- 1Health Science Center, Ningbo University, Ningbo, China
- 2Department of Gastroenterology, The First Affiliated Hospital of Ningbo University, Ningbo, China
Circular RNAs (circRNAs) lack the 5’-end methylated guanine cap structure and 3’ polyadenylate tail structure, classifying it as a non-coding RNA. With the extensive investigation of circRNA, its role in regulating cell death has garnered significant attention in recent years, establishing it as a recognized participant in cancer’s biological processes. Autophagy, an essential pathway in programmed cell death (PCD), involves the formation of autophagosomes using lysosomes to degrade cellular contents under the regulation of various autophagy-related (ATG) genes. Numerous studies have demonstrated that circRNA can modulate the biological activity of cancer cells by influencing the autophagy pathway, exhibiting a dualistic role in suppressing or promoting carcinogenesis. In this review, we comprehensively analyze how autophagy-related circRNA impacts the progression of gastrointestinal cancer (GIC). Additionally, we discuss drug resistance phenomena associated with autophagy regulation in GIC. This review offers valuable insights into exploring potential biological targets for prognosis and treatment strategies related to GIC.
1 Introduction
1.1 CircRNA
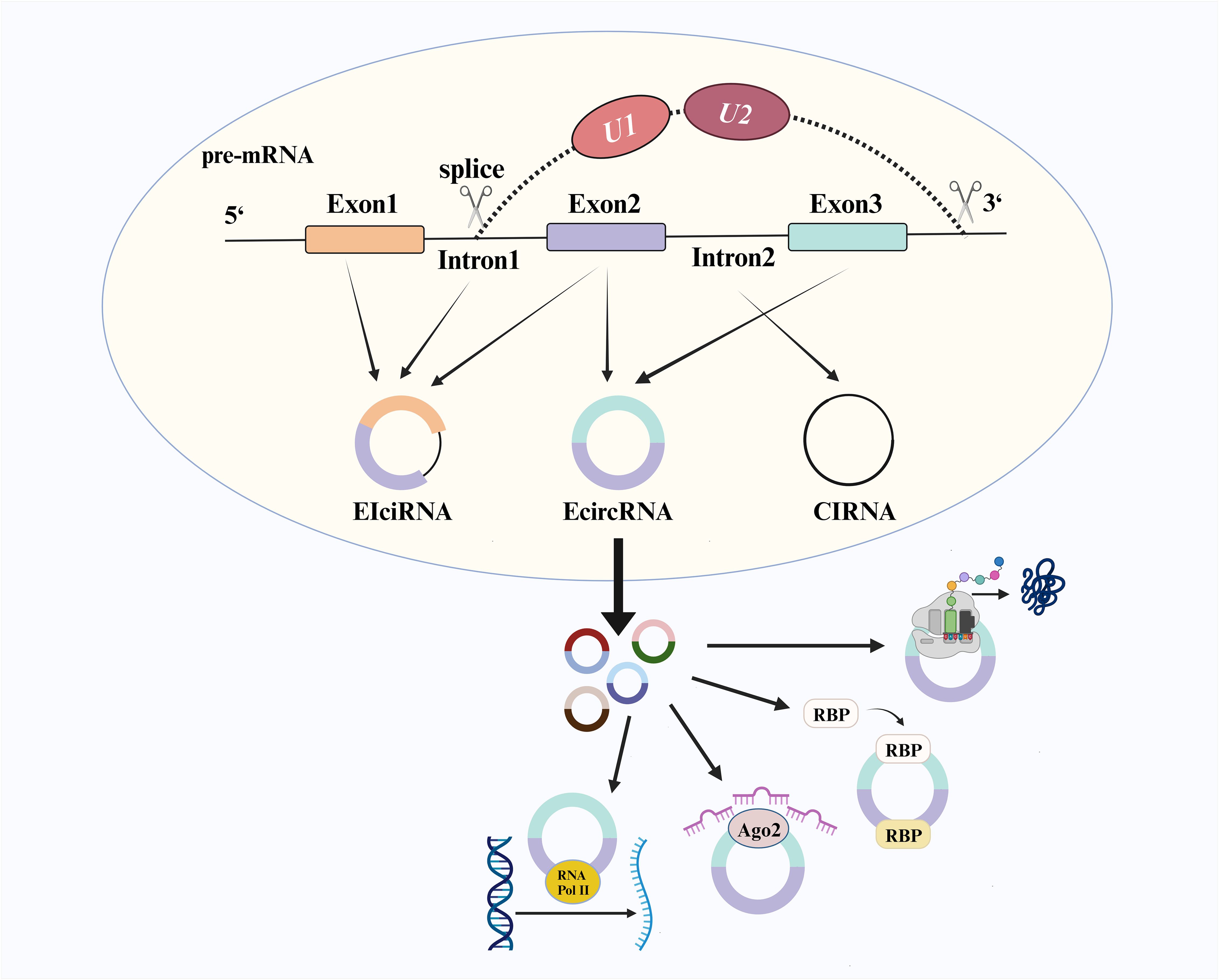
CircRNA is a covalently connected closed-loop structure formed by the reverse splicing protein-coding genes in eukaryotic cells (1). Most circRNA consists only of exons, known as exonic circRNA (EcircRNA). At the same time, some retain intron composition and are referred to as circular intronic RNA (CiRNA) and exon-intron circRNA (EIciRNA) (2) (Figure 1). The splicing process begins with the selection of splice sites on pre-mRNA under the influence of the canonical splicing mechanism, where U1snRNP (U1) and U2snRNP (U2) play crucial roles in mutual regulation during site selection (3, 4). Previous studies have demonstrated that changes in U2/U1 levels generate different splicing products. Depletion of U2 can inhibit the splicing factor and increase the production level of circRNA (5, 6). Initially considered a non-functional product of pre-mRNA (7), further research has revealed various functional mechanisms associated with circRNA, including transcriptional regulation, formation of circRNA-RNA binding protein (RBP) complex, microRNA (miRNA) sponge activity and translation function. CircRNA with an open reading frame was initially overlooked due to the absence of a canonical start codon and the challenges in detecting encoded peptides. However, recent research has revealed that circFNDC3B exists in colon cancer and encodes circFNDC3B-218aa, which induces FBP1 expression and inhibits the growth and metastasis of colon cancer (8). Simultaneously, global social and economic development has shifted the disease epidemiology model from acute infectious diseases to chronic non-communicable diseases, particularly with an increasing burden of cancer worldwide (9, 10). CircRNA has been found to exhibit differential expression levels in various types of cancers, such as gastric cancer (GC), colorectal cancer (CRC), lung cancer, and leukemia (2). Combining different circRNA may enhance early detection stability for cancers. Compared to linear RNA molecules, circRNA possesses a more stable covalent ring structure that avoids exonuclease degradation and can be widely detected in blood samples, tissues, and saliva samples, among others. Henceforth, circRNA holds significant potential as a novel biomarker for cancer (7, 11).
1.2 Autophagy
Autophagy is a crucial form of PCD that maintains cellular homeostasis by degrading or utilizing intracellular harmful or accumulated substances (12). Researchers have gained a more comprehensive understanding of how autophagy impacts human disease progression in recent years through molecular mechanism studies involving knockdowns of ATG genes and analysis of ATG protein products (13).
1.2.1 The biological process of autophagy
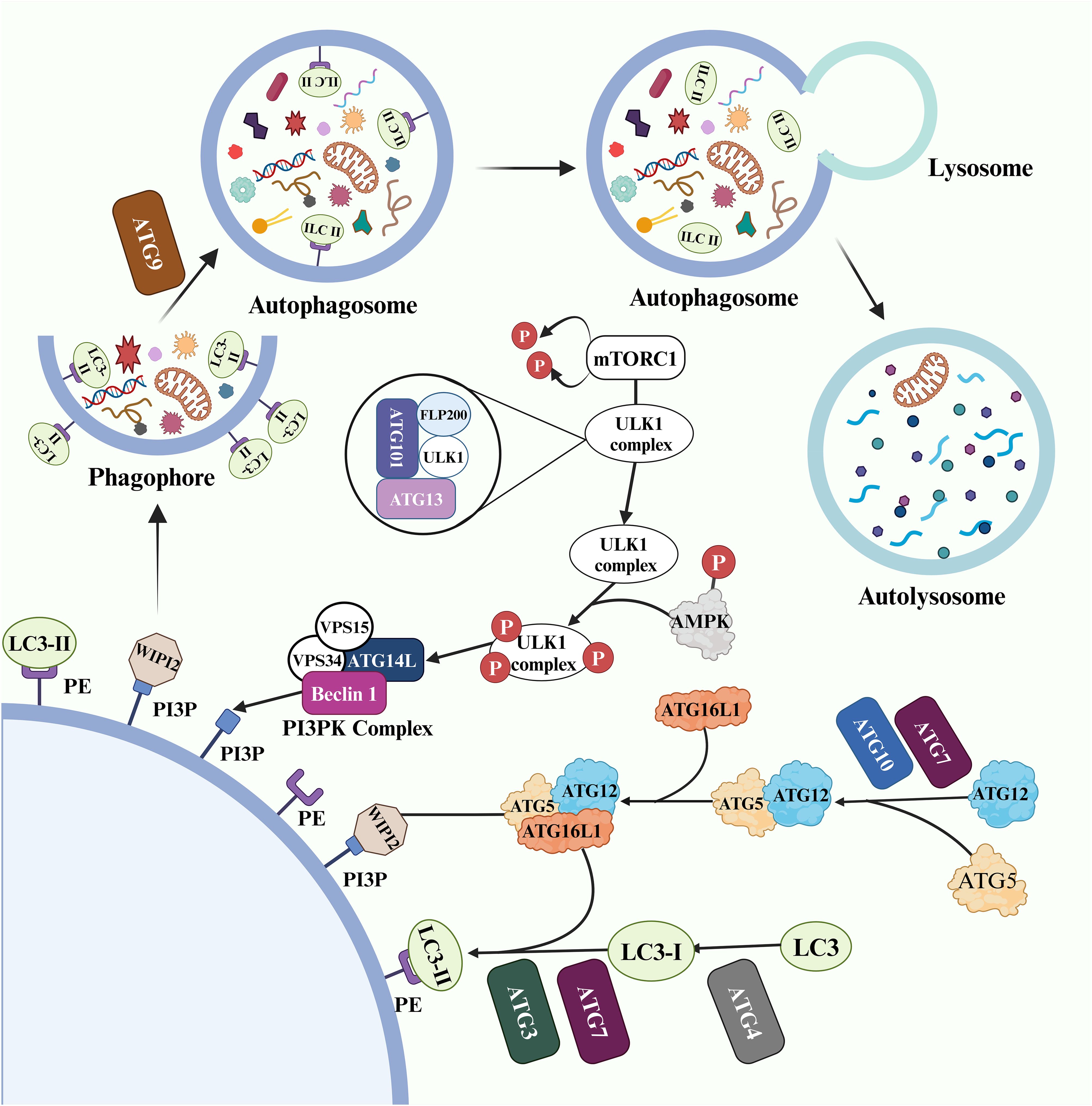
Three main types of autophagy have been identified: macroautophagy, microautophagy, and chaperone-mediated autophagy. Among these, macroautophagy is a highly regulated multi-stage process (14) (Figure 2). ATGs coordinate with each other and selectively recruit core autophagy components to establish double-membrane vesicle structures called autophagosomes. Subsequently, under the influence of effector factors such as soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins, GTPase, and motor proteins, the outer membrane of the vesicles fuses with lysosome to form autolysosome (15).
In this process, the physiological process of macroautophagy is regulated by a series of ATG (16). The UNC-52-like kinase 1 (ULK1) complex, consisting of ATG101, ATG13, FLP200 (focal adhesion kinase family interacting protein of 200kD) and ULK1, serves as a critical initiator of autophagy and its activity is controlled through mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) phosphorylation and dephosphorylation (17). During starvation conditions, the ULK1 complex dissociates from dephosphorylated mTORC1, increasing activity in the free ULK1 complex (18, 19). Additionally, glucose deficiency activates AMP-activated protein kinase (AMPK), which provides phosphate groups to the ULK1 complex for initiating autophagy (19). The activated ULK1 complex then regulates the phosphoinositide 3-kinase (PI3K) complex composed of ATG14L, Beclin-1 (BECN1), VPS34 and VPS15 that participates in phosphatidylinositol 3-phosphate (PI3P) production from PI on the phagophore membrane to promote autophagy (20, 21). WD repeat domain, phosphoinositide interacting 2 (WIPI2) is recruited by PI3P (22).
Two ubiquitin-like cascade modules modify the early stages of autophagy (23). Firstly, ATG5 and ATG12 bind to ATG16L1 with assistance from two ubiquitinating enzymes (ATG7 and ATG10), forming an ATG5-ATG12-ATG16L1 complex. This complex subsequently binds to WIPI2 and localizes on the surface of the phagosome membrane (24). Moreover, ATG4 cleaves the microtubule associated protein 1 light chain 3 (LC3) termini to generate LC3-I, which participates in the production of LC3-II by co-coupling with ubiquitinating enzymes ATG3, ATG7, and the ATG5-ATG12-ATG16L1 complex on the phagophore membrane (25). Under regulation by ATG9, both the inner and outer membranes of the phagophore expand simultaneously to form a fully closed autophagosome. The cargo to be processed enters the autophagosome through selective or non-selective recognition pathways (26).
