- 1Biofidelity Inc., Morrisville, NC, United States
- 2Biofidelity Ltd., Cambridge, United Kingdom
Introduction: Genomic variant testing of tumors is a critical gateway for patients to access the full potential of personalized oncology therapeutics. Current methods such as next-generation sequencing are costly and challenging to interpret, while PCR assays are limited in the number of variants they can cover. We developed ASPYRE® (Allele-Specific PYrophosphorolysis REaction) technology to address the urgent need for rapid, accessible and affordable diagnostics informing actionable genomic target variants of a given cancer. The targeted ASPYRE-Lung panel for non-small cell carcinoma covers 114 variants in 11 genes (ALK, BRAF, EGFR, ERBB2, KRAS, RET, ROS1, MET & NTRK1/2/3) to robustly inform clinical management. The assay detects single nucleotide variants, insertions, deletions, and gene fusions from tissue-derived DNA and RNA simultaneously.
Methods: We tested the limit of detection, specificity, analytical accuracy and analytical precision of ASPYRE-Lung using FFPE lung tissue samples from patients with non-small cell lung carcinoma, variant-negative FFPE tissue from healthy donors, and FFPE-based contrived samples with controllable variant allele fractions.
Results: The sensitivity of ASPYRE-Lung was determined to be ≤ 3% variant allele fraction for single nucleotide variants and insertions or deletions, 100 copies for fusions, and 200 copies for MET exon 14 skipping. The specificity was 100% with no false positive results. The analytical accuracy test yielded no discordant calls between ASPYRE-Lung and expected results for clinical samples (via orthogonal testing) or contrived samples, and results were replicable across operators, reagent lots, runs, and real-time PCR instruments with a high degree of precision.
Conclusions: The technology is simple and fast, requiring only four reagent transfer steps using standard laboratory equipment (PCR and qPCR instruments) with analysis via a cloud-based algorithm. The ASPYRE-Lung assay has the potential to be transformative in facilitating access to rapid, actionable molecular profiling of tissue for patients with non-small cell carcinoma.
Introduction
Worldwide, over 2 million people are diagnosed with lung cancer annually, which has the highest mortality rate of any cancer (1, 2). Non-small cell lung carcinoma (NSCLC) has a five-year survival rate of just 16% when patients with metastatic NSCLC are treated with chemotherapy alone (3). Historically, standard of care treatment for NSCLC included platinum-based cytotoxic chemotherapy. Prognosis has significantly improved following the emergence of targeted therapies, which typically inactivate oncogenic growth factors and their receptors or inhibit oncogenic tyrosine kinase pathways (4, 5). In addition to higher therapeutic success rates, targeted therapies are often better tolerated, with reduced side effects in patients thus improving quality of life (6). There are now over 30 FDA-approved targeted therapies for NSCLC (7), each targeting specific drivers of this disease, making treatment personalized to the genomic variants of a patient’s tumor.
In order to identify patients who are most likely to benefit from specific targeted therapeutics, tools that enable the detection of multiple variants from a single small quantity patient sample are required (6, 8). Small core needle-biopsies yielding limited material are becoming increasingly common, and repeated invasive specimen collection from patients for comprehensive genomic testing is not feasible for many reasons (including safety and access to tissue) (8, 9). PCR and fluorescence in-situ hybridization are commonly utilized diagnostic tools. However, both methods test only a limited number of mutations, quickly leading to sample exhaustion, and thus limiting the opportunity to identify the most appropriate targeted therapeutic option. Conversely, next-generation sequencing (NGS) can analyze multiple variations from a single sample but is a costly diagnostic tool, with complex laboratory processes, a suboptimal turnaround time (TAT), and the need for extensive bioinformatics analysis, which can be difficult to interpret even in a comprehensive cancer care setting (10). Moreover, as only a small subset of the mutations detected by NGS are clinically actionable, detection of such a broad panel (including variants of unknown significance) lends little benefit and added complexity to clinical decision making. While patients wait for diagnostic results from molecular testing, non-targeted standard cytotoxic chemotherapies are frequently administered to mitigate tumor progression and address patient safety, which can compromise outcomes even if a prelude to targeted therapy (6). Similarly, guidelines indicate that appropriate targeted therapy should take precedence over treatment with an immune checkpoint inhibitor as concurrent or sequential immunotherapy and targeted therapy can lead to toxicity (11). Overall, fewer than 50% of patients with NSCLC receive appropriate therapy due to either not being tested, not receiving molecular variant testing results in a timely manner, or not receiving appropriate treatment (12, 13). Life-saving targeted therapies thus remain underutilized. Taken together, it is clear that access to rapid, robust actionable genomic testing is critical in enabling patients to access the appropriate treatment in a timely manner.
To address this genomic testing gap, we have recently described ASPYRE, a new technology for the detection of DNA variants and RNA fusions (14, 15). Previously, we have shown that ASPYRE technology is highly sensitive, allowing the detection of specific DNA sequences to single-molecule level (14) and RNA fusion detection of under six copies (15), alongside high specificity and robustness against interference from carryover contaminants from formalin fixed paraffin embedded (FFPE) samples (14, 15).
The ASPYRE assay comprises four sequential enzymatic stages with reagent transfer between each stage (Figure 1). Briefly, mutant and wild-type molecules are amplified by PCR (or RT-PCR), and the remaining DNA polymerase is enzymatically digested. Amplicons are made single-stranded via exonuclease digestion. Probes that perfectly match target variants hybridize to mutant targets and are subject to pyrophosphorolysis using a DNA polymerase with no exonuclease activity. Conversely, probes that hybridize to wild-type targets are mismatched, and the mismatch prevents pyrophosphorolysis past the variant site. Probes that have been subject to pyrophosphorolysis beyond the variant site are circularized by ligation and amplified by an isothermal hyper-branched rolling-circle amplification reaction that is multiplexed to detect amplification in four fluorescence channels. The ASPYRE assay is run on thermal cyclers and quantitative real-time PCR (qPCR) instruments, which are readily available in most clinical laboratories.
