- 1Department of Laboratory Medicine and Genetics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Republic of Korea
- 2Department of Pathology and Translational Genomics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Republic of Korea
- 3Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Republic of Korea
Introduction: Numerous studies have suggested high concordance between tissue and circulating tumor DNA (ctDNA) comprehensive genomic profiling (CGP) tests but only few of them focused on fusions. In addition, atypical breakpoints occasionally detected from DNA-based fusion detection make interpretation difficult, and their clinical significance remains unclear. This study evaluated the clinical utility of ctDNA CGP for fusion detection.
Methods: The results of ctDNA CGP tests performed on patients with stage IV non-small cell lung cancer during routine clinical care were retrospectively reviewed. The concordance between ctDNA CGP and combined tissue test results was analyzed using CGP, immunohistochemistry, fluorescence in situ hybridization, and reverse transcription polymerase chain reaction. The clinical significance of fusions detected by ctDNA CGP, including those with atypical breakpoints at the DNA level, was assessed.
Results: In total, 264 patients were tested with ctDNA CGP. Fusions were detected in 27 patients (10.2%), and the fusion drivers were RET (n=12, 4.6%), ALK (n=9, 3.4%), ROS1 (n=4, 1.5%), and FGFR2 (n=2, 0.8%). The overall prevalence of fusion in tissue CGP was comparable to that in ctDNA CGP. A total of 371 ctDNA-tissue test pairs were available, and the overall positive and negative percent agreement rates were 92.9% (13/14) and 100.0% (357/357), respectively. One ALK IHC-positive and ctDNA CGP-negative case did not respond to ALK-targeted therapy. Response to targeted therapy was assessed in 16 patients, and a partial response was achieved in all patients, including four with atypical breakpoints.
Conclusion: Fusion detection using ctDNA CGP showed high concordance with tissue tests and accuracy in predicting therapeutic responses in patients with non-small cell lung cancer. ctDNA CGP may provide an important diagnostic tool for fusion detection.
1 Introduction
Owing to the various genetic alterations that can guide therapy selection, molecular testing of tumor specimen has become essential in non-small cell lung cancer (NSCLC) treatment (1, 2). Among the different types of genetic alterations, fusions or rearrangements resulting in the activation of kinases such as ALK, ROS1, and RET are preferred treatment targets based on their superior efficacy and tolerability when treated with appropriate target agents (3–7). In addition to the conventional methods for fusion detection such as fluorescence in situ hybridization (FISH) and reverse transcription polymerase chain reaction (RT-PCR), high expression of immunohistochemistry (IHC) has demonstrated high concordance rate with genomic alteration especially in ALK and ROS1 fusion. Recently, next-generation sequencing (NGS)-based comprehensive genomic profiling (CGP) tests have been increasingly adopted in clinical practice, and NGS of DNA and/or RNA has become the diagnostic method of choice owing to its superior accuracy and versatility (5, 8). CGP tests are particularly useful when multiple molecular targets need to be evaluated from a limited amount of specimen by detecting various types of genetic alterations, including fusions and amplifications.
Until recently, the tissue CGP test was considered the gold standard method of diagnosis because of its high accuracy and low false negative rates. However, owing to the clinical limitation in the acquisition of tissue samples and difficulties in repetitive approach due to the high invasiveness especially in lung cancer, plasma circulating tumor DNA (ctDNA) has emerged as an alternative to tissue specimen as input material for CGP (1, 9). In ctDNA CGP, fraction of tumor-derived DNA in circulation (tumor fraction, TF) is a key parameter that affects assay sensitivity (10). To detect variants with low allele fraction, as low as 0.1–0.5%, ctDNA CGP is normally performed with ultra-high sequencing volume targeting higher depth-of-coverage (DOC) than in tissue CGP (11). As a result, given that TF is sufficiently high, variants can be reliably detected using ctDNA CGP. The sensitivity of ctDNA CGP in detecting variants, including fusions, is nearly 100% when TF is above 10% (10). Large-scale retrospective review of ctDNA CGP tests performed as routine clinical testing revealed that the prevalence of fusions involving major drivers such as ALK, ROS1, RET, and FGFR2 detected by ctDNA CGP was comparable to that detected using tissue CGP when TF was higher than 1% (12, 13).
In addition to TF, the major challenge in detecting fusions using plasma ctDNA is the lack of utilization of RNA. Fusions are subdivided into direct and composite, depending on the number of rearrangement events (14). RNA sequencing is considered superior to DNA sequencing in identifying fusions because RNA reflects the final product of composite fusion events. In contrast, DNA sequencing captures a snapshot of the fusion composite formation process. Interpreting gene fusions detected from tissue DNA sequencing can be challenging, especially when atypical configurations are involved, such as fusions with intergenic regions, fusions in the antisense direction, or fusions where only the reciprocal fusion is detected (15, 16). Although some of these fusions identified through DNA sequencing form canonical fusions at RNA level, others resulted in nonproductive rearrangements that do not produce fusion transcripts or proteins (16). For example, majority of the RET fusions observed to have out-of-frame configurations in DNA sequencing turned out to be in-frame in RNA sequencing (17). Owing to this indirect nature of DNA sequencing in detecting fusions, in the analysis of tissue, RNA has been widely used for fusion detection in addition to DNA (18–22). However, unlike tissue RNA, the utilization of circulating tumor RNA (ctRNA) is still under investigation (13).
This study aimed to validate the clinical utility of fusion detection using plasma ctDNA CGP. ctDNA CGP tests performed during routine clinical care of patients with NSCLC, along with tissue test results and clinical responses, were retrospectively reviewed.
2 Methods
2.1 Participants
Patients with stage IV NSCLC and tested with ctDNA CGP in routine clinical practice at the Samsung Medical Center (SMC) from December 2022 to April 2024 were included. The results of ctDNA CGP tests were retrospectively reviewed, and for patients with ctDNA CGP test results at multiple time points, the first test was used to prioritize pretreatment specimens with a higher tumor burden. The tissue test results were reviewed for concordance. For patients who underwent targeted therapy based on the fusions detected from ctDNA CGP, including those with atypical configurations, the responses were reviewed from the electronic medical records. This study was approved by the Institutional Review Board (IRB) of the SMC, Seoul, Korea (approval numbers 2024-01-133 and 2024-05-063).
