
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 16 December 2024
Sec. Cancer Molecular Targets and Therapeutics
Volume 14 - 2024 | https://doi.org/10.3389/fonc.2024.1436576
Interferon-induced protein 44-like (IFI44L) is regarded as an immune-related gene and is a member of interferon-stimulated genes (ISGs). They participate in network transduction, and its own epigenetic modifications, apoptosis, cell-matrix formation, and many other pathways in tumors, autoimmune diseases, and viral infections. The current review provides a comprehensive overview of the onset and biological mechanisms of IFI44L and its potential clinical applications in malignant tumors and non-neoplastic diseases.
Interferon-induced protein 44-like (IFI44L) is an identified gene that is involved in various cancerous and non-cancerous diseases including oral cancer (OC), renal cell carcinoma (RCC), Sjogren’s syndrome (SS), and systemic lupus erythematosus (SLE) (1–4). It participates in numerous biological events, such as innate immune response, inflammation, cell death, phosphorylation, tumor cell proliferation, and cell signaling. It plays a crucial role in defending organisms against bacteria, viruses, and tumors (5–7). It is frequently used as a biomarker for identifying and diagnosing diseases and also serves as a possible target for treating many diseases (8, 9). For instance, different levels of DNA methylation in IFI44L can distinguish between SLE patients and healthy individuals (8), as well as between patients with viral infections and those with bacterial infections (9). Therefore, IFI44L shows a growing significance as a potential biomarker for the rapid and non-invasive detection of diseases.
The prognosis for tumors remains poor despite the implementation of many innovative therapeutic approaches; they continue to present a significant global health risk (10). In cancer research, the need for biomarkers to aid in identifying and treating diseases has become increasingly significant. The implementation of precise therapeutic targets can substantially enhance patients’ survival rates and significantly improve the rate at which diseases are detected in clinical practice (11). A recently identified gene, IFI44L is present in various types of tumors, including pancreatic ductal adenocarcinoma (PDAC), laryngeal cancer, hepatocellular carcinoma (HCC), and lung cancer (LC). Its expression levels have shown significant differences in these tumor types (6, 12–14). In lung cancer cells, it can activate the JAK/STAT1 signaling pathway to promote apoptosis (14), and upregulation in head and neck squamous cell carcinoma cells can promote cancer cell proliferation and invasion (15). Among non-tumor diseases, IFI44L was found to be strongly associated with immune disorders, especially SLE, SS, rheumatoid arthritis (RA) and various viral infectious diseases (8, 16–19). This gene contributes to immune functions in different immune cells, including T cells, monocytes, and B cells. It inhibits tumor growth, prevents viral and bacterial replication, and reduces inflammation progression via its involvement in signal transduction, and genetic, and DNA methylation level of IFI44L (5, 16, 20, 21).
Thus, this review provides a detailed overview of IFI44L, covering its biological functions and mechanisms, as well as its correlation with tumor and non-tumor diseases. It highlights the potential of IFI44L as a biomarker in the diagnosis and treatment of human diseases.
IFI44L is one of the ISGs and belongs to the IFI44 family (22). This protein is situated on chromosome 1p31.1 and consists of 452 amino acids (23). Its molecular weight is approximately 47 kDa. Based on a study on Crassostrea gigas, IFI44L (also known as CgIFI44L-1) is the most abundant protein in blood cells, primarily localized in the cytoplasm, and its cDNA has an Open Reading Frame length of 1437 base pairs. It encodes a 479 amino acid peptide with the TLDc and MMR_HSR1 domains and it has the highest content in blood cells, mainly distributed in the cytoplasm (24).
It always participates in congenital immune responses (25), and has a strong anti-virus specificity (26). It is also essential for antiviral, antibacterial, and antitumor activity, in other biological processes, and is associated with several diseases (14, 27). Its involvement in innate immunity, inflammation, histone modification, methylation, programmed cell death, phosphorylation, cell proliferation, extracellular matrix (ECM) formation, and cell signaling has been supported by accumulating evidence (5, 28). Here, IFI44L’s biological functions and mechanisms, including genetic epigenetic modification, specific gene regulation and signal transduction, and cell-matrix formation, are summarized in this review.
Commonly, IFI44L exerts its important biological functions as a downstream target gene and signal transduction molecule as shown in Figure 1.
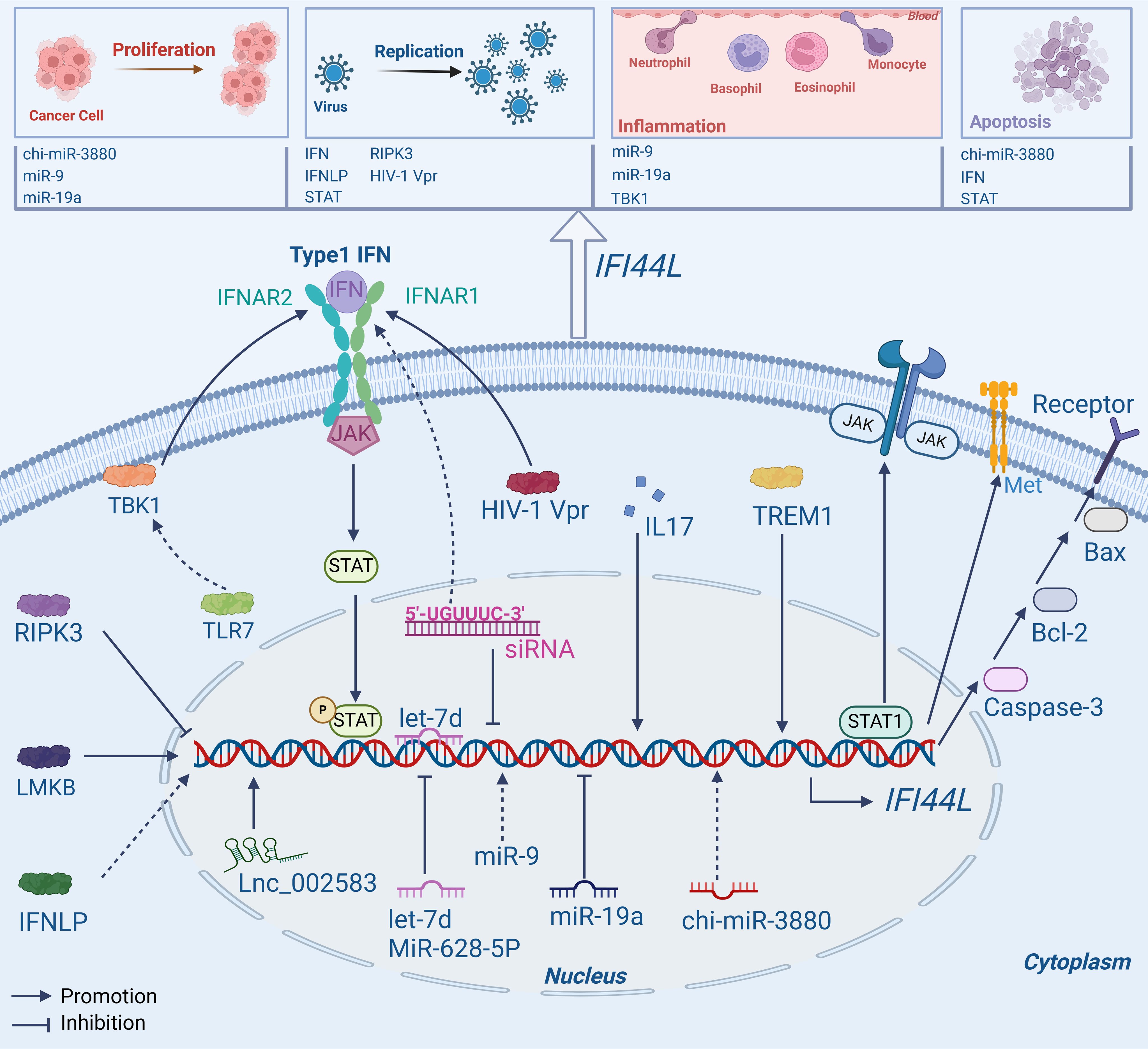
Figure 1. Mechanisms of IFI44L. IFI44L is not only a target gene for upstream molecules like RNA and protein, but can also affect some pathways such as JAK/STAT1, Type1 IFN, and Caspase-3/Bcl-2/Bax signaling pathway, playing a vital role in human diseases.
Various studies have shown that IFI44L has the potential to function as a downstream target gene, binding with some specific proteins or genes. For example, in a recent study, researchers used RNA sequencing to investigate the underlying mechanisms. The findings revealed a connection between Lnc_002583, a type of Long non-coding RNAs, and the expression of IFI44L. It was observed that Lnc_002583 may have a positive impact on IFI44L expression, suggesting a potential synergistic regulatory function in the growth and development of Leqiong cattle (29).
In plasmacytoid dendritic cells (pDCs), IFI44L expression was reduced following BX795 (TBK1 inhibitor) therapy. TANK-binding kinase (TBK1) can produce IFN-I and then promote the production of ISGs including IFI44L, thereby contributing to the suppression of viral replication (30). It has also been observed that human B and T lymphocyte cells express Limkain B (LMKB). A decrease in LMKB expression may stimulate the expression of IFI44L, IFI44L and LMKB can interact to control mRNA stability and reduce the inflammatory response, indicating that LMKB and IFI44L may interact to have an anti-inflammatory effect on B cells and T cells (31). In a recent study, Ji, X., et al. used human umbilical vein endothelial cells (HUVECs) to investigate the effects of a mimic control and let-7d, a specific type of microRNAs. Interestingly, the findings revealed that the transfection of let-7d led to a decrease in the expression of IFI44L. Further validations revealed that let-7d could directly bind to the 3’ untranslated region (3′-UTR) of IFI44L and cause a negative regulatory effect on its expression. This inhibition of IFI44L activity resulted in the suppression of endothelial cell propagation and migration, which in turn affects the development of inflammation and consequently influences the progression of diseases associated with vascular inflammation (32).
The expression of IFI44L was substantially downregulated in A549, HCC827, and CNE2 cells in the miR-19a (a type of microRNA) overexpression group. However, the inhibition of miR-19a facilitates the immuno-oncological activity of IFI44L within cellular compartments. The differential expression patterns of miR-19a and IFI44L may be harnessed as biomarkers for the prognostication and diagnostic evaluation of malignancies, thereby providing insights into the trajectory of disease progression and the efficacy of therapeutic interventions (33). In CNE2 cells, miR-9 was overexpressed to investigate its downstream target gene, and IFI44L emerged as the most significantly upregulated gene. Following the application of miR-9 inhibitors, IFI44L expression was downregulated, indicating that miR-9 can possibley target and regulate IFI44L, thus affecting the relationship between nasopharyngeal carcinoma (NPC) and inflammation (34). To promote the propagation, migration, and invasion of osteosarcoma (OS) cells, miR-9 can bind to the 3′-UTR of IFI44L, thereby targeting and inhibiting its expression (35). Recent studies have indicated that in head and neck squamous cell carcinoma (HNSCC) cells, the knockdown of IRF1 can reduce the expression levels of IFI44L and suppress cell proliferation and invasiveness. The overexpression of ACSL4 disrupts interferon signaling, enhances the expression of IFI44L, and promotes the proliferation and invasiveness of HNSCC cells. However, the specific mechanisms of their roles in tumor development require further investigation (15).
