- Department of Hematology, the Quzhou Affiliated Hospital of Wenzhou Medical University, Quzhou People’s Hospital, Quzhou, China
Background: Recent studies have reported an association between Cytotoxic T-lymphocyte antigen-4 (CTLA-4) polymorphisms and hematologic malignancy susceptibility, while the results remain inconsistent. Hence, we performed a meta-analysis to investigate the association between CTLA-4 polymorphisms with hematologic malignancy susceptibility.
Methods: A comprehensive and systematic search of Cochrane Library, PubMed, Embase databases was performed up to Sep. 20, 2024. The pooled odds ratio (OR) and its 95% confidence interval (CI) were used to determine the strength of the association between CTLA-4 polymorphisms and hematologic malignancy susceptibility. Statistical analysis was performed in STATA 12.0.
Results: A total of 13 studies concerning the CTLA-4 49A/G, CTLA-4 60A/G, CTLA-4 318T/C, CTLA-4 1661A/G, and CTLA-4 319C/T polymorphisms were included in the meta-analysis. The pooled results suggested the CTLA-4 49A/G polymorphism was significantly associated with an increased hematologic malignancy risk (AA vs. GA+GG: OR = 1.77, 95% CI = 1.56-2.02), especially in NHL, multiple myeloma, and leukemia. Similarly, CTLA-4 319C/T polymorphism was found to be associated with decreased chronic lymphocytic leukemia risk. There was no significant association between the CTLA-4 60A/G, 318T/C, and 1661A/G polymorphism and hematologic malignancy risk.
Conclusion: CTLA-4 49A/G and 319C/T polymorphisms were associated with hematologic malignancy susceptibility.
Introduction
Hematologic malignancy originates from bone marrow and lymph nodes, and the heterogeneity of this disease drives the dysregulation of hematopoietic stem cells, leading to blockage and malignant proliferation of the hematopoietic differentiation system (1, 2). Malignant hematopoietic tumors are one of the most common cancers worldwide, and the incidence of these tumors has increased in recent years. According to global cancer statistics, approximately 176,200 people were diagnosed with hematologic malignancy in 2019 and more than 57,000 died of it (3, 4). Although the specific mechanisms of malignant hematopoietic tumors are not yet fully understood, increasing evidence suggests that genetic variations also play a role in influencing the risk of the disease, indicating that genetic factors play an important role in the development of hematologic malignancy.
The cytotoxic T-lymphocyte antigen-4 (CTLA-4) gene, encoding a transmembrane type 1 T-cell inhibitory receptor, plays a critical role as an immune checkpoint (5, 6). CTLA-4 is an important inhibitory factor for T-cell proliferation and activation by inducing Fas-independent cell apoptosis in activated T cells, which is crucial for regulating antitumor immune responses (7). Studies have shown that CTLA-4 inhibitors can block the binding of CTLA-4 to B7, inhibit the generation of T-cell inhibitory signals, and enhance specific antitumor immune responses (8). However, the CTLA-4 rs231775 genetic mutation can also affect the ability of CTLA-4 to bind to B7, thereby affecting T-cell activation (9–11). These results suggest that CTLA-4 genetic polymorphisms may be associated with the occurrence of cancer. The CTLA-4 gene is located on chromosome 2q33 and consists of 4 exons, which have multiple important single nucleotide polymorphisms (12). Recent studies have explored the relationship between CTLA-4 gene polymorphisms and susceptibility to hematologic malignancy (13–16). However, the results of different studies are contradictory. Considering the importance of CTLA-4 in tumor development, the purpose of this meta-analysis was to comprehensively analyze published studies to further clarify the relationship between CTLA-4 gene polymorphisms and hematologic malignancy.
Methods
Literature search
The study was performed according to the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-analyses statement (17). A comprehensive and systematic search of Cochrane Library, PubMed, Embase databases was performed up to Sep. 20, 2024. The following keywords we utilized: cytotoxic t-lymphocyte antigen 4, CTLA-4, polymorphism, variant, variation, mutation, SNP, leukemia, lymphoma, myeloma, and hematologic malignancy. There are no language restrictions on the search for articles. The references of the relevant potential articles on this topic were also manually searched to search for potentially relevant publications.
Inclusion and exclusion criteria
Studies were included when met the following criteria: (1) case-control or cohort design in human subjects; (2) the associations between CTLA-4 gene polymorphisms and hematologic malignancy susceptibility; (3) completed data to calculate the odds ratio (OR) with the 95% confidence interval (95% CI); (4) control subjects satisfied Hardy–Weinberg equilibrium (HWE). The major exclusion criteria were: (1) duplicate publication; (2) non-human trials; and (3) lack of the full text or main genotyping data.
Data extraction and quality assessment
Two investigators conducted literature screening, data extraction, and quality evaluation independently, and any discrepancies were resolved through consensus. The data extracted from the eligible articles included the first author, publication year, country, ethnicity, sample size, number of cases and controls for each genotype, P value of HWE, and score of quality assessment. The Newcastle-Ottawa scale (NOS) was adopted to assess the quality of included studies based on queue selection, comparability of queues, and evaluation of results.
Statistical analysis
The statistical analysis was conducted using STATA 12.0 software. We used the χ² test to assess the HWE for each study. Pooled ORs and 95% CI were utilized to assess the association between the CTLA-4 polymorphisms and hematologic malignancy susceptibility in the dominant, recessive, homozygous, heterozygous, and allelic models. We assessed the heterogeneity between studies using the Cochrane Q-statistic test, and the unreliability was quantified using the I2 statistic. When there was significant heterogeneity (I2 > 50 or P < 0.05), the random-effects model was used, and otherwise, the fixed-effects model was used. Subgroup meta-analysis was performed by ethnicity and the cancer type. Sensitivity analysis was conducted to assess the stability of the results by omitting 1 study each time to exclude studies. The Begg funnel plots and Egger tests were used to assess the existence of publication bias. A 2-tailed P <.05 was considered significantly significant.
