- 1Children’s Medical Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China
- 3Department of Laboratory Medicine, Heyuan People’s Hospital, Heyuan, China
Background: Pediatric AML prognosis research has advanced significantly, yet gaps in understanding genetic and molecular interactions persist. Despite improved outcomes, relapse/refractory cases and personalized treatment integration remain critical clinical challenges.
Objective: To analyze the global research landscape on pediatric AML prognosis, highlight influential components and collaborations, and identify major potential research trends.
Methods: Publications on pediatric AML prognosis research from 1999 to 2023 were retrieved from the Clarivate Analytics Web of Science Core Collection (WoSCC) database. Bibliometric analysis was conducted using CiteSpace and VOSviewer to identify leading countries, prominent institutions, high-impact journals, key research categories, influential authors, and emerging research topics.
Results: The bibliometric analysis encompassed 924 publications, with St. Jude Children’s Research Hospital emerging as the most prolific institution. The United States leads globally in terms of countries, institutions, journals, and authors. Todd A. Alonzo ranks highest in publication volume, while U. Creutzig leads in citations. The top research categories were Oncology, Hematology, and Pediatrics. Key research topics included genomics, transcriptomics, epigenomics, targeted therapies, immune therapy, and integrative diagnostic approaches.
Conclusion: This bibliometric analysis highlights significant advancements in pediatric AML prognosis over the past 25 years, driven by the integration of genetic markers, immunological insights, transcriptomics, and epigenomics, which have collectively transformed risk stratification and treatment strategies. Overcoming challenges, such as discovering new therapeutic targets and enhancing treatment combinations, will depend on global collaboration and advanced technologies to propel the field forward.
1 Introduction
Pediatric acute myeloid leukemia (AML) is a malignant hematological disorder characterized by the clonal proliferation of myeloid precursors in the bone marrow (1). Despite significant advances in the understanding and treatment of pediatric AML, it remains a leading cause of cancer-related mortality in children (2). Over the past few decades, considerable progress has been made in identifying genetic and molecular markers that play crucial roles in predicting prognosis and guiding treatment outcomes (3–6). These advancements have led to the development of targeted therapies and more precise risk stratification methods, significantly improving patient outcomes (7–9). However, high rates of relapse and refractory cases continue to underscore the need for ongoing research and innovation in this field (10).
A critical challenge in pediatric AML research is the disease’s inherent heterogeneity, which complicates both treatment decisions and prognosis (11, 12). While key genetic mutations and molecular abnormalities—such as FLT3-ITD, WT1, CEBPA mutations, and KMT2A rearrangements—have been identified as important prognostic markers (13–16), there remain significant gaps in our understanding of how these markers interact and contribute to disease progression and therapeutic resistance (17). Addressing these gaps is essential to overcoming the persistent challenges of relapse and refractory disease, and to better integrating personalized treatment strategies into clinical practice.
This analysis provides a comprehensive bibliometric overview of global research trends in pediatric AML prognosis from 1999 to 2023. By examining key aspects such as publication trends, leading countries, prestigious institutions, influential journals, major research categories, notable contributors, seminal references, and emerging trends, it aims to offer a clear snapshot of the current landscape and future directions of the field. The analysis specifically focuses on how advancements in genomics, transcriptomics, and epigenomics have shaped prognosis and treatment outcomes. Furthermore, it explores the pivotal role of clinical trials in developing treatment protocols and highlights emerging therapies that show promise for improving patient outcomes.
This bibliometric approach synthesizes extensive research data, providing a detailed and systematic overview of the field. By identifying significant trends and key studies, it offers valuable insights for researchers, clinicians, and policymakers alike. The findings are expected to contribute to the development of more effective, personalized treatment strategies, ultimately improving the prognosis for pediatric AML patients. Notably, this is the first bibliometric study to comprehensively explore prognosis-related research in pediatric AML, marking a significant contribution to the field.
2 Data sources and search methodology
We utilized the Clarivate Analytics Web of Science Core Collection database (WoSCC) to identify and gather publications on prognosis-related pediatric AML research. The search algorithm used was as follows: Topic search #1 included “pediatric acute myeloid leukemia”,”childhood acute myeloid leukemia”, “children acute myeloid leukemia,”,”pediatric AML”, “childhood AML”, “children AML” and Topic search #2 included “prognosis”, “outcome”, “survival”, “predictive factors”, “risk factors”, “long-term outcomes”, “prognostic factors”, “treatment response”, “relapse” and “remission” with a publication date range of 1999-2023. This search yielded 1160 articles in the WoSCC database.
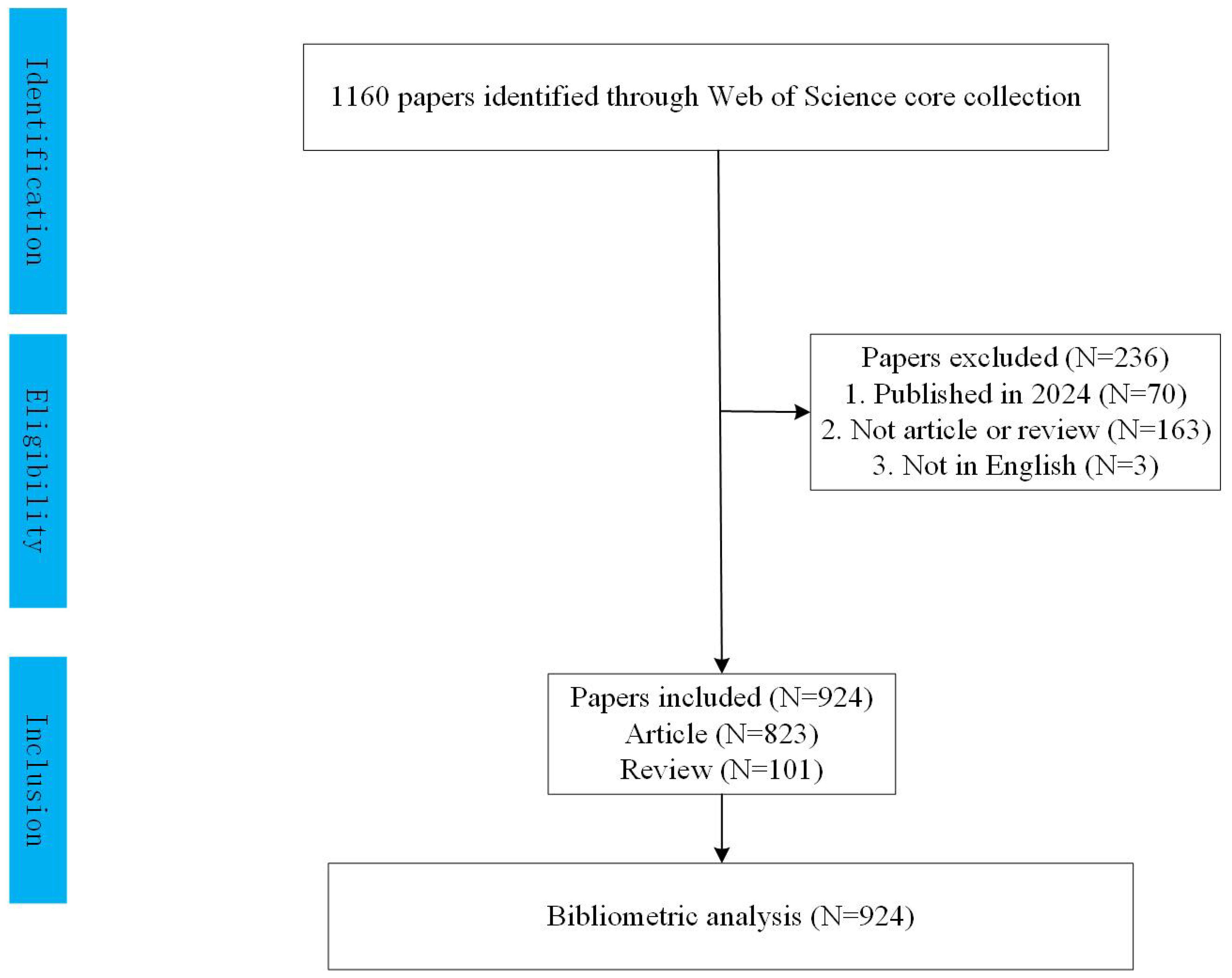
We excluded conference abstracts, editorials, and letters to focus on original research and reviews. To minimize bias, two independent researchers conducted the search together, completing it on February 3, 2024. Titles and abstracts were manually reviewed to select studies specifically addressing prognosis in pediatric AML. Irrelevant articles or those offering limited insights were excluded. We further refined our selection to include only English-language articles and reviews within the specified date range, resulting in 924 publications for analysis, as shown in Figure 1. All records were then imported into CiteSpace v.5.8.R2.
This study analyzed publicly available bibliometric data on pediatric AML prognosis, focusing on published titles and abstracts, with no involvement of human participants or primary data collection. Ethical approval was not required, as the study adhered to international guidelines for bibliometric research, which do not necessitate ethical review for publicly available data without personal identifiers. We followed established ethical standards for data handling, including proper citation and transparency, in line with frameworks from the ISSI and OECD Guidelines for the Responsible Use of Bibliometrics.
2.1 Tools for bibliometric analysis
For our bibliometric analyses, we employed Citespace (version 6.3.R2) and VOSviewer (version 1.6.20).
2.2 Data visualization and presentation
In our bibliometric analysis, we applied several mathematical techniques to uncover significant patterns and trends. Key methods included frequency and co-occurrence analysis, which identified the most discussed topics and their interrelationships. We also used cluster analysis to group related articles, authors, or keywords into distinct clusters, revealing thematic concentrations and research activity patterns. Network analysis played a crucial role in mapping citation and co-authorship networks, highlighting influential authors and publications while visualizing the collaborative landscape. Temporal analysis tracked the evolution of research themes, allowing us to identify both persistent and emerging trends over time (18). For reader clarity, we have provided a brief tutorial on interpreting these visualizations. In CiteSpace, nodes represent articles, authors, or keywords, with size indicating citation count or frequency, while connecting lines show co-citations or co-occurrences (19). In VOSviewer, network diagrams highlight related groups, with proximity indicating stronger connections and colors representing distinct clusters.
Furthermore, We provide illustrative examples to demonstrate the practical application of our results. One network map highlights a key research cluster focused on FLT3 inhibitors and their impact on survival rates in high-risk pediatric AML patients, featuring significant publications and leading authors. Another example is our temporal analysis, which shows the shift in pediatric AML treatment from traditional chemotherapy to combined strategies involving chemotherapy, transplantation, immunotherapy, and targeted therapies. As treatment approaches have become more personalized, patient prognosis has significantly improved.
CiteSpace enabled the extraction of publication and citation graphs, country collaboration networks, and the 25 most prominent keywords based on citation bursts. VOSviewer was essential for visualizing collaborative networks among countries, journals, and authors, as well as conducting keyword analyses. Microsoft Excel 2019 (Microsoft, Redmond, WA, USA) was used to illustrate publication trends and predict next year’s output. The H-index of journals was calculated following Hirsch’s method, which ranks articles by citation count and identifies the number “H” where “H” articles have been cited at least “H” times (20).
3 Results
3.1 Annual trends in global publications
Our bibliometric analysis incorporated 924 publications from 1999 to 2023, with 89% being original research articles and 11% comprehensive reviews. The analysis reveals a steady increase in the number of publications and citations related to prognosis in pediatric AML research from 1999 to 2021, reaching a peak in 2021. Following this peak, a slight decline is observed from 2021 to 2023. Notably, the annual publication output doubled between 2013 and 2021 (Figure 2A). To elucidate this trend, a polynomial fitting curve was utilized. The analysis yielded a coefficient of determination (R²=0.9173), underscoring the statistical significance of the observed pattern (Figure 2B). These data highlight the growing interest and investment in prognosis-related pediatric acute myeloid leukemia (AML) research.
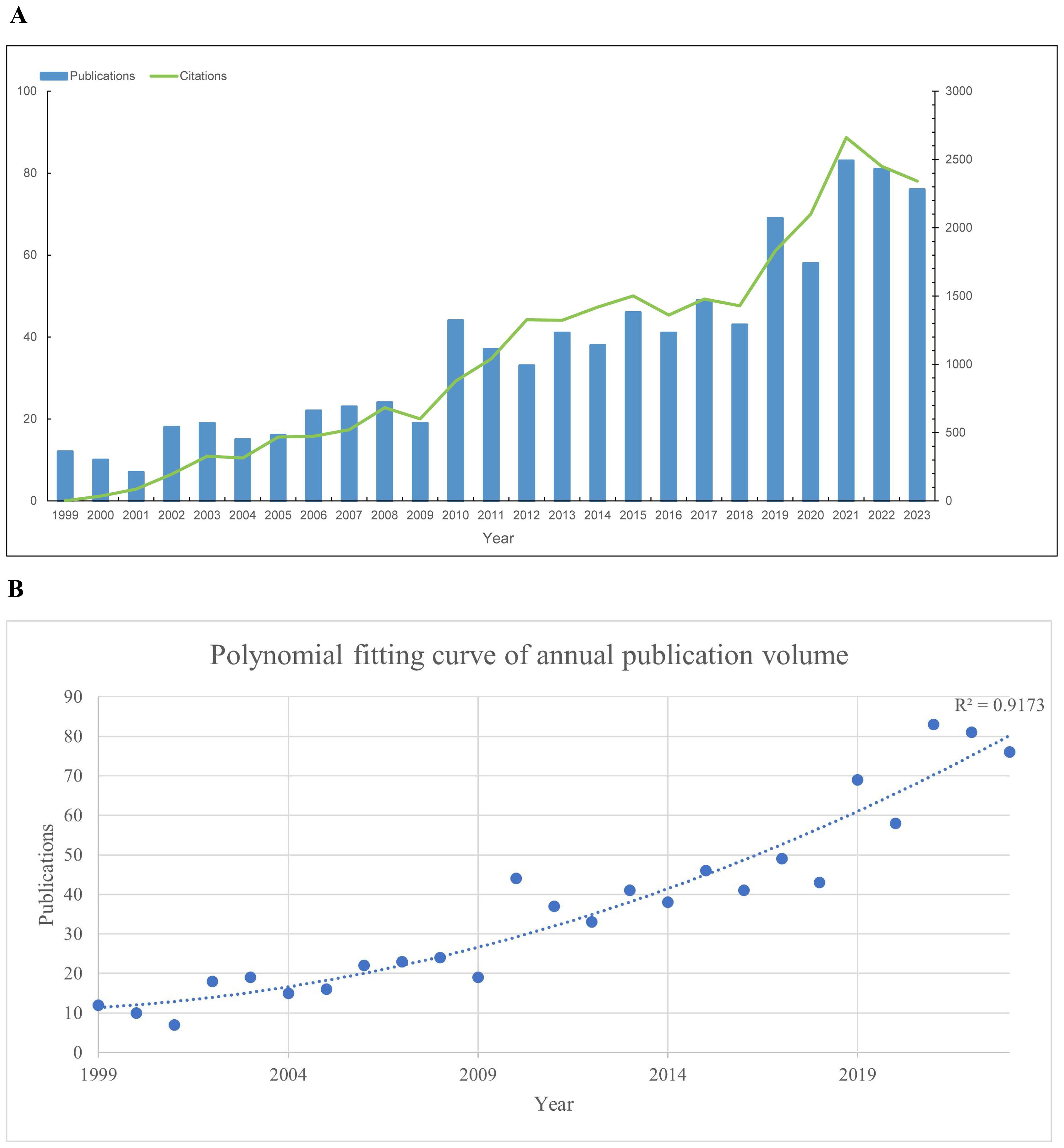
Figure 2. Annual trends in global publications and citations on prognosis-related pediatric AML literatures. (A) Annual trends in global publications and citations on prognosis-related pediatric acute myeloid leukemia (AML) research from 1999 to 2023. (B) Polynomial fitting curve of annual trends in global publications.
