- Haematology-Oncology Unit, Department of Paediatrics, Postgraduate Institute of Medical Education and Research, Chandigarh, India
RNA modification is the critical mechanism for regulating post-transcriptional processes. There are more than 150 RNA modifications reported so far, among which N6-Methyladenosine is the most prevalent one. M6A RNA modification complex consists of ‘writers’, ‘readers’ and ‘erasers’ which together in a group catalyze, recognize and regulate the methylation process of RNA and thereby regulate the stability and translation of mRNA. The discovery of erasers also known as demethylases, revolutionized the research on RNA modifications as it revealed that this modification is reversible. Since then, various studies have focused on discovering the role of m6A modification in various diseases especially cancers. Aberrant expression of these ‘readers’, ‘writers’, and ‘erasers’ is found to be altered in various cancers resulting in disturbance of cellular homeostasis. Acute leukemias are the most common cancer found in pediatric patients and account for 20% of adult cases. Dysregulation of the RNA modifying complex have been reported in development and progression of hematopoietic malignancies. Further, targeting m6A modification is the new approach for cancer immunotherapy and is being explored extensively. This review provides detailed information about current information on the role of m6A RNA modification in acute leukemia and their therapeutic potential.
Introduction
Recent studies have identified RNA modifications as one of the key post-transcriptional regulatory mechanisms in cancer. There are more than 150 types of RNA modifications reported till date. The most abundant RNA modification is the N6-Methyladenosine (m6A) where methylation takes place at the 6th N of adenylate of RNA on the consensus sequence DRACH (D = G, A, or U; R = G or A; H = A, C, or U) (1). This modification is found in the non-coding RNA, messenger RNA, transfer RNA, small nucleolar RNA, microRNA, and ribosomal RNA. It is most abundantly found at the exons, stop codons, and 3’ UTR regions of mRNA (2, 3). M6A modification is catalyzed by methyltransferase complex and is erased to maintain cellular homeostasis. A set of proteins recognized as ‘writers’, ‘erasers’, and ‘readers’ are involved in methylation, demethylation, and signaling pathways respectively (4). Writers consist of methyltransferase complex’s which contains Methyltransferase-like 3 (METTL3), Methyltransferase-like 14 (METTL14), Wilms tumor-1 associated protein (WTAP), Vir-like m6A methyltransferase associated (VIRMA), Zinc finger CCCH-type containing 13 (ZC3H13), RNA-binding protein 15/15B (RBM15/15B), HAKAI (CBLL1) (5). They help to achieve the methylation of RNA molecule. Readers recognize the methylation to regulate the translation and stability of RNA and play an important role in various signaling pathways. YTH domain-containing family protein 1/2/3 (YTHDF1/2/3), YTH domain-containing protein 1/2 (YTHDC1/2), Insulin-like growth factor-binding protein 1/2/3 (IGF2BP1/2/3), Heteronuclear ribonucleoproteins (including HNRNPC, HNRNPG and HNRNPA2B1) are various readers reported so far. Protein categorized under the Erasers category reverses the methylation modification i.e., carry out the demethylation reaction. Fat mass and obesity-associated protein (FTO/ALKBH9) and AlkB homolog 5 (ALKBH5) are two demethylase proteins identified so far (6). These writers, readers and erasers work in a coordinated manner to achieve homeostasis in the body. Any alteration in the function of these may result in the disruption of biological processes causing various diseases including cancers.
There are multiple reports on the development of cancer due to dysregulation of RNA modifications and their aberrant expression is linked to the initiation, progression, and drug resistance in various cancers. Acute leukemia is the most common pediatric malignancy and accounts for 20% adult cancers. Acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) are the two major types of acute leukemia depending on the lineage of cells involved and ALL is further categorized as T-cell and B-cell ALL. With the advancement of various molecular and genetic techniques, the outcome of ALL is significantly improved over the years, leading to better survival rates and more effective treatment regimens. However, despite these advancements, there is still a pressing need for new therapies to tackle relapse, chemoresistance, and to improve outcomes for high-risk patients who do not respond well to standard chemotherapy protocols. The challenge lies in the heterogeneity of the disease, with different genetic mutations and molecular pathways contributing to its aggressiveness and resistance to treatment.
Further, targeting m6A modification regulators is a new approach to cancer immunotherapy and is being widely explored in various cancers. Targeting FTO with inhibitors such as Meclofenamic acid has shown to have inhibitory effects on cell proliferation and metastasis in breast cancer and glioblastoma cells (7). Similar results are also seen on targeting ALKBH5 in AML (8). Moreover, targeting methyltransferase complex with STM2457 has been shown to inhibit the growth of AML cells (9). Thus, targeting the RNA modification regulatory pathways may pave the way for the development of novel therapeutic strategies for the treatment of hematopoietic malignancies. This review summarizes the recent information on the role of m6A RNA modification in pathogenesis of acute leukemia and its therapeutic potential.
Key players of m6A modification: discovery and functions
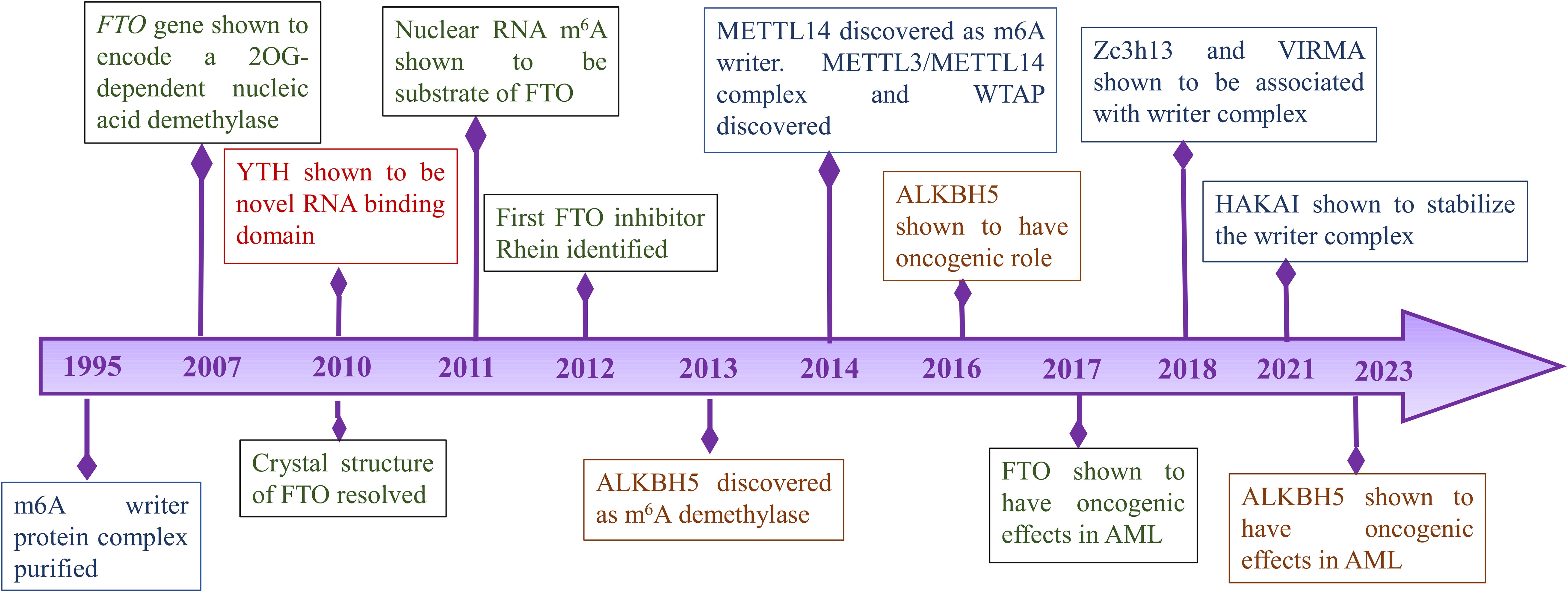
The first report of methylation on mRNA was published in 1974 by Desrosiers et al. in Novikoff hematoma cells (10). However, the writer complex was first purified after two decades in 1994 from HeLa cell extracts which provided insights into the mechanisms of formation of m6A modification. The latter study showed that methyltransferase contains three subunits MT-A1 (30 kDa), MT-A2 (200 kDa) and MT-B (875 kDa) (11). Further studies by the same research group showed that MT-A contains the AdoMet binding site on a 70-kDa subunit. Following that, expression studies in bacteria and antisera production from rabbits revealed that MT-A70 is a critical subunit of methyltransferase complex and Northern blotting studies showed that MT-A70 mRNA undergoes alternative splicing and is present abundantly in human tissues (12). Further components of methyltransferase complex were not known earlier due to the lack of advanced approaches. It was in 2014 when Schwartz et al. using a proteomic approach identified WTAP, KIAA1429 and METTL14 as important components of methyltransferase complex (13). Simultaneously, WTAP and METTL14 were published by two other groups while KIAA1429 could not be identified in any subsequent studies (14, 15). WTAP was shown to act as a regulatory subunit that interacts with METTL3-METTL14 heterodimer and was required for the localization of complex to nuclear speckles (15). The crystal structure of the METTL3-METTL14 complex was explained in 2016 by Wang et al. which revealed METTL3 is the catalytic component (16). The other writer VIRMA was reported as a part of the methyltransferase complex in 2018 which helps in mediating mRNA methylation at 3′UTR and near the stop codon (17). The writer ZC3H13 was reported by Wen et al., as a critical regulator of methyltransferase complex (18). Another recently identified writer HAKAI was shown to stabilize the core components of the methyltransferase complex (19).
FTO gene was identified in association with obesity and body mass regulation (20). It was in 2007 when studies by Gerken et al. revealed that the FTO encodes for a 2-Oxoglutarate-dependent Nucleic Acid Demethylase while the substrates were not known yet (21). In 2011, m6A was first shown to be the substrate of FTO in nuclear RNA and it was speculated that it plays a role in pre-RNA processing, transport or splicing (22). FTO was the first demethylase to be reported indicating m6A is a reversible modification. These findings brought a new dimension to the biological significance of m6A modifications. The other demethylase AlkBH5 was reported in 2013 and was shown to affect mRNA export and RNA metabolism (23).
