
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 19 August 2024
Sec. Pediatric Oncology
Volume 14 - 2024 | https://doi.org/10.3389/fonc.2024.1399442
This article is part of the Research Topic Global Approaches to Molecular Diagnostics for Pediatric Cancer View all 8 articles
 Trevor A. Burrow1,2
Trevor A. Burrow1,2 Balakrishna Koneru1
Balakrishna Koneru1 Shawn J. Macha1,3
Shawn J. Macha1,3 Wenyue Sun4
Wenyue Sun4 Frederic G. Barr4
Frederic G. Barr4 Timothy J. Triche5
Timothy J. Triche5 C. Patrick Reynolds1,2,3*
C. Patrick Reynolds1,2,3*Introduction: Alternative lengthening of telomeres (ALT) occurs in sarcomas and ALT cancers share common mechanisms of therapy resistance or sensitivity. Telomeric DNA C-circles are self-primed circular telomeric repeats detected with a PCR assay that provide a sensitive and specific biomarker exclusive to ALT cancers. We have previously shown that 23% of high-risk neuroblastomas are of the ALT phenotype. Here, we investigate the frequency of ALT in Ewing’s family sarcoma (EFS), rhabdomyosarcoma (RMS), and osteosarcoma (OS) by analyzing DNA from fresh frozen primary tumor samples utilizing the real-time PCR C-circle Assay (CCA).
Methods: We reviewed prior publications on ALT detection in pediatric sarcomas. DNA was extracted from fresh frozen primary tumors, fluorometrically quantified, C-circles were selectively enriched by isothermal rolling cycle amplification and detected by real-time PCR.
Results: The sample cohort consisted of DNA from 95 EFS, 191 RMS, and 87 OS primary tumors. One EFS and 4 RMS samples were inevaluable. Using C-circle positive (CC+) cutoffs previously defined for high-risk neuroblastoma, we observed 0 of 94 EFS, 5 of 187 RMS, and 62 of 87 OS CC+ tumors.
Conclusions: Utilizing the ALT-specific CCA we observed ALT in 0% of EFS, 2.7% of RMS, and 71% of OS. These data are comparable to prior studies in EFS and OS using less specific ALT markers. The CCA can provide a robust and sensitive means of identifying ALT in sarcomas and has potential as a companion diagnostic for ALT targeted therapeutics.
Telomeres are nucleoprotein structures at the ends of chromosomes (1) that contain 5-10 kilobases of the canonical hexanucleotide (5’-TTAGGG-3’) repeat sequence encased in sheltering proteins (2). This complex protects genomic DNA from replicative erosion (3, 4), shields the ends of chromosomes from aberrant fusion (5), and prevents DNA damage response (DDR) elements from errantly recognizing genomic DNA (2, 6, 7). Approximately 85-90% of all cancers achieve replicative immortality by utilizing the telomere maintenance mechanism (TMM) telomerase (TA), a ribonucleotide reverse transcriptase (8, 9). The remaining 10-15% of cancer cases (~250,000 U.S. patients annually) use a non-telomerase TMM called alternative lengthening of telomeres (ALT) (10, 11).
Incidence of ALT varies amongst sarcomas (Tables 1, 2), with the majority of cases arising from tissues of mesenchymal or neuroepithelial origin (10, 22). Cancers with an estimated ALT frequency >40% include osteosarcoma (OS), diffuse and anaplastic astrocytomas, undifferentiated pleomorphic sarcomas, and pediatric grade 4 glioblastoma multiforme (10, 14). Previously reported patient sample screenings have demonstrated a broad range of ALT frequency amongst pediatric cancers, from 0% in Ewing’s Family Sarcoma (EFS), up to 85% in OS (10, 12, 14). Recently, there have been calls for assessing patient samples with currently available ALT biomarkers to confirm historically reported ALT frequencies, especially for OS (23).
The hallmarks of the ALT phenotype include absence of TA activity (TERT mRNA expression provides a suitable surrogate for TA activity) (24) with the presence of high telomere content and heterogenous telomere length (25, 26), non-canonical telomere variant repeats (27), extra-chromosomal telomeric repeats (28), ALT-associated PML bodies (APBs) (14, 29–31), ultrabright telomere foci by FISH (10), and telomeric DNA C-circles (Tables 1, 2) (32). These characteristic markers have been used to screen tumor sample cohorts to determine the frequency of ALT among various tumor histologies (10, 13, 14, 18, 33, 34). Each of the methods has advantages and disadvantages (Table 3).