Results
Process of study selection and study characteristics
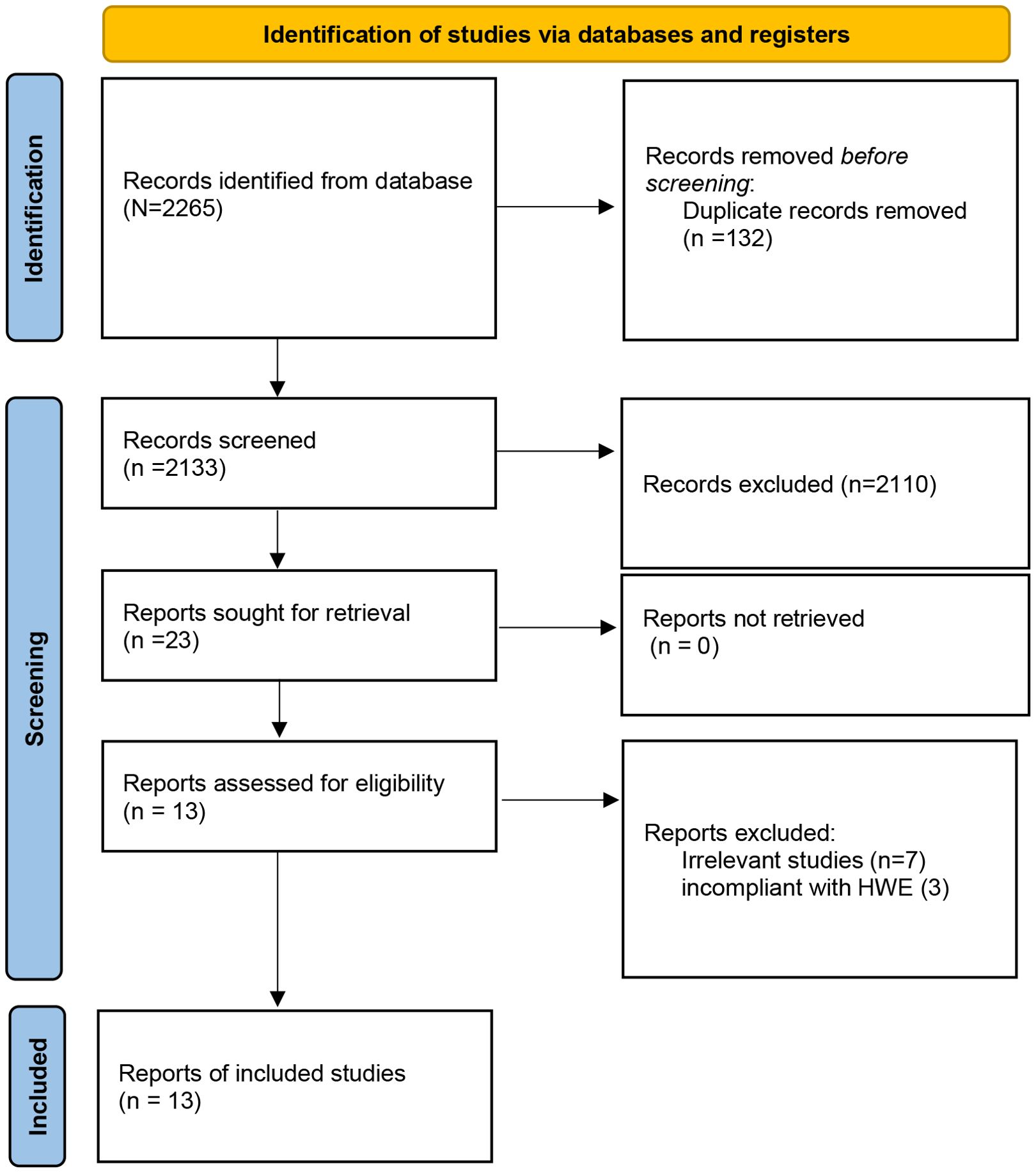
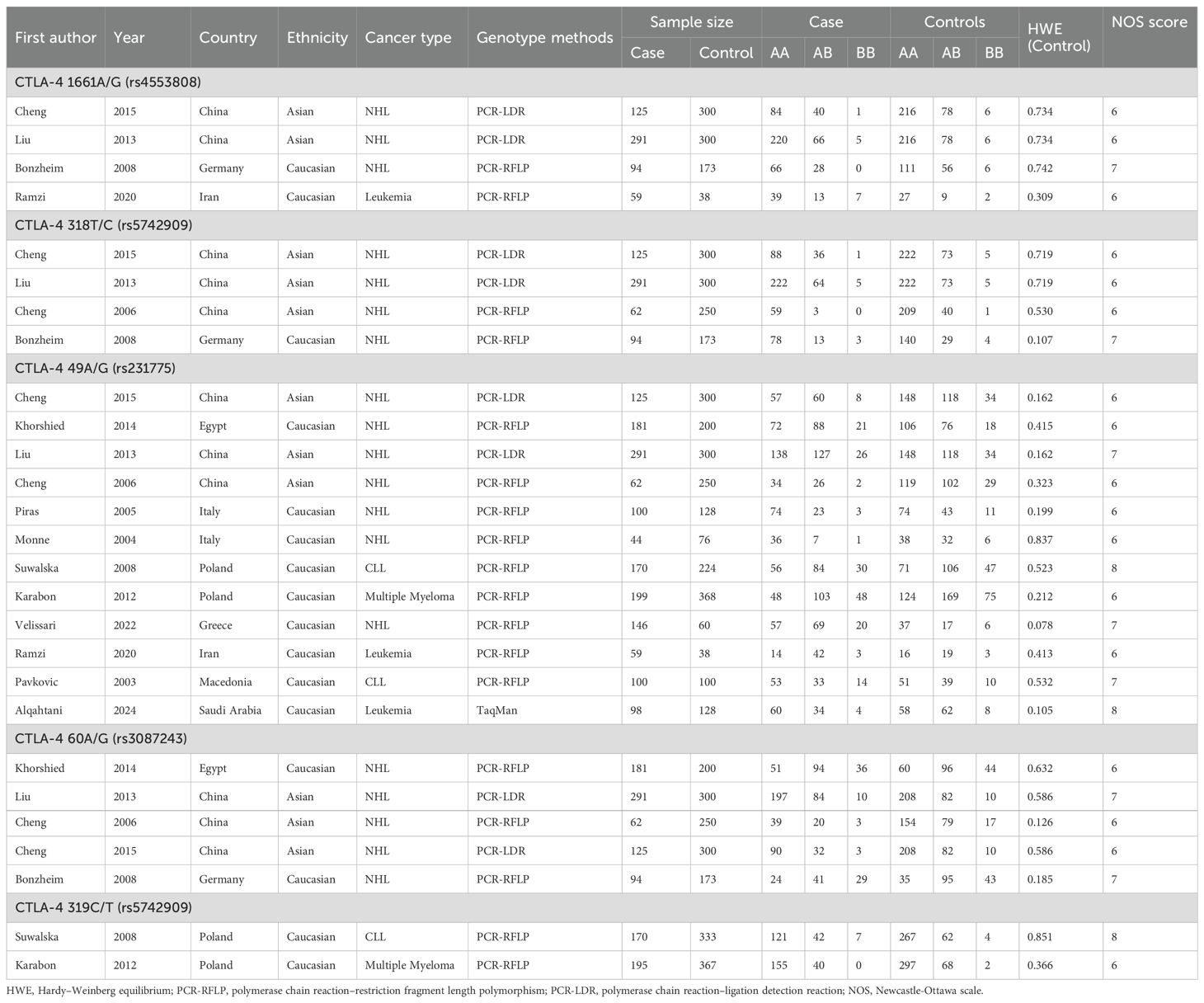
Figure 1 shows a study selection process. Through searching the Embase, PubMed, and Cochrane Library databases, 2265 studies were yielded from the database searches. After eliminating 132 duplicate articles, 2110 publications were further removed by screening titles and abstracts. After reading those articles, 13 articles were identified for this meta-analysis (13–16, 18–26), which included 12 studies for CTLA-4 49A/G (rs231775; located in exon 1 region), 5 studies for CTLA-4 60A/G (rs3087243; located in 3’-UTR region), 4 studies for CTLA-4 318T/C (rs5742909; located in promoter region), 4 studies for CTLA-4 1661A/G (rs4553808; located in promoter region), and 2 studies for CTLA-4 319C/T (rs5742909; located in promoter region) polymorphisms. The articles were published from 2004 to 2024, and the sample size ranged from 105 to 591. Among the included studies, 3 studies were from China, 2 studies from Italy, 2 studies from Poland, 2 studies from Egypt, and one study from Iran, Greece, Germany, Saudi Arabia, and Macedonia. The NOS score of all articles ranged from 6 to 8, implying that all included studies were of high quality. The selected study characteristics were summarized in Table 1.
Correlation between CTLA-4 49A/G polymorphism and hematologic malignancy risk
Twelve relevant studies with 1575 cancer patients and 2172 controls were examined for the association between the CTLA-4 49A/G polymorphism and hematologic malignancy risk. The main results of the meta-analysis about CTLA-4 49A/G polymorphism were shown in Table 2. The pooled results indicated that CTLA-4 49A/G polymorphism was associated with hematologic malignancy risk under dominant model (AA vs. GA+GG: OR = 1.77, 95% CI = 1.56-2.02, P = 0.001) (Figure 2). There was no significant association with hematologic malignancy risk under the other four genetic models. However, in the stratification analysis by ethnicity, we observed that the Asian population was significantly related to an increased hematologic malignancy risk in the recessive model (GG vs. GA+AA: OR = 1.67, 95% CI = 1.10-2.53, P = 0.017), and dominant model (AA vs. GA+GG: OR = 1.85, 95% CI = 1.49-2.30, P = 0.001). The Caucasian population was significantly related to an increased hematologic malignancy risk in dominant model (AA vs. GA+GG: OR = 1.73, 95% CI = 1.47-2.04, P = 0.001). We then performed the subgroup analyses stratified by cancer types. The pooled ORs suggested that the CTLA-4 49A/G polymorphism was significantly associated with NHL risk (AA vs. GA+GG: OR = 1.98, 95% CI = 1.68-2.32, P = 0.001), multiple myeloma risk (AA vs. GA+GG: OR = 1.60, 95% CI = 1.08-2.36, P = 0.018; GG vs. AA: OR = 0.60, 95% CI = 0.37-0.99, P = 0.045; GA vs. AA: OR = 0.64, 95% CI = 0.42-0.96, P = 0.031; G vs. A: OR = 0.76, 95% CI = 0.60-0.98, P = 0.032), and leukemia risk (AA vs. GA+GG: OR = 2.91, 95% CI = 1.22-6.95, P = 0.016).
