
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 01 August 2024
Sec. Cancer Genetics
Volume 14 - 2024 | https://doi.org/10.3389/fonc.2024.1419808
This article is part of the Research Topic Non-coding RNAs in Breast Cancer, volume II View all 13 articles
Breast cancer is the most common cancer in women and the leading cause of female deaths by cancer in the world worldwide. Hence, understanding the molecular mechanisms associated with breast cancer development and progression, including drug resistance and breast cancer metastasis, is essential for achieving the best management of breast cancer patients. Cancer-related long noncoding RNAs have been shown to be involved in the regulation of each stage of breast cancer progression. Additionally, exosomes are extracellular microvesicles that are central to intercellular communication and play an important role in tumorigenesis. Exosomes can be released from primary tumor cells into the bloodstream and transmit cellular signals to distant body sites. In this work, we review the findings regarding the cellular mechanisms regulated by exosomal lncRNAs that are essentials to chemoresistance development and metastasis of breast cancer. Likewise, we evaluate the outcomes of the potential clinical use of exosomal lncRNAs as breast cancer biomarkers to achieve personalized management of the patients. This finding highlights the importance of transcriptomic analysis of exosomal lncRNAs to understand the breast cancer tumorigenesis as well as to improve the clinical tests available for this disease.
Breast cancer (BC) represents a worldwide public health challenge due to its high associated mortality and morbidity rates. The 5-year survival rate of patients with metastatic BC after treatment with adjuvant therapy is less than 30% (1). According to GLOBOCAN 2020 statistics, the number of new cases of BC worldwide was 2.3 million (11.5%), and the number of deaths was 666 103 (6.8%) (2). In women, the BC is the most diagnosed cancer in 159 of 185 countries and is the leading cause of cancer death in 110 of 185 countries (3). BC incidence is highly correlated with human development (4). Thus, incidence rates are 88% higher in developed than in developing countries (55.9 and 29.7 per 100,000, respectively). However, women living in developing countries have 17% higher mortality rates compared with women in developed countries (15.0 and 12.8 per 100,000, respectively) (3). The elevated incidence rates in higher Human Development Index (HDI) countries are associated with BC risk factors (4). The HDI promote a longstanding higher prevalence of reproductive and hormonal risk factors and lifestyle risk factors, as well as increased detection through organized or opportunistic mammographic screening (3).
According with the data, the average age of women deaths associated to BC is lower in developing countries than in countries with high in-come. That is associated to several factor including the late diagnostic, the poor health services and lack of treatments. Thus, in developing countries, more than half of BC is in women under 50, shortening life expectancy in those countries by a decade (4). Almost two-thirds of the deaths in 2020 were recorded in less-developed regions. In developed countries, over 80% of BC patients present an overall survival of 5-year, in contrast to developing regions with 5 years survival less than 50% (3, 4). The observed BC survival profile is associated with later diagnostics that are common in less developed regions of the world, with over half of breast cancers being locally advanced or metastatic at diagnostic (4). Thereby, establishing primary prevention programs for BC remains as a challenge and all approaches to screening tests and early diagnoses are essential, mainly in countries with developing economies (3).
BC usually refers to a group of diseases with biological subtypes that reflect different molecular profiles and clinicopathological characteristics. In addition to histological subtypes, immunohistochemical classification has divided BC into 5 main molecular subtypes: luminal A (estrogen receptor [ER] +, progesterone receptor [PR] +, human epidermal growth factor receptor 2 [HER2] –, Ki-67 low); luminal B HER2 – (ER+, PR+, HER2-, Ki-67 high); luminal B HER2+ (ER+, PR+, HER2+, Ki-67 high); HER2 (ER-, PR-, HER2+); and basal (triple negative [TNBC], ER-, PR-, HER2-), which are related to clinical outcome (5). Additionally, gene expression profiling and molecular diagnosis have significantly impacted the management of BC. Groups of genes have been identified as biomarkers that help predict disease prognosis and estimate the risk of metastasis, tumor recurrence, response to therapy, and clinical decision making. Furthermore, they have helped guide clinical test development for patient follow-up (5, 6).
Exosomes are a class of lipid bilayer-enclosed extracellular vesicles (EVs) that are devoid of intracellular organelles but contain all known molecular constituents within a cell (7, 8). The exosome size ranges from 30 nm to 150 nm (9–11), and exosomes are constantly released by most eukaryotic cells, including platelets, mast cells, dendritic cells, astrocytes, B and T cells, and cancer cells. In vivo, exosomes are broadly observed in numerous body fluids, such as blood, serum, saliva, amniotic fluid, and breast milk (12). In particular, exosomes enclose a wide range of molecules, including proteins, lipids, and other metabolites (7, 13). Furthermore, they also contain single-stranded (14) and double-stranded DNA (15) and different kinds of RNA, such as mRNAs, tRNAs, rRNAs, miRNAs, siRNAs, circRNAs (16), lncRNAs (17), snoRNAs, snRNAs (18) and piRNAs (19). In addition, genomic, mitochondrial, and plasmid DNA have all been identified within exosomes (20–22).
Exosomes are important mediators of intercellular communication and are involved in several physiological and pathological processes (10). Intriguingly, tumor cells secrete approximately 10-fold more exosomes than do normal cells (23), and tumor-derived exosomes (TDEs) play important roles in different stages of cancer progression (24, 25). TDEs can enhance angiogenesis, invasion, and migration, promote premetastatic niche establishment and confer chemoresistance (26–29). The high heterogeneity of TDEs likely reflects the phenotypic state of tumor cells that generate exosomes (30, 31). Thus, TDE analysis provides a robust method for monitoring cancer progression and further guiding clinical decisions and treatment strategies (32). The stability of exosomes in most body fluids and the diversity of their cargo, which reflects the status of the parental cells, make them promising candidates for developing new approaches for cancer diagnosis (33, 34). An advantage of TDE analysis is that their lipid bilayers stabilize and protect macromolecules against enzymatic activity existing in biofluids, unlike other biomarker assays requiring fresh biofluid (35, 36). This allows them to be stored for an extended period (35, 36), thereby greatly increasing their clinical applications while reducing the cost of short-term sample storage (12, 37). TDEs provide a promising platform for cancer prognosis, diagnosis, and treatment follow-up in precision and personalized medicine (12). EVs can be purified, and their isolated materials can be further analyzed using next-generation sequencing (NGS), real-time PCR, digital PCR, and bioinformatics for screening and early detection of cancer (38). The previous describe the advantages that exosomes offer in the development of new clinical tests for the diagnosis and monitoring of breast cancer, however it cannot be ignored that there is still a long way to go in the sense of reaching standardized methods for the purification, characterization, and analysis of exosomes with adequate quality for use in the clinic.
Noncoding RNAs (ncRNAs) are classified by their size, the longest of which are long noncoding RNAs (lncRNAs) with more than 200 nucleotides. LncRNAs are transcribed by RNA polymerase II, and similar to mRNAs, they have a 5’-cap and 3’ poly-A tail (39, 40). According to their chromosomal positions, lncRNAs are classified as antisense, intronic, divergent, intergenic, promoter associated, transcription start site associated, or enhancer (39). LncRNAs have different functions according with their cellular localization. In the nucleus, they are involved in epigenetic and transcriptional regulation, whereas in the cytoplasm, they are associated with posttranscriptional regulation, including mRNA stability and protein translation, and can act as competitive endogenous RNAs (ceRNAs) (39).
It is widely known that lncRNAs play pivotal roles in each state and process of cancer development (39, 40). Since the discovery of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) in 2003 (41), many other cancer-associated lncRNAs have been identified, and their expression can be deregulated. Among the most characterized lncRNAs associated with cancer development and drug resistance are H19 (42), X-inactive-specific transcript (XIST) (43), and homeobox (HOX) transcript antisense RNA (HOTAIR) (44). The role of lncRNAs in cancer development has been established for a number of these molecules, and four main mechanisms of action have been proposed, including acting as signals for transcriptional regulation, acting as decoys that recruit binding partners away from their other targets, acting as scaffolds bringing together multiple biomolecules, and acting as guides directing the targeting of molecular complexes (39, 40).
In this context, due to the lncRNAs are regulators of diverse oncogenic processes, they could be cancer biomarkers. Particularly in BC, the current screening and diagnostic methods for BC are mammography, ultrasound, MRI, and biopsy. However, the prognostic potential of these methods to predict the BC course, including metastasis, is still very limited (45, 46). Likewise, it is important to identify screening biomarkers to measure the BC risk before the tumor onset (45). BC conventional serum biomarkers are carcinoembryonic antigen (CEA) and cancer antigen 15–3 (CA15–3), however their clinical use is limited by their low sensitivity and specificity (45). In this context, aberrant expression of lncRNAs has been observed in many diseases including cancer and lncRNAs possess a high degree of specificity for tissue type and disease, becoming ideal candidates for cancer diagnosis (45). There are some approaches that analyze the use of levels of lncRNAs in circulation as biomarkers of BC. Furthermore, they analyze the potential of measuring the levels of exosomal lncRNAs. In many of these studies, the certainty of using exosome lncRNAs has been compared versus the use of CEA and CA15–3, with promising results that indicate greater sensitivity and specificity when the 3 biomarkers are used (47). However, the current studies investigating the use of EV ncRNA as biomarkers in BC have been focused on discovery and initial technical validation (46) and the clinical implementation for the use of exosomal lncRNAs needs more research to develop standardized methods that reduce the variability between results. It is necessary to resolve issues such as pre-analytical variables (sample type, storage methods, environmental factors, etc); the methods to be used for exosomal RNA isolation and measurements; and standardization of methodologies to normalize the procedures and reduce inter-laboratory and inter-user variability (45).
RNA-seq technology has revealed that all forms of RNA can be detected in exosomes and can be useful for cancer analysis (48, 49). Using high-throughput sequencing technologies, researchers have found that exosomes contain different RNA populations, including circRNAs (34), lncRNAs (35, 50), mRNAs (51, 52), miRNAs (53, 54), mRNA fragments (55), piRNAs, and fragments of numerous noncoding RNAs, including tRNAs and rRNAs (35, 56, 57). Specifically, several exosomal miRNAs were recently described as being diagnostic for lung cancer (58, 59). Additionally, increasing evidence has shown that circRNAs are highly enriched and stable in exosomes (49). Compared with circulating tumor DNA (ctDNA) assays, circulating nucleic acids from exosomal sources could increase the number of mutant copies accessible for sampling (34, 60). This suggests that exosomal RNA may increase the potential for detecting mutations in blood samples, particularly when very few copies of ctDNA are available during the early stages of disease (34, 60). For clinically reliable and suitable NGS analyses in the future, standardization and clinical verification are necessary. Currently, exploring biomarkers in TDEs has shown great potential for the diagnosis, monitoring, and treatment of cancer patients but still has obvious limitations (32). However, the development of standardized methods for the isolation and identification of TDEs, as well as the use of NGS techniques, could be an excellent tool for diagnosing, monitoring, and guiding cancer therapy.
