- Department of Pathology, Hospital for Skin Diseases, Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College, Nanjing, China
Intratumoral microbiota (IM) has emerged as a significant component of the previously thought sterile tumor microenvironment (TME), exerting diverse functions in tumorigenesis and immune modulation. This review outlines the historical background, classification, and diversity of IM, elucidating its pivotal roles in oncogenicity, cancer development, and progression, alongside its influence on anti-tumor immunity. The signaling pathways through which IM impacts tumorigenesis and immunity, including reactive oxygen species (ROS), β-catenin, stimulator of interferon genes (STING), and other pathways [NF-κB, Toll-like receptor (TLR), complement, RhoA/ROCK, PKR-like ER kinase (PERK)], are discussed comprehensively. Furthermore, we briefly introduce the clinical implications of IM, emphasizing its potential as a target for novel cancer therapies, diagnostic biomarkers, and prognostic indicators. Notably, microbe-based therapeutic strategies such as fecal microbiome transplantation (FMT), probiotics regulation, bacteriotherapy, bacteriophage therapy, and oncolytic virotherapy are highlighted. These strategies hold promise for enhancing the efficacy of current cancer treatments and warrant further exploration in clinical settings.
1 Introduction
The tumor microenvironment (TME), encompassing cancer cells, stromal cells, immune cells, and soluble factors such as cytokines and chemokines, plays a pivotal role in cancer progression and therapeutic outcomes. Understanding the regulatory mechanisms governing the immune compartment within the TME is crucial for advancing cancer treatment strategies. Recently, the discovery of intratumoral microbiota (IM) has added a new dimension to our understanding of the TME (1–3). Despite its low biomass ranging from 0.1% to 10%, IMs have emerged as significant contributors to the tumor ecosystem, profoundly influencing tumorigenesis, cancer progression, and responses to cancer therapy (4).
Over the past decades, propelled by technological advancements, researches into IMs have revealed common characteristics and highlighted their complex interplay within the TME. The implications of IM extend beyond mere association; they have been implicated in tumor diagnosis, prognosis, and treatment guidance (1–3). As the field rapidly evolves, an updated review is warranted to consolidate current knowledge and identify emerging trends.
n this review, we first provide a concise overview of the historical context, classification, and diversity of IM. We then introduce their roles in carcinogenesis, cancer development, and progression, emphasizing their influence on anti-tumor immunity and the underlying signaling pathways. Furthermore, we discuss the clinical implications of IM, including their potential as targets for cancer therapy, diagnostic biomarkers, and prognostic indicators. Finally, we propose innovative microbe-based strategies for cancer treatment aimed at broad clinical applications. By synthesizing these insights, this review aims to underscore the transformation potential of IM research in reshaping our approach to cancer management.
2 The history, classification, and diversity of intratumoral microbiota
The concept of IMs as integral components of the TME has evolved significantly over time, tracing its roots back to discoveries made in the mid-19th century. Early microbiologists identified live microorganisms within tumors, yet due to technological limitations and prevailing skepticism, these findings initially faced resistance in the scientific community. A pivotal breakthrough occurred in 1911 with the discovery of the Rous Sarcoma virus, marking one of the first recognized examples of oncogenic viruses within tumors (5). Subsequent decades witnessed the identification of various viruses linked to cancer, such as Epstein-Barr virus (EBV), Hepatitis B virus (HBV), Hepatitis C virus (HCV), and Human papillomavirus (HPV). These discoveries underscored the diverse viral contributions to tumorigenesis. The identification of bacteria as oncobacteria gained prominence following the groundbreaking revelation in 1983 of Helicobacter pylori (H. pylori) as a causative agent in gastric cancer (3, 6). Beyond bacteria, Chlamydia trachomatis (C. trachomatis) emerged as another microbial species implicated in the pathogenesis of cervical cancer, squamous cell carcinoma, and ovarian cancer (7). Recent advancements, exemplified by Nejman et al.’s comprehensive analysis in 2020, have expanded the understanding of IMs to include a wide spectrum of bacterial species across various human tumor types (8). Concurrent studies by Narunsky-Haziza and Dohlman et al. further elucidated the role of fungi within tumors, highlighting their potential impacts on cancer diagnosis and prognosis (9, 10).
In addition to viruses and bacteria, the IM spectrum encompasses bacteriophages and protozoa, though these are not extensively discussed in this review. Notably, the composition and abundance of IMs exhibit significant variability within different cancer types, subtypes, and stages. For instance, lung cancer exhibits a distinct microbial colonization influenced by environmental factors, whereas breast cancer harbors a remarkably diverse microbiome (8, 11). Huang et al. highlighted higher levels of Gammaproteobacteria in cancerous tissues compared to normal tissues, along with significant increases in Streptococcaceae and Lactococcus in cirrhotic hepatocellular carcinoma (HCC), suggesting their potential as biomarkers (12). Fusobacterium and enterotoxigenic Bacteroides fragilis (ETBF) have implications in colorectal cancer (CRC) progression. CRC tissues exhibit higher abundances of Lactococcus, Bacteroides, Fusobacterium, Prevotella, and Streptococcus, while Pseudomonas and E. coli-Shigella are enriched in adjacent normal tissues (13). Moreover, studies have indicated varying intratumor microbiome profiles in cancer recurrence, emphasizing the dynamic nature of IMs in cancer progression. In conclusion, understanding the historical context, diverse classifications, and dynamic nature of intratumoral microbiota is crucial for unraveling their intricate roles in carcinogenesis and disease progression across different cancer types.
3 The roles of IMs in tumorigenesis, cancer development and progression
Bacteria associated with cancer have long been regarded as being opportunistic, but it became clear in the late 20th century that bacteria can directly cause cancer (5). An oncogenic bacterial “driver-passenger” model has been proposed to explain the role of IMs in tumorigenesis (14–16). Oncoviruses such as HPV, HBV, and HCV can also act as “drivers,” exerting various effects on cancer immunosurveillance and cancer progression. Herein, we focus on bacteria as examples to elucidate this model.
Endogenous “driver bacteria,” such as Escherichia coli group B2 strains capable of producing enterotoxin-induced single-stranded DNA breaks, can aggregate into non-anchor colonies within infected cells, thereby increasing mutation frequencies (17). Certain microbial metabolites, like hydrogen sulfide (H2S) produced by resident sulfate-reducing bacteria such as Fusobacterium nucleatum and E. coli, generate reactive oxygen species (ROS) that disrupt host DNA (18). Pathogens like Chlamydia trachomatis and Helicobacter pylori, associated with gynecological and gastric cancers respectively, inhibit homologous recombination and induce error-prone non-homologous end-joining, thereby compromising host DNA repair processes. Other bacteria such as Hungatella hathewayi and Fusobacterium nucleatum induce epigenetic modifications of the host genome by upregulating DNA methyltransferases through mechanisms that are not fully understood, leading to hypermethylation and silencing of tumor-suppressor genes (including CDX2 and MLH1), thereby promoting tumorigenesis (19–21). Enterotoxigenic Pseudomonas fragilis (ETPF) can sustain persistent Th-17-type inflammatory stimulation, gradually decreasing in number due to this stimulation and being replaced by opportunistic “passenger bacteria” that can dominate the existing ecological niche, disrupt local innate immunity, and ultimately contribute to tumor progression (22). Staphylococcus haemolyticus (S. haemolyticus) is considered a “passenger” bacterium, promoting epithelial cell differentiation and forming biofilms on exposed basement membrane collagen fibers within disrupted colonic wall structures (23). Models explain the occurrence of Clostridium-induced CRC, a silent disease affecting various age groups worldwide, with heightened mucosal biofilm levels harboring specific microbial species interacting with mucosal immune and microbial communities, imparting partial oncogenic properties (24). 5-fluorouracil (5-FU), a first-line chemotherapeutic agent for CRC, can serve as an inhibitor of F. nucleatum isolates from CRC but does not work for CRC-isolated E. coli with the capacity to modify 5-FU. When E. coli exists, it attenuates 5-FU virulence against F. nucleatum and human CRC epithelial cells which are otherwise sensitive to 5-FU, thereby leading to resistance to chemotherapeutic agents and promoting tumor progression (25). Collectively, these studies underscore a robust link between intratumoral microbiota and carcinogenesis, as well as cancer progression.
Tumor metastasis remains the leading cause of cancer-related mortality. IM contributes to cancer metastasis through multiple mechanisms across various cancer types: ① Co-migration with primary cancer cells: Fusobacterium has been detected in both primary tumors and matched distant metastases of colorectal cancer (26). ② Disruption of the gut vascular barrier (GVB): pathogens such as Salmonella typhimurium can disrupt the GVB, facilitating translocation and toxin transmission. For instance, VirF+ E. coli within tumors can degrade the GVB, allowing gut microbes to translocate to the liver and establish a “premetastatic niche” (PMN), enhancing chemotactic factor expression and extracellular matrix deposition, and upregulating plasmalemma vesicle-associated protein 1 (PV1) (27). ③ Modification of the TME to promote distant metastasis: Fusobacterium nucleatum infection induces colorectal cancer cells to release exosomes enriched with CXCL16/RhoA/IL-8 and miR-1246/92b-3p/27a-3p, which are transferred to uninfected CRC cells, thereby promoting liver metastasis (28). Additionally, Fusobacterium nucleatum induces M2 macrophage polarization within the TME, promoting CRC liver metastasis by upregulating chemokine CCL20 and ICAM1 expression (29). Most recurrences of lung cancer (LC) may be due to an abundance of butyrate-producing bacteria which promotes LC metastasis by inhibiting HDAC2 and increasing H3K27 acetylation at the H19 promoter and inducing M2 macrophage polarization (30). Altogether, IM profoundly influences cancer initiation, development, progression, and metastasis through diverse mechanisms (Figure 1), while also impacting anti-tumor immunity.
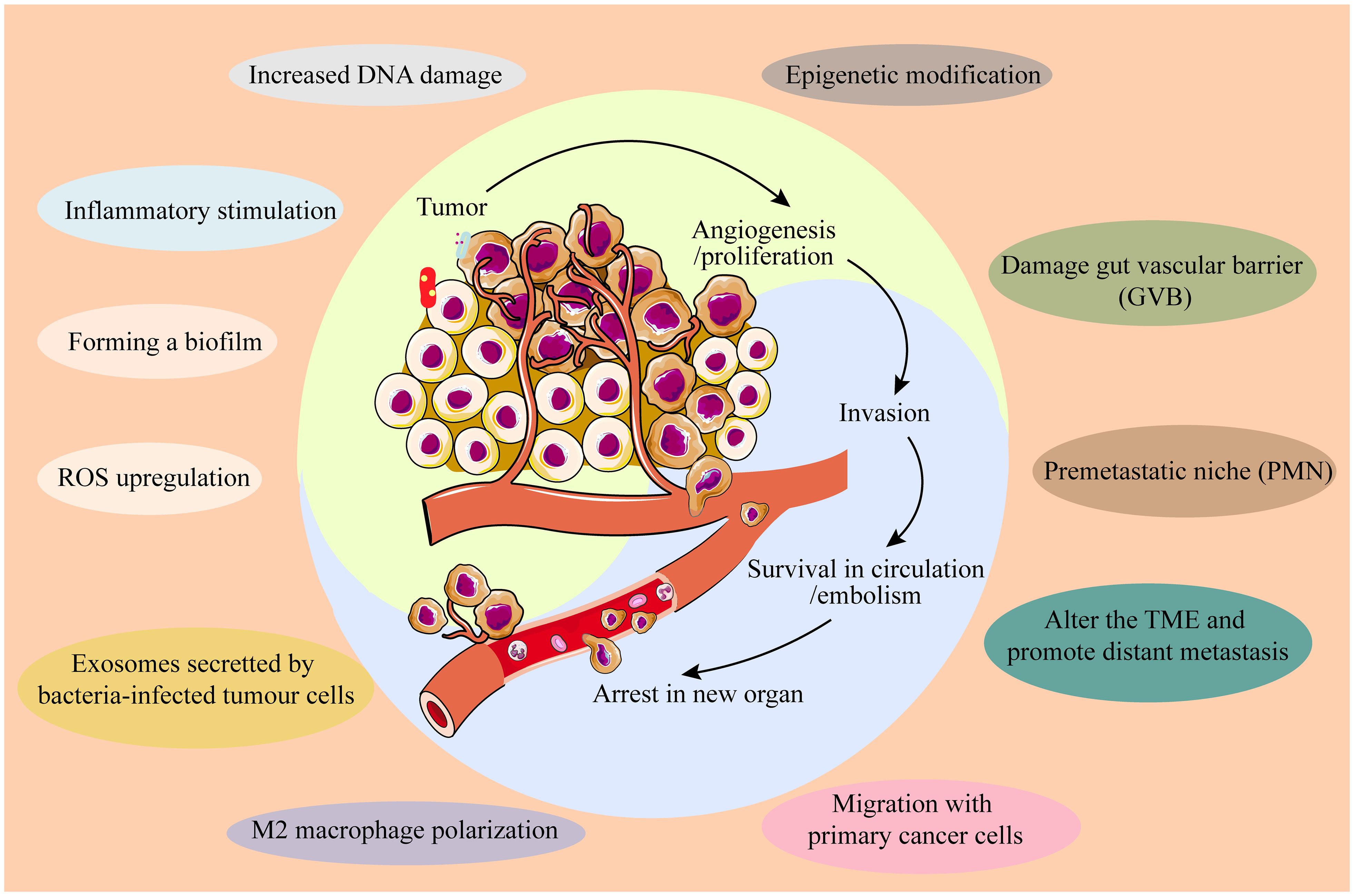
Figure 1. IM influences cancer initiation, development, progression, and metastasis through diverse mechanisms.
4 The impacts of IMs on anti-tumor immunity
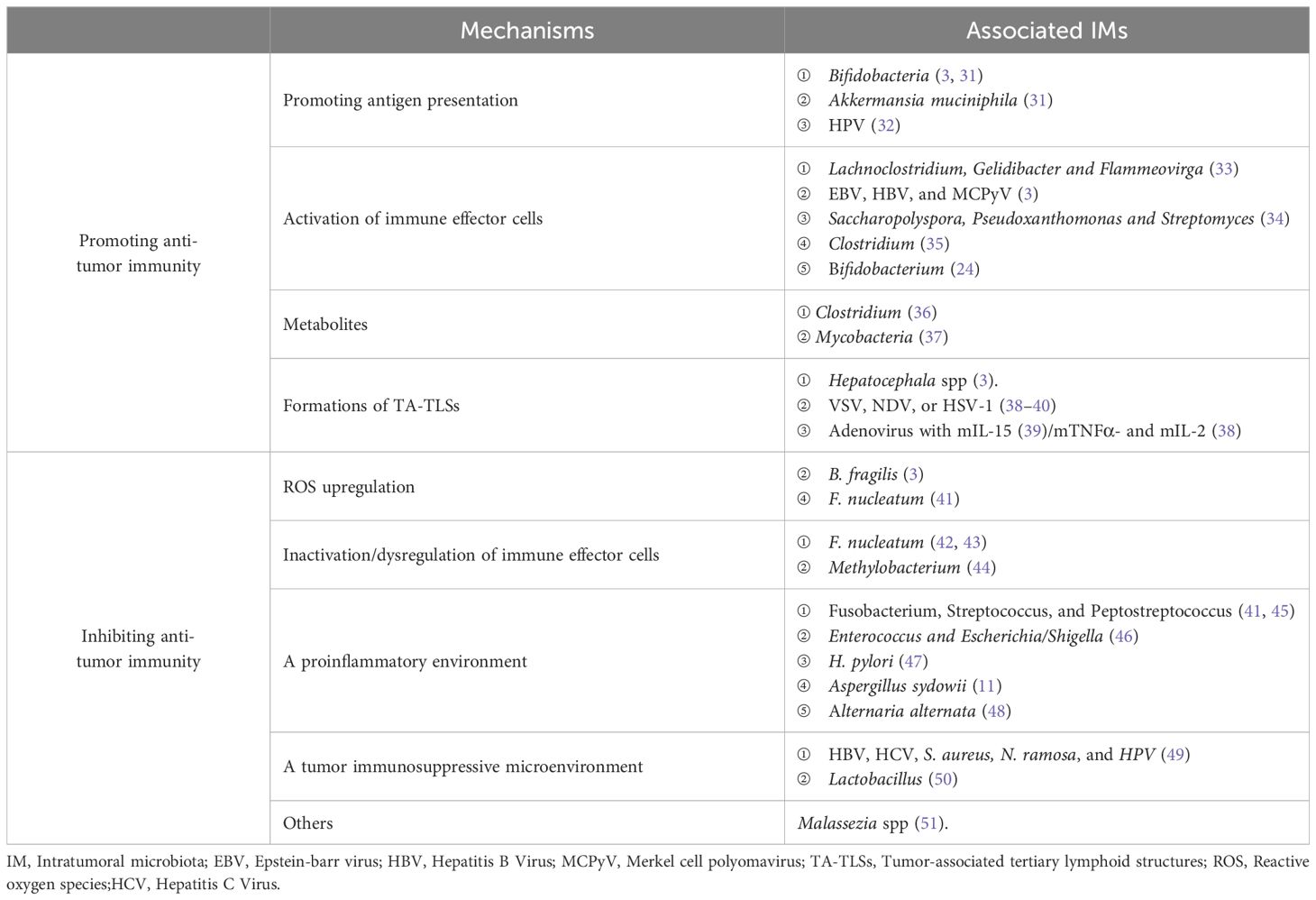
Intratumoral microbiota has different effects on the tumor immune response, some with cancer-promoting effects and others with tumor suppressors (Table 1).
