- The First Affiliated Hospital of Guangxi University Of Chinese Medicine, Nanning, Guangxi, China
Gastric cancer (GC) is one of the leading causes of cancer-related death worldwide. N6-methyladenosine (m6A) modification is the most prominent epigenetic modification of eukaryotic mRNAs, and methyltransferase-like 3 (METTL3), a core component of the methyltransferase complex, catalyzes m6A modification. The results of previous studies indicate that the expression level of METTL3 is significantly elevated in gastric cancer tissues and cells. In addition, fluctuations in m6A levels induced by METTL3 are closely associated with the malignant progression of tumors as well as the poor prognosis of patients with gastric cancer. In this review, we focus on the potential mechanism of METTL3 in gastric cancer, and through our analysis, we suggest that targeting METTL3 could be a new therapeutic tool for treating GC.
Introduction
Gastric cancer (GC) is one of the top five malignant tumors worldwide and is very likely to cause death in cancer patients (1). The cause of stomach cancer varies by country. Asia, especially Japan and Mongolia, has the highest age-standardized rates of incidence and mortality, followed by Central and Eastern Europe and South America (2). In recent years, the incidence of GC in adults aged < 50 years has been gradually increasing in both low-risk and high-risk countries, with an estimated > 1 million new cases per year (3). Poor lifestyle habits and medical history, such as Hp infection, smoking status, smoking history, alcohol consumption status, alcohol consumption history, and pickled food intake, may contribute to the development of GC (4). N6-methyladenosine (m6A) is a form of mRNA modification that works by methylating the sixth nitrogen atom of RNA adenosine, thereby regulating RNA metabolism, which mostly occurs in eukaryotes. The methyltransferase complex is responsible for catalyzing m6A modification and relies on a subunit called methyltransferase-like 3 (METTL3) The core component of the methyltransferase complex is composed of METTL3, METTL4, and WTAP. METTL3 and METTL14 form a heterodimer in which WTAP plays a regulatory role. METTL3 has catalytic activity, and adenosylmethionine (SAM) binds to the catalytic domain of METTL3 through a gate loop to provide methyl. METTL14 provides structural support near the active site to help maintain the stability and integrity of the complex. METTL3 and METTL14 jointly recognize the substrate RNA (5). m6A is composed of 580 amino acids and contains two domains that are important for enzymatic activity: the zinc finger domain and the methyltransferase domain (6). METTL3 has been reported to be important in promoting tumorigenesis, accelerating the proliferation and metastasis of tumor cells, and promoting stem cell differentiation in a variety of cancers (7). METTL3 regulates RNA processing, splicing, metabolism, and ribonucleoprotein complex biogenesis through m6A modification (8). m6A in the nucleus promotes the maturation of pre-mRNA by regulating the splicing process. The transcribed mRNA is exported to the cytoplasm for translation under the action of m6A. m6A in the cytoplasm promotes the translation of mRNA in many ways, and can also maintain mRNA stability or promote mRNA degradation (9). Several studies have shown that abnormal changes in genes involved in RNA methylation may induce gastric cancer in healthy individuals or have adverse consequences in patients with gastric cancer (10). METTL3 promotes drug resistance and proliferation, invasion, migration, and metastasis in GC cells; inhibits apoptosis; and affects the expression of key components of various signaling pathways that regulate GC cell proliferation. In addition, METTL3 can be used as a reliable marker for clinicians to evaluate disease development trends in patients, and targeting METTL3 has gradually become a treatment modality for GC patients (11–13). Therefore, we summarize the research progress on METTL3 in GC tumorigenesis and propose that interfering with METTL3 expression may be a novel approach for treating patients with GC.
Results
METTL3 inhibits downstream gene expression in gastric cancer
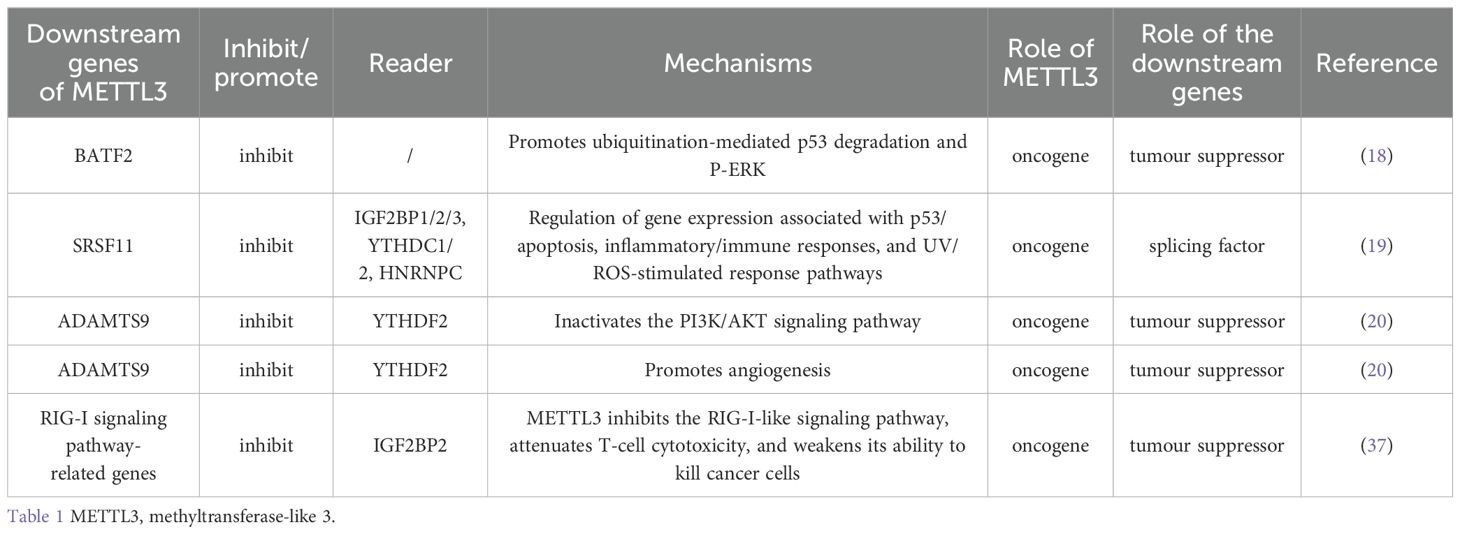
BATF2 contains a structural domain called the basic Leu zipper protein and is encoded by a gene located on human chromosome 11q (14). BATF2 blocks tumor progression through multiple mechanisms, such as the inhibition of epithelial–mesenchymal transition (EMT), CCN1 transcription, c-Jun expression, and angiogenesis (15–17). By catalyzing m6A modification of the MUT3 site in the 3’ untranslated region (3’UTR) of BATF2, METTL3 maintains BATF2 mRNA stability and inhibits BATF2 expression. BATF2 has been shown to inhibit GC cell growth, metastasis, and proliferation by prolonging the cell cycle both in vivo and in vitro. Reduced BATF2 expression is usually associated with poor prognosis in patients with GC. Human phospho-kinase microarray assays revealed that BATF2 overexpression decreased ERK phosphorylation (Thr202/Tyr204); conversely, BATF2 knockdown increased ERK phosphorylation. Western blotting (WB) and immunohistochemistry confirmed that high p-ERK levels were detected in patients with low BATF2 expression. WB demonstrated that the cell cycle proteins D1 (cyclin D1), MMP2, and MMP9 are targets of ERK signaling. These proteins and p-ERK were decreased and increased in BATF2-overexpressing and BATF2-knockdown cells, respectively. Overall survival of GC patients in an internal cohort classified according to BATF2 and p-ERK expression revealed that the level of p-ERK determines the prognostic value of BATF2. BATF2 regulates p53 protein levels via posttranslational modifications. BATF2 deletion promotes ubiquitylation-mediated p53 degradation, which can activate ERK signaling and promote ERK phosphorylation and the expression of MMP2, MMP9, and cyclin D1, accelerating GC cell proliferation, invasion, and metastasis (18).
RNA processing includes transcription initiation, splicing, and termination. Pre-mRNAs are spliced to generate mature mRNAs in one of the most important steps in eukaryotic RNA processing, and the splicing process is controlled by splicing factors. METTL3 downregulates the expression of splicing factors and is overexpressed in cancer tissue. Reduced SRSF11 and TRA2A expression is positively correlated with poor survival in patients with stomach adenocarcinoma. GSEA demonstrated that downregulation of the splicing factor RSF11 affects oncogenesis by regulating the expression of genes associated with the p53/apoptosis, inflammation/immune response, and UV/ROS stimulus response pathways. According to the m6A target database and eCLIP-seq data, several m6A-reading proteins, including IGF2BP1/2/3, YTHDC1/2, and HNRNPC, interact with SRSF11 mRNA. At the molecular level, the specific mechanism of SRSF11 expression related to the regulatory role of METTL3 needs to be elucidated in further studies (19). As a downstream effector of METTL3, ADAMTS9 is regulated by METTL3. METTL3 inhibits ADAMTS9 expression via YTHDF2-dependent mRNA degradation. When this occurs, the PI3K/AKT pathway is noticeably inactivated, and ADAMTS9-mediated downregulation of AKT and PI3K phosphorylation is attenuated, thereby promoting GC progression (20). METTL3 targets ANGPTL3 and downregulates its expression, which is conducive to tumor growth, proliferation, migration, and invasion (21) (Table 1). In this study, we noted that genes downstream of METTL3 are closely related to the stability of p53. The specific mechanism by which these genes regulate p53 is still unclear and needs to be further explored.