As a target gene, IFI44L is differentially expressed in a substantial number of virus-infected cells and, in certain circumstances, displays antiviral activity (36–39). Studies indicated that high levels of IFI44L have been observed to impede the replication and dissemination of the Japanese encephalitis virus (JEV), the expression of IFI44L can be inhibited by interacting with serine/threonine-protein kinase 3 (RIPK3) receptors. Therefore, inhibiting the expression of IFI44L would lead to the propagation of JEV in neurons (40). The expression of IFI44L was upregulated in IFN-like protein (IFNLP) treated haemocytes, but significantly downregulated after the deletion of IFI44L and STAT, suggesting that IFNLP and STAT participate in the antiviral immune response of oysters by targeting IFI44L (24). Therefore, inhibiting viral replication may be possible through the functions of IFI44L.
In addition to being influenced by upstream molecules, this gene can also be affected by LTR-retrotransposons. The 011052702-MALR1044uL locus has been identified in the 3′-UTR of IFI44L and has the potential to impact the expression of IFI44L in specific circumstances (41). However, these studies only identified the molecules that interact with IFI44L, they laid the groundwork for a deeper mechanistic study of this gene.
This IFI44L gene can interact with proteins and also functions as a signaling molecule, contributing to various signaling pathways and influencing the progression of diseases. It is a significant molecular biomarker that has a crucial role in regulating the Interferon (IFN) signaling pathway. Its impact extends to influencing viral replication and bacterial growth (42).
Studies have shown that in dendritic cells (DCs) infected with human immunodeficiency virus type 1 (HIV-1), a protein called Vpr can regulate IFI44L. This pleiotropic protein can stimulate IFI44L, leading to the production and activation of the type I IFN signaling pathway in human DCs (25). This offers new perspectives on the therapeutic management of HIV-1 infection. Wallen, A.J., et al. discovered that specific short interfering RNA (siRNA) contains the hexamer region 5′-UGUUUC-3′; mutation of this region inhibits the expression of IFI44L, which is essential for the IFN signaling pathway (43). During an in vitro infection system with hepatitis B virus (HBV), IFN-γ and IFN-α can remarkably regulate IFI44L expression. It has the potential to resist the HBV virus by suppressing certain pathways involved in nuclear factor-κβand signal transducers and transcription 1 (44). In a study focusing on Leishmania raziliensis, the researchers classified the patients into two groups (high and low expressions) based on their levels of IFN-γ secretion. The findings indicated that IFI44L displayed significant expression in the group characterized by elevated levels of IFN-γ, and showed a correlation with the signaling pathways of IL17 and TREM1 (45).In a recent study conducted by Wahadat, M.J. et al., it was revealed that the IFI44L expression increased with TLR7, RNA, and DNA binding receptors in CD14+ monocytes of patients with Childhood-onset SLE (cSLE). These receptors were found to have a role in promoting IFI44L sensitization via the TBK1 signaling pathway (46).
Many interferon genes may be modulated by chi-miR-3880, which is classified as a microRNA, in goat mammary epithelial cells (GMECs). Furthermore, chi-miR-3880 and IFNγ (including IFI44L) can modulate cell apoptosis via the Caspase-3/Bcl-2/Bax pathways, in addition to regulating lipid forfmation and cell growth via the PI3K/AKT/mTOR pathway (47). In human laryngeal carcinoma HEp2 cells, the expression of IFI44L is upregulated and can ultimately suppress the progression of laryngeal carcinoma by triggering apoptosis of laryngeal carcinoma cells through interaction with IFN-α (6). Nevertheless, the specific mode of interaction between them has not been studied. In LC cells, IFI44L can directly bind to STAT1 (a type of protein), promote STAT1 phosphorylation and activate JAK/STAT1 signaling pathway, causing cell apoptosis, the overexpression of IFI44L exerted a significant inhibitory effect on cellular proliferation and concomitantly induced apoptosis. Conversely, the depletion of IFI44L resulted in an enhancement of cellular proliferative capacity and a correspondingly substantial decrease in apoptotic cell death (14). The ability of IFI44L to stimulate cellular apoptosis indicates its considerable promise as a therapeutic agent for cancer.
Substantially, IFI44L, as a downstream target gene, has the potential to play a key role in different signaling cascades. Some specific proteins and non-coding RNAs, such as TBK1, LMKB, Lnc_002583, and miR-19a, may interact with IFI44L or influence its expression (29–31, 33). As a signal transduction molecule, it can regulate signaling pathways including IFN signaling, Caspase-3/Bcl-2/Bax apoptosis-related pathways, and JAK/STAT1 signaling (14, 44, 47). Unfortunately, the specific chemical mechanism has not been adequately investigated, posing a new avenue for future research.
Besides, IFI44L can also alter its own expression through methylation in its promoter region, thereby affecting biological processes such as matrix formation as displayed Figure 2.

Figure 2. Other mechanisms of action of IFI44L. IFI44L mainly participates in biological activity by altering expression through methylation of its own promoter region, and can participate in the formation of ECM.
The IFI44L gene has multiple biological functions, serving as both a target gene and a signal transduction molecule. It has a significant role in immune diseases, particularly concerning methylation and histone modification. These processes are frequently linked to the development of SLE (20, 21, 48).
For example, a high number of hypomethylated immune genes have a significant role in the development of SLE, as evidenced by the low methylation levels of IFI44L at CpG sites in the DNA of SLE patients, Yeung, K.S., et al. performed a genome-wide DNA methylation analysis of patients with systemic lupus erythematosus and found that the IFI44L gene was undermethylated compared to healthy controls (48). Luo, S., et al. found an interesting finding regarding the promoter region of IFI44L in naive CD4+ T cells of SLE patients. They observed a lack of methylation at the CG site, which aligns with the SLE Disease Activity Index (SLEDAI) scores. Conversely, it is interesting that IFI44L revealed upregulation in CD4+ T cells, indicating that the SLE had no impact on the hypomethylation of naive CD4+ T cells in patients (20). This may be due to the lack of transcription factors required for the IFI44L in naive CD4+ T cells. In epigenetics, the existence of IFI44L with low methylation induces naive CD4+ T cells to activate each other for a prompt type I IFN response (20). The results indicate that some ISGs such as IFI44L may react to IFN-I during T cell activation. This provides evidence for hyper-responsiveness to IFN-I in T cells (20). Similarly, IFI44L was found to be hypomethylated at several CpG sites in CD19+ B cells, researchers performed RNA sequencing on whole blood and CD19+ B cells in a separate investigation of primary SS and found that interferon (IFN) -induced genes were significantly hypomethylated (16). It is markedly elevated in mononuclear cells in SLE patients, it can also be regulated by STAT3 in monocytes (21). Based on the ChIP-qPCR assay, it was observed that the binding of the IFI44L promoter to STAT3 was increased, while the DNA methylation level of IFI44L decreased substantially in STAT3 overexpressed monocytes (21). Therefore, to explore the significance of IFI44L upregulation in monocytes associated with SLE, researchers transfected and overexpressed IFI44L into monocyte-derived DCs (Mo-DCs) to induce differentiation. The results indicated an increase in the mRNA levels of CD40, CD80, CD83, and CD86. The co-culture of naive CD4+ T cells and Mo-DCs cells led to a remarkable upregulation of IFN-γ and IL-17α This finding suggests that IFI44L may be required to maintain the Th1/Th17-related cytokines (21). This study showed that STAT3 can cause DNA demethylation of the IFI44L promoter, thereby regulating its expression. Overexpression of IFI44L can sustain the upregulation of Th1/Th17-related cytokines (21). Zhao, L., et al. investigated the methylation status of the IFI44L gene in SLE patients’ peripheral blood cells using flow cytometry. In resting naive B cells (rNAV) of SLE patients, it was noted that the promoter of IFI44L had hypomethylation of two sites (57.4 vs.27.1 and 93.6 vs.82.02), which suggested that the B lymphocytes of SLE patients suffer epigenetic alterations early in their growth process (49).
Moreover, Zhao, L., et al. found that IVRPIE (a kind of lncRNA) could promote the transcription of IFI44L by affecting the methylation of its transcription start site (50). Following that, heterogeneous nuclear ribonuclear protein U (hnRNP U) was silenced, and IFI44L expression was markedly reduced in IVRPIE overexpressing cells, as shown by RT-qPCR analysis. This finding suggests that hnRNP U is involved in the IVRPIE modulation of IFI44L transcripts (50). HBV carriers are early-onset types of liver cancer, and their genomes exhibit high enrichment of genes on immune-related pathways. In the peripheral leukocytes of patients with early-onset HCC, the cg06872964 probe was found to be located in the proximal promoter of IFI44L and detected methylation in this region (13). In contrast to the healthy control group, liver cancer patients show a reduced expression of IFI44L. This gene not only impedes the metastasis, invasion, and resistance of tumor cells to doxorubicin but also restricts their migration and invasion via the met/Src signaling pathway (51). Collectively, the progression of numerous diseases has been linked to the methylation of IFI44L.
Multiple studies have demonstrated that IFI44L has functions in the formation and modulation of microenvironments. A recent study by Woeckel, V.J., et al. discovered that human osteoblasts reveal sensitivity to the IFN (52). Osteoblasts can stimulate the production of the IFN target gene IFI44L. The ECM is an extremely complex environment that includes several chemicals such as fibrin, which play a vital role in regulating cell growth and differentiation (53). In the early stages of human osteoblast differentiation, the formation of ECM is influenced by the immune cytokine IFN-β, leading to a decrease in mineralization. Therefore, IFI44L may have a certain therapeutic effect in the early stage of osteoblast differentiation (52). Furthermore, IFI44L expression was substantially upregulated in fibroblasts damaged in the heart, indicating that type I INFs may be involved in the damage to heart fibroblasts (54). In a high-throughput sequencing study of Channel Catfish (Ictalurus punctatus), the researchers found that IFI44L might have a significant impact on immune activity and ion transport. However, the precise mechanism underlying this remains unexplored (55).
This gene has significant implications and shows promising potential as both a diagnostic marker and therapeutic target for various human diseases. Next, a detailed analysis was carried out about two areas of malignant tumors and non-neoplastic diseases.
Researchers identified IFI44L as the Hub gene in the protein-protein interaction (PPI) network and a substantially elevated gene in peripheral blood mononuclear cells (PBMCs) by analyzing microarray expression data of all solid tumors (3). When IFI44L is overexpressed, patients’ survival rates are higher, thusthis gene has the potential to function as an immune-related suppressor in tumor therapy (3).
Skin cancer is a very common tumor and its prevention is very crucial. Current scientific research primarily centers around developing effective strategies for its prevention (56, 57). The CDKN2A gene mutation is linked to an increased vulnerability to skin cancer. Compared with normal control cells, in CDKN2A mutant cell lines, the expression of several immune genes, including IFI44L, is significantly increased. This suggests that dysregulation of the transcriptome serves as an important marker for the early progression of skin cancer (58).
Oral cancer (OC) ranks as the 11th most prevalent tumor globally, with approximately 50% of all head and neck tumors originating from the mouth (59, 60). Reyimu, A., et al. observed that IFI44L was highly expressed in OC in contrast to normal tissue (2). Based on the results of COX regression analysis, IFI44L was identified as an independent prognostic marker of overall survival (OS). Furthermore, receiver operating characteristic (ROC) analysis yielded an area under the curve (AUC) of 0.802 for IFI44L, which differentiated OC tissue from normal tissue, indicating that it may serve as a potential predictive biomarker for OC (2).