4.1 Strengthening anti-tumor immune response
IMs boost anti-tumor immunity by augmenting stimulator of interferon genes (STING) signal and antigen presentation, activating immune effector cells and forming a tumor-suppressive microenvironment (Figure 2).
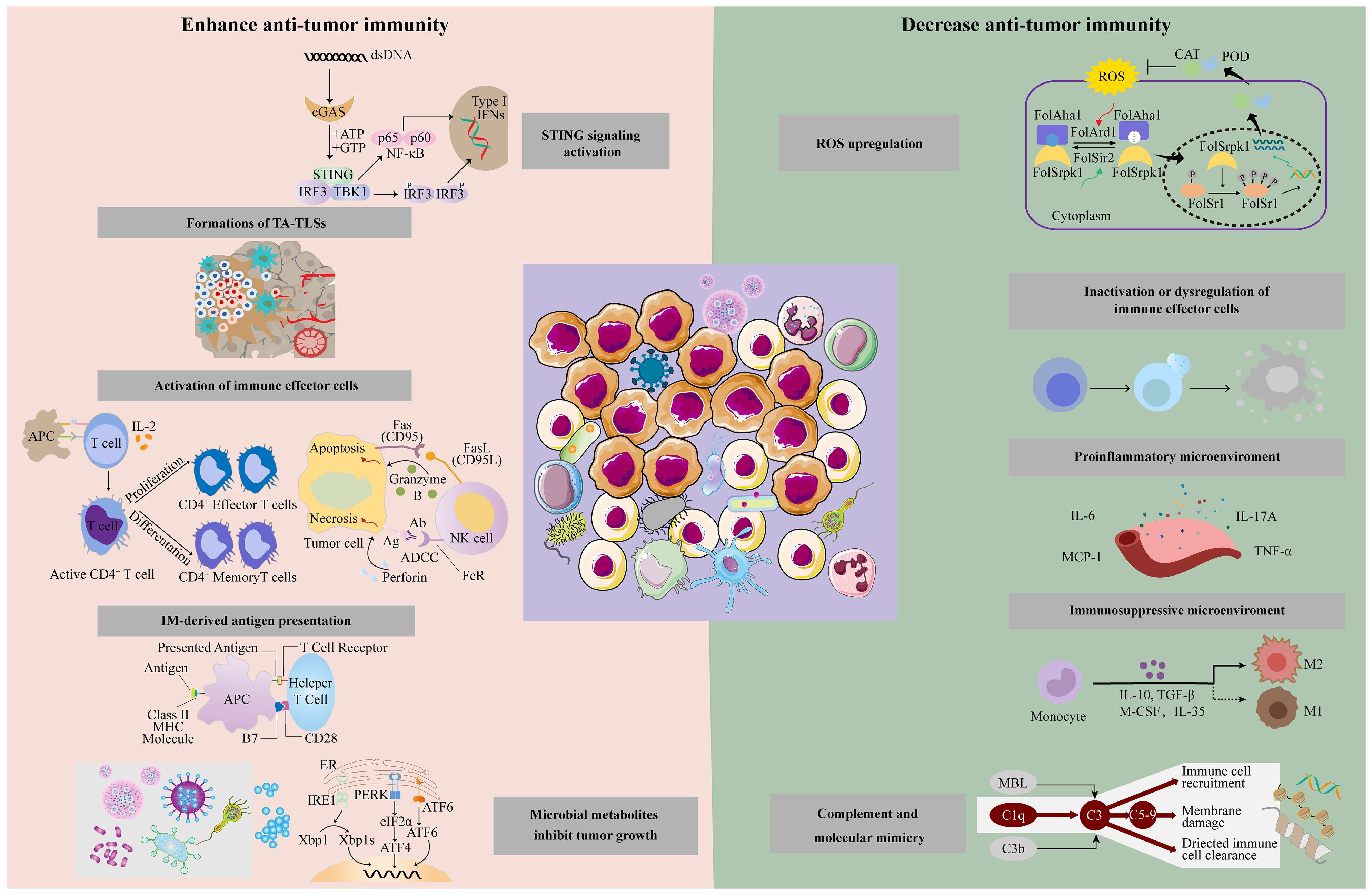
Figure 2. Intratumoral microbiota has different effects on the tumor immune response through different mechanisms, including enhanced anti-tumor immune response and suppression of anti-tumor immunity.
4.1.1 Promoting STING signal and IM-derived antigen presentation
Microbe-associated molecular patterns may stimulate pattern recognition receptors (PRRs) and intensify immune responses based on cross-reactivity between tumors and microbial antigens. Microbiota can activate the STING signaling pathway, which improves the cross-priming of dendric cells (DCs) and further induces naive T cell proliferation and differentiation to promote anti-tumor responses (52). Bifidobacteria can migrate to and invade intestinal cancer cells. Following anti-CD47 targeted immunotherapy, topically and systematically administered Bifidobacteria initiate STING signaling to coordinately kill tumor cells (3). Analogously, STING activators derived from Akkermansia muciniphila induce the release of interferon-I (IFN-I) from intratumoral mononuclear cells, polarize macrophages, and promote the cross-talk with natural killer (NK) cells and DCs (3, 31).
In addition to triggering more potent host antigen presentation through the aforementioned mechanisms, the antigen-presenting cell (APC)-presented peptides of intratumoral bacteria provoke specific T cell responses targeting tumor cells for the microbes mostly located within tumor cells and immune cells. Intratumoral injection of bacteria or their antigens provides immunostimulatory effects to combat tumors, similar to Coley toxin, a heat-inactivated mixture of Serattia marcescence and Streptococcus pyogenes. HPV E2 and E5 are major targets of the intratumoral CD8+ T cell response in patients with HPV-positive head and neck squamous cell carcinoma (HNSCC). Thus, therapeutic vaccination strategies for HPV-positive patients should also consider including E2 and E5 as vaccine antigens to elicit tumor-reactive CD8+ T cell responses of maximal breadth (32).
4.1.2 Activation of immune effector cells
Some IMs are positively correlated with activated and infiltrated immune effector cells and promote anti-tumor immunity through mechanisms such as activation of STING and IFN signaling pathway, regulation of cytokines or chemokines, initiation of endoplasmic reticulum (ER) stress, and other potential mechanisms.
In cutaneous malignant melanoma (MM), intratumoral Lachnoclostridium, Gelidibacter, and Flammeovirga were the top three genera positively associated with CD8+ T cell infiltration. They are also positively correlated with the secretion of chemokines such as CXCL9, CXCL10, and CCL5 (33). Oncoviruses such as EBV, HBV, and Merkel cell polyomavirus (MCPyV) can also induce chemokine production in tumor tissues, and then increase CD8+T cell infiltration to prolong patient survival (3). In addition, a higher α diversity in Ims and density of CD8 + T cells and granzyme + B cells in the TME are associated with long-term survival (LTS) in pancreaticductal adenocarcinoma (PDAC) patients, although the levels of macrophages, Tregs, and MDSC cells exhibited no significant difference from those in short-term survival patients, indicating Saccharopolyspora, Pseudoxanthomonas, and Streptomyces can recruit and activate CD8+ T cells in PDAC tissues (34). The exogenous administration of microbes has also been found to affect immune cell infiltration. For example, bifidobacterium incorporation into cancers facilitates NK cell activation (35), and four commensal Clostridiales strains (CC4) orally administered to mice can accumulate more CD8+ T cells (24).
4.1.3 Microbial metabolites inhibit tumor growth
Trimethylamine-N-oxide (TMAO) released by Clostridium initiates PKR-like ER kinase (PERK)-mediated ER stress, induces pyroptosis, and augments CD8 + T cell-mediated tumor killing (36). A significantly increased fungal mass in HCC was negatively associated with carbohydrate antigen199 (CA199) levels. Metabolomic and transcriptomic analyses have deduced that mycobacteria interfere with tumor progression via amino acid metabolism (37).
4.1.4 Formations of tumor-associated tertiary lymphoid structures
Tertiary lymphoid structures (TLSs) are ectopic lymphoid aggregates developing within non-lymphoid tissues, which exhibit phenotypic and functional similarities with conventional secondary lymphoid organs (SLOs). TLSs are not typically present under normal physiological conditions but have been noted in various pathological states, such as autoimmune diseases, chronic infections, and cancers. Tumor-associated TLSs (TA-TLSs) are associated with improved patient survival by promoting lymphocyte infiltration and activation in the TME to arouse anti-tumor immunity during maturation (53, 54). Some IMs or exogenous microorganisms contributed to TA-TLS formation. Hepatocephala spp. enhance TLS development around tumors and inhibit intestinal cancer (3). Natural oncolytic viruses (OVs) can also induce TLSs. For instance, after infecting tumor-bearing mice with vesicular stomatitis virus (VSV), Newcastle disease virus (NDV), or herpes simplex virus (HSV-1), antiviral memory T cells were reactivated after being immunized secondly, favoring mutual reinforcement of antiviral and anti-tumor activities to form TLSs, thus eliciting larger-range adaptive immunity compared to controls (38–40). In immune checkpoint inhibitor (ICI)-naïve mice with refractory oral carcinoma, the therapeutic efficacy of anti-PD-1/PD-L1 combined with the mTNFα- and mIL-2-carrying non-replicative adenovirus was improved along with the upregulated expression of a TLS relative gene signature (38). A mIL-15 armed-oncolytic adenovirus recruited activated T and NK cells through STING-TBK1-IRF3-mediated DC activation, prompting vascular normalization and TLS formation (39). These studies showed that microbe-induced TA-TLSs could be a new strategy for cancer immunotherapy.
4.2 Inhibiting antitumor immune responses
Some IMs can counteract effective immunotherapy via releasing ROS, inactivating immune effector cells, promoting inflammation, and forming immunosuppressive microenvironments (Figure 2).
4.2.1 ROS upregulation
Symbiotic microorganisms can produce ROS, contributing to tumor progression in multiple ways (55), including the regulation of immune responses. The bacterium enterotoxigenic Bacteroides fragilis (ETBF) causes pre-inflammation or immunosuppression via ROS and DNA damage in the TME, resulting in colorectal cancer (3). In the gastrointestinal tract, activation of the Toll-like receptor (TLR4)-ROS and nitric oxide dioxygenase (NOD)-like receptor (NOD1/2)-dependent signaling by F. nucleatum enriches neutrophil extracellular trap (NET) formation, thus, tumors progress and metastasize through angiogenesis, Epithelial-Mesenchymal Transition (EMT), matrix metalloproteinase (MMP)-mediated basement membrane protein degradation, and the capture of CRC cells (41).
4.2.2 Inactivation or dysregulation of immune effector cells
Some IMs have a negative correlation with tumor-infiltrating T cell density and can cause T cell dysfunction and favor tumor growth and metastasis. Transcriptome and digital pathological analyses revealed that the bacterial load within breast cancer (BC) hampered T cell infiltration, with F. nucleatum being negatively correlated, especially with CD3+T cells (42). F. nucleatum inoculation inhibited the aggregation of tumor-infiltrating T cells and recruited tumor-associated macrophages (TAMs) and fibroblasts (43). Conversely, antibiotic treatment counteracts F. nucleatum-induced side effects, thereby confirming its detrimental effects on immune effector cells (42). Intratumoral Methylobacterium induces dysfunction of the memory CD8+ T cells residing in tumor tissues (44). Altogether, these studies suggest that IMs influence the behavior of immune effector cells and thus impact tumor progression and prognosis.
4.2.3 A proinflammatory microenvironment
Oncobacteria, such as Fusobacterium nucleatum (Fn), Streptococcus gallolyticus, and Prevotella anaerobius, induce the tumor inflammatory microenvironment by regulating inflammatory factor release and forming local inflammatory environments, such as NETs (41), thereby stimulating cancer cells to proliferate (45). In a liver metastasis murine model of CRC, orally administered F. nucleatum exerted deleterious effects on the distant tumor, along with substantially increasing plasma levels of proinflammatory cytokines, such as IL6, IL12, IL9, IL17A, CXCL1, monocyte chemoattractant protein-1(MCP-1), TNF-α, and IFN-γ as well as monocyte myeloid-derived suppressor cell (MDSC) accumulation, abridgment in NK and Th17, and a decrease in the α diversity of the gut microbiome, but elevating Enterococcus and Escherichia/Shigella levels (46). H. pylori induced-NF-κB activates the PIEZO1-YAP1-CTGF axis to reshape the gastric cancer microenvironment by promoting CAF infiltration, therefore targeting PIEZO1-YAP1-CTGF might be a potential therapeutic option to prevent gastric cancer (GC) progression and peritoneal metastasis (47).
As for fungi, Aspergillus sydowii (A. sydowii), enriched in lung adenocarcinoma (LUAD) tissue, provokes IL-1β secretion and its associated MDSC accumulation and expansion through the β-glucan/Dectin-1/CARD9 pathway, which suppresses cytotoxic T cells and ICI therapy (11). Likewise, microbiota residing in the lung mucosa spur myeloid cells release Myd88-dependent IL-1β and IL-23, resulting in Vγ6+Vδ1+γδ T cell proliferation and secretion of IL-17 and other cytokines to form an inflammatory microenvironment, thus accelerating the development of lung cancer due to KRAS mutation and p53 loss. This evidence seems to imply that ablating microbiota through antibiotic treatment is a good candidate. Interestingly, another study shared a different perspective. Mice treated with antibiotics are more susceptible to Lewis lung carcinoma because of defects in inducing the γδT17 cell response and subsequent IL-6 and IL-23 release. This may be a result of the differences in the pathogenesis of different lung cancers (48). In another example, fungi in PDAC elevate IL-33 secretion by tumor cells, further recruiting Th2 and innate lymphoid cells 2 (ILC-2), which then promotes tumor progression by releasing pro-tumorigenic cytokines such as IL-4/5/13 (56).