3.2 Countries and regions
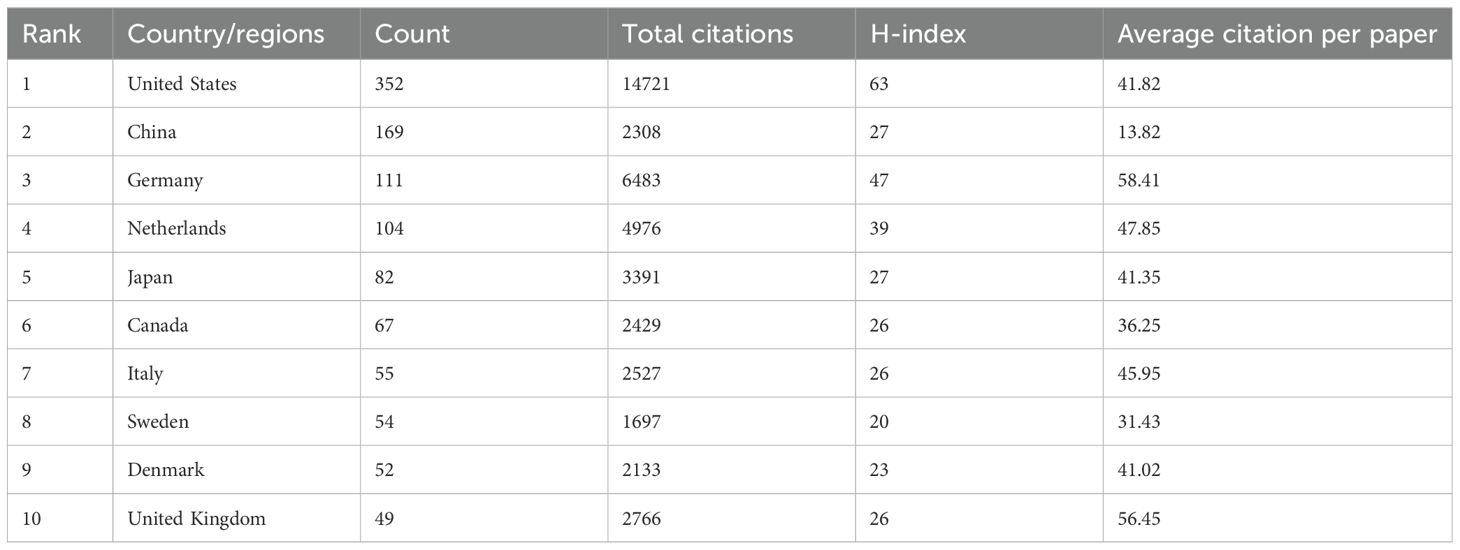
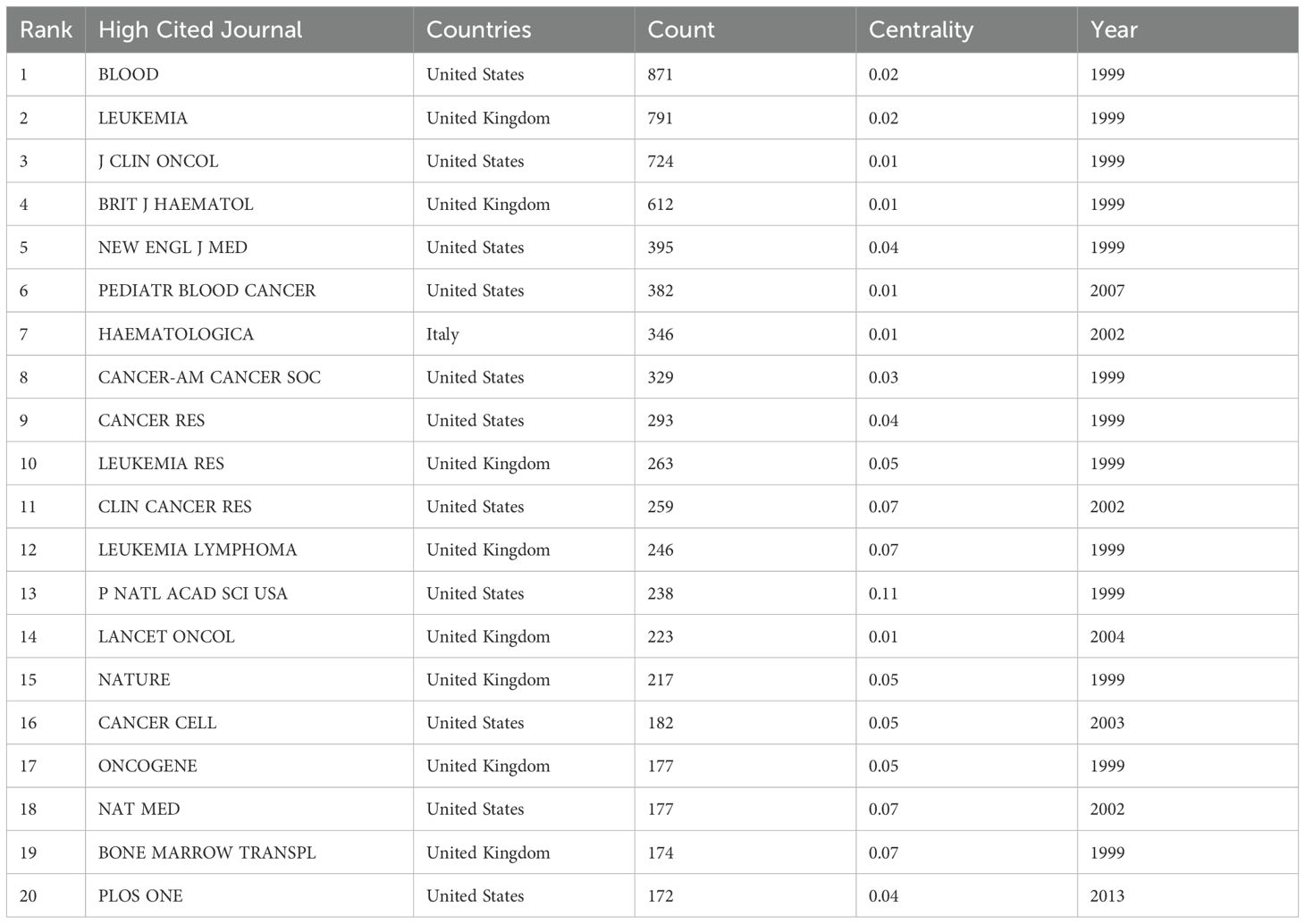
Investigators from 65 countries or regions have contributed to the study of prognosis-related pediatric AML. Table 1 and Figure 3C highlight the top 10 most productive nations in this research field. Europe emerges as a dominant contributor with five countries, while North America and Asia each have two, and Oceania has one. The United States stands out prominently, having the highest number of publications, total citations, and an impressive H-index of 63, indicating both the quantity and quality of its academic output. Moreover, the network view and heatmap illustrate the USA as a major hub, represented by the brightest (red) color, demonstrating extensive collaborative links with numerous countries, with significant participation from North America and Europe (Figures 3A, B). These visualizations effectively identify significant international research networks and highlights areas for potential growth in collaboration, underscoring the United States’ leadership in the study of prognosis-related pediatric AML. Furthermore, Germany is distinguished by achieving the highest average citation per paper, reflecting the nation’s production of high-quality studies with significant influence. Notably, China, the only developing country among the top 10, has demonstrated remarkable collaboration in this field, with high-density collaborative links with surrounding countries (Figure 3A) and a prominent red area (Figure 3B).
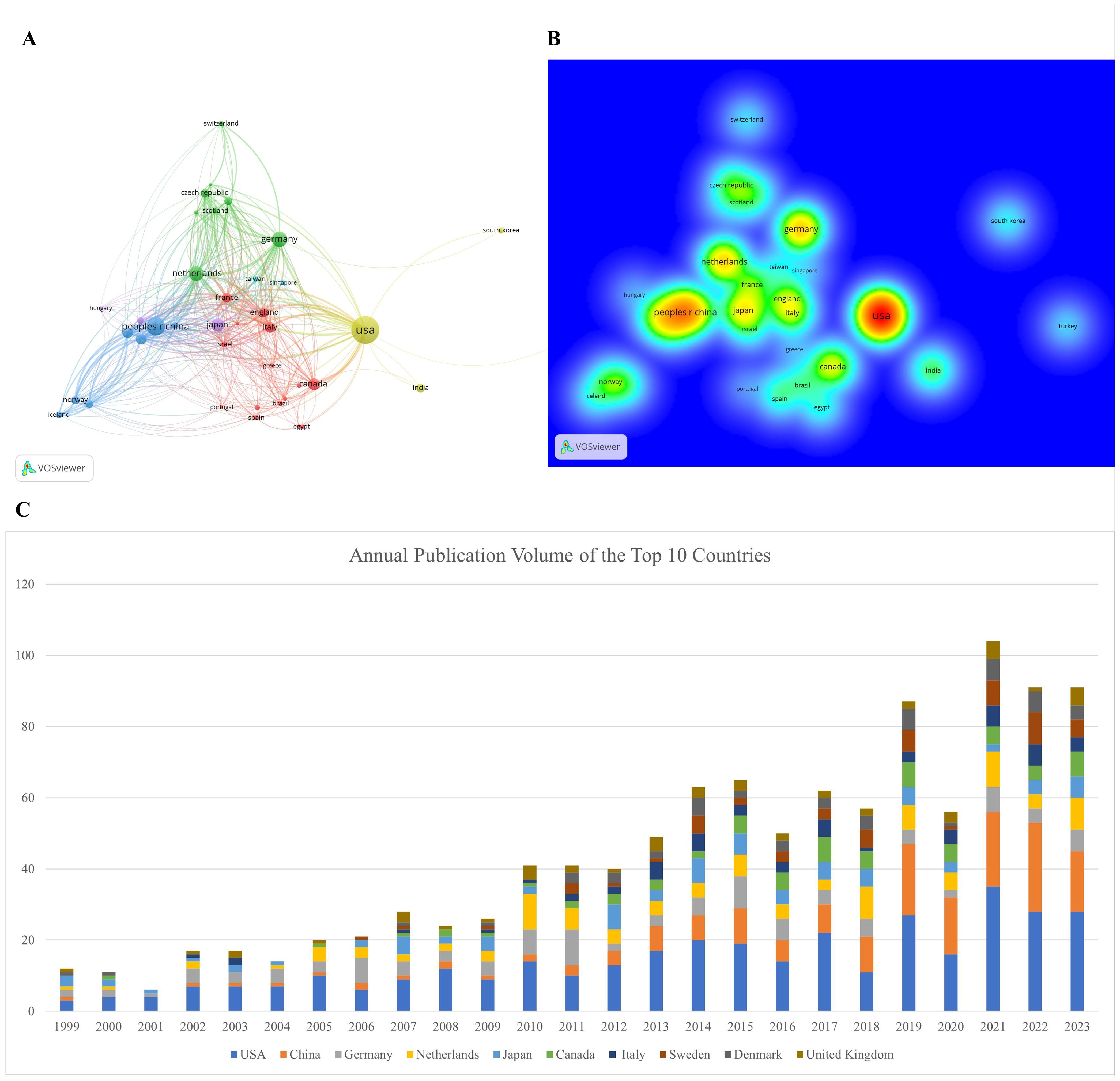
Figure 3. Visualization of international collaborations in prognosis-related pediatric AML research. (A) The graph illustrates collaborations between countries in prognosis-related pediatric AML research. The size of each bubble corresponds to the number of publications from each country, while the thickness of the connecting lines between nodes represents the strength of collaborative ties, indicating the extent of their collaborative efforts. Connection lines between countries show the cooperation relationships, highlighting the global network of research partnerships. (B) Density visualization of collaborations between countries. This visualization displays the density and intensity of collaborative research activities between countries in the field of pediatric AML prognosis. The heatmap is employed to highlight regions of varying collaborative strength, with warmer colors representing areas of higher activity and stronger connections. (C) Annual publication volume of the top 10 countries.
