- 1The First Clinical Medical College of Shandong University of Traditional Chinese Medicine, Jinan, Shandong, China
- 2Department of Oncology, Zibo Hospital of Traditional Chinese Medicine, Zibo, China
- 3Department of Minimally Invasive Comprehensive Treatment of Cancer, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, Shandong, China
- 4Department of Radiotherapy, The Affiliated Yantai Yuhuangding Hospital of Qingdao University, Yantai, China
Colorectal cancer (CRC) ranks among the most prevalent gastrointestinal tumors globally and poses a significant threat to human health. In recent years, tumor organoids have emerged as ideal models for clinical disease research owing to their ability to closely mimic the original tumor tissue and maintain a stable phenotypic structure. Organoid technology has found widespread application in basic tumor research, precision therapy, and new drug development, establishing itself as a reliable preclinical model in CRC research. This has significantly advanced individualized and precise tumor therapies. Additionally, the integration of single-cell technology has enhanced the precision of organoid studies, offering deeper insights into tumor heterogeneity and treatment response, thereby contributing to the development of personalized treatment approaches. This review outlines the evolution of colorectal cancer organoid technology and highlights its strengths in modeling colorectal malignancies. This review also summarizes the progress made in precision tumor medicine and addresses the challenges in organoid research, particularly when organoid research is combined with single-cell technology. Furthermore, this review explores the future potential of organoid technology in the standardization of culture techniques, high-throughput screening applications, and single-cell multi-omics integration, offering novel directions for future colorectal cancer research.
1 Introduction
1.1 Background
1.1.1 Incidence of colorectal cancer and its global implications
Colorectal cancer (CRC) is the third most common malignancy and the second leading cause of cancer-related deaths worldwide, with approximately 1.926 million new cases and 904,000 deaths reported in 2022 (1). Despite advances in treatment, 30-50% of early-stage patients still experience recurrence (2). The global incidence of CRC continues to rise, with projections suggesting 3.2 million new cases and 1.6 million deaths by 2040. Developed countries, such as the United States and Western European nations, face particularly high incidence rates, likely due to dietary habits, lifestyle, and aging populations. Current treatments include surgery, radiotherapy, and chemotherapy, supplemented by emerging immunotherapy and targeted therapies (1). However, challenges in targeting efficiency, drug resistance, and treatment efficacy persist, partly due to the lack of models that accurately replicate the in vivo disease environment. Organoid technology, which closely mimics in vivo tumor characteristics, offers significant potential for both research and clinical applications (3–5).
1.1.2 Concept and importance of precision medicine
Precision medicine is a healthcare model that customizes medical treatment based on an individual’s genetic, environmental, and lifestyle factors. The core of precision medicine lies in using individualized information to prevent, diagnose, and treat diseases, aiming to provide personalized healthcare. This concept emphasizes the application of advanced biotechnologies, particularly genomics, proteomics, metabolomics, and other multi-omics, to achieve more accurate disease diagnosis and treatment (6, 7). Especially in CRC, precision medicine predicts patient responses to specific treatments by identifying particularly genetic mutations, such as KRAS, BRAF, and PIK3CA, while also reducing unnecessary treatment side effects (8).
Precision medicine not only focuses on effective disease treatment but also prioritizes prevention and health management, with the goal of improve treatment outcomes, reduce side effects, and lower healthcare costs (1). Unlike traditional healthcare models that rely on generalized population data, precision medicine seeks to deliver “customized” solutions for each patient (9).
The significance of precision medicine can be summarized in five key areas: 1) Improvement of treatment efficacy: by analyzing individual patients’ genomes and biomarkers, more accurate drug dosages and combinations can be determined, enhancing treatment effectiveness. For example, exome sequencing can identify specific tumor mutations, guiding the selection of optimal targeted therapies (10). 2) Reduction of side effects: by predicting patient responses to medications, physicians can minimize adverse reactions by avoiding ineffective or harmful drugs. 3) Disease prevention: precision medicine facilitates early disease detection and prevention through genetic screenings, enabling the identification or prediction of specific disease risks and allowing for lifestyle adjustments to mitigate morbidity risk (4). 4) Optimization of healthcare resource allocation: precision medicine reduces trial-and-error approaches, expedites diagnostic and treatment decisions, and conserves healthcare resources, thereby lowering costs. This is particularly crucial for optimizing resource allocation (11). 5) Promotion of medical research: precision medicine drives research into disease mechanisms, aids in discovering new therapeutic targets, and supports the development of personalized diagnostic and treatment strategies (12).
1.2 Overview of organoid technology
1.2.1 Definition and application of organoids
Organoids are typically derived from adult stem cells, embryonic stem cells, or induced pluripotent stem cells and are cultured in vitro under specific signaling and environmental conditions. These structures replicate the multicellular composition and physiological functions of their original organs in a laboratory setting, offering a more physiologically relevant model than traditional two-dimensional cell cultures.
Organoid technology has vast potential in cancer research, genetic disease modeling, and the exploration of infectious pathogenesis (13). In cancer research, organoids can recreate the tumor microenvironment, serving as an effective tool for targeted drug screening and personalized therapy. In genetic disease research, patient-derived organoids can mimic the effects of gene mutations, advancing the study and treatment of rare diseases. Patient-derived organoids (PDOs) are invaluable for predicting patient treatment responses in vitro and have been successfully applied in various cancers, including colorectal (14), gastric (15), lung (8), and liver cancers (16). The development of organoid technology, particularly the construction of PDOs from tumor tissues, has significantly enhanced its value in precision medicine. By recreating the tumor microenvironment, organoid technology preserves the structural and functional characteristics of donor tissues, making it a crucial platform for studying tumor biology and developing personalized therapies (Figure 1).
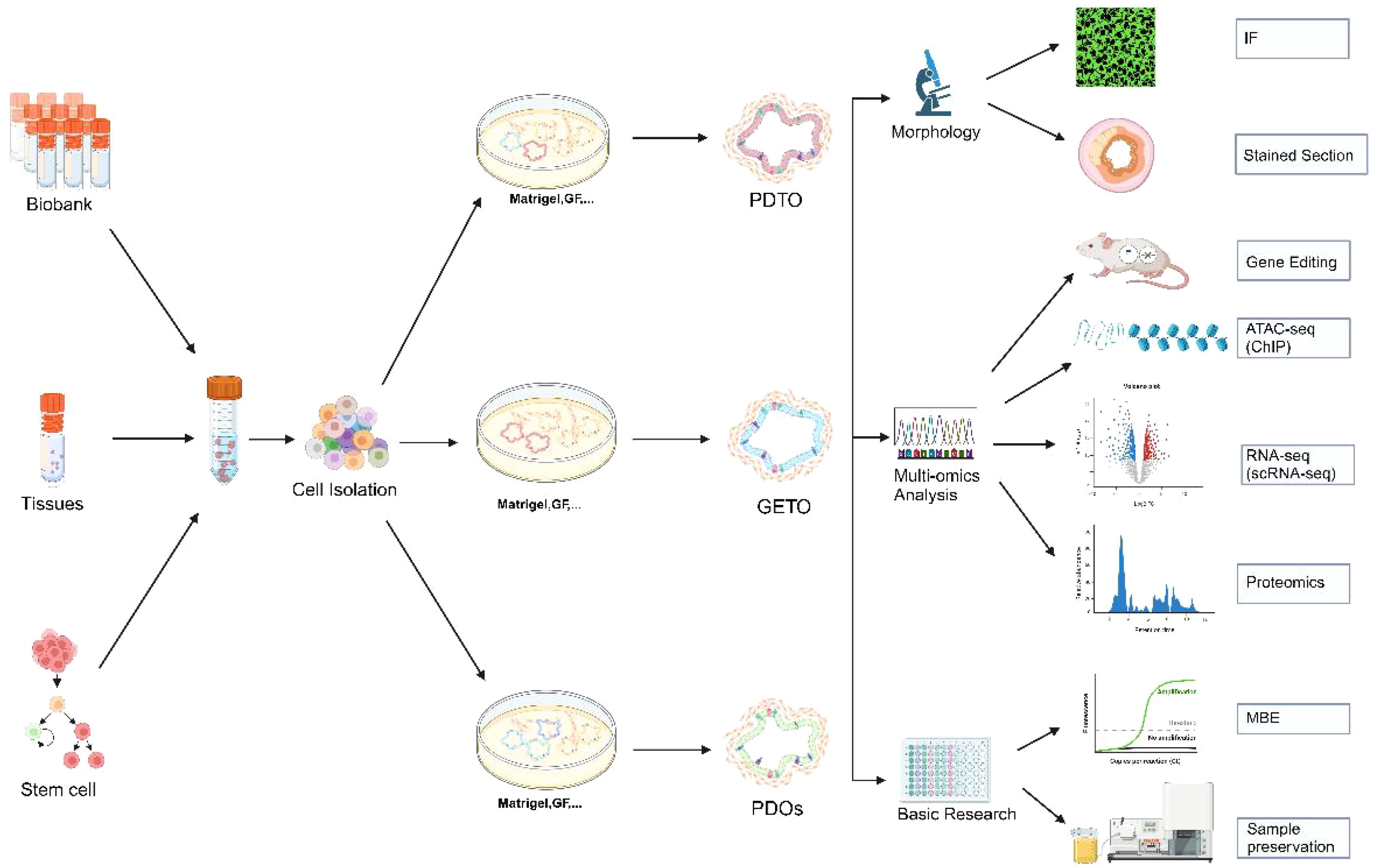
Figure 1. Establishment of colorectal cancer organoids and its application in morphology, multi-omics analysis and basic research. Biobank, human tissue-derived, and stem cell-derived cells can be cultured into colorectal organoids. These organoids are applicable for morphological observation, multi-omics analysis (including gene editing, ATAC-seq analysis, single-cell transcriptome analysis, and proteomic analysis), and basic research. PDTX, Patient-derived tumor xenograft model; GETO, Genetically modified tumor organoids; PDTO, Patient-derived organoids. Created with Biorender.com (accessed on 15 August 2024).
1.2.2 Potential and advantages in colorectal cancer research
Intestinal tumor organoids can differentiate into almost all intestinal cell types, including stem cells, Paneth cells, goblet cells, endocrine cells, and epithelial cells (17). Currently, there are two primary methods for constructing patient-derived tumor organoids (PDTOs): differentiation of induced pluripotent stem cells (iPSCs) or direct derivation from tumor tissues. However, the success rate of iPSC-derived PDOs varies significantly depending on the tumor type, and the complexity of the procedure can reduce efficiency. Additionally, PDOs generated from iPSCs often lack the tumor microenvironment’s complexity, being predominantly epithelial, whereas iPSC-derived organoids may include both epithelial and mesenchymal cells (18). Therefore, the future trend in organoid research lies in the culture techniques of PDTOs directly derived from tumor tissues, supplemented with cytokines and extracellular matrices. Directly deriving organoid cultures from tumor tissues is relatively straightforward, as both surgical specimens and biopsy samples can be used for colorectal cancer organoid cultures.
The culture process involves isolating cancer stem cells from colorectal cancer(CRC) tissues, embedding these cells in stromal gels, and adding specific media for expansion and passage, followed by experimental studies (19)(Figure 1). Although organoids are not fully developed organs, embedding intestinal crypts or tumor cells in an extracellular matrix (ECM) containing growth factors and inhibitors successfully replicates the in vivo stem cell niche at the base of intestinal crypts, enabling long-term proliferation and differentiation without genetic modifications. Consequently, organoids have become an ideal model system for studying intestinal homeostasis and CRC, preserving the molecular characteristics of donor tissues in terms of structure and function (10). Organoids exhibit a high degree of congruence with their source tissues in various biological characteristics, including gene composition, protein expression, biomechanical properties, and biochemical properties (17, 20). Early and late passage organoids maintain essentially identical gene mutations, demonstrating their genetic stability in long-term culture (10). By constructing organoids with APC, KRAS, and TP53 mutations, researchers can simulate the progression of CRC and evaluate the effectiveness of various treatment strategies (21). Organoids also offer advantages such as short cultivation time, ease of operation, high culture efficiency, transmissibility, freezing capability, and ease of genetic modification, rendering them ideal models for achieving precision tumor therapy.