4.2.4 A tumor immunosuppressive microenvironment
IMs and their metabolites can fail ICI or other therapeutics through immune escape and immune effector cell anergy. Previous studies have reported that IMs, such as HBV, HCV, S. aureus, and N. ramosa, enhance Treg immunosuppression in the TME, thereby mediating prostatic cancer (PCA) and HCC growth. As a result of inflammation and antiviral responses (49), advanced forms of HPV (+) tumors have a high infiltration of immune cells in the TME, however, the main content is myeloid cells instead of DCs and cytotoxic T cells, weakening anti-tumor activity (50). Microbiota within pancreatic cancer causes T cell disability through selective Toll-like receptor ligation, thereby leading to increased levels of MDSC and M2 macrophages to form a suppressive TME (57). Commensal fungi in breast cancer and melanoma can reduce T cells and increase M2-like TAMs by conjugation with Dectin-1, thus offsetting anti-tumor immunity (58).
4.2.5 Complement and molecular mimicry
Glycans on the wall of intratumoral Malassezia spp. ligating with mannose-binding lectin (MBL) elicits complement cascades that promote tumor progression (51). Molecular mimicry is another mechanism for fungi to promote cancer. For instance, Candida albicans express complement receptor 3-related protein (CR3-RP), structurally similar to CR3 on the leukocytes, thereby disturbing the immune response in the TME (59).
Taken together, intratumoral microbes modulate anti-tumor immunity in multiple ways. Their functions depend on various factors, far beyond the IM composition, tumor type and status, and external factors including antibiotics and diet, which is worth exploring in clinical practice.
5 The signaling pathway of intratumoral microbiota in tumorigenesis and immunity
As discussed previously, intratumoral microorganisms play a significant role in tumorigenesis and the regulation of anti-tumor immunity. The signaling pathways through which these microorganisms influence tumor progression and immune responses include but are not limited to, ROS, β-catenin, STING, and others (Figure 3).
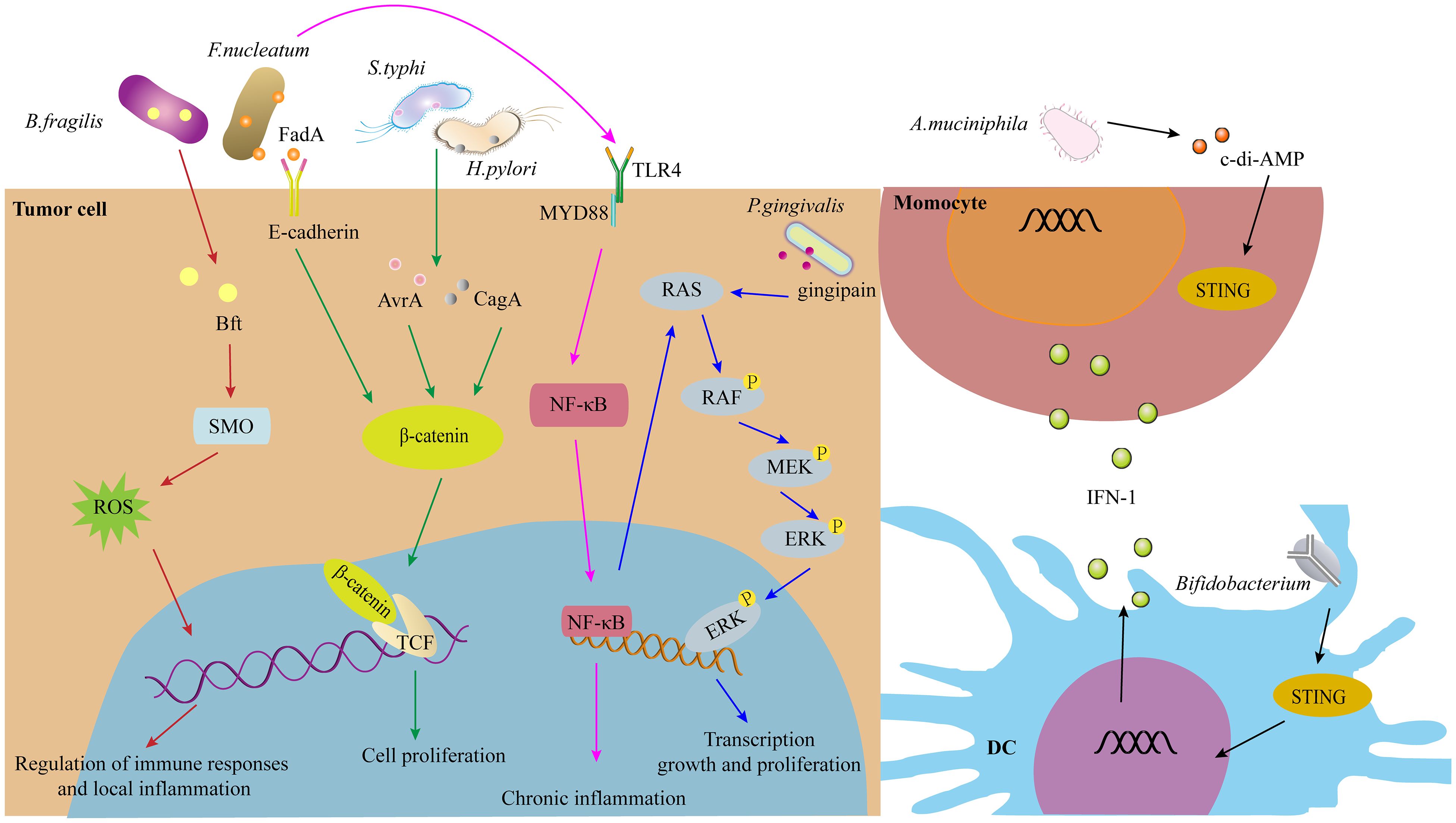
Figure 3. The signaling pathways of IMs in tumorigenesis and immunity, such as ROS, β-catenin, STING signaling pathway, and others.
5.1 ROS signaling pathway
ROS are highly bioactive molecules that, at low to moderate levels, serve as signal transducers that activate cell proliferation, migration, invasion, and angiogenesis. However, high levels of ROS can lead to cellular damage, including protein, nucleic acid, lipid, membrane, and organelle injuries, culminating in cell death, EMT, and metastasis (60). For instance, Enterotoxigenic Bacteroides fragilis (ETBF) induces DNA damage via ROS (3). In the gastrointestinal tract, F. nucleatum activates Toll-like receptor 4 (TLR4)-ROS signaling, promoting NET formation and thereby enhancing tumor progression and metastasis through angiogenesis, EMT, and matrix metalloproteinase (MMP)-mediated degradation of basement membrane proteins (41).
5.2 β-catenin signaling pathway
The Wnt/β-catenin signaling pathway plays crucial roles in embryonic development and adult tissue homeostasis. Dysregulation of this pathway contributes to various diseases, including cancer. H. pylori infection significantly increases cyclin-dependent kinase 1 (CDK1), which phosphorylates and inhibits GSK-3β activity, resulting in the accumulation and activation of β-catenin and NF-κB in gastric tumors (61). Additionally, β-catenin and yes-associated protein (YAP) synergistically promote H. pylori-induced gastric carcinogenesis by regulating downstream genes such as CDX2, LGR5, and RUVBL1 (62). Furthermore, FadA adhesin from F. nucleatum enhances Annexin A1 expression in Wnt/β-catenin signaling, forming a positive feedback loop through E-cadherin and activating Cyclin D1 specifically in cancerous cells, not in non-cancerous cells (63). F. nucleatum interaction with CDH1 triggers phosphorylation events that upregulate downstream β-catenin, Cyclin D1, and Myc to promote squamous cell carcinoma proliferation (64). In another study, the F. nucleatum group showed significantly increased expression of TLR4, PAK1, p-PAK1, p-β-catenin S675, and cyclin D1 compared to the control group, suggesting TLR4 as a potential therapeutic target for F. nucleatum-related colorectal cancer prevention and therapy.
5.3 STING signaling pathway
Cyclic GMP-AMP synthase (cGAS) functions by detecting misplaced genomic, mitochondrial, and microbial double-stranded DNA (dsDNA), leading to the synthesis of 2’3’-cGAMP. This molecule activates STING, thereby initiating innate immune responses. This mechanism serves as a pervasive and effective surveillance system against tissue damage and pathogen invasion. However, dysregulated cGAS-STING signaling plays a significant role in infectious, autoimmune, malignant, fibrotic, and neurodegenerative diseases (65). The cGAS/STING pathway can promote macrophage polarization, contributing to potent anti-tumor immunity (65). In tumor models, both systemic delivery and local administration of Bifidobacterium result in its accumulation within tumors, converting non-responders into responders in a STING- and interferon-dependent manner. Furthermore, Bifidobacterium enhances dendritic cell cross-priming following anti-CD47 treatment (66). In cancer immunotherapy, Bifidobacterium improves the efficacy of immune checkpoint inhibitors through STING and adenosine 2A receptor (A2AR) signaling pathways (67).
5.4 Other signaling pathways (NF-κB, TLR, complement, RhoA/ROCK, PERK)
In addition to the aforementioned pathways, intratumoral microbes may activate other signaling pathways such as NF-κB, TLR, complement, RhoA/ROCK, and PERK. For instance, E. coli colonization in colorectal cancer liver metastasis (CRLM) enhances lactate production, promoting M2-like macrophage polarization via NF-κB signaling facilitated by retinoic acid-inducible gene 1 (RIG-I) lactylation, thereby contributing to colorectal cancer liver metastasis (68). In both immunocompetent and immunodeficient melanoma mouse models, flagellum-deficient Salmonella fails to induce significant anti-tumor effects, despite higher bacterial inoculation, due to the absence of the Flagellin/Toll-like receptor 5 (TLR5) signaling pathway. Mannose-binding lectin (MBL) binds fungal glycans and activates the complement cascade, potentially mediating tumor progression. Intratumoral microorganisms also impact tumorigenesis and anti-tumor immune responses through other signaling pathways such as RhoA/ROCK and PERK (69–71).
6 Clinical implications of intratumoral microbiota
The intratumoral microbiota not only plays a role in tumor initiation, progression, and metastasis but also possesses significant clinical implications. These include targeting intratumoral bacteria for cancer therapy and utilizing intratumoral microbiota as diagnostic and prognostic tools.
6.1 Targeting the microbiota for cancer therapy
Microbe-based cancer therapies have shown efficacy, yet some IMs can induce an immunosuppressive microenvironment, leading to therapeutic failure and adverse effects. Therefore, strategies aimed at eliminating specific tumor-associated IMs are highly desirable. For instance, the administration of bismuth colloidal pectin granules to gastric cancer patients infected with H. pylori reduced side effects and improved symptoms (ClinicalTrials.gov Identifier: NCT05049902) (72). Additionally, the efficacy of itraconazole in treating various cancers has been explored in preclinical and clinical trials (ClinicalTrials.gov Identifier: NCT02749513) (73). Nevertheless, the use of antibiotics may result in flora imbalance and drug resistance. Future advancements may include the use of phage-based or engineered phage-based therapies targeting oncobacteria for effective tumor treatment.
6.2 Diagnostic and prognostic roles of intratumoral microbiota
The abundance of intratumoral microbes in different tumors, along with the presence of tumor type- and subtype-specific microbial profiles, suggests that IMs can serve as valuable biomarkers in cancer diagnostics and prognostics. For example, the intratumoral microbiome characteristics of HNSCC are associated with clinicopathological features such as tumor stage and histological grade. A recent study revealed that the microbiome in GC can be classified into three microbial subtypes (MS1, MS2, and MS3), each with distinct characteristics that are linked to immunotherapy and prognosis (74). Another study found that the relative abundance of viruses in soft tissue sarcomas (STS) is positively correlated with higher NK cell infiltration, which is associated with improved metastasis-free and overall survival (75). In pancreatic cancer, the presence of F. nucleatum and P. gingivalis is associated with a higher risk and worse prognosis (76). High levels of F. nucleatum are particularly correlated with the stage and recurrence of esophageal squamous cell carcinoma (ESCC) (77). These findings underscore the potential of intratumoral microbiota as biomarkers in cancer diagnostics and prognostics.
7 Applications in therapeutics
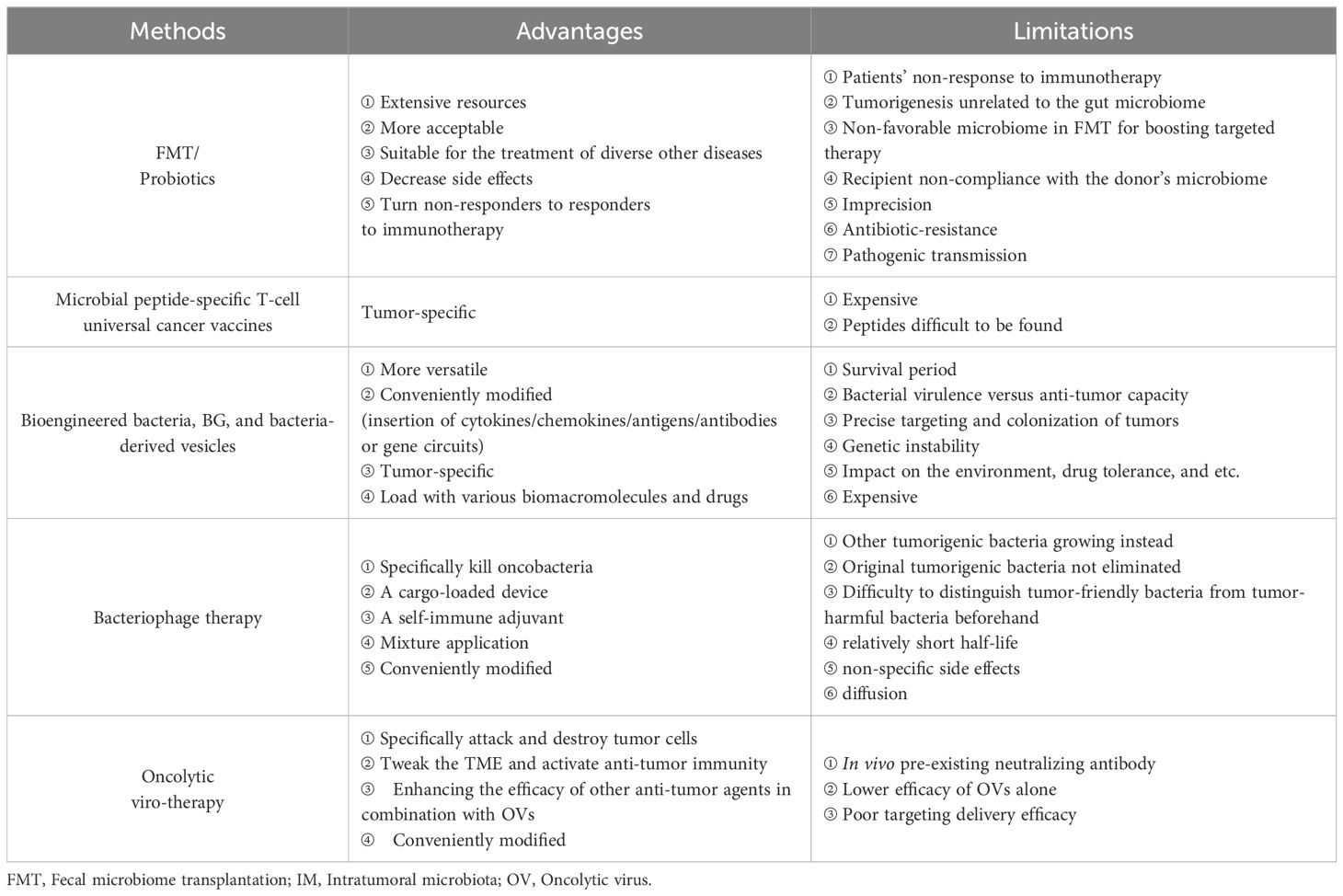
Based on the roles of intratumoral microbiota in tumors, the ablation of pro-tumoral IMs and supplementation with tumor-hostile IMs are feasible approaches therapeutically (78), but fecal microbiome transplantation (FMT), probiotic regulation, microbial peptide-specific T cell universal cancer vaccines, and engineered microbes may have more prospects for cancer therapy (Figure 4).