3.3 Institutions
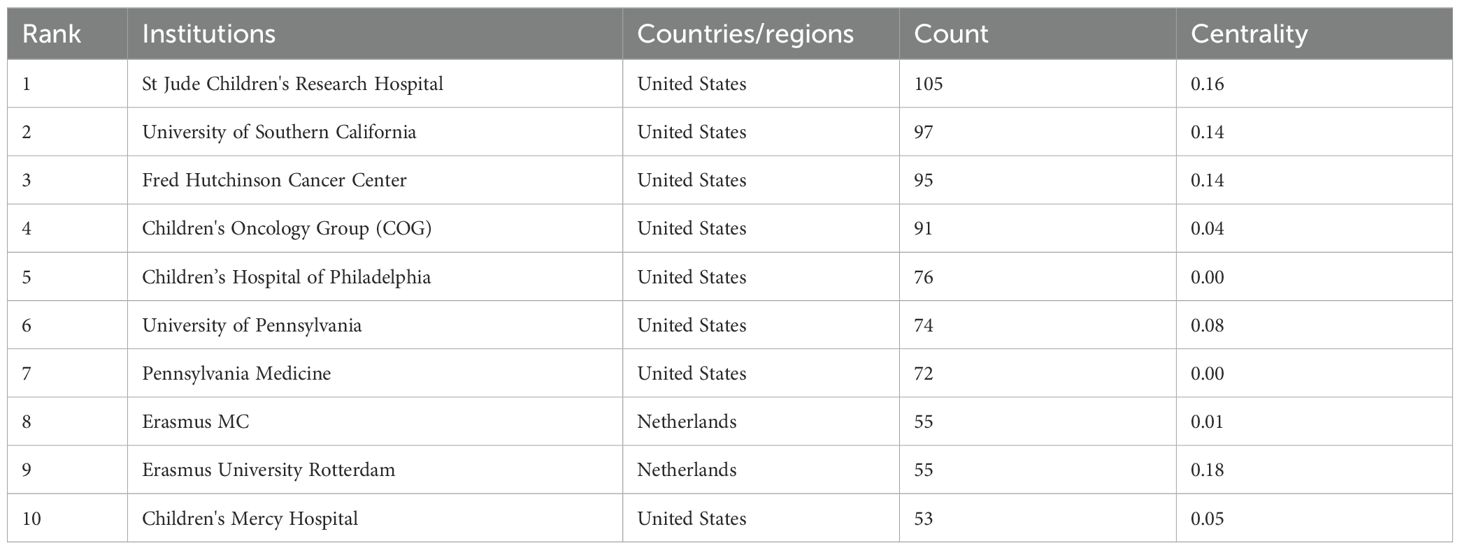
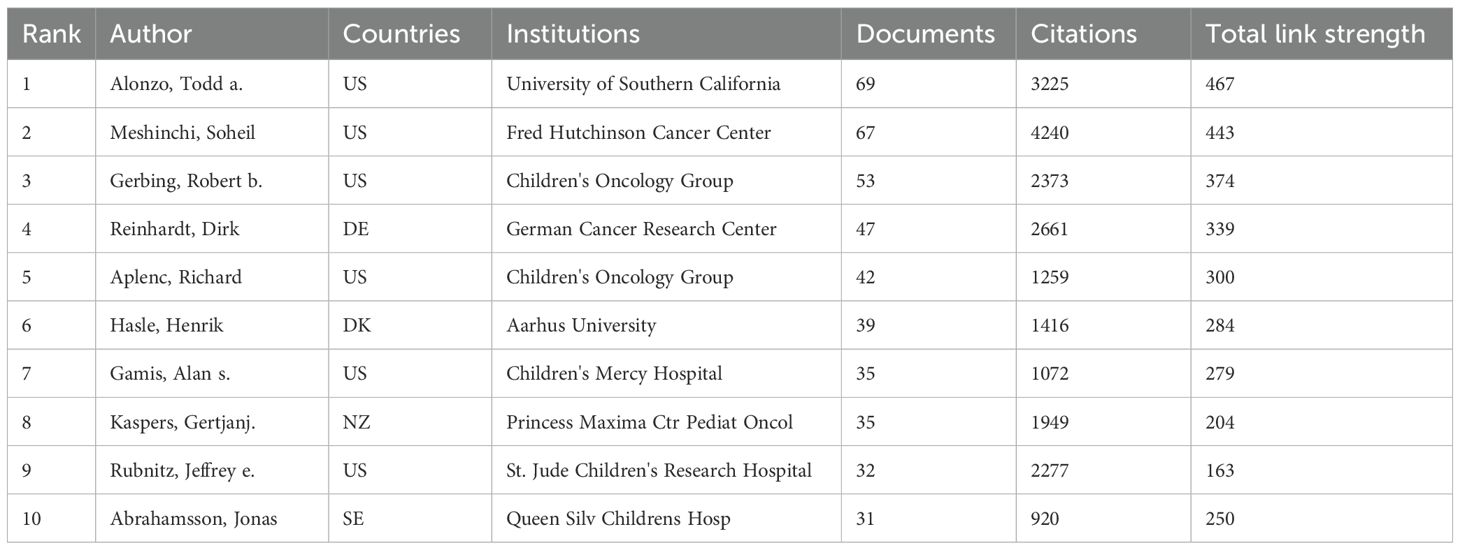
Table 2 shows that the top 10 institutions in pediatric AML prognosis research are mainly from North America and Europe, with eight based in the U.S. and two in the Netherlands. St. Jude Children’s Research Hospital is the most prolific, while Erasmus University Rotterdam leads in centrality (0.18), highlighting its key role in the research network. The Children’s Oncology Group (COG) has been pivotal in advancing pediatric AML prognosis through influential clinical trials and risk stratification strategies, contributing significantly to the field from 1999 to 2023. Figure 4A illustrates the strong institutional collaborations, primarily between U.S. and European institutions, emphasizing the global nature of this research. The density of connections in the figure reflects the frequency and strength of these partnerships, with COG acting as a central collaborative hub, linking with institutions like the University of Southern California and St. Jude Children’s Research Hospital.
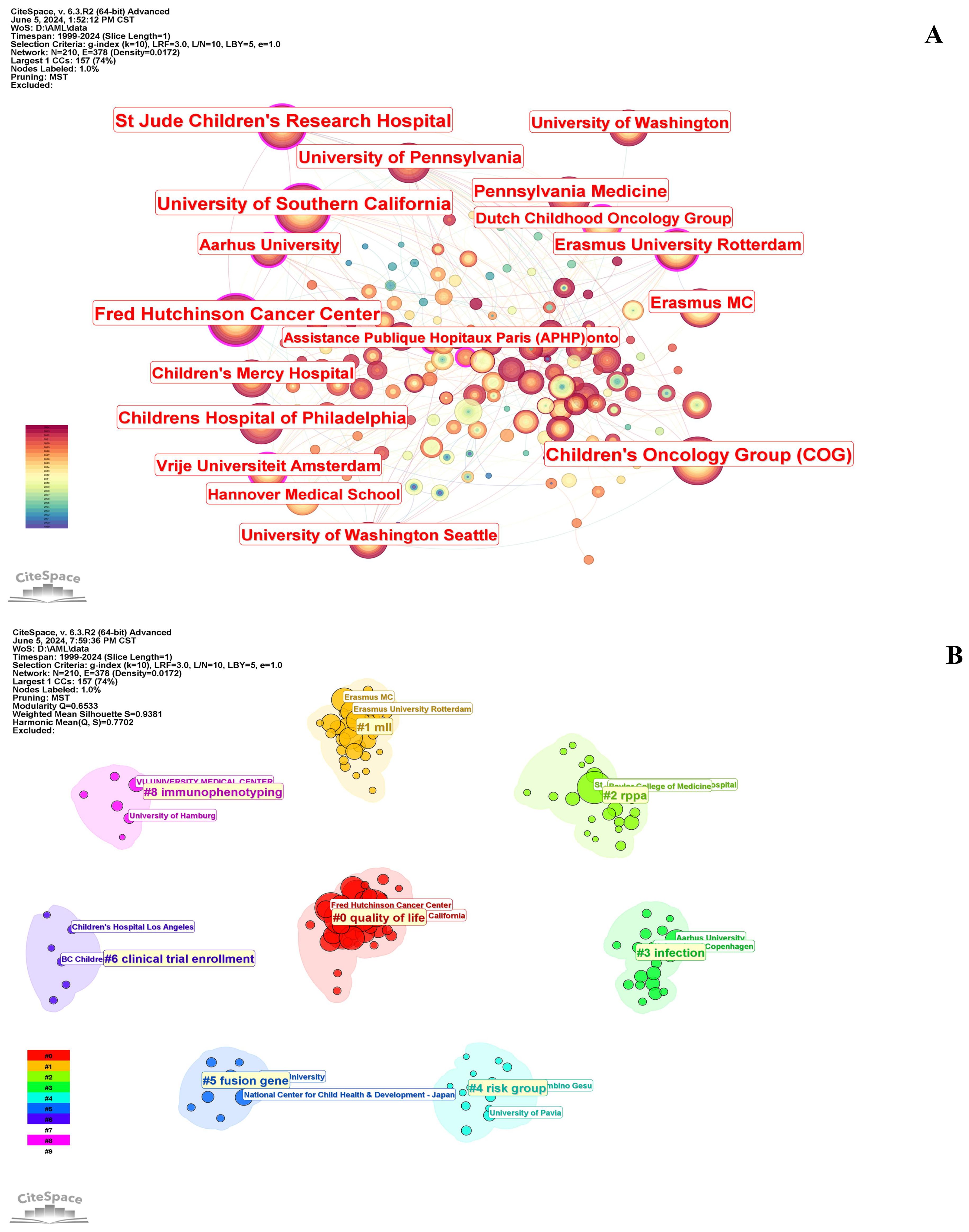
Figure 4. Collaborations between institutions in pediatric AML prognosis research. (A) Visualization of collaborations between institutions in pediatric AML prognosis research with a minimum of 5 publications. The graph illustrates collaborations between institutes involved in prognosis-related pediatric AML research. Each node represents an institute, with the node size indicating the volume of their publications. The lines between the nodes represent collaborative relationships between the institutions. (B) Circular cluster view of institutions involved in pediatric AML prognosis research (LLR). This visualization presents a circular cluster view of institutions engaged in this field. Each cluster is color-coded and spatially organized to reflect the density and strength of research activities. This circular cluster view facilitates the identification of leading institutions and their collaborative networks, providing insights into the institutional landscape of pediatric AML prognosis research.
Figure 4B depicts a circular cluster view categorizing institutions into eight distinct research clusters, each represented by a color and corresponding to specific pediatric AML prognosis themes, highlighting the thematic diversity of institutional research and their collaborative networks. For example, Cluster #5 (blue) focuses on fusion genes, with notable contributions from the National Center for Child Health and Development in Japan. Similarly, Cluster #8 (pink) emphasizes immunophenotyping, led by institutions such as the University Medical Center Hamburg.
3.4 Leading journals, co-cited journals and categories
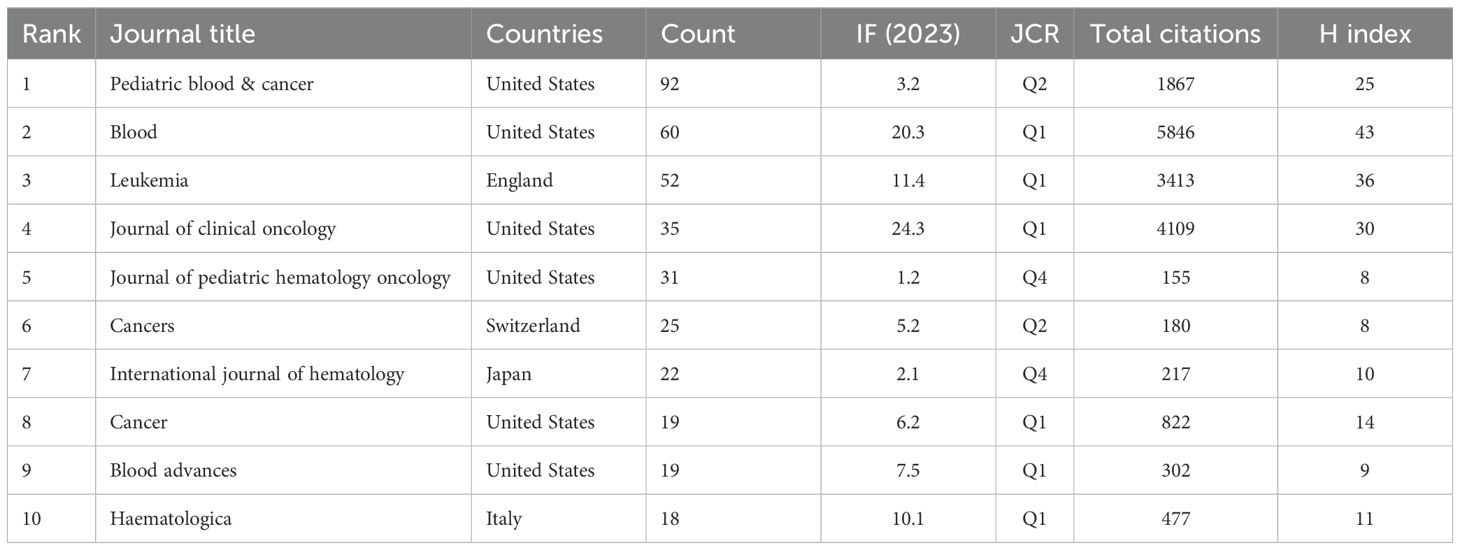
Tables 3 and 4 present the top 10 journals by publication volume and citation impact in pediatric AML prognosis research. The leading co-cited and high-publication journals are primarily based in the United States. While publication volume and impact factor indicate a journal’s reach, citation count and H-index more accurately reflect its significance. Key contributors to the field include Blood, New England Journal of Medicine (NEJM), Lancet Oncology, Nature, and Nature Medicine, which drive advancements in pediatric AML prognosis research (Tables 3, 4, Figures 5A–C).
