- 1Institute of Head and Neck Studies and Education, Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham, United Kingdom
- 2Stanford Center for Biomedical Informatics Research (BMIR), Department of Medicine, and Department of Biomedical Data Science, Stanford University, Stanford, CA, United States
- 3Propath, Hereford, United Kingdom
- 4Institute of Immunology and Immunotherapy, College of Medical and Dental Sciences, University of Birmingham, Birmingham, United Kingdom
- 5Warwick Clinical Trials Unit, University of Warwick, Coventry, United Kingdom
- 6National Institute for Health and Care Research (NIHR) Birmingham Biomedical Research Centre, University of Birmingham, Birmingham, United Kingdom
Background: The incidence of oropharyngeal cancer (OPC) is increasing, due mainly to a rise in Human Papilloma Virus (HPV)-mediated disease. HPV-mediated OPC has significantly better prognosis compared with HPV-negative OPC, stimulating interest in treatment de-intensification approaches to reduce long-term sequelae. Routine clinical testing frequently utilises immunohistochemistry to detect upregulation of p16 as a surrogate marker of HPV-mediation. However, this does not detect discordant p16-/HPV+ cases and incorrectly assigns p16+/HPV- cases, which, given their inferior prognosis compared to p16+/HPV+, may have important clinical implications. The biology underlying poorer prognosis of p16/HPV discordant OPC requires exploration.
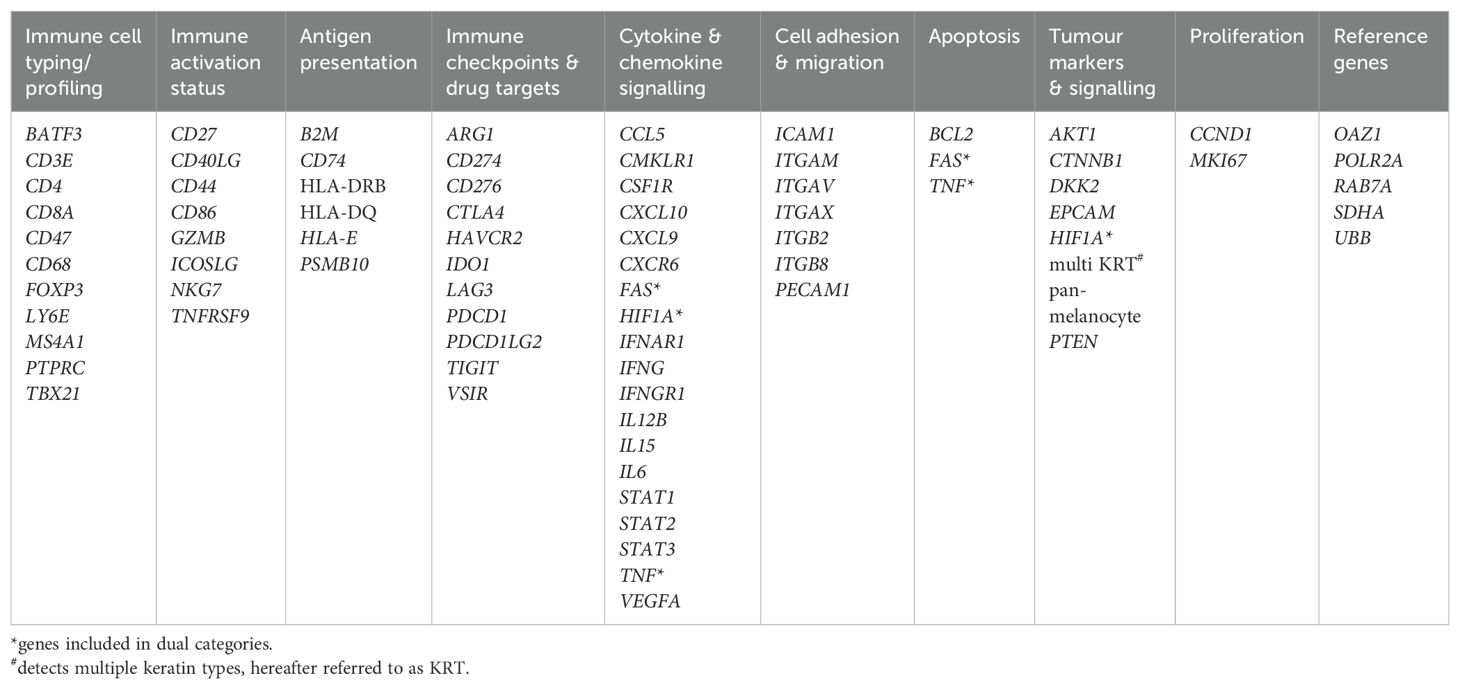
Methods: GeoMx digital spatial profiling was used to compare the expression patterns of selected immuno-oncology-related genes/gene families (n=73) within the tumour and stromal compartments of formalin-fixed, paraffin-embedded OPC tumour tissues (n=12) representing the three subgroups, p16+/HPV+, p16+/HPV- and p16-/HPV-.
Results: Keratin (multi KRT) and HIF1A, a key regulator of hypoxia adaptation, were upregulated in both p16+/HPV- and p16-/HPV- tumours relative to p16+/HPV+. Several genes associated with tumour cell proliferation and survival (CCND1, AKT1 and CD44) were more highly expressed in p16-/HPV- tumours relative to p16+/HPV+. Conversely, multiple genes with potential roles in anti-tumour immune responses (immune cell recruitment/trafficking, antigen processing and presentation), such as CXCL9, CXCL10, ITGB2, PSMB10, CD74, HLA-DRB and B2M, were more highly expressed in the tumour and stromal compartments of p16+/HPV+ OPC versus p16-/HPV- and p16+/HPV-. CXCL9 was the only gene showing significant differential expression between p16+/HPV- and p16-/HPV- tumours being upregulated within the stromal compartment of the former.
Conclusions: In terms of immune-oncology-related gene expression, discordant p16+/HPV- OPCs are much more closely aligned with p16-/HPV-OPCs and quite distinct from p16+/HPV+ tumours. This is consistent with previously described prognostic patterns (p16+/HPV+ >> p16+/HPV- > p16-/HPV-) and underlines the need for dual p16 and HPV testing to guide clinical decision making.
Background
Head and neck cancer (HNC) is the seventh most common cancer worldwide (1). Incidence rates are rising; mostly driven by a rapid increase in oropharyngeal cancer (OPC) incidence within certain global regions including the United States (US), Europe, New Zealand, and parts of Asia (2–4). The main risk factors for OPC include smoking, excessive alcohol intake and infection with high-risk Human Papilloma Virus (HPV). It is the increase in the latter which underpins rising incidence rates. In the US and United Kingdom, HPV-mediated OPC is now more prevalent than HPV-mediated cervical cancer (3, 5). It is estimated that incidence will continue to rise for the next ~20 years before the impact of gender-neutral prophylactic vaccination is felt (6, 7).
HPV-mediated OPC has distinct epidemiological, molecular, and immunological features compared with HPV-negative disease and is associated with better treatment response and outcomes (8, 9). This has led to separate classifications in the latest UICC/AJCC TNM staging system (TNM8) (10). Given the improved prognosis (8) and younger age of HPV-positive OPC patients, there is considerable interest in approaches to de-intensify treatment and reduce long-term morbidity and quality-of-life impact. Unfortunately, clinical trials to date have reported limited or no success (11, 12). One potential explanation for this relates to the determination and definition of HPV status.