BECN1 and LC3-II play critical roles in the process of autophagy and serve as important markers. BECN1 promotes PI3P activation, which leads to the expansion of phagosome membranes and the formation of double membrane vesicles (27). LC3-II recruits ATG2 to interact with WIPI to transport essential lipids for autophagosome formation (28).
1.2.2 Autophagy and cell death
The leading roles of autophagy in dying cells can be categorized into two types: autophagy-dependent cell death (ADCD) and autophagy-mediated cell death (AMCD). ADCD relies solely on components related to autophagy to induce cell death. On the other hand, AMCD triggers various modes of cell death through autophagy, and these two processes can occur independently or simultaneously (29).
Based on specificity, ADCD can be classified into non-selective and selective autophagy (30). When the internal environment detects a lack of nutrients, it initiates the formation of autophagosomes that indiscriminately engulf organelles to ensure sufficient cellular functionality under nutrient-deprived conditions (30). However, current research primarily focuses on selective forms of autophagy, such as mitophagy, ER-phagy, and xenophagy (31). Selective autophagic processes involve receptor proteins with distinct features like LC3-interacting region (LIR) domains and ubiquitin-binding sites that differentiate them from typical selective-autophagy-related proteins (32). For instance, PTEN-induced putative kinase 1 (PINK1) restricts mitochondrial damage by accumulating at damaged mitochondria’s outer membrane through E3 ubiquitin ligase Parkin-mediated ubiquitination; subsequently identified by ubiquitin-proteasome system for degradation leading to PINK1/Parkin pathway-induced mitochondrial-autophagic response (33). Additionally, targeting LIR regions instead of relying on the ubiquitin-proteasome pathway also initiates selective forms of autophagy, an example being TEX264 (an ER-phagic receptor), where its LIR domain interacts with the phagosome, resulting in endoplasmic reticulum clearance under damaged conditions (34).
The emerging evidence substantiates the intricate interplay between autophagy and apoptosis pathways (35); for instance, the crosstalk between PINK1-BECN1 regulates the interplay of autophagy and apoptosis pathways, thereby influencing cancer cell proliferation (36). The convergence of these two signaling cascades is contingent upon the involvement of the second messenger Ca2+. Anthraquinone derivatives have been discovered to concurrently induce apoptosis and autophagy in reproductive system tumors as well as digestive tract tumor cell lines (37). Autophagy can also expedite ferroptosis initiation for anti-tumor programming. Erianin, a natural compound derived from Dendrobium officinale, interacts with KRASG13D mutation to trigger autophagy-dependent ferroptosis while inhibiting the growth and migration of CRC (38). The process of anoikis represents a specific form of PCD that actively contributes to maintaining homeostasis by eliminating cancer cells exhibiting metastatic behavior in the absence of cellular adhesion and loss of extracellular matrix (39). Wu et al. demonstrated that ELAVL1 exerts inhibitory effects on circspecc1 in GC, suppressing of autophagy through ubiquitination and subsequent degradation of ATG4B. Moreover, reduced resistance to anoikis was observed, thereby promoting increased metastasis in GC (40). The mechanism of AMCD, however, remains elusive, and further investigation is warranted for elucidation.
1.2.3 The regulatory factors of autophagy
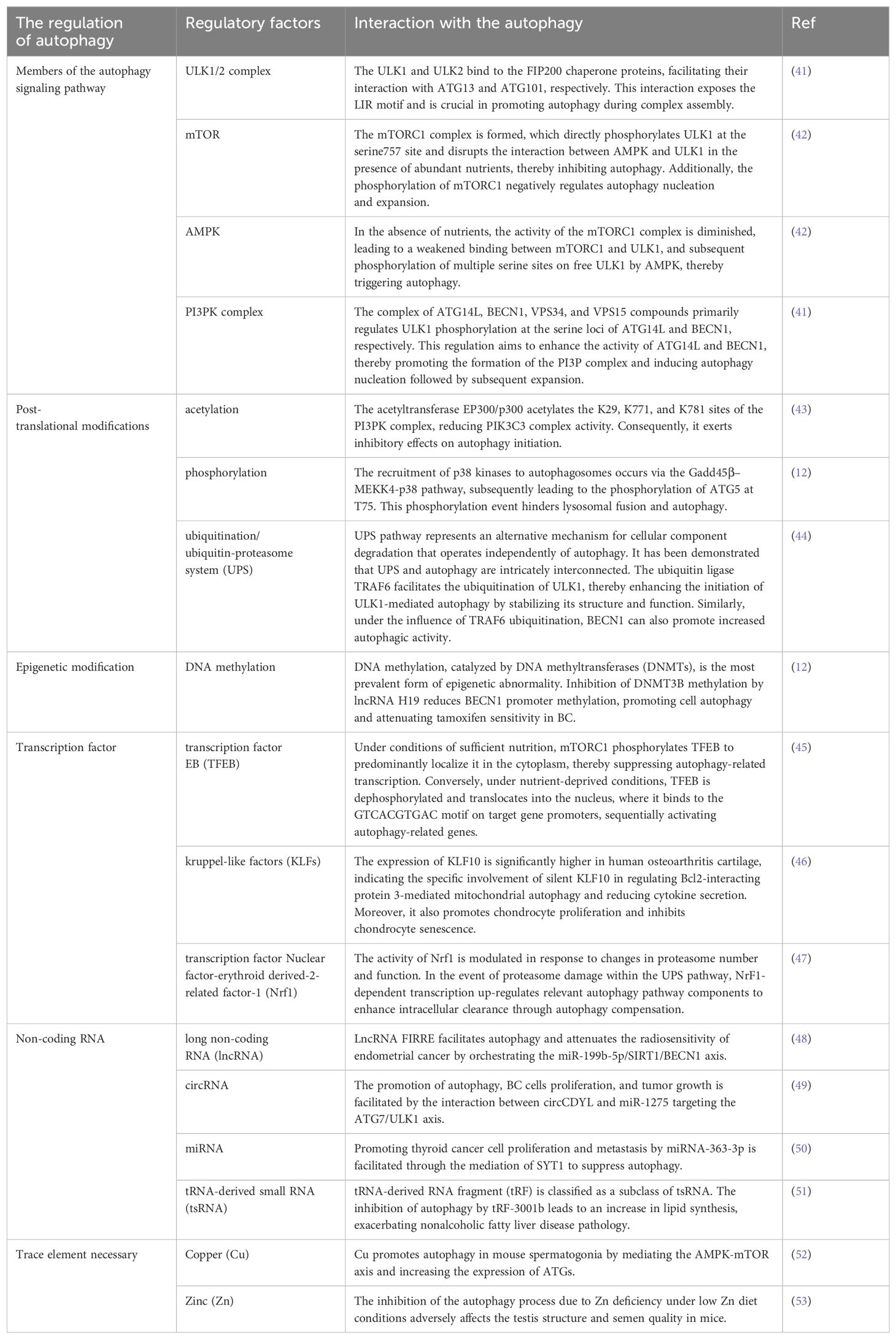
The increasing body of research has unveiled that autophagy is subject to the influence of diverse regulatory factors (Table 1); however, a comprehensive understanding of the precise molecular mechanisms governing autophagy remains elusive (54). Currently, extensive research is being conducted on circRNA’s interference with target gene expression through competitive binding with mRNA via Ago2 sponge miRNA, and its direct or indirect modulation of autophagy efficiency (55). Liang et al. demonstrated that circCDYL acts as a molecular sponge for miR-1275-ATG7, thereby enhancing autophagy in breast cancer (BC) cells by regulating ULK1 expression and ultimately exacerbating the poor prognosis of BC (49). Furthermore, the circRNA-autophagy axis can also regulate chemoresistance to various drugs, thus determining the efficacy of cancer treatment (56). In oral squamous cell carcinoma (OSCC), it was discovered that the circ-PKD2/miR-646/ATG13 axis promotes autophagy and enhances therapeutic sensitivity to cisplatin (DPP) in OSCC (57). Recent studies have also revealed that exosome-loaded circRNA can be transferred between cancer cells, activating WNT-CTNNB1/β-catenin signaling while augmenting autophagy and reducing chemosensitivity to accelerate tumor metastasis (58).
1.2.4 Autophagy is a double-edged sword
Autophagy is a crucial biological process that profoundly impacts cancer. An increasing body of research has established a strong association between autophagy and the initiation, progression, and drug resistance to cancer. The dual effects of autophagy activation on cancer are captivating (59). On the one hand, within the tumor microenvironment, malignant cells experience rapid proliferation and often encounter hypoxia and malnutrition conditions. Autophagy eliminates intolerable cells while promoting the survival of cancer cells under stressful circumstances, including enhancing their tolerance to drug therapy environments (56). Emerging evidence suggests that ISG15, a protein involved in post-translational modifications, stabilizes ATG7 structure and function while augmenting autophagic flux to counteract gemcitabine (GEM) sensitivity in pancreatic cancer (PC) therapy—a factor associated with poor prognosis in PC (60).
On the other hand, when toxic or malignant substances invade normal cells, autophagy inhibits inflammation, oxidation, and necrosis at the primary site while releasing immune regulatory factors to reduce infiltration by foreign harmful cancer cells (61). Lodder et al. simulated autophagy inhibition by knocking out ATG5 and found that mouse liver cells recruit more monocytes to aggravate liver fibrosis (62). Additionally, autophagy can enhance chemotherapy-induced cytotoxicity, improving drug sensitivity in cancer cells (63). Up-regulation of miR-519a expression was found to increase glioblastoma cell sensitivity towards temozolomide—an effect achieved through promoting autophagy via dissociation of Bcl-2/BECN1 complex as well as inhibiting STAT3/Bcl-2 axis activity (64). The dichotomous nature of autophagy reminds for clinicians to carefully consider whether to inhibit or activate this process during targeted therapies since each scenario yields distinct outcomes.
1.2.5 Therapies targeting autophagy
As the understanding of autophagy in cancer progression mechanisms grows, more autophagy compounds are being developed for future clinical trials and drug screening. WJ460, C150, Oroxylin A, and GNS561 have shown potential in selectively targeting autophagy and in anti-tumor effects in PC and hepatocellular carcinoma (HCC) (31). Tigecycline, commonly used for multidrug-resistant bacterial infections, has also been found to induce selective autophagy and inhibit GC cells activity (31).
The only clinically approved autophagy inhibitor is chloroquine (CQ) derivatives. Clinical trials have reported that CQ and hydroxychloroquine (HCQ) act as autophagy inhibitors by disrupting lysosomes and impairing Golgi apparatus function, particularly interfering with autophagosome-lysosome fusion, thus enhancing the anti-tumor efficacy of chemotherapy drugs (65). However, clinical trials using CQ derivatives for GIC, such as PC and CRC, have yielded mixed results (66, 67).
In recent years, a new class of protein-protein interaction (PPI) autophagy inhibitors based on ATG4 and ATG7 proteins has been developed (68). Compound 189 is one such inhibitor that targets the ATG12-ATG3 PPI interaction; it significantly reduces the number of autophagosomes and inhibits tumor growth in PC cells (69). Unfortunately, these inhibitors targeting autophagy-related proteins have not yet undergone clinical testing.
Interestingly enough, though, there are ongoing clinical trials combining traditional chemotherapy drugs or targeted therapy with autophagy inhibitors like HCQ plus FOLFOX (a CRC chemotherapy regimen) or bevacizumab bead sheet resistance, which may improve the anti-tumor effect on CRC patients (67, 70, 71). Additionally, HCQ plus atezolizumab combination therapy may also provide benefits for GIC precisely (70). In clinical practice for PC, the researchers did not observe any significant clinical significance of HCQ monotherapy. However, when combined with GEM therapy, HCQ treatment effectively improved median survival and disease-free survival in patients with PC (66). Radiotherapy is a crucial method in tumor treatment, and further exploration is needed to understand the therapeutic effect of autophagy inhibitors combined with radiotherapy in patients with cancer. Targeting biological molecules to regulate autophagy, such as the circRNA-autophagy axis described in this paper, represents a bold attempt to treat tumors.
The current field of autophagy is just a tiny fraction of the iceberg, with many issues still needing clarification. The same signaling pathway-mediated autophagy may play different roles in different cells in diseases such as cancer, inflammation, and neurodegeneration (72). CircRNA also mediates signaling pathways in this way (73). Studies have shown that elevated levels of circ_0032821 in GC cells inhibit autophagy, suggesting its significance as a critical regulator (74). Compared to other still emerging PCD, research on the impact of circRNA on autophagy is relatively mature, with numerous reports on the regulation of autophagy by circRNAs in GIC emerging in the past five years. There is currently no comprehensive discussion on the circRNA-autophagy axis in GIC, so it is necessary to study further the impact of autophagy on GIC under the regulation of circRNA.
