells in the collagen matrix. The lesion comprised spindle cells with mild atypia, fusiform nuclei, inconspicuous nucleoli, and indistinct cell borders (Figure 3A). In some cells, intracytoplasmic vacuoles were present with lumina formation, exhibiting signet-ring like appearance (Figure 3B). This case also showed nuclear staining (2+) for TFE3 and TFE3-rearrangement (Figures 3C, D). The mitotic figures ranged from 0 to 2 per 10 high-power fields (HPFs) in the 4 cases. Significant necrosis was not identified in four cases.
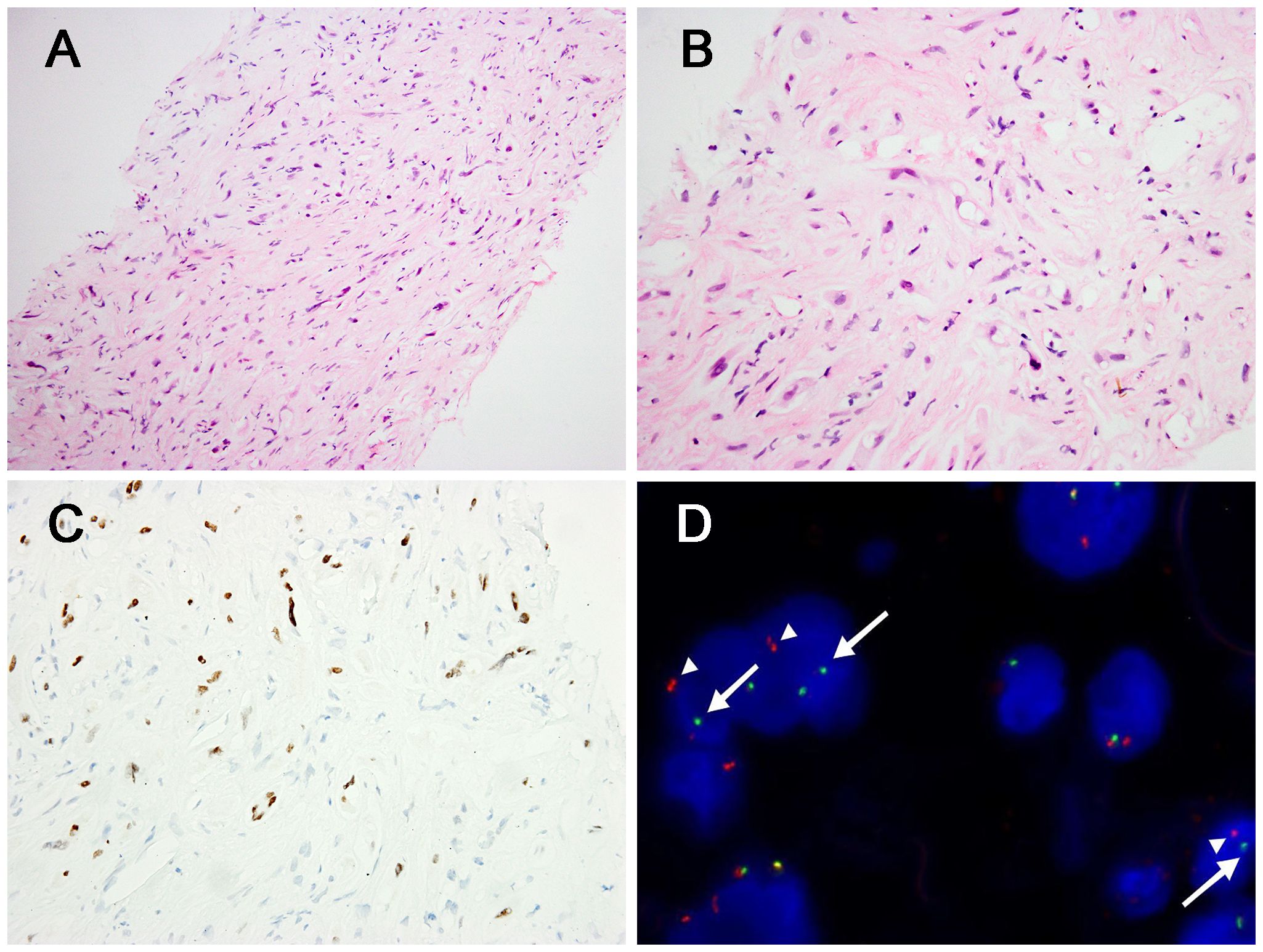
Figure 3. Morphological features of TFE3 rearranged (case4) and corresponding immunohistochemical and genetic results. (A) The tumor cells arranged in discohesive strands or single cells in the collagen matrix, with indistinct cell borders (H&E, ×200). (B) Some tumors cells had intracytoplasmic vacuoles and the presence of signet-ring lumen (H&E, ×400). (C) The case showed positivity for TFE3 (×400). (D) FISH demonstrated the presence of TFE3 gene rearrangement in the neoplastic cells [separation of the red (white arrowhead) and green (white arrow) signals].
In the three CAMTA1 rearranged cases, case 5 and case 6 showed discohesive strands or single epithelioid or spindled cells in a myxohyaline stroma (Figure 4A). Tumor cells had fusiform and oval nuclei with mild atypia, and intracytoplasmic vacuoles, imparting a signet-ring like appearance (Figure 4B), with CAMTA1-immunonegativity but WWTR1::CAMTA1 fusion (Figures 4C, D). Another CAMTA1 rearranged case (case 7) had higher atypia and the tumor exhibited infiltrative growth into liver parenchyma (Figure 5A). The tumor showed solid growth pattern in a sclerotic matrix with lymphocyte infiltration (Figure 5B). Medium-sized epithelioid cells had round nuclei with moderate atypia and conspicuous nucleoli, with moderate amounts of eosinophilic cytoplasm (Figure 5C). In some area, the tumor showed vascular lumens that contain erythrocytes (Figure 5D). Nuclear expression of CAMTA1 was negative while WWTR1::CAMTA1 fusion was identified by FISH and NGS (Figures 5E–G). Nuclear pleomorphism can be seen in case 7 and necrosis can be seen in case 5, but significant mitotic activity was not identified in three cases.
