- 1Department of Oncology, Affiliated Hospital of Youjiang Medical University for Nationalities, Baise, China
- 2Faculty of Medicine, MAHSA University, Jenjarom, Selangor, Malaysia
- 3Department of Preclinical Sciences, Faculty of Dentistry, MAHSA University, Jenjarom, Selangor, Malaysia
- 4Department of Metabolism, Digestion and Reproduction, Imperial College London, Chelsea and Westminster Hospital, London, United Kingdom
- 5Life Science and Clinical Research Center, Youjiang Medical University for Nationalities, Baise, China
Breast cancer (BC) is associated with malignant tumors in women worldwide with persistently high incidence and mortality rates. The traditional therapies including surgery, chemotherapy, radiotherapy and targeted therapy have certain therapeutic effects on BC patients, but acquired drug resistance can lead to tumor recurrence and metastasis. This remains a clinical challenge that is difficult to solve during treatment. Therefore, continued research is needed to identify effective targets and treatment methods, to ultimately implement personalized treatment strategies. Several studies have implicated that the long non-coding RNA LINC00511 is closely linked to the occurrence, development and drug resistance of BC. Here we will review the structure and the mechanisms of action of lnc RNA LINC00511 in various cancers, and then explore its expression and its related regulatory mechanisms during BC. In addition, we will discuss the biological functions and the potential clinical applications of LINC00511 in BC.
1 Introduction
Although governments and health organizations around the world have taken a series of measures and made great efforts with regards to cancer prevention and treatment, the incidence and mortality rates for breast cancer (BC) have not been effectively controlled. These continue to show an increasing trend (1). Cancer data statistics show that in 2020, there were about 2.3 million newly diagnosed cases of BC. As a result, BC has surpassed lung cancer (LC) for the first time, and has become the leading cancer among women (2). Currently, BC remains one of the main causes of death in women worldwide, posing a serious threat to the health and lives (3). There are currently several ways to classify BC. Based on genetic and epigenetic characteristics, breast cancer cell lines can be classified as luminal A, luminal B, HER2 positive, triple negative A and triple negative B (4) Clinically, BC can be classified into estrogen receptor (ER)-positive and ER-negative as well as HER2 (human epidermal growth factor receptor 2)-positive and HER2-negative subtypes. This classification is based on the levels of ER, progesterone receptor (PR) and HER2 present (5, 6).
Although medical professionals have conducted extensive and long-term research on the diagnosis and treatment of BC, as yet no significant breakthroughs have been achieved. Therefore, the therapeutic effects for BC patients have not improved significantly, and the survival times have not been significantly extended. To date, the conventional treatment methods for BC are primarily surgery and targeted therapies using drugs, radiography, hormones and antibodies (7). Although the emerging targeted therapies have brought hope to BC patients in recent years, cellular drug resistance leading to tumor recurrence and metastasis has posed a severe challenge to its treatment. Therefore, continued research to identify effective targets and treatment methods are in demand from BC patients as well as health professionals. In this context, the role of long non-coding RNAs (lncRNAs) in cancers has been the subject of intense research from scholars, worldwide.
LncRNAs are transcripts that contain more than 200 nucleotides. Due to their lack of long open reading frames, these molecules were previously assumed to have no protein-coding ability. However, with the continuous development of medical testing techniques, there is now a substantial body of evidence to show that some lncRNAs have these abilities (8–11). The discovery that lncRNAs can encode proteins has drawn widespread attention to their biological roles and have made them a research hotspot in recent years.
Studies have shown that lncRNAs can regulate the expression and function of various genes through multiple mechanisms. These include regulation of chromatin remodeling, splicing, mRNA transcription, DNA methylation, mRNA stability and translation and post-transcriptional regulation (12–17). In addition, mature lncRNAs can also bind to RNA/DNA binding proteins, transcription factors (TFs), chromatin modification complexes, RNA transcripts, mature mRNAs, microRNAs, DNA and chromatin) to form supramolecular structures (18). This can lead to regulation of the expression and function of target genes. Numerous studies implicate lncRNAs in the regulation of essential biological and cellular processes. These include proliferation, differentiation, invasion, migration, angiogenesis, stemness, epithelial-mesenchymal transition, cell apoptosis, immune responses and tumor treatment resistance in malignant tumors (19–21).
Therefore, dysregulation of either their expression or function can lead to pathological and physiological changes in the human body. These can result in the development of abnormalities and malignancy resulting in diseases. In recent years, the functional roles of lncRNAs in malignant tumors have gradually been revealed. Several studies have shown that lncRNAs are abnormally expressed in malignant tumors such as renal cell carcinoma, gastric cancer, liver cancer, non-small cell LC, colorectal cancer, glioblastoma, osteosarcoma and ovarian cancer (22–31). They have been shown to be implicated in the regulation and development of cancers.
Long intergenic non-coding RNAs (lincRNAs) can be considered to be lncRNAs as they share similarities of structure and function. Although lincRNAs do not participate in encoding specific proteins, they can act as regulatory factors to regulate the expression of target genes and thus play roles in various cellular and biological processes (32). Existing studies have shown that dysregulation of lincRNA expression and function can also lead to the occurrence and progression of tumors (33–36).
LINC00511 is a newly discovered lncRNA and it is known to be associated with LC (37, 38). It has also been reported to be abnormally expressed in many types of malignant tumors where it can accelerate tumor progression. It inhibits malignant cell apoptosis and promotes the proliferation, migration, invasion, metastasis and chemotherapy resistance of tumor cells (39–41). Therefore, LINC00511 is a potential cancer biomarker and a promising therapeutic target. Currently, there have been multiple reports on the link between LINC00511 and BC, but the existing research directions are scattered and there is a lack systematic summarization. Therefore, we reviewed the expression, structural characteristics, mechanisms and functional roles of LINC00511 in BC with a view to clarifying its clinical significance and therapeutic potential.
2 Structural characteristics of LINC00511
Cabanski et al. first reported LINC00511, which is also known as onco-LncRNA-12, in 2015 after a pan-cancer transcriptome analysis (42). It is located on the negative strand of 17q24.3 region, 72,290,091-72,640,472 (GRCh38/hg38), and it has been allocated the transcript number, ENST00000453722.6 (Figure 1A). It is 1716nt in length and consists of 5 exons (43). Studies have identified 107 alternative splice variants of LINC00511. Except for the two splicing variants, LINC00511-279 and LINC00511-278, which retain the introns, the others belong to the lncRNA transcriptome (https://asia.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000227036;r=17:72290091-72640472). The NIH Genotype-Tissue Expression (GTEx) project (https://commonfund.nih.gov/GTEx, accessed on April 9, 2024) suggested that LINC00511 is widely expressed in normal human tissues (Figure 1B). However, its expression is highest in the skin, vagina and esophagus, suggesting that its presence in these organs has biological significance.
