
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 21 March 2025
Sec. Radiation Oncology
Volume 14 - 2024 | https://doi.org/10.3389/fonc.2024.1478078
This article is part of the Research TopicUse of Radiation Therapy for Hematological MalignanciesView all 6 articles
It is well established that hematologic malignancies are often considerably radiosensitive, which enables usage of far lower doses of therapeutic radiotherapy. This review summarizes the currently known genomic landscape of hematologic malignancies, particularly as it relates to radiosensitivity and the field of radiation oncology. By tracing the historical development of the modern understanding of radiosensitivity, we focus on the discovery and implications of pivotal mutated genes in hematologic malignancies such as TP53, ATM, and other genes critical to DNA repair pathways. These genetic insights have contributed significantly to the advancement of personalized medicine, aiming to enhance treatment precision and outcomes, and there is an opportunity to extend these insights to personalized radiotherapy. We explore the transition from early discoveries to the current efforts in integrating comprehensive genomic data into clinical practice. Specific examples from Hodgkin lymphoma, non-Hodgkin lymphoma, and plasma cell neoplasms illustrate how genetic mutations could influence radiosensitivity and impact subsequent radiotherapeutic response. Despite the advancements, challenges remain in translating these genetic insights into routine clinical practice, particularly due to the heterogeneity of alterations and the complex interactions within cancer signaling pathways. We emphasize the potential of radiogenomics to address these challenges by identifying genetic markers that predict radiotherapy response and toxicity, thereby refining treatment strategies. The need for robust decision support systems, standardized protocols, and ongoing education for healthcare providers is critical to the successful integration of genomic data into radiation therapy. As research continues to validate genetic markers and explore novel therapeutic combinations, the promise of personalized radiotherapy becomes increasingly attainable, offering the potential to significantly improve outcomes for patients with hematologic malignancies.
“Know your enemy and know yourself, and you can fight a hundred battles without disaster.”
-The Art of War, Sun Tzu
Oncologists must confront cancer at both the macroscopic level of the patient and the microscopic level of the cancer cell. The modern battle against cancer continues to rely on both systemic therapies, such as chemotherapy, immunotherapies and targeted agents, and local treatments, including surgery and radiation therapy (RT). While therapeutic decision making was historically driven largely by histology and stage, contemporary planning is increasingly reliant on molecular insights. Specifically, since the development of Sanger sequencing in the 1970s, scientists have meticulously mapped out somatic and germline DNA mutations that have already demonstrated strong potential to enhance and refine treatment approaches. In radiation oncology, however, there are currently no widely implemented examples of molecular traits determining appropriate RT utilization, RT targets and/or optimal RT doses. We now stand at a pivotal moment, with the opportunity to leverage this accumulated genetic intelligence to revolutionize our approach and take the offensive against cancer.
In this review, we will first describe the classical understanding of cancer radiosensitivity as it relates to DNA damage and the cell cycle. Next, we will review key studies that have identified specific genes which are related to radiation response. We will then discuss current efforts to develop genetic signatures of therapeutic sensitivity in hematologic malignancies, highlighting recent advancements and research findings. Lastly, we will comment on the potential future of personalized medicine within the field of RT for hematologic malignancies, highlighting potential developments and the implications for patient care.
“Attack him where he is unprepared, appear where you are not expected.”
-The Art of War, Sun Tzu
RT exerts its lethal effects on cancer cells primarily through DNA damage. This damage is not uniformly distributed; it preferentially affects DNA in open chromatin regions compared to heterochromatin (1). Open chromatin is less densely packed and more transcriptionally active, making it more accessible to radiation-induced damage. Direct damage by radiation causes single-strand breaks (SSBs) and double-strand breaks (DSBs) in DNA. However, approximately two-thirds of the DNA damage caused by radiation is indirect, resulting from the generation of free radicals (Figure 1). These reactive oxygen species (ROS) are produced when radiation ionizes water molecules within the cell. The free radicals then diffuse through the cell, causing widespread damage to DNA, lipids, and proteins, resulting in base damage, SSBs, and DSBs. Hypoxic tumors, which have low oxygen levels, are therefore less susceptible to this indirect damage because the production of ROS is oxygen-dependent, leading to lower radiosensitivity (2).
“In war, the way is to avoid what is strong, and strike at what is weak.”
-The Art of War, Sun Tzu
The terms radiosensitive and radioresponsive are used in different contexts in clinical practice. They are sometimes referenced as a state of disease control in a clinical trial testing a radiotherapeutic intervention; for example, if the RT arm shows improved outcomes, patients treated in this study can be referred to as more radioresponsive. In a lesion-specific context, these terms may also reflect a local observation of a specific tumor decreasing in diameter, volume, or PET SUV following RT treatment. One may even use the term radiosensitivity to characterize the toxicity observed in a patient, referring to patients with more toxicity after RT as being more radiosensitive. However, in its most straightforward form, the concept of radiosensitivity can be defined by the observation that different phases of the cell cycle display varying levels of cell death in response to RT (3). For example, in the G0, early G1, and late S phases, cells are generally resistant to RT (4). Conversely, the most radiosensitive parts of the cell cycle are the late G1, G2, and M phases (4). Variability in radiation sensitivity throughout the cell cycle is thought to be related to the biological characteristics of each phase. For example, in the S phase, DNA synthesis leads to more nucleic acid content, a higher probability of DNA damage repair enzyme activity, and intrinsic free radical scavenging via glutathione (5). Tumor cells that proliferate at higher rates are generally seen as more radiosensitive, likely because a larger proportion of these cells are in the radiosensitive phases of the cell cycle. Hematologic malignancies can sometimes exhibit faster or more pronounced responses to relatively lower doses of radiation when compared to treated solid tumors, due to their rapid proliferation rates and the characteristics of the cells in these malignancies (6). To understand the observed inherent radiosensitivity of hematologic malignancies, one must first understand how DNA damage caused by radiation is repaired.
“What is of supreme importance in war is to attack the enemy’s strategy”
-The Art of War, Sun Tzu
Cells have evolved sophisticated strategies to halt the cell cycle and repair DNA damage, and cancer cells have developed methods to overcome these checks and balances (Figure 2). Normal cells have DNA damage repair mechanisms that maintain genomic integrity, involving various pathways to address different types of DNA damage. The MRN complex, consisting of MRE11, RAD50, and NBS1, plays a critical role in detecting and signaling DSBs (7). Upon recognizing damage, the MRN complex recruits ATM (ataxia-telangiectasia mutated kinase), which phosphorylates several key proteins, including p53 and H2AX, to initiate the DNA damage response (DDR). There are two major pathways to repair DSBs: homologous recombination (HR) and non-homologous end joining (NHEJ). HR, active during the S and G2 phases of the cell cycle, uses a sister chromatid as a template for accurate repair, involving proteins such as BRCA1, BRCA2, and RAD51. In contrast, NHEJ, which operates throughout the cell cycle, directly ligates the broken DNA ends but is more error-prone. NHEJ has core components including Ku70/80, DNA protein kinases, LIG4, XRCC4, and XLF. In addition to DSB repair, cells employ mismatch repair (MMR) to correct replication errors, nucleotide excision repair (NER) to remove bulky DNA adducts caused by UV radiation, and base excision repair (BER) to fix small base lesions induced by oxidative stress. The coordination of these pathways and others ensures comprehensive maintenance of DNA integrity and is crucial for preventing mutations that could lead to cancer.
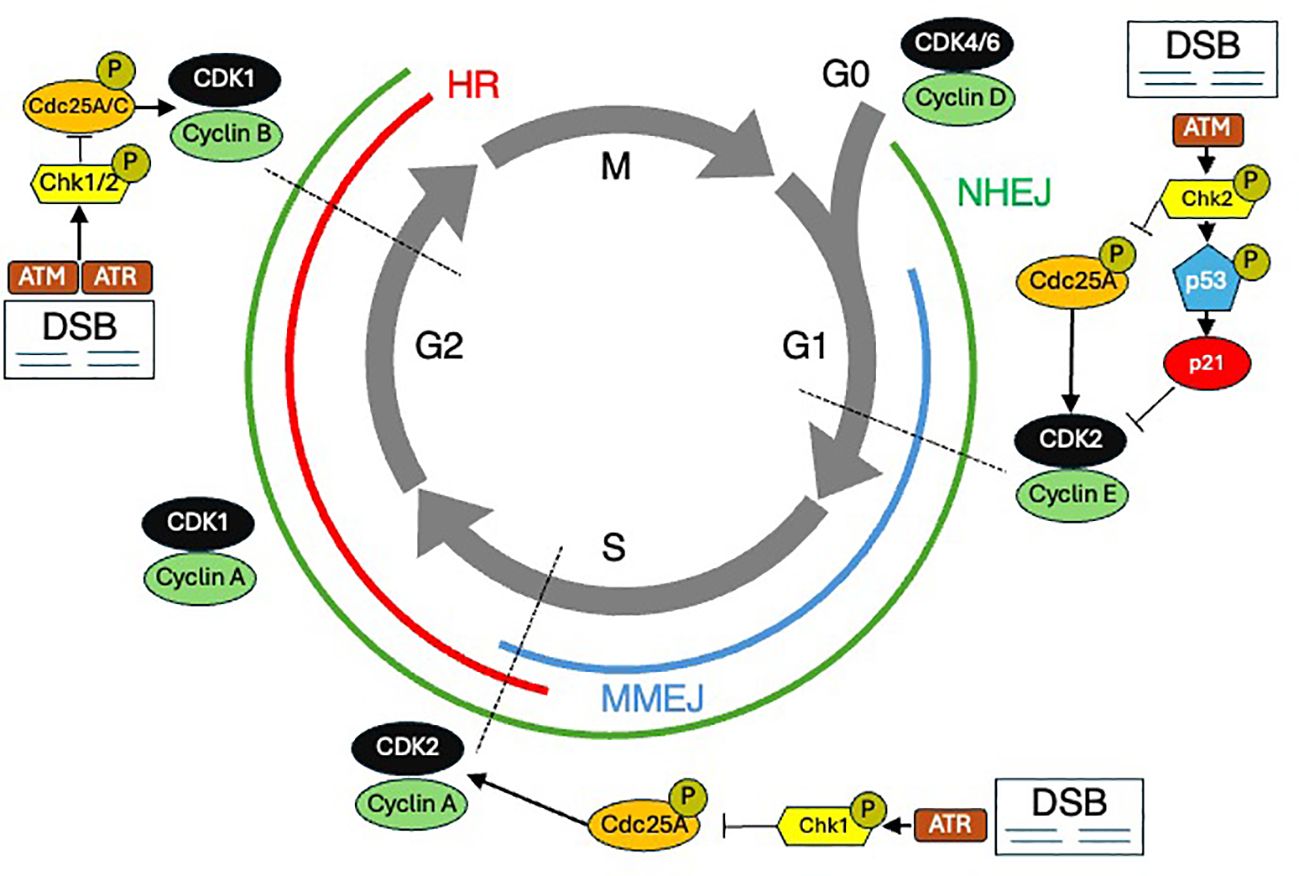
Figure 2. The cell cycle and DNA damage repair (DDR). The cell cycle is shown with various DDR mechanisms shown at the stages during which they predominate. DSB Double strand breaks. HR Homologous recombination. MMEJ Microhomology-mediated end joining. NHEJ Nonhomologous end joining.
Relative resistance to radiation-induced damage in the S phase is thought to be due in part to an elevated amount of DNA synthesis and repair enzymes, as well as increased intracellular levels of glutathione (a free radical scavenger) (8). Cell cycle blockage in the G1 phase after ionizing radiation is believed to allow time for the recognition and repair of DNA damage prior to the initiation of DNA synthesis. Cells are most sensitive in the G2/M phase of the cell cycle, possibly because there is no time for adequate repair before chromosome segregation takes place. Several gene products have been identified which increase expression or are post-translationally altered following DNA damage, and these are thought to participate in halting cell cycle progression (9). These damage-responsive genes and their protein products include p53, p21, growth arrest and damage-delay (GADD45), X-ray induced protein (XIP269), retinoblastoma protein (Rb), and a group of retinoblastoma control proteins (RCPs), which bind to Sp1 sites in DNA promoters and may act to further alter gene transcription in response to DNA damage (10). Understanding the mechanisms that underlie radiation effects on DNA and the cell’s ability to counter these effects highlights that tumors with highly deregulated genomes will exhibit abnormal cell cycle biology, a potentially advantageous observation for inducing radiosensitivity.
“In war, the victorious strategist only seeks battle after the victory has been won, whereas he who is destined to defeat first fights and afterwards looks for victory.”
-The Art of War, Sun Tzu
A rational starting point to identify potential somatic molecular alterations in cancer cells which might be associated with increased radiosensitivity is to review early key observations linking individual genetic mutations to syndromes characterized by heightened radiation toxicity. These putative genes are summarized in Table 1. One of the first significant discoveries was the identification of the ATM gene, which is crucial in Ataxia-Telangiectasia, a disorder marked by increased sensitivity to ionizing radiation (14, 15). Similarly, mutations in the NBS1 gene, part of the MRN complex, cause Nijmegen Breakage Syndrome (NBS), another autosomal recessive disorder with increased radiosensitivity and cancer predisposition (16). The discovery of the TP53 gene, often referred to as the “guardian of the genome,” highlighted its critical role in the cellular response to DNA damage (17–19). Li-Fraumeni Syndrome, caused by germline mutations in TP53, underscored the impact of genetic mutations on radiosensitivity (20). Additionally, Xeroderma Pigmentosum (XP) is marked by extreme sensitivity to UV radiation due to defects in the NER pathway (21), while Fanconi Anemia (FA), characterized by mutations in FANCA and other FA genes, increases susceptibility to DNA crosslinking agents and radiation, demonstrating the role of DNA repair pathways in maintaining genomic stability (22).
