- School of Basic Medicine, Yangtze University, Health Science Center, Yangtze University, Jingzhou, Hubei, China
As global population ageing accelerates, cancer emerges as a predominant cause of mortality. Long non-coding RNAs (lncRNAs) play crucial roles in cancer cell growth and death, given their involvement in regulating downstream gene expression levels and numerous cellular processes. Cell death, especially non-apoptotic regulated cell death (RCD), such as ferroptosis, pyroptosis and necroptosis, significantly impacts cancer proliferation, invasion and metastasis. Understanding the interplay between lncRNAs and the diverse forms of cell death in cancer is imperative. Modulating lncRNA expression can regulate cancer onset and progression, offering promising therapeutic avenues. This review discusses the mechanisms by which lncRNAs modulate non-apoptotic RCDs in cancer, highlighting their potential as biomarkers for various cancer types. Elucidating the role of lncRNAs in cell death pathways provides valuable insights for personalised cancer interventions.
1 Introduction
Cell death can be classified into two groups based on its rate and susceptibility to influence by drugs or genes: accidental cell death and regulated cell death (RCD) (1, 2). Accidental cell death arises from biological processes, whereas RCD is orchestrated by signalling pathways and underlying mechanisms, maintaining homeostasis and influencing the development of various diseases. RCD encompasses apoptotic and non-apoptotic subsets, each with unique signalling induction and molecular regulatory characteristics, as well as implications for disease (3). Currently, non-apoptotic RCDs, including ferroptosis, pyroptosis and necroptosis, plays pivotal roles in cancer progression (4). Ferroptosis, an iron-dependent type of cell death, is characterised by necrotic changes in cells such as cell swelling and plasma membrane rupture, stemming from lipid hydroperoxide accumulation (5). Ferroptosis is widely acknowledged as a critical process influencing the development and advancement of various cancers (6). Pyroptosis, an inflammatory form of cell death, involves cell swelling, lysis, release of pro-inflammatory mediators, ATP production and expression of high mobility group box 1, among other features (7). Necroptosis, another form of cell death, shares morphological similarities with necrosis, such as translucent cytoplasm and organelle swelling, which can be triggered by various mechanisms (8). Cancer is a complex disease that involves the dysregulation of cell death. In cancer initiation and progression, cell death is regulated by various molecules, including long non-coding RNAs (lncRNAs).
LncRNAs, ranging from 200 to 10,000 nucleotides, lack a full open reading frame (ORF) and seldom produce small functional peptides. They are typically found in the nucleus or cytoplasm (9). LncRNAs participate in essential physiological processes such as cell cycle, tissue differentiation, metabolism and immunity (10–12). Their abnormal expression or dysfunction is frequently associated with various human diseases, including cancer (13). Previous studies have demonstrated that lncRNAs possess tissue-specific, cell-type-specific and cell developmental stage-specific properties (14–17). Moreover, the dysregulation of lncRNAs has been demonstrated to be associated with various cancer-related phenotypes, including malignancy proliferation, epithelial-mesenchymal transition (EMT), invasion and metastasis. Furthermore, lncRNAs modulate cancer progression through influencing various non-apoptotic RCDs signalling pathways.
Recent scholarly reviews have highlighted that abnormally expressed ncRNAs exert a essential role on RCDs in the progression of multiple diseases (18–20). Although the integrated roles and mechanisms of lncRNAs in non-apoptotic RCDs remain understudied, an increasing number of studies have demonstrated that lncRNAs can mediate intracellular signalling pathways, subsequently influencing physiological and pathological processes such as cancer progression (21). Herein, this comprehensive review elucidates the diverse functions and underlying molecular mechanisms of lncRNAs in modulating non-apoptotic RCDs processes during the initiation and progression of various cancers. Moreover, we also highlights the potential of these lncRNAs in cancer diagnosis and therapeutics, which have great significance for translational medicine and clinical practice.
2 LncRNAs in cancers
LncRNAs are transcribed molecules longer than 200 nucleotides, which can be located in either the same or opposite direction to protein-coding genes, or within regions between genes (22). They play crucial roles in gene regulation, including the modulation of gene activation and silencing. LncRNAs perform diverse functions, including transcriptional regulation in cis or trans, regulation of gene expression and modulation of proteins or RNA molecules (23). Their mechanisms of action include chromosome looping, chromatin modification, transcription inhibitor/activator, miRNA sponging, protein interaction and translation modulation (Figure 1). While the majority of lncRNAs are localised in the nucleus, some also function in the cytoplasm (24). LncRNAs are transcribed by RNA polymerase II, and some of these transcripts are often polyadenylated or 7-methylguanosine capped and spliced (25–27). Typically, lncRNA functions are divided into cis and trans, including the target is near the genomic location of the lncRNA or is located on other chromosomal loci (28, 29). Within the nucleus, lncRNAs mainly regulate gene expression through interactions with transcription factors (TFs), resulting in various epigenetic chromatin modifications or DNA architecture alterations (chromosome looping) (30). In the cytoplasm, lncRNAs predominantly govern gene expression post-transcriptionally, coordinating various RNA or protein modifications to influence their activation and stability (31–33). Moreover, they can impact the stability and translation of mRNA, as well as mRNA decay, through the formation of regions with complementary sequences to mRNA (34, 35). Additionally, lncRNAs function as ‘sponges’ or ‘decoys’, competing with other genes for miRNA binding and consequently diminishing the regulatory impact of miRNAs on targeted mRNAs (36). Moreover, recent studies suggest that the translation of lncRNAs is initiated by ORFs, with certain lncRNAs carrying out their functions through their coding peptides (37). Furthermore, the dysregulation of lncRNAs plays multifaceted roles in diseases, including cancer.
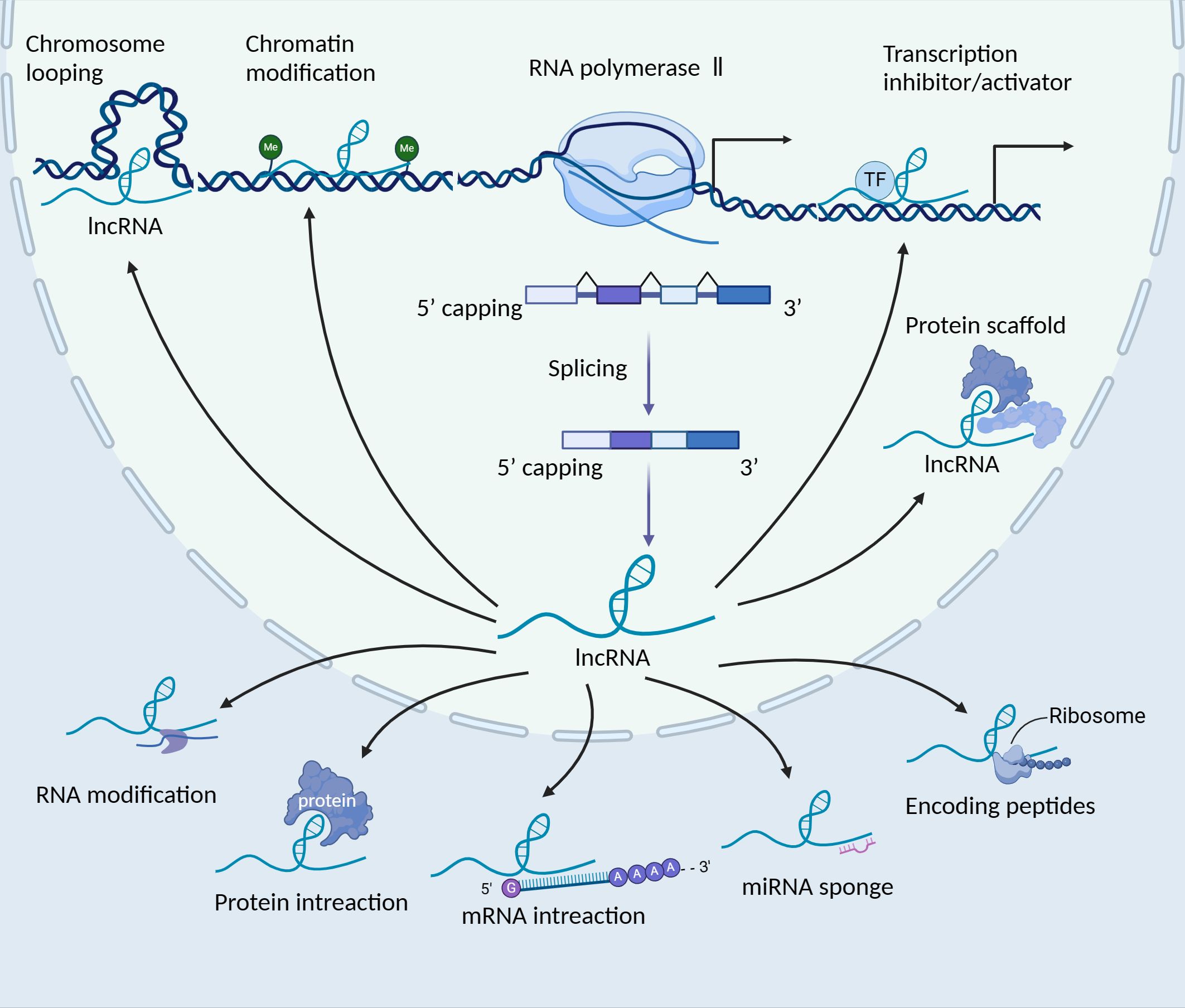
Figure 1. The biogenesis and function of long non-coding RNAs (lncRNAs). LncRNAs are transcribed by RNA polymerase II, localising in the nucleus or cytoplasm. In the nucleus, lncRNAs play various roles, such as the regulation of gene expression in cis or trans, splicing regulation and the formation of subnuclear domains. Certain lncRNAs, which possess 3’ cleavage and polyadenylation features resembling the consensus sequence of mRNAs, can be transported to the cytoplasm. In the cytoplasm, these lncRNAs serve various functions, such as acting as sponges for miRNAs, interacting with signalling proteins, modulating the translation of specific mRNAs and even encoding peptides. TF, transcription factor; lncRNA, long non-coding RNA.
Cancer is a complex disease characterised by genetic mutations, epigenetic alterations, chromosomal translocations, deletions and amplification (38, 39). In addition to mutations or abnormal expression in protein-coding genes, mutations and dysregulation of non-coding RNAs, specifically lncRNAs, are increasingly recognised for their pivotal roles in cancer (40). LncRNAs drive the acquisition of hallmark cancer characteristics, including proliferation, survival, metabolism modulation and interactions with the tumour microenvironment (TME). Recent studies have demonstrated the roles of various lncRNAs in the TME, which is a complex ecosystem involving diverse immune cells, cancer-associated fibroblasts (CAFs), endothelial cells, and the extracellular matrix (ECM), among others (41–44). It have illustrated that the roles of lncRNAs within the TME which is involved in diverse molecular mechanisms including interaction with DNAs, RNAs and proteins. Early indications of lncRNA involvement in cancer came from their transcriptional regulation by prominent oncogenic or tumour-suppressive TFs such as p53, MYC and various signalling pathways, shaping oncogenic or tumour-suppressive responses (45). Previous research has highlighted lncRNA’s participation in multiple signalling pathways, such as Wnt/β-catenin, TGF-β/Smad, STAT3 and VEGFA/VEGFR2, consequently influencing tumour invasion and metastasis, tumour angiogenesis (46–49). While the precise mechanisms through which lncRNAs participate in tumour pathogenesis remain elusive, mounting evidence suggests their key regulatory role in ferroptosis, pyroptosis and necroptosis (50, 51). Consequently, dysregulation of specific lncRNAs may exhibit anti-tumour or pro-carcinogenic functions, impacting various forms of cancer cell death.
3 Ferroptosis, pyroptosis and necroptosis
Research has unveiled significant crosstalk among initiators, executors and implementers of ferroptosis, pyroptosis and necroptosis (52). These three forms of non-apoptotic RCDs are extensively studied, each characterised by distinct molecular traits (53). This section offers a concise overview of the mechanisms and roles of these key forms of non-apoptotic RCDs (Figure 2).