Discovery of demethylases highlighted the importance of m6A modification in various diseases. Further studies revealed ‘readers’ that are involved in various signaling pathways. Wang et al. discovered YTH domain family 2 (YTHDF2) as a reader protein that play a role in RNA metabolism (24). In 2016, another member of the YTH family protein YTHDF2 was identified as an alternative reader. YTHDF2 recruits the CCR4-NOT complex which is essential for the deadenylation of m6A-containing RNAs by CAF1 and CCR4 (25). Further, YTHDC1 and its role in mRNA splicing was identified in 2016 (26). In 2018, Insulin-like growth factor 2 (IGF2) mRNA binding proteins 1,2,3 (IGF2BP1/2/3) which promote stability of mRNA targets were identified as a new family of m6A readers (27). Important timelines of discoveries of different sub-units of RNA modifications are shown in Figure 1.
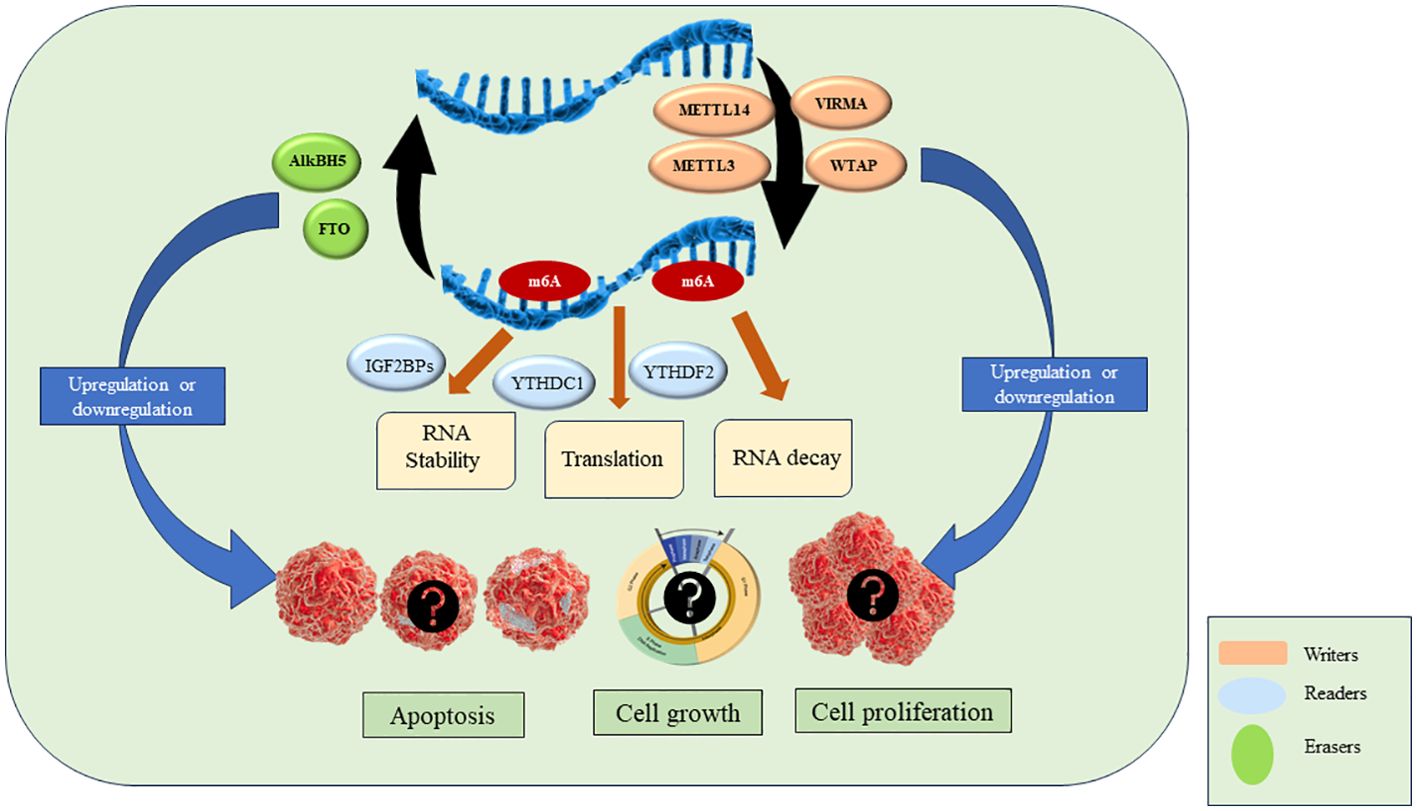
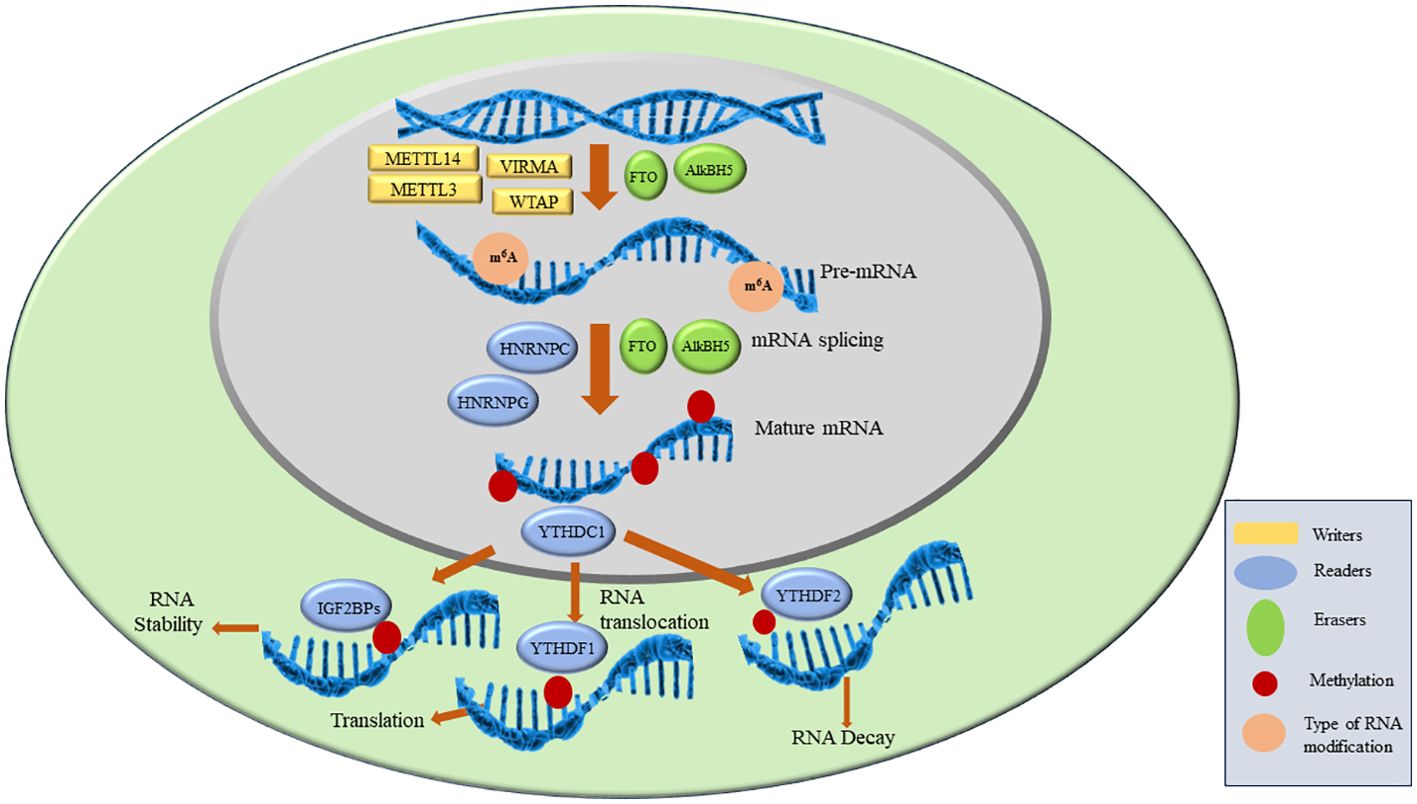
As discussed above, the key players in m6A modifications are ‘writers’, ‘readers’ and ‘erasers’. Writers are methyltransferase complex proteins that help in the methylation of RNA and consist of METTL3, METTL14, WTAP, VIRMA, ZC3H13, RBM15/15B, HAKAI. Readers are proteins that recognize the m6A modification and regulate RNA stability and translation by binding to m6A. YTHDF1/2/3, YTHDC1/2, IGF2BP1/2/3, HNRNPC, HNRNPG and HNRNPA2B1 are reader proteins. Erasers are demethylase proteins that reverse the m6A modification. FTO and ALKBH5 are two demethylases reported so far (4, 5). The details of each of the above component of RNA modification is described in the next section and are shown in Figure 2.
Writers
The writer complex consists of multiple subunits which add m6A to mRNA. This complex achieves methylation in a highly specific manner as only certain mRNAs are methylated and among those only certain consensus sites are methylated. However, the basis of this specificity remains poorly understood. After the discovery of the first methyltransferase METTL3 in 1994 more members are being added to date (11). The already reported and known members of Methyltransferase complex (MTC) includes METTL3, METTL14, VIRMA, WTAP, ZC3H13, RBM15/15B, HAKAI. METTL3 is the only catalytic subunit in MTC while others are associated proteins. METTL3 binds to METTL14 in 1:1 to form a dimer after which METTL3 becomes catalytically active and convert ‘A’ (adenosine) to m6A on mRNA. METTL14 is catalytically inactive as it lacks SAM binding site but it helps in activation of METTL3. This complex is localized in the nucleus but some reports show that stress can cause them to redistribute (28). Inactivation or deletion of METTL3 or METTL14 results in a 99% loss of total m6A modification in RNA indicating they play a potential role in achieving methylation (1). Associated proteins such as VIRMA, WTAP, ZC3H13, RBM15/15B, HAKAI have their specific functions. ZC3H13 helps in the nuclear localization of MTC and enhances m6A by bridging WTAP to the mRNA-binding factor Nito (29). Wilms tumor 1-associated protein (WTAP) is the adaptor protein of MTC which also lacks catalytic methylation. It interacts with METTL3 and METTL14 and helps in their recruitment of nuclear speckle. VIRMA is responsible for preferential m6A modification on mRNA at 3’UTR regions and stop codons. VIRMA mediates preferential m6A mRNA methylation in the 3’UTR and near the stop codon, while HAKAI affects m6A modification distributed in the 5’UTR and around the start codon (17). HAKAI also known as CBLL1, is a RING-finger type E3 ubiquitin ligase. It mediates the ubiquitination and endocytosis of the E-catherin complex which leads to cell-cell adhesion loss and increased cell mobility. It is also shown to regulate cell proliferation by affecting the binding of RNA targets to the PTB-associated splicing factors. Apart from these functions, HAKAI is the component of the methyltransferase complex in mammalian as well as plant cells. HAKAI is the component of MTC which is conserved in both drosophila and human and is a strong interactor of WTAP in mammalian cells (19). ZC3H13 enhances m6A processing through bridging WTAP to the mRNA-binding factor Nito while VIRMA directs m6A in 3′ UTR and near stop codon by recruiting the MTC to modulate region-selective methylation (30).