There are several examples in which RNA-seq technology has been used to identify and describe the role of exosomal ncRNAs associated with BC. Jenjaroenpun and collaborators (2013) were the first to analyze the transcriptomes of exosomes derived from the human BC cell lines MDA-MB-231 and MDA-MB-436 (61). Exosomes contain many classes of RNA, the most common of which is fragmented rRNA. Importantly, the analysis of exosomal RNA reflected the RNA of the donor cells, and several noncoding transcripts were unique to the MDA-MB-231 and MDA-MB-436 cells. In this case, RNA-seq analysis was able to distinguish exosomal RNA delivered by highly metastatic cells (MDA-MB-231) from that delivered by less metastatic cells (MDA-MB-436) (61). In a subsequent exploratory study, RNA sequencing analysis was carried out on serum exosomes derived from one healthy female and two BC patients, followed by comparative analysis with reference data, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Based on these methodologies, they identified five upregulated and six downregulated exosomal lncRNAs as potential biomarkers (62). They proposed the lncRNAs VIM-AS1 (upregulated; with 35 predicted target miRNAs), SNHG8 (downregulated; with 12 predicted target miRNAs), and ELDR (downregulated; with 24 predicted target miRNAs) (62) as possible diagnostic biomarkers. In a more recent study, with the analysis of the Cancer Genome Atlas (TCGA) database and the use of RNA-seq technology, the expression profiles of lncRNAs in EVs from BC patient plasma were analyzed. In this work, the authors identified five lncRNAs in tissue and plasma EVs that could be developed as biomarkers for BC. Four of these lncRNAs (C15orf54, AL157935.1, LINC01117, and SNHG3) were proposed as diagnostic markers for BC lesions, although the plasma EVs from patients were not significantly different. The last lncRNA, AL355974.2, was proposed to be an independent protective prognostic factor after survival analysis (63). These three studies previously described are excellent approaches in the use of NGS techniques and bioinformatic analysis for the massive search of new exosomal biomarkers for BC. However, the results of these studies require further validation with a significant number of exosome samples from BC patients. The study of Jenjaroenpun and cols. was carried out with BC cell lines (61). In the second study, only the exosomal samples of serum from two patients were sequenced (62) and in the third, the results of the levels in EVs of the molecules under study, did not show significant differences between BC patients and healthy ones (63), so they proposed the lncRNAs founded as diagnostic biomarkers of BC only for tissue.
Similarly, there have been several initiatives to develop exosome-based databases for biomarker discovery. These include ExoBCD (64) and exoRBase (65). Using exoRBase, the exosomal sequencing data of BC patients and normal controls were analyzed to identify a ceRNA regulatory network with differential expression profiles from exosomes. Overall, 42 mRNAs, 43 circRNAs, and 26 lncRNAs were found to be differentially expressed (65). In another study using the ExoBCD database, the authors analyzed exosomal lncRNAs and microenvironment interactions in BC. They identified 15 exosome-related differentially expressed lncRNAs that correlated with BC prognosis, and further bioinformatic analysis allowed them to construct a risk model to predict survival outcome. This exosome-related lncRNA risk model could provide a tool to estimate prognosis and immune cell infiltration in BC patients, providing important information for immunotherapeutic decisions (66). The last is an excellent example of the potential of using data stored in platforms such as ExoBCD and exoRBase, so it is necessary to increase the data from these platforms and thus increase their potential in the bioinformatic analysis of the content of exosomes, their function and their possible use as diagnostic and monitoring markers for BC.
One of the main obstacles in cancer management is the development of drug resistance during chemotherapy treatment. Basically, there are two types of drug resistance: intrinsic or natural resistance and acquired resistance, in which therapeutic effectiveness is attenuated over time (67). However, Vasan and colleagues (2019) explained that many tumors are resistant or become resistant due to combinations of each type of resistance (68). Specifically, in BC, the use of different chemotherapies for each of the molecular types of tumors results in drug resistance in a high percentage of BCs (69). Approximately 30%-40% of BC patients are resistant to endocrine therapies and develop metastatic conditions. In addition, modifications of HER receptor signaling have been reported to play a substantial role in the development of BC drug resistance (69). In this context, the role of exosomal ncRNAs in the development of drug resistance has already been reviewed (69); here, we focus only on exosomal lncRNA functions in chemoresistance, including the most current data and summarized in the Table 1.
Tamoxifen is a nonsteroidal synthetic selective estrogen receptor modulator (SERM) that inhibits estrogen receptor (ER) activity in the breast and is a widely used therapeutic agent for BC patients with ER-positive tumors (79). However, endocrine therapy resistance occurs in a significant number of patients. The oncogenic function of the lncRNA urothelial carcinoma-associated 1 (UCA1) in BC has been described previously (80, 81). In an in vitro study in the ER-positive human BC cell line MCF-7 and a tamoxifen-resistant derivative of these cells (LCC2), Xu and collaborators (2016) explored the role of UCA1 in tamoxifen resistance (70). They showed that UCA1 is highly expressed not only in LCC2 cells but also in exosomes released from LCC2 cells compared with exosomes from tamoxifen-sensitive MCF-7 cells. Moreover, incubation with exosomes derived from LCC2 cells increased the viability of MCF-7 cells treated with tamoxifen, reducing apoptosis by decreasing cleaved caspase-3 activity (Figure 1). However, the capacity of LCC2 exosomes to induce tamoxifen resistance was inhibited when their cargo contained impaired UCA1. Thus, UCA1 transfer mediated by exosomes can significantly increase tamoxifen resistance in ER-positive MCF-7 cells (70).
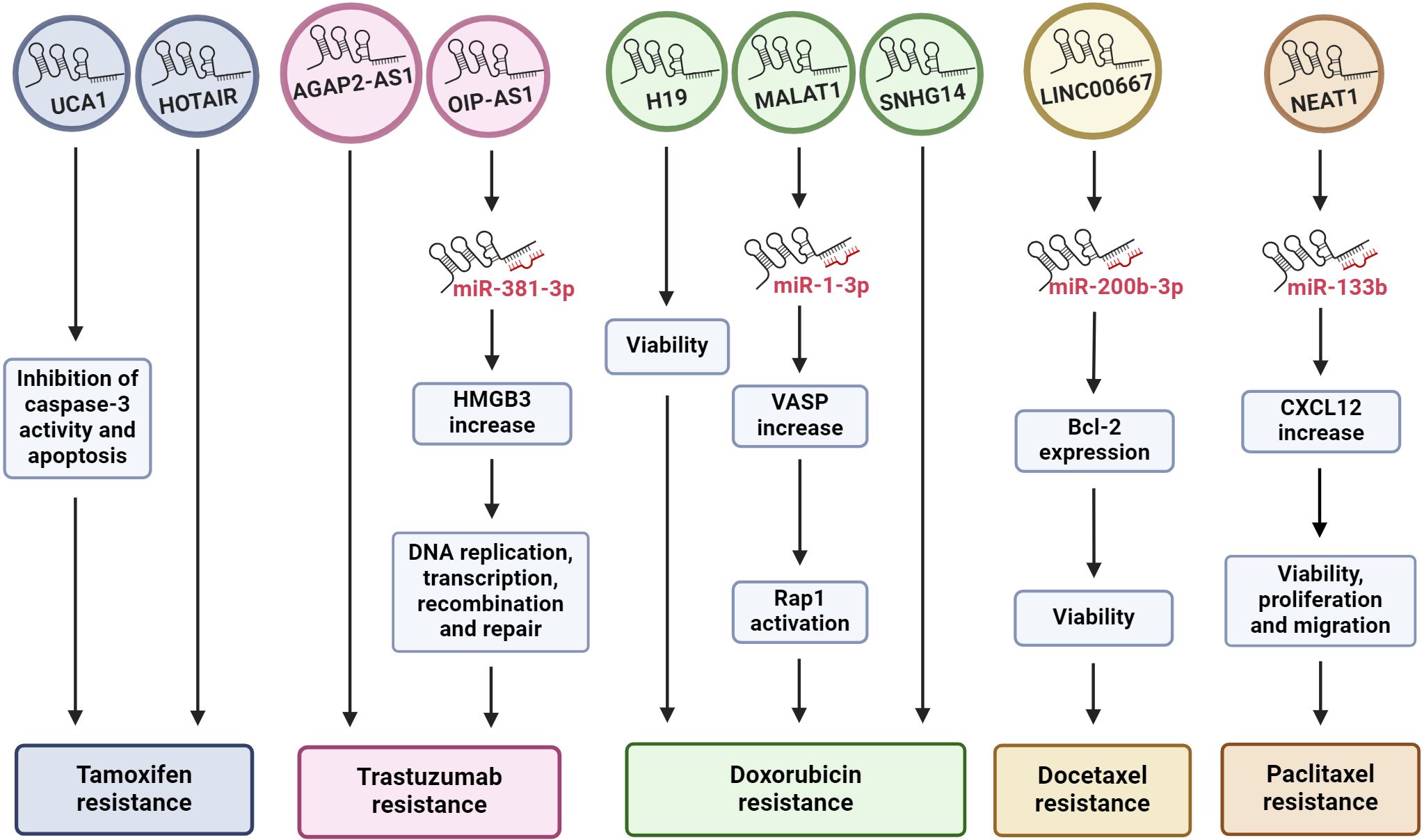
Figure 1 Exosomal lncRNAs regulate drug resistance. The diagram shows how the effect of some lncRNAs released by BC cells, mediated by exosomes, can promote drug resistance through different cell pathways, including in many cases the sponging of miRNAs. The chemoresistance to each specific drug can be modulated by more than one exosomal lncRNA. HMGB3, high mobility group box 3; VASP, vasodilator-stimulated phosphoprotein; Rap 1, Ras-related protein 1; CXCL12, C-X-C motif chemokine ligand 12.