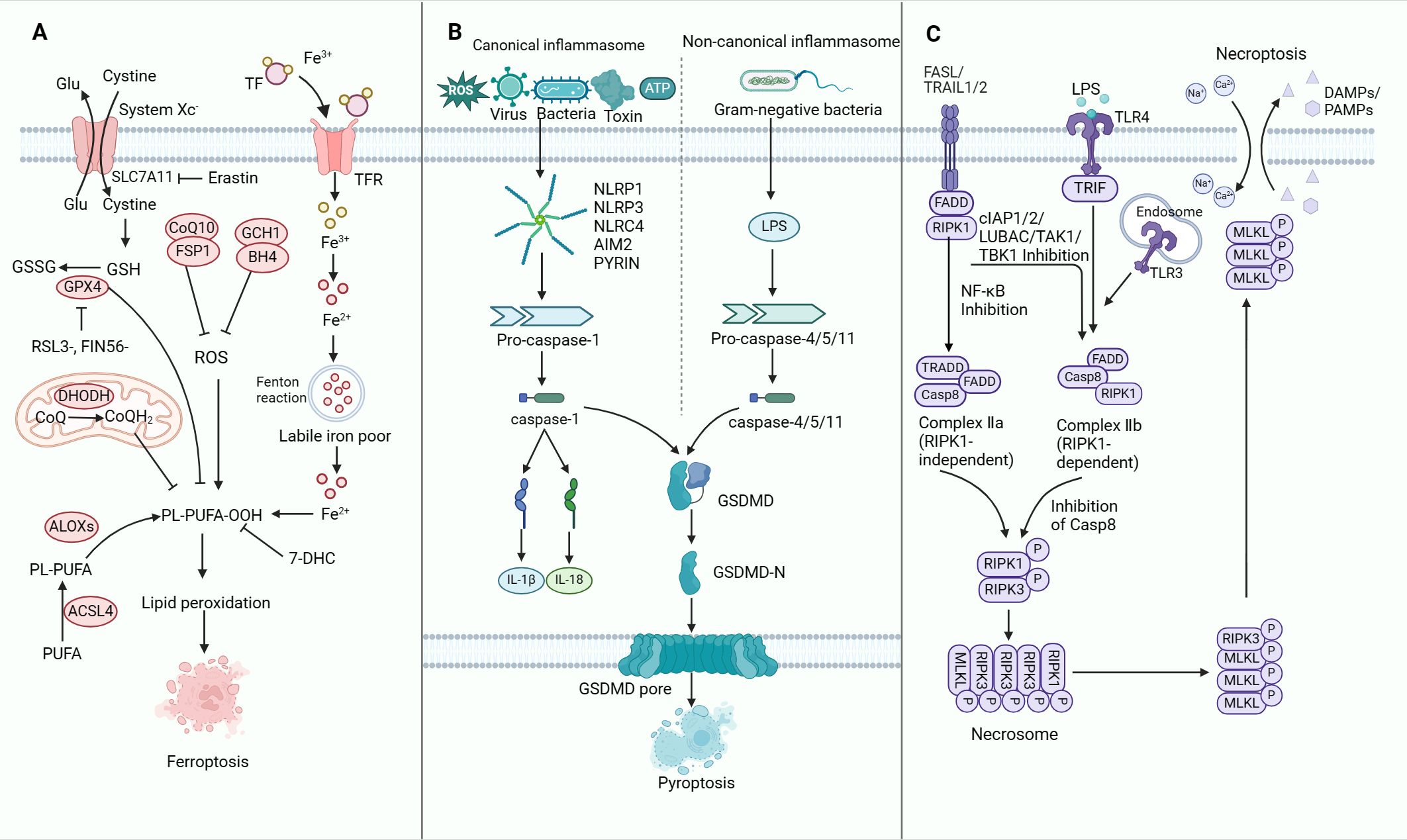
Figure 2. The molecular mechanisms of ferroptosis, pyroptosis and necroptosis. (A) Mechanisms of ferroptosis. The synthesis of PUFA, production of intracellular ROS, upregulation of iron levels and inhibition of the GPX4-dependent defence system collectively promote ferroptosis in tumour cells. (B) Mechanisms of pyroptosis. Upon encountering PAMPs or DAMPs, activated CASP1 or CASP11 triggers the cleavage and generation of GSDMD-N. This process plays a central role in driving pyroptosis through the activation of both canonical and non-canonical inflammasomes. The induction of pyroptosis is further modulated by signalling pathways involving ROS as well as calcium and potassium efflux. Inhibition of pyroptosis is mediated by GPX4, which counteracts ROS-mediated pyroptosis. Notably, the gasdermin-N pore-forming domains and the gasdermin-C repressor domains are distinctly separated. The gasdermin-N pore-forming domains subsequently undergo oligomerisation to form pores within the cell membrane, resulting in the disruption of membrane integrity and the initiation of cell pyroptosis. (C) Mechanisms of necroptosis. Activation of the death receptor results in the activation of RIPK1 and recruitment of intracellular RIPK3 to form a complex called the necrosome. Subsequently, RIPK3 is activated. Activated RIPK3 then phosphorylates MLKL, which is co-trafficked with tight junction proteins to the cell periphery, where it binds to the plasma membrane. The binding of MLKL at the plasma membrane triggers cell necroptosis. TFR, transferrin receptor; ROS, reactive oxygen species; PUFA, polyunsaturated fatty acid; GSDMD, gasdermin D; LPS, lipopolysaccharide; TLR, toll-like receptor; DAMPs, damage-associated molecular patterns; PAMPs, pathogen-associated molecular patterns; RIPK, receptor-interacting serine-threonine kinase; MLKL, mixed lineage kinase domain-like.
3.1 Ferroptosis
Ferroptosis is a unique iron-dependent and oxidative form of cell death (6). Compared to other types of cell death, ferroptosis exhibits marked variations in genetic, biochemical, morphological and metabolic traits (1, 54). Notably, ferroptosis can rapidly propagate within cell populations in a wave-like fashion. Morphologically, cells undergoing ferroptosis display mitochondrial abnormalities including swelling, altered density and rupture of the outer membrane. Uncontrolled lipid peroxidation, driven by iron and Fenton-like reactions, disrupts lipid membranes, a hallmark of ferroptosis. Ultimately, the pathways related to iron, glutathione (GSH) and lipid metabolism intersect to regulate ferroptosis execution (55).
Several cellular processes regulate ferroptosis (Figure 2A). Ferroptosis is triggered by the excessive oxidative damage (peroxidation) of lipids in the cell membranes, dependent on iron, reactive oxygen species (ROS) and phospholipids containing polyunsaturated fatty acids (PUFAs) (56–58). Intracellular PUFAs are enzymatically converted into PUFA-phospholipid-peroxides (PUFA-PL-OOH), leading to lipid peroxide buildup within cellular membranes (59, 60). Iron-mediated lipid peroxidation is regulated by four antioxidant pathways linked to ferroptosis: System Xc–GPX4, FSP1-CoQ10, DHODH/CoQH2 and GCH1-BH4. The antiporter System Xc-, encompassing SLC7A11 and SLC3A2, facilitates cystine absorption, essential for GSH production (61–65). Notably, erastin, a common ferroptosis inducer, can suppress the expression of SLC7A11, leading to the dysfunction of System Xc-. This leads to inhibited cystine uptake, reduced GSH production and ultimately ferroptosis activation (66). Excessive iron, stored as ferritin is released upon cellular stimulation and undergoes degradation, resulting in Fe3+ release in large quantities. This Fe3+ is then converted to Fe2+ by the transferrin receptor-1 (TFR1) and ultimately discharged into the cytoplasmic iron pool. Consequently, ROS levels increase, leading to detrimental effects (67–69). GPX4 assumes a crucial role in safeguarding against ferroptosis by reducing ROS cellular levels and proficiently mending lipid oxidation-induced cellular damage. Nevertheless, GPX4 inactivation due to GSH depletion and direct inhibition via RSL3 or FIN56 comprises the antioxidant capacity and subsequent lipid ROS overproduction, consequently triggering ferroptosis through uncontrolled lipid peroxidation (70–72). Ferroptosis can be initiated by disrupting the System Xc- cystine/glutamate antiporter or GPX4, resulting in impaired redox balance within the GSH system. Additionally, ferroptosis suppressor protein 1 (FSP1) serves as another inhibitor of ferroptosis (73, 74). The FSP1-CoQ10-NAD(P)H pathway operates independently of GPX4 and GSH to suppress phospholipid peroxidation and ferroptosis (61). In the DHODH/CoQH2 pathway, DHODH functions concomitantly with mitochondrial GPX4 (yet not aligned with cytosolic GPX4 or FSP1) in hindering ferroptosis within the inner mitochondrial membrane. This inhibition is achieved through the reduction of ubiquinone to ubiquinol, which serves as a radical-trapping antioxidant possessing anti-ferroptosis properties (63). In another ferroptosis pathway, GTP cyclohydrolase-1 (GCH1) serves as the primary enzyme regulating the production of BH4. Furthermore, by genetically or pharmacologically inhibiting GCH1, levels of BH4 can be depleted, thereby facilitating erastin-induced cell death, enhancing lipid peroxidation and promoting ferrous iron accumulation, ultimately resulting in ferroptosis (75). A recent research found that 7-DHC attenuates ferroptosis by diverting the peroxidation pathway from phospholipids and protects cells from phospholipid peroxidation at the cell membrane and mitochondria. However, high levels of 7-DHC also lead to more aggressive cancer manifestations and promote cancer metastasis (76, 77). Notably, ferroptosis governs the growth and proliferation of various tumour cell types, including lymphocytoma, pancreatic ductal cell carcinoma, renal cell carcinoma and hepatocellular carcinoma (HCC) (78–81).
3.2 Pyroptosis
Pyroptosis, a programmed inflammatory cell death mechanism, involves the cleavage and activation of gasdermin, resulting in compromised cell membrane integrity and the activation of inflammasome sensors, releasing cellular proteins as danger signals (82, 83). Despite certain similarities with apoptosis, such as DNA damage and chromatin state changes, pyroptosis exhibits distinct morphological traits (82, 84). Cells undergoing pyroptosis exhibit swelling and bubble-shaped protrusions on the cellular membrane before eventual rupture, contrasting with the regulated and non-inflammatory nature of apoptosis (85). Notably, pyroptosis elicits inflammation in response to various extrinsic and intrinsic stimuli, including microbial pathogens, toxins and chemotherapeutic agents (86). Compared to the abrupt rupture seen in necrosis, pyroptosis results in cytoplasmic flattening due to the leakage of the plasma membrane (7).
Previously, researchers speculated that pyroptosis arises as a response to bacterial infection, which was primarily mediated by caspase-1 in monocytes. However, recently, caspase-11/4/5 has been demonstrated to play a role in sensing intracellular lipopolysaccharide (LPS) and expanding the range of pyroptosis mediators. This finding indicates that pyroptosis is not specific to a particular cell type (87). Recent research has revealed that gasdermin D (GSDMD), a substrate for caspase-1 and caspase-11/4/5, acts as the executioner of pyroptosis by forming membrane pores (83, 85). During pyroptosis, the pore-forming domains of gasdermin-N are separated from the repressor domains of gasdermin-C. The gasdermin-N domains subsequently oligomerise and create pores in the cell membrane, resulting in compromised membrane integrity and the induction of cell pyroptosis (7, 88).
Pyroptotic cells are orchestrated through two primary molecular pathways: the classical Caspase-1-dependent route and the non-Caspase-1-dependent pathway. Caspase 1 governs the canonical pyroptosis pathway, while caspases 4, 5 and 11 mediate the non-canonical pyroptosis pathway (Figure 2B). In the canonical pathway, various stimuli such as viruses, bacteria, toxins, ATP or ROS, which are known as pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), trigger the activation of specific inflammasome sensors, including NOD-like receptor family pyrin domain-containing 1B (NLRP1b), NOD-like receptor family pyrin domain-containing 3 (NLRP3), NOD-like receptor family CARD domain-containing protein 4 (NLRC4), AIM2 and pyrin (89–91). Upon activation, the sensors of the inflammasome engage in direct or indirect interactions that activate caspase-1. Subsequently, caspase-1 cleaves the N-terminal end of GSDMD, thereby forming pores on the cell membrane (87). This process ultimately culminates in the release of cytoplasmic contents and the onset of cellular pyroptosis. Furthermore, caspase-1 catalyses the dissociation of pro-IL-1β and pro-IL-18, resulting in mature IL-1β and IL-18 (92). In the non-canonical pathway, the LPS derived from Gram-negative bacteria interacts with either CASP11 or CASP4/5, leading to the activation of the inflammasome (93). Subsequently, the inflammasome activation induces the cleavage and generation of the N-terminal fragment of GSDMD (GSDMD-N), which facilitates pyroptotic cell death by forming pores on the plasma membrane through its pore-forming activity (94–97). Pyroptosis is considered pro-inflammatory due to its ability to release DAMPs from expired cells and promote the maturation and secretion of interleukin-1 family members, such as IL-1β and IL-18, through inflammasome activation (98). A novel discovery has shown that GSDMD activation during pyroptosis could be spatiotemporally modulated by a palmitoylation-depalmitoylation relay model (99). Some studies have demonstrated that caspase-3 can modulate and activate GSDME, leading to pyroptosis (86). Recent research has found that USP48 can facilitate pyroptosis by stabilising GSDME and sensitising cancer cells to pyroptosis (100). Current studies have uncovered that pyroptosis is linked with the proliferation and migration of multiple cancers, including colon cancer, osteosarcoma, head and neck squamous cell carcinoma and breast cancer (101–104).
3.3 Necroptosis
Necroptosis is another type of immunogenic cell death (ICD), which is a caspase-independent RCD process triggered by infection. It is characterised by the phosphorylation of pseudokinase mixed lineage kinase domain-like (MLKL) by receptor-interacting serine-threonine kinase 3 (RIPK3) (105–107). Morphologically, necroptosis involves organelle swelling, plasma membrane rupture, cytoplasm and nucleus breakdown, and the release of intracellular components known as DAMPs, which propagate secondary inflammation. This pro-inflammatory nature suggests that necroptosis may have evolved as an innate immune process that supplements apoptosis in pathogen clearance (74, 108, 109). Under conditions of insufficient apoptosis, necroptosis is regulated by distinct death receptors (DRs) like FAS and tumour necrosis factor receptor 1 (TNFR1) or pattern recognition receptors (PRRs) such as toll-like receptor 3 (TLR3), which sense detrimental cues from the intracellular and extracellular environments to trigger necroptosis (110–112) (Figure 2C).
In the necroptosis process, death-inducing molecules such as FasL and TRAIL stimulate necrosome complex assembly. NF-κB inhibition induces Complex II a (RIPK1-independent) formation, which is composed of TRADD, FADD and caspase-8 (113). Moreover, the induction of complex IIb, which comprises RIPK1, FADD and caspase-8, is observed when RIPK1 ubiquitination is inhibited or when cytotoxicity induces phosphorylation, which indicates that the initial stages of TNF signalling are hindered. Inhibition of caspase-8 leads to necrosome complex formation through RIPK1 and RIPK3 homotypic interactions, involving the RIP homotypic interaction motif and consequently activating MLKL through phosphorylation (109). Intracellular adaptor molecules, such as FADD, recruit RIPK1 and subsequently RIPK3, forming the necrosome through phosphorylation processes. The necrosome is a protein complex comprising core components such as RIPK1, RIPK3 and MLKL (105, 112, 114). The translocation of MLKL to the membrane is imperative for facilitating the influx of Ca2+, an early occurrence in TNF-induced necroptosis (115). During TNFR1 stimulation, a critical component of necroptosis, known as the necrosome, assembles. In this process, CASP8, which is typically involved in apoptosis, is suppressed. Upon TNFα stimulation, the intracellular tails of TNFR1 recruit various proteins that collectively form a signalling complex called ‘Complex I’. In general, the activation of RIPK1 kinase by TNFα, FASL (FAS ligand) and TRAIL (tumour necrosis factor-associated apoptosis-inducing ligand) initiates the formation of a complex known as inhibitors of apoptosis (cIAPs) at the cell membrane. This cascade reaction ultimately triggers the activation of NF-κB, thereby promoting cell survival (8, 116, 117). Necroptosis plays indispensable roles in various physiological functions, but dysregulation is associated with multiple human diseases (118). In addition to their essential roles in physiological processes, molecules associated with necroptosis contribute to the development of various cancers including breast cancer, liver cancer, PAAD and CRC (106, 118–121). Recent findings highlight the crucial involvement of necroptosis in tumour development and metastasis, suggesting promising prospects for leveraging necroptosis in cancer treatment (8).