METTL3 promotes the expression of downstream genes
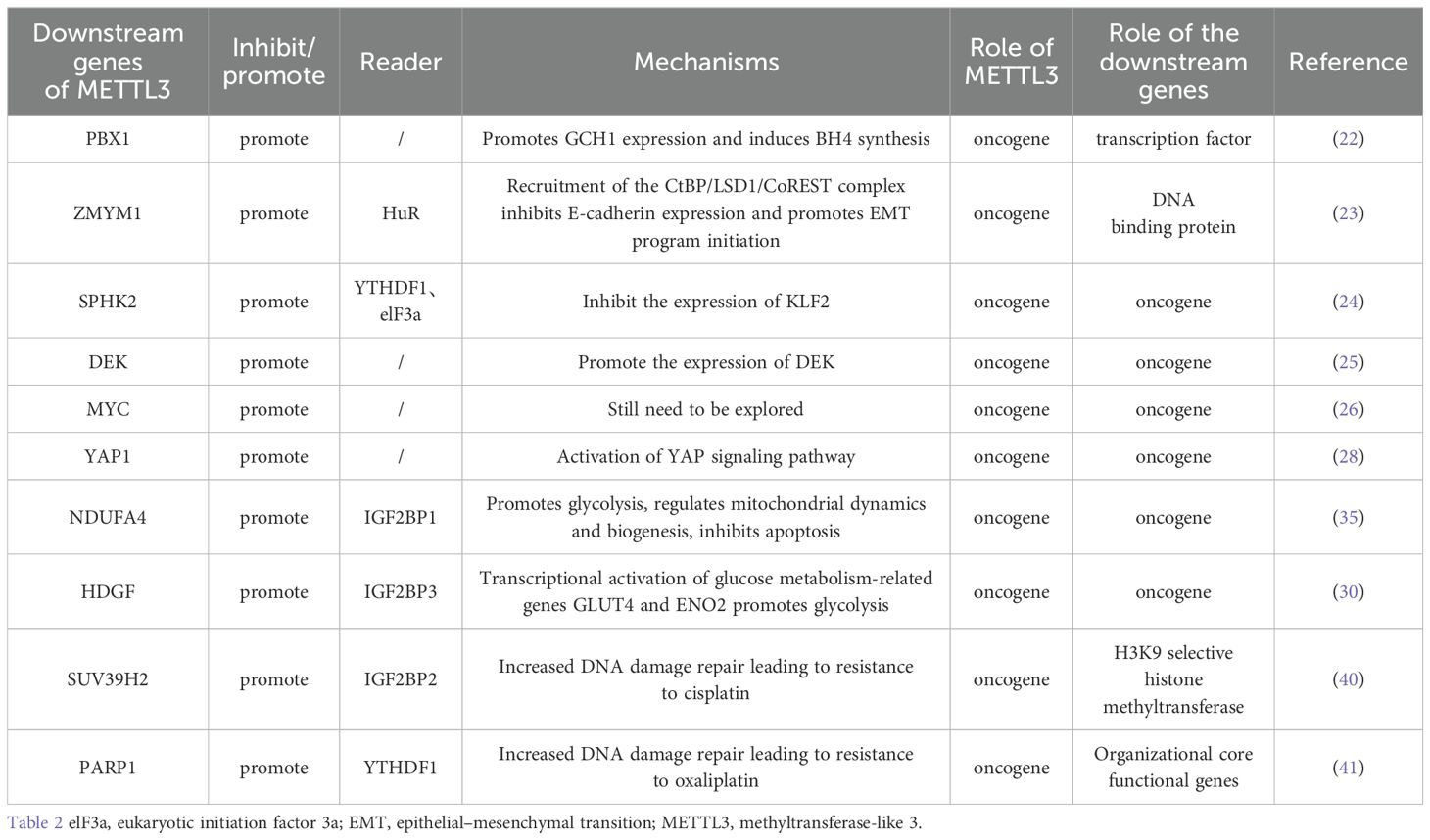
MeRIP-seq, RNA-seq, and TCGA analyses revealed that PBX1 is a downstream target of METTL3 methylation. METTL3-mediated m6A modification of the 5’UTR and 3’UTR of PBX1 mRNA promotes the expression of PBX1 mRNA by maintaining its stability, subsequently promoting the malignant progression of GC. GCH1 is among the genes with higher expression levels in GC tissues than in normal tissues, and GCH1 is the rate-limiting enzyme for BH4 biosynthesis. High PBX1 antibody enrichment was detected in the GCH1 promoter region, and METTL3-overexpressing GC cells with high GCH1 expression also presented high GCH1 expression. However, the expression of GCH1 decreased significantly as a result of PBX1 knockdown. In conclusion, GCH1 functions downstream of the METTL3-PBX1 axis. PBX1 binds to a site 500 bp away from the transcription start site of GCH1 and induces GCH1 synthesis. The GCH1 protein regulates BH4 biosynthesis in GC cells, leading to the proliferation, invasion, migration, and metastasis of GC. BH4 can regulate cellular metabolism and has antioxidant effects, which is why it can progress to gastric cancer, but its specific function remains to be explored (22). The mechanism by which METTL3 induces an increase in BH4 levels is well understood. However, further research is needed to explore the specific mechanism by which BH4 affects gastric cancer cells.
METTL3-mediated m6A methylation occurs at the stop codon of ZMYM1 mRNA and enhances ZMYM1 expression via a m6A-HuR-dependent pathway. ZMYM1 binds to E-cadherin by recruiting the CRS site of the CtBP/LSD1/CoREST complex to the promoter, thereby inhibiting E-cadherin expression. It also promotes EMT and metastasis in GC cells (23). The results of existing studies suggest that the expression level of E-cadherin decreases when the EMT program is initiated. At present, it is only inferred from this conclusion that the decrease in E-cadherin expression indicates that the EMT program is initiated, but the mechanism that causes the change in EMT marker levels is still unclear. Further research is needed to confirm this hypothesis.
SPHK is a protein-regulated cellular metabolic process in which phosphorylated sphingosine catalyzes the synthesis of sphingosine 1-phosphate. SPHK2 is associated with tumorigenesis, cancer progression, and chemoresistance. KLFs are TFs with Cys-2/His-2 zinc-finger-like structures that bind to the CACC or GT box. Abnormal expression of KLF2 has been detected in many tumors and has an important impact on tumorigenesis and development. METTL3 promotes m6A modification of SPHK2 target genes, and YTHDF1 binds to m6A-containing SPHK2 mRNA to form a translation initiation complex by interacting with eukaryotic initiation factor 3a (elF3a) to promote the translation of SPHK2 mRNA. These results suggest that the translation of SPHK2 is promoted by METTL3 and YTHDF1. SPHK2 promotes the ubiquitination of KLF2, which is facilitated by SPHK2-mediated KLF2 phosphorylation. Therefore, SPHK2 induced KLF2 degradation. K122 is the main ubiquitinated residue of KLF2, and Thr173 is the main phosphorylated residue of KLF2. Therefore, SPHK2 promotes the migration, proliferation, and invasion of GC cells (24). METTL3 binds to the 3’UTR of DEK to induce m6A modifications. METTL3 stabilizes DEK mRNAs to promote DEK expression. Thus, METTL3 promotes GC cell proliferation, migration, and metastasis (25). HBXIP can upregulate the expression of METTL3 and promote MYC expression (26). MYC is a downstream target of METTL3. MCM5 and MCM6 are key components of MYC. They assemble into complexes that initiate eukaryotic genome replication. High expression of MCM5 is correlated with poor clinicopathological parameters and survival METTL3 may regulate MYC through directly. For instance, promoting mRNA expression by maintaining the stability of mRNA. Besides, METTL3 may also indirectly regulate MYC. Therefore, the relationship between METTL3 and MYC in gastric cancer is still unclear and still needs further exploration (27). METTL3 modifies YAP1 to promote GC proliferation and metastasis. YAP1 is involved in the YAP signaling pathway through m6A. A new direction and target for GC treatment was developed with the discovery of the METTL3–YAP1 pathway (28) (Table 2). The mechanism by which METTL3 regulates DEK and SPHK2 expression has been elucidated. METTL3 maintains the stability of DEK mRNAs and enriches m6A-modified SPHK2 mRNAs by binding to YTHDF1 and elF3a. The transcript stability of MYC-associated genes decreased because of the depletion of METTL3. This conclusion is speculative, and the mechanism of action of METTL3 and MCM5/MCM6 should be investigated in the future.