The presence of HPV in OPC can be assessed directly using PCR-based methods or in situ hybridisation to detect viral DNA/RNA or indirectly using immunohistochemistry (IHC) to assess overexpression of the protein p16INK4a (p16) (13, 14). p16 overexpression is the indirect result of HPV early protein 7 (E7)-mediated inactivation of retinoblastoma protein. In contrast, frequent loss, mutation, or epigenetic silencing of the CDKN2A gene encoding p16 results in low/absent expression in HPV-negative OPC. Detection of p16 overexpression is therefore a good surrogate marker for HPV status and – being simple and cost-effective to implement – is routinely used in clinical practice. However, dual p16 and HPV-DNA/RNA testing has recently shown that while the majority of p16-positive tumours are HPV-positive (p16+/HPV+), a subset (~10%) are HPV-negative (p16+/HPV-). Likewise, a small subset of HPV-positive tumours do not overexpress p16 (p16-/HPV+). An important issue is whether these two discordant subsets – particularly those that are p16+/HPV- and therefore assigned as ‘HPV-positive’ by p16 routine testing – share the improved treatment response and survival outcomes of p16+/HPV+ cases, or whether outcomes are more closely aligned with p16-/HPV- tumours. Multiple studies suggested differential prognosis but were limited by sample size or restricted geographical sampling (15–19). A recent large multicentre study (n = 7,654) provided strong evidence that patients with discordant OPC (p16+/HPV- or p16-/HPV+) have significantly worse prognosis compared with p16+/HPV+ OPC patients, although significantly better than p16-/HPV-patients (20).
The biology underpinning this intermediate outcome requires exploration to develop understanding, guide development of novel therapies and inform clinical decision making. Here, we utilised NanoString’s GeoMx Digital Spatial Profiling (DSP) platform to explore in situ differences in gene expression between p16+/HPV+, p16+/HPV- and p16-/HPV- oropharyngeal tumours. This approach enables spatially-resolved analysis of gene expression within defined regions of interest, selected based on expression of morphology markers; for example, pan-cytokeratin expression to identify tumour versus stroma in epithelial tumours and/or CD3 to identify T cell rich areas.
Methods
Cohort
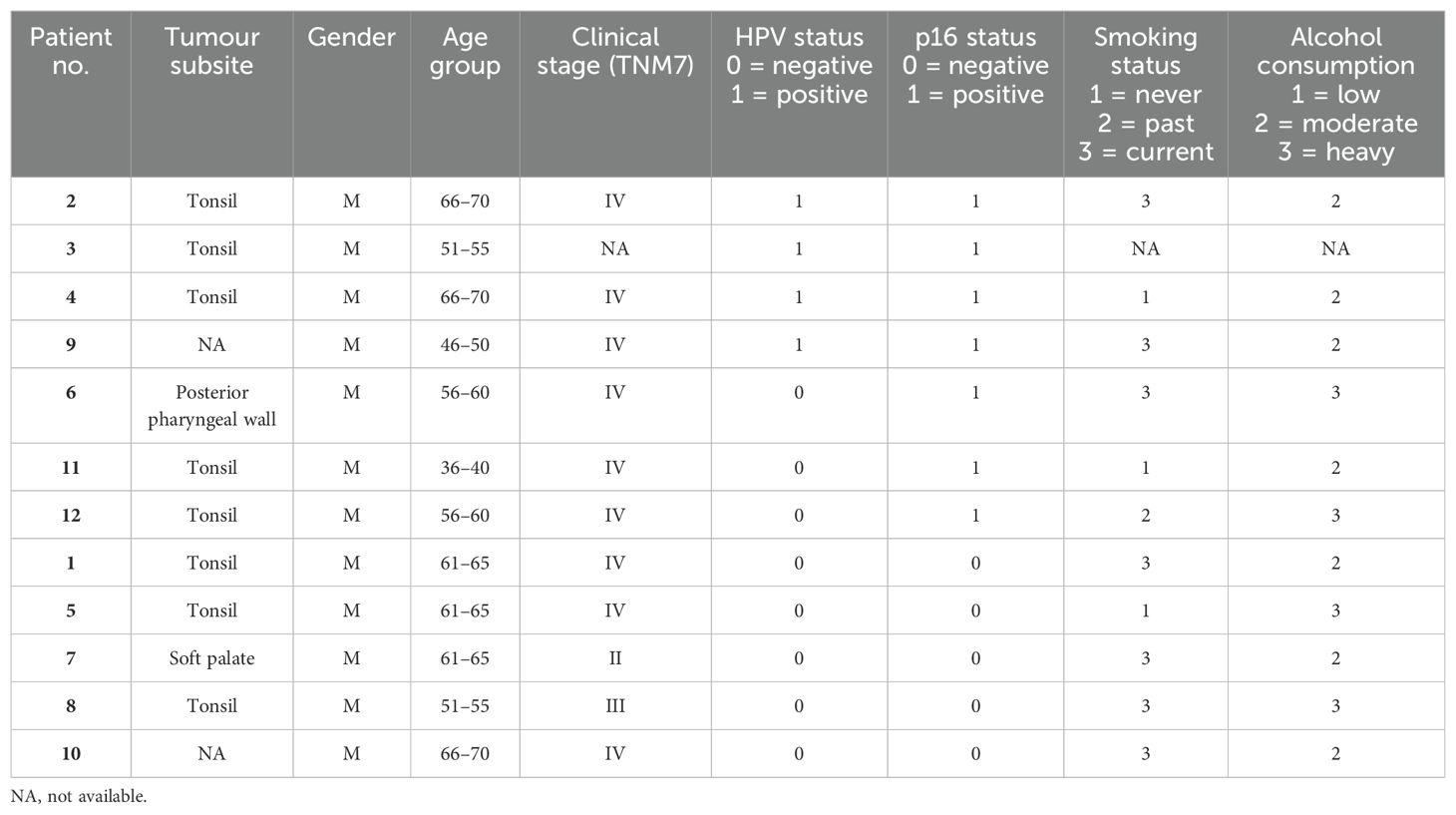
The study utilised formalin-fixed, paraffin-embedded (FPPE) diagnostic biopsy samples (primary tumour) from 12 OPC patients (four p16+/HPV+, three p16+/HPV- and five p16-/HPV-) recruited to the PET-NECK (21) or Predictr (22) clinical studies between 2000 and 2012. Patients were treated with curative intent, with either platinum-based chemoradiotherapy or surgery followed by adjuvant radiotherapy/chemoradiotherapy. Ethical approval for use of tissue samples in translational research was granted by the North West - Preston Research Ethics Committee (Reference: 16/NW/0265).
p16 status was assessed by immunohistochemical staining using the CINtec Histology kit (Roche). Samples with strong nuclear and cytoplasmic staining in >70% of the tumour were considered positive. HPV status was determined by DNA in situ hybridisation using the Ventana INFORM HPV III Family 16 probe or HPV DNA PCR.
