
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 03 April 2024
Sec. Molecular and Cellular Oncology
Volume 14 - 2024 | https://doi.org/10.3389/fonc.2024.1381467
The Keap1-Nrf2 signaling pathway is a major regulator of the cytoprotective response, participating in endogenous and exogenous stress caused by ROS (reactive oxygen species). Nrf2 is the core of this pathway. We summarized the literature on Keap1-Nrf2 signaling pathway and summarized the following three aspects: structure, function pathway, and cancer and clinical application status. This signaling pathway is similar to a double-edged sword: on the one hand, Nrf2 activity can protect cells from oxidative and electrophilic stress; on the other hand, increasing Nrf2 activity can enhance the survival and proliferation of cancer cells. Notably, oxidative stress is also considered a marker of cancer in humans. Keap1-Nrf2 signaling pathway, as a typical antioxidant stress pathway, is abnormal in a variety of human malignant tumor diseases (such as lung cancer, liver cancer, and thyroid cancer). In recent years, research on the Keap1-Nrf2 signaling pathway has become increasingly in-depth and detailed. Therefore, it is of great significance for cancer prevention and treatment to explore the molecular mechanism of the occurrence and development of this pathway.
NRF2 (nuclear factor erythroid 2-related factor 2) is tightly regulated by different mechanisms at transcriptional, epigenetic, or ARE‐binding level; however, its key regulation is interceded by proteasome degradation mainly mediated by the repressor protein Kelch‐like ECH‐associated protein 1 (Keap1) (1). There are differences in the number of cysteine residues between human- and mouse-derived Keap1. There are 25 cysteine residues in mice and 27 in human-derived Keap1. Most can be modified by different oxidants and electrophilic reagents in vitro (2). Nrf2 is an important redox-sensitive transcription factor that is conducive to improving the oxidative stress state of the body, promoting cell survival, and maintaining the redox homeostasis of cells by inducing and regulating the constitutive and inducible expression of phase II detoxification enzymes and antioxidant enzymes in cells (3–5). The Keap1-Nrf2 signaling pathway is a major regulator of the cell protection response, which is involved in endogenous and exogenous stress caused by reactive oxygen species (ROS) (6). This pathway is regulated with Nrf2 as the core. First, Nrf2 and small Maf protein bind to an ARE (antioxidant response element) in the regulatory region of the target gene. Second, Nrf2 binds with Keap1 to promote its degradation through the ubiquitin proteasome pathway (7). At the same time, this pathway also acts as a double-edged sword: Nrf2 activity protects cells against oxidative and electrophilic stress, while increasing Nrf2 activity contributes to cancer cell survival and proliferation.
In view of these remarkable findings, research on the Keap1-Nrf2 signaling pathway has become increasingly in-depth and detailed in recent years. Therefore, it is of great significance to constantly explore the molecular mechanism of the occurrence and development of this pathway for cancer prevention and treatment. In this review, the key molecular mechanisms of the Keap1-Nrf2 signaling pathway in carcinogenesis and development are summarized.
Keap1 was found and reported in 1999, and it belongs to the BTB Kelch protein family (8, 9). Keap1 is a negative regulator of Nrf2, which mainly binds with it in the cytoplasm to form a homodimer. Under normal circumstances, Keap1 interacts with Cullin3 (Cul3) and Rbx1 to ubiquitinate the Nrf2 protein and induce the proteasome to degrade it, thus preventing Nrf2 from being translocated to the nucleus and binding to the ARE site in DNA (10). The protein molecular weight of Keap1 is 69 kDa, which is located at position 19q13.2 of the human chromosome. Under normal circumstances, it is anchored to the actin skeleton in the cytoplasm.