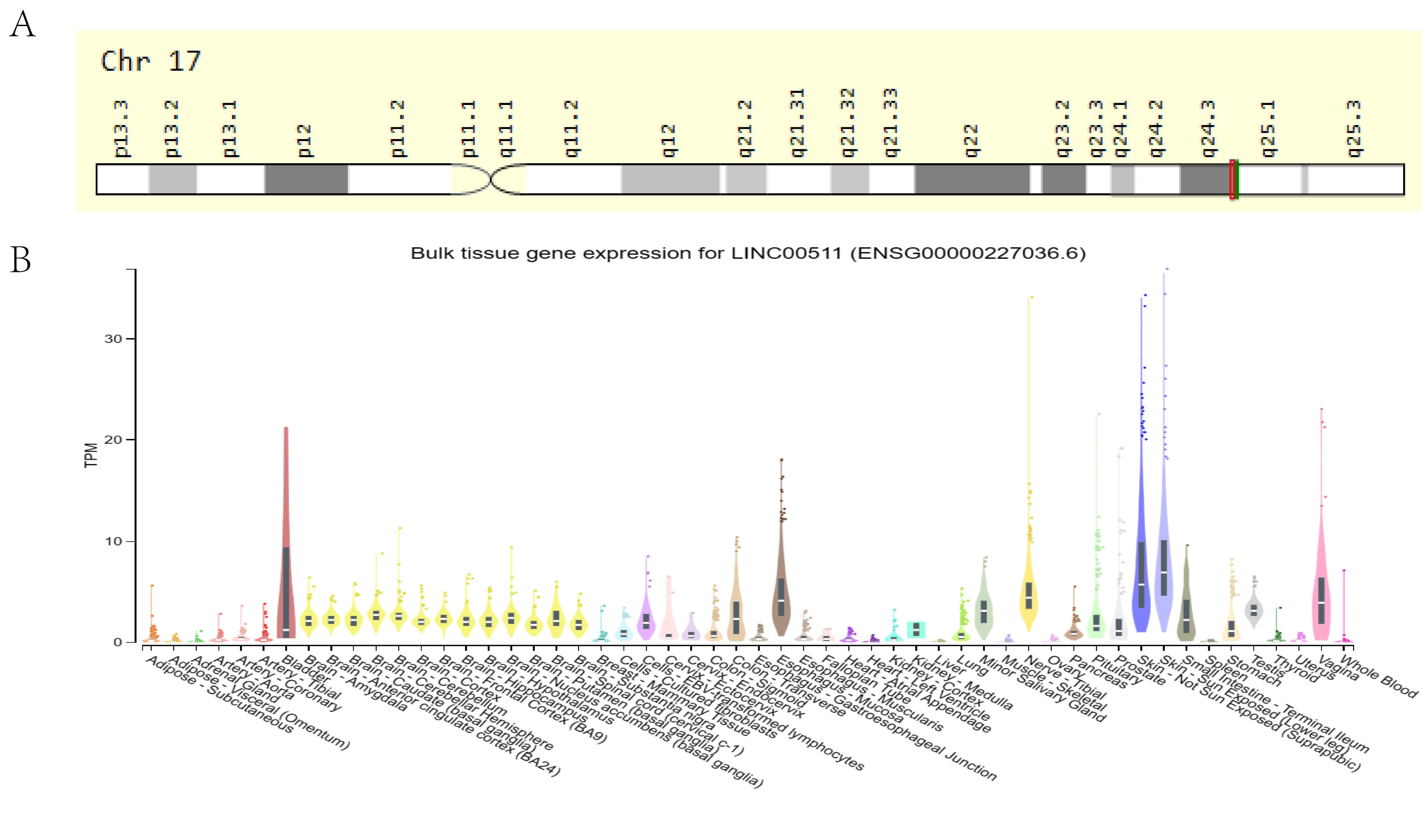
Figure 1. (A) Localization of LINC00511 in the human chromosome. (B) The expression of LINC00511 in normal human tissues from information obtained from the GTEx database.
3 Functional roles of LINC00511
Studies have shown that the biochemical functions and regulatory mechanisms of lncRNAs are closely related to their genomic locations and subcellular localizations (44). For example, lncRNAs that are located in the cell nuclei are mainly associated with regulation in epigenetic modifications and transcriptional processes. However, cytoplasmic lncRNAs typically exert their regulatory effects through post-transcriptional mechanisms such as the regulation of mRNA stability, protein translation and competition with endogenous RNA (ceRNA) networks (9, 45, 46) (Figure 2).
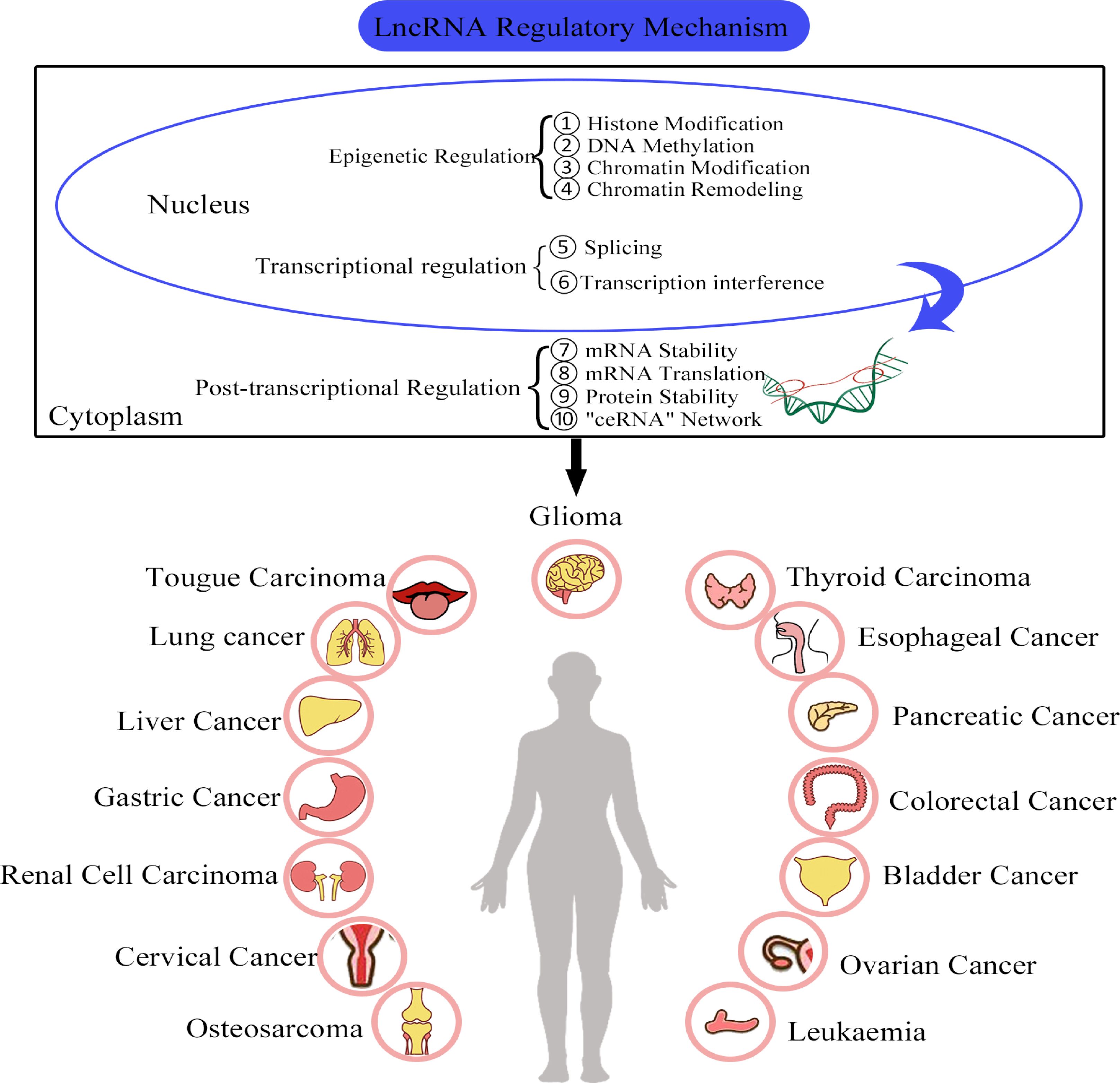
Figure 2. The functional roles and regulatory mechanisms of lncRNAs in various cancers. ① LncRNAs are involved in the regulation of histone modification. ② LncRNAs are involved in DNA methylation. ③ LncRNAs can regulate chromatin modification. ④ LncRNAs can bind to chromatin re-modelling factors and regulate gene expression. ⑤ LncRNAs can interact with splicing factors. ⑥ LncRNAs can cause transcription factors to interfere with gene transcription. ⑦ LncRNAs can regulate RNA stability. ⑧ LncRNAs can participate in the regulation of RNA translation. ⑨ LncRNAs are involved in the regulation protein stability. ⑩ LncRNAs can act as miRNA sponges.
With respect to LINC00511, studies have shown that its intracellular localization varies in different cancers. Wu et al. used subcellular fractionation and FISH assays and they localized LINC00511 primarily in the cytoplasm in lung squamous cell carcinoma (46). However, in ovarian cancer it was localized in the nuclei of ovarian cancer cells (47). Zhao et al. found that LINC00511 was abundant in both the cytoplasm and nuclei of pancreatic cancer cells by using FISH (48). The widespread subcellular localization of LINC00511 differs in different types of tumor cells and this is likely to reflect on its biological functions and regulatory mechanisms (Table 1).

Table 1. The functions and regulatory mechanisms associated with LINC00511 in various malignant tumors.
Although it has a relatively complex role in malignant tumors, the regulatory effects LINC00511 can be exerted in three main ways. Firstly, LINC00511 can act as a ceRNA, where it regulates the expression of target genes by interacting with microRNAs (miRNAs) and thereby affecting malignant tumor cells. Secondly, LINC00511 can regulate its own or target gene expression via epigenetic modifications thereby promoting tumor occurrence. Thirdly, LINC00511 can exert effects at the transcriptional level, such as the regulation of target genes and related signaling pathways, thereby influencing tumor progression.
4 Expression of LINC00511 in BC
LINC00511 is known to be dysregulated in various human malignant tumors. In addition, its expression levels is known to be associated with the age patients and their tumor size, stage and subtype as well as their lymph node status. These factors can affect the diagnosis and prognosis of tumor patients, which suggests that it has a role as a potential molecular biomarker (90). By using the publicly available online databases, GEPIA (91), UALCAN (92) and ENCORI (93), LINC00511 was found to be upregulated in BC tissues. The expression levels of this lncRNA were found to be closely correlated to the clinical stage, subtype and subclass of BC (Figure 3).