2 Biological behavior of GIC
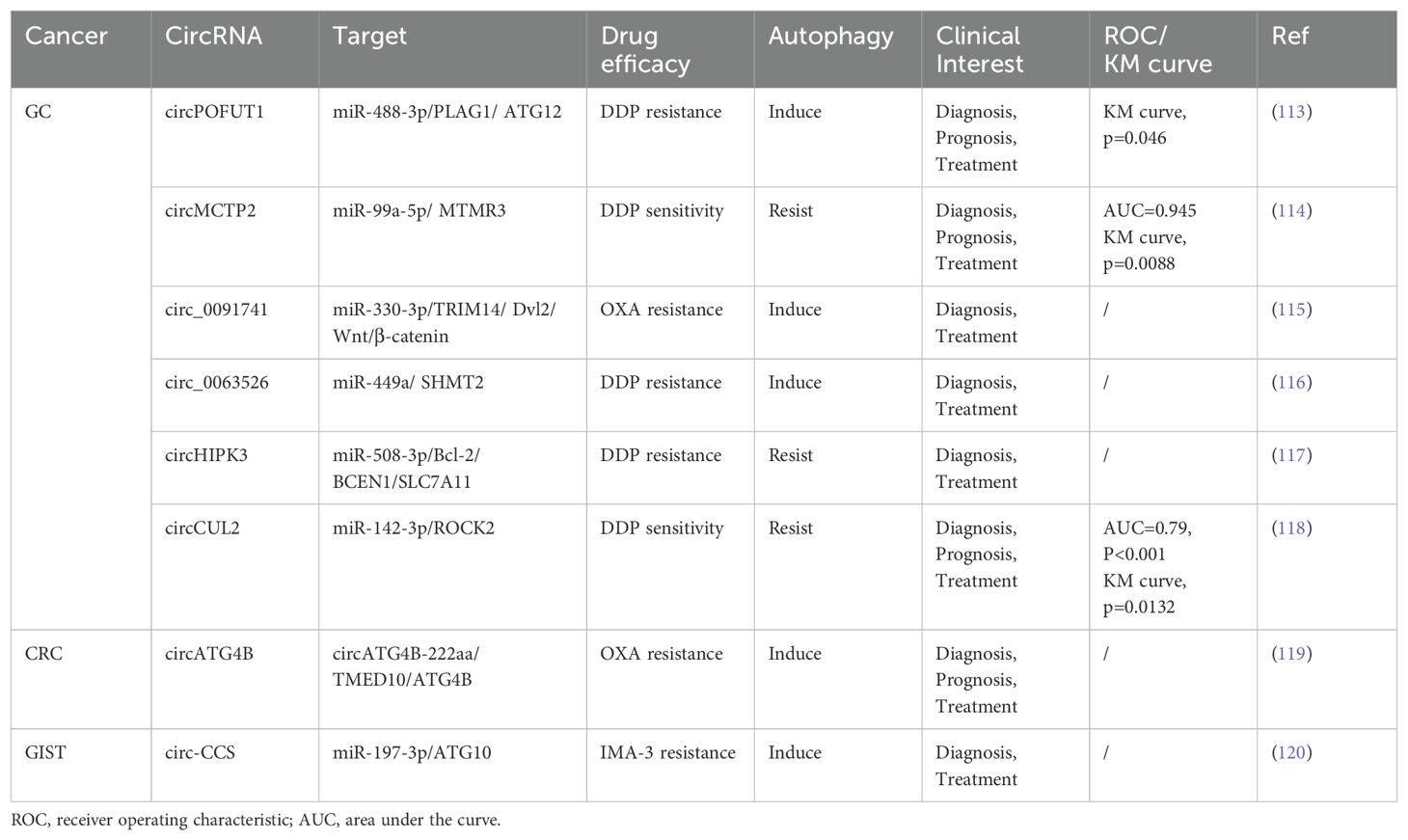
In this context, we emphasize the role of autophagy-related circRNA in GIC to better understand the functional roles of the circRNA-autophagy axis in different situations regarding the biological activity of GIC, such as proliferation, invasion, and metastasis (Table 2; Figure 3).
Figure 3. The mechanism of the circRNA-autophagy axis on biological activity. The mechanism by which circRNA mediates autophagy and impacts the progression of GIC is predominantly through the classical ceRNA pathway, thereby regulating the biological behavior of tumors. A minority of circRNA have been identified to interact with RBP or participate in translation processes, leading to autophagy-related alterations in tumor biology.
2.1 GC
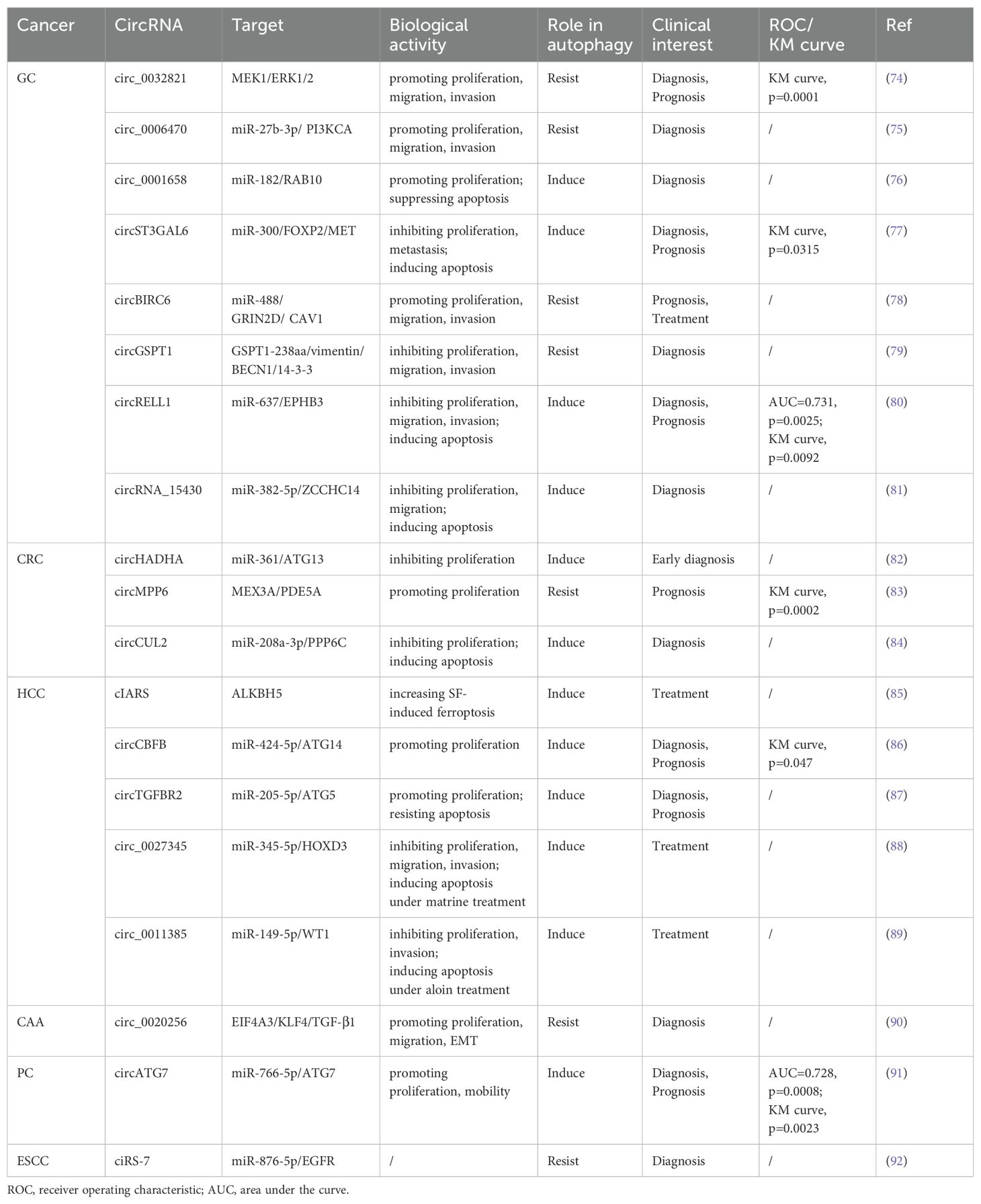
GC has consistently remained one of the most significant types of cancer worldwide, ranking as the fifth most prevalent malignant tumor with the fourth mortality rate globally (93). Due to the absence of specific early clinical indicators for GC, many patients are diagnosed at an advanced stage during endoscopy or imaging procedures. For individuals with advanced GC or distant metastasis, the 5-year survival rate is less than 5% (94). Urgent investigation into novel biomarkers and therapeutic targets for the early diagnosis and treatment of GC is essential. Numerous studies have revealed a significant correlation between circRNA and autophagy in the progression of GC (95).
Cui et al. proved that the circ_0006470/miR-27b-3p/PI3KCA axis regulates autophagy, proliferation, and invasion of GC cells. Specifically, circ_0006470 and PI3KCA act as negative regulators of autophagy levels while functioning as positive regulators of GC malignancy (75). Duan and colleagues discovered that circ_0001658 is highly expressed in GC cells and clinical specimens, promoting GC progression and autophagy by targeting miR-182/RAB10 (76). Silencing this circRNA inhibited the proliferation of GC cells and reduced IL3 site expression on GC cells through immunofluorescence analysis. Additionally, overexpression of circST3GAL6 was observed in both GC tissues and cells. This circRNA regulated downstream transcription factor FOXP2 through sponge miR-300 to inhibit the invasion of GC cells while promoting autophagy (77). Further investigation revealed that FOXP2 promotes autophagy by mediating the binding between circST3GAL6 and MET promoter to activate the AKT/mTOR pathway. The ultimate consequence of this mechanism is to augment the process of autophagy and thereby optimize the prognostic quality for patients diagnosed with GC. Tang et al. discovered that the circBIRC6-miR-488/GRIN2D axis synergistically promotes multiple malignant behaviors in GC. Specifically, circBIRC6 and GRIN2D exhibit concordant effects, as their upregulation reduces caveolin-1 (CAV1)-mediated autophagy (78).
Based on the latest findings, proteins derived from the translation of circRNA play a pivotal role in regulating various biological behaviors of tumors (96). The polypeptide GSPT1-238aa is encoded by circGSPT1. Hu et al. demonstrated that the circGSPT1 inhibits GC cells proliferation and lymph node metastasis, while its product GSPT1-238aa acts as an independent regulator to suppress malignant behavior in GC (79). Vimentin/BECN1/14-3-3 has been identified as an autophagy inhibitory complex, and GSPT1-238aa directly binds to Vimentin. Hu’s team extensively investigated the interaction between GSPT1-238aa and the autophagy inhibitory complex, revealing its efficacy in suppressing autophagy through the PI3K/AKT/mTOR pathway. The combined significance of circRNA translation products with autophagy in GC prognosis warrants further consideration.
Exosomes, secreted by cells into body fluids within organisms, encapsulate genetic materials from parental cells within multivesicular bodies and deliver them to recipient cells, including circRNA (97). Upon reaching recipient cells, exosomal circRNA can assume various functions. For instance, plasma exosomal circRELL1 derived from GC tissues has been confirmed to inhibit the proliferation and invasion of GC cells by mediating the miR-637/EPHB3 axis, in line with findings from cell and animal experiments involving circRELL1 (80). Inversely, the downregulation of exosomal circRELL1 led to a decrease in IL3 levels, suggesting that the circRELL1-autophagy axis also contributes to the biological behavior of GC cells. The implications of exosomal circRNA in cancer progression offer an additional avenue for early cancer diagnosis.
Liu et al. discovered that circ_0015430 exhibited downregulation in GC, facilitating GC cells proliferation and migration. Researchers found that circ_0015430 was also reduced in Helicobacter pylori (H. pylori)-infected chronic gastritis tissues and GC cells, accompanied by an increase in the abundance of autophagosome (81). Circ_0015430 regulated the dissemination process of GC cells and associated autophagy by acting as a sponge for miR-382-5p/ZCCHC14. The inhibition of circ_0015430 reversed the impact of H. pylori infection on autophagy. It is widely acknowledged that H. pylori infection induces chronic gastritis, serving as a common carcinogenic factor leading to a series of malignant pathologies in the gastric mucosa, according to the Correa cascade model (98). Therefore, circ_0015430 may be an effective therapeutic target for treating GC complicated with H. pylori infection.
2.2 CRC
The increasing prevalence of early-onset CRC has become a matter of growing concern. As per the data, approximately 11% of colon cancer patients and 23% of rectal cancer patients globally are anticipated to be under the age of 50 by 2030 (99). In chemotaxis in young individuals with CRC, unraveling more potent and dependable molecular biology markers, such as circRNA associated with the autophagy pathway, which is involved in the initiation and development of CRC (100).