Upstream regulators of METTL3
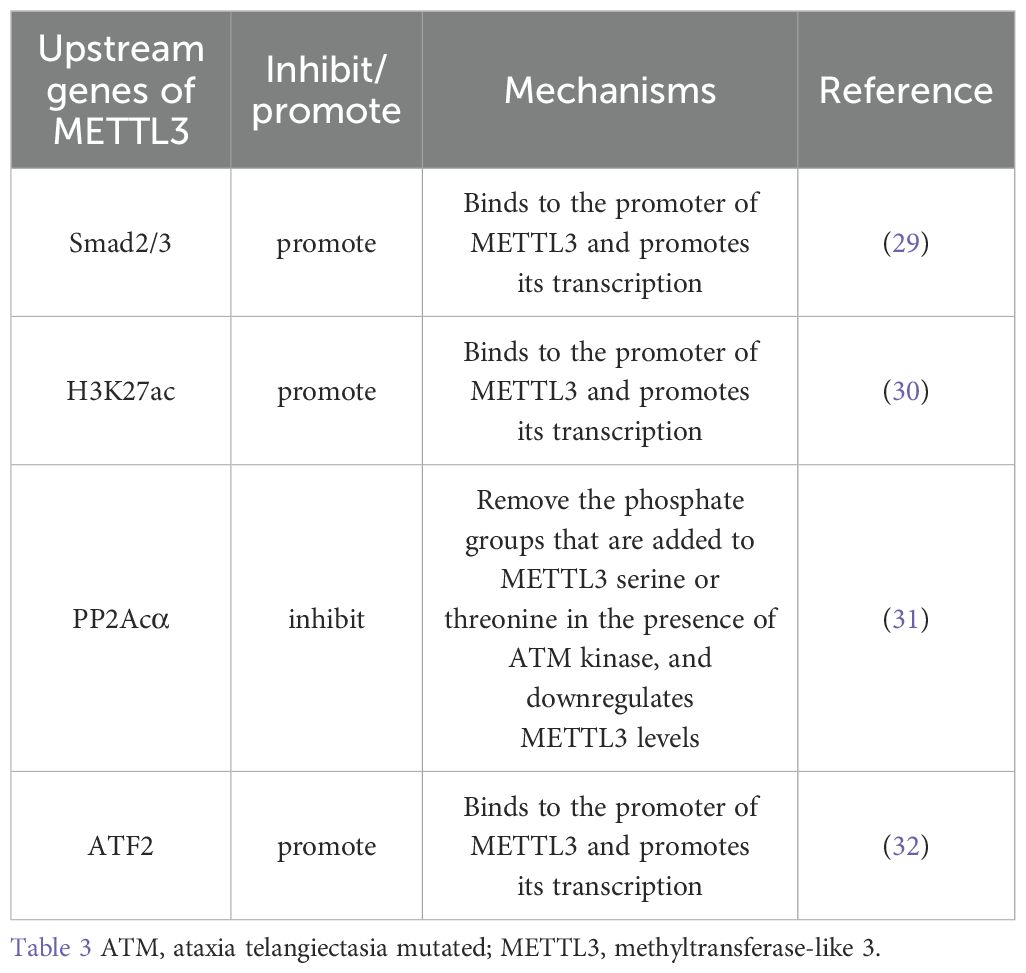
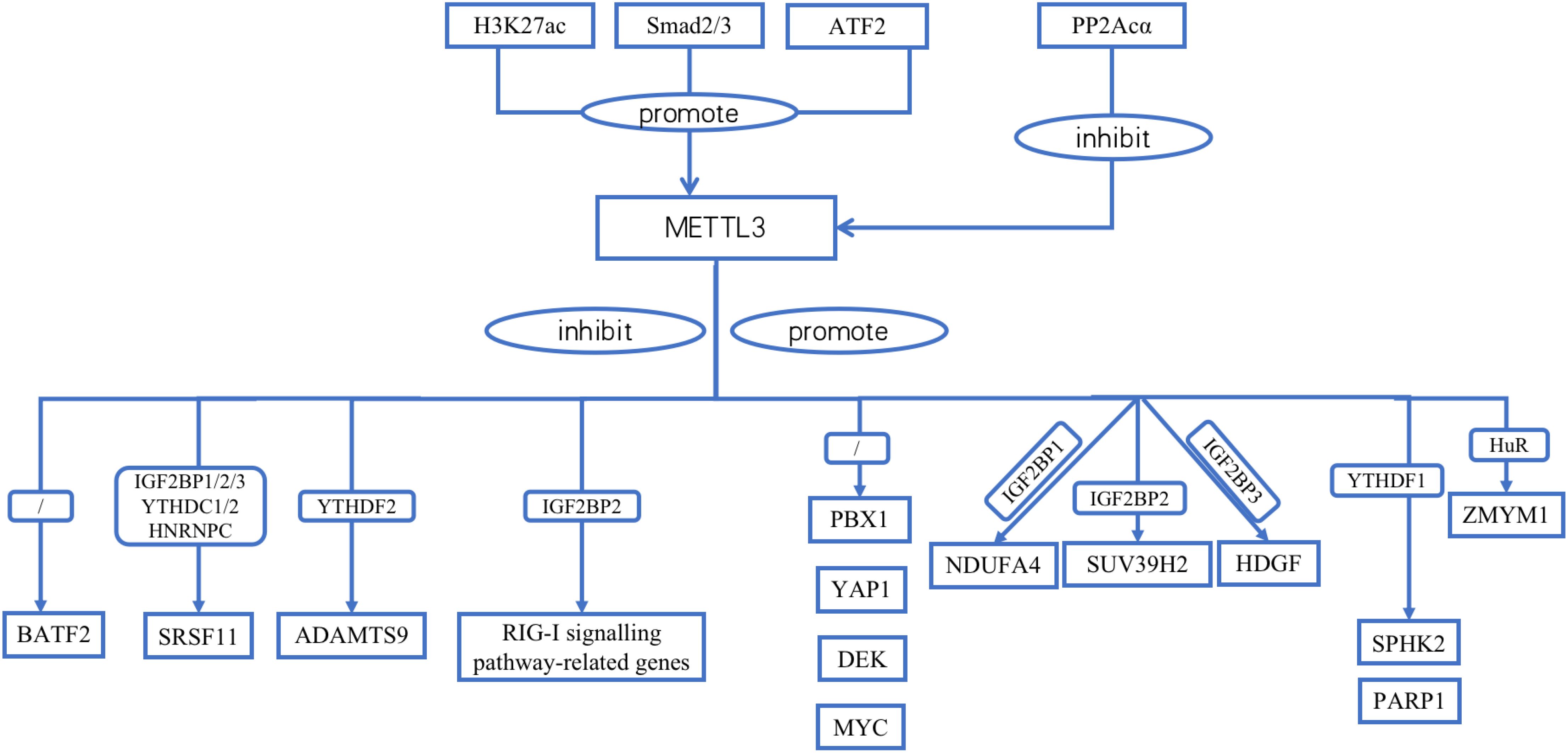
METTL3 contains many upstream regulators. Homologous box (HOX) genes are involved in cell proliferation, migration, metabolism, and apoptosis. HOXA10 binds to the promoter, increases the activity of the TGF-β2 promoter and activates the expression of TGF-β2 through transcriptional regulation. This, in turn, increases the secretion of TGF-β2 in the extracellular space, activates TGF-β/Smad2/3 signaling, and leads to a significant increase in Smad2/3 expression in the nucleus. Smad2/3 bind directly to the METTL3 promoter region in the nucleus. Smad2/3 is involved in the transcriptional regulation of METTL3, promotes METTL3 expression, and promotes m6A RNA modification in GC cells (29). H3K27 acetylation in GC cells is catalyzed by P300. P300 binds to acetylated H3K27, together with the METTL3 promoter region, positively regulating METTL3 expression (30). PP2Acα can remove the phosphoryl group added to the METTL3 serine 43 site (S43) via ataxia telangiectasia mutated (ATM) kinase and inhibits p-ATM. Moreover, PP2Acα can increase the activity of ATM kinases, resulting in the downregulation of METTL3 (31). The transcription factor ATF2 promotes the transcription of METTL3 by binding to the METTL3 promoter region. WB revealed that ATF2 overexpression increased cyclin D1 expression. However, METTL3 knockdown dampened this effect. The downregulation of ATF2 led to a decrease in cyclin D1 expression. This effect was counteracted by the upregulation of METTL3. Therefore, METTL3 is associated with cell cycle progression. Moreover, METTL3 activated cyclin D1 expression. ATF2 activates the METTL3/cyclin D1 signaling pathway to promote GC cell proliferation and inhibit GC cell apoptosis (32). PLAGL2 attaches to the upstream promoter of UCA1 to aid its transcription. UCA1 mRNA may function as a microRNA (miRNA) sponge that adsorbs miR-145-5p bound to YTHDF1 mRNA, upregulates YTHDF1 expression, promotes snail expression through METTL3 methylation, activates EMT, and promotes GC metastasis (33) (Table 3; Figure 1).
Effects of METTL3 on angiogenesis in gastric cancer
Angiogenesis is an important factor in tumor formation, rapid multiplication, and cancer progression. Vascular endothelial growth factor is at the forefront of this process. Vascular endothelial growth factor is secreted by tumor cells and lymphocytes and is an important component of tumor angiogenesis (34).
ADAMTS9 has been shown to be a downstream effector of METTL3. The degradation of ADAMTS9 is usually due to the action of the YTHDF2-dependent pathway. METTL3 represses ADAMTS9 expression by degrading YTHDF2-dependent mRNAs, limiting the ability of ADAMTS9 to promote GC progression via the PI3K/AKT pathway. ADAMTS9 can induce notable inactivation of the PI3K/AKT signaling pathway in gastric cancer cells by inhibiting its phosphorylation (20). The results of this study suggest that METTL3 acts as an oncogene to promote angiogenesis by targeting gastric cancer-related enzymes.
Effects of METTL3 on glycolysis in gastric cancer
Cell survival and other functional activities are energy-intensive processes. Glycolysis and oxidative metabolism release large amounts of energy for cellular use. Aerobic glycolysis, which involves the oxidative phosphorylation of mitochondria, affects the growth of tumor cells. METTL3 promotes m6A methylation of NDUFA4 through a reader called IGF2BP1. This mechanism allows IGF2BP1 to attach to the 3’-UTR of NDUFA4 and then promote NDUFA4 expression. In GC cells, NDUFA4 may promote glycolysis and oxidative metabolism. NDUFA4 can increase oxygen consumption and extracellular acidification rates, which are important indicators of glycolysis for assessing the metabolic state of the cell and energy metabolism. ENO1 and LDHA, two enzymes involved in glycolysis, also increase the expression of NDUFA4 and improve glycolysis and oxidative metabolism in GCs.
