
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 14 June 2024
Sec. Cancer Cell Signaling
Volume 14 - 2024 | https://doi.org/10.3389/fonc.2024.1384928
Sirtuins are pivotal in orchestrating numerous cellular pathways, critically influencing cell metabolism, DNA repair, aging processes, and oxidative stress. In recent years, the involvement of sirtuins in tumor biology has garnered substantial attention, with a growing body of evidence underscoring their regulatory roles in various aberrant cellular processes within tumor environments. This article delves into the sirtuin family and its biological functions, shedding light on their dual roles—either as promoters or inhibitors—in various cancers including oral, breast, hepatocellular, lung, and gastric cancers. It further explores potential anti-tumor agents targeting sirtuins, unraveling the complex interplay between sirtuins, miRNAs, and chemotherapeutic drugs. The dual roles of sirtuins in cancer biology reflect the complexity of targeting these enzymes but also highlight the immense therapeutic potential. These advancements hold significant promise for enhancing clinical outcomes, marking a pivotal step forward in the ongoing battle against cancer.
Epigenetics, encompassing DNA modifications, histone alterations, and chromatin remodeling, has emerged as a focal point in oncological research over recent years (1–3). Among the myriad epigenetic mechanisms, the acetylation and deacetylation of histones stand out as a pivotal mode of covalent histone modification (4, 5). Central to this process are the Sirtuin (Sirt) enzymes (6, 7). Originating from the discovery of the Sir2 gene in yeast, Frye identified five human homologs in 1999, dubbed Sirt1 through Sirt5. Subsequent research expanded the family with the cloning of Sirt6 and Sirt7 from a human spleen cDNA library (8). The sirtuin family’s extensive involvement in a range of biological functions and pathologies, particularly cancer, underscores their significance. Sirtuins critically modulate the metabolic adaptations of cancer cells that favor growth and division and are implicated in the dysregulation of DNA repair and apoptotic pathways that contribute to genomic instability, mutation accumulation, and ultimately, tumor development and progression (9, 10). The intricate role of sirtuins extends to key cellular processes such as the cell cycle, DNA repair, cell survival, and apoptosis, positioning them as pivotal players in the cellular response to genomic instability and tumor dynamics. Given their broad biological impact, sirtuins and their inhibitors have become focal points in the quest for novel cancer therapies. Investigations have shown that sirtuins regulate Base Excision Repair (BER), Nucleotide Excision Repair (NER), and DNA double-strand break (DSB) repair pathways, with Sirt1, in particular, controlling Forkhead Box O (FoxO) protein deacetylation and apoptosis induction (11, 12). SIRT3 localizes mainly to the mitochondrial matrix and plays an important role in regulating mitochondrial metabolism, including the tricarboxylic acid (TCA) cycle, the urea cycle, amino acid metabolism, fatty acid oxidation, ETC/oxidative phosphorylation (OXPHOS), ROS detoxification, mitochondrial dynamics and the mitochondrial unfolded protein response (13).The regulation of various signaling pathways by sirtuins through acetylation processes is crucial in tumor biology, influencing outcomes such as tumor initiation, metastasis, and angiogenesis. The differential effects of sirtuin mutations, overexpression, up-regulation, and activation on cancer further underscore the complexity of their roles (9, 10).
Given the pivotal function of histone deacetylases like sirtuins in the malignant progression of tumors, especially during invasion and metastasis, their study offers critical insights for the development of new anti-cancer drugs and strategies for clinical cancer treatment.
The Sirtuin family, integral to class III histone deacetylase (HDACs), exhibits inhibition by nicotinamide and spans a broad evolutionary spectrum from bacteria to humans. Characterized by a conserved catalytic core, sirtuins engage in the deacetylation of diverse substrates. In mammals, this family comprises seven distinct members, SIRT1–7, each occupying unique genetic loci across chromosomes 10q22.1, 19q13, 11p15, 5, 12q, 5p23, 19p13.3, and 17q25, with gene lengths varying from 6 to 37 (14, 15) (Figure 1). These sirtuins possess specific domains, underlining their structural and functional diversity.
A typical sirtuin structure includes about 270 amino acids, featuring a large domain with Rossmann folds and a smaller, less conserved domain with a zinc ribbon and helical segments, facilitating the catalytic process (16). Subcellular localization differentiates sirtuins’ functions (Table 1): SIRT1 is mainly nuclear, but a cytoplasmic localization has also been reported (17). Heterokaryon assay shows that SIRT1 has nucleocytoplasmic shuttle phenomenon (18). SIRT6, and SIRT7 are nuclear. SIRT6 is mainly found in chromatin and SIRT7 is mainly found in nucleoli, regulating genetic and metabolic activities (19, 20). SIRT2 is cytoplasmic, but it is also found in the nucleus of the G2 to M phase transition of the cell cycle (21). SIRT1 involves a variety of physiological and pathological processes, including cell proliferation, apoptosis, DNA repair, metabolic regulation, and antioxidant defense. It is also involved in the regulation of the aging process and longevity (22). SIRT2 regulates microtubule stability and cell cycle progression. In addition, SIRT2 has also been implicated in the pathogenesis of neurodegenerative diseases (23). SIRT3, SIRT4, and SIRT5 are mitochondrial, overseeing oxygen and energy supply (24). SIRT3 promotes cell metabolism and deacetylates specific substrates to play a carcinogenic role. SIRT3 plays a unique function by mediating interactions between mitochondria and intracellular signaling. SIRT3 regulates oxidative stress, amino acid metabolism, fatty acid oxidation, electron transport, and the TCA cycle through deacetylating various substrate proteins (25). Beyond their deacetylase activity, SIRT4–6 exhibit additional enzymatic functions. In addition to their deacetylase activities, the sirtuins SIRT4–6 possess unique enzymatic functions that are crucial for cellular metabolism and stress responses. SIRT4, for example, has strong Adenosine diphosphate (ADP)-ribosyltransferase activity, allowing it to modify other proteins and influence cellular processes such as insulin secretion and amino acid catabolism (26). SIRT5 distinguishes itself by targeting and removing malonyl and succinyl groups from proteins, which regulates the urea cycle and mitochondrial function, demonstrating its critical role in metabolic regulation (27). SIRT6, on the other hand, is involved in DNA repair, telomere maintenance, and also has deacetylase and long-chain deacylase activity, making it a pivotal player in genomic stability and aging (28). These multifaceted activities underscore the sirtuins’ crucial roles in not just metabolism and energy production, but also in maintaining cellular homeostasis under stress conditions, providing potential targets for therapeutic interventions in metabolic disorders and age-related diseases.
Sirtuins intersect with multiple biological pathways, engaging with proteins such as Tumor protein p53 (P53), FOXO/Peroxisome proliferator-activated receptor-γ coactivator-1 (PGC-1), Nuclear Factor Kappa B (NF-kB), and Ku70 to modulate stress responses, metabolism, aging, and apoptosis. Their pivotal role in cellular processes extends to DNA repair, oxidative stress, and aging (29). Given their critical involvement in tumorigenesis, sirtuins have attracted significant interest as potential targets in cancer research. The dual nature of SIRTs’ function, as either tumor suppressors or promoters, often depends on the cellular context and experimental conditions, with SIRT1 displaying bifunctionality and SIRT2 and SIRT6 predominantly acting as tumor suppressors (19, 30). Explorations into sirtuins’ roles have unveiled their potential in influencing the epigenome, offering insights into how these enzymes affect gene expression and cellular fate without altering the DNA sequence. This epigenetic regulation presents a nuanced layer of control over cellular processes, highlighting sirtuins’ significance in both health and disease states, particularly in cancer where they can dictate cell survival or death. Moreover, the therapeutic implications of targeting sirtuins in cancer treatment have gained momentum, driven by the understanding that modulating sirtuin activity can alter the trajectory of cancer progression. As research delves deeper into the mechanistic pathways governed by sirtuins, their capacity to serve as biomarkers for cancer prognosis or as direct targets for novel anticancer therapies becomes increasingly apparent. This burgeoning area of study not only promises to enhance our grasp of cancer biology but also to usher in a new era of targeted treatments that leverage the unique properties of the sirtuin family.
Sirtuin proteins, commonly referred to as SIRTs, are essential components in the regulation of cellular metabolism, impacting the processing of sugars and fats at various biological levels. These enzymes are pivotal in the management of critical metabolic pathways, such as those involved in the metabolism of glucose—encompassing aerobic glycolysis, the tricarboxylic acid (TCA) cycle, and glutamine metabolism—and fat metabolism, which includes the breakdown of fatty acids, cholesterol, and the production of ketone bodies. Such metabolic activities are crucial in the pathogenesis and progression of cancerous tumors (31). Sirtuins exert their regulatory influence over these metabolic processes by modulating the activity of vital enzymes and transport systems through mechanisms that include transcriptional and post-translational modifications, enzyme function adjustments, and the manipulation of signaling pathways.
SIRT1, for instance, is known to enhance the expression of FOXO1 and PGC-1α or to suppress the activity of transcription factors like sterol regulatory element binding protein-1c (SREBP1c), thereby reducing the cellular fat content by lowering the expression of genes targeted by SREBP1c (32–35). This action underscores SIRT1’s significant role in maintaining metabolic health across various tissues, including muscle, liver, and fat tissue, through the deacetylation of numerous target proteins. Similarly, SIRT3 is instrumental in metabolic regulation by deacetylating and activating key enzymes such as acetyl-CoA synthetase 2 (ACSS2) and glutamate dehydrogenase (GDH), thus facilitating an enhanced Krebs cycle (36). It also plays a role in managing cellular reactive oxygen species (ROS) levels and mitigating ROS-induced activation of iron regulatory protein 1 (IRP 1), which in turn moderates glycolysis and glutamine breakdown (37) (Figure 2). The activity of SIRT3 is intricately linked to the metabolic state of the cell, modulated by the intracellular Nicotinamide Adenine Dinucleotide (NAD+)/Nicotinamide Adenine Dinucleotide Hydrogen (NADH) ratio (38–40). SIRT3 can maintain mitochondrial homeostasis, which is necessary for survival Therefore, SIRT3 is regulated in numerous ways. As a major repair enzyme in the base excision repair pathway, 8-oxoguanine DNA glycosylase (OGG1) is a newly discovered target of SIRT3. Cheng et al. found that SIRT3 deacetylates OGG1 to regulate its incision activity and decrease its degradation. Mitochondrial DNA damage repair, mitochondrial integrity, and apoptotic cell death induced by oxidative stress are closely related to the acetylation- and turnover-related regulation of OGG1 by SIRT3. SIRT3 can regulate different enzymes including manganese dismutase, SOD2, and catalase, which are important in regulating ROS levels (41). On the other hand, SIRT4, with its limited deacetylase but notable ADP-ribosyltransferase activity, modulates glutamine metabolism post-DNA damage and inhibits it through the ADP ribosylation of Glutamate Dehydrogenase (GDH). Furthermore, it has been discovered that mammalian target of rapamycin complex 1 (mTORC1) influences the expression of SIRT4 by promoting the degradation of its transcriptional activator, CAMP response element-binding protein 2 (CREB2), thereby affecting SIRT4 gene expression negatively (42, 43). SIRT5, possessing lysine deacetylase activity, plays a part in the regulation of glycolysis, the TCA cycle, fatty acid oxidation, and the formation of ketone bodies among other processes (44, 45). Additionally, SIRT6 and SIRT7, both of which exhibit ADP ribosyltransferase activity, contribute to the regulation of cellular metabolism with SIRT6 directly inhibiting the expression of various glycolysis-related genes and impacting iron metabolism (46–49). The reprogramming of cellular metabolism is a hallmark of cancer, with cancer cells adapting their metabolic processes to meet energy demands, a phenomenon in which sirtuins play a significant role by regulating metabolic pathways, thus influencing cancer initiation and progression (50–53).