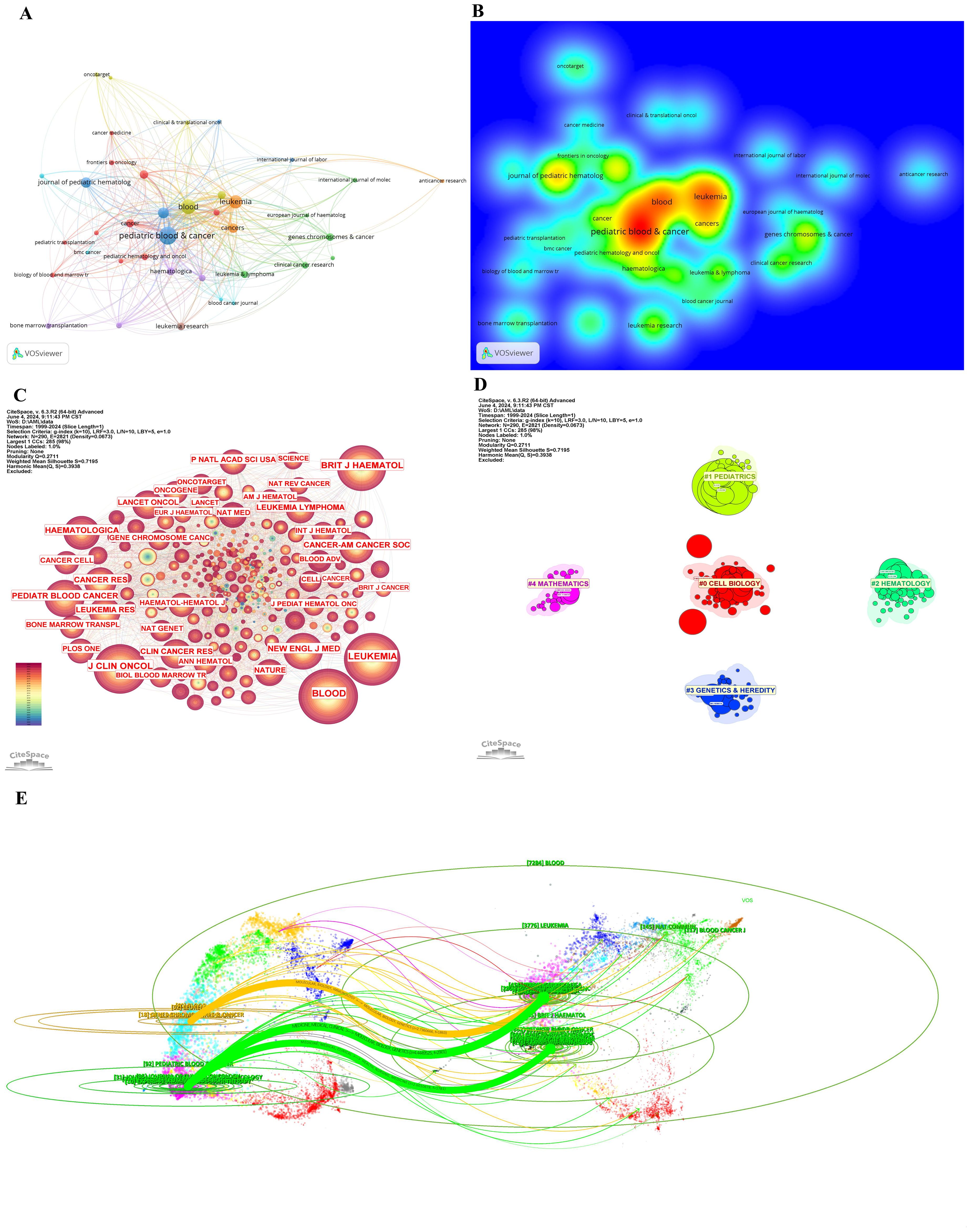
Figure 5. Journals and categories in pediatric AML prognosis research. (A) Visualization of collaborations between journals in pediatric prognosis AML research with a minimum of 5 publications. The graph illustrates collaborations between journals in prognosis-related pediatric AML research. Each node represents a journal, with the size of the node indicating the volume of their publications. The lines between nodes represent the collaborative relationships between the journals, while the thickness of the connecting lines between nodes represents the strength of collaborative ties. (B) Density visualization of journals featuring co-authorship. (C) The cited journals view in pediatric prognosis AML research. The graph illustrates collaborations between cited journals in this field. Each node represents a journal, with the node size indicating the citation count. The lines between the nodes represent collaborative relationships between the cited ones. (D) Circular cluster view of WoSCC Categories in pediatric prognosis AML research (LLR). This visualization presents a circular cluster view of categories engaged in this field. Each cluster is color-coded and spatially organized to reflect the density and strength of research activities. This circular cluster view facilitates the identification of leading categories and their collaborative networks, providing insights into the categories landscape of pediatric AML prognosis. (E) Dual-map overlay of journals in pediatric AML prognosis research. This dual-map overlay depicts the citation relationships between journals in the field of pediatric AML prognosis research. The base map shows citing journals on the left and cited journals on the right, with colored lines indicating citation pathways.
Figures 5A, B visualize the collaboration networks and density of journal relationships, highlighting Blood as the most influential journal, with strong collaborative ties denoted by warm colors. Figure 5C further illustrates connections among the most cited journals, with Blood, Nature, NEJM, Journal of Clinical Oncology, and Leukemia emerging as central hubs due to their high citation counts and dense collaborations.
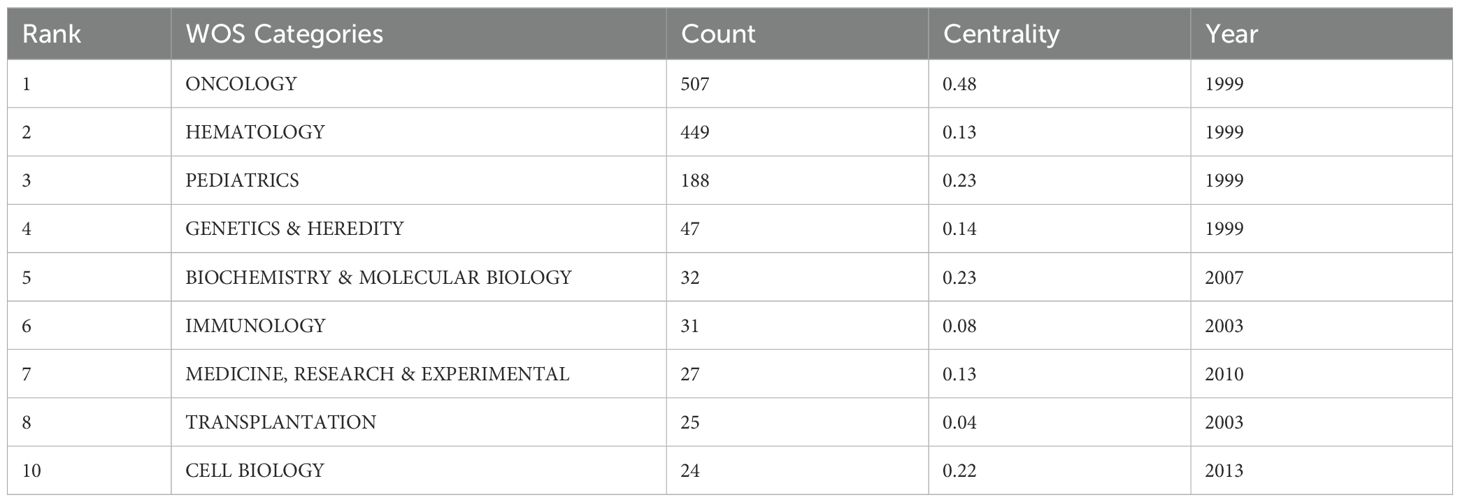
Figure 5D categorizes journals into five clusters—Pediatrics, Mathematics, Hematology, Genetics and Heredity, and Cell Biology—each represented by a distinct color corresponding to specific WoSCC categories. These clusters reflect thematic concentrations within pediatric AML prognosis research. Table 5 identifies Oncology, Hematology, and Pediatrics as the top three research categories, with citation counts and centrality measures providing insights into their relative influence.
The dual-map overlay of journals (Figure 5E) reveals three major citation pathways. Two green citation lines indicate that journals in MEDICINE, MEDICAL, and CLINICAL categories frequently cite those in HEALTH, NURSING, MEDICINE (Citing frequency = 1781) and MOLECULAR, BIOLOGY, GENETICS (Citing frequency = 2901). For instance, journals like Pediatric Blood Cancer often cite Blood and NEJM. A yellow citation line shows frequent citations from MEDICINE, MEDICAL, and IMMUNOLOGY journals to MOLECULAR, BIOLOGY, and GENETICS journals (Citing frequency = 1863), exemplified by journals like Leukemia citing Nature.
3.5 Co-authors, co-cited authors, and leading references
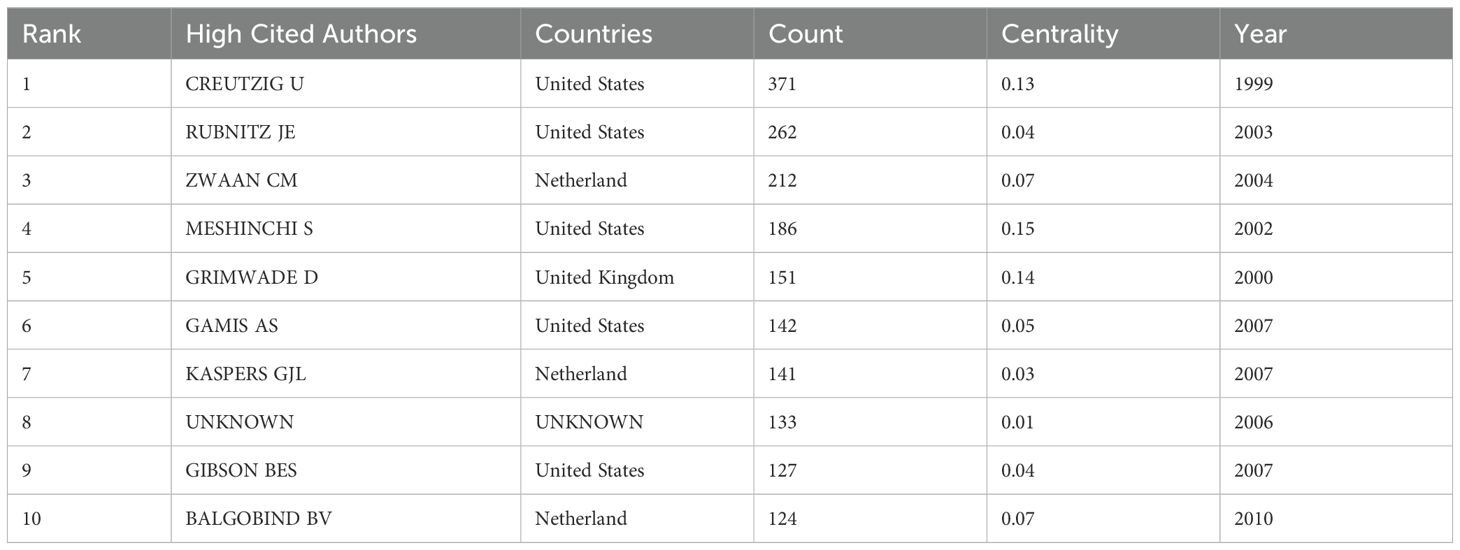
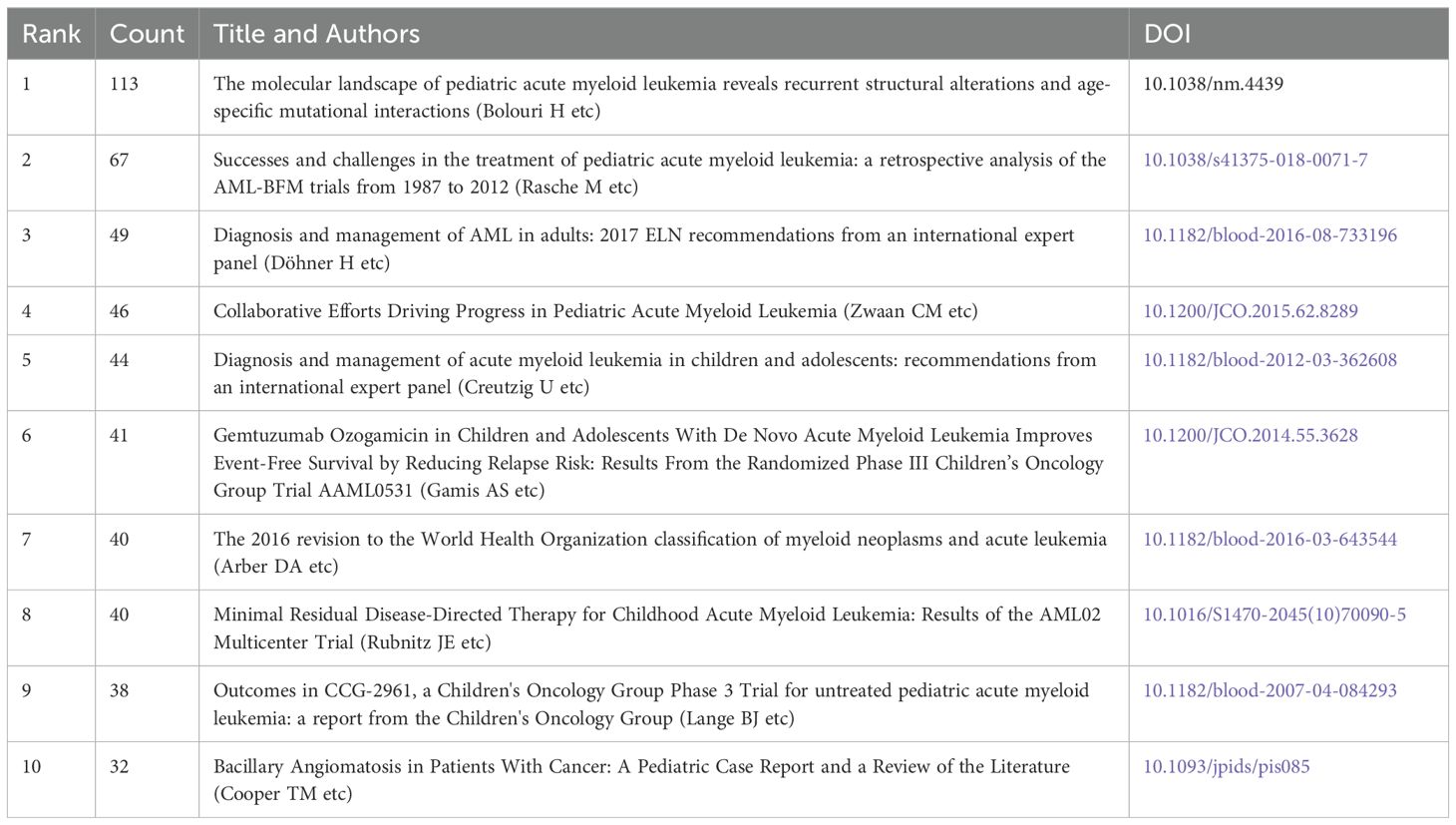
Tables 6-8 highlight the top 10 prolific authors, leading cited authors, and most cited references in pediatric AML prognosis research, with corresponding visualizations in Figures 6A-C. Six prolific authors and five leading cited authors are from the United States, underscoring the country’s leading role in this field, as evident across countries, institutions, journals, and authors. Core literature in this domain is identified by citation frequency, with high-impact publications becoming focal points for researchers. Among the top 10 cited authors, Creutzig U, Rubnitz JE, Zwaan CM, and Gamis AS stand out for contributing influential references that have significantly advanced the field. Figure 6B and Table 7 illustrate their extensive collaboration networks. Notably, four of the most cited references are published in Blood, reaffirming its leadership in this area (Table 8).
