
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Oncol. , 21 February 2025
Sec. Gynecological Oncology
Volume 15 - 2025 | https://doi.org/10.3389/fonc.2025.1461737
Background: The association of the p53 rs1042522 and rs17878362 polymorphisms with cervical cancer risk has been reported in several published original studies and meta-analyses. However, the conclusions of these studies were contradictory. Consequently, we conducted an updated meta-analysis to further validate these debates.
Objective: To evaluate the association between the p53 rs1042522 and rs17878362 polymorphisms and cervical cancer risk.
Materials and Methods: PubMed, Medline, Ovid, Embase, CNKI, and China Wanfang databases were searched. Association was assessed using odds ratio (OR) with 95% confidence interval (CI). Moreover, the false-positive reporting probability (FPRP), Bayesian false-finding probability (BFDP), and Venice criteria were used to assess the credibility of statistically significant association.
Results: A significantly decreased cervical cancer risk was revealed for the p53 rs1042522 polymorphism (Pro/Pro +Arg/Pro vs. Arg/Arg: OR = 0.79, 95% CI = 0.71-0.87; Pro/Pro vs. Arg/Arg: OR = 0.80, 95% CI = 0.70-0.91; Arg/Pro vs. Arg/Arg: OR = 0.78, 95% CI = 0.71-0.86; Pro vs. Arg: OR = 0.87, 95% CI = 0.81-0.93) in overall analysis and several subgroup analyses, such as in Caucasians, Asians, Indians, and so on. However, no significant association was found between the p53 rs17878362 polymorphism and cervical cancer risk. Despite these statistically significant results, reliability analysis using FPRP, BFDP, and Venice criteria deemed all associations “unreliable”.
Conclusions: After considering the reliability of the results, this study indicates that the p53 rs1042522 polymorphism is not associated with the cervical cancer risk.
According to global cancer statistics, cervical cancer is classified by World Health Organization (WHO) as the second most prevalent malignant tumor of the female reproductive system, following breast cancer (1). In many developing countries, there continues to be a rise in the prevalence of cervical cancer. The latest statistics reveal that approximately 3.11 million new cases of cervical cancer occur worldwide each year, with around 570,000 cases being diagnosed annually (2, 3). Furthermore, there is an increasing trend in the occurrence of cervical cancer among young women. The p53 gene plays a crucial role as a tumor suppressor gene and possesses various biological functions such as inhibiting tumor cell growth and inducing cell cycle arrest at G1 phase. It also promotes programmed cell death after DNA damage and safeguards genetic stability.
The p53 gene, situated on the short arm of chromosome 17, holds a pivotal position as a tumor suppressor gene. Its structure encompasses multiple functional domains, including those for transcription activation and DNA binding. The p53 exerts its regulatory influence on the expression of specific genes in response to a variety of stimuli, operating through both transcriptional and non-transcriptional mechanisms. Mutations in p53 have the potential to disrupt its vital functions, encompassing cell cycle regulation, DNA repair, and the induction of apoptosis, thereby facilitating the onset and progression of tumorigenesis (4). The most common locus for variation is the p53 codon 72 (rs1042522). This mutation leads to functional inactivation of coding proteins p53 Arg and p53 Pro and may contribute to tumorigenesis through various mechanisms. Recent investigations on cervical cancer have revealed that mutations in host p53 gene polymorphisms play a significant role in its onset and progression. Furthermore, research suggests that individuals carrying the Arg form of p53 are more susceptible to cervical cancer compared to those carrying Pro (5, 6, 15). Therefore, understanding these genetic variations can provide valuable insights into the development and management strategies for this disease.
Many studies reported the association between the p53 codon 72 (rs1042522) and IVS3 16 bp (rs17878362) and cervical cancer risk. However, this association remained a subject of controversy. One hundred and twenty-three articles (7–129) evaluated the relationship between the p53 codon 72 (rs1042522) and IVS3 16 bp (rs17878362) and cervical cancer risk, yet these findings were inconsistent. Furthermore, previously published meta-analyses did not use the false positive reporting probability (FPRP) (137), Bayesian error detection probability (BFDP) (138), and Venice criteria (139) to assess the credibility of the pooled results (7–15). Therefore, we conducted an updated meta-analysis to further evaluate the above issues.
This meta-analysis was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (130).
PubMed, Medline, Embase, China National Knowledge Network (CNKI), and China Wanfang Databases were used for literature retrieval. The search strategies are as follows (“p53” OR “ tp53 “or” tp-53 “or” p-53 “) and (“ polymorphism “or” variability “or” mutation “or” gene “or” NP “) and (“ cervical “or” cervix “). Literature searches were conducted until October 31, 2023. In addition, a careful review of the reference list of published meta-analyses was conducted to spot all eligible studies.
Inclusion criteria were as follows: (1) case-control or cohort studies, (2) associations were evaluated between p53 rs1042522 and rs17878362 polymorphisms and risk of cervical cancer; (3) detailed genotype data or odds ratios (OR) and their corresponding 95% confidence intervals (CI). Exclusion criteria are as follows: (1) animal experiments or overlapping studies; (2) case reports, abstracts, reviews, letters, and meta-analyses; (3) insufficient genotype data or unavailable for studies.
Two researchers screened all the literatures according to the inclusion and exclusion criteria. Once variations exist and no accord are often reached once discussion, the other author collected the data once more, and at last the three authors can check and ensure along. The following data was extracted: year of publication, first author, country, region, source of case p53 genotyping materials, recruitment source, genotype management cluster, total sample size, matching, genotype distribution, etc.
After comprehensively considering the characteristics of the articles, the quality evaluation of all the included literatures was conducted according to some criteria (such as HWE, control matching, certainty, sample size, etc.), as shown in Supplementary Table S1. In the control group, we applied the goodness-fit Chi-square test to analyze the Hardy-Weinberg balance (HWE) for eligible studies with complete genotype data. P ≥ 0.05 was defined as HWE, and P < 0.05 was considered as Hardy–Weinberg disequilibrium (HWD) (131). The highest score was 23, and the eligible studies that met both scoring ≥16 and HWE compliant were considered as high-quality (Supplementary Table S6). If there is a disagreement on the score, it is assessed again by a superior author.
Association was evaluated applying the following five genetic models: (1) dominant model (rs1042522: Pro/Pro + Arg/Pro vs Arg/Arg, rs17878362: A2/A2+ A1/A2 vs. A1/A1); (2) recessive model (rs1042522: Pro/Pro vs Arg/Arg + Arg/Pro, rs17878362: A2/A2 vs. A1/A1+ A1/A2); (3) homozygous model (rs1042522: Pro/Pro vs Arg/Arg, rs17878362: A2/A2 vs. A1/A1; (4) codominance model (rs1042522: Arg/Pro vs Arg/Arg, rs17878362: A1/A2 vs. A1/A1); (5) allele model (rs1042522: Pro vs Arg, rs17878362: A1 vs. A2). If the P < 0.05 and/or I2 > 50%, indicating significant heterogeneity, a random-effects model was used (132). Instead, a fixed-effects model was used. The sources of heterogeneity were assessed using meta-regression analysis (133). Subgroups were created based on race, region, matching situation, and source of controls. Sensitivity analyses were conducted by individually excluding each study or by excluding studies with both low quality and HWD. Egger’s test (134) and Begg’s test (135) were performed to evaluate potential publication bias. In case of publication bias, a non-parametric “trim and fill” approach (136) was employed to estimate and supplement the number of missing studies. All statistical analyses for this meta-analysis were calculated using STATA code version 12.0 (STATA Corp, College Station, TX, USA).
FPRP, BFDP, and Venetian criteria (139) were utilized to assess the confidence levels for statistically significant associations. Associations meeting the following criteria were considered as highly credible: 1) statistically significant associations observed in at least two genetic models; 2) I2 < 50%; 3) FPRP < 0.2 and BFDP < 0.8; 4) statistical power >80%.
According to the pre-search methodology employed in this study (Figure 1), a total of 5,223 relevant articles were initially identified. After eliminating duplicates from these records, a final set of 3,378 unique publications remained. Subsequently, during the title and abstract screening process, a further 3,212 papers were excluded. Following a thorough full-text review, 22 additional articles were removed due to duplicate or unavailable data, and 30 papers were discarded because of poor quality control. Thus, the final analysis included 114 studies (supplementary Table S4-S5, Figure 1) comprising 125 independent investigations, encompassing a total combined sample size of 13,319 cases and 19,959 controls. As shown in Supplementary Tables S4-S5, p53 rs1042522 was reported in 118 studies (12,655 cases and 19,272 controls), while p53 rs17878362 was reported in seven studies (664 cases and 687 controls). Furthermore, among these studies, there were 37 articles of low quality and 77 articles of high quality for p53 rs1042522; whereas for p53 rs17878362, one article was classified as low quality and five articles as high quality (Supplementary Table S6). The complete characteristics and genotype frequencies of the literature included are presented in Supplementary Table S4-S5.