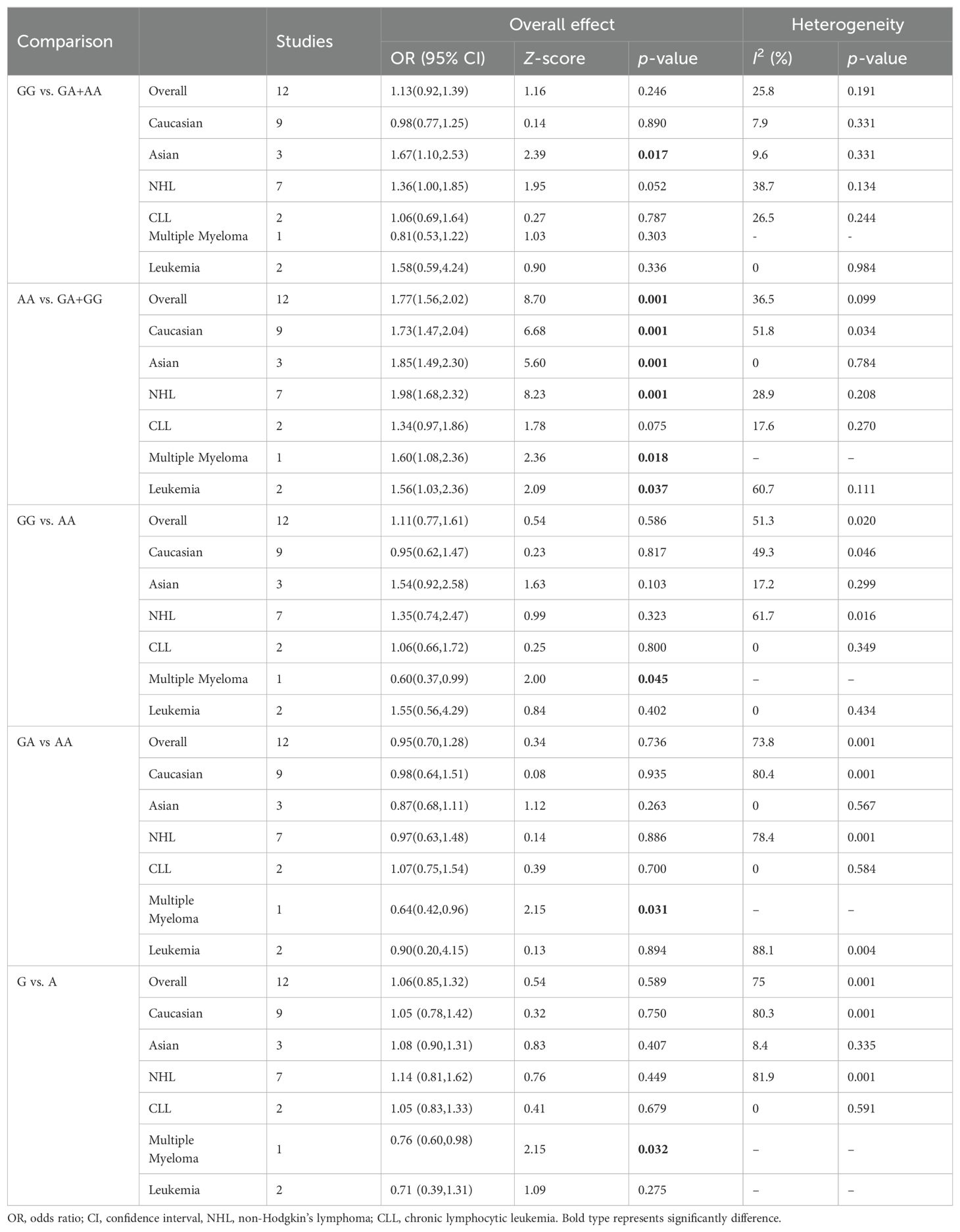
Table 2. Summary of meta-analysis of association of CTLA-4 49A/G (rs231775) polymorphism and hematologic malignancy risk.
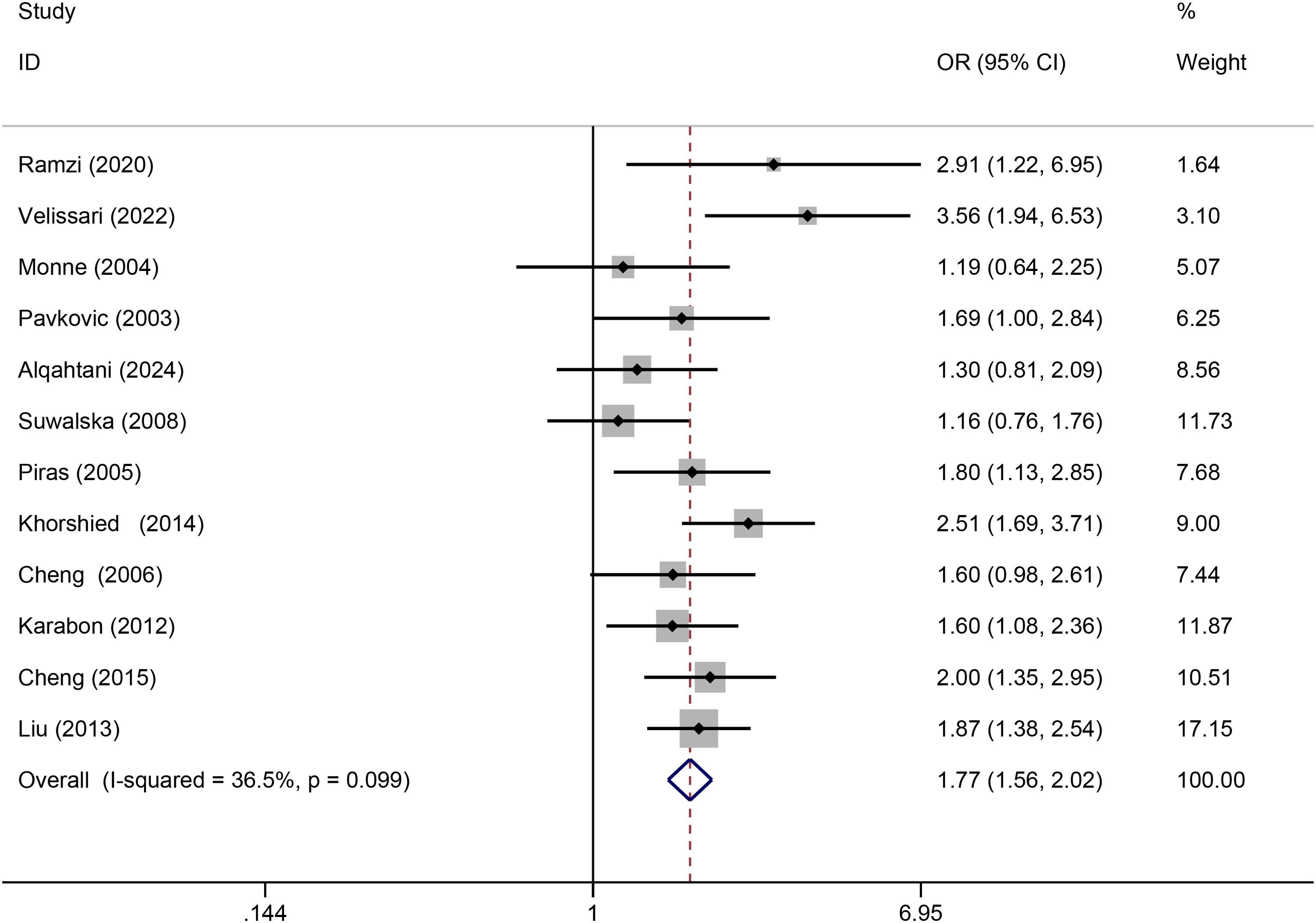
Figure 2. Forrest plot for association between CTLA-4 49A/G polymorphism and hematologic malignancy risk (AA vs. GA+GG).