Digital spatial profiling
5μm tissue sections were cut and mounted on SuperFrost plus microscope slides. Digital Spatial profiling (DSP) was carried out at NanoString Technologies Inc. Seattle, USA as part of their Technology Access Programme and according to their standard protocol (23). Briefly, slides were hybridised/stained with oligo-conjugated RNA detection probes [immuno-oncology panel comprising 78 genes, 73 target genes and five controls (Table 1)], plus three fluorescent conjugated antibodies and nuclear stain for characterisation of tissue compartments to facilitate Region of Interest (ROI) selection. ROIs were segmented into multiple regions representing tumour and stromal (non-tumour) tissue, using a threshold classifier on a fluorescently labelled pan-cytokeratin (PanCK) stain (clone AE1/AE3 594 Novus, 1:200 dilution). Fluorescent antibodies to CD3 (UMAB54 647 Origene, 1:100), to identify T cells, and CA-IX (EPR1451(2) 488, AbCam, 1:100), as a marker of hypoxia, were included to aid ROI selection, with nuclear stain (Syto 83 532, ThermoFisher, 1:25). Slides were imaged and a total of 141 ROIs across the 12 samples selected for analysis. ROIs were then sequentially illuminated with UV light to cleave oligo-probes which were aspirated and dispensed into 96-well plates. Probes were then hybridised to optical barcodes and counted using the nCounter platform (NanoString Technologies). Digital raw counts were exported for analysis.
Data analysis
Quality control and normalisation were performed in accordance with NanoString’s Gene Expression Data Analysis Guidelines MAN-C0011–04, 2017 (https://nanostring.com/wp-content/uploads/Gene_Expression_Data_Analysis_Guidelines.pdf). nCounter readout performance was assessed by evaluating the imaging and binding density QC metrics. All imaging segments demonstrated a high percentage of successfully scanned subsections, with a registered Fields of View (FOV) above 88%. The binding density was below 0.58, indicating low competition for binding the flow cell. Gene expression counts for the target genes were normalised to a set of house-keeping genes (RAB7A, UBB, SDHA, POLR2A, OAZ1), and a normalisation factor was calculated for each segment by comparing the geometric means of house-keeping genes across all segments. The normalised gene expression data were imported into Anndata objects (v0.8.0), and the gene expression values log2 transformed. Differential gene expression analysis was performed using the Kruskal-Wallis test, and pairwise comparisons were made using the Wilcoxon rank-sum test using the Scanpy library (v1.9.6). Raw p-values were adjusted using the Benjamini-Hochberg procedure. To generate the heatmaps, we selected genes that were significantly upregulated in one group compared to the other groups. For this, we required the raw p-values to be smaller than 0.05 and the adjusted p-values to be smaller than 0.2. To generate the volcano plots, we performed pairwise comparisons between each pair of groups, and the significant genes were determined using adjusted p-values below 0.05. Volcano plots were generated using the ggplot2 R library (v3.5.1).
Results
The cohort of 12 FFPE diagnostic biopsy samples from patients with OPC included four patients whose tumours were p16+/HPV+, three discordant (p16+/HPV-), and five p16-/HPV-. Patient characteristics are summarised in Table 2. Gene expression was analysed separately within the tumour (PanCK+) and stromal regions (PanCK-). ROIs were placed to capture gene expression within the tumour core, at the tumour-stroma interface and within the peritumoural stroma. Additional morphology markers were used to identify regions with high versus low T cell density (identified by CD3e) and high versus low hypoxia (as determined by CA-IX staining). Morphology marker staining, ROI placement and segmentation strategy is illustrated for one sample in Figures 1A, B. Comparison of keratin gene expression within the tumour and stromal compartments (Figure 1C), confirmed the efficacy of the segmentation approach.
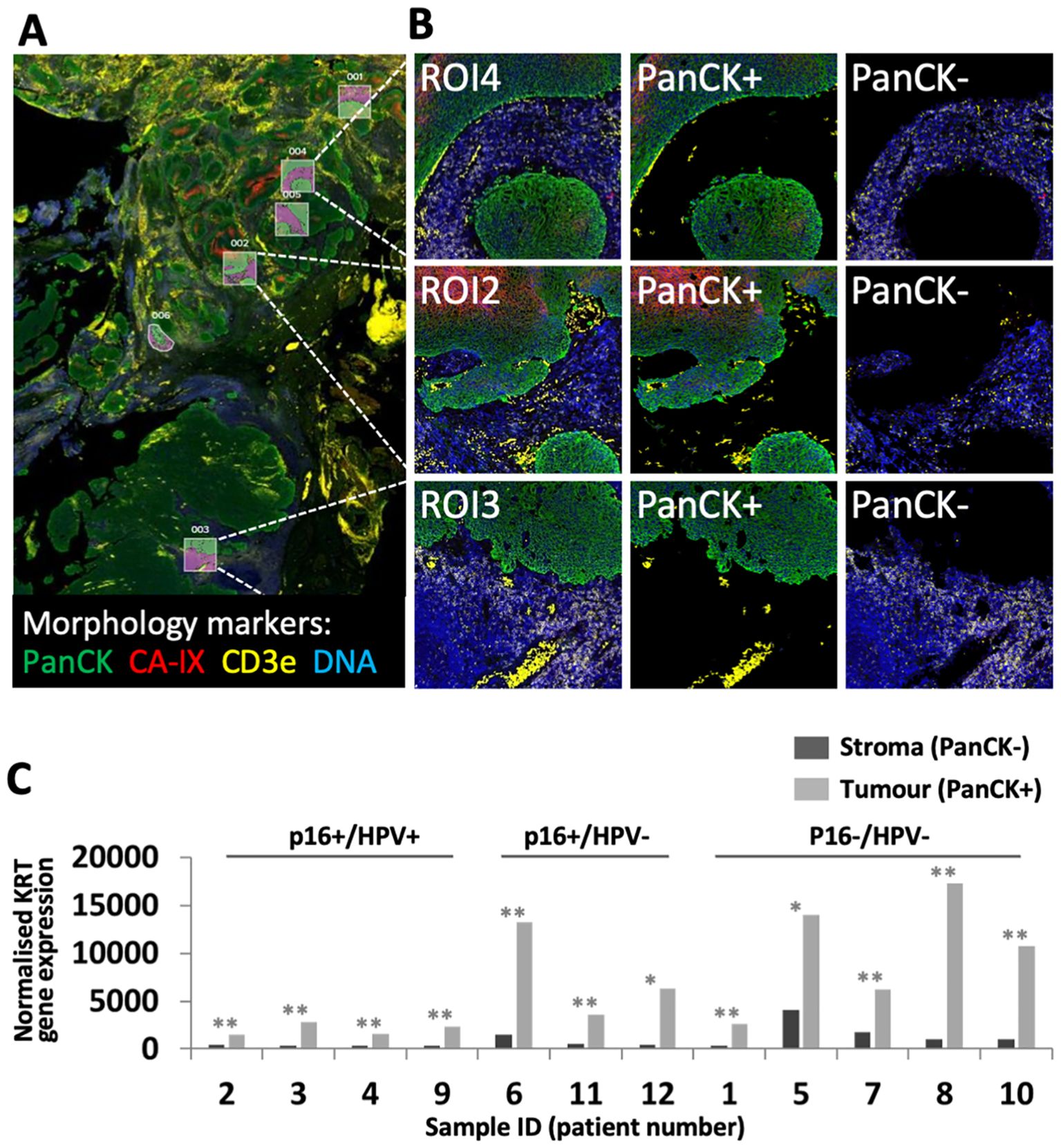
Figure 1. GeoMx DSP ROI selection and segmentation approach. (A) Tumour sections were stained with three morphology markers: PanCK (tumour; green), CD3e (T cell marker; yellow), CA-IX (surrogate marker for regions of hypoxia; red), plus a DNA stain to identify all cells (blue). ROIs were placed to identify regions within tumour nests, tumour-stroma interface and peri-tumoural stroma. In this illustrative example from patient 11, ROIs were placed to capture tumour and adjacent stroma in regions of high (ROIs 1, 2, 4, 5) and low (ROIs 3, 6) hypoxia. (B) Segmentation strategy – PanCK staining was used for identification of tumour (PanCK+) and stromal regions (PanCK-), enabling separate analysis. (C) Comparison of keratin gene expression between stromal and tumour compartments. Graph shows combined normalised KRT gene expression for all PanCK- versus PanCK+ segments within each of the 12 patient samples. P values for comparison of PanCK- versus PanCK+ were calculated using unpaired t test, adjusted p values reported as * <0.05, ** <0.005.