The ratio of the mitochondrial membrane potential (MMP) to reactive oxygen species (ROS) is an important signal for normal physiological functions of the cell and for cellular damage caused by environmental factors. Once the ROS level exceeds the capacity of endogenous antioxidant defenses, redox homeostasis is disrupted, which leads to structural or conformational changes in DNA, lipids, and proteins, ultimately leading to cell death. OPA1, p-Drp1, and PGC1α are associated with mitochondrial fusion, fission, and biogenesis. NDUFA4 overexpression increases the MMP; decreases ROS levels; increases OPA1, p-Drp1, and PGC1α levels; promotes mitochondrial fusion, fission, and biogenesis; and inhibits apoptosis by regulating mitochondrial dynamics and biogenesis in gastric cancer cells (35). Wang et al. reported that in METTL3-overexpressing BGC823 cells, the expression of the proliferation biomarker Ki-67 and the angiogenesis marker CD31 was significantly increased. Moreover, angiogenesis, human umbilical vein endothelial cell growth, and tube formation were significantly promoted by METTL3 overexpression. In experiments with tumor xenografts of the HGC-27 and NCI-N87 cell lines, METTL3 overexpression promoted increases in tumor volume and weight in vivo. In summary, METTL3 acts as an oncogene that helps GC cells generate blood vessels, proliferate, and metastasize. Its function is dependent on m6A catalytic activity (the catalytic mutant of METTL3 does not have any regulatory effects). Analysis of TCGA data revealed that H3K27 acetylation in GC cells is catalyzed by P300. The METTL3 promoter region is rich in P300 binding and H3K27ac signaling, and P300 positively regulates METTL3 transcription and expression. In summary, P300 binds to the promoter region of METTL3 and acetylates H3K27 to positively regulate METTL3 expression. HDGF was identified as a downstream target of METTL3, and its expression was increased by m6A modification. This process is mediated by METTL3 and is inseparable from the involvement of IGF2BP3. When an HDGF overexpression plasmid was transfected into METTL3 knockout cells via plasmid transfection, glycolytic activity was significantly increased (30). Additionally, a greater amount of lactic acid was produced. Mechanistically, HDGF binds to the promoter regions of genes related to glucose metabolism, including GLUT4 and ENO2. HDGF transcription subsequently activates GLUT4 and ENO2 expression, inducing glycolysis in GC cells. This ultimately facilitates GC progression and hepatic metastasis (30). Cellular energy metabolism requires the participation of proteins. Gene ontology enrichment revealed that these proteins were composed mainly of proteins whose expression was reduced under the influence of METTL3 overexpression. These proteins trigger glycolysis in GC cells and promote tumor progression by mediating mitochondria-associated oxidative phosphorylation (36). METTL3 addresses the energy and biosynthetic needs of the ongoing proliferation and metastasis of malignant tumors by regulating glycolysis-related enzymes. Additionally, NDUFA4 inhibits apoptosis by promoting mitochondrial fission.
Effects of METTL3 on the tumour microenvironment of gastric cancer
Exosomes are tiny vesicles released into the extracellular space by almost all cell types and contain DNA, mRNAs, proteins, lipids, miRNAs, and other bioactive substances that are unique to the original cell. These cargoes can build bridges for cells to communicate with each other and participate in various physiological and pathological processes. In the tumour microenvironment (TME), many interactions and communications occur between cells. Recently, increasing evidence has shown that xenobiotics play key roles in this process. THBS1, an angiogenesis inhibitor, is negatively associated with cancer progression. Cytokines are small-molecule proteins. When stimulated, immune and non-immune cells can synthesize and secrete cytokines. They are biologically active, can promote cell growth, and can regulate adaptive immunity. Cytokines can also be used to repair damaged tissues.
Interleukins, interferons, tumor necrosis factors, colony-stimulating factors, and growth factors are cytokines. Exosomes derived from GC cells are more likely to produce IFN-γ and TNF-α. THBS1 was found to stimulate cytotoxicity and cytokine production in Vγ9Vδ2 T cells. Thus, the toxicity of Vγ9Vδ2 T cells to cancer cells increases. Differentially expressed genes identified by RNA sequencing were associated with THBS1 expression. These genes are abundantly expressed in the RIG-I-like receptor signaling pathway. Similarly, in THBS1-OE-treated T cells, factors related to the RIG-I-like receptor signaling pathway were significantly increased in Vγ9Vδ2 T cells. Subsequent rescue experiments revealed that THBS1 regulated the function of Vγ9Vδ2 T cells, causing them to produce more cytokines. In addition, THBS1 can increase the toxicity of Vγ9Vδ2 T cells, thereby increasing their ability to kill GC cells. All the above is accomplished by THBS1 with the help of the RIG-I-like receptor signaling pathway. The SRAMP database was analyzed to confirm that m6A methylation occurs in genes involved in RIG-I signaling. THBS1 downregulated METTL3 in Vγ9Vδ2 T cells at the posttranscriptional level, resulting in increased expression of the reader IGF2BP2 and eraser FTO. The amount of METTL3 expressed in Vγ9Vδ2 T cells was approximately 4-fold greater than the amount of METTL3 expressed in FTOs. As a result, genes involved in the RIG-I signaling pathway are often methylated. THBS1 directly binds to METTL3 to reduce its expression level and the methylation of RIG-I signaling pathway genes. Genes related to the RIG-I signaling pathway are methylated by binding to the IGF2BP2 protein, which increases mRNA stability, promotes expression, activates the RIG-I signaling pathway, and regulates the function of Vγ9Vδ2 T cells. More cytokines subsequently increase the toxicity of Vγ9Vδ2 T cells, further promoting the synthesis and secretion of cytokines such as IFN-γ and TNF-α, thereby inhibiting cancer progression (37).
METTL3-mediated m6A modification and immunological infiltration are also strongly associated with the TME. The expression of PD-1 and PD-L1 is related to m6A regulator expression, according to Spearman correlation analysis. Using consensus clustering analysis, the GC samples were divided into two groups. These two groups of samples are involved in key signaling pathways, such as base excision, nucleotide excision and repair, and the cell cycle. Further experiments revealed that PD-L1 was more highly expressed in Cluster 1 than in Cluster 2. In addition, the infiltration of immune cells was greater in cluster 1. Thus, the number of m6A methylation regulators significantly affects the number of immune cells that infiltrate the TME. Genes that can affect the prognosis of patients with gastric cancer were first identified, and then a risk scoring model was established. This model was sufficiently convincing to predict the disease development status of patients with gastric cancer. WB demonstrated that METTL3 can increase the expression of PD-L1 in GC cells, but bioinformatics data suggest that the link between PD-L1 and METTL3 is weak; thus, m6A regulators may also be involved. The interactions between the TME and other variables need to be further explored (38). Different molecular subtypes of GC patients overexpress different core genes for METTL3 methylation, and the tumor mutational burden (TMB) and infiltration of immune cells are also not static. For patients with higher TMB, immunotherapy allows them to obtain greater therapeutic benefits and are more sensitive to the immune checkpoint inhibitor paclitaxel. Therefore, METTL3-mediated m6A methylation can influence the responsiveness of patients with GC to immunotherapy (39). Overall, METTL3 is a promising new therapeutic target for gastric cancer and may improve the efficacy of immunotherapy.
Effects of METTL3 on the drug resistance of gastric cancer
DNA double-strand breaks are a type of DNA damage, and chemotherapeutic drugs exert their cytotoxic effects through this process. Cisplatin (CDDP) causes DNA double-strand breaks by forming platinum−DNA adducts, which is the mechanism by which CDDP induces cancer cell death. Yang et al. demonstrated that METTL3 increases the expression of SUV39H2 through methylation. SUV39H2 modifies the promoter region of DUSP6 by trimethylation of histone H3K9 to inhibit its transcription, reverse the dephosphorylation of DUSP6, activate homologous recombination repair, and enhance DNA double-strand break repair. In summary, SUV39H-induced DNA damage repair is the main cause of CDDP resistance. Targeting SUV39H2 can increase the sensitivity of tumor cells to CDDP and could become a new chemotherapy adjuvant (40). Cancer stem cells constitute one of the main causes of cancer recurrence. CD133+ tumor cells with high PARP1 expression presented oxaliplatin resistance. Studies have shown that METTL3 promotes stable PARP1 expression through m6A modification. PARP1 activates the base excision repair pathway to promote DNA damage repair, resulting in resistance of CD133+ gastric cancer stem cells to oxaliplatin. These findings provide a new approach for the clinical treatment of oxaliplatin-resistant patients with high METTL3 levels (41). XRCC1, PARP1, and RAD51 are important components of DNA repair. Wang et al. reported that these genes are positively correlated with METTL3 expression. Therefore, METTL3 promotes oxaliplatin resistance in gastric cancer cells via the DNA damage repair process (42). These findings suggest that chemotherapy resistance in gastric cancer is closely related to DNA double-strand break repair. These findings provide a new direction for the treatment of GC. In addition to SUV39H2 and PARP1, METTL3 is a promising new therapeutic target in GC.