Correlation between CTLA-4 60A/G polymorphism and hematologic malignancy risk
Five relevant studies with 753 cancer patients and 1223 controls were examined for the association between the CTLA-4 60A/G polymorphism and hematologic malignancy risk. The overall analyses of 5 genetic models did not have any significant correlation between the CTLA-4 60A/G polymorphism and hematologic malignancy risk. As shown in Table 3, no association between CTLA-4 60A/G polymorphism and hematologic malignancy risk was detected in the ethnicity and cancer types subgroup analyses.
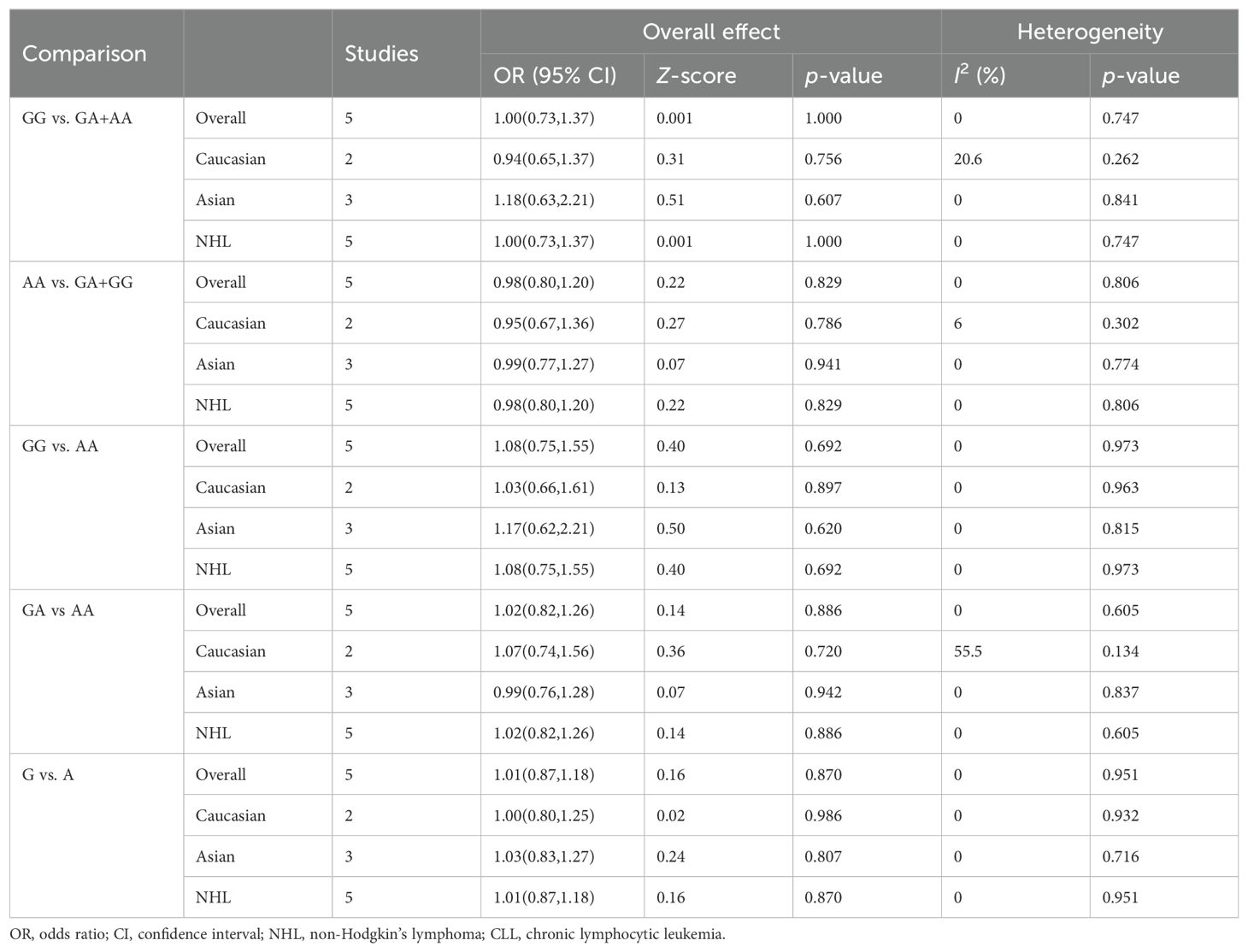
Table 3. Summary of meta-analysis of association of CTLA-4 60A/G (rs3087243) polymorphism and hematologic malignancy risk.
Correlation between CTLA-4 318T/C polymorphism and hematologic malignancy risk
Four relevant studies with 572 cancer patients and 1023 controls were examined for the association between the CTLA-4 318T/C polymorphism and hematologic malignancy risk. The pooled results indicated that CTLA-4 318T/C was not significantly related to hematologic malignancy risk in all models. Then in the stratification analysis by ethnicity and cancer types, no significant association between CTLA-4 318T/C polymorphism and hematologic malignancy risk was discovered (Table 4).
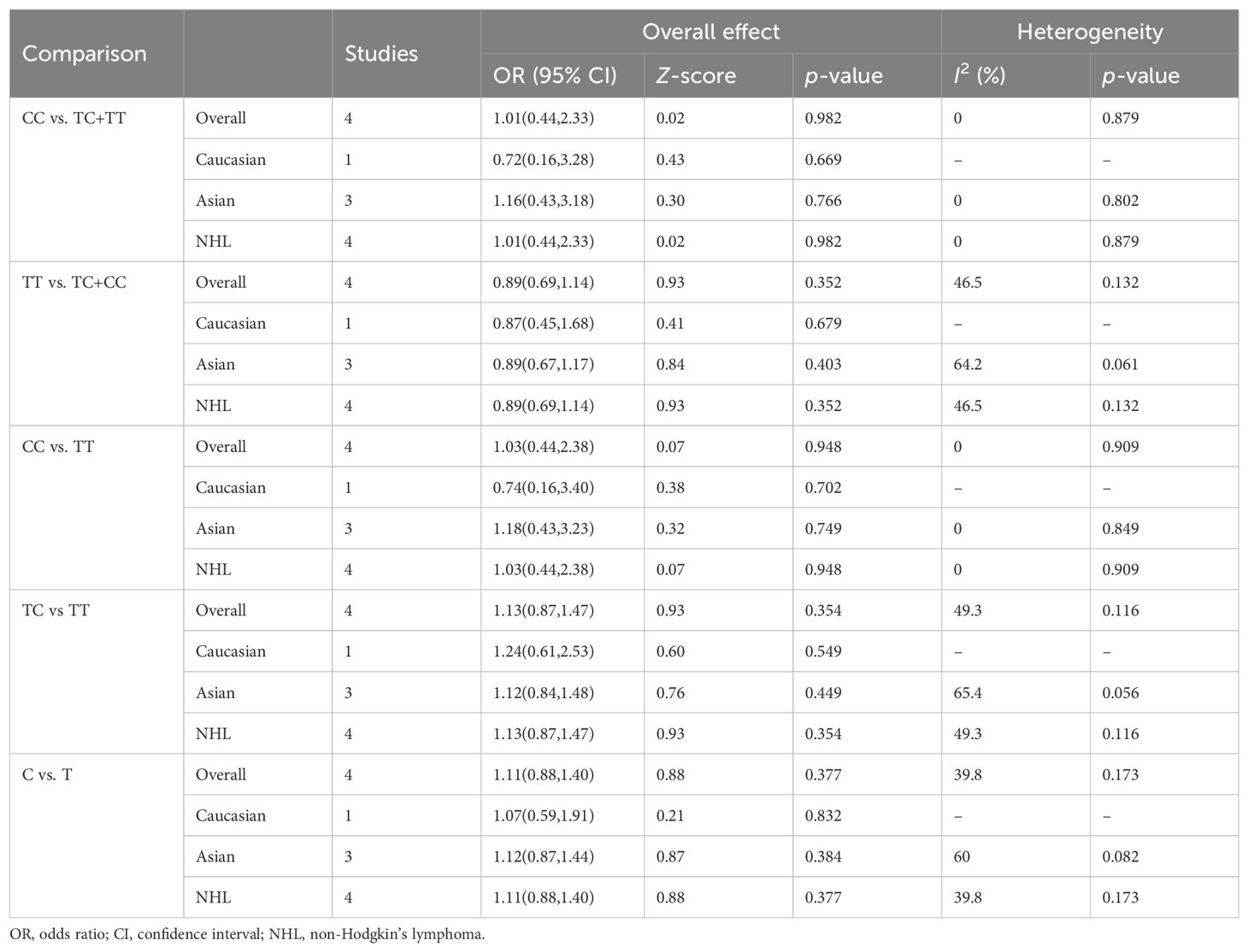
Table 4. Summary of meta-analysis of association of CTLA-4 318T/C (rs5742909) polymorphism and hematologic malignancy risk.