Numerous research has confirmed that colorectal cancer organoids, as a novel tool for in-depth tumor research, drug screening, and therapeutic sensitivity prediction, hold immense potential in tumor precision medicine. This review summarizes the development of colorectal cancer organoids and recent advances in precision medicine, while also discussing current challenges.
2 Construction and culture of colorectal organoids
2.1 Sources of organoids
2.1.1 Human tissue sources
The sources of colorectal cancer organoids primarily include two categories: human tissue and stem cells. Direct cell isolation from human tissues is a common method for constructing colorectal cancer organoids. This approach typically involves extracting cells from a patient’s tumor tissue and forming organoid models through in vitro three-dimensional culture (Figure 1). These organoids retain the genomic features and morphological characteristics of the original tumor, positioning them an essential tool for studying tumor heterogeneity and treatment response (20, 22).
Tumor heterogeneity research: Organoids derived from human tissues can reflect the heterogeneity of different cell populations within a tumor, aiding researchers in understanding the differential responses of various cell types to treatment (20). For instance, different tumor cell subtypes may exhibit varying sensitivities to the same drugs, which is crucial for developing personalized treatment strategies (5).
Drug screening and resistance studies: Using patient-derived organoids allows for the testing of multiple drugs under laboratory conditions and the identification of resistance mechanisms (3, 5). This not only accelerates new drug development but also provides a method for optimizing drug combinations tailored to specific patients (4, 12).
2.1.2 Stem cell sources
Stem cells, particularly induced pluripotent stem cells (iPSCs) and adult stem cells, offer another significant pathway for constructing colorectal cancer organoids. Especially with intestinal stem cells, organoids can be formed under appropriate culture conditions (23). By adding specific growth factors, such as Wnt, Noggin, and R-spondin, to the culture medium, stem cell-derived organoids can maintain long-term proliferation and develop structures resembling the intestine (24, 25). These stem cells can be induced by specific culture conditions and signaling molecules to differentiate into organoids with tumor characteristics in vitro (26).
Application of iPSCs: iPSCs can be reprogrammed into any cell type, and specific tumor mutations can be introduced through gene editing, generating organoids with distinct genomic features. This is vital for examining the role of genetic mutations in tumorigenesis (23).
Utilization of adult stem cells: Adult stem cells isolated from normal or cancerous tissues can also be used to generate organoids. These organoids can be maintained in vitro for extended periods, exhibiting growth and differentiation properties resembling those of in vivo tissues, thereby positioning them an ideal model for studying tumor development and treatment (11, 27, 28).
2.2 Organoid culture techniques
2.2.1 3D culture technology
Culture technology for colorectal cancer organoids is a core aspect of organoid research, providing a more physiologically relevant model by mimicking the in vivo environment for three-dimensional cell growth and differentiation. Three-dimensional (3D) culture technology forms the foundation for organoid construction, utilizing biological scaffolds or matrix gels that place cells in a three-dimensional environment to support tissue characterization (9, 29).
Matrigel culture: The most used 3D culture technique is Matrigel or similar matrix gel-based cultures. This method allows cells to self-assemble into organoid structures by providing a support analogous to the extracellular matrix (ECM). The application of Matrigel mimics the natural microenvironment of cells in vivo, enhancing cell-matrix interactions to better replicate in vivo conditions (9).
Bioscaffolding technology: In addition to matrix gels, bioscaffolding materials are frequently used in 3D culture. These scaffold materials, typically made of natural or synthetic polymers, can be tailored in terms of porosity, rigidity, and other properties to accommodate the growth requirements of various cell types. This approach has broad applications in tissue engineering and regenerative medicine (30).
Self-assembling organoids: Cells can self-assemble into organoids under scaffold-free or low-scaffold conditions by precisely controlling cell density and culture conditions. This self-assembly process relies on cell-to-cell signaling and interactions, generating tissues that more closely resemble physiological states (31). Table 1 provides a detailed comparison of the various models used in cancer research, demonstrating the strengths and limitations of each.
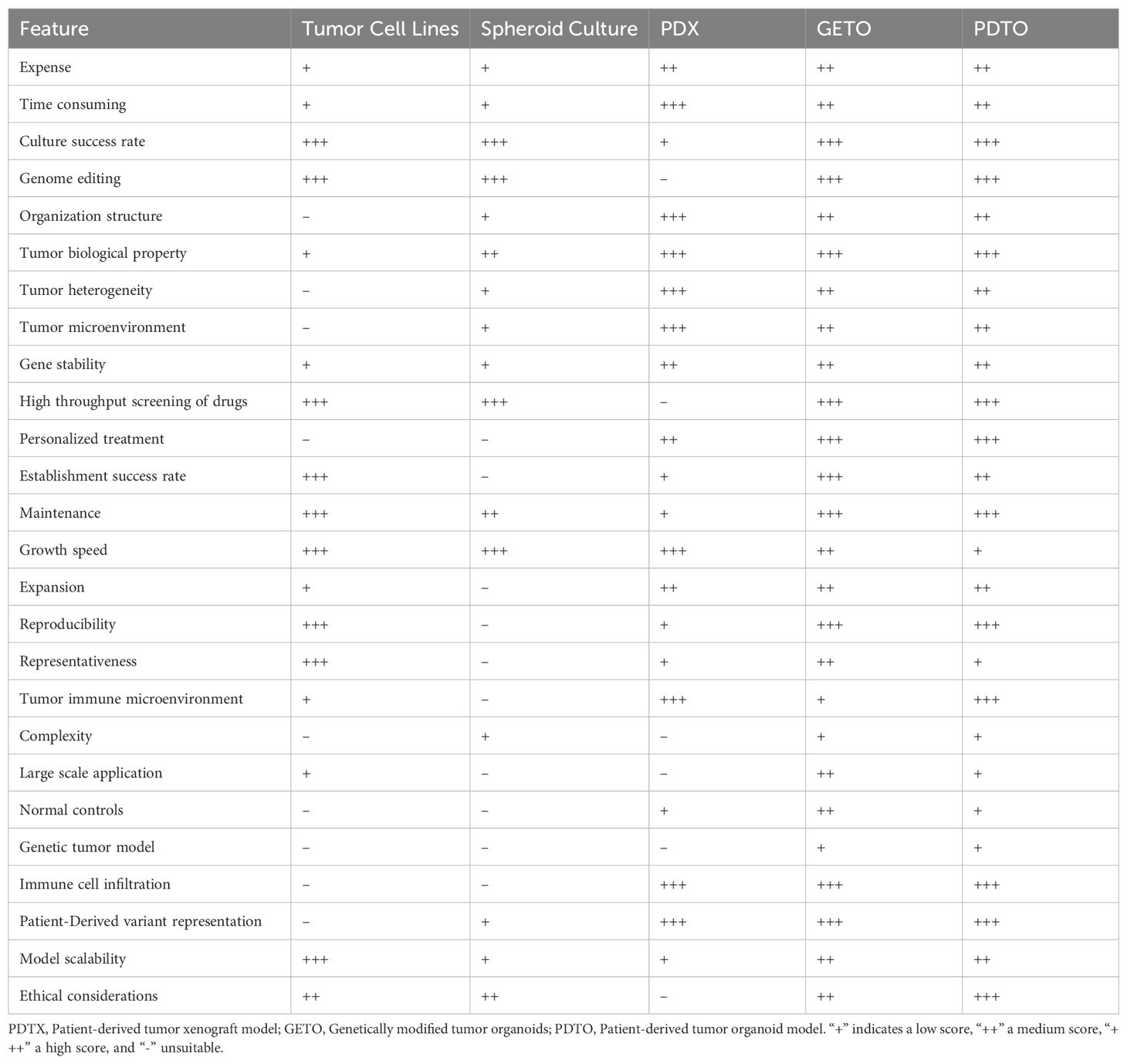
Table 1. Comparison of tumor cell lines, spheroid culture, PDX, GETO, and PDTO models based on key metrics.
2.2.2 Microenvironmental simulation
The success of organoid technology relies heavily on the effective simulation of the in vivo microenvironment, which includes the combined modulation of physical, chemical, and biological factors to sustain organoid growth, differentiation, and functionality (32).
Physical environment simulation: In organoid cultures, physical parameters such as oxygen concentration, shear stress, and mechanical pressure are precisely modulated to mimic the real environment cells encounter in vivo. As an illustration, hypoxic conditions are often used to replicate the hypoxic microenvironment of tumor tissues to study tumor growth and metabolic properties (33).
Control of the chemical environment: The regulation of media composition and growth factors is crucial for organoid growth. By adding or removing specific growth factors and chemical signals, the differentiation and maturation of organoids can be directed. This method has been widely used to investigate cell signaling pathways and drug responses (34, 35).
Reproduction of biological interactions: Microenvironmental simulations also involve the interactions of immune cells, stromal cells, and microbial communities with organoids. By co-culturing with other cell types or introducing microbial communities around organoids, researchers can more comprehensively explore cellular interactions and their impact on disease processes (6, 36, 37).
2.3 Characterization and validation of organoids
2.3.1 Morphological analysis
Characterization and validation of organoids are essential to ensuring they accurately mimic physiological and pathological states in vivo. Morphological analysis is one of the fundamental methods for characterizing organoids, providing detailed observations of their structural and organizational features through microscopy. The formation of typical crypt-villus structures in organoids can be observed, allowing for the assessment of growth and differentiation under various conditions (38).
Microscopic observation: Light and electron microscopy are commonly used to assess the overall morphology and cellular substructures of organoids. Light microscopy enables observation of the three-dimensional structure and growth of organoids, while transmission electron microscopy provides higher resolution details of cellular structures (39).
Tissue sections and staining: Organoid sections, combined with HE staining and immunohistochemistry, can reveal internal tissue structures and cell distribution.
In vivo imaging technology: In vivo imaging, through fluorescence labeling and confocal microscopy, allows real-time observation of organoid growth and development without destruction, making it particularly suitable for studying cell migration and differentiation dynamics.
2.3.2 Multi-omics analysis including genome and transcriptome
Multi-omics analyses offer a comprehensive view of the molecular characteristics of organoids, covering genomic, transcriptomic, proteomic, and metabolomic dimensions.
Genomic analysis: Whole genome sequencing (WGS) and targeted gene sequencing can confirm whether the genetic characteristics of organoids align with the original tissues, validating the genetic accuracy of organoids, especially in cancer research and genetic disease modeling (5). Whole-genome sequencing of organoids derived from 30 patients revealed that these organoids retained key mutations in APC, KRAS, and TP53, and exhibited abnormal activation of the Wnt and PI3K signaling pathways, further validating the effectiveness of the organoid model (40–42).
Transcriptome analysis: RNA sequencing (RNA-seq) reveals the full spectrum of gene expression in organoids, enabling in the identification of key genes and signaling pathways associated with disease states or therapeutic responses (43, 44). Transcriptomic data comparison enables assessment of the similarity between organoids and in vivo tissues in terms of gene expression levels.
Proteomic and metabolomic analyses: Techniques like mass spectrometry identify and quantify proteins and metabolites in organoids, providing essential insights into the functional state and metabolic activity of cells. These data are vital for evaluating the functional and metabolic integrity of organoids. The application of single-cell technology in organoid research will be discussed later (45, 46).
2.4 The role of biospecimen banking in organoid research
2.4.1 Establishment and maintenance of biospecimen banks
Biospecimen repositories are vital in organoid research, offering essential resources and technical support for tumor biology research and personalized medicine. By systematically collecting and managing biological samples from various patients, sample banks not only enhance the scope of organoid research but also contribute to the advancement of precision medicine. Establishing a biospecimen repository requires careful consideration of sample quality and availability, encompassing key aspects such as collection, storage, management, and quality control (5, 22) (Figure 1).