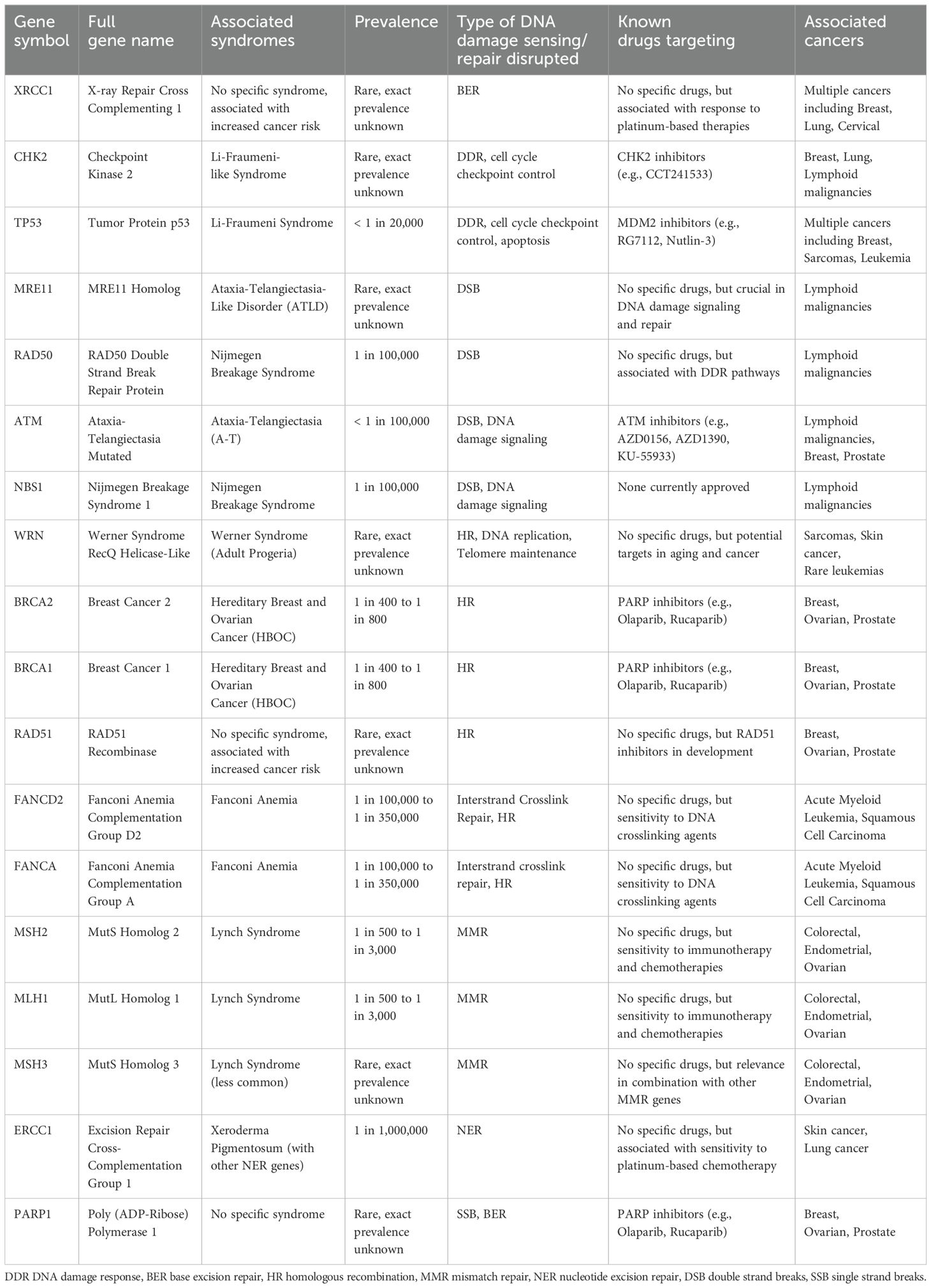
Table 1. Summary of major DNA damage repair genes, associated syndromes, prevalence, known targeted agents, and associated cancers (11–13).
In addition to syndromes that were directly linked to radiation sensitivity by a single altered gene, other DNA damage sensing and repair gene alterations are also associated with increased risk of cancer and radiation sensitivity, though with reduced penetrance. The identification of mutations in the BRCA1 and BRCA2 genes, pivotal in HR repair of DSBs, was associated with a breast cancer risk as high as 70% and emphasized the importance of intact DDR in mitigating cancer risk (23, 24). RAD51 is an essential component for fixing radiation-induced DSBs as part of HR, and germline RAD51 mutations are associated with a 10-20 percent lifetime risk in women for ovarian, fallopian tube, or primary peritoneal cancer and a 30% risk for breast cancer (25). Alterations in XRCC1, involved in the BER pathway crucial for repairing SSBs induced by ionizing radiation, have been associated with increased risk for several cancers (26). Similarly, the elucidation of the CHK2 gene as a key checkpoint kinase in DDR, particularly to ionizing radiation, marked another important discovery. The presence of CHK2 mutations can double the lifetime risk of breast cancer and increase colorectal and prostate cancer risks (27–29). Each of these genetic insights has deepened our understanding of DNA repair mechanisms, and as RT exerts its effect on cells through DNA damage, the presence of germline and somatic DDR alterations has implications for both cancer risk and radiosensitivity. For example, identifying a single altered copy of a cancer-associated gene has allowed clinicians to begin augmenting cancer treatments through exploiting synthetic lethality (30), and agents are already being used to increase radiosensitivity of cells with impaired DDR (31).
Given the effects that radiation has on normal and cancer cells, it is not surprising that cell signaling and transcription factor pathways have also been increasingly implicated in mitigating aspects of radiation sensitivity. The JAK-STAT pathway allows extracellular signals including cytokines such as interferons and growth factors to quickly influence nuclear processes. Many types of cancer have shown that STAT3 can mediate resistance to chemoradioimmunotherapy (32), and targeting STAT3 may overcome radioresistance (33). The Notch pathway involves 4 short-range cell-cell signaling receptors regulating genes involved in cell cycle regulation, cellular differentiation, and stem cell maintenance. Of note, NOTCH1 inhibits the kinase activity of ATM, and blocking Notch in the presence of DNA damage leads to increased radiation sensitivity in an ATM-dependent manner (34). Additionally, inactivation of HR in Notch-driven cancers is shown to cause radiosensitization (35). The Nuclear factor (NF)-κB transcription factor regulates immunity, cellular survival and apoptosis. DSBs, like those resulting from radiation therapy, activate the NF-κB pathway (36). Many other cell signaling pathways are implicated in radiation sensitivity and are active areas of research. They may yield mechanistic insight into radiation response when they are altered or may be targeted to induce radiation sensitivity.
While there are no guideline-approved variations in radiation therapy indications based on clinicogenetic factors, recent insights from solid tumors offer promising directions. For instance, solid tumors with ATM or BRCA mutations exhibit increased radiosensitivity compared to matched controls (37, 38). Similarly, the radiosensitivity of human papillomavirus (HPV)-associated oropharyngeal cancer (OPC) is partly attributed to deficient DNA repair caused by E6 and E7 viral oncoproteins, which degrade p53 and inactivate Rb, disrupting DNA repair pathways and enhancing susceptibility to radiation-induced DSBs (39, 40). This intrinsic radiosensitivity of HPV-positive OPC has facilitated dose de-escalation studies, demonstrating that reduced radiation doses (e.g., 30-60 Gy versus 70 Gy) can achieve comparable local control while minimizing treatment-related toxicity (41). Furthermore, HPV-positive tumors are less likely to harbor hypoxic microenvironments—an important determinant of radioresistance—further enhancing their radiation responsiveness (42). These findings underscore opportunities for tailored radiation dosing and emphasize the importance of considering tumor microenvironmental factors when optimizing radiotherapy for other cancers. Lastly, the Oncotype DX DCIS score was retrospectively correlated with local recurrence risk after lumpectomy for DCIS, and it can guide the use of adjuvant RT (43). However, there are no large-scale prospective trials randomizing DCIS patients to omit radiation solely using this score, and it is primarily used in risk-stratification and shared decision-making contexts with questionable clinical utility and cost-effectiveness (44).
“He who wishes to fight must first count the cost.”
-The Art of War, Sun Tzu
Radiation therapy is a powerful tool in cancer treatment, but no therapy comes without a price: adequate radiation dose to effectively treat a tumor and radiation-associated toxicity to surrounding normal tissues are two sides of the same coin. As discussed above, there are several genetic variants especially those involved with DDR that have been associated with augmented toxicity to RT; this emphasizes a potential to identify patients who may be less able to adequately repair DNA damage to normal cells (45). However, the results of studies trying to associate DDR genes and cell cycle genetic aberration to radiation toxicity are not always straightforward. For example, in a proof-of-principle study, the ATM gene was sequenced in 20 patients with severe late radiation side effects, but no ATM mutations were found (46). Since that time, there has been inconsistent data in gene-level and population-based studies, though, more recently, ATM sequence variants were shown to predict adverse RT response in prostate cancer patients (47, 48). Alternative mechanisms of radiation toxicity are also implicated such as nucleoshuttling of ATM (49). A recent study introduced the PROSTOX assay, a microRNA-based test that may be able predict the risk of long-term genitourinary toxicity in prostate cancer patients undergoing radiation therapy by looking for specific germline microRNA single-nucleotide polymorphisms (SNPs) (50). SNPs and TP53 polymorphisms correlate with severe late adverse effects and clinical outcomes in cancer patients undergoing RT (51, 52). Specifically, certain TP53 mutations predict normal tissue toxicity following RT in head and neck cancer (53). Similarly, the presence of BRCA1/BRCA2 mutations in breast cancer patients may influence the risk of complications after radiation, like brachial plexopathy, though the data is conflicting (54, 55). The Radiosensitivity Index (RSI) predicts a tumor’s response to radiation by analyzing a 10-gene signature related to DDR and cell cycle regulation (56), with specific studies showing its utility in reducing breast cancer treatment toxicity (57). Building on RSI, a dose-adjustment algorithm termed GARD (genomic adjusted radiation dose) assayed multiple solid tumors and integrated the radiosensitivity score with RT dose, optimizing therapeutic outcomes while significantly minimizing the risk of radiation-induced toxicities (58). GARD was associated with risk of local recurrence in breast cancer and has the potential to be used to make decisions on radiation dose adjustment (59). Therefore, it is possible that a general test such as RSI and GARD, or a novel disease-specific genetic signature may be used to optimize radiation doses through personalized escalation or de-escalation.
“In the midst of chaos, there is also opportunity.”
-The Art of War, Sun Tzu
Decades of research into molecular biomarkers has yielded an undeniable truth: the genome is incredibly intricate and its complexity is often difficult to translate into clinically relevant information. We are no longer in an era where single genes establish new syndromes with easily observable phenotypes. While certain single nucleotide variants (SNVs) are associated with targeted therapies (60), the associations with radiosensitivity are more complicated, often involving defects in multiple genes or entire pathways. This complexity necessitates a more nuanced understanding of the mutational landscape and its impact on therapeutic sensitivity.
Preclinical models such as cell lines and transgenic mice have contributed our earliest observations about radiation sensitivity in hematologic malignancies, indicating that lymphomas are at one extreme of the spectrum of radiosensitivity relative to solid tumors like melanoma and glioblastoma (61, 62). In Mantle cell lymphoma (MCL), cell lines derived from patients indicated that observed radiosensitivity may be due to distinct mechanisms in subtypes of MCL such as telomere shortening, loss of heterozygosity at the ATM locus, and the functionality of TP53 mutants (63). Paradoxically, in mice with lymphomas arising in a background of altered ATM, TP53, ARF, or NBS1, showed that mice with an intact DDR had the most durable remission after irradiation (64). In Burkitt lymphoma (BL) cell lines, mutant TP53 abrogated the ability of G-phase arrest following radiation (65). Additional studies of lymphoma cell lines have shown that in the presence of wild type TP53, having mutations in downstream proteins can result in similar radiosensitivity (66). Using exogenous agents to inhibit NF-KB signaling has been shown to radiosensitize BL cell lines (67). Studies of mouse lymphoma cell lines show that radiation resistance correlates to Bcl-2 expression and suggest that Bcl-2 blocks apoptosis by the antioxidant pathway (68). The transition from candidate gene studies to genome-wide association studies (GWAS) in identifying genetic markers of RT toxicity and response will likely involve genomic profiling (69). Through target sequencing panels, whole exome sequencing (WES), and whole genome sequencing (WGS) approaches, a vast amount of molecular information can inform clinical radiation oncology practice. Studies that establish models in which genes are associated with radiation sensitivity or predict patient response could be translated into a framework for personalized RT (70).
A thorough understanding of the genetic underpinnings of hematologic malignancies have already led to advancements in targeted systemic therapy options. For example, Tazemetostat inhibits the EZH2 protein, frequently altered in lymphomas, and phase II trials have demonstrated benefit in patients with EZH2 mutations (71). Moving forward, we foresee that these same insights could facilitate personalized consideration and integration of RT. Specifically, these opportunities include a better understanding of how these aforementioned targeted therapies could be optimally used in conjunction with RT. Furthermore, these insights may highlight opportunities for rational RT dose alterations or even RT inclusion/omission, treatment-response prediction, and RT modality selection (Figure 3). There may also be opportunity to better predict risk of normal tissue toxicity post-RT.

Figure 3. The potential uses of genetic information in radiotherapy (RT). Addition addresses the identification of mutations such as those implicated in synthetic lethality that may be targeted and used in conjunction with RT. Avoidance concerns the germline mutations that may indicate that a patient has greater radiosensitivity of their normal tissue. Dose can be reduced or escalated depending on observed genetic phenotypes. The potential for tumor genetics to provide insight into the behavior of different lesions within the same patient or the aggressiveness of a particular lesion has implications for targeting radiation (i.e., only targeting the largest lesion out of several). Tumor genetics may indicate that RT is not the preferential mode of treatment and provide insight into omission of RT. Genetics can be used to predict the likelihood of RT eradicating the tumor. Lastly, the RT modality (i.e. photons, protons, brachytherapy, etc.) may be chosen by understanding the tumor genetics.
In the following sections, we will review the current landscape of common genetic mutations in hematologic malignancies, focusing on their implications for treatment sensitivity and resistance.