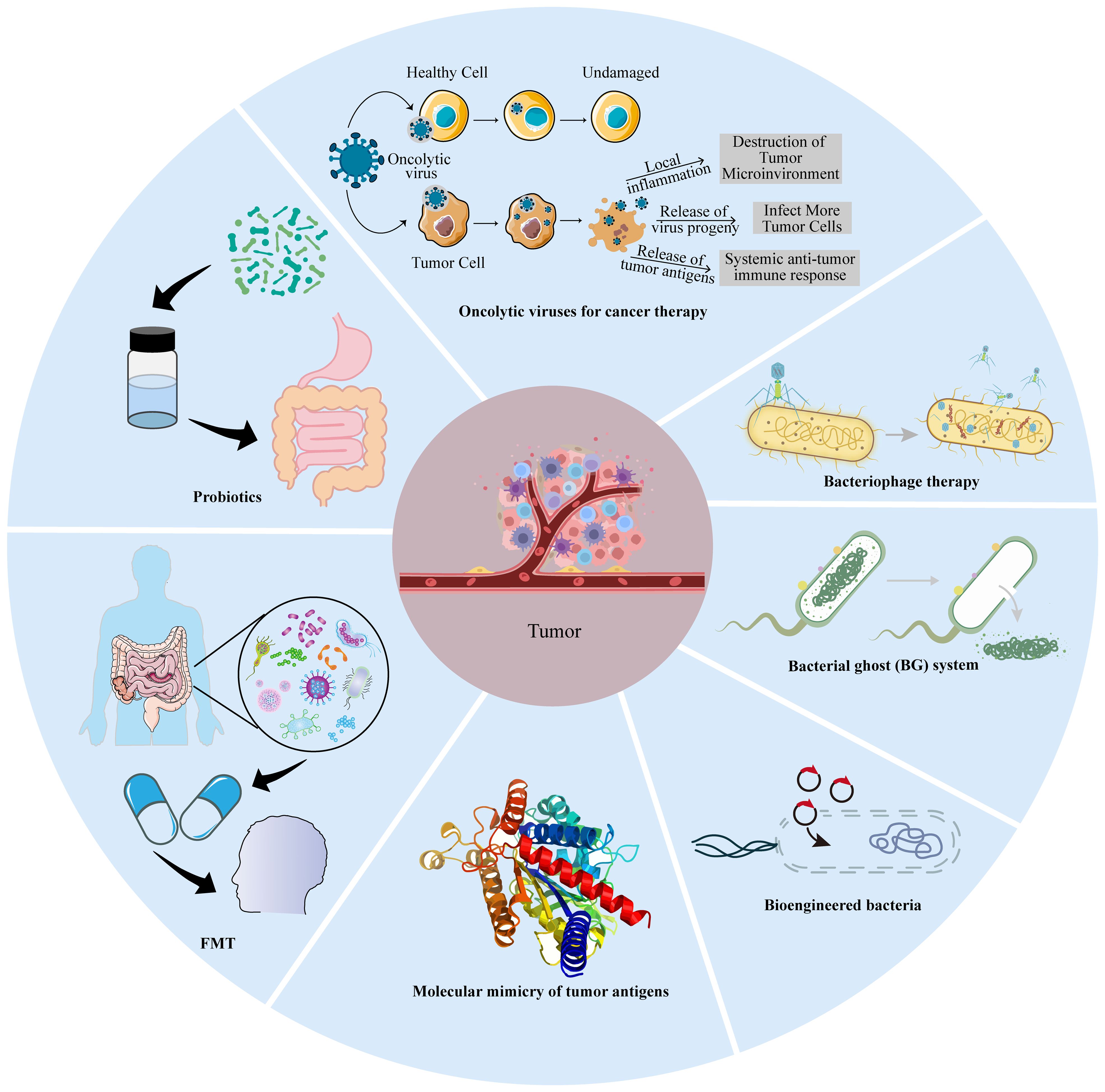
Figure 4. Microbial-based cancer treatment strategies. These include oncolytic virotherapy (OVT), bacteriophage therapy, bacterial ghost (BG), bioengineered bacteria, molecular mimicry, fecal microbiota transplantation (FMT), and probiotics.
7.1 Fecal microbiome transplantation
FMT can effectively treat recurrent antibiotic-resistant infections caused by C. difficile, which has been evaluated in inflammatory bowel disease (IBD) (79). Baruch and coworkers reported that 10 patients with metastatic melanoma who were resistant to ICIs were treated with vancomycin and neomycin for 3 days to ablate native microbiota before FMT via both colonoscopy and administration of oral stool capsules. FMT products were extracted from two donors with a complete response to nivolumab for metastatic melanoma for over 1 year. Three patients (3/10)overcame ICI resistance after FMT, one with a complete response, and two with partial responses (NCT03353402) (80). In two independent clinical trials (NCT03341143 and NCT03772899), FMT from long-term responder donors with ICI-refractory advanced melanoma alone showed effective clinical responses or prolonged survival without serious side effects. FMT ameliorates the dysbiosis of bacterial flora associated with anti-PD-1 clinical responses and increases anti-tumor immunity (81, 82). Although favorable and unfavorable species of bacteria have been identified, the ingredients of an “ideal gut microbiota” remain elusive. Thus, there is not a one-size-fits-all gut microbiota. FMT faces several main challenges: ① The suitable microbiota composition for donors is not yet established at present. ② There are no available screening methods or prognostic factors that can be harnessed to select candidate recipient patients for FMT plus ICI treatment. ③ FMT failure is likely attributed to patients’ non-response to immunotherapy, tumorigenesis unrelated to the gut microbiome, a non-favorable microbiome in FMT for boosting targeted therapy, or recipient non-compliance with the donor’s microbiome. ④ The incidence of antibiotic resistance and pathogenic transmission in practical applications are further challenges. For this reason, FMT is not an approved treatment by either the Food and Drug Administration (FDA) or European Medicines Agency (EMA). Together, these early findings have important implications for modulating the gut microbiota in cancer treatment. In the future, the modulation of the gut microbiota, particularly with FMT regimens (delivery routes, frequency, duration, antibiotic pre-treatment) should select donors and recipients to improve the likelihood of success.
7.2 Probiotic regulation
Probiotics can survive or temporarily colonize the intestine, which is an important mechanism to manipulate the gut microbiota to increase the number of “beneficial bacteria.” Probiotic-based manipulation of the gut microbiota inevitably affects host metabolism, immune function, and digestion (83). At present, some species of bacteria have been utilized as probiotics, such as lactic acid bacteria, Bifidobacterium, and Clostridiales (83), to treat various diseases, including diarrhea, IBD, rheumatoid arthritis, chronic sinusitis, and cancer (84, 85). Mice administered with cisplatin in combination with Lactobacillus exhibited superior outcomes, including reduced tumor size and increased survival rates, compared to mice receiving cisplatin alone or cisplatin plus antibiotics. This suggests that Lactobacillus may mitigate the adverse effects commonly associated with chemotherapy (86). Radiation-induced intestinal injury is a common complication following abdominal or pelvic radiotherapy. The intervention with Lactobacillus rhamnosus GG (LGG) improves intestinal structure, reduces jejunal DNA damage, and inhibits the inflammatory cGAS/STING pathway, suggesting a potential therapeutic radioprotective effect (87). Lactobacillus acidophilus combined with Bifidobacterium bifidum could reduce the incidence of radiation-induced diarrhea and the necessity for anti-diarrheal medication in cervical cancer patients undergoing radiotherapy, with significant improvements in stool consistency (trial registration number: TCTR20170314001) (88). Members of the Clostridiales order within the gut microbiome, which have been linked to a reduced tumor burden, were found in a mouse model of CRC. Furthermore, these beneficial species were observed to be sharply diminished in CRC patients compared to their healthy counterparts. Notably, treatment with four strains of Clostridiales (CC4) or individual strains demonstrated an ability to impede or effectively treat CRC as a monotherapy. In a head-to-head comparison with anti-PD-1 therapy, supplementation with a mixture containing CC4 strains exhibited superior efficacy in mouse models of CRC and MM (24). Accumulation of Bifidobacterium within tumors reversed anti-CD47 treated mice from non-responsive to responsive via potently stimulating STING and IFN-I signaling as aforementioned (66). These studies provide not only pre-clinical evidence for harnessing FMT and probiotics as a monotherapy for solid tumors but also some limitations in practical applications (89). One study revealed that the dietary tryptophan catabolite indole-3-aldehyde (I3A), produced by the colonizing probiotic Lactobacillus reuteri (Lr), locally promotes IFN-γ-producing CD8+ T cells, thereby sustaining the efficacy of ICIs. The absence of Lr-secreted I3A or impaired aryl hydrocarbon receptor (AhR) signaling within CD8+ T cells negates the anti-cancer effects of Lr. Chimeric antigen receptor (CAR)-T cells have demonstrated remarkable success in treating hematological malignancies but have encountered limited efficacy against solid tumors (90). A recent study unveiled that tumor-colonizing probiotics release synthetic targets that mark tumor tissue for CAR-T-mediated cytolysis in situ. The innovative probiotic-guided CAR-T cells (ProCARs) system can activate and direct CAR-T cell activity for antigen-agnostic cell lysis across various cancer models (91). These breakthroughs highlight the potential of intratumoral microbiota manipulation as a complementary strategy to conventional and immunotherapeutic approaches, offering a beacon of hope for improved therapeutic outcomes in cancer treatment.
7.3 Microbial peptide-specific T cell universal cancer vaccines
Molecular mimicry (92–94) of tumor antigens homologous to microbial epitopes supplies cross-reactive T cells with ideal targets (95). Besides this paradigm, tumor cells are also capable of presenting microbial peptides directly to induce microbial-specific T cells, such as in MM (96). These peptides partially overlap with the HLA of tumor cells infected with coequal microbial strains.
These studies could usher in the development of microbial peptide-specific T cell universal cancer vaccines. Although the limited presence (0.1-10%) of bacteria in tumor cells (8, 69) may restrict its therapeutic efficacy, targeting bacterial peptides elicits greater anti-tumor immunity (by antigen dissemination) and facilitates a more precise tumor-specific T cell migration to the tumor site that is difficult for other methods to achieve. For instance, F. nucleatum-specific T cells could elicit anti-tumor immunity at the site of primary and metastatic tumors (43). In another example, one bacterial peptide called azurin-28 showed acceptable toxicity and tumor-inhibiting effects in pediatric brain tumor patients (97, 98).
The concept mentioned above is not novel, as previous attempts have been made to inject attenuated Salmonella directly into tumors over the past two decades in multiple cancer types (99–101). Recently, Salmonella infection in MM induced the unfolded protein response (UPR), leading to the release of peptides from tumor cells to generate anti-tumor responses in vivo (102). This new antigenic peptide can be further applied as a universal vaccine platform. Although Salmonella is not regarded as an intratumoral microorganism, this work provides new approaches for cancer therapy.
7.4 Bioengineered bacteria
Although bacteria can display immunostimulatory features and possess the potential for cancer therapy, bioengineered bacteria can acquire more versatile functions to achieve better efficacy through synthetic biology, which selectively and preferentially colonize in hypoxic and necrotic TMEs. These bioengineered bacteria, including S. typhimurium, Listeria Monocytogenes, Bifidobacterium longum, Lactobacillus species, and Clostridium species, have been utilized as delivery vectors with no serious adverse effects. In S.typhimurium, two chromosomal genes were deleted, namely, purI and msbB, to reduce toxicity and increase safety (103). Recently, attenuated Salmonella typhimurium VNP20009 was engineered to exploit its tumor-targeting capabilities and induce the expression of GM-CSF and IL-7 in response to L-arabinose (named as GM-CSF-IL-7-VNP20009). Intravenous inoculation of GM-CSF-IL-7-VNP20009 in mouse models of subcutaneous and lung metastatic melanoma resulted in significant inhibition of melanoma growth and metastasis and prolonged the survival of the mice by significantly increasing the number of tumor-infiltrating macrophages and mature DCs, and elevated the proportion of CD4+ T cells as well as their proliferation in cancers (104). Currently, an L.monocytogenes-based cancer vaccine named ADXS11-001 is in several Phase II clinical trials to evaluate its efficacy for HPV-associated cancers (NCT02853604) (105). Additionally, different probiotic bacteria including Bifidobacterium longum and Lactobacillus plantarum are regarded as anticancer-targeting vectors. Clostridia strains such as C. acetobutylicum, C. beijerinckii, and C. sporogenes could effectively be modified to express some bacterial enzymes (nitroreductase, cytosine deaminase) or murine-TNFα to enhance the efficacy of cancer treatment (106). Clostridium perfringens exhibits antitumor activity due to its enterotoxin to damage the tight junction in the epithelial cells (107). In a clinical trial (NCT01924689), a single intratumoral injection of C. novyi-NT, which lacked the alpha toxin, for patients with injectable, treatment-refractory solid tumors can generate bacterial spore germination in the tumor, and leads to the killing of tumor cells and an inflammatory response with significant but controllable toxicities (108). An engineered E. coli Nissle 1917 strain can colonize tumors and convert ammonia to increase intratumoral L-arginine levels which increases the number of tumor-infiltrating T cells and has significant synergistic effects with PD-L1 blocking antibodies to clear tumors (109). Bacteria engineered to synthesize CD47 nanobodies could avoid adverse side effects caused by an unwanted CD47 blockade of red blood cells and platelets (110). Intratumoral injection of engineered Lactococcus lactis strains expressing Fms-like tyrosine kinase 3 (Flt3) ligand and OX40 ligand into one flank in mice bearing tumors bilaterally caused regression of tumors on both sides and increased anti-PD1 efficacy by 40% when the two methods were combined, although on the opposite side, after draining lymph nodes and the whole body, viable bacteria were not detected by the culture method (111). The results of these engineered bacteria to improve the response rates are exciting. However, one question remains: can certain bacteria survive for a period of time in a certain TME? Strange infiltration and proliferation behavior of IMs can occur in xenograft tumor models. Salmonella typhimurium is capable of colonizing BXPC-3 cells and slowly proliferating in the HCT116 model, whereas E. coli cannot in BXPC-3 cell-bearing tumors (112). In 1990, the FDA approved BCG for perfusion treatment of superficial bladder cancer, and BCG perfusion treatment remains the standard first-line treatment for bladder cancer (113).
Additionally, the bacterial ghost (BG) system is another innovative bacteria-based cancer therapy, which lacks ribosomes, nucleic acids, and some more intracellular constituents. The drugs, DNA, proteins, enzymes, and other therapeutic agents can fill the inner space of the BGs (114). For instance, cancer cell lysate (oncolysate)-loaded BGs induced by IFN-α and GM-CSF enhances DC maturation to increase the expression of maturation markers and co-stimulatory molecules, produce higher IL-12 levels, and significantly increase the proliferation of CD4+ and CD8+T cells compared to DCs in the presence of LPS (115). Bacteria-derived vesicles are also utilized in cancer therapy. Liu and coworkers constructed a bacteria-derived outer membrane vesicle (OMV)-coated nanoplatform that reached the dual-target kill of pre-tumor F. nucleatum and tumor cells, thus converting intratumoral bacteria into immunopotentiators in immunotherapy of triple-negative breast cancer (TNBC) (116).