A tumor known as NPC has been linked to EBV infection (61). Correlation with INF signaling pathways and regulation by miR-9 (34, 62, 63) led to the discovery that IFI44L was substantially upregulated in NPC cells and tissues; Thus, it may represent a possible target for modulating the advancement of NPC. Moreover, IFI44L was also highly expressed in laryngeal carcinoma HEp2 cells (6). Increased IFI44L expression might facilitate apoptosis mediated by IFN α-1a (6).
Early diagnosis of lung cancer (LC) is challenging due to its typically poor prognosis. Some new treatment methods or diagnostic markers are urgently needed to solve this problem (64). Zeng, Y., et al. found a strong association between IFI44L and LC tumor immune cells (7). A prognostic model was then developed to assess the prognosis of lung cancer patients using ten IFI44L-associated immune modulators. This model yielded an accurate AUC, with all results exceeding 0.75 in a three-year ROC analysis, and was capable of predicting the OS rate for patients with lung adenocarcinoma (LUAD). The prognosis and survival of patients are positively correlated with IFI44L expression. Further, cell function experiments showed that high expression of IFI44L inhibited the propagation, migration, and penetration of LC cells (7). In addition, IFI44L was significantly downexpressed in lung tumorigenesis, and its low expression may be associated with DNA methylation. It was found to facilitate tumor cell apoptosis and may be a tumor suppressor gene through involving the JAK/STAT1 pathway (14). Based on these findings, IFI44L is a potent diagnostic marker for LC and may have immunotherapeutic effects.
The HCC is the most prevalent primary malignant tumor (65). Based on the studies, methylation of the proximal promoter of IFI44L is strongly associated with the early advancement of LC (13). Huang, W.C., et al. revealed that HCC cells display low levels of IFI44L and that low expression is associated with a poorer prognosis for the tumor. Low IFI44L expression may facilitate the invasion and metastasis of LC cells via the Met/Src signaling pathway (51). These studies suggest that IFI44L is a promising therapeutic target and biomarker.
The prognosis for pancreatic cancer is unfortunately very poor, which makes it the primary cause of cancer-related deaths (66). There is a significant correlation between IFI44L overexpression and poorer prognosis among patients with pancreatic ductal adenocarcinoma (PDAC) (p = 0.0153) (12). Furthermore, it may affect pancreatic cancer growth through mechanisms of cell-matrix adhesion and extracellular matrix. These results suggest that IFI44L could potentially serve as a new biomarker for PDAC.
Osteosarcoma (OS) is the most prevalent primary malignant bone tumor, characterized by its high aggression and the tendency for metastasis (67). Bone has a highly specific immune environment, and many immune signal molecules play a significant role in bone homeostasis (68). For example, IFI44L was found to be decreased in OS and be related to the higher survival rate (p = 0.022) (35). The miR-628-5p, classified as a microRNA, inhibits the expression of IFI44L, thereby promoting the propagation, infiltration, and migration of MG-63 cells (35). This result suggests that miR-628-5p may function as a tumor suppressor in OS.
The survival rate of patients with primary central nervous system lymphoma (PCNSL) is another factor associated with IFI44L (69). Takashima, Y., et al. developed a prognosis prediction formula that could effectively differentiate between patients with poor and favorable prognoses. The formula including IFI44L, showed promising results with an AUC of 0.76 for 2 years and 0.69 for 5-year survival, as evaluated by ROC analysis (69). These results demonstrated that IFI44L may hold potential as a therapeutic target for PCNSL.
Acute lymphoblastic leukemia (ALL) is a hematological malignancy that predominantly affects children (70). A prognostic risk model was developed using IFI44L, which displays a commendable predictive value ((AUC) for predicted survival at 1, 3, and 5 years is ≥ 0.7 in the datasets examined). In the high-risk group and early relapse patients, IFI44L expression was reduced (71). Thus, this model may serve as a novel tool for predicting the prognosis and early detection of ALL recurrence.
Myelodysplastic syndrome (MDS) is a hematologic diease with a high probability of developing into acute leukemia (72). They observed that patients with high IFI44L levels in MDS had a more unfavorable prognosis (73), additionally, it may affect pancreatic cancer growth through mechanisms of cell-matrix adhesion and extracellular matrix, suggesting that IFI44L could serve as a valuable and significant survival indicator in MDS. It is evident that IFI44L exhibits varying expression patterns in different types of cancer. This is because different types of cancer exhibit genomic instability changes at both the early and late stages of tumor development (74). Gene expression is regulated at multiple levels, such as transcription, mRNA stability, mRNA translation, and post-translational modifications (75). Consequently, how these regulations affect protein levels and cancer progression remains a focal point of research in each cancer field. Although gene expression may differ, in certain tumors, integrating other diagnostic methods (such as imaging studies, pathological examinations, and multi-gene panel testing) for a comprehensive assessment can still enhance the accuracy of diagnosis.
In general, IFI44L has a significant involvement in malignant tumors. A huge number of genome sequencing studies have revealed that it is associated with the formation of tumors, can alter tumor growth and migration, and is associated with patient prognosis. The expression level of the IFI44L gene can be used to predict patient survival rates, and it has the potential to be a valuable diagnostic and prognostic marker.
Besides the involvement of IFI44L in tumors, it is frequently observed in non-tumor conditions, primarily immune disorders, viral infections, and bacterial infectious diseases.
It is an autoimmune disease with a very complex pathogenesis, mostly caused by autoimmune defects (76). Multiple studies have demonstrated the significant role of IFN-α in SLE. Among these studies, IFI44L has emerged as a key gene with promising potential as a biomarker for diagnosis and treatment (1, 8, 28, 77–79). Zhao, X., et al. found that IFI44L had AUC values ≥ 0.8 in ROC analysis when distinguishing SLE from healthy patients (80). Fan, H., et al. validated that IFI44L expression in SLE patients trended higher in comparison to the healthy control group (81). More interestingly, IFI44L can also be induced by estrogen, suggesting that gender differences should be considered when diagnosing SLE (81). It has been revealed that low methylation genes comprise the majority of the type I INF pathway, with IFI44L being particularly significant due to its close association with the development of SLE (48, 82–84). It was found that in the immune cells of SLE patients, including T cells, monocytes (85), granulocytes, or B cells, the CpGs site of IFI44L display different degrees of methylation (49, 86, 87). DNA demethylation of the IFI44L promoter can be induced by TET2 (which is recruited by STAT3), leading to increased IFI44L expression and SLE via stimulation of Mo-DC maturation (21). Different IFI44L methylation levels show significant differences between SLE patients and healthy controls or other immune diseases like rheumatoid arthritis (RA). This distinction is highly accurate and reliable, with a sensitivity of 0.81 and 0.84 for SLE and RA patients, respectively (83). The results suggest that IFI44L methylation could serve as a valuable epigenetic diagnostic biomarker for SLE (8, 88). Moreover, the methylation of IFI44L can also distinguish between discoid lupus erythematosus (DLE) and SLE (89). It was demonstrated by Zhang, B., et al. that IFI44L was less methylated than the healthy control and the DLE groups. It can distinguish between SLE from healthy individuals (specificity; 0.98 and sensitivity; 0.96) and SLE from DLE (specificity; 0.7255 and sensitivity; 0.96) when the methylation threshold is set to 25% (89). When the methylation level of IFI44L reaches 0.329, its promoter can distinguish SLE from primary antiphospholipid syndrome (APS) (sensitivity; 0.93 and specificity; 0.8) (90). The identification of IFI44L has major implications for the diagnosis of SLE and provides significant possibilities for the advancement of targeted therapies in the treatment of SLE patients (91).
Sjogren’s Syndrome (SS) is an autoimmune disease with an unclear pathogenesis (92). Studies have shown that SS is also associated with some IFN genes such as IFI44L (93–95), the hyperactivation of the IFN system is recognized as a crucial factor in the development of disease (96). Similar to SLE, IFI44L is also found to be hypomethylated in naive CD4+ T cells of SS patients, indicating its involvement in the development of SS (4). Strong associations between IFI44L, a component gene of the IFN type I signature, and myxovirus-resistance protein A (MxA), which can evaluate the activity of SS, suggest that IFI44L may also be capable of measuring the activity of SS (97). Chen, S., et al. found that low methylation of IFN-associated genes such as IFI44L is a common feature of T cells and monocytes in many immune systems diseases such as SLE, RA, and systemic sclerosis (SSc), and may function as biomarkers to differentiate and diagnose these diseases (98, 99). Significant hypomethylation of IFI44L was also observed in whole blood and CD19+ B cells of SS patients (16). Salivary samples and salivary gland biopsy samples from patients diagnosed with SS showed significant differences in IFI44L expression. These findings indicate that IFI44L is present not only in peripheral blood but also in saliva and salivary glands. Thus, IFI44L has the potential to serve as a potent diagnostic biomarker for SS (100).
DNA methylation sites that are connected with RA are present in ISGs (101). Cooles, F.A.H., et al. found that the IFN gene signature (IGS) may influence the epigenetic regulation of lymphocytes in RA patients (19). In B cells and T cells, the levels of IGS and IFN-α are correlated with varying degrees of methylation site abundance; IGS serves as an indicator of IFN-α protein expression. When the IGS score is increased, the prognosis of patients is worse, suggesting that IGS may be a good prognostic biomarker in RA (19). Thousands of molecules were screened by Yadalam, P.K., et al. to determine that IFI44L is a drug target for RA. Vemurafenib is the most suitable drug for the IFI44L target, and the combination of the two has good stability. Vemurafenib may be an anti-inflammatory drug for treating this disease (18). In RA patients, IFI44L primarily regulates the IFN signaling pathway via DCs, resulting in a crucial molecular target (42). Moreover, the IFN signature (including IFI44L and other genes) can predict the sensitivity of RA patients to rituximab (RTX) with excellent ROC analysis (an AUC of 0.87) (102).
In addition to its primary association with SLE, RA, and SS, IFI44L also has a significant impact on various other autoimmune diseases. Different levels of methylation were also found at the CpGs site of IFI44L in monocytes and macrophages of patients with SSc, suggesting that IFI44L contributes to the development of this disease (103–105). Considerable expression of IFI44L was observed in lung microvascular endothelial cells (MVECs) during the study of SSc-induced vascular lesions, this result may indicate the role of IFI44L in the vasculopathy caused by SSc (106). However, very few studies have investigated the molecular mechanism of mixed connective tissue disease (MCTD) and the researchers found the epigenetic signal of IFI44L and confirmed that it is related to the heritability of MCTD. The diagnostic AUC value reached 0.97, 0.9, and 0.84 when distinguishing this disease from healthy people, RA, and SSc patients, suggesting the potential of IFI44L as an epigenetic biomarker for the MCTD (107). Lerkvaleekul, B., et al. found that IFI44L was also associated with clinical activity in juvenile dermatomyositis (JDM) (108). The IFI44L gene has substantial upregulation in CD4+ and CD8+ T cells of patients with large-vessel involvement in giant cell arteritis (LV-GCA). This upregulation may potentially be linked to the pathogenesis of the disease (109). The IFI44L gene is differentially expressed in both membranous nephropathy (MN) and lupus nephritis (LN). It is a member of a group of differentially expressed genes (DEGs) that has a high diagnostic value in differentiating patients with MN from those with LN (110).