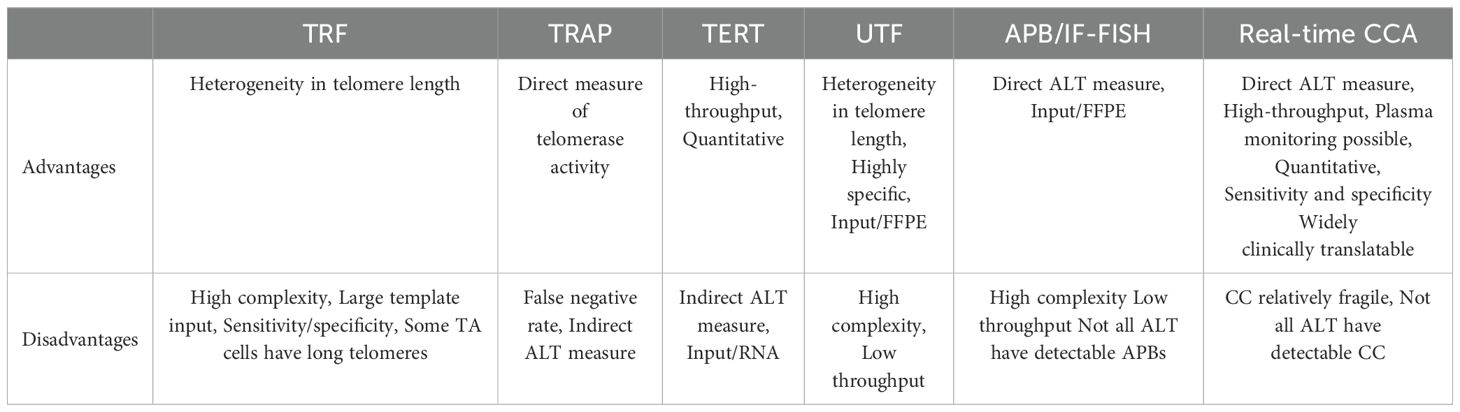
Table 3. Advantages and disadvantages of assays used to determine telomere maintenance mechanisms (TMM).
Historically, ALT has been identified by the telomerase repeated amplification protocol (TRAP) assay to demonstrate low TA activity (35) and/or low TERT mRNA expression, since TA is mutually exclusive to ALT (36). Telomere content and heterogeneity have been evaluated by telomere restriction fragment (TRF) analysis (37) and telomere fluorescence in situ hybridization for ultra-bright telomeric foci (UTF). UTF was combined with immunofluorescence (IF) of the PML protein, which was discovered to co-localize with telomeres in ALT samples, to detect APBs, yielding an additional ALT feature (38). Recently C-circles, circular self-primed telomeric DNA repeats, have been shown to be a sensitive and specific biomarker for ALT in tumors (32) that also circulate in patient plasma, potentially increasing the clinical utility of C-circles as a biomarker (39, 40).
After genomic DNA is extracted (Figure 1) from fresh frozen tumor, or plasma, C-circles can be enriched and subsequently detected by blot or real-time PCR (41, 42). First, C-circles are selectively amplified by ϕ-29 DNA polymerase via isothermal rolling-circle amplification (Figure 1) (41, 43, 44), which enriches the partially double-stranded telomeric DNA, termed C-circles, when compared to a reaction without ϕ-29. Subsequently, the telomeric signals can be compared by real-time PCR (Figure 1) for the ϕ-29 and no ϕ-29 reactions (Supplementary Figures S1A, C), which is then normalized to a single copy gene (e.g. VAV2) for the same ϕ-29 and no ϕ-29 reactions (Supplementary Figures S1B, D) (41, 42). This unique molecular diagnostic assay allows for high-throughput screening of DNA from fresh frozen tumor and plasma samples with as little as 1 ng of template input (14, 41). Herein, we sought to assess the frequency of ALT in pediatric sarcomas using the real-time PCR CCA on DNA samples extracted from fresh frozen tumor.
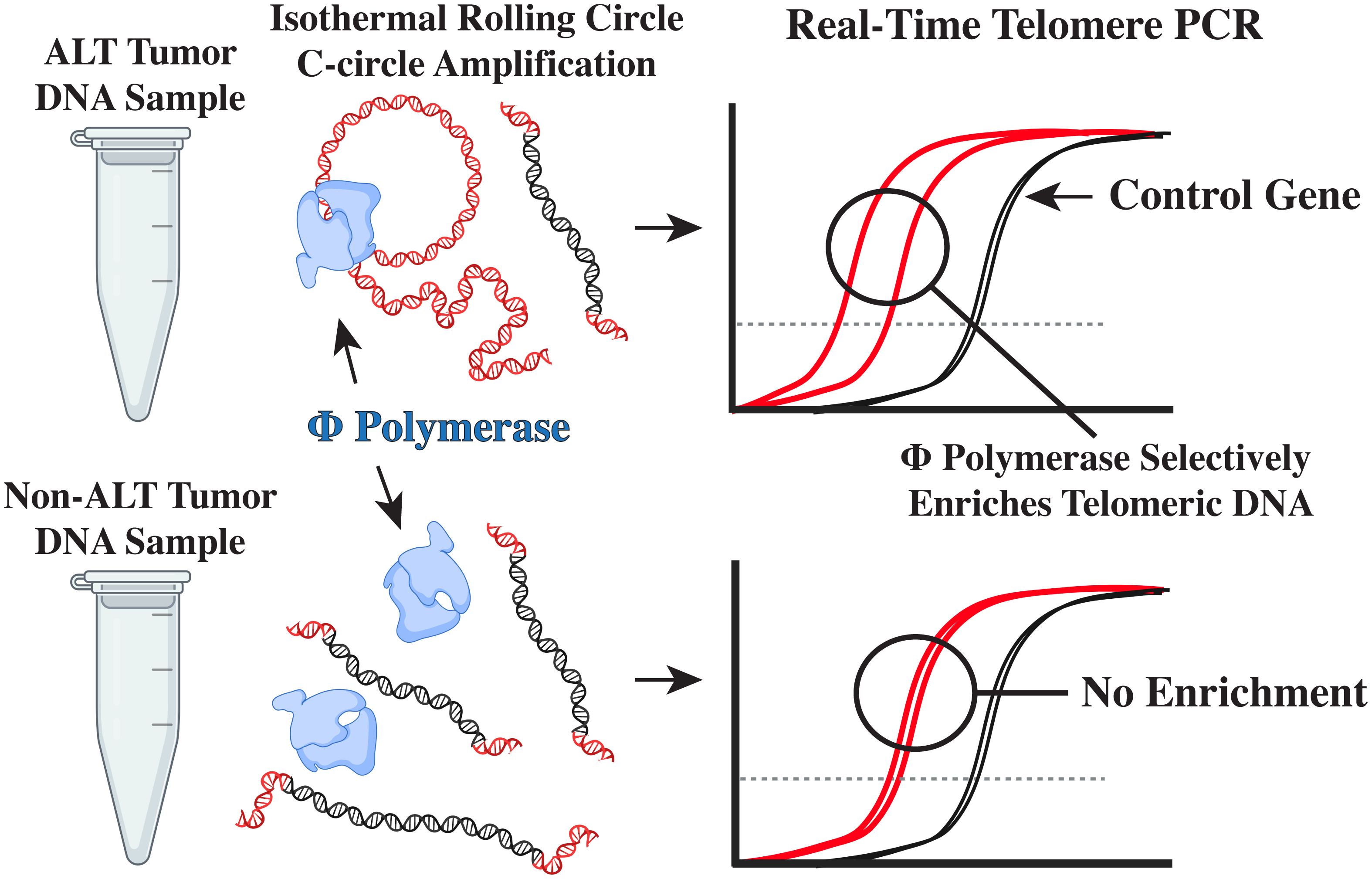
Figure 1. The real-time PCR CCA. Self-primed telomeric C-circles are selectively amplified by ϕ-29 polymerase via rolling circle amplification. Subsequent real-time PCR detection of telomere content reveals an enriched telomeric signal, indicating the presence of C-circles.
