
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 18 June 2024
Sec. Head and Neck Cancer
Volume 14 - 2024 | https://doi.org/10.3389/fonc.2024.1412212
This article is part of the Research TopicAging and Oncology: Molecular Insights and Therapeutic AdvancesView all 3 articles
 Leonard E. Estephan1*†
Leonard E. Estephan1*† Gaurav Kumar2,3†
Gaurav Kumar2,3† Matthew Stewart1,4
Matthew Stewart1,4 Raphael Banoub1
Raphael Banoub1 Alban Linnenbach2,5
Alban Linnenbach2,5 Larry A. Harshyne2,6
Larry A. Harshyne2,6 Ubaldo E. Martinez-Outschoorn2,6
Ubaldo E. Martinez-Outschoorn2,6 My G. Mahoney1,5
My G. Mahoney1,5 Joseph M. Curry1,2
Joseph M. Curry1,2 Jennifer Johnson1,2,6
Jennifer Johnson1,2,6 Andrew P. South2,5
Andrew P. South2,5 Adam J. Luginbuhl1,2
Adam J. Luginbuhl1,2Introduction: Oral cavity squamous cell carcinoma (OSCC) occurs most frequently in patients >60 years old with a history of tobacco and alcohol use. Epidemiological studies describe increased incidence of OSCC in younger adults (<45 years). Despite its poor prognosis, knowledge of OSCC tumor microenvironment (TME) characteristics in younger adults is scarce and could help inform possible resistance to emerging treatment options.
Methods: Patients with OSCC were evaluated using TCGA-HNSC (n=121) and a stage and subsite-matched institutional cohort (n=8) to identify differential gene expression focusing on the extracellular matrix (ECM) and epithelial-mesenchymal transition (EMT) processes in younger (≤45 years) vs. older adults (≥60 years). NanoString nCounter analysis was performed using isolated total RNA from formalin-fixed paraffin-embedded (FFPE) tumor samples. Stained tumor slides from young and old OSCC patients were evaluated for CD8+ T-cell counts using immunohistochemistry.
Results: Younger OSCC patients demonstrated significantly increased expression of ECM remodeling and EMT process genes, as well as TME immunosuppression. Gene set enrichment analyses demonstrated increased ECM pathways and concurrent decreased immune pathways in young relative to old patients. Transcripts per million of genetic markers involved in ECM remodeling including LAMB3, VCAN, S100A9, COL5A1, and ITGB2 were significantly increased in tumors of younger vs. older patients (adjusted p-value < 0.10). Young patient TMEs demonstrated a 2.5-fold reduction in CD8+ T-cells as compared to older patients (p < 0.05).
Conclusion: Differential gene expression impacting ECM remodeling and TME immunosuppression may contribute to disease progression in younger adult OSCC and has implications on response to evolving treatment modalities, such as immune checkpoint inhibitor therapy.
Arising mostly in the mucosal linings of the oral cavity, pharynx, and larynx, head and neck squamous cell carcinoma (HNSCC) represents an aggressive multi-factorial disease that accounts for over 650,000 new cases annually, with a mortality rate of approximately 50–60% per year. Specifically, oral cavity squamous cell carcinoma (OSCC) is primarily a disease of older adults, occurring mostly in patients older than age 60 with a demonstrated history of tobacco and/or alcohol use (1–4). Younger cancer patients, by virtue of their age, have typically had a shorter durations of toxin and chemical exposure than their older counterparts raising the question of other etiologies of disease. Although human papillomavirus (HPV) has a well-described role in squamous cell carcinoma of the oropharynx in younger and middle-aged adults, it is not currently considered a driver of OSCC and data from HPV-positive patients is not included in this manuscript (2, 5).
Whether younger OSCC patients display a more aggressive phenotype with a poorer prognosis and a lower 3–5-year survival rate as compared to older OSCC patients remains controversial. A meta-analysis of 23,382 collective patients with mostly T1/T2 disease demonstrated no difference in disease-specific survival for patients aged 18–40 compared with those >40 years of age, yet multiple single-center studies report worsened survival in younger patients (4, 6–8). Oral cavity cancer is aggressive regardless of age, and includes a similar 5-year overall survival rate to several other head and neck cancers with HPV-negative status (9). This disease may confer an added complexity and severity when presenting in younger patients with no antecedent risk factors. In younger patients that lack extensive exposure to alcohol and tobacco, unique genetic predispositions may contribute to more severe OSCC disease. Notably, previous studies have found local recurrences and distant metastases are more prevalent in younger as compared to older patients (2). Based on its unclear etiology and potentially more aggressive clinical course, OSCC in younger patients may even be considered a distinct clinical entity compared to OSCC in older adults.
Treatment for OSCC has focused on multimodal therapy comprised of surgery, radiation, and chemotherapy. The poor overall survival of this cancer has prompted the search for new treatment strategies and paradigms. Recent success has been found with immune checkpoint inhibitors (ICI), specifically monoclonal antibodies targeting PD-1, which have been approved in the setting of recurrent or metastatic disease. There is intense interest in incorporating these agents into the care of locally advanced disease both to improve patient survival and potentially decrease toxicity. Patients who achieve immune engagement with ICIs can experience prolonged responses, though this occurs in less than 20% of patients. The toxicity profile is favorable and thus far does not appear to be additive with other modalities used in the locally advanced setting (10, 11). In order to optimize the use of novel immunotherapeutic agents we must better understand the enormous diversity of HNSCC. Tumors present with varying degrees of fibrosis, immune infiltrate, and alterations in the tumor microenvironment (TME) that may impact primary response to ICI and facilitate secondary resistance to these agents (12, 13).
Collectively, younger adults with or without risk factors who develop OSCC pose difficult questions of disease course and sequelae of treatment. Oral cavity tumors in this age group have been relatively understudied. Earlier reports have mainly focused on the epidemiological association of younger OSCC patients with prognosis and survival, but lack the mechanistic investigations necessary to understand the etiology of OSCC in this specific population (1–4, 14, 15). To address this, the present study utilizes multiple differential gene expression analysis methods to investigate genetic characteristics of younger and older OSCC cohorts that we hypothesize may contribute tumor aggressiveness and progression in the younger OSCC subset. Aggressiveness is defined in this manuscript as carrying elements of tumor infiltration and known histopathologic features that contribute to worsened disease-specific survival as it pertains to OSCC.