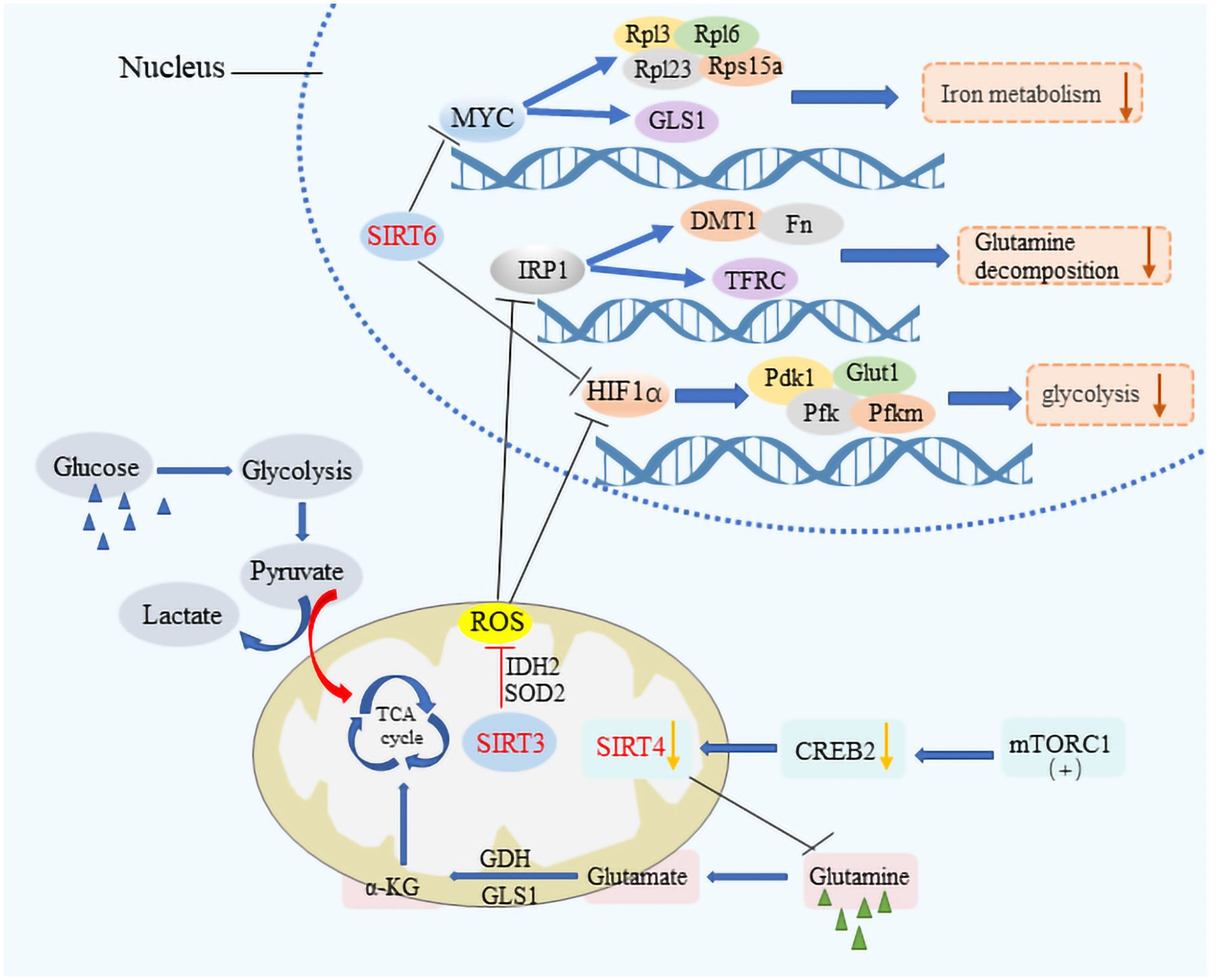
Figure 2 The metabolic modulation by SIRTs in cancer cells. SIRT3, SIRT4, and SIRT6 significantly impact cancer cell metabolism by downregulating key pathways crucial for tumor growth. These sirtuins suppress HIF-1α and IRP1 target genes, affecting glycolytic processes by inhibiting genes such as PDK1, PFK, and Pfkm, alongside Glut1, which is vital for glucose transport. They also regulate iron metabolism by modulating the expression of DMT1 and other bivalent metal transporters, as well as ferritin and the transferrin receptor. The influence extends to MYC target genes, including ribosomal proteins Rpl3, Rpl6, Rpl23, and Rps15a, thereby reducing the metabolic rate encompassing aerobic glycolysis, the tricarboxylic acid (TCA) cycle, and glutamine decomposition. Additionally, the activation of the mTORC1 pathway leads to CREB2 degradation, which results in decreased SIRT4 expression, illustrating a comprehensive mechanism by which these sirtuins suppress metabolic pathways to hinder cancer progression.
Sirtuins are critical for maintaining genome integrity, participating actively in the repair of DNA within both the nucleus and mitochondria. Their role in DNA repair involves the deacetylation of DNA repair components, thereby enhancing the efficiency of repair enzymes and improving the cellular response to damage (54–56). SIRT6, in particular, is highlighted for its pivotal role in DNA repair, with deficiencies in SIRT6 leading to genomic instability in mice (57). In cells under oxidative stress, SIRT6 promotes DSB repair via non-homologous end joining (NHEJ) and homologous recombination (HR) at DSB sites (58, 59). SIRT6 has been shown to form complexes with Myosin Heavy Chain (MYH), Apurinic/apyrimidinic Endonuclease 1 (APE1), and the 9–1-1 complex (RAD9-RAD1-HUS1 Checkpoint Clamp Component, HUS1), thus safeguarding genomic and telomere integrity and enhancing cellular resilience against oxidative stress (60) (Figure 3). Additionally, SIRT6 facilitates DSB repair by augmenting the activity of PARP1 and recruiting SNF2H at DNA damage sites for proper DSB repair (61). SIRT6’s interaction with CtIP and DNA-Protein kinase C (PKC) further underscores its role in DNA damage response (62, 63). SIRT1 also contributes to genome stability by deacetylating DNA damage repair factors, thereby facilitating the repair process. It enhances the BER pathway by promoting APE1 deacetylation and stimulating the activity of Thymine DNA glycosylase (TDG), which mediates DNA repair. In the NER pathway, SIRT1 plays a role by complementing XPA phosphorylation through interactions with ATR Serine/Threonine Kinase (ATR) and replication protein A 32 (RPA32), while in the DSB pathway, it aids in cell cycle regulation and DNA repair assessment through NBS1 deacetylation (64–68). Moreover, SIRT3 has been recognized for its role in repairing mitochondrial DNA (mtDNA) damage and preventing apoptotic cell death under oxidative stress conditions by regulating the acetylation and activity of 8-oxoG glycosylase1 (OGG1) (69).SIRT3 targets different enzymes which regulate mitochondrial metabolism and participate in ROS detoxification, such as the complexes of the respiratory chain, the isocitrate dehydrogenase, or the manganese superoxide dismutase (25). SIRT7 enhances DNA repair by boosting upstream signaling and inducing chromatin condensation, thus inhibiting transcription near DSB sites and limiting DSB mobility during the initial response phase (70, 71). This intricate network of sirtuin activities highlights the close relationship between sirtuin function and DNA damage repair, underscoring their vital role in maintaining cellular health and stability.
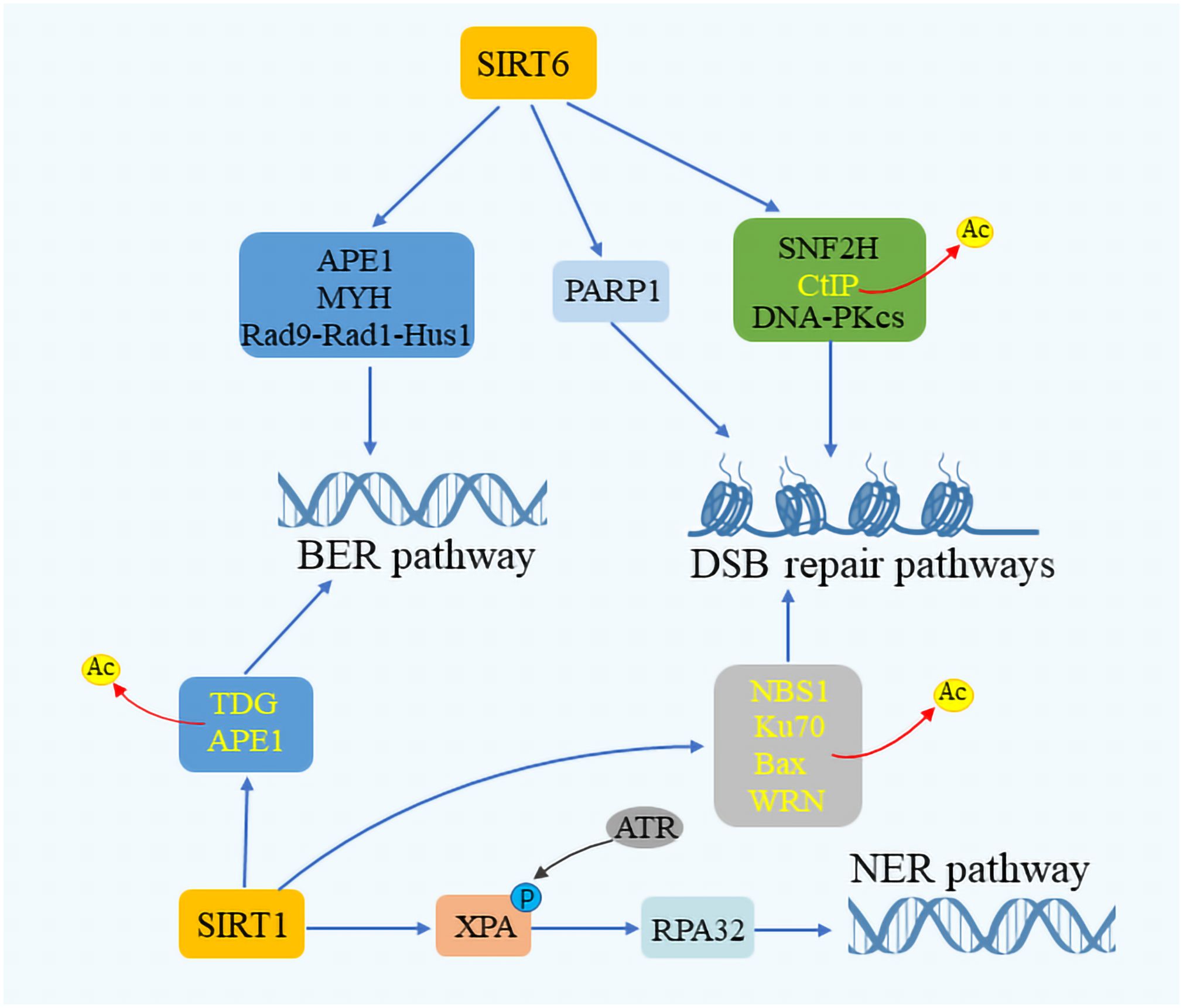
Figure 3 SIRTuins’ involvement in DNA repair mechanisms. SIRT6 plays a crucial role in the base excision repair (BER) pathway by interacting with key components such as Myosin heavy chain (MYH), Apurinic/Apyrimidinic endonuclease 1 (APE1), and the Rad 9-RAD 1-Hus 1 (9–1–1) complex. SIRT1 enhances the DNA repair process by promoting the deacetylation of APE1 and Thymine DNA glycosylase (TDG). In the context of double-strand break (DSB) repair, SIRT6 boosts the activity of ADP-Ribose polymerase 1 (PARP1) and facilitates the deacetylation of CtIP. It also interacts with the DNA-dependent protein kinase Catalytic subunit (DNA-PKcs) and Sucrose Nonfermenter2 Homolog (SNF2H), playing a vital role in responding to DNA damage. Moreover, SIRT1 is instrumental in augmenting DNA repair by promoting the acetylation of Nijmegen breakage syndrome 1 (NBS1), Ku70, Bax, and Werner syndrome protein (WRN). In the nucleotide excision repair (NER) pathway, SIRT1 increases the efficacy of DNA repair by enhancing the interaction between Rad3-related phosphorylation of Xeroderma Pigmentosum (XPA) and Replication protein A32 (RPA32), showcasing the comprehensive involvement of SIRTuins in maintaining genomic stability through various DNA repair mechanisms.