The p53 rs1042522 polymorphism was significantly associated with a reduced risk of cervical cancer (Pro/Pro +Arg/Pro vs. Arg/Arg: OR = 0.79, 95% CI = 0.71-0.87; Pro/Pro vs. Arg/Arg: OR = 0.80, 95% CI = 0.70-0.91; Arg/Pro vs. Arg/Arg: OR = 0.78, 95% CI = 0.71-0.86; Pro vs. Arg: OR = 0.87, 95% CI = 0.81-0.93, Table 1, Figure 2) in overall analysis. Moreover, a significantly reduced cervical cancer risk was also observed in Caucasians (Pro/Pro +Arg/Pro vs. Arg/Arg: OR = 0.81, 95% CI = 0.70-0.94; Pro/Pro vs. Arg/Arg: OR = 0.84, 95% CI = 0.73-0.98; Arg/Pro vs. Arg/Arg: OR = 0.81, 95% CI = 0.70-0.94; Pro vs. Arg: OR = 0.86, 95% CI = 0.77-0.96, Table 1, Figure 3), Asians (Pro/Pro +Arg/Pro vs. Arg/Arg: OR = 0.80, 95% CI = 0.67-0.95; Arg/Pro vs. Arg/Arg: OR = 0.78, 95% CI = 0.66-0.93; Pro vs. Arg: OR = 0.89, 95% CI = 0.79-0.99, Table 1, Figure 3), Indians (Pro/Pro +Arg/Pro vs. Arg/Arg: OR = 0.57, 95% CI = 0.47-0.70; Arg/Pro vs. Arg/Arg: OR = 0.60, 95% CI = 0.48-0.73, Table 1, Figure 3), and mixed population (Pro/Pro vs. Arg/Pro + Arg/Arg: OR = 0.81, 95% CI = 0.68-0.98; Pro/Pro vs. Arg/Arg: OR = 0.73, 95% CI = 0.57-0.92; Pro vs. Arg: OR = 0.88, 95% CI = 0.79-0.98, Table 1, Figure 3). However, no significant association was found between p53 rs1042522 polymorphism and cervical cancer risk in Africans. Furthermore, significantly reduced risk of cervical cancer was observed in Europe (Pro/Pro +Arg/Pro vs. Arg/Arg: OR = 0.77, 95% CI = 0.65-0.92; Pro/Pro vs. Arg/Arg: OR = 0.84, 95% CI = 0.7-0.99; Arg/Pro vs. Arg/Arg: OR = 0.76, 95% CI = 0.64-0.91; Pro vs. Arg: OR = 0.84, 95% CI = 0.74-0.96, Table 1), East Asians (Pro/Pro +Arg/Pro vs. Arg/Arg: OR = 0.74, 95% CI = 0.61-0.90; Pro/Pro vs. Arg/Arg: OR = 0.76, 95% CI = 0.62-0.94; Arg/Pro vs. Arg/Arg: OR = 0.72, 95% CI = 0.59-0.88; Pro vs. Arg: OR = 0.84, 95% CI = 0.75-0.95, Table 1), and Africa (Pro/Pro +Arg/Pro vs. Arg/Arg: OR = 0.75, 95% CI = 0.59-0.95; Pro/Pro vs. Arg/Arg: OR = 0.69, 95% CI = 0.48-0.98, Table 1). Then, we observed that the p53 rs1042522 polymorphism reduced the risk of cervical cancer in the matching studies (Pro/Pro +Arg/Pro vs. Arg/Arg: OR = 0.78, 95% CI = 0.68-0.90; Pro/Pro vs. Arg/Arg: OR = 0.75, 95% CI = 0.63-0.90; Arg/Pro vs. Arg/Arg: OR = 0.79, 95% CI = 0.68-0.91; Pro vs. Arg: OR = 0.88, 95% CI = 0.80-0.97, Table 1) and non-matching studies (Pro/Pro +Arg/Pro vs. Arg/Arg: OR = 0.79, 95% CI = 0.68-0.91; Pro/Pro vs. Arg/Arg: OR = 0.83, 95% CI = 0.74-0.94; Arg/Pro vs. Arg/Arg: OR = 0.78, 95% CI = 0.68-0.90; Pro vs. Arg: OR = 0.86, 95% CI =0.77-0.96, Table 1). Finally, we obtained a significant association in health control population (Pro/Pro +Arg/Pro vs. Arg/Arg: OR = 0.80, 95% CI = 0.69-0.92; Pro/Pro vs. Arg/Arg: OR = 0.80, 95% CI = 0.67-0.95; Arg/Pro vs. Arg/Arg: OR = 0.81, 95% CI = 0.70-0.93; Pro vs. Arg: OR = 0.88, 95% CI =0.8-0.98, Table 1) and non-cancer control population (Pro/Pro +Arg/Pro vs. Arg/Arg: OR = 0.77, 95% CI = 0.68-0.88; Pro/Pro vs. Arg/Arg: OR = 0.8, 95% CI = 0.66-0.97; Arg/Pro vs. Arg/Arg: OR = 0.76, 95% CI = 0.66-0.87; Pro vs. Arg: OR = 0.86, 95% CI =0.76-0.95, Table 1). The results of sensitivity analysis showed no significant changes in this study. Furthermore, Egger’s test and Begg’s funnel plot confirmed the absence of publication bias (Pro/Pro + Arg/Pro vs. Arg/Arg: P = 0.06; Pro/Pro vs. Arg/Pro + Arg/Arg: P = 0.386; Pro/Pro vs. Arg/Arg: P = 0.673; Arg/Pro vs. Arg/Arg: p=0.091; Pro vs. Arg: P = 0.91). In the overall analysis, the results for the Pro Pro +Arg Pro vs. Arg Arg models did not change (data not shown), suggesting that more studies could not change the pooled results (Figure 5).
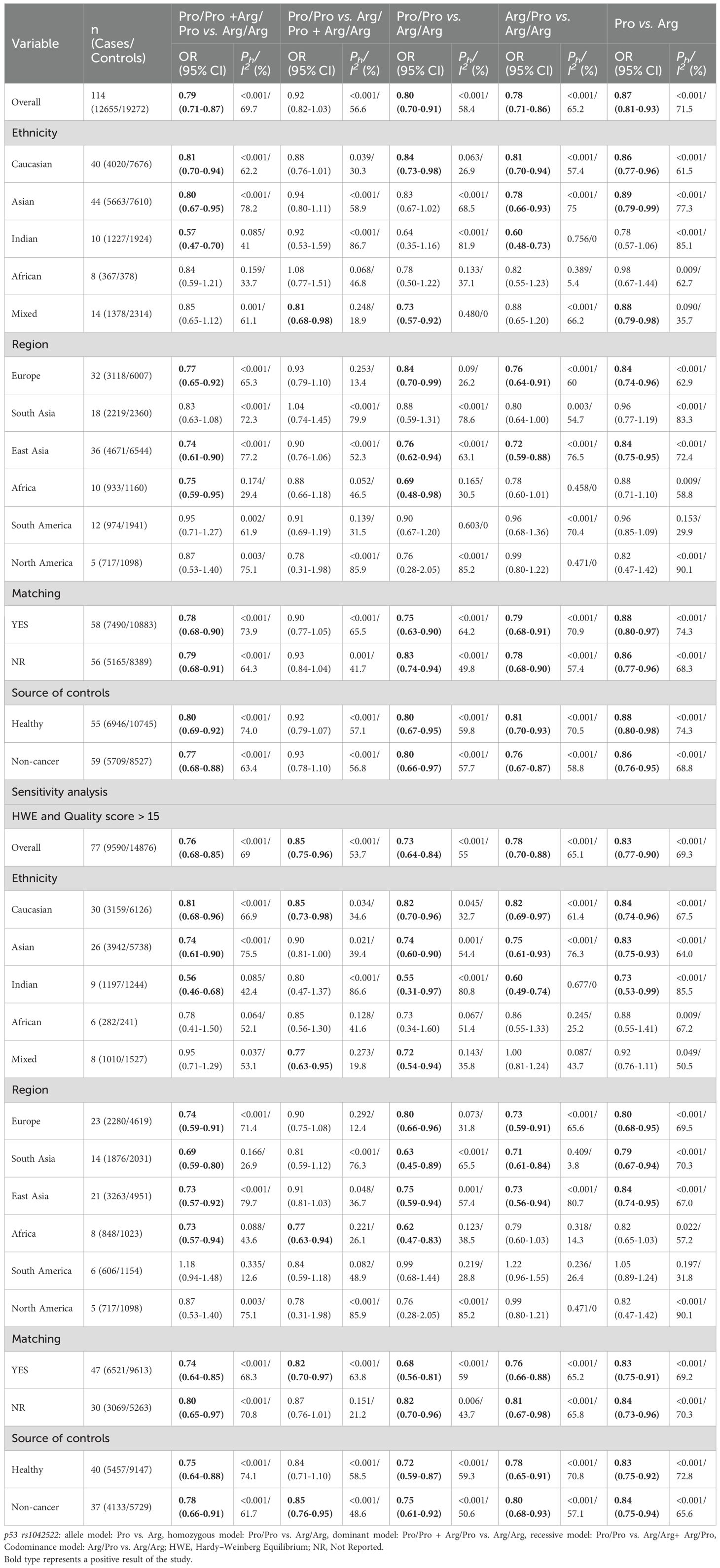
Table 1. Meta-analysis of the association of p53 rs1042522 polymorphism with risk of cervical cancer.