Following data QC and normalisation, genes differentially expressed between the three p16/HPV subgroups were identified. Heatmaps illustrating significantly differentially expressed genes in each subgroup relative to the other two subgroups are shown in Figures 2A, B, for the tumour and stromal compartments respectively. Pairwise comparisons of expression of all immuno-oncology-related genes between the three different p16/HPV subgroups (p16+/HPV+ versus p16-/HPV-, p16+/HPV+ versus p16+/HPV- and p16+/HPV- versus p16-/HPV-) are illustrated in Figure 2C and listed in Supplementary Table 1. Genes whose expression is significantly down-regulated within the tumour compartment of p16+/HPV+ versus p16+/HPV- and/or p16-/HPV- tumours include keratins (KRT), HIF1A, CD44, CCND1, AKT1 and CD276 (Figure 2C top left and centre panels). The expression levels of HIF1A, CCND1 and CD276 are also downregulated in the stromal compartment when comparing the p16+/HPV+ group versus the p16-/HPV- group (Figure 2C bottom left panel). Other genes displaying reduced expression in the stromal compartment include PTEN, ITGAV, ITGB8 and PDCD1 (Figure 2C left and centre panels). Multiple genes are up-regulated within the tumour and stromal compartments of p16+/HPV+ tumours relative to p16+/HPV- and p16-/HPV- (Figure 2C left and centre panels), including several whose function relates to anti-tumoural immune responses (CXCL9, CXCL10, CCL5, STAT1, ITGB2, PSMB10, B2M, CD74 and HLA-DRB). Of note, in these pairwise comparisons only one gene (CXCL9) showed significantly different expression between p16+/HPV- and p16-/HPV- tumours, being overexpressed in the stromal compartment of p16+/HPV- OPC (Figure 2C right hand panels). Three additional genes, CCND1, AKT1 and CD44 displayed a trend for reduced expression in the tumoural compartment of p16+/HPV- OPCs relative to p16-/HPV- (adjusted p values <0.06) see Supplementary Table 1.
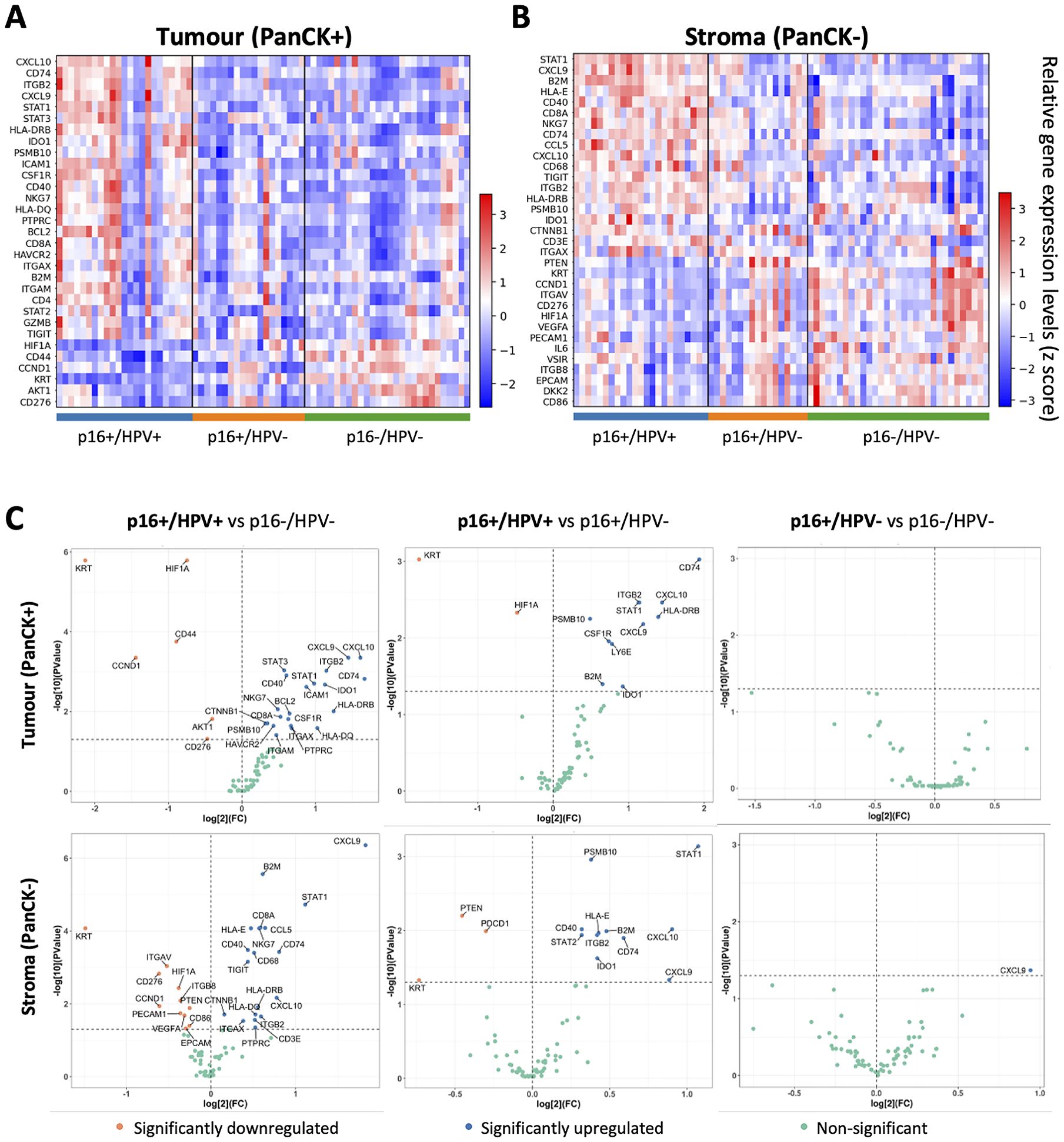
Figure 2. Identification of genes differentially expressed between p16+/HPV+, p16+/HPV- and p16-/HPV- oropharyngeal tumours. (A, B). Heatmaps showing relative gene expression levels for significantly differentially expressed genes across p16/HPV subgroups within the tumour and stromal compartments respectively. Gene names are indicated for rows, ordered by the “scores” generated from the Wilcoxon rank-sum test statistics. Each column represents an individual ROI (p16+/HPV+: n = 46; p16+/HPV-: n = 36; p16-/HPV-: n = 59. (C) Volcano plots showing pairwise comparisons of gene expression between the three groups for the tumour (top) and stromal (bottom) compartments. The p16/HPV subgroup listed first is the primary group, while the subgroup listed second is the reference group. Significantly down regulated genes in the primary group are shown in pink, significantly upregulated genes in blue and non-significantly altered genes in green. P values for pair-wise comparison between any two groups were calculated using the Wilcoxon rank-sum test and adjusted by the Benjamini-Hochberg method across all genes.