As mentioned above, HOTAIR is a widely studied lncRNA due to its pivotal role in many types of malignant tumors (82–84), including BC, where HOTAIR is highly expressed (85, 86). Additionally, studies have shown that there are greater levels of HOTAIR in exosomes from patients with bladder (87) and cervical (88) cancer than in those from healthy individuals. Moreover, exosomal HOTAIR has been suggested to be a good prognostic and diagnostic biomarker in laryngeal squamous cell carcinoma (89). Tang and coworkers (2019) (71) carried out a study on BC patients who were followed for approximately 6 years to evaluate the diagnostic and prognostic value of serum exosomal HOTAIR. Serum samples from 15 healthy individuals, 15 BC patients treated surgically, 25 patients who received neoadjuvant chemotherapy (anthracycline + taxane + cyclophosphamide regimen) before surgery, and another 25 patients who received tamoxifen treatment after surgery were analyzed. Additionally, 20 BC tissue samples were collected. They found that BC patients had high serum exosomal HOTAIR levels, which decreased 3 months after surgery; thus, exosomal HOTAIR is produced by primary tumor cells and released by exosomes into the bloodstream (71). Furthermore, exosomal HOTAIR levels increased with culture time in the human BC cell lines MDA-MB-231 (triple negative) and MCF-7. In a xenograft assay in which nude mice were injected with BC cells, the expression of serum exosomal HOTAIR in the mice was notably greater than that in the mock control group. The authors also explored the diagnostic and prognostic value of serum exosomal HOTAIR and found that high expression of exosomal HOTAIR led to worse disease-free survival and overall survival, indicating that exosomal HOTAIR could be a good diagnostic and prognostic biomarker. When they analyzed the response to chemotherapy, they observed a correlation between poor response and the overexpression of exosomal HOTAIR. In the HOTAIR high-expression group (n=14), 6 patients achieved a partial response (PR), and 8 achieved stable disease (SD). Conversely, in the low-expression group (n=11), 9 patients achieved a PR, and 2 achieved SD. Similarly, there was a better response to tamoxifen in the low exosomal HOTAIR expression group, as only 1 patient (of 11) experienced BC recurrence, while in the high-expression group, 6 (of 13) experienced recurrence (71). This finding implies that high HOTAIR expression is associated with a poor response to endocrine therapy with tamoxifen and that this effect could be modulated by increased levels of exosomal HOTAIR in serum derived from primary BC tumors and released into the bloodstream (Figure 1). Therefore, the authors suggest that serum exosomal HOTAIR is a prognostic and diagnostic biomarker for BC patients and may be useful for making therapeutic decisions in terms of endocrine therapy (71). Furthermore, the expression of exosomal HOTAIR in plasma was positively correlated with the HER2 status of BC patients, supporting the possible use of HOTAIR as a BC prognostic biomarker (90).
Trastuzumab is an antibody against HER2 that is commonly used as a treatment for HER-2-positive BC. Trastuzumab treatment improves the clinical prognosis of patients, prolonging overall survival in adjuvant and metastatic settings (91). Recently, the role of AGAP2 antisense RNA 1 (AGAP2-AS1) in trastuzumab resistance in vitro was explored. The lncRNA AGAP2-AS1 has an oncogenic function in human non-small cell lung cancer (92, 93) and gastric cancer (94). Zheng and colleagues (2019) (72) induced trastuzumab resistance in the HER2-positive BC cell lines SKBR-3 and BT474. AGAP2-AS1 is upregulated in trastuzumab-resistant SKBR-3 and BT474 cells (SKBR3-TR and BT474-TR, respectively), but silencing AGAP2-AS1 reversed trastuzumab resistance. The authors demonstrated that trastuzumab-resistant cell lines release exosomes loaded with AGAP2-AS1 in a manner dependent on RNA-binding hnRNPA2B1, which mediates the packaging of RNAs into exosomes. Moreover, these exosomes can induce trastuzumab resistance in sensitive cells, an effect that is dependent on exosomal AGAP2-AS1 (Figure 1). The authors suggested that knockdown of AGAP2-AS1 may be helpful for improving the clinical outcome of HER2-positive BC patients and could serve as a therapeutic target (72).
Moreover, the lncRNA OPA-interacting protein 5 antisense transcript 1 (OIP5-AS1) plays many oncogenic roles in multiple cancers (95), including BC (96). In an in vitro assay using the same trastuzumab-resistant cells, SKBR3-TR and BT474-TR, OIP5-AS1 was elevated, and resistance was dependent on OIP5-AS1 (73). Like AGAP2-AS1, OIP5-AS1 is released from resistant cells via exosomes, and these exosomes can be absorbed by trastuzumab-sensitive cells and induce cellular resistance. Mechanistically, OIP-AS1 acts as a sponge for miR-381–3p. miR-381–3p targets high mobility group box 3 (HMGB3), which can regulate DNA replication, transcription, recombination, and repair (Figure 1) (97). Therefore, HMGB3 silencing can inhibit cell growth and progression in BC (98). The ability of OIP5-AS1 to induce trastuzumab resistance was established in a murine xenograft model in which the transfer of exosomal OIP5-AS1 induced trastuzumab resistance in vivo. Moreover, exosomal OIP5-AS1 was dysregulated in the serum of BC patients and might be a promising diagnostic biomarker for trastuzumab resistance (73).
LncRNA H19 functions as an oncogene in numerous cancer types, including gastric (99), colorectal (100), pancreatic (101) and BC, to the extent that plasma H19 has been proposed as a diagnostic and prognostic biomarker for BC (47, 102). The role of H19 in the development of doxorubicin resistance in BC cells has recently been explored. Doxorubicin (DOX) is an anthracycline used as a broad-spectrum anti-neoplastic drug and is included in first-line adjuvant BC treatment (103). As mentioned above, the human BC cell lines MCF-7 and MDA-MB-231 are molecularly different. MCF-7 is a hormone-responsive human invasive breast adenocarcinoma that represents a luminal A subtype, while MDA-MB-231 is a basal subtype that does not express hormone receptors and contains the mutant p53 protein. Wang and collaborators (2020) (74) induced doxorubicin resistance in both cell types (MCF-7/DOX and MDA-MB231/DOX). They observed an increase in H19 expression in DOX-resistant BC cells compared with the corresponding parental cells. Additionally, they established that DOX resistance was an H19-dependent event since H19 suppression significantly lowered DOX resistance by decreasing cell viability and inducing apoptosis. Furthermore, DOX resistance is induced in sensitive cells by exosomal H19 (Figure 1). In BC patients, serum exosomal H19 levels are elevated in DOX-resistant patients (46 of 82 total patients). The sensitivity and specificity of exosomal H19 were 75% and 65.2%, respectively, for the prediction of BC resistance to therapy (74). Moreover, doxorubicin resistance occurred in both the basal and hormone receptor-positive subtypes.
Small nucleolar RNA host gene 14 (SNHG14) plays a pivotal role in the carcinogenesis of several malignant tumors, such as BC, by regulating cell proliferation, migration, invasion, and chemoresistance (104). Using RNAseq technology, the differential expression profiles of lncRNAs in DOX-resistant MCF7 cells were determined. These results and the analysis of gene expression omnibus (GEO) datasets allowed researchers to identify the altered expression of SNHG14, which was increased in BC tissues and in MCF7/DOX cells (76). In addition, SNHG14 levels were greater in purified exosomes from MCF7/DOX cells than in those from parental cells. Additionally, exosomal SNHG14 could be transmitted from drug-resistant cells to drug-sensitive cells after coincubation of DOX-sensitive cells with purified MCF7/DOX exosomes (Figure 1). However, the role of SNHG14 in cellular DOX resistance must be further explored in detail (76).
Interestingly, although the MCF-7 and MDA-MB-231 cell lines have distinct molecular profiles, they show similar tamoxifen and doxorubicin resistance responses. In both cell lines, tamoxifen resistance is modulated by HOTAIR, and doxorubicin resistance is modulated by H19. These data imply a molecular mechanism of chemoresistance that is dependent on the treatment and not the molecular profile of BC cells, at least in terms of the effects associated with HOTAIR and H19.
Docetaxel and paclitaxel are two taxanes widely used in clinical cancer treatment. Both drugs bind to the β-subunit of tubulin, stabilizing microtubules and interfering with natural cytoskeleton dynamics, thereby inhibiting mitosis. Paclitaxel acts at the M/A-phase transition, whereas docetaxel is primarily active in G2/M-phase. Therefore, these drugs inhibit proliferation and cell viability, induce apoptosis, and have antiangiogenic effects (105). Docetaxel has been used for treating various cancers, including triple-negative breast cancer (TNBC). Although docetaxel arrests the cell cycle in cancer cells, acquired resistance is an important obstacle for the treatment of TNBC (105, 106). Docetaxel resistance appears to be regulated by lncRNAs, such as LINC00667. Studies have shown that LINC00667 expression is elevated in BC tissues (107) and that this lncRNA promotes the proliferation and migration of BC cells (108). LINC00667 expression is elevated in exosomes derived from MDA-MB-231 cells resistant to docetaxel compared to exosomes from docetaxel-sensitive cells. In addition, LINC00667 can be transmitted via exosomes from docetaxel-resistant to docetaxel-sensitive cells. Furthermore, exosomal LINC00667 induced docetaxel resistance in sensitive cells through the upregulation of antiapoptotic Bcl-2. In this regard, LINC00667 seems to function as a ceRNA to sponge miR-200b-3p, resulting in increased expression of Bcl-2 (Figure 1) (77).
Similarly, high expression of the lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) in SKBR3 BC cells promotes migration, proliferation, and paclitaxel resistance (78). NEAT1 downregulation decreased paclitaxel resistance, cell migration and cell proliferation. This effect is mediated by the release of miR-133b, which in turn downregulates C-X-C motif chemokine ligand 12 (CXCL12), a well-known inducer of cell survival, proliferation, migration, and drug resistance (Figure 1) (109). Moreover, paclitaxel-resistant SKBR3 cells produce exosomes loaded with high levels of NEAT1, and these exosomes are able to induce paclitaxel resistance, cell migration and growth in paclitaxel-sensitive BC cells. In a xenograft mouse model, knockdown of NEAT1 decreased cancer progression and improved the response to paclitaxel (78).