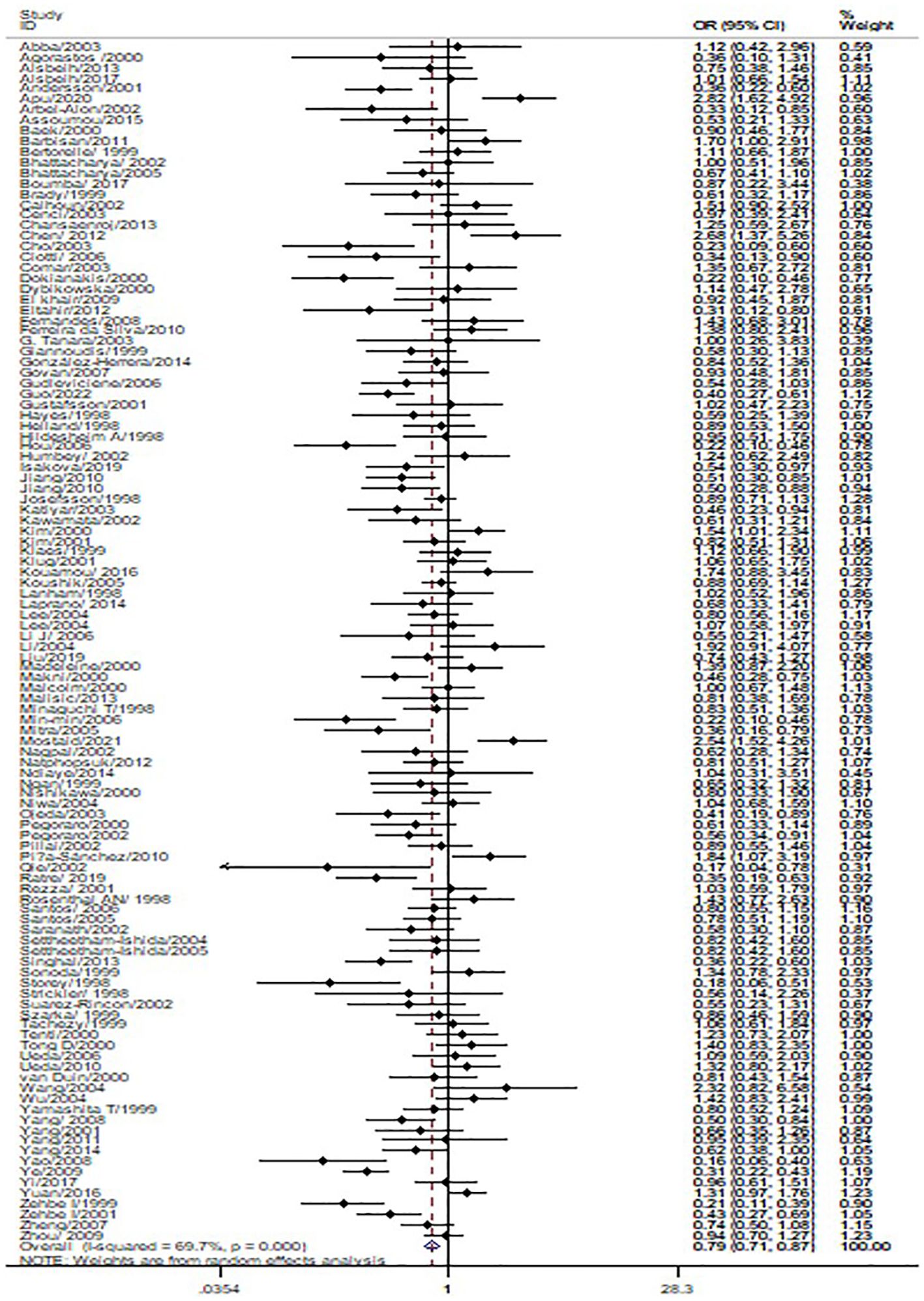
Figure 2. Forest map of the correlation between p53 rs1042522 polymorphism and cervical cancer in overall analysis (Pro Pro + Arg Pro vs. Arg Arg).

Figure 3. Forest map of the correlation of between p53 rs1042522 polymorphism and cervical cancer in the ethnicity group analysis forest map (Pro Pro + Arg Pro vs. Arg Arg).
No significant association was observed between the p53 rs17878362 polymorphism and risk of cervical cancer in the overall population (Table 2, Figure 4). Sensitivity analysis revealed consistent results without significant changes. Additionally, no publication bias was detected based on Egger’s test and Begg’s funnel plot (A2/A2+ A1/A2 vs. A1/A1: P = 0.48; A2/A2 vs. A1/A1+ A1/A2: P = 0.59; A2/A2 vs. A1/A1: P = 0.60; A1/A2 vs. A1/A1: p=0.48; A1 vs. A2: P = 0.65). In the overall analysis, the results for the Pro Pro +Arg Pro vs. Arg Arg models did not change (data not shown), suggesting that more studies could not change the pooled results (Figure 5).
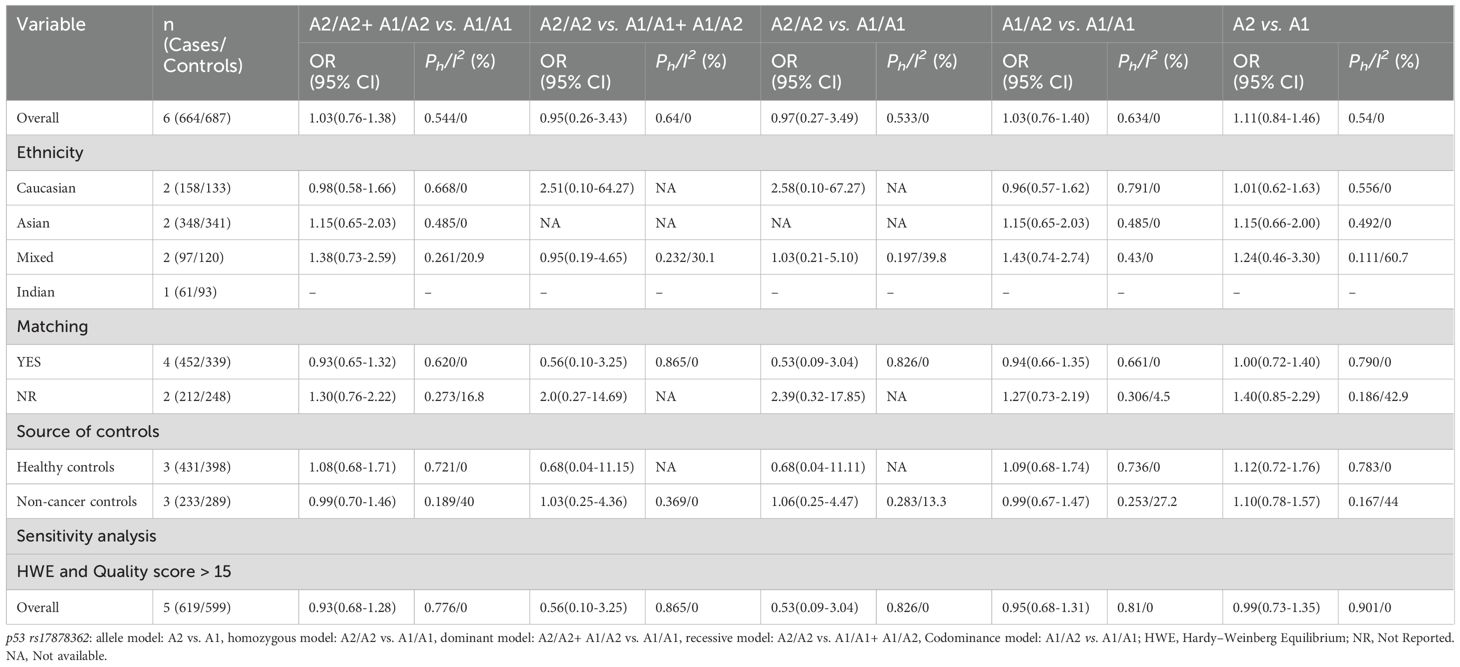
Table 2. Meta-analysis of the association of p53 rs17878362 polymorphism with risk of cervical cancer.
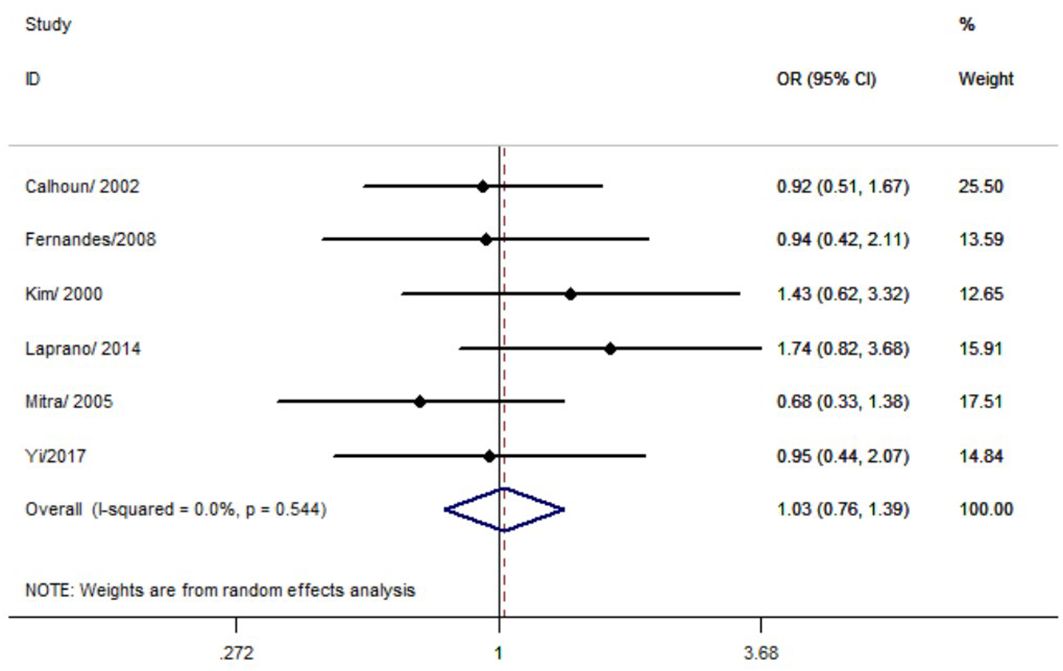
Figure 4. Forest map of the correlation between p53 rs17878362 polymorphism and cervical cancer in overall analysis (A2/A2+ A1/A2 vs. A1/A1).