The RNA sequencing data as raw count matrix, and clinical data for 506 HNSCC patients from The Cancer Genome Atlas (TCGA) was obtained using the ‘TCGAbiolinks’ package in R/Bioconductor (16). Clinical data was stratified (HPV-negative and oral cavity subsite) resulting in 121 patients (n=17 [ ≤ 45 years old], n=104 [≥60 years old]) used for subsequent analysis. Upper and lower age cutoffs were selected to isolate distinct OSCC cohorts for gene expression analysis. The ages of <45 years for the young cohort and >60 years for the older cohort were selected as cutoff criteria based on epidemiological reports of OSCC disease incidence and criteria used in prior studies pertaining to this topic (1–4).
This study was approved by the Thomas Jefferson University Institutional Review Board. Eight subjects from our institutional biobank were identified (4 younger and 4 older adults), and RNA from their formalin-fixed paraffin-embedded (FFPE) tumor samples was isolated using an miRNeasy FFPE kit (Qiagen, Hilden, Germany). Age at the time of surgery was used to define our young (≤45 years) and old (≥60 years) cohorts (Table 1). Briefly, after excess paraffin was trimmed from each sample, the residual was removed by using deparaffinization solution and incubation at 56°C for 3 minutes. At room temperature, buffer PKD was added; samples were transferred to bead mill tubes containing 2.8 mm ceramic beads. Tissue was disrupted using a Bead Ruptor at 5 m/sec for 15 seconds, followed by icing for 45 seconds (Omni International, GA, USA) three separate times. Lysates were centrifuged at 11,000 xg 1 min. Proteinase K was added to the clear phase (~24 mAnson Units/ml), followed by serial incubations at 56°C and 80°C with intermittent vortexing. After centrifugation at 11,000 xg 1 minute, the lower phase was recovered, further clarified by centrifugation, then treated with DNaseI. Buffer RBC and ethanol were added for optimal binding conditions, and samples were applied to RNeasy MinElute spin columns. After rinsing with buffer RPE, total RNA was eluted in 20–30 µl of RNase-free water. FFPE RNA sample QC was determined using an Agilent TapeStation (Agilent Technoligies, CA, USA). DV200 determinations for percentage of total RNA fragments >200 nt, averaged 42 (range 18–63). An nCounter analysis system was utilized to run the PanCancer IO 360 Panel for 16 immuno-oncology pathways and bioprocesses (NanoString, WA, USA). NanoString technology is considered advantageous over next-generation sequencing and polymerase chain reaction as it is rapid, technically simple, and does not require nucleic acid amplification. Additionally, this tool has recently been implicated in diagnostic methods in cancers of the breast and lung, leukemia, and lymphoma (17).
Raw counts were utilized from the TCGA-HNSC cohort and the NanoString nCounter PanCancer IO 360 Panel was applied to our institutional biobank samples. Consisting of 770 genes, the PanCancer IO 360 Panel combines vital components involved in the complex interplay between the tumor microenvironment and the immune response in cancer. Before differential expression, batch effects or sample heterogeneity was tested using iSeqQC (18). Differential gene expression was performed between young and older adults using the DESeq2 package in R/Bioconductor (19). Genes were considered differentially expressed if they had an adjusted p-value ≤ 0.10. All plots were constructed using R/Bioconductor.
The DESeq2 test statistic was used as a ranking metric to perform Gene Set Enrichment Analysis (GSEA) in pre-ranked mode, with genes having zero base mean or “NA” test statistic values filtered out to avoid providing numerous duplicate values. The GSEA was performed against hallmark gene sets from MSigDB collections (20). Additionally, an analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) was performed on our list of differentially expressed genes between younger vs. older patients to identify biological processes modulated between these cohorts (21). Further, IPA software was used (Ingenuity system, Qiagen, CA, USA) to evaluate functional changes in the young adults and network analysis was performed using Cytoscape (22).
Ten additional patients (n=5 each of younger and older patients) with HPV-negative OSCC and available FFPE samples were utilized. Patients were matched based on site and stage of their tumor. Tissue preserved in FFPE was obtained from surgically resected specimens and underwent staining with monoclonal antibodies directed against CD8 (anti-CD8 (SP57) rabbit monoclonal primary antibody; Ventana Medical Systems, AZ, USA). The slides were digitally scanned at 20x magnification using the iScan HT whole-slide image scanner (Roche, Switzerland). Fully automated detection was performed on a Ventana Discovery Ultra System (Ventana Medical Systems, AZ, USA).
To interrogate sequencing data for a signal of cancer aggressiveness in young vs. older OSCC patients, we first utilized the TCGA-HNSC cohort. Differential RNA expression analysis was performed between OSCC from younger patients (n=17) and older patients (n=104) using DESeq2 (19). Statistical power was determined to be adequate for subsequent analysis with the size of each respective cohort. Here, we observed 1856 genes to be differentially expressed, where 214 genes were up-regulated, and 1642 were down-regulated in young patients (Figure 1A). Next, using the differentially expressed genes, we performed gene ontology analysis to evaluate the biological processes implicated in the younger patients using DAVID (21). Here, we observed up-regulation of ECM structural organization (Figure 1B) in the young oral cavity patients when compared to the older patients. Furthermore, GSEA analysis using ranked gene lists showed epithelial-mesenchymal transition (EMT) processes to be among the most up-regulated hallmarks in the younger cohort (Figure 1C). These results implicate the ECM and EMT processes in promoting disease progression in the OSCC tumors of young adults. Patient identifiers from TCGA and a list of differentially expressed genes can be found in the Supplementary File.