The IFI44L gene can participate in virus and bacterial replication (39, 111). It is frequently observed as a DEG in various diseases caused by bacteria and viruses. Its presence is significant in the progression of these diseases (17, 112). It can also distinguish some diseases infected by bacteria and viruses (113), such as acute febrile illness caused by viruses or bacterial infections (114), and thus, IFI44L can significantly contribute to the differential treatment of febrile diseases.
To differentiate between bacterial and viral infections, Herberg, J.A., et al. identified the IFI44L gene for the first time in 2016 (115). In a Disease Risk Score (DRS) composed of IFI44L and FAM89A, IFI44L was highly expressed in patients with bacterial infections. The AUC of this DRS exceeded 0.89 when distinguishing bacterial infections, viral infections, and inflammatory diseases (115). Following this, Pennisi, I., et al. used a disposable diagnostic cartridge for clinical detection using semiconductor sensing technology and the DRS; this represents a significant breakthrough and invention in the detection of viral and bacterial infections (116). Moreover, they observed that IFI44L could differentiate between bacterial and viral infections with an AUC of 0.97. Further, IFI44L expression was reduced during the acute infection phase compared to the recovery phase, indicating that the dynamic change of IFI44L could serve as a prognostic indicator for the therapeutic efficacy of the disease (114). Experimental studies revealed that when IFI44L was knocked out, the survival rate of cells infected with Mycobacterium Tuberculosis increased. Similarly, when the gene was highly expressed, the survival rate of cells increased; these results suggest that IFI44L has effective antibacterial activity and may clear the burden of Mycobacterium tuberculosis from macrophages (27).
Overexpression of IFI44L can affect the activity of many viruses. For instance, when lung epithelial cells are infected with respiratory syncytial virus (RSV), the mRNA level of IFI44L is substantially upregulated. This upregulation in IFI44L can effectively hinder the replication of the virus and lower the stimulation of the IFN pathway caused by respiratory viruses. These results suggest possible possibilities for therapeutic interventions in RSV (117, 118). In germ cells, overexpression of IFI44L can inhibit the production of Zika virus (ZIKV) (119). The COVID-19 pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has had a significant impact globally (120). In PBMCs infected by SARS-CoV-2, IFI44L is upregulated and involved in some immune responses (121–123). Patients infected with COVID-19 showed varying levels of methylation in IFI44L (124). Expression of IFI44L from endothelial cells was also increased in SARS-CoV-2-infected heart tissue, and this upregulation was found to be related to the INF pathway (125). These outcomes indicate that IFI44L could be a promising target for treating SARS-CoV-2 infections. Moreover, differential upregulation of IFI44L has been found in cells or tissues infected by many other viruses such as Hepatitis E virus (HEV), Influenza A virus (IAV), and rhinoviruses (RV), indicating that IFI44L holds significant promise for treating diseases caused by viral infections (126–128). Another interesting finding of Brochado et al. revealed that the IFI44L showed a pattern of overexpression in most antiviral responses; however, it was one of the most downregulated genes in patients co-infected with HIV and HCV (17, 129). Similarly, they also found that IFI44L expression was downregulated in HIV-infected macrophages, possibly due to HIV’s strong resistance to IFN-mediated immune responses (130).
In addition to the above-mentioned diseases, IFI44L also has a significant impact on various other system diseases, such as those affecting the nervous, cardiovascular, and endocrine systems. Many inflammation-related genes, including IFI44L, are downregulated in the brain tissue of patients with Major Depressive Disorder (MDD), underscoring the role of neuroinflammation in MDD (131). Significant associations were also observed between a risk score model (including IFI44L) and the clinical symptoms of patients diagnosed with bipolar disorder and schizophrenia (132). Feenstra et al. identified IFI44L on chromosome 1p31.1 as the initial locus for febrile seizures related to the MMR vaccine. They recognized IFI44L as a gene linked to febrile seizures unrelated to the MMR vaccine (p = 1.2 × 10-9 versus controls and p = 5.9 × 10-12 versus controls, respectively) (133). In diabetes and diabetes-related complications such as diabetic foot ulcer (DFU) and proliferative diabetic retinopathy (FDR), IFI44L has been listed as an immune-related marker and a possible therapeutic target for metformin (134–136). Analysis of the whole genome of leukocytes in obese patients revealed an association between IFI44L and body mass index (BMI). This suggests that IFI44L may serve as a potential biomarker in the diagnosis of obesity (137). The IFI44L was found to be strongly associated with cardiovascular disease. Its expressions were found to be greatly increased and strongly associated with myocardial infarction (MI) in the heart tissues of rat MI models. The expression of IFI44L was also effectively suppressed by metoprolol, indicating that it could serve as a potential marker for human ischemic cardiomyopathy (ISCM) (138). It can mediate fibroblast damage of the fetal heart in SSA/Ro autoantibody-associated congenital heart block (CHB) (54), indicating that IFI44L can serve as a potential clinical intervention target. When circulating endothelial progenitor cells (EPCs) were exposed to monomeric C-reactive protein (mCRP), there was a significant upregulation of interferon-responsive genes, including IFI44L (139). It is also differentially expressed in ischemic stroke and pulmonary arterial hypertension (PAH), revealing its potential as a therapeutic target in PAH (140, 141). It is highly expressed or functions as a hub gene in the PPI network (5, 142–146), in several chronic inflammatory diseases, including airway inflammation, periodontitis, neuritis, psoriasis, necrosis of the femoral head, and parasitic diarrhea. This gene association may have implications for innate immune response pathways and inflammation. It is also involved in the pathogenesis of premature ovarian failure (POF) as a necrotic gene (147).
When bacteria, viruses, and tumor cells infiltrate the body, IFN can trigger the activation of multiple ISGs within the host cell. These ISGs then produce proteins that serve specific biological functions, and these proteins can independently or synergistically participate in a wide range of vital processes (148, 149). They are very important immune-related regulatory genes (150). The functions of these ISG products are highly varied to impeding viral replication, growth and migration of tumor cells, stimulating the propagation of immune cells, enhancing cytotoxicity via regulation of multiple signaling pathways, facilitating protein synthesis and modification (151–155). Among them, the antiviral effect stands out as the most crucial and extensively studied. However, limited research has been conducted on tumors and autoimmune diseases. Recently, an increasing number of studies have revealed the significant impact of ISG in the field of tumor and immune function.
As an ISG, IFI44L has antiviral, antibacterial, and antitumor properties comparable to those of other ISGs. This review also provided a summary of the other crucial functions of IFI44L, including its involvement in ECM formation, conduction of cardiac electrical signals, and induction of cell apoptosis, along with signal transduction and methylation modification (6, 52, 54, 55). Unfortunately, many investigators have only discovered that IFI44L influences the growth, migration, and apoptosis of tumor cells. The precise mechanism by which it acts in the tumor immune environment has not been fully investigated. The clinical development of anti-tumor drugs has resulted in a degree of resistance. Several genes have been extensively researched as potential targets for drug development. However, IFI44L stands out due to its ability to evaluate a patient’s response to anti-tumor drugs like doxorubicin (156). It is a key gene in SLE, SS and RA. When it relates to exploring autoimmune diseases, the IFI44L has proven to be incredibly valuable. It has a crucial role in diagnosing diseases, differentiating patients, and assessing prognosis. As a result, it has become a promising candidate for both diagnostic markers and therapeutic targets in various diseases (30). However, in many studies, IFI44L is only one of the genes that constitute a prognostic risk model. It evaluates disease prognosis along with other immune-related genes, which is not very accurate in assessing the effect of IFI44L independently. Furthermore, IFI44L manifests a favorable impact on resistance to bacteria and viruses. While HIV acquires a certain degree of resistance to it, it is still capable of inhibiting the replication of other viruses. Further study is needed to better understand its inhibitory effect on HIV (130). Moreover, IFI44L has been linked to certain neurological, cardiovascular, chronic inflammatory, and endocrine diseases (131, 134, 138, 142).
In the application of clinical diagnosis, IFI44L can be used as a potential target for tumor drug development, as well as a differential target for bacterial and viral diagnosis. In addition, IFI44L can also be used as a tumor prognostic model to estimate the survival rate of patients, playing an important role in disease diagnosis and treatment. However, the molecular mechanisms underlying IFI44L’s role in disease development have not been extensively studied. Most current research focuses on the surface-level relationship between expression levels and disease progression, failing to fully uncover its intrinsic biological processes and regulatory networks. Although a few studies have explored the regulatory pathways of IFI44L and provided some valuable insights, these findings have yet to be effectively translated into clinically practical diagnostic markers, limiting their potential application in medical practice. Additionally, the significant specificity of IFI44L expression in various types of tumors and immune diseases makes it a promising tool for distinguishing patients from healthy individuals. However, due to the lack of tissue specificity, accurate clinical diagnosis often requires the combination of other auxiliary diagnostic methods. Furthermore, there are limited reports in the existing literature on the sensitivity value of IFI44L in early diagnosis, suggesting that future research should pay more attention to its sensitivity performance at different stages of disease. This would better assess its potential as an early diagnostic marker. Through further research and technological advancements, IFI44L has the potential to be developed into an efficient and reliable clinical diagnostic tool, providing robust support for the early detection and treatment of diseases.
In summary, IFI44L emerged as a promising biomarker in studies examining a range of diseases. A limited number of studies investigated the relationship between IFI44L and diseases. Its application in clinical practice was also uncommon. Few studies developed IFI44L into a disposable diagnostic cartridge for use in clinical settings. Despite this illustrating that IFI44L has the potential to yield results for clinical applications, the current pool of research is quite limited. In the future, more research is required regarding the therapeutic and diagnostic potential of IFI44L, not only for its application in drug development but also in clinical diagnosis.
JD: Writing – original draft. HL: Writing – original draft. SY: Writing – original draft. HZ: Writing – original draft. ZZ: Writing – original draft. KL: Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the Project of NINGBO Leading Medical & Health Discipline (No. 2022-F01); Medical and Health Research Project of Zhejiang Province, Grant/Award (2024KY1469).