Keap1 is mainly composed of the following five domains (Figure 1A): NTR domain (1–49 amino acid residues), BTB domain (50–179 amino acid residues) that can interact with Cul3, IVR domain (180–314 amino acid residues), six repetitive DGR domains (315–598 amino acid residues), and the final CTR domain (599–624 amino acid residues) (11, 12). The details are as follows: the BTB domain, also known as the POZ domain, has diverse functions and can participate in the polymer formation process of the Keap1 protein; for example, it can mediate the mutual recognition of Keap1 homodimers in the cytoplasm and bind to the E3 ubiquitination ligase complex dependent on Cul3 (13). At C151, a cysteine residue was found that is necessary for Keap1 to reduce E3 activity under electrophilic stimulation (14). The IVR domain (also known as the BACK domain) connects the BTB domain with the Kelch/DGR domain on the C-terminal side. Because this domain is rich in cysteine amino acid residues, it can regulate the activity of Keap1 protein. At the same time, Keap1 can interact with the 3-box double helix motif region near the N-terminal of Cul3 through the IVR domain (15). The Kelch/DGR domain of Keap1 is composed of six repetitive Kelch sequences, which contain tyrosine, tryptophan, diglycine, and other repetitive conserved amino acid residues; Keap1 has a homodimer that can interact with ETGE (high affinity) and DLG (low affinity) motifs in the Neh2 domain of the Nrf2 protein (16).
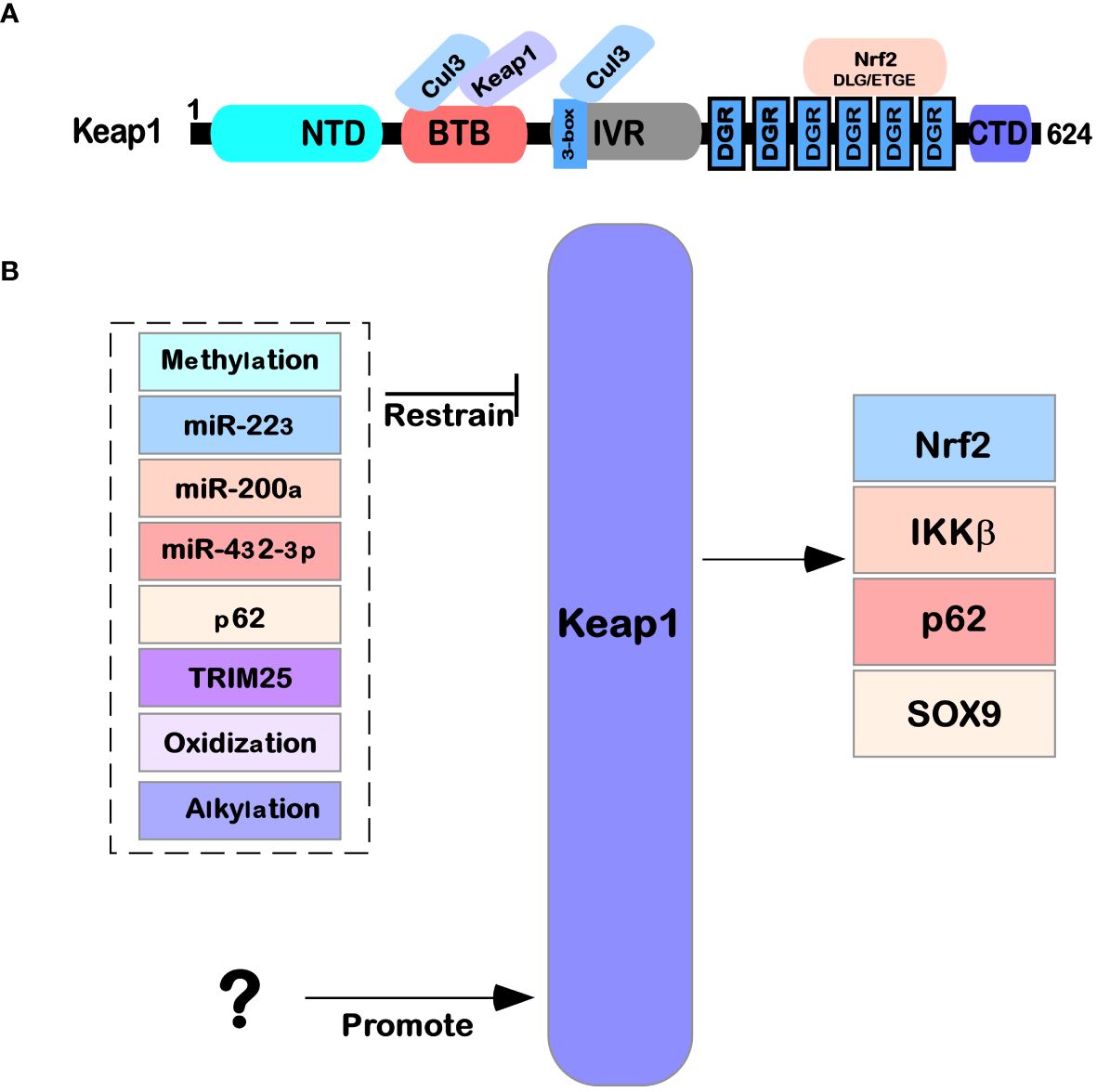
Figure 1 Structure and function of Keap1. (A) The protein structure of Keap1 and the functions of its domains. (B) The regulatory network of Keap1. Several microRNAs (miR-223, miR-200a, and miR-432-39) also affect Keap1 translation levels. The Keap1 protein is also regulated by various modifications (such as methylation, oxidation, glycosylation, and alkylation) after translation. At the same time, it is also affected by other factors (such as P62 and TRIM25) that regulate its expression.