Apart from the above advantages, engineered bacteria also face some obstacles in clinical applications, including bacterial virulence, anti-tumor capacity, precise targeting and colonization of tumors, genetic instability, impacts on the environment, and drug tolerance.
7.5 Bacteriophage therapy
Tumors are non-responsive to immunotherapy (117, 118), which may be due to the effects of human microbiota on inflammation and immune dysregulation (119). Although antibiotic therapy (ABT) can control intratumoral bacteria, it has detrimental effects on the microbiome and leads to antibiotic resistance (26). Moreover, some types of antibiotics increase the incidence of CRC, and ABT and tumor location are significantly correlated (120). Additionally, the broad-spectrum activity of antibiotics may not be able to distinguish whether bacteria are cancer-promoting or cancer-suppressive within the TME.
Bacteriophages are an attractive biomedical tool for therapeutic applications owing to their nanosize, polyvalent surface properties, non-pathogenic nature, and specifically infecting particular bacteria species and are receptive to desirable chemical or genetic modifications. Bacteriophages are able to serve as both a cargo-loaded device and self-immune adjuvant (45, 121), which makes targeting oncobacteria possible. For example, a specific phage may eliminate S. gallolyticus from cancer patients while retaining other anti-tumorigenic Streptococci (122). More H. pylori strictly virulent phages need to be isolated for therapy of chronic gastritis, peptic ulcer disease, and gastric cancer (123).
In the TME, phages can eliminate specific oncobacteria, leading to other tumorigenic bacteria growing (124). Furthermore, due to their adhesive and invasive toxicity, pro-tumoral bacteria such as Fusobacterium are present in biofilms or cells and evade the immune system (125). When utilizing phages as a therapy to restore microbiota from deviation, it is crucial to ascertain how they interact with each other within the niche.
Similar to engineered bacteria, bacteriophages can be genetically manipulated to improve cancer therapy (126, 127). For instance, the filamentous bacteriophage fUSE5-ZZ has been modified genetically and chemically to deliver anti-ErbB2 and anti-ERGR antibodies or the anti-tumor agents hygromycin and doxorubicin, for anti-tumor treatment (128). Likewise, engineered phages targeting F. nucleatum enhanced the effects of chemotherapy and improved survival in mouse and piglet CRC models by modifying them to covalently bind and optimize irinotecan delivery (124, 129).
Multiple phage mixtures have more potential than individual phages and can be prepared from a phage library, formulated, and administrated to some patients to modulate IMs and enhance cancer therapy (130–132). Recently, eight phages were engineered with tail fibers and clustered regularly interspaced short palindromic repeats (CRISPR)-Cas machinery to specifically target E. coli in biofilms. Furthermore, SNIPR001, composed of the four most complementary bacteriophages, is well tolerated and reduces E. coli load in the mouse gut better than its individual components. SNIPR001 can selectively kill E. coli which may lead to fatal infections associated with hematological cancer (133). Of note, before using phage therapy, it is necessary to accurately distinguish pro-tumoral stains from anti-neoplastic ones.
Though bacteriophages for cancer therapy mentioned above have been discussed, there are a few obstacles yet to be addressed during circulation and after reaching the tumor. Once injected into the bloodstream, bacteriophages could be cleared from circulation by the reticuloendothelial system (RES), which impacts their half-life, bio-distribution, and pharmacokinetics, which compromises the efficacy and may result in non-specific side effects. More importantly, biochemical and biophysical barriers prevent phages from entering into tumor sites. The major obstacle encountered by phages upon reaching the tumor site is related to diffusion. In the future, it is necessary to screen phages to target tumorigenic bacteria. Phages are modified to avoid clearance by the RES, prolong their half-life, and target tumors. It is necessary to better understand how to reduce the immune response-mediated phage clearance from the system. In addition, it is also important to optimize the use of bacteriophages for cancer therapy.
7.6 Oncolytic viruses for cancer therapy
Oncolytic viruses (OVs) are natural or genetically modified viruses that can specifically attack and destroy tumor cells. Generally, compared with “cold” tumors, “hot” tumors feature a high number of mutations, increased expression of tumor-infiltrating lymphocytes (TILs) and PD-L1, and response well to PD-1/PD-L1 inhibitors. Currently, there are no accurate methods to screen “cold” and “hot” tumors. Fortunately, oncolytic viruses, as a way to tweak the TME, can enhance the immune response, in combination with other treatments such as immune checkpoint blockade (ICB) (134–138). The most oncolytic viruses are those armed with targeting or functional factors, depending on the inserted gene sequences (139–143). Talimogene laherparepvec (T-Vec), a genetically modified attenuated HSV1 containing granulocyte-macrophage colony-stimulating factor (GM-CSF), is the first FDA-approved OV product to be administrated via intratumoral injection for postoperative recurrent unresectable MM. Several Phase II-III clinical trials of T-Vec for melanoma and other cancers are currently underway(NCT02779855, NCT00769704) (144, 145). There are also some oncolytic viruses being trialed alone or in combination with other treatments(NCT03152318; UMIN-CTR Clinical Trial Registry UMIN000015995; NCT03178032;NCT03072134) (136, 146–148). Various attempts have been made to modify OVs, including multi-gene, microRNA, and gene circuit insertion. Intratumorally inoculating with OVs arms a PD-L1 inhibitor and GM-CSF and fights against both virus-injected and distant tumors by reversing PD-L1-mediated resistance, priming T cell immune responses against mutation-derived dominant and subdominant neoantigen epitopes (149). The engineered measles virus expressing the synthesized microRNA universal cassette expressed the virally encoded microRNA within the endogenous microRNA transcripts and successfully inhibited the target protein (150). The replication and release of an adenovirus selectively in HCC cells can be regulated via programmable and modular synthetic gene circuits responding to multiple promoter and microRNA inputs, and it has superior cytolytic efficacy compared to non-replicable ones (151). Unlocking the potential of oncolytic virotherapy (OVT) appears to have bright prospects for cancer immunotherapy.
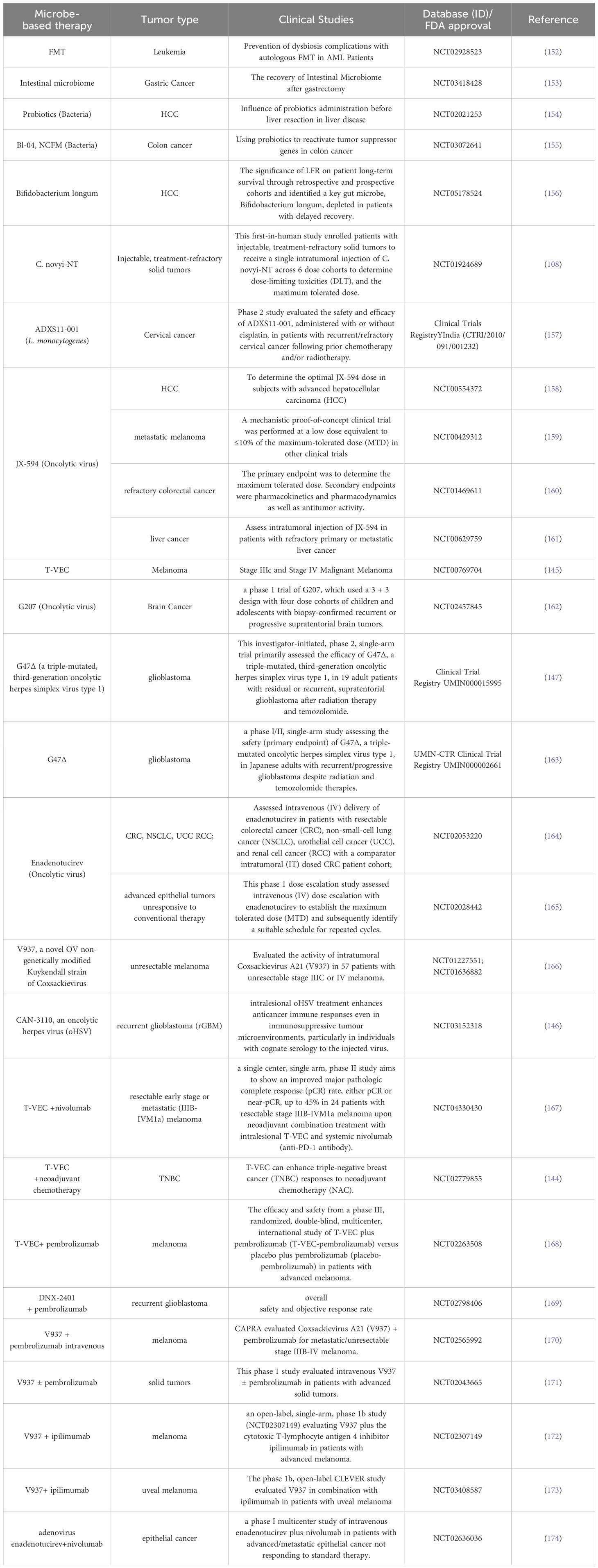
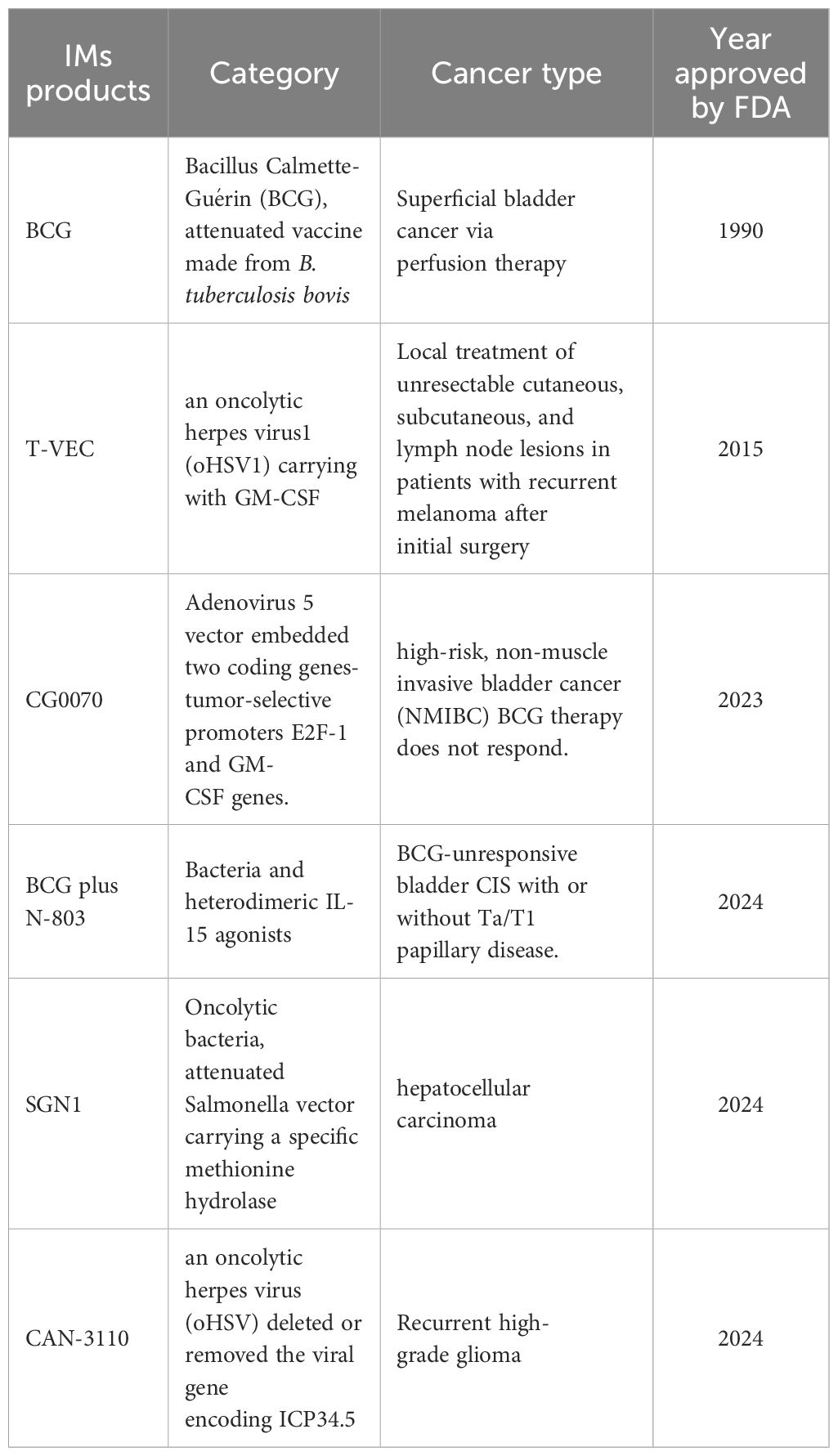
Table 2 gives a comparison of the advantages and limitations of the therapeutic methods mentioned above. Dozens of IM products are being tested in clinical trials (Table 3), some of which have been approved by the FDA for clinical cancer therapy (Table 4). Additionally, microbe-based therapy combined with conventional therapies, and immunotherapy such as ICI or CAR-T, can increase efficacy or mitigate side effects. Clinical trials of microbe-based or combined therapy are listed in Table 4.
8 Conclusions and perspectives
In this review, we have outlined how the composition and diversity of intratumoral microbiota influence tumor initiation, development, and metastasis, as well as anti-tumor immunity through signaling pathways including ROS, Wnt/β-catenin, and STING, with significant clinical implications. We have also proposed strategies for microbe-based cancer therapy. Despite being recognized as a “hallmark” of cancer, research in this field remains nascent, with numerous unresolved issues. For example, the role of intratumoral phages remains largely unexplored. How do bacteria, fungi, viruses, and other microbes within the TME interact to influence tumor initiation, progression, and anti-tumor immune responses? Determining whether specific intratumoral microorganisms act as cancer promoters or tumor suppressors remains a challenge. Additionally, issues surrounding the precision and resistance of FMT or bacterial transplantation need resolution. The systemic administration of engineered bacteria or oncolytic viruses to target tumor tissues also presents considerable challenges.
Further research is needed to elucidate how intratumoral microbes can be leveraged to enhance the efficacy of immunotherapy and mitigate the cytokine storm and neurotoxicity induced by CAR-T cell therapy. With advancements in technologies such as multi-omics, synthetic biology, and artificial intelligence (AI), our understanding of the roles of intratumoral microbiota will improve, leading to the emergence of new concepts and innovative cancer strategies and therapeutic agents. Microbe-based therapies can be used independently or in combination with current immunotherapeutic approaches to enhance efficacy, predict treatment outcomes, or mitigate the toxicity associated with treatments such as CAR-T cell therapy and ICB, thereby advancing effective cancer therapy.
Author contributions
JZ: Formal analysis, Writing – original draft, Writing – review & editing. HC: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The review was supported by Medical Research Project of Jiangsu Provincial Health Commission(M2021045); Clinical Transformation Project of Medical and Health Technology Innovation Project of Chinese Academy of Medical Sciences (2023-2M-C&T-B-110).