Correlation between CTLA-4 1661A/G polymorphism and hematologic malignancy risk
Four relevant studies with 569 cancer patients and 811 controls were examined for the association between the CTLA-4 1661A/G polymorphism and hematologic malignancy risk. The pooled results indicated that no significant association between CTLA-4 1661A/G polymorphism and hematologic malignancy risk was found in overall and subgroup analyses. Table 5 summarizes the evaluation results of the association between CTLA-4 1661A/G polymorphism and hematologic malignancy risk.
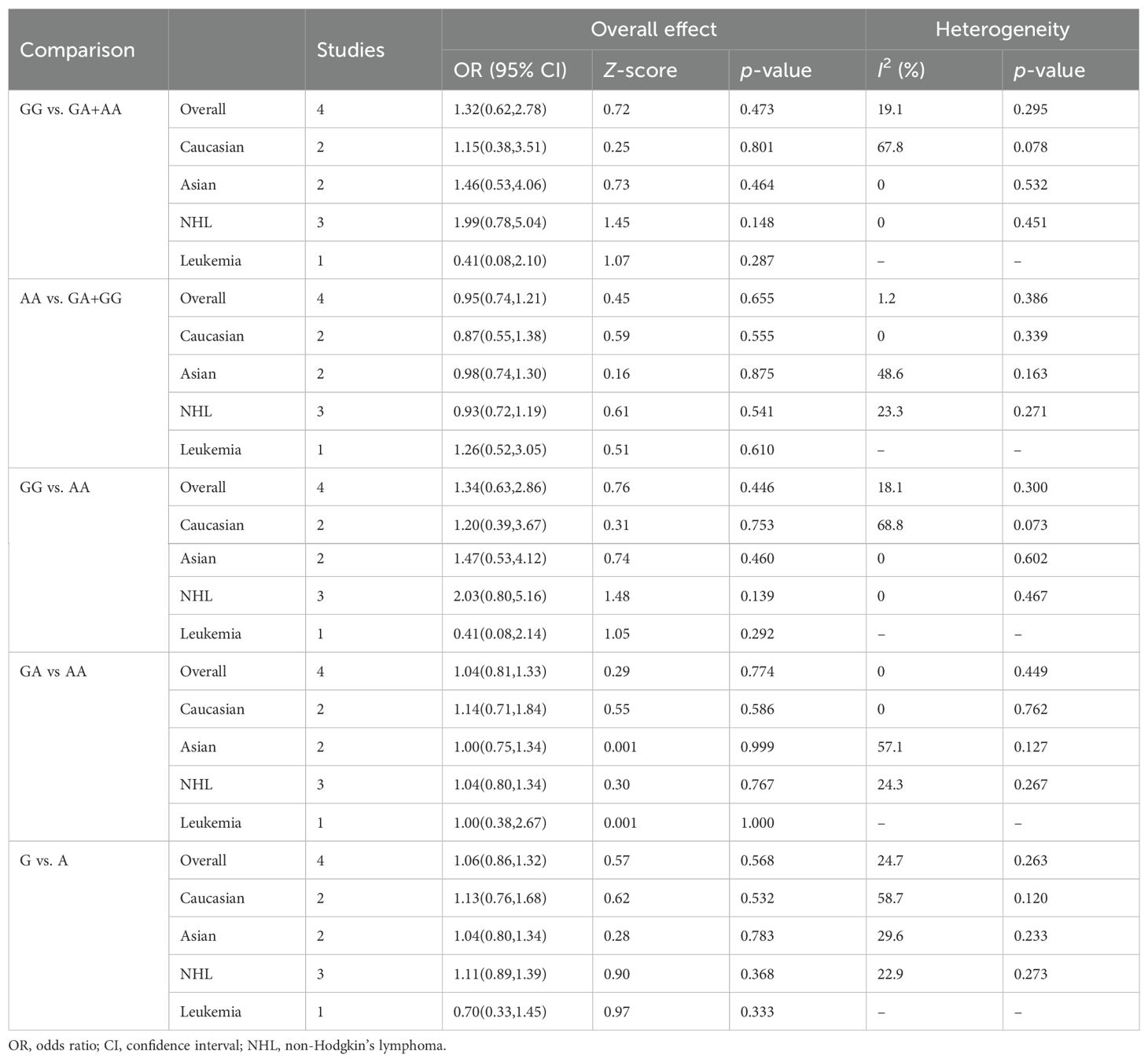
Table 5. Summary of meta-analysis of association of CTLA-4 1661A/G (rs4553808) polymorphism and hematologic malignancy risk.
Correlation between CTLA-4 319C/T polymorphism and hematologic malignancy risk
Two relevant studies with 365 cancer patients and 700 controls were examined for the association between the CTLA-4 319C/T polymorphism and hematologic malignancy risk. Overall, CTLA-4 319C/T was not associated with hematologic malignancy susceptibility in all genetic models (Table 6). When subgroup analysis was conducted according to ethnicity, no significant association between CTLA-4 319 C/T polymorphism and hematologic malignancy risk was discovered. Using cancer types subgroup analyses, we observed that CTLA-4 319C/T polymorphism was significantly associated with the CLL risk in the recessive model (TT vs. TC+CC: OR: 0.28, 95%CI:0.08-0.98, P = 0.047), dominant model (CC vs. TC+TT: OR: 1.64, 95%CI:1.07-2.51, P = 0.024), homozygous model (TT vs. CC: OR: 0.26, 95%CI:0.07-0.90, P = 0.034), and allelic model (T vs C: OR: 0.60, 95%CI:0.42-0.87, P = 0.007).
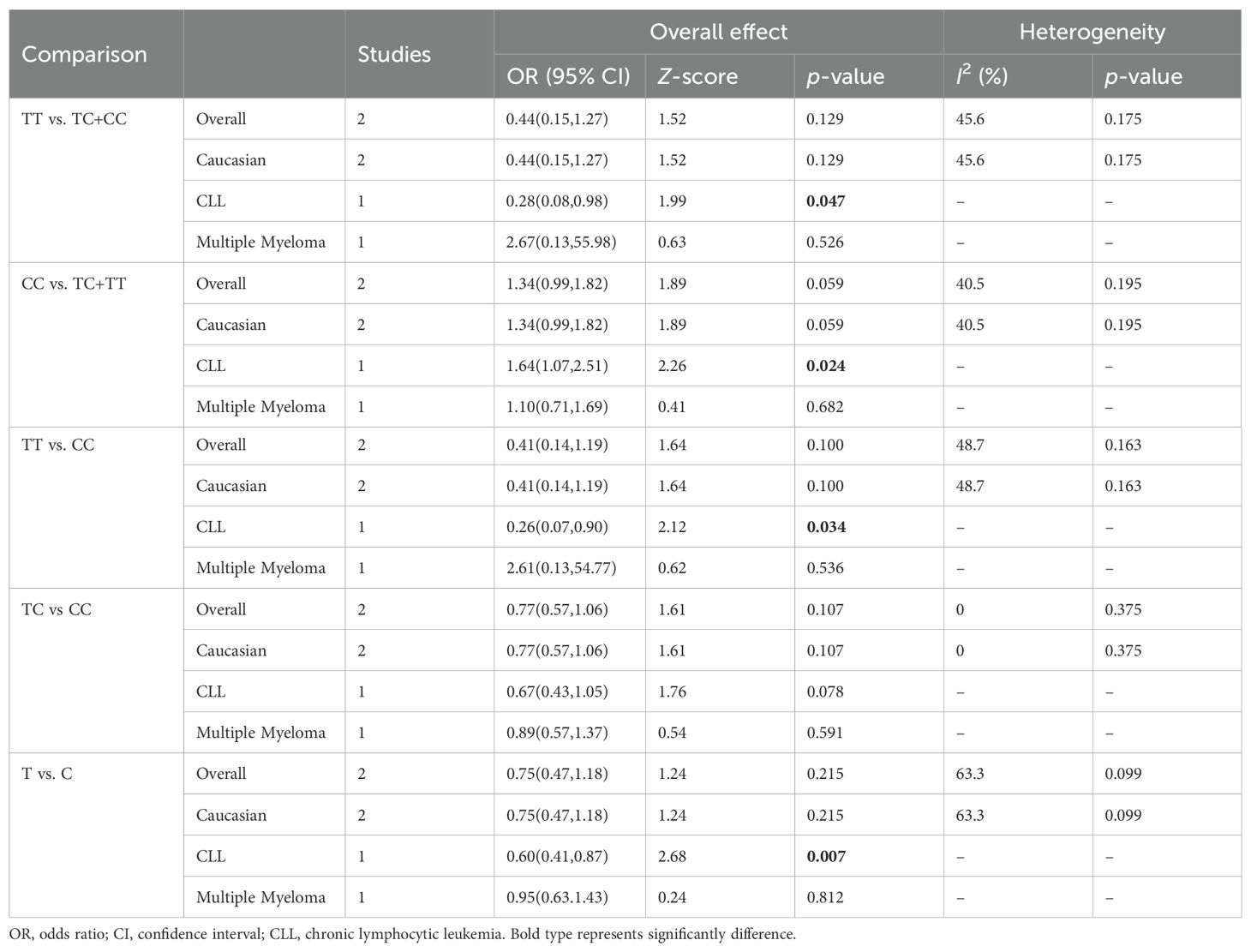
Table 6. Summary of meta-analysis of association of CTLA-4 319C/T (rs5742909) polymorphism and hematologic malignancy risk.
