- Clinical Biochemistry Unit, Department of Pathology, College of Medicine, King Saud University, Riyadh, Saudi Arabia
Thyroid cancer (TC) is the most common endocrine cancer, which contributes to more than 43,600 deaths and 586,000 cases worldwide every year. Among the TC types, PTC and FTC comprise 90% of all TCs. Genetic modifications in genes are responsible for encoding proteins of mitogen-associated protein kinase cascade, which is closely related with numerous cellular mechanisms, including controlling programmed cell death, differentiation, proliferation, gene expression, as well as in genes encoding the PI3K (phosphatidylinositol 3-kinase)/protein kinase B (AKT) cascade, which has contribution in controlling cell motility, adhesion, survival, and glucose metabolism, have been associated with the TC pathogenesis. Various genetic modifications including BRAF mutations, RAS mutations, RET mutations, paired-box gene 8/peroxisome proliferator-activated receptor-gamma fusion oncogene, RET/PTC rearrangements, telomerase reverse transcriptase mutations, neurotrophic tyrosine receptor kinase fusion genes, TP53 mutations, and eukaryotic translation initiation factor 1A X-linked mutations can effectively serve as potential biomarkers in both diagnosis and prognosis of TC. On the other hand, epigenetic modifications can lead to aberrant functions or suppression of a range of signalling cascades, which can ultimately result in cancer. Various studies have observed the link between epigenetic modification and multiple cancers including TC. It has been reported that several epigenetic alterations including histone modifications, aberrant DNA methylation, and epigenetic modulations of non-coding RNAs can play significant roles as potential biomarkers in the diagnosis and prognosis of TC. Therefore, a good understanding regarding the genetic and epigenetic modifications is not only essential for the diagnosis and prognosis of TC, but also for the development of novel therapeutics. In this review, most of the major TC-related genetic and epigenetic modifications and their potential as biomarkers for TC diagnosis and prognosis have been extensively discussed.
1 Introduction
Thyroid cancer (TC) is the most common endocrine cancer, which contributes to more than 43,600 deaths and 586,000 cases worldwide every year (1). In the past 30 years, the occurrence of TC has elevated in multiple developed countries (2, 3). The occurrence of TC varies by up to 15-20-fold according to geographical regions, where it is more commonly diagnosed in developed countries. High-risk regions for TC include Southern Europe, North America, New Zealand, Australia, Eastern Asia, and Polynesia. As per the histological type, TC can be classified into 5 types including anaplastic TC (ATC), medullary TC (MTC), poorly differentiated TC (PDTC), follicular TC (FTC), and papillary TC (PTC). Around 10% of TC patients contain tumour metastasis including lung and bone (20%), lung (50%), bone (25%), and other sites (5%) (Figure 1). PTC and FTC comprise 90% of all TCs, which generally affect people aged between 50 and 60 years (1). A range of risk factors have already been identified that can contribute to TC development including genetic predisposition, increased concentrations of thyroid-stimulating hormone, iodine excess or deficiency, and ionising radiation (5). Common diagnostic techniques of TC include histopathological evaluation of the thyroid gland tissue, fine needle aspiration cytology (FNAC), ultrasonography, and various laboratory examinations with the likelihood of estimating the tumour markers calcitonin and thyroglobulin (6). There is a growing interest in molecular genetic analysis of FNAC samples (6). In addition, there are several mutations that are explicit for specific types of carcinomas and can thus play a role in molecular testing in the preoperative period along with alteration of the diagnosis in the case of cytologically ambiguous results (6). Decreased effectiveness of radioiodine therapy or a propensity to dedifferentiate, involvement of metastatic lymph node, and signs of elevated level of tumour aggressiveness have already been linked along with some mutations (7).
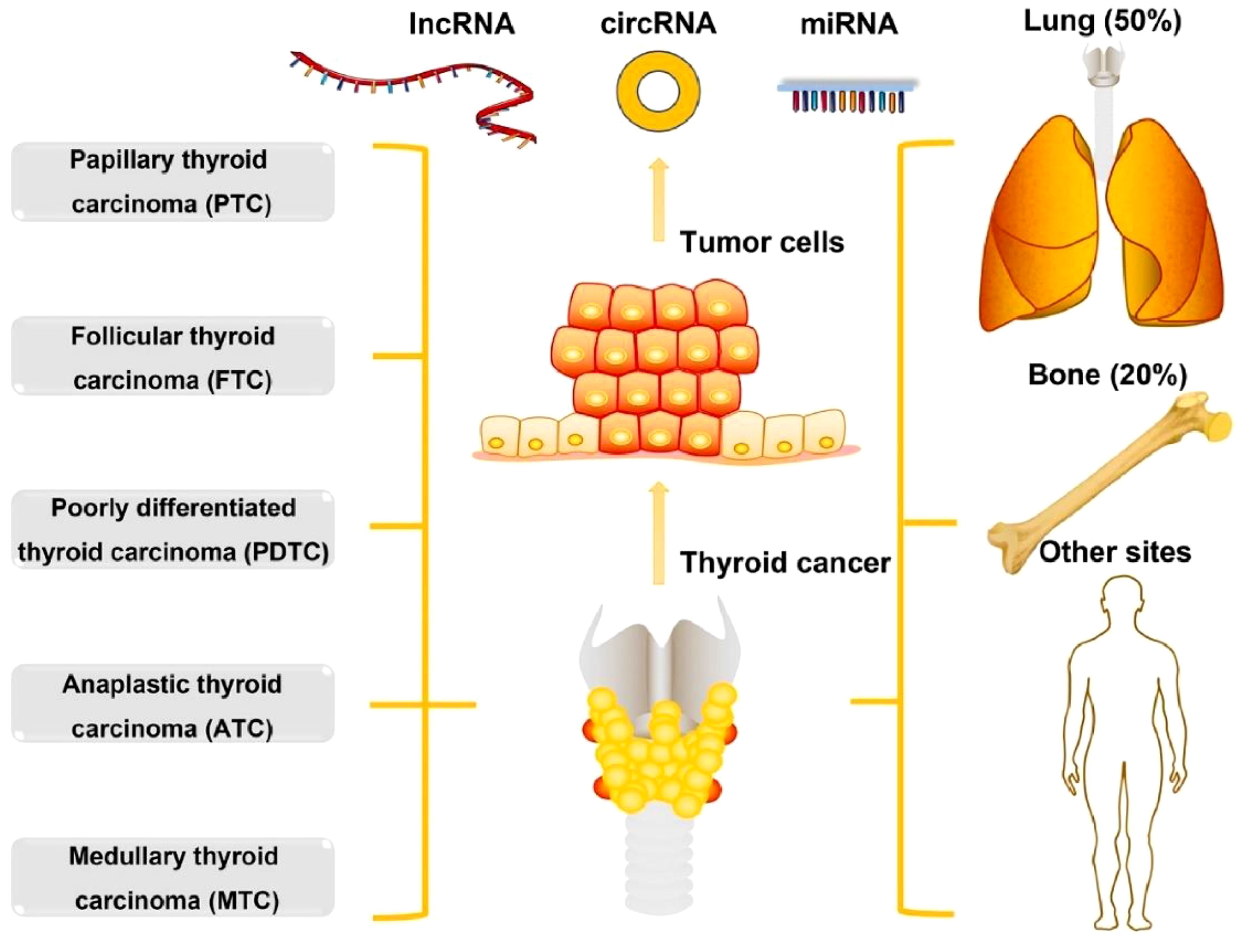
Figure 1. The types and metastasis of thyroid cancer. Reproduced with permission from Elsevier, (4).
MTC arises from neural crest and most of the MTC cases are sporadic (75%) while the remaining are hereditary (25%) (8). Indeed, this wide range of progression is meticulously associated with the pattern of accumulated genetic and epigenetic changes, which are associated with tumour invasion, metastasis, and differentiation. Most of the genetic changes in TC start their activities via causing the activation of metabolic pathways. It has been observed that constitutive activation of the extracellular signal-regulated kinase (ERK)/mitogen-associated protein kinase (MAPK) cascades can result in tumorigenesis and can further mediate cell division. ERK/MAPK cascade activation is an important and common process involved with the human cancer progression and initiation. Genetic abnormalities in the RAS gene, BRAF (B-Raf Proto-Oncogene, Serine/Threonine Kinase) gene, and rearranged in translation (RET)/PTC are also linked with TC. Interestingly, the occurrence of RAS gene-associated activating mutations is reliant on the tumour histology. It was observed that RAS mutations are more commonly seen in FTC as compared to PTC. A cell membrane receptor tyrosine kinase was found to be encoded by the RET proto-oncogene. This kinase’s ligands belong to the glial-cell-line derived neurotropic factor family that results in receptor dimerisation following binding, which further results in tyrosine residue autophosphorylation and starts the ERK/MAPK cascade. Functional deficiency of RET can lead to Hirschsprung’s disease, while increased activities of RET have been linked with a range of cancer types, such as MTC. Simultaneous mutations of BRAF as well as RET/PTC have been linked with PTC (9). Interestingly, BRAF V600E mutation is limited to anaplastic, papillary, and poorly differentiated TC (10). In this review, the genetic and epigenetic basis of TC as well as the main TC-associated genetic and epigenetic modifications and their potential as biomarkers for both diagnosis and prognosis of TC have been extensively covered.
2 Genetic and epigenetic basis of thyroid cancer
Tumour progression and transformation involve the disturbance of cell signal mechanisms that control the balance between apoptosis and cell proliferation (11). Genetic modifications in genes are responsible for encoding proteins of MAPK cascade (Figure 2), which is closely linked with numerous cellular mechanisms, including controlling programmed cell death, differentiation, proliferation, gene expression, as well as in genes encoding the PI3K (phosphatidylinositol 3-kinase)/protein kinase B (AKT) cascade, which has contribution in the regulation of cell motility, survival, adhesion, and glucose metabolism, have been associated with the TC pathogenesis (11, 12). Chromosomal rearrangements (fusion genes) and point mutations are the two main molecular mechanisms associated with TC. A single nucleotide is altered in case of a point mutation, while 2 different genes are fused in case of chromosomal rearrangement. In most of the cases, these genetic alterations are somatic or nonhereditary in nature. Several endocrine neoplasia syndromes (such as- MEN2A, MEN2B) and familial forms of MTC involve hereditary germline mutations (12). Suspected thyroid tissues are analysed for potential somatic mutations. On the other hand, peripheral blood collected from the patients and perhaps their relatives are used to identify germline mutations (11, 13).