2.2 CGP of ctDNA
The TruSight Oncology 500 (TSO500) ctDNA assay (Illumina, San Diego, CA, USA), 1.94 Mb panel covering 523 genes, was used for ctDNA CGP (Supplementary Table S1). The TSO500 ctDNA assay provides hybrid capture-based fusion detection, similar to most other ctDNA CGP assays. Whole blood was collected from each patient into two Cell-Free DNA Collection Tubes (Roche, Basel, Switzerland). DNA was extracted manually from 8 mL of plasma with a QIAamp DSP Circulating NA Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Libraries were prepared using 30 ng of input DNA, according to the TSO500 ctDNA assay protocol provided by the manufacturer. Subsequently, the library was enriched for 523 genes using a pool of target-specific oligos during two rounds of hybridization and target capture, followed by a final library amplification and cleanup. Quality control procedures before and after library preparation were performed using a Qubit (Thermo Fisher Scientific, Waltham, MA, USA) and a Cell-free DNA ScreenTape Assay (Agilent, Santa Clara, CA, USA). Sequencing was performed using an Illumina NovaSeq 6000 instrument with a read length of 2 × 150 bp and a targeted sequencing depth of 800 million reads (120 Gb) per sample. Bioinformatic analyses were performed using the DRAGEN TSO500 ctDNA Analysis Software version 1.1.0 on an Illumina DRAGEN Server, according to the user guide provided by the manufacturer. Within the software, fusion calling step used the Manta fusion caller, considering candidate fusions with at least 3 unique supporting reads, one of which must be a split read (a single read crossing the fusion breakpoint).
2.3 TF estimation
TF can be quantified either using aneuploidy- or maximum somatic allele frequency (MSAF)-based methods (12, 23). As the aneuploidy-based method is applicable only to cases with relatively high TF, at least 3% according to a landmark research, (24) MSAF-based methods have been used to complement aneuploidy-based methods for cases with low TF (12, 13, 23). In this study, to differentiate between cases with sufficient TF [≥ 1% following the previous studies (12, 13)] and those with limited TF (< 1%), MSAF of 0.5%, which is equivalent to TF of 1%, was used as the threshold. MSAF was determined as the highest VAF observed from the list of somatic variants generated by the TSO500 DRAGEN software for tumor mutation burden (TMB) calculation. The TMB variant list was generated by filtering out potential germline variants and variants of genes commonly associated with clonal hematopoiesis from all the variants detected in the coding region.
2.4 Acquisition of tissue-based test results
Tissue-based molecular testing results, including ALK IHC, ALK FISH, ROS1 IHC, ROS1 RT-PCR, and tissue CGP, were reviewed from electronic medical records. When multiple tissue test items were evaluated for the same fusion target, combined tissue results were used. When a discrepancy was observed between the results of IHC and RT-PCR, the results from the latter were used as true values, considering the higher accuracy of RT-PCR compared to that of IHC (5, 8).
To compare the overall prevalence rate of fusion between tissue and ctDNA CGP, the results of tissue CGP performed on patients with NSCLC in SMC during the study period, from December 2022 to April 2024, were used. Tissue CGP tests were performed using the tissue version of the TSO500 assay, and fusion detection was performed by hybridization capture-based enrichment of target regions using tissue RNA. Although the fusion drivers targeted by the two assays were different (55 genes in the tissue and 23 genes in the ctDNA versions, Supplementary Table S1), the most important fusion drivers in NSCLC, namely ALK, ROS1, and RET, were commonly targeted in both assays, and a comparison was performed for these three genes. For cases in which fusion was detected from both ctDNA and tissue CGP, their raw data (BAM files) were compared to observe the difference in fusion configurations in DNA and RNA.
2.5 Statistical analysis
Concordance between the combined tissue results and ctDNA CGP was analyzed in terms of positive percent agreement (PPA) and negative percent agreement (NPA) using combined tissue results as a reference. The positive predictive value (PPV) of ctDNA CGP was also assessed using combined tissue results. Fisher’s exact test was used to compare categorical variables between groups, and a binomial test was used to compare proportions. Statistical significance was set at P < 0.05. Statistical analyses were performed using the R software (version 4.2.2).
3 Results
3.1 Fusions detected from ctDNA CGP
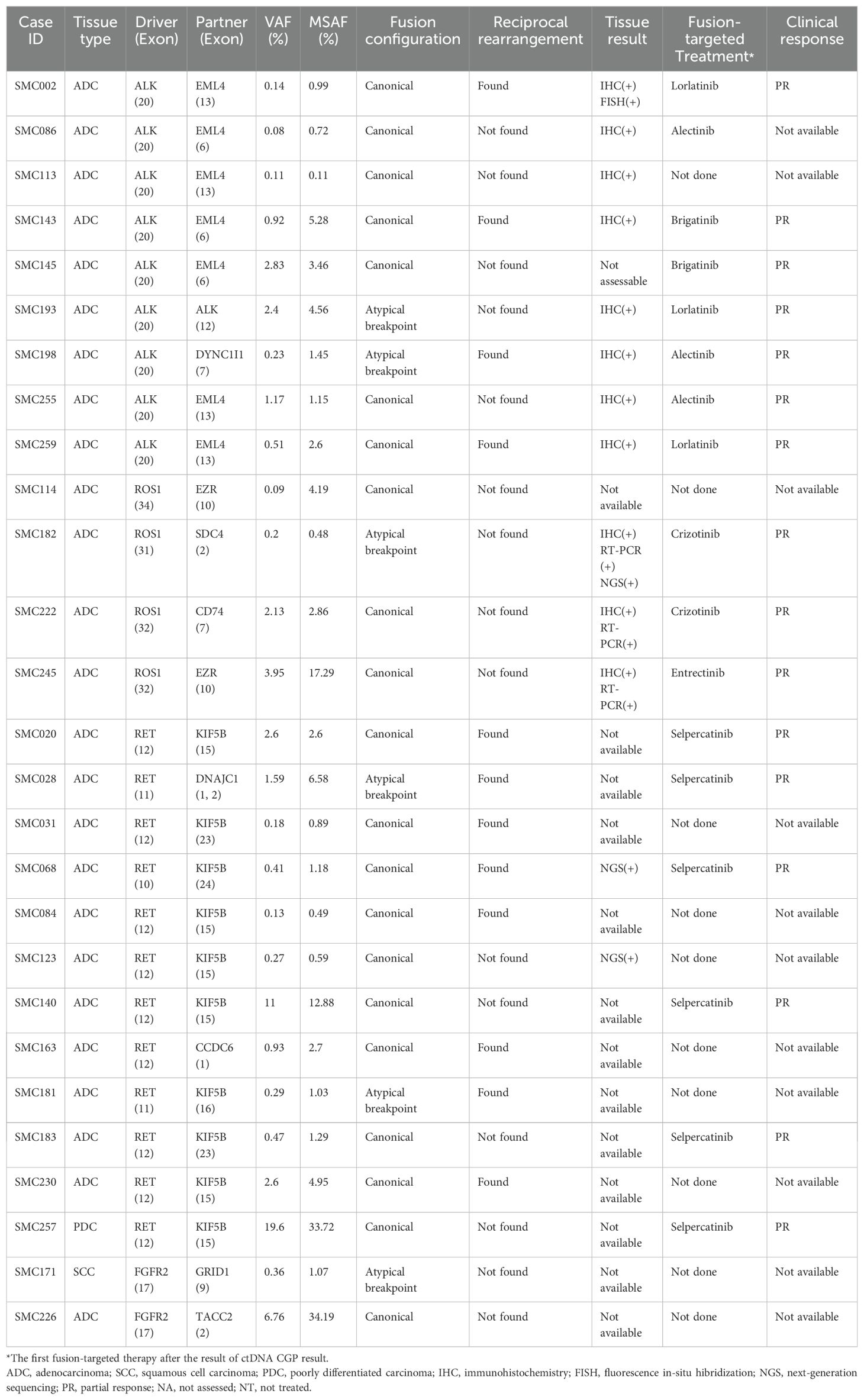
A total of 264 patients were tested for ctDNA CGP during the study period, and their histological subtypes included adenocarcinoma (n=227, 86.0%), squamous cell carcinoma (n=20, 7.6%), and poorly differentiated carcinoma (n=7, 2.7%) (Figure 1). Fusions were detected in 27 patients (10.2%), including RET (n=12; 4.6%), ALK (n=9; 3.4%), ROS1 (n=4; 1.5%), and FGFR2 (n=2; 0.8%) (Table 1). All ALK and ROS1 fusions were detected in adenocarcinomas, and RET fusions were detected mostly in adenocarcinomas (11/12, 91.7%), except in one case of poorly differentiated carcinoma. FGFR2 fusions were detected in one case each of adenocarcinoma and squamous cell carcinoma.