Acknowledgments
We thanks to the guidance and support of the supervisor and colleagues in the process of manuscript preparation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Zitvogel L, Straussman R, Sepich-Poore GD, Knight R, Iliev ID, Sears CL, et al. Cancer microbiome. Cancer Cell. (2022) 40:1443–7. doi: 10.1016/j.ccell.2022.11.007
2. Ji J, Ji F, Bayarsaikhan E. Intratumoral microbiota in HCC: A new kid on the block? Hepatology. (2023) 78:1012–4. doi: 10.1097/HEP.0000000000000454
3. Yang L, Li A, Wang Y, Zhang Y. Intratumoral microbiota: roles in cancer initiation, development and therapeutic efficacy. Signal Transduct Target Ther. (2023) 8:35. doi: 10.1038/s41392-022-01304-4
4. Ferrari V, Rescigno M. The intratumoral microbiota: friend or foe? Trends Cancer. (2023) 9:472–9. doi: 10.1016/j.trecan.2023.03.005
5. Lax AJ. Opinion: Bacterial toxins and cancer–a case to answer? Nat Rev Microbiol. (2005) 3:343–9. doi: 10.1038/nrmicro1130
6. Xie Y, Xie F, Zhou X, Zhang L, Yang B, Huang J, et al. Microbiota in tumors: from understanding to application. Adv Sci (Weinh). (2022) 9:e2200470. doi: 10.1002/advs.202200470
7. Gagnaire A, Nadel B, Raoult D, Neefjes J, Gorvel J-P. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol. (2017) 15:109–28. doi: 10.1038/nrmicro.2016.171
8. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. (2020) 368:973–80. doi: 10.1126/science.aay9189
9. Dohlman AB, Klug J, Mesko M, Gao IH, Lipkin SM, Shen X, et al. A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell. (2022) 185:3807–22. doi: 10.1016/j.cell.2022.09.015
10. Narunsky-Haziza L, Sepich-Poore GD, Livyatan I, Asraf O, Martino C, Nejman D, et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell. (2022) 185:3789–806. doi: 10.1016/j.cell.2022.09.005
11. Liu N-N, Yi C-X, Wei L-Q, Zhou J-A, Jiang T, Hu C-C, et al. The intratumor mycobiome promotes lung cancer progression via myeloid-derived suppressor cells. Cancer Cell. (2023) 41:1927–44. doi: 10.1016/j.ccell.2023.08.012
12. Huang J-H, Wang J, Chai X-Q, Li Z-C, Jiang Y-H, Li J, et al. The intratumoral bacterial metataxonomic signature of hepatocellular carcinoma. Microbiol Spectr. (2022) 10:e0098322. doi: 10.1128/spectrum.00983-22
13. Irrazabal T, Thakur BK, Kang M, Malaise Y, Streutker C, Wong EOY, et al. Limiting oxidative DNA damage reduces microbe-induced colitis-associated colorectal cancer. Nat Commun. (2020) 11:1802. doi: 10.1038/s41467-020-15549-6
14. Avril M, DePaolo RW. Driver-passenger" bacteria and their metabolites in the pathogenesis of colorectal cancer. Gut Microbes. (2021) 13:1941710. doi: 10.1080/19490976.2021.1941710
15. Cogdill AP, Gaudreau PO, Arora R, Gopalakrishnan V, Wargo JA. The impact of intratumoral and gastrointestinal microbiota on systemic cancer therapy. Trends Immunol. (2018) 39:900–20. doi: 10.1016/j.it.2018.09.007
16. Wang Y, Zhang C, Hou S, Wu X, Liu J, Wan X. Analyses of potential driver and passenger bacteria in human colorectal cancer. Cancer Manag Res. (2020) 12:11553–61. doi: 10.2147/CMAR.S275316
17. Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède J-P. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci USA. (2010) 107:11537–42. doi: 10.1073/pnas.1001261107
18. Wang G, He X, Wang Q. Intratumoral bacteria are an important "accomplice" in tumor development and metastasis. Biochim Biophys Acta Rev Cancer. (2023) 1878:188846. doi: 10.1016/j.bbcan.2022.188846
19. Xia X, Wu WKK, Wong SH, Liu D, Kwong TNY, Nakatsu G, et al. Bacteria pathogens drive host colonic epithelial cell promoter hypermethylation of tumor suppressor genes in colorectal cancer. Microbiome. (2020) 8:108. doi: 10.1186/s40168-020-00847-4
20. Gutierrez-Angulo M, Ayala-Madrigal M, Moreno-Ortiz JM, Peregrina-Sandoval J, Garcia-Ayala FD. Microbiota composition and its impact on DNA methylation in colorectal cancer. Front Genet. (2023) 14:1037406. doi: 10.3389/fgene.2023.1037406
21. Kwon S-Y, Thi-Thu Ngo H, Son J, Hong Y, Min J-J. Exploiting bacteria for cancer immunotherapy. Nat Rev Clin Oncol. (2024) 21:569–89. doi: 10.1038/s41571-024-00908-9
22. Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. (2012) 10:575–82. doi: 10.1038/nrmicro2819
23. Boleij A, Muytjens CMJ, Bukhari SI, Cayet N, Glaser P, Hermans PWM, et al. Novel clues on the specific association of Streptococcus gallolyticus subsp gallolyticus with colorectal cancer. J Infect Dis. (2011) 203:1101–9. doi: 10.1093/infdis/jiq169
24. Montalban-Arques A, Katkeviciute E, Busenhart P, Bircher A, Wirbel J, Zeller G, et al. Commensal Clostridiales strains mediate effective anti-cancer immune response against solid tumors. Cell Host Microbe. (2021) 29:1573–88. doi: 10.1016/j.chom.2021.08.001
25. LaCourse KD, Zepeda-Rivera M, Kempchinsky AG, Baryiames A, Minot SS, Johnston CD, et al. The cancer chemotherapeutic 5-fluorouracil is a potent Fusobacterium nucleatum inhibitor and its activity is modified by intratumoral microbiota. Cell Rep. (2022) 41:111625. doi: 10.1016/j.celrep.2022.111625
26. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. (2017) 358:1443–8. doi: 10.1126/science.aal5240
27. Bertocchi A, Carloni S, Ravenda PS, Bertalot G, Spadoni I, Lo Cascio A, et al. Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer Cell. (2021) 39:708–24. doi: 10.1016/j.ccell.2021.03.004
28. Guo S, Chen J, Chen F, Zeng Q, Liu W, Zhang G. Exosomes derived from Fusobacterium nucleatum-infected colorectal cancer cells facilitate tumour metastasis by selectively carrying miR-1246/92b-3p/27a-3p and CXCL16. Gut. (2022) 71:e1–3. doi: 10.1136/gutjnl-2020-321187corr1
29. Xu C, Fan L, Lin Y, Shen W, Qi Y, Zhang Y, et al. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes. (2021) 13:1980347. doi: 10.1080/19490976.2021.1980347
30. Ma Y, Chen H, Li H, Zheng M, Zuo X, Wang W, et al. Intratumor microbiome-derived butyrate promotes lung cancer metastasis. Cell Rep Med. (2024) 5:101488. doi: 10.1016/j.xcrm.2024.101488
31. Lam KC, Araya RE, Huang A, Chen Q, Di Modica M, Rodrigues RR, et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. (2021) 184:5338–56. doi: 10.1016/j.cell.2021.09.019
32. Eberhardt CS, Kissick HT, Patel MR, Cardenas MA, Prokhnevska N, Obeng RC, et al. Functional HPV-specific PD-1+ stem-like CD8 T cells in head and neck cancer. Nature. (2021) 597:279–84. doi: 10.1038/s41586-021-03862-z
33. Zhu G, Su H, Johnson CH, Khan SA, Kluger H, Lu L. Intratumour microbiome associated with the infiltration of cytotoxic CD8+ T cells and patient survival in cutaneous melanoma. Eur J Cancer. (2021) 151:25–34. doi: 10.1016/j.ejca.2021.03.053
34. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. (2019) 178:795–806. doi: 10.1016/j.cell.2019.07.008
35. Rizvi ZA, Dalal R, Sadhu S, Kumar Y, Kumar S, Gupta SK, et al. High-salt diet mediates interplay between NK cells and gut microbiota to induce potent tumor immunity. Sci Adv. (2021) 7:eabg5016. doi: 10.1126/sciadv.abg5016
36. Wang H, Rong X, Zhao G, Zhou Y, Xiao Y, Ma D, et al. The microbial metabolite trimethylamine N-oxide promotes antitumor immunity in triple-negative breast cancer. Cell Metab. (2022) 34:581–94. doi: 10.1016/j.cmet.2022.02.010
37. Zhu Z, Cai J, Hou W, Xu K, Wu X, Song Y, et al. Microbiome and spatially resolved metabolomics analysis reveal the anticancer role of gut Akkermansia muciniphila by crosstalk with intratumoral microbiota and reprogramming tumoral metabolism in mice. Gut Microbes. (2023) 15:2166700. doi: 10.1080/19490976.2023.2166700
38. Clubb JHA, Kudling TV, Heiniö C, Basnet S, Pakola S, Cervera Carrascón V, et al. Adenovirus encoding tumor necrosis factor alpha and interleukin 2 induces a tertiary lymphoid structure signature in immune checkpoint inhibitor refractory head and neck cancer. Front Immunol. (2022) 13:794251. doi: 10.3389/fimmu.2022.794251
39. He T, Hao Z, Lin M, Xin Z, Chen Y, Ouyang W, et al. Oncolytic adenovirus promotes vascular normalization and nonclassical tertiary lymphoid structure formation through STING-mediated DC activation. Oncoimmunology. (2022) 11:2093054. doi: 10.1080/2162402X.2022.2093054
40. Houel A, Foloppe J, Dieu-Nosjean M-C. Harnessing the power of oncolytic virotherapy and tertiary lymphoid structures to amplify antitumor immune responses in cancer patients. Semin Immunol. (2023) 69:101796. doi: 10.1016/j.smim.2023.101796
41. Kong X, Zhang Y, Xiang L, You Y, Duan Y, Zhao Y, et al. Fusobacterium nucleatum-triggered neutrophil extracellular traps facilitate colorectal carcinoma progression. J Exp Clin Cancer Res. (2023) 42:236. doi: 10.1186/s13046-023-02817-8
42. Parhi L, Alon-Maimon T, Sol A, Nejman D, Shhadeh A, Fainsod-Levi T, et al. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun. (2020) 11:3259. doi: 10.1038/s41467-020-16967-2
43. Casasanta MA, Yoo CC, Udayasuryan B, Sanders BE, Umaña A, Zhang Y, et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci Signal. (2020) 13:eaba9157. doi: 10.1126/scisignal.aba9157
44. Peng R, Liu S, You W, Huang Y, Hu C, Gao Y, et al. Gastric microbiome alterations are associated with decreased CD8+ Tissue-resident memory T cells in the tumor microenvironment of gastric cancer. Cancer Immunol Res. (2022) 10:1224–40. doi: 10.1158/2326-6066.CIR-22-0107
45. Kabwe M, Dashper S, Bachrach G, Tucci J. Bacteriophage manipulation of the microbiome associated with tumour microenvironments-can this improve cancer therapeutic response? FEMS Microbiol Rev. (2021) 45:fuab017. doi: 10.1093/femsre/fuab017
46. Yin H, Miao Z, Wang L, Su B, Liu C, Jin Y, et al. Fusobacterium nucleatum promotes liver metastasis in colorectal cancer by regulating the hepatic immune niche and altering gut microbiota. Aging (Albany NY). (2022) 14:1941–58. doi: 10.18632/aging.203914
47. Chen B, Liu X, Yu P, Xie F, Kwan JSH, Chan WN, et al. H. pylori-induced NF-κB-PIEZO1-YAP1-CTGF axis drives gastric cancer progression and cancer-associated fibroblast-mediated tumour microenvironment remodelling. Clin Transl Med. (2023) 13:e1481. doi: 10.1002/ctm2.1481
48. Cheng M, Qian L, Shen G, Bian G, Xu T, Xu W, et al. Microbiota modulate tumoral immune surveillance in lung through a γδT17 immune cell-dependent mechanism. Cancer Res. (2014) 74:4030–41. doi: 10.1158/0008-5472.CAN-13-2462
49. Gorvel L, Olive D. Tumor associated macrophage in HPV+ tumors: Between immunosuppression and inflammation. Semin Immunol. (2023) 65:101671. doi: 10.1016/j.smim.2022.101671
50. Galliverti G, Wullschleger S, Tichet M, Murugan D, Zangger N, Horton W, et al. Myeloid cells orchestrate systemic immunosuppression, impairing the efficacy of immunotherapy against HPV+ Cancers. Cancer Immunol Res. (2020) 8:131–45. doi: 10.1158/2326-6066.CIR-19-0315
51. Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. (2019) 574:264–7. doi: 10.1038/s41586-019-1608-2
52. Mender I, Zhang A, Ren Z, Han C, Deng Y, Siteni S, et al. Telomere stress potentiates STING-dependent anti-tumor immunity. Cancer Cell. (2020) 38:400–11. doi: 10.1016/j.ccell.2020.05.020
53. Munoz-Erazo L, Rhodes JL, Marion VC, Kemp RA. Tertiary lymphoid structures in cancer - considerations for patient prognosis. Cell Mol Immunol. (2020) 17:570–5. doi: 10.1038/s41423-020-0457-0
54. Rodriguez AB, Engelhard VH. Insights into tumor-associated tertiary lymphoid structures: novel targets for antitumor immunity and cancer immunotherapy. Cancer Immunol Res. (2020) 8:1338–45. doi: 10.1158/2326-6066.CIR-20-0432
55. Cheung EC, Vousden KH. The role of ROS in tumour development and progression. Nat Rev Cancer. (2022) 22:280–97. doi: 10.1038/s41568-021-00435-0
56. Alam A, Levanduski E, Denz P, Villavicencio HS, Bhatta M, Alhorebi L, et al. Fungal mycobiome drives IL-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell. (2022) 40:153–67. doi: 10.1016/j.ccell.2022.01.003
57. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discovery. (2018) 8:403–16. doi: 10.1158/2159-8290.CD-17-1134
58. Shiao SL, Kershaw KM, Limon JJ, You S, Yoon J, Ko EY, et al. Commensal bacteria and fungi differentially regulate tumor responses to radiation therapy. Cancer Cell. (2021) 39:1202–13. doi: 10.1016/j.ccell.2021.07.002
59. Dekkerová J, Lopez-Ribot JL, Bujdáková H. Activity of anti-CR3-RP polyclonal antibody against biofilms formed by Candida auris, a multidrug-resistant emerging fungal pathogen. Eur J Clin Microbiol Infect Dis. (2019) 38:101–8. doi: 10.1007/s10096-018-3400-x
60. Nakamura H, Takada K. Reactive oxygen species in cancer: Current findings and future directions. Cancer Sci. (2021) 112:3945–52. doi: 10.1111/cas.v112.10
61. Zhu S, Al-Mathkour M, Cao L, Khalafi S, Chen Z, Poveda J, et al. CDK1 bridges NF-κB and β-catenin signaling in response to H. pylori infection in gastric tumorigenesis. Cell Rep. (2023) 42:112005. doi: 10.1016/j.celrep.2023.112005
62. Li N, Xu X, Zhan Y, Fei X, Ouyang Y, Zheng P, et al. YAP and β-catenin cooperate to drive H. pylori-induced gastric tumorigenesis. Gut Microbes. (2023) 15:2192501. doi: 10.1080/19490976.2023.2192501
63. Rubinstein MR, Baik JE, Lagana SM, Han RP, Raab WJ, Sahoo D, et al. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β-catenin modulator Annexin A1. EMBO Rep. (2019) 20:e47638. doi: 10.15252/embr.201847638