The most common sites of BC metastasis are the lung, bone, lymph nodes, liver, and pleura, with the highest incidence in the lung (110). Therefore, Feng and collaborators (2019) (111) used high-throughput sequencing to analyze lncRNA expression profiles in lung fibroblasts treated with exosomes derived from MDA-MB-231 BC cells compared with exosomes from normal epithelial MCF-10A cells. Exosomes from BC cells increase the proliferation and migration of lung fibroblasts (the WI-38 and HFL1 cell lines). They identified many lncRNA expression abnormalities that could be associated with alterations in the lung microenvironment and metastasis, including 141 upregulated lncRNAs and 98 downregulated lncRNAs in both lung fibroblast lines (111).
More than 30% of patients with metastatic BC develop brain metastasis (112). Additionally, BC is the second most common cause of brain metastases, following lung cancer, with the highest incidence in BC patients with HER2-positive and TNBC subtypes (113). In this context, the lncRNA expression profile associated with brain metastasis from BC patients showed that XIST was downregulated in brain tumors (114). Moreover, XIST is downregulated in the BC cell lines MDA-MB-231-BrM2a and SKBrM3 (derived from MDA-MB-231 and SKBR3, respectively), which preferentially metastasize to the brain. Thus, XIST plays a protective role in brain metastasis development. This finding was supported in a mouse xenograft model in which XIST silencing promoted brain metastasis. Specific knockout of XIST in mouse mammary glands accelerated primary tumor growth as well as metastasis to the brain. Additionally, XIST downregulation stimulated epithelial–mesenchymal transition (EMT) and activated the c-Met pathway by upregulating the MSN gene. Loss of XIST also augmented the secretion of exosomal miRNA-503, leading to reprogramming of microglia in the brain and triggering M1–M2 polarization of microglia. M1-M2 macrophage conversion upregulated the expression of immunosuppressive cytokines in microglia that suppressed T-cell proliferation. The authors searched an FDA-approved drug library and showed that fludarabine selectively inhibits the growth of XIST-low tumor cells. Additionally, they observed that fludarabine blocked brain metastasis in a mouse model. Furthermore, the loss of XIST promoted brain metastasis in BC by altering the tumor cell microenvironment and stimulating EMT. This suggested that fludarabine could be a good therapeutic agent for the prevention and treatment of brain metastasis (114).
Another central element for brain metastasis is the integrity of the blood-brain barrier (BBB). Lu and collaborators (2020) (115) determined that exosomes from BC cells disrupt the BBB system, promoting brain metastasis. They established a highly brain metastatic BC cell line (MDABR3, derived from MDA-MB-231 cells) by in vivo and in vitro selection after 3 rounds of intracardiac injection in mice and subsequent recovery of brain tumors. MDABR3 cells exhibited high levels of migration and invasion in vitro and metastatic capacity in vivo. Exosomes derived from MDABR3 cells can be taken up by brain microvascular endothelial cells (BMECs), reducing transepithelial/transendothelial electrical resistance (TEER) and increasing permeability in a BBB model, thus promoting invasion of BC cells in the BBB model (Figure 2). Using the GEO dataset, the authors selected exosomal lncRNAs associated with the BBB and verified that GS1–600G8.5 was highly expressed in MDABR3 cells and their exosomes than in samples with reduced metastatic behavior. GS1–600G8.5 silencing decreased the BBB permeability induced by MDABR3 exosomes and the infiltration of cancer cells through the BBB. In addition, BMECs treated with GS1–600G8.5-derived exosomes expressed higher levels of tight junction proteins than those treated with exosomes containing GS1–600G8.5 (Figure 2) (115).
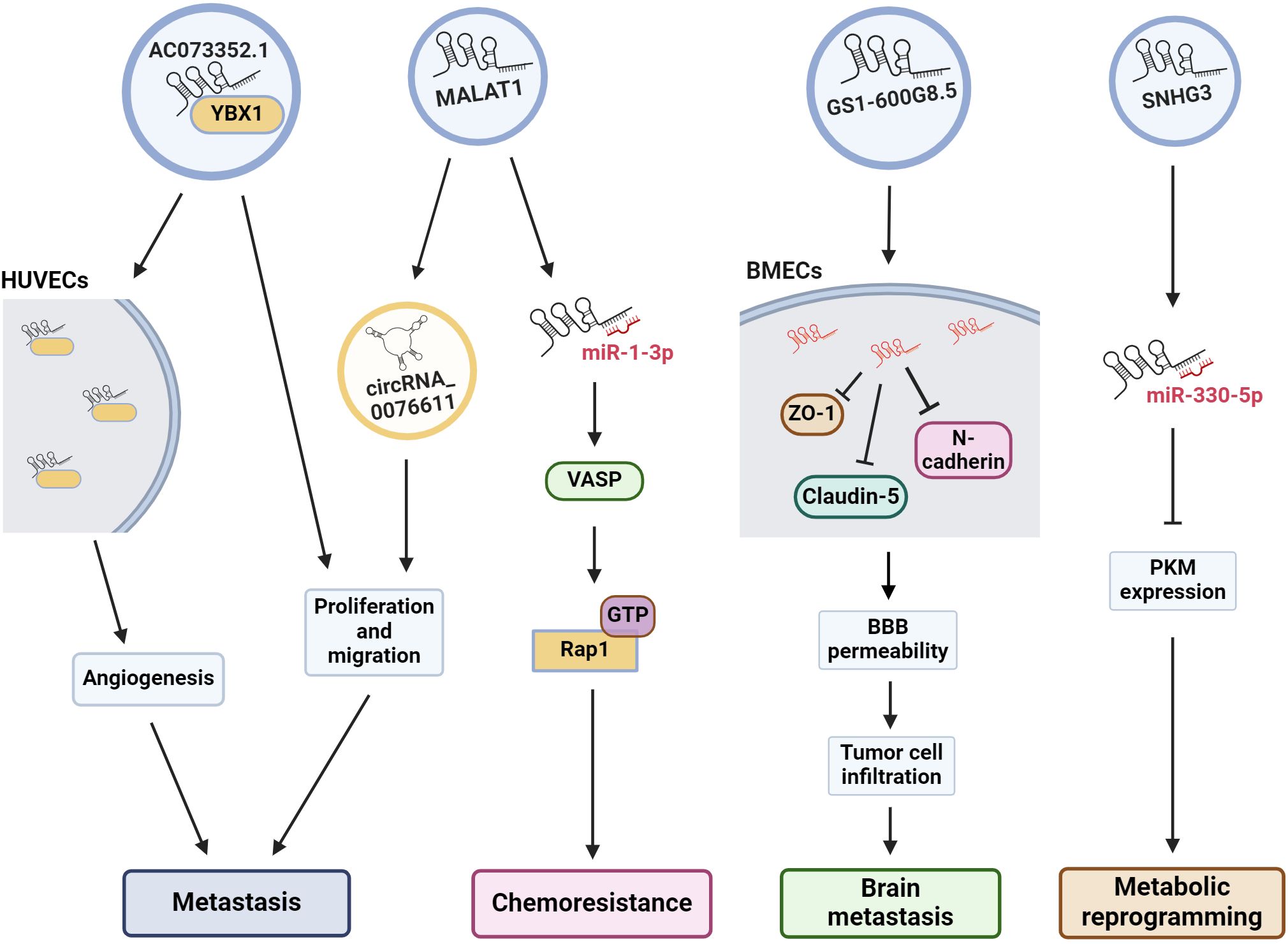
Figure 2 Exosomal lncRNAs are associated to many oncogenic pathways. The illustration shows the effect of some exosomal lncRNAs in the regulation of several cell pathways to induce angiogenesis, metastasis, drug resistance, etc. HUVECs, human umbilical vein endothelial cells; VASP, vasodilator-stimulated phosphoprotein; Rap 1, Ras-related protein 1; BMECs, brain microvascular endothelial cells; BBB, blood-brain barrier; PKM, pyruvate kinase muscle M1/M2.
Angiogenesis is essential for the development of metastasis. Therefore, angiogenesis during cancer progression is another widely studied mechanism. In a recent study, the lncRNA AC073352.1 was identified from a microarray and TCGA database analysis as a novel lncRNA involved in BC metastasis (116). In this study, the authors determined that AC073352.1 was upregulated in BC tissue and was associated with advanced tumor node metastasis (TNM) stage and poor prognosis in BC patients. The overexpression of this lncRNA in MDA-MB-231 and BT549 BC cells (both cell lines with low AC073352.1 expression levels) increased in vitro migration and invasion and augmented in vivo metastasis in a mouse model. However, AC073352.1 suppression in MCF-7 and MDA-MB-468 cell lines (both with high endogenous AC073352.1 levels) reduced their migration and invasion capabilities (116). They also showed that AC073352.1 promoted BC metastasis and angiogenesis through its binding to YBX1, a transcriptional activator and well-known promoter of metastasis (117). The AC073352.1-YBX1 interaction increases YBX1 stability, and in turn, YBX1 promotes the exosomal internalization of AC073352.1. Moreover, exosomes purified from MDA-MB-231 cells overexpressing AC073352.1 promoted angiogenesis in an in vitro tube-formation model using endothelial human umbilical vein endothelial cells (HUVECs) (Figure 2) (116).
MALAT1 is another lncRNA found to be important for BC metastasis and doxorubicin resistance (75). Recently, MALAT1 was shown to be highly expressed in MCF-7 cells and their exosomes. Exosomal MALAT1 increases the malignant properties and chemoresistance of BC cells. This effect is mediated by the downregulation of miR-1–3p, which in turn upregulates vasodilator-stimulated phosphoprotein (VASP), resulting in the activation of the Ras-related protein 1 (Rap 1, a member of the RAS oncogene family) signaling pathway (Figures 1, 2) (75), which is well known for promoting invasion, metastasis and chemoresistance (118, 119). MALAT1 is highly expressed in the tissues and serum of BC patients, and higher MALAT1 expression is positively associated with metastasis and TNM stage but negatively associated with overall patient survival (120). Furthermore, MALAT1 promoted the proliferation of MDA-MB-231 and ZR-75–1 BC cells and promoted tumor growth in a xenograft mouse model. Additionally, exosomal MALAT1 from BC cells was able to induce cell proliferation in vitro (120). Moreover, MALAT1 induces the expression of circRNA_0076611 and promotes the release of circRNA_0076611 from exosomes of MDA-MB-231 cells (Figure 2) (121). CircRNA_0076611 is associated with cell proliferation and migration, and BC patients have been shown to have high levels of this circRNA in serum (121). Another lncRNA involved in metastasis is SNHG3, which has been associated with bone metastasis in BC patients (122). Exosomes from MDA-MB-231 cells containing SNHG3 positively regulate BMP3 expression and bone marrow mesenchymal stem cells (BMSCs), thereby regulating osteogenic differentiation in bone metastasis. Thus, the overexpression of SNHG3 in BC cells may be important for the regulation of osteolytic metastasis (122). All the data described above and summarized in Table 2, shows us that lncRNA present several cellular effects that can be both protective and oncogenic in the regulation of BC. The protective effect of XIST expression in various cancers is already well recognized, as well the oncogenic effect of MALAT1. In this way, exosomes are the vehicle for lncRNAs to reach and modulate molecular mechanisms and cellular changes in in distant places.