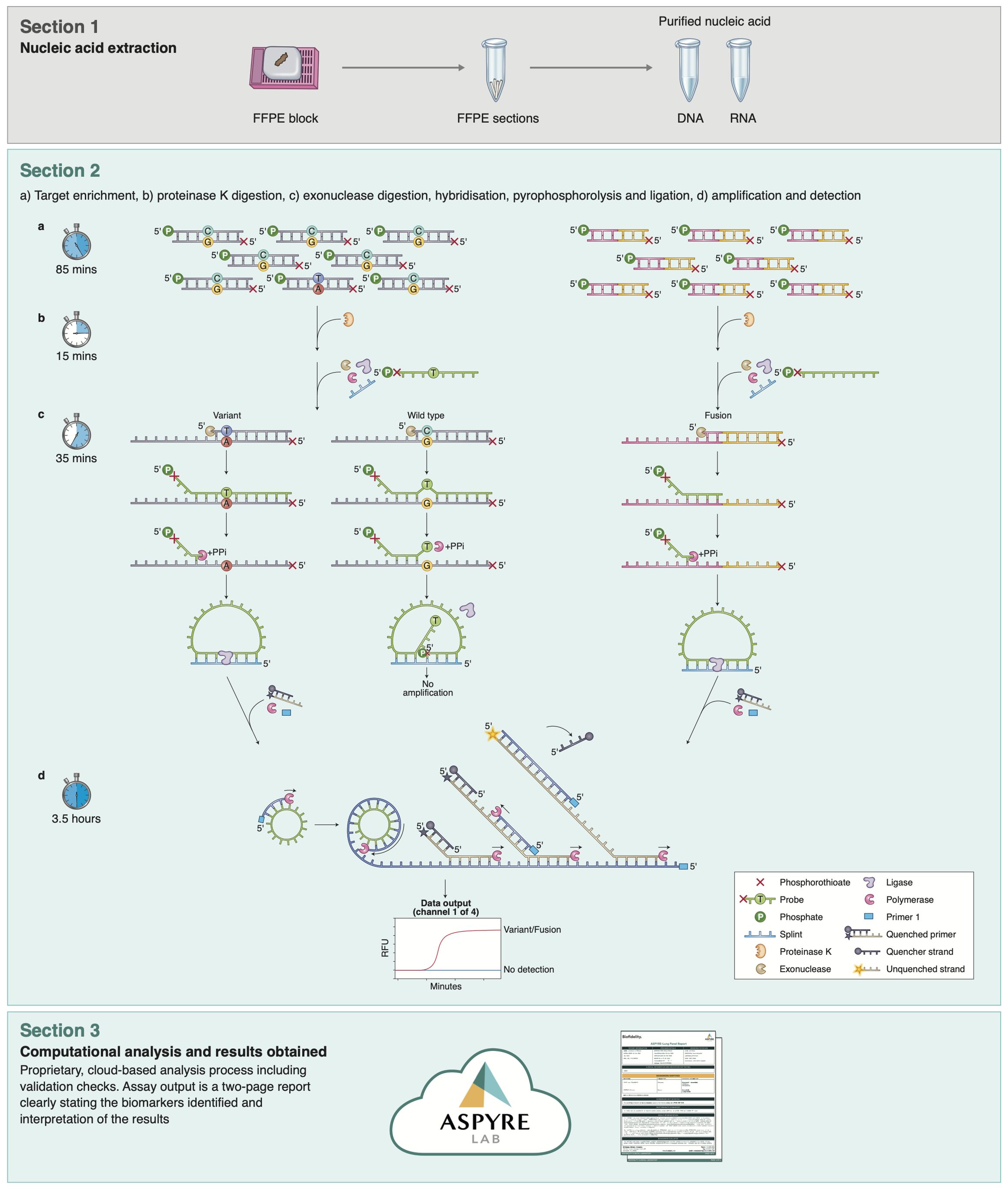
Figure 1. Parallelized workflow of the ASPYRE-Lung targeted mutation panel. Workflow schematic, showing the four separate steps of the assay for DNA (Section 2, left) and RNA (Section 2, right). While steps shown in section 2 differ technically between DNA and RNA, there is no difference to the user in handling the assay, facilitating ease of use.
Herein we describe the ASPYRE-Lung Tissue assay and its performance in our Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory. ASPYRE-Lung enables the concurrent detection of 77 DNA variants, 36 RNA fusions and MET exon 14 skipping (Supplementary Table S1). Importantly, the variants detected within the assay are clinically actionable and recommended by the National Comprehensive Cancer Network (NCCN), College of American Pathologists (CAP), Association for Molecular Pathology (AMP) and European Society of Medical Oncology (ESMO) advanced NSCLC treatment guidelines. The assay uses DNA and RNA extracted from FFPE tissue samples. The TAT from sample extraction to analysis can be completed within two days. A single patient sample is analyzed in a total of 24 wells, allowing 16 patient samples to be run on a 384-well instrument.
We assessed the performance of the ASPYRE-Lung assay across a range of parameters using both contrived samples and FFPE patient lung tissue. The assay limit of detection (LoD) was determined using serially diluted contrived DNA and RNA samples, and assay specificity (Limit of Blank, LoB) was established using FFPE variant-negative tonsil tissue samples from patients without a known cancer diagnosis. Analytical accuracy was assessed by determining variant calls from FFPE NSCLC lung tumor tissue samples, and analytical precision assessed using FFPE NSCLC lung tissue and variant-negative FFPE tissue. Additionally, common exogenous contaminants (potential interfering substances) from nucleic acid extraction were added to DNA and RNA samples to determine the effect on assay performance. Overall, this study successfully validated the performance of the ASPYRE-Lung assay on FFPE lung tissue samples in our CLIA-certified laboratory. Importantly, ASPYRE-Lung will address the gap clinicians face in obtaining sensitive and actionable mutation testing for patients with NSCLC, at a lower cost, faster TAT, and with minimal sample requirements.
Results
The ASPYRE-Lung assay analyzes DNA and RNA derived from tissue
The ASPYRE-Lung assay assesses the status of 114 actionable variants across 11 genes from paired DNA (20 ng) and RNA (6 ng) derived from FFPE lung tissue in a single workflow. Multiplexed targeted PCR amplification of nine exons of DNA (KRAS exons 2 & 3, BRAF exon 15, ERBB2 exons 17 & 20, EGFR exons 18, 19, 20, and 21) occurs alongside a separate but parallel RT-PCR reaction to amplify any of 36 RNA fusion targets and one exon skipping event that are present in the sample. The digestion, hybridization, pyrophosphorolysis and isothermal amplification steps are common to both DNA and RNA. The result for each variant detected by the ASPYRE-Lung panel is interpreted from one of four fluorescent channel signals within each of the 20 DNA and 2 RNA wells (these also incorporate a positive control well for DNA and RNA respectively). An additional two wells are used as negative controls for the DNA and RNA detection reactions. The fluorescent output from the qPCR instrument is analyzed through ASPYRELab software to provide variant call results for each sample, including assay and sample quality checks. Combined with patient information, the parsed results are used to generate a clinical report that indicates presence or absence of a specific mutation (e.g. BRAF exon 15 p.V600E) or class of mutation (e.g. ALK fusion) and the potential therapeutic options available, without manual bioinformatic interpretation. An assay schematic is illustrated in Figure 1.