Sample collection and processing: Tumor tissues obtained through clinical surgery or biopsy are processed quickly to prevent degradation. Samples are typically stored frozen in liquid nitrogen or at -80°C.
Storage and management systems: Modern biobanks utilize advanced database systems to manage and track sample information, recording each sample’s origin, pathological characteristics, genomic data, and clinical information to ensure accurate and prompt access to data.
Quality control and ethical considerations: Regular quality checks, such as DNA/RNA integrity and viability testing, are essential for maintaining sample quality. Additionally, obtaining informed consent from patients is necessary for the use of samples to ensure the ethical legitimacy of the research.
2.4.2 Sample banks in organoid studies
Sample repositories provide various applications for organoid research, facilitating the exploration of disease mechanisms and the development of therapeutic strategies (Figure 1).
Disease modeling: Patient tumor tissues from sample banks can be used to construct multiple organ models that accurately reflect the biological characteristics of a patient’s tumor, serving as crucial tools for studying the mechanisms of various cancer types.
Drug screening and development: Organoid models in sample banks enable high-throughput drug screening to evaluate the efficacy and safety of new drugs and identify potential resistance mechanisms (22).
Multi-omics analysis: Organoid models can be subjected to multi-omics analyses, including genome, transcriptome, and proteome, contributing to the identification of disease biomarkers and therapeutic targets, thereby providing valuable data for precision medicine (Figure 2).
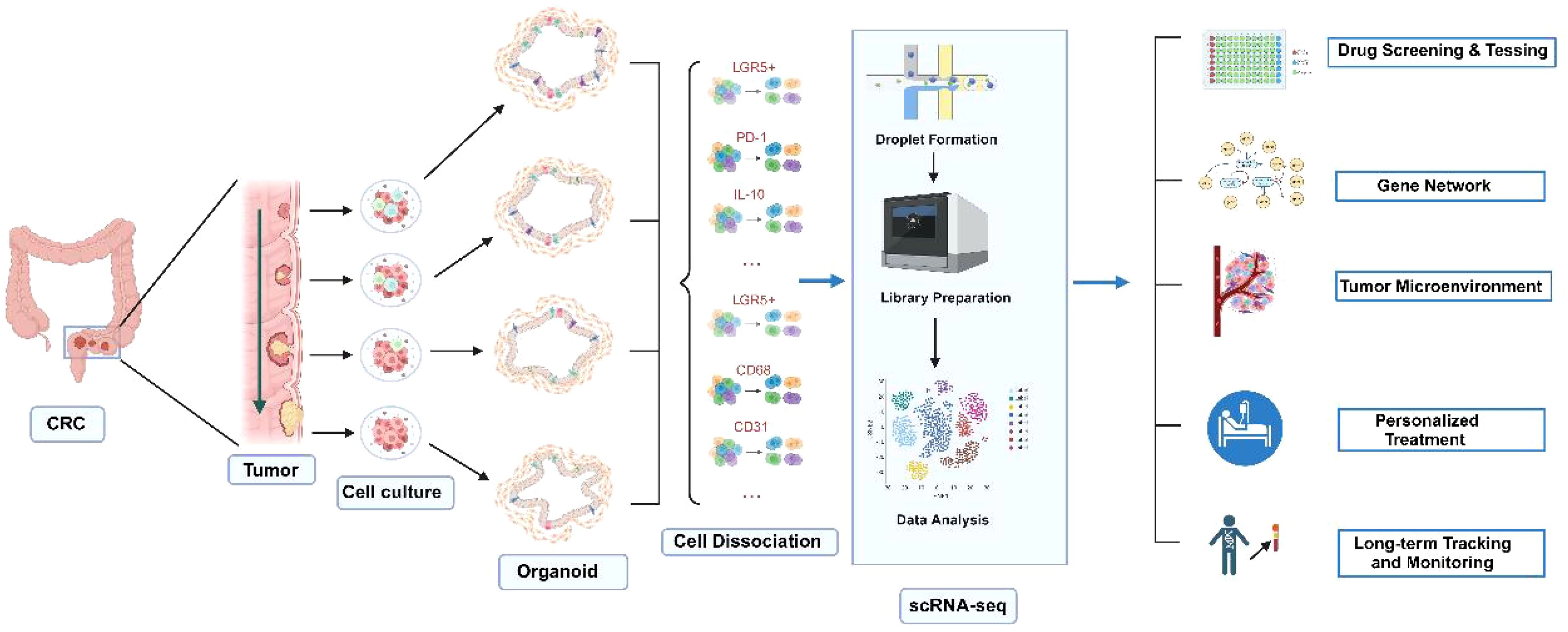
Figure 2. Applications of CRC organoids combined with single-cell sequencing technology. Normal and tumor tissues are used to generate corresponding organoids. By applying single-cell sequencing technology, the roles of various cell types in CRC development can be elucidated. This approach is valuable for drug screening and testing, gene function studies through gene editing, personalized treatment, as well as long-term tracking and monitoring. Created with Biorender.com (accessed on 15 August 2024).
2.4.3 Integration of sample banking and organoid studies
The integration of sample banking and organoid research is a key driver of personalized and precision medicine.
Personalized medicine: The diversity of patient samples provided by sample banks makes personalized medicine feasible. By simulating a patient’s pathological characteristics through organoid models, researchers can validate the effectiveness and safety of individualized treatment plans, advancing the realization of personalized treatment.
Collaborative innovation and resource sharing: Sample banks facilitate collaboration and resource sharing among research institutions, enabling scientists to access a broader range of sample types and data, fostering interdisciplinary research and innovation. This synergistic model can also improve the efficiency of research translation.
Clinical translation and application: Organoid research supported by sample banks accelerates the translation of basic research into clinical applications. The application of organoid modeling in clinical trials enhances the success rate of new drug development and reduces R&D costs.
A comparison of various metrics for the construction of tumor cell lines, spheroid culture, PDX, GETO, and PDTO models is presented in Table 1.
3 Application of organoids in colorectal cancer research
3.1 Disease modeling
3.1.1 Simulation of pathological features of colorectal cancer
Organoids not only retain the heterogeneity of patient tumors by mimicking key pathological features of colorectal cancer, such as multiple cell types, varying differentiation states, and complex tumor microenvironments, but also exhibit biological behaviors and drug responses in vitro that closely resemble those observed in vivo. By decellularizing human colonic tissue and reintroducing organoid cells carrying APC, KRAS, and TP53 mutations, researchers successfully recapitulated the development of colorectal cancer (42). Further transcriptomic analysis revealed significant activation of the Wnt/β-catenin and PI3K/AKT signaling pathways in the organoids, which closely matched the gene expression profiles observed in actual patient samples (21, 47, 48).
Cellular heterogeneity: Colorectal cancer organoids effectively reproduce the cellular heterogeneity of tumors, which is crucial for understanding tumor progression and treatment response (49). These organoid models facilitate in-depth studies of the interactions among different cell types during cancer progression and provide essential data for developing personalized treatment strategies (50, 51).
Tumor microenvironment: Organoid technology also enables the replication of the tumor microenvironment’s complexity, including key factors such as angiogenesis and immune cell infiltration. This capability allows researchers to gain a better understanding of how microenvironmental factors influence tumor growth and treatment response, leading to the development of more effective therapeutic strategies (52).
3.1.2 Application of genome editing in disease modeling
Creation of mutation models: Using CRISPR-Cas9 technology, researchers can precisely introduce specific gene mutations (knock-in) into organoids to study their roles in colorectal cancer initiation and progression (38). This approach is pivotal for deepening our understanding of cancer’s genetic underpinnings and for developing novel targeted therapies (Figure 3).
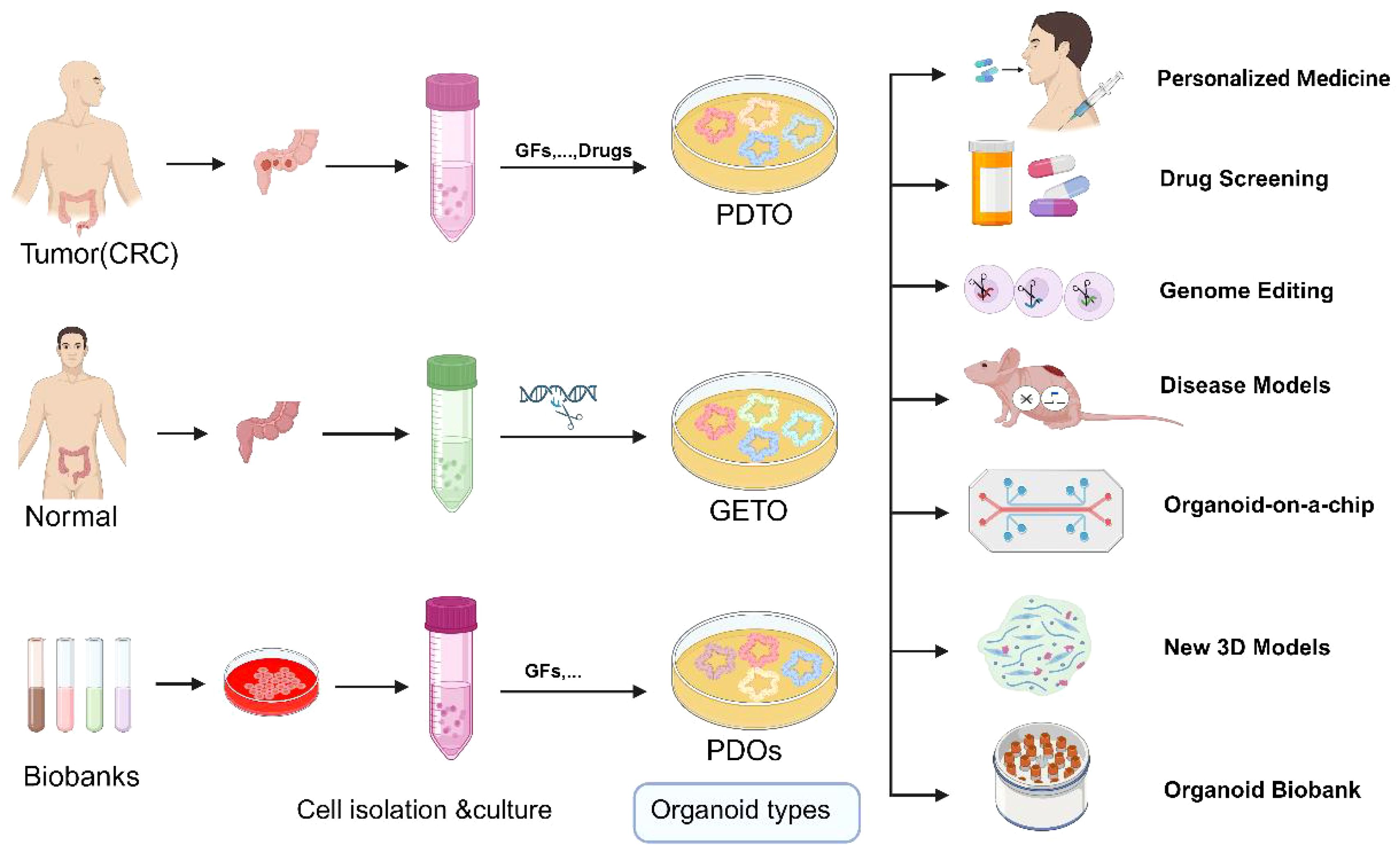
Figure 3. Schematic representation of the application of normal, tumor organoids and Biobank. GETO and PDTO organoid models play a pivotal role in advancing personalized medicine, drug screening, gene editing, disease modeling, organoid-on-a-chip platforms, the development of novel 3D models, and the establishment of organoid biobanks. PDTX, Patient-derived tumor xenograft model; GETO, Genetically modified tumor organoids; PDTO, Patient-derived organoids. Created with Biorender.com (accessed on 15 August 2024).