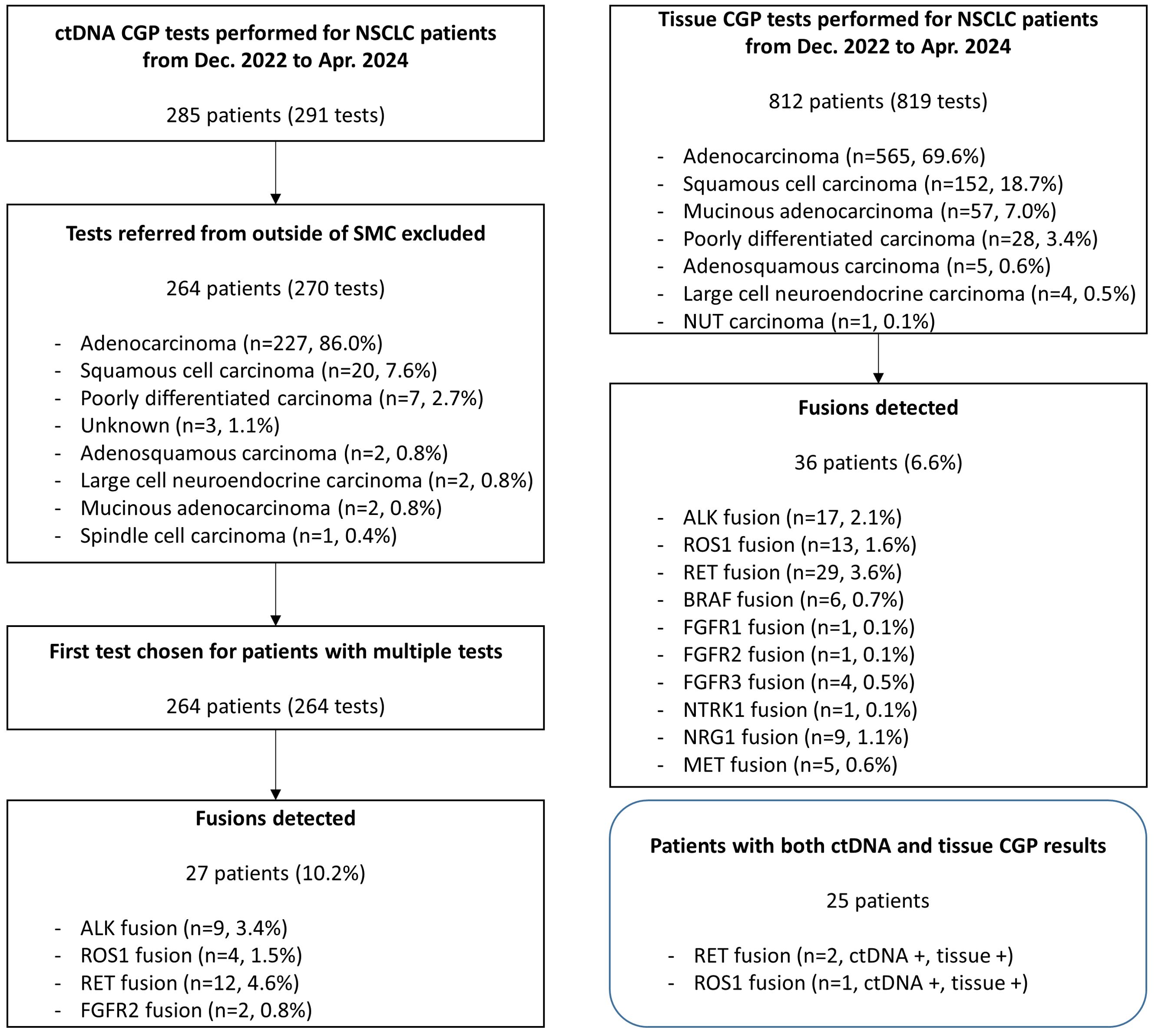
Figure 1. Flowchart of study inclusion and summary of the detected fusions. During the study period, 291 ctDNA CGP tests were performed on samples from 285 patients with NSCLC. After exclusion of tests referred from outside of SMC, 264 patients were finally included, and fusions were detected from 27 cases (10.2%). Tissue CGP was performed in 812 patients with NSCLC during the same period and fusion was detected from 86 patients (10.6%). Twenty five patients had both ctDNA and tissue CGP results and fusions were detected from three of them, commonly from ctDNA and tissue CGP. ctDNA, circulating tumor; CGP, comprehensive genomic profiling; MSAF, maximum somatic allele frequency; NSCLC, non-small cell lung cancer; SMC, Samsung Medical Center.
The prevalence rate of ALK, ROS1, and RET fusions were compared between ctDNA CGP (MSAF ≥ 0.5 group and MSAF < 0.5 group) and tissue CGP (Figure 2). As tissue CGP had a higher proportion of squamous cell carcinoma, in which fusions were rarely detected, the prevalence rate was also derived from non-squamous (NS)-NSCLC subsets (Figure 2B). However, the significant difference in the fusion prevalence was not observed across groups in all fusion drivers.