We acknowledge the Project of NINGBO Leading Medical & Health Discipline (No. 2022-F01); Medical and Health Research Project of Zhejiang Province, Grant/Award (2024KY1469).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
IFI44L: Interferon-induced protein 44-like
ISGs: interferon-stimulated genes
OC: oral cancer
RCC: renal cell carcinoma
SS: Sjogren’s syndrome
SLE: systemic lupus erythematosus
PDAC: pancreatic ductal adenocarcinoma
HCC: hepatocellular carcinoma
LC: lung cancer
ECM: extracellular matrix
pDCs: plasmacytoid dendritic cells
TBK1: TANK-binding kinase
LMKB: Limkain B
HUVECs: human umbilical vein endothelial cells
3′-UTR: 3’ untranslated region
NPC: nasopharyngeal carcinoma
OS: osteosarcoma
RIPK3: serine/threonine-protein kinase 3
JEV: Japanese encephalitis virus
IFNLP: IFN-like protein
IFN: Interferon
DCs: dendritic cells
HIV-1: human immunodeficiency virus type 1
siRNA: short interfering RNA
HBV: hepatitis B virus
cSLE: Childhood-onset SLE
GMECs: goat mammary epithelial cells
SLEDAI: SLE Disease Activity Index
Mo-DCs: monocyte-derived DCs
rNAV: resting naive B cells
hnRNP U: heterogeneous nuclear ribonuclear protein U
PPI: protein-protein interaction
PBMCs: peripheral blood mononuclear cells
OC: Oral cancer
OS: overall survival
ROC: receiver operating characteristic
AUC: area under the curve
LC: lung cancer
LUAD: lung adenocarcinoma
PCNSL: primary central nervous system lymphoma
ALL: Acute lymphoblastic leukemia
MDS: Myelodysplastic syndrome
RA: rheumatoid arthritis
DLE: discoid lupus erythematosus
APS: antiphospholipid syndrome
MxA: myxovirus-resistance protein A
SSc: systemic sclerosis
IGS: IFN gene signature
RTX: rituximab
MVECs: microvascular endothelial cells
MCTD: mixed connective tissue disease
LV-GCA: large-vessel involvement in giant cell arteritis
MN: membranous nephropathy
DEGs: differentially expressed genes
DRS: Disease Risk Score
RSV: respiratory syncytial virus
ZIKV: Zika virus
SARS-CoV-2: severe acute respiratory syndrome coronavirus 2
HEV: Hepatitis E virus
IAV: Influenza A virus
RV: rhinoviruses
MDD: Major Depressive Disorder
DFU: diabetic foot ulcer
FDR: proliferative diabetic retinopathy
BMI: body mass index
MI: myocardial infarction
ISCM: ischemic cardiomyopathy
CHB: congenital heart block
EPCs: endothelial progenitor cells
mCRP: C-reactive protein
PAH: pulmonary arterial hypertension
POF: premature ovarian failure
1. Zhang Q, Wu R, Tan Y, Huang J, Luo S. Novel polymorphism of IFI44L associated with the susceptibility and clinical characteristics of systemic lupus erythematosus in a Chinese population. Int Immunopharmacol. (2023) 117:109979. doi: 10.1016/j.intimp.2023.109979
2. Reyimu A, Chen Y, Song X, Zhou W, Dai J, Jiang F. Identification of latent biomarkers in connection with progression and prognosis in oral cancer by comprehensive bioinformatics analysis. World J Surg Oncol. (2021) 19:240. doi: 10.1186/s12957-021-02360-w
3. Chen S, Liu M, Liang B, Ge S, Peng J, Huang H. Identification of human peripheral blood monocyte gene markers for early screening of solid tumors. PloS One. (2020) 15:e0230905. doi: 10.1371/journal.pone.0230905
4. Altorok N, Coit P, Hughes T, Koelsch KA, Stone DU, Rasmussen A. Genome-wide DNA methylation patterns in naive CD4+ T cells from patients with primary Sjögren’s syndrome. Arthritis Rheumatol. (2014) 66:731–9. doi: 10.1002/art.38264
5. Sabir MJ, Low R, Hall N, Kamli MR, Malik MZ. A bioinformatics approach to identifying potential biomarkers for cryptosporidium parvum: A coccidian parasite associated with fetal diarrhea. Vaccines (Basel). (2021) 9(12):1427. doi: 10.3390/vaccines9121427
6. Xin XL, Zhang R, Yuan XM, Liu L. Mechanisms of IFNalpha-1a-induced apoptosis in a laryngeal cancer cell line. Med Sci Monit. (2019) 25:7100–14. doi: 10.12659/MSM.917097
7. Zeng Y, Zhang Z, Chen H, Fan J, Yuan W, Li J. Comprehensive analysis of immune implication and prognostic value of IFI44L in non-small cell lung cancer. Front Oncol. (2021) 11:798425. doi: 10.3389/fonc.2021.798425
8. Teruel M, Sawalha AH. Epigenetic variability in systemic lupus erythematosus: what we learned from genome-wide DNA methylation studies. Curr Rheumatol Rep. (2017) 19:32. doi: 10.1007/s11926-017-0657-5
9. Gómez-Carballa A, Cebey-López M, Pardo-Seco J, Barral-Arca R, Rivero-Calle I, Pischedda S. A qPCR expression assay of IFI44L gene differentiates viral from bacterial infections in febrile children. Sci Rep. (2019) 9:11780. doi: 10.1038/s41598-019-48162-9
10. Du J, Huang T, Zheng Z, Fang S, Deng H, Liu K. Biological function and clinical application prospect of tsRNAs in digestive system biology and pathology. Cell Commun Signal. (2023) 21:302. doi: 10.1186/s12964-023-01341-8
11. Huang T, Du J, Chen C, Zheng Z, Fang S, Liu K. Dysregulation of transfer RNA-derived small RNAs that regulate cell activity and its related signaling pathways in human cancers. Curr Gene Ther. (2023) 23:291–303. doi: 10.2174/1566523223666230601102506
12. Li H, Wang X, Fang Y, Huo Z, Lu X, Zhan X. Integrated expression profiles analysis reveals novel predictive biomarker in pancreatic ductal adenocarcinoma. Oncotarget. (2017) 8:52571–83. doi: 10.18632/oncotarget.16732
13. Kao WY, Yang SH, Liu WJ, Yeh MY, Lin CL, Liu CJ. Genome-wide identification of blood DNA methylation patterns associated with early-onset hepatocellular carcinoma development in hepatitis B carriers. Mol Carcinog. (2017) 56:425–35. doi: 10.1002/mc.22505
14. Zeng Y, Chen HQ, Zhang Z, Fan J, Li JZ, Zhou SM. IFI44L as a novel epigenetic silencing tumor suppressor promotes apoptosis through JAK/STAT1 pathway during lung carcinogenesis. Environ pollut. (2023) 319:120943. doi: 10.1016/j.envpol.2022.120943
15. Rupa D, Chuang HW, Hu CE, Su WM, Wu SR, Lee HS. ACSL4 upregulates IFI44 and IFI44L expression and promotes the proliferation and invasiveness of head and neck squamous cell carcinoma cells. Cancer Sci. (2024) 115:3026–40. doi: 10.1111/cas.v115.9
16. Imgenberg-Kreuz J, Sandling JK, Almlöf JC, Nordlund J, Signér L, Norheim KB. Genome-wide DNA methylation analysis in multiple tissues in primary Sjögren’s syndrome reveals regulatory effects at interferon-induced genes. Ann Rheum Dis. (2016) 75:2029–36. doi: 10.1136/annrheumdis-2015-208659
17. BroChado Ó, Martínez I, Berenguer J, Medrano L, González-García J, Jiménez-Sousa M. HCV eradication with IFN-based therapy does not completely restore gene expression in PBMCs from HIV/HCV-coinfected patients. J BioMed Sci. (2021) 28:23. doi: 10.1186/s12929-021-00718-6
18. Yadalam PK, Sivasankari T, Rengaraj S, Mugri MH, Sayed M, Khan SS. Gene interaction network analysis reveals IFI44L as a drug target in rheumatoid arthritis and periodontitis. Molecules. (2022) 27(9):2749. doi: 10.3390/molecules27092749
19. Cooles FAH, Tarn J, Lendrem DW, Naamane N, Lin CM, Millar B. Interferon-α-mediated therapeutic resistance in early rheumatoid arthritis implicates epigenetic reprogramming. Ann Rheum Dis. (2022) 81:1214–23. doi: 10.1136/annrheumdis-2022-222370
20. Coit P, Jeffries M, Altorok N, Dozmorov MG, Koelsch KA, Wren JD. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naïve CD4+ T cells from lupus patients. J Autoimmun. (2013) 43:78–84. doi: 10.1016/j.jaut.2013.04.003
21. Luo S, Wu R, Li Q, Zhang G. Epigenetic regulation of IFI44L expression in monocytes affects the functions of monocyte-derived dendritic cells in systemic lupus erythematosus. J Immunol Res. (2022) 2022:4053038. doi: 10.1155/2022/4053038
22. McDowell IC, Modak TH, Lane CE, Gomez-Chiarri M. Multi-species protein similarity clustering reveals novel expanded immune gene families in the eastern oyster Crassostrea virginica. Fish Shellfish Immunol. (2016) 53:13–23. doi: 10.1016/j.fsi.2016.03.157
23. Meng X, Yang D, Yu R, Zhu H. EPSTI1 is involved in IL-28A-mediated inhibition of HCV infection. Mediators Inflammation. (2015) 2015:716315. doi: 10.1155/2015/716315
24. Qiao X, Li Y, Jin Y, Wang S, Hou L, Wang L. The involvement of an interferon-induced protein 44-like (CgIFI44L) in the antiviral immune response of Crassostrea gigas. Fish Shellfish Immunol. (2022) 129:96–105. doi: 10.1016/j.fsi.2022.08.064
25. Zahoor MA, Xue G, Sato H, Aida Y. Genome-wide transcriptional profiling reveals that HIV-1 Vpr differentially regulates interferon-stimulated genes in human monocyte-derived dendritic cells. Virus Res. (2015) 208:156–63. doi: 10.1016/j.virusres.2015.06.017
26. Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. (2011) 472:481–5. doi: 10.1038/nature09907
27. Jiang H, Tsang L, Wang H, Liu C. IFI44L as a forward regulator enhancing host antituberculosis responses. J Immunol Res. (2021) 2021:5599408. doi: 10.1155/2021/5599408
28. Bing PF, Xia W, Wang L, Zhang YH, Lei SF, Deng FY. Common marker genes identified from various sample types for systemic lupus erythematosus. PloS One. (2016) 11:e0156234. doi: 10.1371/journal.pone.0156234
29. Kong L, Liu G, Deng M, Lian Z, Han Y, Sun B. Growth retardation-responsive analysis of mRNAs and long noncoding RNAs in the liver tissue of Leiqiong cattle. Sci Rep. (2020) 10:14254. doi: 10.1038/s41598-020-71206-4
30. Bodewes ILA, Huijser E, van Helden-Meeuwsen CG, Tas L, Huizinga R, Dalm V. TBK1: A key regulator and potential treatment target for interferon positive Sjögren’s syndrome, systemic lupus erythematosus and systemic sclerosis. J Autoimmun. (2018) 91:97–102. doi: 10.1016/j.jaut.2018.02.001
31. Bloch DB, Li P, Bloch EG, Berenson DF, Galdos RL, Arora P. LMKB/MARF1 localizes to mRNA processing bodies, interacts with Ge-1, and regulates IFI44L gene expression. PloS One. (2014) 9:e94784. doi: 10.1371/journal.pone.0094784
32. Ji X, Hua H, Shen Y, Bu S, Yi S. Let-7d modulates the proliferation, migration, tubulogenesis of endothelial cells. Mol Cell Biochem. (2019) 462:75–83. doi: 10.1007/s11010-019-03611-x
33. Li J, Lin TY, Chen L, Liu Y, Dian MJ, Hao WC. miR-19 regulates the expression of interferon-induced genes and MHC class I genes in human cancer cells. Int J Med Sci. (2020) 17:953–64. doi: 10.7150/ijms.44377
34. Gao F, Zhao ZL, Zhao WT, Fan QR, Wang SC, Li J. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Commun. (2013) 431:610–6. doi: 10.1016/j.bbrc.2012.12.097
35. Wang JY, Wang JQ, Lu SB. miR-628-5p promotes growth and migration of osteosarcoma by targeting IFI44L. Biochem Cell Biol. (2020) 98:99–105. doi: 10.1139/bcb-2019-0001
36. Vishnubalaji R, Shaath H, Alajez NM. Protein coding and long noncoding RNA (lncRNA) transcriptional landscape in SARS-CoV-2 infected bronchial epithelial cells highlight a role for interferon and inflammatory response. Genes (Basel). (2020) 11(7):760. doi: 10.3390/genes11070760
37. Chan JCM, Mohammad KN, Zhang LY, Wong SH, Chan MC. Targeted profiling of immunological genes during norovirus replication in human intestinal enteroids. Viruses. (2021) 13(2):155. doi: 10.3390/v13020155
38. Bui TT, Moi ML, Nabeshima T, Takemura T, Nguyen TT, Nguyen LN, et al. A single amino acid substitution in the NS4B protein of Dengue virus confers enhanced virus growth and fitness in human cells in vitro through IFN-dependent host response. J Gen Virol. (2018) 99:1044–57. doi: 10.1099/jgv.0.001092