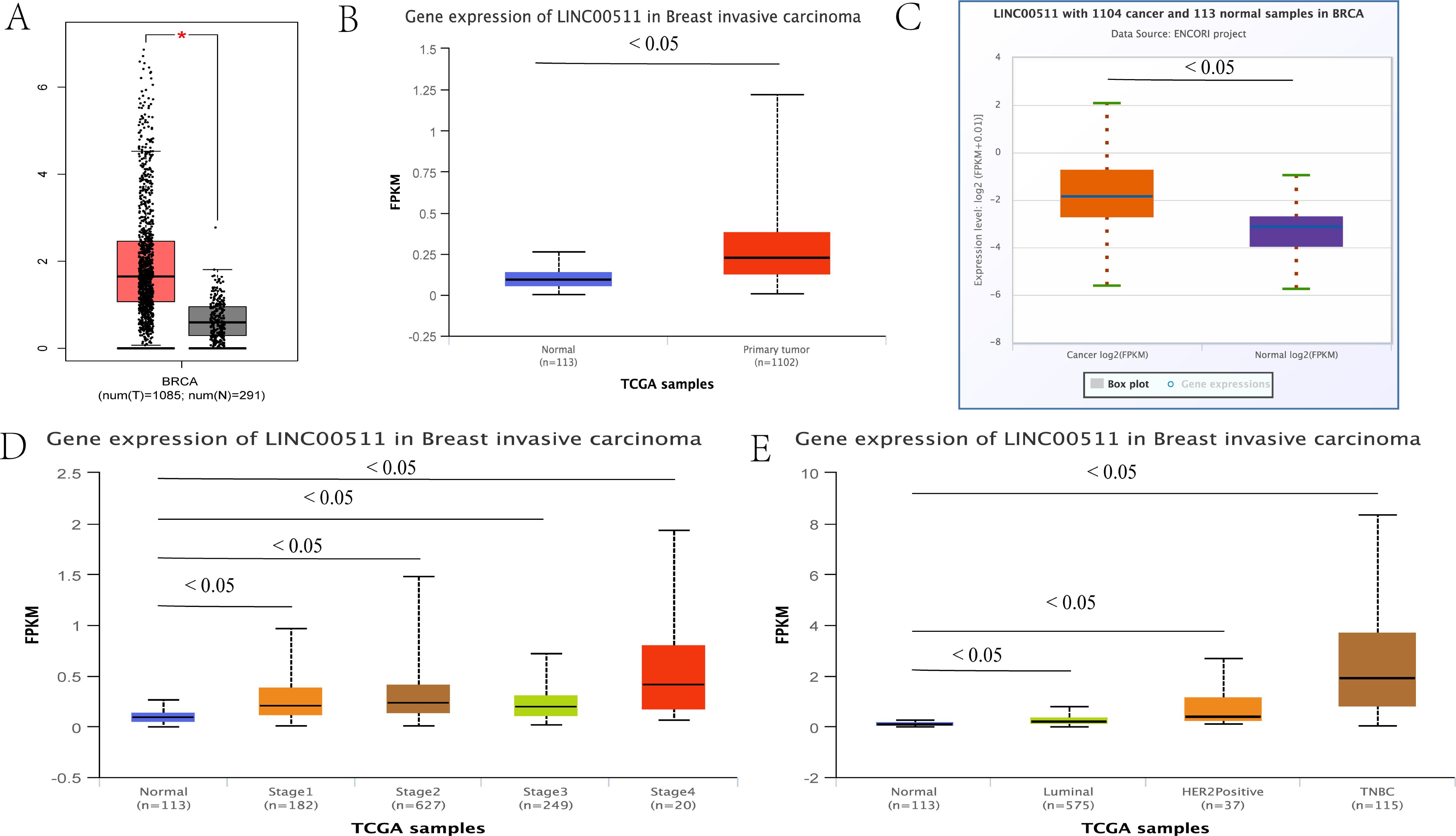
Figure 3. The relationship between the expression levels of LINC00511 in breast cancer and tumor stage and the breast cancer subtype. (A) GEPIA, (B) UALCAN, (C) ENCORI and other online websites were used to show the LINC00511 expression levels in breast cancer. (D) The UALCAN online website showed that LINC00511 expression levels were closely related to tumor stage of cancer progression.(E) The UALCAN online website showed that LINC00511 expression levels were closely related to the breast cancer subtype.
A subsequent study of BC tissue samples confirmed the results obtained from the above online databases. Lu et al. measured LINC00511 expression levels in BC and non-cancerous tissues by qRT-PCR and found that LINC00511 was significantly upregulated in the cancer samples (94). This was confirmed by Tan et al. These authors showed that LINC00511 expression was closely associated with tumor size, clinical stage and lymph node status by using in vitro cell cultures (95). Interestingly, another study using serum samples from BC patients also showed that LINC00511 was overexpressed in BC patients where it was found that this lncRNA was significantly associated with the patients’ PR status, ER status, HER2 status, tumor stage, tumor size as well as lymph node status (43).
In summary, several lines of evidence obtained from bioinformatics, blood, tissue and cell experiments, would suggest that there is an overexpression of LINC00511 in BC. Therefore, further research on the regulatory mechanisms of LINC00511 dysregulation in BC would be beneficial for revealing the molecular nature this cancer and this exercise may point to potential new therapeutic targets to combat this disease.
5 Related regulatory mechanisms of LINC00511 dysregulation in BC
Currently, the specific regulatory mechanisms underlying the upregulation of LINC00511 expression in BC remain unclear. Studies have shown that the abnormal levels of LINC00511 may affect regulatory mechanisms such as epigenetic modifications. DNA methylation is an epigenetic mechanism that has been proven to play a crucial role in the occurrence and development of BC (96). Liu et al. also confirmed that DNA hypo-methylation in the CpG island region of the gene promoter region was positively and significantly correlated with the levels of LINC00511 expression (97).
The estrogen-signaling pathway is another factor that can affect LINC00511 expression. Zhang et al. explored the relationship between LINC00511 and ER-negative status in a study on ER-negative BC and found that silencing ER expression in cancer cells could induce its expression. Surprisingly, after stimulating BC cells with tamoxifen, an anti-estrogen drug, LINC00511 expression was significantly upregulated. Further studies revealed that estrogen deficiency could directly activate the TF, AP-2 (TFAP-2), thereby upregulating LINC00511 expression in BC cells (98). Currently, only DNA methylation and the estrogen-signaling pathway have been reported to be associated with the regulation of LINC00511 expression in BC. Further research is needed to elucidate the precise mechanisms of action of LINC00511 in BC.
6 Molecular regulatory mechanisms of LINC00511
6.1 CeRNA networks
LncRNAs often exert their biological functions as ceRNAs. LINC00511 is a typical ceRNA molecule that is capable of base pairing in a complementary fashion with various miRNAs, by competitively binding to them. This prevents the miRNAs from binding to and degrading target genes, thereby regulating target gene expression. Among them, the LINC00511/miR-185-3p/E2F1 axis is a classic ceRNA interaction network. Lu et al. showed that LINC00511 can bind to miR-185-3p through molecular complementarity. This prevents miR-185-3p from binding to its target gene, E2F1, leading to upregulation of E2F1 protein expression and activation of the expression of the downstream TF, Nanog. These interactions promotes the stem-like state and malignant phenotype of BC cells (94). Additionally, LINC00511 can also promote tumor cell proliferation by alleviating the inhibitory effect of miR-150 on matrix metalloproteinase 13 (MMP13) (99). Such typical ceRNA networks can play important roles in the oncogenic functions of LINC00511.
6.2 Involvement in transcriptional regulation
Numerous studies have shown that LINC00511 may either directly or indirectly affect the transcription and expression of downstream genes through interactions with TFs, regulatory factors as well as other molecules. Blasiak et al. found that LINC00511 could interact with the vitamin D receptor (VDR), thereby affecting the transcriptional activation process and interfering with the expression of genes related to the vitamin D signaling pathway. This can lead to an anti-BC effect (100). Additionally, Liu et al.’s study also confirmed that LINC00511 could alter the radio-sensitivity of BC cells by regulating the expression levels of STXBP4 (101). This suggests that LINC00511 achieves its biological functions at multiple levels, including gene expression regulation.
6.3 Encoding small peptides to regulate signaling pathways
In addition to acting as a classic lncRNA, recent studies have found that LINC00511 may also encode a 133-amino acid small peptide (LINC00511-133aa) resulting in unique biological functions. Tan et al. found that the small peptide encoded by LINC00511 is capable of activating the Wnt/β-catenin signaling pathway. This can promote the invasive capacity and the stem cell state of BC cells (95). This finding provides a novel molecular mechanism for the oncogenic effects of LINC00511 and suggest additional roles for lncRNAs in tumor progression.