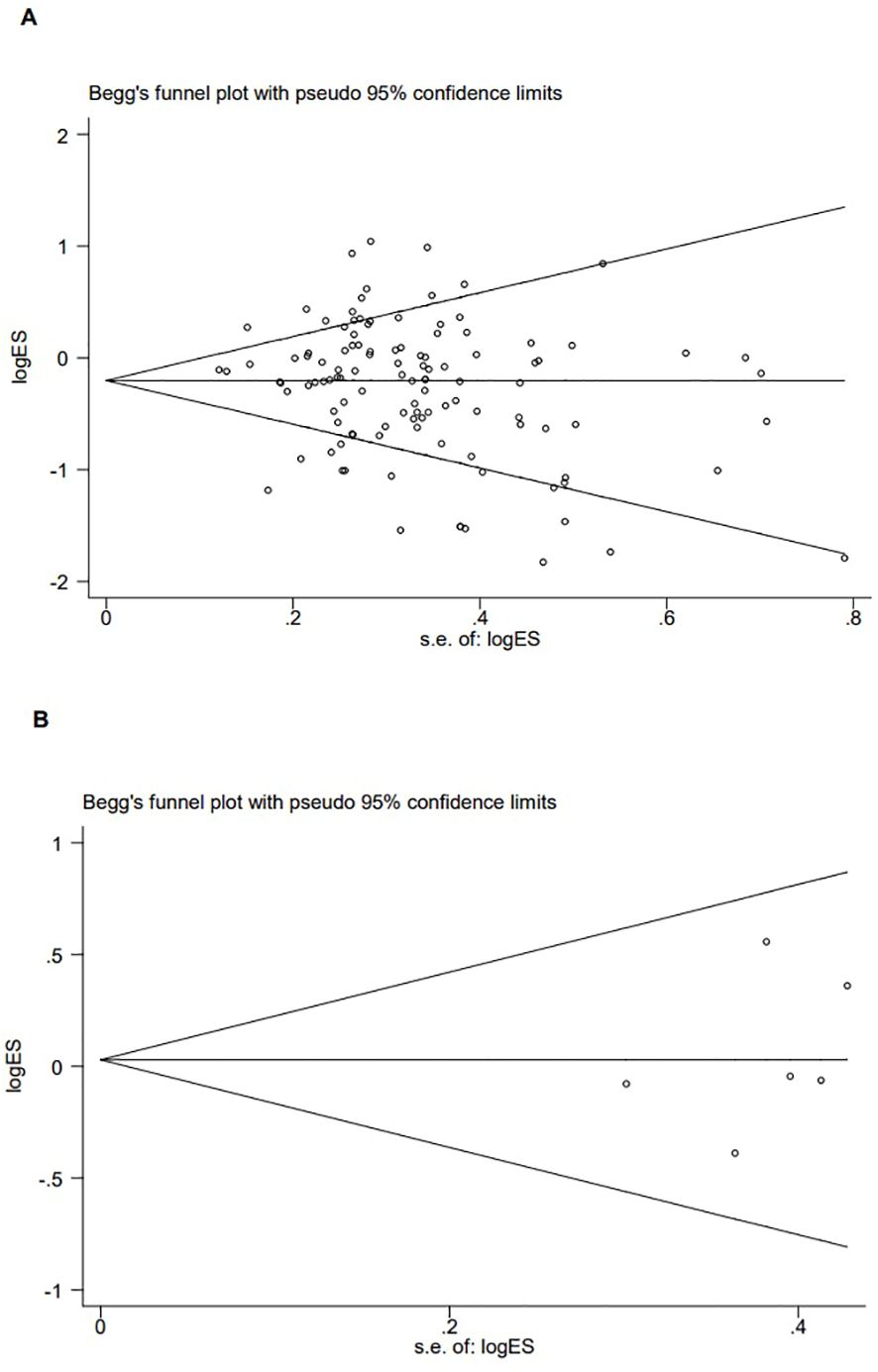
Figure 5. Publication bias of the combined effect of Begg funnel plot assessment of p53 rs1042522 [(A) Pro Pro +Arg Pro vs. Arg Arg) and rs17878362 [(B) Pro Pro +Arg Pro vs. Arg Arg) polymorphisms and cervical cancer.
In our study, the credibility of all significant associations was evaluated using FPRP, BFDP, and Venice criteria; however, they were deemed as having lower credibility (Table 3).
This meta-analysis comprised a total of 125 studies from 114 articles. The application of genetic models in meta-analysis can help us to better reveal the true association between genes and diseases, based on previous research, we chose five genetic models (dominant model; recessive model; homozygous model; codominance model; allele model). Moreover, excluding low-quality studies would provide a more accurate representation of this relationship. Additionally, our findings indicated that p53 rs1042522 polymorphism significantly influenced cervical cancer risk in both matched and control subgroups, suggesting that matching factors and control variables did not affect its association with cervical cancer. However, after considering the reliability of the results, this study indicates that the p53 rs1042522 polymorphism is not associated with the cervical cancer risk. Furthermore, no significant association was found between the p53 rs17878362 polymorphism and cervical cancer risk, these results were consistent with those obtained from sensitivity analysis.
It is important to note that meta-analysis of gene polymorphisms involves aggregation of extensive genomic data which may lead to false positive results; therefor credibility assessment using FPRP, BFDP, and Venice criteria is commonly employed. Based on analytical evaluation using these criteria, we concluded that the confidence intervals for the associations between p53 rs1042522 polymorphism with cervical cancer risk were relatively unreliable. Up to now, a total of nine meta-analyses have investigated the association between p53 rs1042522 polymorphism and the risk of cervical cancer. Francisco et al. (7) and Yu et (14) al found that the p53 rs1042522 was correlated with an increased risk of cervical cancer in whole population. Koushik et al. (11) found was same conclusion, but the number of deviations from Hardy-Weinberg equilibrium in the control group of the included studies was large, which led to an inevitable decrease in the reliability of the conclusions. Kamiza et al. (9) and Li et al. (12) observed that the p53 rs1042522 was associated with an increased risk of cervical cancer in Africans and Chinese population, respectively. Zhou et al. (15) study also found the same results in Asians. Habbous et al. (8) found that the Arg variant is associated with progression of Squamous Intraepithelial Lesion to cervical cancer only in the presence of Human Papillomavirus positivity. Sousa et al. (13) found that p53 codon 72 polymorphism in countries with low incidence rates of cervical cancer, this polymorphism might represent a significant genetic marker. Hower, Klug et al. (10) found that the p53 rs1042522 was not association with risk of cervical cancer. Inconsistencies in the existence of previous studies may be due to differences in the number of studies included in the studies and differences in the study populations. The cases and controls of Klug et al. (10) study most were white women, this can lead to pooling bias. There exist contradictory conclusions among these studies. Moreover, some articles with weak associations were included in the meta-analysis without strict evaluation of their quality. Additionally, none of them accounted for potential false positive results.
To address these conflicting conclusions and determine the precise association between p53 rs1042522 and p53 rs17878362 with cervical cancer, an updated meta-analysis is deemed necessary. The strengths of this updated meta-analysis are as follows: (1) It includes a larger sample size comprising 114 articles compared to previous studies; (2) HWE was assessed in control group; (3) Credibility evaluation was conducted on significant results; (4) Ethnic differences were thoroughly analyzed. However, our study also has certain limitations. Firstly, we only considered eligible studies from specific databases without exploring alternative sources for eligible studies. Secondly, our search was limited to English and Chinese languages while excluding articles published in other languages. Lastly, the genotype data we included were unadjusted. Because of study limitations, we did not adjust for miscarriage, presence or absence of HPV infection, and other factors. Hence, future research should aim to include more comprehensive adjustments for confounding factors in order to obtain accurate conclusions.
In conclusion, the significant association between p53 rs1042522 polymorphism and the risk of cervical cancer may be false positive results. More research is needed to confirm this association.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author/s.
XZ: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Writing – original draft. XB: Data curation, Methodology, Software, Writing – original draft. HZ: Supervision, Writing – review & editing. XH: Supervision, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1461737/full#supplementary-material
BFDP, Bayesian false discovery probability; CI, confidence interval; FPRP, false-positive report probabilities; HWE, Hardy–Weinberg equilibrium; OR, odds ratio; PRISMA, Preferred Reporting Items for Systematic Review and Meta-Analyses.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global cancer in women: burden and trends. Cancer Epidemiol Biomarkers Prev. (2017) 26:444–57. doi: 10.1158/1055-9965.EPI-16-0858
3. Schubert M, Bauerschlag DO, Muallem MZ, Maass N, Alkatout I. Challenges in the diagnosis and individualized treatment of cervical cancer. Med (Kaunas Lithuania). (2023) 59:925. doi: 10.3390/medicina59050925
4. Mao Y, Jiang P. The crisscross between p53 and metabolism in cancer. Acta Biochim Biophys Sin. (2023) 55:914–22. doi: 10.3724/abbs.2023109
5. Haws ALF, Woeber S, Gomez M, Garza N, Gomez Y, Rady P, et al. Human papillomavirus infection and P53 codon 72 genotypes in a hispanic population at high-risk for cervical cancer. J Med Virol. (2005) 77:265–72. doi: 10.1002/jmv.20446
6. Hao X, Wang RS, Li JH. The relationship between the polymorphism of the p53 gene codon 72 and Hp susceptibility and prognosis in patients with gastric ulcers. Shandong Med J. (2019) 59:16–9.