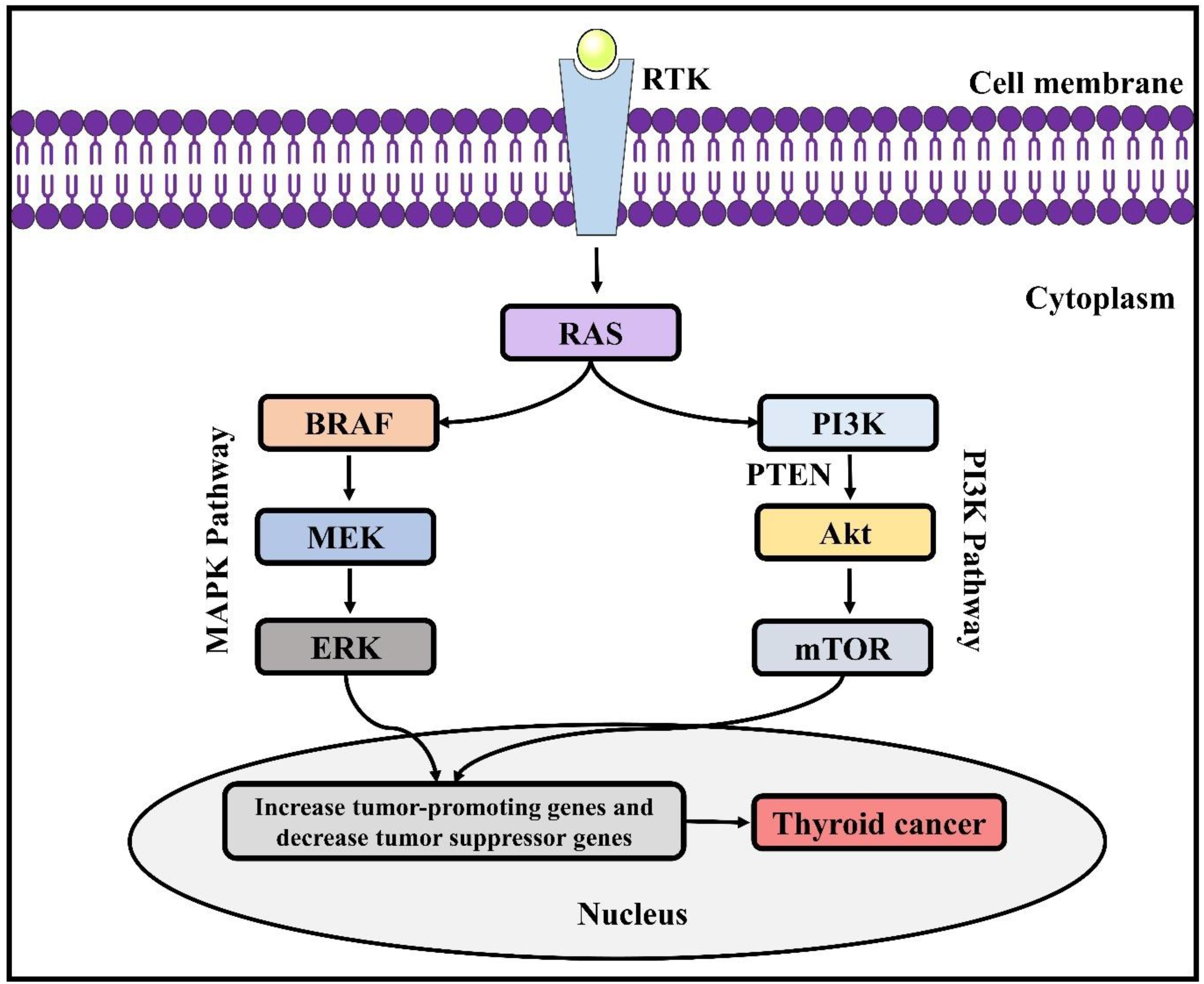
Figure 2. Genetic alterations involved in thyroid cancer. RAS, Rat sarcoma virus; ERK, extracellular signal-regulated kinase; BRAF, B-Raf proto-oncogene; PI3K, phosphatidylinositol 3-kinase; MAPK, mitogen-associated protein kinase; PTEN, phosphatase and tensin homolog; AKT, protein kinase B; mTOR, mammalian target of rapamycin.
Epigenetic processes are vital for the maintenance of tissue-specific gene expression patterns and normal development of cells in mammals (14). Nonetheless, epigenetic changes can lead to abnormal functions or suppression of several signalling cascades, which can eventually result in cancer including TC (Figure 3). Several studies have observed the link between epigenetic modification and a range of cancers, along with several genetic variations (16, 17). Epigenetic processes involve nucleosome remodelling, non-coding RNA expressions, DNA cytosine methylation, and covalent chromatin modification. Abnormal DNA methylation is linked with gene expression and has a contribution in tumorigenesis. Hypomethylation through several mechanisms can result in proto-oncogene activations and genomic instability, which can play a role in cancer progression and development. Nevertheless, hypermethylation is linked with gene silencing (predominantly tumour suppressor genes) and is regarded as a cancer hallmark. More studies regarding how certain genomic areas are targeted for hypermethylation are likely to lead to the development of more therapeutic areas. In addition, miRNA expression profile is another feature of epigenetic modification. Previously, miRNA expression profiles in tumours were compared to the related normal tissues, which indicated extensive expression level alterations. As microRNAs (miRNAs or miRs) control the expression levels of many genes that are associated with apoptosis, cell proliferation, and transcriptional regulation, therefore changes in their expression levels can mediate tumorigenesis. Indeed, miRNAs have the capacity to play a role as oncogenes or tumour suppressors, according to their role on the target genes.
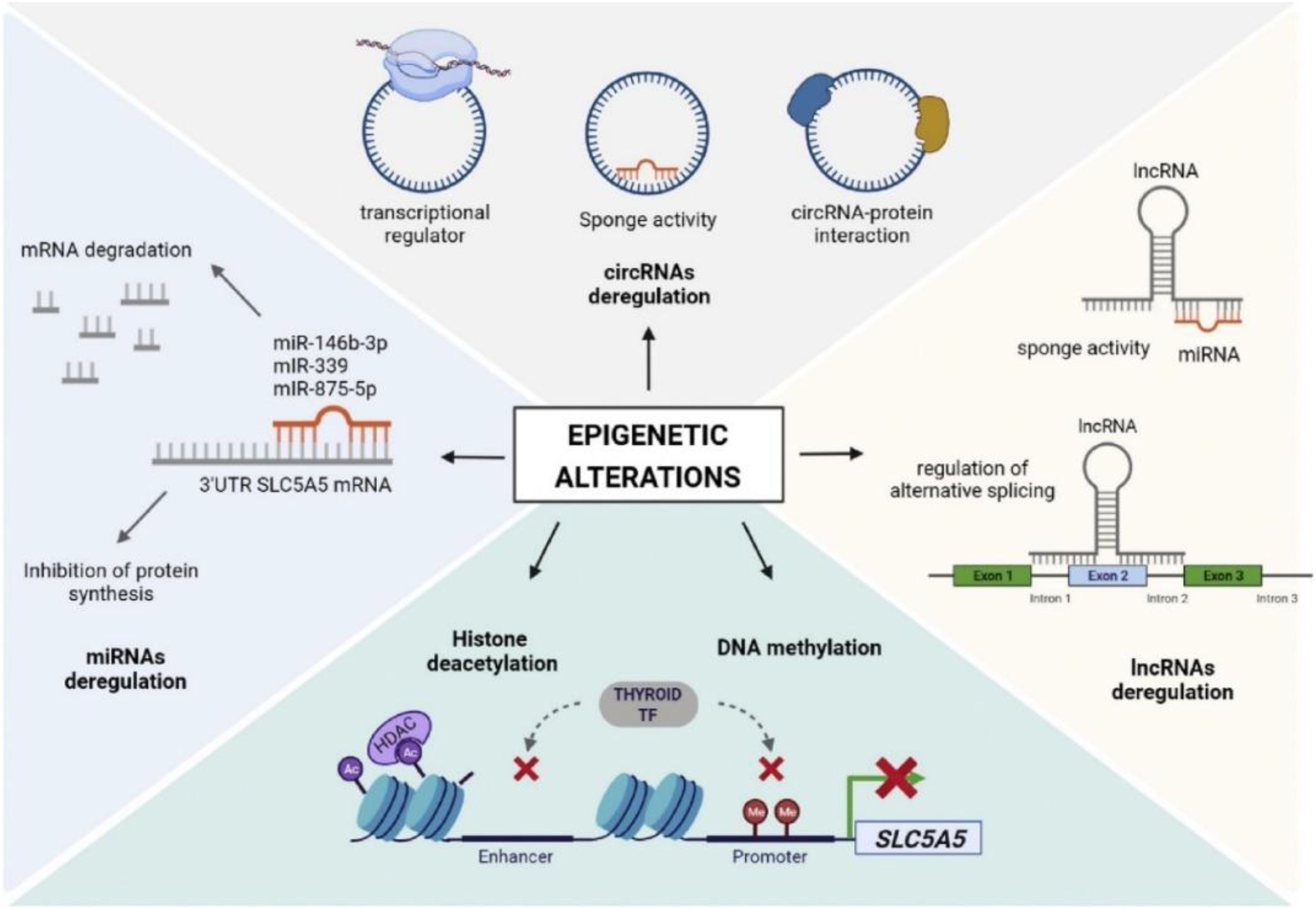
Figure 3. The role of epigenetic modifications in thyroid cancer. Reproduced with permission from Elsevier, (15).
3 Genetic modifications that can be considered as biomarkers in the diagnosis and prognosis of thyroid cancer
3.1 BRAF mutations
BRAF (7q24) is a proto-oncogene, which is responsible for encoding serine/threonine kinase belonging to the RAF-kinase family that has an important contribution in the signal transduction along the RAS/RAF/MEK/ERK cascade controlling apoptosis, differentiation, and cell growth. Among the 3 functional RAF proteins identified in humans including c-RAF, BRAF, and ARAF; BRAF exhibits the maximum basal kinase function and is the most strong MAPK cascade activator. Mutations in BRAF have also been linked with human carcinogenesis, where an increased level of BRAF mutations was observed in ovarian carcinoma, colorectal carcinoma, and melanomas. All such mutations were observed in the kinase domain of the protein, linking either the ATP binding site or activation loop, which further contributes in the activation of BRAF. Subsequently, they were also detected in PTCs and might be a target for potential therapy development against aggressive lesions (18). In the case of PTC, BRAF mutations are the most commonly detected mutations, along with the highest frequencies in classical PTC and tall-cell. BRAF mutations are exclusively observed in PTC and PTC-derived-ATC. So far, over 40 BRAF mutations have been detected, among them V600E is most commonly observed and responsible for around 95% of cases. BRAF K601E is most commonly seen in the case of follicular variant PTC (FV-PTC). Upon radiation exposure, a chromosomal rearrangement leading to the fusion gene AKAP9/BRAF and small in-frame deletions or insertions surrounding codon 600 were detected in PTCs. As compared to children, adults are more commonly affected by BRAF mutations. Fluorescence in situ hybridisation study revealed that BRAF activation includes copy number gain and is present in 35% of FTCs, 25% of follicular adenomas (FAs), and 3% of PTCs (18).
3.2 RAS mutations
Ras proteins belong to the guanosine triphosphate (GTP)-binding protein family that control cell growth via phosphoinositide-3-kinase (PIK3) and MAPK signalling pathways. In TC, 3 of RAS family including members NRAS (located at chromosome position 1p13), KRAS (located at chromosome position 12p12), and HRAS (located at chromosome position 11p11) were found to be mutated, wherein they mainly become activated either through mutations that reduce their intrinsic GTPase action (codon 61) or increase their GTP-binding affinity (codons12/13). Since their discovery, RAS mutations have been detected in 18–27% of poorly differentiated TCs (PDTCs), 10–20% of PTCs (particularly FV-PTC), up to 60% of ATCs, 40–50% of FTCs, and 20–40% of FAs. In general, the most common mutations are less common in codon 61 of HRAS and most common in codon 61 of NRAS. Furthermore, they are rarely observed in radiation-mediated TCs of Chernobyl and more commonly observed in iodine-deficient regions (18). RAS mutations were initially regarded as early events in thyroid carcinogenesis because of their occurrence in both FTCs and FAs, nonetheless this might be elucidated through the high extent of interobserver variability regarding the difference of FAs from FV-PTCs. It has been recently revealed that there is an increased level of RAS mutations in ATCs and PDTCs as well as lower levels of RAS mutations in well-differentiated TCs, which indicates the influence of RAS in the progression of the tumour, instead of the initiation. It has been demonstrated by in vitro studies that elevated levels of genomic instability mediated via RAS-genomic instability can mediate the progression of the tumour through permitting tumour cells to collect mutations that mediate elevated levels of invasiveness and survival. In addition, thyroid-targeted RAS mutation led to follicular tumours that further resulted in PDTCs in mice. RAS mutations might also be prognostic of poor prognosis in the case of PDTCs and well-differentiated TCs (18).
3.3 RET mutations
It is now well-known that the RET gene encodes a receptor tyrosine kinase and has a significant contribution in cell survival, differentiation, and growth (12). Typically, RET gene-associated point mutations are observed in MTC (19). Interestingly, 95% of MEN2B and MEN2A individuals contain germline mutations, while the occurrence of somatic mutations in sporadic MTC and germline mutations in familiar MTC is lower (50%) (20). For relatives of patients, genetic screening is suggested with an identified germline mutation in the RET gene (21). There is a high risk of MTC in case of an inherited RET mutation, where it is suggested to go through a preventive total thyroidectomy (22). Individual suggestions can be provided as per the identified genotype-phenotype correlations, particularly about the timing of prophylactic total thyroidectomy in childhood to avert the disease development (13, 23).