Sensitivity and publication bias tests
Significant heterogeneity was observed in some comparison models; thus, sensitivity analysis was performed to evaluate the influence of each separate study, and the result indicated the omission of any single study did not significantly change the direction of estimates (Figure 3). In addition, there was no publication bias was detected according to Begg rank correlation test (P = 0.312) and Egger linear regression test (P = 0.218) (Figure 4).
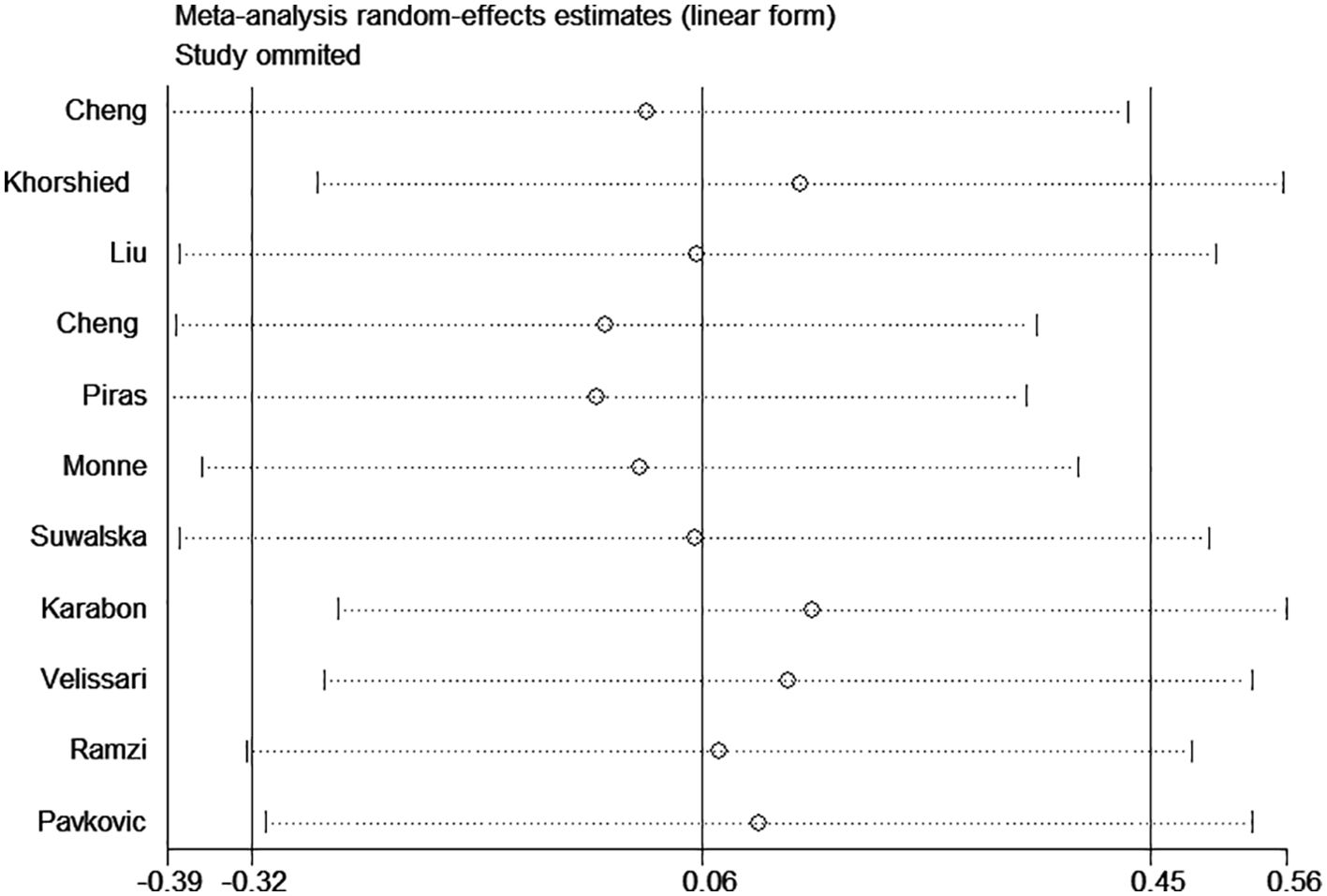
Figure 3. Sensitivity analysis for association between CTLA-4 49A/G polymorphism and hematologic malignancy risk (GG vs. AA).
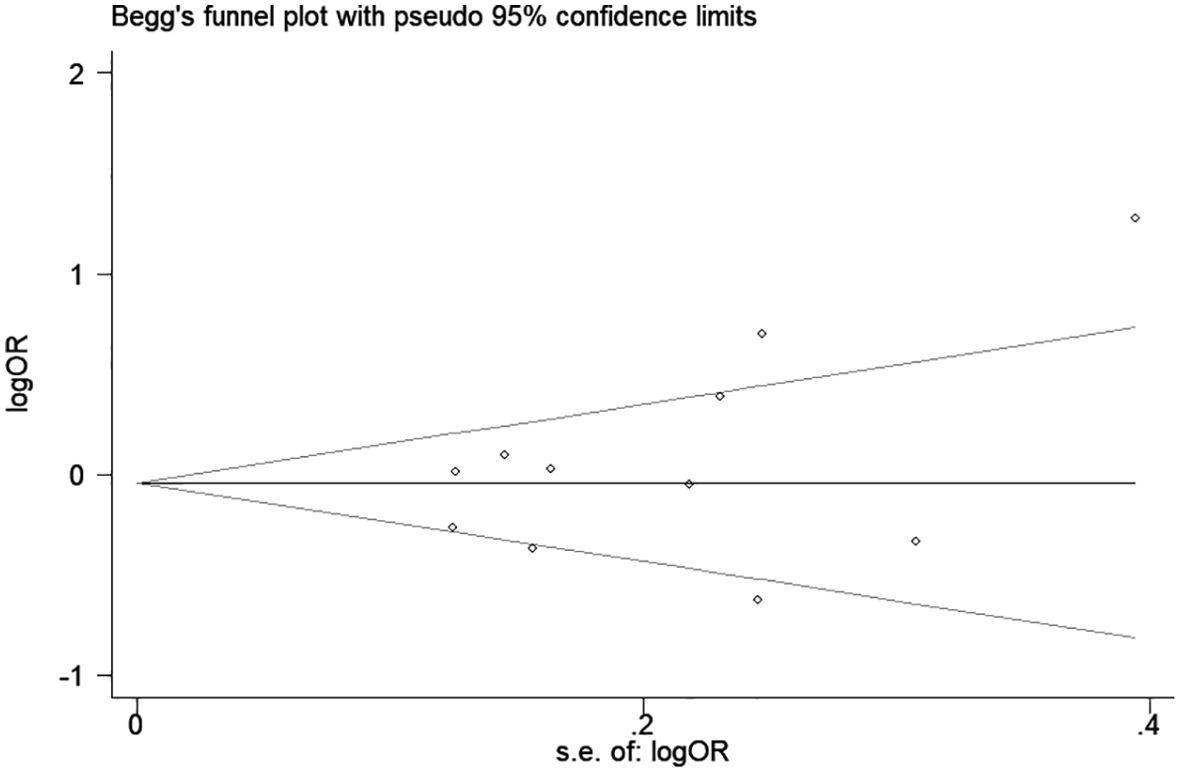
Figure 4. Begg’s funnel plot for association between CTLA-4 49A/G polymorphism and hematologic malignancy risk (G vs. A).