By analyzing the RNA-seq data of 27 different human tissues (from NCBI), it was found that Keap1 was distributed in most human tissues and organs, such as the brain, adrenal gland, bone marrow, gallbladder, kidney, liver, spleen, and pancreas. Interestingly, the expression level of Keap1 RNA in different organs and tissues is quite different. For example, the expression level of Keap1 is high in the brain, kidney, and prostate but low in the bone marrow, pancreas, and salivary gland. The above data come from the National Center for Biotechnology Information (17).
Keap1 is part of a ubiquitin ligase (cul3-rbx1 E3) complex that recruits pgam5, Nrf2, SLK, IKK β, P62, Sox9, Bcl-2 MIRO2, MAD2L1, and MYO9B, which are ubiquitinated and degraded and are involved in the regulation of multiple signaling pathways in cells (18) (Figure 1B). For example, in the Keap1-Nrf2 pathway in oxidative stress and metabolic processes, Nrf2 often causes corresponding case changes after aberrant expression occurs (19, 20). An increasing number of studies have also been used to demonstrate that Keap1 is a shuttling competent protein, i.e., shuttling back and forth in the nucleus and cytosol under specific conditions, whereas a nuclear export signal (leucine) is found in Keap1, and in large amounts of the nuclear protein prothymosin α that binds with Keap1, revealing that Keap1 is able to translocate from the cytoplasm into the nucleus (21). Later in the state of oxidative stress, Keap1 is also able to enter the nucleus and bind with Nrf2 again, allowing Nrf2 to translocate from the nucleus to the cytoplasm again and thus allowing Nrf2, under the mediation of Keap1, to be degraded by ubiquitination (22–25). Keap1 can act as an E3 ubiquitination ligase for p62 (a.k.a. SQSTM1), allowing p62 to be ubiquitinated for degradation and reducing cell death in disease (26) (Figure 2). Keap1 can also function as an IKK β, the role of E3 ligases that inhibit NF-κB expression of the pathway, which in turn inhibits cancer initiation (18).