Previously, we mentioned the many roles of lncRNAs and TDEs in cancer. Several actions of exosomal lncRNAs have been described, particularly in BC. In addition to their role in chemoresistance and metastasis, they can also modulate other cellular processes associated with cellular transformation, including metabolic changes. In this context, MCF-7 and MD-MBA-453 BC cells were treated with exosomes secreted from cultured cancer-associated fibroblasts (CAFs) derived from BC patients. In these assays, CAF exosomes induced the reprogramming of metabolic pathways in BC cells in culture, promoting cell proliferation, decreasing mitochondrial function, and increasing glycolysis, thus modulating metabolic pathways in BC cells (123). This effect is regulated by the lncRNA SNHG3, which is expressed by CAFs and released in exosomes. SNHG3 acts as a molecular sponge for miR-330–5p in cultured BC cells. The suppression of SNHG3 decreases glycolysis and cell proliferation by increasing miR-330–5p, which, in turn, inhibits pyruvate kinase muscle M1/M2 (PKM) expression in cancer cells (Figure 2). Accordingly, the inhibition of miR-330–5p by SNHG3 promotes PKM expression, resulting in the inhibition of mitochondrial oxidative phosphorylation and increased glycolytic carboxylation (123). Thus, SNHG3 has been shown to be an important regulator of metastasis, not only by promoting bone metastasis but also by mediating the metabolic transformation of tumor cells. Importantly, the SNHG3 that is released into exosomes from BC cells and CAFs can have cellular effects on numerous types of cells.
In cancer research, there is a constant search for biomarkers that will help develop screening tests to achieve more timely diagnoses and better follow-up of BC patients to achieve personalized precision medicine, we summarized the assays made in BC patients in the search for exosomal lncRNAs as biomarkers of BC in the Table 3 and we outline the use of exosomal lncRNAs as BC biomarkers (Figure 3). In this context, the possible use of lncRNA H19 has been studied as a diagnostic biomarker of BC (47). Serum exosomal H19 levels were measured in BC patients, and higher levels were detected in BC patients than in benign breast disease (BBD) patients and healthy controls. Furthermore, when serum exosomal H19 levels were compared between BC patients before and 7 days after surgical removal of the tumor mass, H19 levels were significantly lower in postoperative patients than in preoperative patients. In this manner, the analysis of exosomal H19 levels in serum was shown to be more sensitive and specific than the analysis of cancer antigen 15.3 (CA15.3) and carcinoembryonic antigen (CEA) levels in blood. Moreover, exosomal H19 expression levels are positively associated with lymph node metastasis, distant metastasis, TNM stage, ER status, PR status, and HER2 status (47).
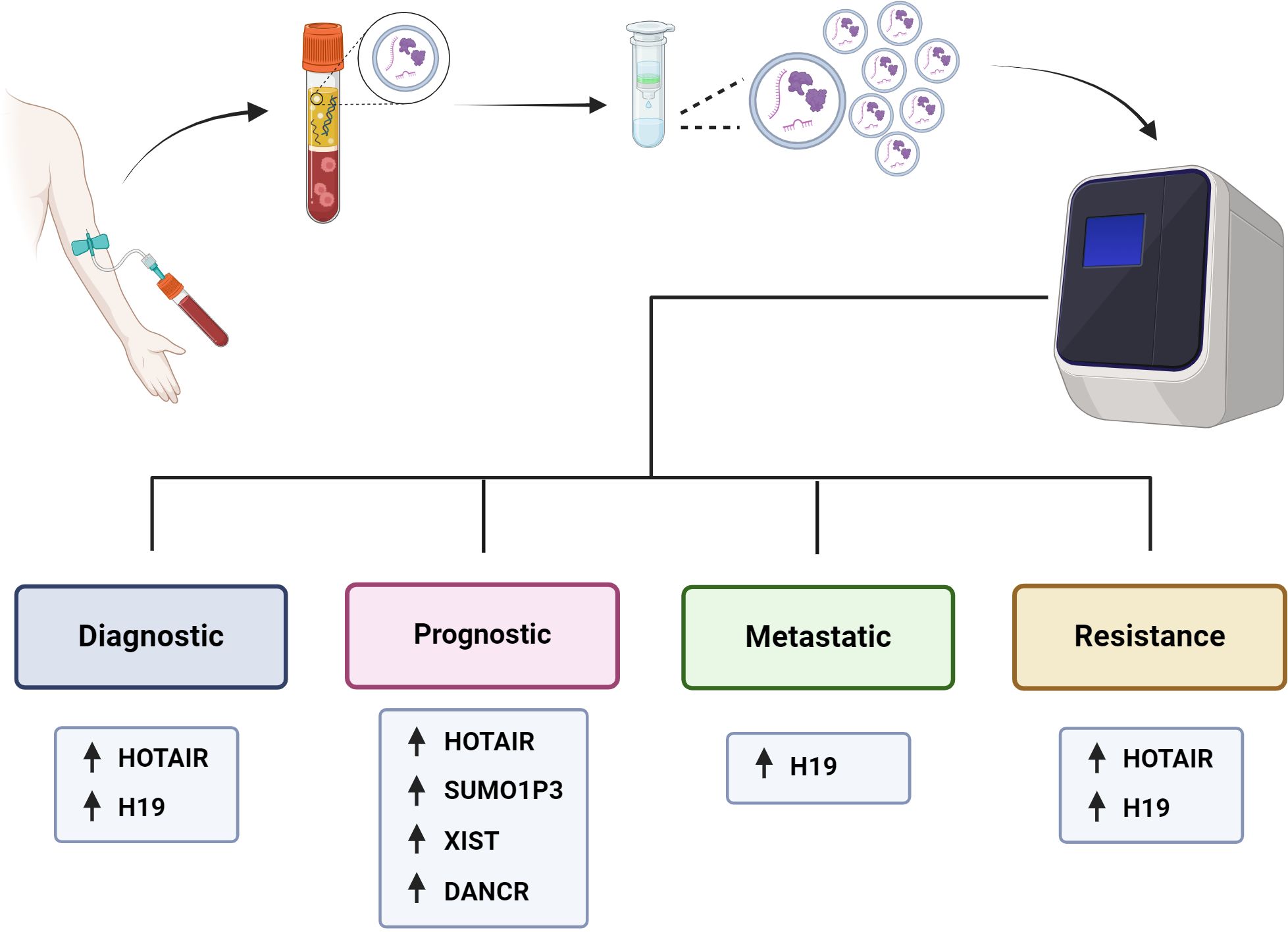
Figure 3 Utility of exosomal lncRNAs as biomarkers of BC. It is outlined how patients with BC would have a peripheral blood sample taken, from which the plasma would be isolated, and the exosomes purified to be analyzed and measure the levels of their components, such as proteins or nucleic acids, including lncRNAs. The levels of exosomal lncRNAs can be measured and used as diagnostic, prognostic, metastatic and drug resistance biomarkers for BC patients.
Na-Er and collaborators (2021) (124) examined the prognostic significance of the serum exosomal lncRNA small ubiquitin-like modifier 1 pseudogene 3 (SUMO1P3) in TNBC. The authors analyzed SUMOP3 levels in BC tissue using the TCGA database. SUMO1P3 was upregulated in BC and correlated with poor survival. Moreover, exosomal SUMO1P3 levels were greater in TNBC patients than in non-TNBC patients, patients with benign breast disease, and healthy people. High exosomal SUMO1P3 levels were correlated with lymphovascular invasion, lymph node metastasis, histological grade and chemoresistance. In addition, patients with high levels of exosomal SUMO1P3 had worse overall survival than patients with low levels of exosomal SUMO1P3. Thus, the authors suggest that exosomal SUMO1P3 might be a prognostic biomarker for TNBC (124). Similarly, Lan and collaborators (2021) (125) proposed exosomal XIST as a biomarker for possible recurrence in TNBC patients. They observed that XIST levels in tumor tissue and exosomal XIST in serum were greater in TNBC patients than in healthy controls. Like those of other lncRNAs, the exosomal levels of XIST decreased after surgical removal of the tumor mass. Furthermore, 5-year follow-up studies revealed greater levels of exosomal XIST in recurrent TNBC patients than in nonrecurrent patients. There was an association between high exosomal XIST levels and poor overall survival, but there was no association with other clinicopathological parameters, such as Karnofsky Performance Status (an assessment tool for functional impairment), sex or age (125).
The lncRNA differentiation antagonizing nonprotein coding RNA (DANCR or ANCR) has been postulated to be a good prognostic biomarker for BC. DANCR has been shown to be involved in the initiation and progression of BC (127, 128). The levels of exosomal DANCR were greater in BC patients than in benign breast disease patients or normal controls, and as expected, exosomal DANCR levels were markedly downregulated in postoperative samples. However, although the diagnostic performance of exosomal DANCR was good, the diagnostic accuracy for BC was better when a combination of exosomal DANCR, CA15.3 and CEA was assessed. In this manner, high exosomal DANCR levels were associated with clinicopathological parameters, including lymph node metastasis, ER status, HER2 status, and TNM stage. Patients with high exosomal DANCR levels had shorter 5-year overall survival, and multivariate analysis of exosomal DANCR levels revealed that this parameter was an independent risk factor for BC (126).
In this review, we describe the importance of exosomal lncRNAs in intercellular communication associated with BC development. The role of lncRNAs in cancer development is well established and the significance of exosomes as mediators of intercellular communication in tumorigenesis has been demonstrated. Particularly, exosomal lncRNAs promote processes such as angiogenesis, invasion, migration, metastasis and chemoresistance, etc, in numerous cancer types, including BC. In this way, the transcriptomic analysis of exosomal lncRNAs is a useful tool in the search for lncRNAs associated with BC tumorigenesis as well in the identification of biomarkers with potential clinical use for the disease.