64. Li Z, Liu Y, Huang X, Wang Q, Fu R, Wen X, et al. F. Nucleatum enhances oral squamous cell carcinoma proliferation via E-cadherin/β-Catenin pathway. BMC Oral Health. (2024) 24:518. doi: 10.1186/s12903-024-04252-3
65. Chen C, Xu P. Cellular functions of cGAS-STING signaling. Trends Cell Biol. (2023) 33:630–48. doi: 10.1016/j.tcb.2022.11.001
66. Shi Y, Zheng W, Yang K, Harris KG, Ni K, Xue L, et al. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J Exp Med. (2020) 217:e20192282. doi: 10.1084/jem.20192282
67. Luo B, Zhang Y, Zhang C, Liu X, Shi C. Intestinal microbiota: A potential target for enhancing the antitumor efficacy and reducing the toxicity of immune checkpoint inhibitors. Cancer Lett. (2021) 509:53–62. doi: 10.1016/j.canlet.2021.04.001
68. Gu J, Xu X, Li X, Yue L, Zhu X, Chen Q, et al. Tumor-resident microbiota contributes to colorectal cancer liver metastasis by lactylation and immune modulation. Oncogene. (2024) 43:2389–404. doi: 10.1038/s41388-024-03080-7
69. Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. (2022) 185:1356–72. doi: 10.1016/j.cell.2022.02.027
70. Mohamed E, Sierra RA, Trillo-Tinoco J, Cao Y, Innamarato P, Payne KK, et al. The unfolded protein response mediator PERK governs myeloid cell-driven immunosuppression in tumors through inhibition of STING signaling. Immunity. (2020) 52:668–82. doi: 10.1016/j.immuni.2020.03.004
71. Andrews AM, Tennant MD, Thaxton JE. Stress relief for cancer immunotherapy: implications for the ER stress response in tumor immunity. Cancer Immunol Immunother. (2021) 70:1165–75. doi: 10.1007/s00262-020-02740-3
72. Ding Y-M, Duan M, Han Z-X, Song X-H, Zhang F-L, Wang Z, et al. Bismuth-containing quadruple therapy for helicobacter pylori eradication: A randomized clinical trial of 10 and 14 days. Dig Dis Sci. (2024) 69:2540–7. doi: 10.1007/s10620-024-08460-3
73. Zhang W, Bhagwath AS, Ramzan Z, Williams TA, Subramaniyan I, Edpuganti V, et al. Itraconazole exerts its antitumor effect in esophageal cancer by suppressing the HER2/AKT signaling pathway. Mol Cancer Ther. (2021) 20:1904–15. doi: 10.1158/1535-7163.MCT-20-0638
74. Wang G, Wang H, Ji X, Wang T, Zhang Y, Jiang W, et al. Intratumoral microbiome is associated with gastric cancer prognosis and therapy efficacy. Gut Microbes. (2024) 16:2369336. doi: 10.1080/19490976.2024.2369336
75. Perry LM, Cruz SM, Kleber KT, Judge SJ, Darrow MA, Jones LB, et al. Human soft tissue sarcomas harbor an intratumoral viral microbiome which is linked with natural killer cell infiltrate and prognosis. J Immunother Cancer. (2023) 11:e004285. doi: 10.1136/jitc-2021-004285
76. Hayashi M, Ikenaga N, Nakata K, Luo H, Zhong P, Date S, et al. Intratumor Fusobacterium nucleatum promotes the progression of pancreatic cancer via the CXCL1-CXCR2 axis. Cancer Sci. (2023) 114:3666–78. doi: 10.1111/cas.v114.9
77. Li Z, Shi C, Zheng J, Guo Y, Fan T, Zhao H, et al. Fusobacterium nucleatum predicts a high risk of metastasis for esophageal squamous cell carcinoma. BMC Microbiol. (2021) 21:301. doi: 10.1186/s12866-021-02352-6
78. Fu A, Yao B, Dong T, Cai S. Emerging roles of intratumor microbiota in cancer metastasis. Trends Cell Biol. (2023) 33:583–93. doi: 10.1016/j.tcb.2022.11.007
79. Porcari S, Baunwall SMD, Occhionero AS, Ingrosso MR, Ford AC, Hvas CL, et al. Fecal microbiota transplantation for recurrent C. difficile infection in patients with inflammatory bowel disease: A systematic review and meta-analysis. J Autoimmun. (2023) 141:103036. doi: 10.1016/j.jaut.2023.103036
80. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. (2021) 371:602–9. doi: 10.1126/science.abb5920
81. Routy B, Lenehan JG, Miller WH, Jamal R, Messaoudene M, Daisley BA, et al. Fecal microbiota transplantation plus anti-PD-1 immunotherapy in advanced melanoma: a phase I trial. Nat Med. (2023) 29:2121–32. doi: 10.1038/s41591-023-02453-x
82. Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin J-M, Morrison RM, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. (2021) 371:595–602. doi: 10.1126/science.abf3363
83. Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. (2017) 28:1368–79. doi: 10.1093/annonc/mdx108
84. Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia. (2017) 19:848–55. doi: 10.1016/j.neo.2017.08.004
85. Ji H, Jiang Z, Wei C, Ma Y, Zhao J, Wang F, et al. Intratumoural microbiota: from theory to clinical application. Cell Commun Signal. (2023) 21:164. doi: 10.1186/s12964-023-01134-z
86. Gui QF, Lu HF, Zhang CX, Xu ZR, Yang YH. Well-balanced commensal microbiota contributes to anti-cancer response in a lung cancer mouse model. Genet Mol Res. (2015) 14:5642–51. doi: 10.4238/2015.May.25.16
87. Zhang L-L, Xu J-Y, Xing Y, Wu P, Jin Y-W, Wei W, et al. Lactobacillus rhamnosus GG alleviates radiation-induced intestinal injury by modulating intestinal immunity and remodeling gut microbiota. Microbiol Res. (2024) 286:127821. doi: 10.1016/j.micres.2024.127821
88. Linn YH, Thu KK, Win NHH. Effect of probiotics for the prevention of acute radiation-induced diarrhoea among cervical cancer patients: a randomized double-blind placebo-controlled study. Probiotics Antimicrob Proteins. (2019) 11:638–47. doi: 10.1007/s12602-018-9408-9
89. Crits-Christoph A, Hallowell HA, Koutouvalis K, Suez J. Good microbes, bad genes? The dissemination of antimicrobial resistance in the human microbiome. Gut Microbes. (2022) 14:2055944. doi: 10.1080/19490976.2022.2055944
90. Bender MJ, McPherson AC, Phelps CM, Pandey SP, Laughlin CR, Shapira JH, et al. Dietary tryptophan metabolite released by intratumoral Lactobacillus reuteri facilitates immune checkpoint inhibitor treatment. Cell. (2023) 186:1846–62. doi: 10.1016/j.cell.2023.03.011
91. Vincent RL, Gurbatri CR, Li F, Vardoshvili A, Coker C, Im J, et al. Probiotic-guided CAR-T cells for solid tumor targeting. Science. (2023) 382:211–8. doi: 10.1126/science.add7034
92. Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. (2017) 551:512–6. doi: 10.1038/nature24462
93. Bessell CA, Isser A, Havel JJ, Lee S, Bell DR, Hickey JW, et al. Commensal bacteria stimulate antitumor responses via T cell cross-reactivity. JCI Insight. (2020) 5:e135597. doi: 10.1172/jci.insight.135597
94. Ragone C, Manolio C, Mauriello A, Cavalluzzo B, Buonaguro FM, Tornesello ML, et al. Molecular mimicry between tumor associated antigens and microbiota-derived epitopes. J Transl Med. (2022) 20:316. doi: 10.1186/s12967-022-03512-6
95. Fluckiger A, Daillère R, Sassi M, Sixt BS, Liu P, Loos F, et al. Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science. (2020) 369:936–42. doi: 10.1126/science.aax0701
96. Kalaora S, Nagler A, Nejman D, Alon M, Barbolin C, Barnea E, et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature. (2021) 592:138–43. doi: 10.1038/s41586-021-03368-8
97. Warso MA, Richards JM, Mehta D, Christov K, Schaeffer C, Rae Bressler L, et al. A first-in-class, first-in-human, phase I trial of p28, a non-HDM2-mediated peptide inhibitor of p53 ubiquitination in patients with advanced solid tumours. Br J Cancer. (2013) 108:1061–70. doi: 10.1038/bjc.2013.74
98. Lulla RR, Goldman S, Yamada T, Beattie CW, Bressler L, Pacini M, et al. Phase I trial of p28 (NSC745104), a non-HDM2-mediated peptide inhibitor of p53 ubiquitination in pediatric patients with recurrent or progressive central nervous system tumors: A Pediatric Brain Tumor Consortium Study. Neuro Oncol. (2016) 18:1319–25. doi: 10.1093/neuonc/now047
99. Yang Z, Zou L, Yue B, Hu M. Salmonella typhimurium may support cancer treatment: a review. Acta Biochim Biophys Sin (Shanghai). (2023) 55:331–42. doi: 10.3724/abbs.2023007
100. Chen W, Zhu Y, Zhang Z, Sun X. Advances in Salmonella Typhimurium-based drug delivery system for cancer therapy. Adv Drug Delivery Rev. (2022) 185:114295. doi: 10.1016/j.addr.2022.114295
101. Avogadri F, Mittal D, Saccheri F, Sarrafiore M, Ciocca M, Larghi P, et al. Intra-tumoral Salmonella typhimurium induces a systemic anti-tumor immune response that is directed by low-dose radiation to treat distal disease. Eur J Immunol. (2008) 38:1937–47. doi: 10.1002/eji.200738035
102. Melacarne A, Ferrari V, Tiraboschi L, Mishto M, Liepe J, Aralla M, et al. Identification of a class of non-conventional ER-stress-response-derived immunogenic peptides. Cell Rep. (2021) 36:109312. doi: 10.1016/j.celrep.2021.109312
103. Zheng JH, Min J-J. Targeted cancer therapy using engineered salmonella typhimurium. Chonnam Med J. (2016) 52:173–84. doi: 10.4068/cmj.2016.52.3.173
104. Lin Z, Meng F, Ma Y, Zhang C, Zhang Z, Yang Z, et al. In situ immunomodulation of tumors with biosynthetic bacteria promote anti-tumor immunity. Bioact Mater. (2024) 32:12–27. doi: 10.1016/j.bioactmat.2023.09.007
105. Galicia-Carmona T, Arango-Bravo E, Serrano-Olvera JA, Flores-de La Torre C, Cruz-Esquivel I, Villalobos-Valencia R, et al. ADXS11-001 LM-LLO as specific immunotherapy in cervical cancer. Hum Vaccin Immunother. (2021) 17:2617–25. doi: 10.1080/21645515.2021.1893036
106. Górska A, Przystupski D, Niemczura MJ, Kulbacka J. Probiotic bacteria: A promising tool in cancer prevention and therapy. Curr Microbiol. (2019) 76:939–49. doi: 10.1007/s00284-019-01679-8
107. Piontek A, Eichner M, Zwanziger D, Beier L-S, Protze J, Walther W, et al. Targeting claudin-overexpressing thyroid and lung cancer by modified Clostridium perfringens enterotoxin. Mol Oncol. (2020) 14:261–76. doi: 10.1002/1878-0261.12615
108. Janku F, Zhang HH, Pezeshki A, Goel S, Murthy R, Wang-Gillam A, et al. Intratumoral injection of clostridium novyi-NT spores in patients with treatment-refractory advanced solid tumors. Clin Cancer Res. (2021) 27:96–106. doi: 10.1158/1078-0432.CCR-20-2065
109. Canale FP, Basso C, Antonini G, Perotti M, Li N, Sokolovska A, et al. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature. (2021) 598:662–6. doi: 10.1038/s41586-021-04003-2
110. Chowdhury S, Castro S, Coker C, Hinchliffe TE, Arpaia N, Danino T. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat Med. (2019) 25:1057–63. doi: 10.1038/s41591-019-0498-z
111. Zhu J, Ke Y, Liu Q, Yang J, Liu F, Xu R, et al. Engineered Lactococcus lactis secreting Flt3L and OX40 ligand for in situ vaccination-based cancer immunotherapy. Nat Commun. (2022) 13:7466. doi: 10.1038/s41467-022-35130-7
112. Takahashi M, Sukowati EW, Nomura S, Kato A, Mizuseki K, Watanabe Y, et al. Impact of tumoral structure and bacterial species on growth and biodistribution of live bacterial therapeutics in xenografted tumours. J Drug Targeting. (2023) 31:194–205. doi: 10.1080/1061186X.2022.2122477
113. Hannouneh ZA, Hijazi A, Alsaleem AA, Hami S, Kheyrbek N, Tanous F, et al. Novel immunotherapeutic options for BCG-unresponsive high-risk non-muscle-invasive bladder cancer. Cancer Med. (2023) 12:21944–68. doi: 10.1002/cam4.v12.24
114. Alanazi FK, Alsuwyeh AA, Haq N, Salem-Bekhit MM, Al-Dhfyan A, Shakeel F. Vision of bacterial ghosts as drug carriers mandates accepting the effect of cell membrane on drug loading. Drug Dev Ind Pharm. (2020) 46:1716–25. doi: 10.1080/03639045.2020.1820039
115. Michalek J, Hezova R, Turanek-Knötigova P, Gabkova J, Strioga M, Lubitz W, et al. Oncolysate-loaded Escherichia coli bacterial ghosts enhance the stimulatory capacity of human dendritic cells. Cancer Immunol Immunother. (2017) 66:149–59. doi: 10.1007/s00262-016-1932-4
116. Liu X, Sun M, Pu F, Ren J, Qu X. Transforming intratumor bacteria into immunopotentiators to reverse cold tumors for enhanced immuno-chemodynamic therapy of triple-negative breast cancer. J Am Chem Soc. (2023) 145:26296–307. doi: 10.1021/jacs.3c09472
117. Bang YJ, Ruiz EY, Van Cutsem E, Lee KW, Wyrwicz L, Schenker M, et al. randomised trial of avelumab versus physician's choice of chemotherapy as third-line treatment of patients with advanced gastric or gastro-oesophageal junction cancer: primary analysis of JAVELIN Gastric 300. Ann Oncol. (2018) 29:2052–60. doi: 10.1093/annonc/mdy264
118. Shitara K, Özgüroğlu M, Bang Y-J, Di Bartolomeo M, Mandalà M, Ryu M-H, et al. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): a randomised, open-label, controlled, phase 3 trial. Lancet. (2018) 392:123–33. doi: 10.1016/S0140-6736(18)31257-1
119. Huang J, Jiang Z, Wang Y, Fan X, Cai J, Yao X, et al. Modulation of gut microbiota to overcome resistance to immune checkpoint blockade in cancer immunotherapy. Curr Opin Pharmacol. (2020) 54:1–10. doi: 10.1016/j.coph.2020.06.004
120. Zhang J, Haines C, Watson AJM, Hart AR, Platt MJ, Pardoll DM, et al. Oral antibiotic use and risk of colorectal cancer in the United Kingdom, 1989-2012: a matched case-control study. Gut. (2019) 68:1971–8. doi: 10.1136/gutjnl-2019-318593
121. Dong X, Pan P, Zhang Q, Ye J-J, Zhang X-Z. Engineered living bacteriophage-enabled self-adjuvanting hydrogel for remodeling tumor microenvironment and cancer therapy. Nano Lett. (2023) 23:1219–28. doi: 10.1021/acs.nanolett.2c04279