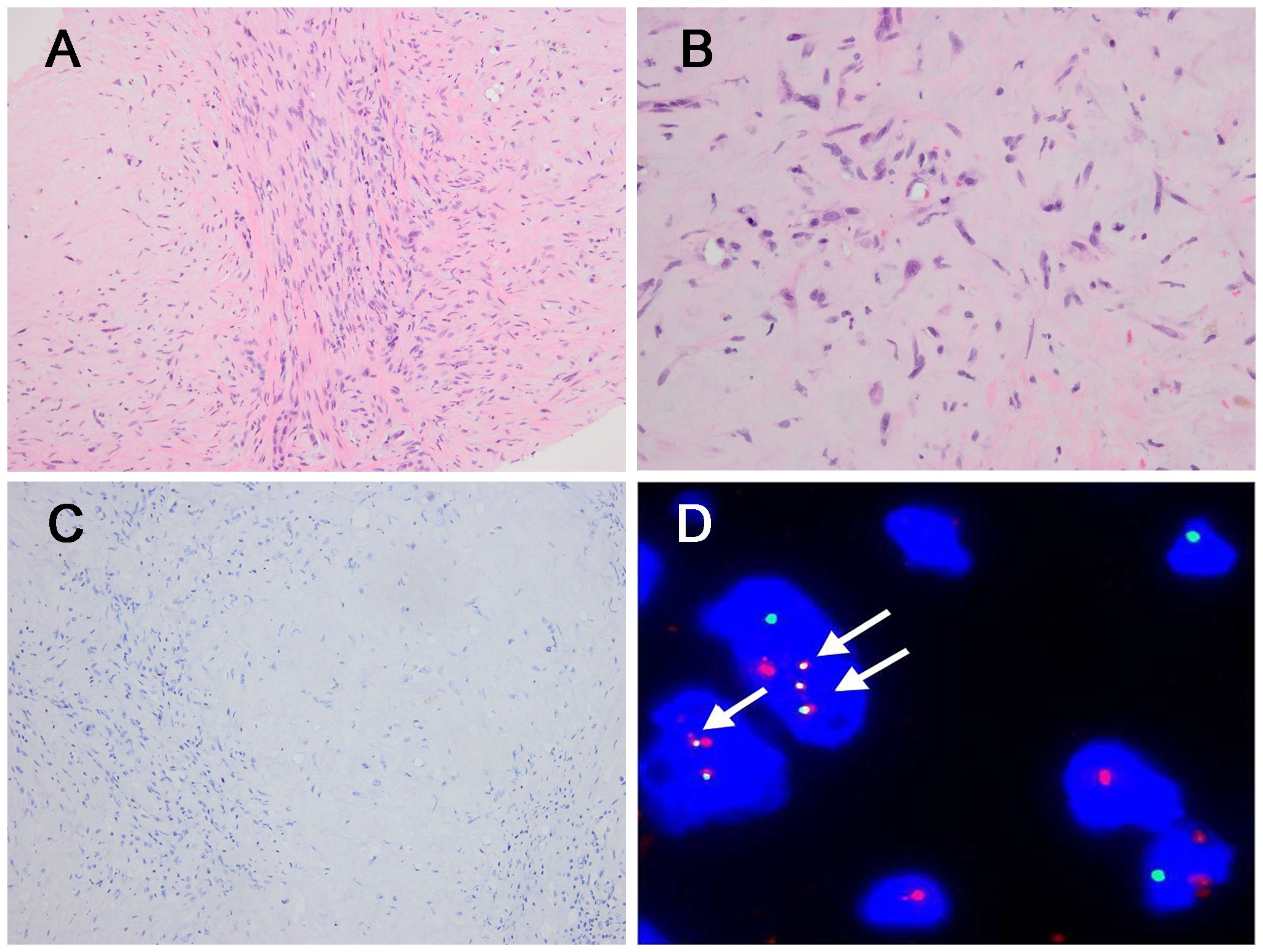
Figure 4. Morphological features of CAMTA1 rearranged and immunohistochemical and genetic results. (A) In some areas, strands or single epithelioid and spindled cells in a myxohyaline stroma (H&E, ×200). (B) Tumor cells had fusiform and oval nuclei with mild atypia, and intracytoplasmic vacuoles, imparting a signet-ring like appearance (H&E, ×400). (C) The tumor was immunonegative for CAMTA1 (×200). (D) FISH revealed WWTR1::CAMTA1 fusion (arrow).
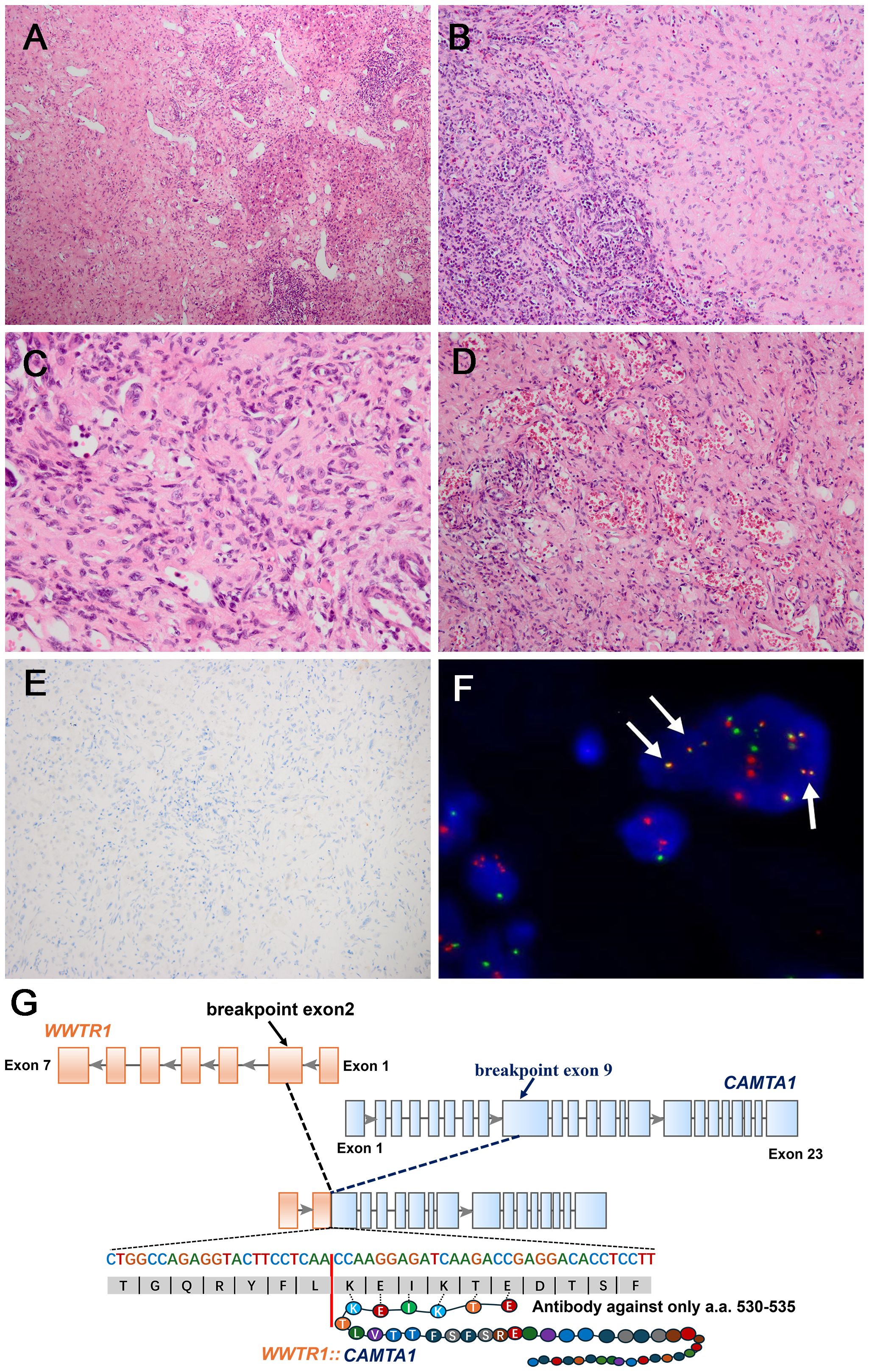
Figure 5. Morphological features of one CAMTA1 rearranged (case7) and immunohistochemical and genetic results. (A) At low magnification, infiltrative growth into liver parenchyma can be seen (H&E, ×100). (B) The tumor showed solid growth in a sclerotic matrix with lymphocyte infiltration (H&E, ×200). (C) Tumor cells had round nuclei with moderate atypia and conspicuous nucleoli, with moderate amounts of eosinophilic cytoplasm (H&E, ×400). (D) Vascular channels containing red blood cells can be seen in some areas (H&E, ×200). (E) Tumor cells were immunonegative for CAMTA1 (×200). (F) FISH revealed WWTR1::CAMTA1 fusion, (arrow). (G) Schematic diagram of the WWTR1::CAMTA1 fusion identified by NGS. The breakpoints were in the WWTR1 (exon2) and CAMTA1(exon9) and the fusion transcript only had 6 amino acids that could be recognized by the CAMTA1 antibody.