We have described the significance of exosomal lncRNAs in promoting resistance to drugs such as tamoxifen, trastuzumab, doxorubicin, docetaxel and paclitaxel. Interestingly, it has been observed that resistance to a specific drug can be regulated by more than one exosomal lncRNA, and apparently through different molecular pathways, as we can see in the development of doxorubicin resistance. However, in many cases the intracellular pathways by which some of the lncRNAs induce chemoresistance are not fully described, as in the case of SNHG14. Curiously, the evidence about the chemoresistance induction in sensitive BC cells suggests that the mechanism of drug resistance is dependent on the treatment and not on the molecular profile of the BC cells. For example, the development of chemoresistance to tamoxifen and doxorubicin, in the MCF-7 and MDA-MB-231 cell lines, representatives of different BC subtypes, both cell lines develop resistance to tamoxifen mediated by exosomal HOTAIR and doxorubicin resistance modulated by exosomal H19. Likewise, exosomal lncRNAs not only participate in the development of chemoresistance, but they also modulate metabolic changes in cancer cells as well as several processes associated with metastasis such as proliferation, invasion and angiogenesis. However, the information available regarding the importance of exosomal lncRNAs in the development and progression of BC is still insufficient. There are still many lncRNAs for which the molecular pathways they promote have not been described, and there are surely many more that have not even been identified. Finally, another important aspect is the identification of exosomal lncRNAs and their potential clinical use both as therapeutic targets and as biomarkers of the disease. In this way, there are already approaches and some exosomal lncRNAs have been proposed as diagnostic, prognostic, metastatic and chemoresistance biomarkers. The transfer of lncRNAs mediated by exosomes provides advantages compared with that mediated by cell-free circulating RNA since exosomes provide a more stable environment for measuring macromolecules that reflect tumor characteristics, and these advantages can be exploited to measure different biomarkers that could guide and improve the management of breast cancer. However, exosomal lncRNAs still must be validated for use in clinical trials. Likewise, the most appropriate method for the isolation and analysis of exosomes remains to be validated.
SB-Z: Writing – review & editing, Writing – original draft, Investigation, Conceptualization. EL: Writing – review & editing. AH-M: Supervision, Writing – review & editing, Resources.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Riggio AI, Varley KE, Welm AL. The lingering mysteries of metastatic recurrence in breast cancer. Br J Cancer. (2021) 124:13–26. doi: 10.1038/s41416-020-01161-4
2. Global cancer observatory. GLOBOCAN (2022). Available at: https://gco.iarc.fr/.
3. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
4. Wilkinson L, Gathani T. Understanding breast cancer as a global health concern. Br J Radiol. (2022) 95:20211033. doi: 10.1259/bjr.20211033
5. Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Primer. (2019) 5:66. doi: 10.1038/s41572-019-0111-2
6. Eliyatkin N, Yalcin E, Zengel B, Aktaş S, Vardar E. Molecular classification of breast carcinoma: from traditional, old-fashioned way to A new age, and A new way. J Breast Health. (2015) 11:59–66. doi: 10.5152/tjbh.
7. Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. (2014) 29:116–25. doi: 10.1016/j.ceb.2014.05.004
8. Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. (2002) 2:569–79. doi: 10.1038/nri855
9. Doyle LM, Wang MZ. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. (2019) 8. doi: 10.3390/cells8070727
10. Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. (2016) 164:1226–32. doi: 10.1016/j.cell.2016.01.043
11. Yáñez-Mó M, Siljander PRM, Andreu Z, Zavec AB, Borràs FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. (2015) 4:27066. doi: 10.3402/jev.v4.27066
12. Hosseini K, Ranjbar M, Pirpour Tazehkand A, Asgharian P, Montazersaheb S, Tarhriz V, et al. Evaluation of exosomal non-coding RNAs in cancer using high-throughput sequencing. J Transl Med. (2022) 20:30. doi: 10.1186/s12967-022-03231-y
13. Kim JH, Kim E, Lee MY. Exosomes as diagnostic biomarkers in cancer. Mol Cell Toxicol. (2018) 14:113–22. doi: 10.1007/s13273-018-0014-4
14. Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. (2011) 2:180. doi: 10.1038/ncomms1180
15. Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch M, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. (2014) 289:3869–75. doi: 10.1074/jbc.C113.532267
16. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. (2019) 20:675–91. doi: 10.1038/s41576-019-0158-7
17. Zhao W, Shan B, He D, Cheng Y, Li B, Zhang C, et al. Recent progress in characterizing long noncoding RNAs in cancer drug resistance. J Cancer. (2019) 10:6693–702. doi: 10.7150/jca.30877
18. Vihinen M. Systematics for types and effects of RNA variations. RNA Biol. (2021) 18:481–98. doi: 10.1080/15476286.2020.1817266
19. Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. (2011) 12:246–58. doi: 10.1038/nrm3089
20. Erdmann S, Tschitschko B, Zhong L, Raftery MJ, Cavicchioli R. A plasmid from an Antarctic haloarchaeon uses specialized membrane vesicles to disseminate and infect plasmid-free cells. Nat Microbiol. (2017) 2:1446–55. doi: 10.1038/s41564-017-0009-2
21. Grüll MP, Mulligan ME, Lang AS. Small extracellular particles with big potential for horizontal gene transfer: membrane vesicles and gene transfer agents. FEMS Microbiol Lett. (2018) 365. doi: 10.1093/femsle/fny192
22. Lázaro-Ibáñez E, Lässer C, Shelke GV, Crescitelli R, Jang SC, Cvjetkovic A, et al. DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J Extracell Vesicles. (2019) 8(1):1656993. doi: 10.1080/20013078.2019.1656993
23. Li W, Li C, Zhou T, Liu X, Liu X, Li X, et al. Role of exosomal proteins in cancer diagnosis. Mol Cancer. (2017) 16:145. doi: 10.1186/s12943-017-0706-8
24. Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. (2016) 30:836–48. doi: 10.1016/j.ccell.2016.10.009
25. Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. (2015) 161:1046–57. doi: 10.1016/j.cell.2015.04.042
26. Boelens MC, Wu TJ, Nabet BY, Xu B, Qiu Y, Yoon T, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. (2014) 159:499–513. doi: 10.1016/j.cell.2014.09.051
27. Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. (2018) 560:382–6. doi: 10.1038/s41586-018-0392-8
28. Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. (2015) 527:329–35. doi: 10.1038/nature15756
29. Le MTN, Hamar P, Guo C, Basar E, Perdigão-Henriques R, Balaj L, et al. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest. (2014) 124:5109–28. doi: 10.1172/JCI75695
30. Kucharzewska P, Belting M. Emerging roles of extracellular vesicles in the adaptive response of tumour cells to microenvironmental stress. J Extracell Vesicles. (2013) 2. doi: 10.3402/jev.v2i0.20304
31. Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. (2009) 284:34211–22. doi: 10.1074/jbc.M109.041152
32. Cheng J, Wang X, Yuan X, Liu G, Chu Q. Emerging roles of exosome-derived biomarkers in cancer theranostics: messages from novel protein targets. Am J Cancer Res. (2022) 12:2226–48.
33. Hornick NI, Huan J, Doron B, Goloviznina NA, Lapidus J, Chang BH, et al. Serum exosome microRNA as a minimally-invasive early biomarker of AML. Sci Rep. (2015) 5:11295. doi: 10.1038/srep11295
34. Yu W, Hurley J, Roberts D, Chakrabortty SK, Enderle D, Noerholm M, et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol Off J Eur Soc Med Oncol. (2021) 32:466–77. doi: 10.1016/j.annonc.2021.01.074
35. Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. (2013) 14:319. doi: 10.1186/1471-2164-14-319
36. Kalra H, Adda CG, Liem M, Ang CS, Mechler A, Simpson RJ, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. (2013) 13:3354–64. doi: 10.1002/pmic.201300282
37. Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y, et al. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther. (2020) 5:144. doi: 10.1038/s41392-020-00258-9
38. Hur JY, Lee KY. Characteristics and clinical application of extracellular vesicle-derived DNA. Cancers. (2021) 13. doi: 10.3390/cancers13153827
39. Gao N, Li Y, Li J, Gao Z, Yang Z, Li Y, et al. Long non-coding RNAs: the regulatory mechanisms, research strategies, and future directions in cancers. Front Oncol. (2020) 10:598817. doi: 10.3389/fonc.2020.598817
40. Carter JM, Ang DA, Sim N, Budiman A, Li Y. Approaches to identify and characterise the post-transcriptional roles of lncRNAs in cancer. Non-Coding RNA. (2021) 7:19. doi: 10.3390/ncrna7010019
41. Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. (2003) 22:8031–41. doi: 10.1038/sj.onc.1206928
42. Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, et al. The H19 non-coding RNA is essential for human tumor growth. PloS One. (2007) 2:e845. doi: 10.1371/journal.pone.0000845
43. Sirchia SM, Tabano S, Monti L, Recalcati MP, Gariboldi M, Grati FR, et al. Misbehaviour of XIST RNA in breast cancer cells. PloS One. (2009) 4:e5559. doi: 10.1371/journal.pone.0005559
44. Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. (2016) 35:2746–55. doi: 10.1038/onc.2015.340
45. Maroni P, Gomarasca M, Lombardi G. Long non-coding RNAs in bone metastasis: progresses and perspectives as potential diagnostic and prognostic biomarkers. Front Endocrinol. (2023) 14:1156494. doi: 10.3389/fendo.2023.1156494
46. Samuels M, Jones W, Towler B, Turner C, Robinson S, Giamas G. The role of non-coding RNAs in extracellular vesicles in breast cancer and their diagnostic implications. Oncogene. (2023) 42:3017–34. doi: 10.1038/s41388-023-02827-y