The Sirtuin family of proteins plays a crucial role in sustaining cellular equilibrium in the face of stress. It has been documented that SIRT1 engages with stress-induced transcription factors including nuclear factor κB (NF-κB), heat shock transcription factor 1 (HSF1), and peroxisome proliferator-activated receptor gamma coactivator 1α (PGC-1α), exerting a regulatory influence (72–74). SIRT1’s interaction with these factors can both elevate cancer risk by hindering P53-induced apoptosis and, paradoxically, stimulate P53 activation through the modulation of P19 alternative reading frame (p19ARF), particularly under conditions of high intracellular ROS (75) (Figure 4). By promoting the deacetylation of P53, SIRT1 facilitates the accumulation of this protein in mitochondria, enhancing cellular survival mechanisms (76). Furthermore, SIRT1 modulates the deacetylation of FOXO proteins, diminishing the transcription of pro-apoptotic genes (77). Intriguingly, FOXO1, when disassociated and acetylated away from SIRT2, can complex with Autophagy Related 7 (Atg7) to trigger autophagy (78). SIRT1 is also known to augment apoptosis in cells exposed to tumor necrosis factor-alpha (TNF-α) by suppressing nuclear NF-κB activity (79). This suppression of NF-κB function is a trait shared with SIRT2 and SIRT6. In the mitochondrial environment, SIRT3, akin to SIRT1, possesses deacetylase activity that shields cells from oxidative stress-induced mortality (80). SIRT3 not only boosts the expression of Forkhead box class O3a (FoxO3A)-dependent genes but also interacts with P53 and BAG Cochaperone 2 (BAG2) to modulate growth and cellular stress response by restraining P53 activity (81). Additionally, SIRT3 plays a vital role in managing ROS levels through the modulation of superoxide dismutase 2 (SOD2), manganese dismutase, catalase, and mitochondrial isocitrate dehydrogenase (IDH2), crucial for mitigating oxidative stress-induced DNA damage, including DSBs (82–84). SIRT6, specifically, is lauded for its effectiveness in facilitating the repair of DSBs, especially under oxidative stress conditions, marking it as a key player in cellular stress management (85).
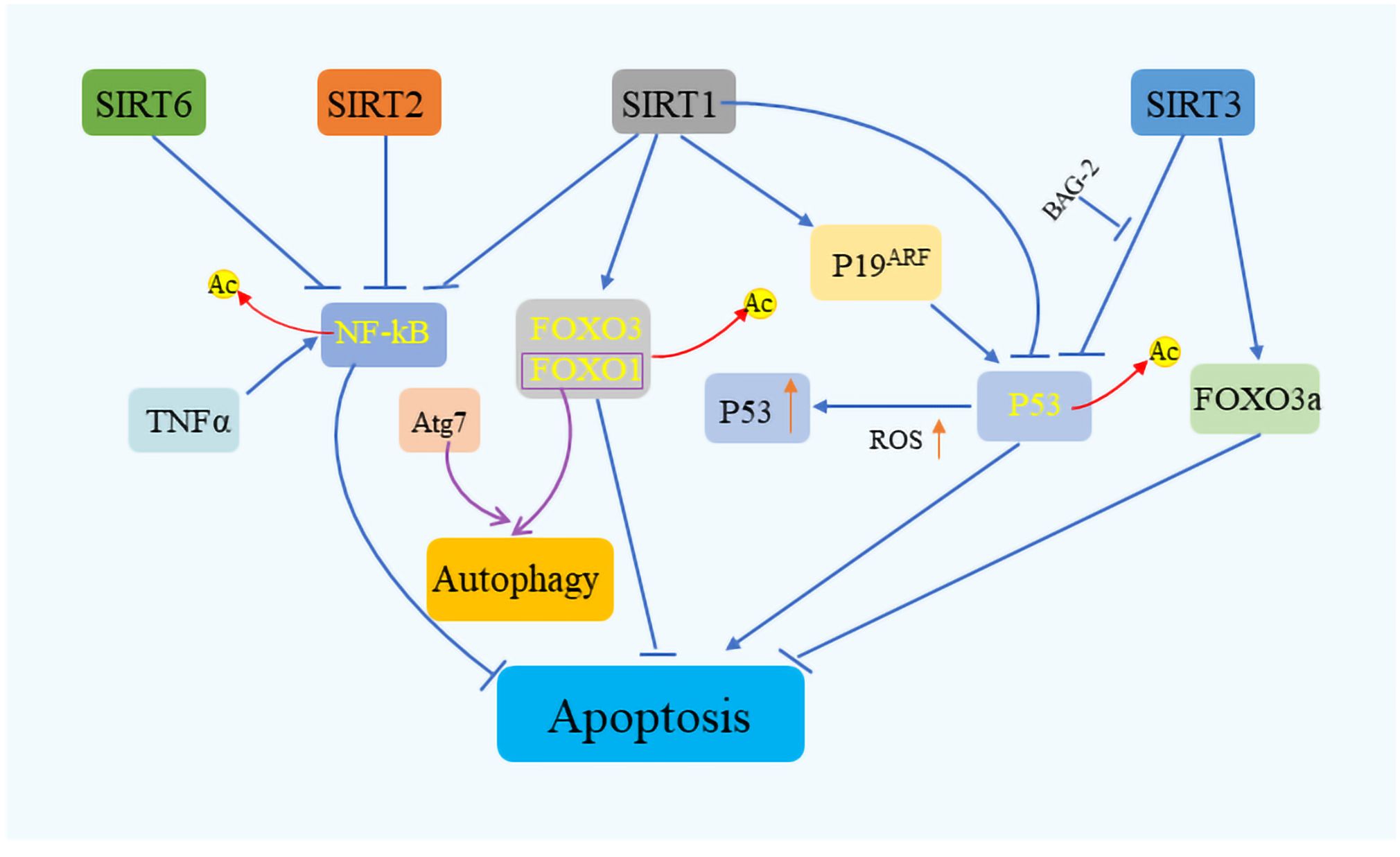
Figure 4 Sirtuins’ role in cell survival and stress response. Sirtuins orchestrate cell fate under various stress conditions by modulating key apoptotic and survival pathways. SIRT1 plays a pivotal role in cell survival by interacting with critical regulators such as P53, FOXO, and NF-κB, while SIRT2 and SIRT6 similarly influence NF-κB signaling. SIRT3, on the other hand, contributes to apoptosis regulation by targeting P53 and FOXO3a. Through these interactions, sirtuins serve as vital mediators of cellular stress response, ensuring cell survival by fine-tuning the balance.
The impact of the Sirtuin family on mitigating cellular aging and extending lifespan has become a focal point of scientific inquiry, with particular emphasis on the roles of SIRT1 and SIRT6. Aging is categorized into replicative and premature forms, the former being attributed to telomere attrition, and the latter to the depletion of critical cellular factors such as oncogenes and tumor suppressors (86). By modulating various cellular mechanisms, Sirtuins contribute to delaying cellular senescence and extending biological longevity. This includes maintaining genomic stability, enhancing DNA repair processes, and protecting against age-related telomere degradation (87). Moreover, Sirtuins influence organismal lifespan through interactions with key regulatory pathways, including the insulin/Insulin-like growth factor-1 (IGF-1) signaling pathway and the AMP-activated protein kinase pathway (88, 89). Research highlights how pathways mediated by SIRT1, alongside Nk2 homeobox 1 (Nkx2–1), and orexin type 2 receptor (Ox2r) within the hypothalamus’ dorsal medial and lateral nuclei, can invigorate bone and muscle functions via the sympathetic nervous system, thereby fostering a physiologically youthful state that aids in delaying the aging process and prolonging life (90, 91). Furthermore, Sirt6 is actively engaged in suppressing age-associated gene expression related to cellular aging, in addition to maintaining the structural integrity of telomere chromatin, thus preventing genomic instability and cellular senescence (92).
The Sirtuin family, known for its deacetylase properties, plays a significant role in safeguarding against a variety of diseases, including atherosclerotic thrombosis, hepatic steatosis, obesity induced by diet, type 2 diabetes, and myocardial infarction (93). Within the realm of cardiovascular diseases, SIRT1 stands out for its ability to mitigate atherosclerosis through the activation of endothelial nitric oxide synthase (eNOS) and the reduction of NF-κB activity in endothelial cells and macrophages (94–96). Similarly, SIRT3 has been identified as beneficial in addressing left ventricular hypertrophy, cardiomyopathy, and dyslipidemia. It achieves this by enhancing the activity of superoxide dismutase 2 (SOD2), manganese superoxide dismutase (MnSOD), and catalase (Cat), thereby preventing cardiac hypertrophy (97). Additionally, SIRT6 contributes to the improved survival of cardiomyocytes under hypoxic conditions by inhibiting necrosis or apoptosis pathways and ameliorating dyslipidemia and left ventricular hypertrophy (98).
In the context of kidney diseases, SIRT1’s presence is widespread in renal tubular cells and podocytes, where it reduces the reabsorption of epithelial sodium through interaction with methyltransferases, ultimately suppressing transcription (99). This function of SIRT1 in regulating renal sodium and water reabsorption has implications for blood pressure control. SIRT3 is pivotal in managing the mitochondrial dynamics within renal cells, regulating the microtubule network-dependent transport of functional mitochondria among renal tubular epithelial cells to maintain cellular bioenergetics (100). The role of SIRT6 in maintaining renal homeostasis has also been acknowledged (101). Moreover, the involvement of Sirtuins in neurological diseases has garnered attention in recent years, particularly given the connection between aging and Alzheimer’s disease (AD). SIRT6, by preserving telomere integrity and correcting abnormal DNA repair mechanisms, plays a role in delaying cellular senescence, which is pivotal in the context of AD (102). This multifaceted involvement of Sirtuins in disease prevention and management underscores their potential as therapeutic targets in various pathological conditions, highlighting the breadth of their influence on human health.