3.4 RET/PTC rearrangements
In children and adolescents, the most commonly observed mutation is the chromosomal rearrangement of RET/PTC (24). This kind of rearrangement is more common in PTC and is linked with frequent metastatic dissemination and more aggressive tumour behaviour (25). Carcinoma cases are observed following previous exposure to radiation (25). Identification of RET/PTC rearrangement serves as a strong indicator of PTC and might also facilitate the molecular diagnosis of FNAC, particularly in uncertain cytological findings. Total thyroidectomy is typically suggested following identification of RET/PTC rearrangements (13, 26).
3.5 Paired-box gene 8/Peroxisome proliferator-activated receptor-gamma fusion oncogene
A balanced translocation, t(2;3)(q13;p25), can lead to PAX8/PPARγ fusion oncogene that fuses PPARγ and PAX8. A translocation t(2;3;6;15) was first detected in an FA, which was then confirmed in FTCs and FAs. Subsequently, the presence of PPARγ and PAX8 was confirmed in this translocation and it was observed that fusion protein expression is induced by the PAX8 promoter. PAX8 plays an important role in regulating the terminal differentiation in thyroid cells, which regulates the expressions of thyroid-stimulating hormone (TSH) receptor, thyroglobulin, and sodium iodide symporter (NIS). Therefore, it is expected that the expression pattern of PAX8/PPARγ fusion protein is associated with the differentiation behaviour of thyroid tumours, where absent or low level is expected in poorly differentiated tumours and high level in well-differentiated tumours. On average, it has been reported that PAX8/PPARγ present in 13% of FV-PTC (0–50%), 11% of FAs (0–55%), and 36% of FTCs (0–63%). The aforementioned findings are based on the RT-PCR, however it should also consider the fact that promoters that drive expression of PAX8/PPARγ mRNA might be missing in the cells that are present in the dedifferentiated tumours, nonetheless still might harbour the fusion at the DNA level. In an attachment-independent manner, PAX8/PPARγ mediated in vitro thyroid cell growth, which decreased apoptosis and elevated soft agar colony formation (27). These effects might take place because of the fusion protein’s dominant negative suppressive action as compared to wild-type PPARγ, which is thought to possess tumour-suppressive activities and has been identified as a possible target for the development of therapeutics against various cancer types. The role of PPARγ in TC has been further revealed by the identification of another fusion protein in FTC, CREB3L2-PPARγ, which might arise a chromosomal rearrangement, t(3;7)(p25;q34). However, more studies are required to find out whether PAX8/PPARγ alone can mediate TC or whether additional epigenetic or genetic processes are needed to activate the full phenotypic expression of follicular TC (18).
3.6 Telomerase Reverse Transcriptase mutations
The rate-limiting catalytic subunit of telomerase is encoded by the TERT gene, which is accountable for the elongation of telomere during the replication of DNA (12). C250T and C228T are the two main point mutations that have been identified in TERT gene (28, 29). An increased level of telomerase expressions have been observed in cancer cells positive for C250T and C228T mutations, where these cancer cells can maintain chromosomal telomere length and can continue proliferation indefinitely (29). It has been observed that there is a link between TERT mutations and tumour aggressiveness as well as distant or local metastasis (28). TERT mutations are found in ATC, more aggressive forms of PTC, and poorly differentiated TCs (28, 30). Co-occurrence of BRAF V600E mutation and TERT mutations is linked with increased level of tumour aggressiveness in case of PTC as compared to BRAF and TERT mutations occurring alone (31). Region VI elective neck dissection and total thyroidectomy are recommended for the preoperative detection of TERT mutations in nodules bigger than 1 cm as per the European Thyroid Association (13, 26).
3.7 Neurotrophic tyrosine receptor kinase fusion genes
The occurrence of NTRK fusion genes in PTC is 5–10% in adolescent and pediatric patients (32). If a NTRK fusion gene is detected in a thyroid sample, the risk of malignancy is considered as 100% (21, 32). The follicular arrangement is observed in NTRK fusion-positive carcinomas, along with the incidence of frequent lymph node metastasis and chronic lymphocytic thyroiditis (13, 19).
3.8 TP53 mutations
TP53 gene (a tumour suppressor) starts apoptosis in the case of nonrepairable DNA and controls the cell growth by regulating the cell division (12). Increased levels of TP53 expressions and mutations are identified in over 75% of undifferentiated and invasive carcinomas (21). In differentiated carcinomas, the occurrence of TP53 mutations is considered as an indication of subsequent ATC dedifferentiation (19).
3.9 Eukaryotic translation initiation factor 1A X-linked mutations
The EIF1AX gene encodes a vital eukaryotic translation initiation factor and EIF1AX mutations have been linked to several cancers. Mutations in the EIF1AX gene have been detected in tumours that typically be deficient in other common drivers and identified in 1.5% of The Cancer Genome Atlas (TCGA) cohort, which are indicating that EIF1AX gene may play a role a novel PTC oncogene (33). RAS and EIF1AX mutations often co-occur, however the precise mechanism of action is yet to be discovered. These mutations are observed in benign neoplasms and in around 30% of PTC, which are closely linked with TERT and RAS mutations and indicate individuals in advanced TC with a reduced rate of disease-specific survival (34).
4 Epigenetic modifications that can be considered as biomarkers in the diagnosis and prognosis of thyroid cancer
4.1 Aberrant DNA methylation
Aberrant DNA methylation (ADM) of tumour suppressor genes and proto-oncogenes are found in TC and other human cancers. Tumour suppressor genes present in the thyroid include tissue inhibitor of metalloproteinase 3 (TIMP3), solute carrier family 5 member 8 (SLC5A8), Ras association domain family member 1, isoform A (RASSF1A), RAP1 GTPase activating protein (RAP1GAP), RAPβ2, phosphatase and tensin homolog (PTEN), and death associated protein kinase (DAPK). A class of GTPase-activating proteins is encoded by the RAP1GAP gene, which is responsible for the deactivation of RAS-related protein. This gene also controls mitogenic and oncogenic mechanisms in thyroid cells. On the other hand, RAP1 has a significant contribution in the ERK-dependent cascade regulation and BRAF-MEK-ERK cascade activation. In thyroid tumour cell lines, the immunohistochemistry studies showed RAP1GAP gene downregulation in PTC along with its invasion and proliferation. PTEN is a tumour-suppressive gene, which is responsible for encoding phosphatidylinositol-3, 4, 5-triphosphate 3-phosphatase protein. PTEN mutations have been detected in several cancer types. PTEN gene is also responsible for the negative regulation of AKT/protein kinase B (PKB) signalling cascade. Moreover, this gene has a significant contribution in controlling cell cycle and opposing rapid cell division and growth. ADM of PTEN is commonly observed in both PTC and FTC.
The TIMP3 gene is responsible for the suppression of cell development, angiogenesis, infiltration, and metastasis of many tumours. A hypermethylation of TIMP3 gene has been detected in the case of TC. This gene is also linked with extrathyroidal invasion and lymph node metastasis (35). A protein analogue to the RAS effectors protein is encoded by the RASSF1A gene. A link has been reported between the cancer and RASSF1A mRNA expression deregulations, where ADM plays an important in the inactivation of RASSF1A gene. Unlike FTC, ADM of RASSF1A is present in a small proportion in PTC, which might have a significant contribution in thyroid carcinogenesis. DNA hypomethylation also has a significant contribution in carcinogenesis, however its exact role is yet to be fully revealed. Nonetheless, international patterns of ADM have been revealed in subtypes of thyroid malignancy through DNA methylation arrays. Collectively, 13 and 21 hypomethylated genes in FTC as well as 262 and 352 hypermethylated genes in PTC have been identified. Moreover, 86 and 131 hypermethylated as well as 280 and 393 hypomethylated genes were detected in MTC and ATC, respectively. Out of these genes, 4 oncogenes including TCL1B, NOTCH4, INSL4, and DPPA2 were found to be commonly controlled by hypomethylation (35).
4.2 Histone modifications
The link between the behaviour of thyroid tumours and histone modifications has been demonstrated. It is well-established that the gene transcription is dependent on the chromatin accessibility and conformation. Chromatin remains “open” (euchromatin) in a transcriptionally active state, which permits the transcriptional machinery interaction with DNA to initiate the transcription of genes, while DNA remains tightly wrapped around the closed state of chromatin (heterochromatin). On the other hand, DNA tight packaging is caused by its coiling around histone proteins, which provide structural support to a chromatin. Through de-acetylation and acetylation of lysine residues, the extent of DNA condensation around histones regulates gene transcription, which involves histone post-translational modification. Histone deacetylases and acetyltransferases are the enzymes that cause such reversible acetylation-deacetylation alterations. The interaction of DNA and histones is hindered by histone acetylation of lysine residues through the removal of the positive charge on the histones that is responsible for the interaction with the negative charge containing phosphate groups of DNA. The role of histone acetylation in cancer has been widely evaluated, where this acetylation has a significant contribution in tumorigenesis.
Histone acetylation in TC plays a role from the early stages of thyroid carcinogenesis. Increased concentrations of H3K9–K14ac and H3K18ac have been reported in FTC and PTC than in control tissues, while histone H3K9–K14ac was only identified in ATC tissues, further indicates that the deficiency of the expression of H3K18Ac in case of ATC might have contribution during the progression of TC (36). It was observed that ectopic induction of major driver oncogenes including RAS, BRAF or RET/PTC resulted in elevated concentrations of acetylated histones in thyroid cell lines, which is in contrast with the events that take place in undifferentiated and advanced tumours where acetylation decreases expressions of a range of thyroid differentiation genes including NKX2.1 (NK2 Homeobox 1), TPO (thyroid peroxidase), TG (Thyroglobulin), and SLC5A5 (solute carrier family 5 member 5) (37). HRAS and BRAFV600E mutations (unlike other genetic modifications) exhibit less response to histone deacetylase (HDAC) inhibitor therapies regardless of thyroid tumour subtypes. An improved antitumour action has been observed in some tumour cell lines with the combined treatment of PI3K/Akt or MAPK inhibitors and HDAC inhibitors. Panobinostat is a strong inhibitor of HDAC that exerted in vitro and in vivo cytotoxic actions on ATC cell lines, which resulted in apoptosis as well as cell cycle arrest and blocked growth of tumours in a xenograft mouse model. Treatment with panobinostat also re-stimulated mRNA expression of SLC5A5 along with an increased level of NIS, these effects were also demonstrated with two other inhibitors of HDAC trichostatin and suberoylanilide hydroxamic acid (38). Several clinical trials were carried out utilising various HDAC inhibitors including depsipeptide, valproic acid, romidepsin, vorinostat, and suberoylanilide hydroxamic acid (39, 40). Nonetheless, these clinical trials failed to show promising outcomes, in spite of the great potential of these HDAC inhibitors (15).