47. Zhong G, Wang K, Li J, Xiao S, Wei W, Liu J. Determination of serum exosomal H19 as a noninvasive biomarker for breast cancer diagnosis. OncoTargets Ther. (2020) 13:2563–71. doi: 10.2147/OTT.S243601
48. Mjelle R, Dima SO, Bacalbasa N, Chawla K, Sorop A, Cucu D, et al. Comprehensive transcriptomic analyses of tissue, serum, and serum exosomes from hepatocellular carcinoma patients. BMC Cancer. (2019) 19:1007. doi: 10.1186/s12885-019-6249-1
49. Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. (2019) 18:116. doi: 10.1186/s12943-019-1041-z
50. Kogure T, Yan IK, Lin WL, Patel T. Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: A mechanism of intercellular signaling in human hepatocellular cancer. Genes Cancer. (2013) 4:261–72. doi: 10.1177/1947601913499020
51. Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, Baran J, Urbanowicz B, Brański P, et al. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol Immunother CII. (2006) 55:808–18. doi: 10.1007/s00262-005-0075-9
52. Deregibus MC, Cantaluppi V, Calogero R, Lo Iacono M, Tetta C, Biancone L, et al. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. (2007) 110:2440–8. doi: 10.1182/blood-2007-03-078709
53. Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. (2012) 40:10937–49. doi: 10.1093/nar/gks832
54. Huang Q, Yang J, Zheng J, Hsueh C, Guo Y, Zhou L. Characterization of selective exosomal microRNA expression profile derived from laryngeal squamous cell carcinoma detected by next generation sequencing. Oncol Rep. 40:2584–94. doi: 10.3892/or.2018.6672
55. Batagov AO, Kurochkin IV. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3’-untranslated regions. Biol Direct. (2013) 8:12. doi: 10.1186/1745-6150-8-12
56. Dhahbi JM, Spindler SR, Atamna H, Boffelli D, Martin DI. Deep sequencing of serum small RNAs identifies patterns of 5’ tRNA half and YRNA fragment expression associated with breast cancer. biomark Cancer. (2014) 6:37–47. doi: 10.4137/BIC.S20764
57. Miranda KC, Bond DT, Levin JZ, Adiconis X, Sivachenko A, Russ C, et al. Massively parallel sequencing of human urinary exosome/microvesicle RNA reveals a predominance of non-coding RNA. PloS One. (2014) 9:e96094. doi: 10.1371/journal.pone.0096094
58. Grimolizzi F, Monaco F, Leoni F, Bracci M, Staffolani S, Bersaglieri C, et al. Exosomal miR-126 as a circulating biomarker in non-small-cell lung cancer regulating cancer progression. Sci Rep. (2017) 7:15277. doi: 10.1038/s41598-017-15475-6
59. Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X, et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res Off J Am Assoc Cancer Res. (2017) 23:5311–9. doi: 10.1158/1078-0432.CCR-17-0577
60. Möhrmann L, Huang HJ, Hong DS, Tsimberidou AM, Fu S, Piha-Paul SA, et al. Liquid biopsies using plasma exosomal nucleic acids and plasma cell-free DNA compared with clinical outcomes of patients with advanced cancers. Clin Cancer Res Off J Am Assoc Cancer Res. (2018) 24:181–8. doi: 10.1158/1078-0432.CCR-17-2007
61. Jenjaroenpun P, Kremenska Y, Nair VM, Kremenskoy M, Joseph B, Kurochkin IV. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ. (2013) 1:e201. doi: 10.7717/peerj.201
62. Zhao X, Guo X, Jiao D, Zhu J, Xiao H, Yang Y, et al. Analysis of the expression profile of serum exosomal lncRNA in breast cancer patients. Ann Transl Med. (2021) 9:1382. doi: 10.21037/atm
63. Wang H, Shu L, Niu N, Zhao C, Lu S, Li Y, et al. Novel lncRNAs with diagnostic or prognostic value screened out from breast cancer via bioinformatics analyses. PeerJ. (2022) 10:e13641. doi: 10.7717/peerj.13641
64. Wang X, Chai Z, Pan G, Hao Y, Li B, Ye T, et al. ExoBCD: a comprehensive database for exosomal biomarker discovery in breast cancer. Brief Bioinform. (2021) 22:bbaa088. doi: 10.1093/bib/bbaa088
65. Zhu K, Wang Q, Wang L. Analysis of competitive endogenous RNA regulatory network of exosomal breast cancer based on exoRBase. Evol Bioinforma Online. (2022) 18:11769343221113286. doi: 10.1177/11769343221113286
66. Qiu P, Guo Q, Lin J, Pan K, Chen J, Ding M. An exosome-related long non-coding RNAs risk model could predict survival outcomes in patients with breast cancer. Sci Rep. (2022) 12:22322. doi: 10.1038/s41598-022-26894-5
67. Cosentino G, Plantamura I, Tagliabue E, Iorio MV, Cataldo A. Breast cancer drug resistance: overcoming the challenge by capitalizing on microRNA and tumor microenvironment interplay. Cancers. (2021) 13:3691. doi: 10.3390/cancers13153691
68. Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature. (2019) 575:299–309. doi: 10.1038/s41586-019-1730-1
69. Rezaee M, Mohammadi F, Keshavarzmotamed A, Yahyazadeh S, Vakili O, Milasi YE, et al. The landscape of exosomal non-coding RNAs in breast cancer drug resistance, focusing on underlying molecular mechanisms. Front Pharmacol. (2023) 14:1152672. doi: 10.3389/fphar.2023.1152672
70. Xu CG, Yang MF, Ren YQ, Wu CH, Wang LQ. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur Rev Med Pharmacol Sci. (2016) 20:4362–8.
71. Tang S, Zheng K, Tang Y, Li Z, Zou T, Liu D. Overexpression of serum exosomal HOTAIR is correlated with poor survival and poor response to chemotherapy in breast cancer patients. J Biosci. (2019) 44:37. doi: 10.1007/s12038-019-9861-y
72. Zheng Z, Chen M, Xing P, Yan X, Xie B. Increased expression of exosomal AGAP2-AS1 (AGAP2 antisense RNA 1) in breast cancer cells inhibits trastuzumab-induced cell cytotoxicity. Med Sci Monit Int Med J Exp Clin Res. (2019) 25:2211–20. doi: 10.12659/MSM.915419
73. Yu Q, Li Y, Peng S, Li J, Qin X. Exosomal-mediated transfer of OIP5-AS1 enhanced cell chemoresistance to trastuzumab in breast cancer via up-regulating HMGB3 by sponging miR-381-3p. Open Med Wars Pol. (2021) 16:512–25. doi: 10.1515/med-2021-0249
74. Wang X, Pei X, Guo G, Qian X, Dou D, Zhang Z, et al. Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J Cell Physiol. (2020) 235:6896–904. doi: 10.1002/jcp.29585
75. Tao S, Bai Z, Liu Y, Gao Y, Zhou J, Zhang Y, et al. Exosomes derived from tumor cells initiate breast cancer cell metastasis and chemoresistance through a MALAT1-dependent mechanism. J Oncol. (2022) 2022:5483523. doi: 10.1155/2022/5483523
76. Wan J, Feng Z, Shi J, Li Q. Identification of key lncRNAs in exosomes with doxorubicin resistance in the MCF7 cells. Asian J Surg. (2023) 46:3187–9. doi: 10.1016/j.asjsur.2023.02.113
77. Li J, Kang J, Liu W, Liu J, Pan G, Mao A, et al. Docetaxel-resistant triple-negative breast cancer cell-derived exosomal lncRNA LINC00667 reduces the chemosensitivity of breast cancer cells to docetaxel via targeting miR-200b-3p/Bcl-2 axis. Eur J Histochem EJH. (2022) 66. doi: 10.4081/ejh.2022.3529
78. Wei X, Tao S, Mao H, Zhu H, Mao L, Pei W, et al. Exosomal lncRNA NEAT1 induces paclitaxel resistance in breast cancer cells and promotes cell migration by targeting miR-133b. Gene. (2023) 860:147230. doi: 10.1016/j.gene.2023.147230
79. Lumachi F, Brunello A, Maruzzo M, Basso U, Basso SMM. Treatment of estrogen receptor-positive breast cancer. Curr Med Chem. (2013) 20:596–604. doi: 10.2174/092986713804999303
80. Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. (2014) 5:e1008. doi: 10.1038/cddis.2013.541
81. Tuo YL, Li XM, Luo J. Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur Rev Med Pharmacol Sci. (2015) 19:3403–11.
82. Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. (2011) 18:1243–50. doi: 10.1245/s10434-011-1581-y
83. Wu Y, Liu J, Zheng Y, You L, Kuang D, Liu T. Suppressed expression of long non-coding RNA HOTAIR inhibits proliferation and tumourigenicity of renal carcinoma cells. Tumour Biol J Int Soc Oncodevelopmental Biol Med. (2014) 35:11887–94. doi: 10.1007/s13277-014-2453-4
84. Nakayama I, Shibazaki M, Yashima-Abo A, Miura F, Sugiyama T, Masuda T, et al. Loss of HOXD10 expression induced by upregulation of miR-10b accelerates the migration and invasion activities of ovarian cancer cells. Int J Oncol. (2013) 43:63–71. doi: 10.3892/ijo.2013.1935
85. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. (2010) 464:1071–6. doi: 10.1038/nature08975
86. Sørensen KP, Thomassen M, Tan Q, Bak M, Cold S, Burton M, et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat. (2013) 142:529–36. doi: 10.1007/s10549-013-2776-7
87. Berrondo C, Flax J, Kucherov V, Siebert A, Osinski T, Rosenberg A, et al. Expression of the long non-coding RNA HOTAIR correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PloS One. (2016) 11:e0147236. doi: 10.1371/journal.pone.0147236
88. Zhang J, Liu SC, Luo XH, Tao GX, Guan M, Yuan H, et al. Exosomal long noncoding RNAs are differentially expressed in the cervicovaginal lavage samples of cervical cancer patients. J Clin Lab Anal. (2016) 30:1116–21. doi: 10.1002/jcla.21990
89. Wang J, Zhou Y, Lu J, Sun Y, Xiao H, Liu M, et al. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol Northwood Lond Engl. (2014) 31:148. doi: 10.1007/s12032-014-0148-8
90. Wang YL, Liu LC, Hung Y, Chen CJ, Lin YZ, Wu WR, et al. Long non-coding RNA HOTAIR in circulatory exosomes is correlated with ErbB2/HER2 positivity in breast cancer. Breast Edinb Scotl. (2019) 46:64–9. doi: 10.1016/j.breast.2019.05.003
91. Doss S, Robertson J, Adam J. Lapatinib or trastuzumab in combination with an aromatase inhibitor for first-line treatment of metastatic hormone-receptor-positive breast cancer that overexpresses HER2. Lancet Oncol. (2012) 13:766–7. doi: 10.1016/S1470-2045(12)70290-5
92. Li W, Sun M, Zang C, Ma P, He J, Zhang M, et al. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. (2016) 7:e2225. doi: 10.1038/cddis.2016.126
93. Fan KJ, Liu Y, Yang B, Tian XD, Li CR, Wang B. Prognostic and diagnostic significance of long non-coding RNA AGAP2-AS1 levels in patients with non-small cell lung cancer. Eur Rev Med Pharmacol Sci. (2017) 21:2392–6.