Functional genomics research: Systematic knockdown or activation of specific genes using CRISPR technology allows the exploration of gene functions in cancer. This approach not only aids in identifying potential therapeutic targets and biomarkers but also provides molecular data to support precision medicine. Using CRISPR-Cas9 technology, human intestinal organoids were sequentially edited to introduce APC, TP53, KRAS, and SMAD4 mutations, successfully modeling the multistep progression of colorectal cancer (52). More than 50 gene-edited organoids were transplanted into mice, resulting in the formation of invasive adenocarcinomas (12). Genomic sequencing indicated that these organoids exhibited typical chromosomal instability (CIN) and highlighted the synergistic action of key genes during tumor progression (38) (Figure 2).
Research on drug resistance mechanisms: Genome editing can also be utilized to investigate drug resistance mechanisms. By simulating various drug exposure environments in organoids, researchers can elucidate how genetic changes lead to resistance to chemotherapy and targeted therapies, thus providing new directions and strategies to overcome drug resistance (11, 53) (Figure 2).
Gene editing in organoids using adenoviral and lentiviral transduction, combined with Cre-lox and CRISPR-Cas9 technologies, enables the investigation of specific genetic events in cancer development. Additionally, the integration of CRISPR technology with single-cell RNA sequencing techniques, such as Perturb-seq and CROP-seq, allows the elucidation of gene functions at the single-cell level and the study of multicellular interactions within the tumor microenvironment during tumorigenesis and development (54).
3.2 Drug screening
3.2.1 High-throughput drug screening
Organoids have emerged as an indispensable tool for drug screening due to their genetic and epigenetic stability and their highly similar gene expression profiles compared to primary tissues. Tumor organoids effectively capture tumor heterogeneity, meeting the demand for large-scale drug screening within short timeframes (55, 56) (Figure 3).
The Clevers Laboratory conducted the first medium-scale drug screening using its colorectal cancer (CRC) organoid biobank, demonstrating the feasibility of using patient-derived organoids (PDOs) for such tests and highlighting their potential as clinically relevant biomarkers of therapeutic response (57). Van De Wetering pioneered preclinical high-throughput drug screening with organoid libraries, successfully establishing PDOs from both primary and metastatic colorectal cancer lesions (22). They performed extensive comparisons of the genomic profiles of these organoids with their original tumors, showing that the organoids closely resembled the genetic change profiles of the tumors and enabling the identification of gene-drug correlations. For instance, CRC organoids with RNF43 mutations were sensitive to Porcupine inhibitors, while those with Kras mutations exhibited resistance to tyrosine kinase receptor inhibitors (22).
High-throughput drug screening was performed on 22 tumor organoids derived from 20 colorectal cancer patients, testing over 200 compounds. The results revealed that organoids harboring the KRASG13D mutation exhibited significant sensitivity to MEK inhibitors, while their response to standard chemotherapy drugs, such as 5-FU and irinotecan, was highly dependent on specific genetic backgrounds (41).
Kondo et al. screened 2,427 drugs using CRC patient-derived organoids, discovering that the organoids displayed varying sensitivities to different drugs (58). Additionally, Du et al. developed a higher-throughput organoid drug screening platform capable of efficiently completing screening tests for thousands of drugs, further validating the enormous potential of organoids in developing new therapeutic regimens (59). Shen et al. identified the Kruppel-like factor 5 inhibitor ML264 by conducting in vitro drug screening with patient-derived CRC organoids. ML264 overcame drug resistance by restoring apoptosis in CRC organoids and re-sensitizing them to oxaliplatin (60). USUI et al. constructed organoids expressing high levels of LGR5 and CD44 to study tumor cell drug resistance mechanisms and screen colorectal cancer drugs. Their findings indicated that combining Hedgehog signaling inhibitors (AY9944 and GANT61) with anticancer drugs could inhibit tumor cell stemness, reduce organoid colony formation, and reverse drug resistance, significantly improving patient outcomes (61).
The OncoTrack Collaboration established a large biobank comprising 106 tumors, 35 organoids, and 59 xenografts, testing 16 clinical drugs on both ex vivo and in vivo models (62). The study by Shen et al. reaffirms the potential of organoid models for high-throughput drug screening and contributes to advancing precision therapies.
3.2.2 New drug development and personalized treatment
Human tumor cell lines remain the primary targets for anticancer drug research. However, one of the significant obstacles in new drug development is translating laboratory results into clinical trials, largely because traditional 2D tumor cell lines do not accurately reflect the physiological characteristics of primary tumors. Organoids facilitate this process and reduce the costs associated with new drug development. Tumor tissues and normal tissues from the same patient can be cultured into organoids, enabling simultaneous drug sensitivity and toxicity tests. This dual approach allows for the identification of drugs that specifically kill tumor cells without being toxic to normal cells, thereby reducing adverse drug reactions (Figure 3). For instance, in drug development, tumor organoids can be used to test a drug’s target-specific killing ability, while organoids derived from liver, heart, and kidney tissues can assess hepatotoxicity, cardiotoxicity, and nephrotoxicity, respectively. This strategy ensures the selection of drugs that target tumor cells without harming normal tissues. Furthermore, by simulating APC, KRAS, TP53, and PIK3CA mutations in organoids, researchers have successfully screened and validated several targeted therapies specific to different genotypes. Genomic and transcriptomic analyses revealed that these drugs significantly inhibited the activation of MYC and Wnt signaling pathways, thereby suppressing organoid growth. This paper demonstrates that integrating patient-specific organoids with genomic data can effectively optimize personalized treatment strategies and expedite the development of new drugs (41). Researchers have developed a powerful drug screening system that identified 34 drugs with anti-CRC properties among 335 drugs. Drugs were classified into five modes of action through transcriptome analysis: differentiation induction, growth inhibition, metabolic inhibition, immune promotion response, and inhibition of cell cycle (3).
3.3 Research on drug resistance mechanisms
3.3.1 Mechanisms of chemotherapy resistance
Drug resistance remains one of the major challenges in colorectal cancer treatment, highlighting the importance of understanding the underlying mechanisms to develop more effective therapeutic strategies. Chemotherapy is a standard treatment for colorectal cancer, but many patients develop resistance during therapy, resulting in diminished efficacy.
Expression of multidrug resistance pumps: Cancer cells often excrete chemotherapeutic drugs by upregulating multidrug resistance proteins such as P-glycoprotein (P-gp) and multidrug resistance-associated proteins (MRP), thereby reducing intracellular drug concentrations (63). Organoid models can accurately mimic changes in the expression of these proteins, providing valuable insights into their role in drug resistance (53). The study found that organoids harboring the KRASG12D mutation were resistant to standard chemotherapy regimens, whereas those with the KRASG13D mutation exhibited varying degrees of drug sensitivity (64).
Apoptotic escape: Chemotherapeutic drugs typically induce cancer cell death by triggering apoptosis, but some cancer cells evade this process by regulating apoptotic signaling pathways, such as by upregulating Bcl-2 family proteins (65). Organoids retain the apoptotic regulatory properties of tumors, thereby serving as an ideal platform for the investigation of this escape mechanism (66).
Enhanced DNA repair: Cancer cells can resist chemotherapy-induced DNA damage by enhancing their DNA repair capabilities, often through upregulating key DNA repair enzymes like PARP (67). Organoid models are valuable for elucidating the role of these repair pathways in drug resistance and for devising novel strategies to overcome resistance (68).
3.3.2 Analysis of resistance to targeted therapy
Targeted therapies inhibit tumor growth by interfering with specific molecular pathways, but drug resistance remains a significant challenge.
Genetic mutations and altered therapeutic targets: Targeted drugs often focus on specific genetic mutations, but cancer cells can acquire new mutations or alter existing targets, leading to resistance. As an example, EGFR and KRAS mutations render targeted therapies ineffective in some colorectal cancer patients (69). The combined inhibition of PD-1, BRAF, and MEK has been used to treat colorectal cancer patients and organoid models with the BRAFV600E mutation (63, 70). Although the initial treatment achieved significant antitumor effects, some patients and organoids developed resistance during the later stages of therapy. Molecular analysis indicated that upregulation of EGFR signaling was the primary cause of this resistance (70).
Activation of paracrine signaling pathways: When primary signaling pathways in cancer cells are inhibited, these cells may continue to grow by activating alternative pathways. When the EGFR pathway is blocked, cancer cells may maintain growth by activating the MET or HER2 pathways (71). Organoid technology can be employed to study the redundancy of these signaling pathways and their role in drug resistance (52, 72).
Impact of the tumor microenvironment: The tumor microenvironment, including interactions between immune cells and stromal cells, significantly affects the efficacy of targeted drugs. Organoids can mimic these complex microenvironmental interactions, enabling researchers to investigate how drug resistance might be overcome by modulating the microenvironment (32).
3.4 Organoid chip technology
3.4.1 Principles of organoid chips
Organochips are miniature cell culture systems derived from the application of microfluidic devices in cell culture and bioanalysis. By utilizing micro-nanofluidic processing technology, a multifactorial and complex reaction system is integrated onto a chip, where the entire process of culture, experimentation, and analysis is completed under precise control. The objective is to replicate essential cellular structures and functions of human organs on a chip. Organoids on a chip serve as a platform to investigate the processes and mechanisms driving tumor progression and treatment resistance, including tumor cell migration and invasion, tumor heterogeneity, and interactions with the tumor microenvironment (TME), encompassing stromal components, immune cells, endothelial cells, and chemokines (27).
Microarrays are widely used for 3D high-throughput organoid culture. Microwell structures facilitate the rapid aggregation of stem cells, a crucial step in generating uniformly sized organoids. These structures not only organize the culture system into regular arrays, enhancing throughput, but also provide geometric constraints and microenvironmental cues that guide organoid development and maturation. Recent studies have aimed to optimize microwell substrate materials to further improve culture efficiency.
3.4.2 Applications in colorectal cancer research
Organoid microarray models have been developed to explore the interaction between tumor cells and blood vessels in colorectal cancer. Research on the activin-ALK 7 pathway revealed its key role in insufficient vascularization of tumors, affecting drug delivery and chemotherapy efficacy (73). Hu et al. combined organoid technology with microfluidics to develop a chip with integrated superhydrophobic microwell arrays, achieving drug sensitivity predictions in just one week (74). Maxim et al (75). developed a targeted organoid sequencing technology (TORNADO-seq) for monitoring the expression of large genes to assess cellular phenotypes in organoids. TORNADO-seq enables the analysis of cell mixtures and their differentiation status within the intestinal system, providing valuable insights into drug mechanisms of action. A gut-on-a-chip integrated with transepithelial electrical resistance (TEER) sensors and electrochemical sensors was successfully used to simulate the human intestinal microenvironment and monitor cellular physiology (76).
3.4.3 Advantages of organoid chips
A significant advancement in organoid microarray technology is the “fusion” of normal tissue organoids with tumor organoids to form “organ microchips.” This technology allows not only the observation of cell-cell interactions within the tumor but also the examination of overall organ-to-organ correlations. Such a system simulates the process of tumor metastasis in vivo and enables the visual detection of drug pharmacokinetics and pharmacodynamics, thereby enhancing drug screening success rates and mitigating ethical concerns associated with animal models. A GOC integrated with a carcinoembryonic antigen (CEA) biosensor was developed, it can detect CEA through antigen antibody specific binding, with high selectivity, excellent stability, and good reproducibility (77).Furthermore, the development of high-throughput automated organoid microarrays will contribute to the standardization of organoid culture processes.