Among 7 cases, 5 cases underwent surgical resection, one of which was followed by adjuvant chemotherapy. Two cases were biopsied and treated with chemotherapy. (case 4 with Epirubicin and case 5 with Doxorubicin and Sirolimus). Follow-up information was available for 6 patients (6/7, 85.7%) with a median follow-up duration of 46 months (range 16-86 months). Among the 6 patients, 1 patient had centrum metastases at initial diagnosis. Four patients (4/6, 66.7%) were alive with no evidence of disease, and 2 patients (2/6, 33.3%) were alive with disease.
EHE is a rare malignant vascular neoplasm which was first described by Weiss and Enzinger in 1982 (13). The majority of EHEs (>90%) are characterized by WWTR1::CAMTA1 fusions, therefore, CAMTA1 IHC is commonly utilized as a diagnostic tool for EHE. Subsequently, a small subset EHE cases were found to harbor YAP1::TFE3 fusion gene and negative for CAMTA1 IHC but showed positivity to TFE3. Most CAMTA1-immunonegative cases are composed of the TFE3 subtype, however, in our clinical practice, CAMTA1-immunonegative but CAMTA1 gene rearrangements cases were identified, and further literature review found more such cases (8, 9). Moreover, with the development of NGS, a few EHE cases were found to be negative for CAMTA1 rearrangement, but with variant WWTR1 fusion, including WWTR1::MAML2 (n=2), WWTR1::ACTL6A (n=2), WWTR1::TFE3 (n=1) (Table 2) (8, 10, 11). Furthermore, there existed EHE cases with WWTR1 rearrangement, but NGS failed to identify fusion gene candidate (11). So far, no studies have systematically reported these peculiar cases. Hence, we focused on liver, one of the most common sites of EHE, to further explore the clinicopathological and molecular features of these CAMTA1-immunonegative tumors.