The significance of Sirtuins in the realm of oncology has been increasingly recognized, with a growing body of evidence suggesting their involvement in regulating numerous aberrant cellular processes in cancer. These include cell cycle modulation, DNA repair mechanisms, cell survival and apoptosis, and the cellular response to genomic instability, which are pivotal in the onset and progression of cancer (Table 2).
The role of SIRT1 in oral cancer presents a dichotomy. Research indicates that SIRT1’s expression levels critically influence the pathophysiology of oral cancer. Stable SIRT1 expression contributes to epithelial integrity by upregulating E-cadherin expression, thus mitigating the risk of invasion and metastasis in oral cancer (103, 104). These findings suggest a tumor-suppressive role for SIRT1 in oral cancer. However, conflicting evidence exists, with some studies highlighting SIRT1’s tumor-promoting aspects. For instance, SIRT1 overexpression has been linked to cisplatin resistance in oral cancer via the upregulation of annexin A4 (105, 106). Similarly, the involvement of SIRT3 in oral cancer has been scrutinized. SIRT3 is vital for regulating cellular ROS levels, exerting anti-tumor effects in oral cancer cells by managing ROS and diminishing its activity. The absence of SIRT3 leads to increased ROS, which escalates genomic instability and fosters tumorigenesis (107–109). Furthermore, SIRT7 has been identified as a metastasis inhibitor in oral squamous cell carcinoma (OSCC), where its downregulation promotes invasion and migration of OSCC cells, whereas its overexpression hampers metastasis through the negative regulation of epithelial-mesenchymal transition (EMT) (110–112).
SIRT7 has emerged as a critical modulator of the transforming growth factor-β (TGF-β) signaling pathway and an antagonist of breast cancer metastasis. It is markedly reduced in lung metastases from breast cancer in both humans and mice, indicating that a deficiency in SIRT7 propels the metastasis of breast cancer cells. The overexpression of SIRT7, particularly in early stages of breast cancer, appears crucial for countering malignancy progression (113). On the other hand, SIRT1 has been pinpointed as a key player in cell growth and survival. The miR-211–5p is known to directly target SIRT1, reducing both its gene and protein expression levels, which correlates with decreased P53 acetylation, reduced cell viability, and increased apoptosis in breast cancer cells (114–116). SIRT3, the principal mitochondrial deacetylase, safeguards key proteins by regulating mitochondrial homeostasis and antioxidants, playing a significant role in ROS production and clearance (117). A decrease in SIRT3 expression can elevate oxidative stress, leading to cell death (118). Thus, SIRT3 supports cell proliferation and survival by maintaining ROS balance and preventing apoptosis pathway activation (119). SIRT3’s role as an adjunct in breast cancer treatment, enhancing the effectiveness of chemotherapy and hormonal therapy through its diverse mechanisms, is also noteworthy. Zhang’s team showed that SIRT3 expression was higher in Tam resistant breast cancer cells. Knockdown of SIRT3 increased the sensitivity of Tam-resistant cells and induced apoptosis, with an increase in mitochondrial ROS and ERβ levels (13).In studies focusing on SIRT4, an elevated expression level in breast tissue compared to non-tumor tissues was observed, suggesting a carcinogenic role for SIRT4 at the cellular level and implicating its involvement in breast cancer development (120).
Mitochondrial ROS production has been linked to the progression and metastasis of liver cancer, contributing to DNA damage and alterations in mitochondrial pathways. SIRT3 has been shown to counter tumor progression by suppressing HIF-1α and mitochondrial ROS production (121, 122), and by elevating SOD2 activity, a key mitochondrial antioxidant, thus bolstering resistance to oxidative stress (123). Liu et al. found that SIRT3 is underexpressed or absent in hepatocellular carcinoma (HCC) tissues (124). Compared to patients with low SIRT3 expression, those with high SIRT3 expression exhibit longer survival, better tumor differentiation, and smaller tumor size in HCC (125). Wang et al. discovered that in patients with Barcelona Clinic Liver Cancer (BCLC) stage A or without vascular invasion, SIRT3 can serve as a biomarker for post-hepatectomy recurrence (123). This suggests that SIRT3 plays a crucial role in the development of liver cancer and can serve as a reliable prognostic predictor. In HCC cells, the expression of mitochondrial Ca2+ uniporter (MCU) complex is upregulated, increasing mitochondrial Ca2+ uptake, reducing NAD+ levels, and decreasing NAD+-dependent SIRT3 deacetylase activity (126). This inhibits MnSOD activity, leading to increased ROS levels and activation of c-Jun N-terminal kinase (JNK) signaling, inducing Matrix Metallopeptidase 2 (MMP-2) activity, and promoting HCC cell migration (126). Additionally, research has shown that SIRT3 can inhibit the development and proliferation of HepG2 cells and induce apoptosis by upregulating P53 and MnSOD, increasing BCL2-Associated X (Bax) and Factor Related Apoptosis (Fas) (124). Song et al. found that SIRT3 deacetylates and activates Glycogen synthase kinase 3β (GSK-3β), inhibiting cell growth through the GSK-3β/Bax-dependent apoptotic pathway (127). Similarly, SIRT4 levels are found reduced in liver cancer cell lines and patient samples, with significant deletions at the SIRT4 locus in hepatocellular carcinoma (HCC), suggesting its protective role against liver cancer. SIRT4 impedes glutamine metabolism, enhancing ADP/AMP levels through LKB1 activation and subsequent phosphorylation of AMP-activated protein kinase (AMPK), showcasing its anticancer efficacy in liver cancer (128–130). Conversely, most liver cancer tissues exhibit elevated SIRT7 mRNA and protein levels compared to adjacent non-tumor liver tissues, playing a crucial role in negating p53-dependent cytotoxic effects in liver cancer. SIRT7’s expression, regulated by tumor suppressors miR-125a and miR-125b, underscores its upregulation in liver cancer (131, 132). SIRT1 remains consistently overexpressed in liver cancer, deemed essential at all cancer stages, with its heightened expression necessary for liver cancer persistence (133–135). SIRT5 promotes liver cancer cell proliferation; its downregulation significantly hampers this proliferation, indicating SIRT5’s role in inhibiting mitochondrial apoptosis in hepatoma cells via cytochrome c deacetylation (136). SIRT6, associated with hepatoma cell proliferation and reduced apoptosis, regulates these processes through the extracellular signal-related kinases 1 and 2 (ERK1/2) pathway, influencing the activation of endogenous apoptosis pathways (137).
SIRT1’s ability to deacetylate lysine residues at core histones’ N-terminus reduces transcription by tightening DNA. It negatively regulates pro-apoptotic protein Smac and NF-κB, which delivers anti-apoptotic signals, thus protecting cells from apoptosis. SIRT1 modulates lung cancer cell apoptosis through the SIRT1-NF-κB-Smac signaling pathway (138–140). SIRT3 deacetylates the tumor suppressor Phosphatase And Tensin Homolog (PTEN), inhibiting murine double minute 2 (mdm2) transcription and stabilizing p53, yet its overexpression in non-small cell lung cancer (NSCLC) acts carcinogenically. In PTEN-deficient cancers, SIRT3’s unilateral deacetylation of p53 may contribute to NSCLC malignancies (141, 142). SIRT6, identified as a driver of NSCLC metastasis, promotes cancer cell apoptosis, suggesting its downregulation could be linked to NSCLC’s malignant progression, positioning SIRT6 as a potential therapeutic target (143).
SIRT1’s inhibition through NF-κB-cyclin D1 signaling plays a role in gastric cancer (GC) formation. It also impacts GC invasion and metastasis, with both in vitro and in vivo studies showing that SIRT1 reduction diminishes GC cell migration, invasion, and lung metastasis by targeting Rho GTPase Activating Protein 5 (ARHGAP5) through deacetylation and inhibition of c-JUN (144–146). SIRT2 appears to facilitate GC cell migration and invasion, possibly acting as a carcinogen in GC by activating the RAS/ERK/JNK/MMP-9 pathway and enhancing cytoplasmic PEPCK1 levels and mitochondrial activity, thus promoting cell migration and invasion. Inhibiting SIRT2 activity is crucial for mitigating GC cell migration and invasion (147, 148). Conversely, SIRT5 suppresses GC progression by inhibiting cell proliferation and glycolysis, with its downregulation or overexpression significantly impacting GC growth. SIRT5’s tumor-suppressive role in GC is mediated by its inhibition of the Oxoglutarate Dehydrogenase (OGDH) complex and interference with mitochondrial function (149, 150). A decrease in SIRT3 elevates hypoxia-inducible factor-1α (HIF-1α) expression in MGC-803 gastric cancer cells, suggesting SIRT3’s involvement in GC progression through the regulation of cellular ROS and HIF-1α stability, offering a mitochondrial tumor-suppressive mechanism (151–153).
The Sirtuin family has been implicated in a wide range of cancers, showcasing a spectrum of expressions that influence tumor behavior. In acute myeloid leukemia (AML), heightened SIRT1 expression has been observed (154), whereas reductions in SIRT1 are linked to the onset of cancers in tissues such as the brain (glioma), bladder, ovary, and others (155–157). Prostate cancer presents a unique scenario where SIRT1 levels vary, sometimes inhibiting FOXO1 transcription and enhancing FOXO1 deacetylation to prevent apoptosis, while also capable of decreasing androgen-induced proliferation (158). Skin cancer and melanoma studies reveal a significant drop in SIRT2 expression, with mutations in SIRT2 (P199L) leading to loss of enzymatic activity and tumor proliferation. SIRT2’s knockout resulted in altered expression of keratin markers and increased stem cell markers, promoting tumor growth (159). Additionally, SIRT2 has been proposed as a tumor suppressor in glioma development, offering new molecular markers for treatment (160).
The critical role of the Sirtuin family in cancer biology has illuminated the pathway for developing novel therapeutics. This ongoing exploration into Sirtuins’ function in tumor genesis, coupled with a deeper understanding of their regulatory mechanisms, presents a promising frontier in cancer treatment with the advent of small molecule drugs targeting these enzymes (Table 3).
Resveratrol, a naturally occurring polyphenolic compound, has demonstrated significant potential in cancer prevention and treatment. As a SIRT1 activator, it enhances the protein’s expression and enzymatic action within HepG2 cells, leading to a reduction in the phosphorylation levels of the Phosphoinositide 3-Kinase (PI3K)/Protein Kinase B (PKB, AKT) pathway components. This suggests that Resveratrol’s inhibition of the PI3K/AKT pathway, a critical signaling pathway in cancer progression, is mediated through SIRT1 activation (161). Further evidence indicates that Resveratrol can impede the proliferation and migration of liver cancer cells by modulating the PI3K/AKT pathway through SIRT1’s post-translational modifications (162).
Curcumin, the primary active compound found in turmeric, exerts its anticancer effects through multiple mechanisms, including the activation of the Ataxia telangiectasia mutated (ATM)/checkpoint kinase2 (CHK2) pathway and suppression of NF-κB signaling. Research has revealed that Curcumin’s induction of DNA damage and apoptosis in tumor cells is partly facilitated by its activation of SIRT1, highlighting a dual-pathway mechanism towards exerting its anticancer effects (163).
Additional research into Resveratrol’s effects on tumor cells with constitutively active signal transducer and activator of transcription 3 (STAT3) demonstrates that this SIRT1 activator can lead to the deacetylation and subsequent inactivation of STAT3, reducing surviving gene transcription and promoting tumor cell apoptosis (164).