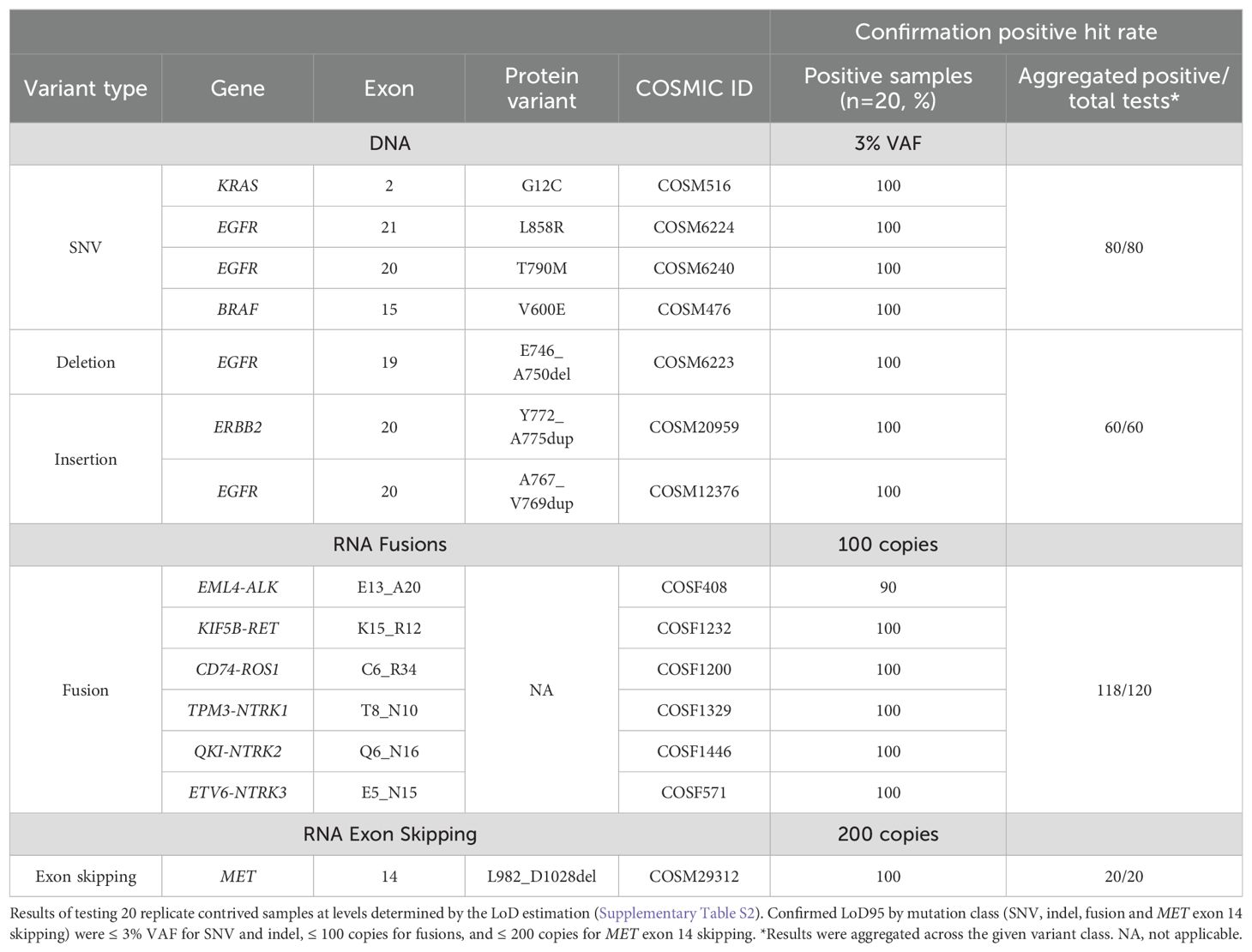
The limit of detection of ASPYRE-Lung is ≤ 3% VAF for SNV and indels, ≤ 100 copies for fusions, and ≤ 200 copies for MET exon 14 skipping
The LoD95 was established as the lowest test level with at least a 95% hit rate. This was determined in two stages: first by estimating the value within a wide range of variant allele fractions (VAFs) or copies for each class-representative variant in the panel (DNA single nucleotide variations (SNV), DNA insertions or deletions (indel), RNA fusions, and MET exon 14 skipping), and second by confirming the estimated value with greater replicate testing power. During the estimation stage all replicates were positive across all SNV and indel samples at 3% VAF, all fusion samples at 100 copies, and the MET exon 14 skipping mutation at 200 copies, although one false negative was observed for a single exon 14 skipping replicate at 400 copies (Supplementary Table S2). The levels chosen for the estimation phase may therefore have been conservative, and the LoD95 likely lies below, or considerably below, these levels.
Secondly, the LoD95 was confirmed by testing 20 replicates of each variant at the estimated LoD95 using a new reagent lot. A minimum of 17/20 (85%) positive results was required for confirmation per variant as per CLSI EP17-A2 specifications with 20 replicates (an upper one-sided 95% confidence limit of 93.8%). At this confirmation stage, 100% of replicates were positive across all SNV and indel samples at 3% VAF, 100% of RET, ROS1 and NTRK1/2/3 fusions were positive at 100 copies, and 100% of the MET exon 14 skipping mutation at 200 copies (Table 1). 90% (18/20) of replicates of the ALK fusion were positive at 100 copies. Therefore, the confirmed LoD95 for SNV and indels was ≤ 3%, for fusions ≤ 100 copies, and for MET exon 14 skipping mutation ≤ 200 copies (Table 1). The LoD of each variant was confirmed at the lowest tested level, which suggests that the true LoD95 likely lies below (and possibly considerably below) 3% VAF for SNVs and indels, 100 copies for fusions, and 200 copies for MET exon 14 skipping.