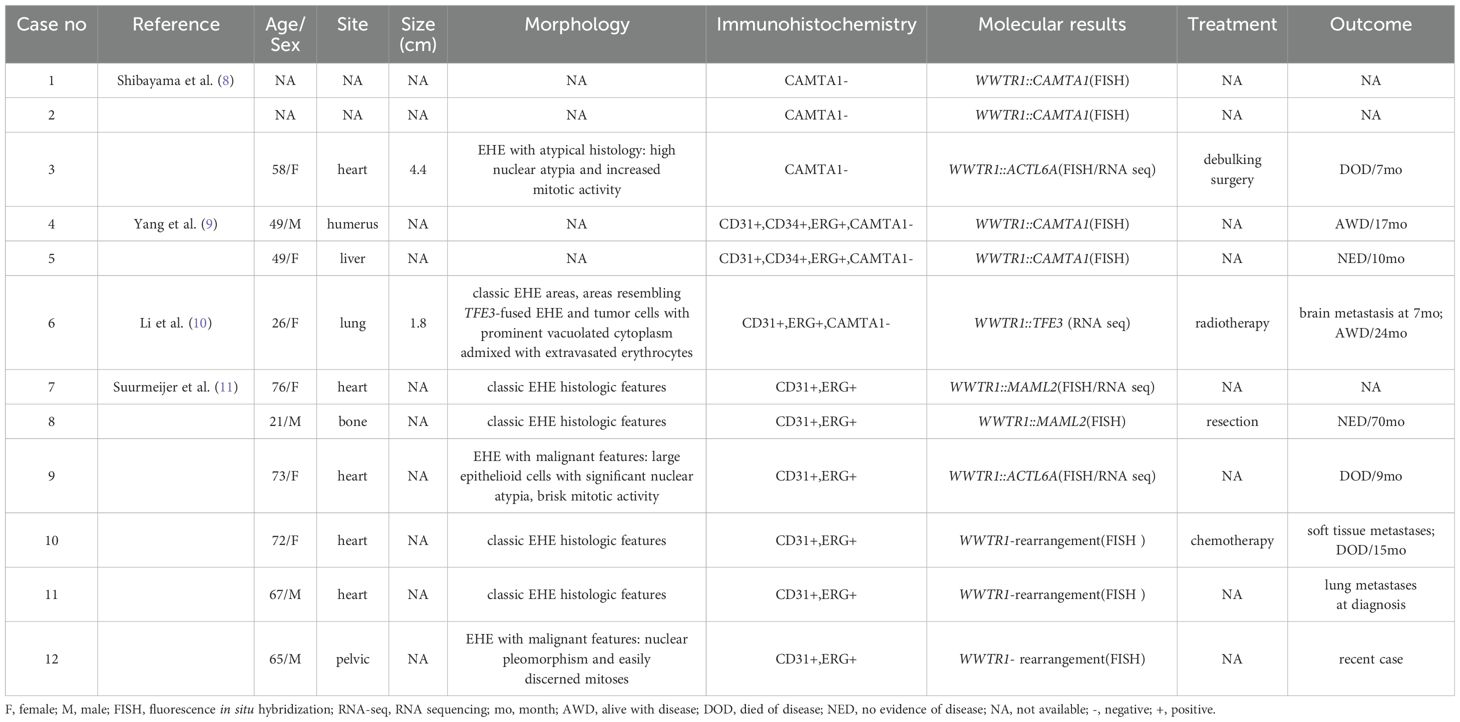
Table 2. Clinicopathological and molecular features of historical EHE cases with CAMTA1-immunonegativity or variant WWTR1 gene rearrangement.
In our series, 4 CATMA1-immunonegative hepatic cases were proved to be TFE3-subtype EHE, including 2 males and 2 females with a median age of 60 years. There were 17 TFE3-subtype hepatic EHE cases that had been reported in English literatures (Table 3) (14–23). Sixteen (including our study) historical, hepatic TFE3-subtype EHEs had clinical information, including 8 males and 8 females. Histologically, 2 TFE3-rearranged cases (case1 and 2) in our cohort displayed classic TFE3-subtype morphologic change, and case 3 also showed TFE3-subtype morphology with a certain degree variation, harboring a lower degree of atypia, smaller tumor cells and moderate cytoplasm. Furthermore, case 4 only displayed mild atypia that could potentially be misdiagnosed as CAMTA1-subtype EHE morphologically, harboring cells with intracytoplasmic vacuoles and lumina formation, presenting a signet-ring like appearance. In the hepatic TFE3-subtype cases we reviewed, morphological manifestations were available in 7 cases, of which 4 showed classical TFE3-type morphology and only 1 case had similar morphology to case 4, with morphologic change as CAMTA1 subtype EHE (23). However, unlike our case, this case was not genetically confirmed. Besides, the morphology of the other 2 historical TFE3-subtype cases were uncommon, with dual growth pattern. One case exhibited pseudo-alveolar architecture and cords, nests, and single neoplastic cells in myxoid area simultaneously (14). Another case had classic TFE3-subytpe area and focal area resembling nodular hyperplasia (17). Both in our series and historical cases, the hepatic TFE3-subtype EHE harbored unusual morphologic change, and great care with ancillary analysis should be taken to avoid misdiagnosis.
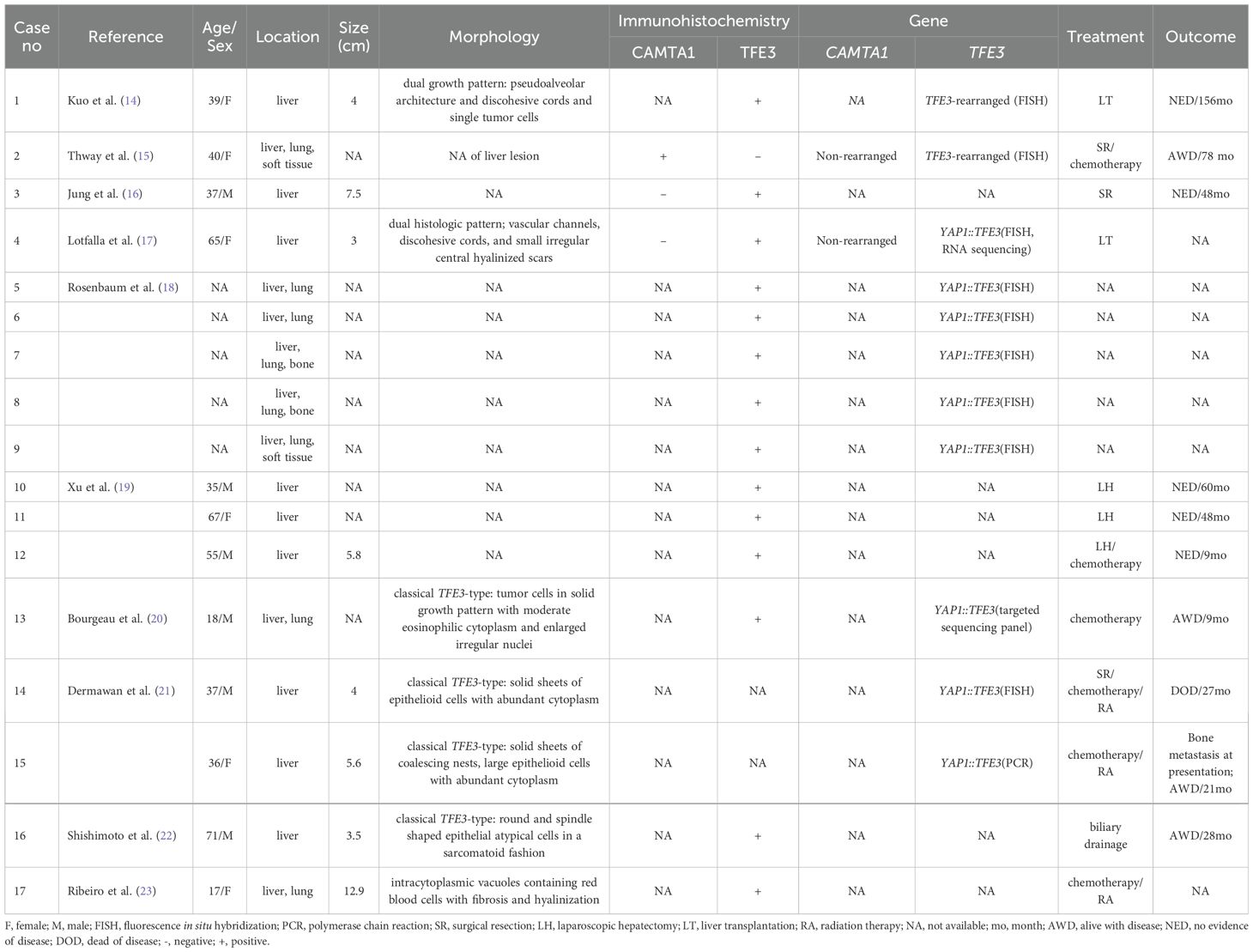
Table 3. Clinicopathological and molecular features of hepatic YAP1::TFE3-fused EHEs in the published literature.