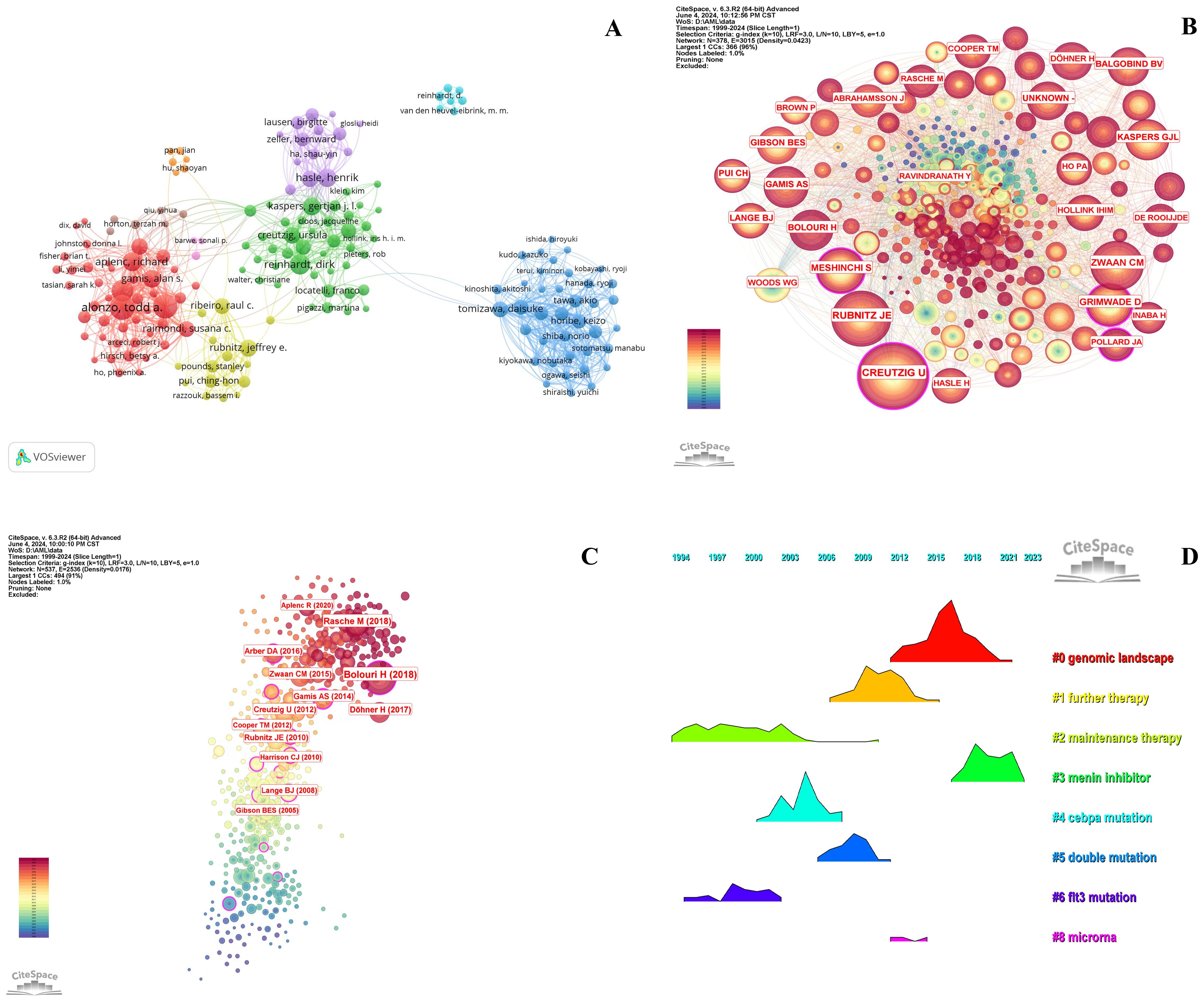
Figure 6. Analysis of authors and references in prognosis-related pediatric AML research. (A) The visualization map shows the analysis of authors in prognosis-related pediatric AML research. Each node represents an author, with the node size indicating their number of citations or publications. The lines between the nodes represent collaborative relationships among the authors. (B) The cited authors view in pediatric prognosis AML research. The graph illustrates collaborations between cited authors in this field. Each node represents a author, with the node size indicating the citation count. The lines between the nodes represent collaborative relationships between the cited ones. (C) The cited references view in pediatric prognosis AML research. The node illustration is similar with Figure 5B. (D) Ridgeline plot of the cited references in pediatric AML prognosis research. This ridgeline plot visualizes the temporal trends of key cited references in this field. Each colored line represents a distinct research focus, illustrating the evolution of influential topics over time.
The highly cited references fall into three main themes: guidelines and recommendations (e.g., 2017 ELN and 2016 WHO), genomic and epigenetic landscapes (e.g., structural alterations and age-specific mutations), and targeted therapies and clinical trials (e.g., Phase III Children’s Oncology Group Trial AAML0531). These categories reflect pivotal advancements and research priorities in pediatric AML prognosis (Figure 6C, Table 8). Temporal trends of influential research topics are captured in the ridgeline plot, highlighting key themes such as “genomic landscape” and “further therapy,” which show increasing attention in recent years. Earlier peaks in topics like “menin inhibitor,” “CEBPA mutation,” and “microRNA” indicate prior research focus shifts (Figure 6D).
3.6 Keywords co-occurrence analysis
Figures 7A, B highlight central themes in pediatric AML prognosis research, including “acute myeloid leukemia,” “pediatric AML,” “prognosis,” “relapse,” “chemotherapy,” “bone marrow transplantation,” “stem cell transplantation,” “immune therapy,” and “survival.” These keywords demonstrate strong interconnections, reflecting their critical role in the research community. The density visualization reveals intense research activity around these themes, with warmer colors indicating high-focus areas. Key genetic factors such as “CEBPA,” “KMT2A,” “WT1,” and “FLT3-ITD,” along with targeted therapies like “sorafenib,” emerge as significant research hotspots, critical to understanding prognostic factors and treatment outcomes.
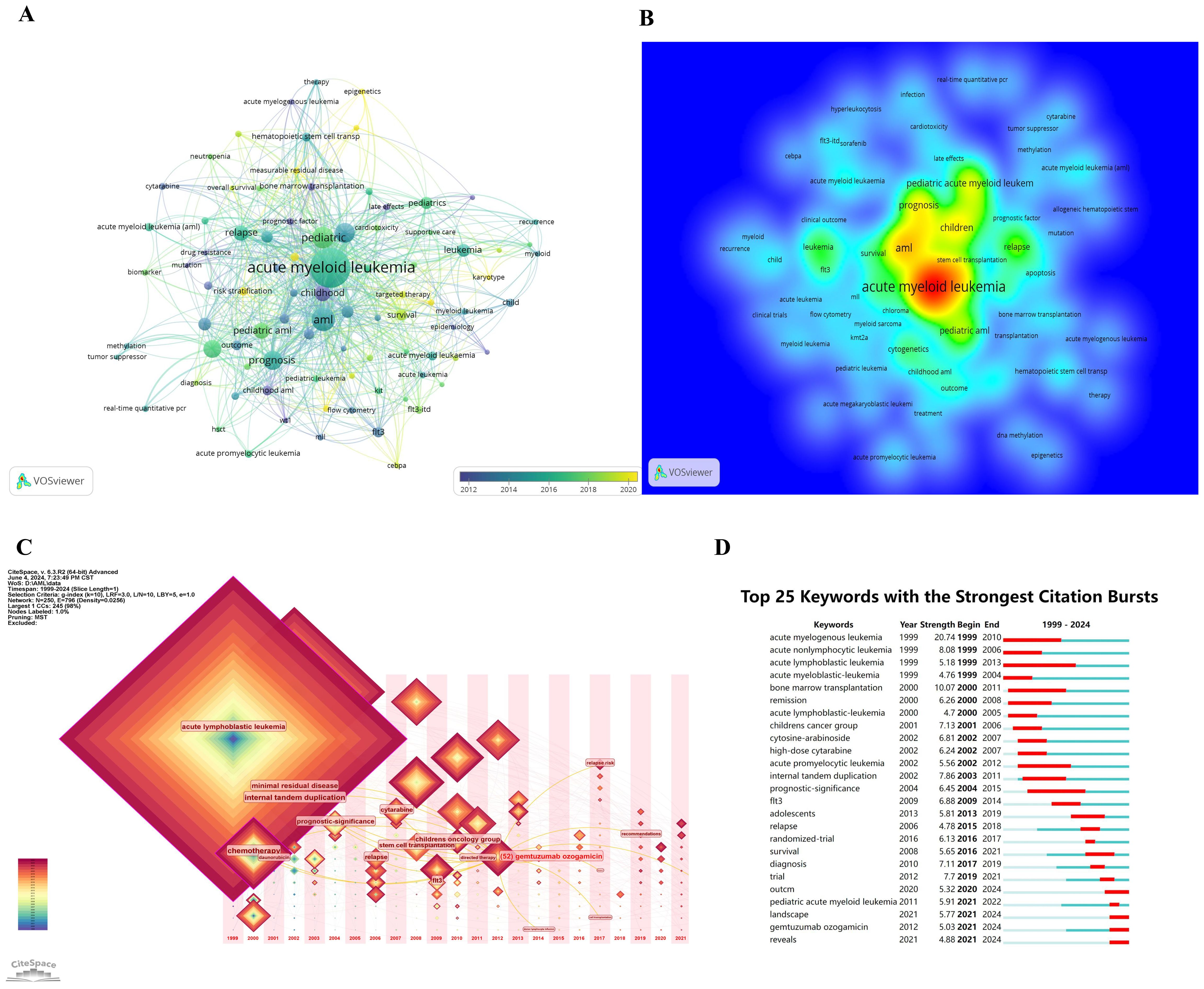
Figure 7. Keywords co-occurrence analysis in prognosis-related pediatric AML research. (A) Network visualization of keyword co-occurrence analysis with a minimum of 5 occurrences. Each node represents a keyword, with the node size indicating the number of occurrences. Lines between nodes represent collaborations between keywords; the larger the size of the node, the higher the frequency of the keyword. (B) Density visualization of keyword co-occurrence analysis. This visualization displays the density and intensity of research themes in this field. The heatmap is employed to highlight regions of varying study strength, with warmer colors representing areas of higher activity and stronger connections. (C) Time zone chart of keyword co-occurrence analysis. This time zone chart visualizes the temporal evolution of key research topics in this field. Each keyword’s prominence is depicted over time, with larger and more central nodes representing higher frequency and significance. (D) The 25 keywords with the strongest citation bursts are shown. The blue line represents the time axis, while the red segments indicate the start year, end year, and duration of each burst.
Figures 7C, D illustrate the temporal evolution of research topics. Early focus areas included “chemotherapy” and “acute lymphoblastic leukemia,” while recent trends highlight “minimal residual disease,” “stem cell transplantation,” and “FLT3,” indicating a shift toward advanced therapies and molecular research. Figure 7D further identifies the top 25 keywords with the strongest citation bursts, showcasing evolving priorities. Keywords like “acute myelogenous leukemia” (burst period: 1999–2010, strength: 20.74) and “acute lymphoblastic leukemia” (1999–2013, strength: 5.18) demonstrate lasting importance. More recent bursts include “landscape” and “GO,” emerging after 2021, reflecting shifting research interests toward novel directions.
The combined results from Figures 6D and 7 underscore the growing prominence of genomics, transcriptomics, epigenomics, targeted therapies, immune therapy, and integrative diagnostic approaches in pediatric AML prognosis research. These findings emphasize the field’s increasing focus on molecular and personalized medicine to improve outcomes.
4 Discussion
4.1 General information
Research on pediatric AML prognosis has grown substantially over the past 25 years, as reflected by the increasing number of publications and citations. This growth highlights the heightened global focus on improving prognostic outcomes for pediatric AML patients. Our bibliometric analysis, encompassing 924 publications from 1999 to 2023, shows a significant peak in publication and citation activity in 2021, driven by advancements in multi-omics and precision medicine.
Initially, research in this field centered on clinical outcomes and standard chemotherapy regimens (21). Over time, the focus has evolved toward genetic and molecular insights, leading to the discovery of key prognostic markers and novel therapeutic targets (22, 23). This shift underscores the dynamic nature of the field, which continuously adapts to scientific breakthroughs and technological progress (24). High-frequency keywords such as “CEBPA,” “WT1,” and “FLT3-ITD” emphasize the pivotal role of genetic mutations in disease progression and treatment response. Multi-omics approaches—encompassing genomics, transcriptomics, and epigenomics—have further refined prognostic assessments and enabled personalized treatment strategies (23, 25). For example, Pollard and Alonzo demonstrated that adding sorafenib to standard chemotherapy significantly improved event-free survival (EFS) and reduced relapse risk in FLT3/ITD+ pediatric AML patients (26). Similarly, Menin inhibitors demonstrate substantial therapeutic effectiveness in treating KMT2A-rearranged and NPM1-mutated pediatric acute leukemias (27).
Our analysis of contributions from 65 countries highlights a diverse and collaborative research landscape. The United States leads in publication volume, institutional output, journal contributions, categories, and collaborations, underscoring its dominant role in this field (Tables 1-7, Figures 3-6). European countries, particularly Germany, have produced high-quality and influential studies, while China, as a developing nation, has shown rapid growth in publication output, reflecting increasing investment in this area (28). Collaboration across countries with varying levels of development and across continents remains vital for fostering innovation and advancing knowledge in pediatric AML prognosis research.
Prominent institutions such as St. Jude Children’s Research Hospital, Erasmus University Rotterdam, and the Children’s Oncology Group (COG) have played instrumental roles in shaping the field. St. Jude leads in publication output, while Erasmus University Rotterdam’s high centrality score reflects its pivotal role in global research networks. COG has contributed groundbreaking clinical trials and comprehensive risk stratification methods, demonstrating the value of multidisciplinary and collaborative approaches. Leading journals, including Blood and Journal of Clinical Oncology, play a critical role in disseminating high-impact research. These journals’ high impact factors, citation counts, and H-index scores underscore their authority in the field (Tables 2, 4, 8; Figures 4, 6C, D). Encouraging cross-institutional, cross-journal, and interdisciplinary collaborations could further accelerate knowledge dissemination and foster innovative research.
Pioneering researchers, such as Creutzig U, Rubnitz JE, Zwaan CM, Bolouri H, and Gamis AS, have significantly influenced pediatric AML prognosis research through highly cited publications and seminal references (Table 8; Figures 6B–D). For instance, Gamis AS demonstrated that GO, a CD33-targeted immunoconjugate, improves three years event-free survival (53.1% vs. 46.9%) and reduces relapse risk (32.8% vs. 41.3%) in children and adolescents (29). Bolouri H’s study on nearly 1,000 participants revealed that somatic structural variants are more prevalent in younger AML patients, with genetic mutations such as GATA2, FLT3, and CBL highlighting the need for age-specific therapeutic strategies (22).
Keyword analysis offers valuable insights into research trends and emerging foci. Terms such as “survival,” “chemotherapy,” “bone marrow transplantation,” “stem cell transplantation,” “immune therapy,” and “relapse” reflect ongoing efforts to improve outcomes. The increasing prominence of genetic and targeted therapies underscores the shift toward precision medicine. Emerging keywords post-2021 reflect the dynamic nature of the field, as researchers continue exploring novel avenues to enhance prognosis and treatment.