Hodgkin lymphoma (HL) is challenging to characterize from a genomic perspective given that the main neoplastic cells, Hodgkin and Reed-Sternberg (HRS) cells, comprise a minority of the tumor microenvironment (<5%). Only recently has it become realistic to isolate HRS cells for better characterization; whereas, most existing work relied on bulk sequencing with admixed cell populations. The observed mutations in HL are shown in Table 2. From targeted sequencing panels of FFPE biopsy samples, TP53 is the most frequently mutated gene in HL (approximately 20% of patients) (78). BCL2, an antiapoptotic factor, is thought to be involved in the pathogenesis of HL, and overexpression of BCL2 in HL is correlated with poorer response to chemotherapy (79). Microdissected HL samples indicated that EP300 and CREBBP, epigenetic regulators, are mutated in up to 40% of samples (80). No studies have identified specific genetic markers that can determine the radiosensitivity of HL; however, genetic aberrations are found in the immune system, JAK-STAT, NF-κB, DNA repair and cell cycle pathways, indicating that there are potentially radiosensitive HLs depending on the underlying molecular characteristics (81).
Though ATM is not mutated at high frequencies in HL, ataxia-telangiectasia and Rad3-related (ATR), which is also essential for proliferation, is shown to be mutated in some HL cell lines (82). Previous studies suggest that ATR might participate in the signaling of ionizing radiation (IR)- and ultraviolet (UV)-induced DNA damage (83). ATR is activated not only by UV-induced SSBs but also other forms of DNA damage and replication blocks, with mutations believed to cause abrogation of ATR in HL (82). Other cell cycle genes like KLHDC8B61 (84) are NPAT (85) (nuclear protein, ataxia-telangiectasia), are also implicated in HL, and may convey similar radiosensitivity to ATM/ATR mutations. Finally, mutations in POT1 in HL are associated with increased chromosomal instability (CIN) which may associate with increased radiosensitivity (86). Copy number alterations (CNA), which involve the gain or loss of DNA segments directly affecting gene dosage, are present in more than 20% of HL cases and are enriched in genes related to NF-κB signaling, such as REL, IKBKB, CD40, and MAP3K14 (87). The presence of specific recurrent CNAs in HL was shown to be related to chemotherapy resistance (88).
Non-Hodgkin lymphoma (NHL) encompasses a diverse group of lymphoid malignancies, each characterized by distinct genetic profiles that may influence radiosensitivity and treatment outcomes (Figure 4). Indolent lymphomas, such as follicular lymphoma (FL) and marginal zone lymphoma (MZL), generally exhibit slow disease progression and relatively good responses to treatment, while other lymphomas like MCL, BL and diffuse large B-cell lymphoma (DLBCL) can have more aggressive courses (Figure 5). Of note, depending on histology and clinical situation, guideline-supported RT doses acceptable for treatment of NHL range from 4-54 Gy (89). This wide spectrum underscores a critical need to personalize RT dosing.
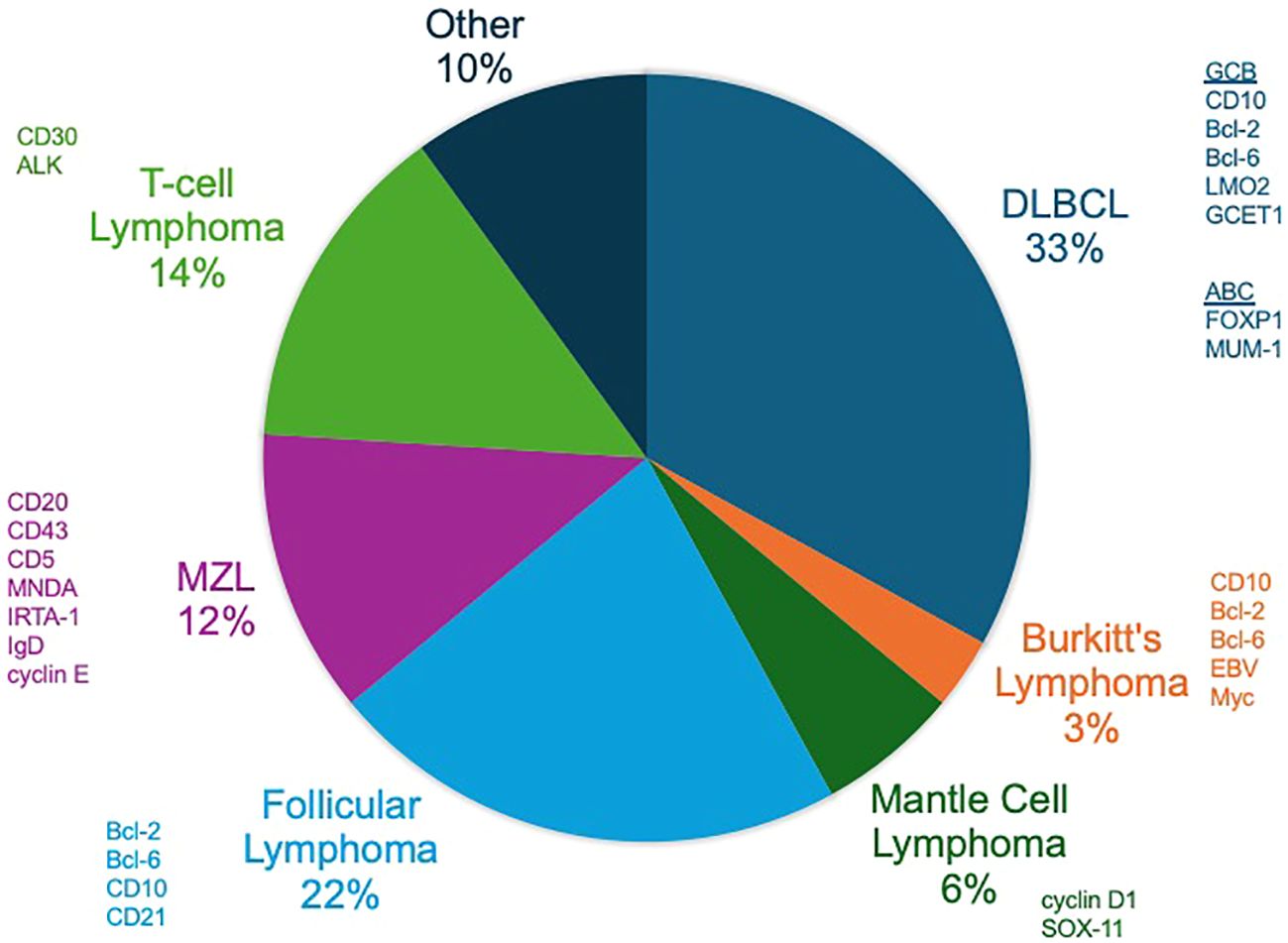
Figure 4. Non-Hodgkin lymphomas (NHLs). The distribution of the prevalence of the most common NHLs with commonly expressed factors on histopathology shown.
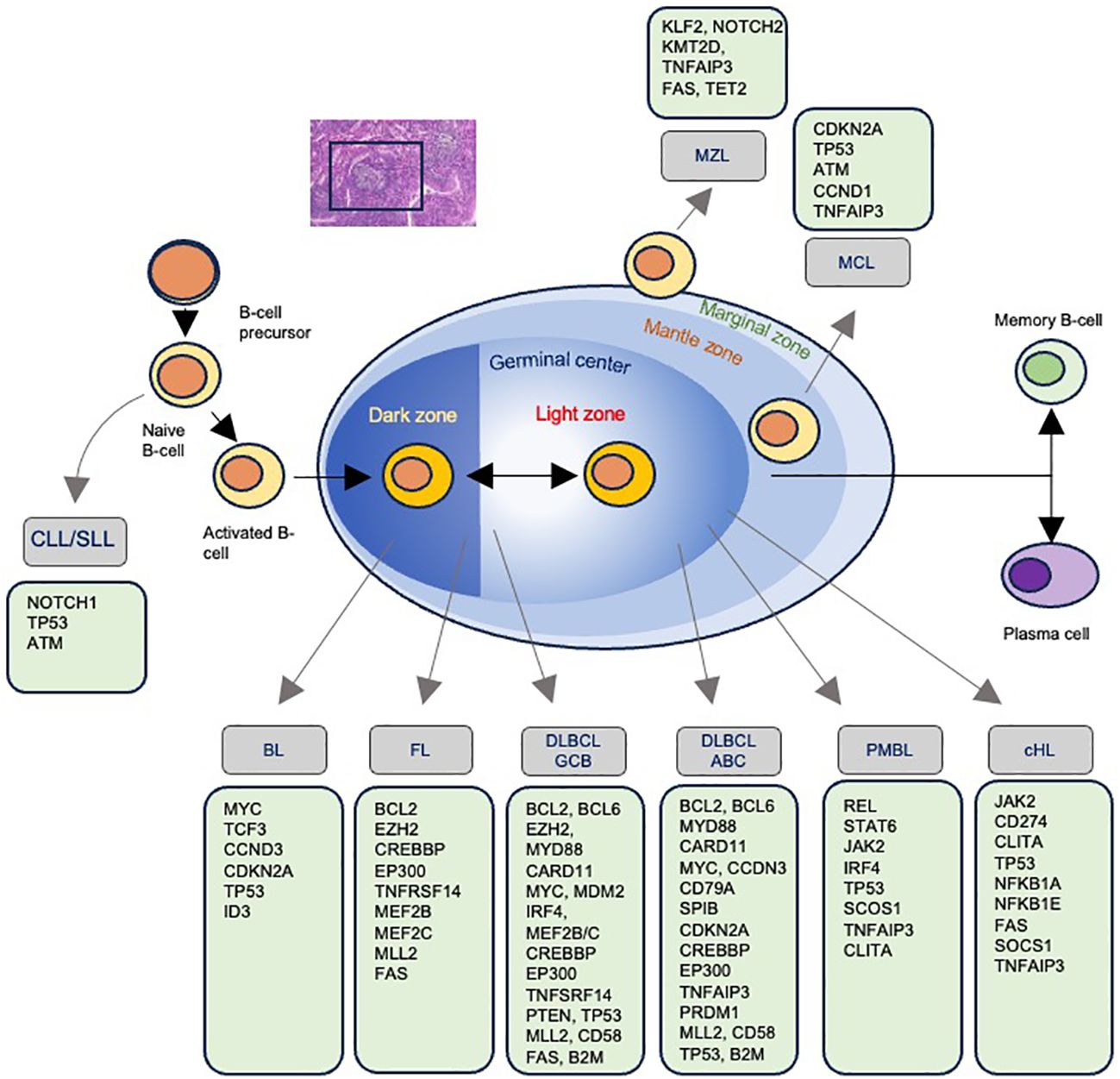
Figure 5. Mutations associated with each type of non-Hodgkin lymphoma. The progression of B-cell maturation and the associated development of lymphoma with the most commonly identified mutations.
“If he is taking his ease, give him no rest.”
-The Art of War, Sun Tzu
Indolent lymphomas are mature B-cell neoplasms that often involve translocations which bring oncogenes in proximity to the immunoglobulin heavy chain (IGH) locus at chromosome band 14q32, resulting in overexpression of the oncogene (Table 3). For example, FL is commonly defined by t(14;18)(q32;q21) occurring in up to 90% of cases and resulting in overexpression of BCL2 by placing it under control of the IGH promoter. Extranodal marginal zone lymphomas (ENMZL) often arise in the gastrointestinal tract, salivary glands, and other extranodal sites. ENMZLs often have rearrangement involving the MALT1 gene, such as t(11;18)(q21;q21), t(14;18)(q32;q21), and t(1;14)(p22;q32), which result in the activation of the NF-κB pathway (97). Chromosomal translocations involving the BIRC3 gene, such as t(1;14)(p22;q32), have also been implicated in MALT lymphomas of the intestine and lung (98). BIRC3 (also known as cIAP2) is an inhibitor of apoptosis, and its dysregulation can contribute to radioresistance by preventing radiation-induced cell death (99). In ENMZLs of the GI system, BCL10-IGH translocation t(1;14) is a driving feature. In ocular ENMZLs, t(3;14)(p14.1;q32) resulting in FOXP1-IGH can occur. Other indolent lymphomas, such as nodal MZL and lymphoplasmacytic lymphoma, can also harbor distinct genetic alterations such as MYD88 and L265P mutations, implicated in promoting survival through constitutive NF-κB activation (100). Understanding the genetic landscape of indolent lymphomas provides critical insights into their biology and may inform personalized therapeutic approaches, including the potential for targeted radiosensitization strategies.
In FL, genetic mutations occur in epigenetic regulators such as KMT2D or CREBBP in roughly two-thirds of patients (101). Given the high frequency of epigenetic dysregulation, epigenetic targets are hypothesized to be cancer-driving and involve cell-cycle regulation. If a clear genetic phenotype is identified, it is possible that there may be an observable and exploitable relationship with radiation sensitivity (102). Although no specific mutations have been identified that are directly associated with radiation sensitivity per se, there are already some examples of incorporating genetic information into tools to guide clinical decision making with respect to chemoimmunotherapy. For example, the m7-FLIPI (Follicular Lymphoma International Prognostic Index) score combines clinical factors with genetic mutations in EZH2, ARID1A, MEF2B, EP300, FOXO1, CREBBP, and CARD11, providing a more nuanced risk stratification that guides therapeutic decisions and identifies patients who may benefit from more aggressive treatment strategies (103); of note, the utility of m7-FLIPL for prediction of response to chemotherapy is inconsistent (104, 105). A study of the m7-FLIPI on patients from the GALLIUM trial, a clinical study which evaluated rituximab vs. obinutuzumab frontline treatment (106), found that EZH2 mutations were associated with more benefit from cyclophosphamide, adriamycin, vincristine and prednisone (CHOP)/cyclophosphamide, vincristine and prednisone (CVP) chemotherapy, whereas EZH2 wild-type patients had superior outcomes with bendamustine-based regimens (107).