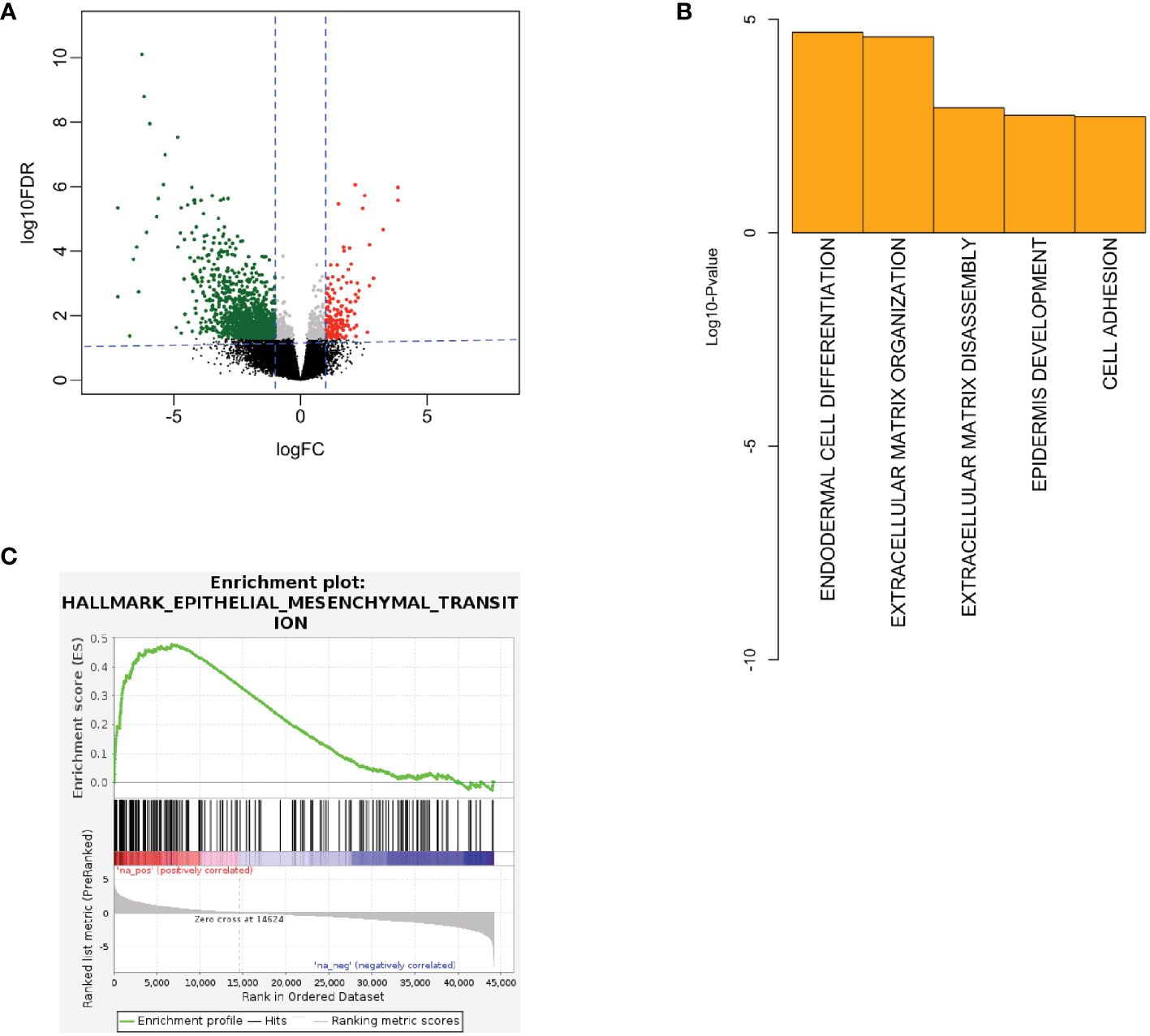
Figure 1 (A) Volcano plot showing RNA expression between young and old patients in TCGA. Here, red color denotes up-regulated genes in young and green denotes down-regulated genes in young patients. (B) Biological processes being up-regulated in the young patients in TCGA. (C) GSEA showing EMT processes to be up-regulated in the young TCGA patients (FDR q-value < 0.01, NES = 2.49). EMT, epithelial-mesenchymal transition, FC, fold-change, FDR, false discovery rate, GSEA, gene set enrichment analysis, IHC, immunohistochemistry, NES, normalized enrichment score, TCGA, the cancer genome atlas.
Next, to validate our findings from the TCGA-HNSC cohort, we utilized an institutional OSCC cohort. Here, FFPE tumor blocks were obtained for 8 site and stage-matched patients (n=4 each for young and old patients) and RNA was extracted to perform NanoString nCounter gene expression assays using the PanCancer IO 360 Panel. A summary of patient age (at time of surgery), self-reported race, sex, tumor subsite within the oral cavity, HPV status, TNM staging, evidence of perineural invasion, evidence of lymphovascular invasion, disease recurrence, reported alcohol use, reported smoking history, and current status of patients from the institutional cohort can be found in Table 1. Patient RNA expression for the 770 genes of the PanCancer IO 360 Panel was obtained and differential expression was performed between young and old OSCC patients using DESeq2. Here, we found 40 genes to be differentially expressed in the young patients compared to older patients (34 up-regulated, 6 down-regulated) (Figure 2A). Next, functional annotations were performed on the differentially expressed genes, confirming our TCGA results. We found genes involved in cell adhesion and ECM processes to be up-regulated in young vs. older patients. These data correspond to our TCGA-HNSC observations, and strongly suggest that ECM remodeling is a key biological process contributing to aggressiveness in the OSCC tumors of young patients. NanoString outputs are available in the public repository FigShare or are available upon request.