This study underscores significant advancements in AML prognosis research, highlighting leading countries, key institutions, prominent journals, major categories, influential contributors, seminal references, and emerging trends. By providing valuable insights into future research directions, this work emphasizes the importance of addressing existing challenges and fostering global collaborations to further advance the field and improve outcomes for pediatric AML prognosis.
In conclusion, this bibliometric analysis provides a comprehensive overview of prognosis-related pediatric AML research, highlighting significant advancements, key contributors, and emerging trends. By identifying influential components, this study offers valuable insights into future research directions. Additionally, this analysis has highlighted several key subtopics within the research on pediatric AML prognosis.
4.2 Research progress on pediatric AML prognosis
4.2.1 Genetic insights and risk stratification
Prognostic factors and risk stratification are paramount in guiding treatment decisions and improving outcomes for pediatric AML patients (30). The identification and validation of key genetic and molecular markers have revolutionized our understanding of disease mechanisms and patient prognosis. For instance, FLT3-ITD mutations, especially when combined with WT1 mutations, significantly worsen prognosis and treatment response, necessitating more aggressive treatment protocols (31). Independently, WT1 mutations are associated with poor outcomes and higher relapse rates, highlighting the need for intensified therapeutic strategies (32). The combination of FLT3-ITD and CEBPA mutations further impacts patient survival, with a 3-year overall survival (OS) of only 44%, underscoring the importance of tailored treatment approaches in pediatric AML (33). Conversely, NPM1 mutations, whether accompanied by FLT3-ITD or not, allow for less intensive regimens, reducing treatment-related toxicity while maintaining high survival rates (34, 35).
The discovery of cytogenetic abnormalities has been equally transformative. Chromosomal translocations involving the KMT2A gene are strongly linked to poor outcomes and elevated relapse rates (15). Studies on 11q23/MLL rearrangements reveal significant variations in prognosis depending on specific translocation partners, offering critical insights for risk stratification and treatment planning (36–38). Recent advancements, such as Optical Genome Mapping, have identified novel structural aberrations, enhancing diagnostic accuracy and enabling refined minimal residual disease (MRD) monitoring (39). Additionally, the integration of multi-omics data with machine learning has pinpointed robust molecular markers that predict treatment responses, paving the way for precision medicine in AML (40).
4.2.2 Gene expression profiling
Gene expression profiling is a powerful prognostic tool in pediatric AML (41). Elevated expression of specific genes increases relapse risk, prompting strategies like early stem cell transplantation (42).
Real-time PCR and flow cytometry aid in identifying high-risk patients (43, 44), while next-generation sequencing (NGS) and transcriptome analysis uncover gene fusions such as NPM1-CCDC28A, TRIP12-NPM1, and MLLT10-DNAJC1, enabling precise diagnosis and insights into disease mechanisms (17).
Single-cell sequencing further enhances understanding. scDNA-seq techniques like Phertilizer identify tumor clonal structures via copy number aberrations (45), while scRNA-seq, exemplified by SMART-seq, reveals cellular heterogeneity and gene regulation (46, 47). The NanoString GeoMx Digital Spatial Profiler integrates spatial analysis with single-cell sequencing, mapping tumor microenvironments to identify distinct gene expression patterns critical for understanding disease progression (48). These approaches are vital for advancing tumor biology and targeted therapy.
Epigenetic modifications, especially DNA methylation, predict treatment response and disease progression. DNA methylation clusters linked to cytogenetic abnormalities enhance classification (49), while methylation at bivalent promoters and Polycomb-repressed regions guides therapeutic decisions (50). Specific miRNA profiles also serve as biomarkers for prognosis (51).
Single-cell epigenome sequencing, including scATAC-seq and scChIP-seq, reveals chromatin accessibility and transcription factor binding, advancing gene regulation research. Integrating scRNA-seq and scATAC-seq across treatment time points uncovers transcriptional changes and chromatin dynamics, shedding light on disease progression and treatment resistance (52).
Gene expression profiling drives personalized medicine by identifying genetic abnormalities and molecular signatures, improving risk stratification and enabling precise predictions of disease progression and treatment response, aligning with the rise of precision medicine in pediatric AML.
4.2.3 Multi-omics integration and its influence on prognosis
The integration of molecular genetics, gene expression profiling, and epigenetic modifications has advanced precise and personalized treatment approaches. Patients with favorable genetic profiles can receive fewer intensive therapies, reducing treatment-related toxicity while maintaining high survival rates. In contrast, high-risk patients with adverse genetic markers require more aggressive protocols. As shown in Figure 7D, the keyword “landscape” has exhibited the strongest citation burst since 2021, highlighting the growing focus on integrating genomic, transcriptomic, epigenomic, metabolomic, and pharmacogenomic landscapes to provide a comprehensive understanding of pediatric AML pathophysiology (53). The depth and degree of personalization enabled by multi-omics integration are closely interlinked (54), paving the way for more refined therapeutic strategies and improved patient outcomes.
4.3 Outcomes in pediatric AML
4.3.1 Remission, induction failure, and transplantation failure
Achieving remission is a crucial objective in pediatric AML treatment. Induction therapy aims to achieve complete remission (CR) by eradicating leukemic cells (55). However, 10-20% of patients fail to reach CR following initial induction therapy (56), often due to factors such as a high initial leukemic burden and adverse genetic profiles.
For patients who achieve CR, sustaining remission and preventing relapse are critical priorities. Allogeneic HSCT is commonly used for high-risk patients in CR1 or those who relapse (57), Nevertheless, transplantation failure and transplant-related mortality remain significant challenge (58, 59). To address these issues, current research focuses on developing less toxic conditioning regimens and improving donor selection to reduce failure rates and enhance overall outcomes.
4.3.2 Relapse and cumulative incidence of relapse
Relapse after remission remains a major challenge in pediatric AML, often resulting in poorer outcomes (60). Key causes of relapse include residual leukemic cells that escape initial treatment, genetic mutations, and chemotherapy resistance. The cumulative incidence of relapse (CIR) varies by risk group, with high-risk patients experiencing significantly higher relapse rates. Studies indicate that approximately 30-40% of pediatric AML patients relapse, highlighting the urgent need for effective post-relapse treatment strategies (60). Minimal residual disease (MRD) is a critical predictor of relapse, with leading references emphasizing its role in guiding therapeutic decisions and improving patient outcomes (8).
4.3.3 Event-free survival and overall survival
Event-free survival (EFS) and overall survival (OS) are critical endpoints for evaluating the efficacy of pediatric AML treatments. EFS measures the duration after treatment during which patients remain free of complications or events, such as relapse or treatment-related mortality. Recent advances have significantly improved EFS rates in pediatric AML, with COG trials reporting 3-year and 5-year EFS rates exceeding 50% for high-risk groups, reflecting notable progress in treatment protocols (61, 62).
OS, defined as the time from diagnosis to death from any cause, has also improved due to enhanced therapeutic strategies. Over the past two decades, 5-year OS rates for pediatric AML have risen to nearly 70% (55). These gains are attributed to better risk stratification, targeted therapies, and advancements in supportive care, all of which contribute to improved patient outcomes. Compared to adult AML, where OS improvements have been slower, pediatric AML has achieved more pronounced advancements, underscoring the effectiveness of tailored pediatric treatment protocols (63).
4.4 Clinical trials advancing pediatric AML prognosis
Clinical trials have been pivotal in establishing current treatment standards for pediatric AML, driving advancements in chemotherapy, immunotherapy, targeted therapy, and transplantation. Our bibliometric analysis highlights the importance of clinical trials, as evidenced by the frequent occurrence of keywords such as “treatment,” “clinical trials,” and “stem cell transplantation.” Landmark trials have defined contemporary protocols, such as the COG studies, which consistently demonstrate the efficacy of intensive chemotherapy combined with transplantation, resulting in significant improvements in EFS and OS (61, 64). Similarly, the AML-BFM 98 trial emphasized the effectiveness of risk-adapted therapy, tailoring treatment intensity to individual risk profiles to maximize benefits while minimizing toxicity, reinforcing the value of personalized treatment approaches (65).
Beyond validating existing strategies, clinical trials have paved the way for innovative therapies. The incorporation of FLT3 inhibitors, such as midostaurin and gilteritinib, into treatment protocols has significantly improved survival outcomes for FLT3-ITD-positive patients (66). Additionally, the introduction of GO has enhanced the ability to target CD33-positive leukemic cells, demonstrating efficacy in both frontline and relapsed settings (9, 61, 67). Ongoing trials are further exploring the potential of immunotherapy, including monoclonal antibodies and CAR T-cell therapies, which hold promise for advancing treatment options in pediatric AML (65).
Building on current advancements, new trials are developing strategies to improve outcomes for relapsed and refractory pediatric AML. For example, studies are evaluating the safety and efficacy of Talazoparib, a DNA repair inhibitor, in combination with chemotherapy for relapsed pediatric AML (NCT05101551). Similarly, trials are exploring JK500 cell injections in relapsed/refractory pediatric AML to assess safety and preliminary efficacy (NCT05519384). These efforts, alongside additional studies (NCT03173612, NCT06262438, NCT03164057), are crucial for identifying new therapeutic avenues and optimizing treatment regimens to improve patient outcomes.
4.5 Treatment advances and prognosis research in childhood AML
Over the past 25 years, pediatric AML treatment has progressed through evolving strategies, refined risk stratification, and novel therapies, as evidenced by our bibliometric analysis of key trends (Figure 7, Table 9).
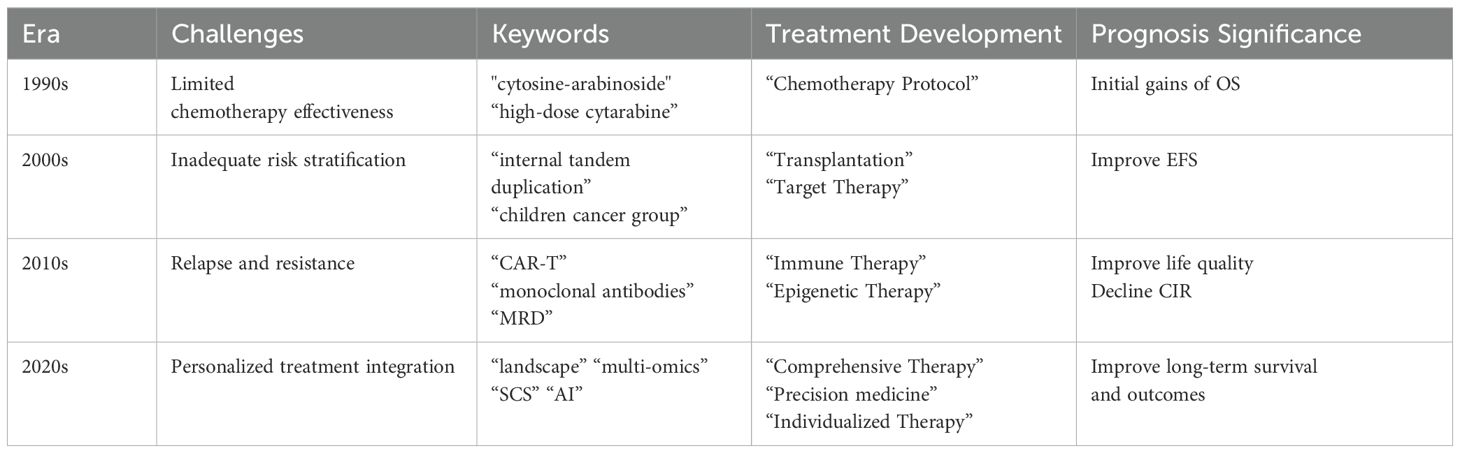
4.5.1 1990s: chemotherapy regimens and initial gains
In the 1990s, pediatric AML treatment relied on intensive chemotherapy with cytarabine and anthracyclines as cornerstone agents. Optimized regimens and improved supportive care significantly enhanced overall survival (OS). Keywords like “cytosine-arabinoside” and “high-dose cytarabine” showed strong citation bursts from 2002 to 2007, reflecting foundational chemotherapy research. Multi-agent chemotherapy during this era notably improved 5-year OS, laying the groundwork for future advancements (68).
4.5.2 2000s: risk stratification and targeted therapies
The early 2000s marked a pivotal shift in pediatric AML treatment, driven by the integration of genetic and molecular insights into risk stratification and the emergence of targeted therapies. Key genetic mutations such as FLT3-ITD, NPM1, and CEBPA were identified, enabling more precise tailoring of treatment protocols to individual patient profiles. Keywords like “internal tandem duplication” and “prognostic-significance” experienced citation bursts during this period, reflecting the growing focus on targeted approaches (69). COG played a critical role in incorporating molecular diagnostics into clinical protocols, with FLT3 inhibitors like midostaurin significantly improving event-free survival (EFS) for high-risk patients (70). This period also saw increased use of bone marrow transplantation for high-risk cases, further enhancing survival outcomes (71).
4.5.3 2010s: progress in immunotherapy and epigenetic therapies
The 2010s marked significant progress in immunotherapy and epigenetic therapies as novel treatment approaches for pediatric AML. Immunotherapy, including monoclonal antibodies and CAR T-cell therapies, enables precise targeting of leukemic cells with fewer side effects than conventional chemotherapy. For example, Anti-CLL1 CAR T-cell therapy targets CLL1 expressed on AML blasts and has shown promise in treating pediatric AML (72). Similarly, PD-1/PD-L1 inhibitors are under investigation for their potential to enhance immune responses against AML cells (73).
Epigenetic therapies, such as azacitidine and decitabine, target DNA methylation and histone modifications, offering new hope for relapsed or refractory AML by reversing aberrant methylation and restoring normal gene expression (74, 75). Additionally, miRNA-based therapies, including mimics and inhibitors, show potential in targeting dysregulated miRNA pathways to improve treatment response and overcome drug resistance (76). Exploring the epigenomic landscape in pediatric AML not only complements traditional therapies but also enhances risk stratification and treatment personalization.
4.5.4 Current challenges in treatment beyond 2020
Since 2020, pediatric AML treatment has increasingly focused on integrating emerging therapies such as CAR-T, GO, and checkpoint inhibitors, alongside a stronger emphasis on precision medicine and individualized care driven by multi-omics insights. Despite these advancements, high relapse and refractory rates, along with challenges in personalized integration, remain significant barriers that impact long-term survival and outcomes. Relapsed or refractory AML is particularly resistant to standard therapies, underscoring the need for innovative treatment strategies (77). Additionally, the toxicity associated with intensive chemotherapy and HSCT, including cardiotoxicity, continues to be a major concern, especially for young patients (78).