6.4 Mediating target gene regulation of the immune microenvironment
Recently, it was shown that there is a connection between LINC00511 and tumor immunogenicity and the tumor microenvironment. Sun et al. reported that LINC00511 could regulate the activation of inflammasomes through the LINC00511/miR-573/GSDMC axis, thereby affecting immune cell infiltration and tumor immunogenicity (102). Lian et al. also provided evidence through bioinformatics analysis combined with tissue and cell experiments that LINC00511 participated in regulating immune cell infiltration by modulating the related signaling pathways. They found that LINC00511 could target miR-29-3p, thereby promoting the upregulation of SLC31A1 expression. This molecule then promoted BC progression by regulating tumor immune infiltration (103). These findings suggest that LINC00511 may be involved in remodeling of the immune microenvironment leading to tumor evasion by immune system. This may provide another potential target for tumor immunotherapy.
7 Biological functions of LINC00511 in BC
7.1 Mediation of tumor cell apoptosis, proliferation, migration and invasion
In several studies, LINC00511 has been shown to significantly promote the proliferation, migration and invasion capabilities of BC cells. Liu et al. found that DNA hypo-methylation could lead to upregulation of LINC00511, thereby promoting BC cell proliferation, invasion and migration (97). LINC00511 could also inhibit cell apoptosis and promote cell proliferation in some in vivo experiments where it was found that knocking down LINC00511 could inhibit tumor growth (98). Another study revealed that LINC00511 could positively regulate the expression of MMP13, thereby promoting the migration and invasion of BC cells (99). Therefore, LINC00511 can promote the progression of malignancy in BC by regulating various downstream molecular targets.
7.2 Maintenance the tumor stem cell state
Cancer stem cells belong to a small subgroup of tumor cells, which possess self-renewal and tumor-initiating abilities, playing a crucial role in various stages of tumorigenesis, drug resistance, recurrence as well as metastasis. Studies have shown that LINC00511 could positively regulate Nanog expression, thereby maintaining the tumor stem cell-like state and characteristics. Knocking down LINC00511 can significantly downregulate Nanog expression and its downstream genes, inhibiting the formation and self-renewal capacity of tumor stem cells. There is also evidence that LINC00511 may encode a 133-amino acid small peptide (LINC00511-133aa), which can activate the Wnt/β-catenin signaling pathway, and thus maintaining the stem-like state of BC cells (95). Hence, LINC00511 has the ability to regulate the existence and activity of tumor stem cells.
7.3 Involvement of LINC00511 in tumor chemotherapy and radiotherapy resistance
Tumor cell resistance to chemotherapeutic drugs and radiation can lead to failure in cancer treatment, recurrence and metastasis. Multiple studies have found that LINC00511 can cause resistance in BC patients. Liu et al. confirmed that LINC00511 could regulate miR-185 expression, thereby affecting the levels of its target gene, STXBP4, ultimately leading to enhanced radio-resistance in BC cells (101). Zhang et al. showed that LINC00511 could enhance cellular resistance to paclitaxel by upregulating the expression levels of CDK6, while knocking down LINC00511 could significantly increase tumor cell sensitivity to this chemotherapeutic drug (104). Wu et al. found that nanomaterial-mediated LINC00511 siRNA delivery technology could significantly improve the chemo-sensitivity of triple negative BC (TNBC) cell lines to cisplatin (105). These findings reveal a key regulatory role of LINC00511 in processes such as chemotherapeutic drug- and radiation-induced tumor cell apoptosis and DNA damage repair.
8 Potential clinical applications of LINC00511 in BC
8.1 LINC00511 as a molecular biomarker for diagnosis and prognosis of BC
Extensive preclinical and retrospective clinical data have shown that LINC00511 may be used as a potential molecular biomarker for the diagnosis and prognosis of BC. Currently, the main blood biomarkers used in the clinical screening of BC are CEA and CA15-3. However, these have limited diagnostic efficacy for early-stage BC. To find new blood biomarkers in order to improve the diagnostic efficacy of BC, Mahmoud et al. conducted relevant studies on the sera of Egyptian female BC patients (43). They found that, compared to using CEA and CA15-3 alone for diagnosis, a combination screening with either LINC00511/CA15-3 or LINC00511/CEA had higher diagnostic efficacy in distinguishing BC patients from healthy individuals. In addition, LINC00511 also had a certain diagnostic value in the staging and metastasis of BC patients. This study found that the expression pattern of LINC00511 also differed among different molecular subtypes of BC, with a relatively higher expression in TNBC, suggesting its intrinsic association with the occurrence and development of specific subtypes.
LINC00511 has also shown to have a potential value in predicting the survival of BC patients. A systematic review and meta-analysis of LINC00511 in BC patients showed that those with high LINC00511 expression had a shorter overall survival (106). Chen et al. also reached a similar conclusion (107). These findings suggest that LINC00511 may be a potentially valuable molecular marker for BC. Further in-depth studies on LINC00511 will help to optimize the molecular subtyping, prognostic assessment and personalized treatment strategies for BC.
8.2 LINC00511 as a therapeutic target for BC
High expression levels of LINC00511 in BC patients precedes a worse survival prognosis with the lncRNA acting as an oncogene in BC. Therefore, knocking out LINC00511 expression in order to inhibit its effects on oncogenes and their related signaling pathways may be an effective new strategy for treatment of BC. Currently, the main targeting strategies for lncRNAs include antisense oligonucleotides, siRNAs (99, 104) and shRNAs (94, 101), which can act as small inhibitor molecules. SiRNA silencing studies by Yuan et al. have involved the construction of a carrier by combining siRNA with cationic nano-bubbles (CNBs). This significantly improved the silencing efficiency of LINC00511 by employing ultrasound-mediated nano-bubble destruction and forming siRNA-CNBs, which were ideal carriers for treating BC (107). Additionally, Wu et al. constructed a novel type of therapeutic diagnostic agent, by using a complex of low-frequency ultrasound irradiation and nano-bubbles, which appeared to be an efficient and safe siRNA transfection strategy (105).
Another method that could be used to inhibit the effects of LINC00511 on oncogenes, involved CRISPR-Cas9-mediated gene editing (108). Azadbakht et al. showed that CRISPR/Cas9 was another potential method for knocking out LINC00511, and subsequent studies have shown that this technique could specifically knock out this lncRNA gene (109). These studies indicate that LINC00511 may a potential novel therapeutic target for BC, and many scholars have proposed and implemented various methods for targeting LINC00511 (38, 110). However, up to now, these practices have only yielded results in cell and animal models, and clinical application studies have not produced satisfactory results. Therefore, further in-depth understanding and elucidation of the functional mechanisms of LINC00511 are needed, followed by the development of targeted drugs, to bring new treatment options for BC patients.
9 Prospects and challenges
LINC00511 is a functionally diverse and broad acting oncogenic lncRNA in BC, participating in multiple key biological processes in these cells (Figure 4). Its mechanisms of action involve ceRNA networks, transcriptional regulation, signaling pathway activation as well as small peptide encoding at multiple levels. New functions and regulatory mechanisms of LINC00511 in BC are being discovered. However, there are currently several gaps in our understanding of the overall mechanism of action of LINC00511 in tumor occurrence and development, as well as its associated networks with other molecular events. Additionally, the precise mechanisms by which LINC00511 regulates downstream targets and signaling pathways, and its differential roles at different time points in BC remain to be further explored. With respect to its clinical applications, LINC00511 may be used to improve prognostic assessment for BC, although further studies are urgently needed.