The relationship between METTL3 and non-coding RNAs in gastric cancer
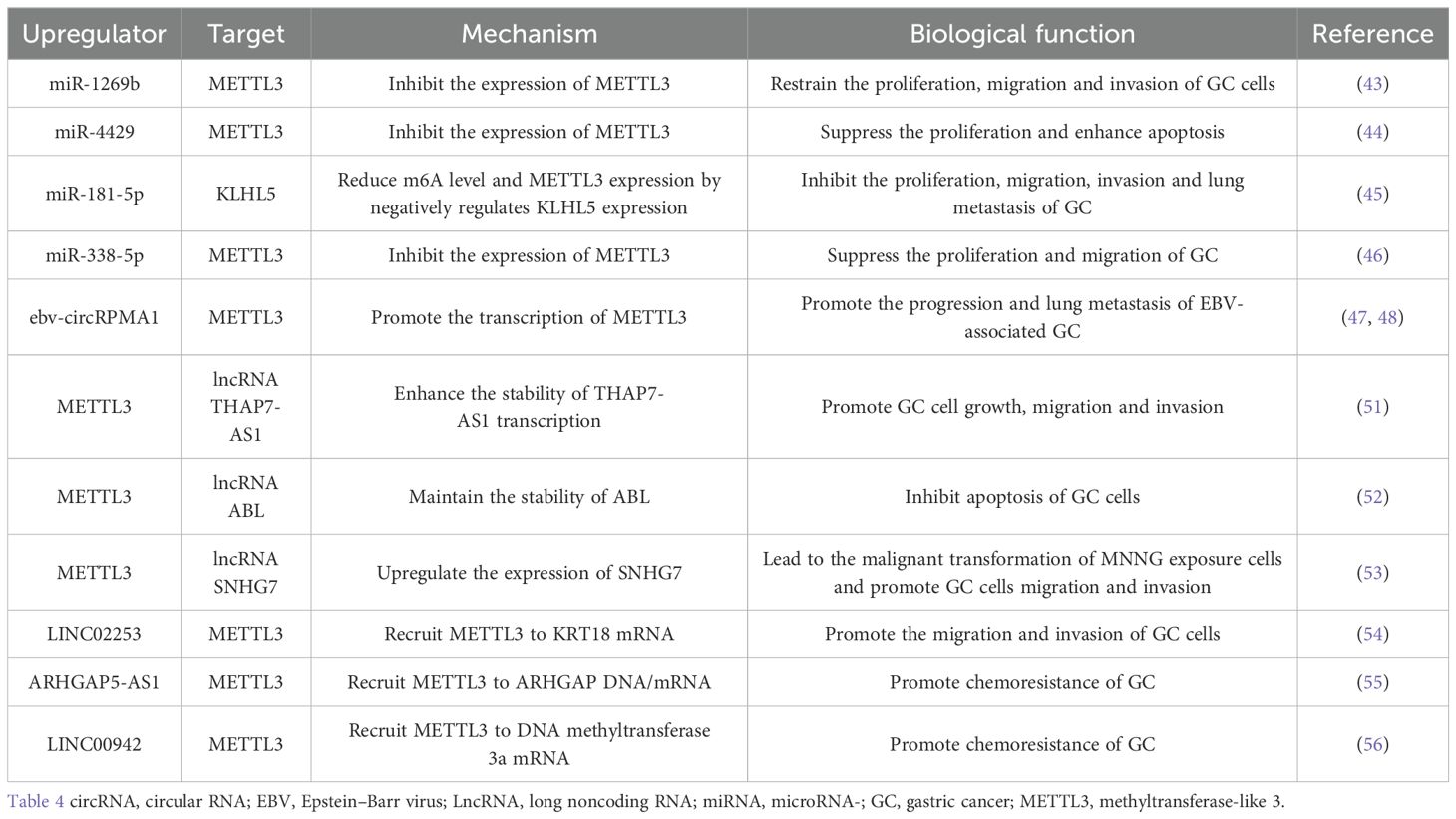
miRNAs are upstream regulators of METTL3 expression. miR-1269b can target the METTL3 3’-UTR and negatively regulate the expression of METTL3 in GC cells (43). miR-4429 inhibits the expression of METTL3 by targeting METTL3 and reducing the methylation level of SEC62 mRNA, thus playing a role in inhibiting GC cell proliferation and enhancing GC cell apoptosis (44). miR-181-5p negatively regulates KLHL5 mRNA expression by binding to the 3’-UTR of KLHL5, reducing m6A levels and METTL3 expression (45). EED reversed the low expression of METTL3 by inhibiting the expression of miR-338-5p (46).
In addition to miRNAs, METTL3 contains several other upstream regulators. Circular RNAs (circRNAs) are important regulators of tumorigenesis that modulate the malignant behavior of tumor cells. Small noncoding RNAs encoded by Epstein–Barr virus (EBV) are found in Epstein–Barr virus (EBV)-associated GC (EBVaGC). Moreover, EBV also produces circular RNAs (ebv-circRNAs). The complex formed by circRPMS1 with Sam68 and P53 is enriched in the promoter of METTL3 and regulates the malignant behavior of EBVaGC cells by promoting the transcription of METTL3 (47, 48). The Epstein–Barr virus circRNA does not function as a miRNA sponge, and it directly interacts with the Sam68 protein. Therefore, Sam68 is expected to become a new target for the treatment of gastric cancer. However, P53 is a cofactor that interacts with Sam68, and the relationship between METTL3 and p53 still needs to be studied.
The relationship between METTL3 and long non-coding RNAs in gastric cancer
Long noncoding RNAs (lncRNAs) are a class of noncoding RNAs that are more than 200 nucleotides long and do not have the ability to encode proteins. lncRNAs can bind to mRNAs, miRNAs, or proteins to regulate chromatin modification, transcription, and posttranscriptional and posttranslational regulation (49). lncRNAs have been shown to play critical roles in disease, viral infections, cell cycle differentiation, and metabolic processes (50).
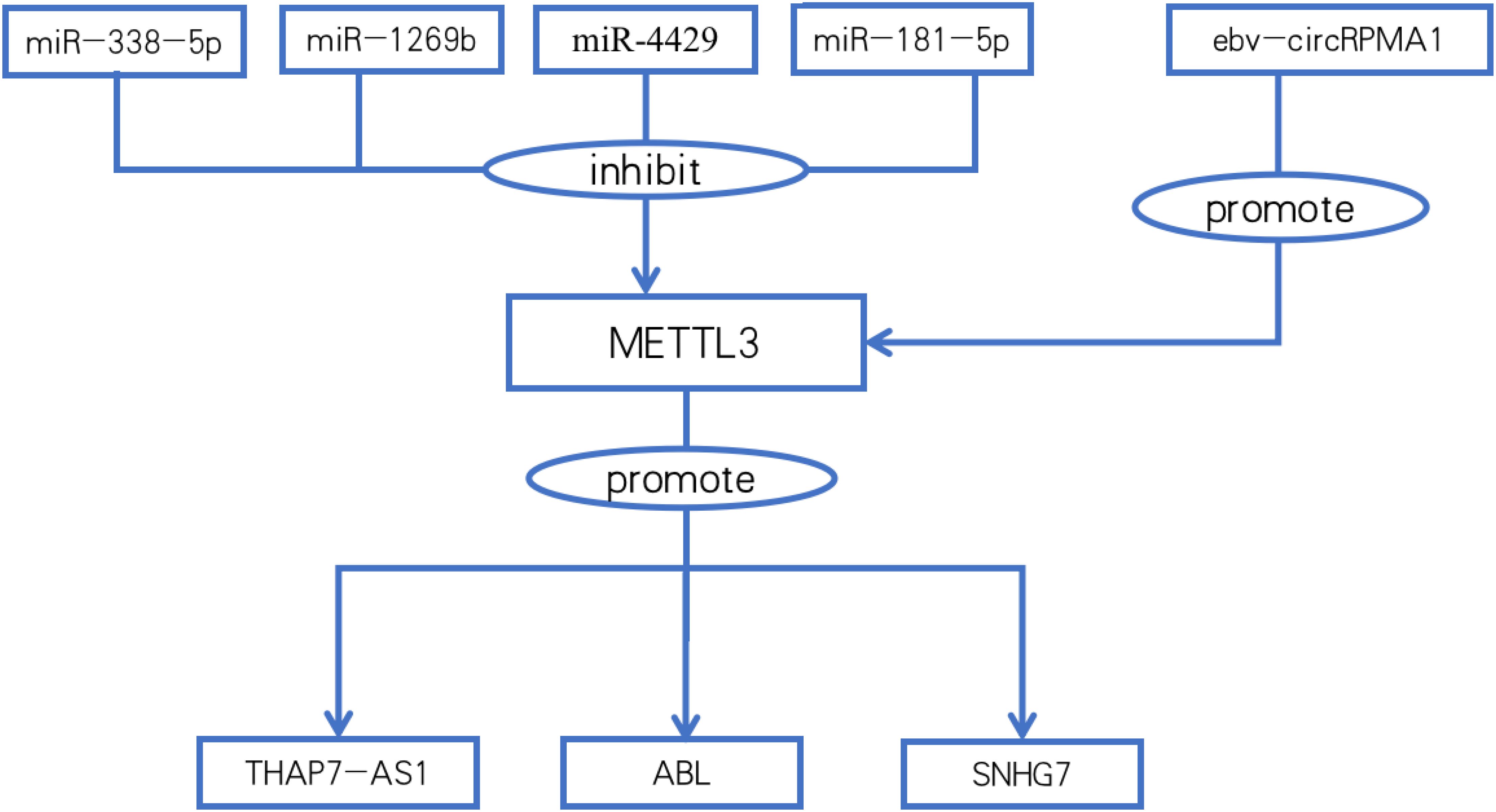
lncRNAs are genes downstream of METTL3. METTL3 maintains the stability of THAP7-AS1 through posttranscriptional modifications (51). IGF2BP1 directly binds to and recognizes m6A modifications in METTL3-mediated ABLs and maintains their stability. ABLs in GC tissues promote gastric cancer cell survival and inhibit the apoptosis induced by various drugs (52). In GC induced by exposure to environmental chemical carcinogens, METTL3 promotes the expression of SNHG7 by increasing the m6A methylation level of the lncRNA SNHG7 and promotes the malignant progression of gastric cancer (53).
lncRNAs play a key role in the recruitment of METTL3. LINC02253 stabilizes KRT18 mRNA and upregulates KRT18 expression by promoting the recruitment of METTL3 to KRT18 mRNA, activating the MAPK/ERK signaling pathway, and promoting tumor growth and GC cell migration and invasion (54).