RBM 15 belongs to the split-end protein family. RBM15 and its paralog RBM15B were identified to interact with WTAP which showed that they might be a part of the methyltransferase complex (31). Later studies showed that they bind U-rich RNA consensus motifs and thus facilitate the recruitment of the MTC to specific sites in mRNA. RBM15 plays an important role in X chromosome inactivation by regulating m6A modification (32). Various studies show that RBM15 plays an important role in the recurrence of phyllodes tumour, laryngeal squamous cell carcinoma and hepatocellular carcinoma (33–35).
Readers
Readers recognize the m6A and bind to them leading to different signaling pathways. They comprise of YTH domain-containing family protein 1/2/3 (YTHDF1/2/3), YTH domain-containing protein 1/2 (YTHDC1/2), insulin-like growth factor-binding protein 1/2/3 (IGF2BP1/2/3), heteronuclear ribonucleoproteins (including HNRNPC, HNRNPG and HNRNPA2B1 (6).
YTH domain family have a conserved m6A-binding domain. They bind at the RRm6ACH consensus sequence. YTHDF2 is the first characterized reader which is shown to accelerate the decay of m6A-modified transcripts by recruiting the CCR4-NOT deadenylase complex directly. YTHDF-1 interacts with the translation initiation factors which results in enhanced mRNA stability and protein synthesis. Further, interaction of YTHDF3 with YTHDF1 results in the enhanced RNA translation or RNA degradation. YTHDC1 also plays an important part in RNA splicing and export (26). Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) including IGF2BP1–3 is recently identified as reader which bind to the m6A. They enhance the mRNA stability thus enhancing the RNA expressions. Another reader HNRNP A2/B1 (heterogeneous nuclear ribonucleoprotein A2/B1) binds to the RGAC motif on the RNA sites which results in m6A-dependent pre-miRNA processing events. It plays an important role in the mRNA stability by regulating pre-mRNA splicing in m6A dependent manner. It is shown to be upregulated in various cancers and is often associated with poor prognosis (36–38).
Erasers
The m6A erasers are the demethylases that convert m6A to A. Though writers and readers were identified several decades ago, demethylases were discovered in 2011 when FTO was shown to have demethylase activity (22). ALKBH5 was discovered in 2013 and shows tissue-specific expression. Discovery of demethylases showed the first evidence of reversible post-transcriptional modification in mRNA (23). FTO was first defined as demethylase and belongs to the non-heme Fe (II)-2-oxoglutarate (2OG)-dependent dioxygenase AlkB family proteins that repair N-alkylated nucleobases by oxidative demethylation. It is present on chromosome 16q12.2 and contains 9 exons and 8 introns. It contains two domains namely the N-terminal domain and the C-terminal domain containing 32-326 residues and 327-498 residues, respectively. Both these domains interact with each other to destabilize the N-terminal domain to achieve demethylation. The unique structure of FTO helps in recognition of double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) or single-stranded RNA (ssRNA) (4). FTO shows binding to multiple types of RNAs, including mRNA, snRNA, and tRNA, and can demethylate m6A and N6, 2'-O-dimethyladenosine (m6Am) in mRNA, m6A in U6RNA, m6Am in snRNAs, and N1-methyladenosine (m1A) in tRNA. However, m6A is the most favourable nucleobase substrate of FTO (39).
FTO is differentially expressed in different age groups and also varies with body mass index. Its expression is high in foetal and adult tissues especially in brain. Single nucleotide polymorphisms in the FTO gene are shown to be associated with obesity and various cancers such as breast, prostate and lung cancer (40). Various studies indicate the role of FTO in the susceptibility to obesity as it increases adipogenesis by regulation of various adipogenic pathways and adipocyte differentiation (41). There are emerging studies that define the role of FTO in tumorigenesis and progression. Various molecular studies on the correlation between obesity and tumorigenesis yield contradictory results as most of the single nucleotide polymorphisms lie in the intronic regions of the FTO gene and intron 1 and 2 show the strongest association with obesity. These results can be explained by two hypotheses that either FTO introns act as auto-regulators or as cis-regulatory sites for adjacent genes. Knockout of the FTO gene in mice models shows reduced adipose tissues and lean body mass, suppressed mitochondria biogenesis and energy production by positively regulating mammalian target of rapamycin-peroxisome proliferator-activated receptor-g coactivator-1a (mTOR-PGC-1a) pathway. FTO is also linked to heart failure as its deficiency decreases cardiomyocytes contractile functions by accelerating Ca2+ decay and increasing sarcomere shortening during cardiac remodeling and repair (42). Thus, wide cellular role of FTO with major involvement in RNA demethylation process suggests its immense potential as candidate of research in various biological processes.
ALKBH5 is the second demethylase that was discovered in 2013 and is a member of the AlkB family. It also plays an important role in demethylation along with FTO and influences RNA stability, translation, splicing and export (23). ALKBH5 contains 395 amino acids. The catalytic core of ALKBH5 contains a DSBH domain consisting of 11 b strands (b1−b11) and 5 a helix (a1− a5). ALKBH5 binds to 2OG and metal ions in a conserved manner (4). Colocalization studies of ALKBH5 show that it is colocalized with mRNA-processing factors15 and thus regulates the translocation of mRNA from nuclear to cytoplasm. ALKBH5 can decrease the synthesis rate of nascent RNA and maintain global RNA stability by affecting RNA metabolism. ALKBH5 also shows differential expression in tissues and is located in nuclear speckles similar to FTO indicating different roles in the methylation. Knockout of ALKBH5 in cell lines showed increased export of target RNAs to the cytoplasm from the nucleus. Further, functional studies of ALKBH5 show that it mediates demethylation at 3’ UTR m6A. This demethylation results in cancer progression in various cancers such as breast cancers and glioblastoma. FOXM1 which plays an important role in cell cycle and genomic stability is the downstream target of ALKBH5 in various cancers (43, 44). ALKBH5 also modulates the stability of 3’ UTR mRNAs in male germ cells (45). Recent studies have shown that ALKBH5 is expressed in the nucleus of adult mouse brain neurons, and is abundant in the cerebellum and olfactory bulb. The expression of ALKBH5 is usually high in the embryonic stages and decreases during the development of the brain indicating a probable role in brain development.
Role of m6A RNA modification in leukemia
M6A modifications play an important role in progression, metastasis and chemoresistance in various cancers such as breast cancer, lung cancer, etc. The studies on leukemia are limited but they show that m6A plays a significant role in leukemia by influencing various cellular signaling and gene expression. A list of studies on expression of readers, writers and erasers in leukemia is listed in Table 1.
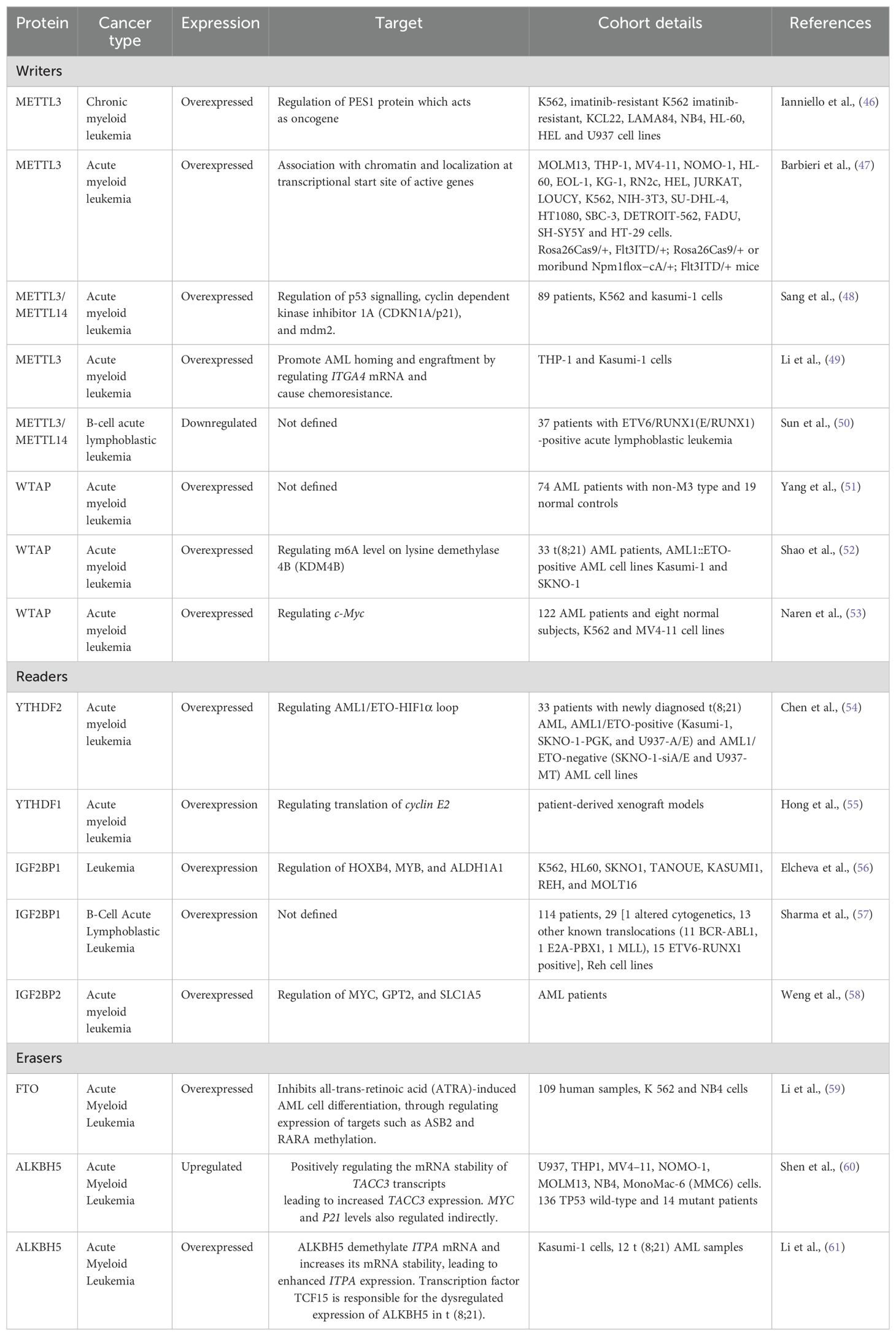
Table 1. Role of proteins involved in RNA modification in different leukemia and their target genes.