More recently, genomically distinct subtypes of FL have been identified with different, often more favorable, treatment response (108). Translocation t(14;18) negative FL is noted to arise predominantly in inguinopelvic sites, is enriched for STAT6 and CREBBP mutations, and usually has a good response to chemotherapy and RT (109). Additionally, pediatric-type FL, which occurs in young patients and adolescents, is recognized for its excellent prognosis and high responsiveness to treatment with roughly 50% of cases harbor mutation of IRF8 and frequent MAPK mutations with a surprising absence of epigenetic modifier mutations (110). This subtype is often localized and exhibits a low-grade histology, resulting in long-term remission with standard treatment protocols (111).
In MZL, most ENMZLs show trisomy of chromosomes 3 and 18. The t(11;18)(q21;q21) translocation resulting an API2–MLT/MALT1 is associated with resistance to antibiotic therapy in gastric MALT lymphoma, suggesting a more advanced and therapy-resistant disease (51). In lung cancer cell lines, MALT1 loss is associated with increased radiosensitivity (112). In MALT1 rearrangement-negative gastric MALTs, TRAF3, TNFAIP3, and NOTCH1 are commonly altered (113). Targeted mutations of TNFAIP3 are seen in ocular adnexal MALT, possibly associated with more DDR errors and radiosensitivity (114); whereas salivary MALTs have mutations in TBL1XR1 and GPR34, which have been implicated in lung cancer radiation resistance (115). In a study of ocular ENMZL patients using whole genome and targeted sequencing, JAK3 mutation occurred in 11% of cases and was associated with reduced PFS after chemotherapy relative to wild-type (116). Studies also showed deletions of TNFAIP3, and amplifications of NOTCH targets and the CEBP transcription factor family (116). Mutations of TBL1XR1 leads to increased NCoR degradation and activation of NF-kB and JUN signaling pathways (117). Lastly, MYD88 mutations in ocular ENMZL have been associated with inferior disease-free survival after chemotherapy (DFS) (118).
“If your opponent is in superior strength, evade him.”
-The Art of War, Sun Tzu
Aggressive lymphomas encompass a subset of NHLs characterized by increased proliferation, more genetic complexity, and higher clinical acuity than indolent lymphomas. Subtypes such as MCL, DLBCL, and BL can exhibit brisk disease progression and are often driven by distinct genetic mutations or chromosomal rearrangements that influence both prognosis and treatment responsiveness (Table 4). Despite their aggressive nature, these malignancies can be highly responsive to therapy, especially when their specific molecular features are targeted.
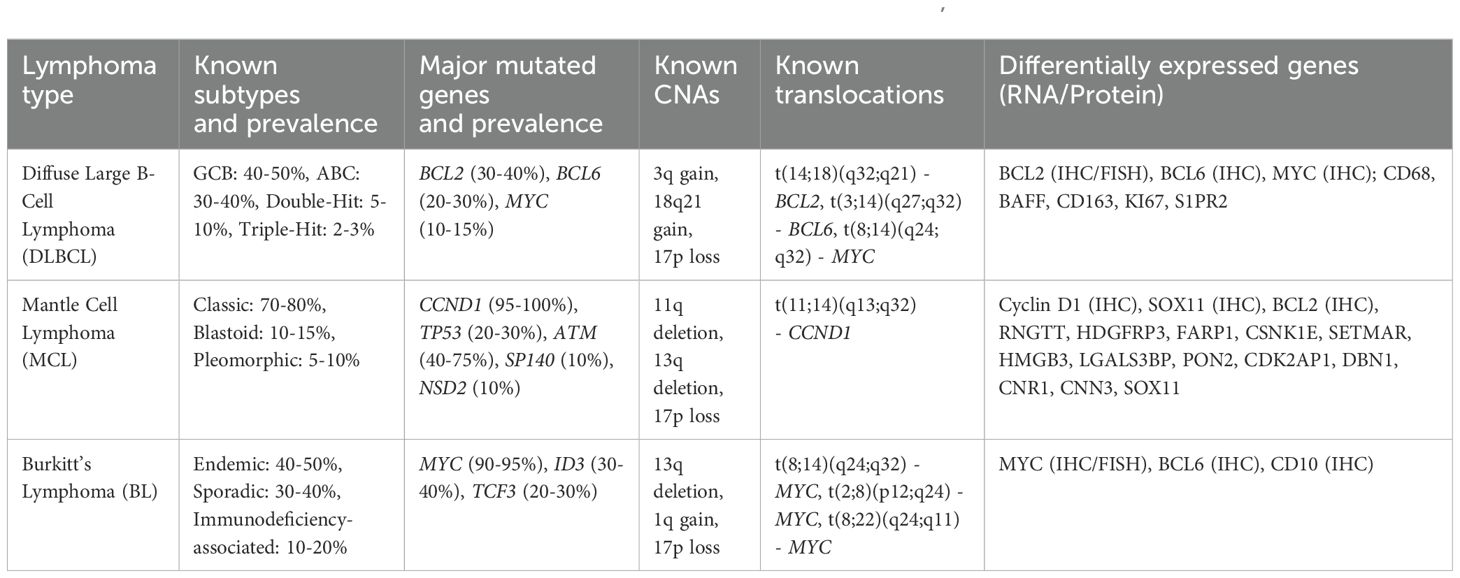
Table 4. Summary of aggressive non-Hodgkin lymphoma and associated genetic aberrations (92, 119–123).
MCL is a subtype of NHL characterized by the presence of characteristic rearrangement t(11;14) involving cyclin D1 (CCND1), a cell cycle signaling factor. Cyclin D1 can form a complex with CDK4 or CDK6, both of which are overexpressed in MCL (124). The downregulation of specific microRNAs has been linked to CDK6 upregulation and poorer survival (125). Studies have shown that there are also frequent mutations in the TP53 and ATM genes, which have implications for therapeutic sensitivity. In MCL, mutations in TP53 are associated with poor prognosis and resistance to conventional therapies (126). TP53 mutation is associated with MCL blastoid morphology, and its presence informs chemotherapy decision making. ATM is mutated in roughly 70% of MCL, and since they are thought to result in defective DNA repair mechanisms, they implicate ATM mutation as a likely MCL defining event. The high frequency of ATM mutations make it ideal for further study as a clinical biomarker (127). At present, neither TP53 nor ATM alteration are used routinely to guide RT utilization; however, MCL can be extremely sensitive to very low dose radiation (128), and therefore, a relationship between MCL’s altered DDR and radiosensitivity may exist.
Another interesting finding is that MCL has a relatively high degree of CNA compared to other lymphomas, perhaps attributed to mechanisms allowing a bypass of the normal cell cycle checks through cyclin D1/CDK4 (129). CNA in MCL has also been linked to altered MAP2 and MAP6, microtubule genes that could contribute to genomic instability (130). From WES of MCL, recurrent mutations were identified including WHSC1, RB1, POT1, and SMARCA4 (131). Another study noted 4 mutational signatures in MCL with different overall survival probabilities: mutated IGH variable, CCND1 mutation, amplified 11q13, and active B cell receptor signaling (122). The CCND1 signature was also associated with del(11q), ATM mutations, and upregulation of NF-κB and DNA repair pathways (122). Understanding these genetic mutations may allow for more precise decision making with respect to radiation doses, potentially improving patient outcomes by enhancing the therapeutic ratio.
BL is a highly aggressive form of NHL characterized by the rapid growth of malignant B-cells. There are three distinct clinical subtypes: sporadic BL, endemic BL predominantly in sub-Saharan Africa, and immunodeficiency-associated BL. Genetic studies have demonstrated both common mutations across subtypes (e.g., mutations in ID3), and distinct alterations for each subtype (132). BL is most commonly associated with translocation t(8;14)(q24;q32), which juxtaposes the MYC gene with IGH locus (121). This translocation leads to the overexpression of the MYC oncogene, a critical regulator of cell proliferation, apoptosis, and metabolism. The aberrant expression of MYC drives the uncontrolled growth and rapid proliferation of lymphoma cells, contributing to BL’s aggressive clinical behavior. The translocation involving MYC can also occur with other immunoglobulin loci, such as t(2;8)(p12;q24) involving the kappa light chain or t(8;22)(q24;q11) involving the lambda light chain, although these are less common. MYC dysregulation is a key driver of oncogenesis in BL, and its detection is critical for diagnosis and therapeutic decision making. Studies have shown that MYC overexpression is associated with poor prognosis and resistance to conventional therapies, making it a target of interest for novel therapeutic approaches (133). Sporadic and immunodeficiency-associated BLs were shown to be genetically similar with mutations in TCF3, CCND3, and SMARCA4 (134); whereas, endemic BL has more frequent mutations in BCL7A and BCL6 and fewer genetic alterations in DNMT1, SNTB2, and CTCF (132).
DLBCL is the most common form of NHL and is characterized by significant genetic and phenotypic heterogeneity, which influences treatment response and prognosis. DLBCL can be classified into two major phenotypes based on gene expression profiles: activated B-cell-like (ABC) and germinal center B-cell-like (GCB), with distinct molecular characteristics and clinical behaviors. ABC DLBCL is often associated with poorer prognosis and resistance to standard therapies, including radiation, due to constitutive activation of the NF-κB pathway (135). In contrast, GCB DLBCL generally has a better prognosis and is more responsive to chemotherapy (136). Despite efforts to classify the cell of origin of DLBCL, precision oncology based on these subtypes has not consistently translated into clinical benefit (137), although upfront Polatuzumab was recently demonstrated to have more benefit for ABC DLBCL than GCB DLBCL (138, 139). Rearrangements, mutations, and overexpression of BCL2, BCL6, and MYC are critical genetic alterations in DLBCL that further influence treatment outcomes. BCL2 rearrangements are more common in GCB DLBCL and are associated with resistance to apoptosis, which can impact the effectiveness of radiation therapy (140). BCL6 rearrangements, which are found in both ABC and GCB subtypes, can inhibit differentiation and promote survival of lymphoma cells, potentially affecting their radiosensitivity (141). MYC rearrangements, often associated with a more aggressive disease course, lead to overexpression of the MYC protein, driving rapid cell proliferation and metabolic activity. The co-occurrence of MYC rearrangements with BCL2 and/or BCL6 rearrangements, known previously as “double-hit” or “triple-hit” lymphomas, now known as High-Grade B-Cell Lymphoma (HGBCL), is associated with very poor prognosis and resistance to conventional treatments (142). MYC-positive cells are shown to have more oxidative stress and replication errors which lead to DNA damage and genomic instability (143). DDR activation in DLBCL correlates with MYC expression and predicts poor prognosis (144). In fact, inhibition of ATR-CHK1/2 mediated DDR was linked to chemotherapy resistance in MYC-positive DLBCL (145).
Early WES studies of DLBCL, showed recurrent mutations in MYD88, CARD11, EZH2, and CREBBP, which were known to be altered in DLBCL and somatic mutations in novel genes like MEF2B, MLL2, BTG1, GNA13, ACTB, P2RY8, PCLO, and TNFRSF14 (146). Later studies compared ABC and GCB subtypes to show GCB type preferentially mutated in EZH2, SGK1, GNA13, SOCS1, STAT6, and TNFRSF14, and ABC type biased toward mutations ETV6, MYD88, PIM1, and TBL1XR1 (147). Loss of CDKN2A is associated shorter survival after Rituximab-CHOP through dysregulation of the RB/E2F pathway, activation of cellular metabolism, and decreased immune and inflammatory responses (148). Shipp et al. identified five DLBCL genetic phenotypes including transformed from indolent lymphoma, two subsets of GCB-DLBCLs and group with biallelic inactivation of TP53, CDKN2A loss, and associated genomic instability (142). The LymphGen classification further refines the molecular categorization of DLBCL into distinct genetic subgroups with specific phenotypic and clinical characteristics (149). This classification includes subtypes MCD (MYD88L265P and CD79B mutations), BN2 (BCL6 fusions and NOTCH2 mutations), N1 (NOTCH1 mutations), EZB (EZH2 mutations and BCL2 translocations), ST2 (PI3K signaling and JAK2 signaling), A53 (low p53 target genes), and TP53Mut (p53 signaling dysregulation, immune deficiency, and PI3K activation), each associated with different therapeutic responses and outcomes (150). Lastly, primary central nervous system lymphoma (PCNSL) is an aggressive large B-cell lymphoma that also displays CDKN2A loss and mutations in MYD88, CD79B, and TBL1XR1, thus with potentially definable genetic subtypes (151). Understanding these genetic and phenotypic subtypes may allow for more precise tailoring of radiation therapy protocols. For instance, patients with MYC, BCL2, and/or BCL6 rearrangements may benefit from more aggressive treatment regimens or novel therapeutic combinations that enhance radiosensitivity and overcome resistance mechanisms.
T-cell lymphomas are a heterogeneous group of malignancies characterized by various genetic alterations that influence their response to treatments. Among the key genetic mutations associated with T-cell lymphomas are those in the TP53 and anaplastic lymphoma kinase (ALK) genes, which significantly impact disease behavior and treatment outcomes (152). Early genetic studies exploring mutations in ATM and TP53 showed that mutant mice developed T-cell lymphoma with mice having both genes knocked out displaying resistance to radiation (153). In T-cell lymphomas, TP53 mutations can lead to impaired apoptosis and increased survival of malignant cells despite radiation-induced DNA damage, suggesting a potential for reduced radiation sensitivity (154). Conversely, ALK mutations or translocations, particularly in ALK-positive anaplastic large cell lymphoma (ALCL), are associated with a distinct clinical and biological profile (155). ALK-positive T-cell lymphomas generally exhibit better responses to chemotherapy and RT compared to ALK-negative variants, likely due to the oncogenic driver role of ALK mutations that render the cells more susceptible to targeted treatments, though no clear relationship with radiation is known (156). The potential to understand variability in radiation sensitivity based on genetic alterations underscores the importance of molecular profiling in T-cell lymphomas to optimize treatment strategies and improve patient outcomes.