Autophagy is essential for maintaining optimal cellular function during energy metabolism and helps inhibit the progression of various diseases, such as aging and cancer. Autophagy also degrades cancer-promoting proteins. Dysregulation of autophagy can lead to a variety of pathological processes, such as cancer chemotherapy resistance. ARHGAP5-AS1 enhances the interaction between ARHGAP5 mRNA and HuR (reader) by recruiting METTL3, maintaining ARHGAP5 mRNA stability, and promoting its expression. High ARHGAP5 expression is associated with chemoresistance and shorter overall and progression-free survival in GC cells (55). Similarly, DNA modification is DNA methylation catalyzed by DNA methyltransferase 3a (DNMT3a). Linc00942 recruits METTL3 to DNMT3a mRNA and stabilizes DNMT3a mRNA through “IGF2BP3/HuR-dependent” m6A modification, resulting in an increase in the level of DNA modification and chemotherapy resistance. Linc00942 autophagy, which is mediated by p62, restores gastric cancer cell sensitivity to chemotherapeutic agents by decreasing DNA methylation levels (56). (Table 4; Figure 2). These results suggest that in addition to inducing lncRNA autophagy, DNA methylation-targeted therapy provides a new way to solve the problem of chemoresistance (Figure 2).
METTL3 functions independent of enzyme activity
The subcellular localization of METTL3 demonstrated that cytoplasmic METTL3 regulates translation initiation by interacting with cytoplasmic METTL3-related proteins, which in turn promotes gastric carcinogenesis independent of m6A modification. The translation initiation factor PABPC1 binds to the 3 poly(A) tail. PABPC1 interacts with eIF4F to form a closed-loop conformation of RNA, in which METTL3 contributes to PABPC1 translation initiation by directly binding to PABPC1 to promote the interaction between PABPC1 and eIF4F to maintain the loop RNA structure and regulate translation. The METTL3-PABPC1 complex promotes the translation of histone modification genes to deactivate histones to regulate epigenetic inheritance in GC cells. METTL3-PABPC1 does not promote methylation (57). Studies have shown that acetylation of METTL3 is associated with the progression of gastric cancer. Acetylated METTL3 cannot effectively promote RNA translation. The acetyl group blocks the binding of METTL3 to EIF3H and destroys the ring structure. Acetylation of METTL3 occurs on amino acids related to mRNA translation and has nothing to do with the methyltransferase activity of METTL3 (58). Ianniello Z et al. demonstrated that in cells from patients with chronic myeloid leukemia, METTL3 in the nucleus is only responsible for transferring methyl groups to PES1 mRNA, while the enhancement of PES1 protein synthesis is the result of the action of METTL3 in the cytoplasm (59).
Discussion
The occurrence and progression of GC are closely related to high METTL3 expression levels, as shown in recent studies. METTL3 oncogene mRNA expression was significantly higher in GC tissues than in normal tissues. Tumor suppression can be achieved by downregulating METTL3 expression. Consequently, a novel strategy for the treatment of GC is to target METTL3. YANKOVA et al. identified a compound called STM2457 that binds directly to the SAM binding pocket. It inhibits the catalytic activity of METTL3 by competing with SAM but does not destroy the enzyme structure (60). In addition, some natural substances, such as quercetin and coptisine chloride, are antitumor small-molecule drugs. These compounds inhibited the enzymatic activity of METTL3. However, the mechanism by which these natural substances bind to METTL3 remains unclear (61). Manna et al. discovered several natural products that can bind SAM-binding pockets via methods such as docking and molecular dynamic simulation. These natural products have stronger binding properties than STM2457 does (62). These results suggest that the role of METTL3 small-molecule drugs in tumor treatment involves competing with SAM as well as disrupting the methyltransferase activity of METTL3. However, methylation is not the only mechanism by which METTL3 promotes gastric cancer progression. It has been demonstrated that KH12, formed by PROTAC technology, degrades METTL3 via the ubiquitin−proteasome pathway (63). WD6305 is a newly developed PROTAC degrader that degrades METTL3 by ubiquitination. METTL14, as a protein that interacts with METTL3, is also degraded at the same time (64). These discoveries provide a new theoretical basis for targeted therapy of GC unrelated to the m6A methylation function of METTL3.
METTL3 leads to the upregulation of oncogenes or the downregulation of tumor suppressors, which affects the malignant progression of GC. However, the molecular mechanisms regulating gastric carcinogenesis and progression are complex, and existing findings are just the tip of the iceberg. Notably, METTL3 is the only methyltransferase with enzymatic activity among m6A regulators, such as methyltransferases (writers), demethylases (erasers), and m6A recognition proteins (readers) (65). Stable knockdown of the demethylase FTO can lead to upregulation of E-calmodulin and downregulation of waveform proteins in a variety of GC cell lines, suggesting that FTO may participate in generating stable and overexpressed EMT-related genes, thereby exerting carcinogenic effects (66). FTO can demethylate HOXB13 mRNA, resulting in high HOXB13 expression. This is a deterioration in the condition of patients with stomach cancer. HOXB13 promotes IGF-1R transcription and activates the PI3K/AKT/mTOR pathway. In contrast, when FTO levels are reduced, the malignant development of cancer cells is inhibited (67). Therefore, METTL3 is not the only factor that determines m6A levels, and the factors and mechanisms that regulate m6A levels need to be further investigated in more comprehensive experiments.
METTL3 mostly causes GC progression and drug resistance in gastric cancer. Targeting METTL3 can improve the efficacy of immunotherapy, alleviate chemotherapy resistance, and inhibit tumor growth, which is an advantage of using METTL3 as a target for gastric cancer. However, their disadvantages cannot be ignored. According to Zhang et al., METTL3 may act as a cancer suppressor in GC cell lines and other tumors. Low m6A signaling is strongly associated with the malignant progression of GC. Decreased m6A levels mediate the activation of signals such as Wnt and PI3K-Ak, whereas increased m6A levels mediate the activation of tumor suppressor signals (68). Sun et al. reported that METTL3 promoted the maturation of pri-miR-17-92 and that miR-17-92 activated the AKT/mTOR pathway and improved the sensitivity of GC to chemotherapy (69).
In addition to targeting METTL3 for the treatment of GC, many upstream regulators of METTL3 can also be targeted. For example, H3K27ac binds to the METTL3 promoter to regulate METTL3 expression, and HOXA10 activates the expression of TGFB2 through transcriptional regulation. Increased Smad2/3 expression results from activation of the TGF-β-Smad2/3 pathway. Smad2/3 bind directly to the METTL3 promoter, participate in regulating METTL3 transcription, increase the m6A levels of Slug and Snail in GC cells, accelerate the translation of Snail and Slug, and induce EMT progression (29). In addition, molecules such as Linc00942 and ARHGAP5-AS1 can promote METTL3 recruitment, and this effect can be attenuated by inducing autophagy. In addition, in the immunotherapy of tumors, the methylation of core molecules regulated by METTL3 determines the TMB of different GC subtypes. In cells with high METTL3 expression, the level of PD-L1 expression is also greater, and patients with a high TMB have greater sensitivity to immune checkpoint inhibitors, such as paclitaxel. Some retrospective studies have demonstrated that METTL3 in GC is the main cause of poor overall survival and is correlated with advanced PT, PN, and TNM stages; a tumor size greater than 5 cm; and vascular infiltration (70, 71). Therefore, METTL3 and METTL3-related molecules can be used as clinical targets for patients with GC and as predictive and prognostic biomarkers for GC.
Author contributions
ZY: Writing – original draft, Writing – review & editing. YY: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
GC, Gastric cancer; m6A, N6-methyladenosine; METTL3, methyltransferase-like 3; EMT, epithelial–mesenchymal transition; 3’UTR, 3’ untranslated region; WB, western blotting; cyclin D1, cell cycle protein D1; elF3a, eukaryotic initiation factor 3a; HOX, Homologous box; S43, serine 43 site; ATM, ataxia telangiectasia mutated; miRNA, MicroRNA; MMP, mitochondrial membrane potential; ROS, reactive oxygen species; TME, tumour microenvironment; TMB, tumour mutational burden; CDDP, Cisplatin; circRNAs, Circular RNAs; EBV, Epstein–Barr virus; EBVaGC, EBV-associated GC; LncRNAs, Long noncoding RNAs; DNMT3a, DNA methyltransferase 3; SAM, S-adenosyl methionine.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Lopez MJ, Carbajal J, Alfaro AL, Saravia LG, Zanabria D, Araujo JM, et al. Characteristics of gastric cancer around the world. Crit Rev Oncol Hematol. (2023) 181:103841. doi: 10.1016/j.critrevonc.2022.103841
3. Lordick F, Carneiro F, Cascinu S, Fleitas T, Haustermans K, Piessen G, et al. Gastric cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. (2022) 33:1005–20. doi: 10.1016/j.annonc.2022.07.004
4. Poorolajal J, Moradi L, Mohammadi Y, Cheraghi Z, Gohari-Ensaf F. Risk factors for stomach cancer: a systematic review and meta-analysis. Epidemiol Health. (2020) 42:e2020004. doi: 10.4178/epih.e2020004
5. Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of mettl3 and mettl14 methyltransferases. Mol Cell. (2016) 63:306–17. doi: 10.1016/j.molcel.2016.05.041
6. Zeng C, Huang W, Li Y, Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol. (2020) 13:117. doi: 10.1186/s13045-020-00951-w
7. Liu S, Zhuo L, Wang J, Zhang Q, Li Q, Li G, et al. METTL3 plays multiple functions in biological processes. Am J Cancer Res. (2020) 10:1631–46.