39. DeDiego ML, Martinez-Sobrido L, Topham DJ. Novel functions of IFI44L as a feedback regulator of host antiviral responses. J Virol. (2019) 93. doi: 10.1128/JVI.01159-19
40. Bian P, Ye C, Zheng X, Luo C, Yang J, Li M. RIPK3 promotes JEV replication in neurons via downregulation of IFI44L. Front Microbiol. (2020) 11:368. doi: 10.3389/fmicb.2020.00368
41. Mommert M, Tabone O, Oriol G, Cerrato E, Guichard A, Naville M. LTR-retrotransposon transcriptome modulation in response to endotoxin-induced stress in PBMCs. BMC Genomics. (2018) 19:522. doi: 10.1186/s12864-018-4901-9
42. Zheng Q, Wang D, Lin R, Lv Q, Wang W. IFI44 is an immune evasion biomarker for SARS-CoV-2 and Staphylococcus aureus infection in patients with RA. Front Immunol. (2022) 13:1013322. doi: 10.3389/fimmu.2022.1013322
43. Wallen AJ, Barker GA, Fein DE, Jing H, Diamond SL. Enhancers of adeno-associated virus AAV2 transduction via high throughput siRNA screening. Mol Ther. (2011) 19:1152–60. doi: 10.1038/mt.2011.4
44. Nosaka T, Naito T, Murata Y, Matsuda H, Ohtani M, Hiramatsu K. Regulatory function of interferon-inducible 44-like for hepatitis B virus covalently closed circular DNA in primary human hepatocytes. Hepatol Res. (2022) 52:141–52. doi: 10.1111/hepr.13722
45. Carneiro MW, Fukutani KF, Andrade BB, Curvelo RP, Cristal JR, Carvalho AM. Gene expression profile of high IFN-γ Producers stimulated with leishmania Braziliensis identifies genes associated with cutaneous leishmaniasis. PloS Negl Trop Dis. (2016) 10:e0005116. doi: 10.1371/journal.pntd.0005116
46. Wahadat MJ, Bodewes ILA, Maria NI, van Helden-Meeuwsen CG, van Dijk-Hummelman A. Type I IFN signature in childhood-onset systemic lupus erythematosus: a conspiracy of DNA- and RNA-sensing receptors? Arthritis Res Ther. (2018) 20:4. doi: 10.1186/s13075-017-1501-z
47. Zhang Y, Liu J, Niu G, Wu Q, Cao B. Chi-miR-3880 mediates the regulatory role of interferon gamma in goat mammary gland. Dev Biol. (2023) 501:104–10. doi: 10.1016/j.ydbio.2023.04.004
48. Yeung KS, Chung BH, Choufani S, Mok MY, Wong WL, Mak CC. Genome-wide DNA methylation analysis of chinese patients with systemic lupus erythematosus identified hypomethylation in genes related to the type I interferon pathway. PloS One. (2017) 12:e0169553. doi: 10.1371/journal.pone.0169553
49. Hurtado C, Rojas-Gualdrón DF, Urrego R, Cashman K, Vásquez-Trespalacios EM, Díaz-Coronado JC. Altered B cell phenotype and CD27+ memory B cells are associated with clinical features and environmental exposure in Colombian systemic lupus erythematosus patients. Front Med (Lausanne). (2022) 9:950452. doi: 10.3389/fmed.2022.950452
50. Zhao L, Xia M, Wang K, Lai C, Fan H, Gu H. A long non-coding RNA IVRPIE promotes host antiviral immune responses through regulating interferon β1 and ISG expression. Front Microbiol. (2020) 11:260. doi: 10.3389/fmicb.2020.00260
51. Huang WC, Tung SL, Chen YL, Chen PM, Chu PY. IFI44L is a novel tumor suppressor in human hepatocellular carcinoma affecting cancer stemness, metastasis, and drug resistance via regulating met/Src signaling pathway. BMC Cancer. (2018) 18:609. doi: 10.1186/s12885-018-4529-9
52. Woeckel VJ, Eijken M, van Peppel J, Chiba H, van der Eerden BC, van Leeuwen JP. IFNβ impairs extracellular matrix formation leading to inhibition of mineralization by effects in the early stage of human osteoblast differentiation. J Cell Physiol. (2012) 227:2668–76. doi: 10.1002/jcp.v227.6
53. Theocharis AD, Skandalis SS, Gialeli C, Karamanos NK. Extracellular matrix structure. Advanced Drug Delivery Rev. (2016) 97:4–27. doi: 10.1016/j.addr.2015.11.001
54. Clancy RM, Markham AJ, Jackson T, Rasmussen SE, Blumenberg M, Buyon JP. Cardiac fibroblast transcriptome analyses support a role for interferogenic, profibrotic, and inflammatory genes in anti-SSA/Ro-associated congenital heart block. Am J Physiol Heart Circ Physiol. (2017) 313:H631–h640. doi: 10.1152/ajpheart.00256.2017
55. Ma X, Su B, Tian Y, Backenstose NJC, Ye Z, Moss A. Deep Transcriptomic Analysis Reveals the Dynamic Developmental Progression during Early Development of Channel Catfish (Ictalurus punctatus). Int J Mol Sci. (2020) 21(55):5535. doi: 10.3390/ijms21155535
56. Geller AC, Dickerman BA, Taber JM, Dwyer LA, Hartman AM, Perna FM. Skin cancer interventions across the cancer control continuum: A review of experimental evidence (1/1/2000-6/30/2015) and future research directions. Prev Med. (2018) 111:442–50. doi: 10.1016/j.ypmed.2018.01.018
57. Rojas KD, Perez ME, Marchetti MA, Nichols AJ, Penedo FJ, Jaimes N. Skin cancer: Primary, secondary, and tertiary prevention. Part II. J Am Acad Dermatol. (2022) 87:271–88. doi: 10.1016/j.jaad.2022.01.053
58. Puig-Butille JA, Escámez MJ, Garcia-Garcia F, Tell-Marti G, Fabra À, Martínez-Santamaría L. Capturing the biological impact of CDKN2A and MC1R genes as an early predisposing event in melanoma and non melanoma skin cancer. Oncotarget. (2014) 5:1439–51. doi: 10.18632/oncotarget.1444
60. D’Souza S, Addepalli V. Preventive measures in oral cancer: An overview. BioMed Pharmacother. (2018) 107:72–80. doi: 10.1016/j.biopha.2018.07.114
61. Wong KCW, Hui EP, Lo KW, Lam WKJ, Johnson D, Li L. Nasopharyngeal carcinoma: an evolving paradigm. Nat Rev Clin Oncol. (2021) 18:679–95. doi: 10.1038/s41571-021-00524-x
62. Hsu YB, Huang CF, Lin KT, Kuo YL, Lan MC, Lan MY. Podoplanin, a potential therapeutic target for nasopharyngeal carcinoma. BioMed Res Int. (2019) 2019:7457013. doi: 10.1155/2019/7457013
63. Wu X, Lin L, Zhou F, Yu S, Chen M, Wang S. The highly expressed IFIT1 in nasopharyngeal carcinoma enhances proliferation, migration, and invasion of nasopharyngeal carcinoma cells. Mol Biotechnol. (2022) 64:621–36. doi: 10.1007/s12033-021-00439-z
64. Li MY, Liu LZ, Dong M. Progress on pivotal role and application of exosome in lung cancer carcinogenesis, diagnosis, therapy and prognosis. Mol Cancer. (2021) 20:22. doi: 10.1186/s12943-021-01312-y
65. Nagaraju GP, Dariya B, Kasa P, Peela S, El-Rayes BF. Epigenetics in hepatocellular carcinoma. Semin Cancer Biol. (2022) 86:622–32. doi: 10.1016/j.semcancer.2021.07.017
66. Klein AP. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastroenterol Hepatol. (2021) 18:493–502. doi: 10.1038/s41575-021-00457-x
67. Chen C, Xie L, Ren T, Huang Y, Xu J, Guo W. Immunotherapy for osteosarcoma: Fundamental mechanism, rationale, and recent breakthroughs. Cancer Lett. (2021) 500:1–10. doi: 10.1016/j.canlet.2020.12.024
68. Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. (2014) 14:722–35. doi: 10.1038/nrc3838
69. Takashima Y, Hamano M, Yoshii K, Hayano A, Fukai J, Iwadate Y. Reciprocal expression of the immune response genes CXCR3 and IFI44L as module hubs are associated with patient survivals in primary central nervous system lymphoma. Int J Clin Oncol. (2023) 28:468–81. doi: 10.1007/s10147-022-02285-8
70. Stanulla M, Cavé H, Moorman AV. IKZF1 deletions in pediatric acute lymphoblastic leukemia: still a poor prognostic marker? Blood. (2020) 135:252–60. doi: 10.1182/blood.2019000813
71. Huang Y, Li J, Chen Y, Jiang P, Wang L, Hu J. Identification of early recurrence factors in childhood and adolescent B-cell acute lymphoblastic leukemia based on integrated bioinformatics analysis. Front Oncol. (2020) 10:565455. doi: 10.3389/fonc.2020.565455
72. Zeidan AM, Shallis RM, Wang R, Davidoff A, Ma X. Epidemiology of myelodysplastic syndromes: Why characterizing the beast is a prerequisite to taming it. Blood Rev. (2019) 34:1–15. doi: 10.1016/j.blre.2018.09.001
73. Le Y. Screening and identification of key candidate genes and pathways in myelodysplastic syndrome by bioinformatic analysis. PeerJ. (2019) 7:e8162. doi: 10.7717/peerj.8162
74. Martínez-Jiménez F, Movasati A, Brunner SR, Nguyen L, Priestley P, Cuppen E. Pan-cancer whole-genome comparison of primary and metastatic solid tumours. Nature. (2023) 618:333–41. doi: 10.1038/s41586-023-06054-z
75. Ghoshdastider U, Sendoel A. Exploring the pan-cancer landscape of posttranscriptional regulation. Cell Rep. (2023) 42:113172. doi: 10.1016/j.celrep.2023.113172
76. Pan L, Lu MP, Wang JH, Xu M, Yang SR. Immunological pathogenesis and treatment of systemic lupus erythematosus. World J Pediatr. (2020) 16:19–30. doi: 10.1007/s12519-019-00229-3
77. Liu W, Zhang S, Wang J. IFN-γ, should not be ignored in SLE. Front Immunol. (2022) 13:954706. doi: 10.3389/fimmu.2022.954706
78. Siddiqi KZ, Wilhelm TR, Ulff-Møller CJ, Jacobsen S. Cluster of highly expressed interferon-stimulated genes associate more with African ancestry than disease activity in patients with systemic lupus erythematosus. A systematic review of cross-sectional studies. Transl Res. (2021) 238:63–75. doi: 10.1016/j.trsl.2021.07.006
79. He Z, Zhou S, Yang M, Zhao Z, Mei Y, Xin Y. Comprehensive analysis of epigenetic modifications and immune-cell infiltration in tissues from patients with systemic lupus erythematosus. Epigenomics. (2022) 14:81–100. doi: 10.2217/epi-2021-0318
80. Zhao X, Zhang L, Wang J, Zhang M, Song Z, Ni B. Identification of key biomarkers and immune infiltration in systemic lupus erythematosus by integrated bioinformatics analysis. J Transl Med. (2021) 19:35. doi: 10.1186/s12967-020-02698-x
81. Fan H, Zhao G, Ren D, Liu F, Dong G, Hou Y. Gender differences of B cell signature related to estrogen-induced IFI44L/BAFF in systemic lupus erythematosus. Immunol Lett. (2017) 181:71–8. doi: 10.1016/j.imlet.2016.12.002
82. Ehtesham N, Habibi Kavashkohie MR, Mazhari SA, Azhdari S, Ranjbar H, Mosallaei M. DNA methylation alterations in systemic lupus erythematosus: A systematic review of case-control studies. Lupus. (2023) 32:363–79. doi: 10.1177/09612033221148099
83. Salesi M, Dehabadi MH, Salehi R, Salehi A, Pakzad B. Differentially methylation of IFI44L gene promoter in Iranian patients with systemic lupus erythematosus and rheumatoid arthritis. Mol Biol Rep. (2022) 49:3065–72. doi: 10.1007/s11033-022-07134-5
84. Joseph S, George NI, Green-Knox B, Treadwell EL, Word B, Yim S. Epigenome-wide association study of peripheral blood mononuclear cells in systemic lupus erythematosus: Identifying DNA methylation signatures associated with interferon-related genes based on ethnicity and SLEDAI. J Autoimmun. (2019) 96:147–57. doi: 10.1016/j.jaut.2018.09.007
85. Karimifar M, Pakzad B, Karimzadeh H, Mousavi M, Kazemi M, Salehi A. Interferon-induced protein 44-like gene promoter is differentially methylated in peripheral blood mononuclear cells of systemic lupus erythematosus patients. J Res Med Sci. (2019) 24:99. doi: 10.4103/jrms.JRMS_83_19
86. Ulff-Møller CJ, Asmar F, Liu Y, Svendsen AJ, Busato F, Grønbaek K. Twin DNA methylation profiling reveals flare-dependent interferon signature and B cell promoter hypermethylation in systemic lupus erythematosus. Arthritis Rheumatol. (2018) 70:878–90. doi: 10.1002/art.40422
87. Xie S, Zeng Q, Ouyang S, Liang Y, Xiao C. Bioinformatics analysis of epigenetic and SNP-related molecular markers in systemic lupus erythematosus. Am J Transl Res. (2021) 13:6312–29.