7. Francisco G, Menezes PR, Eluf-Neto J, Chammas R. Arg72Pro TP53 polymorphism and cancer susceptibility: A comprehensive meta-analysis of 302 case-control studies. Int J Cancer. (2011) 129:920–30. doi: 10.1002/ijc.25710
8. Habbous S, Pang V, Eng L, Xu W, Kurtz G, Liu F-F, et al. p53 arg72Pro polymorphism, HPV status and initiation, progression, and development of cervical cancer: A systematic review and meta-analysis. Clin Cancer Res. (2012) 18:6407–15. doi: 10.1158/1078-0432.CCR-12-1983
9. Kamiza AB, Kamiza S, Singini MG, Mathew CG. Association of TP53 rs1042522 with cervical cancer in the sub-Saharan African population: A meta-analysis. Trop Med Int Health. (2020) 25:666–72. doi: 10.1111/tmi.13397
10. Klug SJ, Ressing M, Koenig J, Abba MC, Agorastos T, Brenna SM, et al. TP53 codon 72 polymorphism and cervical cancer: A pooled analysis of individual data from 49 studies. Lancet Oncol. (2009) 10:772–84. doi: 10.1016/S1470-2045(09)70187-1
11. Koushik A, Platt RW, Franco EL. p53 codon 72 polymorphism and cervical neoplasia: A meta-analysis review. Cancer Epidemiol Biomarkers Prev. (2004) 13(1):11–22. doi: 10.1158/1055-9965.epi-083-3
12. Li B, Wang X, Chen H, Shang L-X, Wu N. TP53 codon 72 polymorphism and susceptibility to cervical cancer in the Chinese population: An update meta-analysis. Int J Clin Exp Med. (2015) 8(6):9055–62.
13. Sousa H, Santos AM, Pinto D, Medeiros R. Is the p53 codon 72 polymorphism a key biomarker for cervical cancer development? A meta-analysis review within European populations. Int J Mol Med. (2007) 20(5):731–41. doi: 10.3892/ijmm.20.5.731
14. Yu M, Zhang Q, Zhao X. Associations of MDM2 rs2279744 and TP53 rs1042522 polymorphisms with cervical cancer risk: A meta-analysis and systematic review. Front Oncol. (2022) 12:973077. doi: 10.3389/fonc.2022.973077
15. Zhou X, Gu Y, Zhang S-L. Association Between p53 codon 72 Polymorphism and Cervical Cancer Risk Among Asians: A Huge Review and Meta-analysis. Asian Pacific J Cancer Prev. (2012) 13:4909–14. doi: 10.7314/APJCP.2012.13.10.4909
16. Abba M. P53 codon 72 genotypes in HPV infection and cervical disease. Eur J Obstetrics Gynecol Reprod Biol. (2003) 109:63–6. doi: 10.1016/S0301-2115(02)00474-8
17. Agorastos T, Lambropoulos AF, Constantinidis TC, Kotsis A, Bontis JN. P53 codon 72 polymorphism and risk of intra-epithelial and invasive cervical neoplasia in Greek women. Eur J Cancer Prevention: Off J Eur Cancer Prev Organisation (ECP). (2000) 9:113–8. doi: 10.1097/00008469-200004000-00007
18. Alsbeih GA, Al-Harbi NM, Bin Judia SS, Khoja HA, Shoukri MM, Tulbah AM. Reduced rate of human papillomavirus infection and genetic overtransmission of TP53 72C polymorphic variant lower cervical cancer incidence. Cancer. (2017) 123:2459–66. doi: 10.1002/cncr.30635
19. Alsbeih G, Al-Harbi N, El-Sebaie M, Al-Badawi I. HPV prevalence and genetic predisposition to cervical cancer in Saudi Arabia. Infect Agents Cancer. (2013) 8:15. doi: 10.1186/1750-9378-8-15
20. Andersson S, Rylander E, Strand A, Sällström J, Wilander E. The significance of p53 codon 72 polymorphism for the development of cervical adenocarcinomas. Br J Cancer. (2001) 85:1153–6. doi: 10.1054/bjoc.2001.2085
21. Apu MNH, Rashed AZM, Bashar T, Rahman MM, Mostaid MS. TP53 genetic polymorphisms and susceptibility to cervical cancer in Bangladeshi women: A case–control study. Mol Biol Rep. (2020) 47:4357–64. doi: 10.1007/s11033-020-05523-2
22. Arbel-Alon S, Menczer J, Feldman N, Glezerman M, Yeremin L, Friedman E. Codon 72 polymorphism of p53 in Israeli Jewish cervical cancer patients and healthy women. Int J Gynecol Cancer. (2002) 12:741–4. doi: 10.1046/j.1525-1438.2002.01124.x
23. Assoumou SZ, Boumba ALM, Ndjoyi-Mbiguino A, Khattabi A, Ennaji MM. The preliminary study of p53 codon 72 polymorphism and risk of cervical carcinoma in Gabonese women. Med Oncol. (2015) 32:281. doi: 10.1007/s12032-014-0281-4
24. Baek WK, Cho JW, Suh SI, Suh MH, Shin DH, Cho CH, et al. P53 codon 72 polymorphism and risk of cervical carcinoma in Korean women. J Korean Med Sci. (2000) 15:65–7. doi: 10.3346/jkms.2000.15.1.65
25. Barbisan G, Contreras A, Pérez LO, Difranza L, Golijow CD. The effect of TP53 codon 72 and RNASEL codon 462 polymorphisms on the development of cervical cancer in Argentine women. Cancer Genet. (2011) 204:270–7. doi: 10.1016/j.cancergen.2011.04.001
26. Bertorelle R, Chieco-Bianchi L, Del Mistro A. Papillomavirus andp53 codon 72 polymorphism. Int J Cancer. (1999) 82:616–7. doi: 10.1002/(SICI)1097-0215(19990812)82:4<616::AID-IJC24>3.0.CO;2-7
27. Bhattacharya P, Sengupta S. Lack of evidence that proline homozygosity at codon 72 of p53 and rare arginine allele at codon 31 of p21, jointly mediate cervical cancer susceptibility among Indian women. Gynecol Oncol. (2005) 99:176–82. doi: 10.1016/j.ygyno.2005.06.007
28. Bhattacharya P, Duttagupta C, Sengupta S. Proline homozygosity in codon 72 of p53: A risk genotype for human papillomavirus related cervical cancer in Indian women. Cancer Lett. (2002) 188:207–11. doi: 10.1016/S0304-3835(02)00430-5
29. Boumba LMA, Moukassa D, Hilali L, Ennaji MM. Molecular Analysis of P53 Codon 72 polymorphism and risk of cervical carcinoma among women in southwest of the Republic of Congo: a case-control study. Int J Virol. (2017) 2:021–4.
30. Brady CS, Duggan-Keen MF, Davidson JA, Varley JM, Stern PL. Human papillomavirus type 16 E6 variants in cervical carcinoma: Relationship to host genetic factors and clinical parameters. J Gen Virol. (1999) 80:3233–40. doi: 10.1099/0022-1317-80-12-3233
31. Calhoun ES, McGovern RM, Janney CA, Cerhan JR, Iturria SJ, Smith DI, et al. Host genetic polymorphism analysis in cervical cancer. Clin Chem. (2002) 48:1218–24. doi: 10.1093/clinchem/48.8.1218
32. Cenci M, French D, Pisani T, Alderisio M, Lombardi AM, Marchese R, et al. P53 polymorphism at codon 72 is not a risk factor for cervical carcinogenesis in central Italy. Anticancer Res. (2003) 23:1385–7.
33. Chansaenroj J, Theamboonlers A, Junyangdikul P, Swangvaree S, Karalak A, Chinchai T, et al. Polymorphisms in TP53 (rs1042522), p16 (rs11515 and rs3088440) and NQO1 (rs1800566) Genes in Thai Cervical Cancer Patients with HPV 16 Infection. Asian Pacific J Cancer Prev. (2013) 14:341–6. doi: 10.7314/APJCP.2013.14.1.341
34. Chen CC, Ding XH, Cai HB. HPV16 infection and p53 codon72 polymorphism in cervical cancer and recursor lesions. Chin Modern Med. (2012) 19:9–10.
35. Cho NH, Lim SY, Kim YT, Kim D, Kim YS, Kim JW. G2 checkpoint in uterine cervical cancer with HPV 16 E6 according to p53 polymorphism and its screening value. Gynecol Oncol. (2003) 90:15–22. doi: 10.1016/S0090-8258(03)00198-7
36. Ciotti M, Coletti A, Giuliani L, Cappiello G, Syrjanen K, Favalli C. The p53 Codon 72 arg/arg Homozygous Women in Central Italy are at Increased Risk for HPV Infections. Anticancer Res. (2006) 26(5B):3745–8.
37. Comar M, Molin GD, Guaschino S, Campello C. p53 at codon 72 polymorphism, human papillomavirus infection and cervical lesions: A cross-sectional study from northeastern Italy. Eur J Obstetrics Gynecol Reprod Biol. (2004) 114:210–4. doi: 10.1016/j.ejogrb.2003.10.021
38. Dokianakis DN, Spandidos DA. P53 codon 72 polymorphism as a risk factor in the development of HPV-associated cervical cancer. Mol Cell Biol Res Commun. (2000) 3:111–4. doi: 10.1006/mcbr.2000.0196
39. Dybikowska A, Dettlaff A, Konopa K, Podhajska A. P53 codon 72 polymorphism in cervical cancer patients and healthy women from Poland. Acta Biochim Polonica. (2000) 47:1179–82. doi: 10.18388/abp.2000_3970
40. El Khair MM, Ennaji MM, El Kebbaj R, Mhand RA, Attaleb M, El Mzibri M. P53 codon 72 polymorphism and risk of cervical carcinoma in Moroccan women. Med Oncol. (2010) 27:861–6. doi: 10.1007/s12032-009-9297-6
41. Eltahir HA, Ibrahim M. Contribution of retinoblastoma LOH and the p53 Arg/Pro polymorphism to cervical cancer. Mol Med Rep. (2012) 6(3):473–6. doi: 10.3892/mmr.2012.942
42. Fernandes T, Lima GLF, De Souza FCG, Fernandes JV, Meissner RV. Evaluation of the polymorphisms in the exons 2 to 4 of the TP53 in cervical carcinoma patients from a Brazilian population. Cell Mol Biol (Noisy-Le-Grand France). (2008) 54 Suppl:OL1025–1031.