8. Ge Y, Zhang Y, Yang S, Liu T, Zhang T, Feng Y, et al. The comprehensive analysis of N6-methyadenosine writer METTL3 and METTL14 in gastric cancer. J Oncol. (2023) 2023:9822995. doi: 10.1155/2023/9822995
9. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. (2019) 18:176. doi: 10.1186/s12943-019-1109-9
10. Jing JJ, Zhao X, Li H, Sun LP, Yuan Y. Expression profiles and prognostic roles of m6A writers, erasers and readers in gastric cancer. Future Oncol. (2021) 17:2605–20. doi: 10.2217/fon-2020-0630
11. Jiang L, Chen T, Xiong L, Xu JH, Gong AY, Dai B, et al. Knockdown of m6A methyltransferase METTL3 in gastric cancer cells results in suppression of cell proliferation. Oncol Lett. (2020) 20:2191–8. doi: 10.3892/ol.2020.11794
12. Okugawa Y, Toiyama Y, Yin C, Ruiya M, Goel A, Ichikawa T, et al. Prognostic potential of METTL3 expression in patients with gastric cancer. Oncol Lett. (2023) 25:64. doi: 10.3892/ol.2022.13651
13. Lin S, Liu J, Jiang W, Wang P, Sun C, Wang X, et al. METTL3 promotes the proliferation and mobility of gastric cancer cells. Open Med (Wars). (2019) 14:25–31. doi: 10.1515/med-2019-0005
14. Su ZZ, Lee SG, Emdad L, Lebdeva IV, Gupta P, Valerie K, et al. Cloning and characterization of SARI (suppressor of AP-1, regulated by IFN). Proc Natl Acad Sci U S A. (2008) 105:20906–11. doi: 10.1073/pnas.0807975106
15. Wang C, Su Y, Zhang L, Wang M, You J, Zhao X, et al. The function of SARI in modulating epithelial-mesenchymal transition and lung adenocarcinoma metastasis. PloS One. (2012) 7:e38046. doi: 10.1371/journal.pone.0038046
16. Dash R, Su ZZ, Lee SG, Azab B, Boukerche H, Sarkar D, et al. Inhibition of AP-1 by SARI negatively regulates transformation progression mediated by CCN1. Oncogene. (2010) 29:4412–23. doi: 10.1038/onc.2010.194
17. Dai L, Cui X, Zhang X, Cheng L, Liu Y, Yang Y, et al. SARI inhibits angiogenesis and tumour growth of human colon cancer through directly targeting ceruloplasmin. Nat Commun. (2016) 7:11996. doi: 10.1038/ncomms11996
18. Xie JW, Huang XB, Chen QY, Ma YB, Zhao YJ, Liu LC, et al. m(6)A modification-mediated BATF2 acts as a tumor suppressor in gastric cancer through inhibition of ERK signaling. Mol Cancer. (2020) 19:114. doi: 10.1186/s12943-020-01223-4
19. Oh J, Oh JM, Cho SY. METTL3-mediated downregulation of splicing factor SRSF11 is associated with carcinogenesis and poor survival of cancer patients. Eur Rev Med Pharmacol Sci. (2023) 27:2561–70. doi: 10.26355/eurrev_202303_31793
20. Wang N, Huo X, Zhang B, Chen X, Zhao S, Shi X, et al. METTL3-mediated ADAMTS9 suppression facilitates angiogenesis and carcinogenesis in gastric cancer. Front Oncol. (2022) 12:861807. doi: 10.3389/fonc.2022.861807
21. Zhang Z, Fu J, Zhang Y, Qin X, Wang Y, Xing C. METTL3 regulates N6-methyladenosine modification of ANGPTL3 mRNA and potentiates Malignant progression of stomach adenocarcinoma. BMC Gastroenterol. (2023) 23:217. doi: 10.1186/s12876-023-02844-x
22. Liu Y, Zhai E, Chen J, Qian Y, Zhao R, Ma Y, et al. m(6) A-mediated regulation of PBX1-GCH1 axis promotes gastric cancer proliferation and metastasis by elevating tetrahydrobiopterin levels. Cancer Commun (Lond). (2022) 42:327–44. doi: 10.1002/cac2.12281
23. Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, et al. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. (2019) 18:142. doi: 10.1186/s12943-019-1065-4
24. Huo FC, Zhu ZM, Zhu WT, Du QY, Liang J, Mou J. METTL3-mediated m(6)A methylation of SPHK2 promotes gastric cancer progression by targeting KLF2. Oncogene. (2021) 40:2968–81. doi: 10.1038/s41388-021-01753-1
25. Zhang HM, Qi FF, Wang J, Duan YY, Zhao LL, Wang YD, et al. The m6A Methyltransferase METTL3-Mediated N6-Methyladenosine Modification of DEK mRNA to Promote Gastric Cancer Cell Growth and Metastasis. Int J Mol Sci. (2022) 23:6451. doi: 10.3390/ijms23126451
26. Yang Z, Jiang X, Li D, Jiang X. HBXIP promotes gastric cancer via METTL3-mediated MYC mRNA m6A modification. Aging (Albany NY). (2020) 12:24967–82. doi: 10.18632/aging.103767
27. Yang DD, Chen ZH, Yu K, Lu JH, Wu QN, Wang Y, et al. METTL3 promotes the progression of gastric cancer via targeting the MYC pathway. Front Oncol. (2020) 10:115. doi: 10.3389/fonc.2020.00115
28. Zhou W, Xian Q, Wang Q, Wu C, Yan H, Li X, et al. m6A Methyltransferase 3 Promotes the Proliferation and Migration of Gastric Cancer Cells through the m6A Modification of YAP1. J Oncol. (2021) 2021:8875424. doi: 10.1155/2021/8875424
29. Song C, Zhou C. HOXA10 mediates epithelial-mesenchymal transition to promote gastric cancer metastasis partly via modulation of TGFB2/Smad/METTL3 signaling axis. J Exp Clin Cancer Res. (2021) 40:62. doi: 10.1186/s13046-021-01859-0
30. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. (2020) 69:1193–205. doi: 10.1136/gutjnl-2019-319639
31. Cheng Z, Gao S, Liang X, Lian C, Chen J, Fang C. Inhibiting PP2Acalpha promotes the Malignant phenotype of gastric cancer cells through the ATM/METTL3 axis. BioMed Res Int. (2021) 2021:1015293. doi: 10.1155/2021/1015293
32. Meng H, Li J, Sun H, Lin Y, Xu H, Zhang N. The transcription factor ATF2 promotes gastric cancer progression by activating the METTL3/cyclin D1 pathway. Drug Dev Res. (2023) 84:1325–34. doi: 10.1002/ddr.22092
33. Chen W, He Q, Liu J, Li N, Xiao K, Chen H. PLAGL2 promotes Snail expression and gastric cancer progression via UCA1/miR-145-5p/YTHDF1 axis. Carcinogenesis. (2023) 44:328–40. doi: 10.1093/carcin/bgad016
34. Zhang N, Zuo Y, Peng Y, Zuo L. Function of N6-methyladenosine modification in tumors. J Oncol. (2021) 2021:6461552. doi: 10.1155/2021/6461552
35. Xu W, Lai Y, Pan Y, Tan M, Ma Y, Sheng H, et al. m6A RNA methylation-mediated NDUFA4 promotes cell proliferation and metabolism in gastric cancer. Cell Death Dis. (2022) 13:715. doi: 10.1038/s41419-022-05132-w
36. Peng G, Chen S, Zheng N, Tang Y, Su X, Wang J, et al. Integrative proteomics and m6A microarray analyses of the signatures induced by METTL3 reveals prognostically significant in gastric cancer by affecting cellular metabolism. Front Oncol. (2022) 12:996329. doi: 10.3389/fonc.2022.996329
37. Li J, Feng H, Zhu J, Yang K, Zhang G, Gu Y, et al. Gastric cancer derived exosomal THBS1 enhanced Vgamma9Vdelta2 T-cell function through activating RIG-I-like receptor signaling pathway in a N6-methyladenosine methylation dependent manner. Cancer Lett. (2023) 576:216410. doi: 10.1016/j.canlet.2023.216410
38. Xu Z, Chen Q, Shu L, Zhang C, Liu W, Wang P. Expression profiles of m6A RNA methylation regulators, PD-L1 and immune infiltrates in gastric cancer. Front Oncol. (2022) 12:970367. doi: 10.3389/fonc.2022.970367
39. Chen S, Su X, Wang J, Zheng N, Tang Y, Peng G, et al. Identification and validation of METTL3-related molecules for predicting prognosis and efficacy of immunotherapy in gastric cancer based on m6A methylome and transcriptome sequencing analysis. Front Oncol. (2022) 12:935239. doi: 10.3389/fonc.2022.935239
40. Yang J, Xu P, Chen Z, Zhang X, Xia Y, Fang L, et al. N6-methyadenosine modified SUV39H2 regulates homologous recombination through epigenetic repression of DUSP6 in gastric cancer. Cancer Lett. (2023) 558:216092. doi: 10.1016/j.canlet.2023.216092
41. Li H, Wang C, Lan L, Yan L, Li W, Evans I, et al. METTL3 promotes oxaliplatin resistance of gastric cancer CD133+ stem cells by promoting PARP1 mRNA stability. Cell Mol Life Sci. (2022) 79:135. doi: 10.1007/s00018-022-04129-0