4 Organoid applications in precision medicine
Organoids offer substantial potential in precision medicine, especially in the formulation of individualized treatment strategies. By accurately replicating the 3D architecture and microenvironment of a patient’s tumor, organoid technology provides a robust in vitro model for personalized therapy. Notable applications include:
Individualized drug screening: leveraging high-throughput screening of patient-derived organoids to identify the most effective therapeutic regimens.
Predicting treatment response: utilizing organoid-based assays to evaluate various drug combinations, enabling the prediction of patient responses and the optimization of treatment plan.
Disease modeling and gene editing: applying gene editing techniques and disease modeling in organoids to investigate the functional roles of specific genes in disease pathogenesis.
4.1 Development of individualized treatment strategies
4.1.1 Application of patient-specific organoids
Immunotherapy research is progressing rapidly, and the integration of organoid technology offers a powerful tool to advance our understanding of tumor immunity and escape mechanisms. This approach has the potential to significantly accelerate the development of immunotherapies and enhance the efficacy of precision cancer treatments. It is well established that bidirectional communication between tumor cells and both cellular components (e.g., fibroblasts, endothelial cells, immune cells) and non-cellular components (e.g., extracellular matrix) within the tumor microenvironment (TME) plays a crucial role in tumor initiation and progression (Figures 1, 2).
However, conventional organoid cultures typically contain only tumor cells and lack stromal and immune cells, necessitating the exogenous addition of immune cells to study tumor-immune cell interactions. Co-culture of organoids and immune cells reconstructs the tumor immune microenvironment, overcoming this limitation (36, 78). As an illustration, researchers constructed an organoid biobank from colorectal cancer patient tumor tissues, with these organoids retaining the genomic characteristics of the original tumors, including mutations in APC, TP53, and KRAS. High-throughput screening revealed that these patient-specific organoids could accurately predict responses to standard chemotherapy and targeted therapies. For instance, organoids carrying PIK3CA mutations showed significant sensitivity to PI3K inhibitors, providing strong support for the development of individualized treatment plans (22).
Organoid models simulating the tumor microenvironment (TME) can be categorized into initial TME models and reconstructed TME models, depending on the inclusion of exogenous immune and stromal cells. Initial TME models encompass microfluidic culture and air-liquid interface (ALI) culture. Microfluidic culture can preserve endogenous immune cell subpopulations from tumor tissues while allowing the addition of exogenous immune cells, such as CAR-T cells. In contrast, ALI culture is more practical due to its simplicity and the absence of specialized equipment requirements. They found that these cells could specifically kill EGFRvIII+ colorectal cancer organoids without affecting normal tissues, suggesting that the organoid-immune cell co-culture system could assess CAR-mediated tumor-specific cytotoxicity. Additionally, studies have demonstrated that peripheral T cells can acquire intraepithelial lymphocyte characteristics—including morphology, markers, and motility—when co-cultured with intestinal organoids (79, 80). However, further research is needed to fully align the immune microenvironment of organoids with in vivo conditions (81).
4.1.2 Predicting treatment response and optimizing treatment regimens
Tumor heterogeneity is a hallmark of cancer biology and a significant contributor to treatment failure. As a highly heterogeneous solid tumor, colorectal cancer presents a challenge in that only a small fraction of patients responds favorably to treatment (32). Colorectal cancer organoids offer a more accurate reflection of patient treatment sensitivity, providing reliable experimental data and new insights for clinical precision therapy. While organoids are an ideal platform for antitumor drug testing and personalized treatment, a standardized protocol for drug sensitivity testing has yet to be established. The most common approach involves assessing organoid cell viability at therapeutic doses (Figure 2).
In 2015, Weeber et al. (82) demonstrated that organoids from metastatic colorectal cancer patients retain 90% of somatic mutations with a DNA copy number correlation coefficient of 0.89, confirming their genetic fidelity to the original tumor. Organoids accurately predicted treatment responses in colorectal and gastroesophageal cancers with a positive predictive value of 88% and a negative predictive value of 100% (83), and they also effectively forecast radiotherapy sensitivity, enabling personalized treatment (84).
Organoids have shown high accuracy in predicting chemotherapy outcomes (85). Tiriac et al. (81) linked chemotherapy responses in pancreatic tumor-derived organoids with patient outcomes, while Ganesh et al. (86) found a significant correlation between organoid sensitivity and progression-free survival. Rectal cancer organoids also predicted chemotherapy and radiotherapy responses, with TP53 and BRAF mutations linked to higher radiotherapy resistance, confirmed clinically (86). However, Ooft et al. (87) faced significant challenges in predicting the efficacy of various chemotherapy regimens. The observed discrepancies between organoid sensitivity and clinical outcomes may be attributed to the low success rate of organoid culture and the suboptimal quality of the samples.
In summary, colorectal cancer (CRC) organoids have demonstrated a robust ability to predict treatment efficacy, underscoring their significant potential for clinical application. By tailoring individualized treatment regimens based on organoid-derived predictions, it is anticipated that drug resistance can be reduced, overtreatment avoided, and adverse effects minimized. This makes organoid technology a promising tool for integration into prospective clinical trials on a broader scale (Figure 2).
4.2 Application of single-cell technology in precision medicine
4.2.1 Cellular-level interpretation of individualized clinics
Single-cell technology reveals heterogeneity and dynamic changes among cells by analyzing genomic, transcriptomic, and epigenetic features at the individual cell level, providing detailed cellular information for disease diagnosis and treatment strategies in precision medicine.
Identify cell types associated with individual therapeutic response: Single-cell technology can identify cell subpopulations closely associated with therapeutic response. By analyzing single-cell RNA sequencing data from tumor samples, key cell types and gene expression patterns influencing drug efficacy can be identified (88) (Figure 3). The presence of immunosuppressive cell subpopulations in tumors has been found to predict the effectiveness of immunotherapy. Tumor-reactive T cells were generated by co-culturing organoids with patient-derived peripheral blood lymphocytes (6). These T cells demonstrated the ability to specifically target and kill colorectal cancer organoids with defective mismatch repair (dMMR). Further single-cell RNA sequencing analysis uncovered the diversity of T-cell subsets and their correlation with tumor-killing efficacy (6) (Figure 2).
Customized treatment plan optimization: Leveraging single-cell data, clinicians can refine and optimize personalized treatment plans. By gaining detailed insights into the responses of various cell types within a patient’s tumor, treatment regimens can be precisely tailored to the tumor’s unique characteristics, thereby enhancing treatment efficacy and minimizing adverse side effects (89).
4.2.2 Early detection of tumor progression and recurrence
Single-cell technology has important applications in early tumor detection. By recognizing specific cancer biomarkers, single-cell technology can diagnose the disease in its early stages, making it particularly useful for detecting tiny lesions and metastases that are difficult to identify using traditional methods. For instance, researchers utilized multivariate single-cell mass spectrometry flow analysis of heterogeneous organoids to uncover the specific signaling networks of different cell types involved in tumor progression. The analysis revealed that cells within the tumor microenvironment possessing certain post-translational modifications (PTMs) exhibited increased invasiveness and a higher potential for recurrence (90). The abnormal expression of microRNA-21 (miR-21) is closely related to the pathogenesis of cancer, but in the early stage of cancer, its serum concentration is very low. To solve this problem, a nano-genosensor based on the free-standing MWCNT electrode was successfully developed for measuring miR-21 (91).
Monitoring the risk of tumor recurrence after treatment: Single-cell analysis can monitor changes in the tumor microenvironment post-treatment and identify early signs of recurrence. By tracking dynamic changes in residual tumor cells, treatment strategies can be adjusted in a timely manner to prevent disease recurrence (92).
4.2.3 Assisted gene therapy and immunotherapy
Single-cell technology enables precise evaluation of gene editing effects, providing in-depth insights into editing efficiency and off-target effects. By analyzing single-cell genomic data from edited cells, researchers can verify the specificity and accuracy of gene editing to optimize gene therapy strategies (88).
Combining single-cell technology with organoids allows for the simulation of the tumor immune microenvironment and the study of immune cell-tumor cell interactions. This approach is crucial for developing and optimizing immunotherapy strategies, such as checkpoint inhibitors and CAR-T cell therapy. The dynamics of immune cells in organoids are closely related to clinical patient responses. A genome-wide CRISPR screen identified the Wnt-FZD5 signaling pathway as a viable drug target for RNF43 mutant pancreatic tumors. Investigators developed antibodies that specifically target FZD5 and validated their anti-tumor efficacy in both organoid and mouse models (93).
By integrating organoid and single-cell technologies, precision medicine is progressively achieving the goal of individualized diagnosis and treatment. This integration not only enhances our understanding of tumor biology but also provides patients with more precise and effective treatment options, laying the foundation for improving overall cancer treatment outcomes.
4.3 Challenges and opportunities for clinical translation
4.3.1 Data integration and analysis
Organoid research generates vast amounts of multi-omics data, including genomic, transcriptomic, proteomic, and metabolomic information. The cellular heterogeneity of colorectal cancer organoids was comprehensively analyzed using single-cell RNA sequencing. The study revealed significant differences in gene expression and drug response across various clones within the organoids (40). Integrating these heterogeneous data types is a complex task requiring standardization and integration to achieve a comprehensive understanding of biological systems (94).
Data Standardization: Given the diversity and heterogeneity of organoid data, it is crucial to develop uniform data standards and formats. This includes standardizing the entire process of data acquisition, storage, and analysis (53).
Data integration platforms: Developing flexible data integration platforms and tools is essential for effectively combining multiple data types and helping researchers identify new biological associations and potential therapeutic targets. Data analysis is a critical step in extracting meaningful insights from large datasets, and advanced analytical tools will accelerate the clinical application of organoid technologies.
Establishing a good data integration platform first requires accurate provision of patient clinical information, and secondly, clear and detailed records of organoid culture, passage, and cryopreservation should be kept for data extraction and further analysis. In addition, accurately recording and analyzing the morphology and structure of organoids is also an important aspect, as they can reflect their growth and development status. For example, the area of colorectal cancer organoids can indicate their survival ability. However, currently there are few automated tools and instruments used to analyze the growth status of organoids, and they still mainly rely on visual analysis or immunofluorescence staining observation after sectioning. With the rapid development of artificial intelligence, a series of excellent algorithms have emerged that can obtain detailed information and decipher image content. Li analyzed organ chips based on deep learning for data analysis, automation, and image digitization (95). More and more researchers are seeking methods to quantify and analyze organoids through AI, in order to achieve three-dimensional imaging, intelligent recognition, and analysis of organoids.
Artificial intelligence and machine learning: The application of Artificial Intelligence (AI) and Machine Learning technologies allows for a degree of automation in processing and analyzing organoid data, helping to identify complex biological patterns and predict drug responses.
Big data analytics: Utilizing big data analytics methods enables researchers to extract patterns and trends from large-scale datasets, which is particularly important for understanding disease mechanisms and developing personalized treatment plans.
4.3.2 Application in clinical trials
Organoid technology offers a physiologically relevant model for drug development and testing, significantly enhancing the efficiency and success of clinical trials. A study evaluated the efficacy of a combination therapy involving PD-1, BRAF, and MEK inhibitors in BRAFV600E mutant colorectal cancer. The clinical efficacy and safety of this therapy were validated by successfully translating findings from organoid studies into clinical trials (70). A clinical trail from Denmark enrolled 90 patients with metastatic colorectal cancer following progression on or after standard therapy and treated with a panel of drugs from organoids models based-drug screening. The results revealed that the primary endpoint was met, as half of the patients were without progression at two months (96). This clinical trial strongly proved that cancer patients may benefit from functional testing using tumor-derived organoids.
Drug screening and toxicity testing: Organoids can be used for high-throughput drug screening and toxicity testing, aiding in the identification of potential drug candidates and the optimization of drug combinations (53).