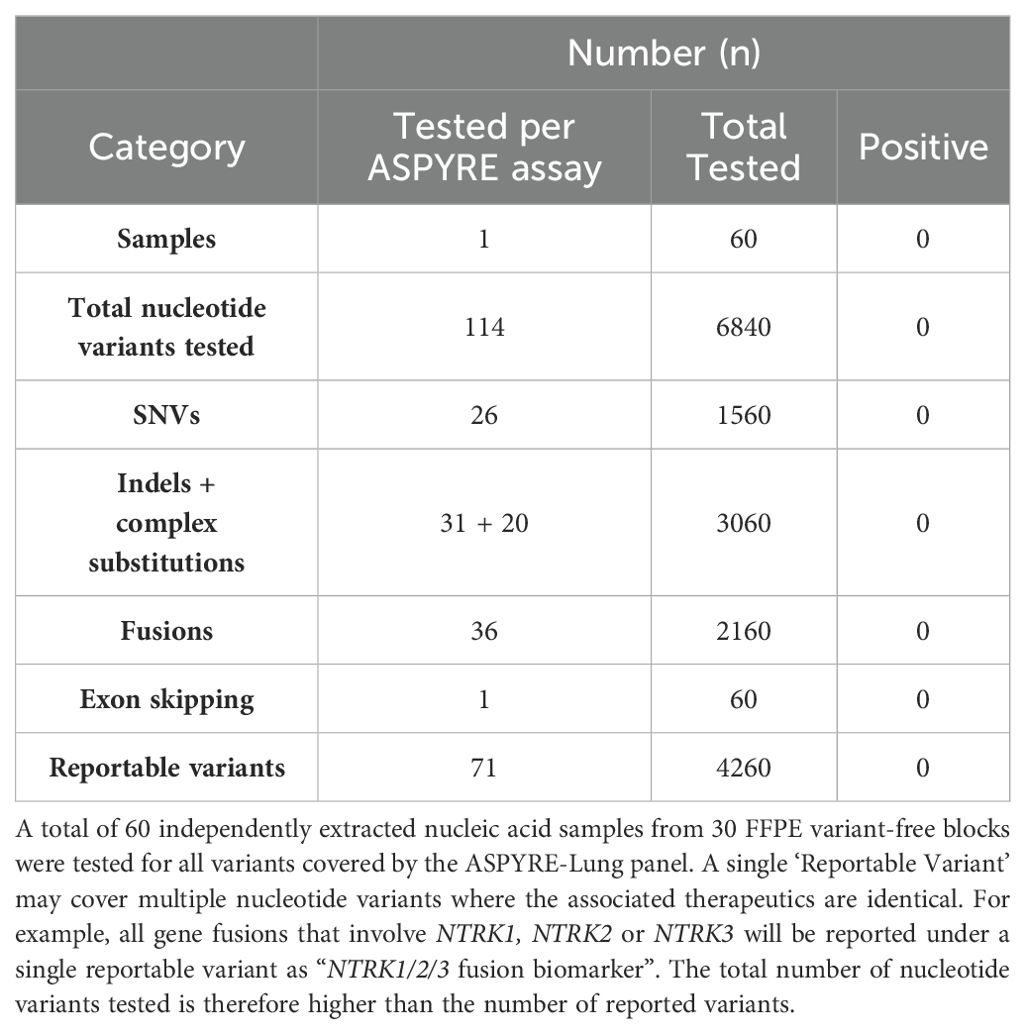
The ASPYRE-Lung assay in FFPE tissue is highly specific
Next, the specificity of the assay was tested using DNA and RNA extracts from 30 FFPE tonsil samples from donors with no history of cancer diagnosis. DNA and RNA from each FFPE sample was extracted, and each nucleic acid tested in duplicate using two different reagent lots for a total of 60 tests. One DNA sample was repeated once to obtain a valid result as the internal positive control failed in the first run. There were no positive calls for any sample at any of the 6840 variants analyzed by ASPYRE-Lung (Table 2); therefore, the false positive rate was 0% (0-6% Clopper-Pearson 95%CI) and the LoB of the test is zero (0-4.1% Jeffreys 95% confidence interval).
ASPYRE-Lung has high analytical accuracy
The analytical accuracy of the ASPYRE-Lung assay was assessed using both contrived specimens (Table 3; Supplementary Table S3) and nucleic acid extracted from 30 FFPE samples from patients with a diagnosis of NSCLC for which targeted enrichment NGS data were available (Table 3; Supplementary Table S4). Each of the contrived samples was tested once at twice the LoD95. Variants were aggregated according to class, and all results were according to expected output with 100% positive percent agreement (PPA) and 100% negative percent agreement (NPA) across all samples (Table 3).
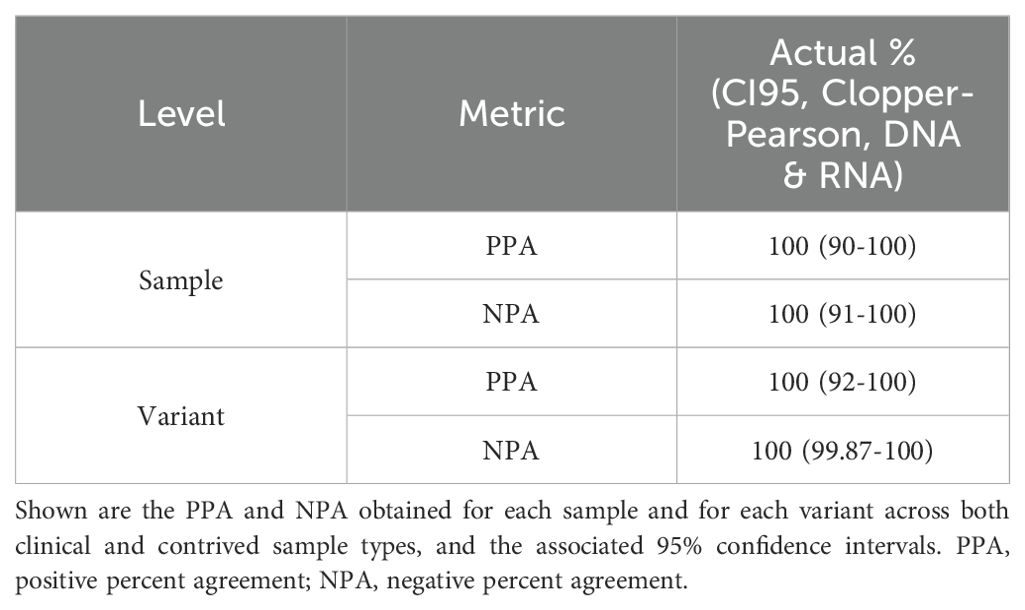
Table 3. Summary of analytical accuracy of ASPYRE-Lung assessed using contrived and clinical samples.