4 LncRNAs regulate ferroptosis, pyroptosis and necroptosis
Emerging studies uncovered that lncRNAs influenced tumour progression by regulating non-apoptotic RCDs. In the following sections, the role of lncRNA-regulated ferroptosis, pyroptosis and necroptosis in different cancer types are discussed, including lung cancer, liver cancer, gastric cancer (GC), breast cancer (BC), bladder cancer (BLCA), colorectal cancer (CRC), endometrial cancer (EC), ovarian cancer (OC), pancreatic adenocarcinoma (PAAD), prostatic cancer (PCa), oral squamous cell carcinoma (OSCC) and glioma (Figure 3, Table 1).
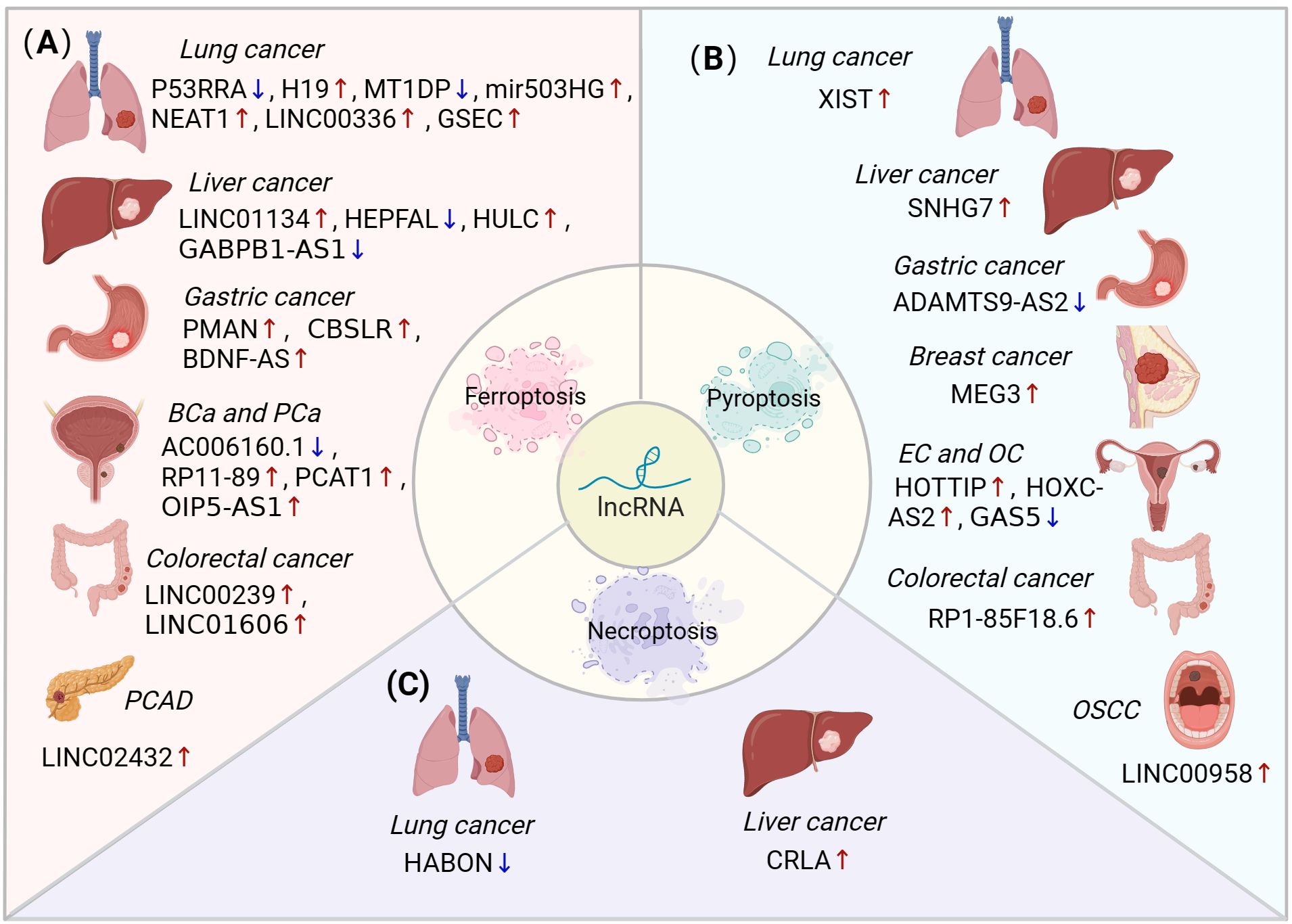
Figure 3. The role of lncRNA-mediated ferroptosis, pyroptosis and necroptosis in different cancer types. (A) LncRNAs can regulate ferroptosis to influence various types of cancers, including lung, liver, stomach, bladder, prostate, colorectal, and pancreatic cancers. (B) LncRNAs mediate pyroptosis in lung, liver, gastric, breast, endometrial, ovarian, colorectal, and oral squamous cell carcinomas. (C) The dysregulation of lncRNAs inhibits necroptosis in lung and liver cancers. BLCA, bladder cancer; PCa, prostatic cancer; PAAD, pancreatic adenocarcinoma; EC, endometrial cancer; OC, ovarian cancer; OSCC, oral squamous cell carcinoma.
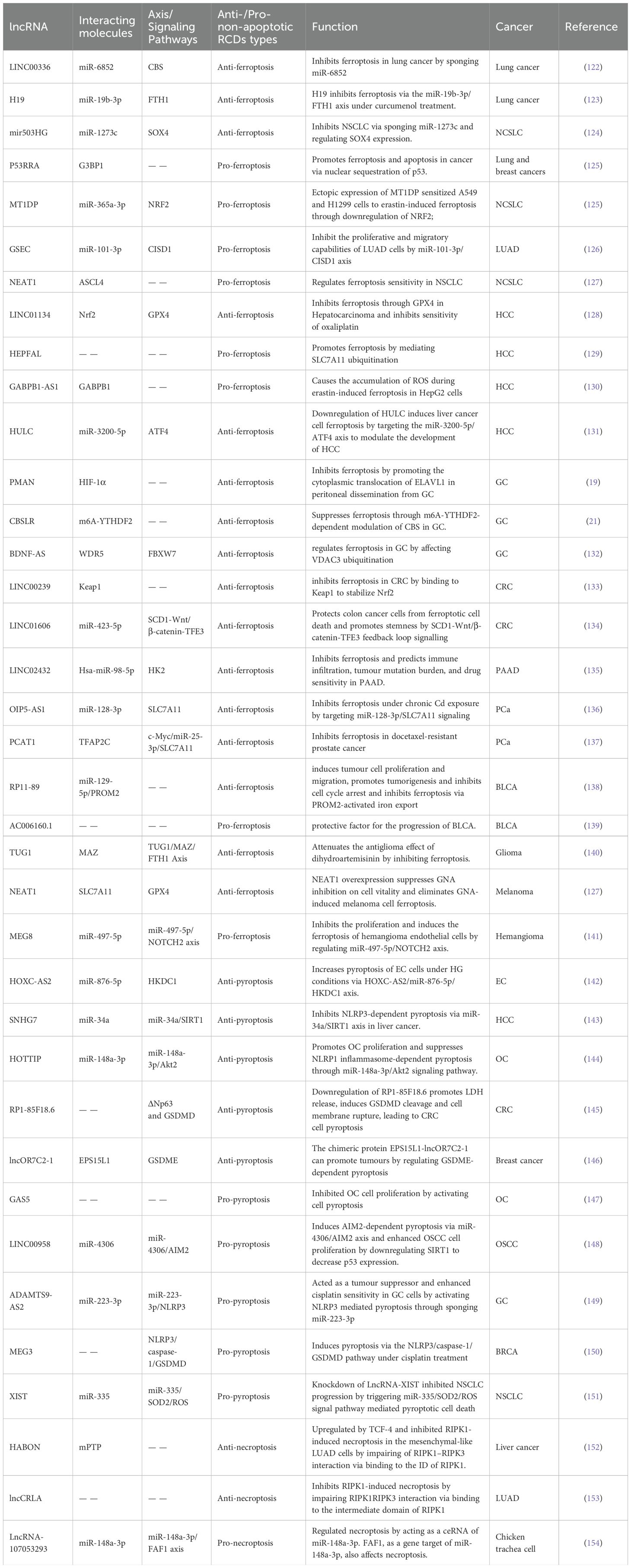
Table 1. Examples of non-apoptotic regulated cell death (RCD) regulated by long non-coding RNAs (lncRNAs) in cancers.
4.1 LncRNAs regulate ferroptosis
Ferroptosis is intricately linked to cancer progression. However, the underlying mechanisms, particularly those involving lncRNA-mediated ferroptosis in oncogenesis, remain insufficiently explored. This section focuses on the lncRNAs that regulate ferroptosis across various cancers through via diverse mechanisms (Figure 3A).
4.1.1 LncRNAs regulate ferroptosis in lung cancer
Lung cancer remains the leading cause of cancer-related mortality globally, with non-small cell lung cancer (NSCLC) representing over 80% of all cases (155). Recent studies have unveiled that lncRNAs can either promote or inhibit ferroptosis, thereby influencing lung cancer progression. Ferroptosis plays a significant role in tumorigenesis depression by selectively eliminating cells experiencing a nutrient deficiency or damage caused by environmental or infection-induced stress (156). For instance, LINC00336 acts as a competing endogenous RNA (ceRNA) to suppress ferroptosis in lung cancer. The inactivation of p53 by lymphoid-specific helicase leads to the induction of ELAV-like RNA binding protein 1 expression, which, in turn, enhances the levels of LINC00336 through transcriptional regulation. Subsequently, LINC00336 acts as a ceRNA by sequestering miR-6852, ultimately resulting in increased CBS mRNA levels. This molecular cascade promotes cell proliferation and tumour growth, while concurrently inhibiting ferroptosis in lung cancer (122). As another example, lncRNA H19 counteracts the anticancer properties of curcumenol via the miR-19b-3p/FTH1 axis in lung cancer. H19 suppresses lipid ROS generation while promoting GSH production, thereby impeding ferroptosis (123). The reduction in lncRNA MIR503HG levels, influenced by XAV939, may suppress NSCLC by serving as a sponge for miR-1273c and controlling SOX4 expression. Additionally, the decrease in SLC7A11 expression, which is triggered by XAV939, may hinder NSCLC progression through the ferroptosis pathway (124).
Moreover, lncRNA can also facilitate ferroptosis in lung cancer. For example, the tumour-suppressive lncRNA P53RRA, also referred to as LINC00472, is downregulated in various cancers, such as lung, liver, breast and colon cancers. In lung cancer, P53RRA interacts with Ras GTPase-activating protein-binding protein 1 (G3BP1) within the cytosol, resulting in the displacement of p53 from the G3BP1 complex. As a result, p53 is localised to the nucleus, resulting in cell cycle arrest, apoptosis activation and ferroptosis promotion. P53RRA facilitates ferroptosis by influencing the transcription of various metabolic genes, including the inhibition of SCL7A11. Furthermore, P53RRA enhances erastin-induced ferroptosis and increases lipid ROS and iron levels in cells (125). A recent study reported a new approach wherein targeting the MT1DP/miR-365a-3p/NRF2 pathway facilitated erastin-induced ferroptosis. The use of erastin/MT1DP@FA-LPs (E/M@FA-LPs) effectively intensified ferroptosis, leading to reduced cellular GSH levels and increased lipid ROS. Moreover, the ectopic expression of the metallothionein 1D pseudogene (MT1DP) enhanced the susceptibility of A549 and H1299 cells to erastin-induced ferroptosis by suppressing NRF2. Additionally, MT1DP overexpression resulted in elevated levels of malondialdehyde (MDA) and ROS. Notably, in instances of cancer cells being exposed to erastin, an evident increase in intracellular ferrous iron levels has been noted along with a decline in GSH concentrations. Conversely, downregulating MT1DP demonstrated the opposite effect (126). In lung adenocarcinoma (LUAD), ferroptosis-related lncRNA GSEC expression was increased, while miR-101-3p expression was decreased. Moreover, GSEC can influence ferroptosis by sequestering miR-101-3p, thereby influencing LUAD progression (157). Another study investigated the involvement of nuclear-enriched transcript 1 (NEAT1) has been reported to influence ferroptosis, exhibiting susceptibility to erastin-induced ferroptosis (127).
4.1.2 LncRNAs regulate ferroptosis in liver cancer
Liver cancer is the leading cause of death from malignancies globally, underscoring the urgent need for new treatment options for patients (158, 159). Numerous studies have shown that lncRNAs can either promote or inhibit ferroptosis in liver cancer through various mechanisms, including transcriptional regulation, protein modification and sequestering miRNA. For instance, Kang et al. revealed that LINC01134 suppresses ferroptosis by facilitating Nrf2 protein recruitment to the promoter region of the GPX4 gene, thereby promoting GPX4 transcription and enhancing liver cancer resistance to OXA (128). Another study indicated that lncRNA HEPFAL contributes to ferroptosis in hepatoma cells by facilitating the ubiquitination of SLC7A11. Thereby, HEPFAL emerges as a promising candidate for the diagnosis and therapeutic intervention of HCC (129). A recent study has revealed that URB1-AS1 attenuates sorafenib-induced ferroptosis by facilitating ferritin phase separation and decreasing the intracellular free iron level. Moreover, it was discovered that specifically silencing the expression of URB1-AS1 with N-acetylgalactosamine (GalNAc)-small interfering URB1-AS1 effectively potentiated the sensitivity of HCC cells to sorafenib in an in vivo tumour model. This suggests that URB1-AS1 targeting may represent a potential therapeutic approach to overcome sorafenib resistance in HCC (160).