Based on the evidence that serrated polyps can progress to CRC, researchers are increasingly persuaded that polyps play a significant role as precursor lesions in the development of CRC (101). The relevance of colonic polyps concerning colon cancer has become increasingly evident. Shi’s research team confirmed differential expression of circHADHA in the plasma samples from healthy individuals, patients with colonic polyps, and patients with colon cancer. Compared to healthy individuals, circHADHA was upregulated in the plasma of patients with colonic polyps and downregulated in the plasma of patients with colon cancer (82). Cell experiments demonstrated that circHADHA could inhibit the proliferation of colon cancer cells by enhancing autophagy. Dual-luciferase assay revealed that miR-361 could bind to circHADHA/ATG13, and overexpression of circHADHA could competitively bind to miR-361 to increase ATG13 and LC3-II levels for restoring autophagy. In summary, the dynamic changes observed in plasma circHADHA among patients with colon polyps and colon cancer provide valuable insights for the early detection of malignant tendencies associated with polyps. Additionally, targeting the circHADHA/miR-361/ATG13 axis may offer a novel biological approach to treating colon cancer.
As a biomolecule in RBP, MEX3A is significantly up-regulated in CRC, but its expression level is opposite to the autophagy marker LC3-II, indicating that MEX3A is an autophagy inhibitor. In subsequent experiments, researchers have further proved circMPP6, after forming complexes with MEX3A biological accumulation, can accelerate PDE5A mRNA degradation, MEX3A level and PDE5A levels of the rise of synergistic inhibition of autophagy and accelerate CRC malignant behavior (83). The competitive endogenous RNA (ceRNA) network represents a classical pathway through which circRNA regulates autophagy (102). In light of this, studies involving the circRNA-RBP complex are instrumental in comprehensively understanding the mechanism underlying the circRNA-autophagy axis.
To delve into the regulatory mechanism of circCUL2 in CRC, Yang et al. initially detected the downregulation of circCUL2 expression in collected CRC tissues. Subsequently, they ascertained that increased circCUL2 expression robustly inhibited CRC cell proliferation and arrested tumor growth in both Cell Counting Kit-8 (CCK-8) and tumor allograft experiments (84). Further experimental validation corroborated that circCUL2 induced autophagy through its interaction with miR-208a-3p, thereby exerting a tumor-suppressive role in CRC.
2.3 HCC
The emergence of HCC has garnered significant attention in the global healthcare community. At present, the primary etiology of HCC is transitioning from viral hepatitis to non-alcoholic fatty liver disease, a condition intimately linked with obesity (103). With the increasing burden of HCC, it is vital to identify novel biomarkers with enhanced sensitivity and specificity in addition to alpha fetoprotein (AFP), including circRNA, to guide the diagnosis and systemic treatment of HCC (104).
Sorafenib (SF) is the preferred systemic treatment for HCC and confers a survival benefit across different subtypes (105). CIARS, a highly expressed circRNA in HCC following SF treatment, interacts with ALKBH5 to antagonize the inhibitory effects on autophagy. Moreover, cIARS/ALKBH5 promotes ferroptosis in HCC cells after SF treatment (85).
Zhao et al. transfected circCBFB mimic into HCC cells and observed enhanced proliferation ability using CCK-8 assays, reversing the growth inhibition caused by an autophagy inhibitor called 3-methyladenine (3-MA) treatment (86). Furthermore, transfection of miR-424-5p mimic or ATG14 mimic led to decreased cell viability and inhibited autophagy in HCC cells. The investigator confirmed that circCBFB/miR-424/ATG14 enhances HCC proliferation activity by promoting autophagy via dual-luciferase assay. Therefore, there are promising prospects for the clinical application of circ-SPECC1 and circCBFB in improving the survival rate of patients with HCC. Wang et al. revealed that exosomal circTGFBR2, in combination with miR-205-5p, targets ATG5 downstream to enhance the autophagy process in HCC cells, thereby increasing their resistance to starvation stress (87).
Natural chemical products derived from Chinese herbal medicine exhibit anti-inflammatory, anti-tumor, and other therapeutic properties. Due to their relatively low toxicity and natural compounds, researchers have progressively investigated them. The mechanisms by which certain natural compounds, such as terpenoids, alkaloids, phenols, and flavonoids, inhibit tumor growth have been elucidated (106). Lin et al. experimented with Matrine to treat HCC and demonstrated its efficacy in inhibiting HCC progression. By observing epigenetic changes in the treatment group, they identified down-regulated circ_0027345 (88). Subsequent experiments revealed that Matrine utilized the circ_0027345/miR-345-5p/HOXD3 axis to promote autophagy while attenuating its anti-tumor effectiveness. On the other hand, Fu et al. identified an oncogene, circ_0011385, in aloin-treated HCC that targeted miR-149-5p/WT1. The administration of aloin effectively inhibited HCC cells proliferation and tumor growth. Inhibition of circ_0011385 expression was observed alongside WT1 inhibition, accompanied by increased autophagy markers LC3-II and BECN1 expression. Conversely, overexpression of circ_0011385 reversed the effects induced by aloin (89). Long-term regulation of circRNA can enhance the therapeutic effect of natural chemicals in treating HCC while also providing novel pharmacological mechanisms for future drug research and development.
2.4 Cholangiocarcinoma
The CAA can be classified into three types based on the location of the lesion: intrahepatic CAA, hilar CAA, and extrahepatic CAA. Although the incidence of CAA in GIC is relatively low, early detection through conventional tumor markers remains essential due to its high mortality rate (107). In recent years, dysregulated expression of circRNA has been implicated in the pathogenesis of CAA (108). EIF4A3 functions as an RBP that interacts with circ_0020256 to consolidate the expression of transcription factor KLF4 in CAA. This interaction further promotes TGF-β1 secretion (90), which plays a role in epithelial-mesenchymal transition (EMT) mediated by TGF-β1/Smad pathway activation (109). Furthermore, CAA cells activate cancer-associated fibroblasts (CAFs) via the TGF-β1/Smad pathway, while IL-6 secreted by CAF inhibits autophagy and accelerates CAA growth and infiltration. In future studies, targeting the circ_0020256/EIF4A3/KLF4/TGF-β1 axis could provide a novel approach for intervention in cholangiocarcinoma.
2.5 PC
PC is a markedly aggressive tumor, with pancreatic ductal adenocarcinoma being the most prevalent pathological type. The majority of patients are ineligible for surgical intervention upon diagnosis, and even after surgery, nearly three-quarters of patients experience recurrence within three years (110). He et al. identified a negative correlation between circATG7 and miR-766-5p but a positive correlation with ATG7 in PC. This circRNA was found to enhance invasion and metastasis in PC cells, upregulate ATG7 expression through miR-766-5p sequestration, and provide human antigen R protein (HuR) scaffold to stabilize ATG7 mRNA. Consequently, it can facilitate autophagy in PC cells (91). Recognizing contemporary medicine’s substantial challenges in diagnosing and treating PC is paramount. Consequently, circRNA is garnering increasing acknowledgment for its role in elucidating the pathogenesis of PC.
2.6 Esophageal carcinoma
Esophageal cancer is a prevalent tumor of the digestive tract, with squamous cell carcinoma (ESCC) and adenocarcinoma being the primary histological subtypes. As revealed by GLOBOCAN’s data, a projected increase of 58.4% in new cases and 61.7% in deaths due to esophageal cancer is anticipated from 2020 to 2040 (111). Meng et al. demonstrated that ciRS-7 exhibits abnormal expression in ESCC and functions as an oncogenic circRNA that regulates the miR-1299/EGFR axis, thereby impacting the AKT/mTOR pathway (92). CiRS-7 acts as a negative regulator of autophagy, suggesting that the integrated ciRS-7-autophagy axis may contribute to enhancing the malignant behavior of ESCC.
3 Drug resistance
Pharmaceuticals assume a vital function in cancer management, acting as indispensable instruments to mitigate preoperative lesions, forestall postoperative recurrence, and offer palliative care for advanced stages. The conventional agents employed in GIC therapy primarily encompass DPP, carboplatin, oxaliplatin (OXA), and GEM. The advent of targeted drugs and anti-programmed death ligand 1 (PD-L1) drugs has opened up new opportunities for patients with GIC. However, drug resistance is a common challenge encountered in GIC treatment regardless of the specific antineoplastic agent employed. Scientists have elucidated various mechanisms underlying tumor drug resistance, including reduced intracellular drug concentration, disrupted drug metabolism processes, target mutations or difficulties in forming effective drug-target complexes, and alterations in signaling pathways (112). We actively explore new biomarkers that may regulate drug resistance and have identified the research potential of the circRNA-autophagy axis (Table 3; Figure 4), as well as circRNA and autophagy-related factors as promising biological targets to enhance the therapeutic efficacy of GIC medications.
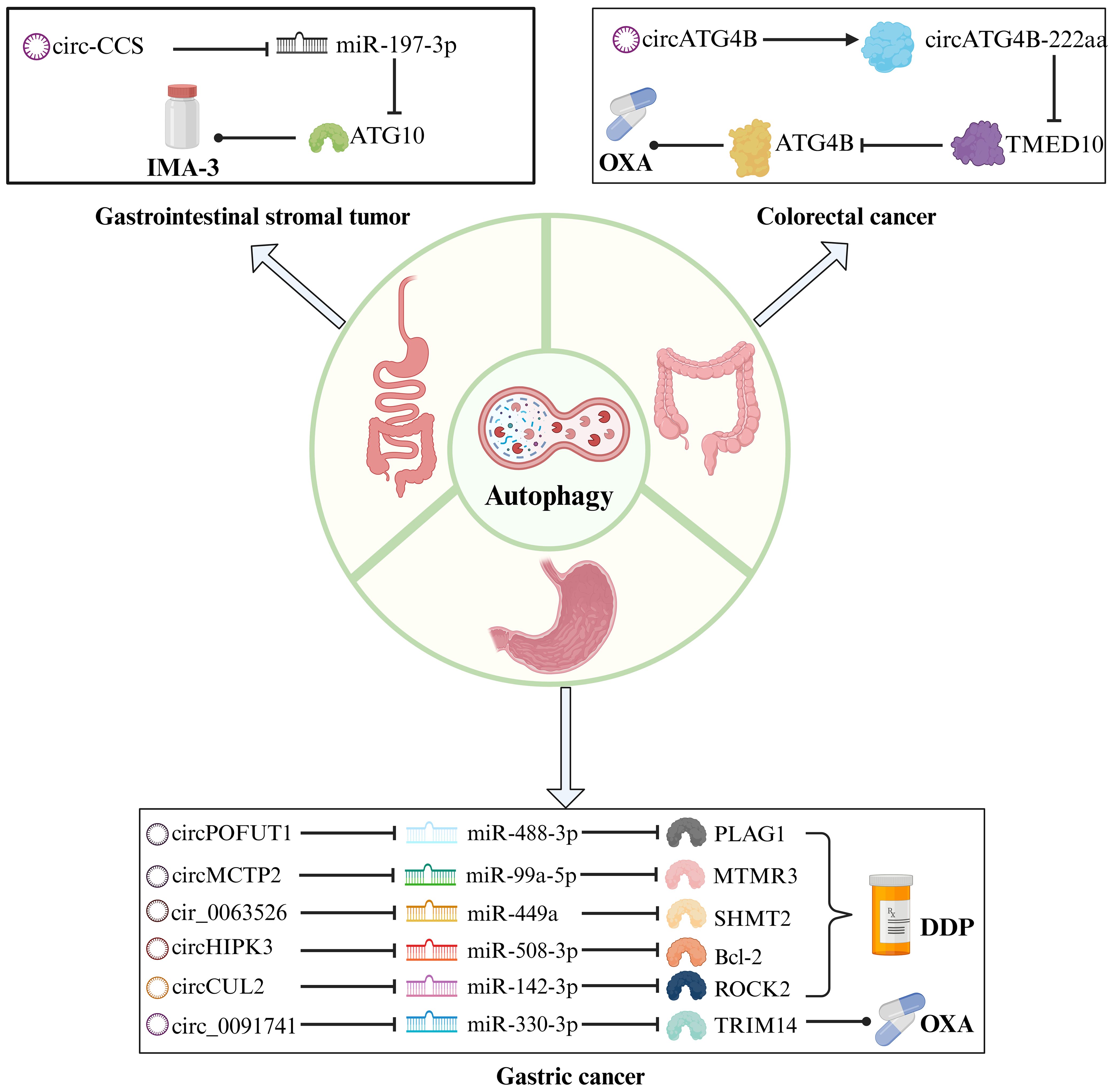
Figure 4. The mechanism of the circRNA-autophagy axis on drug efficacy. CircRNA-mediated autophagy regulates the signaling pathway of chemoresistance. Platinum drugs such as DDP and OXA are traditional chemotherapy drugs for the treatment of GC and CRC, and IMA-3 is a common drug for the treatment of GIST.
3.1 GC
Luo et al. examined the function of circPOFUT1 as a ceRNA sponge for miR-488-3p, and identified PLAG1 as a protooncogene that directly interacts with miR-488-3p to regulate ATG12 expression in GC indirectly (113). The upregulation of circPOFUT1 in GC promotes tumor progression, autophagy progression, and confers resistance to DPP through autophagy-related mechanisms. Numerous studies have centered on DPP resistance. Sun et al. also demonstrated that circMCTP2, which modulates DPP resistance, collaborates with miR-99a-5p/MTMR3 to suppress the autophagic behavior of DPP-resistant GC cells (114). The functional significance of circMCTP2 in GC is dissimilar to that of circPOFUT1, as it serves to restrain DPP resistance.