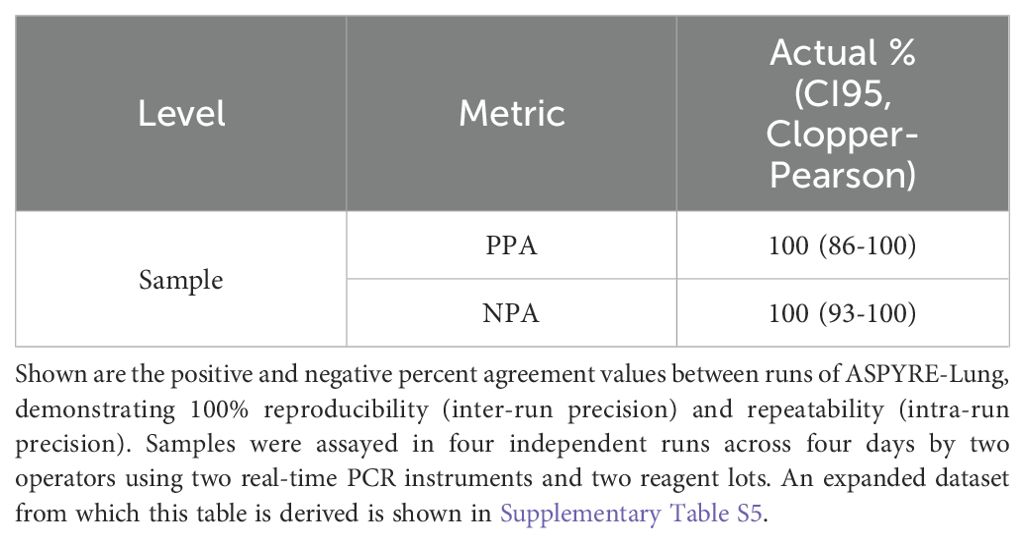
The ASPYRE-Lung assay has excellent analytical precision
The reproducibility and repeatability of the assay were tested using FFPE tissue samples (DNA and RNA from variant-positive and variant-negative patient samples) tested in duplicate across four runs with two instruments by two operators over four days (Table 4). The three FFPE NSCLC variant-positive tissue samples were sequenced by targeted enrichment NGS (Supplementary Table S4). All replicate runs were matched with each other and expected results, giving 100% PPA and NPA, and 100% reproducibility and repeatability.
ASPYRE-Lung is not affected by common interfering substances present in nucleic acid extracts
As ASPYRE-Lung utilizes isolated nucleic acids as sample input, it is potentially susceptible to interfering substances carried over during DNA and RNA extraction. Common substances used in nucleic acid extraction that often impact molecular assays include guanidinium salts and ethanol. We tested whether the performance of the ASPYRE-Lung assay was affected by the presence of carry-over chemicals by spiking two DNA and two RNA samples with guanidinium thiocyanate and ethanol, with five replicates per sample and spike-in condition (Table 5). The ethanol test level was the same as used by the Foundation Medicine FoundationOne CDx FDA submission (16), while guanidine thiocyanate levels were chosen from in-house preliminary testing of levels found in samples with incomplete removal of contaminants. None of the contaminants tested showed any effect on assay results at the levels tested and are therefore considered non-interfering (Table 5).
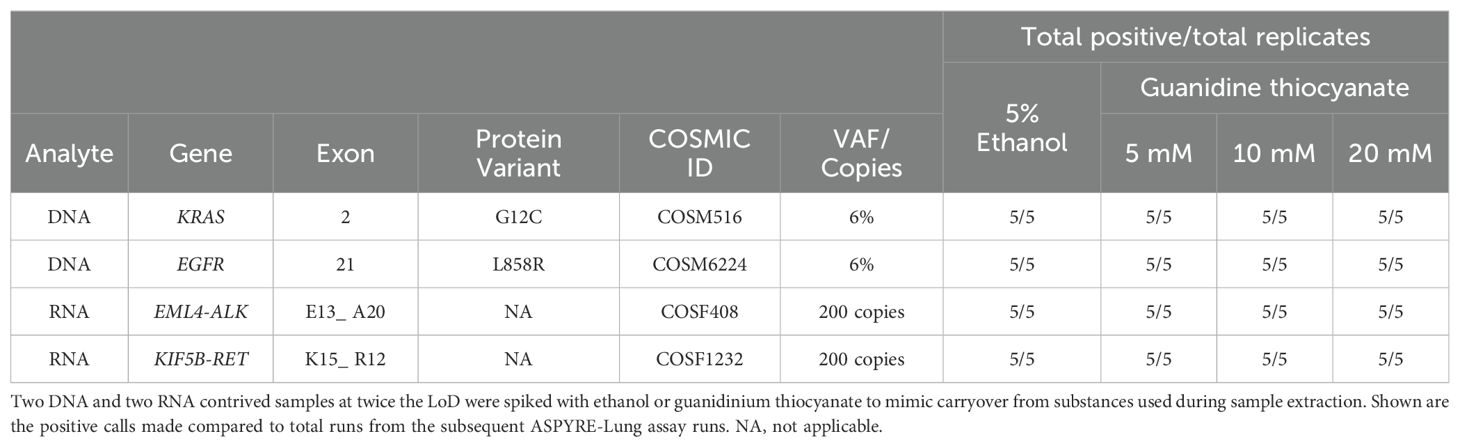
Table 5. Contaminants carried over from FFPE sample extraction do not interfere with the ASPYRE-Lung assay.
Discussion
In an ideal scenario, patients newly diagnosed with NSCLC would receive results from genomic tests that identify all actionable driver mutations within a short timeframe after confirmed diagnosis to access the appropriate targeted therapy as soon as possible (6). While practice guidelines recommend molecular testing at the outset for all actionable biomarkers (17, 18), around 90% of patients are tested for at least one actionable biomarker, only just under half are tested for five or more biomarkers, with many starting chemotherapy before results from biomarker testing are returned (19, 20). Stratification of patients according to the mutation in their tumor can be achieved through a variety of methods, including single-target and multiplex PCR which look for specific alterations, and NGS which captures specific genomic regions and analyzes changes therein. There are advantages and disadvantages to each method. Targeted methods of detection are well established, faster, and more affordable, but can require running multiple sequential assays to capture all possible actionable variants, with the potential for tissue exhaustion and extended laboratory time (21). NGS has a long TAT and yields large amounts of data that require expert bioinformatic interpretation and may generate technically unclear results for some targets (for example, the prediction of gene fusions at the RNA level from DNA sequence in introns) (12, 19, 22).