42. Wang Y, Hong Z, Song J, Zhong P, Lin L. METTL3 promotes drug resistance to oxaliplatin in gastric cancer cells through DNA repair pathway. Front Pharmacol. (2023) 14:1257410. doi: 10.3389/fphar.2023.1257410
43. Kang J, Huang X, Dong W, Zhu X, Li M, Cui N. MicroRNA-1269b inhibits gastric cancer development through regulating methyltransferase-like 3 (METTL3). Bioengineered. (2021) 12:1150–60. doi: 10.1080/21655979.2021.1909951
44. He H, Wu W, Sun Z, Chai L. MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m(6)A-caused stabilization of SEC62. Biochem Biophys Res Commun. (2019) 517:581–7. doi: 10.1016/j.bbrc.2019.07.058
45. Li R, Li Y, Wang Z, Suo R, Ma R, Zhang J. miR-181-5p/KLHL5 promoted proliferation and migration of gastric cancer through activating METTL3-mediated m6A process. Mol Biotechnol. (2024) 66:2415–25. doi: 10.1007/s12033-023-00877-x
46. Zhang F, Yan Y, Cao X, Zhang J, Li Y, Guo C. Methylation of microRNA-338-5p by EED promotes METTL3-mediated translation of oncogene CDCP1 in gastric cancer. Aging (Albany NY). (2021) 13:12224–38. doi: 10.18632/aging.103822
47. Zhang JY, Du Y, Gong LP, Shao YT, Pan LJ, Feng ZY, et al. ebv-circRPMS1 promotes the progression of EBV-associated gastric carcinoma via Sam68-dependent activation of METTL3. Cancer Lett. (2022) 535:215646. doi: 10.1016/j.canlet.2022.215646
48. Zhang JY, Du Y, Gong LP, Shao YT, Pan LJ, Feng ZY, et al. Corrigendum to “ebv-circRPMS1 promotes the progression of EBV-associated gastric carcinoma via Sam68-dependent activation of METTL3 [Cancer Letters 535 (2022) 215646]’’. Cancer Lett. (2022) 545:215824. doi: 10.1016/j.canlet.2022.215646
49. Chi Y, Wang D, Wang J, Yu W, Yang J. Long non-coding RNA in the pathogenesis of cancers. Cells. (2019) 8:1015. doi: 10.3390/cells8091015
50. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. (2021) 220:e202009045. doi: 10.1083/jcb.202009045
51. Liu HT, Zou YX, Zhu WJ, Sen L, Zhang GH, Ma RR, et al. lncRNA THAP7-AS1, transcriptionally activated by SP1 and post-transcriptionally stabilized by METTL3-mediated m6A modification, exerts oncogenic properties by improving CUL4B entry into the nucleus. Cell Death Differ. (2022) 29:627–41. doi: 10.1038/s41418-021-00879-9
52. Wang Q, Chen C, Xu X, Shu C, Cao C, Wang Z, et al. APAF1-binding long noncoding RNA promotes tumor growth and multidrug resistance in gastric cancer by blocking apoptosome assembly. Adv Sci (Weinh). (2022) 9:e2201889. doi: 10.1002/advs.202201889
53. Liu T, Feng Y, Yang S, Ge Y, Zhang T, Li J, et al. Depicting the Profile of METTL3-Mediated lncRNA m6A Modification Variants and Identified SNHG7 as a Prognostic Indicator of MNNG-Induced Gastric Cancer. Toxics. (2023) 11:944. doi: 10.3390/toxics11110944
54. Gao Z, Long Y, Wu Y, Pu Y, Xue F. LncRNA LINC02253 activates KRT18/MAPK/ERK pathway by mediating N6-methyladenosine modification of KRT18 mRNA in gastric cancer. Carcinogenesis. (2022) 43:419–29. doi: 10.1093/carcin/bgac018
55. Zhu L, Zhu Y, Han S, Chen M, Song P, Dai D, et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. (2019) 10:383. doi: 10.1038/s41419-019-1585-2
56. Zhu L, Zhu Y, Li F, Meng Y, Wang H, Xu W, et al. RNautophagic regulation of DNMT3a-dependent DNA methylation by Linc00942 enhances chemoresistance in gastric cancer. Clin Transl Med. (2023) 13:e1337. doi: 10.1002/ctm2.v13.7
57. Wei X, Huo Y, Pi J, Gao Y, Rao S, He M, et al. METTL3 preferentially enhances non-m(6)A translation of epigenetic factors and promotes tumourigenesis. Nat Cell Biol. (2022) 24:1278–90. doi: 10.1038/s41556-022-00968-y
58. Liu C, Yu M, Wang M, Yang S, Fu Y, Zhang L, et al. PCAF-mediated acetylation of METTL3 impairs mRNA translation efficiency in response to oxidative stress. Sci China Life Sci. (2024) 67(10):2157–68. doi: 10.1007/s11427-023-2535-x
59. Ianniello Z, Sorci M, Ceci Ginistrelli L, Iaiza A, Marchioni M, Tito C, et al. New insight into the catalytic -dependent and -independent roles of METTL3 in sustaining aberrant translation in chronic myeloid leukemia. Cell Death Dis. (2021) 12:870. doi: 10.1038/s41419-021-04169-7
60. Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. (2021) 593:597–601. doi: 10.1038/s41586-021-03536-w
61. Li H, Zhang Q, Feng Q, You Q, Guo X. The development of small molecules targeting methyltransferase-like 3. Drug Discovery Today. (2023) 28:103513. doi: 10.1016/j.drudis.2023.103513
62. Manna S, Samal P, Basak R, Mitra A, Roy AK, Kundu R, et al. Amentoflavone and methyl hesperidin, novel lead molecules targeting epitranscriptomic modulator in acute myeloid leukemia: in silico drug screening and molecular dynamics simulation approach. J Mol Model. (2022) 29:9. doi: 10.1007/s00894-022-05407-1
63. Hwang K, Bae J, Jhe YL, Kim J, Cheong JH, Choi HS, et al. Targeted degradation of METTL3 against acute myeloid leukemia and gastric cancer. Eur J Med Chem. (2024) 279:116843. doi: 10.1016/j.ejmech.2024.116843
64. Du W, Huang Y, Chen X, Deng Y, Sun Y, Yang H, et al. Discovery of a PROTAC degrader for METTL3-METTL14 complex. Cell Chem Biol. (2024) 31:177–83 e17. doi: 10.1016/j.chembiol.2023.12.009
65. Xu P, Ge R. Roles and drug development of METTL3 (methyltransferase-like 3) in anti-tumor therapy. Eur J Med Chem. (2022) 230:114118. doi: 10.1016/j.ejmech.2022.114118
66. Shimura T, Kandimalla R, Okugawa Y, Ohi M, Toiyama Y, He C, et al. Novel evidence for m(6)A methylation regulators as prognostic biomarkers and FTO as a potential therapeutic target in gastric cancer. Br J Cancer. (2022) 126:228–37. doi: 10.1038/s41416-021-01581-w
67. Guo C, Chu H, Gong Z, Zhang B, Li C, Chen J, et al. HOXB13 promotes gastric cancer cell migration and invasion via IGF-1R upregulation and subsequent activation of PI3K/AKT/mTOR signaling pathway. Life Sci. (2021) 278:119522. doi: 10.1016/j.lfs.2021.119522
68. Zhang C, Zhang M, Ge S, Huang W, Lin X, Gao J, et al. Reduced m6A modification predicts Malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. (2019) 8:4766–81. doi: 10.1002/cam4.v8.10
69. Sun Y, Li S, Yu W, Zhao Z, Gao J, Chen C, et al. N(6)-methyladenosine-dependent pri-miR-17-92 maturation suppresses PTEN/TMEM127 and promotes sensitivity to everolimus in gastric cancer. Cell Death Dis. (2020) 11:836. doi: 10.1038/s41419-020-03049-w
70. Zhu C, Wu Q, Xu Y, Ma J, Hu Y, Wang J, et al. Prognostic significance of N6-methyladenosine-modified related chemotransferase METTL3 in gastric carcinoma: Evidence from meta-analysis. Int J Biol Markers. (2023) 38:185–93. doi: 10.1177/03936155231184908
Keywords: METTL3, m6A, gastric cancer, angiogenesis, glycolysis, tumour microenvironment, drug resistance
Citation: Yu Z and Yang Y (2024) METTL3 as a potential therapeutic target in gastric cancer. Front. Oncol. 14:1483435. doi: 10.3389/fonc.2024.1483435
Received: 20 August 2024; Accepted: 28 October 2024;
Published: 29 November 2024.
Edited by:
Dunfa Peng, University of Miami, United StatesReviewed by:
Qiang Wang, First Affiliated Hospital of Anhui Medical University, ChinaKrishnapriya Thangaretnam, University of Miami, United States
Copyright © 2024 Yu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Yang, ZG9jb2xpZGFAc29odS5jb20=
 Zhefei Yu
Zhefei Yu Yang Yang*
Yang Yang*