Writers
A study by Barbieri et al. using CRISPR screens showed METTL3 plays an essential role in the growth of leukemic cells in AML. METTL3 was shown to be associated with chromatin by localizing to the transcriptional start site of genes. In vivo studies show that downregulation of METTL3 results in suppression of leukemic cell progression and cell cycle arrest (47). Further in 2019, METTL3 and METTL14 expression was reported to be downregulated in ETV6/RUNX1-positive acute lymphoblastic leukemia as compared to the controls. METTL3 and METTL14 expression in relapse patients was also reported to be lower than the non-relapsed patients while the associated pathways were not studied (50). METTL3/METTL14 is also shown to be overexpressed in chronic myeloid leukemia and play an important role in the proliferation of leukemic cells. METTL3 plays an important role in ribosome biogenesis and translation. It also regulates the level of PES1 protein which is shown to have oncogenic role in breast cancer (46, 62). METTL3/METTL14 also regulates the expression of p53, CDKN1a/p21 and mdm2 thus having oncogenic effects in AML patients (48). METTL3 also plays a role in chemoresistance in AML by enhancing the half-life of ITGA4 mRNA leading to increased expression of ITGA4 protein. This chemoresistance is shown to be reversed when cells are treated with METTL3 inhibitors (49).
Expression of WTAP is shown to be altered in AML patients and AML cell lines (63). WTAP expression is reported to be significantly higher in non-M3 Acute myeloid leukemia patients. WTAP is also shown to be highly expressed in FLT3-ITD mutated AML patients than in FLT3-ITD unmutated AML patients. WTAP mRNA expression is positively correlated with WT1 expression (51). Further, WTAP has shown to be involved in proliferation, tumorigenesis, differentiation, cell cycle, and chemoresistance of AML cells. Moreover, WTAP has shown to regulate c-Myc expression which is oncogenic in nature (53). Overexpression of WTAP in t(8;21) AML patients is also linked to poor prognosis. Its activation by HIF1α results in reduced m6A levels on KDM4B transcripts which increase its degradation, thus enhancing the malignancy of leukemic cells (52). Further, studies in xenograft models show that growth rates of tumours in WTAP-knockdown cells are significantly reduced indicating WTAP plays an oncogenic role in AML.
Readers
YTHDF2 is reported to be expressed across all AML samples but is significantly upregulated in t(8;21) acute myeloid leukemia patients as compared to non-t(8;21) controls. High expression of YTHDF2 is correlated to higher chances of relapse and high mortality in patients (54). It is shown to increase cell proliferation by regulating AML1/ETO-HIF1α loop. Differential expression of YTHDF2 in t(8;21) indicates to its potential to be used as a prognostic marker after more elaborative studies (54). Studies also show that YTHDF1 is also upregulated in AML and is required for the progression of leukemia. It is achieved by promoting the translation of cyclin E2. YTHDF2 is also being explored as a therapeutic target in AML (55). Apart from YTH family proteins, IGF2BP1 is shown to be highly expressed in various leukemic cell lines. Low levels of IGF2BP1 are correlated with better survival in AML patients as it results in reduced tumorgenicity in mice models. It regulates the expression of ALDH1A1, HOXB4, and MYB at the post-transcriptional level thus plays a role in leukemia stem cell phenotype (56). IGF2BP2 is also upregulated in AML and B-ALL. In AML, IGF2BP2 is shown to control the expression of MYC, GPT2 and SLC1A5, although the specific signalling mechanisms in B-ALL are yet unidentified (57, 58).
Erasers
FTO is shown to be abnormally regulated in t(11q23)/MLL-rearranged, t(15;17)/PML-RARA, FLT3-ITD, and/or NPM1-mutated AML phenotypes. It enhances cellular viability, and proliferation and inhibits apoptosis thus helping in leukemogenesis. FTO downregulation has been shown to result in an increase of all-trans-retinoic acid (ATRA)-induced AML cell differentiation. This is achieved by methylation regulation of two genes by FTO: Ankyrin repeat and SOCS box protein 2 (ASB2) and retinoic acid receptor (RARA). This process also involves the interaction of FTO with YTHDF2 and YTHDF1; which indicates that the role of FTO is multifactorial (41, 64).
In studies involving the use of FTO inhibitors, it is reported that use of R-2-hydroxyglutarate (R-2HG) which was previously reported small-molecular inhibitor of FTO shows antileukemic effects in AML cell lines. R-2HG inhibits the FTO and thus increases the m6A levels on FTO oncogenic mRNA target MYC/CEBPA. These studies are also verified in the synergistic combination with first-line chemotherapy drugs such as daunorubicin or decitabine in murine models and further preclinical studies are required to check the therapeutic potentials (65).
ALKBH5 plays a critical role in autophagy, stem-cell renewal, cell proliferation, metastasis, and radioresistance in various cancers. Copy number alterations in the ALKBH5 are often reported in AML patients and are associated with inferior outcomes (66). MLST8, a common member of mTORC1 and mTORC2 protein complexes of the mTor pathway. mTor pathway is overexpressed in more than 70% of cancers. Studies show that targeting ALKBH5 by bioactive peptides results in decreased AML cell proliferation by repressing the demethylation of m6a in MLST8/EIF4EBP1 mRNA axis. These findings suggest that RNA demethylase could be a potential therapeutic targets in leukemia (67).
Distribution of m6A levels in cells of acute leukemia
The location of m6A sites on transcripts regulates m6A functions. Previous studies on transcriptome-wide m6A mapping of polyadenylated RNAs have shown that m6A modifications are enriched at 3′UTR region and near stop codon. Methylated RNA immunoprecipitation sequencing (MeRIP-seq) is one of the most commonly used tools for quantifying m6A modification at a specific site. Utilizing MeRIP-seq, studies have shown that that total m6A level are high in AML cells as compared to the normal hematopoietic progenitors. Deletion of METTL3 and METTL14 in embryonic cells results in reduced m6A levels affecting the differentiation of the cells (47, 68, 69). The reduction in m6A RNA by downregulating the expression of writers has shown to induce apoptosis in leukemic cells while having no effect on the normal hematopoietic progenitors. However, the detailed pathways studies associated with these observations are still awaited (70).
Methylation pattern of RNA is also shown to vary during the leukemia treatment. A study by Zhang et al. have shown that the number of m6A sites were decreased after azacytidine plus venetoclax treatment in patients bone marrow who achieved complete remission as compared to the AML bone marrows. Similar results were also obtained when HL-60 cell lines were treated with azacytidine. Further, downstream analysis has shown that most of the downregulated genes were protein coding indicating that they might play an important role in disease proliferation. In addition, HPRT1, ANP32B, and SNRPC were found to be linked to favourable outcomes in AML. These genes also showed a notable decrease in both m6A modification and expression (71).
Role of m6A in various metabolic pathways in acute leukemia
Dysregulation of m6A modification can impact various metabolic pathways, contributing to the development and progression of acute leukemia. A study by Liu et al. has shown that m6A modifications are more prevalent on short-term hematopoietic stem and progenitor cells (ST-HSPC) than long-term hematopoietic stem (LT-HSPCs) and progenitor cells indicating its role in cell development. Further analysis of m6A methylome in AML cells has shown that the genes associated with fatty acid catabolic process-related genes shows higher m6A modification in leukemia initiating cells as compared to normal HSPCs. ATP-binding cassette subfamily D member 2 (ABCD2, also called ALDRP) which is involved in lipid metabolism was shown to be significantly high in AML patients as compared to healthy controls. Also, the increased expression of ABCD2 was shown to be correlated with poor prognosis. Further studies on AML cell lines MOLM13 and MV4-11 showed that ABCD2 plays a role in proliferation of AML cells (72). Similar study in AML has shown that IGFBP2 is upregulated in AML patients as well as in cell lines MV4-11, KASUMI, MOLM13, and THP1. Further in-vivo studies have shown that IGFBP2 is required for AML cell growth and PRMT6 is downstream target. The expression of PMRT6 was also shown to be higher in AML patients and was correlated with poor overall survival in the patients. Targeting the expression of PMRT using inhibitor EPZ020411 resulted in decreased AML cell proliferation. MFSD2A which is sodium-dependent lysophosphatidylcholine symporter that is responsible for the uptake of docosahexaenoic acid (DHA) was found to be the downstream target of PMRT6. PMRT6 was shown to inhibit the MFSD2A by catalyzing H3R2me2a modification (73). IGFBP2 is shown to play a role in regulation of glutamine metabolism by stabilizing the associated mRNA transcripts MYC, GPT2, or SLC1A5 (58). ALKBH5 is also shown to modulate energy metabolism by m6A dependent manner. It controls OGDH RNA stability which is a key metabolic enzyme of TCA cycle (74).
Role of m6A in various signalling pathways
Malignant cells are often characterized by the abrupt signalling pathways. M6A modification is associated with different signalling pathways in various cancers. PI3K/AKT pathway which is associated with cell growth, is shown to be regulated by m6A in many cancers. The alterations in PI3K or AKT genes or upstream signalling molecules result in uncontrolled cell growth and chemoresistance. Dysregulation of METTL3/14 is linked with PTEN expression which can activate PI3K/AKT pathway in pancreatic cancer and renal cell cancer (75, 76). The Janus kinase (JAK)-signal transducer and activator of transcription (JAK-STAT) is also associated with cell proliferation, differentiation and migration. SOCS protein negatively regulate the JAK-STAT signalling. SOCS2 is shown to be regulated by METTL3 and YTHDF2 in various cancers (77–79). This signalling is also shown to play an important role in T-cell homeostasis by regulating STAT signalling inhibitory proteins SOCS1, SOCS3 and CISH which further play a role in IL-2 signalling pathways in m6A dependent manner (64). p53 pathway plays an important role in cellular stress signalling. Aberrant expression of p53 is linked to various cancers. M6A modification regulate the genes such as Mdm2, Mdm4 and p21 which are involved in p53 signalling. Dysregulation of METTL3, METTL14, ALKBH5 and FTO is shown to affect the p53 signalling in various cancers leading to cancer progression (80–82).