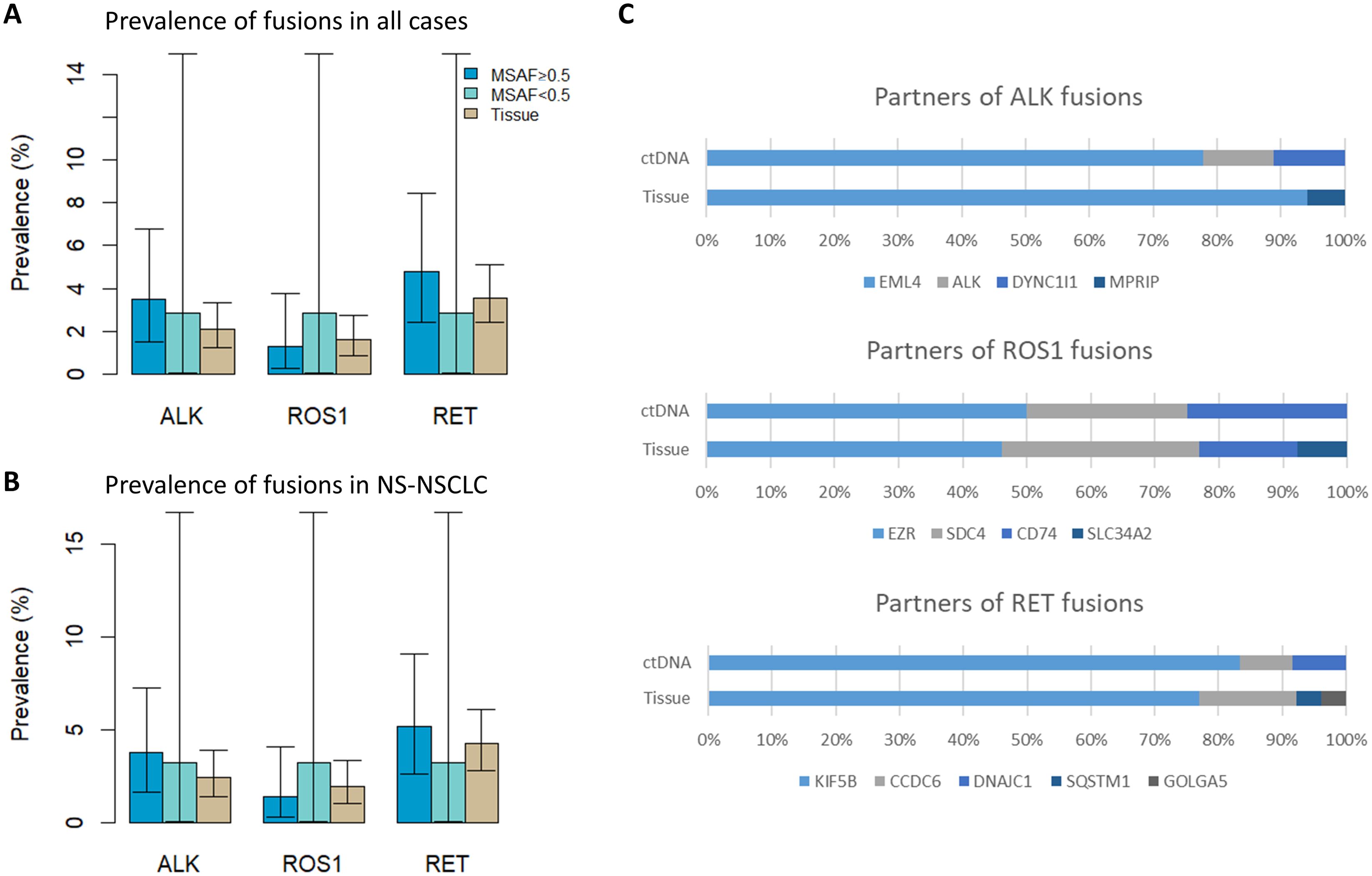
Figure 2. Detection of fusions from ctDNA and tissue. (A) Prevalence of fusions from ctDNA (MSAF ≥ 0.5 and <0.5 groups) and tissue (all cases) (B) Prevalence of fusions when only NS-NSCLC cases were considered (C) Distribution of fusion partners. ctDNA, circulating tumor; MSAF, maximum somatic allele frequency; NS-NSCLC, non-squamous non-small cell lung cancer.
The distribution of fusion partners is shown in Figure 2C. The majority of fusion partners for ALK were EML4 in ctDNA CGP (7/9, 77.8%), while two cases had atypical configuration, and the exact partners forming in-frame fusions were not detected (SMC193 and SMC198, as shown in Supplementary Figure S1, S2). The fusion partners of ROS1 were EZR (2/4, 50.0%), CD74(1/4, 25.0%), and SDC4(1/4, 25.0%), with the breakpoint observed in SDC4 being non-canonical (SMC182; Figure 3). Fusion partners of RET were KIF5B (10/12, 83.3%), CCDC6 (1/12, 8.3%), and DNAJC1 (1/12, 8.3%), with two cases showing RET fusions involving only the 5′ region of RET (SMC028 and SMC181, shown in Figure 4; Supplementary Figure S3). Breakpoints of FGFR2 commonly involved the known hotspot of FGFR2 intron 18, and the partners were GRID1 and TACC2 (Table 1). Although TACC2 is a common fusion partner of FGFR2, GRID1 has not yet been reported.