The role of Aspirin in cancer chemoprevention has gained attention, particularly in colorectal cancer. Studies have shown that Aspirin can induce senescence in CRC cells by upregulating SIRT1 and activating the AMPK pathway, marking a significant step forward in understanding the molecular mechanisms underlying Aspirin’s anticancer effects (165).
The development of MHY2256, a novel SIRT inhibitor, has shown promise in reducing the viability of breast cancer cells. This compound’s ability to decrease SIRT1, 2, and 3 protein levels and increase the acetylation of p53 in MCF-7 cells signifies its potential for inducing apoptosis through p53 acetylation, offering a new therapeutic option in breast cancer treatment (166).
EX527, specifically targeting SIRT1, has been studied for its impact on glioma cell proliferation and survival. The inhibitor’s capacity to upregulate p53, acetylated p53, and the downstream target p21, thereby inducing apoptosis, presents a viable strategy for glioma therapy (167).
Salermide, a reverse amide compound, exhibits potent inhibitory effects against SIRT1 and SIRT2 in vitro. Its unique ability to induce p53-mediated apoptosis in breast cancer cells without affecting normal cells underscores its therapeutic potential (168).
A comprehensive analysis comparing Sirtinol, Salermide, and EX527 revealed their effectiveness as SIRT1/2 inhibitors, with EX527 showing the highest specificity towards SIRT1. This comparative study suggests the necessity of targeting both SIRT1 and SIRT2 to achieve optimal pro-apoptotic effects in cancer therapy (169).
The role of microRNAs (miRNAs) as key regulators in cancer has gained prominence, with their dual capacity to act as either oncogenes or tumor suppressors. This dual functionality allows miRNAs to finely tune the expression of crucial genes involved in tumorigenesis, including those related to the Sirtuin family, thereby offering a nuanced approach to cancer therapy (Table 4).
One pivotal example is MicroRNA-34a (miR-34a), which is activated by p53 and targets SIRT1 along with other critical oncogenes. The modulation of SIRT1 by miR-34a leads to cell cycle arrest and apoptosis, making miR-34a a potential ally in chemotherapy regimens aimed at enhancing cancer cell susceptibility to treatment. This synergy between miRNA-34a and chemotherapy could lead to more effective therapeutic strategies, enhancing treatment efficacy while minimizing side effects (170, 171).
Further, the miRNAs miRNA-204–5p and miR-22 have been identified as potent regulators of SIRT1, showcasing the capability of miRNAs to inhibit cancer progression by modulating key molecular pathways. These miRNAs can suppress SIRT1 activity, influencing cellular processes such as cell survival, proliferation, and apoptosis. By altering SIRT1 levels, miRNA-204–5p and miR-22 can potentially reshape the tumor microenvironment, thereby stalling cancer progression and providing new avenues for targeted cancer therapies (172, 173). Furthermore, miRNA-217 could target SIRT1 to induce epithelial-mesenchymal transition (EMT), and lead to poor prognosis of pancreatic cancer (174). MiR-125a-5p has been reported to increase radioresistance in NSCLC via upregulation of SIRT7 (175). The expanding research into miRNAs offers promising insights into their potential to manipulate Sirtuin activity, revealing a complex network of genetic regulation that could be harnessed to develop innovative cancer treatments. These findings underscore the need for continued exploration into miRNA-mediated pathways as integral components of cancer therapeutics, where the strategic targeting of Sirtuins could lead to groundbreaking advancements in cancer treatment.
The multifaceted roles of Sirtuins in cancer biology have become increasingly evident, revealing their profound influence on tumorigenesis and cancer progression through diverse mechanisms. These proteins, with their capacity to either suppress or promote tumor growth, embody the complex nature of cancer regulation at the molecular level. While Sirtuins can act as guardians by inhibiting tumor development, they can also play a role in facilitating cancer progression under certain conditions. This dual functionality underscores the nuanced understanding required to harness their potential in cancer therapy effectively. Despite the strides made in elucidating the functions of Sirtuins in cancer, numerous questions remain. The intricate balance between their protective and oncogenic roles presents a significant challenge in developing therapeutic strategies. However, the growing body of evidence underscores the significant impact these enzymes have on the cancer landscape. Their ability to modulate key cancer pathways offers a promising avenue for targeted therapy, making them critical components in the fight against cancer.
Moreover, the potential of Sirtuins to synergize with existing anticancer drugs opens new possibilities for combination therapies. By integrating Sirtuin-targeted treatments with conventional chemotherapy, targeted therapy, or immunotherapy, we can potentially enhance therapeutic efficacy and overcome resistance mechanisms. This approach not only amplifies the anti-tumor effects but also provides a strategy to mitigate the adverse side effects associated with high-dose monotherapies. As research continues to advance, it is anticipated that novel Sirtuin modulators will emerge, offering more precise and personalized treatment options for cancer patients. The development of these modulators, guided by an in-depth understanding of Sirtuin biology and cancer-specific contexts, could lead to breakthroughs in cancer care. Furthermore, exploring the interplay between Sirtuins and the tumor microenvironment may unveil additional therapeutic targets and strategies to prevent tumor metastasis and recurrence.
In conclusion, Sirtuins represent a pivotal frontier in cancer research, with the potential to revolutionize cancer treatment paradigms. Their dual roles in cancer biology reflect the complexity of targeting these enzymes but also highlight the immense therapeutic potential they hold. As we continue to unravel the intricacies of Sirtuin function in cancer, the future holds promise for innovative therapies that could significantly impact patient outcomes in the battle against cancer.
LY: Writing – original draft. YL: Writing – original draft. SS: Writing – original draft. YZ: Writing – original draft. YipW: Funding acquisition, Writing – original draft. HW: Funding acquisition, Writing – original draft. ZY: Funding acquisition, Writing – review & editing. YiW: Conceptualization, Funding acquisition, Investigation, Project administration, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China (81802504), grants from the Sichuan Science and Technology Department 2021YFS0380 and 2023YFH0010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Garcia-Martinez L, Zhang Y, Nakata Y, Chan HL, Morey L. Epigenetic mechanisms in breast cancer therapy and resistance. Nat Commun. (2021) 12:1786. doi: 10.1038/s41467–021-22024–3
2. Hogg SJ, Beavis PA, Dawson MA, Johnstone RW. Targeting the epigenetic regulation of antitumour immunity. Nat Rev Drug Discovery. (2020) 19:776–800. doi: 10.1038/s41573–020-0077–5
3. Miranda Furtado CL, Dos Santos Luciano MC, Silva Santos RD, Furtado GP, Moraes MO, Pessoa C. Epidrugs: targeting epigenetic marks in cancer treatment. Epigenetics. (2019) 14:1164–76. doi: 10.1080/15592294.2019.1640546
4. Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb Perspect Biol. (2016) 8:a019521. doi: 10.1101/cshperspect.a019521
5. Zaib S, Rana N, Khan I. Histone modifications and their role in epigenetics of cancer. Curr Med Chem. (2022) 29:2399–411. doi: 10.2174/0929867328666211108105214
6. Damodaran S, Damaschke N, Gawdzik J, Yang B, Shi C, Allen GO, et al. Dysregulation of Sirtuin 2 (SIRT2) and histone H3K18 acetylation pathways associates with adverse prostate cancer outcomes. BMC Cancer. (2017) 17:874. doi: 10.1186/s12885–017-3853–9
7. Carafa V, Poziello A, Della Torre L, Giovannelli P, Di Donato M, Safadeh E, et al. Enzymatic and biological characterization of novel sirtuin modulators against cancer. Int J Mol Sci. (2019) 20(22)5654. doi: 10.3390/ijms20225654
8. Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. (1999) 260:273–9. doi: 10.1006/bbrc.1999.0897
9. Voelter-Mahlknecht S, Mahlknecht U. Cloning, chromosomal characterization and mapping of the NAD-dependent histone deacetylases gene sirtuin 1. Int J Mol Med. (2006) 17:59–67. doi: 10.3892/ijmm
10. Hsu CC, Shi J, Yuan C, Zhao D, Jiang S, Lyu J, et al. Recognition of histone acetylation by the GAS41 YEATS domain promotes H2A.Z deposition in non-small cell lung cancer. Genes Dev. (2018) 32:58–69. doi: 10.1101/gad.303784.117
11. Pan Z, Dong H, Huang N, Fang J. Oxidative stress and inflammation regulation of sirtuins: New insights into common oral diseases. Front Physiol. (2022) 13:953078. doi: 10.3389/fphys.2022.953078
12. Jalgaonkar MP, Parmar UM, Kulkarni YA, Oza MJ. SIRT1-FOXOs activity regulates diabetic complications. Pharmacol Res. (2022) 175:106014. doi: 10.1016/j.phrs.2021.106014
13. Ouyang S, Zhang Q, Lou L, Zhu K, Li Z, Liu P, et al. The double-edged sword of SIRT3 in cancer and its therapeutic applications. Front Pharmacol. (2022) 13:871560. doi: 10.3389/fphar.2022.871560
14. Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. (2009) 460:587–91. doi: 10.1038/nature08197