Differential expression of selected genes (those showing the most significant differences in the pairwise comparisons) between all three groups is presented in Figure 3. The violin plots illustrate both inter-group and within-group differences in gene expression. In respect of inter-group differences, Figure 3 again highlights the similarity between the p16+/HPV- and p16-/HPV- groups and differential expression relative to the p16+/HPV+ group. With respect to the within-group distribution, some genes have a narrow expression range (for example, KRT, HIF1A in both tumour and stromal compartments of p16+/HPV+ tumours, and PSMB10, B2M in the stromal compartment for all p16/HPV subgroups). Conversely, many genes have broad, often bi-/multimodal distribution (for example CD44, CCND1 and CD74 in tumour and STAT1 in both compartments for all p16/HPV subgroups), potentially reflecting spatial context – tumour core versus tumour periphery or tumour-adjacent versus more distant stroma.
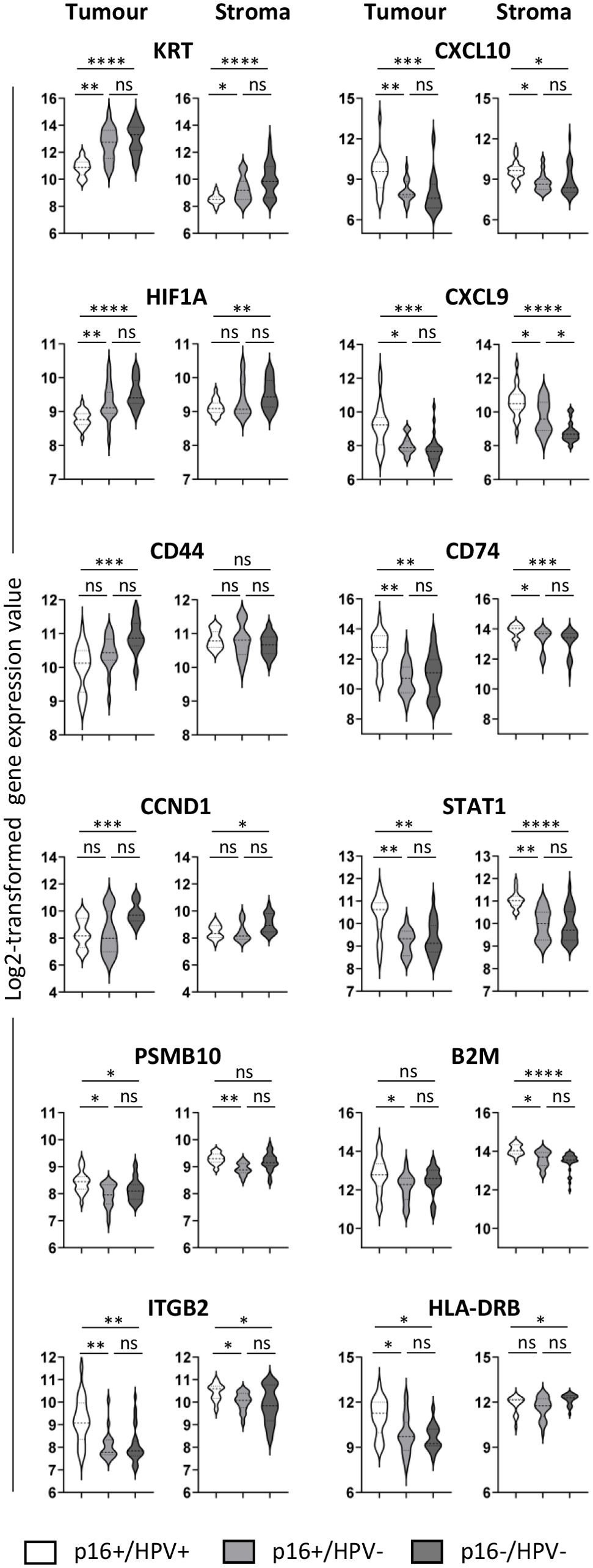
Figure 3. Expression of selected immuno-oncology-related genes within the three p16/HPV subgroups. Violin plots showing log2-transformed normalised gene expression for p16+/HPV+ (white), p16+/HPV- (pale grey) and p16-/HPV- (dark grey) OPC samples. First and third columns represent tumour (PanCK+), second and fourth columns represent stroma (PanCK-). Dashed and dotted lines represent the median and quartiles respectively. Data were analysed using Kruskal–Wallis test, and pair-wise comparisons were conducted with the Wilcoxon rank-sum test. Adjusted P values (see Supplementary Table 1) are reported as: ns, nonsignificant; *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0001.
Discussion
Spatially resolved analysis of gene (or protein) expression within distinct tumour regions offers valuable insights for unravelling tumour composition and microenvironmental phenotypes (23, 24). Here, we employed GeoMx DSP to partition OPC tissues into tumour (PanCK+) and stromal (PanCK-) compartments and quantitatively assessed gene expression within each segmented region. This approach provides valuable information that is lost in bulk RNA-sequencing. Previous studies in HNC have successfully utilised this platform to analyse the tumour and microenvironment in patients with recurrent/metastatic HNC treated with immune checkpoint inhibitors (25–27). Here, we analysed expression of immuno-oncology-related genes in whole tissue sections from locally-advanced OPC patients treated with chemoradiotherapy +/- surgery in the curative setting, in relation to their p16 and HPV status. Overall, p16+/HPV- and p16-/HPV- tumours showed highly similar gene expression profiles – in both tumour and stromal compartments – which are quite distinct from the pattern of gene expression displayed by p16+/HPV+ tumours.
The ‘gene family’ displaying most consistent differential expression between the three groups was KRT, being overexpressed in both p16-/HPV- and p16+/HPV- tumours relative to p16+/HPV+ tumours. This is consistent with histologic classification of OPC, whereby HPV-related tumours are described as non-keratinising (28, 29). Elevated keratin expression, particularly KRT17, is associated with poor prognosis and decreased survival in multiple cancer types (30) including OPC (31), potentially due to inhibition of T cell infiltration (32). Previous studies in HNC link cytokeratin upregulation (including KRT17) with HPV-negative status (33) or an ‘immune low’ subset of HPV-positive tumours (34). In these studies, HPV status was assigned based on detection of HPV DNA/RNA/gene signature and the potential effects of discordant p16/HPV status were not explored. The differential expression of KRT (albeit at very low levels) within the stromal compartment of OPCs mirrors expression patterns in the tumoural compartment, probably reflecting imperfect tumour-stroma segmentation – especially where tumours are discontinuous.