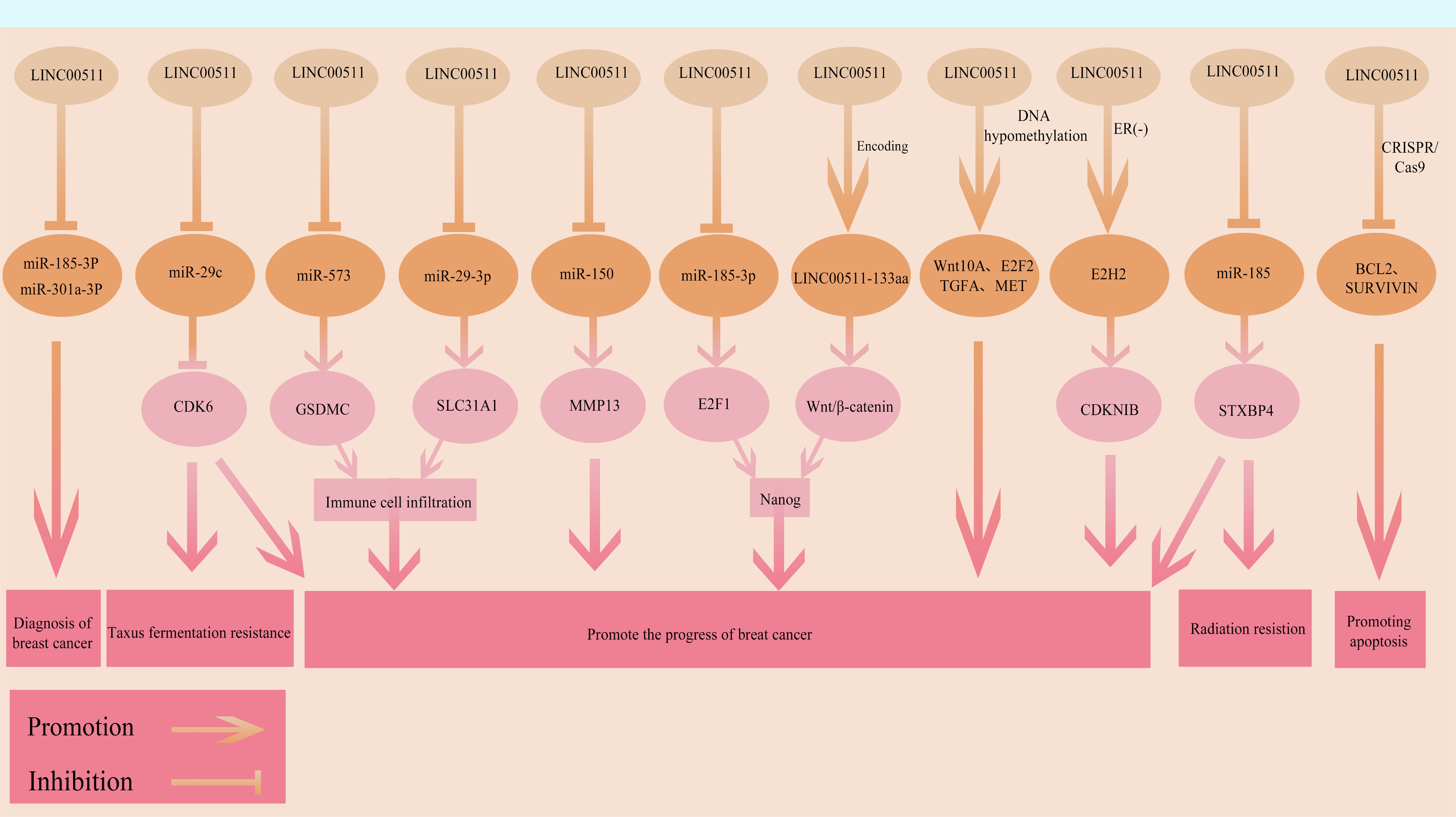
Figure 4. The functional roles and regulatory mechanisms of LINC00511 in BRCA. The arrow and blocking symbols represent promotion and inhibition of molecules/events, respectively.
In conclusion, in-depth elucidation of the functions and mechanisms of action LINC00511 will not only expand our understanding of the molecular networks underlying tumor occurrence and development, but will also provide new insights for early diagnosis, molecular subtyping and clinical treatment of BC. The study of lncRNAs and in particular, LINC00511 and its regulatory networks, has the potential to deliver novel breakthroughs in the precise and personalize diagnosis and treatment of all cancers.
Author contributions
LZ: Data curation, Visualization, Writing – original draft, Writing – review & editing. SB: Conceptualization, Writing – original draft, Writing – review & editing. YL: Conceptualization, Funding acquisition, Resources, Writing – original draft, Writing – review & editing. SS: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The Young and Middle-aged Talent Program of the Affiliated Hospital of Youjiang Medical University for Nationalities (Y202210316) supported this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. (2023) 73:17–48. doi: 10.3322/caac.21763
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
3. Kunkler IH, Williams LJ, Jack W, Cameron DA, Dixon JM. Breast-conserving surgery with or without irradiation in early breast cancer. N Engl J Med. (2023) 388:585–94. doi: 10.1056/NEJMoa2207586
4. Dai X, Cheng H, Bai Z, Li J. Breast cancer cell line classification and its relevance with breast tumor subtyping. J Cancer. (2017) 8:3131–41. doi: 10.7150/jca.18457
5. Shi Y, Huang Q, Kong X, Zhao R, Chen X, Zhai Y, et al. Current knowledge of long non-coding rna hotair in breast cancer progression andits application. Life (Basel). (2021) 11:483. doi: 10.3390/life11060483
6. Singh A, Mishra R, Mazumder A. Breast cancer and its therapeutic targets: a comprehensive review. Chem Biol Drug Des. (2024) 103:e14384. doi: 10.1111/cbdd.14384
7. Isaac-Lam MF, DeMichael KM. Calorie restriction and breast cancer treatment: a mini-review. J Mol Med (Berl). (2022) 100:1095–109. doi: 10.1007/s00109-022-02226-y
8. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding rnas and its biological functions. Nat Rev Mol Cell Biol. (2021) 22:96–118. doi: 10.1038/s41580-020-00315-9
9. Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding rnas. Nat Cell Biol. (2019) 21:542–51. doi: 10.1038/s41556-019-0311-8
10. Zhao Y, Teng H, Yao F, Yap S, Sun Y, Ma L. Challenges and strategies in ascribing functions to long noncoding rnas. Cancers (Basel). (2020) 12:1458. doi: 10.3390/cancers12061458
11. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding rnas. Cell. (2018) 172:393–407. doi: 10.1016/j.cell.2018.01.011
12. Islam F, Zhou Y, Lam AK. Long non-coding rnas profiling using microarray in papillary thyroid carcinoma. Methods Mol Biol. (2022) 2534:135–48. doi: 10.1007/978-1-0716-2505-7_10
13. Singh D, Assaraf YG, Gacche RN. Long non-coding rna mediated drug resistance in breast cancer. Drug Resist Updat. (2022) 63:100851. doi: 10.1016/j.drup.2022.100851
14. Bohosova J, Kubickova A, Slaby O. Lncrna pvt1 in the pathogenesis and clinical management of renal cell carcinoma. Biomolecules. (2021) 11:664. doi: 10.3390/biom11050664
15. Anastasiadou E, Jacob LS, Slack FJ. Non-coding rna networks in cancer. Nat Rev Cancer. (2018) 18:5–18. doi: 10.1038/nrc.2017.99
16. Martone J, Mariani D, Santini T, Setti A, Shamloo S, Colantoni A, et al. Smart lncrna controls translation of a g-quadruplex-containing mrna antagonizingthe dhx36 helicase. EMBO Rep. (2020) 21:e49942. doi: 10.15252/embr.201949942
17. Fernandes J, Acuna SM, Aoki JI, Floeter-Winter LM, Muxel SM. Long non-coding rnas in the regulation of gene expression: physiology anddisease. Noncoding Rna. (2019) 5:17. doi: 10.3390/ncrna5010017
18. Sun Y, Xu C, Wu Q, Zhang L, Wang P. Long noncoding rna kcnq1ot1 promotes proliferation, migration, and invasion inmaxillary sinus squamous cell carcinoma by regulating mir-204/epha7 axis. J Cell Biochem. (2020) 121:2962–9. doi: 10.1002/jcb.29548
19. Cong Z, Diao Y, Xu Y, Li X, Jiang Z, Shao C, et al. Long non-coding rna linc00665 promotes lung adenocarcinoma progression and functions as cerna to regulate akr1b10-erk signaling by sponging mir-98. Cell Death Dis. (2019) 10:84. doi: 10.1038/s41419-019-1361-3
20. Bin X, Hongjian Y, Xiping Z, Bo C, Shifeng Y, Binbin T. Research progresses in roles of lncrna and its relationships with breast cancer. Cancer Cell Int. (2018) 18:179. doi: 10.1186/s12935-018-0674-0
21. Wu Y, Shao A, Wang L, Hu K, Yu C, Pan C, et al. The role of lncrnas in the distant metastasis of breast cancer. Front Oncol. (2019) 9:407. doi: 10.3389/fonc.2019.00407
22. Yang Y, Dong MH, Hu HM, Min QH, Xiao L. Lncrna fgd5-as1/mir-5590-3p axis facilitates the proliferation and metastasis ofrenal cell carcinoma through erk/akt signalling. Eur Rev Med Pharmacol Sci. (2020) 24:8756–66. doi: 10.26355/eurrev_202009_22814
23. Yang W, Zhou J, Zhang K, Li L, Xu Y, Ma K, et al. Identification and validation of the clinical roles of the vhl-related lncrnas inclear cell renal cell carcinoma. J Cancer. (2021) 12:2702–14. doi: 10.7150/jca.55113
24. Gao Y, Xie M, Guo Y, Yang Q, Hu S, Li Z. Long non-coding rna fgd5-as1 regulates cancer cell proliferation andchemoresistance in gastric cancer through mir-153-3p/cited2 axis. Front Genet. (2020) 11:715. doi: 10.3389/fgene.2020.00715
25. Zhang J, Lou W. A key mrna-mirna-lncrna competing endogenous rna triple sub-network linked to diagnosis and prognosis of hepatocellular carcinoma. Front Oncol. (2020) 10:340. doi: 10.3389/fonc.2020.00340
26. Fan Y, Li H, Yu Z, Dong W, Cui X, Ma J, et al. Long non-coding rna fgd5-as1 promotes non-small cell lung cancer cellproliferation through sponging hsa-mir-107 to up-regulate fgfrl1. Biosci Rep. (2020) 40:BSR20193309. doi: 10.1042/BSR20193309
27. Gao SJ, Ren SN, Liu YT, Yan HW, Chen XB. Targeting egfr sensitizes 5-fu-resistant colon cancer cells through modification of the lncrna-fgd5-as1-mir-330-3p-hexokinase 2 axis. Mol Ther Oncolytics. (2021) 23:14–25. doi: 10.1016/j.omto.2021.06.012
28. Su D, Ji Z, Xue P, Guo S, Jia Q, Sun H. Long-noncoding rna fgd5-as1 enhances the viability, migration, and invasion ofglioblastoma cells by regulating the mir-103a-3p/tpd52 axis. Cancer Manag Res. (2020) 12:6317–29. doi: 10.2147/CMAR.S253467