Immunohistochemically, nuclear TFE3 was uniformly expressed, while CAMTA1 was negative in the 4 cases. In terms of genetics, all 4 cases harbored YAP1::TFE3 fusion and had gene fusions between YAP1 exon 1 and TFE3 exon 4. Of the 17 historical cases, 11 had genetic information, harboring YAP1::TFE3 fusions gene or TFE3 rearrangement. Three of the 11 cases had exon information, involving YAP1 exon 1 and TFE3 exon 4, like our report. The above data was also similar to the previous largest TFE3-subtype EHEs study, among which the majority (14/16, 88%) cases had YAP1 exon 1 fused to TFE3 exon 4 (21). These results suggested the TFE3 immunohistochemistry could aid in the EHE diagnosis and further genetic analysis could help to establish the final diagnosis.
In our study, another 3 hepatic CAMTA1-immunonegative cases were proved to be positive for WWTR1::CAMTA1 gene fusion. Four historical cases harbored similar immunohistochemical and genetic change, with CAMTA1 rearrangement and negative CAMTA1 immunoreactivity (8, 9). However, the 4 cases lacked the description of their morphology. In our study, 2 of the 3 cases exhibited classic histology of CAMTA1-subtype, showing nests of epithelioid tumor cells with small, oval nuclei and intracytoplasmic vacuoles. It needs to be pointed out, another 1 case (case 7), unlike characteristic EHE, showing the morphology analogous to TFE3-subtype, with medium-sized epithelioid cells having moderate amounts of eosinophilic cytoplasm and vascular lumen, containing erythrocytes. More cases and further studies and needed to clarify the pathological feature of these peculiar cases. More importantly, when meeting such peculiar and challenging cases with unusual morphologic and immunohistochemical results, molecular studies such as FISH and furthermore, NGS are needed to establish the diagnosis and reveal the reason for these changes.
In this study, we identified a novel fusion between WWTR1 exon 2 and CAMTA1 exon 9 in one case (case 7) through NGS. This breakpoint is located at the 1663 nucleotide of CAMTA1 gene (NM_015215.4), a previously unreported site, and the breakpoint of WWTR1 gene is identical to previous studies. The current CAMTA1 antibody only recognizes 84 amino acids encoded by nucleotides 1428 to 1682 of the CAMTA1 gene, which is located downstream of all previously reported CAMTA1 breakpoints (4, 6, 24–27). However, the antibody recognition sequence is located upstream of the breakpoint of case 7, with only 20bp nucleotide overlapped and corresponding to 6 amino acids (Figure 5G). This result suggested that the uncommon fusion point might result in the negative IHC result. This finding underscores the importance of understanding the precise location of fusion points, as breakpoint could impact the gene translation and antibody recognition and may lead to unexpected results. Additionally, the CAMTA1 immuno-negativity may also occurred in TFE3-rearranged EHE cases and more rarely, in cases with variant WWTR1 gene rearrangements. Hence, when meeting such challenging cases with uncommon IHC results, the FISH and NGS is valuable to assist the establishment of correct diagnosis.
The histopathologic morphology of hepatic EHE is diverse and heterogeneous, particularly in small biopsy samples, and it is usually too difficult to confirm endothelial differentiation and to exclude histologic mimics. EHE includes a wide range of differential diagnoses, including poorly differentiated adenocarcinoma, perivascular epithelioid cell tumor (PEComa), epithelioid angiosarcoma and so on.
Poorly differentiated adenocarcinoma can also have distinct signet-ring cell components and morphologically overlapped with EHE. The typical features of adenocarcinoma are a large number of cells with intracytoplasmic vacuoles containing mucus, readily confirmed by epithelial mucinous histochemical staining such as PAS (28). Immunohistochemically, adenocarcinoma expresses epithelial markers such as CK and EMA, but negative for CD31, CD34 and especially CAMTA1 and TFE3. Genetically, CAMTA1 or TFE3-rearrangement is present in EHE, but not in poorly differentiated adenocarcinomas.
PEComa also has spindle or epithelioid cells arranged in fascicular or nested distribution, like EHE, but the former lacks evident vascular differentiation. PEComa exhibit expression of melanocyte markers such as HMB45 and Melan-A, as well as myogenic markers including SMA and desmin, while they do not express the endothelial marker CD34 (29). Generally, specific immunohistochemical markers can be utilized for distinguishing between these two entities. Since both have TFE3 gene break, they can be identified by gene sequencing if necessary to prevent misdiagnosis or missed diagnosis. Unlike YAP1::TFE3 EHEs, PEComa is frequently accompanied by SFPQ::TFE3 fusion.
The distinction between EHE and epithelioid angiosarcoma can be challenging due to their similar histological change with large epithelioid cells, eosinophilic cytoplasm, distinct nucleoli, and occasional intracytoplasmic vacuoles containing erythrocytes. However, the latter exhibits higher atypia and marked mitosis. Furthermore, CAMTA1 or TFE3 immunohistochemistry may provide valuable insights. Moreover, confirming the presence of WWTR1::CAMTA1 or YAP1::TFE3 gene fusion in the tumor would offer more convincing evidence.