The characteristics of the ASPYRE-Lung assay make it eminently suitable for all patients with NSCLC to undergo mutational profiling, as it is simple and rapid to perform using standard laboratory techniques and equipment (14, 15). The panel covers the guideline recommended 77 mutations from DNA and 36 gene fusions and MET exon 14 skipping events in RNA. In this study, we have estimated and confirmed an LoD95 of ASPYRE-Lung in FFPE tissue at ≤ 3% VAF for SNV and indels found in DNA, ≤ 100 copies for RNA fusions and ≤ 200 copies for MET exon 14 skipping. The specificity of the assay was 100%, with no false positive results yielded from 60 independent tests of 30 variant-free FFPE tissue samples using two lots of reagents. The accuracy of the assay compared with targeted enrichment NGS was 100% PPA and NPA, with no false negative calls, including for samples with more than one mutation present. Repeated testing of six samples (three positive and three negative) across different days, operators, reagent lots, instruments and assay runs gave 100% consistent results. Finally, spiking samples with potential interfering substances that are commonly found in extraction kits, and can be incompletely removed during pre-analytical procedures, did not affect whether correct calls were made.
ASPYRE-Lung, like any molecular diagnostic assay, has some limitations. The ASPYRE platform is predicated on the prior identification of target sequences, which constrains ASPYRE-Lung’s ability to detect low-prevalence genomic alterations and novel fusion breakpoints that have not been previously characterized. In such cases, RNA-based next-generation sequencing (NGS) theoretically provides a broader capability to capture these fusion events, whereas DNA-based NGS would also miss these fusion events (22). Moreover, ASPYRE-Lung is a qualitative assay and does not quantify tumor VAF. Although VAF can provide additional insights into tumor biology, current clinical guidelines do not endorse its use in therapeutic decision-making. Another potential limitation is that ASPYRE-Lung does not measure tumor mutational burden (TMB), a biomarker that has recently been excluded from lung cancer guidelines by the NCCN, as well as from joint guidelines issued by the College of American Pathologists, Association for Molecular Pathology, International Association for the Study of Lung Cancer, Pulmonary Pathology Society, and LUNGevity Foundation. This exclusion is due to inconsistent evidence regarding the clinical utility of TMB. While initial findings from the CheckMate-227 trial suggested that patients with advanced NSCLC and TMB ≥10 mutations/megabase benefited from first-line treatment with nivolumab plus ipilimumab, subsequent data failed to demonstrate a survival advantage stratified by TMB levels, leading to the withdrawal of the FDA’s supplemental application for TMB as a predictive biomarker in this context (23, 24). Finally, the ASPYRE-Lung assay is designed to target genes with established clinical relevance as per current NSCLC guidelines (18). Consequently, it does not include variants associated with therapeutics approved for indications outside of NSCLC, but which may be prescribed off-label (e.g., FGFR, PIK3CA, NRAS). In clinical scenarios where such variants are of interest, ASPYRE-Lung’s rapid turnaround time and cost-effectiveness position it as a practical initial screening tool for guideline-recommended biomarkers, with the potential for reflex testing using comprehensive NGS panels if no actionable variants are detected.
While these results demonstrate excellent assay performance, work is ongoing to expand the remit of the assay by further testing and validating performance characteristics on a wide range of samples to address unmet clinical need. Lung tissue biopsies can take different forms, including core needle biopsies and samples taken for cytology (pleural effusions, bronchial washings) which do not meet tissue requirements for NGS or sequential PCR testing. Previous work has shown that the ASPYRE assay has robust results down to 1 ng extracted RNA from clinical lung FFPE tissue samples (15), and this work will be updated and expanded to include DNA and a lower range of sample input levels. Further goals include validating the assay and analysis algorithm for circulating nucleic acid extracted from plasma. As new targeted therapies become available and guidelines change, the panel can be expanded to include detection of additional biomarkers, allowing more patients to access the appropriate targeted therapy for which they may best respond.
Conclusions
There are now many targeted therapies available, but biomarker testing remains a crucial roadblock to accessing these highly efficacious and well-tolerated treatments in a timely manner. This is largely due to the limitations of existing genomic testing technologies. To address these limitations, we have developed ASPYRE, a new class of diagnostic technology that leverages a unique series of exquisitely sensitive and specific enzymatic reactions to give the benefits of multi-gene testing yet with rapid TAT, simple bioinformatics, and easily interpretable clinical decision making (as only actionable markers are tested). In this study, we demonstrate that the ASPYRE-Lung Tissue assay has excellent analytical sensitivity at ≤ 3% VAF for SNV and indels from DNA, ≤ 100 copies for gene fusions from RNA, and ≤ 200 copies MET exon 14 skipping from RNA. The assay has high specificity with no false positive results out of 6,840 calls made from variant-free samples. The assay is also highly reproducible and repeatable across different operators, reagent lots, runs, days and qPCR instruments. As obtaining sufficient quantity and quality of tissue samples from lung biopsies for robust genomic analysis can be challenging, future plans for ASPYRE-Lung include validating the assay on small biopsy and cytopathology samples. Finally, we are developing a version of the assay that will analyze liquid (plasma) biopsy samples as input to provide results for individuals where tissue specimens are not available.
Methods
Reference samples
DNA and RNA variants were selected to represent the most common variant in each target exon from BRAF exon 15; EGFR exons 18, 19, 20, 21; KRAS exons 2, 3; and ERBB2 exons 17, 20; and the most common ALK, RET, ROS1, NTRK1/2/3 fusions alongside MET exon 14 exon skipping (selected variants are listed in Supplementary Table S2). Contrived control oligonucleotides made from DNA (SNV, indel) or RNA (fusions, MET exon 14 skipping) were manufactured (DNA from Eurofins, Wolverhampton, UK, RNA from IDT, Leuven, Belgium), quantified by digital PCR (QIAcuity Digital PCR system, Qiagen) at the Biofidelity R&D facility (Cambridge, UK), and spiked into DNA or RNA extracted from FFPE variant-free tonsil tissue quantified by dPCR (DNA) or Qubit (RNA), serially diluted to the appropriate concentrations, and immediately frozen at -20°C (DNA) or -80°C (RNA).