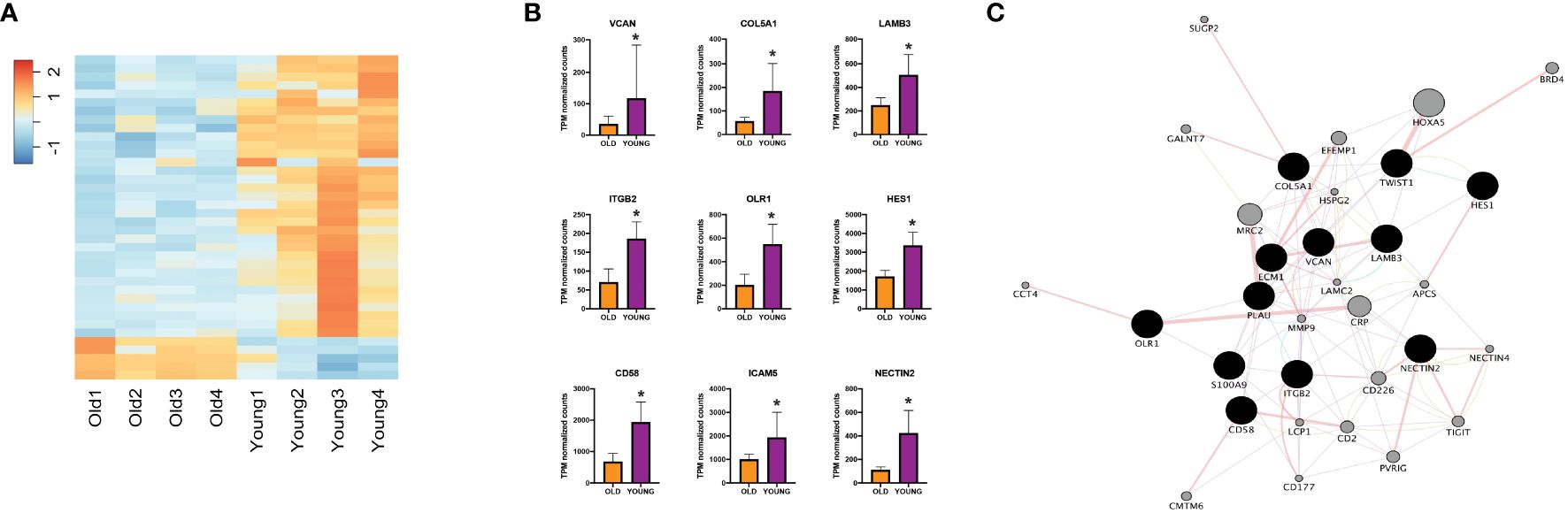
Figure 2 (A) Heatmap showing expression of differentially expressed genes between young and old patients using nCounter. (B) NanoString nCounter gene expression profiles of different genes of interest. (C) Network analysis including genes of interest showing physical interaction with ECM-related genes. *Adjusted p-value < 0.10 for young compared to old. ECM, extracellular matrix, TPM, transcripts per million.
To characterize the key mediators that might be promoting ECM remodeling processes in young OSCC patients, we performed functional annotation analysis using IPA software. We identified key genes including VCAN, S100A9, COL5A1, LAMB3, ITGB2, OLR1, HES1, CD58, TWIST1, and NECTIN2 to be of high-interest due to their described role in functions of disease progression in multiple cancers (Figure 2B) (23–32). To evaluate their interaction in ECM remodeling, we constructed a network map using Cytoscape (Figure 2C), which showed physical interaction of LAMB3 with key ECM-regulating genes such as COL5A1, PLAU, ITGB2, and ECM1 (33–36).
Our analysis of the TCGA-HNSC and nCounter gene expression panel showed down-regulation of immune-related processes in the young OSCC patients. Here, functional annotation analysis of the TCGA-HNSC cohort demonstrated down-regulation of complement and B-cell activations, as well as the interferon gamma response processes in the young OSCC patients (Figures 3A, B). To assess the immune cell presence in younger patients, we performed immunohistochemistry (IHC) of CD8+ T-cells on FFPE samples of younger and older OSCC patients (n=10; 5 each for younger and older patients). Here, we observed a significant reduction (2.5-fold change; p<0.05) of CD8+ T-cell infiltration in the young OSCC tumors (Figure 3C). Collectively, these data suggest that younger OSCC patients have a more immunosuppressive TME, where the stromal cells may dynamically modulate the microenvironment to favor tumor aggressiveness in this age group.
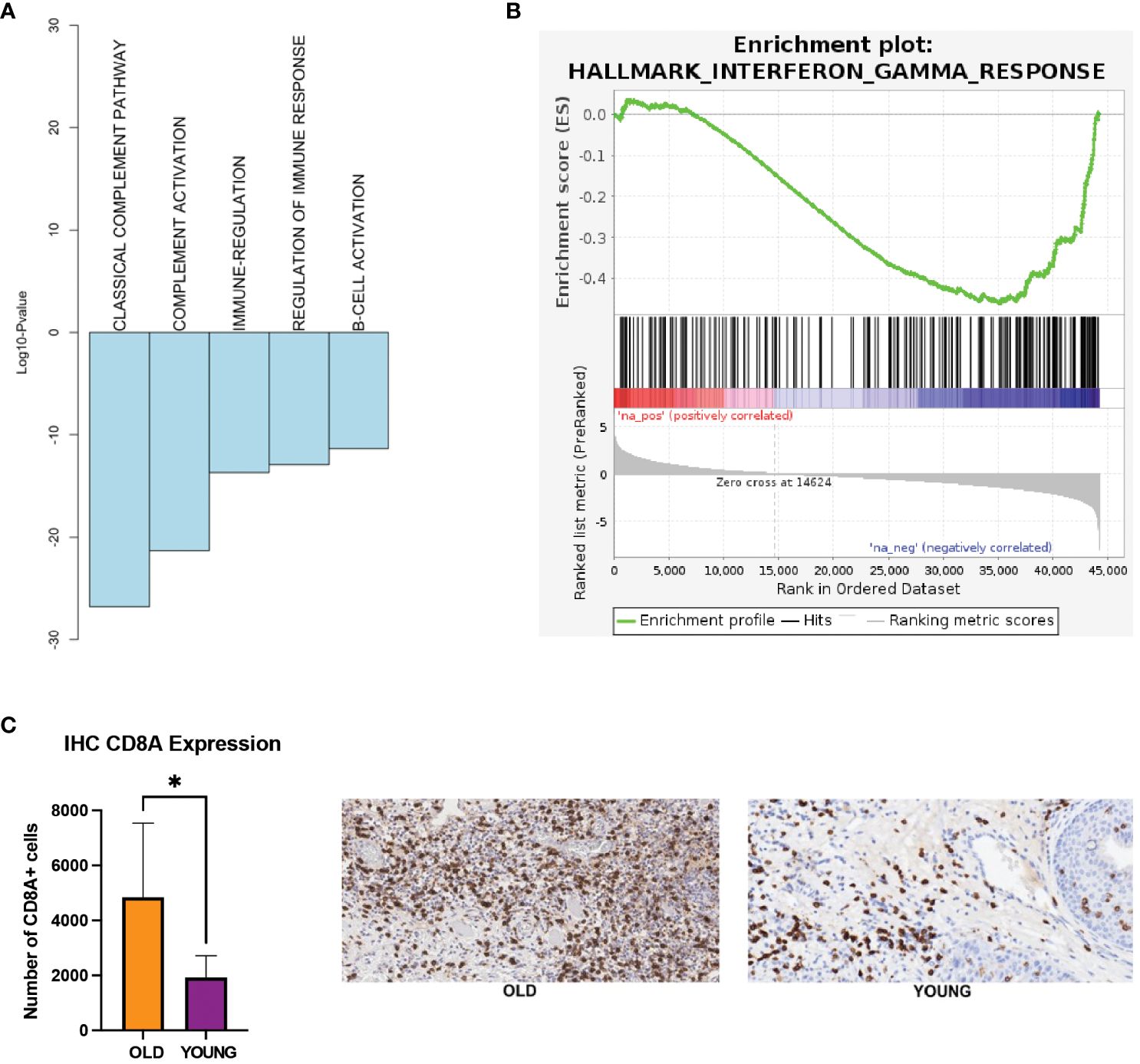
Figure 3 (A) Biological processes being down-regulated in the young patients in TCGA. (B) GSEA showing interferon gamma response processes to be down-regulated in the young patients in TCGA (FDR q-value = 0.01, NES = -1.61). (C) IHC expression of CD8 T-cells. *p-value < 0.05 for young compared to old patients. EMT, epithelial-mesenchymal transition; FDR, false discovery rate; GSEA, gene set enrichment analysis; IHC, immunohistochemistry; NES, normalized enrichment score; TCGA, the cancer genome atlas.