Erastin-induced upregulation of lncRNA GABPB1-AS1 promotes the formation of RNA duplexes with GABPB1 mRNA, leading to the inhibition of GABPB1 translation. This inhibition ultimately results in reduced expression of PRDX5, leading to ROS accumulation. Therefore, GABPB1-AS1 lncRNA could potentially contribute significantly to erastin-induced ferroptosis in HCC (130). Another study found that liver cancer cell ferroptosis can be induced by suppressing HULC, mediated by the miR-3200-5p/ATF4 axis. This regulatory pathway emerges as a crucial determinant in the pathogenesis of HCC (131).
4.1.3 LncRNAs regulate ferroptosis in gastrointestinal tumours
GC, the fifth most prevalent cancer worldwide, is the third highest cause of cancer-related deaths (161, 162). LncRNAs can influence ferroptosis by interacting with different regulators, thereby catalysing the initiation and progression of gastrointestinal tumours. For example, in GC, hypoxia-induced HIF-1α and lncRNA-PMAN facilitate the cytoplasmic translocation of ELAVL1 during peritoneal dissemination. This orchestration effectively inhibits ferroptosis, as PMAN, upregulated by HIF-1α, stabilises SLC7A11 mRNA through ELAVL1-mediated cytoplasmic distribution. The resulting accumulation of SLC7A11 elevates l-GSH levels, inhibiting ROS and iron accumulation, thereby shielding GC cells from ferroptosis induced by agents like erastin and RSL3 (19). Additionally, hypoxia-induced signalling pathways, including CBSLR, CBS and ACSL4, modulate the metabolism of PUFAs, conferring resistance to ferroptosis in GC cells. Under hypoxic conditions, CBSLR is transactivated by HIF-1α, leading to the modulation of ferroptosis in GC cells. Moreover, the increased expression of CBSLR in GC tissues correlates with an unfavourable prognosis and reduced responsiveness to chemotherapy, underscoring its potential as a prognostic biomarker and a determinant of chemotherapy efficacy (21). As another example, the BDNF-AS/WDR5/FBXW7 axis regulates ferroptosis in GC by influencing the ubiquitination of VDAC3. Furthermore, BDNF-AS demonstrates promising prospects as both a prognostic biomarker and a therapeutic target in GC (132).
In CRC, which ranks as the second leading cause of cancer-related deaths globally, lncRNAs like LINC00239 fosters CRC cell proliferation by interacting with Keap1, thereby disrupting the Keap1/Nrf2 complex and consequently bolstering Nrf2 protein stability, which results in its translocation to the nucleus (163, 164). The overexpression of LINC00239 suppresses ferroptosis by interacting with the Kelch domain (Nrf2-binding site) of Keap1, thereby blocking Nrf2 ubiquitination and augmenting its protein stability. LINC00239 promotes CRC tumorigenesis in vitro and in vivo (133). Similarly, LINC01606 inhibits ferroptosis and promotes CRC stemness via the SCD1-Wnt/β-catenin-IGHM enhancer 3 (TFE3) positive feedback loop signalling. Thus, LINC01606 emerges as a pivotal therapeutic target for CRC treatment (134).
Furthermore, LINC02432 exert an oncogenic role in PAAD by inhibiting ferroptosis through the miR-98-5p/HK2 axis. Moreover, its potential as a predictive marker for immune infiltration, drug responsiveness and tumour mutation burden underscores its significance. Further research is warranted to elucidate the precise mechanism underlying LINC02432 impact on PAAD prognosis (135).
4.1.4 LncRNAs regulate ferroptosis in other cancers
In other cancers, lncRNAs also play crucial roles in initiating and progressing tumours by affecting ferroptosis. For instance, in PCa cells exposed to chronic Cd, the expression of lncRNA OIP5-AS1 is significantly elevated. OIP5-AS1 enhances cell proliferation and inhibits ferroptosis upon chronic Cd exposure by modulating the miR-128-3p/SLC7A11 pathway. Furthermore, targeting OIP5-AS1 holds promise as a viable therapeutic approach for managing Cd-induced progression of PCa (136). Another example is TFAP2C-regulated lncRNA PCAT1, which suppresses ferroptosis in docetaxel-resistant PCa via c-Myc/miR-25-3p/SLC7A11 pathway (137).
In BLCA, the lncRNA RP11-89 exerts an inhibitory effect on ferroptosis by facilitating PROM2-mediated iron export through its interaction with miR-129-5p. Moreover, the miR-129-5p/PROM2 axis is utilised by RP11-89 to promote tumour cell proliferation and migration, boost tumorigenesis and hinder cell cycle arrest. RP11-89 upregulates PROM2 expression and functions as a ceRNA targeting miR-129-5p (138). Additionally, the lncRNA AC006160.1 is a ferroptosis-related lncRNA that demonstrates significant potential in accurately predicting survival outcomes, clinical stages, tumour grades, immune cell infiltration and immune checkpoint expression in BLCA. Moreover, AC006160.1 exhibits a protective role in hindering BLCA progression (139).
In glioma, the lncRNA TUG1 undermines the antiglioma efficacy of dihydroartemisinin (DHA) by suppressing ferroptosis through the MAZ/FTH1 axis. Modulating TUG1 expression or inhibiting FTH1 can augment the antiglioma effects of DHA, presenting a promising strategy to enhance DHA’s effectiveness against glioma (140). As another example, SNAI3-AS1 interacted with SND1 in a competitive manner, disrupting the m6A-dependent recognition of Nrf2 mRNA 3’UTR by SND1, thereby reducing the mRNA stability of Nrf2. This, in turn, enhanced the anti-tumour efficacy of erastin in vitro and in vivo by facilitating ferroptosis, providing a theoretical basis for inducing ferroptosis to improve the treatment of glioma (165). Furthermore, the downregulation of NEAT1 inhibits the direct interaction between SLC7A11, indirectly inhibiting GPX-4 activity and facilitating ferroptosis. Moreover, NEAT1 overexpression counteracts GNA-mediated suppression of cell viability and prevents GNA-induced ferroptosis in melanoma cells (166). Another study investigated the impact of lncRNA MEG8 downregulation on haemangioma endothelial cells and found that decreased expression of lncRNA MEG8 inhibited cell proliferation and stimulated the occurrence of ferroptosis. Additionally, they observed that these effects were mediated through the modulation of the miR-497-5p/NOTCH2 signal pathway (141).
In conclusion, lncRNAs can affect the development of cancers by regulating the ferroptosis inducer ROS, the key regulatory factor Nrf2, SLC7A11, and the NF-κB signalling pathway. Therefore, an in-depth exploration of the molecular mechanism of lncRNA-regulated ferroptosis in tumours will illuminate new insight on the prevention and treatment of cancers.
4.2 LncRNAs regulate pyroptosis
Pyroptosis can suppress tumour initiation and progression, it also has the potential to create a microenvironment that favours tumour growth. LncRNAs can influence the initiation and progression of various cancers by interacting with various regulators to modulate pyroptosis (Figure 3B). This section describes the signalling pathways associated with lncRNA-mediated pyroptosis, which may represent a potential approach for regulating pyroptosis in cancers.
4.2.1 LncRNAs regulate pyroptosis in gastrointestinal tumours
In CRC, silencing of lncRNA RP1-85F18.6 facilitates the induction of pyroptosis in CRC cells by escalating the discharge of LDH, promoting the fragmentation of GSDMD and disrupting the cell membrane, thereby impeding CRC cell proliferation and invasion. Additionally, lncRNA RP1-85F18.6 can inhibit GSDMD activity by enhancing ΔNp63 expression, thereby inhibiting the pyroptosis of CRC cells and facilitating the progression of cancer (145). The upregulation of ADAMTS9-AS2 suppresses GC advancement and enhances the sensitivity of cisplatin-resistant GC cells to cisplatin by mediating the miR-223-3p/NLRP3 axis, thereby inducing pyroptosis (149).
4.2.2 LncRNAs regulate pyroptosis in OC
It has been demonstrated that HOTTIP is highly expressed in OC tissue samples and cell lines. The inhibition of HOTTIP inhibits the progression of OC and the initiation of pyroptosis regulated by the NLRP1 inflammasome through the miR-148a-3p/Akt2 signalling pathway (144). Likewise, lncRNA growth arrest-specific transcript 5 (GAS5) induces the formation of the inflammasome and promotes the inflammatory process by interfering with the glucocorticoid receptor. LncRNA GAS5 acts as a tumour suppressor and could be used as a potential therapeutic target for the diagnosis and treatment of OC (147).
4.2.3 LncRNAs regulate pyroptosis in other cancers
In triple-negative breast cancer (TNBC), the inhibition of MEG3 not only partially mitigated the stimulatory effect of DDP on the NLRP3/caspase-1/GSDMD-mediated pyroptosis pathways, but also counteracted DDP’s inhibitory effect on tumour growth and metastasis, highlighting new avenues for the development of innovative therapeutic approaches for TNBC (150). HOXC‐AS2, a ceRNA, promotes pyroptosis and glycolysis through the miR‐876‐5p/HKDC1 signalling pathway, thereby stimulating the generation of inflammatory factors and the release of lactic acid into the TME, ultimately enhancing the proliferation and migration of EC cells (142). In liver cancer, lncRNA small nucleolar RNA host gene 7 (SNHG7) suppresses NLRP3-triggered pyroptosis through the miR-34a/SIRT1 pathway (143). LINC00958 also induces AIM2-dependent pyroptosis via the miR-4306/AIM2 axis and enhances OSCC cell growth by reducing SIRT1 levels, resulting in decreased p53 expression (148). Silencing of lncRNA-XIST hinders the proliferation of NSCLC via triggering the miR-335/SOD2/ROS axis-regulated cell pyroptosis (151). Moreover, emerging research indicates a potential correlation between the fusion of mRNA-lncRNA, leading to the production of the chimeric protein EPS15L1-lncOR7C2-1, which facilitates tumour growth by potentially controlling GSDME-related pyroptosis. These findings underscore the significance of lncRNA fusions in the regulation of tumour immunity via pyroptosis (146). Recent research has substantially refined our knowledge of the interplay between lncRNAs and pyroptosis in various cancers, offering significant insights that could enhance clinical strategies for the diagnosis and treatment of cancers.
4.3 LncRNAs regulate necroptosis
Mounting evidence underscores the role of lncRNA in either promoting or inhibiting necroptosis, thereby influencing tumour progression. In this section, we listed some lncRNAs linked with necroptosis in cancers (Figure 3C). For instance, lncCRLA, exhibits significant upregulation mediated by TCF-4 and suppresses RIPK1-induced necroptosis in mesenchymal-like LUAD cells. By impairing the interaction between RIPK1 and RIPK3, lncCRLA effectively obstructs necroptosis (153). Furthermore, HABON, a hypoxia-activated lncRNA, exerts a suppressive effect on necroptosis in liver cancer by binding with the mitochondria-related protein VDAC1. This interaction modulates the opening of the mitochondrial permeability transition pore, thereby regulating necroptosis (152). In addition, lncRNA-107053293 inhibits necroptosis induced by ammonia in trachea cells and LMH cells via miR-148a-3p/FAF1 axis. FAF1, as a gene target of miR-148a-3p, also affects necroptosis via its interaction with the necrosis-related genes RIPK1 and RIPK3 (154). Together, lncRNAs can affect necroptosis to influence the development of cancers by interacting with the molecular effectors RIPK3 and MLKL. Although lncRNA-mediated necroptosis are still limited to molecular biology and disease model experiments, the regulatory role of lncRNA-mediated necroptosis in cancers, as well as its participation in the pathophysiological process of various cancers reflected by lncRNA, are bound to be applied and reflected in future clinical studies. The scene will be set for lncRNA-mediated necroptosis to emerge prominently through early screening of high-risk groups for tumours and diagnosis and treatment of patients.
Taken together, lncRNAs exhibit oncogenic or anti-carcinogenic effects by various mechanisms to regulate non-apoptotic RCD genes and relative pathways involved in GSH, NLRP3 and RIPK3/MLKL. The above mentioned that a large number of lncRNAs have been implicated in hallmarks of cancer by promoting or inhibiting non-apoptotic RCDs. Therefore, non-apoptotic RCD associated lncRNAs could be used as promising diagnostic biomarkers and therapeutic targets in cancer treatment.
5 Potential clinical applications of lncRNAs in non-apoptotic RCDs
Recent studies have highlighted the close association between lncRNAs and non-apoptotic RCDs processes, including ferroptosis, pyroptosis and necroptosis. These lncRNAs play pivotal roles in modulating RCD and related cell death pathways, thereby influencing cancer progression and the efficacy of clinical therapies (134, 167). Given their prevalence, functional significance, and expression specificity, there is growing interest in evaluating lncRNAs as novel biomarkers and therapeutic targets in clinical settings. In the following sections, we will delve into the involvement of lncRNAs in pathways leading to RCD across various tumour types. Additionally, we will explore the therapeutic potential of targeting lncRNAs involved in RCD as a promising strategy for cancer treatment (Figure 4).