88. Weeding E, Sawalha AH. Deoxyribonucleic acid methylation in systemic lupus erythematosus: implications for future clinical practice. Front Immunol. (2018) 9:875. doi: 10.3389/fimmu.2018.00875
89. Zhang B, Zhou T, Wu H, Zhao M, Lu Q. Difference of IFI44L methylation and serum IFN-a1 level among patients with discoid and systemic lupus erythematosus and healthy individuals. J Transl Autoimmun. (2021) 4:100092. doi: 10.1016/j.jtauto.2021.100092
90. Weeding E, Coit P, Yalavarthi S, Kaplan MJ, Knight JS, Sawalha AH. Genome-wide DNA methylation analysis in primary antiphospholipid syndrome neutrophils. Clin Immunol. (2018) 196:110–6. doi: 10.1016/j.clim.2018.11.011
91. Jiang Z, Shao M, Dai X, Pan Z, Liu D. Identification of diagnostic biomarkers in systemic lupus erythematosus based on bioinformatics analysis and machine learning. Front Genet. (2022) 13:865559. doi: 10.3389/fgene.2022.865559
93. Yao Q, Song Z, Wang B, Qin Q, Zhang JA. Identifying key genes and functionally enriched pathways in sjögren’s syndrome by weighted gene co-expression network analysis. Front Genet. (2019) 10:1142. doi: 10.3389/fgene.2019.01142
94. Jara D, Carvajal P, Castro I, Barrera MJ, Aguilera S, González S. Type I interferon dependent hsa-miR-145-5p downregulation modulates MUC1 and TLR4 overexpression in salivary glands from sjögren’s syndrome patients. Front Immunol. (2021) 12:685837. doi: 10.3389/fimmu.2021.685837
95. Björk A, Richardsdotter Andersson E, Imgenberg-Kreuz J, Thorlacius GE, Mofors J, Syvänen AC. Protein and DNA methylation-based scores as surrogate markers for interferon system activation in patients with primary Sjögren’s syndrome. RMD Open. (2020) 6(1):e000995. doi: 10.1136/rmdopen-2019-000995
96. Imgenberg-Kreuz J, Sandling JK, Norheim KB, Johnsen SJA, Omdal R, Syvänen AC. DNA methylation-based interferon scores associate with sub-phenotypes in primary sjögren’s syndrome. Front Immunol. (2021) 12:702037. doi: 10.3389/fimmu.2021.702037
97. Maria NI, Brkic Z, Waris M, van Helden-Meeuwsen CG, Heezen K, van de Merwe JP. MxA as a clinically applicable biomarker for identifying systemic interferon type I in primary Sjogren’s syndrome. Ann Rheum Dis. (2014) 73:1052–9. doi: 10.1136/annrheumdis-2012-202552
98. Chen S, Pu W, Guo S, Jin L, He D, Wang J. Genome-wide DNA methylation profiles reveal common epigenetic patterns of interferon-related genes in multiple autoimmune diseases. Front Genet. (2019) 10:223. doi: 10.3389/fgene.2019.00223
99. Luo X, Peng Y, Chen YY, Wang AQ, Deng CW, Peng LY. Genome-wide DNA methylation patterns in monocytes derived from patients with primary Sjogren syndrome. Chin Med J (Engl). (2021) 134:1310–6. doi: 10.1097/CM9.0000000000001451
100. Khuder SA, Al-Hashimi I, Mutgi AB, Altorok N. Identification of potential genomic biomarkers for Sjögren’s syndrome using data pooling of gene expression microarrays. Rheumatol Int. (2015) 35:829–36. doi: 10.1007/s00296-014-3152-6
101. Zhu H, Wu LF, Mo XB, Lu X, Tang H, Zhu XW. Rheumatoid arthritis-associated DNA methylation sites in peripheral blood mononuclear cells. Ann Rheum Dis. (2019) 78:36–42. doi: 10.1136/annrheumdis-2018-213970
102. Raterman HG, Vosslamber S, Ridder S, Nurmohamed MT, Lems WF, Boers M. The interferon type I signature towards prediction of non-response to rituximab in rheumatoid arthritis patients. Arthritis Res Ther. (2012) 14:R95. doi: 10.1186/ar3819
103. Ramos PS, Zimmerman KD, Haddad S, Langefeld CD, Medsger TA Jr, Feghali-Bostwick CA. Integrative analysis of DNA methylation in discordant twins unveils distinct architectures of systemic sclerosis subsets. Clin Epigenet. (2019) 11:58. doi: 10.1186/s13148-019-0652-y
104. Martire S, Navone ND, Montarolo F, Perga S, Bertolotto A. A gene expression study denies the ability of 25 candidate biomarkers to predict the interferon-beta treatment response in multiple sclerosis patients. J Neuroimmunol. (2016) 292:34–9. doi: 10.1016/j.jneuroim.2016.01.010
105. Malhotra S, Bustamante MF, Pérez-Miralles F, Rio J, Ruiz de Villa MC, Vegas E. Search for specific biomarkers of IFNβ bioactivity in patients with multiple sclerosis. PloS One. (2011) 6:e23634. doi: 10.1371/journal.pone.0023634
106. Piera-Velazquez S, Mendoza FA, Addya S, Pomante D, Jimenez SA. Increased expression of interferon regulated and antiviral response genes in CD31+/CD102+ lung microvascular endothelial cells from systemic sclerosis patients with end-stage interstitial lung disease. Clin Exp Rheumatol. (2021) 39:1298–306. doi: 10.55563/clinexprheumatol/ret1kg
107. Carnero-Montoro E, Barturen G, Povedano E, Kerick M, Martinez-Bueno M, Ballestar E. Epigenome-wide comparative study reveals key differences between mixed connective tissue disease and related systemic autoimmune diseases. Front Immunol. (2019) 10:1880. doi: 10.3389/fimmu.2019.01880
108. Lerkvaleekul B, Veldkamp SR, van der Wal MM, Schatorjé EJH, Kamphuis SSM, van den Berg JM. Siglec-1 expression on monocytes is associated with the interferon signature in juvenile dermatomyositis and can predict treatment response. Rheumatol (Oxford). (2022) 61:2144–55. doi: 10.1093/rheumatology/keab601
109. Vieira M, Régnier P, Maciejewski-Duval A, Le Joncour A, Darasse-Jèze G, Rosenzwajg M. Interferon signature in giant cell arteritis aortitis. J Autoimmun. (2022) 127:102796. doi: 10.1016/j.jaut.2022.102796
110. Dong Z, Dai H, Liu W, Jiang H, Feng Z, Liu F. Exploring the differences in molecular mechanisms and key biomarkers between membranous nephropathy and lupus nephritis using integrated bioinformatics analysis. Front Genet. (2021) 12:770902. doi: 10.3389/fgene.2021.770902
111. Mutua F, Su RC, Mesa C, Lopez C, Ball TB, Kiazyk S. Type I interferons and Mycobacterium tuberculosis whole cell lysate induce distinct transcriptional responses in M. tuberculosis infection. Tuberculosis (Edinb). (2023) 143:102409. doi: 10.1016/j.tube.2023.102409
112. Shi T, Huang L, Zhou Y, Tian J. Role of GBP1 in innate immunity and potential as a tuberculosis biomarker. Sci Rep. (2022) 12:11097. doi: 10.1038/s41598-022-15482-2
113. Tian S, Deng J, Huang W, Liu L, Chen Y, Jiang Y. FAM89A and IFI44L for distinguishing between viral and bacterial infections in children with febrile illness. Pediatr Investig. (2021) 5:195–202. doi: 10.1002/ped4.12295
114. Xu N, Hao F, Dong X, Yao Y, Guan Y, Yang L. A two-transcript biomarker of host classifier genes for discrimination of bacterial from viral infection in acute febrile illness: a multicentre discovery and validation study. Lancet Digit Health. (2021) 3:e507–16. doi: 10.1016/S2589-7500(21)00102-3
115. Herberg JA, Kaforou M, Wright VJ, Shailes H, Eleftherohorinou H, Hoggart CJ. Diagnostic test accuracy of a 2-transcript host RNA signature for discriminating bacterial vs viral infection in febrile children. Jama. (2016) 316:835–45. doi: 10.1001/jama.2016.11236
116. Pennisi I, Rodriguez-Manzano J, Moniri A, Kaforou M, Herberg JA, Levin M. Translation of a host blood RNA signature distinguishing bacterial from viral infection into a platform suitable for development as a point-of-care test. JAMA Pediatr. (2021) 175:417–9. doi: 10.1001/jamapediatrics.2020.5227
117. Busse DC, Habgood-Coote D, Clare S, Brandt C, Bassano I, Kaforou M. Interferon-induced protein 44 and interferon-induced protein 44-like restrict replication of respiratory syncytial virus. J Virol. (2020) 94(18):e00297-20. doi: 10.1128/JVI.00297-20
118. Lempainen J, Korhonen LS, Kantojärvi K, Heinonen S, Toivonen L, Räty P. Associations between IFI44L gene variants and rates of respiratory tract infections during early childhood. J Infect Dis. (2021) 223:157–65. doi: 10.1093/infdis/jiaa341
119. Robinson CL, Chong ACN, Ashbrook AW, Jeng G, Jin J, Chen H. Male germ cells support long-term propagation of Zika virus. Nat Commun. (2018) 9:2090. doi: 10.1038/s41467-018-04444-w
120. To KK, Sridhar S, Chiu KH, Hung DL, Li X, Hung IF. Lessons learned 1 year after SARS-CoV-2 emergence leading to COVID-19 pandemic. Emerg Microbes Infect. (2021) 10:507–35. doi: 10.1080/22221751.2021.1898291
121. Li H, Huang F, Liao H, Li Z, Feng K, Huang T. Identification of COVID-19-specific immune markers using a machine learning method. Front Mol Biosci. (2022) 9:952626. doi: 10.3389/fmolb.2022.952626