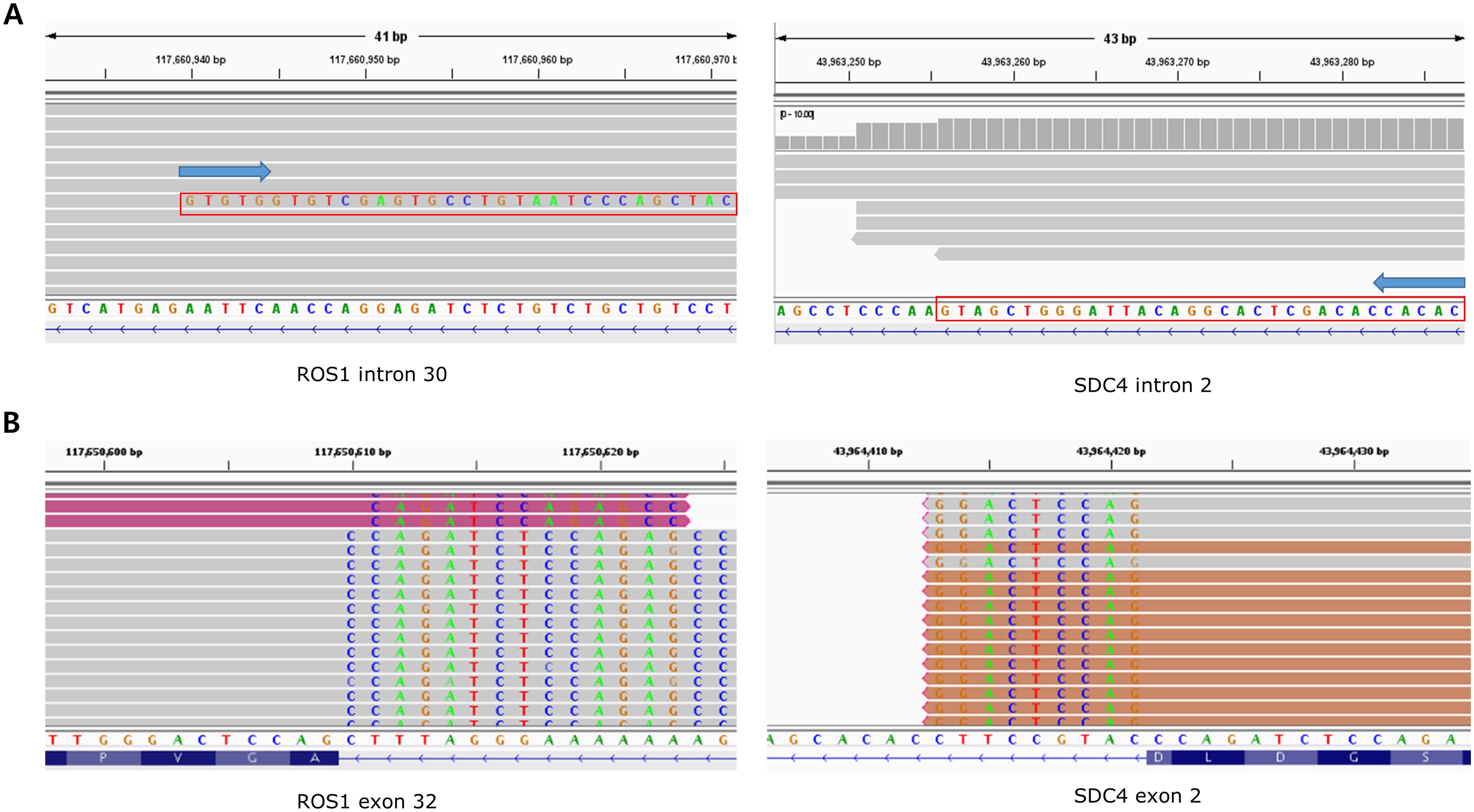
Figure 3. Comparison of BAM file findings of a fusion detected from SMC182 between ctDNA and tissue RNA. (A) From ctDNA, downstream part of ROS1 intron 30 was fused to SDC4 intron 2. However, the downstream part, rather than upstream part, of SDC4 intron 2 constituted the fusion. (B) From tissue RNA, ROS1 exon 32 was fused to SDC4 exon 2, which was in-frame.
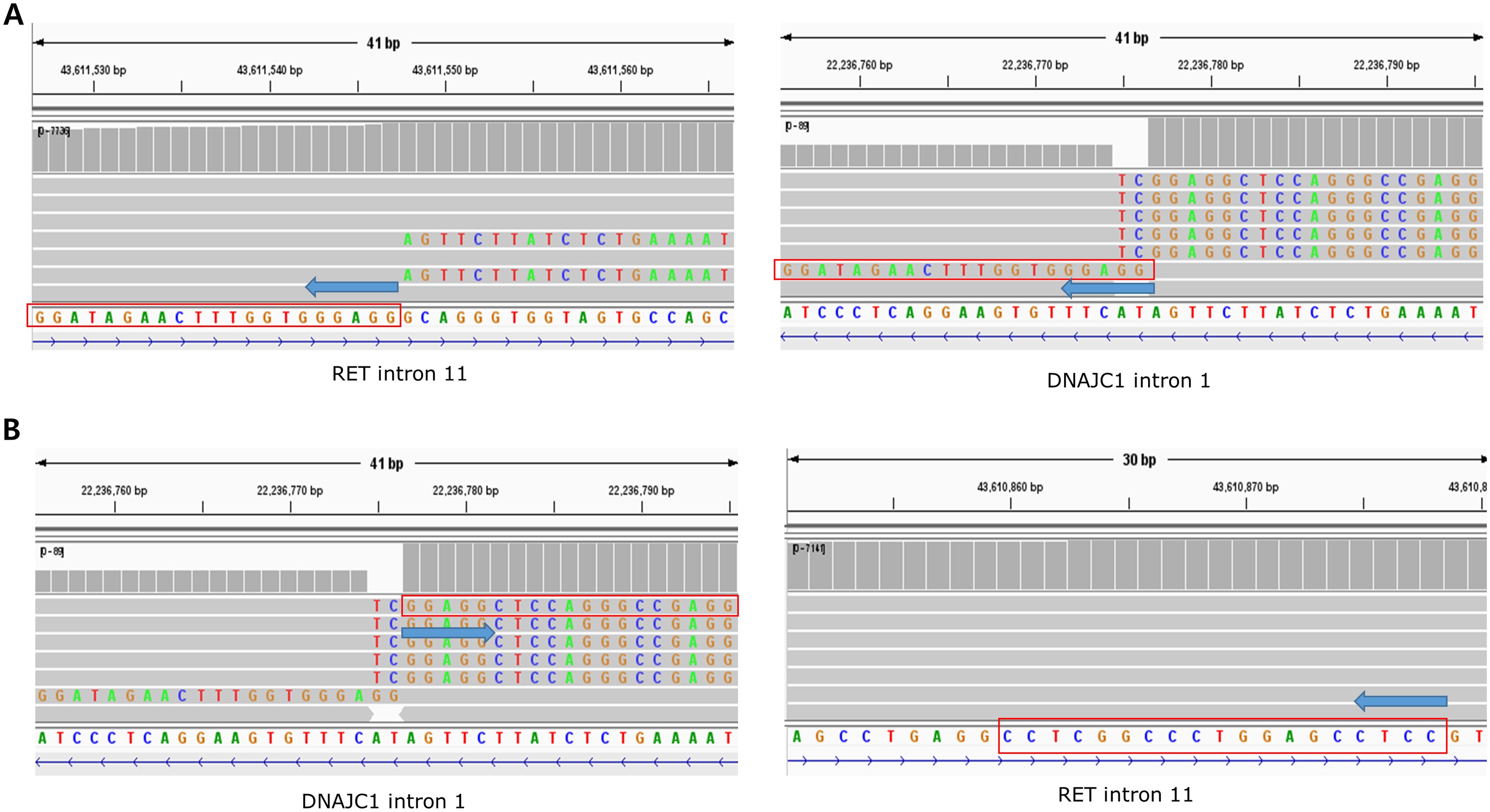
Figure 4. A fusion of reversed partner gene direction detected from ctDNA of SMC028. (A) Upstream part of DNAJC1 and upstream part of RET are fused. (B) The downstream part of DNAJC1, rather than upstream part, is fused to the upstream part of the RET.