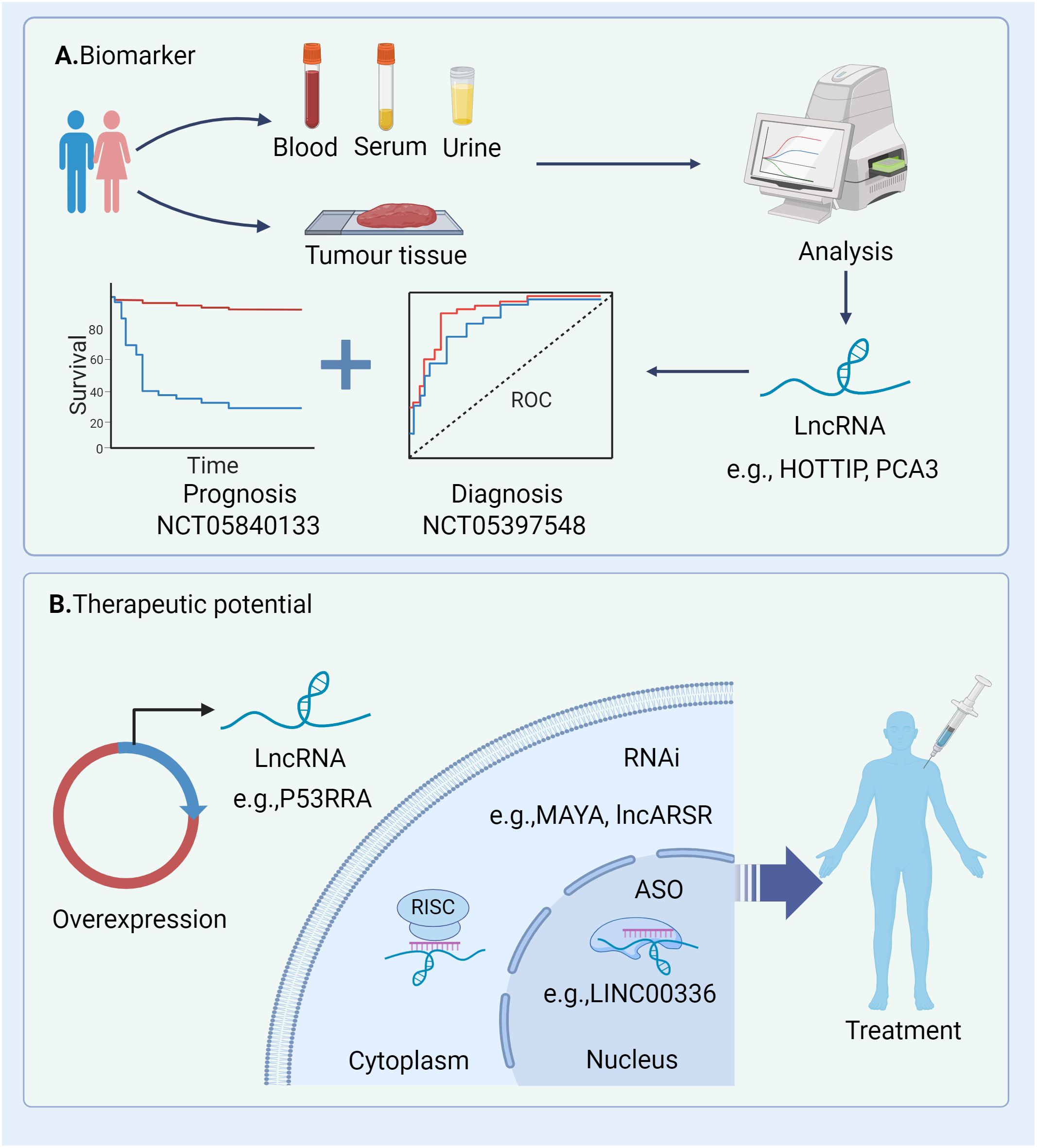
Figure 4. Potential clinical applications of lncRNAs in cancers. (A) These lncRNAs can be quantified and analysed from tumour tissue and liquid biopsy specimens (e.g. serum and urine) and thus have great potential as diagnostic and prognostic biomarkers; (B) Strategies for modulating lncRNAs within the cytoplasm and nucleus encompass knockdown and overexpression techniques. Knockdown methodologies comprise RNA interference (RNAi) and antisense oligonucleotides (ASOs). RNAi can be achieved through chemically synthesized siRNAs or pooled via lentivirally delivered short hairpin RNAs (shRNAs). RNAi, RNA interference; ASO, antisense oligonucleotides; RISC, RNA-induced silencing complex; ROC, receiver operating characteristic.
5.1 The diagnostic and prognostic value of lncRNAs
A considerable number of lncRNAs is detectable in both traditional tumour biopsies and liquid biopsies (e.g. blood, serum, and urine), presenting potential as diagnostic and prognostic biomarkers (Figure 4A). The current clinical applications of lncRNAs functioning as biomarkers in different tumours are outlined in Table 2. These lncRNA biomarkers offer distinct advantages over traditional diagnostic markers in clinical settings. For instance, lncRNA prostate cancer antigen 3 (PCA3) has been established as a non-invasive initial diagnostic indicator for prostate cancer, demonstrating reliable test characteristics and clinical utility (NCT01632930) (168). Additionally, several clinical trials are currently underway to explore the diagnostic and prognostic potential of lncRNAs in various cancer types (e.g., NCT04269746, NCT05397548, NCT04729855).
In preclinical studies, lncRNAs have shown promise as valuable biomarkers for diagnosing and predicting the prognosis of different cancer types. For example, recent research has identified C5orf66 antisense RNA 1 as a diagnostic marker for early GC, with an area under the curve (AUC) value of 0.789 (169). Combining multiple lncRNAs often enhances biomarker performance. For instance, a combination of lncRNA PANDAR, FOXD2-AS1 and SMARCC2 increases the AUC value to 0.84, indicating improved diagnostic accuracy (170). Similarly, an 11-lncRNA signature has been proposed as a prognostic indicator for BC, independent of various clinicopathological parameters (171). While numerous studies await validation in clinical settings, the utilisation of lncRNA expression profiles in cancer holds promise for future applications in early detection and prognosis, offering high accuracy and specificity.
As lncRNAs exhibit dysregulated expression in cancer and can modulate key tumorigenic processes by promoting or inhibiting different RCDs, they hold potential as diagnostic and prognostic biomarkers. For instance, plasma-derived ZFAS1, SNHG11, LINC00909 and LINC00654 together demonstrated a strong diagnostic capability for CRC with an AUC of 0.937, particularly in early-stage disease (AUC: 0.935) (172). Additionally, a nine-pyroptosis-associated lncRNA signature has been independently validated as a prognostic indicator for patients with BC. The AUC of this lncRNA signature associated with pyroptosis reached 0.880 in the training dataset and 0.799 in the validation dataset (173). As their regulatory roles in non-apoptotic RCDs are gradually being unveiled, lncRNAs are emerging as effective diagnostic and prognosis biomarkers in diseases. For instance, a clinical trial reveals that lncRNA NBR2 regulates septic endothelial pyroptosis, thereby underscoring the prognostic significance of pyroptosis levels in patients with sepsis (NCT04427371). Furthermore, lncRNAs regulating non-apoptotic RCDs pathways hold promise as tumour diagnostic markers. Integrating specific lncRNAs with other biomarkers has the potential to enhance diagnostic sensitivity and specificity across various cancer types. In conclusion, the integration of lncRNAs with traditional biomarkers will enhance diagnostic sensitivity and specificity, particularly in specific diseases or cellular subtypes.
5.2 Therapeutic potential of lncRNAs
Recent studies have unveiled the capacity of lncRNAs to influence RCD processes in cancer cells, presenting a potential avenue for suppressing tumour development. Understanding the mechanisms underlying gene overexpression or knockdown could provide new insight for targeting lncRNAs in treatment strategies. Currently, lncRNA-focused therapeutic strategies primarily involve RNA interference (RNAi) and antisense oligonucleotides (ASOs), offering customisable options for targeting a diverse range of transcripts (174) (Figure 4B). Preclinical models of human tumours have demonstrated the potential of lncRNAs as targets for cancer therapy. For example, the targeting of lncRNAs (e.g. LINC00336) using shRNA has shown promise in reducing cancer cell growth by promoting ferroptosis, suggesting these lncRNAs may represent novel therapeutic targets for human cancer (19, 122). In other studies, silencing of lncRNAs MAYA, MALAT1 and lncARSR using ASO has been shown to attenuate metastasis in mouse models, underscoring the therapeutic potential of ASOs targeting lncRNAs in cancer (175–178). Despite the emergence of preclinical studies highlighting the therapeutic potential of lncRNAs in cancer, further clinical trials are warranted to elucidate the targeting of lncRNAs as novel therapeutic options in clinical settings. Several ongoing clinical trials focus on anticancer therapy targeting non-apoptotic RCDs processes in cancers such as GC and HCC (e.g., NCT05334849, NCT04767750). Additionally, targeting mediators of non-apoptotic RCDs holds promise as candidates to enhance the efficacy and safety of therapeutic interventions. For instance, clinical trials evaluating the potential efficacy of targeting necroptosis in metastatic solid cancer (NCT04739618) and assessing the clinical efficacy of Aurora kinase A (AURKA), a negative regulator of necrosome activation, inhibitors in pancreatic ductal adenocarcinoma (NCT04479306, NCT04555837, NCT04085315 and NCT01924260) are underway. However, the clinical application of targeting lncRNAs is still in its early stages, necessitating extensive research to develop advanced strategies and effective agents for clinical settings. Furthermore, emerging studies have revealed that targeting specific lncRNAs (e.g. H19) is associated with various types of cell death in human cells, suggesting that targeting these lncRNAs could be a promising strategy for cancer therapy (123, 179). However, the precise mechanism by which lncRNAs facilitate or impede various RCD processes in cancer cells by targeting shared key molecules remains to be elucidated tumour prognosis by initiating multiple RCD mechanisms, offering novel insights for both diagnosis and treatment strategies.
Nonetheless, the current research on the role of lncRNAs as biomarkers or therapeutic targets has some specific limitations that require further study in cancer therapy. Although non-apoptotic RCD is a significant programmed cell death process that moderates the inflammatory response and cell demise in cancer, the investigation into how lncRNAs affect cancer therapy by modulating non-apoptotic RCD has not been thoroughly investigated in clinical trials, and this represents a potential area of exploration.
6 Conclusion and perspectives
Recent studies have highlighted the association between dysregulated expression of lncRNAs and the onset and progression of various cancers. Dysregulation of non-apoptotic RCDs, including necroptosis, pyroptosis and ferroptosis, has been shown to play a significant role in the etiopathogenesis of cancer (180). This review provides an overview of the functions and mechanisms of lncRNA-mediated ferroptosis, pyroptosis and necroptosis in tumour initiation and progression, emphasising the diagnostic, prognostic and therapeutic implications of lncRNAs in cancers. Thus, gaining a comprehensive understanding of the correlation between lncRNAs and non-apoptotic RCDs could offer valuable insights into the underlying molecular mechanisms of cancer pathogenesis. Moreover, inducing cancer cell death by targeting theses lncRNAs presents a promising strategy for cancer therapy.
However, several critical questions remain unanswered. Firstly, how can lncRNAs be used as reliable biomarkers for cancer diagnosis or prognosis, and how can they be explored further as therapeutic targets? While lncRNAs have shown promise as biomarkers and therapeutic targets, further research is needed to fully exploit their potential in clinical applications (181). A large quantity of lncRNAs modulate non-apoptotic RCDs and related pathways in tumour cells to exert oncogenic or anticancer effects, indicating potential value as predictive biomarkers of cancer chemotherapy responsiveness and clinical prognosis (74, 182, 183). Secondly, it is crucial to understand how to specifically target cancer cells to promote non-apoptotic RCDs and overcome treatment resistance. It is also important to understand the common signalling pathways (e.g., caspase-8 and RIPK1) between non-apoptotic RCDs and apoptosis, which are critical for targeting non-apoptotic RCDs regulated by lncRNAs in cancer therapy (184). Thirdly, targeting non-apoptotic RCD associated lncRNAs to reshape the TME could accelerate the development of therapeutic strategies for cancer therapy. Recent researches have highlighted that abnormally expressed lncRNAs can contribute to the modulation of the TME via non-apoptotic RCDs (185–187). Currently, the integration of novel technologies, such as CRISPR screening and single-cell sequencing to accurately identify genetic perturbations in different components of the TME will aid in the development of lncRNA-based therapeutics for clinical practice.
To date, only a limited number of chemical drugs and herbal remedies have been identified to target lncRNAs in non-apoptotic RCDs for cancer treatment, primarily focusing on understanding the regulatory mechanisms of non-apoptotic RCDs. Therefore, the development of lncRNA-targeted anticancer strategy, particularly involving herbal remedies, holds significant promise and may contribute to a more comprehensive clinical approach to enhancing cancer therapeutic outcomes.
Author contributions
KH: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Investigation, Visualization. LY: Writing – original draft, Writing – review & editing, Visualization. DL: Writing – review & editing, Supervision. ZZ: Writing – review & editing, Supervision. MS: Writing – review & editing, Supervision. ZM: Writing – review & editing, Conceptualization, Funding acquisition, Resources, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the following research grants from the National Foreign Experts Program of China: Belt and Road Initiative Innovative Talent Exchange Foreign Expert Project (DL2023027001L); Yangtze University Science and Technology Aid to Tibet Medical Talent Training Program Project (2023YZ18).