Individualized drug testing: With patient-derived organoids, clinical trials can more accurately mimic an individual patient’s tumor response, providing data to support personalized therapy. In a phase 2, single-center, open-label, non-comparative study (ClinicalTrials.gov, register NCT03251612), 90 patients with metastatic tumors who progressed after treatment were subjected to organoid culture, of which 44 were successfully established and drugs with anticancer activity were screened in vitro, and at least one drug was used for treatment. The results showed that 17 patients (50%) achieved a progression free survival of two months, exceeding the expected level (14 of 45;31%). This strongly suggests that in vitro sensitivity testing of organoids can effectively guide the selection of clinical anti-tumor drugs and improve patient benefits (97).
Biomarker identification: Organoids can be used to identify and validate biomarkers that predict therapeutic response, thereby optimizing patient selection strategies and improving the success of clinical trials (4, 98).
Simulation in early trials: By simulating the efficacy and toxicity of a drug in an organoid model, potential issues can be identified, reducing the risk of failure in the early stages of clinical trials (99).
The use of organoid technology allows for faster and more efficient clinical trials while significantly improving the success rate of new drug development. These advantages make organoids an indispensable tool in precision medicine research. However, despite their potential, challenges remain in translating organoid applications to clinical settings.
5 Limitations and future directions of organoid technologies
5.1 Technical limitations
5.1.1 Complexity and reproducibility of models
Complexity: The complex structure of organoids makes their culture and maintenance challenging. Variations in experimental conditions can lead to significant differences in organoid growth and differentiation, affecting the reliability of experimental results (12, 32).
Reproducibility: Inter-laboratory reproducibility is a critical issue due to the diversity of organoid sources and culture conditions. Different laboratories may use varying media, scaffold materials, and growth factors, resulting in greater variability in outcomes (96).
5.1.2 Differences from the internal environment
Simulation of the microenvironment: Organoids cannot fully replicate the complex microenvironment present in vivo, including interactions with the vascular, nervous, and immune systems. This limitation hinders the application of organoids in studying multicellular interactions and systemic diseases (32).
Physiological dynamic changes: Organoids are typically static models that lack the dynamic physiological changes observed in vivo, such as blood flow, mechanical stress, and metabolic activity. This deficiency limits their ability to model certain physiological processes (6, 14). ssQuadruple mutant organoids (APC, KRAS, TP53, SMAD4) did not exhibit metastatic behavior despite forming aggressive tumors in mice. This finding suggests that cancer development and progression are not solely driven by genetic mutations but also require the interplay of additional molecular and environmental factors (100).
To summarize, the limitations of organoid technologies are fourfold: (1) the success rate of organoid construction is often compromised by bacterial contamination, tumor characteristics, and specific culture media, making it difficult to establish organoids for some types; (2) organoids established from a single time-point sample only represent the state at that moment, whereas tumors in vivo are dynamically evolving; (3) organoids are inadequate at fully modeling the tumor microenvironment; and (4) organoids do not replicate the collaborative interactions between multiple organs in vivo, limiting their ability to provide insights into systemic biological processes.
5.2 Future directions
Although organoid models offer new perspectives for understanding carcinogenesis and developing therapeutic strategies, they still have inherent limitations. Compared to animal models, organoids lack functional vascular, nervous, and immune systems, which restricts their use in simulating the in vivo environment. Cancer development and therapeutic response involve complex processes such as vascularization, local hypoxia, and immune system activity, which organoids cannot fully replicate. For example, the standardization of culture techniques is crucial for the growth of tumor tissue-derived organoids and the accuracy of experimental results (101). Challenges such as preventing “contamination” in normal tissue-derived organoids and successfully introducing vascular systems into organoids remain unresolved. Additionally, extracellular matrix gels (e.g., Matrigel) may impede drug penetration, affecting the application of organoids in drug screening. These gels also suffer from batch-to-batch variability and lack components of the tumor microenvironment (TME) (102, 103).
5.2.1 Integration with other cutting-edge technologies
Microfluidics: The combination of organoids with microfluidics can provide a dynamic fluidic environment that mimics blood flow and nutrient transport in vivo. This integration makes organoids more physiologically relevant for drug screening and toxicity testing (104).
Artificial intelligence: The use of artificial intelligence and machine learning to analyze organoid-generated data enables the identification of complex biological patterns and drug responses, thereby accelerating the development of new drugs and personalized treatment regimens.
5.2.2 Standardization and large-scale production of organoids
Standardization: Developing uniform culture standards and protocols will improve the reproducibility of organoid experiments. This includes standardizing medium composition, scaffold materials, and culture conditions, which will help reduce inter-laboratory variation. To achieve standardization in organoid culture, it is necessary to clarify the sampling requirements for each type of organ, and standardize the composition of the culture medium and culture conditions as much as possible; And after successful cultivation, the consistency between the properties of the organoid and the parent tissue was determined through methods such as IHC and NGS. Taking colon cancer organoids as an example, for surgical removal of sample tissue, it is recommended that the size of fava beans be appropriate; For microscopic examination and puncture sample tissues, it is recommended to take at least 3 tissue samples with forceps or 3 tissue samples with coarse needles. The collected tumor tissue samples must be quickly stored in sterile tubes containing specialized preservation solution for tumor samples, and quickly transported to the laboratory at around 4°C for preservation. Growth factors such as Y-27632 to the culture medium should be added in the culture medium to maintain cell viability. It is recommended to use immunohistochemistry methods to identify tumor organoids, such as HE,CDX-2,STAB-2,CK8/18,etc (78, 105).
Scale-up production: High-throughput organoid culture technology needs to be developed to achieve large-scale production, meeting the demands of drug screening and clinical research. This advancement will not only enhance efficiency but also reduce research costs.
By addressing the limitations of current technologies and exploring new development directions, organoid technologies will continue to play a pivotal role in biomedical research, driving precision medicine toward broader clinical applications.
6 Conclusion
The application of colorectal cancer organoid technology in precision medicine has shown great potential. Organoid models can highly mimic multiple biological features of the original tumor, including gene expression, protein profile, metabolic activities and other aspects of the characteristics, thus providing a physiologically relevant in vitro model for disease research and personalized treatment. Through organoid culture, researchers can gain a deeper understanding of the pathophysiological characteristics of colorectal cancer and study key issues such as tumor heterogeneity, drug screening, and drug resistance mechanisms. These models can not only simulate the three-dimensional structure of tumors, but also reproduce the complex interactions of the tumor microenvironment, thus providing a solid foundation for new drug development and treatment optimization. In recent years, the combination of organoid technology with genome editing, single-cell technology, microfluidic technology and other cutting-edge technologies has further promoted the development of organoid research and enhanced the breadth and depth of its application in precision medicine.
However, despite the remarkable progress of organoid technology in colorectal cancer research, its application still faces many challenges. Technical complexity, standardization issues, and inter-laboratory reproducibility issues are all key factors affecting the widespread application of organoid research. In addition, organoids still have some limitations in mimicking the in vivo environment, such as the lack of a complete vascular system, nervous system, and immune system functions, which limits their potential application in studying systemic diseases. Therefore, future research should focus on solving these technical difficulties and enhancing the standardized and scaled production capacity of organoids by further combining cutting-edge technologies such as artificial intelligence and machine learning, so as to make their role in precision medicine more comprehensive and effective.
Overall, the development of organoid technology in colorectal cancer research is promising. By overcoming the existing technological limitations, organoids are expected to become an important tool for precision medicine, providing reliable support for the development of individualized treatment plans and new drug development. This process requires not only in-depth advancement of scientific research, but also multidisciplinary synergistic innovation to accelerate the clinical translational application of organoid technology.
Author contributions
YZ: Writing – original draft, Writing – review & editing. RM: Writing – original draft, Writing – review & editing. DS: Conceptualization, Data curation, Writing – original draft. HG: Conceptualization, Data curation, Formal analysis, Writing – original draft. SW: Writing – original draft. JZ: Data curation, Writing – original draft. XW: Conceptualization, Data curation, Writing – original draft. FL: Writing – review & editing, Conceptualization. XL: Writing – review & editing. WS: Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Beijing Medical Award Foundation (YXJL-2020-0785-1016) to WS and Beijing Science and Technology Innovation Medical Development Foundation to DS (KC2023-JX-0186-FM052).
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-a Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Argiles G, Tabernero J, Labianca R, Hochhauser D, Salazar R, Iveson T, et al. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2020) 31:1291–305. doi: 10.1016/j.annonc.2020.06.022