Plasma cell neoplasms like solitary plasmacytoma and multiple myeloma (MM) have also shown associations with mutations in the TP53 and ATM genes. Initial reports showed roughly a third of extramedullary MM reported t(4;14), deletion of 13q the RB1 locus, and deletion 17p the TP53 locus. Recently, WES of extramedullary MM showed most patients had 1q21 amplification including the CKS1B gene and at least one mutated gene in the MAPK signaling pathway, with KRAS as the most frequently mutated gene (157). NBS1 mutations in myeloma may contribute to carcinogenesis and may be targetable (158). Additionally, MM polymorphisms in DNA repair genes such as XRCC1 have been identified and could potentially influence radiosensitivity and treatment responses across myeloma and various lymphoid malignancies (159).
While chronic lymphocytic leukemia (CLL) is not thought to have a large amount of genetic dysregulation, Richter’s syndrome (RS), a transformation to aggressive disease, has recently been characterized through WGS with some recurrent mutations observed (160). Mutations were found in the DDR pathway, but also other genes like PTPRD and TRAF3. Immune genes were also implicated including BTG2, CXCR4, NFATC1, PAX5, and NOTCH. Transformed FL may be associated with CDKN2A/B deletions which are also associated with inferior PFS and OS after Rituximab (161).
“The rule is, not to besiege walled cities if it can possibly be avoided”
-The Art of War, Sun Tzu
While successful efforts in non-hematologic malignancies have linked radiation sensitivity to genomic classifiers for solid tumors, implicating genes like KEAP1 and CTNNB1 and pathways such as ROS reduction and cell-cycle deregulation, these studies have often explicitly omitted hematologic malignancies (162). In the absence of clear molecular markers of radiosensitivity, radiation oncologists determine radiation candidates, targets, and doses, based on factors including existing retrospective and prospective trials, lymphoma aggressiveness, disease burden, disease location, and chemotherapy response, though practice patterns may vary. As different hematologic cancer subtypes exhibit both distinct and common molecular traits that influence disease behavior and treatment response, we are now in an era of where integrating genomic information into treatment decision-making is possible. The prospect of incorporating genetic information into a radiation treatment plans has given rise to the exciting, emerging field of radiogenomics. Radiogenomics promises to identify genetic markers that can predict RT toxicity (163), and better predict and prognosticate RT efficacy (164). By leveraging genomic data, oncologists can already tailor treatment plans to individual patients in medical oncology, enhancing the precision and efficacy of chemoimmunotherapy, and similar techniques should exist in radiation oncology.
Genomic tools may offer a powerful method to help determine which patients are most likely to respond well to RT. While chemotherapy alone can effectively control malignancies, there are specific situations where RT after chemotherapy may be necessary, such as a bridging strategy before CAR T-cell therapy, cytoreduction prior to transplant, or consolidation therapy for high-risk disease. Identifying specific genomic markers linked to radiosensitivity can guide these decisions, particularly in cases like MCL and HL, where combined modality therapy may offer advantages, potentially reducing the need for extensive chemotherapy. In the pre-rituximab era, efforts like SWOG 8736 established that fewer cycles of chemotherapy combined with radiation were superior in treating DLBCL compared to more cycles of chemotherapy alone (165). Future efforts are needed to determine which genetic markers reliably identify patients who can tolerate reduced-intensity chemotherapy when additional cycles are substituted for radiotherapy.
Currently, it is unknown whether signals of radiosensitivity exist in hematologic malignancies. Recently, in an abstract, CREBBP alterations in FL were shown to predict response to very low dose RT, offering a significant advancement in RT personalization (166). However, in a study of higher RT doses in early-stage FL, no gene alterations were associated with outcomes (167). Similarly, Ma et al. explored genomic correlates of radiosensitivity in DLBCL, focusing on the LymphGen classification (168). Studies like these could stratify patients from the outset based on whether certain subtypes will likely benefit from lower-dose regimens. For example, ATM-mutant MCL could be preferentially treated with 4 Gy in an adaptive RT approach, potentially minimizing RT exposure for specific MCL patients. Additionally, genomic profiling opens opportunities for novel treatment combinations with lower RT doses. For example, incorporating EZH2 inhibitors with low-dose RT could be explored for specific LymphGen subtypes in relapsed/refractory DLBCL. Furthermore, radiopharmaceuticals could be utilized for patients with systemic disease predicted to have high RT sensitivity based on their genomic profile. However, challenges in accessing radiopharmaceutical agents and regulatory hurdles limit their use. Personalized genomic profiling may justify broader adoption of radiopharmaceuticals, which could also be combined with immune checkpoint inhibitors to unlock new treatment avenues for systemic lymphomas. By moving beyond standard approaches, these novel strategies have the potential to improve outcomes in patients with challenging lymphoma cases.
Bulky tumors, which are often less sensitive to radiation, might benefit from the addition of radiosensitizers based on specific genomic vulnerabilities. Conversely, identifying patients at lower risk of long-term RT toxicities—through germline or somatic mutations affecting DNA repair pathways—could lead to better-informed decisions about dose and modality, allowing oncologists to balance efficacy and safety. These personalized approaches align with the overarching goal of optimizing radiation oncology through genomics, ensuring that each patient receives the most appropriate treatment for their unique cancer profile.
Future directions in this field are promising, with ongoing research aimed at refining genomic profiling techniques and expanding our understanding of the molecular mechanisms underlying radiosensitivity. Studies are needed to investigate the significance of mutations in commonly mutated cell cycle-regulating genes like TP53, MYC, BCL2, and BCL6 and DDR genes like ATM. Additionally, associations between lymphoma and epigenetic modifiers offer clinically useful biomarkers given their high frequency and connection to disease development. As genomic technologies continue to evolve, they hold the potential to transform radiation oncology, making personalized treatment a standard practice. Collaborative efforts like the Radiogenomics Consortium will be essential in overcoming current obstacles and realizing the full potential of genomics in optimizing radiation therapy for lymphoma patients (169).
Recognizing the transformative potential of genomics in radiation oncology within the framework of precision medicine, the American Society for Radiation Oncology, American Association of Physicists in Medicine, and the National Cancer Institute convened a Precision Medicine in Radiation Oncology Workshop (170). The group emphasized the urgent need to develop and validate genomic markers for predicting radiosensitivity in tumors and normal tissues, with a focus on creating polygenic risk scores. They also called for deeper exploration of molecular mechanisms underlying radiosensitivity and resistance, including hypoxia, HPV status, and alterations in DNA damage response pathways, while investigating synergies between radiation therapy and systemic treatments like immune checkpoint inhibitors and targeted molecular therapies.
To support these goals, the workshop advocated for multi-institutional consortia such as ORIEN to pool resources for genomic research (171), cooperative group trials that integrate genomics into radiation therapy protocols, and the expansion of genomic repositories to include detailed radiation therapy data such as dose-volume histograms and locoregional recurrence rates. The group stressed the importance of designing randomized trials to assess the benefits of genomically personalized radiation therapy, including selective dose escalation and chemoradiotherapy combinations tailored to genetic profiles. Practical recommendations included enhancing radiation oncologists’ genomic literacy through residency curricula, symposia at national meetings, and dedicated study sections within funding agencies like the NIH to prioritize precision radiation oncology research. Lastly, the group highlighted the importance of fostering academic-industry collaborations to advance radiation-guided precision oncology platforms, drawing inspiration from the pharmaceutical industry’s success in integrating precision medicine.
“He will win who knows when to fight and when not to fight.”
-The Art of War, Sun Tzu
Translating genetic research into the clinic poses numerous challenges, largely due to the complexity and variability of genetic data. The transition from candidate gene studies to GWAS necessitates robust validation and reproducibility (172), and large, well-designed studies to confirm associations before clinical use (173). Ethical, legal, and social implications also play a significant role (174), as incorporating genetic information into cancer care issues of consent, privacy, and potential discrimination. Integrating genetic data into clinical workflows presents practical challenges, requiring comprehensive clinical decision support systems, timely availability during clinical decision-making, and extensive training for healthcare providers for interpretation (70). Technical and methodological barriers, such as the need for standardized protocols and the complexity of polygenic models, further complicate the clinical translation of genetic research (172). Economic and resource constraints are additional obstacles resulting in financial and infrastructural requirements for implementing genetic testing and genomic data analysis in clinical practice (175). Moreover, the acceptance of genetic testing by both patients and physicians remains a significant barrier, and education is needed to facilitate the adoption of genetic research in clinical settings (176). Regulatory and policy issues also need to be addressed, highlighting the necessity for clear guidelines and policies to support the integration of genetic research into clinical practice (177). Future research should focus on addressing these challenges, developing more precise and comprehensive genomic profiles, and validating predictive models in large, diverse patient populations.
“Ponder and deliberate before you make a move.”
-The Art of War, Sun Tzu
The intersection of genetic research and radiation therapy in hematologic malignancies represents a promising frontier for personalized medicine. Over the years, substantial progress has been made in understanding the molecular underpinnings of radiosensitivity through the study of key genes involved in DNA repair and cell cycle regulation. Syndromes such as Ataxia-Telangiectasia, Nijmegen Breakage Syndrome, and Li-Fraumeni Syndrome have provided critical insights into the role of ATM, NBS1, and TP53 mutations in radiation response. Additionally, advances in genomic profiling have revealed complex mutational landscapes in various types of non-Hodgkin lymphomas, including FL, MCL, and MALT. These discoveries have paved the way for the development of predictive tools which combines clinical and genetic information to guide therapeutic decisions and optimize treatment outcomes.
The war on cancer needs to be fought with smarter, more deliberate tactics, and future research should focus on validating genetic markers of radiosensitivity through large, well-designed clinical trials and exploring novel therapeutic combinations to overcome resistance mechanisms. The emerging field of radiogenomics holds great potential to enhance the precision of radiation therapy, minimize adverse effects, and improve patient outcomes. By bridging the gap between genetic research and clinical application, we can move closer to realizing the full potential of personalized RT in the treatment of hematologic malignancies.
NW: Conceptualization, Data curation, Investigation, Writing – original draft, Writing – review & editing. JY: Writing – review & editing. BI: Conceptualization, Writing – review & editing.