Genomic DNA was extracted using the QIAamp DNA mini kit (Qiagen, 51104), in accordance with the manufacturer’s instructions, and stored at -20°C, or in liquid nitrogen vapor, until aliquoted and sent to TTUHSC on dry ice for use in the C-circle assay. PAX3-FOXO1 or PAX7-FOXO1 fusion status was previously determined on all samples without unambiguous embryonal RMS histology. Fusion status was determined by reverse transcriptase-polymerase chain reaction assays (45) of RNA isolated using RNA STAT-60 (Tel-Test, Friendswood, TX).
Fluorometric quantification of DNA samples was carried out on a Qubit 2.0 system with the Qubit dsDNA Broad Range Assay Kit (Invitrogen Cat. No. Q32853).
The isothermal rolling circle amplification reactions were performed on an Eppendorf Vapo.Protect thermocycler at 30°C for 8 hrs, 65°C for 20 min, and held at 4°C. Reactions were comprised of: 32 ng of template DNA, 2 µL BSA (2 µg/µL), 2 µL of 1% Tween, 0.8 µL DTT (100µM), 2 µL of 10 mM dNTPs (NEB, Ipswich, MA, N0447L), 2 µL of ϕ-29 Buffer, 0.8 µL of ϕ-29 DNA polymerase (NEB, Ipswich, MA, M0269L), and nuclease-free water up to 20 µL. No ϕ-29 control reactions consisted of the aforementioned reagents with nuclease-free water in place of ϕ-29 DNA polymerase. After isothermal rolling circle amplification, all reactions were diluted with 20 µL nuclease-free water to a final volume of 40 µL.
Subsequent real-time PCR amplification of telomere DNA (Forward Primer: 5’ - CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT - 3’, Reverse Primer: 5’ - GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT - 3’) and VAV2 DNA (Forward Primer: 5’ - TGGGCATGACTGAAGATGAC - 3’, Reverse Primer: 5’ - ATCTGCCCTCACCTTCTCAA - 3’) (IDT, Coralville, IA) was performed using a 96-well Thermo-Fisher Quantstudio 3 Real-Time PCR System with the following cycling conditions: Telomere reaction: 95°C for 15 min, 33 cycles of 95°C for 15 sec and 56°C for 2 min, and VAV2 Reaction: 95°C for 15 min, 40 cycles of 95°C for 15 sec, 57°C for 30 sec, and 72°C for 1 min. Real-time PCR reactions consisted of: 5 µL of diluted isothermal reaction product, 12.5 µL QuantiTect SYBR Green PCR Master Mix (Qiagen, 204445), 1 µL DTT (100 µM), 0.5 µL DMSO, 1 µL nuclease-free water, and 2.5 µL of primers (5 µM Tel, or 2 µM VAV2). All real-time reactions (Telomere ϕ, Telomere No-ϕ, VAV2 ϕ, VAV2 No-ϕ) were carried out in triplicate and assessed via arbitrary unit (AU) calculations. DNA from CHLA-90 and CHLA-20 cell lines were used for positive and negative controls, respectively. Samples were considered CC+ if they had ≥5 AU, after normalization to CHLA-90, as previously described (46–48).
The relationship between clinical characteristics and C-circle status (Table 4) was evaluated by Chi-square, or Fisher’s exact test, when appropriate. The Mann-Whitney U Test was used to analyze telomere content. Two-tailed statistical tests with P values ≤ 0.05 were considered significant. All analyses were performed in GraphPad Prism v10.2.2.