43. Ferreira Da Silva I, Koifman RJ, Quinto Santos Souza C, Ferreira De Almeida Neto O, Koifman S. TP53 genetic polymorphisms and environmental risk factors associated with cervical carcinogenesis in A cohort of Brazilian women with cervical lesions. J Toxicol Environ Health Part A. (2010) 73:888–900. doi: 10.1080/15287391003744823
44. Tanara G, Falugi C, Cesario A, Margaritora S, Russo P, Cosimi A. TP53 codon 72 polymorphism does not affect risk of cervical cancer in patients from The Gambia. Int J Biol Markers. (2003) 18:280–3. doi: 10.1177/172460080301800405
45. Giannoudis A, Graham DA, Southern SA, Herrington CS. P53 codon 72 ARG/PRO polymorphism is not related to HPV type or lesion grade in low- and high-grade squamous intra-epithelial lesions and invasive squamous carcinoma of the cervix. Int J Cancer. (1999) 83:66–9. doi: 10.1002/(SICI)1097-0215(19990924)83:1<66::AID-IJC13>3.0.CO;2-K
46. González-Herrera L, Rodríguez-Morales P, Del-Refugio-González-Losa M, Pérez-Mendoza G, Canul-Canché J, Rosado-López I, et al. MTHFR/p53 polymorphisms as genetic factors for cervical intraepithelial neoplasia and cervical cancer in HPV-infected mexican women. Int J Biol Markers. (2014) 29:142–9. doi: 10.5301/jbm.5000070
47. Govan VA, Loubser S, Saleh D, Hoffman M, Williamson A. No relationship observed between human p53 codon-72 genotype and HPV-associated cervical cancer in a population group with a low arginine-72 allele frequency. Int J Immunogenetics. (2007) 34:213–7. doi: 10.1111/j.1744-313X.2007.00678.x
48. Gudleviciene Z, Didziapetriene J, Ramael M, Uleckiene S, Valuckas KP. Human papillomavirus and p53 polymorphism in Lithuanian cervical cancer patients. Gynecol Oncol. (2006) 102:530–3. doi: 10.1016/j.ygyno.2006.01.019
49. Guo H, Wen Z, Yang S, Qi H. Association of p73 G4C14-A4T14 and p53 codon 72 polymorphism with cervical cancer in Chinese population. Indian J Cancer. (2021). doi: 10.4103/ijc.IJC_538_19
50. Gustafsson AC, Zhongmin G, Xinron A. HPV-related cancer susceptibility and p53 codon 72 polymorphism. Acta Dermato-Venereol. (2001) 81:125–9. doi: 10.1080/00015550152384272
51. Hayes VM, Hofstra RM, Buys CH, Hollema H, van der Zee AG. Homozygous arginine-72 in wild type p53 and risk of cervical cancer. Lancet (London England). (1998) 352:1756. doi: 10.1016/S0140-6736(05)79829-9
52. Helland A, Langerod A, Johnsen H, Olsen AO, Skovlund E, Borresen-Dale AL. p53 olymorphism and risk of cervical cancer. Nature. (1998) 396:530–31. doi: 10.1038/25034
53. Hildesheim A, Schiff man M, Brinton LA, Fraumeni JF Jr, Herrero R, Bratti MC, et al. p53 polymorphism and risk of cervical cancer. Nature. (1998) 396:531–32. doi: 10.1038/25040
54. Hou MM, Qie MR, Cao ZY. A study on the relationship between the 72nd codon polymorphism of p53 gene and cervical cancer, HPV-16, 18E6. J Sichuan Univ (Medical Edition). (2006) 3:404–7.
55. Humbey O, Aubin F, Cairey-Remonnay S, Riethmuller D, Pretet JL, Fest T, et al. TP53 polymorphism at exon 4 in caucasian women from eastern France: Lack of correlation with HPV status and grade of cervical precancerous lesions. Eur J Obstetrics Gynecol Reprod Biol. (2002) 103:60–4. doi: 10.1016/S0301-2115(02)00006-4
56. Isakova J, Vinnikov D, Bukuev N, Talaibekova E, Adasheva N. [amp]]Tcy;Р53 codon 72 polymorphism and human papilloma virus-associated cervical cancer in kyrgyz women. Asian Pacific J Cancer Prev. (2019) 20:1057–62. doi: 10.31557/APJCP.2019.20.4.1057
57. Jiang P, Liu J, Li W, Zeng X, Tang J. Role of p53 and p51 polymorphisms in the risk of cervical cancer among Chinese women. Acta Biochim Biophys Sin. (2010) 42:671–6. doi: 10.1093/abbs/gmq069
58. Jiang P, Liu J, Zeng X, Li W, Tang J. Association of TP53 codon 72 polymorphism with cervical cancer risk in Chinese women. Cancer Genet Cytogenetics. (2010) 197:174–8. doi: 10.1016/j.cancergencyto.2009.11.011
59. Josefsson AM, Magnusson PK, Ylitalo N, Quarforth-Tubbin P, Pontén J, Adami HO, et al. p53 polymorphism and risk of cervical cancer. Nature. (1998) 396:531. doi: 10.1038/25037
60. Katiyar S, Thelma BK, Murthy NS, Hedau S, Jain N, Gopalkrishna V, et al. Polymorphism of the p53 codon 72 Arg/Pro and the risk of HPV type 16/18-associated cervical and oral cancer in India. Mol Cell Biochem. (2003) 252:117–24. doi: 10.1023/a:1025546610920
61. Kawamata Y, Mitsuhashi A, Unno Y, Kado S, Shino Y, Uesugi K, et al. HPV 16-E6-mediated degradation of intrinsic p53 is compensated by upregulation of p53 gene expression in normal cervical keratinocytes. Int J Oncol. (2002) 6(3):473–6. doi: 10.3892/ijo.21.3.561
62. Kim JW, Lee CG, Park YG, Kim KS, Kim I-K, Sohn YW, et al. Combined analysis of germline polymorphisms of p53, GSTM1, GSTT1, CYP1a1, and CYP2e1: Relation to the incidence rate of cervical carcinoma. Cancer. (2000) 88:2082–91. doi: 10.1002/(SICI)1097-0142(20000501)88:9<2082::AID-CNCR14>3.0.CO;2-D
63. Kim JW, Roh JW, Park NH, Song YS, Kang SB, Lee HP. Polymorphism of TP53 codon 72 and the risk of cervical cancer among Korean women. Am J Obstetrics Gynecol. (2001) 184:55–8. doi: 10.1067/mob.2001.108329
64. Klaes R, Ridder R, Schaefer U, Benner A, Von Knebel Doeberitz M. No evidence of p53 allele-specific predisposition in human papillomavirus-associated cervical cancer. J Mol Med. (1999) 77:299–302. doi: 10.1007/s001090050353
65. Klug SJ, Wilmotte R, Santos C, Almonte M, Herrero R, Guerrero I, et al. TP53 polymorphism, HPV infection, and risk of cervical cancer. Cancer Epidemiol Biomarkers Prevention: A Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. (2001) 10:1009–12.
66. Kouamou V, Chin’ombe N, Matimba A, Kadzatsa W, Nyandoro G, Musarurwa C. P53 Codon 72 polymorphism and the risk of cervical cancer in Zimbabwean women. Int J Trop Dis Health. (2016) 15:1–6. doi: 10.9734/IJTDH/2016/24676
67. Koushik A, Ghosh A, Duarte-Franco E, Forest P, Voyer H, Matlashewski G, et al. The p53 codon 72 polymorphism and risk of high-grade cervical intraepithelial neoplasia. Cancer Detection Prev. (2005) 29:307–16. doi: 10.1016/j.cdp.2005.06.007
68. Lanham S, Campbell I, Watt P, Gornall R. P53 polymorphism and risk of cervical cancer. Lancet (London, England). (1998) 352(9140):1631. doi: 10.1016/S0140-6736(05)61083-5
69. Laprano TDR, Lemos ÉH, Cunha LMP, Júnior JE, De SousaTeles RA, Rabenhorst SHB. Association of TP53 codon 72 and intron 3 16-bp Ins/Del polymorphisms with cervical cancer risk. Tumor Biol. (2014) 35:7435–40. doi: 10.1007/s13277-014-1988-8
70. Lee JE, Lee SJ, Namkoong SE, Um SJ, Sull JW, Jee SH, et al. Gene—Gene and gene—Environmental interactions of p53, p21, and IRF-1 polymorphisms in Korean women with cervix cancer. Int J Gynecol Cancer. (2004) 14(1):118–25. doi: 10.1136/ijgc-00009577-200401000-00016
71. Lee S-A, Kim JW, Roh JW, Choi JY, Lee K-M, Yoo K-Y, et al. Genetic polymorphisms of GSTM1, p21, p53 and HPV infection with cervical cancer in Korean women. Gynecol Oncol. (2004) 93:14–8. doi: 10.1016/j.ygyno.2003.11.045
72. Li J, Zheng QQ, Wang P. The relationship between p53 codon 72 gene polymorphism and susceptibility to cervical squamous cell carcinoma in Shanxi Han population: a case-control study. J Fourth Military Med Univ. (2006) 16:1499–501.