HIF1A gene expression is upregulated in the tumour compartment of both p16+/HPV- and p16-/HPV- OPC relative to p16+/HPV+, and also in the stromal compartment of p16-/HPV-. HIF-1α comprises one subunit of the heterodimeric HIF1 protein, a key transcriptional regulator of cellular adaptation to low oxygen levels. While control of HIF-1α expression is mostly achieved through post-translational regulation (reduced degradation leading to protein accumulation under hypoxic conditions), transcriptional regulation also plays a role (35, 36). High tumoural hypoxia is associated with poor prognosis (37, 38), mediated by multiple factors including radiotherapy/chemotherapy resistance, heightened immunosuppressive nature of the tumour microenvironment, and increased epithelial-to-mesenchymal transition facilitating metastasis (39, 40). Although the HPV E7 oncogene has been shown to upregulate HIF1A expression (41), previous literature associates highest HIF1A expression with HPV-negative status (33). Here, we identified that this association extends to include p16+/HPV- discordant tumours and not just classical p16-/HPV-, a finding that could have important implications for future treatment strategies.
CCND1, AKT1 and CD44 were all expressed at significantly higher levels in the tumour compartment of p16-/HPV- relative to p16+/HPV+ OPC, with CCND1 also being upregulated in the stromal compartment. CCND1 (Cyclin D1) gene amplification, a relatively frequent event in head and neck cancer, has been associated with HPV-negative status and poor survival (33, 42). Mechanistically, this is linked to enhanced tumour cell proliferation and invasion, with recent evidence supporting a role for stromal expression in promoting inflammation and immunosuppression (43). CD44 has been widely implicated as a marker of cancer stem cells in multiple cancer types including OPC (44). High expression is associated with treatment resistance, epithelial-to-mesenchymal transition, and poor survival in most studies (45). Higher expression of CCND1 and CD44 in p16-/HPV- tumours relative to p16+/HPV+ aligns with their poorer prognosis. The trend for reduced expression of all three genes in discordant p16+/HPV- tumours compared with p16-/HPV- is consistent with intermediate prognosis (20).
Phenotypic and transcriptomic studies have characterised HPV-positive tumours as ‘immune hot’ (46–48), with greater numbers of peritumoural immune cells and increased intratumoural T cell infiltration relative to HPV-negative tumours. Many of the genes upregulated in p16+/HPV+ tumours (tumoural and stromal compartments) have potential roles in anti-tumoural immunity. These include, for example, key chemokines controlling T cell trafficking (CXCL9, CXCL10), adhesion molecules involved in T cell recruitment (ITGB2) and components of antigen processing and presentation pathways for CD4 or CD8 T cell recognition (B2M, PBSM10, CD74, HLA-DRB). STAT1, a key transcription factor shaping the tumour microenvironment, is also upregulated in both the tumour and stromal compartments of p16+/HPV+ tumours. Although STAT1 expression is essential for effective antitumor T cell responses (49, 50), tumour cell-specific expression may have negative consequences (50, 51). Of note, expression of anti-tumour immune response-related genes is down-regulated in both p16+/HPV- and HPV-/p16- tumours, consistent with the poorer survival outcomes of both these subgroups relative to p16+/HPV+. CXCL9 upregulation in the stromal compartment of p16+/HPV- tumours relative to p16-/HPV- is suggestive of subtle amelioration of immune responses consistent with intermediate prognosis.
Our study has several limitations. Firstly, the sample size is small, reflecting the capacity of the GeoMx DSP platform which provides in depth analysis rather than high throughput screening. Secondly, the cohort does not include any examples of the second discordant group (p16-/HPV+). This subgroup is much less clinically relevant as it is smaller and also not misdiagnosed (i.e. treated as p16+/HPV+, unlike the p16+/HPV- discordant subgroup) if p16 IHC is used on its own. Thirdly, we here present analysis of 73 immuno-oncology-related genes; GeoMx DSP technology has now progressed to enable whole transcriptome analysis, which we are currently employing in an extended cohort to provide further information on mechanistic differences. For example, this might further define an intermediate microenvironmental p16+/HPV- discordant phenotype, as hinted by differential CXCL9 expression, which aligns with the observed intermediate clinical outcome.
In summary, our study demonstrates that, in terms of immuno-oncology-related gene expression, p16+/HPV- OPC are much more closely aligned with p16-/HPV- than p16+/HPV+, although some subtle differences exist between p16+/HPV- and p16-/HPV- tumours. This is consistent with prognostic patterns described in a large pivotal study, p16+/HPV+ >> p16+/HPV- > p16-/HPV- and underlines the need for dual testing to support informed clinical decision making.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by North West - Preston Research Ethics Committee (Reference: 16/NW/0265). The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from previous clinical trials/studies where participants gave their informed consent for samples to be used in future ethically-approved research. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
JB: Conceptualization, Funding acquisition, Investigation, Visualization, Writing – original draft, Writing – review & editing. YZ: Data curation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. KH: Conceptualization, Data curation, Funding acquisition, Investigation, Writing – review & editing, Writing – original draft. BW: Conceptualization, Funding acquisition, Writing – review & editing, Writing – original draft, Investigation. JD: Resources, Writing – review & editing, Writing – original draft. PN: Resources, Writing – review & editing, Writing – original draft. OG: Resources, Writing – review & editing, Writing – original draft, Conceptualization, Formal analysis. HM: Conceptualization, Funding acquisition, Resources, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by Cancer Research UK (CRUK Centre funding) and National Institute for Health and Care Research (NIHR) UK.
Acknowledgments
A version of this work was published as a preprint on MedRxiv doi: https://doi.org/10.1101/2023.11.17.23298621. The copyright holder for this preprint is the author/funder, who has granted medRxiv a licence to display the preprint in perpetuity.