122. Kabwe M, Meehan-Andrews T, Ku H, Petrovski S, Batinovic S, Chan HT, et al. Lytic bacteriophage EFA1 modulates HCT116 colon cancer cell growth and upregulates ROS production in an enterococcus faecalis co-culture system. Front Microbiol. (2021) 12:650849. doi: 10.3389/fmicb.2021.650849
123. Sousa C, Ferreira R, Azevedo NF, Oleastro M, Azeredo J, Figueiredo C, et al. Helicobacter pylori infection: from standard to alternative treatment strategies. Crit Rev Microbiol. (2022) 48:376–96. doi: 10.1080/1040841X.2021.1975643
124. Zheng D-W, Dong X, Pan P, Chen K-W, Fan J-X, Cheng S-X, et al. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat BioMed Eng. (2019) 3:717–28. doi: 10.1038/s41551-019-0423-2
125. Hashemi Goradel N, Heidarzadeh S, Jahangiri S, Farhood B, Mortezaee K, Khanlarkhani N, et al. Fusobacterium nucleatum and colorectal cancer: A mechanistic overview. J Cell Physiol. (2019) 234:2337–44. doi: 10.1002/jcp.v234.3
126. Dong X, Pan P, Zheng D-W, Bao P, Zeng X, Zhang X-Z. Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor-immune microenvironment against colorectal cancer. Sci Adv. (2020) 6:eaba1590. doi: 10.1126/sciadv.aba1590
127. Mahler M, Costa AR, van Beljouw SPB, Fineran PC, Brouns SJJ. Approaches for bacteriophage genome engineering. Trends Biotechnol. (2023) 41:669–85. doi: 10.1016/j.tibtech.2022.08.008
128. Bar H, Yacoby I, Benhar I. Killing cancer cells by targeted drug-carrying phage nanomedicines. BMC Biotechnol. (2008) 8:37. doi: 10.1186/1472-6750-8-37
129. Kannen V, Parry L, Martin FL. Phages enter the fight against colorectal cancer. Trends Cancer. (2019) 5:577–9. doi: 10.1016/j.trecan.2019.08.002
130. Malik DJ, Sokolov IJ, Vinner GK, Mancuso F, Cinquerrui S, Vladisavljevic GT, et al. Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv Colloid Interface Sci. (2017) 249:100–33. doi: 10.1016/j.cis.2017.05.014
131. Vandenheuvel D, Lavigne R, Brüssow H. Bacteriophage therapy: advances in formulation strategies and human clinical trials. Annu Rev Virol. (2015) 2:599–618. doi: 10.1146/annurev-virology-100114-054915
132. Liu N, Lewis C, Zheng W, Fu ZQ. Phage cocktail therapy: multiple ways to suppress pathogenicity. Trends Plant Sci. (2020) 25:315–7. doi: 10.1016/j.tplants.2020.01.013
133. Gencay YE, Jasinskytė D, Robert C, Semsey S, Martínez V, Petersen AØ, et al. Engineered phage with antibacterial CRISPR-Cas selectively reduce E. coli burden in mice. Nat Biotechnol. (2024) 42:265–74. doi: 10.1038/s41587-023-01759-y
134. Ramelyte E, Tastanova A, Balázs Z, Ignatova D, Turko P, Menzel U, et al. Oncolytic virotherapy-mediated anti-tumor response: a single-cell perspective. Cancer Cell. (2021) 39:394–406. doi: 10.1016/j.ccell.2020.12.022
135. Wang G, Zhang Z, Zhong K, Wang Z, Yang N, Tang X, et al. CXCL11-armed oncolytic adenoviruses enhance CAR-T cell therapeutic efficacy and reprogram tumor microenvironment in glioblastoma. Mol Ther. (2023) 31:134–53. doi: 10.1016/j.ymthe.2022.08.021
136. Gállego Pérez-Larraya J, Garcia-Moure M, Labiano S, Patiño-García A, Dobbs J, Gonzalez-Huarriz M, et al. Oncolytic DNX-2401 virus for pediatric diffuse intrinsic pontine glioma. N Engl J Med. (2022) 386:2471–81. doi: 10.1056/NEJMoa2202028
137. Sasso E, D'Alise AM, Zambrano N, Scarselli E, Folgori A, Nicosia A. New viral vectors for infectious diseases and cancer. Semin Immunol. (2020) 50:101430. doi: 10.1016/j.smim.2020.101430
138. Duan Q, Zhang H, Zheng J, Zhang L. Turning Cold into Hot: Firing up the Tumor Microenvironment. Trends Cancer. (2020) 6:605–18. doi: 10.1016/j.trecan.2020.02.022
139. Ma R, Li Z, Chiocca EA, Caligiuri MA, Yu J. The emerging field of oncolytic virus-based cancer immunotherapy. Trends Cancer. (2023) 9:122–39. doi: 10.1016/j.trecan.2022.10.003
140. Bommareddy PK, Shettigar M, Kaufman HL. Integrating oncolytic viruses in combination cancer immunotherapy. Nat Rev Immunol. (2018) 18:498–513. doi: 10.1038/s41577-018-0014-6
141. Peters C, Rabkin SD. Designing herpes viruses as oncolytics. Mol Ther Oncolytics. (2015) 2:15010–. doi: 10.1038/mto.2015.10
142. Mondal M, Guo J, He P, Zhou D. Recent advances of oncolytic virus in cancer therapy. Hum Vaccin Immunother. (2020) 16:2389–402. doi: 10.1080/21645515.2020.1723363
143. Greig SL. Talimogene laherparepvec: first global approval. Drugs. (2016) 76:147–54. doi: 10.1007/s40265-015-0522-7
144. Soliman H, Hogue D, Han H, Mooney B, Costa R, Lee MC, et al. Oncolytic T-VEC virotherapy plus neoadjuvant chemotherapy in nonmetastatic triple-negative breast cancer: a phase 2 trial. Nat Med. (2023) 29:450–7. doi: 10.1038/s41591-023-02309-4
145. Andtbacka RHI, Kaufman HL, Collichio F, Amatruda T, Senzer N, Chesney J, et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. (2015) 33:2780–8. doi: 10.1200/JCO.2014.58.3377
146. Ling AL, Solomon IH, Landivar AM, Nakashima H, Woods JK, Santos A, et al. Clinical trial links oncolytic immunoactivation to survival in glioblastoma. Nature. (2023) 623:157–66. doi: 10.1038/s41586-023-06623-2
147. Todo T, Ito H, Ino Y, Ohtsu H, Ota Y, Shibahara J, et al. Intratumoral oncolytic herpes virus G47∆ for residual or recurrent glioblastoma: a phase 2 trial. Nat Med. (2022) 28:1630–9. doi: 10.1038/s41591-022-01897-x
148. Fares J, Ahmed AU, Ulasov IV, Sonabend AM, Miska J, Lee-Chang C, et al. Neural stem cell delivery of an oncolytic adenovirus in newly diagnosed Malignant glioma: a first-in-human, phase 1, dose-escalation trial. Lancet Oncol. (2021) 22:1103–14. doi: 10.1016/S1470-2045(21)00245-X
149. Wang G, Kang X, Chen KS, Jehng T, Jones L, Chen J, et al. An engineered oncolytic virus expressing PD-L1 inhibitors activates tumor neoantigen-specific T cell responses. Nat Commun. (2020) 11:1395. doi: 10.1038/s41467-020-15229-5
150. Anker SC, Szczeponik MG, Dessila J, Dittus K, Engeland CE, Jäger D, et al. Oncolytic measles virus encoding microRNA for targeted RNA interference. Viruses. (2023) 15:308. doi: 10.3390/v15020308
151. Huang H, Liu Y, Liao W, Cao Y, Liu Q, Guo Y, et al. Oncolytic adenovirus programmed by synthetic gene circuit for cancer immunotherapy. Nat Commun. (2019) 10:4801. doi: 10.1038/s41467-019-12794-2
152. Malard F, Vekhoff A, Lapusan S, Isnard F, D'Incan-Corda E, Rey J, et al. Gut microbiota diversity after autologous fecal microbiota transfer in acute myeloid leukemia patients. Nat Commun. (2021) 12:3084. doi: 10.1038/s41467-021-23376-6
153. Horvath A, Bausys A, Sabaliauskaite R, Stratilatovas E, Jarmalaite S, Schuetz B, et al. Distal gastrectomy with billroth II reconstruction is associated with oralization of gut microbiome and intestinal inflammation: A proof-of-concept study. Ann Surg Oncol. (2021) 28:1198–208. doi: 10.1245/s10434-020-08678-1
154. Roussel E, Brasse-Lagnel C, Tuech J-J, Montialoux H, Papet E, Tortajada P, et al. Influence of probiotics administration before liver resection in patients with liver disease: A randomized controlled trial. World J Surg. (2022) 46:656–65. doi: 10.1007/s00268-021-06388-7
155. Hibberd AA, Lyra A, Ouwehand AC, Rolny P, Lindegren H, Cedgård L, et al. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. (2017) 4:e000145. doi: 10.1136/bmjgast-2017-000145
156. Yu J, Zhu P, Shi L, Gao N, Li Y, Shu C, et al. Bifidobacterium longum promotes postoperative liver function recovery in patients with hepatocellular carcinoma. Cell Host Microbe. (2024) 32:131–44. doi: 10.1016/j.chom.2023.11.011
157. Basu P, Mehta A, Jain M, Gupta S, Nagarkar RV, John S, et al. A randomized phase 2 study of ADXS11-001 listeria monocytogenes-listeriolysin O immunotherapy with or without cisplatin in treatment of advanced cervical cancer. Int J Gynecol Cancer. (2018) 28:764–72. doi: 10.1097/IGC.0000000000001235
158. Heo J, Reid T, Ruo L, Breitbach CJ, Rose S, Bloomston M, et al. Randomized dose-finding clinical trial of oncolytic immunotherapeutic vaccinia JX-594 in liver cancer. Nat Med. (2013) 19:329–36. doi: 10.1038/nm.3089
159. Hwang T-H, Moon A, Burke J, Ribas A, Stephenson J, Breitbach CJ, et al. A mechanistic proof-of-concept clinical trial with JX-594, a targeted multi-mechanistic oncolytic poxvirus, in patients with metastatic melanoma. Mol Ther. (2011) 19:1913–22. doi: 10.1038/mt.2011.132
160. Park SH, Breitbach CJ, Lee J, Park JO, Lim HY, Kang WK, et al. Phase 1b trial of biweekly intravenous pexa-vec (JX-594), an oncolytic and immunotherapeutic vaccinia virus in colorectal cancer. Mol Ther. (2015) 23:1532–40. doi: 10.1038/mt.2015.109
161. Park B-H, Hwang T, Liu T-C, Sze DY, Kim J-S, Kwon H-C, et al. Use of a targeted oncolytic poxvirus, JX-594, in patients with refractory primary or metastatic liver cancer: a phase I trial. Lancet Oncol. (2008) 9:533–42. doi: 10.1016/S1470-2045(08)70107-4
162. Friedman GK, Johnston JM, Bag AK, Bernstock JD, Li R, Aban I, et al. Oncolytic HSV-1 G207 immunovirotherapy for pediatric high-grade gliomas. N Engl J Med. (2021) 384:1613–22. doi: 10.1056/NEJMoa2024947
163. Todo T, Ino Y, Ohtsu H, Shibahara J, Tanaka M. A phase I/II study of triple-mutated oncolytic herpes virus G47∆ in patients with progressive glioblastoma. Nat Commun. (2022) 13:4119. doi: 10.1038/s41467-022-31262-y
164. Garcia-Carbonero R, Salazar R, Duran I, Osman-Garcia I, Paz-Ares L, Bozada JM, et al. Phase 1 study of intravenous administration of the chimeric adenovirus enadenotucirev in patients undergoing primary tumor resection. J Immunother Cancer. (2017) 5:71. doi: 10.1186/s40425-017-0277-7
165. Machiels J-P, Salazar R, Rottey S, Duran I, Dirix L, Geboes K, et al. A phase 1 dose escalation study of the oncolytic adenovirus enadenotucirev, administered intravenously to patients with epithelial solid tumors (EVOLVE). J Immunother Cancer. (2019) 7:20. doi: 10.1186/s40425-019-0510-7
166. Andtbacka RHI, Curti B, Daniels GA, Hallmeyer S, Whitman ED, Lutzky J, et al. Clinical responses of oncolytic coxsackievirus A21 (V937) in patients with unresectable melanoma. J Clin Oncol. (2021) 39:3829–38. doi: 10.1200/JCO.20.03246
167. Rohaan MW, Stahlie EHA, Franke V, Zijlker LP, Wilgenhof S, van der Noort V, et al. Neoadjuvant nivolumab + T-VEC combination therapy for resectable early stage or metastatic (IIIB-IVM1a) melanoma with injectable disease: study protocol of the NIVEC trial. BMC Cancer. (2022) 22:851. doi: 10.1186/s12885-022-09896-4
168. Chesney JA, Ribas A, Long GV, Kirkwood JM, Dummer R, Puzanov I, et al. Randomized, double-blind, placebo-controlled, global phase III trial of talimogene laherparepvec combined with pembrolizumab for advanced melanoma. J Clin Oncol. (2023) 41:528–40. doi: 10.1200/JCO.22.00343
169. Nassiri F, Patil V, Yefet LS, Singh O, Liu J, Dang RMA, et al. Oncolytic DNX-2401 virotherapy plus pembrolizumab in recurrent glioblastoma: a phase 1/2 trial. Nat Med. (2023) 29:1370–8. doi: 10.1038/s41591-023-02347-y
170. Silk AW, O'Day SJ, Kaufman HL, Bryan J, Norrell JT, Imbergamo C, et al. A phase 1b single-arm trial of intratumoral oncolytic virus V937 in combination with pembrolizumab in patients with advanced melanoma: results from the CAPRA study. Cancer Immunol Immunother. (2023) 72:1405–15. doi: 10.1007/s00262-022-03314-1
171. Rudin CM, Pandha HS, Zibelman M, Akerley WL, Harrington KJ, Day D, et al. Phase 1, open-label, dose-escalation study on the safety, pharmacokinetics, and preliminary efficacy of intravenous Coxsackievirus A21 (V937), with or without pembrolizumab, in patients with advanced solid tumors. J Immunother Cancer. (2023) 11:e005007. doi: 10.1136/jitc-2022-005007
172. Curti BD, Richards J, Hyngstrom JR, Daniels GA, Faries M, Feun L, et al. Intratumoral oncolytic virus V937 plus ipilimumab in patients with advanced melanoma: the phase 1b MITCI study. J Immunother Cancer. (2022) 10:e005224. doi: 10.1136/jitc-2022-005224
173. Lutzky J, Sullivan RJ, Cohen JV, Ren Y, Li A, Haq R. Phase 1b study of intravenous coxsackievirus A21 (V937) and ipilimumab for patients with metastatic uveal melanoma. J Cancer Res Clin Oncol. (2023) 149:6059–66. doi: 10.1007/s00432-022-04510-3
174. Fakih M, Harb W, Mahadevan D, Babiker H, Berlin J, Lillie T, et al. Safety and efficacy of the tumor-selective adenovirus enadenotucirev, in combination with nivolumab, in patients with advanced/metastatic epithelial cancer: a phase I clinical trial (SPICE). J Immunother Cancer. (2023) 11:e006561. doi: 10.1136/jitc-2022-006561
Keywords: intratumoral microbiota (IM), tumor microenvironment (TME), fecal microbiome transplantation (FMT), engineered bacteria, bacteriophage, oncolytic virus
Citation: Zheng J and Chen H (2024) Effects of intratumoral microbiota on tumorigenesis, anti-tumor immunity, and microbe-based cancer therapy. Front. Oncol. 14:1429722. doi: 10.3389/fonc.2024.1429722
Received: 08 May 2024; Accepted: 26 August 2024;
Published: 26 September 2024.
Edited by:
Khashayarsha Khazaie, Mayo Clinic Arizona, United StatesReviewed by:
Abdullah Saeed, City of Hope National Medical Center, United StatesAmit Kumar, Virginia Commonwealth University, United States
Copyright © 2024 Zheng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao Chen, Y2g3NmNoQDE2My5jb20=
 Jingwei Zheng
Jingwei Zheng Hao Chen
Hao Chen