Acknowledgments
Figures in the article were drawn using BioRender.com (accessed on 21 May 2024).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. (2018) 25:486–541. doi: 10.1038/s41418-017-0012-4
2. Newton K, Strasser A, Kayagaki N, Dixit VM. Cell death. Cell. (2024) 187:235–56. doi: 10.1016/j.cell.2023.11.044
3. Tang D, Kang R, Berghe TV, Vandenabeele P, Kroemer G. The molecular machinery of regulated cell death. Cell Res. (2019) 29:347–64. doi: 10.1038/s41422-019-0164-5
4. Koren E, Fuchs Y. Modes of regulated cell death in cancer. Cancer Discovery. (2021) 11:245–65. doi: 10.1158/2159-8290.CD-20-0789
5. Jin X, Tang J, Qiu X, Nie X, Ou S, Wu G, et al. Ferroptosis: Emerging mechanisms, biological function, and therapeutic potential in cancer and inflammation. Cell Death Discovery. (2024) 10:1–10. doi: 10.1038/s41420-024-01825-7
6. Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. (2012) 149:1060–72. doi: 10.1016/j.cell.2012.03.042
7. Yu P, Zhang X, Liu N, Tang L, Peng C, Chen X. Pyroptosis: mechanisms and diseases. Signal Transduct Target Ther. (2021) 6:128. doi: 10.1038/s41392-021-00507-5
8. Yan J, Wan P, Choksi S, Liu Z-G. Necroptosis and tumor progression. Trends Cancer. (2022) 8:21–7. doi: 10.1016/j.trecan.2021.09.003
9. Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigó R, Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet. (2018) 19:535–48. doi: 10.1038/s41576-018-0017-y
10. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. (2016) 73:2491–509. doi: 10.1007/s00018-016-2174-5
11. Sirey TM, Roberts K, Haerty W, Bedoya-Reina O, Rogatti-Granados S, Tan JY, et al. The long non-coding RNA Cerox1 is a post transcriptional regulator of mitochondrial complex I catalytic activity. Elife. (2019) 8:e45051. doi: 10.7554/eLife.45051.043
12. Li S, Ran M-Y, Qiao H. A cell cycle-related lncRNA signature predicts the progression-free interval in papillary thyroid carcinoma. Front Endocrinol (Lausanne). (2023) 14:1110987. doi: 10.3389/fendo.2023.1110987
13. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. (2015) 47:199–208. doi: 10.1038/ng.3192
14. Seifuddin F, Singh K, Suresh A, Judy JT, Chen Y-C, Chaitankar V, et al. lncRNAKB, a knowledgebase of tissue-specific functional annotation and trait association of long noncoding RNA. Sci Data. (2020) 7:326. doi: 10.1038/s41597-020-00659-z
15. Mattioli K, Volders P-J, Gerhardinger C, Lee JC, Maass PG, Melé M, et al. High-throughput functional analysis of lncRNA core promoters elucidates rules governing tissue specificity. Genome Res. (2019) 29:344–55. doi: 10.1101/gr.242222.118
16. Bjørklund SS, Aure MR, Häkkinen J, Vallon-Christersson J, Kumar S, Evensen KB, et al. Subtype and cell type specific expression of lncRNAs provide insight into breast cancer. Commun Biol. (2022) 5:834. doi: 10.1038/s42003-022-03559-7
17. Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. (2013) 14:1190–8. doi: 10.1038/ni.2712
18. Zhang M, Dang P, Liu Y, Qiao B, Sun Z. Noncoding RNAs in pyroptosis and cancer progression: Effect, mechanism, and clinical application. Front Immunol. (2022) 13:982040. doi: 10.3389/fimmu.2022.982040
19. Lin Z, Song J, Gao Y, Huang S, Dou R, Zhong P, et al. Hypoxia-induced HIF-1α/lncRNA-PMAN inhibits ferroptosis by promoting the cytoplasmic translocation of ELAVL1 in peritoneal dissemination from gastric cancer. Redox Biol. (2022) 52:102312. doi: 10.1016/j.redox.2022.102312
20. Zuo Y-B, Zhang Y-F, Zhang R, Tian J-W, Lv X-B, Li R, et al. Ferroptosis in cancer progression: role of noncoding RNAs. Int J Biol Sci. (2022) 18:1829–43. doi: 10.7150/ijbs.66917
21. Yang H, Hu Y, Weng M, Liu X, Wan P, Hu Y, et al. Hypoxia inducible lncRNA-CBSLR modulates ferroptosis through m6A-YTHDF2-dependent modulation of CBS in gastric cancer. J Adv Res. (2022) 37:91–106. doi: 10.1016/j.jare.2021.10.001
22. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen L-L, et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. (2023) 24:430–47. doi: 10.1038/s41580-022-00566-8
23. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. (2018) 172:393–407. doi: 10.1016/j.cell.2018.01.011
24. Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. (2015) 16:20. doi: 10.1186/s13059-015-0586-4
25. Wilusz JE, Freier SM, Spector DL. 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. (2008) 135:919–32. doi: 10.1016/j.cell.2008.10.012
26. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. (2012) 22:1775–89. doi: 10.1101/gr.132159.111
27. Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. (2005) 308:1149–54. doi: 10.1126/science.1108625
28. Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. (2012) 22:885–98. doi: 10.1101/gr.131037.111
29. Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. (2016) 17:756–70. doi: 10.1038/nrm.2016.126
30. Wu H, Yang L, Chen L-L. The diversity of long noncoding RNAs and their generation. Trends Genet. (2017) 33:540–52. doi: 10.1016/j.tig.2017.05.004
31. Huang H, Weng H, Chen J. m6A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. (2020) 37:270–88. doi: 10.1016/j.ccell.2020.02.004
32. Zhang K, Shi Z-M, Chang Y-N, Hu Z-M, Qi H-X, Hong W. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene. (2014) 547:1–9. doi: 10.1016/j.gene.2014.06.043
33. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. (2014) 344:310–3. doi: 10.1126/science.1251456
34. Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. (2013) 493:231–5. doi: 10.1038/nature11661
35. Herman AB, Tsitsipatis D, Gorospe M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol Cell. (2022) 82:2252–66. doi: 10.1016/j.molcel.2022.05.027
36. Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing miRNA-lncRNA interactions. Methods Mol Biol. (2016) 1402:271–86. doi: 10.1007/978-1-4939-3378-5_21
37. Yin X, Jing Y, Xu H. Mining for missed sORF-encoded peptides. Expert Rev Proteomics. (2019) 16:257–66. doi: 10.1080/14789450.2019.1571919
38. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
39. Glassman ML, de Groot N, Hochberg A. Relaxation of imprinting in carcinogenesis. Cancer Genet Cytogenet. (1996) 89:69–73. doi: 10.1016/0165-4608(95)00364-9
40. Tan Y-T, Lin J-F, Li T, Li J-J, Xu R-H, Ju H-Q. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond). (2021) 41:109–20. doi: 10.1002/cac2.12108
41. Bejarano L, Jordāo MJC, Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Discovery. (2021) 11:933–59. doi: 10.1158/2159-8290.CD-20-1808
42. Liu C-G, Chen J, Goh RMW-J, Liu Y-X, Wang L, Ma Z. The role of tumor-derived extracellular vesicles containing noncoding RNAs in mediating immune cell function and its implications from bench to bedside. Pharmacol Res. (2023) 191:106756. doi: 10.1016/j.phrs.2023.106756
43. Park E-G, Pyo S-J, Cui Y, Yoon S-H, Nam J-W. Tumor immune microenvironment lncRNAs. Brief Bioinform. (2022) 23:bbab504. doi: 10.1093/bib/bbab504
44. de Visser KE, Joyce JA. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell. (2023) 41:374–403. doi: 10.1016/j.ccell.2023.02.016
45. Statello L, Guo C-J, Chen L-L, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. (2021) 22:96–118. doi: 10.1038/s41580-020-00315-9
46. Li Z, Qin X, Bian W, Li Y, Shan B, Yao Z, et al. Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA-124/STAT3 axis. J Exp Clin Cancer Res. (2019) 38:477. doi: 10.1186/s13046-019-1473-8
47. Deng X, Ruan H, Zhang X, Xu X, Zhu Y, Peng H, et al. Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int J Cancer. (2020) 146:1700–16. doi: 10.1002/ijc.32608
48. Wu D-M, Deng S-H, Liu T, Han R, Zhang T, Xu Y. TGF-β-mediated exosomal lnc-MMP2-2 regulates migration and invasion of lung cancer cells to the vasculature by promoting MMP2 expression. Cancer Med. (2018) 7:5118–29. doi: 10.1002/cam4.1758
49. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discovery. (2022) 12:31–46. doi: 10.1158/2159-8290.CD-21-1059
50. Balihodzic A, Prinz F, Dengler MA, Calin GA, Jost PJ, Pichler M. Non-coding RNAs and ferroptosis: potential implications for cancer therapy. Cell Death Differ. (2022) 29:1094–106. doi: 10.1038/s41418-022-00998-x
51. Liu Y, Chen Q, Zhu Y, Wang T, Ye L, Han L, et al. Non-coding RNAs in necroptosis, pyroptosis and ferroptosis in cancer metastasis. Cell Death Discovery. (2021) 7:210. doi: 10.1038/s41420-021-00596-9
52. Yang Y-C, Jiang Q, Yang K-P, Wang L, Sethi G, Ma Z. Extracellular vesicle-mediated ferroptosis, pyroptosis, and necroptosis: potential clinical applications in cancer therapy. Cell Death Discovery. (2024) 10:23. doi: 10.1038/s41420-024-01799-6
53. Tang R, Xu J, Zhang B, Liu J, Liang C, Hua J, et al. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J Hematol Oncol. (2020) 13:110. doi: 10.1186/s13045-020-00946-7
54. Galluzzi L, Maiuri MC, Vitale I, Zischka H, Castedo M, Zitvogel L, et al. Cell death modalities: classification and pathophysiological implications. Cell Death Differ. (2007) 14:1237–43. doi: 10.1038/sj.cdd.4402148
55. Fang X, Ardehali H, Min J, Wang F. The molecular and metabolic landscape of iron and ferroptosis in cardiovascular disease. Nat Rev Cardiol. (2023) 20:7–23. doi: 10.1038/s41569-022-00735-4
56. Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. (2021) 22:266–82. doi: 10.1038/s41580-020-00324-8
57. Kuang F, Liu J, Tang D, Kang R. Oxidative damage and antioxidant defense in ferroptosis. Front Cell Dev Biol. (2020) 8:586578. doi: 10.3389/fcell.2020.586578
58. Li J, Cao F, Yin H-L, Huang Z-J, Lin Z-T, Mao N, et al. Ferroptosis: past, present and future. Cell Death Dis. (2020) 11:88. doi: 10.1038/s41419-020-2298-2
59. Tang D, Chen X, Kang R, Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. (2021) 31:107–25. doi: 10.1038/s41422-020-00441-1
60. Lei G, Zhuang L, Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer. (2022) 22:381–96. doi: 10.1038/s41568-022-00459-0
61. Doll S, Freitas FP, Shah R, Aldrovandi M, da Silva MC, Ingold I, et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. (2019) 575:693–8. doi: 10.1038/s41586-019-1707-0
62. Bersuker K, Hendricks JM, Li Z, Magtanong L, Ford B, Tang PH, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. (2019) 575:688–92. doi: 10.1038/s41586-019-1705-2
63. Mao C, Liu X, Zhang Y, Lei G, Yan Y, Lee H, et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. (2021) 593:586–90. doi: 10.1038/s41586-021-03539-7
64. Zheng J, Conrad M. The metabolic underpinnings of ferroptosis. Cell Metab. (2020) 32:920–37. doi: 10.1016/j.cmet.2020.10.011
65. Liang D, Feng Y, Zandkarimi F, Wang H, Zhang Z, Kim J, et al. Ferroptosis surveillance independent of GPX4 and differentially regulated by sex hormones. Cell. (2023) 186:2748–2764.e22. doi: 10.1016/j.cell.2023.05.003
66. Wang L, Liu Y, Du T, Yang H, Lei L, Guo M, et al. ATF3 promotes erastin-induced ferroptosis by suppressing system Xc. Cell Death Differ. (2020) 27:662–75. doi: 10.1038/s41418-019-0380-z
67. Roemhild K, von Maltzahn F, Weiskirchen R, Knüchel R, von Stillfried S, Lammers T. Iron metabolism: pathophysiology and pharmacology. Trends Pharmacol Sci. (2021) 42:640–56. doi: 10.1016/j.tips.2021.05.001
68. Park E, Chung SW. ROS-mediated autophagy increases intracellular iron levels and ferroptosis by ferritin and transferrin receptor regulation. Cell Death Dis. (2019) 10:822. doi: 10.1038/s41419-019-2064-5
69. Mancardi D, Mezzanotte M, Arrigo E, Barinotti A, Roetto A. Iron overload, oxidative stress, and ferroptosis in the failing heart and liver. Antioxid (Basel). (2021) 10:1864. doi: 10.3390/antiox10121864
70. Stockwell BR, Jiang X, Gu W. Emerging mechanisms and disease relevance of ferroptosis. Trends Cell Biol. (2020) 30:478–90. doi: 10.1016/j.tcb.2020.02.009
71. Xu T, Ding W, Ji X, Ao X, Liu Y, Yu W, et al. Molecular mechanisms of ferroptosis and its role in cancer therapy. J Cell Mol Med. (2019) 23:4900–12. doi: 10.1111/jcmm.14511
72. Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. (2014) 156:317–31. doi: 10.1016/j.cell.2013.12.010