3.2 Concordance between tissue and ctDNA CGP
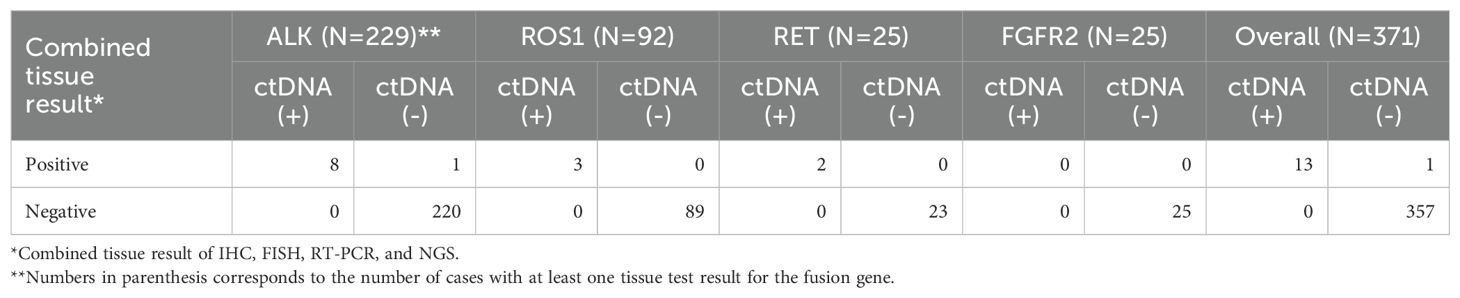
Among the 264 cases available for ctDNA CGP, the number of cases in which combined tissue test results were available was 229 for ALK, 92 for ROS1, 25 for RET, and 25 for FGFR2 fusion (Table 2). When all four genes were considered together, 371 ctDNA-tissue test pairs were available, and the overall PPA and NPA were 92.9% (13/14) and 100.0% (357/357), respectively. PPV was 100.0% (13/13). The only discrepant case, SMC063, was ALK IHC-positive and ctDNA CGP-negative (Supplementary Tables S2, S3). The MSAF of this case was 3.19% and the median unique molecular DOC was 3,075 ×, which was higher than the 2,500 × recommended by the manufacturer to ensure a 0.5% limit of detection. Based on the ALK IHC results, only approximately 10% of the tumor cells were positive, whereas the remaining tumor cells showed no immunoreactivity. The patient was treated with alectinib based on the positive ALK-IHC result. However, a PET-CT performed 2 months later revealed aggravated or newly appeared multiple hypermetabolic lesions in the right pleura, suggesting progressive disease. At the time of disease progression, ctDNA CGP was performed, which showed no alterations in ALK. T his finding raised the possibility of either a false positive IHC result or primary resistance caused by another oncogenic driver.
Both tissue and ctDNA CGP were performed in 25 cases, and fusion was detected in three cases (SMC068, SMC123, and SMC182). The driver and partner gene configurations observed in the ctDNA and RNA were identical for SMC068 and SMC123 (Supplementary Figure S4, S5). However, the observed breakpoint of the ROS1 fusion in SMC182 cells differed between ctDNA and tissue RNA (Figure 4). In this case, while the fusion observed from ctDNA was expected to produce an out-of-frame product, a fusion between ROS1 exon 31 and SDC4 exon, the one observed from tissue RNA, was a fusion between ROS1 exon 32 and SDC4 exon 2, which was in-frame.
3.3 Clinical significance of fusions detected from ctDNA CGP
Among the 27 patients with fusions detected in ctDNA CGP, 17 (63.0%) underwent targeted therapy for the fusions, and a partial response was observed in all patients whose responses were assessed (16/16) (Table 1). Five patients (SMC020, SMC028, SMC140, SMC145, and SMC183) underwent targeted therapy based solely on the ctDNA CGP results without tissue test results, and four of them had RET fusions for which IHC/FISH/RT-PCR was not available.
Among the six patients whose fusion configuration was non-canonical, four were treated with targeted agents (SMC028, SMC182, SMC193, and SMC198), and all showed a partial response (Table 1). The objective response ratio (ORR) did not differ between patients with canonical fusions and those with atypical breakpoints (10/10 vs. 4/4, P=1.000). Specifically, SMC028 cells were treated with selpercatinib based solely on the non-canonical findings of DNAJC1-RET fusion derived from ctDNA GCP (Figure 4), which showed a significant response that persisted for 14 months.
Among the 16 cases in which the response was assessed, reciprocal rearrangement was observed in seven cases. The ORR was not affected by reciprocal rearrangement (7/7 vs. 9/9, P=1.000).
4 Discussion
Owing to the expansion of targetable biomarkers, testing for molecular markers has become an essential part of NSCLC treatment. Genetic evolutions encountered during treatment have made repeated biopsies a common practice (25). Considering the cost and complications associated with repeated biopsy procedures, ctDNA CGP is expected to play an increasingly important role (26). Although numerous studies have suggested high concordance between tissue and ctDNA CGP tests, only a few of them focused on fusions (12, 13). Owing to the perceived inferior sensitivity of ctDNA CGP and the perception that DNA is a suboptimal specimen for fusion detection compared to RNA, the clinical utility of ctDNA CGP in terms of fusion detection has not yet been fully recognized.
In this study, the results of ctDNA CGP and combined tissue tests were retrospectively analyzed, and the results were perfectly concordant, except for one case in which false positivity of immunohistochemistry (IHC) was suspected based on the response to targeted therapy. In this case, only a small proportion of the tumor cells were positive. Typically, any percentage of positive tumor cells was considered a positive result (27). However, this case suggests that a confirmatory test may be necessary only when focal positivity is observed.
The prevalence of fusions was comparable between ctDNA and tissue CGP, supporting the concordance observed between the two methods. RET fusions have been reported to occur in 1–2% of patients with NSCLC, and the prevalence was reported to be higher in the Korean population (28–31). The prevalence rate of RET fusion observed in this study from ctDNA CGP was 4.5% (12/264), which was similar to that previously reported in Korea (30). Considering the high prevalence of RET fusions in specific populations and low availability of other tests such as RET IHC, (31) CGP tests should be considered a priority, and ctDNA could be an appropriate option based on the high clinical utility observed in this study.