Clinical samples
FFPE variant-free tonsil tissue blocks from patients without any known cancer diagnosis were procured from a commercial biobank retrospectively, sample selection and data access July 1 2022 (Reprocell, Maryland, USA). FFPE lung tissue blocks with a confirmed NSCLC diagnosis were obtained prospectively from commercial biobanks (Geneticist, Tissue Solutions, Reprocell, BocaBio, Cureline, VitroVivo); patients were recruited between August 2020 and August 2021.
Nucleic acid extraction
FFPE blocks were manually sectioned with a microtome (Shandon Finesse, ThermoFisher), producing three 12 μM thick curls, at the Biofidelity R&D facility (Cambridge, UK). DNA and RNA were extracted from specimens in parallel using the Quick-DNA/RNA™ FFPE miniprep kit (Zymo Research). Nucleic acid concentration was determined with a Qubit™ 1X dsDNA or RNA high sensitivity kit (ThermoFisher) and stored at -80°C until further usage.
ASPYRE reaction
The ASPYRE reaction has been described for DNA detection (14) and RNA detection (15) and was performed as carried out previously with input levels of 20 ng DNA per PCR reaction and 6 ng RNA per RT-PCR reaction.
Orthogonal testing of clinical samples
DNA extracted from FFPE lung tissue samples was sequenced through an orthogonal method by targeted enrichment (Roche Avenio Targeted Assay) and sequencing (NextSeq 500, Illumina) by Glasgow Polyomics (University of Glasgow, UK), according to the manufacturer’s guidelines. Analysis was performed by the Roche Sequencing Solutions team.
Interfering substances
Four contrived reference samples of COSM516 at 6% VAF, COSM6224 at 6% VAF, COSF408 at 200 copies or COSF1232 at 200 copies prepared in background DNA or RNA (extracted from FFPE variant-free tissue) was spiked with molecular biology-grade ethanol (Sigma-Aldrich) or guanidinium thiocyanate (Sigma-Aldrich) at concentrations mimicking potential carryover during the extraction process (ethanol at 5%, guanidine thiocyanate at 5, 10 or 20 mM).
Data analysis
Data were downloaded from QuantStudio 5 RealTime PCR System (Thermofisher) instruments running Design and Analysis 2 software. The Raw Data CSV produced by this software was analyzed using custom ASPYRELab v1.0.0 software. This cloud-based web application takes the Raw Data CSV as input and generates CSm values (25) that are translated to variant calls and control statuses as output. All variant calling was blinded to results from orthogonal analyses. The ASPYRELab results were then further collated and analyzed using standalone Python scripts.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra/PRJNA1103989.
Ethics statement
Institutional Review Board (IRB) or equivalent approval was obtained for the use of samples in diagnostics development by biobanks through participating collection sites. All patients provided written informed consent to the collection sites and data were de-identified so no patients could be identified by study personnel outside of the clinical trial site including the biobanks and the study authors.
Author contributions
RE: Conceptualization, Formal analysis, Investigation, Writing – review & editing. EG-Z: Investigation, Writing – review & editing. JB: Investigation, Writing – review & editing. KK: Investigation, Writing – review & editing. CK: Investigation, Writing – review & editing. AG: Conceptualization, Formal analysis, Project administration, Writing – review & editing. A-LS: Investigation, Methodology, Writing – review & editing. JM: Investigation, Writing – review & editing. RP: Formal analysis, Investigation, Methodology, Writing – review & editing. AT: Formal analysis, Investigation, Writing – review & editing. ALC: Investigation, Writing – review & editing. CX: Investigation, Writing – review & editing. IT: Investigation, Writing – review & editing. CH: Investigation, Writing – review & editing. DN: Investigation, Writing – review & editing. JJ: Investigation, Writing – review & editing. SA: Investigation, Writing – review & editing. NP: Investigation, Writing – review & editing. KB: Investigation, Writing – review & editing. EG: Investigation, Writing – original draft, Writing – review & editing. MS-J: Investigation, Methodology, Supervision, Writing – review & editing. AIC: Software, Supervision, Writing – review & editing. HR: Conceptualization, Methodology, Project administration, Supervision, Writing – review & editing. BB: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing. RO: Conceptualization, Formal analysis, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding for this work was provided by Biofidelity Ltd.
Acknowledgments
We thank Kasia Anton, Maksym Artomenko, Ethan A. Clark, Seb Dangerfield, Timon Heide, Donald McAusland, Paulina Powalowska for assistance in the development of ASPYRE-Lung. We acknowledge the work of the Roche Sequencing Solutions team in Mannheim, Germany in assisting with analyses of NGS data.
Conflict of interest
Authors RE, EG-Z, JB, KK, CK, AG, and BB are employed by the company Biofidelity Inc. Authors A-LS, JM, RP, AT, AC, CX, IT, CH, DN, JJ, SA, NP, KB, EG, MS-J, AC and RO are employees of Biofidelity Ltd, a privately held company. All authors may hold stock or stock options. Author HR was a consultant of Biofidelity Inc at the time of the study. Biofidelity has filed patent applications on aspects of this research WO2021130494A1.
The author HR declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The authors declare that this study received funding from Biofidelity Ltd. The following had the following involvement in the study: study design, data collection, analysis, decision to publish, and preparation of the manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1420162/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, et al. Cancer statistics for the year 2020: An overview. Int J Cancer. (2021) 149:778–89. doi: 10.1002/ijc.33588
3. Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, et al. Five-year outcomes with pembrolizumab versus chemotherapy for metastatic non–small-cell lung cancer with PD-L1 tumor proportion score ≥ 50%. J Clin Oncol. (2021) 39:2339–49. doi: 10.1200/jco.21.00174
4. Ciavarella S, Milano A, Dammacco F, Silvestris F. Targeted therapies in cancer. BioDrugs. (2010) 24:77–88. doi: 10.2165/11530830-000000000-00000
5. Wang M, Herbst RS, Boshoff C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat Med. (2021) 27:1345–56. doi: 10.1038/s41591-021-01450-2
6. Scott JA, Lennerz J, Johnson ML, Gordan LN, Dumanois RH, Quagliata L, et al. Compromised outcomes in stage IV non small-cell lung cancer with actionable mutations initially treated without tyrosine kinase inhibitors: A retrospective analysis of real-world data. JCO Oncol Pr. (2023) 20(1):145–53. doi: 10.1200/op.22.00611
7. Targeted therapy drug list by cancer type - NCI. Available online at: https://www.cancer.gov/about-cancer/treatment/types/targeted-therapies/approved-drug-list#targeted-therapy-approved-for-lung-cancer (Accessed September 21, 2023).