73. Li CY, Liu JH, Huang BJ. Preliminary study on the relationship between P53 gene polymorphism and cervical cancer. Cancer. (2004) S1:1396–9.
74. Liu G-C, Zhou Y-F, Su X-C, Zhang J. Interaction between TP53 and XRCC1 increases susceptibility to cervical cancer development: A case control study. BMC Cancer. (2019) 19:24. doi: 10.1186/s12885-018-5149-0
75. Madeleine MM, Shera K, Schwartz SM, Daling JR, Galloway DA, Wipf GC, et al. The p53 Arg72Pro polymorphism, human papillomavirus, and invasive squamous cell cervical cancer. Cancer Epidemiol Biomarkers Prevention: A Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. (2000) 9:225–7.
76. Makni H, Franco EL, Kaiano J, Villa LL, Labrecque S, Dudley R, et al. p53 polymorphism in codon 72 and risk of human papillomavirus-induced cervical cancer: Effect of inter-laboratory variation. Int J Cancer. (2000) 87:528–33. doi: 10.1002/1097-0215(20000815)87:4<528::AID-IJC11>3.0.CO;2-O
77. Malcolm EK, Baber GB, Boyd JC, Stoler MH. Polymorphism at Codon 72 of p53 Is Not Associated with Cervical Cancer Risk. Modern Pathol. (2000) 13:373–8. doi: 10.1038/modpathol.3880061
78. Malisic E, Jankovic R, Brotto K, Radulovic S. TP53 codon 72 polymorphism and risk of cervical carcinoma in Serbian women. Arch Gynecol Obstetrics. (2013) 288:621–5. doi: 10.1007/s00404-013-2783-2
79. Minaguchi T, Kanamori Y, Matsushima M, Yoshikawa H, Taketani Y, Nakamura Y. No evidence of correlation between polymorphism at codon 72 of p53 and risk of cervical cancer in Japanese patients with human papillomavirus 16/18 infection. Cancer Res. (1998) 58:4585–6.
80. Min-Min H, Ming-Rong X, Ze-Yi C, Kai-Xuan Y, Zhi-Lin S. Analysis of p53 codon 72 polymorphism and its association with human papillomavirus 16 and 18 E6 in Chinese cervical lesions. Int J Gynecol Cancer. (2006) 16:2004–8. doi: 10.1111/j.1525-1438.2006.00733.x
81. Mitra S. Association of specific genotype and haplotype of p53 gene with cervical cancer in India. J Clin Pathol. (2005) 58:26–31. doi: 10.1136/jcp.2004.019315
82. Mostaid MS, Mumu SB, Haque MA, Sharmin S, Jamiruddin MR, Sayedur Rahman GM, et al. Elevated serum expression of p53 and association of TP53 codon 72 polymorphisms with risk of cervical cancer in Bangladeshi women. PloS One. (2021) 16:e0261984. doi: 10.1371/journal.pone.0261984
83. Nagpal JK, Sahni S, Das BR. P53 BlackwellScience,Ltd codon 72 polymorphism and susceptibility to development of human papilloma virus-associated cervical cancer in Indian women. Eur J Clin Invest. (2002) 32(12):943–8. doi: 10.1046/j.1365-2362.2002.01096.x
84. Natphopsuk S, Settheetham-Ishida W, Sinawat S, Pientong C, Yuenyao P, Ishida T. Risk factors for cervical cancer in northeastern Thailand: detailed analyses of sexual and smoking behavior. Asian Pacific J Cancer Prev. (2012) 13:5489–95. doi: 10.7314/APJCP.2012.13.11.5489
85. Ndiaye R, Dem A, Mbaye PM, Guèye PM, Diop G, Diop PA, et al. [amp]]Eacute;tude du codon 72 du gène p53 dans la prédisposition au cancer du col de l’utérus au Sénégal. Bull Du Cancer. (2014) 101:789–94. doi: 10.1684/bdc.2014.1911
86. Ngan HYS, Liu VWS, Liu SS. Risk of cervical cancer is not increased in Chinese carrying homozygous arginine at codon 72 of p53. Br J Cancer. (1999) 80:1828–9. doi: 10.1038/sj.bjc.6690606
87. Nishikawa A, Fujimoto T, Akutagawa N, Iwasaki M, Takeuchi M, Fujinaga K, et al. P53 Polymorphism (codon-72) has no correlation with the development and the clinical features of cervical cancer. Int J Gynecol Cancer. (2000) 10:402–7. doi: 10.1046/j.1525-1438.2000.010005402.x
88. Niwa Y, Hamajima N, Atsuta Y, Yamamoto K, Tamakoshi A, Saito T, et al. Genetic polymorphisms of p73 G4C14-to-A4T14 at exon 2 and p53 Arg72Pro and the risk of cervical cancer in Japanese. Cancer Lett. (2004) 205:55–60. doi: 10.1016/j.canlet.2003.11.014
89. Ojeda JM, Ampuero S, Rojas P, Prado R, Allende JE, Barton SA, et al. P53 codon 72 polymorphism and risk of cervical cancer. Biol Res. (2003) 36(2):279–83. doi: 10.4067/S0716-97602003000200017
90. Pegoraro R, Moodley J, Naiker S, Lanning P, Rom L. The p53 codon 72 polymorphism in black South African women and the risk of cervical cancer. BJOG: Int J Obstetrics Gynaecol. (2000) 107:1164–5. doi: 10.1111/j.1471-0528.2000.tb11118.x
91. Pegoraro RJ, Rom L, Lanning PA, Moodley M, Naiker S, Moodley J. P53 codon 72 polymorphism and human papillomavirus type in relation to cervical cancer in South African women. Int J Gynecol Cancer. (2002) 12:383–8. doi: 10.1046/j.1525-1438.2002.01109.x
92. Pillai R, S. S, Pollock B, P. J, Herman B. Polymorphism at codon 72 of p53, human papillomavirus, and cervical cancer in South India. J Cancer Res Clin Oncol. (2002) 128:627–31. doi: 10.1007/s00432-002-0383-9
93. Piña-Sánchez P, Hernández-Hernández DM, Taja-Chayeb L, Cerda-Flores RM, González-Herrera AL, Rodea-Avila C, et al. Polymorphism in exon 4 of TP53 gene associated to HPV 16 and 18 in Mexican women with cervical cancer. Med Oncol. (2011) 28:1507–13. doi: 10.1007/s12032-010-9599-8
94. Qie MR, Zhang YH, Wu JM. A study on the relationship between P53 Codon 72 polymorphism and cervical cancer. J West China Univ Med Sci. (2002) 2:274–5.
95. Ratre YK, Jain V, Amle D, Patra PK, Mishra PK. Association of TP53 gene codon 72 polymorphism with incidence of cervical cancer in chhattisgarh. Indian J Exp Biol. (2019) 57:580–5.
96. Rezza G, Giuliani M, Garbuglia AR, Serraino D, Cappiello G, Migliore G, et al. Lack of association between p53 codon-72 polymorphism and squamous intraepithelial lesions in women with, or at risk for, human immunodeficiency virus and/or human papillomavirus infections. Cancer Epidemiol Biomarkers Prevention: A Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. (2001) 10:565–6.
97. Rosenthal AN, Ryan A, Al-Jehani RM, Storey A, Harwood CA, Jacobs IJ. P53 codon 72 polymorphism and risk of cervical cancer in UK. Lancet. (1998) 352:871–2. doi: 10.1016/S0140-6736(98)07357-7
98. Santos AM, Sousa H, Catarino R, Pinto D, Pereira D, Vasconcelos A, et al. TP53 codon 72 polymorphism and risk for cervical cancer in Portugal. Cancer Genet Cytogenetics. (2005) 159:143–7. doi: 10.1016/j.cancergencyto.2004.10.005
99. Santos AM, Sousa H, Pinto D, Portela C, Pereira D, Catarino R, et al. Linking TP53 codon 72 and P21 nt590 genotypes to the development of cervical and ovarian cancer. Eur J Cancer. (2006) 42:958–63. doi: 10.1016/j.ejca.2006.01.015
100. Saranath D, Khan Z, Tandle AT, Dedhia P, Sharma B, Contractor R, et al. HPV16/18 prevalence in cervical lesions/cancers and p53 genotypes in cervical cancer patients from India. Gynecol Oncol. (2002) 86:157–62. doi: 10.1006/gyno.2002.6735
101. Settheetham-Ishida W, Singto Y, Yuenyao P, Tassaneeyakul W, Kanjanavirojkul N, Ishida T. Contribution of epigenetic risk factors but not p53 codon 72 polymorphism to the development of cervical cancer in Northeastern Thailand. Cancer Lett. (2004) 210:205–11. doi: 10.1016/j.canlet.2004.03.039
102. Settheetham-Ishida W, Kanjanavirojkul N, Kularbkaew C, Ishida T. Human papillomavirus genotypes and the p53 codon 72 polymorphism in cervical cancer of northeastern Thailand. Microbiol Immunol. (2005) 49:417–21. doi: 10.1111/j.1348-0421.2005.tb03745.x
103. Singhal P, Hussain S, Thakur N, Batra S, Salhan S, Bhambani S, et al. Association of MDM2 and p53 polymorphisms with the advancement of cervical carcinoma. DNA Cell Biol. (2013) 32:19–27. doi: 10.1089/dna.2012.1718
104. Sonoda Y, Saigo PE, Boyd J. P53 and genetic susceptibility to cervical cancer. JNCI J Natl Cancer Institute. (1999) 91:557–7. doi: 10.1093/jnci/91.6.557
105. Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, et al. Role of a p53 polymorphism in the development of human papilloma-virus-associated cancer. Nature. (1998) 393:229–34. doi: 10.1038/30400
106. Strickler H, Manns A, Escoffery C, Rattray C, Brown C, Vellucci V, et al. P53 polymorphisms at position 72 and development of cervical cancer. Int J Gynecol Cancer. (1998) 8:439–9. doi: 10.1046/j.1525-1438.1998.0LE05.x
107. Suárez-Rincón AE, Morán-Moguel MC, Montoya-Fuentes H, Gallegos-Arreola MP, Sánchez-Corona J. Polymorphism in codon 72 of the p53 gene and cervico-uterine cancer risk in Mexico. Ginecol Y Obstetricia Mexico. (2002) 70:344–8.