94. Qi F, Liu X, Wu H, Yu X, Wei C, Huang X, et al. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J Hematol OncolJ Hematol Oncol. (2017) 10:48. doi: 10.1186/s13045-017-0420-4
95. Li Y, Han X, Feng H, Han J. Long noncoding RNA OIP5-AS1 in cancer. Clin Chim Acta Int J Clin Chem. (2019) 499:75–80. doi: 10.1016/j.cca.2019.08.031
96. Zeng H, Wang J, Chen T, Zhang K, Chen J, Wang L, et al. Downregulation of long non-coding RNA Opa interacting protein 5-antisense RNA 1 inhibits breast cancer progression by targeting sex-determining region Y-box 2 by microRNA-129-5p upregulation. Cancer Sci. (2019) 110:289–302. doi: 10.1111/cas.13879
97. Nemeth MJ, Curtis DJ, Kirby MR, Garrett-Beal LJ, Seidel NE, Cline AP, et al. Hmgb3: an HMG-box family member expressed in primitive hematopoietic cells that inhibits myeloid and B-cell differentiation. Blood. (2003) 102:1298–306. doi: 10.1182/blood-2002-11-3541
98. Gu J, Xu T, Huang QH, Zhang CM, Chen HY. HMGB3 silence inhibits breast cancer cell proliferation and tumor growth by interacting with hypoxia-inducible factor 1α. Cancer Manag Res. (2019) 11:5075–89. doi: 10.2147/CMAR
99. Li H, Yu B, Li J, Su L, Yan M, Zhu Z, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. (2014) 5:2318–29. doi: 10.18632/oncotarget.v5i8
100. Yang W, Ning N, Jin X. The lncRNA H19 Promotes Cell Proliferation by Competitively Binding to miR-200a and Derepressing β-Catenin Expression in Colorectal Cancer. BioMed Res Int. (2017) 2017:2767484. doi: 10.1155/2017/2767484
101. Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, et al. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumour Biol J Int Soc Oncodevelopmental Biol Med. (2014) 35:9163–9. doi: 10.1007/s13277-014-2185-5
102. Zhang K, Luo Z, Zhang Y, Zhang L, Wu L, Liu L, et al. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer biomark Sect Dis Markers. (2016) 17:187–94. doi: 10.3233/CBM-160630
103. Rivankar S. An overview of doxorubicin formulations in cancer therapy. J Cancer Res Ther. (2014) 10:853–8. doi: 10.4103/0973-1482.139267
104. Shen S, Wang Y, Zhang Y, Dong Z, Xing J. Long non-coding RNA small nucleolar RNA host gene 14, a promising biomarker and therapeutic target in Malignancy. Front Cell Dev Biol. (2021) 9:746714. doi: 10.3389/fcell.2021.746714
105. Ashrafizadeh M, Mirzaei S, Hashemi F, Zarrabi A, Zabolian A, Saleki H, et al. New insight towards development of paclitaxel and docetaxel resistance in cancer cells: EMT as a novel molecular mechanism and therapeutic possibilities. BioMed Pharmacother. (2021) 141:111824. doi: 10.1016/j.biopha.2021.111824
106. Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol Off J Am Soc Clin Oncol. (2023) 41:1809–15. doi: 10.1200/JCO.22.02572
107. Zhu M, Lv Q, Huang H, Sun C, Pang D, Wu J. Identification of a four-long non-coding RNA signature in predicting breast cancer survival. Oncol Lett. (2020) 19(1):221–8. doi: 10.3892/ol.2019.11063
108. Ren S, Zhang Y, Yang X, Li X, Zheng Y, Liu Y, et al. N6-methyladenine- induced LINC00667 promoted breast cancer progression through m6A/KIAA1429 positive feedback loop. Bioengineered. (2022) 13:13462–73. doi: 10.1080/21655979.2022.2077893
109. Gonçalves TL, de Araújo LP, Pereira Ferrer V. Tamoxifen as a modulator of CXCL12-CXCR4-CXCR7 chemokine axis: A breast cancer and glioblastoma view. Cytokine. (2023) 170:156344. doi: 10.1016/j.cyto.2023.156344
110. Echeverria GV, Powell E, Seth S, Ge Z, Carugo A, Bristow C, et al. High-resolution clonal mapping of multi-organ metastasis in triple negative breast cancer. Nat Commun. (2018) 9:5079. doi: 10.1038/s41467-018-07406-4
111. Feng T, Zhang P, Sun Y, Wang Y, Tong J, Dai H, et al. High throughput sequencing identifies breast cancer-secreted exosomal LncRNAs initiating pulmonary pre-metastatic niche formation. Gene. (2019) 710:258–64. doi: 10.1016/j.gene.2019.06.004
112. Weigelt B, Peterse JL, van ‘t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. (2005) 5:591–602. doi: 10.1038/nrc1670
113. Venur VA, Leone JP. Targeted therapies for brain metastases from breast cancer. Int J Mol Sci. (2016) 17:1543. doi: 10.3390/ijms17091543
114. Xing F, Liu Y, Wu SY, Wu K, Sharma S, Mo YY, et al. Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal miRNA to Promote Brain Metastasis. Cancer Res. (2018) 78:4316–30. doi: 10.1158/0008-5472.CAN-18-1102
115. Lu Y, Chen L, Li L, Cao Y. Exosomes derived from brain metastatic breast cancer cells destroy the blood-brain barrier by carrying lncRNA GS1-600G8. 5. BioMed Res Int. (2020) 2020:7461727. doi: 10.1155/2020/7461727
116. Kong X, Li J, Li Y, Duan W, Qi Q, Wang T, et al. A novel long non-coding RNA AC073352.1 promotes metastasis and angiogenesis via interacting with YBX1 in breast cancer. Cell Death Dis. (2021) 12:670. doi: 10.1038/s41419-021-03943-x
117. Lim JP, Nair S, Shyamasundar S, Chua PJ, Muniasamy U, Matsumoto K, et al. Silencing Y-box binding protein-1 inhibits triple-negative breast cancer cell invasiveness via regulation of MMP1 and beta-catenin expression. Cancer Lett. (2019) 452:119–31. doi: 10.1016/j.canlet.2019.03.014
118. McSherry EA, Brennan K, Hudson L, Hill ADK, Hopkins AM. Breast cancer cell migration is regulated through junctional adhesion molecule-A-mediated activation of Rap1 GTPase. Breast Cancer Res BCR. (2011) 13:R31. doi: 10.1186/bcr2853
119. Khattar E, Maung KZY, Chew CL, Ghosh A, Mok MMH, Lee P, et al. Rap1 regulates hematopoietic stem cell survival and affects oncogenesis and response to chemotherapy. Nat Commun. (2019) 10:5349. doi: 10.1038/s41467-019-13082-9
120. Zhang P, Zhou H, Lu K, Lu Y, Wang Y, Feng T. Exosome-mediated delivery of MALAT1 induces cell proliferation in breast cancer. OncoTargets Ther. (2018) 11:291–9. doi: 10.2147/OTT
121. Turco C, Esposito G, Iaiza A, Goeman F, Benedetti A, Gallo E, et al. MALAT1-dependent hsa_circ_0076611 regulates translation rate in triple-negative breast cancer. Commun Biol. (2022) 5:598. doi: 10.1038/s42003-022-03539-x
122. Sun Z, Hu J, Ren W, Fang Y, Hu K, Yu H, et al. LncRNA SNHG3 regulates the BMSC osteogenic differentiation in bone metastasis of breast cancer by modulating the miR-1273g-3p/BMP3 axis. Biochem Biophys Res Commun. (2022) 594:117–23. doi: 10.1016/j.bbrc.2021.12.075
123. Li Y, Zhao Z, Liu W, Li X. SNHG3 functions as miRNA sponge to promote breast cancer cells growth through the metabolic reprogramming. Appl Biochem Biotechnol. (2020) 191:1084–99. doi: 10.1007/s12010-020-03244-7
124. Na-Er A, Xu YY, Liu YH, Gan YJ. Upregulation of serum exosomal SUMO1P3 predicts unfavorable prognosis in triple negative breast cancer. Eur Rev Med Pharmacol Sci. (2021) 25(1):154–60. doi: 10.26355/eurrev_202101_24379
125. Lan F, Zhang X, Li H, Yue X, Sun Q. Serum exosomal lncRNA XIST is a potential non-invasive biomarker to diagnose recurrence of triple-negative breast cancer. J Cell Mol Med. (2021) 25:7602–7. doi: 10.1111/jcmm.16009
126. Shi W, Jin X, Wang Y, Zhang Q, Yang L. High serum exosomal long non-coding RNA DANCR expression confers poor prognosis in patients with breast cancer. J Clin Lab Anal. (2022) 36:e24186. doi: 10.1002/jcla.24186
127. Zhang KJ, Tan XL, Guo L. The long non-coding RNA DANCR regulates the inflammatory phenotype of breast cancer cells and promotes breast cancer progression via EZH2-dependent suppression of SOCS3 transcription. Mol Oncol. (2020) 14:309–28. doi: 10.1002/1878-0261.12622
Keywords: breast cancer, lncRNA, exosomes, metastasis, chemoresistance, cancer biomarkers
Citation: Blancas-Zugarazo SS, Langley E and Hidalgo-Miranda A (2024) Exosomal lncRNAs as regulators of breast cancer chemoresistance and metastasis and their potential use as biomarkers. Front. Oncol. 14:1419808. doi: 10.3389/fonc.2024.1419808
Received: 18 April 2024; Accepted: 16 July 2024;
Published: 01 August 2024.
Edited by:
Naoyuki Kataoka, The University of Tokyo, JapanReviewed by:
Jessian Munoz, Texas Children’s Hospital, United StatesCopyright © 2024 Blancas-Zugarazo, Langley and Hidalgo-Miranda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sugela Susana Blancas-Zugarazo, c2JsYW5jYXN6QGlubWVnZW4uZWR1Lm14; Alfredo Hidalgo-Miranda, YWhpZGFsZ29AaW5tZWdlbi5nb2IubXg=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.