73. Jiang L, Kon N, Li T, Wang S-J, Su T, Hibshoosh H, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. (2015) 520:57–62. doi: 10.1038/nature14344
74. Niu X, Chen L, Li Y, Hu Z, He F. Ferroptosis, necroptosis, and pyroptosis in the tumor microenvironment: Perspectives for immunotherapy of SCLC. Semin Cancer Biol. (2022) 86:273–85. doi: 10.1016/j.semcancer.2022.03.009
75. Hu Q, Wei W, Wu D, Huang F, Li M, Li W, et al. Blockade of GCH1/BH4 axis activates ferritinophagy to mitigate the resistance of colorectal cancer to erastin-induced ferroptosis. Front Cell Dev Biol. (2022) 10:810327. doi: 10.3389/fcell.2022.810327
76. Li Y, Ran Q, Duan Q, Jin J, Wang Y, Yu L, et al. 7-Dehydrocholesterol dictates ferroptosis sensitivity. Nature. (2024) 626:411–8. doi: 10.1038/s41586-023-06983-9
77. Freitas FP, Alborzinia H, Dos Santos AF, Nepachalovich P, Pedrera L, Zilka O, et al. 7-Dehydrocholesterol is an endogenous suppressor of ferroptosis. Nature. (2024) 626:401–10. doi: 10.1038/s41586-023-06878-9
78. Yu H, Guo P, Xie X, Wang Y, Chen G. Ferroptosis, a new form of cell death, and its relationships with tumourous diseases. J Cell Mol Med. (2017) 21:648–57. doi: 10.1111/jcmm.13008
79. Yang W-H, Ding C-KC, Sun T, Rupprecht G, Lin C-C, Hsu D, et al. The hippo pathway effector TAZ regulates ferroptosis in renal cell carcinoma. Cell Rep. (2019) 28:2501–2508.e4. doi: 10.1016/j.celrep.2019.07.107
80. Linkermann A, Skouta R, Himmerkus N, Mulay SR, Dewitz C, De Zen F, et al. Synchronized renal tubular cell death involves ferroptosis. Proc Natl Acad Sci U.S.A. (2014) 111:16836–41. doi: 10.1073/pnas.1415518111
81. Chen Q, Zheng W, Guan J, Liu H, Dan Y, Zhu L, et al. SOCS2-enhanced ubiquitination of SLC7A11 promotes ferroptosis and radiosensitization in hepatocellular carcinoma. Cell Death Differ. (2023) 30:137–51. doi: 10.1038/s41418-022-01051-7
82. Maltez VI, Tubbs AL, Cook KD, Aachoui Y, Falcone EL, Holland SM, et al. Inflammasomes coordinate pyroptosis and natural killer cell cytotoxicity to clear infection by a ubiquitous environmental bacterium. Immunity. (2015) 43:987–97. doi: 10.1016/j.immuni.2015.10.010
83. Liu X, Xia S, Zhang Z, Wu H, Lieberman J. Channelling inflammation: gasdermins in physiology and disease. Nat Rev Drug Discovery. (2021) 20:384–405. doi: 10.1038/s41573-021-00154-z
84. Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. (1972) 26:239–57. doi: 10.1038/bjc.1972.33
85. Chen X, He W-T, Hu L, Li J, Fang Y, Wang X, et al. Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Res. (2016) 26:1007–20. doi: 10.1038/cr.2016.100
86. Wang Y, Gao W, Shi X, Ding J, Liu W, He H, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. (2017) 547:99–103. doi: 10.1038/nature22393
87. Kovacs SB, Miao EA. Gasdermins: effectors of pyroptosis. Trends Cell Biol. (2017) 27:673–84. doi: 10.1016/j.tcb.2017.05.005
88. Hou J, Zhao R, Xia W, Chang C-W, You Y, Hsu J-M, et al. PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis. Nat Cell Biol. (2020) 22:1264–75. doi: 10.1038/s41556-020-0575-z
89. Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. (2012) 481:278–86. doi: 10.1038/nature10759
90. Chen GY, Nuñez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. (2010) 10:826–37. doi: 10.1038/nri2873
91. He Y, Hara H, Núñez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci. (2016) 41:1012–21. doi: 10.1016/j.tibs.2016.09.002
92. Wang Y, Wang Y, Pan J, Gan L, Xue J. Ferroptosis, necroptosis, and pyroptosis in cancer: Crucial cell death types in radiotherapy and post-radiotherapy immune activation. Radiother Oncol. (2023) 184:109689. doi: 10.1016/j.radonc.2023.109689
93. Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, et al. Non-canonical inflammasome activation targets caspase-11. Nature. (2011) 479:117–21. doi: 10.1038/nature10558
94. Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. (2015) 526:666–71. doi: 10.1038/nature15541
95. Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. (2015) 526:660–5. doi: 10.1038/nature15514
96. Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. (2016) 535:153–8. doi: 10.1038/nature18629
97. Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. (2016) 535:111–6. doi: 10.1038/nature18590
98. Shi J, Gao W, Shao F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem Sci. (2017) 42:245–54. doi: 10.1016/j.tibs.2016.10.004
99. Zhang N, Zhang J, Yang Y, Shan H, Hou S, Fang H, et al. A palmitoylation-depalmitoylation relay spatiotemporally controls GSDMD activation in pyroptosis. Nat Cell Biol. (2024) 26:757–69. doi: 10.1038/s41556-024-01397-9
100. Ren Y, Feng M, Hao X, Liu X, Li J, Li P, et al. USP48 stabilizes gasdermin E to promote pyroptosis in cancer. Cancer Res. (2023) 83:1074–93. doi: 10.1158/0008-5472.CAN-22-1812
101. Yu J, Li S, Qi J, Chen Z, Wu Y, Guo J, et al. Cleavage of GSDME by caspase-3 determines lobaplatin-induced pyroptosis in colon cancer cells. Cell Death Dis. (2019) 10:193. doi: 10.1038/s41419-019-1441-4
102. Ding Q, Zhang W, Cheng C, Mo F, Chen L, Peng G, et al. Dioscin inhibits the growth of human osteosarcoma by inducing G2/M-phase arrest, apoptosis, and GSDME-dependent cell death in vitro and in vivo. J Cell Physiol. (2020) 235:2911–24. doi: 10.1002/jcp.29197
103. Wang S, Wu Z-Z, Zhu S-W, Wan S-C, Zhang M-J, Zhang B-X, et al. CTLA-4 blockade induces tumor pyroptosis via CD8+ T cells in head and neck squamous cell carcinoma. Mol Ther. (2023) 31:2154–68. doi: 10.1016/j.ymthe.2023.02.023
104. Ji X, Huang X, Li C, Guan N, Pan T, Dong J, et al. Effect of tumor-associated macrophages on the pyroptosis of breast cancer tumor cells. Cell Commun Signal. (2023) 21:197. doi: 10.1186/s12964-023-01208-y
105. He S, Wang L, Miao L, Wang T, Du F, Zhao L, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. (2009) 137:1100–11. doi: 10.1016/j.cell.2009.05.021
106. Galluzzi L, Kepp O, Chan FK-M, Kroemer G. Necroptosis: mechanisms and relevance to disease. Annu Rev Pathol. (2017) 12:103–30. doi: 10.1146/annurev-pathol-052016-100247
107. Shan B, Pan H, Najafov A, Yuan J. Necroptosis in development and diseases. Genes Dev. (2018) 32:327–40. doi: 10.1101/gad.312561.118
108. Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature. (2015) 517:311–20. doi: 10.1038/nature14191
109. Sun L, Wang H, Wang Z, He S, Chen S, Liao D, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. (2012) 148:213–27. doi: 10.1016/j.cell.2011.11.031
110. Takemura R, Takaki H, Okada S, Shime H, Akazawa T, Oshiumi H, et al. PolyI:C-induced, TLR3/RIP3-dependent necroptosis backs up immune effector-mediated tumor elimination. In Vivo Cancer Immunol Res. (2015) 3:902–14. doi: 10.1158/2326-6066.CIR-14-0219
111. Dondelinger Y, Darding M, Bertrand MJM, Walczak H. Poly-ubiquitination in TNFR1-mediated necroptosis. Cell Mol Life Sci. (2016) 73:2165–76. doi: 10.1007/s00018-016-2191-4
112. Zhang D-W, Shao J, Lin J, Zhang N, Lu B-J, Lin S-C, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. (2009) 325:332–6. doi: 10.1126/science.1172308
113. Seo J, Nam YW, Kim S, Oh D-B, Song J. Necroptosis molecular mechanisms: Recent findings regarding novel necroptosis regulators. Exp Mol Med. (2021) 53:1007–17. doi: 10.1038/s12276-021-00634-7
114. Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. (2009) 137:1112–23. doi: 10.1016/j.cell.2009.05.037
115. Cai Z, Jitkaew S, Zhao J, Chiang H-C, Choksi S, Liu J, et al. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat Cell Biol. (2014) 16:55–65. doi: 10.1038/ncb2883
116. Tong X, Tang R, Xiao M, Xu J, Wang W, Zhang B, et al. Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research. J Hematol Oncol. (2022) 15:174. doi: 10.1186/s13045-022-01392-3
117. Gao W, Wang X, Zhou Y, Wang X, Yu Y. Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor immunotherapy. Signal Transduct Target Ther. (2022) 7:196. doi: 10.1038/s41392-022-01046-3
118. Gong Y, Fan Z, Luo G, Yang C, Huang Q, Fan K, et al. The role of necroptosis in cancer biology and therapy. Mol Cancer. (2019) 18:100. doi: 10.1186/s12943-019-1029-8
119. Zhou P, Zhang S, Wang M, Zhou J. The induction mechanism of ferroptosis, necroptosis, and pyroptosis in inflammatory bowel disease, colorectal cancer, and intestinal injury. Biomolecules. (2023) 13:820. doi: 10.3390/biom13050820
120. Chen L, Zhang X, Zhang Q, Zhang T, Xie J, Wei W, et al. A necroptosis related prognostic model of pancreatic cancer based on single cell sequencing analysis and transcriptome analysis. Front Immunol. (2022) 13:1022420. doi: 10.3389/fimmu.2022.1022420
121. Baik JY, Liu Z, Jiao D, Kwon H-J, Yan J, Kadigamuwa C, et al. ZBP1 not RIPK1 mediates tumor necroptosis in breast cancer. Nat Commun. (2021) 12:2666. doi: 10.1038/s41467-021-23004-3
122. Wang M, Mao C, Ouyang L, Liu Y, Lai W, Liu N, et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. (2019) 26:2329–43. doi: 10.1038/s41418-019-0304-y
123. Zhang R, Pan T, Xiang Y, Zhang M, Xie H, Liang Z, et al. Curcumenol triggered ferroptosis in lung cancer cells via lncRNA H19/miR-19b-3p/FTH1 axis. Bioact Mater. (2022) 13:23–36. doi: 10.1016/j.bioactmat.2021.11.013
124. Yu H, Han Z, Xu Z, An C, Xu L, Xin H. RNA sequencing uncovers the key long non-coding RNAs and potential molecular mechanism contributing to XAV939-mediated inhibition of non-small cell lung cancer. Oncol Lett. (2019) 17:4994–5004. doi: 10.3892/ol.2019.10191
125. Mao C, Wang X, Liu Y, Wang M, Yan B, Jiang Y, et al. A G3BP1-Interacting lncRNA Promotes Ferroptosis and Apoptosis in Cancer via Nuclear Sequestration of p53. Cancer Res. (2018) 78:3484–96. doi: 10.1158/0008-5472.CAN-17-3454
126. Gai C, Liu C, Wu X, Yu M, Zheng J, Zhang W, et al. MT1DP loaded by folate-modified liposomes sensitizes erastin-induced ferroptosis via regulating miR-365a-3p/NRF2 axis in non-small cell lung cancer cells. Cell Death Dis. (2020) 11:751. doi: 10.1038/s41419-020-02939-3
127. Wu H, Liu A. Long non-coding RNA NEAT1 regulates ferroptosis sensitivity in non-small-cell lung cancer. J Int Med Res. (2021) 49:300060521996183. doi: 10.1177/0300060521996183
128. Kang X, Huo Y, Jia S, He F, Li H, Zhou Q, et al. Silenced LINC01134 enhances oxaliplatin sensitivity by facilitating ferroptosis through GPX4 in hepatocarcinoma. Front Oncol. (2022) 12:939605. doi: 10.3389/fonc.2022.939605
129. Zhang B, Bao W, Zhang S, Chen B, Zhou X, Zhao J, et al. LncRNA HEPFAL accelerates ferroptosis in hepatocellular carcinoma by regulating SLC7A11 ubiquitination. Cell Death Dis. (2022) 13:734. doi: 10.1038/s41419-022-05173-1
130. Qi W, Li Z, Xia L, Dai J, Zhang Q, Wu C, et al. LncRNA GABPB1-AS1 and GABPB1 regulate oxidative stress during erastin-induced ferroptosis in HepG2 hepatocellular carcinoma cells. Sci Rep. (2019) 9:16185. doi: 10.1038/s41598-019-52837-8
131. Guan L, Wang F, Wang M, Han S, Cui Z, Xi S, et al. Downregulation of HULC Induces Ferroptosis in Hepatocellular Carcinoma via Targeting of the miR-3200-5p/ATF4 Axis. Oxid Med Cell Longev. (2022) 2022:9613095. doi: 10.1155/2022/9613095
132. Huang G, Xiang Z, Wu H, He Q, Dou R, Lin Z, et al. The lncRNA BDNF-AS/WDR5/FBXW7 axis mediates ferroptosis in gastric cancer peritoneal metastasis by regulating VDAC3 ubiquitination. Int J Biol Sci. (2022) 18:1415–33. doi: 10.7150/ijbs.69454
133. Han Y, Gao X, Wu N, Jin Y, Zhou H, Wang W, et al. Long noncoding RNA LINC00239 inhibits ferroptosis in colorectal cancer by binding to Keap1 to stabilize Nrf2. Cell Death Dis. (2022) 13:742. doi: 10.1038/s41419-022-05192-y
134. Luo Y, Huang S, Wei J, Zhou H, Wang W, Yang J, et al. Long noncoding RNA LINC01606 protects colon cancer cells from ferroptotic cell death and promotes stemness by SCD1-Wnt/β-catenin-TFE3 feedback loop signalling. Clin Transl Med. (2022) 12:e752. doi: 10.1002/ctm2.752