In clinical practice, the diagnosis of EHE is mainly based on the histological evaluation and IHC results, and for the challenging cases with ambiguous morphology or unexpected IHC profile, the FISH for CAMTA1 and TFE3 should be performed, and furthermore, NGS is recommended for cases which is highly suspected for EHE to make the final diagnosis. We recommend the diagnostic algorithm shown in the figure to minimize misdiagnosis (Figure 6).
Owing to the rarity and heterologous clinical course of hepatic EHE, there is no well-established treatment strategy for such entity. In our series, 4 patients were treated with surgical resection only, 1 patient received surgical resection and chemotherapy and 2 patients underwent chemotherapy. According to previous large series studies of hepatic EHE, the common treatment modalities include hepatic resection, liver transplantation, ablations, chemotherapy, and radiotherapy. Hepatic resection is recommended for resectable intrahepatic lesions, and liver transplantation is considered to be the first choice for the treatment of tumor patients when there is intrahepatic metastasis or the tumor is too large to be resected (30, 31). In addition, adjuvant chemoradiotherapy is recommended when metastasis occurs.
In cases of EHE, the discovery of the WWTR1::CAMTA1 and YAP1::TFE3 fusion genes has provided a new perspective for the treatment of EHE. WWTR1 and YAP1 are key components of the Hippo signaling pathway, playing a central role in a variety of physiological and pathological processes. The WWTR1::CAMTA1 fusion gene can induce endothelial cells to transform into vascular tumors with EHE characteristics. This process involves WWTR1 (TAZ)::CAMTA1 as a continuously activated form of TAZ, which is mainly located in the cell nucleus and activates its pro-tumor transcription program. Similarly, the carcinogenic potential of the YAP1::TFE3 fusion gene also requires interaction with TEAD. The fusion protein utilizes the transcriptional activation domain and nuclear localization sequence of TFE3, binding to DNA through the TEAD binding site of YAP1, forming a continuously activated chimeric transcription factor (32). Additionally, trametinib, an MEK inhibitor, has demonstrated certain therapeutic effects in EHE patients with the WWTR1::CAMTA1 fusion (33). In the future, it is necessary to further collect and analyze cases to gain a deeper understanding of the molecular basis of EHE, in order to provide more precise treatment strategies for patients.
In our series, 4 patients (4/6; 66.7%) were alive with no evidence of disease, and 2 patients (2/6;33.3%) were alive with disease. Previous studies found the 5-year overall survival rate of hepatic EHE ranging from 57% to 67% (18, 34). Pathologic parameters including tumor size >3 cm, mitotic activity >3 mitoses per 50 high-powered fields and histologic grade were associated with worse outcome of EHE patients (18, 35). Moreover, a recent large series study discovered the CAMTA1-subtype EHE correlated with a worse prognosis compared to TFE3 subtype, with the 5-year overall survival (OS) as 59% versus 86%, respectively (18). Hence, we carefully reviewed the historical hepatic EHE cases and found 74 CAMTA1 subtype and 13 TFE3 subtype EHE cases with available follow-up information (including our cases)) (9, 16, 26, 31, 36–45). The survival results showed 10 of 74 (13.5%) CAMTA1 subtype cases died of disease and only 1 of 13 (7.7%) TFE3-subtype EHEs succumbed to disease. Combined with this study and retrospective data analysis, most patients with hepatic EHE have a good prognosis, and TFE3 subtype cases seems to harbor a favorable outcome. While the prognosis difference between the TFE3-subtype and CAMTA1-subtype hepatic EHE still needs to be further explored in a larger study cohort.
In summary, our study reported a series of 7 peculiar, CAMTA1-immunonegative hepatic EHE cases and most of them were TFE3-rearranged EHEs. Moreover, our study showed that there existed the CAMTA1-immunonegative but CAMTA1-rearranged EHE cases. Therefore, the diagnosis of EHE should be based on histological morphology, combined with the dual detection of CAMTA1 and TFE3 IHC, and if necessary, supplemented by comprehensive analysis, including FISH and NGS for the genetic aberration, to ensure correct diagnosis.
The data presented in the study are deposited in the NCBI database, accession number PV083432.
The studies involving humans were approved by West China Hospital Institutional Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
YN: Writing – original draft. WJ: Writing – original draft. XZ: Writing – original draft. XH: Writing – review & editing. MC: Writing – original draft. HZ: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No. 81972520), Natural Science Foundation Project of Science & Technology Department of Sichuan Province (No.2024NSFSC0763) and the Cadre Health Research Project of Sichuan Province (No.2024-113).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Rubin BP, Deyrup AT, Doyle LA. Epithelioid haemangioendothelioma. In: WHO Classification of Tumours Editorial Board, editors. World Health Organization classification of soft tissue and bone tumours, 5th ed. IARC Press, Lyon (2020). p. 172–5.
2. Folpe AL. Vascular tumors of intermediate Malignancy: An update. Hum Pathology. (2024) 147:114–28. doi: 10.1016/j.humpath.2024.01.014
3. Mendlick MR, Nelson M, Pickering D, Johansson SL, Seemayer TA, Neff JR, et al. Translocation t(1;3)(p36.3;q25) is a nonrandom aberration in epithelioid hemangioendothelioma. Am J Surg Pathol. (2001) 25:684–7. doi: 10.1097/00000478-200105000-00019
4. Errani C, Zhang L, Sung YS, Hajdu M, Singer S, Maki RG, et al. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosomes Cancer. (2011) 50:644–53. doi: 10.1002/gcc.20886
5. Doyle LA, Fletcher CD, Hornick JL. Nuclear expression of CAMTA1 distinguishes epithelioid hemangioendothelioma from histologic mimics. Am J Surg Pathol. (2016) 40:94–102. doi: 10.1097/PAS.0000000000000511
6. Shibuya R, Matsuyama A, Shiba E, Harada H, Yabuki K, Hisaoka M. CAMTA1 is a useful immunohistochemical marker for diagnosing epithelioid haemangioendothelioma. Histopathology. (2015) 67:827–35. doi: 10.1111/his.12713
7. Antonescu CR, Le Loarer F, Mosquera JM, Sboner A, Zhang L, Chen CL, et al. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer. (2013) 52:775–84. doi: 10.1002/gcc.22073
8. Shibayama T, Makise N, Motoi T, Mori T, Hiraoka N, Yonemori K, et al. Clinicopathologic characterization of epithelioid hemangioendothelioma in a series of 62 cases: A proposal of risk stratification and identification of a synaptophysin-positive aggressive subset. Am J Surg Pathol. (2021) 45:616–26. doi: 10.1097/pas.0000000000001660
9. Yang P, Zhang S, Yu C, Yan W, Yang N, Li N, et al. Fluorescence in situ hybridization for WWTR1-CAMTA1 has higher sensitivity and specificity for epithelioid hemangioendothelioma diagnosis. Am J Transl Res. (2020) 12:4561–8.