Although there have been reports concerning the reliability of fusions observed to have atypical configurations using DNA-based methods, (15, 16) all cases in this study responded to targeted therapy. This result is in concordance with previous studies, in which the majority of fusions detected to have atypical configurations in DNA sequencing turned out to have canonical fusions in RNA sequencing or IHC (15–17). Validation using tissue RNA or protein assays could be warranted for available cases, as suggested in previous reports (16, 17). However, considering the potential scarcity of tissue specimens from patients who have already chosen ctDNA CGP instead of tissue, a trial of targeted therapy without confirmation could be considered based on risk vs. benefit analysis. Notably, most atypical fusions confirmed to produce canonical fusions had breakpoints located at or slightly upstream of the canonical location.
A limitation of this study was the sample size, especially for MSAF < 0.5 group. In the SMC, only patients with stage IV were eligible for ctDNA CGP, following the results of the Korean Food and Drug Administration accreditation, which might have resulted in the depletion of cases with a low tumor burden. One of the limitations of ctDNA CGP is the low applicability to cases with low tumor burden. However, the number of cases in MSAF < 0.5 group was too small for the statistical analysis and consequently, the difference in the prevalence was not observed between the MSAF ≥ 0.5 and < 0.5 groups as that observed in previous studies (12, 13). Future studies with larger number of cases, including earlier stage NSCLC cases, could help reveal the true performance of ctDNA CGP in fusion detection. On the other hand, the overall number of cases was sufficient for the comparison of prevalence between ctDNA and tissue CGP, which demonstrated that ctDNA is at least comparable to tissue CGP in detecting fusions. It should also be noted that the frequency of ALK fusions in tissue CGP may be underestimated because ALK IHC is prioritized and positive cases may not be tested for CGP. Lastly, the timing of tissue acquisition and blood sampling for ctDNA CGP could not be aligned owing to the retrospective nature of the study. The mutational profile, including fusions, might have been affected by the treatment or clonal evolution during the gap between tissue and blood sampling, affecting the correlation observed in this study.
In conclusion, fusion detection using ctDNA CGP showed high concordance with tissue tests and accuracy in predicting the therapeutic response in patients with NSCLC. ctDNA CGP is expected to provide an important diagnostic tool for fusion detection.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics statement
The studies involving humans were approved by Institutional Review Board (IRB) of the Samsung Medical Cener, Seoul, Korea (approval numbers 2024-01-133 and 2024-05-063). The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because of retrospective nature of study and minimal chance of harm to the participants.
Author contributions
Y-GK: Conceptualization, Writing – original draft. BL: Conceptualization, Writing – original draft. CH: Formal analysis, Investigation, Writing – review & editing. CL: Data curation, Investigation, Writing – review & editing. HJ: Formal analysis, Validation, Writing – review & editing. J-MS: Project administration, Validation, Writing – review & editing. S-HL: Project administration, Validation, Writing – review & editing. M-JA: Supervision, Validation, Writing – review & editing. Y-LC: Project administration, Validation, Writing – review & editing. SP: Supervision, Writing – review & editing. J-WK: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (grant number: HI23C1538).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1463341/full#supplementary-material
References
1. Aggarwal C, Rolfo CD, Oxnard GR, Gray JE, Sholl LM, Gandara DR. Strategies for the successful implementation of plasma-based NSCLC genotyping in clinical practice. Nat Rev Clin Oncol. (2021) 18:56–62. doi: 10.1038/s41571-020-0423-x
2. Bourreau C, Treps L, Faure S, Fradin D, Clere N. Therapeutic strategies for non-small cell lung cancer: Experimental models and emerging biomarkers to monitor drug efficacies. Pharmacol Ther. (2023) 242:108347. doi: 10.1016/j.pharmthera.2023.108347
3. Drilon A, Oxnard GR, Tan DSW, Loong HHF, Johnson M, Gainor J, et al. Efficacy of selpercatinib in RET fusion-positive non-small-cell lung cancer. N Engl J Med. (2020) 383:813–24. doi: 10.1056/NEJMoa2005653
4. Gendarme S, Bylicki O, Chouaid C, Guisier F. ROS-1 fusions in non-small-cell lung cancer: evidence to date. Curr Oncol. (2022) 29:641–58. doi: 10.3390/curroncol29020057
5. Kazdal D, Hofman V, Christopoulos P, Ilie M, Stenzinger A, Hofman P. Fusion-positive non-small cell lung carcinoma: Biological principles, clinical practice, and diagnostic implications. Genes Chromosomes Cancer. (2022) 61:244–60. doi: 10.1002/gcc.23022
6. Lei Y, Lei Y, Shi X, Wang J. EML4-ALK fusion gene in non-small cell lung cancer. Oncol Lett. (2022) 24:277. doi: 10.3892/ol.2022.13397
7. Drilon A, Camidge DR, Lin JJ, Kim SW, Solomon BJ, Dziadziuszko R, et al. Repotrectinib in ROS1 fusion-positive non-small-cell lung cancer. N Engl J Med. (2024) 390:118–31. doi: 10.1056/NEJMoa2302299
8. Lin C, Shi X, Yang S, Zhao J, He Q, Jin Y, et al. Comparison of ALK detection by FISH, IHC and NGS to predict benefit from crizotinib in advanced non-small-cell lung cancer. Lung Cancer. (2019) 131:62–8. doi: 10.1016/j.lungcan.2019.03.018
9. Page RD, Drusbosky LM, Dada H, Raymond VM, Daniel DB, Divers SG, et al. Clinical outcomes for plasma-based comprehensive genomic profiling versus standard-of-care tissue testing in advanced non–small cell lung cancer. Clin Lung Cancer. (2022) 23:72–81. doi: 10.1016/j.cllc.2021.10.001