3. Mao Y, Wang W, Yang J, Zhou X, Lu Y, Gao J, et al. Drug repurposing screening and mechanism analysis based on human colorectal cancer organoids. Protein Cell. (2024) 15:285–304. doi: 10.1093/procel/pwad038
4. Avci CB, Bagca BG, Shademan B, Takanlou LS, Takanlou MS, Nourazarian A. The future of cancer therapy: exploring the potential of patient-derived organoids in drug development. Front Cell Dev Biol. (2024) 12:1401504. doi: 10.3389/fcell.2024.1401504
5. Fang XC, Shu L, Chen TL, Zhao XL, Yang LC, Dou TT, et al. Organoids derived from patients provide a new opportunity for research and individualized treatment of Malignant peritoneal mesothelioma. Mol Cancer. (2024) 23:12. doi: 10.1186/s12943-023-01901-z
6. Yan SC, He YX, Zhu YH, Ye WF, Chen Y, Zhu C, et al. Human patient derived organoids: an emerging precision medicine model for gastrointestinal cancer research. Front Cell Dev Biol. (2024) 12:1384450. doi: 10.3389/fcell.2024.1384450
7. Kiwaki T, Kataoka H. Patient-derived organoids of colorectal cancer: A useful tool for personalized medicine. J Personal Med. (2022) 12(5):695. doi: 10.3390/jpm12050695
8. Li H, Chen Z, Chen N, Fan Y, Xu Y, Xu X. Applications of lung cancer organoids in precision medicine: from bench to bedside. Cell Commun Signaling. (2023) 21(1):350. doi: 10.1186/s12964-023-01332-9
9. Wen J, Liu F, Cheng Q, Weygant N, Liang X, Fan F, et al. Applications of organoid technology to brain tumors. CNS Neurosci Ther. (2023) 29:2725–43. doi: 10.1111/cns.14272
10. Yan HHN, Siu HC, Ho SL, Yue SSK, Gao Y, Tsui WY, et al. Organoid cultures of early-onset colorectal cancers reveal distinct and rare genetic profiles. Gut. (2020) 69:2165–79. doi: 10.1136/gutjnl-2019-320019
11. Patricio D, Santiago J, Mano JF, Fardilha M. Organoids of the male reproductive system: Challenges, opportunities, and their potential use in fertility research. Wires Mech Dis. (2023) 15(2):e1590. doi: 10.1002/wsbm.1590
12. Ma X, Wang Q, Li G, Li H, Xu S, Pang D. Cancer organoids: A platform in basic and translational research. Genes Dis. (2024) 11:614–32. doi: 10.1016/j.gendis.2023.02.052
13. de Vries NL, van de Haar J, Veninga V, Chalabi M, Ijsselsteijn ME, Ploeg Mvd, et al. gammadelta T cells are effectors of immunotherapy in cancers with HLA class I defects. Nature. (2023) 613:743–50. doi: 10.1038/s41586-022-05593-1
14. Liu Y, Wang D, Luan Y, Tao B, Li Q, Feng Q, et al. The application of organoids in colorectal diseases. Front Pharmacol. (2024) 15:1412489. doi: 10.3389/fphar.2024.1412489
15. Fan Y, Bai B, Liang YT, Ren Y, Liu YX, Zhou FL, et al. Proteomic profiling of gastric signet ring cell carcinoma tissues reveals characteristic changes of the complement cascade pathway. Mol Cell Proteom. (2021) 20:100068. doi: 10.1016/j.mcpro.2021.100068
16. Ren XX, Chen WK, Yang QX, Li XX, Xu LX. Patient-derived cancer organoids for drug screening: Basic technology and clinical application. J Gastroenterol Hepatol. (2022) 37:1446–54. doi: 10.1111/jgh.15930
17. Sprangers J, Zaalberg IC, Maurice MM. Organoid-based modeling of intestinal development, regeneration, and repair. Cell Death Differ. (2021) 28:95–107. doi: 10.1038/s41418-020-00665-z
18. Barbachano A, Fernandez-Barral A, Bustamante-Madrid P, Prieto I, Rodriguez-Salas N, Larriba MJ, et al. Organoids and colorectal cancer. Cancers (Basel). (2021) 13(11):2657. doi: 10.3390/cancers13112657
19. Bergin CJ, Benoit YD. Protocol for serial organoid formation assay using primary colorectal cancer tissues to evaluate cancer stem cell activity. STAR Protoc. (2022) 3:101218. doi: 10.1016/j.xpro.2022.101218
20. Kozlowski MT, Crook CJ, Ku HT. Towards organoid culture without Matrigel. Commun Biol. (2021) 4:1387. doi: 10.1038/s42003-021-02910-8
21. Chen HJ, Wei Z, Sun J, Bhattacharya A, Savage DJ, Serda R, et al. A recellularized human colon model identifies cancer driver genes. Nat Biotechnol. (2016) 34:845–51. doi: 10.1038/nbt.3586
22. van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. (2015) 161:933–45. doi: 10.1016/j.cell.2015.03.053
23. Novelli G, Spitalieri P, Murdocca M, Centanini E, Sangiuolo F. Organoid factory: The recent role of the human induced pluripotent stem cells (hiPSCs) in precision medicine. Front Cell Dev Biol. (2022) 10:1059579. doi: 10.3389/fcell.2022.1059579
24. van Husen LS, Katsori AM, Meineke B, Tjernberg LO, Schedin-Weiss S, Elsässer SJ. Engineered human induced pluripotent cells enable genetic code expansion in brain organoids. Chembiochem. (2021) 22:3208–13. doi: 10.1002/cbic.202100399
25. Kyprianou N, Calderon-Gierszal EL, Prins GS. Directed differentiation of human embryonic stem cells into prostate organoids in vitro and its perturbation by low-dose bisphenol A exposure. PloS One. (2015) 10(7):e0133238. doi: 10.1371/journal.pone.0133238
26. Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. (2013) 340:1190–4. doi: 10.1126/science.1234852
27. Teriyapirom I, Batista-Rocha AS, Koo BK. Genetic engineering in organoids. J Mol Med (Berl). (2021) 99:555–68. doi: 10.1007/s00109-020-02029-z
28. Li C, Chiu MC, Yu YF, Liu XJ, Xiao D, Huang JJ, et al. Establishing human lung organoids and proximal differentiation to generate mature airway organoids. Jove-Journal Visual Experiments. (2022) 23(181):e63684. doi: 10.3791/63684
29. Zhao Y, Zhang B, Ma Y, Zhao F, Chen J, Wang B, et al. Colorectal cancer patienttal68436 2D and 3D models efficiently recapitulate interi and intratumoral heterogeneity. Adv Sci. (2022) 9:15. doi: 10.1002/advs.202201539
30. Jung M, Ghamrawi S, Du EY, Gooding JJ, Kavallaris M. Advances in 3D bioprinting for cancer biology and precision medicine: from matrix design to application. Adv Healthc Mater. (2022) 11:e2200690. doi: 10.1002/adhm.202200690
31. Chiu MC, Li C, Liu X, Song W, Wan Z, Yu Y, et al. Human nasal organoids model SARS-coV-2 upper respiratory infection and recapitulate the differential infectivity of emerging variants. mBio. (2022) 13:e0194422. doi: 10.1128/mbio.01944-22
32. Polak R, Zhang ET, Kuo CJ. Cancer organoids 2.0: modelling the complexity of the tumour immune microenvironment. Nat Rev Cancer. (2024) 24:523–39. doi: 10.1038/s41568-024-00706-6
33. Peuker K, Muff S, Wang J, Kunzel S, Bosse E, Zeissig Y, et al. Epithelial calcineurin controls microbiota-dependent intestinal tumor development. Nat Med. (2016) 22:506–15. doi: 10.1038/nm.4072
34. Kato M, Sasaki T, Inoue T. Current experimental human tissue-derived models for prostate cancer research. Int J Urol. (2020) 28:150–62. doi: 10.1111/iju.14441
35. Wang Y, Li S, Li C. Clinical features, immunopathogenesis, and therapeutic strategies in vitiligo. Clin Rev Allergy Immunol. (2021) 61:299–323. doi: 10.1007/s12016-021-08868-z
36. Cattaneo CM, Dijkstra KK, Fanchi LF, Kelderman S, Kaing S, van Rooij N, et al. Tumor organoid-T-cell coculture systems. Nat Protoc. (2020) 15:15–39. doi: 10.1038/s41596-019-0232-9
37. Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell. (2018) 174:1586–98 e12. doi: 10.1016/j.cell.2018.07.009
38. Roper J, Tammela T, Cetinbas NM, Akkad A, Roghanian A, Rickelt S, et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat Biotechnol. (2017) 35:569–76. doi: 10.1038/nbt.3836
39. Yu YF, Zhou TT, Cao L. Use and application of organ-on-a-chip platforms in cancer research. J Cell Commun Signaling. (2023) 17:1163–79. doi: 10.1007/s12079-023-00790-7
40. Roerink SF, Sasaki N, Lee-Six H, Young MD, Alexandrov LB, Behjati S, et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature. (2018) 556:457–62. doi: 10.1038/s41586-018-0024-3
41. Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. (2015) 21:256–62. doi: 10.1038/nm.3802
42. Ping YY, Xu CH, Xu LW, Liao GM, Zhou Y, Deng CY, et al. Prioritizing gene cascading paths to model colorectal cancer through engineered organoids. Front Bioeng Biotechnol. (2020) 8:12. doi: 10.3389/fbioe.2020.00012
43. Li FJ, Wu Y, Yan YJ, Wu SZ, Zhu JY, Zhang GH, et al. Transcriptomic landscape of sodium butyrate-induced growth inhibition of human colorectal cancer organoids. Mol Omics. (2022) 18:754–64. doi: 10.1039/d2mo00127f
44. Wu Y, Li K, Li Y, Sun T, Liu C, Dong C, et al. Grouped-seq for integrated phenotypic and transcriptomic screening of patient-derived tumor organoids. Nucleic Acids Res. (2022) 50:e28. doi: 10.1093/nar/gkab1201
45. Neef SK, Janssen N, Winter S, Wallisch SK, Hofmann U, Dahlke MH, et al. Metabolic drug response phenotyping in colorectal cancer organoids by LC-QTOF-MS. Metabolites. (2020) 10(12):494. doi: 10.3390/metabo10120494
46. Chen L, Dai ZH, Ge CY, Huang D, Zhou X, Pan KL, et al. Specific metabolic response of patient-derived organoids to curcumin of colorectal cancer. J Chromatogr B-Anal Technol Biomed Life Sci. (2022) 1203:12360. doi: 10.1016/j.jchromb.2022.123260
47. Sclafani F, Rimassa L, Colombo P, Destro A, Stinco S, Lutman FR, et al. An exploratory biomarker study in metastatic tumors from colorectal cancer patients treated with bevacizumab. Int J Biol Mark. (2015) 30:E73–80. doi: 10.5301/jbm.5000097
48. Cho Y-H, Ro EJ, Yoon J-S, Mizutani T, Kang D-W, Park J-C, et al. 5-FU promotes stemness of colorectal cancer via p53-mediated WNT/β-catenin pathway activation. Nat Commun. (2020) 11(1):5321. doi: 10.1038/s41467-020-19173-2
49. Zhang R, Hu M, Chen HN, Wang XX, Xia ZL, Liu Y, et al. Phenotypic heterogeneity analysis of APC-mutant colon cancer by proteomics and phosphoproteomics identifies RAI14 as a key prognostic determinant in East Asians and Westerners. Mol Cell Proteom. (2023) 22(5):100532. doi: 10.1016/j.mcpro.2023.100532
50. Schumacher D, Andrieux G, Boehnke K, Keil M, Silvestri A, Silvestrov M, et al. Heterogeneous pathway activation and drug response modelled in colorectal-tumor-derived 3D cultures. PloS Genet. (2019) 15:27. doi: 10.1371/journal.pgen.1008076
51. Yang Q, Li MM, Yang XM, Xiao Z, Tong XY, Tuerdi A, et al. Flourishing tumor organoids: History, emerging technology, and application. Bioeng Trans Med. (2023) 8(5):e10559. doi: 10.1002/btm2.10559
52. Jin HR, Yang Q, Yang J, Wang FY, Feng JY, Lei LJ, et al. Exploring tumor organoids for cancer treatment. Apl Mater. (2024) 12:060602. doi: 10.1063/5.0216185
53. Betge J, Rindtorff N, Sauer J, Rauscher B, Dingert C, Gaitantzi H, et al. The drug-induced phenotypic landscape of colorectal cancer organoids. Nat Commun. (2022) 13:15. doi: 10.1038/s41467-022-30722-9
54. Dijkstra JJ, Neikes HK, Rezaeifard S, Ma XH, Voest EE, Tauriello DVF, et al. Multiomics of colorectal cancer organoids reveals putative mediators of cancer progression resulting from SMAD4 inactivation. J Proteome Res. (2023) 22:138–51. doi: 10.1021/acs.jproteome.2c00551
55. Lewis SK, Nachun D, Martin MG, Horvath S, Coppola G, Jones DL. DNA methylation analysis validates organoids as a viable model for studying human intestinal aging. Cell Mol Gastroenterol Hepatol. (2020) 9:527–41. doi: 10.1016/j.jcmgh.2019.11.013
56. Xiao Y, Kim D, Dura B, Zhang K, Yan R, Li H, et al. Ex vivo dynamics of human glioblastoma cells in a microvasculature-on-a-chip system correlates with tumor heterogeneity and subtypes. Adv Sci (Weinh). (2019) 6:1801531. doi: 10.1002/advs.201801531
57. Geurts MH, Gandhi S, Boretto MG, Akkerman N, Derks LLM, van Son G, et al. One-step generation of tumor models by base editor multiplexing in adult stem cell-derived organoids. Nat Commun. (2023) 14(1):4998. doi: 10.1038/s41467-023-40701-3