10. Li S, Dermawan JK, Seavey CN, Ma S, Antonescu CR, Rubin BP. Epithelioid hemangioendothelioma (EHE) with WWTR1::TFE3 gene fusion, a novel fusion variant. Genes Chromosomes Cancer. (2024) 63:e23226. doi: 10.1002/gcc.23226
11. Suurmeijer AJH, Dickson BC, Swanson D, Sung YS, Zhang L, Antonescu CR. Variant WWTR1 gene fusions in epithelioid hemangioendothelioma-A genetic subset associated with cardiac involvement. Genes Chromosomes Cancer. (2020) 59:389–95. doi: 10.1002/gcc.22839
12. Jing W, Lan T, Chen H, Zhang Z, Chen M, Peng R, et al. Amplification of FRS2 in atypical lipomatous tumour/well-differentiated liposarcoma and de-differentiated liposarcoma: a clinicopathological and genetic study of 146 cases. Histopathology. (2018) 72:1145–55. doi: 10.1111/his.13473
13. Weiss SW, Enzinger FM. Epithelioid hemangioendothelioma: a vascular tumor often mistaken for a carcinoma. Cancer. (1982) 50:970–81. doi: 10.1002/1097-0142(19820901)50:5<970::aid-cncr2820500527>3.0.co;2-z
14. Kuo FY, Huang HY, Chen CL, Eng HL, Huang CC. TFE3-rearranged hepatic epithelioid hemangioendothelioma-a case report with immunohistochemical and molecular study. APMIS. (2017) 125:849–53. doi: 10.1111/apm.12716
15. Thway K, Mentzel T, Perrett CM, Calonje E. Multicentric visceral epithelioid hemangioendothelioma, with extremity dermal deposits, unusual late recurrence on the nasal bridge, and TFE3 gene rearrangement. Hum Pathol. (2018) 72:153–9. doi: 10.1016/j.humpath.2017.08.020
16. Jung H, Kim HN, Jang Y, Park CK, Ha SY. CAMTA-1 expression in 24 cases of hepatic epithelioid hemangioendothelioma in a single institute: diagnostic utility for differential diagnosis from hepatic angiosarcoma. In Vivo. (2019) 33:2293–7. doi: 10.21873/invivo.11736
17. Lotfalla MM, Folpe AL, Fritchie KJ, Greipp PT, Galliano GG, Halling KC, et al. Hepatic YAP1-TFE3 rearranged epithelioid hemangioendothelioma. Case Rep Gastrointest Med. (2019) 2019:7530845. doi: 10.1155/2019/7530845
18. Rosenbaum E, Jadeja B, Xu B, Zhang L, Agaram NP, Travis W, et al. Prognostic stratification of clinical and molecular epithelioid hemangioendothelioma subsets. Mod Pathol. (2020) 33:591–602. doi: 10.1038/s41379-019-0368-8
19. Xu J, Hu S, Li S, Wang W, Zhou X, Wu Y, et al. Laparoscopic resection of hepatic epithelioid hemangioendothelioma: report of eleven rare cases and literature review. World J Surg Oncol. (2020) 18:282. doi: 10.1186/s12957-020-02034-z
20. Bourgeau M, Martinez A, Deeb KK, Reid MD, Lewis M, Point du Jour KS, et al. Cytologic features of hepatic YAP1-TFE3 rearranged epithelioid hemangioendothelioma. Diagn Cytopathol. (2021) 49:E447–E52. doi: 10.1002/dc.24853
21. Dermawan JK, Azzato EM, Billings SD, Fritchie KJ, Aubert S, Bahrami A, et al. YAP1-TFE3-fused hemangioendothelioma: a multi-institutional clinicopathologic study of 24 genetically-confirmed cases. Mod Pathol. (2021) 34:2211–21. doi: 10.1038/s41379-021-00879-7
22. Shishimoto T, Oura S, Motozato K, Tanaka H, Takamatsu S, Ono W. Epithelioid hemangioendothelioma of the liver showing spontaneous complete regression after the cessation of methotrexate intake. Case Rep Oncol. (2023) 16:628–33. doi: 10.1159/000531133
23. Ribeiro MCO, Lemos JVB, de Toledo Moraes MP, Oliver FA, Alvarez M, Silva GF, et al. Hepatic epithelioid hemangioendothelioma: how fast does it grow and which findings could have prevented diagnostic delay?-a case report. Transl Gastroenterol Hepatol. (2023) 8:12. doi: 10.21037/tgh-22-48
24. Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, et al. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med. (2011) 3:98ra82. doi: 10.1126/scitranslmed.3002409
25. Errani C, Sung YS, Zhang L, Healey JH, Antonescu CR. Monoclonality of multifocal epithelioid hemangioendothelioma of the liver by analysis of WWTR1-CAMTA1 breakpoints. Cancer Genet. (2012) 205:12–7. doi: 10.1016/j.cancergen.2011.10.008
26. Flucke U, Vogels RJ, de Saint Aubain Somerhausen N, Creytens DH, Riedl RG, van Gorp JM, et al. Epithelioid Hemangioendothelioma: clinicopathologic, immunhistochemical, and molecular genetic analysis of 39 cases. Diagn Pathol. (2014) 9:131. doi: 10.1186/1746-1596-9-131
27. Patel NR, Salim AA, Sayeed H, Sarabia SF, Hollingsworth F, Warren M, et al. Molecular characterization of epithelioid haemangioendotheliomas identifies novel WWTR1-CAMTA1 fusion variants. Histopathology. (2015) 67:699–708. doi: 10.1111/his.12697
28. Jurczyk M, Zhu B, Laskin W, Lin X. Pitfalls in the diagnosis of hepatic epithelioid hemangioendothelioma by FNA and needle core biopsy. Diagn Cytopathol. (2014) 42:516–20. doi: 10.1002/dc.22943
29. Folpe AL, Kwiatkowski DJ. Perivascular epithelioid cell neoplasms: pathology and pathogenesis. Hum Pathol. (2010) 41:1–15. doi: 10.1016/j.humpath.2009.05.011