To discern whether LAMB3 might be regulating the immunosuppression in the TME of young OSCC adults, we evaluated the correlation of these genes with immune-regulation using an in-silico approach. We did so by performing network analysis of LAMB3 to identify its interactions with the CD8A gene. As demonstrated in Figure 4, LAMB3 is predicted to interact with CD8A, CD3D, and CD3E. Other immune and ECM-specific genes such as ECM1, ITGA2, COl17A1, KRT5, and HLA are also shown to be amongst the node genes.
The present study identifies ECM remodeling and EMT processes as central in promoting increased aggressiveness in younger adult OSCC tumors. The ECM is an essential component of the TME, as increased ECM component deposition and crosslinking contributes to tumor progression and is a major barrier to effective checkpoint immunotherapy treatment (37, 38). Previous reports discuss the pathological role of increased ECM remodeling in HNSCC relative to normal tissue (39–41). However, to our knowledge, this is the first study to describe ECM remodeling processes as having an enhanced differential expression in younger vs. older OSCC patients. Specifically, we report increased LAMB3 activity, suggesting its role as a primary gene impacting the ECM in younger adults. LAMB3 is known to exhibit tumorigenic effects in multiple types of cancer, including HNSCC (42). However, its role in promoting tumorigenesis particularly in young OSCC patients has not been previously well-described.
As it pertains to tumor resistance in immune checkpoint inhibition (ICI) therapy, increased ECM deposition in young OSCC patients may drastically impact prognosis. Previous work has demonstrated the deleterious effects of aberrant ECM remodeling on ICI therapy in several types of cancer. In a study of metastatic melanoma samples, Hugo et al. assessed the transcriptomes and tumor mutanomes of pretreatment melanoma biopsies to better define the role of genetics in resistance to anti-PD-1 therapy. They reported that tumors with innate resistance to immunotherapy displayed transcriptional signatures of upregulated genes implicated in ECM remodeling in addition to cellular adhesion, mesenchymal transition, and angiogenesis (38). Separately, Peng et al. identified the negative consequences of extensive tumor collagen deposition on anti-PD-1/PD-L1 effectiveness in lung tumors. Increased ECM resulted in elevated LAIR1 activity which mechanistically exhausts T-cells by way of tyrosine phosphatase SHP-1, rendering ICI therapy less effective. As a proof-of-concept, when this group reduced collagen deposition in their model through LOXL2 suppression, both T-cell exhaustion and anti-PD-1 resistance were diminished (37). Given these findings, the increased ECM activity described in our young OSCC cohort may contribute to ICI treatment resistance of such patients.
Extracellular matrix remodeling not only contributes to tumor progression and resistance to therapy, but also impacts TME immune cell presence and activity. It is known that the TME is comprised of several cell populations, including cancer and stromal cells, in addition to non-cell ECM components. A complex interplay exists between cellular stromal subpopulations and tumor cells communicated through various secreted cytokines, chemokines, growth factors and ECM proteins (39). Our findings suggest that in young OSCC patients there is a degree of immune suppression relative to older individuals, which we believe is due in part to increased ECM remodeling processes. In assessing our nCounter gene expression panel, we observed up-regulation of genes associated with fibroblasts (LAMB3 and VCAN) in the tumors of our younger patients. In addition, CD8+ T-cells were not as prevalent in younger patients demonstrated through IHC. Mechanistically, several specific effects of ECM processing on immune cell function have been proposed. The ECM dynamically modulates the physical organization, signaling cascades, and cellular constituents in the tumors for both exclusion and inactivation of T-cells (37, 43–47). Exclusion of T-cells might be regulated through haptotaxis signals from altered ECM, where instead of entering the TME, T-cells migrate along ECM-rich encapsulation of tumors due to the gradients of substrate rigidity and adhesion molecules such as aligned collagen fiber. Stiffened ECM can induce poor diffusion that can enhances glycolytic metabolism and acidification, which suppresses the activation of T-lymphocytes through the specific interaction between V-domain immunoglobulin suppressor of T-cell activation and co-inhibitory receptor P-selectin glycoprotein ligand-1 in acidic TMEs. Proteins of the ECM are known to contribute to the direct regulation of T-lymphocytes. As reported, CD8+ T-cells are suppressed by collagen through LAIR1 and tyrosine phosphatase SHP-1, and high molecular weight hyaluronic acid could enhance the activity of regulatory T-cells in vitro (37, 43–47). Concurrent increases in matrix remodeling paralleled by suppressed TME immune activity may promote both disease progression and tumor aggressiveness.
Our results suggest that LAMB3 may be up-regulated in the TME of the younger OSCC patients and act as part of a gene network along with ITGA2 and LAMC2. In a recently published study of pancreatic ductal adenocarcinoma (PDAC), Islam et al. proposed that these three genes potentiate disease progression and severity. Levels of gene mRNA expression were significantly higher in PDAC tissue as compared to normal tissue, and expression levels were associated with increased pathological tumor stage. Disease-specific and overall survival were significantly reduced in patients with high LAMB3, ITGA2, and LAMC2 expression as compared to patients with low expression (31). Similar to PDAC, these ECM-related genes may contribute to disease aggressiveness in the young OSCC population.