4.3 Non-coding RNAs
4.3.1 Long non-coding RNAs
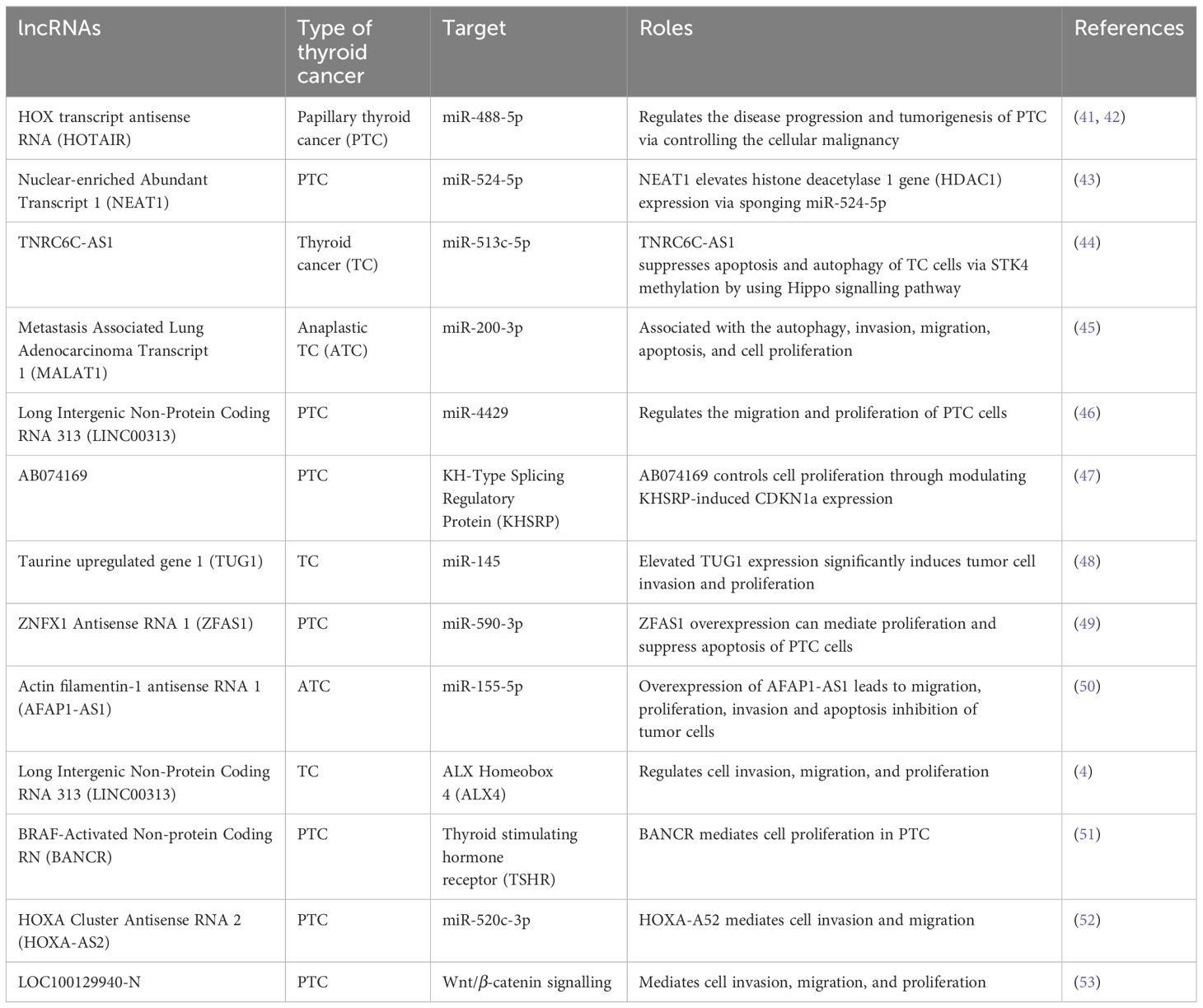
Typically, lncRNAs are described as transcripts that contain over 200 nucleotides that are not generally translated into functional proteins. In addition, they are commonly found in the nucleus, wherein lncRNAs exhibit various activities including gene expression and splicing regulation via various mechanisms. Chromatin structure can be altered due to the interaction of lncRNAs with DNA, which can result in epigenetic modifications and further cause alterations in the target gene expressions. In addition, lncRNAs can interact with miRNAs or mRNAs and play a role as molecular sponges or competing endogenous RNAs (ceRNAs) to control the miRNA interaction with the targets or to regulate the translation and stability of mRNAs. A range of deregulated lncRNAs have already been reported, which can play role as biomarkers in the prognosis and diagnosis of TC (Table 1). For instance, HOX transcript antisense RNA (HOTAIR) is a lncRNA that is commonly overexpressed, which was found to be overexpressed in patients with PTC and TC (41). HOTAIR is commonly linked with survival and it contributes in thyroid carcinogenesis through Wnt signalling (42). In a miR-1-induced manner, silencing of HOTAIR markedly decreased the tumour growth in vivo as well as the growth of FTC-133 and TPC-1 cell lines via controlling the expression of CCND2. Overexpression of nuclear-enriched Abundant Transcript 1 (NEAT1) has been reported in case of PTC and NEAT1 silencing in TPC-1 cells resulted in decreased in vivo tumour growth, motility, and cell survival induced by the downregulation of β-catenin through the regulation of miR-214 (54). Trinucleotide Repeat Containing Adaptor 6C-Anti Sense1 (TNRC6C-AS1) is another lncRNA that plays a role as an oncogene in TC via sponging miR-129-5p as well as mediating invasion, proliferation, and migration of TPC-1 cells (55). TNRC6C overexpression or TNRC6C-AS1 silencing re-induced the expressions of various thyroid genes including TPO, TSHR (thyroid stimulating hormone receptor), SLC26A4, and SLC5A5 (56). Therefore, it is likely that the TNRC6C-AS1–TNRC6C axis has a contribution in iodine metabolism regulation in case of PTC. Overexpression of MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) has also been observed in PTC, where it exerts oncogenic activities (57). Nonetheless, downregulation of MALAT1 has been reported in ATC and PDTC (57). In the case of TC, MALAT1 mediates invasion and proliferation of cells via IQGAP1 upregulation (58), which is a vital MAPK scaffold protein that has significant contribution in TC (59).
The expression of MALAT1 is controlled via TGFβ, which indicates its contribution in TC progression by processes associated with epithelial-mesenchymal transition (60). Interestingly, some lncRNAs act as tumour suppressors via controlling epithelial differentiation and cell homeostasis. For example, in PTC, CASC2 (Cancer Susceptibility Candidate 2) was found to be downregulated in samples and its lower level was linked with poor prognosis (60). CASC2 overexpression markedly decreased in vitro cell proliferation and resulted in ERK1/2 and AKT inactivation (60, 61). Furthermore, CASC2 suppressed the invasion and migration of TC cells via sponging miR-18a-5p and miR-155 (62). GAS5 (Growth Arrest-Specific 5) is another lncRNA and its downregulation was observed in TC cell lines and PTC in contrast with samples containing benign tumour. In patients with PTC, a lower level of GAS5 expression is linked with poor prognosis, tumour nodules metastasis (TNM) staging, multiple cancer foci, and lymph node metastasis (63). In PTC cell lines, GAS5 played a role as a ceRNA of miR-222-3p and inactivated the PI3K signalling caused by the upregulation of PTEN (63). These findings indicate the role as a tumour suppressor in TC. LINC00893 (Long Intergenic Non-Protein Cocing RNA 893) is another lncRNA that is linked with PI3K signalling through PTEN signalling. Reduced expression of LINC00893 has been reported in TC and PTC cells (64). In PTC cell lines, LINC00893 ectopic overexpression diminished cell migration and proliferation via AKT phosphorylation blockage (64).
4.3.2 Circular RNAs
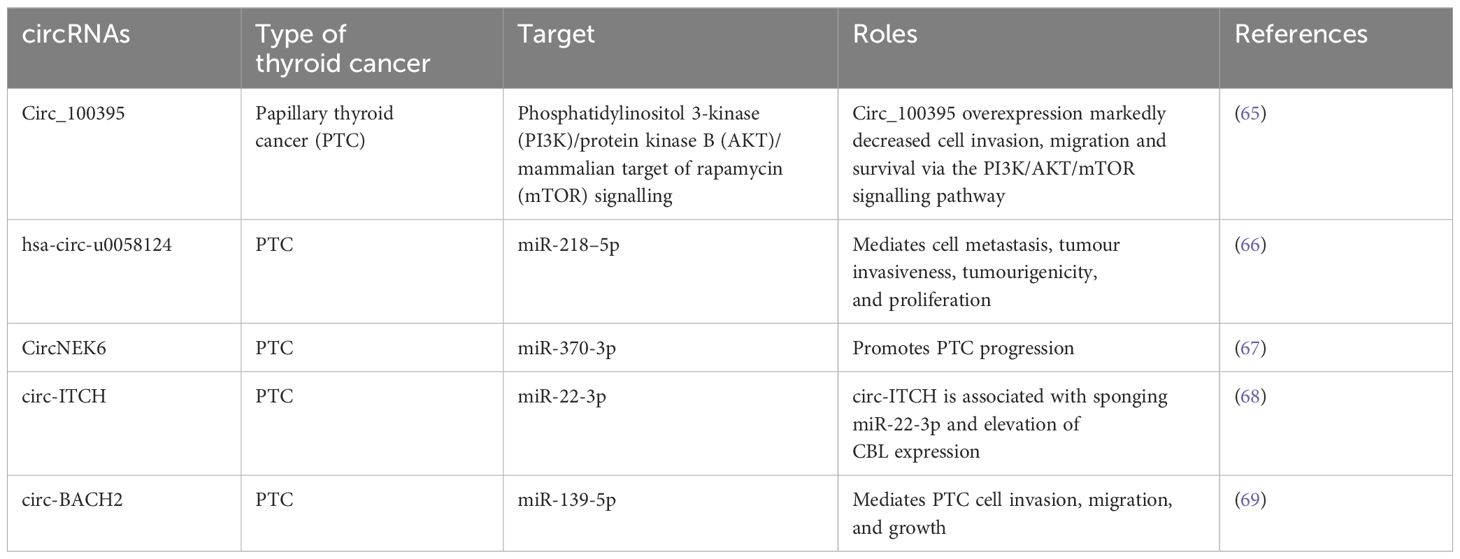
Numerous studies have already discovered the link between circRNAs and tumorigenesis. The circRNAs play a role as a ceRNA to control gene expression via suppressing miRNA activities (Table 2). There is a growing interest regarding the functions and roles of circRNAs. In a study, 98 deregulated circRNAs were detected when comparing six PTC tumours with nearby normal tissues (70). Among them, circRNA-102171 overexpression was observed in 47 PTC samples as compared to contralateral healthy tissues (71). In a different study, 146 deregulated circRNAs were detected after RNA sequencing of 11 PTCs and their heterolateral non-tumour tissues (72). On the other hand, circ_0011058 is another circRNA that mediated in vivo tumour growth and angiogenesis and facilitated cell proliferation in PTC cell lines via controlling YAP1 expression and sponging miR-355-5p (73). An upregulated level of CircRUNX1 was observed in cancer samples to control the miR-296-3p/DDHD2 axis associated with metastasis formation and tumour growth (74). Additional activities of circRNAs include altering protein and gene expression by controlling gene transcription, through their role as a translation template or by interacting with the transcription machinery. Indeed, circRNAs also have the capacity to modify the localisation or functions of the proteins via interactions. For instance, circ-Amotl1 can interact with c-MYC to activate and stabilise its transcriptional function through the regulation of its nuclear translocation (75). Furthermore, circRNA_102171 activates the Wnt/β-catenin signalling through direct interaction with CTNNBIP1, which eventually mediates the invasiveness of PTC cells (71).