Conflict of interest
KH works for Propath. HM reports advisory board fees from AstraZeneca, MSD, Merck, Nanobiotix, and Seagen and is Director of Warwickshire Head and Neck Clinic and Docspert Health.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The views expressed in this article are those of the authors and not necessarily those of the NIHR.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1428741/full#supplementary-material
Abbreviations
AJCC, American Joint Committee on Cancer; CCL, chemokine (C-C motif) ligand; CXCL, chemokine (C-X-C motif) ligand; DSP, Digital Spatial Profiling; HIF, hypoxia inducible factor; HPV, Human Papilloma Virus; KRT, keratin; OPC, Oropharyngeal cancer; p16, CDK4/6 inhibitor p16INK4a; ROI, region of interest; TNM, Tumour, Node, Metastasis; UICC, Union for International Cancer Control.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Chaturvedi AK, Anderson WF, Lortet-Tieulent J, Curado MP, Ferlay J, Franceschi S, et al. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. (2013) 31:4550–9. doi: 10.1200/JCO.2013.50.3870
3. Lechner M, Liu J, Masterson L, Fenton TR. HPV-associated oropharyngeal cancer: epidemiology, molecular biology and clinical management. Nat Rev Clin Oncol. (2022) 19:306–27. doi: 10.1038/s41571-022-00603-7
4. Mehanna H, Beech T, Nicholson T, El-Hariry I, McConkey C, Paleri V, et al. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer–systematic review and meta-analysis of trends by time and region. Head Neck. (2013) 35:747–55. doi: 10.1002/hed.22015
5. Damgacioglu H, Sonawane K, Zhu Y, Li R, Balasubramanian BA, Lairson DR, et al. Oropharyngeal cancer incidence and mortality trends in all 50 states in the US, 2001-2017. JAMA Otolaryngol Head Neck Surg. (2022) 148:155–65. doi: 10.1001/jamaoto.2021.3567
6. Tota JE, Best AF, Zumsteg ZS, Gillison ML, Rosenberg PS, Chaturvedi AK. Evolution of the oropharynx cancer epidemic in the United States: moderation of increasing incidence in younger individuals and shift in the burden to older individuals. J Clin Oncol. (2019) 37:1538–46. doi: 10.1200/JCO.19.00370
7. Zhang Y, Fakhry C, D’Souza G. Projected association of human papillomavirus vaccination with oropharynx cancer incidence in the US, 2020-2045. JAMA Oncol. (2021) 7:e212907. doi: 10.1001/jamaoncol.2021.2907
8. Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. New Engl J Med. (2010) 363:24–35. doi: 10.1056/NEJMoa0912217
9. Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. (2023) 41:3081–8. doi: 10.1200/JCO.22.02625
10. Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, et al. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. (2017) 67:93–9. doi: 10.3322/caac.21388
11. Gillison ML, Trotti AM, Harris J, Eisbruch A, Harari PM, Adelstein DJ, et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG Oncology RTOG 1016): a randomised, multicentre, non-inferiority trial. Lancet. (2019) 393:40–50. doi: 10.1016/S0140-6736(18)32779-X
12. Mehanna H, Robinson M, Hartley A, Kong A, Foran B, Fulton-Lieuw T, et al. Radiotherapy plus cisplatin or cetuximab in low-risk human papillomavirus-positive oropharyngeal cancer (De-ESCALaTE HPV): an open-label randomised controlled phase 3 trial. Lancet. (2019) 393:51–60. doi: 10.1016/S0140-6736(18)32752-1
13. Klussmann JP, Gultekin E, Weissenborn SJ, Wieland U, Dries V, Dienes HP, et al. Expression of p16 protein identifies a distinct entity of tonsillar carcinomas associated with human papillomavirus. Am J Pathol. (2003) 162:747–53. doi: 10.1016/S0002-9440(10)63871-0
14. Larsen CG, Gyldenlove M, Jensen DH, Therkildsen MH, Kiss K, Norrild B, et al. Correlation between human papillomavirus and p16 overexpression in oropharyngeal tumours: a systematic review. Brit J Cancer. (2014) 110:1587–94. doi: 10.1038/bjc.2014.42
15. Mena M, Taberna M, Tous S, Marquez S, Clavero O, Quiros B, et al. Double positivity for HPV-DNA/p16 is the biomarker with strongest diagnostic accuracy and prognostic value for human papillomavirus related oropharyngeal cancer patients. Oral Oncol. (2018) 78:137–44. doi: 10.1016/j.oraloncology.2018.01.010
16. Rietbergen MM, Brakenhoff RH, Bloemena E, Witte BI, Snijders PJ, Heideman DA, et al. Human papillomavirus detection and comorbidity: critical issues in selection of patients with oropharyngeal cancer for treatment De-escalation trials. Ann Oncol. (2013) 24:2740–5. doi: 10.1093/annonc/mdt319
17. Shinn JR, Davis SJ, Lang-Kuhs KA, Rohde S, Wang XW, Liu P, et al. Oropharyngeal Squamous Cell Carcinoma With Discordant p16 and HPV mRNA Results Incidence and Characterization in a Large, Contemporary United States Cohort. Am J Surg Pathol. (2021) 45:951–61. doi: 10.1097/Pas.0000000000001685
18. Wagner S, Prigge ES, Wuerdemann N, Reder H, Bushnak A, Sharma SJ, et al. Evaluation of p16 expression as a single marker to select patients with HPV-driven oropharyngeal cancers for treatment de-escalation (vol 123, pg 1114, 2020). Brit J Cancer. (2021) 125:307–7. doi: 10.1038/s41416-021-01457-z
19. Coordes A, Lenz K, Qian X, Lenarz M, Kaufmann AM, Albers AE. Meta-analysis of survival in patients with HNSCC discriminates risk depending on combined HPV and p16 status. Eur Arch Otorhinolaryngol. (2016) 273:2157–69. doi: 10.1007/s00405-015-3728-0
20. Mehanna H, Taberna M, von Buchwald C, Tous S, Brooks J, Mena M, et al. Prognostic implications of p16 and HPV discordance in oropharyngeal cancer (HNCIG-EPIC-OPC): a multicentre, multinational, individual patient data analysis. Lancet Oncol. (2023) 24:239–51. doi: 10.1016/S1470-2045(23)00013-X
21. Mehanna H, Wong WL, McConkey CC, Rahman JK, Robinson M, Hartley AG, et al. PET-CT surveillance versus neck dissection in advanced head and neck cancer. N Engl J Med. (2016) 374:1444–54. doi: 10.1056/NEJMoa1514493
22. Mehanna H, Rapozo D, von Zeidler SV, Harrington KJ, Winter SC, Hartley A, et al. Developing and validating a multivariable prognostic-predictive classifier for treatment escalation of oropharyngeal squamous cell carcinoma: the PREDICTR-OPC study. Clin Cancer Res. (2024) 30(2):356–67. doi: 10.1158/1078-0432.CCR-23-1013
23. Merritt CR, Ong GT, Church SE, Barker K, Danaher P, Geiss G, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol. (2020) 38:586–99. doi: 10.1038/s41587-020-0472-9
24. Zheng Y, Carrillo-Perez F, Pizurica M, Heiland DH, Gevaert O. Spatial cellular architecture predicts prognosis in glioblastoma. Nat Commun. (2023) 14:4122. doi: 10.1038/s41467-023-39933-0
25. Kulasinghe A, Taheri T, O'Byrne K, Hughes BGM, Kenny L, Punyadeera C. Highly multiplexed digital spatial profiling of the tumor microenvironment of head and neck squamous cell carcinoma patients. Front Oncol. (2020) 10:607349. doi: 10.3389/fonc.2020.607349
26. Gavrielatou N, Vathiotis I, Aung TN, Shafi S, Burela S, Fernandez AI, et al. Digital spatial profiling links beta-2-microglobulin expression with immune checkpoint blockade outcomes in head and neck squamous cell carcinoma. Cancer Res Commun. (2023) 3:558–63. doi: 10.1158/2767-9764.CRC-22-0299