Exosomal circRNA regulates the malignant behavior of tumors and serves as a novel mechanism of cancer drug resistance (121). circ_0091741 (115) and cir_0063526 (116) facilitate intercellular communication through exosomes, promoting autophagy-induced OXA and DPP resistance via the miR-330-3p/TRIM14 axis and the miR-449a/SHMT2 axis, respectively. These exosomal circRNAs effectively propagate drug resistance among cells. Notably, increased expression of TRIM14 activates the Dvl2/Wnt/β-catenin signaling pathway to enhance OXA resistance in GC, while elevated expression of SHMT2 promotes DPP resistance. Surprisingly, Shang et al. confirmed that the miR-508-3p/Bcl-2/BECN1/SLC7A11 axis negatively regulates autophagy and ferroptosis, where autophagy activates ferroptosis (117). Considering that serum levels of exosomal circHIPK3 were lower in DPP-sensitive patients compared to DPP-resistant patients, this circRNA’s role in enhancing DPP resistance was validated. The findings suggest a notable reduction in serum exosomal circHIPK3 levels in GC patients treated with DPP. The exosomal circ_0091741, circ_0063526, and circHIPK3 molecules can serve as robust, non-invasive indicators for monitoring the efficacy of chemotherapy in GC.
3.2 CRC
CircRNA plays a diverse function in CRC, including protein translation (122). ATG4B is a vital signaling molecule that governs the autophagy transduction pathway, and its activity is intricately linked to the high mortality rate and chemoresistance of CRC (123). CircATG4B is a circRNA generated through processing and splicing of the ATG4B gene. Pan et al. contrasted OXA-resistant CRC cells with OXA-sensitive CRC cells, corroborating a significant expression of circATG4B and its protein coding product, cirCATG4B-222aa, in resistant cells (119). Regarding autophagy, circATG4B-222aa functions as a binding platform for TMED10 to augment autophagy. TMED10 is recognized to regulate ATG4B as an autophagy regulator negatively (124). Upon binding to TMED10, circATG4B-222aa dissociates from it and releases more ATG4B, thereby attenuating the anti-tumor effect of OXA in CRC. A principal challenge resides in translating circATG4B-222aa into clinical practice.
3.3 Gastrointestinal stromal tumor
The primary origin of GIST is spontaneous mutations in receptor tyrosine kinase (KIT) or platelet-derived growth factor α (PDGFRα). Tyrosine kinase inhibitors and surgical resection are prevalent treatment methods for GIST (125). Sui et al. detected elevated expression of circ-CCS in patients with imatinib (IMA-3) resistance and recurrence, which promotes the proliferation, dissemination, and autophagy of GIST cells (120).miR-197-3p binds to circ-CCS to regulate ATG10, ultimately resulting in IMA-3 resistance. IMA-3 resistance poses a substantial challenge in the management of GIST. By investigating circ-CCS, potential breakthroughs in drug resistance may be uncovered.
4 The future directions of autophagy-related circRNA
The emergence of immunotherapy (IT), a significant milestone in cancer treatment, encompasses four main types: immune checkpoint inhibitors (ICIs), with PD-L1 being the classic ICIs, peripatetic immunotherapy, tumor vaccines, and chimeric antigen receptor T-cell therapy (126). Autophagy-related molecules stimulate immune responses to engage in anti-tumor therapy and recruit immune cells such as natural killer cells and T lymphocytes to target tumor cells (127). Studies have demonstrated that ATG5 and ATG7 regulate ferroptosis-mediated tumor IT (127). Taraborrelli et al. reported that ATG16L1 protects CRC from immune cell infiltration and cytokine-induced inflammation (128).
In addition, the proteins encoded by circRNA play significant roles in the field of biological vaccines (129). Lipid nanoparticles serve as a delivery medium to transport circRNA into cells, while the protein acts as a specific antigen to initiate systemic immune response (130). A novel engineered circular mRNA vaccine introduces immune cells into tumors and inhibits tumor growth. Utilizing engineered circRNA enhances translation efficiency and protein stability while reducing immunogenicity (131). Although there have been numerous reports on autophagy or circRNA research and their applications in IT, studying the circRNA-autophagy axis in IT is still early. Exploring the connection between the circRNA-autophagy axis and IT will greatly improve cancer treatment.
Immune evasion is a challenge in cancer treatment. When the expression of mitochondrial translocator protein (TPSO) was specifically increased in HCC, autophagy-mediated ferroptosis was inhibited. Mechanistically, upregulated TPSO targeted the p62/KEAP1/Nrf2 pathway and upregulated the expression of PD-L1, thereby promoting the immune evasion of HCC cells (132). Miao et al. reported that circ_0136666 combined with miR-375/PRKDC axis and phosphorylation PD-L1 improved immune evasion GC cells (133). PD-L1 and MHC-I/II regulation are important in immune evasion (134). A comprehensive understanding of the mechanism of autophagy and circRNA in immune evasion can better avoid the treatment risks caused by immune evasion.
5 Discussion
The regulation of autophagy by circRNA has been associated with the modulation of tumor progression, cardiovascular diseases, and neurodegenerative disorders (55). GIC represents a significant form of cancer. In the long run, the circRNA-autophagy axis presents a novel perspective for diagnostic and therapeutic strategies targeting GIC.
Indeed, the same circRNA plays a principal role in various human tumors (135). By carefully comparing the aberrantly expressed circRNA in different tumor types, it has been observed that they not only regulate the progression and drug resistance of multiple tumors, but also participate in the process of autophagy. CircCUL2 and circHIPK3 act as miRNA sponges to target downstream genes, forming a circRNA-autophagy axis that inhibits CRC progression (84) and promotes GC DPP resistance (117), respectively. Previously, Peng et al. demonstrated that circCUL2 mediates the inhibition of autophagy through the miR-142-3p/ROCK2 axis to induce DPP sensitivity in GC (118). Expanding beyond GIC, Wang et al. confirmed that circHIPK3 binds to and inhibits VCP (a kind of RBP), thereby suppressing BECN1-mediated autophagy and impeding bladder cancer progression (136). Consequently, the regulatory roles of both the circCUL2-autophagy axis and circHIPK3-autophagy axis have been elucidated across different tumor types, thus highlighting the need for further exploration of the involvement of the circRNA-autophagy axis in additional malignancies.
In reality, the development of new anti-tumor drugs is imminent. Natural chemicals derived from plants impact HCC progression by modulating autophagy through circRNA (89), which holds great promise. The liver serves as the primary site for anthocyanin metabolism, and Matboli’s team has validated certain anthocyanins like cyanidin-3-glucoside (C3G) to exhibit a dose-dependent anti-liver cancer effect via cell cycle regulation (137). Zabady et al. administered C3G to rats induced with precancerous lesions. Compared to the C3G treatment group, the precancerous lesion group showed increased levels of AFP and liver enzymes and decreased albumin content, consistent with Matboli’s experimental findings (138). Even more striking, circ_0001345 and autophagy-related gene ATG16L1 were concurrently upregulated in the C3G treatment group, while miRNA-106b was downregulated upon C3G treatment. Therefore, it was predicted that C3G could enhance ATG16L1-mediated autophagy through the circ_0001345/miR-106b/ATG16L1 axis identified via database analysis to achieve therapeutic goals for HCC treatment. Yang et al. isolated total flavonoids, luteolin, and apigenin from Scutellaria barbata D.Donc (SB) and Oldenlandia diffusa (Willd.) Roxb (OD) demonstrates their ability to inhibit the invasion of HCC cells, suppress hepatitis B virus replication activity, and hinder autophagy processes. The HCC cells treated with SB extract (SBE) and OD extract (ODE) exhibited a diverse range of differentially expressed circRNA. Researchers constructed a circRNA-miRNA-gene network diagram associated with the SBE and ODE. At the same time, further investigation is required regarding the involvement of the circRNA-autophagy axis in SBE and ODE (139). In future studies, combining traditional chemical drugs with natural compounds will greatly contribute to HCC treatment advancements and expand their application beyond HCC into GIC treatment.
The metabolic microenvironment significantly influences the metastatic dissemination of cancer. Altered metabolic status in the microenvironment leads to changes in nutrient uptake and oxidative stress. Cancer cells employ the autophagy pathway as an adaptive response to their surrounding environment, thereby regulating their ability to metastasize to distant organs (140). Gastrointestinal organs are commonly affected sites of metastasis; for instance, the liver is pivotal in tumor metastasis (141). Wang et al. confirmed that circROBO1 and FUS were highly expressed in BC, with KLF5-FUS promoting circROBO1 back-splicing. This behavior further facilitated BC cells proliferation, metastasis, and tumor growth (142). Subsequent experiments verified that miR-217-5p counteracted the malignant events induced by circROBO1 and formed a signaling pathway with KLF5 to establish the circROBO1/miR-217-5p/KLF5 axis. This axis inhibited afadin-related autophagy while promoting liver metastasis of BC through positive feedback mechanisms. Therefore, the circRNA-autophagy axis also regulates secondary GIC, and understanding its regulatory mechanism may improve prognoses for primary cancers.
The exact contribution of autophagy to the biological behavior and drug resistance of GIC remains to be fully understood. While it has been demonstrated that circFARP1 in PC plays a role in CAF and induces GEM resistance, the interaction between autophagy and circFARP1 remains unclear (143). Previous studies have shown that the TGF-β1/Smad2/3 pathway upregulates ATG4 levels, leading to reduced efficacy of GEM in PC (144). Furthermore, activation of the TGF-β1/Smad pathway induces CAF activation in CAA and promotes the secretion of autophagy-related mediators (90). Therefore, the potential link between circFARP1 and the autophagy pathway in PC warrants further investigation. ULK1 is a known activator of autophagy (145). Wang et al. demonstrated that ULK1, as a downstream target of miR-142-5p, was positively regulated by circTMEM87A to enhance proliferation, metastasis, and invasion of GC cells. However, this study did not assess changes in BECN1, IL3-II, or p65 levels (146). It is hypothesized that circTMEM87A may target ULK1 and participate in regulating autophagy processes. Importantly, it should be noted that ULK1 also possesses non-autophagic functions.
Autophagy-related circRNA accurately target autophagy genes or pathways during the process of autophagy and regulate cancer progression. CircRNA are widely detected in human body fluids (7). Furthermore, its inherent stability aids in validating the clinical significance of more autophagy-related circRNA in cancer (121), establishing a comprehensive database for subsequent clinical applications. The expression of circRNA varies among different patients and disease stages (147). Therefore, personalized circRNA treatments based on individual differences will be more targeted. Biological nanoparticles and viruses serve as carriers to encapsulate autophagy-related circRNA and deliver them to target cells, protecting against degradation caused by external stressors and promoting production (148, 149). The pre-clinical trials should be rigorously regulated under ethical and moral guidelines, with a focus on minimizing drug toxicity and side effects, as well as optimizing research and development costs for the benefit of a larger population of cancer patients.
6 Conclusions
This review summarizes recent studies on the circRNA-autophagy axis in the biological behavior and drug resistance of GIC. CircRNA regulates autophagy through ceRNA, RBP binding, and other mechanisms within multiple signaling pathways. Emerging research highlights the interplay between circRNA translation function and autophagy, influencing the progression and resistance of GIC, including secondary GIC. The complexity of autophagy in GIC necessitates further investigation due to its dual nature. Several circRNA have been identified as regulators of autophagy in various types of GIC and other cancers. The generalizability of the circRNA-autophagy axis beyond individual GIC to encompass a broader range of cancers or cancer screening applications merits exploration. Notably, plant extracts have demonstrated involvement with the circRNA-autophagy axis in treating GIC, highlighting its potential for application in novel drug development. Future research will primarily focus on elucidating the specific mechanism of the circRNA-autophagy axis in regulating the occurrence and progression of GIC. This endeavor aims to utilize the circRNA-autophagy axis as a novel biological marker for early diagnosis and severity evaluation of GIC. It also explores its potential as a therapeutic target to overcome drug resistance and reduce the global disease burden associated with GIC.
Author contributions
BZ: Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. ZL: Methodology, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. GY: Funding acquisition, Resources, Writing – review & editing. KH: Project administration, Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from Ningbo Top Medical and Health Research Program (No.2023020612), the Ningbo Leading Medical & Healthy Discipline (2022-S04), Ningbo Key Technology Research and Development Special Project (no. 2021Z133) and the Medical and Health Research Project of Zhejiang Province (no. 2023RC262).