4.3.3 MicroRNAs
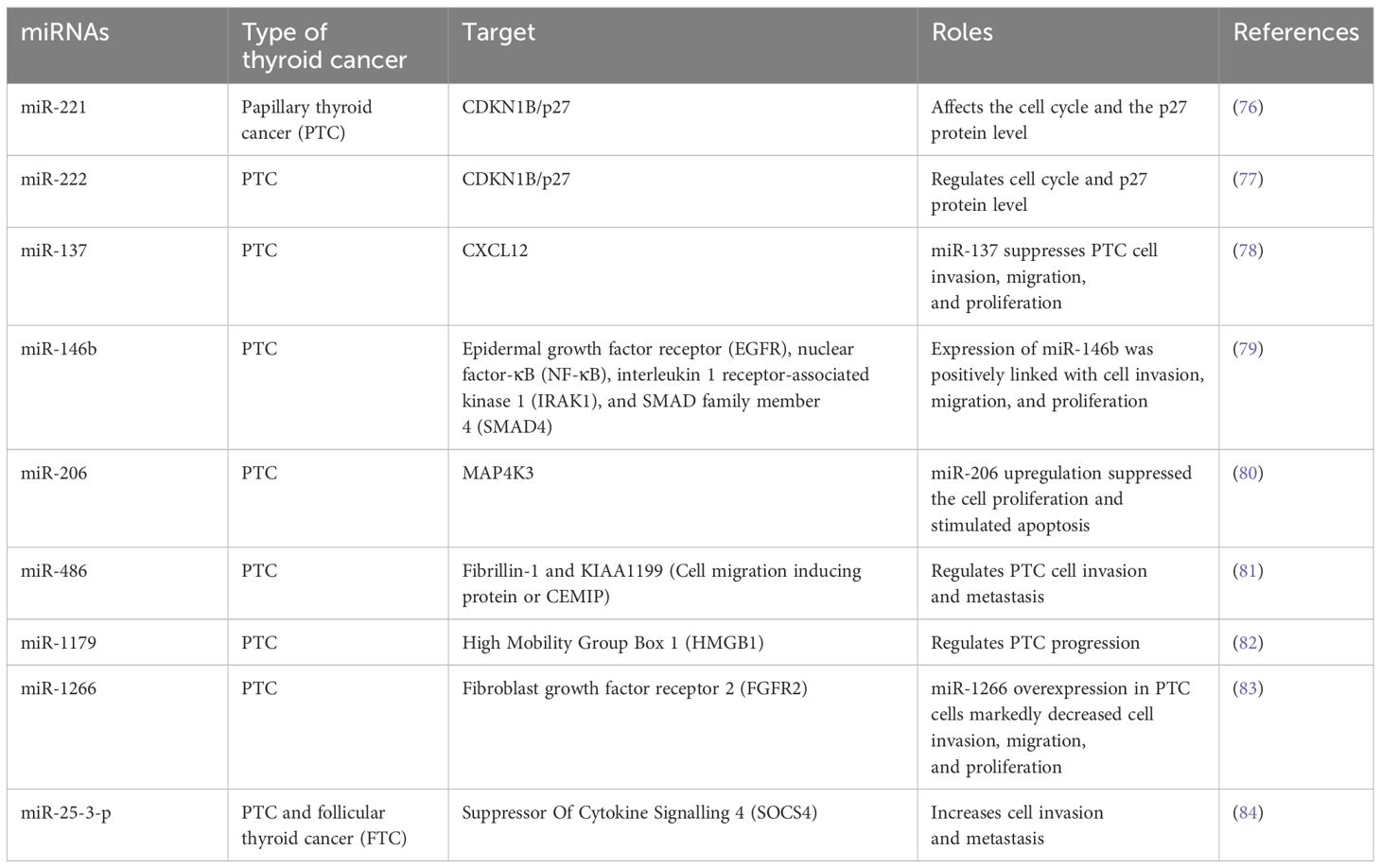
Among the ncRNAs, miRNAs have been best characterised and most studied. These ncRNAs contain RNA transcripts of 18–24 nucleotides that interact with the 3′-UTR of mRNAs to hinder protein translation of target genes. They have a significant role in cancers, where miRNAs play a role in enhancing tumour progression and loss of differentiation. They can be classified as tumour suppressors and oncogenic (oncomiRs) as per their activities on death and proliferation of cells as well as expression patterns in malignant samples in contrast with healthy tissues. Certain profiles of miRNA expressions can be linked with genetic mutations commonly detected in DTCs. Moreover, miRNAs can be easily detected in blood samples and they show resistance to various environmental conditions including room temperature, therefore a miRNA profile can be used as therapeutic targets and prognostic biomarkers (Table 3). In contrast with circulating mRNAs, miRNAs have the ability to remain protected from nucleases in the bloodstream by being encased in exosomes or microvesicles or through interaction with proteins as well as miRNAs have the capacity to remain undamaged in paraffin-fixed tissue samples (85, 86). Numerous studies have already assessed the role of various miRNAs in thyroid carcinogenesis, where they regulate the major cancer-associated signalling mechanisms including transforming growth factor beta (TGFβ), PI3K, MAPK, Hippo and Wnt signalling mechanisms (15, 87). PTC miRNAs are classified into 6 clusters as per TCGA that are markedly different as per the parameters including risk profiles, histological phenotype, driver mutations, and so on. For example, Cluster 1 is closely linked with RAS mutations and FV-PTC, while clusters- 5 and 6 contain the highest level of BRAFV600E mutations and a very high risk.
On the other hand, Cluster 6 possesses most of the tall-cell PTC samples (74%) of TCGA (88). As like FTC tumours, FV-PTC exhibits a RAS-like signature, however some of the most deregulated miRNAs are common in PTC (76, 89), unlike TCGA findings where the expression patterns of miRNA of cluster 1 were markedly dissimilar to the most deregulated miRNA in PTC (15).
Unlike PTC, FTC shows a specific and different pattern of miRNA expression, however there are a minimum 2 shared miRNAs including mir-221 and miR-34 that have significant contributions in well-differentiated tumours in case of thyroid carcinogenesis (76). On the other hand, ATC contains several deregulated miRNAs present in FTC and PTC, however it contains specific deregulated miRNAs that are associated with the malignancy and dedifferentiation (90, 91). ATC also contains heterogenous cell types and a higher level of downregulated miRNAs (91). Collectively, these findings confirm the significance of epigenetics as a diagnostic tool in the case of TC. Numerous studies have already analysed and identified miRNA profiles differentiating benign from malignant thyroid neoplasms, which could be used by clinicians in postoperative monitoring. A combination of quantitative reverse transcription polymerase chain reaction (RT-PCR) and sequencing showed overexpressed levels of miR-222 and miR-151-5p in tissue and serum samples of PTC patients in comparison with healthy controls and goitre patients (77). Levels of miR-222 and miR-151-5p were found to be reduced following thyroidectomy compared to the levels observed in healthy people (92–94). A range of other PTC-linked miRNAs including miRNA-146b, miR-451a, miR-25-3p, miRNA-190, miR-29b, miRNA-95, and miRNA-579 have been identified.
Indeed, miR-146b is a well-studied and most overexpressed miRNAs in TC, which is most commonly seen in PTC. The miR-146b expression is closely linked with the malignant thyroid neoplasm occurrence, which makes this miRNA an important biomarker (79). It was observed that miR-146b-3p can interact with the 3′-UTR of SLC5A5 and PAX8, which further leads to iodide uptake and protein translation inhibition (95). It has been observed that miR-146b-5p interacts with 3′-UTR of PTEN and SMAD4 (SMAD Family Member 4) to decrease the level of mRNA expression and stimulate PI3K/AKT signalling mechanism and TGFβ hyperactivation, which eventually leads to an elevated level of cell aggressiveness (96, 97). In addition, miR-146b-5p decreases the biosynthesis of miRNA via targeting DICER1, which is an important protein required for miRNA maturation that contributes as a tumour suppressor in TC (98). Both cell proliferation as well as invasion are regulated by miR-146b-5p and these processes are up-regulated throughout the epithelial-mesenchymal transition, therefore affects the progression of PTC. Several studies already reported the potential of miRNA profiles (including miR-146b) for the accurate differentiation benign as well as malignant tumours in fine-needle aspiration biopsy (FNAB) specimens (99–102). Levels of circulating miR-146b have been designated as an important and reliable serological marker for differentiating between benign tumours and PTC (103). Identification of the expressions of miR-146b in case of several thyroid nodules via using in situ hybridisation study confirmed its significance in differentiating between PTC from anaplastic TC, FTC, follicular adenomas, or poorly differentiated TC (84, 104). Increased concentrations of miR-21 and miR-146b-5p were also linked with markedly lower survival rates in PTC patients. In PTC, mir-146b-5p has been suggested as a prognostic and diagnostic marker because of its increased expression level in PTC unlike other tissues examined (104).
TCGA carried out a study with TC samples, which observed that miR-146b-5p-induced regulation of the IRAK1 (interleukin-1 receptor-associated kinase 1) gene which is different from the conventional form of PTC (105). NF-κB/IL6/STAT3 signalling cascade was also found to be linked with the regulation of synthesis of miR-146b-5p, whose elevated concentrations downregulate the expressions of various pro-inflammatory mediators including IRAK1 (106). The deregulation of miR-146b was linked with aggressive behaviour of tumours in clinical PTC specimens positive for BRAF and individuals containing BRAF mutations showed elevated expressions of miR-146b as compared to BRAF wild-type controls (107, 108). Various other miRNAs including miR-17-92, miR-339, and miR-875-5p cluster also have the capacity to interact with the 3′UTR of SLC5A5, which further decreases the expressions of NIS. An increased concentration of miR-875-5p expression was detected in the case of PDTC, while miR-339 expression was less commonly observed (109). Elevated levels of miR-17-92 expression decreased PAX8 and other genes responsible for iodine metabolism (110), whereas the miR-29a level is downregulated and targets LOX gene (91). An elevated level of PI3K cascade regulation by microRNAs has been observed in TC. PTEN is also targeted by miR-21, where an increased expression has been observed in various solid tumours including PTC. The expression of miR-21 is controlled through RAS by the activator Protein-1 transcription factor and minimum 2 downstream pathways including PI3K and MAPK (111, 112). Both miR-222 and miR-221 can interact with the 2 target regions in the 3′-UTR area of the p27-kip1 transcript, an established downstream effector of the PI3K cascade and a negative cell cycle regulator, which decreases its protein concentrations (112). The overexpression of miR-221 triggered cell invasion, migration and proliferation in various PTC cell lines (113).
An overexpression of miR-29 and miR-23 has also been reported, which are found to be controlled by TSH and its target SMAD3 (a key regulator of TGFβ activities) (114). Among the TC-related downregulated miRNAs, the let-7 miRNA family has a significant contribution in the development and control of cell fate. Members of the let-7 miRNA family have the capacity to interact with the 3′-UTR of all 3 RAS genes (including KRAS, NRAS and HRAS) via several binding regions, which contribute in decreasing the levels of protein expression. An activation of the RAS-ERK cascade was detected following the downregulation of let-7 miRNAs. A downregulation of let-7f was observed in PTC (115), while its expression was found to be linked with the levels of RAS protein (116). Moreover, a stable let-7f transfection in TPC-1 (the human PTC cell line) containing RET/PTC rearrangement resulted in the decreased cell proliferation and MAPK activation.
As per TCGA findings, expression of miR-137 is not markedly deregulated in TC, however a lower level of expression was observed in PTC as compared to adjacent normal tissues (117). It was observed that miR-137 can directly target the transcript of the tyrosine kinase receptor gene EGFR (epidermal growth factor receptor), which has a significant contribution in the MAPK signalling translocation from the membrane to the nucleus in order to cause activation of the major effectors of MAPK. In addition, miR-137 downregulated cell invasion, colony formation ability, and cell proliferation as well as negatively regulated ERK and Akt signalling pathways in TPC-1 and B-CPAP (the PTC cell lines). The reduction of EGFR repealed the action of miR-137 suppression on these signalling cascades, which indicates that miR-137 contributes on signalling in an EGFR-dependent manner. In a study, Nieto et al. used combined miRNA and mRNA expression as prognostic indicators of TC recurrence (78). They formulated a risk score model as per the comprehensive bioinformatics and experimental miRNA, mRNA, and somatic mutation study in recurrent tumours. Furthermore, they utilised RNA sequencing results of 501 TC samples obtained from TCGA datasets, including 46 recurrent tumours and 455 non-recurrent samples. These researchers also carried out a functional gene analysis in several thyroid cell lines in cell-based assays and evaluated the prognostic values of the genes by utilising the TCGA datasets (78). In total, they detected 59 genetic variants, 39 miRNAs, and 40 mRNAs as important biomarkers of TC recurrence. Among them, miR-1179 and miR-486 showed marked activities in suppressing in vitro TC cell migration, while deletion of miR-1179 and miR-486 elevated cellular migration in vitro (82, 118). A markedly higher level of miR-375 overexpression was observed in the case of MTC in comparison with the normal thyroid tissues. Moreover, there is a close connection between patient outcomes, expression of miR-375, and tumour aggressiveness, which indicates an important role in the pathogenesis of MTC (119, 120). Therefore, levels of circulating miR-375 are regarded as important prognostic markers for advanced MTC (121). In addition to this, serum miR-375 can be used as prognostic and diagnostic markers of MTC, which differentiates between control subjects and MTC patients along with a 97.6% specificity and a 92.6% sensitivity (122).
5 Conclusion and future perspectives
Advances in genetic research and the epigenetics revolution have extensively evaluated Over the past decades to find out whether genetic codes predominately determine gene function or not. Various experiments have also established the involvement of genetic and epigenetic modifications in cancer, which indicates that genome packaging is also vital as like genome in controlling the major cellular mechanisms. Therefore, a comprehension regarding genetic and epigenetic modifications is not only essential for the diagnosis and prognosis of various cancers including TC but also for the development of therapeutics. As like any other cancers, most of the genetic and epigenetic modifications in the case of TC are somatic in nature, thus evaluation of the epigenetic pattern in TC exhibited an important contribution in these modifications in the prognosis and classification of tumours. Interestingly, TC-associated epigenetic alterations are reversible; therefore it is possible to develop an optional epigenetic therapy. As miRNAs have substantial contribution in cell invasion, differentiation and proliferation, thus miRNAs as well as target genes can be used as potential targets for the diagnosis and treatment of tumours. Indeed, whole genome sequencing methods can extraordinarily identify the genetic lesions accountable for the dedifferentiation, progression and onset of TC. The molecular pathogenesis of TC has been changed owing to the growing knowledge of genomics and epigenomics. This improved understanding of TC-associated signalling mechanisms and complex intracellular networks has resulted in clinical trials with small kinase inhibitors.