122. Shaath H, Vishnubalaji R, Elkord E, Alajez NM. Single-cell transcriptome analysis highlights a role for neutrophils and inflammatory macrophages in the pathogenesis of severe COVID-19. Cells. (2020) 9(11):2374. doi: 10.3390/cells9112374
123. Dong Z, Yan Q, Cao W, Liu Z, Wang X. Identification of key molecules in COVID-19 patients significantly correlated with clinical outcomes by analyzing transcriptomic data. Front Immunol. (2022) 13:930866. doi: 10.3389/fimmu.2022.930866
124. Lee Y, Riskedal E, Kalleberg KT, Istre M, Lind A, Lund-Johansen F. EWAS of post-COVID-19 patients shows methylation differences in the immune-response associated gene, IFI44L, three months after COVID-19 infection. Sci Rep. (2022) 12:11478. doi: 10.1038/s41598-022-15467-1
125. Bräuninger H, Stoffers B, Fitzek ADE, Meißner K, Aleshcheva G, Schweizer M. Cardiac SARS-CoV-2 infection is associated with pro-inflammatory transcriptomic alterations within the heart. Cardiovasc Res. (2022) 118:542–55. doi: 10.1093/cvr/cvab322
126. Moal V, Textoris J, Ben Amara A, Mehraj V, Berland Y, Colson P. Chronic hepatitis E virus infection is specifically associated with an interferon-related transcriptional program. J Infect Dis. (2013) 207:125–32. doi: 10.1093/infdis/jis632
127. Zhou A, Dong X, Liu M, Tang B. Comprehensive transcriptomic analysis identifies novel antiviral factors against influenza A virus infection. Front Immunol. (2021) 12:632798. doi: 10.3389/fimmu.2021.632798
128. Jansen K, Wirz OF, van de Veen W, Tan G, Mirer D, Sokolowska M. Loss of regulatory capacity in Treg cells following rhinovirus infection. J Allergy Clin Immunol. (2021) 148:1016–1029.e16. doi: 10.1016/j.jaci.2021.05.045
129. BroChado-Kith Ó, Martínez I, Berenguer J, González-García J, Salgüero S, Sepúlveda-Crespo D. HCV cure with direct-acting antivirals improves liver and immunological markers in HIV/HCV-coinfected patients. Front Immunol. (2021) 12:723196. doi: 10.3389/fimmu.2021.723196
130. Wie SH, Du P, Luong TQ, Rought SE, Beliakova-Bethell N, Lozach J. HIV downregulates interferon-stimulated genes in primary macrophages. J Interferon Cytokine Res. (2013) 33:90–5. doi: 10.1089/jir.2012.0052
131. Mahajan GJ, Vallender EJ, Garrett MR, Challagundla L, Overholser JC, Jurjus G. Altered neuro-inflammatory gene expression in hippocampus in major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. (2018) 82:177–86. doi: 10.1016/j.pnpbp.2017.11.017
132. Ruderfer DM, Fanous AH, Ripke S, McQuillin A, Amdur RL, Gejman PV. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatry. (2014) 19:1017–24. doi: 10.1038/mp.2013.138
133. Feenstra B, Pasternak B, Geller F, Carstensen L, Wang T, Huang F. Common variants associated with general and MMR vaccine-related febrile seizures. Nat Genet. (2014) 46:1274–82. doi: 10.1038/ng.3129
134. Liu S, Ren W, Yu J, Li C, Tang S. Identification of hub genes associated with diabetes mellitus and tuberculosis using bioinformatic analysis. Int J Gen Med. (2021) 14:4061–72. doi: 10.2147/IJGM.S318071
135. Rong Y, Yang H, Xu H, Li S, Wang P, Wang Z. Bioinformatic analysis reveals hub immune-related genes of diabetic foot ulcers. Front Surg. (2022) 9:878965. doi: 10.3389/fsurg.2022.878965
136. Shang M, Zhang Y, Zhang T. IFI44L and C1QTNF5 as promising biomarkers of proliferative diabetic retinopathy. Med (Baltimore). (2022) 101:e31961. doi: 10.1097/MD.0000000000031961
137. Dhana K, Braun KVE, Nano J, Voortman T, Demerath EW, Guan W. An epigenome-wide association study of obesity-related traits. Am J Epidemiol. (2018) 187:1662–9. doi: 10.1093/aje/kwy025
138. Chen C, Tian J, He Z, Xiong W, He Y, Liu S. Identified three interferon induced proteins as novel biomarkers of human ischemic cardiomyopathy. Int J Mol Sci. (2021) 22(23):13116. doi: 10.3390/ijms222313116
139. Ahrens I, Domeij H, Eisenhardt SU, Topcic D, Albrecht M, Leitner E. Opposing effects of monomeric and pentameric C-reactive protein on endothelial progenitor cells. Basic Res Cardiol. (2011) 106:879–95. doi: 10.1007/s00395-011-0191-y
140. He W, Wei D, Cai D, Chen S, Li S, Chen W. Altered long non-coding RNA transcriptomic profiles in ischemic stroke. Hum Gene Ther. (2018) 29:719–32. doi: 10.1089/hum.2017.064
141. Yang H, Lu Y, Yang H, Zhu Y, Tang Y, Li L. Integrated weighted gene co-expression network analysis uncovers STAT1(signal transducer and activator of transcription 1) and IFI44L (interferon-induced protein 44-like) as key genes in pulmonary arterial hypertension. Bioengineered. (2021) 12:6021–34. doi: 10.1080/21655979.2021.1972200
142. Rossios C, Pavlidis S, Gibeon D, Mumby S, Durham A, Ojo O. Impaired innate immune gene profiling in airway smooth muscle cells from chronic cough patients. Biosci Rep. (2017) 37(6):BSR20171090. doi: 10.1042/BSR20171090
143. Zhang J, Zhang C, Yang H, Han X, Fan Z, Hou B. Depletion of PRDM9 enhances proliferation, migration and chemotaxis potentials in human periodontal ligament stem cells. Connect Tissue Res. (2020) 61:498–508. doi: 10.1080/03008207.2019.1620224
144. Garau J, Charras A, Varesio C, Orcesi S, Dragoni F, Galli J. Altered DNA methylation and gene expression predict disease severity in patients with Aicardi-Goutières syndrome. Clin Immunol. (2023) 249:109299. doi: 10.1016/j.clim.2023.109299
145. Lu YW, Chen YJ, Shi N, Yang LH, Wang HM, Dong RJ. L36G is associated with cutaneous antiviral competence in psoriasis. Front Immunol. (2022) 13:971071. doi: 10.3389/fimmu.2022.971071
146. Yan Z, Xu J, Wu G, Zhen Y, Liao X, Zou F. Identification of key genes and pathways associated with gender difference in osteonecrosis of the femoral head based on bioinformatics analysis. J Musculoskelet Neuronal Interact. (2023) 23:122–30.
147. Jin X, Chen W, Wang J, Xu X, Zhang T, Wang L. Unveiling circular RNA-mediated regulatory mechanisms in necroptosis in premature ovarian failure. Front Biosci (Landmark Ed). (2023) 28:314. doi: 10.31083/j.fbl2811314
148. Li Q, Sun B, Zhuo Y, Jiang Z, Li R, Lin C. Interferon and interferon-stimulated genes in HBV treatment. Front Immunol. (2022) 13:1034968. doi: 10.3389/fimmu.2022.1034968
149. Schoggins JW. Interferon-stimulated genes: what do they all do? Annu Rev Virol. (2019) 6:567–84. doi: 10.1146/annurev-virology-092818-015756
150. Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. (2014) 32:513–45. doi: 10.1146/annurev-immunol-032713-120231
151. Pitha-Rowe IF, Pitha PM. Viral defense, carcinogenesis and ISG15: novel roles for an old ISG. Cytokine Growth Factor Rev. (2007) 18:409–17. doi: 10.1016/j.cytogfr.2007.06.017
152. Montano EN, Bose M, Huo L, Tumurkhuu G, De Los Santos G, Simental B. [amp]]alpha;-ketoglutarate-dependent KDM6 histone demethylases regulate Interferon Stimulated Gene expression in Lupus. Arthritis Rheumatol. (2024) 76(3):396–410. doi: 10.1002/art.42724
153. D’Cunha J, Knight E Jr, Haas AL, Truitt RL, Borden EC. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc Natl Acad Sci U.S.A. (1996) 93:211–5. doi: 10.1073/pnas.93.1.211
154. Yap AS, Kovacs EM. Direct cadherin-activated cell signaling: a view from the plasma membrane. J Cell Biol. (2003) 160:11–6. doi: 10.1083/jcb.200208156
155. Loeb KR, Haas AL. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J Biol Chem. (1992) 267:7806–13. doi: 10.1016/S0021-9258(18)42585-9
Keywords: biomarker, diagnosis, immune response, interferon-induced protein 44-like (IFI44L), therapeutic target, tumor
Citation: Du J, Luo H, Ye S, Zhang H, Zheng Z and Liu K (2024) Unraveling IFI44L’s biofunction in human disease. Front. Oncol. 14:1436576. doi: 10.3389/fonc.2024.1436576
Received: 22 May 2024; Accepted: 26 November 2024;
Published: 16 December 2024.
Edited by:
Sharon R. Pine, University of Colorado Anschutz Medical Campus, United StatesReviewed by:
Kenneth Shindler, University of Pennsylvania, United StatesCopyright © 2024 Du, Luo, Ye, Zhang, Zheng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaitai Liu, bGt0MTk4MkAxMjYuY29t
†These authors have contributed equally to this work
‡ORCID: Juan Du, orcid.org/0009-0006-5908-7608
Shuang Ye, orcid.org/0000-0002-5919-5856
Hui Zhang, orcid.org/0000-0003-1223-5141
Kaitai Liu, orcid.org/0000-0001-6441-6417
Hui Luo, orcid.org/0009-0008-2587-7980
Zhen Zheng, orcid.org/0009-0003-0522-8709
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.