15. Vall-Llaura N, Mir N, Garrido L, Vived C, Cabiscol E. Redox control of yeast Sir2 activity is involved in acetic acid resistance and longevity. Redox Biol. (2019) 24:101229. doi: 10.1016/j.redox.2019.101229
16. Vassilopoulos A, Fritz KS, Petersen DR, Gius D. The human sirtuin family: evolutionary divergences and functions. Hum Genomics. (2011) 5:485–96. doi: 10.1186/1479–7364-5–5-485
17. Yanagisawa S, Baker JR, Vuppusetty C, Koga T, Colley T, Fenwick P, et al. The dynamic shuttling of SIRT1 between cytoplasm and nuclei in bronchial epithelial cells by single and repeated cigarette smoke exposure. PloS One. (2018) 13:e0193921. doi: 10.1371/journal.pone.0193921
18. Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. (2007) 282:6823–32. doi: 10.1074/jbc.M609554200
19. Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. (2006) 124:315–29. doi: 10.1016/j.cell.2005.11.044
20. Emmons M, Boulware D, Sullivan DM, Hazlehurst LA. Topoisomerase II beta levels are a determinant of melphalan-induced DNA crosslinks and sensitivity to cell death. Biochem Pharmacol. (2006) 72:11–8. doi: 10.1016/j.bcp.2006.03.017
21. Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. (2006) 20:1256–61. doi: 10.1101/gad.1412706
22. Nandave M, Acharjee R, Bhaduri K, Upadhyay J, Rupanagunta GP, Ansari MN. A pharmacological review on SIRT 1 and SIRT 2 proteins, activators, and inhibitors: Call for further research. Int J Biol Macromol. (2023) 242:124581. doi: 10.1016/j.ijbiomac.2023.124581
23. Wang Y, Yang J, Hong T, Chen X, Cui L. SIRT2: Controversy and multiple roles in disease and physiology. Ageing Res Rev. (2019) 55:100961. doi: 10.1016/j.arr.2019.100961
24. Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. (2005) 16:4623–35. doi: 10.1091/mbc.e05–01-0033
25. Torrens-Mas M, Oliver J, Roca P, Sastre-Serra J. SIRT3: oncogene and tumor suppressor in cancer. Cancers (Basel) (2017) 9(7):90. doi: 10.3390/cancers9070090
26. Wang C, Liu Y, Zhu Y, Kong C. Functions of mammalian SIRT4 in cellular metabolism and research progress in human cancer. Oncol Lett. (2020) 20:11. doi: 10.3892/ol.2020.11872
27. Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, et al. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol Cell. (2015) 59:321–32. doi: 10.1016/j.molcel.2015.05.022
28. Chang AR, Ferrer CM, Mostoslavsky R. SIRT6, a mammalian deacylase with multitasking abilities. Physiol Rev. (2020) 100:145–69. doi: 10.1152/physrev.00030.2018
29. Preyat N, Leo O. Sirtuin deacylases: a molecular link between metabolism and immunity. J Leukocyte Biol. (2013) 93:669–80. doi: 10.1189/jlb.1112557
30. Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. (2008) 14:312–23. doi: 10.1016/j.ccr.2008.09.001
31. Hamanaka RB, Chandel NS. Targeting glucose metabolism for cancer therapy. J Exp Med. (2012) 209:211–5. doi: 10.1084/jem.20120162
32. Simmons GE Jr., Pruitt WM, Pruitt K. Diverse roles of SIRT1 in cancer biology and lipid metabolism. Int J Mol Sci. (2015) 16:950–65. doi: 10.3390/ijms16010950
33. Chakrabarti P, English T, Karki S, Qiang L, Tao R, Kim J, et al. SIRT1 controls lipolysis in adipocytes via FOXO1-mediated expression of ATGL. J Lipid Res. (2011) 52:1693–701. doi: 10.1194/jlr.M014647
34. Choi HK, Cho KB, Phuong NT, Han CY, Han HK, Hien TT, et al. SIRT1-mediated FoxO1 deacetylation is essential for multidrug resistance-associated protein 2 expression in tamoxifen-resistant breast cancer cells. Mol Pharmaceut. (2013) 10:2517–27. doi: 10.1021/mp400287p
35. Ponugoti B, Kim DH, Xiao Z, Smith Z, Miao J, Zang M, et al. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J Biol Chem. (2010) 285:33959–70. doi: 10.1074/jbc.M110.122978
36. Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA. (2006) 103:10230–5. doi: 10.1073/pnas.0604392103
37. Jeong SM, Lee J, Finley LW, Schmidt PJ, Fleming MD, Haigis MC. SIRT3 regulates cellular iron metabolism and cancer growth by repressing iron regulatory protein 1. Oncogene. (2015) 34:2115–24. doi: 10.1038/onc.2014.124
38. Carafa V, Nebbioso A, Altucci L. Sirtuins and disease: the road ahead. Front Pharmacol. (2012) 3:4. doi: 10.3389/fphar.2012.00004
39. Nogueiras R, Habegger KM, Chaudhary N, Finan B, Banks AS, Dietrich MO, et al. Sirtuin 1 and sirtuin 3: physiological modulators of metabolism. Physiol Rev. (2012) 92:1479–514. doi: 10.1152/physrev.00022.2011
40. Peek CB, Affinati AH, Ramsey KM, Kuo HY, Yu W, Sena LA, et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Sci (New York NY). (2013) 342:1243417. doi: 10.1126/science.1243417
41. Zhang J, Xiang H, Liu J, Chen Y, He RR, Liu B. Mitochondrial Sirtuin 3: New emerging biological function and therapeutic target. Theranostics. (2020) 10:8315–42. doi: 10.7150/thno.45922
42. Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. (2013) 23:450–63. doi: 10.1016/j.ccr.2013.02.024
43. Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. (2013) 153:840–54. doi: 10.1016/j.cell.2013.04.023
44. Kumar S, Lombard DB. Functions of the sirtuin deacylase SIRT5 in normal physiology and pathobiology. Crit Rev Biochem Mol Biol. (2018) 53:311–34. doi: 10.1080/10409238.2018.1458071
45. Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Sci (New York NY). (2011) 334:806–9. doi: 10.1126/science.1207861
46. Rack JG, Morra R, Barkauskaite E, Kraehenbuehl R, Ariza A, Qu Y, et al. Identification of a class of protein ADP-ribosylating sirtuins in microbial pathogens. Mol Cell. (2015) 59:309–20. doi: 10.1016/j.molcel.2015.06.013
47. Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature. (2013) 496:110–3. doi: 10.1038/nature12038
48. Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. (2010) 140:280–93. doi: 10.1016/j.cell.2009.12.041
49. Sebastián C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. (2012) 151:1185–99. doi: 10.1016/j.cell.2012.10.047
50. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
51. Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. (2012) 21:297–308. doi: 10.1016/j.ccr.2012.02.014
52. Yi M, Li J, Chen S, Cai J, Ban Y, Peng Q, et al. Correction to: Emerging role of lipid metabolism alterations in Cancer stem cells. J Exp Clin Cancer Res: CR. (2018) 37:155. doi: 10.1186/s13046-018-0826-z
53. Taylor DM, Maxwell MM, Luthi-Carter R, Kazantsev AG. Biological and potential therapeutic roles of sirtuin deacetylases. Cell Mol Life Sci: CMLS. (2008) 65:4000–18. doi: 10.1007/s00018-008-8357-y
54. Agarwal P, Miller KM. The nucleosome: orchestrating DNA damage signaling and repair within chromatin. Biochem Cell Biol Biochim Biol Cellulaire. (2016) 94:381–95. doi: 10.1139/bcb-2016–0017
55. Stadler J, Richly H. Regulation of DNA repair mechanisms: how the chromatin environment regulates the DNA damage response. Int J Mol Sci. (2017) 18(8):1715. doi: 10.3390/ijms18081715
56. Lagunas-Rangel FA. Current role of mammalian sirtuins in DNA repair. DNA Repair. (2019) 80:85–92. doi: 10.1016/j.dnarep.2019.06.009
57. Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell. (2013) 51:454–68. doi: 10.1016/j.molcel.2013.06.018
58. Van Meter M, Simon M, Tombline G, May A, Morello TD, Hubbard BP, et al. JNK phosphorylates SIRT6 to stimulate DNA double-strand break repair in response to oxidative stress by recruiting PARP1 to DNA breaks. Cell Rep. (2016) 16:2641–50. doi: 10.1016/j.celrep.2016.08.006
59. Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Sci (New York NY). (2011) 332:1443–6. doi: 10.1126/science.1202723
60. Hwang BJ, Jin J, Gao Y, Shi G, Madabushi A, Yan A, et al. SIRT6 protein deacetylase interacts with MYH DNA glycosylase, APE1 endonuclease, and Rad9-Rad1-Hus1 checkpoint clamp. BMC Mol Biol. (2015) 16:12. doi: 10.1186/s12867–015-0041–9
61. Gao Y, Tan J, Jin J, Ma H, Chen X, Leger B, et al. SIRT6 facilitates directional telomere movement upon oxidative damage. Sci Rep. (2018) 8:5407. doi: 10.1038/s41598–018-23602–0
62. Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Sci (New York NY). (2010) 329:1348–53. doi: 10.1126/science.1192049
63. McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging. (2009) 1:109–21. doi: 10.18632/aging.100011
64. Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. (2007) 27:149–62. doi: 10.1016/j.molcel.2007.05.029
65. Madabushi A, Hwang BJ, Jin J, Lu AL. Histone deacetylase SIRT1 modulates and deacetylates DNA base excision repair enzyme thymine DNA glycosylase. Biochem J. (2013) 456:89–98. doi: 10.1042/bj20130670
66. Jarrett SG, Carter KM, Bautista RM, He D, Wang C, D’Orazio JA. Sirtuin 1-mediated deacetylation of XPA DNA repair protein enhances its interaction with ATR protein and promotes cAMP-induced DNA repair of UV damage. J Biol Chem. (2018) 293:19025–37. doi: 10.1074/jbc.RA118.003940
67. Li K, Casta A, Wang R, Lozada E, Fan W, Kane S, et al. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J Biol Chem. (2008) 283:7590–8. doi: 10.1074/jbc.M709707200
68. Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, et al. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp Mol Med. (2007) 39:8–13. doi: 10.1038/emm.2007.2
69. Cheng Y, Ren X, Gowda AS, Shan Y, Zhang L, Yuan YS, et al. Interaction of Sirt3 with OGG1 contributes to repair of mitochondrial DNA and protects from apoptotic cell death under oxidative stress. Cell Death Dis. (2013) 4:e731. doi: 10.1038/cddis.2013.254
70. Kiran S, Oddi V, Ramakrishna G. Sirtuin 7 promotes cellular survival following genomic stress by attenuation of DNA damage, SAPK activation and p53 response. Exp Cell Res. (2015) 331:123–41. doi: 10.1016/j.yexcr.2014.11.001
71. Li L, Shi L, Yang S, Yan R, Zhang D, Yang J, et al. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun. (2016) 7:12235. doi: 10.1038/ncomms12235
72. Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. (2005) 434:113–8. doi: 10.1038/nature03354
73. Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. (2008) 582:46–53. doi: 10.1016/j.febslet.2007.11.034
74. Westerheide SD, Anckar J, Stevens SM Jr., Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Sci (New York NY). (2009) 323:1063–6. doi: 10.1126/science.1165946
75. Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. (2005) 123:437–48. doi: 10.1016/j.cell.2005.08.011
76. Chua KF, Mostoslavsky R, Lombard DB, Pang WW, Saito S, Franco S, et al. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. (2005) 2:67–76. doi: 10.1016/j.cmet.2005.06.007
77. Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Sci (New York NY). (2004) 303:2011–5. doi: 10.1126/science.1094637
78. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. (2008) 132:27–42. doi: 10.1016/j.cell.2007.12.018
79. Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. (2004) 23:2369–80. doi: 10.1038/sj.emboj.7600244
80. Kweon KH, Lee CR, Jung SJ, Ban EJ, Kang SW, Jeong JJ, et al. Sirt1 induction confers resistance to etoposide-induced genotoxic apoptosis in thyroid cancers. Int J Oncol. (2014) 45:2065–75. doi: 10.3892/ijo.2014.2585
81. Li S, Banck M, Mujtaba S, Zhou MM, Sugrue MM, Walsh MJ. p53-induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. PloS One. (2010) 5:e10486. doi: 10.1371/journal.pone.0010486
82. Merksamer PI, Liu Y, He W, Hirschey MD, Chen D, Verdin E. The sirtuins, oxidative stress and aging: an emerging link. Aging. (2013) 5:144–50. doi: 10.18632/aging.100544
83. Miao L, St Clair DK. Regulation of superoxide dismutase genes: implications in disease. Free Radical Biol Med. (2009) 47:344–56. doi: 10.1016/j.freeradbiomed.2009.05.018
84. Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. (2010) 143:802–12. doi: 10.1016/j.cell.2010.10.002
85. Van Meter M, Mao Z, Gorbunova V, Seluanov A. Repairing split ends: SIRT6, mono-ADP ribosylation and DNA repair. Aging. (2011) 3:829–35. doi: 10.18632/aging.100389
86. Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. (2010) 24:2463–79. doi: 10.1101/gad.1971610
87. Muñoz-Espín D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol. (2014) 15:482–96. doi: 10.1038/nrm3823
88. Mazucanti CH, Cabral-Costa JV, Vasconcelos AR, Andreotti DZ, Scavone C, Kawamoto EM. Longevity pathways (mTOR, SIRT, insulin/IGF-1) as key modulatory targets on aging and neurodegeneration. Curr Topics Medicinal Chem. (2015) 15:2116–38. doi: 10.2174/1568026615666150610125715
89. Zhao X, Lu L, Qi Y, Li M, Zhou L. Emodin extends lifespan of Caenorhabditis elegans through insulin/IGF-1 signaling pathway depending on DAF-16 and SIR-2.1. Biosci Biotechnol Biochem. (2017) 81:1908–16. doi: 10.1080/09168451.2017.1365592
90. Satoh A, Brace CS, Ben-Josef G, West T, Wozniak DF, Holtzman DM, et al. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J Neurosci: Off J Soc Neurosci. (2010) 30:10220–32. doi: 10.1523/jneurosci.1385–10.2010
91. Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, et al. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. (2013) 18:416–30. doi: 10.1016/j.cmet.2013.07.013
92. Tasselli L, Zheng W, Chua KF. SIRT6: novel mechanisms and links to aging and disease. Trends Endocrinol Metabolism: TEM. (2017) 28:168–85. doi: 10.1016/j.tem.2016.10.002
93. Winnik S, Auwerx J, Sinclair DA, Matter CM. Protective effects of sirtuins in cardiovascular diseases: from bench to bedside. Eur Heart J. (2015) 36:3404–12. doi: 10.1093/eurheartj/ehv290
94. Zhang QJ, Wang Z, Chen HZ, Zhou S, Zheng W, Liu G, et al. Endothelium-specific overexpression of class III deacetylase SIRT1 decreases atherosclerosis in apolipoprotein E-deficient mice. Cardiovasc Res. (2008) 80:191–9. doi: 10.1093/cvr/cvn224
95. Stein S, Lohmann C, Schäfer N, Hofmann J, Rohrer L, Besler C, et al. SIRT1 decreases Lox-1-mediated foam cell formation in atherogenesis. Eur Heart J. (2010) 31:2301–9. doi: 10.1093/eurheartj/ehq107
96. Stein S, Schäfer N, Breitenstein A, Besler C, Winnik S, Lohmann C, et al. SIRT1 reduces endothelial activation without affecting vascular function in ApoE-/- mice. Aging. (2010) 2:353–60. doi: 10.18632/aging.100162
97. Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. (2009) 119:2758–71. doi: 10.1172/jci39162
98. Maksin-Matveev A, Kanfi Y, Hochhauser E, Isak A, Cohen HY, Shainberg A. Sirtuin 6 protects the heart from hypoxic damage. Exp Cell Res. (2015) 330:81–90. doi: 10.1016/j.yexcr.2014.07.013
99. Zhang D, Li S, Cruz P, Kone BC. Sirtuin 1 functionally and physically interacts with disruptor of telomeric silencing-1 to regulate alpha-ENaC transcription in collecting duct. J Biol Chem. (2009) 284:20917–26. doi: 10.1074/jbc.M109.020073
100. Morigi M, Perico L, Rota C, Longaretti L, Conti S, Rottoli D, et al. Sirtuin 3-dependent mitochondrial dynamic improvements protect against acute kidney injury. J Clin Invest. (2015) 125:715–26. doi: 10.1172/jci77632
101. Huang W, Liu H, Zhu S, Woodson M, Liu R, Tilton RG, et al. Sirt6 deficiency results in progression of glomerular injury in the kidney. Aging. (2017) 9:1069–83. doi: 10.18632/aging.101214
102. Mohamad Nasir NF, Zainuddin A, Shamsuddin S. Emerging roles of sirtuin 6 in alzheimer’s disease. J Mol Neurosci: MN. (2018) 64:157–61. doi: 10.1007/s12031-017-1005-y
103. Chen IC, Chiang WF, Huang HH, Chen PF, Shen YY, Chiang HC. Role of SIRT1 in regulation of epithelial-to-mesenchymal transition in oral squamous cell carcinoma metastasis. Mol Cancer. (2014) 13:254. doi: 10.1186/1476–4598-13–254
104. Kang YY, Sun FL, Zhang Y, Wang Z. SIRT1 acts as a potential tumor suppressor in oral squamous cell carcinoma. J Chin Med Association: JCMA. (2018) 81:416–22. doi: 10.1016/j.jcma.2017.09.004
105. Xiong P, Li YX, Tang YT, Chen HG. Proteomic analyses of Sirt1-mediated cisplatin resistance in OSCC cell line. Protein J. (2011) 30:499–508. doi: 10.1007/s10930–011-9354–9
106. Wei B, Guo C, Liu S, Sun MZ. Annexin A4 and cancer. Clin Chim Acta Int J Clin Chem. (2015) 447:72–8. doi: 10.1016/j.cca.2015.05.016
107. Chen IC, Chiang WF, Chen PF, Chiang HC. STRESS-responsive deacetylase SIRT3 is up-regulated by areca nut extract-induced oxidative stress in human oral keratinocytes. J Cell Biochem. (2014) 115:328–39. doi: 10.1002/jcb.24667
108. Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. (2010) 17:41–52. doi: 10.1016/j.ccr.2009.11.023
109. Chen IC, Chiang WF, Liu SY, Chen PF, Chiang HC. Role of SIRT3 in the regulation of redox balance during oral carcinogenesis. Mol Cancer. (2013) 12:68. doi: 10.1186/1476–4598-12–68
110. Shi L, Liang F, Zheng J, Zhou K, Chen S, Yu J, et al. Melatonin regulates apoptosis and autophagy Via ROS-MST1 pathway in subarachnoid hemorrhage. Front Mol Neurosci. (2018) 11:93. doi: 10.3389/fnmol.2018.00093
111. Yang H, Pan L, Xu C, Zhang Y, Li K, Chen S, et al. Overexpression of tumor suppressor gene ZNF750 inhibits oral squamous cell carcinoma metastasis. Oncol Lett. (2017) 14:5591–6. doi: 10.3892/ol.2017.6908
112. Kim SE, Shin SH, Lee JY, Kim CH, Chung IK, Kang HM, et al. Resveratrol induces mitochondrial apoptosis and inhibits epithelial-mesenchymal transition in oral squamous cell carcinoma cells. Nutr Cancer. (2018) 70:125–35. doi: 10.1080/01635581.2018.1397708
113. Aljada A, Saleh AM, Alkathiri M, Shamsa HB, Al-Bawab A, Nasr A. Altered sirtuin 7 expression is associated with early stage breast cancer. Breast Cancer: Basic Clin Res. (2015) 9:3–8. doi: 10.4137/bcbcr.S23156
114. Chen LL, Zhang ZJ, Yi ZB, Li JJ. MicroRNA-211–5p suppresses tumour cell proliferation, invasion, migration and metastasis in triple-negative breast cancer by directly targeting SETBP1. Br J Cancer. (2017) 117:78–88. doi: 10.1038/bjc.2017.150
115. Yarahmadi S, Abdolvahabi Z, Hesari Z, Tavakoli-Yaraki M, Yousefi Z, Seiri P, et al. Inhibition of sirtuin 1 deacetylase by miR-211–5p provides a mechanism for the induction of cell death in breast cancer cells. Gene. (2019) 711:143939. doi: 10.1016/j.gene.2019.06.029
116. Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. (2003) 100:10794–9. doi: 10.1073/pnas.1934713100
117. Torrens-Mas M, Pons DG, Sastre-Serra J, Oliver J, Roca P. SIRT3 silencing sensitizes breast cancer cells to cytotoxic treatments through an increment in ROS production. J Cell Biochem. (2017) 118:397–406. doi: 10.1002/jcb.25653
118. Papa L, Germain D. SirT3 regulates the mitochondrial unfolded protein response. Mol Cell Biol. (2014) 34:699–710. doi: 10.1128/mcb.01337–13
119. Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL. Sirtuin-3 (SIRT3) and the hallmarks of cancer. Genes Cancer. (2013) 4:164–71. doi: 10.1177/1947601913486351
120. Huang G, Lin Y, Zhu G. SIRT4 is upregulated in breast cancer and promotes the proliferation, migration and invasion of breast cancer cells. Int J Clin Exp Pathol. (2017) 10:11849–56.
121. Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. (2011) 19:416–28. doi: 10.1016/j.ccr.2011.02.014
122. Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. (2010) 40:893–904. doi: 10.1016/j.molcel.2010.12.013
123. Wang JX, Yi Y, Li YW, Cai XY, He HW, Ni XC, et al. Down-regulation of sirtuin 3 is associated with poor prognosis in hepatocellular carcinoma after resection. BMC Cancer. (2014) 14:297. doi: 10.1186/1471–2407-14–297
124. Liu Y, Liu YL, Cheng W, Yin XM, Jiang B. The expression of SIRT3 in primary hepatocellular carcinoma and the mechanism of its tumor suppressing effects. Eur Rev Med Pharmacol Sci. (2017) 21:978–98.