Acknowledgments
We thank all colleagues who contributed to this manuscript, and acknowledge using Biorender to create Figures 1–4.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Feng XY, Zhu SX, Pu KJ, Huang HJ, Chen YQ, Wang WT. New insight into circRNAs: characterization, strategies, and biomedical applications. Exp Hematol Oncol. (2023) 12:91. doi: 10.1186/s40164-023-00451-w
2. Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. (2017) 16:94. doi: 10.1186/s12943-017-0663-2
3. Zhang Z, Will CL, Bertram K, Dybkov O, Hartmuth K, Agafonov DE, et al. Molecular architecture of the human 17S U2 snRNP. Nature. (2020) 583:310–3. doi: 10.1038/s41586-020-2344-3
4. Shenasa H, Movassat M, Forouzmand E, Hertel KJ. Allosteric regulation of U1 snRNP by splicing regulatory proteins controls spliceosomal assembly. Rna. (2020) 26:1389–99. doi: 10.1261/rna.075135.120
5. Pervouchine DD. Circular exonic RNAs: When RNA structure meets topology. Biochim Biophys Acta Gene Regul Mech. (2019) 1862:194384. doi: 10.1016/j.bbagrm.2019.05.002
6. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. (2019) 20:675–91. doi: 10.1038/s41576-019-0158-7
7. Wang H, Meng Q, Qian J, Li M, Gu C, Yang Y. Review: RNA-based diagnostic markers discovery and therapeutic targets development in cancer. Pharmacol Ther. (2022) 234:108123. doi: 10.1016/j.pharmthera.2022.108123
8. Zhou B, Yang H, Yang C, Bao YL, Yang SM, Liu J, et al. Translation of noncoding RNAs and cancer. Cancer Lett. (2021) 497:89–99. doi: 10.1016/j.canlet.2020.10.002
9. Kocarnik J. Cancer’s global epidemiological transition and growth. Lancet. (2020) 395:757–8. doi: 10.1016/S0140-6736(19)32046-X
10. Brennan P, Davey-Smith G. Identifying novel causes of cancers to enhance cancer prevention: new strategies are needed. J Natl Cancer Inst. (2022) 114:353–60. doi: 10.1093/jnci/djab204
11. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. (2019) 179:1033–55. doi: 10.1016/j.cell.2019.10.017
12. Shu F, Xiao H, Li QN, Ren XS, Liu ZG, Hu BW, et al. Epigenetic and post-translational modifications in autophagy: biological functions and therapeutic targets. Signal Transduct Target Ther. (2023) 8:32. doi: 10.1038/s41392-022-01300-8
13. Nakatogawa H. Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol. (2020) 21:439–58. doi: 10.1038/s41580-020-0241-0
14. Fleming A, Bourdenx M, Fujimaki M, Karabiyik C, Krause GJ, Lopez A, et al. The different autophagy degradation pathways and neurodegeneration. Neuron. (2022) 110:935–66. doi: 10.1016/j.neuron.2022.01.017
15. Debnath J, Gammoh N, Ryan KM. Autophagy and autophagy-related pathways in cancer. Nat Rev Mol Cell Biol. (2023) 24:560–75. doi: 10.1038/s41580-023-00585-z
16. Shang JN, Yu CG, Li R, Xi Y, Jian YJ, Xu N, et al. The nonautophagic functions of autophagy-related proteins. Autophagy. (2024) 20:720–34. doi: 10.1080/15548627.2023.2254664
17. Pareek G, Kundu M. Physiological functions of ULK1/2. J Mol Biol. (2024) 436:168472. doi: 10.1016/j.jmb.2024.168472
18. Chen X, Cao Y, Guo Y, Liu J, Ye X, Li H, et al. microRNA-125b-1-3p mediates autophagy via the RRAGD/mTOR/ULK1 signaling pathway and mitigates atherosclerosis progression. Cell Signal. (2024) 118:111136. doi: 10.1016/j.cellsig.2024.111136
19. Alers S, Löffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. (2012) 32:2–11. doi: 10.1128/MCB.06159-11
20. Adriaenssens E, Ferrari L, Martens S. Orchestration of selective autophagy by cargo receptors. Curr Biol. (2022) 32:R1357–r1371. doi: 10.1016/j.cub.2022.11.002
21. Safaroghli-Azar A, Sanaei MJ, Pourbagheri-Sigaroodi A, Bashash D. Phosphoinositide 3-kinase (PI3K) classes: From cell signaling to endocytic recycling and autophagy. Eur J Pharmacol. (2023) 953:175827. doi: 10.1016/j.ejphar.2023.175827
22. Fracchiolla D, Chang C, Hurley JH, Martens S. A PI3K-WIPI2 positive feedback loop allosterically activates LC3 lipidation in autophagy. J Cell Biol. (2020) 219:e201912098. doi: 10.1083/jcb.201912098
23. Wei S, Leng B, Yan G. Targeting autophagy process in center nervous trauma. Front Neurosci. (2023) 17:1128087. doi: 10.3389/fnins.2023.1128087
24. Mohamud Y, Shi J, Tang H, Xiang P, Xue YC, Liu H, et al. Coxsackievirus infection induces a non-canonical autophagy independent of the ULK and PI3K complexes. Sci Rep. (2020) 10:19068. doi: 10.1038/s41598-020-76227-7
25. Zhou Y, Wang Z, Huang Y, Bai C, Zhang X, Fang M, et al. Membrane dynamics of ATG4B and LC3 in autophagosome formation. J Mol Cell Biol. (2022) 13:853–63. doi: 10.1093/jmcb/mjab059
26. Holzer E, Martens S, Tulli S. The role of ATG9 vesicles in autophagosome biogenesis. J Mol Biol. (2024) 436:168489. doi: 10.1016/j.jmb.2024.168489
27. Proikas-Cezanne T, Takacs Z, Dönnes P, Kohlbacher O. WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome. J Cell Sci. (2015) 128:207–17. doi: 10.1242/jcs.146258
28. Yamamoto H, Zhang S, Mizushima N. Autophagy genes in biology and disease. Nat Rev Genet. (2023) 24:382–400. doi: 10.1038/s41576-022-00562-w
29. Liu S, Yao S, Yang H, Liu S, Wang Y. Autophagy: Regulator of cell death. Cell Death Dis. (2023) 14:648. doi: 10.1038/s41419-023-06154-8
30. Vargas JNS, Hamasaki M, Kawabata T, Youle RJ, Yoshimori T. The mechanisms and roles of selective autophagy in mammals. Nat Rev Mol Cell Biol. (2023) 24:167–85. doi: 10.1038/s41580-022-00542-2
31. Liu J, Wu Y, Meng S, Xu P, Li S, Li Y, et al. Selective autophagy in cancer: mechanisms, therapeutic implications, and future perspectives. Mol Cancer. (2024) 23:22. doi: 10.1186/s12943-024-01934-y
32. Ma W, Lu Y, Jin X, Lin N, Zhang L, Song Y. Targeting selective autophagy and beyond: From underlying mechanisms to potential therapies. J Adv Res. (2024) S2090-1232:00199-1. doi: 10.1016/j.jare.2024.05.009
33. Lu Y, Li Z, Zhang S, Zhang T, Liu Y, Zhang L. Cellular mitophagy: Mechanism, roles in diseases and small molecule pharmacological regulation. Theranostics. (2023) 13:736–66. doi: 10.7150/thno.79876
34. Chino H, Mizushima N. ER-phagy: quality and quantity control of the endoplasmic reticulum by autophagy. Cold Spring Harb Perspect Biol. (2023) 15:a041256. doi: 10.1101/cshperspect.a041256
35. Sorice M. Crosstalk of autophagy and apoptosis. Cells. (2022) 11:1479. doi: 10.3390/cells11091479
36. Prerna K, Dubey VK. Beclin1-mediated interplay between autophagy and apoptosis: New understanding. Int J Biol Macromol. (2022) 204:258–73. doi: 10.1016/j.ijbiomac.2022.02.005
37. Tian W, Wang C, Li D, Hou H. Novel anthraquinone compounds as anticancer agents and their potential mechanism. Future Med Chem. (2020) 12:627–44. doi: 10.4155/fmc-2019-0322
38. Miao Q, Deng WQ, Lyu WY, Sun ZT, Fan SR, Qi M, et al. Erianin inhibits the growth and metastasis through autophagy-dependent ferroptosis in KRAS(G13D) colorectal cancer. Free Radic Biol Med. (2023) 204:301–12. doi: 10.1016/j.freeradbiomed.2023.05.008
39. Han YH, Wang Y, Lee SJ, Jin MH, Sun HN, Kwon T. Regulation of anoikis by extrinsic death receptor pathways. Cell Commun Signal. (2023) 21:227. doi: 10.1186/s12964-023-01247-5
40. Wu Y, Chen Y, Yan X, Dai X, Liao Y, Yuan J, et al. Lopinavir enhances anoikis by remodeling autophagy in a circRNA-dependent manner. Autophagy. (2024) 20:1651–72. doi: 10.1080/15548627.2024.2325304
41. Corona Velazquez AF, Jackson WT. So many roads: the multifaceted regulation of autophagy induction. Mol Cell Biol. (2018) 38:e00303-18. doi: 10.1128/MCB.00303-18
42. Han X, Goh KY, Lee WX, Choy SM, Tang HW. The importance of mTORC1-autophagy axis for skeletal muscle diseases. Int J Mol Sci. (2022) 24:297. doi: 10.3390/ijms24010297
43. Xu Y, Wan W. Acetylation in the regulation of autophagy. Autophagy. (2023) 19:379–87. doi: 10.1080/15548627.2022.2062112
44. Chen RH, Chen YH, Huang TY. Ubiquitin-mediated regulation of autophagy. J BioMed Sci. (2019) 26:80. doi: 10.1186/s12929-019-0569-y
45. Abokyi S, Ghartey-Kwansah G, Tse DY. TFEB is a central regulator of the aging process and age-related diseases. Ageing Res Rev. (2023) 89:101985. doi: 10.1016/j.arr.2023.101985
46. Wang H, Han J, Dmitrii G, Ning K, Zhang XA. KLF transcription factors in bone diseases. J Cell Mol Med. (2024) 28:e18278. doi: 10.1111/jcmm.18278
47. Ward MA, Vangala JR, Kamber Kaya HE, Byers HA, Hosseini N, Diaz A, et al. Transcription factor Nrf1 regulates proteotoxic stress-induced autophagy. J Cell Biol. (2024) 223:e202306150. doi: 10.1083/jcb.202306150
48. Cai J, Wang R, Chen Y, Zhang C, Fu L, Fan C. LncRNA FIRRE regulated endometrial cancer radiotherapy sensitivity via the miR-199b-5p/SIRT1/BECN1 axis-mediated autophagy. Genomics. (2024) 116:110750. doi: 10.1016/j.ygeno.2023.110750
49. Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W, et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer. (2020) 19:65. doi: 10.1186/s12943-020-01152-2
50. Zhang J, Ren G, Huang T, Sang Y, Zhong Y, Yi Y. miRNA-363-3p hinders proliferation, migration, invasion and autophagy of thyroid cancer cells by controlling SYT1 transcription to affect NF-κB. Endocr Metab Immune Disord Drug Targets. (2024) 24:153–62. doi: 10.2174/1871530323666230504112553
51. Zhu J, Cheng M, Zhao X. A tRNA-derived fragment (tRF-3001b) aggravates the development of nonalcoholic fatty liver disease by inhibiting autophagy. Life Sci. (2020) 257:118125. doi: 10.1016/j.lfs.2020.118125
52. Guo H, Ouyang Y, Yin H, Cui H, Deng H, Liu H, et al. Induction of autophagy via the ROS-dependent AMPK-mTOR pathway protects copper-induced spermatogenesis disorder. Redox Biol. (2022) 49:102227. doi: 10.1016/j.redox.2021.102227
53. Sun B, Ma J, Te L, Zuo X, Liu J, Li Y, et al. Zinc-deficient diet causes imbalance in zinc homeostasis and impaired autophagy and impairs semen quality in mice. Biol Trace Elem Res. (2023) 201:2396–406. doi: 10.1007/s12011-022-03324-1
54. Ma Q, Long S, Gan Z, Tettamanti G, Li K, Tian L. Transcriptional and post-transcriptional regulation of autophagy. Cells. (2022) 11:441. doi: 10.3390/cells11030441
55. Wang Y, Mo Y, Peng M, Zhang S, Gong Z, Yan Q, et al. The influence of circular RNAs on autophagy and disease progression. Autophagy. (2022) 18:240–53. doi: 10.1080/15548627.2021.1917131
56. Wang T, He M, Zhang X, Guo Z, Wang P, Long F. Deciphering the impact of circRNA-mediated autophagy on tumor therapeutic resistance: a novel perspective. Cell Mol Biol Lett. (2024) 29:60. doi: 10.1186/s11658-024-00571-z
57. Gao L, Zhang Q, Li S, Zheng J, Ren W, Zhi K. Circ-PKD2 promotes Atg13-mediated autophagy by inhibiting miR-646 to increase the sensitivity of cisplatin in oral squamous cell carcinomas. Cell Death Dis. (2022) 13:192. doi: 10.1038/s41419-021-04497-8