M6A as therapeutic target
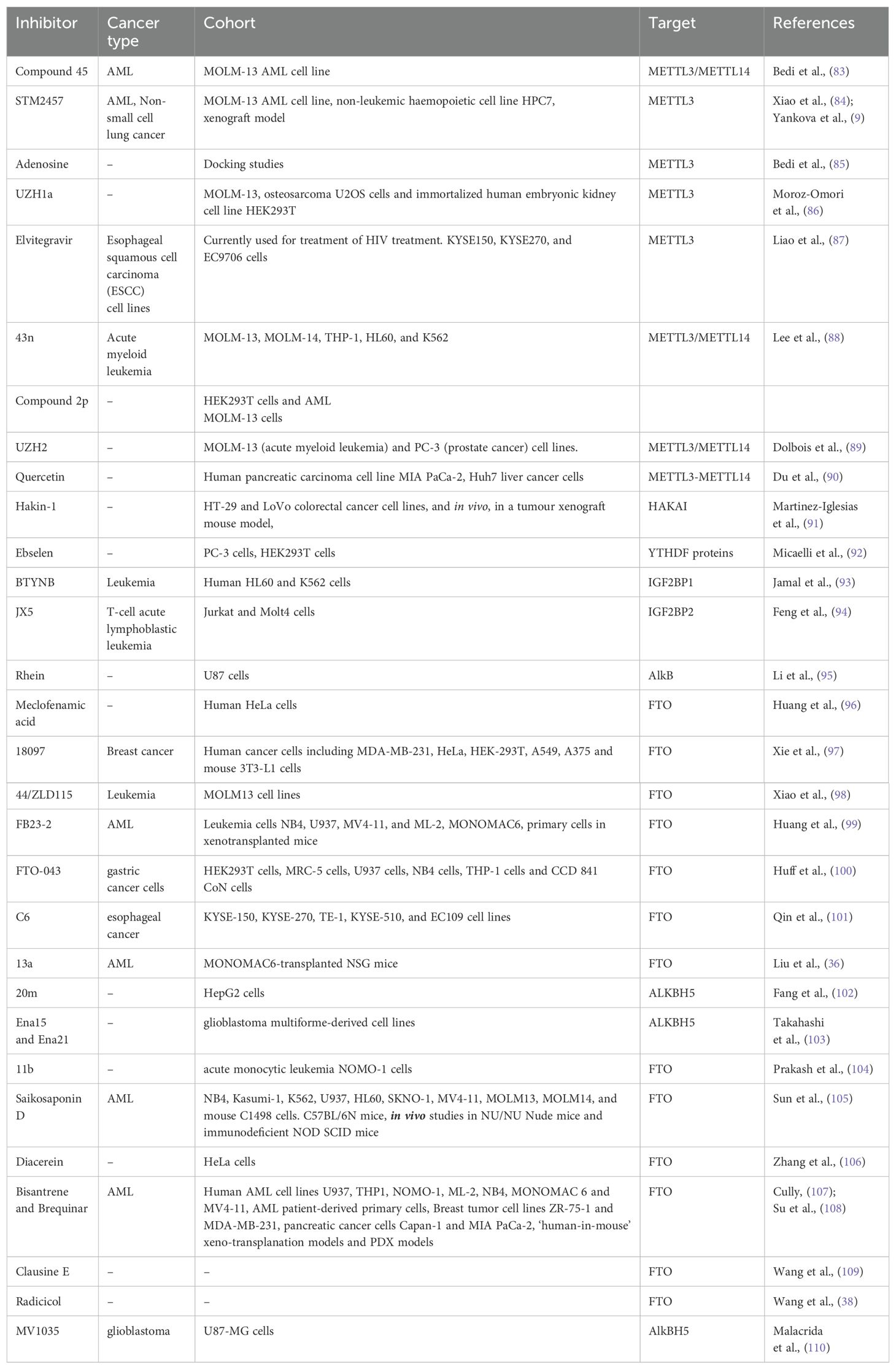
The differential expression of ‘readers’, ‘writers’ and ‘erasers’ make them a suitable candidate for targeted therapy. The discovery of demethylases has revolutionized the research on RNA modifications. After this discovery, writers and readers also grabbed the attention of various researchers and various inhibitors targeting writers and readers were reported which are listed in Table 2 and depicted in Figure 3. One of the first reports of inhibitors of ‘writers’ is that of STM2457 which shows selective inhibition of METTL3 and does not interfere with any other methyltransferases. These findings were confirmed by X-ray crystallography, surface plasmon resonance, and cellular thermal-shift assay. In vitro assays on MOLM-13 cells show decreased proliferation of cells and cell cycle arrest when treated with STM2457 while no such effects were seen on normal human umbilical cells and non-leukemic hematopoietic cell line HPC7. Similar results were seen on primary mice AML cells. Treatment with STM2457 resulted in decreased m6A levels on mRNA. Protein levels of SP1 and BRD4 which are substrates of METTL3 in AML were also reduced after STM2457 treatment. Further to decipher the detailed mode of action, MOLM-13 cells treated with STM2457 were subjected to m6A-specific methylated RNA immunoprecipitation (m6A-meRIP-seq) which revealed m6A associated DRACH motif as the top substrate. Further, in vivo studies using three human AML patient-derived xenograft showed that daily treatment with STM2457 resulted in decreased AML proliferation and prolonged survival in mice. Also, STM2457 did not show any toxicity on hematopoietic cells. Further trials are being done on STM2457 (9). β-elemene extracted from the Chinese medicinal plant Curcuma Wenyujin is also shown to inhibit METTL3 activity and have tumor suppressive effects. Various other studies show that it also targets p21, p53, Myc, HTRA etc. indicating that it has multiple targets of action (111, 112).
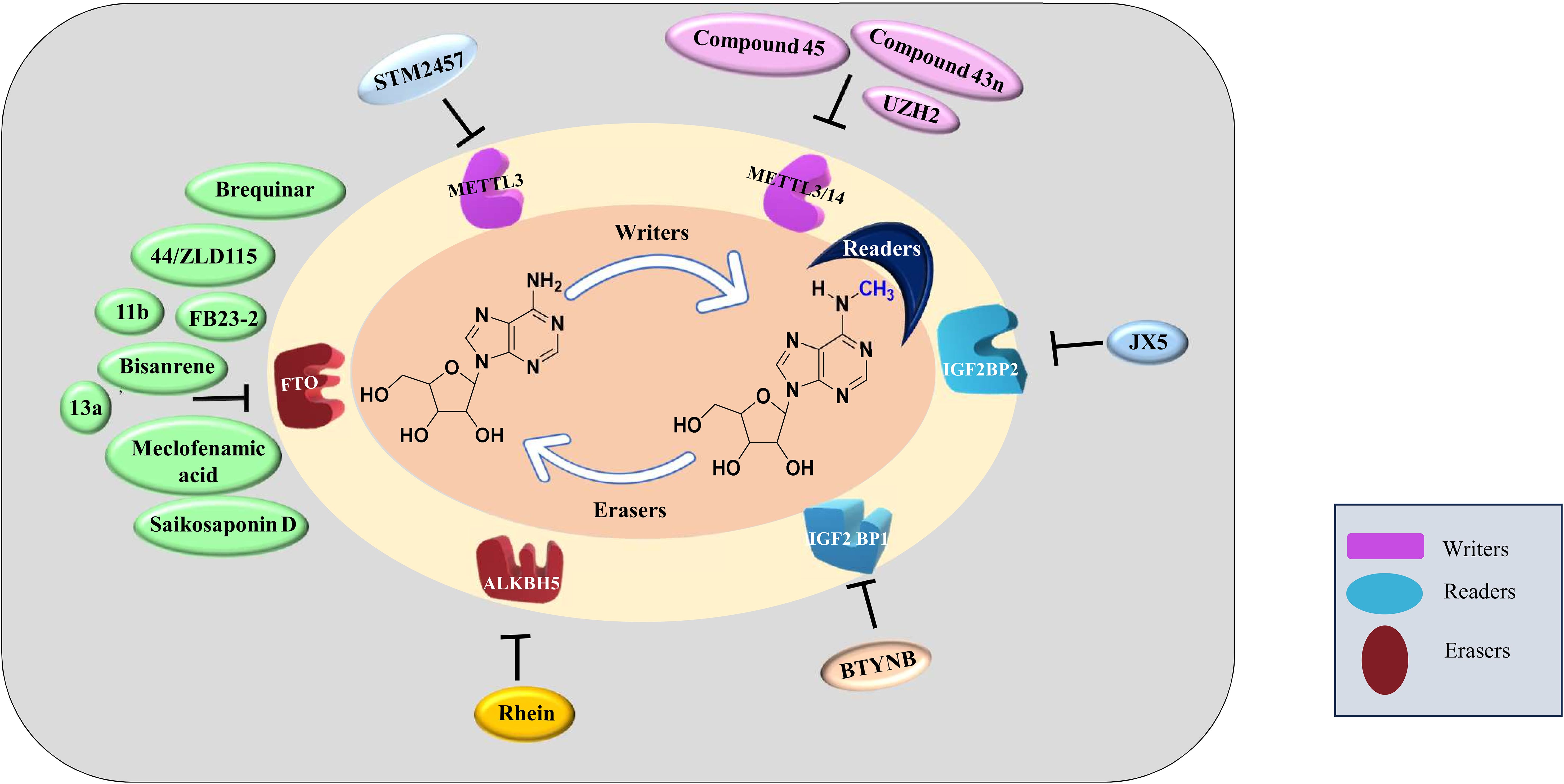
Figure 3. Inhibitors of m6A methylation complex targeting different components of RNA modification reported in different cancers.
Rhein is the first natural molecule reported as an FTO inhibitor identified by using structure-based virtual screening. It competitively binds to the catalytic subunit of FTO (113). Meclofenamic acid is an anti-inflammatory drug that selectively inhibit FTO over ALKBH5. Due to poor pharmacodynamics and low efficacy, their analogues are being synthesized. FB23 and FB23-2 are recently developed analogues of Meclofenamic acid showing high efficacy against primary AML leukemic stem cells. However, their downstream targets are under study (99). Another study identified CS1 and CS2 as inhibitors of FTO by using in silico screening. They show significant inhibition of AML cells with IC50 much lower than FB23 and FB23-2 by activating apoptotic signaling and inhibiting the MYC pathway (108).