Besides LAMB3, prior reports discuss other significantly up-regulated genes from our study as they pertain to various cancers. Mitsui et al. studied VCAN and its potential implications in clear cell renal cell carcinoma (ccRCC). This group concluded that VCAN was associated with poor prognosis through induction of tumor development by reducing TNF-mediated cellular apoptosis (28). Besides ccRCC, VCAN has been described for its potential negative role in malignancy of the prostate, colon, and ovary (48–50). Similarly, COL5A1 was found to negatively contribute to ovarian cancer, as described in a recent study by Zhang et al. This group demonstrated elevated expression of COL5A1 in ovarian cancer cells, while knockdown of this gene reduced cellular capabilities to proliferate and migrate. Additionally, this protein was found to be overexpressed specifically in cells resistant to paclitaxel, which was significantly improved following gene silencing (51). The remainder of genes described in our study have also been implicated in several types of cancer, but an extensive discussion of each is beyond the scope of this manuscript.
Converse to an apparent upregulation of such genes, we observed a diminished CD8+ T-cell presence in the TME of young patients. Natural killer (NK) cells have been shown to secrete FLT3L, which is a potent driver of dendritic cell (DC) activation and antigen presentation ability. Further, DC-CD8 interactions within the TME are vital for proper immune surveillance. The implications of theoretically diminished FLT3L pathway in young patients with OSCC may be drastic. Bickett et al. used the TCGA to describe an increased 5-year overall survival in HNSCC patients with increased FLT3L and FLT3 (ligand receptor) expression, demonstrating the consequences of a depleted NK cell response (52). Taken together, the immune suppression in the TME of our young patients, as evidenced by decreased CD8+ cell presence, may contribute to enhanced tumor aggressiveness through obtunded immune cell signaling, activity, and surveillance. Not only are the implications for aggressive tumor behavior elucidated in these findings, but also the major implications of immunotherapy resistance. As ICI treatment becomes a front-line approach either in the neoadjuvant or definitive setting, we need to be thoughtful about defining patients’ TME to inform us about known resistance patterns. In addition, further work aimed at identifying strategies to improve therapeutic efficacy is vitally important for patients with OSCC.
Our study is not without certain limitations. Tumor tissue utilized for the NanoString nCounter analysis was obtained from a cohort of 4 younger and 4 older patients from our institution. Ideally, this matched cohort could have been enlarged to include additional patients and possibly strengthen our findings. Additionally, the tumor samples available for our IHC quantification of CD8+ T-cells within the TME were of institutional patients of the same age restrictions but were separate from the NanoString group. Under ideal conditions, these samples would have come from the same matched cohort, however this was not possible due to tissue availability. Lastly, this study of young OSCC patients provides evidence of differential expression for genetic markers known to be involved in cancer progression, however future studies are required to further elucidate mechanisms of relatively poor prognosis in this population.
This study suggests that younger adults with OSCC have a TME with depletion of effector immune cells and enhanced ECM remodeling processes, contributing to increased disease aggressiveness. As a downstream effect, this patient population may also suffer from increased resistance to certain therapeutic options, such as immunotherapy. Further work is required to expand upon these initial findings and define specific mechanistic pathways leading to worsened prognosis for OSCC patients of young age.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://figshare.com/, https://figshare.com/s/152292bb44d76d45c2ad.
The studies involving humans were approved by Thomas Jefferson University Hospital Institutional Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
LE: Investigation, Writing – original draft, Writing – review & editing, Data curation. GK: Writing – review & editing, Data curation, Formal analysis, Writing – original draft. MS: Data curation, Investigation, Writing – review & editing. RB: Data curation, Investigation, Writing – review & editing. ALi: Data curation, Writing – review & editing. LH: Writing – review & editing. UM-O: Writing – review & editing. MM: Writing – review & editing. JC: Writing – review & editing. JJ: Visualization, Writing – review & editing. AS: Visualization, Writing – review & editing. ALu: Writing – review & editing, Methodology, Supervision, Visualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Financial support for The Wistar Institute core facilities was provided by Cancer Center Support Grant P30 CA010815. This project utilized the Biostatistics, Bioinformatics and Cancer Genomics Shared Resources at the Sidney Kimmel Cancer Center, supported by the NCI, grant 5P30CA056036–17.