8. Nesline MK, Subbiah V, Previs RA, Strickland KC, Ko H, DePietro P, et al. The impact of prior single-gene testing on comprehensive genomic profiling results for patients with non-small cell lung cancer. Oncol Ther. (2024) 12:329–43. doi: 10.1007/s40487-024-00270-x
9. Harada S, Agosto-Arroyo E, Levesque JA, Alston E, Janowski KM, Coshatt GM, et al. Poor cell block adequacy rate for molecular testing improved with the addition of Diff-Quik–stained smears: Need for better cell block processing. Cancer Cytopathol. (2015) 123:480–7. doi: 10.1002/cncy.21561
10. Malapelle U, Leighl N, Addeo A, Hershkovitz D, Hochmair MJ, Khorshid O, et al. Recommendations for reporting tissue and circulating tumour (ct)DNA next-generation sequencing results in non-small cell lung cancer. Br J Cancer. (2024) 131:212–9. doi: 10.1038/s41416-024-02709-4
11. McCoach CE. A cautionary analysis of immunotherapy prior to targeted therapy. J Thorac Oncol. (2019) 14:8–10. doi: 10.1016/j.jtho.2018.10.009
12. Sadik H, Pritchard D, Keeling D-M, Policht F, Riccelli P, Stone G, et al. Impact of clinical practice gaps on the implementation of personalized medicine in advanced non–small-cell lung cancer. JCO Precis Oncol. (2022) 6:e2200246. doi: 10.1200/po.22.00246
13. Tsimberidou AM, Sireci A, Dumanois R, Pritchard D. Strategies to address the clinical practice gaps affecting the implementation of personalized medicine in cancer care. JCO Oncol Pr. (2024) 20:761–6. doi: 10.1200/op.23.00601
14. Silva A-L, Powalowska PK, Stolarek M, Gray ER, Palmer RN, Herman B, et al. Single-copy detection of somatic variants from solid and liquid biopsy. Sci Rep. (2021) 11:6068. doi: 10.1038/s41598-021-85545-3
15. Gray ER, Mordaka JM, Christoforou ER, Bargen KV, Potts ND, Xyrafaki C, et al. Ultra-sensitive molecular detection of gene fusions from RNA using ASPYRE. BMC Med Genomics. (2022) 15:215. doi: 10.1186/s12920-022-01363-0
16. SUMMARY OF SAFETY AND EFFECTIVENESS DATA (SSED) P170019/S006(2019). Available online at: https://www.accessdata.fda.gov/cdrh_docs/pdf17/P170019S006B.pdf. (Accessed August 13, 2024)
17. Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors guideline from the college of American pathologists, the international association for the study of lung cancer, and the association for molecular pathology. In: Archives of Pathology and Laboratory Medicine. United States: College of American Pathologists (2018). p. 321–46. doi: 10.1016/j.jtho.2017.12.001
18. NCCN. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) Non-Small Cell Lung Cancer v7.2024. Plymouth Meeting, PA, USA: NCCN (2024).
19. Robert NJ, Nwokeji ED, Espirito JL, Chen L, Karhade M, Evangelist MC, et al. Biomarker tissue journey among patients (pts) with untreated metastatic non-small cell lung cancer (mNSCLC) in the U.S. Oncology Network community practices. J Clin Oncol. (2021) 39:9004–4. doi: 10.1200/jco.2021.39.15_suppl.9004
20. Lim C, Tsao MS, Le LW, Shepherd FA, Feld R, Burkes RL, et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann Oncol. (2015) 26:1415–21. doi: 10.1093/annonc/mdv208
21. Zheng Y, Vioix H, Liu FX, Singh B, Sharma S, Sharda D. Diagnostic and economic value of biomarker testing for targetable mutations in non-small-cell lung cancer: a literature review. Futur Oncol. (2022) 18:505–18. doi: 10.2217/fon-2021-1040
22. Li W, Wan R, Guo L, Chang G, Jiang D, Meng L, et al. Reliability analysis of exonic-breakpoint fusions identified by DNA sequencing for predicting the efficacy of targeted therapy in non-small cell lung cancer. BMC Med. (2022) 20:160. doi: 10.1186/s12916-022-02362-9
23. Hellmann MD, Ciuleanu T-E, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. (2018) 378:2093–104. doi: 10.1056/nejmoa1801946
24. Borghaei H, O’Byrne KJ, Paz-Ares L, Ciuleanu T-E, Yu X, Pluzanski A, et al. Nivolumab plus chemotherapy in first-line metastatic non-small-cell lung cancer: results of the phase III CheckMate 227 Part 2 trial. ESMO Open. (2023) 8:102065. doi: 10.1016/j.esmoop.2023.102065
Keywords: molecular diagnosis, NSCLC, assay validation, precision oncology, pyrophosphorolysis, targeted panel
Citation: Evans RT, Gillon-Zhang E, Brown JN, Knudsen KE, King C, Green AS, Silva A-L, Mordaka JM, Palmer RN, Tomassini A, Collazos A, Xyrafaki C, Turner I, Ho CH, Nugent D, Jose J, Andreazza S, Potts ND, von Bargen K, Gray ER, Stolarek-Januszkiewicz M, Cooke A, Reddi HV, Balmforth BW and Osborne RJ (2024) ASPYRE-Lung: validation of a simple, fast, robust and novel method for multi-variant genomic analysis of actionable NSCLC variants in FFPE tissue. Front. Oncol. 14:1420162. doi: 10.3389/fonc.2024.1420162
Received: 19 April 2024; Accepted: 27 August 2024;
Published: 25 September 2024.
Edited by:
Sharon R. Pine, University of Colorado Anschutz Medical Campus, United StatesCopyright © 2024 Evans, Gillon-Zhang, Brown, Knudsen, King, Green, Silva, Mordaka, Palmer, Tomassini, Collazos, Xyrafaki, Turner, Ho, Nugent, Jose, Andreazza, Potts, von Bargen, Gray, Stolarek-Januszkiewicz, Cooke, Reddi, Balmforth and Osborne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barnaby W. Balmforth, Yi5iYWxtZm9ydGhAYmlvZmlkZWxpdHkuY29t
 Ryan T. Evans
Ryan T. Evans Elizabeth Gillon-Zhang1
Elizabeth Gillon-Zhang1 Nicola D. Potts
Nicola D. Potts Eleanor R. Gray
Eleanor R. Gray Honey V. Reddi
Honey V. Reddi Barnaby W. Balmforth
Barnaby W. Balmforth