108. Szarka K, Veress G, Kónya J, Gergely L. Frequency of p53 codon 72 genotypes in human papillomavirus associated squamous intraepithelial lesions and cervical cancer. Anticancer Res. (1999) 19:2377–9.
109. Tachezy R, Mikyšková I, Saláková M, Van Ranst M. Correlation between human papillomavirus-associated cervical cancer and p53 codon 72 arginine/proline polymorphism. Hum Genet. (1999) 105:564–6. doi: 10.1007/s004399900138
110. Tenti P, Vesentini N, Rondo Spaudo M, Zappatore R, Migliora P, Carnevali L, et al. P53 codon 72 polymorphism does not affect the risk of cervical cancer in patients from northern Italy. Cancer Epidemiol Biomarkers Prevention: A Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. (2000) 9:435–8.
111. Tong D, Kucera E, Stimpfl M, Kölbl H, Leodolter S, Zeillinger R. Detection of p53 polymorphism at codon 72 by PCR and allele-specific oligonucleotide hybridization on microtiter plates. Clin Chem. (2000) 46:124–6. doi: 10.1093/clinchem/46.1.124
112. Ueda M, Terai Y, Kanda K, Kanemura M, Takehara M, Yamaguchi H, et al. Germline polymorphism of p53 codon 72 in gynecological cancer. Gynecol Oncol. (2006) 100:173–8. doi: 10.1016/j.ygyno.2005.08.015
113. Ueda M, Toji E, Nunobiki O, Sato N, Izuma S, Torii K, et al. Germline polymorphisms of glutathione-S-transferase GSTM1, GSTT1 and p53 codon 72 in cervical carcinogenesis: GST and p53 polymorphisms in cervical cancer. Hum Cell. (2010) 23:119–25. doi: 10.1111/j.1749-0774.2010.00089.x
114. Van Duin M, Snijders PJF, Vossen MTM, Klaassen E, Voorhorst F, Verheijen RHM, et al. Analysis of human papillomavirus type 16 E6 variants in relation to p53 codon 72 polymorphism genotypes in cervical carcinogenesis. Microbiology. (2000) 81:317–25. doi: 10.1099/0022-1317-81-2-317
115. Wang XL, Pan XL, Gao ZB. The correlation between the 72nd codon polymorphism of P53 gene and cervical cancer. J Clin Exp Pathol. (2004) 5:525–8. doi: 10.13315/j.cnki.cjcep.2004.05.004
116. Wu M-T, Liu C-L, Ho C-K, Wu T-N. Genetic polymorphism of p53 and XRCC1 in cervical intraepithelial neoplasm in Taiwanese women. J Formosan Med Assoc. (2004) 103:337–43.
117. Yamashita T, Yaginuma Y, Saitoh Y, Kawai K, Kurakane T, Hayashi H, et al. Codon 72 polymorphism of p53 as a risk factor for patients with human papillomavirus-associated squamous intraepithelial lesions and invasive cancer of the uterine cervix. Carcinogenesis. (1999) 20:1733–6. doi: 10.1093/carcin/20.9.1733
118. Yang Y-C, Chang C-L, Chen M-L. Effect of p53 polymorphism on the susceptibility of cervical cancer. Gynecol Obstetric Invest. (2001) 51:197–201. doi: 10.1159/000052924
119. Yang M, Xu JJ. Correlation analysis between P53 gene codon 72 gene polymorphism and cervical cancer in Menggu women. Med Recapitulate. (2011) 17:3352–4.
120. Yang S-D, Cai Y-L, Jiang P, Li W, Tang J-X. Association of a miR-502-binding site single nucleotide polymorphism in the 3’-untranslated region of SET8 and the TP53 codon 72 polymorphism with cervical cancer in the Chinese population. Asian Pacific J Cancer Prevention: APJCP. (2014) 15:6505–10. doi: 10.7314/apjcp.2014.15.16.6505
121. Yang AQ, Zheng XZ, Tao L. The correlation between p53 Arg72Pro, p21Ser31 Arg polymorphism and cervical cancer in Xinjiang Uyghur ethnic group. J Shihezi Univ (Natural Sci Edition). (2008) 1:6–11. doi: 10.13880/j.cnki.65-1174/n.2008.01.026
122. Yao N, Zheng H, Lu J. A study on the relationship between P53 gene Codon72 polymorphism and cervical cancer. J Zunyi Med Coll. (2008) 1:3–5.
123. Ye F, Zhang J, Cheng Q, Shen J, Chen H. P53 Codon 72 polymorphism is associated with occurrence of cervical carcinoma in the Chinese population. Cancer Lett. (2010) 287:117–21. doi: 10.1016/j.canlet.2009.06.004
124. Yi C. A study on the correlation between host p53 gene polymorphism and HPV16 infection in cervical cancer. Yunnan Province, China: Master’s degree, Kunming Medical University (2016). Available at: https://kns.cnki.net/kcms2/article/abstract?v=hqt_j-uEELEpqkb1MBpjodgyWvlY3yh1NWbpQ9xUwufHnTiDCSL4m2yHqlfAXH49yUgqT5UFMZT25Mrn25tm8JBcx1dmT52nMLOVhiIksWzZD8xLmxT4bOAruYx4sEufjLW1NVXYpNd8pM7BzqJb6g==&uniplatform=NZKPT&language=CHS (Accessed May 5, 20204).
125. Yuan F, Sun R, Chen P, Liang Y, Ni S, Quan Y, et al. Combined analysis of pri-miR-34b/c rs4938723 and TP53 Arg72Pro with cervical cancer risk. Tumor Biol. (2016) 37:6267–73. doi: 10.1007/s13277-015-4467-y
126. Zehbe I, Voglino G, Wilander E, Genta F, Tommasino M. Codon 72 polymorphism of p53 and its association with cervical cancer. Lancet. (1999) 354:218–9. doi: 10.1016/S0140-6736(99)01914-5
127. Zehbe I, Voglino G, Wilander E, Delius H, Marongiu A, Edler L, et al. P53 codon 72 polymorphism and various human papillomavirus 16 E6 genotypes are risk factors for cervical cancer development. Cancer Res. (2001) 61:608–11.
128. Zheng X-Z, Yang A-Q, Pan X-L, Zheng L-L, Wang X-L, Zhou Q-Y, et al. Ethnicity determines association of p53Arg72Pro alleles with cervical cancer in China. Eur J Cancer Prev. (2008) 17:460–6. doi: 10.1097/CEJ.0b013e3282f75f3e
129. Zhou X, Han S, Wang S, Chen X, Dong J, Shi X, et al. Polymorphisms in HPV E6/E7 protein interacted genes and risk of cervical cancer in Chinese women: A case-control analysis. Gynecol Oncol. (2009) 114:327–31. doi: 10.1016/j.ygyno.2009.05.011
130. Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. J Clin Epidemiol. (2009) 62:1006–12. doi: 10.1016/j.jclinepi.2009.06.005
131. Thakkinstian A, McKay GJ, McEvoy M, Chakravarthy U, Chakrabarti S, Silvestri G, et al. Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: A HuGE review and meta-analysis. Am J Epidemiol. (2011) 173:1365–79. doi: 10.1093/aje/kwr025
132. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Institute. (1959) 22:719–48.
133. DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemp Clin Trials. (2015) 45:139–45. doi: 10.1016/j.cct.2015.09.002
134. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clin Res Ed). (1997) 315:629–34. doi: 10.1136/bmj.315.7109.629
135. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. (1994) 50:1088–101. doi: 10.2307/2533446
136. Duval S, Tweedie R. A nonparametric “Trim and fill” Method of accounting for publication bias in meta-analysis. J Am Stat Assoc. (2000) 95:89–98. doi: 10.1080/01621459.2000.10473905
137. Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: An approach for molecular epidemiology studies. J Natl Cancer Institute. (2004) 96:434–42. doi: 10.1093/jnci/djh075
138. Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet. (2007) 81:208–27. doi: 10.1086/519024
Keywords: p53, polymorphism, risk, cervical cancer, meta-analysis
Citation: Zhang X-Q, Bai X-H, Zhang H-Z and He X-F (2025) Association between the p53 polymorphisms and cervical cancer risk: an updated meta-analysis. Front. Oncol. 15:1461737. doi: 10.3389/fonc.2025.1461737
Received: 12 July 2024; Accepted: 27 January 2025;
Published: 21 February 2025.
Edited by:
Carlos Pérez-Plasencia, National Autonomous University of Mexico, MexicoReviewed by:
Erik Rene Lizárraga-Verdugo, Autonomous University of Sinaloa, MexicoCopyright © 2025 Zhang, Bai, Zhang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui-Zhen Zhang, emhhbmdoejE5OTRAMTYzLmNvbQ==; Xiao-Feng He, MzkzMTIwODIzQHFxLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.