We are grateful to The Wistar Institute’s Genomics Facility for technical support with the NanoString nCounter analysis system.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1412212/full#supplementary-material
1. Majchrzak E, Szybiak B, Wegner A, Pienkowski P, Pazdrowski J, Luczewski L, et al. Oral cavity and oropharyngeal squamous cell carcinoma in young adults: a review of the literature. Radiol Oncol. (2014) 48:1–10. doi: 10.2478/raon-2013-0057
2. Liu X, Gao XL, Liang XH, Tang YL. The etiologic spectrum of head and neck squamous cell carcinoma in young patients. Oncotarget. (2016) 7:66226–38. doi: 10.18632/oncotarget.v7i40
3. Myers JN, Elkins T, Roberts D, Byers RM. Squamous cell carcinoma of the tongue in young adults: increasing incidence and factors that predict treatment outcomes. Otolaryngol Head Neck Surg. (2000) 122:44–51. doi: 10.1016/S0194-5998(00)70142-2
4. Jeon JH, Kim MG, Park JY, Lee JH, Kim MJ, Myoung H, et al. Analysis of the outcome of young age tongue squamous cell carcinoma. Maxillofac Plast Reconstr Surg. (2017) 39:41. doi: 10.1186/s40902-017-0139-8
5. Dos Santos Costa SF, Brennan PA, Gomez RS, Fregnani ER, Santos-Silva AR, Martins MD, et al. Molecular basis of oral squamous cell carcinoma in young patients: Is it any different from older patients? J Oral Pathol Med. (2018) 47:541–6. doi: 10.1111/jop.12642
6. Park JO, Sun DI, Cho KJ, Joo YH, Yoo HJ, Kim MS. Clinical outcome of squamous cell carcinoma of the tongue in young patients: a stage-matched comparative analysis. Clin Exp Otorhinolaryngol. (2010) 3:161–5. doi: 10.3342/ceo.2010.3.3.161
7. Mneimneh WS, Xu B, Ghossein C, Alzumaili B, Sethi S, Ganly I, et al. Clinicopathologic characteristics of young patients with oral squamous cell carcinoma. Head Neck Pathol. (2021) 15:1099–108. doi: 10.1007/s12105-021-01320-w
8. Lee DS, Ramirez RJ, Lee JJ, Valenzuela CV, Zevallos JP, Mazul AL, et al. Survival of young versus old patients with oral cavity squamous cell carcinoma: A meta-analysis. Laryngoscope. (2021) 131:1310–9. doi: 10.1002/lary.29260
9. Li H, Torabi SJ, Yarbrough WG, Mehra S, Osborn HA, Judson B. Association of human papillomavirus status at head and neck carcinoma subsites with overall survival. JAMA Otolaryngol Head Neck Surg. (2018) 144:519–25. doi: 10.1001/jamaoto.2018.0395
10. Tolentino Ede S, Centurion BS, Ferreira LH, Souza AP, Damante JH, Rubira-Bullen IR. Oral adverse effects of head and neck radiotherapy: literature review and suggestion of a clinical oral care guideline for irradiated patients. J Appl Oral Sci. (2011) 19:448–54. doi: 10.1590/s1678-77572011000500003
11. Johnson DB, Nebhan CA, Moslehi JJ, Balko JM. Immune-checkpoint inhibitors: long-term implications of toxicity. Nat Rev Clin Oncol. (2022) 19:254–67. doi: 10.1038/s41571-022-00600-w
12. Zaitsev A, Chelushkin M, Dyikanov D, Cheremushkin I, Shpak B, Nomie K, et al. Precise reconstruction of the TME using bulk RNA-seq and a machine learning algorithm trained on artificial transcriptomes. Cancer Cell. (2022) 40:879–94.e16. doi: 10.1016/j.ccell.2022.07.006
13. Bagaev A, Kotlov N, Nomie K, Svekolkin V, Gafurov A, Isaeva O, et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell. (2021) 39:845–65.e7. doi: 10.1016/j.ccell.2021.04.014
14. Cariati P, Cabello-Serrano A, Perez-de Perceval-Tara M, Monsalve-Iglesias F, Martinez-Lara I. Oral and oropharyngeal squamous cell carcinoma in young adults: A retrospective study in Granada University Hospital. Med Oral Patol Oral Cir Bucal. (2017) 22:e679–e85. doi: 10.4317/medoral.21755
15. Garavello W, Spreafico R, Gaini RM. Oral tongue cancer in young patients: a matched analysis. Oral Oncol. (2007) 43:894–7. doi: 10.1016/j.oraloncology.2006.10.013
16. Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. (2016) 44:e71. doi: 10.1093/nar/gkv1507
17. Goytain A, Ng T. NanoString nCounter technology: high-throughput RNA validation. Methods Mol Biol. (2020) 2079:125–39. doi: 10.1007/978-1-4939-9904-0_10
18. Kumar G, Ertel A, Feldman G, Kupper J, Fortina P. iSeqQC: a tool for expression-based quality control in RNA sequencing. BMC Bioinf. (2020) 21:56. doi: 10.1186/s12859-020-3399-8
19. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
20. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U.S.A. (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
21. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. (2009) 4:44–57. doi: 10.1038/nprot.2008.211
22. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. doi: 10.1101/gr.1239303
23. Zhao Z, Rahman MA, Chen ZG, Shin DM. Multiple biological functions of Twist1 in various cancers. Oncotarget. (2017) 8:20380–93. doi: 10.18632/oncotarget.v8i12
24. Xu S, Wen Z, Jiang Q, Zhu L, Feng S, Zhao Y, et al. CD58, a novel surface marker, promotes self-renewal of tumor-initiating cells in colorectal cancer. Oncogene. (2015) 34:1520–31. doi: 10.1038/onc.2014.95
25. Wu A, Zhang S, Liu J, Huang Y, Deng W, Shu G, et al. Integrated analysis of prognostic and immune associated integrin family in ovarian cancer. Front Genet. (2020) 11:705. doi: 10.3389/fgene.2020.00705
26. Rani A, Greenlaw R, Smith RA, Galustian C. HES1 in immunity and cancer. Cytokine Growth Factor Rev. (2016) 30:113–7. doi: 10.1016/j.cytogfr.2016.03.010
27. Oshima T, Sato S, Kato J, Ito Y, Watanabe T, Tsuji I, et al. Nectin-2 is a potential target for antibody therapy of breast and ovarian cancers. Mol Cancer. (2013) 12:60. doi: 10.1186/1476-4598-12-60
28. Mitsui Y, Shiina H, Kato T, Maekawa S, Hashimoto Y, Shiina M, et al. Versican promotes tumor progression, metastasis and predicts poor prognosis in renal carcinoma. Mol Cancer Res. (2017) 15:884–95. doi: 10.1158/1541-7786.MCR-16-0444
29. Liu W, Wei H, Gao Z, Chen G, Liu Y, Gao X, et al. COL5A1 may contribute the metastasis of lung adenocarcinoma. Gene. (2018) 665:57–66. doi: 10.1016/j.gene.2018.04.066