Furthermore, mutation identifications in novel genes led to the detection of potential and novel molecular markers of TC. An updated classification of thyroid tumours as per the RAS-and BRAF-score or differentiation-score has mediated the development of precise molecular classification of these tumours. Novel findings on histone acetylation and DNA methylation might result in the detection of repressive molecules of both modifications that may further facilitate the thyroid tumour re-differentiation, which can further decrease their aggressive behaviours and their refractoriness to radioactive iodine. Collectively, all these findings have improved the knowledge regarding TC pathogenesis and its causes. Moreover, this knowledge has provided insights regarding the biological mechanisms linked with the progression and initiation of TC, regulatory circuits, new targetable cancer genes, and molecular markers with clinical uses in prognosis as well as diagnosis of TC.
Author contributions
ES: Conceptualization, Formal analysis, Funding acquisition, Investigation, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Huang J, Ngai CH, Deng Y, Pun CN, Lok V, Zhang L, et al. Incidence and mortality of thyroid cancer in 50 countries: a joinpoint regression analysis of global trends. Endocrine. (2023) 80:355–65. doi: 10.1007/S12020-022-03274-7/FIGURES/5
2. Lortet-Tieulent J, Franceschi S, Dal Maso L, Vaccarella S. Thyroid cancer “epidemic” also occurs in low- and middle-income countries. Int J Cancer. (2019) 144:2082–7. doi: 10.1002/IJC.31884
3. Li M, Maso LD, Vaccarella S. Global trends in thyroid cancer incidence and the impact of overdiagnosis. Lancet Diabetes Endocrinol. (2020) 8:468–70. doi: 10.1016/S2213-8587(20)30115-7
4. Cao J, Zhang M, Zhang L, Lou J, Zhou F, Fang M. Non-coding RNA in thyroid cancer - Functions and mechanisms. Cancer Lett. (2021) 496:117–26. doi: 10.1016/J.CANLET.2020.08.021
5. Miranda-Filho A, Lortet-Tieulent J, Bray F, Cao B, Franceschi S, Vaccarella S, et al. Thyroid cancer incidence trends by histology in 25 countries: a population-based study. Lancet Diabetes Endocrinol. (2021) 9:225–34. doi: 10.1016/S2213-8587(21)00027-9
6. Grimmichova T, Pacesova P, Hill M, Pekova B, Vankova M, Moravcova J, et al. Thyroid cancer detection in a routine clinical setting: performance of ACR TI-RADS, FNAC, and molecular testing in prospective cohort study. Biomedicines. (2022) 10:954. doi: 10.3390/BIOMEDICINES10050954
7. Xing M, Alzahrani AS, Carson KA, Shong YK, Kim TY, Viola D, et al. Association between BRAF V600E mutation and recurrence of papillary thyroid cancer. J Clin Oncol. (2015) 33:42. doi: 10.1200/JCO.2014.56.8253
8. Hedayati M, Yeganeh MZ, Eslami SS, Barez SR, Rad LH, Azizi F. Predominant RET germline mutations in exons 10, 11, and 16 in Iranian patients with hereditary medullary thyroid carcinoma. J Thyroid Res. (2011) 2011. doi: 10.4061/2011/264248
9. Musholt TJ, Schönefeld S, Schwarz CH, Watzka FM, Musholt PB, Fottner C, et al. Impact of pathognomonic genetic alterations on the prognosis of papillary thyroid carcinoma: ESES vienna presentation. Langenbecks Arch Surg. (2010) 395:877–83. doi: 10.1007/S00423-010-0682-6/FIGURES/2
10. Faam B, Ali Ghaffari M, Ghadiri A, Azizi F. Epigenetic modifications in human thyroid cancer. BioMed Rep. (2015) 3:3. doi: 10.3892/BR.2014.375
11. Wu C, Schwartz JM, Brabant G, Nenadic G. Molecular profiling of thyroid cancer subtypes using large-scale text mining. BMC Med Genomics. (2014) 7:1–11. doi: 10.1186/1755-8794-7-S3-S3/TABLES/8
12. Prete A, Borges de Souza P, Censi S, Muzza M, Nucci N, Sponziello M. Update on fundamental mechanisms of thyroid cancer. Front Endocrinol (Lausanne). (2020) 11:102/BIBTEX. doi: 10.3389/FENDO.2020.00102/BIBTEX
13. Hlozek J, Pekova B, Rotnagl J, Holy R, Astl J. Genetic changes in thyroid cancers and the importance of their preoperative detection in relation to the general treatment and determination of the extent of surgical intervention—A review. Biomedicines. (2022) 10:1515. doi: 10.3390/BIOMEDICINES10071515
14. Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. (2010) 31:27–36. doi: 10.1093/CARCIN/BGP220
15. Acuña-Ruiz A, Carrasco-López C, Santisteban P. Genomic and epigenomic profile of thyroid cancer. Best Pract Res Clin Endocrinol Metab. (2023) 37:101656. doi: 10.1016/J.BEEM.2022.101656
16. Katz TA, Huang Y, Davidson NE, Jankowitz RC. Epigenetic reprogramming in breast cancer: From new targets to new therapies. Ann Med. (2014) 46:397–408. doi: 10.3109/07853890.2014.923740
17. Moon JW, Lee SK, Lee JO, Kim N, Lee YW, Kim SJ, et al. Identification of novel hypermethylated genes and demethylating effect of vincristine in colorectal cancer. J Exp Clin Cancer Res. (2014) 33:1–10. doi: 10.1186/1756-9966-33-4/FIGURES/4
18. Cassol CA, Asa SL. Molecular pathology of thyroid cancer. Diagn Histopathol. (2011) 17:124–39. doi: 10.1016/J.MPDHP.2010.12.006
19. Liu Y, Marti A, Umbricht C. Molecular diagnostics of thyroid tumors. Arch Pathol Lab Med. (2011) 135:569–77. doi: 10.5858/2010-0664-RAIR.1
20. Laha D, Nilubol N, Boufraqech M. New therapies for advanced thyroid cancer. Front Endocrinol (Lausanne). (2020) 11:82/BIBTEX. doi: 10.3389/FENDO.2020.00082/BIBTEX
21. Nikiforov YE. Role of molecular markers in thyroid nodule management: then and now. Endocr Pract. (2017) 23:979–89. doi: 10.4158/EP171805.RA
22. Haugen BR, Alexander EK, Bible KC, Doherty GM, Mandel SJ, Nikiforov YE, et al. 2015 American thyroid association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: the american thyroid association guidelines task force on thyroid nodules and differentiated thyroid cancer. Thyroid. (2016) 26:1–133. doi: 10.1089/THY.2015.0020/ASSET/IMAGES/MEDIUM/INLINE1.TIF.GIF
23. Machens A, Dralle H. Long-term outcome after DNA-based prophylactic neck surgery in children at risk of hereditary medullary thyroid cancer. Best Pract Res Clin Endocrinol Metab. (2019) 33:101274. doi: 10.1016/J.BEEM.2019.04.008
24. Pekova B, Dvorakova S, Sykorova V, Vacinova G, Vaclavikova E, Moravcova J, et al. Somatic genetic alterations in a large cohort of pediatric thyroid nodules. Endocr Connect. (2019) 8:796–805. doi: 10.1530/EC-19-0069
25. Zitzelsberger H, Bauer V, Thomas G, Unger K. Molecular rearrangements in papillary thyroid carcinomas. Clin Chim Acta. (2010) 411:301–8. doi: 10.1016/J.CCA.2009.11.028
26. Paschke R, Cantara S, Crescenzi A, Jarzab B, Musholt TJ, Sobrinho Simoes M. European thyroid association guidelines regarding thyroid nodule molecular fine-needle aspiration cytology diagnostics. Eur Thyroid J. (2017) 6:115–29. doi: 10.1159/000468519
27. Eberhardt NL, Grebe SKG, McIver B, Reddi HV. The role of the PAX8/PPARγ fusion oncogene in the pathogenesis of follicular thyroid cancer. Mol Cell Endocrinol. (2010) 321:50–6. doi: 10.1016/J.MCE.2009.10.013
28. Liu X, Bishop J, Shan Y, Pai S, Liu D, Murugan AK, et al. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr Relat Cancer. (2013) 20:603. doi: 10.1530/ERC-13-0210
29. McKelvey BA, Umbricht CB, Zeiger MA. Telomerase reverse transcriptase (TERT) regulation in thyroid cancer: A review. Front Endocrinol (Lausanne). (2020) 11:485/BIBTEX. doi: 10.3389/FENDO.2020.00485/BIBTEX
30. Chung JH. BRAF and TERT promoter mutations: clinical application in thyroid cancer. Endocr J. (2020) 67:577–84. doi: 10.1507/ENDOCRJ.EJ20-0063
31. Ren H, Shen Y, Hu D, He W, Zhou J, Cao Y, et al. Co-existence of BRAFv600e and TERT promoter mutations in papillary thyroid carcinoma is associated with tumor aggressiveness, but not with lymph node metastasis. Cancer Manag Res. (2018) 10:1005–13. doi: 10.2147/CMAR.S159583
32. Pekova B, Sykorova V, Mastnikova K, Vaclavikova E, Moravcova J, Vlcek P, et al. NTRK fusion genes in thyroid carcinomas: Clinicopathological characteristics and their impacts on prognosis. Cancers (Basel). (2021) 13:1932. doi: 10.3390/CANCERS13081932/S1
33. Cancer Genome Atlas Research Network T, Agrawal N, Akbani R, Arman Aksoy B, Ally A, Arachchi H, et al. Integrated genomic characterization of papillary thyroid carcinoma. Cell (2014) 159:676–90. doi: 10.1016/j.cell.2014.09.050
34. Wang Y, Zhang L, Liu Z. Molecular classification of thyroid tumors and key molecular features to identify high-grade thyroid carcinomas. Thyroid FNA Cytol. (2023) 139–46. doi: 10.1007/978-981-99-6782-7_21
35. Ahmed AA, Essa MEA. Potential of epigenetic events in human thyroid cancer. Cancer Genet. (2019) 239:13–21. doi: 10.1016/J.CANCERGEN.2019.08.006
36. Puppin C, Passon N, Lavarone E, Di Loreto C, Frasca F, Vella V, et al. Levels of histone acetylation in thyroid tumors. Biochem Biophys Res Commun. (2011) 411:679–83. doi: 10.1016/J.BBRC.2011.06.182
37. Hou P, Bojdani E, Xing M. Induction of thyroid gene expression and radioiodine uptake in thyroid cancer cells by targeting major signaling pathways. J Clin Endocrinol Metab. (2010) 95:820–8. doi: 10.1210/JC.2009-1888
38. Wächter S, Damanakis AI, Elxnat M, Roth S, Wunderlich A, Verburg FA, et al. Epigenetic modifications in thyroid cancer cells restore NIS and radio-iodine uptake and promote cell death. J Clin Med. (2018) 7:61. doi: 10.3390/JCM7040061
39. Nilubol N, Merkel R, Yang L, Patel D, Reynolds JC, Sadowski SM, et al. A phase II trial of valproic acid in patients with advanced, radioiodine-resistant thyroid cancers of follicular cell origin. Clin Endocrinol (Oxf). (2017) 86:128–33. doi: 10.1111/CEN.13154
40. Amiri-Kordestani L, Luchenko V, Peer CJ, Ghafourian K, Reynolds J, Draper D, et al. Phase I trial of a new schedule of romidepsin in patients with advanced cancers. Clin Cancer Res. (2013) 19:4499–507. doi: 10.1158/1078-0432.CCR-13-0095/273189/AM/PHASE-I-TRIAL-OF-A-NEW-SCHEDULE-OF-ROMIDEPSIN-IN
41. Zhang Y, Yu S, Jiang L, Wang X, Song X. HOTAIR is a promising novel biomarker in patients with thyroid cancer. Exp Ther Med. (2017) 13:2274. doi: 10.3892/ETM.2017.4231
42. Li HM, Yang H, Wen DY, Luo YH, Liang CY, Pan DH, et al. Overexpression of lncRNA HOTAIR is associated with poor prognosis in thyroid carcinoma: A study based on TCGA and GEO data. Hormone Metab Res. (2017) 49:388–99. doi: 10.1055/S-0043-103346/ID/R2016-08-0268-0011/BIB