Computational modeling using the crystal structure of ALKBH5 identified 2-[(1-hydroxy-2-oxo-2- phenylethyl)sulfanyl]acetic acid and 4-{[(furan-2-yl)-methyl]amino}-1,2-diazinane-3,6-dione which showed inhibitory effects on leukemic cell lines by inhibiting ALKBH5 (8). A study published by Zhang et al. showed that bioactive peptide (BP) can downregulate ALKBH5 which results in decreased proliferation of leukemic cells in AML. Treatment with BP resulted in decreased ALKBH5 expression that increased apoptosis and inhibition of proliferation of leukemic cells in vitro and in vivo models. Further, MLST8 and EIF4EBP1 expression was also found to be downregulated and these two were identified as downstream targets of ALKBH5 by m6A sequencing. Downregulation of ALKBH5 results in reduced demethylation of these target mRNA resulting in inhibition of cell proliferation. Interestingly, they have also reported that combination therapy of cytarabine and BP demonstrated decreases tumor growth and increases survival rates in in vivo mice model (67).
According to the report published by Megan Cully, various companies are targeting RNA epigenetics. Compounds targeting METTL3 currently being developed by STORM and Gotham Therapeutics are likely to be under trails from 2021. STORM therapeutics was also aiming to put STM2457 in clinical trials in 2022 but the details are awaited (107).
Apart from using various compounds targeting the m6A regulators, CRISPR-Cas-based strategies are also being explored to develop molecular therapies. A study by Liu et al. has reported ‘m6A editing’ approach which uses the engineered CRISPR-Cas9 fused with m6A methyltransferase or erasers that can be programmed with a guide RNA to install or erase m6A modification (114). Further study by Wilson et al. used CRISPR-Cas13 and RNA methyltransferases to develop Targeted RNA methylation (TRM) system which can install m6A modification on endogenous RNA transcripts. TRM is shown to achieve install methylation in nucleus as well as cytoplasmic transcripts by using dCas13–M3nls and dCas13–M3M14nes respectively (115). Methylation pattern alteration of HMBOX1 RNA in LnCAP cells using endonuclease-dead Cas9 protein (also known as dCas9) fused to the RNA demethylase ALKBH5 or METTL3 has shown that m6A modification plays an important role in determining the tumour suppressive function of HMBOX1 (116).
While studies show the efficacy of various m6A modification inhibitors, detailed studies are required to study off-target effects, synergistic studies with various chemotherapy agents and enhanced effectiveness to utilize them in regimens.
Future perspectives
The role of m6A modification has garnered significant attention as a promising therapeutic target for various cancers, including leukemia. Research has shown that the dysregulation of m6A-related proteins—such as m6A writers (e.g., METTL3, METTL14), erasers (e.g., FTO, ALKBH5), and readers (e.g., the YTHDF family)—is crucial in driving cancer cell proliferation, survival, chemoresistance, and relapse. In leukemia, these proteins influence the fate of leukemic stem cells, playing a key role in disease progression and therapy resistance.
One important consideration is the context-dependent expression and function of m6A-associated proteins across different cancers, and even within subtypes of the same malignancy. For instance, while METTL3 overexpression may promote oncogenesis in some leukemias, it could serve a different or even opposing function in other cancers. This variability highlights the need for a advanced approach that examines the tumor-specific expression profiles of m6A regulators, rather than adopting a uniform strategy for targeting m6A modifications across all cancers. A deeper understanding of the role of m6A machinery in different cancer subtypes is essential for the development of effective, personalized therapies.
Compounds targeting m6A regulators can be potential candidates for cancer therapy. These studies have shown high efficacy in in vitro and in vivo studies. M6A modification also influences how cancers respond to various treatments such as chemotherapy, radiotherapy, and immunotherapy. Utilizing m6A regulators in combination with these therapies could potentially enhance treatment outcomes and patient responses. Further, detailed pharmacological studies should be conducted to provide a deeper understanding of off-target effects, tissue distribution and stability.
Although targeting m6A modulators holds considerable potential, it also presents significant challenges. Since m6A modification is crucial in regulating normal physiological processes such as hematopoiesis, immune responses, and cellular differentiation, inhibiting these proteins could lead to unintended off-target effects and toxicity in healthy cells. This underlines the importance of developing selective targeting approaches that can distinguish between cancerous and healthy tissues, possibly by identifying specific vulnerabilities in cancer cells that are dependent on abnormal m6A regulation.
Moreover, the discovery of specific inhibitors or small molecules capable of modulating m6A-related proteins with precision is crucial. Any therapeutic strategy aimed at these proteins must be thoroughly evaluated to minimize interference with normal biological functions and reduce the risk of adverse side effects.
In summary, while targeting m6A-associated proteins represents a promising new direction in cancer treatment, including for leukemia, it is critical to further explore their cancer-specific roles. The development of safe, selective, and effective therapeutic strategies requires a comprehensive understanding of the differential expression and function of m6A regulators across various cancer types and subtypes, ensuring the therapeutic potential of m6A modulation is fully realized while minimizing associated risks.
Author contributions
PK: Data curation, Formal analysis, Investigation, Writing – original draft. PS: Data curation, Investigation, Writing – review & editing. PB: Formal analysis, Supervision, Writing – review & editing. MS: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. (2019) 20:608–24. doi: 10.1038/s41580-019-0168-5
2. Lei K, Lin S, Yuan Q. N6-methyladenosine (m6A) modification of ribosomal RNAs (rRNAs): Critical roles in mRNA translation and diseases. Genes Dis. (2023) 10:126–34. doi: 10.1016/j.gendis.2021.10.005
3. Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. (2019) 12:121. doi: 10.1186/s13045-019-0805-7
4. Shen D, Wang B, Gao Y, Zhao L, Bi Y, Zhang J, et al. Detailed resume of RNA m6A demethylases. Acta Pharm Sin B. (2022) 12:2193–205. doi: 10.1016/j.apsb.2022.01.003
5. Zhou Y, Kong Y, Fan W, Tao T, Xiao Q, Li N, et al. Principles of RNA methylation and their implications for biology and medicine. Biomed Pharmacother. (2020) 131:110731. doi: 10.1016/j.biopha.2020.110731
6. Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. (2019) 74:640–50. doi: 10.1016/j.molcel.2019.04.025
7. Chen H, Jia B, Zhang Q, Zhang Y. Meclofenamic acid restores gefinitib sensitivity by downregulating breast cancer resistance protein and multidrug resistance protein 7 via FTO/m6A-demethylation/c-myc in non-small cell lung cancer. Front Oncol. (2022) 12:870636. doi: 10.3389/fonc.2022.870636
8. Selberg S, Seli N, Kankuri E, Karelson M. Rational design of novel anticancer small-molecule RNA m6A demethylase ALKBH5 inhibitors. ACS Omega. (2021) 6:13310–20. doi: 10.1021/acsomega.1c01289
9. Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. (2021) 593:597–601. doi: 10.1038/s41586-021-03536-w
10. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from novikoff hepatoma cells. Proc Natl Acad Sci. (1974) 71:3971–5. doi: 10.1073/pnas.71.10.3971
11. Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. (1994) 269:17697–704. doi: 10.1016/S0021-9258(17)32497-3
12. Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA (New York N.Y.). (1997) 3:1233–47.
13. Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. Perturbation of m6A Writers Reveals Two Distinct Classes of mRNA Methylation at Internal and 5′ Sites. Cell Rep. (2014) 8:284–96. doi: 10.1016/j.celrep.2014.05.048
14. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. (2014) 10:93–5. doi: 10.1038/nchembio.1432
15. Ping X-L, Sun B-F, Wang L, Xiao W, Yang X, Wang W-J, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. (2014) 24:177–89. doi: 10.1038/cr.2014.3
16. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature. (2016) 534:575–8. doi: 10.1038/nature18298
17. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discovery. (2018) 4:10. doi: 10.1038/s41421-018-0019-0
18. Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol Cell. (2018) 69:1028–1038.e6. doi: 10.1016/j.molcel.2018.02.015
19. Bawankar P, Lence T, Paolantoni C, Haussmann IU, Kazlauskiene M, Jacob D, et al. Hakai is required for stabilization of core components of the m6A mRNA methylation machinery. Nat Commun. (2021) 12:3778. doi: 10.1038/s41467-021-23892-5
20. Loos RJF, Yeo GSH. The bigger picture of FTO—the first GWAS-identified obesity gene. Nat Rev Endocrinol. (2014) 10:51–61. doi: 10.1038/nrendo.2013.227
21. Gerken T, Girard CA, Tung Y-CL, Webby CJ, Saudek V, Hewitson KS, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Sci (New York N.Y.). (2007) 318:1469–72. doi: 10.1126/science.1151710
22. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. (2011) 7:885–7. doi: 10.1038/nchembio.687
23. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. (2013) 49:18–29. doi: 10.1016/j.molcel.2012.10.015
24. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. (2014) 505:117–20. doi: 10.1038/nature12730
25. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat Commun. (2016) 7:12626. doi: 10.1038/ncomms12626
26. Xiao W, Adhikari S, Dahal U, Chen Y-S, Hao Y-J, Sun B-F, et al. Nuclear m 6 A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. (2016) 61:507–19. doi: 10.1016/j.molcel.2016.01.012
27. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. (2018) 20:285–95. doi: 10.1038/s41556-018-0045-z
28. He PC, He C. m 6 A RNA methylation: From mechanisms to therapeutic potential. EMBO J. (2021) 40:e105977. doi: 10.15252/embj.2020105977
29. Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. (2018) 32:415–29. doi: 10.1101/gad.309146.117
30. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. (2019) 18:176. doi: 10.1186/s12943-019-1109-9
31. Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, et al. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. (2013) 288:33292–302. doi: 10.1074/jbc.M113.500397
32. Patil DP, Chen C-K, Pickering BF, Chow A, Jackson C, Guttman M, et al. M(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. (2016) 537:369–73. doi: 10.1038/nature19342
33. Cai X, Chen Y, Man D, Yang B, Feng X, Zhang D, et al. RBM15 promotes hepatocellular carcinoma progression by regulating N6-methyladenosine modification of YES1 mRNA in an IGF2BP1-dependent manner. Cell Death Discovery. (2021) 7:315. doi: 10.1038/s41420-021-00703-w
34. Garcia-Dios DA, Levi D, Shah V, Gillett C, Simpson MA, Hanby A, et al. MED12, TERT promoter and RBM15 mutations in primary and recurrent phyllodes tumours. Br J Cancer. (2018) 118:277–84. doi: 10.1038/bjc.2017.450
35. Wang X, Tian L, Li Y, Wang J, Yan B, Yang L, et al. RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J Exp Clin Cancer Res. (2021) 40:80. doi: 10.1186/s13046-021-01871-4
36. Liu H, Li D, Sun L, Qin H, Fan A, Meng L, et al. Interaction of lncRNA MIR100HG with hnRNPA2B1 facilitates m6A-dependent stabilization of TCF7L2 mRNA and colorectal cancer progression. Mol Cancer. (2022) 21:74. doi: 10.1186/s12943-022-01555-3
37. Tauler J, Zudaire E, Liu H, Shih J, Mulshine JL. hnRNP A2/B1 modulates epithelial-mesenchymal transition in lung cancer cell lines. Cancer Res. (2010) 70:7137–47. doi: 10.1158/0008-5472.CAN-10-0860
38. Wang H, Liang L, Dong Q, Huan L, He J, Li B, et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma. Theranostics. (2018) 8:2814–29. doi: 10.7150/thno.23012
39. Zhang X, Wei L, Wang Y, Xiao Y, Liu J, Zhang W, et al. Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc Natl Acad Sci. (2019) 116(8):2919–24. doi: 10.1073/pnas.1820574116
40. Hernández-Caballero ME, Sierra-Ramírez JA. Single nucleotide polymorphisms of the FTO gene and cancer risk: An overview. Mol Biol Rep. (2015) 42:699–704. doi: 10.1007/s11033-014-3817-y
41. Azzam SK, Alsafar H, Sajini AA. FTO m6A demethylase in obesity and cancer: implications and underlying molecular mechanisms. Int J Mol Sci. (2022) 23:3800. doi: 10.3390/ijms23073800
42. Mathiyalagan P, Adamiak M, Mayourian J, Sassi Y, Liang Y, Agarwal N, et al. FTO-dependent N 6 -methyladenosine regulates cardiac function during remodeling and repair. Circulation. (2019) 139:518–32. doi: 10.1161/CIRCULATIONAHA.118.033794
43. Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6 A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. (2016) 113(14):E2047-56. doi: 10.1073/pnas.1602883113
44. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. M 6 A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. (2017) 31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013
45. Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y, et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc Natl Acad Sci. (2018) 115(2). doi: 10.1073/pnas.1717794115
46. Ianniello Z, Sorci M, Ceci Ginistrelli L, Iaiza A, Marchioni M, Tito C, et al. New insight into the catalytic -dependent and -independent roles of METTL3 in sustaining aberrant translation in chronic myeloid leukemia. Cell Death Dis. (2021) 12:870. doi: 10.1038/s41419-021-04169-7
47. Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millán-Zambrano G, Robson SC, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature. (2017) 552:126–31. doi: 10.1038/nature24678
48. Sang L, Wu X, Yan T, Naren D, Liu X, Zheng X, et al. The m 6 A RNA methyltransferase METTL3/METTL14 promotes leukemogenesis through the mdm2/p53 pathway in acute myeloid leukemia. J Cancer. (2022) 13:1019–30. doi: 10.7150/jca.60381
49. Li M, Ye J, Xia Y, Li M, Li G, Hu X, et al. METTL3 mediates chemoresistance by enhancing AML homing and engraftment via ITGA4. Leukemia. (2022) 36:2586–95. doi: 10.1038/s41375-022-01696-w
50. Sun C, Chang L, Liu C, Chen X, Zhu X. The study of METTL3 and METTL14 expressions in childhood ETV6/RUNX1 -positive acute lymphoblastic leukemia. Mol Genet Genomic Med. (2019) 7(10):e00933. doi: 10.1002/mgg3.933
51. Yang L-L, Zhao R-R, Jiang R-Y, Liu H, Zhou S-Y, Gu B, et al. The expression of WTAP gene in acute myeloid leukemia and its clinical significance. Zhongguo Shi Yan Xue Ye Xue Za Zhi. (2021) 29:653–60. doi: 10.19746/j.cnki.issn.1009-2137.2021.03.001
52. Shao Y-L, Li Y-Q, Li M-Y, Wang L-L, Zhou H-S, Liu D-H, et al. HIF1α-mediated transactivation of WTAP promotes AML cell proliferation via m6A-dependent stabilization of KDM4B mRNA. Leukemia. (2023) 37:1254–67. doi: 10.1038/s41375-023-01904-1
53. Naren D, Yan T, Gong Y, Huang J, Zhang D, Sang L, et al. High Wilms’ tumor 1 associating protein expression predicts poor prognosis in acute myeloid leukemia and regulates m6A methylation of MYC mRNA. J Cancer Res Clin Oncol. (2021) 147:33–47. doi: 10.1007/s00432-020-03373-w
54. Chen Z, Shao Y-L, Wang L-L, Lin J, Zhang J-B, Ding Y, et al. YTHDF2 is a potential target of AML1/ETO-HIF1α loop-mediated cell proliferation in t(8;21) AML. Oncogene. (2021) 40:3786–98. doi: 10.1038/s41388-021-01818-1
55. Hong Y-G, Yang Z, Chen Y, Liu T, Zheng Y, Zhou C, et al. The RNA m6A reader YTHDF1 is required for acute myeloid leukemia progression. Cancer Res. (2023) 83:845–60. doi: 10.1158/0008-5472.CAN-21-4249
56. Elcheva IA, Wood T, Chiarolanzio K, Chim B, Wong M, Singh V, et al. RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia. (2020) 34:1354–63. doi: 10.1038/s41375-019-0656-9
57. Sharma G, Boby E, Nidhi T, Jain A, Singh J, Singh A, et al. Diagnostic utility of IGF2BP1 and its targets as potential biomarkers in ETV6-RUNX1 positive B-cell acute lymphoblastic leukemia. Front Oncol. (2021) 11:588101. doi: 10.3389/fonc.2021.588101
58. Weng H, Huang F, Yu Z, Chen Z, Prince E, Kang Y, et al. The m6A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell. (2022) 40:1566–1582.e10. doi: 10.1016/j.ccell.2022.10.004
59. Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N 6 -methyladenosine RNA demethylase. Cancer Cell. (2017) 31:127–41. doi: 10.1016/j.ccell.2016.11.017
60. Shen C, Sheng Y, Zhu AC, Robinson S, Jiang X, Dong L, et al. RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell. (2020) 27(1):64–80.e9. doi: 10.1016/j.stem.2020.04.009
61. Li R, Wu X, Xue K, Feng D, Li J, Li J. RNA demethylase ALKBH5 promotes tumorigenesis of t (8;21) acute myeloid leukemia via ITPA m6A modification. biomark Res. (2023) 11:30. doi: 10.1186/s40364-023-00464-x
62. Cheng L, Li J, Han Y, Lin J, Niu C, Zhou Z, et al. PES1 promotes breast cancer by differentially regulating ERα and ERβ. J Clin Invest. (2012) 122:2857–70. doi: 10.1172/JCI62676
63. Bansal H, Yihua Q, Iyer SP, Ganapathy S, Proia D, Penalva LO, et al. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia. (2014) 28:1171–4. doi: 10.1038/leu.2014.16
64. Li H-B, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. (2017) 548:338–42. doi: 10.1038/nature23450
65. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. (2018) 172:90–105.e23. doi: 10.1016/j.cell.2017.11.031
66. Kwok C-T, Marshall AD, Rasko JEJ, Wong JJL. Genetic alterations of m6A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol. (2017) 10:39. doi: 10.1186/s13045-017-0410-6
67. Zhang L, Su X. Bioactive peptide inhibits acute myeloid leukemia cell proliferation by downregulating ALKBH5-mediated m6A demethylation of EIF4EBP1 and MLST8 mRNA. Cell Oncol. (2022) 45:355–65. doi: 10.1007/s13402-022-00666-9
68. Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. (2017) 23:1369–76. doi: 10.1038/nm.4416
69. Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell. (2018) 22:191–205.e9. doi: 10.1016/j.stem.2017.11.016
70. Ianniello Z, Paiardini A, Fatica A. N6-methyladenosine (m6A): A promising new molecular target in acute myeloid leukemia. Front Oncol. (2019) 9:251. doi: 10.3389/fonc.2019.00251
71. Zhang Z, Zhang L, Li J, Feng R, Li C, Liu Y, et al. Comprehensive analysis of m6A methylome alterations after azacytidine plus venetoclax treatment for acute myeloid leukemia by nanopore sequencing. Comput Struct Biotechnol J. (2024) 23:1144–53. doi: 10.1016/j.csbj.2024.02.029
72. Wang Y, Wang Y, Gu J, Su T, Gu X, Feng Y. The role of RNA m6A methylation in lipid metabolism. Front Endocrinol. (2022) 13:866116. doi: 10.3389/fendo.2022.866116
73. Cheng Y, Gao Z, Zhang T, Wang Y, Xie X, Han G, et al. Decoding m6A RNA methylome identifies PRMT6-regulated lipid transport promoting AML stem cell maintenance. Cell Stem Cell. (2023) 30:69–85.e7. doi: 10.1016/j.stem.2022.12.003
74. Gao Y, Zimmer JT, Vasic R, Liu C, Gbyli R, Zheng S-J, et al. ALKBH5 modulates hematopoietic stem and progenitor cell energy metabolism through m6A modification-mediated RNA stability control. Cell Rep. (2023) 42:113163. doi: 10.1016/j.celrep.2023.113163