4.6 Future directions and challenges
Despite significant progress, further exploration is needed to improve pediatric AML prognosis. Novel biomarkers, multi-omics integration, and technologies like machine learning and deep learning offer promising insights into disease mechanisms, personalized therapeutic responses, and prognosis assessment. For example, Pamela S. Becker et al. used a multi-omics computational framework to analyze gene expression profiles and drug sensitivity in 30 AML patients, identifying SMARCA4 as a driver of sensitivity to topoisomerase II inhibitors (40). Support vector machines (SVMs) and advanced statistical tools have identified relapse signatures and predicted therapeutic outcomes, improving risk stratification and enabling personalized treatments, surpassing traditional prognostic models (79, 80). Deep learning applications, such as immune cell segmentation, provide insights into T-cell distribution and its impact on immunotherapy, highlighting immune priming as a strategy to enhance outcomes for patients with low T-cell infiltration (81).
Advanced diagnostic technologies, including NGS, transcriptome sequencing, and SCS, are revolutionizing pediatric AML diagnosis and prognosis. These tools provide comprehensive insights into genetic and molecular drivers, identifying actionable mutations and therapeutic targets. For instance, NGS can detect rare genetic variants associated with treatment resistance, while SCS offers detailed cellular landscape insights to inform the development of targeted therapies (47, 82, 83).
Comprehensive treatment strategies, combining chemotherapy, transplantation, targeted therapy, and immunotherapy, remain critical for improving outcomes in pediatric AML. Notably, CAR T-cell therapy offers significant promise for refractory or relapsed cases (72). Future efforts should prioritize optimizing these combinations to minimize toxicity and enhance long-term survival.
Global disparities in resources present significant challenges to pediatric AML research. High-income countries dominate the field, while low- and middle-income countries face barriers such as limited access to diagnostic tools, fewer clinical trial opportunities, and insufficient research funding. For example, while China is the only developing nation among the top contributors, countries in sub-Saharan Africa, Latin America, and South Asia face similar challenges. Addressing these disparities is critical for fostering a more extensive research landscape. Future efforts should prioritize international collaborations, increase funding for research in low-resource settings, and leverage technologies like telemedicine and mobile diagnostics to bridge gaps. Building infrastructure and providing equitable research opportunities are essential to advancing pediatric AML prognosis research globally.
4.7 Limitations
This analysis has several limitations, primarily stemming from its reliance on the Web of Science Core Collection (WOSCC) database, which may exclude regional journals and non-English publications, potentially introducing bias. By focusing solely on English-language articles, the study might overlook valuable research published in other languages. Additionally, while highlighting significant collaborations and contributions from leading countries and institutions, the analysis may miss nuanced contributions from smaller or underrepresented regions, potentially overlooking local peculiarities in global publications. Finally, the emphasis on publication and citation metrics may fail to capture the qualitative aspects of research contributions, such as their practical applicability in clinical settings.
5 Conclusion
This bibliometric analysis provides a comprehensive overview of pediatric AML prognosis research over the past 25 years, highlighting key advancements, emerging trends, and transformative innovations. The integration of genetic markers, immunological insights, transcriptomics, and epigenomics has revolutionized risk stratification and treatment strategies, leading to significant improvements in patient outcomes. Advanced tools such as single-cell sequencing (SCS), multi-omics integration, and AI offer valuable support in identifying molecular interaction and novel therapeutic targets. Together with expanded clinical trials and global collaboration, these advancements are crucial for optimizing treatment combinations and improving outcomes for relapsed and refractory cases.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: 10.6084/m9.figshare.28343714.
Author contributions
MR: Conceptualization, Formal analysis, Investigation, Methodology, Writing – original draft. WL: Data curation, Formal analysis, Funding acquisition, Writing – original draft. CL: Data curation, Formal analysis, Methodology, Writing – review & editing. BW: Funding acquisition, Supervision, Validation, Writing – review & editing. TX: Data curation, Methodology, Writing – review & editing. ZW: Methodology, Writing – review & editing. HD: Methodology, Writing – review & editing. KL: Methodology, Writing – review & editing. DZ: Conceptualization, Funding acquisition, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors acknowledge financial support for the research, authorship, and/or publication of this literature. This work was funded by the Guangdong Basic and Applied Basic Research Foundation Project (2024A1515012445), Guangdong Province Rural Science and Technology Aid to Xinjiang (Special Commissioner) Project (KTPYJ2021010) and Heyuan Municipal Social Development Science and Technology Plan Project (Guiding Project) (No: 220917101607619).
Conflict of interest
The authors affirm that the research was carried out without any commercial or financial affiliations that could be perceived as potential conflicts of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Rubnitz JE, Inaba H, Dahl G, Ribeiro RC, Bowman WP, Taub J, et al. Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: results of the AML02 multicentre trial. Lancet Oncol. (2010) 11:543–52. doi: 10.1016/S1470-2045(10)70090-5
2. Kaspers GJL, Zwaan CM. Pediatric acute myeloid leukemia: towards high-quality cure of all patients. Haematologica. (2007) 92:1519–32. doi: 10.3324/haematol.11203
3. Iwai T, Yokota S, Nakao M, Okamoto T, Taniwaki M, Onodera N, et al. Internal tandem duplication of the FLT3 gene and clinical evaluation in childhood acute myeloid leukemia. Leukemia. (1999) 13:38–43. doi: 10.1038/sj.leu.2401241
4. Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. (2008) 358:1909–18. doi: 10.1056/NEJMoa074306
5. Othus M, Estey E, Gale RP. Assessment of minimal residual disease in standard-risk AML. New Engl J Med. (2016) 375(5):422–33. doi: 10.1056/NEJMc1603847
6. Stankiewicz J, Styczynski J. Improved outcome in pediatric acute myeloid leukemia: progress with hematopoietic cell transplantation. Anticancer Res. (2022) 42:4499–504. doi: 10.21873/anticanres.15951
7. Tomizawa D, Tsujimoto S-I. Risk-stratified therapy for pediatric acute myeloid leukemia. Cancers. (2023) 15(16):4171. doi: 10.3390/cancers15164171
8. Rubnitz J, Inaba H, Dahl G, Bowman WP, Taub J, Pui C-H, et al. Minimal residual disease-directed therapy for childhood acute myeloid leukemia: results of the AML02 multicenter trial. Blood. (2009) 114:12–3. doi: 10.1182/blood.V114.22.16.16
9. Gamis AS, Aplenc R, Alonzo TA, Sung L, Meshinchi S, Gerbing RB, et al. Gemtuzumab ozogamicin (GO) in children with de novo acute myeloid leukemia (AML) improves event-free survival (EFS) by reducing relapse risk - results from the randomized phase III children’s oncology group (COG) trial, AAML0531. Blood. (2013) 32(27):3021–32. doi: 10.1200/JCO.2014.55.3628
10. Hoffman AE, Schoonmade LJ, Kaspers GJL. Pediatric relapsed acute myeloid leukemia: a systematic review. Expert Rev Anticancer Ther. (2021) 21:45–52. doi: 10.1080/14737140.2021.1841640
11. Bertuccio SN, Anselmi L, Masetti R, Lonetti A, Cerasi S, Polidori S, et al. Exploiting clonal evolution to improve the diagnosis and treatment efficacy prediction in pediatric AML. Cancers. (2021) 13(9):1995. doi: 10.3390/cancers13091995
12. Jordan SC, Thomas B, Mumme H, Bhasin S, Bhasin M. Elucidating single-cell transcriptional differences of the MLL subtype in pediatric acute myeloid leukemia using machine learning and gene regulatory analysis. Blood. (2023) 142:6019. doi: 10.1182/blood-2023-189542
13. Zwaan CM, Meshinchi S, Radich JP, Veerman AJP, Huismans DR, Munske L, et al. FLT3 internal tandem duplication in 234 children with acute myeloid leukemia:: prognostic significance and relation to cellular drug resistance. Blood. (2003) 102:2387–94. doi: 10.1182/blood-2002-12-3627
14. Ho PA, Alonzo TA, Gerbing RB, Pollard J, Stirewalt DL, Hurwitz C, et al. Prevalence and prognostic implications of CEBPA mutations in pediatric acute myeloid leukemia (AML): a report from the Children’s Oncology Group. Blood. (2009) 113:6558–66. doi: 10.1182/blood-2008-10-184747
15. Hoffmeister LM, Orhan E, Walter C, Niktoreh N, Hanenberg H, von Neuhoff N, et al. Impact of KMT2A rearrangement and CSPG4 expression in pediatric acute myeloid leukemia. Cancers. (2021) 13(19):4817. doi: 10.3390/cancers13194817
16. van Weelderen RE, Klein K, Harrison CJ, Jiang Y, Abrahamsson J, Arad-Cohen N, et al. Measurable residual disease and fusion partner independently predict survival and relapse risk in childhood KMT2A-rearranged acute myeloid leukemia: A study by the international berlin-frankfurt-munster study group. J Clin Oncol: Off J Am Soc Clin Oncol. (2023) 41:2963–74. doi: 10.1200/JCO.22.02120
17. Shiba N, Yoshida K, Hara Y, Yamato G, Shiraishi Y, Matsuo H, et al. Transcriptome analysis offers a comprehensive illustration of the genetic background of pediatric acute myeloid leukemia. Blood Advances. (2019) 3:3157–69. doi: 10.1182/bloodadvances.2019000404
18. Lim WM, Kumar S. Guidelines for interpreting the results of bibliometric analysis: A sensemaking approach. Global Business Organizational Excellence. (2023) 43:17–26. doi: 10.1002/joe.22229
19. Stalph F, Heravi B. Exploring data visualisations: an analytical framework based on dimensional components of data artefacts in journalism. Digital Journalism. (2023) 11:1641–63. doi: 10.1080/21670811.2021.1957965
20. Hirsch JE. An index to quantify an individual’s scientific research output. Proc Natl Acad Sci United States America. (2005) 102:16569–72. doi: 10.1073/pnas.0507655102
21. Creutzig U, Ritter J, Riehm H, Langermann HJ, Henze G, Kabisch H, et al. Improved treatment results in childhood acute myelogenous leukemia: a report of the German cooperative study AML-BFM-78. Blood. (1985) 65:298–304. doi: 10.1182/blood.V65.2.298.298
22. Bolouri H, Farrar JE, Triche T Jr., Ries RE, Lim EL, Alonzo TA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions (vol 24, pg 103, 2018). Nat Med. (2018) 24:526–. doi: 10.1038/nm0418-526b
23. Tarlock K, Zong S, Rhonda RE, Bailey M, Morley S, Balasubramanian S, et al. Comprehensive clinical genomic profiling defines age-associated molecular targets in pediatric and adult acute myeloid leukemia. Blood. (2016) 128(22):596. doi: 10.1182/blood.V128.22.596.596
24. Collins FS, Lander ES, Rogers J, Waterston RH. Int Human Genome Sequencing C. Finishing the euchromatic sequence of the human genome. Nature. (2004) 431:931–45. doi: 10.1038/nature03001
25. Menyhart O, Gyorffy B. Multi-omics approaches in cancer research with applications in tumor subtyping, prognosis, and diagnosis. Comput Struct Biotechnol J. (2021) 19:949–60. doi: 10.1016/j.csbj.2021.01.009
26. Pollard JA, Alonzo TA, Brown PA, Gerbing RB, Fox E, Choi JK, et al. Sorafenib in combination with standard chemotherapy for children with high allelic ratio FLT3/ITD+ AML improves event-free survival and reduces relapse risk: A report from the children’s oncology group protocol AAML1031. Blood. (2022) 40(18):2023–35. doi: 10.1182/blood-2019-129557
27. Cuglievan B, Kantarjian H, Rubnitz JE, Cooper TM, Zwaan CM, Pollard JA, et al. Menin inhibitors in pediatric acute leukemia: a comprehensive review and recommendations to accelerate progress in collaboration with adult leukemia and the international community. Leukemia. (2024) 38:2073–84. doi: 10.1038/s41375-024-02368-7
28. Li N, Huang H-Y, Wu D-W, Yang Z-M, Wang J, Wang J-S, et al. Changes in clinical trials of cancer drugs in mainland China over the decade 2009-18: a systematic review. Lancet Oncol. (2019) 20:E619–E26. doi: 10.1016/S1470-2045(19)30491-7
29. Gamis AS, Alonzo TA, Meshinchi S, Sung L, Gerbing RB, Raimondi SC, et al. Gemtuzumab ozogamicin in children and adolescents with de novo acute myeloid leukemia improves event-free survival by reducing relapse risk: results from the randomized phase III children’s oncology group trial AAML0531. J Clin Oncol. (2014) 32:3021. doi: 10.1200/JCO.2014.55.3628
30. Qayed M, Ahn KW, Kitko CL, Johnson MH, Shah NN, Dvorak C, et al. A validated pediatric disease risk index for allogeneic hematopoietic cell transplantation. Blood. (2021) 137:983–93. doi: 10.1182/blood.2020009342
31. Tarlock K, Alonzo TA, Gerbing RB, Ries RE, Gibson B, Niktoreh N, et al. Distinct co-occurring mutational profiles in acute myeloid leukemia confers prognostic significance in children and young adults with FLT3/ITD mutations. Blood. (2018) 132:443. doi: 10.1182/blood-2018-99-113676