43. Liao G, Huang Z, Gan T, Wu C, Wang X, Li D. Long non-coding RNA nuclear enriched abundant transcript 1 (NEAT1) modulates inhibitor of DNA binding 1 (ID1) to facilitate papillary thyroid carcinoma development by sponging microRNA-524-5p. Bioengineered. (2022) 13:13201–12. doi: 10.1080/21655979.2022.2076498
44. Peng X, Ji C, Tan L, Lin S, Zhu Y, Long M, et al. Long non-coding RNA TNRC6C-AS1 promotes methylation of STK4 to inhibit thyroid carcinoma cell apoptosis and autophagy via Hippo signalling pathway. J Cell Mol Med. (2020) 24:304. doi: 10.1111/JCMM.14728
45. Gou L, Zou H, Li B. Long noncoding RNA MALAT1 knockdown inhibits progression of anaplastic thyroid carcinoma by regulating miR-200a-3p/FOXA1. Cancer Biol Ther. (2019) 20:1355. doi: 10.1080/15384047.2019.1617567
46. Wu WJ, Yin H, Hu JJ, Wei XZ. Long noncoding RNA LINC00313 modulates papillary thyroid cancer tumorigenesis via sponging miR-4429. Neoplasma. (2018) 65:933–42. doi: 10.4149/NEO_2018_180219N125
47. Gou Q, Gao L, Nie X, Pu W, Zhu J, Wang Y, et al. Long noncoding RNA AB074169 inhibits cell proliferation via modulation of KHSRP-mediated CDKN1a expressionin papillary thyroid carcinoma. Cancer Res. (2018) 78:4163–74. doi: 10.1158/0008-5472.CAN-17-3766/653203/AM/LONG-NONCODING-RNA-AB074169-INHIBITS-CELL
48. Lei H, Gao Y, Xu X. LncRNA TUG1 influences papillary thyroid cancer cell proliferation, migration and EMT formation through targeting miR-145. Acta Biochim Biophys Sin (Shanghai). (2017) 49:588–97. doi: 10.1093/ABBS/GMX047
49. Tong H, Zhuang X, Cai J, Ding Y, Si Y, Zhang H, et al. Long noncoding RNA ZFAS1 promotes progression of papillary thyroid carcinoma by sponging miR-590-3p and upregulating HMGA2 expression. Onco Targets Ther. (2019) 12:7501. doi: 10.2147/OTT.S209138
50. Ning ML, Qin S, Tian J, Wang Y, Liu Q. LncRNA AFAP-AS1 promotes anaplastic thyroid cancer progression by sponging miR-155-5p through ETS1/ERK pathway. Bioengineered. (2021) 12:1543. doi: 10.1080/21655979.2021.1918537
51. Zheng H, Wang M, Jiang L, Chu H, Hu J, Ning J, et al. BRAF-activated long noncoding RNA modulates papillary thyroid carcinoma cell proliferation through regulating thyroid stimulating hormone receptor. Cancer Res Treatment: Off J Korean Cancer Assoc. (2016) 48:698. doi: 10.4143/CRT.2015.118
52. Xia F, Chen Y, Jiang B, Du X, Peng Y, Wang W, et al. Long noncoding RNA HOXA-AS2 promotes papillary thyroid cancer progression by regulating miR-520c-3p/S100A4 pathway. Cell Physiol Biochem. (2018) 50:1659–72. doi: 10.1159/000494786
53. Choy M, Guo Y, Li H, Wei G, Ye R, Liang W, et al. Long noncoding RNA LOC100129940-N is upregulated in papillary thyroid cancer and promotes the invasion and progression. Int J Endocrinol. (2019) 2019. doi: 10.1155/2019/7043509
54. Li J-H, Zhang S-Q, Qiu X-G, Zhang S-J, Zheng S-H, Zhang D-H. Long non-coding RNA NEAT1 promotes Malignant progression of thyroid carcinoma by regulating miRNA-214. Int J Oncol. (2017) 50:708–16. doi: 10.3892/ijo.2016.3803
55. Hou S, Lin Q, Guan F, Lin C. LncRNA TNRC6C-AS1 regulates UNC5B in thyroid cancer to influence cell proliferation, migration, and invasion as a competing endogenous RNA of miR-129-5p. J Cell Biochem. (2018) 119:8304–16. doi: 10.1002/JCB.26868
56. Muhanhali D, Zhai T, Jiang J, Ai Z, Zhu W, Ling Y. Long non-coding antisense RNA TNRC6C-AS1 is activated in papillary thyroid cancer and promotes cancer progression by suppressing TNRC6C expression. Front Endocrinol (Lausanne). (2018) 9:360/BIBTEX. doi: 10.3389/FENDO.2018.00360/BIBTEX
57. Zhang R, Hardin H, Huang W, Chen J, Asioli S, Righi A, et al. MALAT1 long non-coding RNA expression in thyroid tissues: analysis by in situ hybridization and real-time PCR. Endocr Pathol. (2017) 28:7–12. doi: 10.1007/S12022-016-9453-4/FIGURES/4
58. Huang JK, Ma L, Song WH, Lu BY, Huang YB, Dong HM, et al. MALAT1 promotes the proliferation and invasion of thyroid cancer cells via regulating the expression of IQGAP1. Biomed Pharmacother. (2016) 83:1–7. doi: 10.1016/J.BIOPHA.2016.05.039
59. Liu Z, Liu D, Bojdani E, El-Naggar AK, Vasko V, Xing MM. IQGAP1 plays an important role in the invasiveness of thyroid cancer. Clin Cancer Res. (2010) 16:6009–18. doi: 10.1158/1078-0432.CCR-10-1627/83681/AM/IQGAP1-PLAYS-AN-IMPORTANT-ROLE-IN-THE-INVASIVENESS
60. Xiong X, Zhu H, Chen X. Low expression of long noncoding RNA CASC2 indicates a poor prognosis and promotes tumorigenesis in thyroid carcinoma. Biomed Pharmacother. (2017) 93:391–7. doi: 10.1016/J.BIOPHA.2017.06.063
61. Huang F, Zhang Q, Chen W, Zhang H, Lu G, Chen J, et al. Long noncoding RNA cancer susceptibility candidate 2 suppresses papillary thyroid carcinoma growth by inactivating the AKT/ERK1/2 signaling pathway. J Cell Biochem. (2019) 120:10380–90. doi: 10.1002/JCB.28322
62. Liu QY, Gao LY, Xu L, Zhang XL, Zhang LJ, Gong XL, et al. CASC2 inhibits the growth, migration, and invasion of thyroid cancer cells through sponging miR-18a-5p/FIH1 axis. Kaohsiung J Med Sci. (2021) 37:268–75. doi: 10.1002/KJM2.12331
63. Zhang XF, Ye Y, Zhao SJ. LncRNA Gas5 acts as a ceRNA to regulate PTEN expression by sponging miR-222-3p in papillary thyroid carcinoma. Oncotarget. (2018) 9:3519. doi: 10.18632/ONCOTARGET.23336
64. Li S, Zhang Y, Dong J, Li R, Yu B, Zhao W, et al. LINC00893 inhibits papillary thyroid cancer by suppressing AKT pathway via stabilizing PTEN. Cancer Biomarkers. (2021) 30:277–86. doi: 10.3233/CBM-190543
65. Chen LD, Zhuo DQ, Yuan HY. Circ_100395 impedes Malignancy and glycolysis in papillary thyroid cancer: Involvement of PI3K/AKT/mTOR signaling pathway. Immunol Lett. (2022) 246:10–7. doi: 10.1016/J.IMLET.2022.04.004
66. Wang D, Li Z, Wu Y. The research progression and clinical significance of circular RNAs in head and neck cancers. BioMed Res Int. (2020) 2020. doi: 10.1155/2020/2712310
67. Xia F, Zhang Z, Li X. Emerging roles of circular RNAs in thyroid cancer. Front Cell Dev Biol. (2021) 9:636838. doi: 10.3389/FCELL.2021.636838
68. Liu T, Huang T, Shang M, Han G. CircRNA ITCH: insight into its role and clinical application prospect in tumor and non-tumor diseases. Front Genet. (2022) 13:927541. doi: 10.3389/FGENE.2022.927541
69. Cai X, Zhao Z, Dong J, Lv Q, Yun B, Liu J, et al. Circular RNA circBACH2 plays a role in papillary thyroid carcinoma by sponging miR-139-5p and regulating LMO4 expression. Cell Death Dis. (2019) 10. doi: 10.1038/S41419-019-1439-Y
70. Peng N, Shi L, Zhang Q, Hu Y, Wang N, Ye H. Microarray profiling of circular RNAs in human papillary thyroid carcinoma. PloS One. (2017) 12:e0170287. doi: 10.1371/JOURNAL.PONE.0170287
71. Bi W, Huang J, Nie C, Liu B, He G, Han J, et al. CircRNA circRNA-102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway 06 Biological Sciences 0601 Biochemistry and Cell Biology. J Exp Clin Cancer Res. (2018) 37:1–9. doi: 10.1186/S13046-018-0936-7/FIGURES/6
72. Teng H, Mao F, Liang J, Xue M, Wei W, Li X, et al. Transcriptomic signature associated with carcinogenesis and aggressiveness of papillary thyroid carcinoma. Theranostics. (2018) 8:4345. doi: 10.7150/THNO.26862
73. Zhang Z, Wang W, Su Z, Zhang J, Cao H. Circ_0011058 facilitates proliferation, angiogenesis and radioresistance in papillary thyroid cancer cells by positively regulating YAP1 via acting as miR-335-5p sponge. Cell Signal. (2021) 88:110155. doi: 10.1016/J.CELLSIG.2021.110155
74. Chu J, Tao L, Yao T, Chen Z, Lu X, Gao L, et al. Circular RNA circRUNX1 promotes papillary thyroid cancer progression and metastasis by sponging MiR-296-3p and regulating DDHD2 expression. Cell Death Dis. (2021) 12:1–16. doi: 10.1038/s41419-020-03350-8
75. Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differentiation. (2017) 24:1609–20. doi: 10.1038/cdd.2017.86
76. Mancikova V, Castelblanco E, Pineiro-Yanez E, Perales-Paton J, De Cubas AA, Inglada-Perez L, et al. MicroRNA deep-sequencing reveals master regulators of follicular and papillary thyroid tumors. Modern Pathol. (2015) 28:748–57. doi: 10.1038/modpathol.2015.44
77. Yu S, Liu Y, Wang J, Guo Z, Zhang Q, Yu F, et al. Circulating microRNA profiles as potential biomarkers for diagnosis of papillary thyroid carcinoma. J Clin Endocrinol Metab. (2012) 97:2084–92. doi: 10.1210/JC.2011-3059
78. Nieto HR, Thornton CEM, Brookes K, Nobre De Menezes A, Fletcher A, Alshahrani M, et al. Recurrence of papillary thyroid cancer: A systematic appraisal of risk factors. J Clin Endocrinol Metab. (2022) 107:1392–406. doi: 10.1210/CLINEM/DGAB836
79. Chou CK, Liu RT, Kang HY. MicroRNA-146b: A novel biomarker and therapeutic target for human papillary thyroid cancer. Int J Mol Sci. (2017) 18:636. doi: 10.3390/IJMS18030636
80. Ghafouri-Fard S, Shirvani-Farsani Z, Taheri M. The role of microRNAs in the pathogenesis of thyroid cancer. Noncoding RNA Res. (2020) 5:88. doi: 10.1016/J.NCRNA.2020.06.001
81. Jiao X, Ye J, Wang X, Yin X, Zhang G, Cheng X. KIAA1199, a target of micoRNA-486-5p, promotes papillary thyroid cancer invasion by influencing epithelial-mesenchymal transition (EMT). Med Sci Monit. (2019) 25:6788. doi: 10.12659/MSM.918682
82. Wang Y, Zong H, Zhou H. Circular RNA circ_0062389 modulates papillary thyroid carcinoma progression via the miR-1179/high mobility group box 1 axis. Bioengineered. (2021) 12:1484–94. doi: 10.1080/21655979.2021.1914470
83. Tabatabaeian H, Yang SP, Tay Y. Non-coding RNAs: uncharted mediators of thyroid cancer pathogenesis. Cancers (Basel). (2020) 12:1–36. doi: 10.3390/CANCERS12113264
84. Macvanin MT, Gluvic ZM, Zaric BL, Essack M, Gao X, Isenovic ER. New biomarkers: prospect for diagnosis and monitoring of thyroid disease. Front Endocrinol (Lausanne). (2023) 14:1218320. doi: 10.3389/FENDO.2023.1218320
85. Chevillet JR, Lee I, Briggs HA, He Y, Wang K. Issues and prospects of microRNA-based biomarkers in blood and other body fluids. Molecules. (2014) 19:6080–105. doi: 10.3390/MOLECULES19056080
86. Li X, Abdel-Mageed AB, Mondal D, Kandil E. MicroRNA expression profiles in differentiated thyroid cancer, a review. Int J Clin Exp Med. (2013) 6:74.