The author(s) declare that no financial support was received for the research and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Friedman DA, Tait L, Vaughan AT. Influence of nuclear structure on the formation of radiation-induced lethal lesions. Int J Radiat Biol. (2016) 92:229–40. doi: 10.3109/09553002.2016.1144941
2. Harrison L, Blackwell K. Hypoxia and anemia: factors in decreased sensitivity to radiation therapy and chemotherapy? oncologist. (2004) 9:31–40. doi: 10.1634/theoncologist.9-90005-31
3. Pawlik TM, Keyomarsi K. Role of cell cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol Biol Physics. (2004) 59:928–42. doi: 10.1016/j.ijrobp.2004.03.005
4. Deckbar D, Jeggo PA, Löbrich M. Understanding the limitations of radiation-induced cell cycle checkpoints. Crit Rev Biochem Mol Biol. (2011) 46:271–83. doi: 10.3109/10409238.2011.575764
5. Kciuk M, Marciniak B, Mojzych M, Kontek R. Focus on UV-induced DNA damage and repair—disease relevance and protective strategies. Int J Mol Sci. (2020) 21:7264. doi: 10.3390/ijms21197264
6. Lee CK. Evolving role of radiation therapy for hematologic Malignancies. Hematology/Oncol Clinics. (2006) 20:471–503. doi: 10.1016/j.hoc.2006.01.020
7. Adams KE, Medhurst AL, Dart DA, Lakin ND. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. (2006) 25:3894–904. doi: 10.1038/sj.onc.1209426
8. Morse ML, Dahl RH. Cellular glutathione is a key to the oxygen effect in radiation damage. Nature. (1978) 271:660–2. doi: 10.1038/271660a0
9. Bender K, Blattner C, Knebel A, Iordanov M, Herrlich P, Rahmsdorf HJ. UV-induced signal transduction. J Photochem Photobiol B. (1997) 37:1–17. doi: 10.1016/S1011-1344(96)07459-3
10. Benjamin CL, Ananthaswamy HN. p53 and the pathogenesis of skin cancer. Toxicol Appl Pharmacol. (2007) 224:241–8. doi: 10.1016/j.taap.2006.12.006
11. Davalli P, Marverti G, Lauriola A, D’Arca D. Targeting oxidatively induced DNA damage response in cancer: opportunities for novel cancer therapies. Oxid Med Cell longevity. (2018) 2018:2389523. doi: 10.1155/2018/2389523
12. Li Z, Pearlman AH, Hsieh P. DNA mismatch repair and the DNA damage response. DNA repair. (2016) 38:94–101. doi: 10.1016/j.dnarep.2015.11.019
13. Lam FC. The DNA damage response-from cell biology to human disease. J Transl Genet Genom. (2022) 6:204–22. doi: 10.20517/jtgg.2021.61
14. Lavin MF, Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu Rev Immunol. (1997) 15:177–202. doi: 10.1146/annurev.immunol.15.1.177
15. Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. (1995) 268:1749–53. doi: 10.1126/science.7792600
16. Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, et al. A novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. (1998) 93:467–76. doi: 10.1016/S0092-8674(00)81174-5
17. Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. (1989) 244:217–21. doi: 10.1126/science.2649981
18. Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harbor Perspect Biol. (2010) 2:a001008. doi: 10.1101/cshperspect.a001008
19. Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, et al. TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum mutation. (2016) 37:865–76. doi: 10.1002/humu.2016.37.issue-9
20. Varley JM, Chapman P, McGown G, Thorncroft M, White GR, Greaves MJ, et al. Genetic and functional studies of a germline TP53 splicing mutation in a Li–Fraumeni-like family. Oncogene. (1998) 16:3291–8. doi: 10.1038/sj.onc.1201878
21. Dworaczek H, Xiao W. Xeroderma pigmentosum: a glimpse into nucleotide excision repair, genetic instability, and cancer. Crit Reviews™ Oncogenesis. (2007) 13:159–77. doi: 10.1615/CritRevOncog.v13.i2.20
22. Strathdee CA, Gavish H, Shannon WR, Buchwald M. Cloning of cDNAs for Fanconi’s anaemia by functional complementation. Nature. (1992) 356:763–7. doi: 10.1038/356763a0
23. Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. (1994) 266:66–71. doi: 10.1126/science.7545954
24. Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. (1995) 378:789–92. doi: 10.1038/378789a0
25. Shinohara A, Shinohara M. Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenetic Genome Res. (2004) 107:201–7. doi: 10.1159/000080598
26. Thompson LH, Brookman KW, Jones NJ, Allen SA, Carrano AV. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol Cell Biol. (1990) 10:6160–71. doi: 10.1128/mcb.10.12.6160
27. Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci. (2000) 97:10389–94. doi: 10.1073/pnas.190030497
28. Zannini L, Delia D, Buscemi G. CHK2 kinase in the DNA damage response and beyond. J Mol Cell Biol. (2014) 6:442–57. doi: 10.1093/jmcb/mju045
29. Bell DW, Sikdar N, Lee KY, Price JC, Chatterjee R, Park HD, et al. Predisposition to cancer caused by genetic and functional defects of mammalian Atad5. PloS Genet. (2011) 7:e1002245. doi: 10.1371/journal.pgen.1002245
30. O’Neil NJ, Bailey ML, Hieter P. Synthetic lethality and cancer. Nat Rev Genet. (2017) 18:613–23. doi: 10.1038/nrg.2017.47
31. Yap TA, Sandhu SK, Carden CP, De Bono JS. Poly (ADP-Ribose) polymerase (PARP) inhibitors: Exploiting a synthetic lethal strategy in the clinic. CA: Cancer J Clin. (2011) 61:31–49. doi: 10.3322/caac.20095
32. Spitzner M, Ebner R, Wolff HA, Ghadimi BM, Wienands J, Grade M. STAT3: A novel molecular mediator of resistance to chemoradiotherapy. Cancers. (2014) 6:1986–2011. doi: 10.3390/cancers6041986
33. Park SY, Lee CJ, Choi JH, Kim JH, Kim JW, Kim JY, et al. The JAK2/STAT3/CCND2 Axis promotes colorectal Cancer stem cell persistence and radioresistance. J Exp Clin Cancer Res. (2019) 38:399. doi: 10.1186/s13046-019-1405-7
34. Yahyanejad S, Theys J, Vooijs M. Targeting Notch to overcome radiation resistance. Oncotarget. (2016) 7:7610. doi: 10.18632/oncotarget.6714
35. Deng X, Michaelson D, Tchieu J, Cheng J, Rothenstein D, Feldman R, et al. Targeting homologous recombination in notch-driven C. elegans stem cell and human tumors. PloS One. (2015) 10:e0127862. doi: 10.1371/journal.pone.0127862
36. Singh V, Gupta D, Arora R. NF-kB as a key player in regulation of cellular radiation responses and identification of radiation countermeasures. Discoveries. (2015) 3:1–22. doi: 10.15190/d.2015.27
37. Pitter KL, Casey DL, Lu YC, Hannum M, Zhang Z, Song X, et al. Pathogenic ATM mutations in cancer and a genetic basis for radiotherapeutic efficacy. JNCI: J Natl Cancer Institute. (2021) 113:266–73. doi: 10.1093/jnci/djaa095
38. Kim KH, Kim HS, Kim SS, Shim HS, Yang AJ, Lee JJ, et al. Increased radiosensitivity of solid tumors harboring ATM and BRCA1/2 mutations. Cancer Res Treat. (2022) 54:54–64. doi: 10.4143/crt.2020.1247
39. Leeman JE, Li Y, Bell A, Hussain SS, Majumdar R, Rong-Mullins X, et al. Human papillomavirus 16 promotes microhomology-mediated end-joining. Proc Natl Acad Sci. (2019) 116:21573–9. doi: 10.1073/pnas.1906120116
40. Mirghani H, Amen F, Tao Y, Deutsch E, Levy A. Increased radiosensitivity of HPV-positive head and neck cancers: Molecular basis and therapeutic perspectives. Cancer Treat Rev. (2015) 41:844–52. doi: 10.1016/j.ctrv.2015.10.001
41. Tsai CJ, Kim JK, McBride S, Gelblum D, Riaz N, Dutta PR, et al. Prospective personalized elective nodal dose de-escalation in patients with human papillomavirus (HPV)-positive oropharyngeal cancer (OPC) undergoing definitive concurrent chemoradiation (CCRT): acute toxicities and tolerance. Int J Radiat Oncol Biol Physics. (2018) 102:e362. doi: 10.1016/j.ijrobp.2018.07.1088
42. Lee NY, Sherman EJ, Schöder H, Wray R, Boyle JO, Singh B, et al. Hypoxia-directed treatment of human papillomavirus–related oropharyngeal carcinoma. J Clin Oncol. (2024) 42:940–50. doi: 10.1200/JCO.23.01308
43. Solin LJ, Gray R, Baehner FL, Butler SM, Hughes LL, Yoshizawa C, et al. A multigene expression assay to predict local recurrence risk for ductal carcinoma in situ of the breast. J Natl Cancer Institute. (2013) 105:701–10. doi: 10.1093/jnci/djt067
44. Raldow AC, Sher D, Chen AB, Recht A, Punglia RS. Cost effectiveness of the oncotype DX DCIS score for guiding treatment of patients with ductal carcinoma in situ. J Clin Oncol. (2016) 34:3963–8. doi: 10.1200/JCO.2016.67.8532
45. Barnett GC, Coles CE, Elliott RM, Baynes C, Luccarini C, Conroy D, et al. Independent validation of genes and polymorphisms reported to be associated with radiation toxicity: a prospective analysis study. Lancet Oncol. (2012) 13:65–77. doi: 10.1016/S1470-2045(11)70302-3
46. Oppitz U, Bernthaler U, Schindler D, Sobeck A, Hoehn H, Platzer M, et al. Sequence analysis of the ATM gene in 20 patients with RTOG grade 3 or 4 acute and/or late tissue radiation side effects. Int J Radiat Oncol Biol Physics. (1999) 44:981–8. doi: 10.1016/S0360-3016(99)00108-X
47. Pollard JM, Gatti RA. Clinical radiation sensitivity with DNA repair disorders: an overview. Int J Radiat Oncol Biol Physics. (2009) 74:1323–31. doi: 10.1016/j.ijrobp.2009.02.057
48. Cesaretti JA, Stock RG, Lehrer S, Atencio DA, Bernstein JL, Stone NN, et al. ATM sequence variants are predictive of adverse radiotherapy response among patients treated for prostate cancer. Int J Radiat Oncol Biol Physics. (2005) 61:196–202. doi: 10.1016/j.ijrobp.2004.09.031
49. Granzotto A, Benadjaoud MA, Vogin G, Devic C, Ferlazzo ML, Bodgi L, et al. Influence of nucleoshuttling of the ATM protein in the healthy tissues response to radiation therapy: toward a molecular classification of human radiosensitivity. Int J Radiat Oncol Biol Physics. (2016) 94:450–60. doi: 10.1016/j.ijrobp.2015.11.013
50. Weidhaas JB, Marco N, Steinberg ML, Lee A, Xiang M, Valle LF, et al. Early findings from the GARUDA trial: The impact of a genetic signature of late radiation toxicity on prostate cancer treatment decision making. J Clin Oncol. (2023) 41:5089. doi: 10.1200/JCO.2023.41.16_suppl.5089
51. Azria D, Ozsahin M, Kramar A, Peters S, Atencio DP, Crompton NE, et al. Single nucleotide polymorphisms, apoptosis, and the development of severe late adverse effects after radiotherapy. Clin Cancer Res. (2008) 14:6284–8. doi: 10.1158/1078-0432.CCR-08-0700
52. Parliament MB, Murray D. Single nucleotide polymorphisms of DNA repair genes as predictors of radioresponse. Semin Radiat Oncol. (2010) 20:232–40. doi: 10.1016/j.semradonc.2010.05.003
53. Alsner J, Andreassen CN, Overgaard J. Genetic markers for prediction of normal tissue toxicity after radiotherapy. Semin Radiat Oncol. (2008) 18:126–35. doi: 10.1016/j.semradonc.2007.10.004
54. Senkus-Konefka E, Jassem J. Complications of breast-cancer radiotherapy. Clin Oncol. (2006) 18:229–35. doi: 10.1016/j.clon.2005.11.004
55. Bourgier C, Kerns S, Gourgou S, Lemanski C, Gutowski M, Fenoglietto P, et al. Concurrent or sequential letrozole with adjuvant breast radiotherapy: final results of the CO-HO-RT phase II randomized trial. Ann Oncol. (2016) 27:474–80. doi: 10.1093/annonc/mdv602
56. Dai YH, Wang YF, Shen PC, Lo CH, Yang JF, Lin CS, et al. Radiosensitivity index emerges as a potential biomarker for combined radiotherapy and immunotherapy. NPJ genomic Med. (2021) 6:40. doi: 10.1038/s41525-021-00200-0
57. Eschrich SA, Fulp WJ, Pawitan Y, Foekens JA, Smid M, Martens JW, et al. Validation of a radiosensitivity molecular signature in breast cancer. Clin Cancer Res. (2012) 18:5134–43. doi: 10.1158/1078-0432.CCR-12-0891
58. Scott JG, Berglund A, Schell MJ, Mihaylov I, Fulp WJ, Yue B, et al. A genome-based model for adjusting radiotherapy dose (GARD): a retrospective, cohort-based study. Lancet Oncol. (2017) 18:202–11. doi: 10.1016/S1470-2045(16)30648-9
59. Ahmed KA, Liveringhouse CL, Mills MN, Figura NB, Grass GD, Washington IR, et al. Utilizing the genomically adjusted radiation dose (GARD) to personalize adjuvant radiotherapy in triple negative breast cancer management. EBioMedicine. (2019) 47:163–9. doi: 10.1016/j.ebiom.2019.08.019
60. Schmidt KT, Chau CH, Price DK, Figg WD. Precision oncology medicine: the clinical relevance of patient-specific biomarkers used to optimize cancer treatment. J Clin Pharmacol. (2016) 56:1484–99. doi: 10.1002/jcph.v56.12
61. Bush RS, Bruce WR. The radiation sensitivity of transplanted lymphoma cells as determined by the spleen colony method. Radiat Res. (1964) 21:612–21. doi: 10.2307/3571654
62. Malaise EP, Fertil B, Chavaudra N, Guichard M. Distribution of radiation sensitivities for human tumor cells of specific histological types: comparison of in vitro to in vivo data. Int J Radiat Oncol Biol Physics. (1986) 12:617–24. doi: 10.1016/0360-3016(86)90071-4
63. M’kacher R, Bennaceur A, Farace F, Lauge A, Plassa LF, Wittmer E, et al. Multiple molecular mechanisms contribute to radiation sensitivity in mantle cell lymphoma. Oncogene. (2003) 22:7905–12. doi: 10.1038/sj.onc.1206826
64. Reimann M, Loddenkemper C, Rudolph C, Schildhauer I, Teichmann B, Stein H, et al. The Myc-evoked DNA damage response accounts for treatment resistance in primary lymphomas in vivo. Blood. J Am Soc Hematol. (2007) 110:2996–3004. doi: 10.1182/blood-2007-02-075614
65. O’Connor PM, Jackman J, Jondle D, Bhatia K, Magrath I, Kohn KW. Role of the p53 tumor suppressor gene in cell cycle arrest and radiosensitivity of Burkitt’s lymphoma cell lines. Cancer Res. (1993) 53:4776–80.
66. Bae I, Fan S, Bhatia K, Kohn KW, Fornace AJ Jr, O’Connor PM. Relationships between G1 arrest and stability of the p53 and p21Cip1/Waf1 proteins following γ-irradiation of human lymphoma cells. Cancer Res. (1995) 55:2387–93.