Discussion
CTLA-4 is expressed on activated T cells and negatively regulates their activation and proliferation (27). Previous studies have shown that CTLA-4 regulates the duration and intensity of T-cell-mediated immune responses by competitively binding to the costimulatory B7 molecule and activating FAS-dependent cell apoptosis in T cells (27). Recently, abnormal expression of the CTLA-4 gene has been documented in many types of cancers, suggesting that it may contribute to the development and progression of cancer (10, 21, 24). Considering that genetic polymorphisms may affect gene expression and even protein function, the relationship between CTLA-4 gene polymorphisms and different types of malignant diseases has been widely explored. In this meta-analysis, we summarized the results of 13 relevant studies and highlighted the potential relationship between CTLA-4 gene polymorphisms and hematologic malignancies. Our meta-analysis suggested that the CTLA-4 49A/G polymorphism is associated with a decreased NHL, multiple myeloma, and leukemia risk, while the CTLA-4 319C/T polymorphism is significantly associated with a decreased CLL risk.
The difference between lymphoid tumors and other tumors is that this malignant tumor originates from the immune system itself. Most lymphoid tumors originate from mature B and T lymphocytes (28). Therefore, the role of the immune pathway in lymphadenopathy is very complex. As a typical immune regulatory checkpoint, CTLA-4 plays an important role in T-cell anergy and T and B-cell inhibitory responses (27). Therefore, abnormal expression of CTLA-4 may play a role in the pathogenesis of malignant lymphoid tumors. In addition, SNPs in the CTLA-4 gene are widely believed to alter the activity of the promoter and regulate the expression level of CTLA-4. The CTLA4 gene is located on chromosome 2q33 and can exert its negative regulatory effect on T-cell proliferation and activation by competing with CD28 for binding to B7-1/B7-2, downregulating T-cell responses, and peripheral tolerance (29). When CTLA4 is overexpressed, the inhibitory signals produced exceed the immunogenicity provided by tumor cell surface antigens, downregulating or terminating T-cell activation, leading to immune escape of the tumor (30). The abnormal expression of the CTLA-4 gene has been documented in many types of cancers, and it may contribute to the occurrence and progression of cancer (8, 31). Considering that genetic polymorphisms may affect gene expression and even protein function, the relationship between CTLA-4 gene polymorphisms and different types of malignant diseases has been widely explored.
Currently, the pathogenesis of lymphoma is not fully understood, and its occurrence and development involve a multifactorial and multistep process (32). Identifying high-risk populations, clarifying pathogenesis, and increasing treatment options are the most urgent issues to address. Epidemiological studies have shown that the onset of lymphoma is closely related to certain factors. A family history of lymphoma, age, region, race, diet and lifestyle habits, and viral and bacterial infections may all be risk factors for lymphoma (33). Reports also suggest that factors such as tall stature, early immune function, nutritional status, and growth hormone levels play a role in the onset of lymphoma (34). Changes in immune function increase the risk of lymphoma, but the relationship between CTLA4 polymorphisms and susceptibility to hematologic malignancies is inconsistent (21–24). The reasons for these differences are diverse; for example, the origin of the tumor can affect the results of meta-analyses, so we conducted subgroup analyses of CTLA4 polymorphisms by cancer type. The results indicate that the +49A/G polymorphism is associated with an increased risk of NHL, multiple myeloma, and leukemia but not with CLL. In addition, our study results suggest that the 60A/G, 318T/C, and 1661A/G polymorphisms are not associated with hematologic malignancies. However, these results should be interpreted with caution. Due to the inclusion of only 1-2 case-control studies for certain cancer types, the ability to reveal reliable associations may be limited; therefore, further research is needed to validate these associations in the future. In the current meta-analysis, the impact on different racial groups was also analyzed. The +49A/G polymorphism was associated with an increased risk of cancer in Asians but was associated with an decreased cancer risk in Caucasians, while the CTLA-4 319C/T polymorphism was associated with a notable decreased cancer risk in Caucasians. These results strongly suggest that genetic diversity and interactions between genetic variations among different racial groups may lead to different cancer risks. Both racial and environmental factors influence the risk of cancer in different populations. In the future, more research should be conducted to analyze these associations, particularly gene-environment and gene-gene interactions.
Meta-analysis can overcome some of the issues caused by single studies, such as small sample sizes, selection bias, and low-test power; therefore, it is considered a powerful tool for integrating conflicting results from different studies. However, limitations should be noted in our meta-analysis. First, in some subgroups, the number of cases and controls was relatively small, which may have limited the statistical power. Second, significant heterogeneity was observed in some comparisons, which may stem from differences in the histopathological classification of hematologic malignancies and variations in gene detection methods. Third, due to the lack of uniform gene-environment interaction data in the included studies, we were unable to further stratify the data by other factors (such as age, sex, alcohol consumption, smoking status, diet, and other lifestyle factors). Fourth, no available data to examine the association between CTLA-4 polymorphisms and clinical characteristics or comorbidities.
In conclusion, this meta-analysis suggests a significant association between CTLA-4 49A/G and CTLA-4 319C/T with hematologic malignancy risk. However, CTLA-4 60A/G, 318T/C, and 1661A/G polymorphisms showed no significant association with susceptibility to hematologic malignancies. Therefore, further studies with larger sample sizes, different ethnicities, and various types of cancer are needed to confirm these findings.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
XY: Conceptualization, Data curation, Formal analysis, Methodology, Software, Writing – original draft, Writing – review & editing. NZ: Data curation, Formal analysis, Methodology, Software, Writing – review & editing. GW: Data curation, Formal analysis, Methodology, Software, Writing – review & editing. JW: Conceptualization, Formal analysis, Investigation, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Furutani E, Shimamura A. Germline genetic predisposition to hematologic Malignancy. J Clin Oncol. (2017) 35:1018–28. doi: 10.1200/JCO.2016.70.8644
2. Ferreyro BL, Scales DC, Wunsch H, Cheung MC, Gupta V, Saskin R, et al. Critical illness in patients with hematologic Malignancy: a population-based cohort study. Intensive Care Med. (2021) 47:1104–14. doi: 10.1007/s00134-021-06502-2
3. Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Abate D, Abbasi N, Abbastabar H, Abd-Allah F, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups 1990 to 2017: A systematic analysis for the global burden of disease study. JAMA Oncol. (2019) 5:1749–68. doi: 10.1001/jamaoncol.2019.2996