The Children’s Oncology Group (COG) Biopathology Center provided 82 RMS DNA samples from residual stored DNA. These RMS specimens were collected from patients enrolled on a variety of Intergroup Rhabdomyosarcoma Study Group or COG Soft Tissue Sarcoma studies and received as de-identified samples. All Ewing sarcoma cases were part of COG clinical trial AEWS0031. All cases were reviewed by COG pathologists and a EWS-ETS fusion gene was identified in all cases. None were Ewing-like tumors with FET-ETS or CICX-DUX4. All cases expressed an EWS-FLI1 or EWS-ERG fusion gene and were part of the NCI Strategic Partnering to Evaluate Cancer Signatures (SPECS) program for childhood sarcoma gene expression profiling (49). EFS, OS, and additional RMS DNA was isolated from primary tumors obtained under informed consent by COG and processed by the pediatric division of Cooperative Human Tissue Network at Nationwide Children’s Hospital. These anonymized samples were originally used for genomic analyses in the NCI SPECS program, and in the case of OS, also the NCI TARGET program, and, in both cases, exempt from Human Subjects Research per IRB review (49–51).
Of the 373 DNA samples received (n = 95 EFS, n = 191 RMS, and n = 87 OS), five samples (1 EFS and 4 RMS) did not amplify due to poor DNA quality. CCA results are shown in Figure 2A. We observed 0 of 94 (0%) CC+ EFS cases, which is in concordance with previous reports (Table 1) (10, 12, 23). In contrast to EFS, 62 of 87 (71%) of OS tumors were CC+, which fell within the range of prior studies (12, 23). We did not observe statistically significant relationships between C-circle status and the clinicopathological data (Table 4), which aligns with the conclusions of previous studies that identified ALT through methods other than the CCA (14–16). We observed 5 of 187 (2.7%) CC+ RMS in the sample cohort (Table 5), which is lower than the previously reported 6%, which was determined by APB analysis (14). Of the 5 CC+ RMS samples identified, four were fusion negative (FN) embryonal RMS (ERMS) and one was fusion positive (FP) alveolar RMS (ARMS).
Telomere content amongst EFS, OS, and RMS (Figure 2B) ranged from 0.35 - 9.1, 0.56 - 33.73, and 0.93 - 14.42, respectively. Each histology showed a significant difference (p < 0.05) in telomere content, and CC+ OS had a significantly higher (p < 0.05) telomere content than CC- OS samples (Figure 2C), which is in concordance with reports that ALT telomere content is generally higher than non-ALT samples (39, 52).
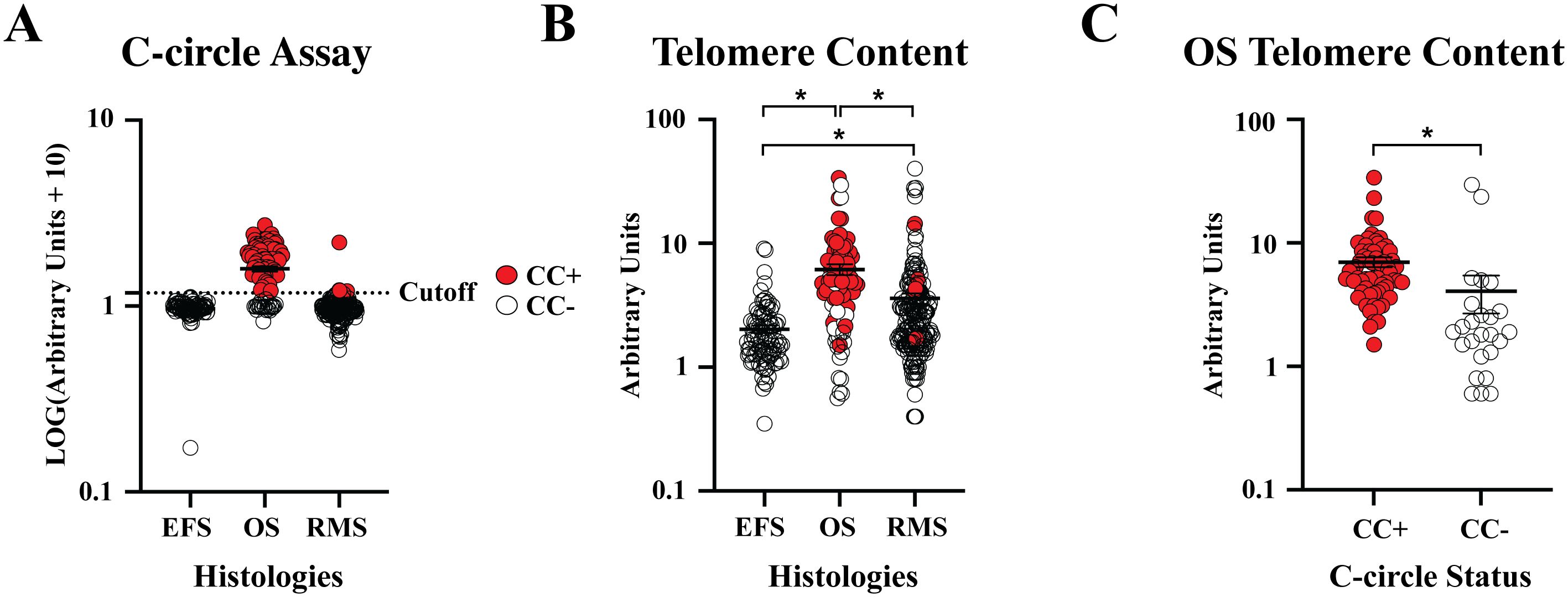
Figure 2. Patient sample CC status and telomere content. (A) Normalized relative CC content was plotted by tumor histology. Samples above the previously established cutoff of 5 arbitrary units (AU) were considered CC+. (B) Telomere content, normalized to CHLA-90 at 5 AU, were plotted by histology. (C) Telomere content was plotted for CC+ and CC- OS samples. * P < 0.05.