67. Qiao Q, Jiang Y, Li G. Inhibition of the PI3K/AKT-NF-κB pathway with curcumin enhanced radiation-induced apoptosis in human Burkitt’s lymphoma. J Pharmacol Sci. (2013) 121:247–56. doi: 10.1254/jphs.12149FP
68. Mirkovic N, Voehringer DW, Story MD, McConkey DJ, McDonnell TJ, Meyn RE. Resistance to radiation-induced apoptosis in Bcl-2-expressing cells is reversed by depleting cellular thiols. Oncogene. (1997) 15:1461–70. doi: 10.1038/sj.onc.1201310
69. Barnett GC, Kerns SL, Noble DJ, Dunning AM, West CM, Burnet NG. Incorporating genetic biomarkers into predictive models of normal tissue toxicity. Clin Oncol. (2015) 27:579–87. doi: 10.1016/j.clon.2015.06.013
70. Overgaard J, Aznar MC, Bacchus C, Coppes RP, Deutsch E, Georg D, et al. Personalised radiation therapy taking both the tumour and patient into consideration. Radiother Oncol. (2022) 166:A1–5. doi: 10.1016/j.radonc.2022.01.010
71. Julia E, Salles G. EZH2 inhibition by tazemetostat: mechanisms of action, safety and efficacy in relapsed/refractory follicular lymphoma. Future Oncol. (2021) 17:2127–40. doi: 10.2217/fon-2020-1244
72. Mata E, Díaz-López A, Martín-Moreno AM, Sánchez-Beato M, Varela I, Mestre MJ, et al. Analysis of the mutational landscape of classic Hodgkin lymphoma identifies disease heterogeneity and potential therapeutic targets. Oncotarget. (2017) 8:111386. doi: 10.18632/oncotarget.22799
73. Brune MM, Juskevicius D, Haslbauer J, Dirnhofer S, Tzankov A. Genomic landscape of Hodgkin lymphoma. Cancers. (2021) 13:682. doi: 10.3390/cancers13040682
74. Alig SK, Shahrokh Esfahani M, Garofalo A, Li MY, Rossi C, Flerlage T, et al. Distinct Hodgkin lymphoma subtypes defined by noninvasive genomic profiling. Nature. (2024) 625:778–87. doi: 10.1038/s41586-023-06903-x
75. Birgersdotter A, Baumforth KR, Porwit A, Sjöberg J, Wei W, Björkholm M, et al. Inflammation and tissue repair markers distinguish the nodular sclerosis and mixed cellularity subtypes of classical Hodgkin’s lymphoma. Br J cancer. (2009) 101:1393–401. doi: 10.1038/sj.bjc.6605238
76. Maura F, Ziccheddu B, Xiang JZ, Bhinder B, Rosiene J, Abascal F, et al. Molecular evolution of classic Hodgkin lymphoma revealed through whole-genome sequencing of Hodgkin and Reed Sternberg cells. Blood Cancer discovery. (2023) 4:208–27. doi: 10.1158/2643-3230.BCD-22-0128
77. Gomez F, Fisk B, McMichael JF, Mosior M, Foltz JA, Skidmore ZL, et al. Ultra-deep sequencing reveals the mutational landscape of classical Hodgkin lymphoma. Cancer Res Commun. (2023) 3:2312–30. doi: 10.1158/2767-9764.CRC-23-0140
78. Maggio EM, Stekelenburg E, Van den Berg A, Poppema S. TP53 gene mutations in Hodgkin lymphoma are infrequent and not associated with absence of Epstein-Barr virus. Int J cancer. (2001) 94:60–6. doi: 10.1002/(ISSN)1097-0215
79. Rassidakis GZ, Medeiros LJ, Vassilakopoulos TP, Viviani S, Bonfante V, Nadali G, et al. BCL-2 expression in Hodgkin and Reed-Sternberg cells of classical Hodgkin disease predicts a poorer prognosis in patients treated with ABVD or equivalent regimens. Blood J Am Soc Hematol. (2002) 100:3935–41. doi: 10.1182/blood.V100.12.3935
80. Mata E, Fernández S, Astudillo A, Fernández R, García-Cosío M, Sánchez-Beato M, et al. Genomic analyses of microdissected Hodgkin and Reed-Sternberg cells: mutations in epigenetic regulators and p53 are frequent in refractory classic Hodgkin lymphoma. Blood Cancer J. (2019) 9:34. doi: 10.1038/s41408-019-0195-7
81. Li Z, Mu W, Xiao M. Genetic lesions and targeted therapy in Hodgkin lymphoma. Ther Adv Hematol. (2023) 14:20406207221149245. doi: 10.1177/20406207221149245
82. Liu A, Takakuwa T, Fujita S, Luo WJ, Tresnasari K, Van den Berg A, et al. ATR alterations in Hodgkin’s lymphoma. Oncol Rep. (2008) 19:999–1005. doi: 10.3892/or.19.4.999
83. Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, et al. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. (2000) 14:2989–3002. doi: 10.1101/gad.851000
84. Salipante SJ, Mealiffe ME, Wechsler J, Krem MM, Liu Y, Namkoong S, et al. Mutations in a gene encoding a midbody kelch protein in familial and sporadic classical Hodgkin lymphoma lead to binucleated cells. Proc Natl Acad Sci. (2009) 106:14920–5. doi: 10.1073/pnas.0904231106
85. Saarinen S, Aavikko M, Aittomäki K, Launonen V, Lehtonen R, Franssila K, et al. Exome sequencing reveals germline NPAT mutation as a candidate risk factor for Hodgkin lymphoma. Blood J Am Soc Hematol. (2011) 118:493–8. doi: 10.1182/blood-2011-03-341560
86. Cosper PF, Copeland SE, Tucker JB, Weaver BA. Chromosome missegregation as a modulator of radiation sensitivity. Semin Radiat Oncol. (2022) 32:54–63. doi: 10.1016/j.semradonc.2021.09.002
87. Steidl C, Telenius A, Shah SP, Farinha P, Barclay L, Boyle M, et al. Genome-wide copy number analysis of Hodgkin Reed-Sternberg cells identifies recurrent imbalances with correlations to treatment outcome. Blood J Am Soc Hematol. (2010) 116:418–27. doi: 10.1182/blood-2009-12-257345
88. Derenzini E, Younes A. Predicting treatment outcome in classical Hodgkin lymphoma: genomic advances. Genome Med. (2011) 3:1–3. doi: 10.1186/gm240
89. National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology (NCCN guidelines): B-cell lymphomas. Version 1.2025 (2025). Available online at: https://www.nccn.org/guidelines/category_1 (Accessed January 01, 2025).
90. Ma MC, Tadros S, Bouska A, Heavican T, Yang H, Deng Q, et al. Subtype-specific and co-occurring genetic alterations in B-cell non-Hodgkin lymphoma. Haematologica. (2021) 107:690. doi: 10.3324/haematol.2020.274258
91. Gaidano G, Rossi D. The mutational landscape of chronic lymphocytic leukemia and its impact on prognosis and treatment. Hematol 2014 Am Soc Hematol Educ Program Book. (2017) 2017:329–37. doi: 10.1182/asheducation-2017.1.329
92. Bobée V, Drieux F, Marchand V, Sater V, Veresezan L, Picquenot JM, et al. Combining gene expression profiling and machine learning to diagnose B-cell non-Hodgkin lymphoma. Blood Cancer J. (2020) 10:59. doi: 10.1038/s41408-020-0322-5
93. Spina V, Khiabanian H, Messina M, Monti S, Cascione L, Bruscaggin A, et al. The genetics of nodal marginal zone lymphoma. Blood J Am Soc Hematol. (2016) 128:1362–73. doi: 10.1182/blood-2016-02-696757
94. van den Brand M, van Krieken JH. Recognizing nodal marginal zone lymphoma: recent advances and pitfalls. A systematic review haematologica. (2013) 98:1003. doi: 10.3324/haematol.2012.083386
95. Staiger AM, Hoster E, Jurinovic V, Winter S, Leich E, Kalla C, et al. Localized-and advanced-stage follicular lymphomas differ in their gene expression profiles. Blood J Am Soc Hematol. (2020) 135:181–90. doi: 10.1182/blood.2019000560
96. Green MR, Kihira S, Liu CL, Nair RV, Salari R, Gentles AJ, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci. (2015) 112:E1116–25. doi: 10.1073/pnas.1501199112
97. Sagaert X, Laurent M, Baens M, Wlodarska I, De-Wolf-Peeters C. MALT1 and BCL10 aberrations in MALT lymphomas and their effect on the expression of BCL10 in the tumour cells. Modern pathol. (2006) 19:225–32. doi: 10.1038/modpathol.3800523
98. Di Rocco A, Petrucci L, Assanto GM, Martelli M, Pulsoni A. Extranodal marginal zone lymphoma: pathogenesis, diagnosis and treatment. Cancers. (2022) 14:1742. doi: 10.3390/cancers14071742
99. Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez JM, Hossfeld DK, et al. The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t (11; 18)(q21; q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood J Am Soc Hematol. (1999) 93:3601–9.
100. Treon SP, Xu L, Yang G, Zhou Y, Liu X, Cao Y, et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. New Engl J Med. (2012) 367:826–33. doi: 10.1056/NEJMoa1200710
101. Green MR. Chromatin modifying gene mutations in follicular lymphoma. Blood J Am Soc Hematol. (2018) 131:595–604. doi: 10.1182/blood-2017-08-737361
102. Ren J, Chu Y, Ma H, Zhang Y, Zhang X, Zhao D, et al. Epigenetic interventions increase the radiation sensitivity of cancer cells. Curr Pharm design. (2014) 20:1857–65. doi: 10.2174/13816128113199990529
103. Pastore A, Jurinovic V, Kridel R, Hoster E, Staiger AM, Szczepanowski M, et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol. (2015) 16:1111–22. doi: 10.1016/S1470-2045(15)00169-2
104. Lockmer S, Ren W, Brodtkorb M, Østenstad B, Wahlin BE, Pan-Hammarström Q, et al. M7-FLIPI is not prognostic in follicular lymphoma patients with first-line rituximab chemo-free therapy. Br J haematol. (2020) 188:259–67. doi: 10.1111/bjh.v188.2
105. Vindi J, Kridel R, Staiger AM, Szczepanowski M, Horn H, Dreyling MH, et al. A clinicogenetic risk model (m7-FLIPI) prospectively identifies one-half of patients with early disease progression of follicular lymphoma after first-line immunochemotherapy. Blood. (2015) 126:333. doi: 10.1182/blood.V126.23.333.333
106. Marcus R, Davies A, Ando K, Klapper W, Opat S, Owen C, et al. Obinutuzumab for the first-line treatment of follicular lymphoma. New Engl J Med. (2017) 377:1331–44. doi: 10.1056/NEJMoa1614598
107. Jurinovic V, Passerini V, Oestergaard MZ, Knapp A, Mundt K, Araf S, et al. Evaluation of the m7-FLIPI in patients with follicular lymphoma treated within the gallium trial: EZH2 mutation status may be a predictive marker for differential efficacy of chemotherapy. Blood. (2019) 134:122. doi: 10.1182/blood-2019-130208
108. Zhou T, Pittaluga S, Jaffe ES. Molecular insights into the pathogenesis of follicular lymphoma. Ann Lymphoma. (2021) 5:1–13. doi: 10.21037/aol-20-49
109. Nann D, Ramis-Zaldivar JE, Müller I, Gonzalez-Farre B, Schmidt J, Egan C, et al. Follicular lymphoma t (14; 18)-negative is genetically a heterogeneous disease. Blood advances. (2020) 4:5652–65. doi: 10.1182/bloodadvances.2020002944
110. Louissaint A Jr., Schafernak KT, Geyer JT, Kovach AE, Ghandi M, Gratzinger D, et al. Pediatric-type nodal follicular lymphoma: a biologically distinct lymphoma with frequent MAPK pathway mutations. Blood J Am Soc Hematol. (2016) 128:1093–100. doi: 10.1182/blood-2015-12-682591
111. Agrawal R, Wang J. Pediatric follicular lymphoma: a rare clinicopathologic entity. Archives of pathology & laboratory medicine. (2009) 133(1):142–6.
112. Wang Y, Deng X, Xie J, Lu T, Qian R, Guo Z, et al. The COP9 signalosome stabilized MALT1 promotes Non-Small Cell Lung Cancer progression through activation of NF-κB pathway. Cell Biol Toxicol. (2024) 40:45. doi: 10.1007/s10565-024-09888-z
113. Hyeon J, Lee B, Shin S-H, Yoo HY, Kim SJ, Kim WS, et al. Targeted deep sequencing of gastric marginal zone lymphoma identified alterations of TRAF3 and TNFAIP3 that were mutually exclusive for MALT1 rearrangement. Mod Pathol. (2018) 31:1418–28. doi: 10.1038/s41379-018-0064-0
114. Yang C, Zang W, Tang Z, Ji Y, Xu R, Yang Y, et al. A20/TNFAIP3 regulates the DNA damage response and mediates tumor cell resistance to DNA-damaging therapy. Cancer Res. (2018) 78:1069–82. doi: 10.1158/0008-5472.CAN-17-2143
115. Wang P, Ke L, Cai C, Dong F. LINC01578 affects the radiation resistance of lung cancer cells through regulating microRNA-216b-5p/TBL1XR1 axis. Bioengineered. (2022) 13:10721–33. doi: 10.1080/21655979.2022.2051881
116. Johansson P, Klein-Hitpass L, Budeus B, Kuhn M, Lauber C, Seifert M, et al. Identifying genetic lesions in ocular adnexal extranodal marginal zone lymphomas of the MALT subtype by whole genome, whole exome and targeted sequencing. Cancers. (2020) 12:986. doi: 10.3390/cancers12040986
117. Jung H, Yoo HY, Lee SH, Shin S, Kim SC, Lee S, et al. The mutational landscape of ocular marginal zone lymphoma identifies frequent alterations in TNFAIP3 followed by mutations in TBL1XR1 and CREBBP. Oncotarget. (2017) 8:17038–49. doi: 10.18632/oncotarget.14928
118. Johansson P, Klein-Hitpass L, Grabellus F, Arnold G, Klapper W, Pförtner R, et al. Recurrent mutations in NF-κB pathway components, KMT2D, and NOTCH1/2 in ocular adnexal MALT-type marginal zone lymphomas. Oncotarget. (2016) 7:62627–39. doi: 10.18632/oncotarget.11548
119. Miao Y, Medeiros LJ, Li Y, Li J, Young KH. Genetic alterations and their clinical implications in DLBCL. Nat Rev Clin Oncol. (2019) 16:634–52. doi: 10.1038/s41571-019-0225-1
120. Royo C, Salaverria I, Hartmann EM, Rosenwald A, Campo E, Beà S. The complex landscape of genetic alterations in mantle cell lymphoma. Semin Cancer Biol. (2011) 21:322–34. doi: 10.1016/j.semcancer.2011.09.007
121. Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. (2012) 44:1321–5. doi: 10.1038/ng.2468
122. Yi S, Yan Y, Jin M, Bhattacharya S, Wang Y, Wu Y, et al. Genomic and transcriptomic profiling reveals distinct molecular subsets associated with outcomes in mantle cell lymphoma. J Clin Invest. (2022) 132:1–18. doi: 10.1172/JCI153283