135. Tan P, Li M, Liu Z, Li T, Zhao L, Fu W. Glycolysis-related LINC02432/Hsa-miR-98-5p/HK2 axis inhibits ferroptosis and predicts immune infiltration, tumor mutation burden, and drug sensitivity in pancreatic adenocarcinoma. Front Pharmacol. (2022) 13:937413. doi: 10.3389/fphar.2022.937413
136. Zhang Y, Guo S, Wang S, Li X, Hou D, Li H, et al. LncRNA OIP5-AS1 inhibits ferroptosis in prostate cancer with long-term cadmium exposure through miR-128-3p/SLC7A11 signaling. Ecotoxicol Environ Saf. (2021) 220:112376. doi: 10.1016/j.ecoenv.2021.112376
137. Jiang X, Guo S, Xu M, Ma B, Liu R, Xu Y, et al. TFAP2C-Mediated lncRNA PCAT1 Inhibits Ferroptosis in Docetaxel-Resistant Prostate Cancer Through c-Myc/miR-25-3p/SLC7A11 Signaling. Front Oncol. (2022) 12:862015. doi: 10.3389/fonc.2022.862015
138. Luo W, Wang J, Xu W, Ma C, Wan F, Huang Y, et al. LncRNA RP11-89 facilitates tumorigenesis and ferroptosis resistance through PROM2-activated iron export by sponging miR-129-5p in bladder cancer. Cell Death Dis. (2021) 12:1043. doi: 10.1038/s41419-021-04296-1
139. Liu J, Cui J, Zhao S, Wu M, Wang J, Zhang Y, et al. Ferroptosis-related long noncoding RNAs have excellent predictive ability for multiomic characteristics of bladder cancer. Oxid Med Cell Longev. (2022) 2022:9316847. doi: 10.1155/2022/9316847
140. Gong H, Gao M, Lin Y, Liu J, Hu Z, Liu J. TUG1/MAZ/FTH1 axis attenuates the antiglioma effect of dihydroartemisinin by inhibiting ferroptosis. Oxid Med Cell Longev. (2022) 2022:7843863. doi: 10.1155/2022/7843863
141. Ma Q, Dai X, Lu W, Qu X, Liu N, Zhu C. Silencing long non-coding RNA MEG8 inhibits the proliferation and induces the ferroptosis of hemangioma endothelial cells by regulating miR-497-5p/NOTCH2 axis. Biochem Biophys Res Commun. (2021) 556:72–8. doi: 10.1016/j.bbrc.2021.03.132
142. Guo J, Ye F, Xie W, Zhang X, Zeng R, Sheng W, et al. The HOXC-AS2/miR-876-5p/HKDC1 axis regulates endometrial cancer progression in a high glucose-related tumor microenvironment. Cancer Sci. (2022) 113:2297–310. doi: 10.1111/cas.15384
143. Chen Z, He M, Chen J, Li C, Zhang Q. Long non-coding RNA SNHG7 inhibits NLRP3-dependent pyroptosis by targeting the miR-34a/SIRT1 axis in liver cancer. Oncol Lett. (2020) 20:893–901. doi: 10.3892/ol.2020.11635
144. Tan C, Liu W, Zheng Z-H, Wan X-G. LncRNA HOTTIP inhibits cell pyroptosis by targeting miR-148a-3p/AKT2 axis in ovarian cancer. Cell Biol Int. (2021) 45:1487–97. doi: 10.1002/cbin.11588
145. Gatti V, Fierro C, Annicchiarico-Petruzzelli M, Melino G, Peschiaroli A. ΔNp63 in squamous cell carcinoma: defining the oncogenic routes affecting epigenetic landscape and tumour microenvironment. Mol Oncol. (2019) 13:981–1001. doi: 10.1002/1878-0261.12473
146. Guo M, Xiao Z-D, Dai Z, Zhu L, Lei H, Diao L-T, et al. The landscape of long noncoding RNA-involved and tumor-specific fusions across various cancers. Nucleic Acids Res. (2020) 48:12618–31. doi: 10.1093/nar/gkaa1119
147. Li J, Yang C, Li Y, Chen A, Li L, You Z. LncRNA GAS5 suppresses ovarian cancer by inducing inflammasome formation. Biosci Rep. (2018) 38:BSR20171150. doi: 10.1042/BSR20171150
148. Jiang L, Ge W, Cui Y, Wang X. The regulation of long non-coding RNA 00958 (LINC00958) for oral squamous cell carcinoma (OSCC) cells death through absent in melanoma 2 (AIM2) depending on microRNA-4306 and Sirtuin1 (SIRT1) in vitro. Bioengineered. (2021) 12:5085–98. doi: 10.1080/21655979.2021.1955561
149. Ren N, Jiang T, Wang C, Xie S, Xing Y, Piao D, et al. LncRNA ADAMTS9-AS2 inhibits gastric cancer (GC) development and sensitizes chemoresistant GC cells to cisplatin by regulating miR-223-3p/NLRP3 axis. Aging (Albany NY). (2020) 12:11025–41. doi: 10.18632/aging.103314
150. Yan H, Luo B, Wu X, Guan F, Yu X, Zhao L, et al. Cisplatin induces pyroptosis via activation of MEG3/NLRP3/caspase-1/GSDMD pathway in triple-negative breast cancer. Int J Biol Sci. (2021) 17:2606–21. doi: 10.7150/ijbs.60292
151. Liu J, Yao L, Zhang M, Jiang J, Yang M, Wang Y. Downregulation of LncRNA-XIST inhibited development of non-small cell lung cancer by activating miR-335/SOD2/ROS signal pathway mediated pyroptotic cell death. Aging (Albany NY). (2019) 11:7830–46. doi: 10.18632/aging.102291
152. Wo L, Zhang X, Ma C, Zhou C, Li J, Hu Z, et al. LncRNA HABON promoted liver cancer cells survival under hypoxia by inhibiting mPTP opening. Cell Death Discovery. (2022) 8:171. doi: 10.1038/s41420-022-00917-6
153. Min W, Sun L, Li B, Gao X, Zhang S, Zhao Y. lncCRLA enhanced chemoresistance in lung adenocarcinoma that underwent epithelialMesenchymal transition. Oncol Res. (2022) 28:857–72. doi: 10.3727/096504021X16203818567367
154. Wang W, Shi Q, Wang S, Zhang H, Xu S. Ammonia regulates chicken tracheal cell necroptosis via the LncRNA-107053293/MiR-148a-3p/FAF1 axis. J Hazard Mater. (2020) 386:121626. doi: 10.1016/j.jhazmat.2019.121626
155. Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am. (2019) 103:463–73. doi: 10.1016/j.mcna.2018.12.006
156. Lin X, Ping J, Wen Y, Wu Y. The mechanism of ferroptosis and applications in tumor treatment. Front Pharmacol. (2020) 11:1061. doi: 10.3389/fphar.2020.01061
157. Jiang X, Yuan Y, Tang L, Wang J, Zhang D, Duan L. Systematic analysis and validation of the prognosis, immunological role and biology function of the ferroptosis-related lncRNA GSEC/miRNA-101-3p/CISD1 axis in lung adenocarcinoma. Front Mol Biosci. (2021) 8:793732. doi: 10.3389/fmolb.2021.793732
158. Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. (2020) 1873:188314. doi: 10.1016/j.bbcan.2019.188314
160. Gao Y, Tong M, Wong T-L, Ng K-Y, Xie Y-N, Wang Z, et al. Long noncoding RNA URB1-antisense RNA 1 (AS1) suppresses sorafenib-induced ferroptosis in hepatocellular carcinoma by driving ferritin phase separation. ACS Nano. (2023) 17:22240–58. doi: 10.1021/acsnano.3c01199
161. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. (2015) 65:87–108. doi: 10.3322/caac.21262
162. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. (2020) 396:635–48. doi: 10.1016/S0140-6736(20)31288-5
163. Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2023. CA Cancer J Clin. (2023) 73:233–54. doi: 10.3322/caac.21772
164. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. (2019) 394:1467–80. doi: 10.1016/S0140-6736(19)32319-0
165. Zheng J, Zhang Q, Zhao Z, Qiu Y, Zhou Y, Wu Z, et al. Epigenetically silenced lncRNA SNAI3-AS1 promotes ferroptosis in glioma via perturbing the m6A-dependent recognition of Nrf2 mRNA mediated by SND1. J Exp Clin Cancer Res. (2023) 42:127. doi: 10.1186/s13046-023-02684-3
166. Wang M, Cheng H, Wu H, Liu C, Li S, Li B, et al. Gambogenic acid antagonizes the expression and effects of long non-coding RNA NEAT1 and triggers autophagy and ferroptosis in melanoma. BioMed Pharmacother. (2022) 154:113636. doi: 10.1016/j.biopha.2022.113636
167. Chen E, Yi J, Jiang J, Zou Z, Mo Y, Ren Q, et al. Identification and validation of a fatty acid metabolism-related lncRNA signature as a predictor for prognosis and immunotherapy in patients with liver cancer. BMC Cancer. (2022) 22:1037. doi: 10.1186/s12885-022-10122-4
168. Roobol MJ, Schröder FH, van Leeuwen P, Wolters T, van den Bergh RCN, van Leenders GJLH, et al. Performance of the prostate cancer antigen 3 (PCA3) gene and prostate-specific antigen in prescreened men: exploring the value of PCA3 for a first-line diagnostic test. Eur Urol. (2010) 58:475–81. doi: 10.1016/j.eururo.2010.06.039
169. Zhou Q, Li H, Jing J, Yuan Y, Sun L. Evaluation of C5orf66-AS1 as a potential biomarker for predicting early gastric cancer and its role in gastric carcinogenesis. Onco Targets Ther. (2020) 13:2795–805. doi: 10.2147/OTT.S239965
170. Yang Z, Sun Y, Liu R, Shi Y, Ding S. Plasma long noncoding RNAs PANDAR, FOXD2-AS1, and SMARCC2 as potential novel diagnostic biomarkers for gastric cancer. Cancer Manag Res. (2019) 11:6175–84. doi: 10.2147/CMAR.S201935
171. Shen Y, Peng X, Shen C. Identification and validation of immune-related lncRNA prognostic signature for breast cancer. Genomics. (2020) 112:2640–6. doi: 10.1016/j.ygeno.2020.02.015
172. Xu W, Zhou G, Wang H, Liu Y, Chen B, Chen W, et al. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int J Cancer. (2020) 146:2901–12. doi: 10.1002/ijc.32747
173. Ping L, Zhang K, Ou X, Qiu X, Xiao X. A novel pyroptosis-associated long non-coding RNA signature predicts prognosis and tumor immune microenvironment of patients with breast cancer. Front Cell Dev Biol. (2021) 9:727183. doi: 10.3389/fcell.2021.727183
174. Coan M, Haefliger S, Ounzain S, Johnson R. Targeting and engineering long non-coding RNAs for cancer therapy. Nat Rev Genet. (2024) 25:578–95. doi: 10.1038/s41576-024-00693-2
175. Arun G, Diermeier S, Akerman M, Chang K-C, Wilkinson JE, Hearn S, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. (2016) 30:34–51. doi: 10.1101/gad.270959.115
176. Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. (2013) 73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850
177. Li C, Wang S, Xing Z, Lin A, Liang K, Song J, et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol. (2017) 19:106–19. doi: 10.1038/ncb3464
178. Qu L, Ding J, Chen C, Wu Z-J, Liu B, Gao Y, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. (2016) 29:653–68. doi: 10.1016/j.ccell.2016.03.004
179. Han Y, Dong B, Chen M, Yao C. LncRNA H19 suppresses pyroptosis of cardiomyocytes to attenuate myocardial infarction in a PBX3/CYP1B1-dependent manner. Mol Cell Biochem. (2021) 476:1387–400. doi: 10.1007/s11010-020-03998-y
180. Yu L, Huang K, Liao Y, Wang L, Sethi G, Ma Z. Targeting novel regulated cell death: Ferroptosis, pyroptosis and necroptosis in anti-PD-1/PD-L1 cancer immunotherapy. Cell Prolif. (2024) 57:e13644. doi: 10.1111/cpr.13644
181. Hashemi M, Moosavi MS, Abed HM, Dehghani M, Aalipour M, Heydari EA, et al. (lncRNA) H19 in human cancer: From proliferation and metastasis to therapy. Pharmacol Res. (2022) 184:106418. doi: 10.1016/j.phrs.2022.106418
182. Xie J, Tian W, Tang Y, Zou Y, Zheng S, Wu L, et al. Establishment of a cell necroptosis index to predict prognosis and drug sensitivity for patients with triple-negative breast cancer. Front Mol Biosci. (2022) 9:834593. doi: 10.3389/fmolb.2022.834593
183. Wu Z, Lu Z, Li L, Ma M, Long F, Wu R, et al. Identification and validation of ferroptosis-related lncRNA signatures as a novel prognostic model for colon cancer. Front Immunol. (2021) 12:783362. doi: 10.3389/fimmu.2021.783362
184. Ai Y, Meng Y, Yan B, Zhou Q, Wang X. The biochemical pathways of apoptotic, necroptotic, pyroptotic, and ferroptotic cell death. Mol Cell. (2024) 84:170–9. doi: 10.1016/j.molcel.2023.11.040
185. Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol. (2018) 19:1112–25. doi: 10.1038/s41590-018-0207-y
186. Zhang Z, Shang J, Hu B, Shi H, Cao Y, Li J, et al. Prognosis and tumour immune microenvironment of patients with hepatocellular carcinoma by a novel pyroptosis-related lncRNA signature. Front Immunol. (2022) 13:836576. doi: 10.3389/fimmu.2022.836576
Keywords: cancers, cancer therapy, long non-coding RNA (IncRNA), ferroptosis, pyroptosis, necroptosis
Citation: Huang K, Yu L, Lu D, Zhu Z, Shu M and Ma Z (2024) Long non-coding RNAs in ferroptosis, pyroptosis and necroptosis: from functions to clinical implications in cancer therapy. Front. Oncol. 14:1437698. doi: 10.3389/fonc.2024.1437698
Received: 24 May 2024; Accepted: 12 August 2024;
Published: 29 August 2024.
Edited by:
Cheng Zhang, Anhui Medical University, ChinaReviewed by:
Giovannino Silvestri, University of Maryland Medical Center, United StatesJinghui Liang, Wistar Institute, United States
Jindong Xie, Sun Yat-sen University Cancer Center (SYSUCC), China
Copyright © 2024 Huang, Yu, Lu, Zhu, Shu and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaowu Ma, bWF6d0B5YW5ndHpldS5lZHUuY24=
†These authors have contributed equally to this work
 Ke Huang
Ke Huang Li Yu†
Li Yu† Zhaowu Ma
Zhaowu Ma