The prognostic value of ALT, and other TMM, is gaining traction (23, 33, 53), including in veterinary care (54). Studies have linked high telomerase expression with exceptionally aggressive tumors that can result in rapid progression and poor clinical outcomes (15, 33). By contrast, ALT has been associated with indolent disease progression; yet, patients with various tumor types have been observed to have a worse overall survival (23, 28, 33, 55). Recently, we observed high amounts of ATM kinase activation (which promotes chemotherapy resistance) in patient-derived neuroblastoma cell lines (PDCLs) and patient-derived xenografts (PDXs) (48), and also in PDCLs of other histologies (rhabdomyosarcoma, osteogenic sarcoma, triple negative breast cancer, and colorectal cancer) that have the ALT phenotype (47). We have also observed that certain clinical stage drugs (an ATM kinase inhibitor (48) and a p53 reactivator (47)) are active in reversing chemotherapy resistance in ALT PDCLs and PDXs. Thus, robust identification of ALT has the potential to be a prognostic biomarker and a companion diagnostic for ALT-targeted therapies.
Generally, ALT is activated by loss-of-function (LOF) genetic alterations in the chromatin remodelers α thalassemia-mental retardation, X linked (ATRX) (33) and death domain-associated protein 6 (DAXX) (56). ATRX inactivating mutations are commonly observed among different tumor types, while DAXX mutations are primarily associated with pancreatic neuroendocrine tumors (PanNETs) (56). ALT is less frequently associated with LOF alterations in H3F3A (57, 58) and SMARCAL1 mutated tumors (59, 60). Previous studies have used these genomic alterations as proxies to identify ALT, but depending on histology, as many as ½ ALT cancers can be wild-type for ATRX or DAXX (41, 61).
C-circles, TERT expression, high telomere content with heterogenous telomere length, and APBs have been used to screen sample sets to establish ALT frequencies amongst sarcomas; however, each of these techniques have their own advantages and disadvantages. Relatively fragile, C-circles can be degraded by excess freeze-thaw cycles, prolonged vortexing, and formalin-fixing; thus, proper sample handling and storage are required (62). Recently, ALT tumors have been shown to protect C-circles from nuclease degradation in the blood by releasing C-circles within exosomes, which may provide a non-invasive blood-based biomarker for the detection and monitoring of ALT tumors in vivo (40). Although there is no standardized method for determining ALT status (28), C-circles are the only known molecule specific to ALT (40), and the molecularly based real-time PCR C-circle assay can utilize DNA that has been isolated for sequencing; thus, it is readily translatable to the clinical laboratory, and it’s for these reasons that we selected this approach (28, 32, 42, 61).
We observed no CC+ EFS cases, which is likely due to the activation of TERT by EFS fusion proteins (39). The ALT phenotype is known to be essentially exclusive to TERT activation (32, 39, 40). In OS patients, expression of TERT has been shown to portend an unfavorable clinical prognosis (15); however, stage and clinical outcomes of ALT cases were shown to be equivalent to TA cases (16), but, the ALT phenotype provides a potentially targetable mechanism present in the majority of OS patients, some of which have poor clinical outcomes (47, 63, 64).
ALT is also known to occur in RMS (14), the most common pediatric soft tissue sarcoma (65). Classically, pediatric RMS cases were generally categorized histologically as ERMS, which was linked with better prognoses, or ARMS, which was associated with poor clinical outcomes (66). Further, molecular identification of PAX3, or PAX7, fusions with forkhead box protein O1 (FOXO1), is currently considered the preferred method of distinguishing the latter from the former (67). Instead of histologic criteria, which are inexact, the fusion status identifies ARMS and ERMS, which are FP and FN, respectivly (68).
The tested RMS samples were from banked DNA extracted from fresh frozen tissue; thus, it is possible that the age of the samples, or excess freeze-thaw cycles could have contributed to the lower ALT frequency, due to the degradation of C-circles (41). APB analysis from a previous study (14) has the advantage of using FFPE material, which enables distinguishing of tumor cells from stromal tissue; however, the APB assay is very labor intensive, not all ALT samples have APBs (33), and C-circles have been postulated to be more specific than other ALT markers (32). Ideally, future studies should evaluate the various methods for detecting ALT in the same histology within the same patient sample cohort, since each ALT marker is not necessarily present in every ALT sample or tumor model (14, 33). However, our data suggests that the real-time PCR CCA can identify ALT in sarcomas, and it has potential as a companion diagnostic assay for ALT targeted therapies in RMS, and especially OS, patient populations.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The studies involving humans were approved by IRB Texas Tech University Health Sciences Center. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired without linkage to patient identifiers from another research group. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
TB: Formal analysis, Investigation, Methodology, Writing – original draft. BK: Investigation, Methodology, Writing – review & editing. SM: Investigation, Writing – review & editing. WS: Resources, Writing – review & editing. FB: Resources, Writing – review & editing. TT: Resources, Writing – review & editing. CR: Conceptualization, Data curation, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by UO1 CA263988 and RO1 CA264949 from the National Cancer Institute and RP220460 and RP210154 from the Cancer Prevention & Research Institute of Texas.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1399442/full#supplementary-material