32. Wang Y, Weng W-J, Zhou D-H, Fang J-P, Mishra S, Li C, et al. Wilms tumor 1 mutations are independent poor prognostic factors in pediatric acute myeloid leukemia. Front Oncol. (2021) 11. doi: 10.3389/fonc.2021.632094
33. Wang H, Li X-Q, Chu T-T, Han S-Y, Qi J-Q, Tang Y-Q, et al. Clinical significance of FLT3-ITD/CEBPA mutations and minimal residual disease in cytogenetically normal acute myeloid leukemia after hematopoietic stem cell transplantation. J Cancer Res Clin Oncol. (2021) 147:2659–70. doi: 10.1007/s00432-021-03530-9
34. Xu L-H, Fang J-P, Liu Y-C, Jones AI, Chai L. Nucleophosmin mutations confer an independent favorable prognostic impact in 869 pediatric patients with acute myeloid leukemia. Blood Cancer J. (2020) 10(1):1. doi: 10.1038/s41408-019-0268-7
35. Minetto P, Candoni A, Guolo F, Clavio M, Zannier ME, Miglino M, et al. Fludarabine, high dose cytarabine and idarubicin-based induction is highly effective in young AML patients with concomitant NPM1 and FLT3-ITD mutation irrespectively of FLT3-ITD allelic burden. Blood. (2019) 134:3828. doi: 10.1182/blood-2019-131303
36. Balgobind BV, Raimondi SC, Harbott J, Zimmermann M, Alonzo TA, Auvrignon A, et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood. (2009) 114:2489–96. doi: 10.1182/blood-2009-04-215152
37. Doehner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. (2022) 140:1345–77. doi: 10.1182/blood.2022016867
38. Doehner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buechner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. (2017) 129:424–47. doi: 10.1182/blood-2016-08-733196
39. Suttorp J, Luehmann JL, Behrens YL, Goehring G, Steinemann D, Reinhardt D, et al. Optical genome mapping as a diagnostic tool in pediatric acute myeloid leukemia. Cancers. (2022) 14(9):2058. doi: 10.3390/cancers14092058
40. Lee S-I, Celik S, Logsdon BA, Lundberg SM, Martins TJ, Oehler VG, et al. A machine learning approach to integrate big data for precision medicine in acute myeloid leukemia. Nat Commun. (2018) 9:42. doi: 10.1038/s41467-017-02465-5
41. Chaudhary S, Ganguly S, Palanichamy JK, Singh A, Pradhan D, Bakhshi R, et al. Mitochondrial gene expression signature predicts prognosis of pediatric acute myeloid leukemia patients. Front Oncol. (2023) 13. doi: 10.3389/fonc.2023.1109518
42. Beck D, Thoms JAI, Palu C, Herold T, Shah A, Olivier J, et al. A four-gene LincRNA expression signature predicts risk in multiple cohorts of acute myeloid leukemia patients. Leukemia. (2018) 32:263–72. doi: 10.1038/leu.2017.210
43. Kunz F, Kontopoulou E, Reinhardt K, Soldierer M, Strachan S, Reinhardt D, et al. Detection of AML-specific mutations in pediatric patient plasma using extracellular vesicle-derived RNA. Ann Hematol. (2019) 98:595–603. doi: 10.1007/s00277-019-03608-y
44. Sasaki K, Uchiyama Y, Ikeda J, Yoshitomi M, Shimosato-Wada Y, Tokumasu M, et al. The detection of minor clones with somatic KIT D816V mutations using droplet digital PCR in pediatric de novo AML: AML-05 trial from the Japanese pediatric leukemia/lymphoma study group. Blood. (2019) 134:1419. doi: 10.1182/blood-2019-127656
45. Weber LL, Zhang C, Ochoa I, El-Kebir M. Phertilizer: Growing a clonal tree from ultra-low coverage single-cell DNA sequencing of tumors. PloS Comput Biol. (2023) 19(10):e1011544. doi: 10.1371/journal.pcbi.1011544
46. Aran D. Single-cell RNA sequencing for studying human cancers. Annu Rev Biomed Data Sci. (2021) 35(10):2799–812. doi: 10.1146/annurev-biodatasci-020722-091857
47. Stetson LC, Balasubramanian D, Ribeiro SP, Stefan T, Gupta K, Xu X, et al. Single cell RNA sequencing of AML initiating cells reveals RNA-based evolution during disease progression. Leukemia. (2021) 35:2799–812. doi: 10.1038/s41375-021-01338-7
48. Zhao C, Wong-Rolle A, Divakar P, Calvo K, Hourigan C. 250 Spatial-transcriptomic analysis of tumor-immune microenvironment in AML patients receiving pembrolizumab and decitabine. Regular Young Investigator Award Abstracts. (2021) 9(Suppl 2):A270. doi: 10.1136/jitc-2021-SITC2021.250
49. Lamba JK, Cao X, Raimondi S, Downing J, Ribeiro R, Gruber TA, et al. DNA methylation clusters and their relation to cytogenetic features in pediatric AML. Cancers. (2020) 12(10):3024. doi: 10.3390/cancers12103024
50. Yamato G, Kawai T, Shiba N, Hara Y, Ohki K, Kaburagi T, et al. Significant features of DNA methylation at bivalent promotor and repressed polycomb regions in pediatric AML-the jccg study, JPLSG AML-05. Blood. (2019) 134:2739. doi: 10.1182/blood-2019-125025
51. Szczepanek J. Role of microRNA dysregulation in childhood acute leukemias: Diagnostics, monitoring and therapeutics: A comprehensive review. World J Clin Oncol. (2020) 11:348. doi: 10.5306/wjco.v11.i6.348
52. Wei L, Trinh D, Ries RE, Jin D, Corbett RD, Smith JL, et al. Integrative analysis of single-cell RNA-seq and ATAC-seq data across treatment time points in pediatric AML. Blood. (2020) 136:29. doi: 10.1182/blood-2020-140473
53. Becker PS, Oehler VG, Blau CA, Martins TS, Curley N, Chien S, et al. A multi-omic precision medicine clinical trial in acute leukemia. Blood. (2019) 134:1269. doi: 10.1182/blood-2019-130996
54. Cantini L, Zakeri P, Hernandez C, Naldi A, Thieffry D, Remy E, et al. Benchmarking joint multi-omics dimensionality reduction approaches for the study of cancer. Nat Commun. (2021) 12(1):124. doi: 10.1038/s41467-020-20430-7
55. Rasche M, Zimmermann M, Borschel L, Bourquin J-P, Dworzak M, Klingebiel T, et al. Successes and challenges in the treatment of pediatric acute myeloid leukemia: a retrospective analysis of the AML-BFM trials from 1987 to 2012. Leukemia. (2018) 32:2167–77. doi: 10.1038/s41375-018-0071-7
56. Miyamura T, Tanaka S, Nakayama H, Moritake H, Tomizawa D, Saito AM, et al. Clinical and biological features of pediatric acute myeloid leukemia with primary induction failure in the Japanese pediatric leukemia/lymphoma study group (JPLSG) AML-05 study. Blood. (2019) 185(2):284–8. doi: 10.1182/blood.V128.22.1610.1610
57. Solh M, Zhang X, Bashey A, Morris LE, Bashey Z, Holland HK, et al. Transplant rates and outcomes of young acute myeloid leukemia (AML) patients with intermediate and poor risk disease referred to a coordinated acute leukemia and hematopoietic stem cell transplant (HSCT) program. Blood. (2018) 132:4797. doi: 10.1182/blood-2018-99-119185
58. Gassas A, Sivaprakasam P, Cummins M, Breslin P, Patrick K, Slatter M, et al. High transplant-related mortality associated with haematopoietic stem cell transplantation for paediatric therapy-related acute myeloid leukaemia (t-AML). A study on behalf of the United Kingdom Paediatric Blood and Bone Marrow Transplant Group. Bone Marrow Transplantation. (2018) 53:1165–9. doi: 10.1038/s41409-018-0157-x
59. Wobma H, Jin Z, Moscoso S, Bhatia M, Broglie L, George D, et al. Risk factors, clinical outcomes, and cost-of-care related to graft failure in pediatric allogeneic hematopoietic cell transplant recipients. Biol Blood Marrow Transplantation. (2020) 26:1318–25. doi: 10.1016/j.bbmt.2020.03.009
60. Rasche M, Steidel E, Zimmermann M, Bourquin J-P, Boztug H, Janotova I, et al. Second relapse of pediatric patients with acute myeloid leukemia: A report on current treatment strategies and outcome of the AML-BFM study group. Blood. (2020) 136. doi: 10.1182/blood-2020-139317
61. Pollard JA, Guest E, Alonzo TA, Gerbing RB, Loken MR, Brodersen LE, et al. Gemtuzumab ozogamicin improves event-free survival and reduces relapse in pediatric KMT2A-rearranged AML: results from the phase III children’s oncology group trial AAML0531. J Clin Oncol. (2021) 39:3149. doi: 10.1200/JCO.20.03048
62. Conneely SE, McAtee CL, Rau RE. Impact of race/ethnicity on pediatric core binding factor AML outcomes and response to gemtuzumab ozogamicin. Blood. (2020) 136:10–1. doi: 10.1182/blood-2020-138890
63. Lancet JE. Is the overall survival for older adults with AML finally improving? Best Pract Res Clin Haematol. (2018) 31:387–90. doi: 10.1016/j.beha.2018.09.005
64. Aplenc R, Meshinchi S, Sung L, Alonzo T, Choi J, Fisher B, et al. Bortezomib with standard chemotherapy for children with acute myeloid leukemia does not improve treatment outcomes: a report from the Children’s Oncology Group. Haematologica. (2020) 105:1879–86. doi: 10.3324/haematol.2019.220962
65. Rasche M, Steidel E, Kondryn D, Von Neuhoff N, Sramkova L, Creutzig U, et al. Impact of a risk-adapted treatment approach in pediatric AML: A report of the AML-BFM registry 2012. Blood. (2019) 134:293. doi: 10.1182/blood-2019-130969
66. Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. New Engl J Med. (2017) 377:454–64. doi: 10.1056/NEJMoa1614359
67. Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini S, et al. Gilteritinib significantly prolongs overall survival in patients with FLT3-mutated (FLT3mut+) relapsed/refractory (R/R) acute myeloid leukemia (AML): Results from the Phase III ADMIRAL trial. Cancer Res. (2019) 3:392–3. doi: 10.1158/1538-7445.AM2019-CT184
68. Ravindranath Y, Steuber CP, Krischer J, Civin CI, Ducore J, Vega R, et al. High-dose cytarabine for intensification of early therapy of childhood acute myeloid leukemia: a Pediatric Oncology Group study. J Clin Oncol: Off J Am Soc Clin Oncol. (1991) 9:572–80. doi: 10.1200/JCO.1991.9.4.572
69. Shen S, Liu Y, Tang J, Gu L-J. FLT3, NPM1 and MLL mutations help risk stratification in pediatric acute myeloid leukemia. Blood. (2009) 114:629–30. doi: 10.1182/blood.V114.22.1574.1574
70. Brown P, Alonzo T, Gerbing R, McLntyre E, Lange B, Small D, et al. High level expression of wild type FLT3 is associated with poor outcome and selective sensitivity to FLT3 inhibitors in childhood acute myeloid leukemia: A children’s oncology group study. Blood. (2008) 112:60–. doi: 10.1182/blood.V112.11.147.147
71. Verhaak RGW, Goudswaard CS, van Putten W, Bijl MA, Sanders MA, Hugens W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. (2005) 106:3747–54. doi: 10.1182/blood-2005-05-2168
72. Zhang H, Wang P, Li Z, He Y, Gan W, Jiang H. Anti-CLL1 chimeric antigen receptor T-cell therapy in children with relapsed/refractory acute myeloid leukemia. Clin Cancer Res. (2021) 27:3549–55. doi: 10.1158/1078-0432.CCR-20-4543
73. Aureli A, Marziani B, Sconocchia T, Del Principe MI, Buzzatti E, Pasqualone G, et al. Immunotherapy as a turning point in the treatment of acute myeloid leukemia. Cancers. (2021) 13(24):6246. doi: 10.3390/cancers13246246
74. Castaigne S, Pautas C, Terre C, Raffoux E, Bordessoule D, Bastie J-N, et al. Effect of gemtuzumab ozogamicin on survival of adult patients with de-novo acute myeloid leukaemia (ALFA-0701): a randomised, open-label, phase 3 study. Lancet. (2012) 379:1508–16. doi: 10.1016/S0140-6736(12)60485-1
75. Wouters BJ, Delwel R. Epigenetics and approaches to targeted epigenetic therapy in acute myeloid leukemia. Blood. (2016) 127:42–52. doi: 10.1182/blood-2015-07-604512
76. Gabra MM, Salmena L. microRNAs and acute myeloid leukemia chemoresistance: a mechanistic overview. Front Oncol. (2017) 7:255. doi: 10.3389/fonc.2017.00255
77. Stratmann S, Yones SA, Mayrhofer M, Norgren N, Skaftason A, Sun J, et al. Genomic characterization of relapsed acute myeloid leukemia reveals novel putative therapeutic targets. Blood Advances. (2021) 5:900–12. doi: 10.1182/bloodadvances.2020003709
78. Getz KD, Sung L, Alonzo TA, Leger KJ, Gerbing RB, Pollard JA, et al. Effect of dexrazoxane on left ventricular systolic function and treatment outcomes in patients with acute myeloid leukemia: A report from the children’s oncology group. J Clin Oncol. (2020) 38:2398. doi: 10.1200/JCO.19.02856
79. Qin Y, Pu X, Hu D, Yang M. Machine learning-based biomarker screening for acute myeloid leukemia prognosis and therapy from diverse cell-death patterns. Sci Rep. (2024) 14:17874. doi: 10.1038/s41598-024-68755-3
80. Wallace LK, Peplinski JH, Ries RE, Huang BJ, Ma X, Furlan SN, et al. Identification of a relapse signature in t (8; 21) pediatric AML. Blood. (2023) 142:4318. doi: 10.1182/blood-2023-189817
81. Koedijk JB, van der Werf I, Vermeulen MA, Nierkens S, Zwaan CM, Heidenreich O. Deep learning-based cell segmentation identifies T cell infiltration and spatial distribution in de novo pediatric AML. Klinische Padiatrie. (2022) 234:186–7. doi: 10.1055/s-0042-1748734
82. Heuser M, Heida B, Buettner K, Wienecke CP, Teich K, Funke C, et al. Posttransplantation MRD monitoring in patients with AML by next-generation sequencing using DTA and non-DTA mutations. Blood Advances. (2021) 5:2294–304. doi: 10.1182/bloodadvances.2021004367
Keywords: pediatric AML, prognosis, bibliometric analysis, genetic markers, immune therapy
Citation: Rao M, Luo W, Luo C, Wu B, Xu T, Wei Z, Deng H, Li K and Zhou D (2025) Prognostic factors and outcomes in pediatric acute myeloid leukemia: a comprehensive bibliometric analysis of global research trends. Front. Oncol. 15:1466818. doi: 10.3389/fonc.2025.1466818
Received: 26 July 2024; Accepted: 21 January 2025;
Published: 17 February 2025.
Edited by:
Teresa de Souza Fernandez, National Cancer Institute (INCA), BrazilReviewed by:
Daniela P. Mendes-de-Almeida, Oswaldo Cruz Foundation (Fiocruz), BrazilNathalia Lopez Duarte, Rio de Janeiro Municipal Health Secretariat (SMS-RJ), Brazil
Copyright © 2025 Rao, Luo, Luo, Wu, Xu, Wei, Deng, Li and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingliang Rao, cmFvbWxpYW5nQG1haWwyLnN5c3UuZWR1LmNu; Dunhua Zhou, emhvdWR1bmhAbWFpbC5zeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
 Mingliang Rao
Mingliang Rao Wenna Luo3†
Wenna Luo3† Caiju Luo
Caiju Luo Ziqian Wei
Ziqian Wei Kejing Li
Kejing Li