58. Qin Y, Ashrafizadeh M, Mongiardini V, Grimaldi B, Crea F, Rietdorf K, et al. Autophagy and cancer drug resistance in dialogue: Pre-clinical and clinical evidence. Cancer Lett. (2023) 570:216307. doi: 10.1016/j.canlet.2023.216307
59. Lim SM, Mohamad Hanif EA, Chin SF. Is targeting autophagy mechanism in cancer a good approach? The possible double-edge sword effect. Cell Biosci. (2021) 11:56. doi: 10.1186/s13578-021-00570-z
60. Meng Y, Bian L, Zhang M, Zhou P, Zhang S, Ying Y, et al. ISG15 promotes progression and gemcitabine resistance of pancreatic cancer cells through ATG7. Int J Biol Sci. (2024) 20:1180–93. doi: 10.7150/ijbs.85424
61. Wang G, Jiang X, Torabian P, Yang Z. Investigating autophagy and intricate cellular mechanisms in hepatocellular carcinoma: Emphasis on cell death mechanism crosstalk. Cancer Lett. (2024) 588:216744. doi: 10.1016/j.canlet.2024.216744
62. Pan W, Zhang J, Zhang L, Zhang Y, Song Y, Han L, et al. Comprehensive view of macrophage autophagy and its application in cardiovascular diseases. Cell Prolif. (2024) 57:e13525. doi: 10.1111/cpr.13525
63. Kenific CM, Thorburn A, Debnath J. Autophagy and metastasis: another double-edged sword. Curr Opin Cell Biol. (2010) 22:241–5. doi: 10.1016/j.ceb.2009.10.008
64. Palizkaran Yazdi M, Barjasteh A, Moghbeli M. MicroRNAs as the pivotal regulators of Temozolomide resistance in glioblastoma. Mol Brain. (2024) 17:42. doi: 10.1186/s13041-024-01113-6
65. Chen JL, Wu X, Yin D, Jia XH, Chen X, Gu ZY, et al. Autophagy inhibitors for cancer therapy: Small molecules and nanomedicines. Pharmacol Ther. (2023) 249:108485. doi: 10.1016/j.pharmthera.2023.108485
66. Ferreira PMP, Sousa RWR, Ferreira JRO, Militão GCG, Bezerra DP. Chloroquine and hydroxychloroquine in antitumor therapies based on autophagy-related mechanisms. Pharmacol Res. (2021) 168:105582. doi: 10.1016/j.phrs.2021.105582
67. Wang X, Zhou L, Wang H, Chen W, Jiang L, Ming G, et al. Metabolic reprogramming, autophagy, and ferroptosis: Novel arsenals to overcome immunotherapy resistance in gastrointestinal cancer. Cancer Med. (2023) 12:20573–89. doi: 10.1002/cam4.6623
68. Hama Y, Ogasawara Y, Noda NN. Autophagy and cancer: Basic mechanisms and inhibitor development. Cancer Sci. (2023) 114:2699–708. doi: 10.1111/cas.15803
69. Nuta GC, Gilad Y, Goldberg N, Meril S, Bahlsen M, Carvalho S, et al. Identifying a selective inhibitor of autophagy that targets ATG12-ATG3 protein-protein interaction. Autophagy. (2023) 19:2372–85. doi: 10.1080/15548627.2023.2178159
70. Gao W, Wang X, Zhou Y, Wang X, Yu Y. Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor immunotherapy. Signal Transduct Target Ther. (2022) 7:196. doi: 10.1038/s41392-022-01046-3
71. Wu Q, Sharma D. Autophagy and breast cancer: connected in growth, progression, and therapy. Cells. (2023) 12:1156. doi: 10.3390/cells12081156
72. Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. (2004) 306:990–5. doi: 10.1126/science.1099993
73. Zhang Z, Gao Z, Fang H, Zhao Y, Xing R. Therapeutic importance and diagnostic function of circRNAs in urological cancers: from metastasis to drug resistance. Cancer Metastasis Rev. (2024) 43:867–88. doi: 10.1007/s10555-023-10152-9
74. Jiang Y, Zhang Y, Chu F, Xu L, Wu H. Circ_0032821 acts as an oncogene in cell proliferation, metastasis and autophagy in human gastric cancer cells in vitro and in vivo through activating MEK1/ERK1/2 signaling pathway. Cancer Cell Int. (2020) 20:74. doi: 10.1186/s12935-020-1151-0
75. Cui Y, Cao J, Huang S, Ye J, Huang H, Liao D, et al. circRNA_0006470 promotes the proliferation and migration of gastric cancer cells by functioning as a sponge of miR-27b-3p. Neoplasma. (2021) 68:1245–56. doi: 10.4149/neo_2021_210222N235
76. Duan X, Yu X, Li Z. Circular RNA hsa_circ_0001658 regulates apoptosis and autophagy in gastric cancer through microRNA-182/Ras-related protein Rab-10 signaling axis. Bioengineered. (2022) 13:2387–97. doi: 10.1080/21655979.2021.2024637
77. Xu P, Zhang X, Cao J, Yang J, Chen Z, Wang W, et al. The novel role of circular RNA ST3GAL6 on blocking gastric cancer Malignant behaviours through autophagy regulated by the FOXP2/MET/mTOR axis. Clin Transl Med. (2022) 12:e707. doi: 10.1002/ctm2.707
78. Tang Z, Li J, Lu B, Zhang X, Yang L, Qi Y, et al. CircBIRC6 facilitates the Malignant progression via miR-488/GRIN2D-mediated CAV1-autophagy signal axis in gastric cancer. Pharmacol Res. (2024) 202:107127. doi: 10.1016/j.phrs.2024.107127
79. Hu F, Peng Y, Chang S, Luo X, Yuan Y, Zhu X, et al. Vimentin binds to a novel tumor suppressor protein, GSPT1-238aa, encoded by circGSPT1 with a selective encoding priority to halt autophagy in gastric carcinoma. Cancer Lett. (2022) 545:215826. doi: 10.1016/j.canlet.2022.215826
80. Sang H, Zhang W, Peng L, Wei S, Zhu X, Huang K, et al. Exosomal circRELL1 serves as a miR-637 sponge to modulate gastric cancer progression via regulating autophagy activation. Cell Death Dis. (2022) 13:56. doi: 10.1038/s41419-021-04364-6
81. Liu Y, Cao J, Yang Q, Zhu L, Zhao W, Wang X, et al. CircRNA_15430 reduced by Helicobacter pylori infection and suppressed gastric cancer progression via miR-382-5p/ZCCHC14 axis. Biol Direct. (2023) 18:51. doi: 10.1186/s13062-023-00402-9
82. Shi Y, Li J, Tang M, Liu J, Zhong Y, Huang W. CircHADHA-augmented autophagy suppresses tumor growth of colon cancer by regulating autophagy-related gene via miR-361. Front Oncol. (2022) 12:937209. doi: 10.3389/fonc.2022.937209
83. Chen RX, Xu SD, Deng MH, Hao SH, Chen JW, Ma XD, et al. Mex-3 RNA binding family member A (MEX3A)/circMPP6 complex promotes colorectal cancer progression by inhibiting autophagy. Signal Transduct Target Ther. (2024) 9:80. doi: 10.1038/s41392-024-01787-3
84. Yang BL, Liu GQ, Li P, Li XH. Circular RNA CUL2 regulates the development of colorectal cancer by modulating apoptosis and autophagy via miR-208a-3p/PPP6C. Aging (Albany NY). (2022) 14:497–508. doi: 10.18632/aging.v14i1
85. Liu Z, Wang Q, Wang X, Xu Z, Wei X, Li J. Circular RNA cIARS regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell Death Discov. (2020) 6:72. doi: 10.1038/s41420-020-00306-x
86. Zhao Z, He J, Feng C. CircCBFB is a mediator of hepatocellular carcinoma cell autophagy and proliferation through miR-424-5p/ATG14 axis. Immunol Res. (2022) 70:341–53. doi: 10.1007/s12026-021-09255-8
87. Wang X, Dong FL, Wang YQ, Wei HL, Li T, Li J. Exosomal circTGFBR2 promotes hepatocellular carcinoma progression via enhancing ATG5 mediated protective autophagy. Cell Death Dis. (2023) 14:451. doi: 10.1038/s41419-023-05989-5
88. Lin S, Zhuang J, Zhu L, Jiang Z. Matrine inhibits cell growth, migration, invasion and promotes autophagy in hepatocellular carcinoma by regulation of circ_0027345/miR-345-5p/HOXD3 axis. Cancer Cell Int. (2020) 20:246. doi: 10.1186/s12935-020-01293-w
89. Fu D, Ji Q, Wang C, Yu L, Yu R. Aloin decelerates the progression of hepatocellular carcinoma through circ_0011385/miR-149-5p/WT1 axis. Cell Cycle. (2021) 20:2476–93. doi: 10.1080/15384101.2021.1988227
90. Li Z, Chen Z, Li S, Qian X, Zhang L, Long G, et al. Circ_0020256 induces fibroblast activation to drive cholangiocarcinoma development via recruitment of EIF4A3 protein to stabilize KLF4 mRNA. Cell Death Discov. (2023) 9:161. doi: 10.1038/s41420-023-01439-5
91. He Z, Cai K, Zeng Z, Lei S, Cao W, Li X. Autophagy-associated circRNA circATG7 facilitates autophagy and promotes pancreatic cancer progression. Cell Death Dis. (2022) 13:233. doi: 10.1038/s41419-022-04677-0
92. Meng L, Liu S, Ding P, Chang S, Sang M. Circular RNA ciRS-7 inhibits autophagy of ESCC cells by functioning as miR-1299 sponge to target EGFR signaling. J Cell Biochem. (2020) 121:1039–49. doi: 10.1002/jcb.29339
93. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
94. Seeneevassen L, Bessède E, Mégraud F, Lehours P, Dubus P, Varon C. Gastric cancer: advances in carcinogenesis research and new therapeutic strategies. Int J Mol Sci. (2021) 22:3418. doi: 10.3390/ijms22073418
95. Yang P, Yang W, Wei Z, Li Y, Yang Y, Wang J. Novel targets for gastric cancer: The tumor microenvironment (TME), N6-methyladenosine (m6A), pyroptosis, autophagy, ferroptosis and cuproptosis. BioMed Pharmacother. (2023) 163:114883. doi: 10.1016/j.biopha.2023.114883
96. Wang Y, Wu C, Du Y, Li Z, Li M, Hou P, et al. Expanding uncapped translation and emerging function of circular RNA in carcinomas and noncarcinomas. Mol Cancer. (2022) 21:13. doi: 10.1186/s12943-021-01484-7
97. Liu J, Ren L, Li S, Li W, Zheng X, Yang Y, et al. The biology, function, and applications of exosomes in cancer. Acta Pharm Sin B. (2021) 11:2783–97. doi: 10.1016/j.apsb.2021.01.001
98. Malfertheiner P, Camargo MC, El-Omar E, Liou JM, Peek R, Schulz C, et al. Helicobacter pylori infection. Nat Rev Dis Primers. (2023) 9:19. doi: 10.1038/s41572-023-00431-8
99. Spaander MCW, Zauber AG, Syngal S, Blaser MJ, Sung JJ, You YN, et al. Young-onset colorectal cancer. Nat Rev Dis Primers. (2023) 9:21. doi: 10.1038/s41572-023-00432-7
100. Chen L, He M, Zhang M, Sun Q, Zeng S, Zhao H, et al. The Role of non-coding RNAs in colorectal cancer, with a focus on its autophagy. Pharmacol Ther. (2021) 226:107868. doi: 10.1016/j.pharmthera.2021.107868
101. He X, Wu K, Ogino S, Giovannucci EL, Chan AT, Song M. Association between risk factors for colorectal cancer and risk of serrated polyps and conventional adenomas. Gastroenterology. (2018) 155:355–73.e18. doi: 10.1053/j.gastro.2018.04.019
102. Bian M, Wang W, Song C, Pan L, Wu Y, Chen L. Autophagy-related genes predict the progression of periodontitis through the ceRNA network. J Inflamm Res. (2022) 15:1811–24. doi: 10.2147/JIR.S353092
103. Toh MR, Wong EYT, Wong SH, Ng AWT, Loo LH, Chow PK, et al. Global epidemiology and genetics of hepatocellular carcinoma. Gastroenterology. (2023) 164:766–82. doi: 10.1053/j.gastro.2023.01.033
104. Meng H, Niu R, Huang C, Li J. Circular RNA as a novel biomarker and therapeutic target for HCC. Cells. (2022) 11:1948. doi: 10.3390/cells11121948
105. Bruix J, Cheng AL, Meinhardt G, Nakajima K, De Sanctis Y, Llovet J. Prognostic factors and predictors of sorafenib benefit in patients with hepatocellular carcinoma: Analysis of two phase III studies. J Hepatol. (2017) 67:999–1008. doi: 10.1016/j.jhep.2017.06.026