1. Blackburn EH. Structure and function of telomeres. Nature. (1991) 350(6319):569–73. doi: 10.1038/350569a0
2. de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. (2005) 19(18):2100–10. doi: 10.1101/gad.1346005
3. Levy MZ, Allsopp RC, Futcher AB, Greider CW, Harley CB. Telomere end-replication problem and cell aging. J Mol Biol. (1992) 225:951–60. doi: 10.1016/0022-2836(92)90096-3
4. Wellinger RJ. In the end, what’s the problem? Mol Cell. (2014) 53:855–6. doi: 10.1016/j.molcel.2014.03.008
5. Markiewicz-Potoczny M, Lobanova A, Loeb AM, Kirak O, Olbrich T, Ruiz S, et al. TRF2-mediated telomere protection is dispensable in pluripotent stem cells. Nature. (2021) 589:110–5. doi: 10.1038/s41586-020-2959-4
6. Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. (2003) 13:1549–56. doi: 10.1016/S0960-9822(03)00542-6
7. d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. (2003) 426:194–8. doi: 10.1038/nature02118
8. Broccoli D, Young JW, de Lange T. Telomerase activity in normal and Malignant hematopoietic cells. Proc Natl Acad Sci USA. (1995) 92:9082–6. doi: 10.1073/pnas.92.20.9082
9. Nugent CI, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. (1998) 12:1073–85. doi: 10.1101/gad.12.8.1073
10. Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA, Gabrielson E, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol. (2011) 179:1608–15. doi: 10.1016/j.ajpath.2011.06.018
11. Zhang JM, Genois MM, Ouyang J, Lan L, Zou L. Alternative lengthening of telomeres is a self-perpetuating process in ALT-associated PML bodies. Mol Cell. (2021) 81:1027–1042 e4. doi: 10.1016/j.molcel.2020.12.030
12. Ulaner GA, Hoffman AR, Otero J, Huang HY, Zhao Z, Mazumdar M, et al. Divergent patterns of telomere maintenance mechanisms among human sarcomas: sharply contrasting prevalence of the alternative lengthening of telomeres mechanism in Ewing’s sarcomas and osteosarcomas. Genes Chromosomes Cancer. (2004) 41:155–62. doi: 10.1002/gcc.20074
13. Liau JY, Lee JC, Tsai JH, Yang CY, Liu TL, Ke ZL, et al. Comprehensive screening of alternative lengthening of telomeres phenotype and loss of ATRX expression in sarcomas. Mod Pathol. (2015) 28:1545–54. doi: 10.1038/modpathol.2015.114
14. Henson JD, Hannay JA, McCarthy SW, Royds JA, Yeager TR, Robinson RA, et al. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin Cancer Res. (2005) 11:217–25. doi: 10.1158/1078-0432.217.11.1
15. Sanders RP, Drissi R, Billups CA, Daw NC, Valentine MB, Dome JS. Telomerase expression predicts unfavorable outcome in osteosarcoma. J Clin Oncol. (2004) 22:3790–7. doi: 10.1200/JCO.2004.03.043
16. Ulaner GA, Huang HY, Otero J, Zhao Z, Ben-Porat L, Satagopan JM, et al. Absence of a telomere maintenance mechanism as a favorable prognostic factor in patients with osteosarcoma. Cancer Res. (2003) 63:1759–63.
17. Yasir S, Thompson S, Chen ZE, Knudson R, Knutson D, Kloft-Nelson S, et al. Alternative lengthening of telomeres in primary hepatic neoplasms. Hum Pathol. (2023) 131:79–86. doi: 10.1016/j.humpath.2022.11.003
18. Chudasama P, Mughal SS, Sanders MA, Hubschmann D, Chung I, Deeg KI, et al. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat Commun. (2018) 9:144. doi: 10.1038/s41467-017-02602-0
19. Liau JY, Tsai JH, Jeng YM, Lee JC, Hsu HH, Yang CY. Leiomyosarcoma with alternative lengthening of telomeres is associated with aggressive histologic features, loss of ATRX expression, and poor clinical outcome. Am J Surg Pathol. (2015) 39:236–44. doi: 10.1097/PAS.0000000000000324
20. Costa A, Daidone MG, Daprai L, Villa R, Cantu S, Pilotti S, et al. Telomere maintenance mechanisms in liposarcomas: association with histologic subtypes and disease progression. Cancer Res. (2006) 66:8918–24. doi: 10.1158/0008-5472.CAN-06-0273
21. Venturini L, Daidone MG, Motta R, Cimino-Reale G, Hoare SF, Gronchi A, et al. Telomere maintenance mechanisms in Malignant peripheral nerve sheath tumors: expression and prognostic relevance. Neuro Oncol. (2012) 14:736–44. doi: 10.1093/neuonc/nos083
22. Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. (2002) 21:598–610. doi: 10.1038/sj.onc.1205058
23. MacKenzie D Jr., Watters AK, To JT, Young MW, Muratori J, Wilkoff MH, et al. ALT positivity in human cancers: prevalence and clinical insights. Cancers (Basel). (2021) 13. doi: 10.3390/cancers13102384
24. Reddel RR, Bryan TM, Colgin LM, Perrem KT, Yeager TR. Alternative lengthening of telomeres in human cells. Radiat Res. (2001) 155:194–200. doi: 10.1667/0033-7587(2001)155[0194:alotih]2.0.co;2
25. Murnane JP, Sabatier L, Marder BA, Morgan WF. Telomere dynamics in an immortal human cell line. EMBO J. (1994) 13:4953–62. doi: 10.1002/embj.1994.13.issue-20
26. Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. (1995) 14:4240–8. doi: 10.1002/embj.1995.14.issue-17
27. Lee M, Hills M, Conomos D, Stutz MD, Dagg RA, Lau LM, et al. Telomere extension by telomerase and ALT generates variant repeats by mechanistically distinct processes. Nucleic Acids Res. (2014) 42:1733–46. doi: 10.1093/nar/gkt1117
28. Dilley RL, Greenberg RA. ALTernative telomere maintenance and cancer. Trends Cancer. (2015) 1:145–56. doi: 10.1016/j.trecan.2015.07.007
29. Nabetani A, Yokoyama O, Ishikawa F. Localization of hRad9, hHus1, hRad1, and hRad17 and caffeine-sensitive DNA replication at the alternative lengthening of telomeres-associated promyelocytic leukemia body. J Biol Chem. (2004) 279:25849–57. doi: 10.1074/jbc.M312652200
30. Wu G, Lee WH, Chen PL. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J Biol Chem. (2000) 275:30618–22. doi: 10.1074/jbc.C000390200
31. Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. (1999) 59:4175–9.
32. Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, et al. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol. (2009) 27:1181–5. doi: 10.1038/nbt.1587
33. Koneru B, Lopez G, Farooqi A, Conkrite KL, Nguyen TH, Macha SJ, et al. Telomere maintenance mechanisms define clinical outcome in high-risk neuroblastoma. Cancer Res. (2020) 80:2663–75. doi: 10.1158/0008-5472.CAN-19-3068
34. Meeser A, Bartenhagen C, Werr L, Hellmann AM, Kahlert Y, Hemstedt N, et al. Reliable assessment of telomere maintenance mechanisms in neuroblastoma. Cell Biosci. (2022) 12:160. doi: 10.1186/s13578-022-00896-2
35. Mender I, Shay JW. Telomerase repeated amplification protocol (TRAP). Bio Protoc. (2015) 5. doi: 10.21769/BioProtoc.1657
36. Reddel RR. Alternative lengthening of telomeres, telomerase, and cancer. Cancer Lett. (2003) 194:155–62. doi: 10.1016/S0304-3835(02)00702-4
37. Mender I, Shay JW. Telomere restriction fragment (TRF) analysis. Bio Protoc. (2015) 5. doi: 10.21769/BioProtoc.1658
38. Grobelny JV, Godwin AK, Broccoli D. ALT-associated PML bodies are present in viable cells and are enriched in cells in the G(2)/M phase of the cell cycle. J Cell Sci. (2000) 113 Pt 24:4577–85. doi: 10.1242/jcs.113.24.4577
39. Hartlieb SA, Sieverling L, Nadler-Holly M, Ziehm M, Toprak UH, Herrmann C, et al. Alternative lengthening of telomeres in childhood neuroblastoma from genome to proteome. Nat Commun. (2021) 12:1269. doi: 10.1038/s41467-021-21247-8
40. Chen YY, Dagg R, Zhang Y, Lee JHY, Lu R, Martin La Rotta N, et al. The C-circle biomarker is secreted by alternative-lengthening-of-telomeres positive cancer cells inside exosomes and provides a blood-based diagnostic for ALT activity. Cancers (Basel). (2021) 13(21). doi: 10.3390/cancers13215369
41. Henson JD, Lau LM, Koch S, Martin La Rotta N, Dagg RA, Reddel RR. The C-Circle Assay for alternative-lengthening-of-telomeres activity. Methods. (2017) 114:74–84. doi: 10.1016/j.ymeth.2016.08.016
42. Lau LM, Dagg RA, Henson JD, Au AY, Royds JA, Reddel RR. Detection of alternative lengthening of telomeres by telomere quantitative PCR. Nucleic Acids Res. (2013) 41:e34. doi: 10.1093/nar/gks781
43. Blanco L, Salas M. Characterization and purification of a phage phi 29-encoded DNA polymerase required for the initiation of replication. Proc Natl Acad Sci USA. (1984) 81:5325–9. doi: 10.1073/pnas.81.17.5325
44. Blanco L, Bernad A, Lazaro JM, Martin G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. (1989) 264:8935–40. doi: 10.1016/S0021-9258(18)81883-X
45. Duan F, Smith LM, Gustafson DM, Zhang C, Dunlevy MJ, Gastier-Foster JM, et al. Genomic and clinical analysis of fusion gene amplification in rhabdomyosarcoma: a report from the Children’s Oncology Group. Genes Chromosomes Cancer. (2012) 51:662–74. doi: 10.1002/gcc.21953
46. Dagg RA, Pickett HA, Neumann AA, Napier CE, Henson JD, Teber ET, et al. Extensive proliferation of human cancer cells with ever-shorter telomeres. Cell Rep. (2017) 19:2544–56. doi: 10.1016/j.celrep.2017.05.087
47. Macha SJ, Koneru B, Burrow TA, Zhu C, Savitski D, Rahman RL, et al. Alternative lengthening of telomeres in cancer confers a vulnerability to reactivation of p53 function. Cancer Res. (2022) 82:3345–58. doi: 10.1158/0008-5472.CAN-22-0125
48. Koneru B, Farooqi A, Nguyen TH, Chen WH, Hindle A, Eslinger C, et al. ALT neuroblastoma chemoresistance due to telomere dysfunction-induced ATM activation is reversible with ATM inhibitor AZD0156. Sci Transl Med. (2021) 13(607). doi: 10.1126/scitranslmed.abd5750
49. Volchenboum SL, Andrade J, Huang L, Barkauskas DA, Krailo M, Womer RB, et al. Gene expression profiling of ewing sarcoma tumors reveals the prognostic importance of tumor-stromal interactions: A report from the children’s oncology group. J Pathol Clin Res. (2015) 1:83–94. doi: 10.1002/cjp2.9
50. Taylor AM, Sun JM, Yu A, Voicu H, Shen J, Barkauskas DA, et al. Integrated DNA copy number and expression profiling identifies IGF1R as a prognostic biomarker in pediatric osteosarcoma. Int J Mol Sci. (2022) 23. doi: 10.3390/ijms23148036
51. Hingorani P, Missiaglia E, Shipley J, Anderson JR, Triche TJ, Delorenzi M, et al. Clinical application of prognostic gene expression signature in fusion gene-negative rhabdomyosarcoma: A report from the children’s oncology group. Clin Cancer Res. (2015) 21:4733–9. doi: 10.1158/1078-0432.CCR-14-3326
52. Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. (2010) 11:319–30. doi: 10.1038/nrg2763
53. Matsuo T, Shimose S, Kubo T, Fujimori J, Yasunaga Y, Ochi M. Telomeres and telomerase in sarcomas. Anticancer Res. (2009) 29:3833–6.