29. Song QH, Guo MJ, Zheng JS, Zheng XH, Ye ZH, Wei P. Study on targeting relationship between mir-320b and fgd5-as1 and its effect onbiological function of osteosarcoma cells. Cancer Manag Res. (2020) 12:13589–98. doi: 10.2147/CMAR.S264682
30. Fang K, Xu ZJ, Jiang SX, Tang DS, Yan CS, Deng YY, et al. Lncrna fgd5−as1 promotes breast cancer progression by regulating thehsa−mir−195−5p/nuak2 axis. Mol Med Rep. (2021) 23:460. doi: 10.3892/mmr.2021.12099
31. Aichen Z, Kun W, Xiaochun S, Lingling T. Lncrna fgd5-as1 promotes the Malignant phenotypes of ovarian cancer cells via targeting mir-142-5p. Apoptosis. (2021) 26:348–60. doi: 10.1007/s10495-021-01674-0
32. Louca M, Gkretsi V. Lincrnas and snornas in breast cancer cell metastasis: the unknown players. Cancers (Basel). (2022) 14:4528. doi: 10.3390/cancers14184528
33. Zhang Y, Qian J, Gu C, Yang Y. Alternative splicing and cancer: a systematic review. Signal Transduct Target Ther. (2021) 6:78. doi: 10.1038/s41392-021-00486-7
34. Xing Z, Zhang M, Liu J, Liu G, Feng K, Wang X. Linc00337 induces tumor development and chemoresistance to paclitaxel of breast cancer by recruiting m2 tumor-associated macrophages. Mol Immunol. (2021) 138:1–9. doi: 10.1016/j.molimm.2021.07.009
35. Zhu Y, Yang L, Chong QY, Yan H, Zhang W, Qian W, et al. Long noncoding rna linc00460 promotes breast cancer progression by regulating themir-489-5p/fgf7/akt axis. Cancer Manag Res. (2019) 11:5983–6001. doi: 10.2147/CMAR.S207084
36. Wang HB, Wei H, Wang JS, Li L, Chen AY, Li ZG. Down-regulated expression of linc00518 prevents epithelial cell growth and metastasis in breast cancer through the inhibition of cdx2 methylation and the wnt signaling pathway. Biochim Biophys Acta Mol Basis Dis. (2019) 1865:708–23. doi: 10.1016/j.bbadis.2019.01.003
37. Ding J, Cao J, Chen Z, He Z. The role of long intergenic noncoding rna 00511 in Malignant tumors: a meta-analysis, database validation and review. Bioengineered. (2020) 11:812–23. doi: 10.1080/21655979.2020.1795384
38. Ghafouri-Fard S, Safarzadeh A, Hussen BM, Taheri M, Ayatollahi SA. A review on the role of linc00511 in cancer. Front Genet. (2023) 14:1116445. doi: 10.3389/fgene.2023.1116445
39. Huang HG, Tang XL, Huang XS, Zhou L, Hao YG, Zheng YF. Long noncoding rna linc00511 promoted cell proliferation and invasion via regulating mir-124-3p/ezh2 pathway in gastric cancer. Eur Rev Med Pharmacol Sci. (2020) 24:4232–45. doi: 10.26355/eurrev_202004_21003
40. Mao BD, Xu P, Xu P, Zhong Y, Ding WW, Meng QZ. Linc00511 knockdown prevents cervical cancer cell proliferation and reduces resistance to paclitaxel. J Biosci. (2019) 44:44. doi: 10.1007/s12038-019-9851-0
41. Hu WY, Wei HY, Li KM, Wang RB, Xu XQ, Feng R. Linc00511 as a cerna promotes cell Malignant behaviors and correlates withprognosis of hepatocellular carcinoma patients by modulating mir-195/eya1 axis. BioMed Pharmacother. (2020) 121:109642. doi: 10.1016/j.biopha.2019.109642
42. Cabanski CR, White NM, Dang HX, Silva-Fisher JM, Rauck CE, Cicka D, et al. Pan-cancer transcriptome analysis reveals long noncoding rnas with conservedfunction. RNA Biol. (2015) 12:628–42. doi: 10.1080/15476286.2015.1038012
43. Mahmoud MM, Sanad EF, Elshimy R, Hamdy NM. Competitive endogenous role of the linc00511/mir-185-3p axis and mir-301a-3p from liquid biopsy as molecular markers for breast cancer diagnosis. Front Oncol. (2021) 11:749753. doi: 10.3389/fonc.2021.749753
44. Yang Q, Al-Hendy A. The regulatory functions and the mechanisms of long non-coding rnas in cervical cancer. Cells. (2022) 11:1149. doi: 10.3390/cells11071149
45. Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, et al. Mechanisms and functions of long non-coding rnas at multiple regulatory levels. Int J Mol Sci. (2019) 20:5573. doi: 10.3390/ijms20225573
46. Wu Y, Li L, Wang Q, Zhang L, He C, Wang X, et al. Linc00511 promotes lung squamous cell carcinoma proliferation and migration viainhibiting mir-150-5p and activating tada1. Transl Lung Cancer Res. (2020) 9:1138–48. doi: 10.21037/tlcr-19-701
47. Wang J, Tian Y, Zheng H, Ding Y, Wang X. An integrated analysis reveals the oncogenic function of lncrna linc00511 in human ovarian cancer. Cancer Med. (2019) 8:3026–35. doi: 10.1002/cam4.2171
48. Zhao X, Liu Y, Li Z, Zheng S, Wang Z, Li W, et al. Linc00511 acts as a competing endogenous rna to regulate vegfa expression throughsponging hsa-mir-29b-3p in pancreatic ductal adenocarcinoma. J Cell Mol Med. (2018) 22:655–67. doi: 10.1111/jcmm.13351
49. Lu Y, Tian M, Liu J, Wang K. Linc00511 facilitates temozolomide resistance of glioblastoma cells via sponging mir-126-5p and activating wnt/beta-catenin signaling. J Biochem Mol Toxicol. (2021) 35:e22848. doi: 10.1002/jbt.22848
50. Liu Z, Tao B, Li L, Liu P, Xia K, Zhong C. Linc00511 knockdown suppresses glioma cell Malignant progression through mir-15a-5p/aebp1 axis. Brain Res Bull. (2021) 173:82–96. doi: 10.1016/j.brainresbull.2021.05.010
51. Du X, Tu Y, Liu S, Zhao P, Bao Z, Li C, et al. Linc00511 contributes to glioblastoma tumorigenesis and epithelial-mesenchymal transition via linc00511/mir-524-5p/yb1/zeb1 positive feedback loop. J Cell Mol Med. (2020) 24:1474–87. doi: 10.1111/jcmm.14829
52. Li C, Liu H, Yang J, Yang J, Yang L, Wang Y, et al. Long noncoding rna linc00511 induced by sp1 accelerates the glioma progression through targeting mir-124-3p/ccnd2 axis. J Cell Mol Med. (2019) 23:4386–94. doi: 10.1111/jcmm.14331
53. Zhu FY, Zhang SR, Wang LH, Wu WD, Zhao H. Linc00511 promotes the progression of non-small cell lung cancer through downregulating lats2 and klf2 by binding to ezh2 and lsd1. Eur Rev Med Pharmacol Sci. (2019) 23:8377–90. doi: 10.26355/eurrev_201910_19149
54. Sun CC, Li SJ, Li G, Hua RX, Zhou XH, Li DJ. Long intergenic noncoding rna 00511 acts as an oncogene in non-small-cell lung cancer by binding to ezh2 and suppressing p57. Mol Ther Nucleic Acids. (2016) 5:e385. doi: 10.1038/mtna.2016.94
55. Xue J, Zhang F. Lncrna linc00511 plays an oncogenic role in lung adenocarcinoma by regulatingpkm2 expression via sponging mir-625-5p. Thorac Cancer. (2020) 11:2570–9. doi: 10.1111/1759-7714.13576
56. Zhang Y, Xiao P, Hu X. Linc00511 enhances luad Malignancy by upregulating gcnt3 via mir-195-5p. BMC Cancer. (2022) 22:389. doi: 10.1186/s12885-022-09459-7
57. Cheng Y, Wang S, Mu X. Long non-coding rna linc00511 promotes proliferation, invasion, and migration of non-small cell lung cancer cells by targeting mir-625-5p/gspt1. Transl Cancer Res. (2021) 10:5159–73. doi: 10.21037/tcr-21-1468