106. Nan Y, Su H, Zhou B, Liu S. The function of natural compounds in important anticancer mechanisms. Front Oncol. (2022) 12:1049888. doi: 10.3389/fonc.2022.1049888
107. Qurashi M, Vithayathil M, Khan SA. Epidemiology of cholangiocarcinoma. Eur J Surg Oncol. (2023), 107064. doi: 10.1016/j.ejso.2023.107064
108. Liao W, Feng Q, Liu H, Du J, Chen X, Zeng Y. Circular RNAs in cholangiocarcinoma. Cancer Lett. (2023) 553:215980. doi: 10.1016/j.canlet.2022.215980
109. Zhang Y, Yuan B, Xu Y, Zhou N, Zhang R, Lu L, et al. MiR-208b/miR-21 Promotes the Progression of Cardiac Fibrosis Through the Activation of the TGF-β1/Smad-3 Signaling Pathway: An in vitro and in vivo Study. Front Cardiovasc Med. (2022) 9:924629. doi: 10.3389/fcvm.2022.924629
110. Halbrook CJ, Lyssiotis CA, Pasca di Magliano M, Maitra A. Pancreatic cancer: Advances and challenges. Cell. (2023) 186:1729–54. doi: 10.1016/j.cell.2023.02.014
111. Morgan E, Soerjomataram I, Rumgay H, Coleman HG, Thrift AP, Vignat J, et al. The global landscape of esophageal squamous cell carcinoma and esophageal adenocarcinoma incidence and mortality in 2020 and projections to 2040: new estimates from GLOBOCAN 2020. Gastroenterology. (2022) 163:649–58.e2. doi: 10.1053/j.gastro.2022.05.054
112. Ward RA, Fawell S, Floc'h N, Flemington V, McKerrecher D, Smith PD. Challenges and opportunities in cancer drug resistance. Chem Rev. (2021) 121:3297–351. doi: 10.1021/acs.chemrev.0c00383
113. Luo M, Deng X, Chen Z, Hu Y. Circular RNA circPOFUT1 enhances Malignant phenotypes and autophagy-associated chemoresistance via sequestrating miR-488-3p to activate the PLAG1-ATG12 axis in gastric cancer. Cell Death Dis. (2023) 14:10. doi: 10.1038/s41419-022-05506-0
114. Sun G, Li Z, He Z, Wang W, Wang S, Zhang X, et al. Circular RNA MCTP2 inhibits cisplatin resistance in gastric cancer by miR-99a-5p-mediated induction of MTMR3 expression. J Exp Clin Cancer Res. (2020) 39:246. doi: 10.1186/s13046-020-01758-w
115. Chen Y, Liu H, Zou J, Cao G, Li Y, Xing C, et al. Exosomal circ_0091741 promotes gastric cancer cell autophagy and chemoresistance via the miR-330-3p/TRIM14/Dvl2/Wnt/β-catenin axis. Hum Cell. (2023) 36:258–75. doi: 10.1007/s13577-022-00790-6
116. Yang G, Tan J, Guo J, Wu Z, Zhan Q. Exosome-mediated transfer of circ_0063526 enhances cisplatin resistance in gastric cancer cells via regulating miR-449a/SHMT2 axis. Anticancer Drugs. (2022) 33:1047–57. doi: 10.1097/CAD.0000000000001386
117. Shang Z, Luo Z, Wang Y, Liu Q, Xin Y, Zhang M, et al. CircHIPK3 contributes to cisplatin resistance in gastric cancer by blocking autophagy-dependent ferroptosis. J Cell Physiol. (2023) 238:2407–24. doi: 10.1002/jcp.31093
118. Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X, et al. circCUL2 regulates gastric cancer Malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol Cancer. (2020) 19:156. doi: 10.1186/s12943-020-01270-x
119. Pan Z, Zheng J, Zhang J, Lin J, Lai J, Lyu Z, et al. A novel protein encoded by exosomal circATG4B induces oxaliplatin resistance in colorectal cancer by promoting autophagy. Adv Sci (Weinh). (2022) 9:e2204513. doi: 10.1002/advs.202204513
120. Sui S, Ma F, Mi L, Gao L, Yu W, Li M, et al. Circ-CCS enhances autophagy during imatinib resistance of gastrointestinal stromal tumor by regulating miR-197-3p/ATG10 signaling. J Cancer Res Ther. (2022) 18:1338–45. doi: 10.4103/jcrt.jcrt_625_22
121. Zhang F, Jiang J, Qian H, Yan Y, Xu W. Exosomal circRNA: emerging insights into cancer progression and clinical application potential. J Hematol Oncol. (2023) 16:67. doi: 10.1186/s13045-023-01452-2
122. Zhang Y, Luo J, Yang W, Ye WC. CircRNAs in colorectal cancer: potential biomarkers and therapeutic targets. Cell Death Dis. (2023) 14:353. doi: 10.1038/s41419-023-05881-2
123. Hu WH, Liu TT, Liu PF, Morgan P, Lin IL, Tsai WL, et al. ATG4B and pS383/392-ATG4B serve as potential biomarkers and therapeutic targets of colorectal cancer. Cancer Cell Int. (2023) 23:63. doi: 10.1186/s12935-023-02909-7
124. Shin JH, Park SJ, Jo DS, Park NY, Kim JB, Bae JE, et al. Down-regulated TMED10 in Alzheimer disease induces autophagy via ATG4B activation. Autophagy. (2019) 15:1495–505. doi: 10.1080/15548627.2019.1586249
125. Serrano C, Martín-Broto J, Asencio-Pascual JM, López-Guerrero JA, Rubió-Casadevall J, Bagué S, et al. 2023 GEIS Guidelines for gastrointestinal stromal tumors. Ther Adv Med Oncol. (2023) 15:17588359231192388. doi: 10.1177/17588359231192388
126. Donisi C, Pretta A, Pusceddu V, Ziranu P, Lai E, Puzzoni M, et al. Immunotherapy and cancer: the multi-omics perspective. Int J Mol Sci. (2024) 25:3563. doi: 10.3390/ijms25063563
127. Jiang M, Wu W, Xiong Z, Yu X, Ye Z, Wu Z. Targeting autophagy drug discovery: Targets, indications and development trends. Eur J Med Chem. (2024) 267:116117. doi: 10.1016/j.ejmech.2023.116117
128. Taraborrelli L, Şenbabaoğlu Y, Wang L, Lim J, Blake K, Kljavin N, et al. Tumor-intrinsic expression of the autophagy gene Atg16l1 suppresses anti-tumor immunity in colorectal cancer. Nat Commun. (2023) 14:5945. doi: 10.1038/s41467-023-41618-7
129. Liu C, Wu X, Gokulnath P, Li G, Xiao J. The functions and mechanisms of translatable circular RNAs. J Pharmacol Exp Ther. (2023) 384:52–60. doi: 10.1124/jpet.122.001085
130. Xu S, Xu Y, Solek NC, Chen J, Gong F, Varley AJ, et al. Tumor-tailored ionizable lipid nanoparticles facilitate IL-12 circular RNA delivery for enhanced lung cancer immunotherapy. Adv Mater. (2024) 36:e2400307. doi: 10.1002/adma.202400307
131. Xie J, Ye F, Deng X, Tang Y, Liang JY, Huang X, et al. Circular RNA: A promising new star of vaccine. J Transl Int Med. (2023) 11:372–81. doi: 10.2478/jtim-2023-0122
132. Zhang D, Man D, Lu J, Jiang Y, Ding B, Su R, et al. Mitochondrial TSPO promotes hepatocellular carcinoma progression through ferroptosis inhibition and immune evasion. Adv Sci (Weinh). (2023) 10:e2206669. doi: 10.1002/advs.202206669
133. Miao Z, Li J, Wang Y, Shi M, Gu X, Zhang X, et al. Hsa_circ_0136666 stimulates gastric cancer progression and tumor immune escape by regulating the miR-375/PRKDC Axis and PD-L1 phosphorylation. Mol Cancer. (2023) 22:205. doi: 10.1186/s12943-023-01883-y
134. Duan Y, Tian X, Liu Q, Jin J, Shi J, Hou Y. Role of autophagy on cancer immune escape. Cell Commun Signal. (2021) 19:91. doi: 10.1186/s12964-021-00769-0
135. Chen J, Yang J, Fei X, Wang X, Wang K. CircRNA ciRS-7: a novel oncogene in multiple cancers. Int J Biol Sci. (2021) 17:379–89. doi: 10.7150/ijbs.54292
136. Wang C, Liu T, Wang J, Cheng C, Zhang Z, Zhang J, et al. CircHIPK3 negatively regulates autophagy by blocking VCP binding to the Beclin 1 complex in bladder cancer. Discov Oncol. (2023) 14:86. doi: 10.1007/s12672-023-00689-0
137. Matboli M, Hasanin AH, Hussein R, El-Nakeep S, Habib EK, Ellackany R, et al. Cyanidin 3-glucoside modulated cell cycle progression in liver precancerous lesion, in vivo study. World J Gastroenterol. (2021) 27:1435–50. doi: 10.3748/wjg.v27.i14.1435
138. Zabady S, Mahran N, Soltan MA, Alaa Eldeen M, Eid RA, Albogami S, et al. Cyanidin-3-Glucoside Modulates hsa_circ_0001345/miRNA106b/ATG16L1 Axis Expression as a Potential Protective Mechanism against Hepatocellular Carcinoma. Curr Issues Mol Biol. (2022) 44:1677–87. doi: 10.3390/cimb44040115
139. Yang PW, Chen TT, Zhao WX, Liu GW, Feng XJ, Wang SM, et al. Scutellaria barbata D.Don and Oldenlandia diffusa (Willd.) Roxb crude extracts inhibit hepatitis-B-virus-associated hepatocellular carcinoma growth through regulating circRNA expression. J Ethnopharmacol. (2021) 275:114110. doi: 10.1016/j.jep.2021.114110
140. Gerstberger S, Jiang Q, Ganesh K. Metastasis. Cell. (2023) 186:1564–79. doi: 10.1016/j.cell.2023.03.003
141. Li X, Ramadori P, Pfister D, Seehawer M, Zender L, Heikenwalder M. The immunological and metabolic landscape in primary and metastatic liver cancer. Nat Rev Cancer. (2021) 21:541–57. doi: 10.1038/s41568-021-00383-9
142. Wang Z, Yang L, Wu P, Li X, Tang Y, Ou X, et al. The circROBO1/KLF5/FUS feedback loop regulates the liver metastasis of breast cancer by inhibiting the selective autophagy of afadin. Mol Cancer. (2022) 21:29. doi: 10.1186/s12943-022-01498-9
143. Hu C, Xia R, Zhang X, Li T, Ye Y, Li G, et al. circFARP1 enables cancer-associated fibroblasts to promote gemcitabine resistance in pancreatic cancer via the LIF/STAT3 axis. Mol Cancer. (2022) 21:24. doi: 10.1186/s12943-022-01501-3
144. Wei L, Lin Q, Lu Y, Li G, Huang L, Fu Z, et al. Cancer-associated fibroblasts-mediated ATF4 expression promotes Malignancy and gemcitabine resistance in pancreatic cancer via the TGF-β1/SMAD2/3 pathway and ABCC1 transactivation. Cell Death Dis. (2021) 12:334. doi: 10.1038/s41419-021-03574-2
145. Zou L, Liao M, Zhen Y, Zhu S, Chen X, Zhang J, et al. Autophagy and beyond: Unraveling the complexity of UNC-51-like kinase 1 (ULK1) from biological functions to therapeutic implications. Acta Pharm Sin B. (2022) 12:3743–82. doi: 10.1016/j.apsb.2022.06.004
146. Wang H, Sun G, Xu P, Lv J, Zhang X, Zhang L, et al. Circular RNA TMEM87A promotes cell proliferation and metastasis of gastric cancer by elevating ULK1 via sponging miR-142-5p. J Gastroenterol. (2021) 56:125–38. doi: 10.1007/s00535-020-01744-1
147. Jagtap U, Anderson ES, Slack FJ. The emerging value of circular noncoding RNA research in cancer diagnosis and treatment. Cancer Res. (2023) 83:809–13. doi: 10.1158/0008-5472.CAN-22-3014
148. Muskan M, Abeysinghe P, Cecchin R, Branscome H, Morris KV, Kashanchi F. Therapeutic potential of RNA-enriched extracellular vesicles: The next generation in RNA delivery via biogenic nanoparticles. Mol Ther. (2024) S1525-0016:00094-7. doi: 10.1016/j.ymthe.2024.02.025
Keywords: autophagy, biological activity, circular RNA, drug resistance, gastrointestinal cancer
Citation: Zhang B, Li Z, Ye G and Hu K (2024) Biologic activity and treatment resistance to gastrointestinal cancer: the role of circular RNA in autophagy regulation. Front. Oncol. 14:1393670. doi: 10.3389/fonc.2024.1393670
Received: 29 February 2024; Accepted: 15 August 2024;
Published: 30 August 2024.
Edited by:
John G Clohessy, Harvard Medical School, United StatesReviewed by:
Xinpei Deng, Sun Yat-sen University Cancer Center (SYSUCC), ChinaShanchun Guo, Xavier University of Louisiana, United States
Copyright © 2024 Zhang, Li, Ye and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kefeng Hu, ZnlodWtlZmVuZ0BuYnUuZWR1LmNu
†These authors have contributed equally to this work
 Bo Zhang1,2†
Bo Zhang1,2† Guoliang Ye
Guoliang Ye Kefeng Hu
Kefeng Hu