58. Kondo J, Ekawa T, Endo H, Yamazaki K, Tanaka N, Kukita Y, et al. High-throughput screening in colorectal cancer tissue-originated spheroids. Cancer Sci. (2019) 110:345–55. doi: 10.1111/cas.13843
59. Du Y, Li X, Niu Q, Mo X, Qui M, Ma T, et al. Development of a miniaturized 3D organoid culture platform for ultra-high-throughput screening. J Mol Cell Biol. (2020) 12:630–43. doi: 10.1093/jmcb/mjaa036
60. Norkin M, Ordonez-Moran P, Huelsken J. High-content, targeted RNA-seq screening in organoids for drug discovery in colorectal cancer. Cell Rep. (2021) 35:109026. doi: 10.1016/j.celrep.2021.109026
61. Auman JT, McLeod HL. Colorectal cancer cell lines lack the molecular heterogeneity of clinical colorectal tumors. Clin Colorectal Cancer. (2010) 9:40–7. doi: 10.3816/CCC.2010.n.005
62. Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol. (2014) 32:40–51. doi: 10.1038/nbt.2786
63. Post JB, Roodhart JML, Snippert HJG. Colorectal cancer modeling with organoids: discriminating between oncogenic RAS and BRAF variants. Trends Cancer. (2020) 6:111–29. doi: 10.1016/j.trecan.2019.12.005
64. van de Haar J, Ma X, Ooft SN, van der Helm PW, Hoes LR, Mainardi S, et al. Codon-specific KRAS mutations predict survival benefit of trifluridine/tipiracil in metastatic colorectal cancer. Nat Med. (2023) 29:605–14. doi: 10.1038/s41591-023-02240-8
65. Shen XH, Zhang YC, Xu ZQ, Gao H, Feng WQ, Li WC, et al. KLF5 inhibition overcomes oxaliplatin resistance in patient-derived colorectal cancer organoids by restoring apoptotic response. Cell Death Dis. (2022) 13:13. doi: 10.1038/s41419-022-04773-1
66. Rong L, Li Z, Leng X, Li H, Ma Y, Chen Y, et al. Salidroside induces apoptosis and protective autophagy in human gastric cancer AGS cells through the PI3K/Akt/mTOR pathway. Biomed Pharmacother. (2020) 122:109726. doi: 10.1016/j.biopha.2019.109726
67. Song YR, Kerr TD, Sanders C, Dai LS, Baxter SS, Somerville B, et al. Organoids and metastatic orthotopic mouse model for mismatch repair-deficient colorectal cancer. Front Oncol. (2023) 13:1223915. doi: 10.3389/fonc.2023.1223915
68. Li X, Krause A, Giardina G, Jiang ZW, Andreana M, Huntosová V, et al. Morpho-molecular functional drug response analysis of patient-derived organoids of colorectal cancer. In: Conference on Optical Biopsy XXII - Toward Real-Time Spectroscopic Imaging and Diagnosis. San Francisco, CA (2024). doi: 10.1117/12.3000978
69. Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, et al. EGFR Blockade Reverts Resistance to KRAS(G12C) Inhibition in Colorectal Cancer. Cancer Discovery. (2020) 10:1129–39. doi: 10.1158/2159-8290.CD-20-0187
70. Tian J, Chen JH, Chao SX, Pelka K, Giannakis M, Hess J, et al. Combined PD-1, BRAF and MEK inhibition in BRAF(V600E) colorectal cancer: a phase 2 trial. Nat Med. (2023) 29:458–66. doi: 10.1038/s41591-022-02181-8
71. Mangiapane LR, Nicotra A, Turdo A, Gaggianesi M, Bianca P, Di Franco S, et al. PI3K-driven HER2 expression is a potential therapeutic target in colorectal cancer stem cells. Gut. (2022) 71:119–28. doi: 10.1136/gutjnl-2020-323553
72. Vintila BI, Arseniu AM, Butuca A, Sava M, Birlutiu V, Rus LL, et al. Adverse Drug Reactions Relevant to Drug Resistance and Ineffectiveness Associated with Meropenem, Linezolid, and Colistin: An Analysis Based on Spontaneous Reports from the European pharmacovigilance database. Antibiotics-Basel. (2023) 12(5):918. doi: 10.3390/antibiotics12050918
73. Artegiani B, Hendriks D, Beumer J, Kok R, Zheng X, Joore I, et al. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat Cell Biol. (2020) 22:321–31. doi: 10.1038/s41556-020-0472-5
74. Hu YW, Sui XZ, Song F, Li YQ, Li KY, Chen ZY, et al. Lung cancer organoids analyzed on microwell arrays predict drug responses of patients within a week. Nat Commun. (2021) 12:14. doi: 10.1038/s41467-021-22676-1
75. Drost J, van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V, et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science. (2017) 358:234–38. doi: 10.1126/science.aao3130
76. Wang L, Han J, Su W, Li A, Zhang W, Li H, et al. Gut-on-a-chip for exploring the transport mechanism of Hg(II). Microsys Nanoeng. (2023) 9:2. doi: 10.1038/s41378-022-00447-2
77. Pan X, Chen J, Han J, Zhang W, Su W, Xu Z, et al. Critical suitability evaluation of caco-2 cells for gut-on-a-chip. ACS Appl Mater Interf. (2024) 16:51139–49. doi: 10.1021/acsami.4c11409
78. Ning RX, Liu CY, Wang SQ, Li WK, Kong X, He ZW. Application status and optimization suggestions of tumor organoids and CAR-T cell co-culture models. Cancer Cell Int. (2024) 24:98. doi: 10.1186/s12935-024-03272-x
79. Tamura H, Higa A, Hoshi H, Hiyama G, Takahashi N, Ryufuku M, et al. Evaluation of anticancer agents using patient-derived tumor organoids characteristically similar to source tissues. Oncol Rep. (2018) 40:635–46. doi: 10.3892/or.2018.6501
80. Hsu KS, Adileh M, Martin ML, Makarov V, Chen J, Wu C, et al. Colorectal cancer develops inherent radiosensitivity that can be predicted using patient-derived organoids. Cancer Res. (2022) 82:2298–312. doi: 10.1158/0008-5472.CAN-21-4128
81. Tiriac H, Belleau P, Engle DD, Plenker D, Deschenes A, Somerville TDD, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discovery. (2018) 8:1112–29. doi: 10.1158/2159-8290.CD-18-0349
82. Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T, et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U S A. (2015) 112:13308–11. doi: 10.1073/pnas.1516689112
83. Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. (2018) 359:920–26. doi: 10.1126/science.aao2774
84. Rogoz A, Reis BS, Karssemeijer RA, Mucida D. A 3-D enteroid-based model to study T-cell and epithelial cell interaction. J Immunol Methods. (2015) 421:89–95. doi: 10.1016/j.jim.2015.03.014
85. Zhao X, Yuan C, Wangmo D, Subramanian S. Tumor-secreted extracellular vesicles regulate T-cell costimulation and can be manipulated to induce tumor-specific T-cell responses. Gastroenterology. (2021) 161:560–74 e11. doi: 10.1053/j.gastro.2021.04.036
86. Ganesh K, Wu C, O'Rourke KP, Szeglin BC, Zheng YY, Sauvé CEG, et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat Med. (2019) 25:1607–14. doi: 10.1038/s41591-019-0584-2
87. Ooft SN, Weeber F, Dijkstra KK, McLean CM, Kaing S, van Werkhoven E, et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Trans Med. (2019) 11(513):eaay2574. doi: 10.1126/scitranslmed.aay2574
88. Wang R, Mao Y, Wang W, Zhou X, Wang W, Gao S, et al. Systematic evaluation of colorectal cancer organoid system by single-cell RNA-Seq analysis. Genome Biol. (2022) 23:106. doi: 10.1186/s13059-022-02673-3
89. Shin YJ, Jo EH, Oh Y, Kim DS, Hyun S, Yu A, et al. Improved drug-response prediction model of APC mutant colon cancer patient-derived organoids for precision medicine. Cancers (Basel). (2023) 15(23):5531. doi: 10.3390/cancers15235531
90. Qin X, Sufi J, Vlckova P, Kyriakidou P, Acton SE, Li VSW, et al. Cell-type-specific signaling networks in heterocellular organoids. Nat Methods. (2020) 17:335–42. doi: 10.1038/s41592-020-0737-8
91. Wang L, Li H, Su W, Zhang W, Xu Z, Wang J, et al. Fabrication of a freeication59 MWCNT electrode by electric field force for an ultrariceon592- microRNAeon nanooRNAeon592-0. Small. (2022) 18(25):e2201791. doi: 10.1002/smll.202201791
92. Shimokawa M, Ohta Y, Nishikori S, Matano M, Takano A, Fujii M, et al. Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature. (2017) 545:187–92. doi: 10.1038/nature22081
93. Steinhart Z, Pavlovic Z, Chandrashekhar M, Hart T, Wang X, Zhang X, et al. Genome-wide CRISPR screens reveal a Wnt-FZD5 signaling circuit as a druggable vulnerability of RNF43-mutant pancreatic tumors. Nat Med. (2017) 23:60–8. doi: 10.1038/nm.4219
94. Kim SC, Park JW, Seo HY, Kim M, Park JH, Kim GH, et al. Multifocal organoid capturing of colon cancer reveals pervasive intratumoral heterogenous drug responses. Adv Sci (Weinh). (2022) 9:e2103360. doi: 10.1002/advs.202103360
95. Li J, Chen J, Bai H, Wang H, Hao S, Ding Y, et al. An overview of organs-on-chips based on deep learning. Research. (2022) 2022:9869518. doi: 10.34133/2022/9869518
96. Wu Y, Ye W, Gao Y, Yi Z, Chen Z, Qu C, et al. Application of organoids in regenerative medicine. Stem Cells. (2023) 41:1101–12. doi: 10.1093/stmcls/sxad072
97. Jensen LH, Rogatto SR, Lindebjerg J, Havelund B, Abildgaard C, do Canto LM, et al. Precision medicine applied to metastatic colorectal cancer using tumor-derived organoids and in-vitro sensitivity testing: a phase 2, single-center, open-label, and non-comparative study. J Exp Clin Cancer Res. (2023) 42(1):115. doi: 10.1186/s13046-023-02683-4
98. Zhu J, Ji L, Chen Y, Li H, Huang M, Dai Z, et al. Organoids and organs-on-chips: insights into predicting the efficacy of systemic treatment in colorectal cancer. Cell Death Discovery. (2023) 9:72. doi: 10.1038/s41420-023-01354-9
99. Schnalzger TE, de Groot MH, Zhang C, Mosa MH, Michels BE, Roder J, et al. 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. (2019) 38(12):e100928. doi: 10.15252/embj.2018100928
100. Salahudeen AA, Kuo CJ. Toward recreating colon cancer in human organoids. Nat Med. (2015) 21:215–6. doi: 10.1038/nm.3818
101. Kopper O, de Witte CJ, Lohmussaar K, Valle-Inclan JE, Hami N, Kester L, et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat Med. (2019) 25:838–49. doi: 10.1038/s41591-019-0422-6
102. Zhao Y, Zhang B, Ma Y, Zhao F, Chen J, Wang B, et al. Colorectal cancer patient-derived 2D and 3D models efficiently recapitulate inter- and intratumoral heterogeneity. Adv Sci (Weinh). (2022) 9:e2201539. doi: 10.1002/advs.202201539
103. Zhou Z, van der Jeught K, Fang Y, Yu T, Li Y, Ao Z, et al. Author Correction: An organoid-based screen for epigenetic inhibitors that stimulate antigen presentation and potentiate T-cell-mediated cytotoxicity. Nat BioMed Eng. (2023). doi: 10.1038/s41551-023-01096-0
104. O'Rourke KP, Loizou E, Livshits G, Schatoff EM, Baslan T, ManChado E, et al. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat Biotechnol. (2017) 35:577–82. doi: 10.1038/nbt.3837
105. Fernando EH, Dicay M, Stahl M, Gordon MH, Vegso A, Baggio C, et al. A simple, cost-effective method for generating murine colonic 3D enteroids and 2D monolayers for studies of primary epithelial cell function. Am J Physiology-Gastrointest Liver Physiol. (2017) 313:G467–G75. doi: 10.1152/ajpgi.00152.2017
Keywords: colorectal, organoid, precision medicine, drug screening, single-cell sequencing, gene editing, CRC
Citation: Zhang Y, Meng R, Sha D, Gao H, Wang S, Zhou J, Wang X, Li F, Li X and Song W (2024) Advances in the application of colorectal cancer organoids in precision medicine. Front. Oncol. 14:1506606. doi: 10.3389/fonc.2024.1506606
Received: 05 October 2024; Accepted: 18 November 2024;
Published: 03 December 2024.
Edited by:
Wenbin Yu, Shandong University, ChinaReviewed by:
Li Wang, Qilu University of Technology, ChinaJing Sun, Charles Sturt University, Australia
Copyright © 2024 Zhang, Meng, Sha, Gao, Wang, Zhou, Wang, Li, Li and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Song, c29uZ3dlaUBzZGZtdS5lZHUuY24=; Xinyu Li, bGl4aW55dUBzZGZtdS5lZHUuY24=; Fuxia Li, bGlmdXhpYTEyM0AxMjYuY29t
 Yanan Zhang1,2
Yanan Zhang1,2 Ruoyu Meng
Ruoyu Meng Dan Sha
Dan Sha Huiquan Gao
Huiquan Gao Xinyu Li
Xinyu Li