54. Bicanova L, Kreilmeier-Berger T, Reifinger M, Holzmann K, Kleiter M. Prevalence and potentially prognostic value of C-circles associated with alternative lengthening of telomeres in canine appendicular osteosarcoma. Vet Comp Oncol. (2021) 19:222–31. doi: 10.1111/vco.12665
55. Lawlor RT, Veronese N, Pea A, Nottegar A, Smith L, Pilati C, et al. Alternative lengthening of telomeres (ALT) influences survival in soft tissue sarcomas: a systematic review with meta-analysis. BMC Cancer. (2019) 19:232. doi: 10.1186/s12885-019-5424-8
56. Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. (2011) 333:425. doi: 10.1126/science.1207313
57. Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. (2012) 22:425–37. doi: 10.1016/j.ccr.2012.08.024
58. Minasi S, Baldi C, Gianno F, Antonelli M, Buccoliero AM, Pietsch T, et al. Alternative lengthening of telomeres in molecular subgroups of pediatric high-grade glioma. Childs Nerv Syst. (2021) 37:809–18. doi: 10.1007/s00381-020-04933-8
59. Zhang JM, Zou L. Alternative lengthening of telomeres: from molecular mechanisms to therapeutic outlooks. Cell Biosci. (2020) 10:30. doi: 10.1186/s13578-020-00391-6
60. Diplas BH, He X, Brosnan-Cashman JA, Liu H, Chen LH, Wang Z, et al. The genomic landscape of TERT promoter wildtype-IDH wildtype glioblastoma. Nat Commun. (2018) 9:2087. doi: 10.1038/s41467-018-04448-6
61. de Nonneville A, Reddel RR. Alternative lengthening of telomeres is not synonymous with mutations in ATRX/DAXX. Nat Commun. (2021) 12:1552. doi: 10.1038/s41467-021-21794-0
62. Henson JD, Reddel RR. Assaying and investigating Alternative Lengthening of Telomeres activity in human cells and cancers. FEBS Lett. (2010) 584:3800–11. doi: 10.1016/j.febslet.2010.06.009
63. Alexander JH, Binitie OT, Letson GD, Joyce DM. Osteosarcoma: an evolving understanding of a complex disease. J Am Acad Orthop Surg. (2021) 29:e993–e1004. doi: 10.5435/JAAOS-D-20-00838
64. Yoshida A. Osteosarcoma: old and new challenges. Surg Pathol Clin. (2021) 14:567–83. doi: 10.1016/j.path.2021.06.003
65. Milgrom SA, Million L, Mandeville H, Safwat A, Ermoian RP, Terezakis S. Non-rhabdomyosarcoma soft-tissue sarcoma. Pediatr Blood Cancer. (2021) 68 Suppl 2:e28279. doi: 10.1002/pbc.28279
66. Hinson AR, Jones R, Crose LE, Belyea BC, Barr FG, Linardic CM. Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: utility and pitfalls. Front Oncol. (2013) 3:183. doi: 10.3389/fonc.2013.00183
67. Skapek SX, Ferrari A, Gupta AA, Lupo PJ, Butler E, Shipley J, et al. Rhabdomyosarcoma. Nat Rev Dis Primers. (2019) 5:1. doi: 10.1038/s41572-018-0051-2
68. Kirk R. Pediatric oncology: Fusion-gene status crucial in RMS. Nat Rev Clin Oncol. (2010) 7:356. doi: 10.1038/nrclinonc.2010.88
Keywords: rhabdomyosarcoma, Ewing sarcoma, osteosarcoma, alternative lengthening of telomeres, telomere
Citation: Burrow TA, Koneru B, Macha SJ, Sun W, Barr FG, Triche TJ and Reynolds CP (2024) Prevalence of alternative lengthening of telomeres in pediatric sarcomas determined by the telomeric DNA C-circle assay. Front. Oncol. 14:1399442. doi: 10.3389/fonc.2024.1399442
Received: 11 March 2024; Accepted: 26 July 2024;
Published: 19 August 2024.
Edited by:
Jeremy Wang, University of North Carolina at Chapel Hill, United StatesReviewed by:
Susana Galli, Georgetown University Medical Center, United StatesCopyright © 2024 Burrow, Koneru, Macha, Sun, Barr, Triche and Reynolds. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: C. Patrick Reynolds, UGF0cmljay5SZXlub2xkc0BUVFVIU0MuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.