27. Sadeghirad H, Liu N, Monkman J, Ma N, Ben Cheikh B, Jhaveri N, et al. Compartmentalized spatial profiling of the tumor microenvironment in head and neck squamous cell carcinoma identifies immune checkpoint molecules and tumor necrosis factor receptor superfamily members as biomarkers of response to immunotherapy. Front Immunol. (2023) 14:1135489. doi: 10.3389/fimmu.2023.1135489
28. El-Mofty SK, Patil S. Human papillomavirus (HPV)-related oropharyngeal nonkeratinizing squamous cell carcinoma: Characterization of a distinct phenotype. Oral Surg Oral Med O. (2006) 101:339–45. doi: 10.1016/j.tripleo.2005.08.001
29. Chernock RD, El-Mofty SK, Thorstad WL, Parvin CA, Lewis JS Jr. HPV-related nonkeratinizing squamous cell carcinoma of the oropharynx: utility of microscopic features in predicting patient outcome. Head Neck Pathol. (2009) 3:186–94. doi: 10.1007/s12105-009-0126-1
30. Lozar T, Wang W, Gavrielatou N, Christensen L, Lambert PF, Harari PM, et al. Emerging prognostic and predictive significance of stress keratin 17 in HPV-associated and non HPV-associated human cancers: A scoping review. Viruses. (2023) 15. doi: 10.3390/v15122320
31. Regenbogen E, Mo M, Romeiser J, Shroyer ALW, Escobar-Hoyos LF, Burke S, et al. Elevated expression of keratin 17 in oropharyngeal squamous cell carcinoma is associated with decreased survival. Head Neck-J Sci Spec. (2018) 40:1788–98. doi: 10.1002/hed.25164
32. Wang W, Lozar T, Golfinos AE, Lee DN, Gronski E, Ward-Shaw E, et al. Stress keratin 17 expression in head and neck cancer contributes to immune evasion and resistance to immune-checkpoint blockade. Clin Cancer Res. (2022) 28:2953–68. doi: 10.1158/1078-0432.Ccr-21-3039
33. Keck MK, Zuo Z, Khattri A, Stricker TP, Brown CD, Imanguli M, et al. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res. (2015) 21:870–81. doi: 10.1158/1078-0432.CCR-14-2481
34. Zhang Y, Koneva LA, Virani S, Arthur AE, Virani A, Hall PB, et al. Subtypes of HPV-positive head and neck cancers are associated with HPV characteristics, copy number alterations, PIK3CA mutation, and pathway signatures. Clin Cancer Res. (2016) 22:4735–45. doi: 10.1158/1078-0432.CCR-16-0323
35. Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. (1992) 12:5447–54. doi: 10.1128/mcb.12.12.5447-5454.1992
36. Belaiba RS, Bonello S, Zahringer C, Schmidt S, Hess J, Kietzmann T, et al. Hypoxia up-regulates hypoxia-inducible factor-1alpha transcription by involving phosphatidylinositol 3-kinase and nuclear factor kappaB in pulmonary artery smooth muscle cells. Mol Biol Cell. (2007) 18:4691–7. doi: 10.1091/mbc.e07-04-0391
37. Aebersold DM, Burri P, Beer KT, Laissue J, Djonov V, Greiner RH, et al. Expression of hypoxia-inducible factor-1alpha: a novel predictive and prognostic parameter in the radiotherapy of oropharyngeal cancer. Cancer Res. (2001) 61:2911–6.
38. Nordsmark M, Bentzen SM, Rudat V, Brizel D, Lartigau E, Stadler P, et al. Prognostic value of tumor oxygenation in 397 head and neck tumors after primary radiation therapy. An international multi-center study. Radiother Oncol. (2005) 77:18–24. doi: 10.1016/j.radonc.2005.06.038
39. Hill RM, Rocha S, Parsons JL. Overcoming the impact of hypoxia in driving radiotherapy resistance in head and neck squamous cell carcinoma. Cancers (Basel). (2022) 14. doi: 10.3390/cancers14174130
40. Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. (2020) 6:92. doi: 10.1038/s41572-020-00224-3
41. Bodily JM, Mehta KP, Laimins LA. Human papillomavirus E7 enhances hypoxia-inducible factor 1-mediated transcription by inhibiting binding of histone deacetylases. Cancer Res. (2011) 71:1187–95. doi: 10.1158/0008-5472.CAN-10-2626
42. Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. (2015) 517:576–82. doi: 10.1038/nature14129
43. Montalto FI, De Amicis F. Cyclin D1 in cancer: A molecular connection for cell cycle control, adhesion and invasion in tumor and stroma. Cells. (2020) 9. doi: 10.3390/cells9122648
44. Mesrati MH, Syafruddin SE, Mohtar MA, Syahir A. CD44: A multifunctional mediator of cancer progression. Biomolecules. (2021) 11:1850. doi: 10.3390/biom11121850
45. Chen JQ, Zhou JD, Lu J, Xiong H, Shi XL, Gong L. Significance of CD44 expression in head and neck cancer: a systemic review and meta-analysis. BMC Cancer. (2014) 14:15. doi: 10.1186/1471-2407-14-15
46. Partlova S, Boucek J, Kloudova K, Lukesova E, Zabrodsky M, Grega M, et al. Distinct patterns of intratumoral immune cell infiltrates in patients with HPV-associated compared to non-virally induced head and neck squamous cell carcinoma. Oncoimmunology. (2015) 4:e965570. doi: 10.4161/21624011.2014.965570
47. Mandal R, Senbabaoglu Y, Desrichard A, Havel JJ, Dalin MG, Riaz N, et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight. (2016) 1:e89829. doi: 10.1172/jci.insight.89829
48. Brooks JM, Menezes AN, Ibrahim M, Archer L, Lal N, Bagnall CJ, et al. Development and validation of a combined hypoxia and immune prognostic classifier for head and neck cancer. Clin Cancer Res. (2019) 25:5315–28. doi: 10.1158/1078-0432.CCR-18-3314
49. Ryan N, Anderson K, Volpedo G, Hamza O, Varikuti S, Satoskar AR, et al. STAT1 inhibits T-cell exhaustion and myeloid derived suppressor cell accumulation to promote antitumor immune responses in head and neck squamous cell carcinoma. Int J Cancer. (2020) 146:1717–29. doi: 10.1002/ijc.32781
50. Anderson K, Ryan N, Nedungadi D, Lamenza F, Swingler M, Siddiqui A, et al. STAT1 is regulated by TRIM24 and promotes immunosuppression in head and neck squamous carcinoma cells, but enhances T cell antitumour immunity in the tumour microenvironment. Brit J Cancer. (2022) 127:624–36. doi: 10.1038/s41416-022-01853-z
Keywords: oropharyngeal cancer, Human Papilloma Virus (HPV), p16, dual p16/HPV testing, treatment de-intensification, Digital Spatial Profiling, tumour microenvironment
Citation: Brooks JM, Zheng Y, Hunter K, Willcox BE, Dunn J, Nankivell P, Gevaert O and Mehanna H (2024) Digital Spatial Profiling identifies distinct patterns of immuno-oncology-related gene expression within oropharyngeal tumours in relation to HPV and p16 status. Front. Oncol. 14:1428741. doi: 10.3389/fonc.2024.1428741
Received: 06 May 2024; Accepted: 08 August 2024;
Published: 12 September 2024.
Edited by:
Natalia Isaeva, University of North Carolina at Chapel Hill, United StatesReviewed by:
Bruce Ashford, Illawarra Shoalhaven Local Health District (ISLHD), AustraliaArun Khattri, Indian Institute of Technology (BHU), India
Ivan Sabol, Rudjer Boskovic Institute, Croatia
Copyright © 2024 Brooks, Zheng, Hunter, Willcox, Dunn, Nankivell, Gevaert and Mehanna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hisham Mehanna, aC5tZWhhbm5hQGJoYW0uYWMudWs=
†These authors share first authorship
‡These authors share senior authorship
 Jill M. Brooks
Jill M. Brooks Yuanning Zheng
Yuanning Zheng Kelly Hunter
Kelly Hunter Benjamin E. Willcox
Benjamin E. Willcox Janet Dunn5
Janet Dunn5 Paul Nankivell
Paul Nankivell Olivier Gevaert
Olivier Gevaert Hisham Mehanna
Hisham Mehanna