10. Tsui DWY, Cheng ML, Shady M, Yang JL, Stephens D, Won H, et al. Tumor fraction-guided cell-free DNA profiling in metastatic solid tumor patients. Genome Med. (2021) 13:96. doi: 10.1186/s13073-021-00898-8
11. Deveson IW, Gong B, Lai K, Lococo JS, Richmond TA, Schageman J, et al. Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology. Nat Biotechnol. (2021) 39:1115–28. doi: 10.1038/s41587-021-00857-z
12. Lee JK, Hazar-Rethinam M, Decker B, Gjoerup O, Madison RW, Lieber DS, et al. The pan-tumor landscape of targetable kinase fusions in circulating tumor DNA. Clin Cancer Res. (2022) 28:728–37. doi: 10.1158/1078-0432.ccr-21-2136
13. Kasi PM, Lee JK, Pasquina LW, Decker B, Vanden Borre P, Pavlick DC, et al. Circulating tumor DNA enables sensitive detection of actionable gene fusions and rearrangements across cancer types. Clin Cancer Res. (2024) 30:836–48. doi: 10.1158/1078-0432.ccr-23-2693
14. Dorney R, Dhungel BP, Rasko JEJ, Hebbard L, Schmitz U. Recent advances in cancer fusion transcript detection. Brief Bioinform. (2023) 24:1–12. doi: 10.1093/bib/bbac519
15. Li W, Liu Y, Li W, Chen L, Ying J. Intergenic breakpoints identified by DNA sequencing confound targetable kinase fusion detection in NSCLC. J Thorac Oncol. (2020) 15:1223–31. doi: 10.1016/j.jtho.2020.02.023
16. Li W, Guo L, Liu Y, Dong L, Yang L, Chen L, et al. Potential unreliability of uncommon ALK, ROS1, and RET genomic breakpoints in predicting the efficacy of targeted therapy in NSCLC. J Thorac Oncol. (2021) 16:404–18. doi: 10.1016/j.jtho.2020.10.156
17. Xiang C, Guo L, Zhao R, Teng H, Wang Y, Xiong L, et al. Identification and validation of noncanonical RET fusions in non-small-cell lung cancer through DNA and RNA sequencing. J Mol Diagn. (2022) 24:374–85. doi: 10.1016/j.jmoldx.2021.12.004
18. Reeser JW, Martin D, Miya J, Kautto EA, Lyon E, Zhu E, et al. Validation of a targeted RNA sequencing assay for kinase fusion detection in solid tumors. J Mol Diagn. (2017) 19:682–96. doi: 10.1016/j.jmoldx.2017.05.006
19. Heyer EE, Deveson IW, Wooi D, Selinger CI, Lyons RJ, Hayes VM, et al. Diagnosis of fusion genes using targeted RNA sequencing. Nat Commun. (2019) 10:1388. doi: 10.1038/s41467-019-09374-9
20. Cohen D, Hondelink LM, Solleveld-Westerink N, Uljee SM, Ruano D, Cleton-Jansen AM, et al. Optimizing mutation and fusion detection in NSCLC by sequential DNA and RNA sequencing. J Thorac Oncol. (2020) 15:1000–14. doi: 10.1016/j.jtho.2020.01.019
21. Moore DA, Benafif S, Poskitt B, Argue S, Lee SM, Ahmad T, et al. Optimising fusion detection through sequential DNA and RNA molecular profiling of non-small cell lung cancer. Lung Cancer. (2021) 161:55–9. doi: 10.1016/j.lungcan.2021.08.008
22. Talebi A, Thiery JP, Kerachian MA. Fusion transcript discovery using RNA sequencing in formalin-fixed paraffin-embedded specimen. Crit Rev Oncol Hematol. (2021) 160:103303. doi: 10.1016/j.critrevonc.2021.103303
23. Husain H, Pavlick DC, Fendler BJ, Madison RW, Decker B, Gjoerup O, et al. Tumor fraction correlates with detection of actionable variants across> 23,000 circulating tumor DNA samples. JCO Precis Oncol. (2022) 6. doi: 10.1200/PO.22.00261
24. Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. (2017) 8:1324. doi: 10.1038/s41467-017-00965-y
25. Zhang Y, Shi L, Simoff MJ, Wagner OJ, Lavin J. Biopsy frequency and complications among lung cancer patients in the United States. Lung Cancer Manag. (2020) 9:LMT40. doi: 10.2217/lmt-2020-0022
26. Chiu YW, Kao YH, Simoff MJ, Ost DE, Wagner O, Lavin J, et al. Costs of biopsy and complications in patients with lung cancer. Clinicoecon Outcomes Res. (2021) 13:191–200. doi: 10.2147/CEOR.S295494
27. Wynes MW, Sholl LM, Dietel M, Schuuring E, Tsao MS, Yatabe Y, et al. An international interpretation study using the ALK IHC antibody D5F3 and a sensitive detection kit demonstrates high concordance between ALK IHC and ALK FISH and between evaluators. J Thorac Oncol. (2014) 9:631–8. doi: 10.1097/jto.0000000000000115
28. Zhou C. Lung cancer molecular epidemiology in China: recent trends. Transl Lung Cancer Res. (2014) 3:270–9. doi: 10.3978/j.issn.2218-6751.2014.09.01
29. Parimi V, Tolba K, Danziger N, Kuang Z, Sun D, Lin DI, et al. Genomic landscape of 891 RET fusions detected across diverse solid tumor types. NPJ Precis Oncol. (2023) 7:10. doi: 10.1038/s41698-023-00347-2
30. Yoon S, Kim K-P, Segall G, Jeon A, Jen M, Kim S, et al. EP12. 02-09 Prevalence and Treatment of Advanced NSCLC patients with REarranged During Transfection [RET]-fusion at a South Korean Centre. J Thorac Oncol. (2023) 18:S677–8. doi: 10.1016/j.jtho.2023.09.1302
Keywords: circulating-tumor DNA, ctDNA, gene fusion, non-small cell lung cancer, comprehensive genomic profiling
Citation: Kim Y-g, Lee B, Ha C, Lee C, Jung HA, Sun J-M, Lee S-H, Ahn M-J, Choi Y-L, Park S and Kim J-W (2024) Clinical utility of circulating tumor DNA profiling in detecting targetable fusions in non-small cell lung cancer. Front. Oncol. 14:1463341. doi: 10.3389/fonc.2024.1463341
Received: 11 July 2024; Accepted: 09 October 2024;
Published: 24 October 2024.
Edited by:
Elisa Frullanti, University of Siena, ItalyReviewed by:
Jiayue Qin, Acornmed Biotechnology Co., Ltd, ChinaDakai Zhang, University of Texas Health Science Center at Houston, United States
Copyright © 2024 Kim, Lee, Ha, Lee, Jung, Sun, Lee, Ahn, Choi, Park and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sehhoon Park, c2VoaG9vbi5wYXJrQGdtYWlsLmNvbQ==; Jong-Won Kim, a2ltandAc2trdS5lZHU=
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
 Young-gon Kim
Young-gon Kim Boram Lee
Boram Lee Changhee Ha
Changhee Ha Cheonghwa Lee1
Cheonghwa Lee1 Se-Hoon Lee
Se-Hoon Lee Myung-Ju Ahn
Myung-Ju Ahn Jong-Won Kim
Jong-Won Kim