125. De Matteis S, Scarpi E, Granato AM, Vespasiani-Gentilucci U, La Barba G, Foschi FG, et al. Role of SIRT-3, p-mTOR and HIF-1alpha in hepatocellular carcinoma patients affected by metabolic dysfunctions and in chronic treatment with metformin. Int J Mol Sci. (2019) 20(6):1503. doi: 10.3390/ijms20061503
126. Ren T, Zhang H, Wang J, Zhu J, Jin M, Wu Y, et al. MCU-dependent mitochondrial Ca(2+) inhibits NAD(+)/SIRT3/SOD2 pathway to promote ROS production and metastasis of HCC cells. Oncogene. (2017) 36:5897–909. doi: 10.1038/onc.2017.167
127. Song CL, Tang H, Ran LK, Ko BC, Zhang ZZ, Chen X, et al. Sirtuin 3 inhibits hepatocellular carcinoma growth through the glycogen synthase kinase-3beta/BCL2-associated X protein-dependent apoptotic pathway. Oncogene. (2016) 35:631–41. doi: 10.1038/onc.2015.121
128. Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat Rev Cancer. (2016) 16:635–49. doi: 10.1038/nrc.2016.77
129. Yu J, Auwerx J. The role of sirtuins in the control of metabolic homeostasis. Ann New York Acad Sci. (2009) 1173 Suppl 1:E10–9. doi: 10.1111/j.1749-6632.2009.04952.x
130. Wang YS, Du L, Liang X, Meng P, Bi L, Wang YL, et al. Sirtuin 4 depletion promotes hepatocellular carcinoma tumorigenesis through regulating adenosine-monophosphate-activated protein kinase alpha/mammalian target of rapamycin axis in mice. Hepatol (Baltimore Md). (2019) 69:1614–31. doi: 10.1002/hep.30421
131. Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ, Kim MG, et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatol (Baltimore Md). (2013) 57:1055–67. doi: 10.1002/hep.26101
132. Zhao J, Wozniak A, Adams A, Cox J, Vittal A, Voss J, et al. SIRT7 regulates hepatocellular carcinoma response to therapy by altering the p53-dependent cell death pathway. J Exp Clin Cancer Res: CR. (2019) 38:252. doi: 10.1186/s13046–019-1246–4
133. Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. (2009) 69:1702–5. doi: 10.1158/0008–5472.Can-08–3365
134. Wang Y, Cui R, Zhang X, Qiao Y, Liu X, Chang Y, et al. SIRT1 increases YAP- and MKK3-dependent p38 phosphorylation in mouse liver and human hepatocellular carcinoma. Oncotarget. (2016) 7:11284–98. doi: 10.18632/oncotarget.7022
135. Farcas M, Gavrea AA, Gulei D, Ionescu C, Irimie A, Catana CS, et al. SIRT1 in the development and treatment of hepatocellular carcinoma. Front Nutr. (2019) 6:148. doi: 10.3389/fnut.2019.00148
136. Zhang R, Wang C, Tian Y, Yao Y, Mao J, Wang H, et al. SIRT5 promotes hepatocellular carcinoma progression by regulating mitochondrial apoptosis. J Cancer. (2019) 10:3871–82. doi: 10.7150/jca.31266
137. Zhang C, Yu Y, Huang Q, Tang K. SIRT6 regulates the proliferation and apoptosis of hepatocellular carcinoma via the ERK1/2 signaling pathway. Mol Med Rep. (2019) 20:1575–82. doi: 10.3892/mmr.2019.10398
138. Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: the ‘magnificent seven’, function, metabolism and longevity. Ann Med. (2007) 39:335–45. doi: 10.1080/07853890701408194
139. Hwang SL, Guo HR, Hsieh WA, Hwang JS, Lee SD, Tang JL, et al. Cancer risks in a population with prolonged low dose-rate gamma-radiation exposure in radiocontaminated buildings, 1983–2002. Int J Radiat Biol. (2006) 82:849–58. doi: 10.1080/09553000601085980
140. Ji K, Sun X, Liu Y, Du L, Wang Y, He N, et al. Regulation of apoptosis and radiation sensitization in lung cancer cells via the sirt1/NF-κB/smac pathway. Cell Physiol Biochem: Int J Exp Cell Physiol Biochem Pharmacol. (2018) 48:304–16. doi: 10.1159/000491730
141. Zhao K, Zhou Y, Qiao C, Ni T, Li Z, Wang X, et al. Oroxylin A promotes PTEN-mediated negative regulation of MDM2 transcription via SIRT3-mediated deacetylation to stabilize p53 and inhibit glycolysis in wt-p53 cancer cells. J Hematol Oncol. (2015) 8:41. doi: 10.1186/s13045–015-0137–1
142. Xiong Y, Wang L, Wang S, Wang M, Zhao J, Zhang Z, et al. SIRT3 deacetylates and promotes degradation of P53 in PTEN-defective non-small cell lung cancer. J Cancer Res Clin Oncol. (2018) 144:189–98. doi: 10.1007/s00432–017-2537–9
143. Zhu B, Yan Y, Shao B, Tian L, Zhou W. Downregulation of SIRT6 is associated with poor prognosis in patients with non-small cell lung cancer. J Int Med Res. (2018) 46:1517–27. doi: 10.1177/0300060517750298
144. Dong G, Wang B, An Y, Li J, Wang X, Jia J, et al. SIRT1 suppresses the migration and invasion of gastric cancer by regulating ARHGAP5 expression. Cell Death Dis. (2018) 9:977. doi: 10.1038/s41419–018-1033–8
145. Shen X, Li P, Xu Y, Chen X, Sun H, Zhao Y, et al. Association of sirtuins with clinicopathological parameters and overall survival in gastric cancer. Oncotarget. (2017) 8:74359–70. doi: 10.18632/oncotarget.20799
146. Li L, Zhang HN, Chen HZ, Gao P, Zhu LH, Li HL, et al. SIRT1 acts as a modulator of neointima formation following vascular injury in mice. Circ Res. (2011) 108:1180–9. doi: 10.1161/circresaha.110.237875
147. Jing H, Hu J, He B, Negrón Abril YL, Stupinski J, Weiser K, et al. A SIRT2-selective inhibitor promotes c-myc oncoprotein degradation and exhibits broad anticancer activity. Cancer Cell. (2016) 29:297–310. doi: 10.1016/j.ccell.2016.02.007
148. Li Y, Zhang M, Dorfman RG, Pan Y, Tang D, Xu L, et al. SIRT2 promotes the migration and invasion of gastric cancer through RAS/ERK/JNK/MMP-9 pathway by increasing PEPCK1-related metabolism. Neoplasia (New York NY). (2018) 20:745–56. doi: 10.1016/j.neo.2018.03.008
149. Fernández-Coto DL, Gil J, Hernández A, Herrera-Goepfert R, Castro-Romero I, Hernández-Márquez E, et al. Quantitative proteomics reveals proteins involved in the progression from non-cancerous lesions to gastric cancer. J Proteomics. (2018) 186:15–27. doi: 10.1016/j.jprot.2018.07.013
150. Lu X, Yang P, Zhao X, Jiang M, Hu S, Ouyang Y, et al. OGDH mediates the inhibition of SIRT5 on cell proliferation and migration of gastric cancer. Exp Cell Res. (2019) 382:111483. doi: 10.1016/j.yexcr.2019.06.028
151. Haigis MC, Deng CX, Finley LW, Kim HS, Gius D. SIRT3 is a mitochondrial tumor suppressor: a scientific tale that connects aberrant cellular ROS, the Warburg effect, and carcinogenesis. Cancer Res. (2012) 72:2468–72. doi: 10.1158/0008–5472.Can-11–3633
152. Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene. (2011) 30:2986–96. doi: 10.1038/onc.2011.37
153. Yang B, Fu X, Shao L, Ding Y, Zeng D. Aberrant expression of SIRT3 is conversely correlated with the progression and prognosis of human gastric cancer. Biochem Biophys Res Commun. (2014) 443:156–60. doi: 10.1016/j.bbrc.2013.11.068
154. Sasca D, Hähnel PS, Szybinski J, Khawaja K, Kriege O, Pante SV, et al. SIRT1 prevents genotoxic stress-induced p53 activation in acute myeloid leukemia. Blood. (2014) 124:121–33. doi: 10.1182/blood-2013–11-538819
155. Sayd S, Thirant C, El-Habr EA, Lipecka J, Dubois LG, Bogeas A, et al. Sirtuin-2 activity is required for glioma stem cell proliferation arrest but not necrosis induced by resveratrol. Stem Cell Rev Rep. (2014) 10:103–13. doi: 10.1007/s12015–013-9465–0
156. Qin J, Liu Y, Lu Y, Liu M, Li M, Li J, et al. Hypoxia-inducible factor 1 alpha promotes cancer stem cells-like properties in human ovarian cancer cells by upregulating SIRT1 expression. Sci Rep. (2017) 7:10592. doi: 10.1038/s41598–017-09244–8
157. Zuo Q, Wu W, Li X, Zhao L, Chen W. HDAC6 and SIRT2 promote bladder cancer cell migration and invasion by targeting cortactin. Oncol Rep. (2012) 27:819–24. doi: 10.3892/or.2011.1553
158. Yang Y, Hou H, Haller EM, Nicosia SV, Bai W. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J. (2005) 24:1021–32. doi: 10.1038/sj.emboj.7600570
159. Ming M, Qiang L, Zhao B, He YY. Mammalian SIRT2 inhibits keratin 19 expression and is a tumor suppressor in skin. Exp Dermatol. (2014) 23:207–9. doi: 10.1111/exd.12323
160. Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, et al. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys Res Commun. (2003) 309:558–66. doi: 10.1016/j.bbrc.2003.08.029
161. Hussain AR, Uddin S, Bu R, Khan OS, Ahmed SO, Ahmed M, et al. Resveratrol suppresses constitutive activation of AKT via generation of ROS and induces apoptosis in diffuse large B cell lymphoma cell lines. PloS One. (2011) 6:e24703. doi: 10.1371/journal.pone.0024703
162. Chai R, Fu H, Zheng Z, Liu T, Ji S, Li G. Resveratrol inhibits proliferation and migration through SIRT1 mediated post-translational modification of PI3K/AKT signaling in hepatocellular carcinoma cells. Mol Med Rep. (2017) 16:8037–44. doi: 10.3892/mmr.2017.7612
163. Hu A, Huang J-J, Li R-L, Lu Z-Y, Duan J-L, Xu W-H, et al. Curcumin as therapeutics for the treatment of head and neck squamous cell carcinoma by activating SIRT1. Sci Rep. (2015) 5:13429. doi: 10.1038/srep13429
164. Speck U, Häckel A, Schellenberger E, Kamann S, Löchel M, Clever YP, et al. Drug distribution and basic pharmacology of paclitaxel/resveratrol-coated balloon catheters. Cardiovasc Interventional Radiol. (2018) 41:1599–610. doi: 10.1007/s00270–018-2018–9
165. Jung YR, Kim EJ, Choi HJ, Park JJ, Kim HS, Lee YJ, et al. Aspirin targets SIRT1 and AMPK to induce senescence of colorectal carcinoma cells. Mol Pharmacol. (2015) 88:708–19. doi: 10.1124/mol.115.098616
166. Park EY, Woo Y, Kim SJ, Kim DH, Lee EK, De U, et al. Anticancer Effects of a New SIRT Inhibitor, MHY2256, against Human Breast Cancer MCF-7 Cells via Regulation of MDM2-p53 Binding. Int J Biol Sci. (2016) 12:1555–67. doi: 10.7150/ijbs.13833
167. Wang T, Li X, Sun SL. EX527, a Sirt-1 inhibitor, induces apoptosis in glioma via activating the p53 signaling pathway. Anti-cancer Drugs. (2020) 31:19–26. doi: 10.1097/cad.0000000000000824
168. Luthi-Carter R, Taylor DM, Pallos J, Lambert E, Amore A, Parker A, et al. SIRT2 inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proc Natl Acad Sci USA. (2010) 107:7927–32. doi: 10.1073/pnas.1002924107
169. Peck B, Chen CY, Ho KK, Di Fruscia P, Myatt SS, Coombes RC, et al. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol Cancer Ther. (2010) 9:844–55. doi: 10.1158/1535–7163.Mct-09–0971
170. Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. (2010) 17:193–9. doi: 10.1038/cdd.2009.56
171. Fujita Y, Kojima K, Hamada N, Ohhashi R, Akao Y, Nozawa Y, et al. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem Biophys Res Commun. (2008) 377:114–9. doi: 10.1016/j.bbrc.2008.09.086
172. Jiang G, Wen L, Zheng H, Jian Z, Deng W. miR-204–5p targeting SIRT1 regulates hepatocellular carcinoma progression. Cell Biochem Funct. (2016) 34:505–10. doi: 10.1002/cbf.3223
173. Chen H, Lu Q, Fei X, Shen L, Jiang D, Dai D. miR-22 inhibits the proliferation, motility, and invasion of human glioblastoma cells by directly targeting SIRT1. Tumor Biol. (2015) 37:6761–8. doi: 10.1007/s13277–015-4575–8
174. Deng S, Zhu S, Wang B, Li X, Liu Y, Qin Q, et al. Chronic pancreatitis and pancreatic cancer demonstrate active epithelial-mesenchymal transition profile, regulated by miR-217-SIRT1 pathway. Cancer Lett. (2014) 355:184–91. doi: 10.1016/j.canlet.2014.08.007
Keywords: sirtuins, histone deacetylase, metabolic functions, tumor, cancer therapy
Citation: Yu L, Li Y, Song S, Zhang Y, Wang Y, Wang H, Yang Z and Wang Y (2024) The dual role of sirtuins in cancer: biological functions and implications. Front. Oncol. 14:1384928. doi: 10.3389/fonc.2024.1384928
Received: 11 February 2024; Accepted: 30 May 2024;
Published: 14 June 2024.
Edited by:
Anurag Kumar Singh, Martin Luther University of Halle-Wittenberg, GermanyReviewed by:
Sabita Saldanha, Alabama State University, United StatesCopyright © 2024 Yu, Li, Song, Zhang, Wang, Wang, Yang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Wang, d195aTIwMjJAMTYzLmNvbQ==; Zhengteng Yang, MzMwNTYwMTY3QHFxLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.