30. Khaidakov M, Mitra S, Kang BY, Wang X, Kadlubar S, Novelli G, et al. Oxidized LDL receptor 1 (OLR1) as a possible link between obesity, dyslipidemia and cancer. PloS One. (2011) 6:e20277. doi: 10.1371/journal.pone.0020277
31. Islam S, Kitagawa T, Baron B, Abiko Y, Chiba I, Kuramitsu Y. ITGA2, LAMB3, and LAMC2 may be the potential therapeutic targets in pancreatic ductal adenocarcinoma: an integrated bioinformatics analysis. Sci Rep. (2021) 11:10563. doi: 10.1038/s41598-021-90077-x
32. Gebhardt C, Németh J, Angel P, Hess J. S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol. (2006) 72:1622–31. doi: 10.1016/j.bcp.2006.05.017
33. Zoppi N, Gardella R, De Paepe A, Barlati S, Colombi M. Human fibroblasts with mutations in COL5A1 and COL3A1 genes do not organize collagens and fibronectin in the extracellular matrix, down-regulate alpha2beta1 integrin, and recruit alphavbeta3 Instead of alpha5beta1 integrin. J Biol Chem. (2004) 279:18157–68. doi: 10.1074/jbc.M312609200
34. Yin H, Wang J, Li H, Yu Y, Wang X, Lu L, et al. Extracellular matrix protein-1 secretory isoform promotes ovarian cancer through increasing alternative mRNA splicing and stemness. Nat Commun. (2021) 12:4230. doi: 10.1038/s41467-021-24315-1
35. Xu D, Li T, Wang R, Mu R. Expression and pathogenic analysis of integrin family genes in systemic sclerosis. Front Med (Lausanne). (2021) 8:674523. doi: 10.3389/fmed.2021.674523
36. Chen G, Sun J, Xie M, Yu S, Tang Q, Chen L. PLAU promotes cell proliferation and epithelial-mesenchymal transition in head and neck squamous cell carcinoma. Front Genet. (2021) 12:651882. doi: 10.3389/fgene.2021.651882
37. Peng DH, Rodriguez BL, Diao L, Chen L, Wang J, Byers LA, et al. Collagen promotes anti-PD-1/PD-L1 resistance in cancer through LAIR1-dependent CD8(+) T cell exhaustion. Nat Commun. (2020) 11:4520. doi: 10.1038/s41467-020-18298-8
38. Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. (2016) 165:35–44. doi: 10.1016/j.cell.2016.02.065
39. Peltanova B, Raudenska M, Masarik M. Effect of tumor microenvironment on pathogenesis of the head and neck squamous cell carcinoma: a systematic review. Mol Cancer. (2019) 18:63. doi: 10.1186/s12943-019-0983-5
40. Plzak J, Boucek J, Bandurova V, Kolar M, Hradilova M, Szabo P, et al. The head and neck squamous cell carcinoma microenvironment as a potential target for cancer therapy. Cancers (Basel). (2019) 11. doi: 10.3390/cancers11040440
41. Kramer RH, Shen X, Zhou H. Tumor cell invasion and survival in head and neck cancer. Cancer Metastasis Rev. (2005) 24:35–45. doi: 10.1007/s10555-005-5046-2
42. Liu L, Jung SN, Oh C, Lee K, Won HR, Chang JW, et al. LAMB3 is associated with disease progression and cisplatin cytotoxic sensitivity in head and neck squamous cell carcinoma. Eur J Surg Oncol. (2019) 45:359–65. doi: 10.1016/j.ejso.2018.10.543
43. Pruitt HC, Lewis D, Ciccaglione M, Connor S, Smith Q, Hickey JW, et al. Collagen fiber structure guides 3D motility of cytotoxic T lymphocytes. Matrix Biol. (2020) 85-86:147–59. doi: 10.1016/j.matbio.2019.02.003
44. Johnston RJ, Su LJ, Pinckney J, Critton D, Boyer E, Krishnakumar A, et al. VISTA is an acidic pH-selective ligand for PSGL-1. Nature. (2019) 574:565–70. doi: 10.1038/s41586-019-1674-5
45. Huang J, Zhang L, Wan D, Zhou L, Zheng S, Lin S, et al. Extracellular matrix and its therapeutic potential for cancer treatment. Signal Transduct Target Ther. (2021) 6:153. doi: 10.1038/s41392-021-00544-0
46. Bougherara H, Mansuet-Lupo A, Alifano M, Ngô C, Damotte D, Le Frère-Belda MA, et al. Real-time imaging of resident T cells in human lung and ovarian carcinomas reveals how different tumor microenvironments control T lymphocyte migration. Front Immunol. (2015) 6:500. doi: 10.3389/fimmu.2015.00500
47. Bollyky PL, Falk BA, Wu RP, Buckner JH, Wight TN, Nepom GT. Intact extracellular matrix and the maintenance of immune tolerance: high molecular weight hyaluronan promotes persistence of induced CD4+CD25+ regulatory T cells. J Leukoc Biol. (2009) 86:567–72. doi: 10.1189/jlb.0109001
48. Suhovskih AV, Aidagulova SV, Kashuba VI, Grigorieva EV. Proteoglycans as potential microenvironmental biomarkers for colon cancer. Cell Tissue Res. (2015) 361:833–44. doi: 10.1007/s00441-015-2141-8
49. Ricciardelli C, Mayne K, Sykes PJ, Raymond WA, McCaul K, Marshall VR, et al. Elevated levels of versican but not decorin predict disease progression in early-stage prostate cancer. Clin Cancer Res. (1998) 4:963–71.
50. Ghosh S, Albitar L, LeBaron R, Welch WR, Samimi G, Birrer MJ, et al. Up-regulation of stromal versican expression in advanced stage serous ovarian cancer. Gynecol Oncol. (2010) 119:114–20. doi: 10.1016/j.ygyno.2010.05.029
51. Zhang J, Zhang J, Wang F, Xu X, Li X, Guan W, et al. Overexpressed COL5A1 is correlated with tumor progression, paclitaxel resistance, and tumor-infiltrating immune cells in ovarian cancer. J Cell Physiol. (2021) 236:6907–19. doi: 10.1002/jcp.30350
Keywords: oral cavity squamous cell carcinoma (OSCC), age-related tumor aggressiveness, tumor microenvironment, immunosuppression, extracellular matrix
Citation: Estephan LE, Kumar G, Stewart M, Banoub R, Linnenbach A, Harshyne LA, Martinez-Outschoorn UE, Mahoney MG, Curry JM, Johnson J, South AP and Luginbuhl AJ (2024) Altered extracellular matrix correlates with an immunosuppressive tumor microenvironment and disease progression in younger adults with oral cavity squamous cell carcinoma. Front. Oncol. 14:1412212. doi: 10.3389/fonc.2024.1412212
Received: 04 April 2024; Accepted: 05 June 2024;
Published: 18 June 2024.
Edited by:
Dechao Feng, University College London, United KingdomReviewed by:
Geza Tamas Szabo, Semmelweis University, HungaryCopyright © 2024 Estephan, Kumar, Stewart, Banoub, Linnenbach, Harshyne, Martinez-Outschoorn, Mahoney, Curry, Johnson, South and Luginbuhl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leonard E. Estephan, TGVvbmFyZC5Fc3RlcGhhbkBqZWZmZXJzb24uZWR1
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.