58. Li C, Fu Y, He Y, Huang N, Yue J, Miao Y, et al. Knockdown of linc00511 enhances radiosensitivity of lung adenocarcinoma via regulating mir-497-5p/smad3. Cancer Biol Ther. (2023) 24:2165896. doi: 10.1080/15384047.2023.2165896
59. Wang Y, Mei X, Song W, Wang C, Qiu X. Lncrna linc00511 promotes col1a1-mediated proliferation and metastasis bysponging mir-126-5p/mir-218-5p in lung adenocarcinoma. BMC Pulm Med. (2022) 22:272. doi: 10.1186/s12890-022-02070-3
60. Zhu Z, Shi Y, Gong X, Li J, Zhang M. Linc00511 knockdown suppresses resistance to cisplatin in lung adenocarcinoma byinteracting with mir-182-3p and birc5. Mol Biotechnol. (2022) 64:252–62. doi: 10.1007/s12033-021-00400-0
61. Wang D, Liu K, Chen E. Linc00511 promotes proliferation and invasion by sponging mir-515-5p in gastric cancer. Cell Mol Biol Lett. (2020) 25:4. doi: 10.1186/s11658-020-0201-x
62. Sun CB, Wang HY, Han XQ, Liu YN, Wang MC, Zhang HX, et al. Linc00511 promotes gastric cancer cell growth by acting as a cerna. World J Gastrointest Oncol. (2020) 12:394–404. doi: 10.4251/wjgo.v12.i4.394
63. Chen Z, Wu H, Zhang Z, Li G, Liu B. Linc00511 accelerated the process of gastric cancer by targeting mir-625-5p/nfix axis. Cancer Cell Int. (2019) 19:351. doi: 10.1186/s12935-019-1070-0
64. Wang Q, Mao X, Luo F, Wang J. Linc00511 promotes gastric cancer progression by regulating sox4 and epigenetically repressing pten to activate pi3k/akt pathway. J Cell Mol Med. (2021) 25:9112–27. doi: 10.1111/jcmm.16656
65. Cui N, Sun Q, Liu H, Li L, Guo X, Shi Y, et al. Long non-coding rna linc00511 regulates the expression of microrna-625-5p and activates signal transducers and activators of transcription 3 (stat3) toaccelerate the progression of gastric cancer. Bioengineered. (2021) 12:2915–27. doi: 10.1080/21655979.2021.1940611
66. Sun S, Xia C, Xu Y. Hif-1alpha induced lncrna linc00511 accelerates the colorectal cancer proliferation through positive feedback loop. BioMed Pharmacother. (2020) 125:110014. doi: 10.1016/j.biopha.2020.110014
67. Lu Y, Yu Y, Liu F, Han Y, Xue H, Sun X, et al. Linc00511-dependent inhibition of il-24 contributes to the oncogenic role of hnf4alpha in colorectal cancer. Am J Physiol Gastrointest Liver Physiol. (2021) 320:G338–50. doi: 10.1152/ajpgi.00243.2020
68. Hu Y, Zhang Y, Ding M, Xu R. Lncrna linc00511 acts as an oncogene in colorectal cancer via sponging mir-29c-3pto upregulate nfia. Onco Targets Ther. (2020) 13:13413–24. doi: 10.2147/OTT.S250377
69. Qian X, Jiang C, Zhu Z, Han G, Xu N, Ye J, et al. Long non-coding rna linc00511 facilitates colon cancer development through regulating microrna-625-5p to target wee1. Cell Death Discovery. (2022) 8:233. doi: 10.1038/s41420-021-00790-9
70. Wang RP, Jiang J, Jiang T, Wang Y, Chen LX. Increased long noncoding rna linc00511 is correlated with poor prognosis and contributes to cell proliferation and metastasis by modulating mir-424 in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. (2019) 23:3291–301. doi: 10.26355/eurrev_201904_17691
71. Hu P, Cui H, Lei T, Li S, Mai E, Jia F. Linc00511 indicates a poor prognosis of liver hepatocellular carcinoma. Onco Targets Ther. (2019) 12:9367–76. doi: 10.2147/OTT.S228231
72. Peng X, Li X, Yang S, Huang M, Wei S, Ma Y, et al. Linc00511 drives invasive behavior in hepatocellular carcinoma by regulating exosome secretion and invadopodia formation. J Exp Clin Cancer Res. (2021) 40:183. doi: 10.1186/s13046-021-01990-y
73. Guo W, Yu Q, Zhang M, Li F, Liu Y, Jiang W, et al. Long intergenic non-protein coding rna 511 promotes the progression ofosteosarcoma cells through sponging microrna 618 to upregulate the expression ofmaelstrom. Aging (Albany Ny). (2019) 11:5351–67. doi: 10.18632/aging.102109
74. Yan L, Wu X, Liu Y, Xian W. Lncrna linc00511 promotes osteosarcoma cell proliferation and migration throughsponging mir-765. J Cell Biochem. (2019) 120:7248–56. doi: 10.1002/jcb.27999
75. Qiao S, Qi K, Liu C, Xu C, Ma J, Xu X, et al. Long intergenic non-coding rna 511 correlates with improved prognosis, andhinders osteosarcoma progression both. Vitro vivo. J Clin Lab Anal. (2020) 34:e23164. doi: 10.1002/jcla.23164
76. Zhang T, Yu H, Bai Y, Song J, Chen J, Li Y, et al. Extracellular vesicle-derived linc00511 promotes glycolysis and mitochondrialoxidative phosphorylation of pancreatic cancer through macrophage polarization bymicrorna-193a-3p-dependent regulation of plasminogen activator urokinase. Immunopharmacol Immunotoxicol. (2023) 45:355–69. doi: 10.1080/08923973.2022.2145968
77. Shi Y, Liu M, Huang Y, Zhang J, Yin L. Promotion of cell autophagy and apoptosis in cervical cancer by inhibition of long noncoding rna linc00511 via transcription factor rxra-regulated pld1. J Cell Physiol. (2020) 235:6592–604. doi: 10.1002/jcp.29529
78. Zhang X, Wang Y, Zhao A, Kong F, Jiang L, Wang J. Long non-coding rna linc00511 accelerates proliferation and invasion in cervical cancer through targeting mir-324-5p/dram1 axis. Onco Targets Ther. (2020) 13:10245–56. doi: 10.2147/OTT.S255067
79. Lu M, Gao Q, Wang Y, Ren J, Zhang T. Linc00511 promotes cervical cancer progression by regulating the mir-497-5p/mapk1 axis. Apoptosis. (2022) 27:800–11. doi: 10.1007/s10495-022-01768-3
80. Li S, Guo W, Geng H, Wang C, Yang S, Xu X. Linc00511 exacerbated t-cell acute lymphoblastic leukemia via mir-195-5p/lrrk1 axis. Biosci Rep. (2020) 40:BSR20193631. doi: 10.1042/BSR20193631
81. Xiang J, Guan Y, Bhandari A, Xia E, Wen J, Wang O. Linc00511 influences cellular proliferation through cyclin-dependent kinases in papillary thyroid carcinoma. J Cancer. (2020) 11:450–9. doi: 10.7150/jca.35364
82. Chen Y, Bao C, Zhang X, Lin X, Fu Y. Knockdown of linc00511 promotes radiosensitivity of thyroid carcinoma cells via suppressing jak2/stat3 signaling pathway. Cancer Biol Ther. (2019) 20:1249–57. doi: 10.1080/15384047.2019.1617569
83. Deng H, Huang C, Wang Y, Jiang H, Peng S, Zhao X. Linc00511 promotes the Malignant phenotype of clear cell renal cell carcinoma by ponging microrna-625 and thereby increasing cyclin d1 expression. Aging (Albany Ny). (2019) 11:5975–91. doi: 10.18632/aging.102156
84. Dong LM, Zhang XL, Mao MH, Li YP, Zhang XY, Xue DW, et al. Linc00511/mirna-143-3p modulates apoptosis and Malignant phenotype of bladdercarcinoma cells via pcmt1. Front Cell Dev Biol. (2021) 9:650999. doi: 10.3389/fcell.2021.650999