123. Fernàndez V, Salamero O, Espinet B, Solé F, Royo C, Navarro A, et al. Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res. (2010) 70:1408–18. doi: 10.1158/0008-5472.CAN-09-3419
124. Pérez-Galán P, Dreyling M, Wiestner A. Mantle cell lymphoma: biology, pathogenesis, and the molecular basis of treatment in the genomic era. Blood J Am Soc Hematol. (2011) 117:26–38. doi: 10.1182/blood-2010-04-189977
125. Zhao JJ, Lin J, Lwin T, Yang H, Guo J, Kong W, et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood J Am Soc Hematol. (2010) 115:2630–9. doi: 10.1182/blood-2009-09-243147
126. Eskelund CW, Dahl C, Hansen JW, Westman M, Kolstad A, Pedersen LB, et al. TP53 mutations identify younger mantle cell lymphoma patients who do not benefit from intensive chemoimmunotherapy. Blood J Am Soc Hematol. (2017) 130:1903–10. doi: 10.1182/blood-2017-04-779736
127. Ahmed M, Li L, Pinnix C, Dabaja B, Nomie K, Lam L, et al. ATM mutation and radiosensitivity: An opportunity in the therapy of mantle cell lymphoma. Crit Rev oncology/hematol. (2016) 107:14–9. doi: 10.1016/j.critrevonc.2016.08.008
128. Ning MS, Pinnix CC, Chapman BV, Gunther JR, Milgrom SA, Khoury JD, et al. Low-dose radiation (4 Gy) with/without concurrent chemotherapy is highly effective for relapsed, refractory mantle cell lymphoma. Blood Advances. (2019) 3:2035–9. doi: 10.1182/bloodadvances.2019030858
129. Kim JK, Diehl JA. Nuclear cyclin D1: an oncogenic driver in human cancer. J Cell Physiol. (2009) 220:292–6. doi: 10.1002/jcp.v220:2
130. Vater I, Wagner F, Kreuz M, Berger H, Martín-Subero JI, Pott C, et al. GeneChip analyses point to novel pathogenetic mechanisms in mantle cell lymphoma. Br J haematol. (2009) 144:317–31. doi: 10.1111/j.1365-2141.2008.07443.x
131. Zhang J, Jima D, Moffitt AB, Liu Q, Czader M, Hsi ED, et al. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood J Am Soc Hematol. (2014) 123:2988–96. doi: 10.1182/blood-2013-07-517177
132. Panea RI, Love CL, Shingleton JR, Reddy A, Bailey JA, Moormann AM, et al. The whole-genome landscape of Burkitt lymphoma subtypes. Blood J Am Soc Hematol. (2019) 134:1598–607. doi: 10.1182/blood.2019001880
133. Dhanasekaran R, Deutzmann A, Mahauad-Fernandez WD, Hansen AS, Gouw AM, Felsher DW. The MYC oncogene—the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol. (2022) 19:23–36. doi: 10.1038/s41571-021-00549-2
134. López C, Kleinheinz K, Aukema SM, Rohde M, Bernhart SH, Hübschmann D, et al. Genomic and transcriptomic changes complement each other in the pathogenesis of sporadic Burkitt lymphoma. Nat Commun. (2019) 10:1459. doi: 10.1038/s41467-019-08578-3
135. Pavan A, Spina M, Canzonieri V, Sansonno S, Toffoli G, De Re V. Recent prognostic factors in diffuse large B-cell lymphoma indicate NF-κB pathway as a target for new therapeutic strategies. Leukemia lymphoma. (2008) 49:2048–58. doi: 10.1080/10428190802444176
136. Cabanillas F, Shah B. Advances in diagnosis and management of diffuse large B-cell lymphoma. Clin Lymphoma Myeloma Leukemia. (2017) 17:783–96. doi: 10.1016/j.clml.2017.10.007
137. Rossi A, Orecchioni S, Falvo P, Tabanelli V, Baiardi E, Agostinelli C, et al. Dual targeting of the DNA damage response pathway and BCL-2 in diffuse large B-cell lymphoma. Leukemia. (2022) 36:197–209. doi: 10.1038/s41375-021-01347-6
138. Tilly H, Morschhauser F, Sehn LH, Friedberg JW, Trněný M, Sharman JP, et al. Polatuzumab vedotin in previously untreated diffuse large B-cell lymphoma. New Engl J Med. (2022) 386:351–63. doi: 10.1056/NEJMoa2115304
139. Russler-Germain DA, Cliff ER, Bartlett NL. Cell-of-origin effect of polatuzumab vedotin in diffuse large B-cell lymphoma: No ordinary subgroup analysis. Blood. (2023) 142:2216–9. doi: 10.1182/blood.2023022048
140. Kapoor I, Bodo J, Hill BT, Hsi ED, Almasan A. Targeting BCL-2 in B-cell Malignancies and overcoming therapeutic resistance. Cell Death disease. (2020) 11:941. doi: 10.1038/s41419-020-03144-y
141. Rosenquist R, Beà S, Du MQ, Nadel B, Pan-Hammarström Q. Genetic landscape and deregulated pathways in B-cell lymphoid Malignancies. J Internal Med. (2017) 282:371–94. doi: 10.1111/joim.2017.282.issue-5
142. Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. (2018) 24:679–90. doi: 10.1038/s41591-018-0016-8
143. Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, et al. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. (2002) 9:1031–44. doi: 10.1016/S1097-2765(02)00520-8
144. Derenzini E, Agostinelli C, Imbrogno E, Iacobucci I, Casadei B, Brighenti E, et al. Constitutive activation of the DNA damage response pathway as a novel therapeutic target in diffuse large B-cell lymphoma. Oncotarget. (2015) 6:6553–69. doi: 10.18632/oncotarget.2720
145. Restelli V, Lupi M, Chilà R, Vagni M, Tarantelli C, Spriano F, et al. DNA damage response inhibitor combinations exert synergistic antitumor activity in aggressive B-cell lymphomas. Mol Cancer Ther. (2019) 18:1255–64. doi: 10.1158/1535-7163.MCT-18-0919
146. Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci. (2012) 109:3879–84. doi: 10.1073/pnas.1121343109
147. Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. (2017) 171:481–94. doi: 10.1016/j.cell.2017.09.027
148. Jardin F, Jais JP, Molina TJ, Parmentier F, Picquenot JM, Ruminy P, et al. Diffuse large B-cell lymphomas with CDKN2A deletion have a distinct gene expression signature and a poor prognosis under R-CHOP treatment: a GELA study. Blood J Am Soc Hematol. (2010) 116:1092–104. doi: 10.1182/blood-2009-10-247122
149. Runge HF, Lacy S, Barrans S, Beer PA, Painter D, Smith A, et al. Application of the LymphGen classification tool to 928 clinically and genetically-characterised cases of diffuse large B cell lymphoma (DLBCL). Br. J. Haematol. (2021) 192:216–20. doi: 10.1111/bjh.17132
150. Wright GW, Phelan JD, Coulibaly ZA, Roulland S, Young RM, Wang JQ, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. (2020) 37:551–68. doi: 10.1016/j.ccell.2020.03.015
151. Nayyar N, White MD, Gill CM, Lastrapes M, Bertalan M, Kaplan A, et al. MYD88 L265P mutation and CDKN2A loss are early mutational events in primary central nervous system diffuse large B-cell lymphomas. Blood advances. (2019) 3:375–83. doi: 10.1182/bloodadvances.2018027672
152. Sakamoto Y, Ishida T, Masaki A, Murase T, Takeshita M, Muto R, et al. Clinical significance of TP53 mutations in adult T-cell leukemia/lymphoma. Br J Haematol. (2021) 195:571–84. doi: 10.1111/bjh.v195.4
153. Westphal CH, Rowan S, Schmaltz C, Elson A, Fisher DE, Leder P. atm and p53 cooperate in apoptosis and suppression of tumorigenesis, but not in resistance to acute radiation toxicity. Nat Genet. (1997) 16:397–401. doi: 10.1038/ng0897-397
154. Johnson WT, Ganesan N, Epstein-Peterson ZD, Moskowitz AJ, Stuver RN, Maccaro CR, et al. TP53 mutations identify high-risk events for peripheral T-cell lymphoma treated with CHOP-based chemotherapy. Blood Advances. (2023) 7:5172–86. doi: 10.1182/bloodadvances.2023009953
155. Ferreri AJ, Govi S, Pileri SA, Savage KJ. Anaplastic large cell lymphoma, ALK-positive. Crit Rev oncology/hematol. (2012) 83:293–302. doi: 10.1016/j.critrevonc.2012.02.005
156. Zhang XR, Chien PN, Nam SY, Heo CY. Anaplastic large cell lymphoma: molecular pathogenesis and treatment. Cancers. (2022) 14:1650. doi: 10.3390/cancers14071650
157. Jelinek T, Zihala D, Sevcikova T, Anilkumar Sithara A, Kapustova V, Sahinbegovic H, et al. Beyond the marrow: insights from comprehensive next-generation sequencing of extramedullary multiple myeloma tumors. Leukemia. (2024) 16:1–1. doi: 10.1038/s41375-024-02206-w
158. Hu Y, Chen W, Wang J. Progress in the identification of gene mutations involved in multiple myeloma. OncoTargets Ther. (2019) 24:4075–80. doi: 10.2147/OTT.S205922
159. Duell EJ, Wiencke JK, Cheng TJ, Varkonyi A, Zuo ZF, Ashok TD, et al. Polymorphisms in the DNA repair genes XRCC1 and ERCC2 and biomarkers of DNA damage in human blood mononuclear cells. Carcinogenesis. (2000) 21:965–71. doi: 10.1093/carcin/21.5.965
160. Klintman J, Appleby N, Stamatopoulos B, Ridout K, Eyre TA, Robbe P, et al. Genomic and transcriptomic correlates of Richter transformation in chronic lymphocytic leukemia. Blood J Am Soc Hematol. (2021) 137:2800–16. doi: 10.1182/blood.2020005650
161. Alhejaily A, Day AG, Feilotter HE, Baetz T, LeBrun DP. Inactivation of the CDKN2A tumor-suppressor gene by deletion or methylation is common at diagnosis in follicular lymphoma and associated with poor clinical outcome. Clin Cancer Res. (2014) 20:1676–86. doi: 10.1158/1078-0432.CCR-13-2175
162. Gopal P, Yard BD, Petty A, Lal JC, Bera TK, Hoang TQ, et al. The mutational landscape of cancer’s vulnerability to ionizing radiation. Clin Cancer Res. (2022) 28:5343–58. doi: 10.1158/1078-0432.CCR-22-1914
163. Kerns SL, West Cm L, Andreassen CN, Barnett GC, Bentzen SR, Burnet NG, et al. Radiogenomics: the search for genetic predictors of radiotherapy response. Future Oncol. (2014) 10:2391–406. doi: 10.2217/fon.14.173
164. Khan S, Naim S, Bilwani R, Salem A, Gorlin D, Muhammad A, et al. Radiogenomics and its role in lymphoma. Curr Hematologic Malignancy Rep. (2020) 15:211–24. doi: 10.1007/s11899-020-00577-2
165. Stephens DM, Li H, LeBlanc ML, Puvvada SD, Persky D, Friedberg JW, et al. Continued risk of relapse independent of treatment modality in limited-stage diffuse large B-cell lymphoma: final and long-term analysis of Southwest Oncology Group Study S8736. J Clin Oncol. (2016) 34:2997–3004. doi: 10.1200/JCO.2015.65.4582
166. Wijetunga NA, Lebow ES, Lee J, Fregonese B, Tringale KR, Hubbeling H, et al. Genetic alterations of crebbp are associated with radiosensitivity in follicular lymphoma. Blood. (2022) 140:6413–4. doi: 10.1182/blood-2022-169501
167. Hershenfeld SA, Tobin JW, Shelton V, Calvente L, Lajkosz K, Liu T, et al. Single gene mutations and prognosis in limited-stage follicular lymphoma treated with radiation therapy. Br J Haematol. (2024) 205:1810–4. doi: 10.1111/bjh.v205.5
168. Ma J, Sarkar RR, Uppal M, Wijetunga N, Salles G, Joffe E, et al. Genomic correlates of radiosensitivity in diffuse large B cell lymphoma. Blood. (2023) 142:6104. doi: 10.1182/blood-2023-190846
169. West C, Rosenstein BS. Establishment of a radiogenomics consortium. Int J Radiat oncol biol physics. (2010) 76:1295–6. doi: 10.1016/j.ijrobp.2009.12.017
170. Hall WA, Bergom C, Thompson RF, Baschnagel AM, Vijayakumar S, Willers H, et al. Precision oncology and genomically guided radiation therapy: a report from the American Society for radiation Oncology/American association of physicists in Medicine/National cancer Institute precision medicine conference. Int J Radiat Oncol Biol Physics. (2018) 101:274–84. doi: 10.1016/j.ijrobp.2017.05.044
171. Caligiuri MA, Dalton WS, Rodriguez L, Sellers T, Willman CL. Orien: reshaping cancer research & treatment. Oncol Issues. (2016) 31:62–6. doi: 10.1080/10463356.2016.11884100
172. Andreassen CN, Alsner J. Genetic variants and normal tissue toxicity after radiotherapy: a systematic review. Radiother Oncol. (2009) 92:299–309. doi: 10.1016/j.radonc.2009.06.015
173. Roberts JS, Gornick MR. Ethical, Legal, and Social Implications of Precision Cancer Medicine. In: Precision Cancer Medicine: Challenges and Opportunities (2019). p. 129–43 Cham, Switzerland: Springer Nature Switzerland AG.
174. Barnett GC, West CM, Dunning AM, Elliott RM, Coles CE, Pharoah PD, et al. Normal tissue reactions to radiotherapy: towards tailoring treatment dose by genotype. Nat Rev Cancer. (2009) 9:134–42. doi: 10.1038/nrc2587
175. Baumann M, Krause M, Overgaard J, Debus J, Bentzen SM, Daartz J, et al. Radiation oncology in the era of precision medicine. Nat Rev Cancer. (2016) 16:234–49. doi: 10.1038/nrc.2016.18
176. Haga SB, O’Daniel JM, Tindall GM, Lipkus IR, Agans R. Survey of US public attitudes toward pharmacogenetic testing. pharmacogenomics J. (2012) 12:197–204. doi: 10.1038/tpj.2011.1
Keywords: hematologic malignances, personalized medicine, radiation therapy, precision oncology, genomics, lymphoma
Citation: Wijetunga NA, Yahalom J and Imber BS (2025) The art of war: using genetic insights to understand and harness radiation sensitivity in hematologic malignancies. Front. Oncol. 14:1478078. doi: 10.3389/fonc.2024.1478078
Received: 09 August 2024; Accepted: 20 November 2024;
Published: 21 March 2025.
Edited by:
Thomas FitzGerald, University of Massachusetts Boston, United StatesReviewed by:
Lara Hilal, American University of Beirut Medical Center, LebanonCopyright © 2025 Wijetunga, Yahalom and Imber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: N. Ari Wijetunga, d2lqQGVtYWlsLnVuYy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.