75. Jiang Z, Song X, Wei Y, Li Y, Kong D, Sun J. N(6)-methyladenosine-mediated miR-380-3p maturation and upregulation promotes cancer aggressiveness in pancreatic cancer. Bioengineered. (2022) 13:14460–71. doi: 10.1080/21655979.2022.2088497
76. Zhang L, Luo X, Qiao S. METTL14-mediated N6-methyladenosine modification of Pten mRNA inhibits tumour progression in clear-cell renal cell carcinoma. Br J Cancer. (2022) 127:30–42. doi: 10.1038/s41416-022-01757-y
77. Chen M, Wei L, Law C, Tsang FH, Shen J, Cheng CL, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. (2018) 67:2254–70. doi: 10.1002/hep.29683
78. Jiang L, Chen T, Xiong L, Xu J, Gong A, Dai B, et al. Knockdown of m6A methyltransferase METTL3 in gastric cancer cells results in suppression of cell proliferation. Oncol Lett. (2020) 20:2191–8. doi: 10.3892/ol.2020.11794
79. Xu J, Chen Q, Tian K, Liang R, Chen T, Gong A, et al. m6A methyltransferase METTL3 maintains colon cancer tumorigenicity by suppressing SOCS2 to promote cell proliferation. Oncol Rep. (2020) 44:973–86. doi: 10.3892/or.2020.7665
80. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. (2012) 485:201–6. doi: 10.1038/nature11112
81. Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, et al. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer. (2020) 19:91. doi: 10.1186/s12943-020-01158-w
82. Zhou J, Wang J, Hong B, Ma K, Xie H, Li L, et al. Gene signatures and prognostic values of m6A regulators in clear cell renal cell carcinoma – a retrospective study using TCGA database. Aging. (2019) 11:1633–47. doi: 10.18632/aging.101856
83. Bedi RK, Huang D, Li Y, Caflisch A. Structure-based design of inhibitors of the m6A-RNA writer enzyme METTL3. ACS Bio Med Chem Au. (2023) 3:359–70. doi: 10.1021/acsbiomedchemau.3c00023
84. Xiao H, Zhao R, Meng W, Liao Y. Effects and translatomics characteristics of a small-molecule inhibitor of METTL3 against non-small cell lung cancer. J Pharm Anal 13:625–39. doi: 10.1016/j.jpha.2023.04.009
85. Bedi RK, Huang D, Eberle SA, Wiedmer L, Śledź P, Caflisch A. Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer. ChemMedChem. (2020) 15:744–8. doi: 10.1002/cmdc.202000011
86. Moroz-Omori EV, Huang D, Kumar Bedi R, Cheriyamkunnel SJ, Bochenkova E, Dolbois A, et al. METTL3 Inhibitors for Epitranscriptomic Modulation of Cellular Processes. ChemMedChem. (2021) 16:3035–43. doi: 10.1002/cmdc.202100291
87. Liao J, Wei Y, Liang J, Wen J, Chen X, Zhang B, et al. Insight into the structure, physiological function, and role in cancer of m6A readers—YTH domain-containing proteins. Cell Death Discov. (2022) 8:137. doi: 10.1038/s41420-022-00947-0
88. Lee J, Kim S, Jin MS, Kim Y. Discovery of substituted indole derivatives as allosteric inhibitors of m 6 A-RNA methyltransferase, METTL3 -14 complex. Drug Dev Res. (2022), ddr.21910. doi: 10.1002/ddr.21910
89. Dolbois A, Bedi RK, Bochenkova E, Müller A, Moroz-Omori EV, Huang D, et al. 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors. J Med Chem. (2021) 64:12738–60. doi: 10.1021/acs.jmedchem.1c00773
90. Du Y, Yuan Y, Xu LX, Zhao F, Wenbin Wang Wenbin W, Xu Y, et al. Discovery of METTL3 Small Molecule Inhibitors by Virtual Screening of Natural Products. Front Pharmacol. (2022) 13:878135. doi: 10.3389/fphar.2022.878135
91. Martinez-Iglesias O, Casas-Pais A, Castosa R, Díaz-Díaz A, Roca-Lema D, Concha Á, et al. Hakin-1, a New Specific Small-Molecule Inhibitor for the E3 Ubiquitin-Ligase Hakai, Inhibits Carcinoma Growth and Progression. Cancers. (2020) 12:1340. doi: 10.3390/cancers12051340
92. Micaelli M, Vedove AD, Cerofolini L, Vigna J, Sighel D, Zaccara S, et al. Small-Molecule Ebselen Binds to YTHDF Proteins Interfering with the Recognition of N 6 -Methyladenosine-Modified RNAs. ACS Pharmacol Transl Sci. (2022) 5:872–91. doi: 10.1021/acsptsci.2c00008
93. Jamal A, Hassan Dalhat M, Jahan S, Choudhry H, Imran Khan M. BTYNB, an inhibitor of RNA binding protein IGF2BP1 reduces proliferation and induces differentiation of leukemic cancer cells. Saudi J Biol Sci. (2023) 30:103569. doi: 10.1016/j.sjbs.2023.103569
94. Feng P, Chen D, Wang X, Li Y, Li Z, Li B, et al. Inhibition of the m6A reader IGF2BP2 as a strategy against T-cell acute lymphoblastic leukemia. Leukemia. (2022) 36:2180–8. doi: 10.1038/s41375-022-01651-9
95. Li Q, Huang Y, Liu X, Gan J, Chen H, Yang C-G. Rhein Inhibits AlkB Repair Enzymes and Sensitizes Cells to Methylated DNA Damage. J Biol Chem. (2016) 291:11083–93. doi: 10.1074/jbc.M115.711895
96. Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. (2015) 43:373–84. doi: 10.1093/nar/gku1276
97. Xie G, Wu X-N, Ling Y, Rui Y, Wu D, Zhou J, et al. A novel inhibitor of N6-methyladenosine demethylase FTO induces mRNA methylation and shows anti-cancer activities. Acta Pharm Sin B. (2022) 12:853–66. doi: 10.1016/j.apsb.2021.08.028
98. Xiao P, Duan Z, Liu Z, Chen L, Zhang D, Liu L, et al. Rational Design of RNA Demethylase FTO Inhibitors with Enhanced Antileukemia Drug-Like Properties. J Med Chem. (2023) 66:9731–52. doi: 10.1021/acs.jmedchem.3c00543
99. Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. (2019) 35:677–691.e10. doi: 10.1016/j.ccell.2019.03.006
100. Huff S, Kummetha IR, Zhang L, Wang L, Bray W, Yin J, et al. Rational Design and Optimization of m 6 A-RNA Demethylase FTO Inhibitors as Anticancer Agents. J Med Chem. (2022) 65:10920–37. doi: 10.1021/acs.jmedchem.1c02075
101. Qin B, Bai Q, Yan D, Yin F, Zhu Z, Xia C, et al. Discovery of novel mRNA demethylase FTO inhibitors against esophageal cancer. J Enzyme Inhib Med Chem. (2022) 37:1995–2003. doi: 10.1080/14756366.2022.2098954
102. Fang Z, Mu B, Liu Y, Guo N, Xiong L, Guo Y, et al. Discovery of a potent, selective and cell active inhibitor of m6A demethylase ALKBH5. Eur J Med Chem. (2022) 238:114446. doi: 10.1016/j.ejmech.2022.114446
103. Takahashi H, Hase H, Yoshida T, Tashiro J, Hirade Y, Kitae K, et al. Discovery of two novel ALKBH5 selective inhibitors that exhibit uncompetitive or competitive type and suppress the growth activity of glioblastoma multiforme. Chem Biol Drug Des. (2022) 100:1–12. doi: 10.1111/cbdd.14051
104. Prakash M, Itoh Y, Fujiwara Y, Takahashi Y, Takada Y, Mellini P, et al. Identification of Potent and Selective Inhibitors of Fat Mass Obesity-Associated Protein Using a Fragment-Merging Approach. J Med Chem. (2021) 64:15810–24. doi: 10.1021/acs.jmedchem.1c01107
105. Sun K, Du Y, Hou Y, Zhao M, Li J, Du Y, et al. Saikosaponin D exhibits anti-leukemic activity by targeting FTO/m 6 A signaling. Theranostics. (2021) 11:5831–46. doi: 10.7150/thno.55574
106. Zhang Y, Li Q, Zhou K, Xu Q, Zhang C. Identification of Specific N 6 -Methyladenosine RNA Demethylase FTO Inhibitors by Single-Quantum-Dot-Based FRET Nanosensors. Anal Chem. (2020) 92:13936–44. doi: 10.1021/acs.analchem.0c02828
107. Cully M. Chemical inhibitors make their RNA epigenetic mark. Nat Rev Drug Discovery. (2019) 18:892–4. doi: 10.1038/d41573-019-00179-5
108. Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, et al. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. (2020) 38:79–96.e11. doi: 10.1016/j.ccell.2020.04.017
109. Wang Y, Li J, Han X, Wang N, Song C, Wang R, et al. Identification of Clausine E as an inhibitor of fat mass and obesity-associated protein (FTO) demethylase activity. J Mol Recognit. (2019) 32. doi: 10.1002/jmr.2800
110. Malacrida A, Rivara M, Di Domizio A, Cislaghi G, Miloso M, Zuliani V, et al. 3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell line. Bioorg Med Chem. (2020) 28:115300. doi: 10.1016/j.bmc.2019.115300
111. Alafnan A, Dogan R, Bender O, Celik I, Mollica A, Malik JA, et al. Beta Elemene induces cytotoxic effects in FLT3 ITD-mutated acute myeloid leukemia by modulating apoptosis. Eur Rev Med Pharmacol Sci. (2023) 27:3270–87. doi: 10.26355/eurrev_202304_32098
112. Liu S, Li Q, Li G, Zhang Q, Zhuo L, Han X, et al. The mechanism of m6A methyltransferase METTL3-mediated autophagy in reversing gefitinib resistance in NSCLC cells by β-elemene. Cell Death Dis. (2020) 11:969. doi: 10.1038/s41419-020-03148-8
113. Chen B, Ye F, Yu L, Jia G, Huang X, Zhang X, et al. Development of cell-active N6 -methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc. (2012) 134:17963–71. doi: 10.1021/ja3064149
114. Liu X-M, Zhou J, Mao Y, Ji Q, Qian S-B. Programmable RNA N6-methyladenosine editing by CRISPR-Cas9 conjugates. Nat Chem Biol. (2019) 15:865–71. doi: 10.1038/s41589-019-0327-1
115. Wilson C, Chen PJ, Miao Z, Liu DR. Programmable m6A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat Biotechnol. (2020) 38:1431–40. doi: 10.1038/s41587-020-0572-6
Keywords: RNA modification “writers”, RNA modification “readers”, RNA modification “erasers”, acute leukemia, N6-methyladenosine (m6A)
Citation: Kaur P, Sharma P, Bhatia P and Singh M (2024) Current insights on m6A RNA modification in acute leukemia: therapeutic targets and future prospects. Front. Oncol. 14:1445794. doi: 10.3389/fonc.2024.1445794
Received: 08 June 2024; Accepted: 08 October 2024;
Published: 12 November 2024.
Edited by:
Ning Liu, Shanghai Ocean University, ChinaReviewed by:
Deobrat Dixit, Columbia University, United StatesJiexin Li, Sun Yat-sen University, China
Copyright © 2024 Kaur, Sharma, Bhatia and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minu Singh, bWVlbnVzaW5naC5kckBnbWFpbC5jb20=
 Parminder Kaur
Parminder Kaur Pankaj Sharma
Pankaj Sharma Prateek Bhatia
Prateek Bhatia Minu Singh
Minu Singh