30. Mehrabi A, Kashfi A, Fonouni H, Schemmer P, Schmied BM, Hallscheidt P, et al. Primary Malignant hepatic epithelioid hemangioendothelioma: a comprehensive review of the literature with emphasis on the surgical therapy. Cancer. (2006) 107:2108–21. doi: 10.1002/cncr.22225
31. Treska V, Daum O, Svajdler M, Liska V, Ferda J, Baxa J. Hepatic epithelioid hemangioendothelioma - a rare tumor and diagnostic dilemma. In Vivo. (2017) 31:763–7. doi: 10.21873/invivo.11128
32. Merritt N, Garcia K, Rajendran D, Lin ZY, Zhang X, Mitchell KA, et al. TAZ-CAMTA1 and YAP-TFE3 alter the TAZ/YAP transcriptome by recruiting the ATAC histone acetyltransferase complex. Elife. (2021) 10:e62857. doi: 10.7554/eLife.62857
33. Schuetze SM, Ballman KV, Heise R, Ganjoo KN, Davis EJ, George S, et al. A single-arm phase 2 trial of trametinib in patients with locally advanced or metastatic epithelioid hemangioendothelioma. Clin Cancer Res. (2024) 30:4584–92. doi: 10.1158/1078-0432.Ccr-23-3817
34. Kaltenmeier C, Stacchiotti S, Gronchi A, Sapisochin G, Liu H, Ashwat E, et al. Treatment modalities and long-term outcomes of hepatic hemangioendothelioma in the United States. HPB (Oxford). (2022) 24:1688–96. doi: 10.1016/j.hpb.2022.03.013
35. Deyrup AT, Tighiouart M, Montag AG, Weiss SW. Epithelioid hemangioendothelioma of soft tissue: a proposal for risk stratification based on 49 cases. Am J Surg Pathol. (2008) 32:924–7. doi: 10.1097/pas.0b013e31815bf8e6
36. Abdelmogod A, Papadopoulos L, Riordan S, Wong M, Weltman M, Lim R, et al. A matched molecular and clinical analysis of the epithelioid haemangioendothelioma cohort in the stafford fox rare cancer program and contextual literature review. Cancers (Basel). (2023) 15:4378. doi: 10.3390/cancers15174378
37. Tomassen T, Versleijen-Jonkers YMH, Hillebrandt-Roeffen MHS, Van Cleef PHJ, van Dalen T, Weidema ME, et al. Prognostic factors in epithelioid hemangioendothelioma: analysis of a nationwide molecularly/immunohistochemically confirmed cohort of 57 cases. Cancers (Basel). (2023) 15:3304. doi: 10.3390/cancers15133304
38. Tang W, Niu Y, Luo Y, Ma X. Characteristic contrast−enhanced ultrasound findings of hepatic epithelioid haemangioendothelioma: A case report and literature review. Oncol Lett. (2023) 25:253. doi: 10.3892/ol.2023.13839
39. Somers N, Creytens D, Van Belle S, Sys G, Lapeire L. Diagnosis of epithelioid hemangioendothelioma eight days postpartum: Is there a link with pregnancy? A case report and review of the literature. Acta Clin Belg. (2022) 77:157–62. doi: 10.1080/17843286.2020.1802146
40. Terasaki F, Yamamoto Y, Sugiura T, Okamura Y, Ito T, Ashida R, et al. Laparoscopic repeat liver resection for hepatic epithelioid hemangioendothelioma. Surg Case Rep. (2020) 6:254. doi: 10.1186/s40792-020-01036-z
41. Taniai T, Onda S, Sato S, Shiba H, Sakamoto T, Yanaga K. Hepatic epithelioid hemangioendothelioma: difficult differential diagnosis from angiosarcoma. Case Rep Gastroenterol. (2020) 14:56–62. doi: 10.1159/000505513
42. Mogler C, Koschny R, Heilig CE, Frohling S, Schirmacher P, Weichert W, et al. Molecular characterization of hepatic epithelioid hemangioendothelioma reveals alterations in various genes involved in DNA repair, epigenetic regulation, signaling pathways, and cell cycle control. Genes Chromosomes Cancer. (2020) 59:106–10. doi: 10.1002/gcc.22795
43. Agostini-Vulaj D, Pehlivanoglu B, Weiss SW, Krasinskas A, Feely MM, Hornick JL, et al. Intrasinusoidal spread of hepatic epithelioid hemangioendothelioma: implications for the diagnosis in minimal samples. Am J Surg Pathol. (2019) 43:573–9. doi: 10.1097/pas.0000000000001221
44. Hettmer S, Andrieux G, Hochrein J, Kurz P, Rössler J, Lassmann S, et al. Epithelioid hemangioendotheliomas of the liver and lung in children and adolescents. Pediatr Blood Cancer. (2017) 64. doi: 10.1002/pbc.26675
Keywords: epithelioid hemangioendothelioma, liver tumor, WWTR1::CAMTA1 fusion gene, YAP1::TFE3 fusion gene, molecular analysis
Citation: Nie Y, Jing W, Zheng X, He X, Chen M and Zhang H (2025) CAMTA1-immunonegative epithelioid hemangioendotheliomas of the liver: a clinicopathological and molecular analysis of seven cases. Front. Oncol. 15:1478036. doi: 10.3389/fonc.2025.1478036
Received: 09 August 2024; Accepted: 27 January 2025;
Published: 18 February 2025.
Edited by:
Gisella Figlioli, Humanitas University, ItalyReviewed by:
Xiaolei Liu, China-Japan Friendship Hospital, ChinaCopyright © 2025 Nie, Jing, Zheng, He, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongying Zhang, aHlfemhhbmdAc2N1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.