87. Ramírez-Moya J, Santisteban P. MiRNA-directed regulation of the main signaling pathways in thyroid cancer. Front Endocrinol (Lausanne). (2019) 10:430/BIBTEX. doi: 10.3389/FENDO.2019.00430/BIBTEX
88. Agrawal N, Akbani R, Aksoy BA, Ally A, Arachchi H, Asa SL, et al. Integrated genomic characterization of papillary thyroid carcinoma. Cell. (2014) 159:676. doi: 10.1016/J.CELL.2014.09.050
89. Dettmer M, Perren A, Moch H, Komminoth P, Nikiforov YE, Nikiforova MN. Comprehensive microRNA expression profiling identifies novel markers in follicular variant of papillary thyroid carcinoma. Thyroid. (2013) 23:1383–9. doi: 10.1089/THY.2012.0632
90. Santiago K, Chen Wongworawat Y, Khan S. Differential microRNA-signatures in thyroid cancer subtypes. J Oncol. (2020) 2020. doi: 10.1155/2020/2052396
91. Heb́rant A, Floor S, Saiselet M, Antoniou A, Desbuleux A, Snyers B, et al. miRNA expression in anaplastic thyroid carcinomas. PloS One. (2014) 9:e103871. doi: 10.1371/JOURNAL.PONE.0103871
92. Lee JC, Zhao JT, Clifton-Bligh RJ, Gill A, Gundara JS, Ip JC, et al. MicroRNA-222 and MicroRNA-146b are tissue and circulating biomarkers of recurrent papillary thyroid cancer. Cancer. (2013) 119:4358–65. doi: 10.1002/CNCR.28254
93. Cantara S, Pilli T, Sebastiani G, Cevenini G, Busonero G, Cardinale S, et al. Circulating miRNA95 and miRNA190 Are Sensitive Markers for the Differential Diagnosis of Thyroid Nodules in a Caucasian Population. J Clin Endocrinol Metab. (2014) 99:4190–8. doi: 10.1210/JC.2014-1923
94. Li M, Song Q, Li H, Lou Y, Wang L. Circulating miR-25-3p and miR-451a May Be Potential Biomarkers for the Diagnosis of Papillary Thyroid Carcinoma. PloS One. (2015) 10:e0132403. doi: 10.1371/JOURNAL.PONE.0132403
95. Riesco-Eizaguirre G, Wert-Lamas L, Perales-Paton J, Sastre-Perona A, Fernandez LP, Santisteban P. The miR-146b-3p/PAX8/NIS regulatory circuit modulates the differentiation phenotype and function of thyroid cells during carcinogenesis. Cancer Res. (2015) 75:4119–30. doi: 10.1158/0008-5472.CAN-14-3547/651817/AM/THE-MIR-146B-3P-PAX8-NIS-REGULATORY-CIRCUIT
96. Ramírez-Moya J, Wert-Lamas L, Santisteban P. MicroRNA-146b promotes PI3K/AKT pathway hyperactivation and thyroid cancer progression by targeting PTEN. Oncogene. (2018) 37:3369–83. doi: 10.1038/s41388-017-0088-9
97. Geraldo MV, Yamashita AS, Kimura ET. MicroRNA miR-146b-5p regulates signal transduction of TGF-β by repressing SMAD4 in thyroid cancer. Oncogene. (2011) 31:1910–22. doi: 10.1038/onc.2011.381
98. Ramírez-Moya J, Wert-Lamas L, Riesco-Eizaguirre G, Santisteban P. Impaired microRNA processing by DICER1 downregulation endows thyroid cancer with increased aggressiveness. Oncogene. (2019) 38:5486–99. doi: 10.1038/s41388-019-0804-8
99. Shen R, Liyanarachchi S, Li W, Wakely PE, Saji M, Huang J, et al. MicroRNA signature in thyroid fine needle aspiration cytology applied to “Atypia of undetermined significance” Cases. Thyroid. (2012) 22:9–16. doi: 10.1089/THY.2011.0081
100. Keutgen XM, Filicori F, Crowley MJ, Wang Y, Scognamiglio T, Hoda R, et al. A panel of four miRNAs accurately differentiates Malignant from benign indeterminate thyroid lesions on fine needle aspiration. Clin Cancer Res. (2012) 18:2032–8. doi: 10.1158/1078-0432.CCR-11-2487/84892/AM/A-PANEL-OF-FOUR-MICRORNAS-ACCURATELY
101. Panebianco F, Mazzanti C, Tomei S, Aretini P, Franceschi S, Lessi F, et al. The combination of four molecular markers improves thyroid cancer cytologic diagnosis and patient management. BMC Cancer. (2015) 15:1–11. doi: 10.1186/S12885-015-1917-2/FIGURES/4
102. Vriens MR, Weng J, Suh I, Huynh N, Guerrero MA, Shen WT, et al. MicroRNA expression profiling is a potential diagnostic tool for thyroid cancer. Cancer. (2012) 118:3426–32. doi: 10.1002/CNCR.26587
103. Lee YS, Lim YS, Lee JC, Wang SG, Park HY, Kim SY, et al. Differential expression levels of plasma-derived miR-146b and miR-155 in papillary thyroid cancer. Oral Oncol. (2015) 51:77–83. doi: 10.1016/J.ORALONCOLOGY.2014.10.006
104. Guo Z, Hardin H, Montemayor-Garcia C, Asioli S, Righi A, Maletta F, et al. In Situ Hybridization Analysis of miR-146b-5p and miR-21 in Thyroid Nodules: Diagnostic Implications. Endocr Pathol. (2015) 26:157–63. doi: 10.1007/S12022-015-9363-X/TABLES/3
105. Stokowy T, Gawel D, Wojtas B. Differences in miRNA and mRNA Profile of Papillary Thyroid Cancer Variants. Int J Endocrinol. (2016) 2016. doi: 10.1155/2016/1427042
106. Chou CK, Chi SY, Huang CH, Chou FF, Huang CC, Liu RT, et al. IRAK1, a target of miR-146b, reduces cell aggressiveness of human papillary thyroid carcinoma. J Clin Endocrinol Metab. (2016) 101:4357–66. doi: 10.1210/JC.2016-2276/SUPPL_FILE/JC-16-2276.PDF
107. Yip L, Kelly L, Shuai Y, Armstrong MJ, Nikiforov YE, Carty SE, et al. MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann Surg Oncol. (2011) 18:2035–41. doi: 10.1245/S10434-011-1733-0/FIGURES/3
108. Chou CK, Chen RF, Chou FF, Chang HW, Chen YJ, Lee YF, et al. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAFV600E mutation. Thyroid. (2010) 20:489–94. doi: 10.1089/THY.2009.0027
109. Lakshmanan A, Wojcicka A, Kotlarek M, Zhang X, Jazdzewski K, Jhiang SM. microRNA-339-5p modulates Na+/I– symporter-mediated radioiodide uptake. Endocr Relat Cancer. (2015) 22:11–21. doi: 10.1530/ERC-14-0439
110. Fuziwara CS, Saito KC, Kimura ET. Thyroid follicular cell loss of differentiation induced by microRNA miR-17-92 cluster is attenuated by CRISPR/cas9n gene silencing in anaplastic thyroid cancer. Thyroid. (2020) 30:81–94. doi: 10.1089/THY.2018.0601/ASSET/IMAGES/LARGE/THY.2018.0601_FIGURE6.JPEG
111. Frezzetti D, De Menna M, Zoppoli P, Guerra C, Ferraro A, Bello AM, et al. Upregulation of miR-21 by Ras in vivo and its role in tumor growth. Oncogene. (2010) 30:275–86. doi: 10.1038/onc.2010.416
112. Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. (2007) 14:791–8. doi: 10.1677/ERC-07-0129
113. Wei ZL, Bin GA, Wang Q, XE L, Zhao J, Lu QJ. MicroRNA-221 promotes papillary thyroid carcinoma cell migration and invasion via targeting RECK and regulating epithelial–mesenchymal transition. Onco Targets Ther. (2019) 12:2323–33. doi: 10.2147/OTT.S190364
114. Leone V, D’Angelo D, Pallante P, Croce CM, Fusco A. Thyrotropin Regulates Thyroid Cell Proliferation by Up-Regulating miR-23b and miR-29b that Target SMAD3. J Clin Endocrinol Metab. (2012) 97:3292–301. doi: 10.1210/JC.2012-1349
115. Braun J, Hoang-Vu C, Dralle H, Hüttelmaier S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene. (2010) 29:4237–44. doi: 10.1038/onc.2010.169
116. Perdas E, Stawski R, Nowak D, Zubrzycka M. The Role of miRNA in Papillary Thyroid Cancer in the Context of miRNA Let-7 Family. Int J Mol Sci. (2016) 17:909. doi: 10.3390/IJMS17060909
117. Luo Y, Li X, Dong J, Sun W. microRNA-137 is downregulated in thyroid cancer and inhibits proliferation and invasion by targeting EGFR. Tumor Biol. (2016) 37:7749–55. doi: 10.1007/S13277-015-4611-8/FIGURES/4
118. ElKhouly AM, Youness RA, Gad MZ. MicroRNA-486-5p and microRNA-486-3p: Multifaceted pleiotropic mediators in oncological and non-oncological conditions. Noncoding RNA Res. (2020) 5:11–21. doi: 10.1016/J.NCRNA.2020.01.001
119. Mian C, Pennelli G, Fassan M, Balistreri M, Barollo S, Cavedon E, et al. MicroRNA profiles in familial and sporadic medullary thyroid carcinoma: preliminary relationships with RET status and outcome. Thyroid. (2012) 22:890–6. doi: 10.1089/THY.2012.0045
120. Galuppini F, Bertazza L, Barollo S, Cavedon E, Rugge M, Guzzardo V, et al. MiR-375 and YAP1 expression profiling in medullary thyroid carcinoma and their correlation with clinical–pathological features and outcome. Virchows Archiv. (2017) 471:651–8. doi: 10.1007/S00428-017-2227-7/FIGURES/5
121. Romeo P, Colombo C, Granata R, Calareso G, Gualeni AV, Dugo M, et al. Circulating miR-375 as a novel prognostic marker for metastatic medullary thyroid cancer patients. Endocr Relat Cancer. (2018) 25:217–31. doi: 10.1530/ERC-17-0389
Keywords: thyroid cancer, genetic modifications, epigenetic modifications, biomarkers, diagnosis, prognosis
Citation: Sabi EM (2024) The role of genetic and epigenetic modifications as potential biomarkers in the diagnosis and prognosis of thyroid cancer. Front. Oncol. 14:1474267. doi: 10.3389/fonc.2024.1474267
Received: 01 August 2024; Accepted: 14 October 2024;
Published: 04 November 2024.
Edited by:
Hua Tan, National Human Genome Research Institute (NIH), United StatesReviewed by:
Hengrui Liu, University of Cambridge, United KingdomIwona Sidorkiewicz, Medical University of Bialystok, Poland
Copyright © 2024 Sabi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Essa M. Sabi, ZXNhYmlAa3N1LmVkdS5zYQ==
 Essa M. Sabi
Essa M. Sabi