4. Gbd Lip O, Pharyngeal Cancer C, Cunha ARD, Compton K, Xu R, Mishra R, et al. The global, regional, and national burden of adult lip, oral, and pharyngeal cancer in 204 countries and territories: A systematic analysis for the global burden of disease study 2019. JAMA Oncol. (2023) 9:1401–16. doi: 10.1001/jamaoncol.2023.2960
5. Fong L, Small EJ. Anti-cytotoxic T-lymphocyte antigen-4 antibody: the first in an emerging class of immunomodulatory antibodies for cancer treatment. J Clin Oncol. (2008) 26:5275–83. doi: 10.1200/JCO.2008.17.8954
6. Gaynor N, Crown J, Collins DM. Immune checkpoint inhibitors: Key trials and an emerging role in breast cancer. Semin Cancer Biol. (2022) 79:44–57. doi: 10.1016/j.semcancer.2020.06.016
7. Xiao W, Du N, Huang T, Guo J, Mo X, Yuan T, et al. TP53 mutation as potential negative predictor for response of anti-CTLA-4 therapy in metastatic melanoma. EBioMedicine. (2018) 32:119–24. doi: 10.1016/j.ebiom.2018.05.019
8. Podlesnykh SV, Abramova KE, Gordeeva A, Khlebnikov AI, Chapoval AI. Peptide blocking CTLA-4 and B7-1 interaction. Molecules. (2021) 26:253. doi: 10.3390/molecules26020253
9. Kim HJ, Jeong KH, Lee SH, Moon JY, Lee TW, Kang SW, et al. Polymorphisms of the CTLA4 gene and kidney transplant rejection in Korean patients. Transpl Immunol. (2010) 24:40–4. doi: 10.1016/j.trim.2010.10.001
10. Charbonneau B, Moysich KB, Kalli KR, Oberg AL, Vierkant RA, Fogarty ZC, et al. Large-scale evaluation of common variation in regulatory T cell-related genes and ovarian cancer outcome. Cancer Immunol Res. (2014) 2:332–40. doi: 10.1158/2326-6066.CIR-13-0136
11. Pan H, Shi Z, Gao L, Zhang L, Wei S, Chen Y, et al. Impact of the cytotoxic T-lymphocyte associated antigen-4 rs231775 A/G polymorphism on cancer risk. Heliyon. (2023) 9:e23164. doi: 10.1016/j.heliyon.2023.e23164
12. Sun L, Niu T, Zhang Y. Association between thyroid cancer and CTLA-4 gene polymorphisms. Cell Mol Biol (Noisy-le-grand). (2023) 69:31–6. doi: 10.14715/cmb/2023.69.4.5
13. Monne M, Piras G, Palmas A, Arru L, Murineddu M, Latte G, et al. Cytotoxic T-lymphocyte antigen-4 (CTLA-4) gene polymorphism and susceptibility to non-Hodgkin's lymphoma. Am J Hematol. (2004) 76:14–8. doi: 10.1002/ajh.20045
14. Piras G, Monne M, Uras A, Palmas A, Murineddu M, Arru L, et al. Genetic analysis of the 2q33 region containing CD28-CTLA4-ICOS genes: association with non-Hodgkin's lymphoma. Br J Haematol. (2005) 129:784–90. doi: 10.1111/j.1365-2141.2005.05525.x
15. Cheng TY, Lin JT, Chen LT, Shun CT, Wang HP, Lin MT, et al. Association of T-cell regulatory gene polymorphisms with susceptibility to gastric mucosa-associated lymphoid tissue lymphoma. J Clin Oncol. (2006) 24:3483–9. doi: 10.1200/JCO.2005.05.5434
16. Bonzheim I, Geissinger E, Chuang WY, Roth S, Strobel P, Marx A, et al. Analysis of single nucleotide polymorphisms in the FAS and CTLA-4 genes of peripheral T-cell lymphomas. J Hematop. (2008) 1:11–21. doi: 10.1007/s12308-008-0003-y
17. Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. (2021) 372:n71. doi: 10.1136/bmj.n71
18. Pavkovic M, Georgievski B, Cevreska L, Spiroski M, Efremov DG. CTLA-4 exon 1 polymorphism in patients with autoimmune blood disorders. Am J Hematol. (2003) 72:147–9. doi: 10.1002/ajh.10278
19. Suwalska K, Pawlak E, Karabon L, Tomkiewicz A, Dobosz T, Urbaniak-Kujda D, et al. Association studies of CTLA-4, CD28, and ICOS gene polymorphisms with B-cell chronic lymphocytic leukemia in the Polish population. Hum Immunol. (2008) 69:193–201. doi: 10.1016/j.humimm.2008.01.014
20. Karabon L, Pawlak-Adamska E, Tomkiewicz A, Jedynak A, Kielbinski M, Woszczyk D, et al. Variations in suppressor molecule ctla-4 gene are related to susceptibility to multiple myeloma in a polish population. Pathol Oncol Res. (2012) 18:219–26. doi: 10.1007/s12253-011-9431-6
21. Liu J, Liu J, Song B, Wang T, Liu Y, Hao J, et al. Genetic variations in CTLA-4, TNF-alpha, and LTA and susceptibility to T-cell lymphoma in a Chinese population. Cancer Epidemiol. (2013) 37:930–4. doi: 10.1016/j.canep.2013.08.011
22. Khorshied MM, Gouda HM, Khorshid OM. Association of cytotoxic T-lymphocyte antigen 4 genetic polymorphism, hepatitis C viral infection and B-cell non-Hodgkin lymphoma: an Egyptian study. Leuk Lymphoma. (2014) 55:1061–6. doi: 10.3109/10428194.2013.820294
23. Cheng S, Li J, Liu W, Liu C, Su L, Liu X, et al. LTA + 252A > G polymorphism is associated with risk of nasal NK/T-cell lymphoma in a Chinese population: a case-control study. BMC Cancer. (2015) 15:480. doi: 10.1186/s12885-015-1506-4
24. Ramzi M, Arandi N, Saadi MI, Yaghobi R, Geramizadeh B. Genetic variation of costimulatory molecules, including cytotoxic T-lymphocyte antigen 4, inducible T-cell costimulator, cluster differentiation 28, and programmed cell death 1 genes, in Iranian patients with leukemia. Exp Clin Transplant. (2020) 18:719–24. doi: 10.6002/ect.2017.0176
25. Velissari A, Vassilakopoulos TP, Angelopoulou MK, Korkolopoulou P, Bamias G, Daikos G, et al. Genetic polymorphisms and risk of MALT lymphoma in Greek population. Curr Res Transl Med. (2022) 70:103330. doi: 10.1016/j.retram.2021.103330
26. Alqahtani M, Aljuaimlani A, Al-Tamimi J, Alomar S, Mansour L. TIM3 and CTLA4 immune checkpoint polymorphisms are associated with acute myeloid leukemia in Saudi Arabia. Hematology. (2024) 29:2329024. doi: 10.1080/16078454.2024.2329024
27. Walker LSK. CTLA-4 and autoimmunity: new twists in the tale. Trends Immunol. (2015) 36:760–2. doi: 10.1016/j.it.2015.11.002
28. Duran-Ferrer M, Martin-Subero JI. Epigenomic characterization of lymphoid neoplasms. Annu Rev Pathol. (2024) 19:371–96. doi: 10.1146/annurev-pathmechdis-051122-100856
29. Zhang H, Dai Z, Wu W, Wang Z, Zhang N, Zhang L, et al. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J Exp Clin Cancer Res. (2021) 40:184. doi: 10.1186/s13046-021-01987-7
30. Lee J, Kim EH. Mechanisms underlying response and resistance to immune checkpoint blockade in cancer immunotherapy. Front Oncol. (2023) 13:1233376. doi: 10.3389/fonc.2023.1233376
31. Zhang C, Chen J, Song Q, Sun X, Xue M, Yang Z, et al. Comprehensive analysis of CTLA-4 in the tumor immune microenvironment of 33 cancer types. Int Immunopharmacol. (2020) 85:106633. doi: 10.1016/j.intimp.2020.106633
32. Lopez C, Burkhardt B, Chan JKC, Leoncini L, Mbulaiteye SM, Ogwang MD, et al. Burkitt lymphoma. Nat Rev Dis Primers. (2022) 8:78. doi: 10.1038/s41572-022-00404-3
33. Luan Y, Li X, Luan Y, Luo J, Dong Q, Ye S, et al. Therapeutic challenges in peripheral T-cell lymphoma. Mol Cancer. (2024) 23:2. doi: 10.1186/s12943-023-01904-w
Keywords: CTLA-4, hematologic malignancy, polymorphism, leukemia, meta-analysis
Citation: Yan X, Zhang N, Wang G and Wang J (2024) Association of CTLA-4 polymorphisms with hematologic malignancy susceptibility: a meta-analysis. Front. Oncol. 14:1467740. doi: 10.3389/fonc.2024.1467740
Received: 20 July 2024; Accepted: 25 September 2024;
Published: 11 October 2024.
Edited by:
Massimo Breccia, Sapienza University of Rome, ItalyReviewed by:
Lucia Guadalupe Taja Chayeb, National Institute of Cancerology (INCAN), MexicoAllyson Guimarães Costa, Federal University of Amazonas, Brazil
Adolfo Martinez, General Hospital of Mexico, Mexico
Copyright © 2024 Yan, Zhang, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiaheng Wang, MTg2NTcwMTU2NjhAMTYzLmNvbQ==
 Xuefen Yan
Xuefen Yan Jiaheng Wang
Jiaheng Wang