85. Li J, Li Y, Meng F, Fu L, Kong C. Knockdown of long non-coding rna linc00511 suppresses proliferation and promotes apoptosis of bladder cancer cells via suppressing wnt/beta-catenin signaling pathway. Biosci Rep. (2018) 38:BSR20171701. doi: 10.1042/BSR20171701
86. Ding J, Yang C, Yang S. Linc00511 interacts with mir-765 and modulates tongue squamous cell carcinoma progression by targeting lamc2. J Oral Pathol Med. (2018) 47:468–76. doi: 10.1111/jop.12677
87. Wang K, Zhu G, Bao S, Chen S. Long non-coding rna linc00511 mediates the effects of esr1 on proliferation andinvasion of ovarian cancer through mir-424-5p and mir-370-5p. Cancer Manag Res. (2019) 11:10807–19. doi: 10.2147/CMAR.S232140
88. Han D, Yuan RX, Su F. Linc00511 can promote the proliferation, migration and invasion of esophageal cancer cells through regulating microrna-150-5p. Eur Rev Med Pharmacol Sci. (2020) 24:2462–9. doi: 10.26355/eurrev_202003_20514
89. Chen YN, Fu XR, Guo H, Fu XY, Shi KS, Gao T, et al. Yy1-induced lncrna00511 promotes melanoma progression via the mir-150-5p/adam19axis. Am J Cancer Res. (2024) 14:809–31. doi: 10.62347/VRBK1334
90. Wang XF, Liang B, Chen C, Zeng DX, Zhao YX, Su N, et al. Long intergenic non-protein coding rna 511 in cancers. Front Genet. (2020) 11:667. doi: 10.3389/fgene.2020.00667
91. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. Gepia: a web server for cancer and normal gene expression profiling andinteractive analyses. Nucleic Acids Res. (2017) 45:W98–102. doi: 10.1093/nar/gkx247
92. Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, et al. Ualcan: an update to the integrated cancer data analysis platform. Neoplasia. (2022) 25:18–27. doi: 10.1016/j.neo.2022.01.001
93. Zhao T, Li X, Li M, Jamil M, Zhang J. Characterization and verification of mmp family members as potential biomarkers in kidney clear cell renal carcinoma. Am J Cancer Res. (2023) 13:3941–62.
94. Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q, et al. Long noncoding rna linc00511 contributes to breast cancer tumourigenesis andstemness by inducing the mir-185-3p/e2f1/nanog axis. J Exp Clin Cancer Res. (2018) 37:289. doi: 10.1186/s13046-018-0945-6
95. Tan Z, Zhao L, Huang S, Jiang Q, Wei Y, Wu JL, et al. Small peptide linc00511-133aa encoded by linc00511 regulates breast cancer cell invasion and stemness through the wnt/beta-catenin pathway. Mol Cell Probes. (2023) 69:101913. doi: 10.1016/j.mcp.2023.101913
96. Yang Y, Wu L, Shu XO, Cai Q, Shu X, Li B, et al. Genetically predicted levels of dna methylation biomarkers and breast cancerrisk: data from 228 951 women of european descent. J Natl Cancer Inst. (2020) 112:295–304. doi: 10.1093/jnci/djz109
97. Liu C, Xu Y, Liu X, Fu Y, Zhu K, Niu Z, et al. Upregulation of linc00511 expression by dna hypomethylation promotes the progression of breast cancer. Gland Surg. (2021) 10:1418–30. doi: 10.21037/gs-21-84
98. Zhang J, Sui S, Wu H, Zhang J, Zhang X, Xu S, et al. The transcriptional landscape of lncrnas reveals the oncogenic function of linc00511 in er-negative breast cancer. Cell Death Dis. (2019) 10:599. doi: 10.1038/s41419-019-1835-3
99. Shi G, Cheng Y, Zhang Y, Guo R, Li S, Hong X. Long non-coding rna linc00511/mir-150/mmp13 axis promotes breast cancerproliferation, migration and invasion. Biochim Biophys Acta Mol Basis Dis. (2021) 1867:165957. doi: 10.1016/j.bbadis.2020.165957
100. Blasiak J, Chojnacki J, Pawlowska E, Jablkowska A, Chojnacki C. Vitamin d may protect against breast cancer through the regulation of longnoncoding rnas by vdr signaling. Int J Mol Sci. (2022) 23:3189. doi: 10.3390/ijms23063189
101. Liu L, Zhu Y, Liu AM, Feng Y, Chen Y. Long noncoding rna linc00511 involves in breast cancer recurrence andradioresistance by regulating stxbp4 expression via mir-185. Eur Rev Med Pharmacol Sci. (2019) 23:7457–68. doi: 10.26355/eurrev_201909_18855
102. Sun K, Chen RX, Li JZ, Luo ZX. Linc00511/hsa-mir-573 axis-mediated high expression of gasdermin c associates with dismal prognosis and tumor immune infiltration of breast cancer. Sci Rep. (2022) 12:14788. doi: 10.1038/s41598-022-19247-9
103. Lian W, Yang P, Li L, Chen D, Wang C. A ceRNA network-mediated over-expression of cuproptosis-related gene slc31a1 correlates with poor prognosis and positive immune infiltration in breast cancer. Front Med (Lausanne). (2023) 10:1194046. doi: 10.3389/fmed.2023.1194046
104. Zhang H, Zhao B, Wang X, Zhang F, Yu W. Linc00511 knockdown enhances paclitaxel cytotoxicity in breast cancer viaregulating mir-29c/cdk6 axis. Life Sci. (2019) 228:135–44. doi: 10.1016/j.lfs.2019.04.063
105. Wu B, Yuan Y, Han X, Wang Q, Shang H, Liang X, et al. Structure of linc00511-sirna-conjugated nanobubbles and improvement of cisplatin sensitivity on triple negative breast cancer. FASEB J. (2020) 34:9713–26. doi: 10.1096/fj.202000481R
106. Agbana YL, Abi ME, Ni Y, Xiong G, Chen J, Yun F, et al. Linc00511 as a prognostic biomarker for human cancers: a systematic review and meta-analysis. BMC Cancer. (2020) 20:682. doi: 10.1186/s12885-020-07188-3
107. Chen M, Qi P, Jiang WW. Prognostic significance of long intergenic non-protein-coding rna 511expression in Malignant tumors: a systematic review and meta-analysis. Med (Baltimore). (2020) 99:e23054. doi: 10.1097/MD.0000000000023054
108. Kansara S, Pandey V, Lobie PE, Sethi G, Garg M, Pandey AK. Mechanistic involvement of long non-coding rnas in oncotherapeutics resistance in triple-negative breast cancer. Cells. (2020) 9:1511. doi: 10.3390/cells9061511
109. Azadbakht N, Doosti A, Jami MS. Crispr/cas9-mediated linc00511 knockout strategies, increased apoptosis of breast cancer cells via suppressing antiapoptotic genes. Biol Proced Online. (2022) 24:8. doi: 10.1186/s12575-022-00171-1
Keywords: lncRNAs, LINC00511, breast cancer, mechanisms, biological functions
Citation: Zhao L, Biswas S, Li Y and Sooranna SR (2024) The emerging roles of LINC00511 in breast cancer development and therapy. Front. Oncol. 14:1429262. doi: 10.3389/fonc.2024.1429262
Received: 07 May 2024; Accepted: 29 July 2024;
Published: 14 August 2024.
Edited by:
Wenwen Zhang, Nanjing Medical University, ChinaReviewed by:
Yuehua Li, University of South China, ChinaJia Li, University of North Carolina at Charlotte, United States
Copyright © 2024 Zhao, Biswas, Li and Sooranna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sangita Biswas, c2FuZ2l0YUBtYWhzYS5lZHUubXk=; Yepeng Li, bGl5ZXBlbmcyNzMyQHltdW4uZWR1LmNu
†These authors have contributed equally to this work
 Lifeng Zhao
Lifeng Zhao Sangita Biswas
Sangita Biswas Yepeng Li1*†
Yepeng Li1*† Suren Rao Sooranna
Suren Rao Sooranna