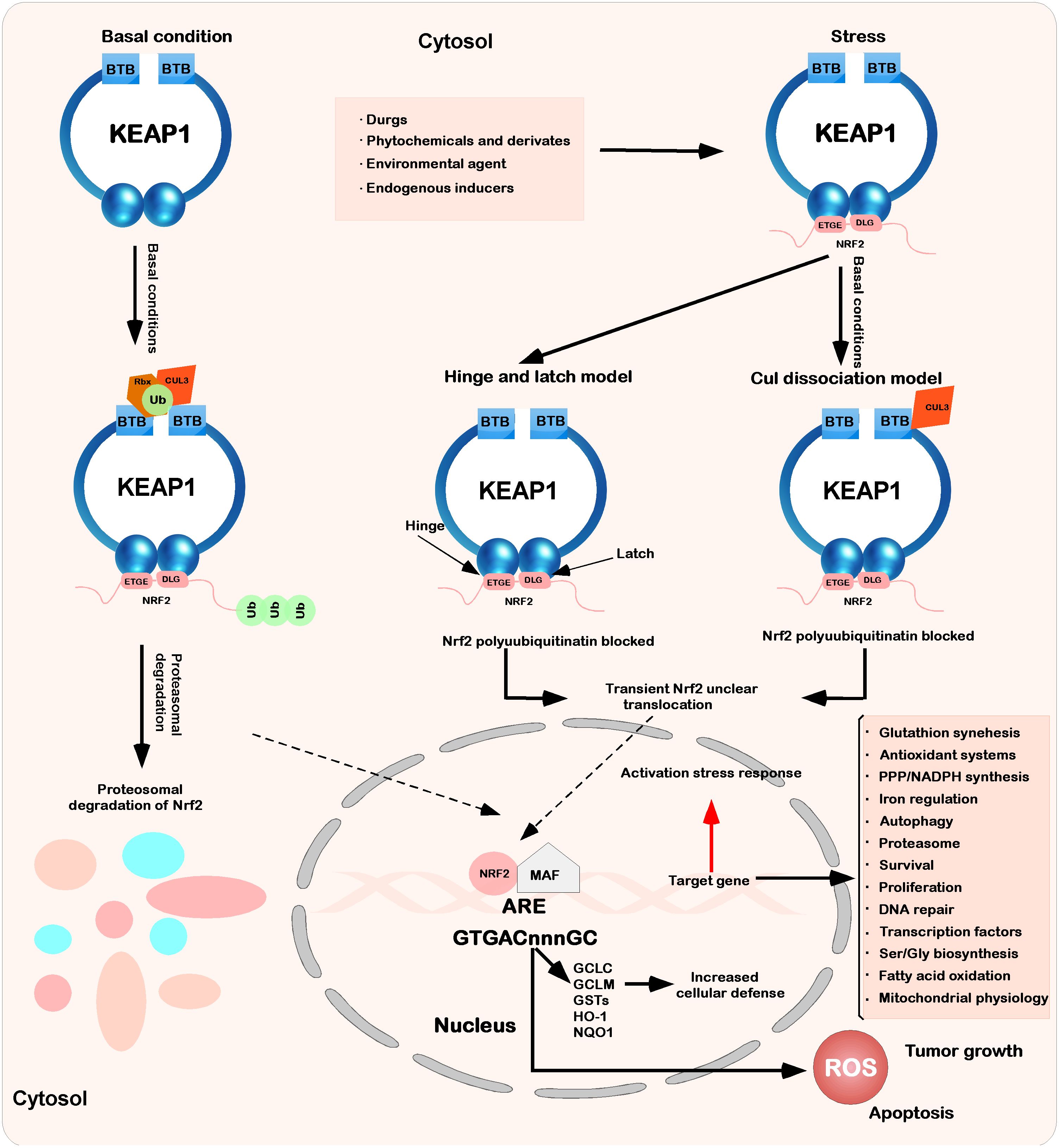
Figure 2 Interaction between Keap1 and Nrf2. Under basic conditions, Keap1 binds to Nrf2 through ETGE and DLEG, and Nrf2 is polyadenylated by the cul3-based E3 ligase complex. This polygeneralization leads to the rapid degradation of Nrf2 by the proteasome. At the same time, a small amount of Nrf2 escapes from the inhibition complex and reaggregates in the nucleus, mediating the expression of basic ARE-dependent genes and thus maintaining intracellular homeostasis. When stimulated by the outside world (drugs, phytochemicals and devivates, environmental agents, and endogenous inducers), the inducer modifies Keap1 cysteine and inhibits Nrf2 ubiquitination by dissociating the inhibition complex. According to the hinge and latch model, the modification of specific Keap1 cysteine residues leads to the conformational change of Keap1, leading to the separation of the Nrf2 DLG motif from Keap1. The ubiquitination of Nrf2 is destroyed, but binding to the ETGE motif still occurs. At the same time, in another model (Keap1-Cul3 dissociation model), the binding of Keap1 and Cul3 is destroyed under the action of electrophilic reagents, which leads to the escape of Nrf2 from the ubiquitination system. In these two models of Keap1-Nrf2, both will induce modification and inactivate Keap1, which will bind Nrf2. Therefore, the newly synthesized Nrf2 protein bypasses Keap1 and enters the nucleus, binds to the antioxidant response element (ARE), and drives the expression of the Nrf2 target genes GCLC, GCLM, NQO1, HO-1, and GST. At the same time, it will also affect other processes (glutathione synthesis, antioxidant systems, PPP/NADPH synthesis, iron regulation, etc.).
It has been shown that Keap1 is not only a tumor suppressor but also a prooncogenic protein. In terms of disease initiation and progression, Keap1 has been implicated in several diseases, such as kidney disease, liver disease, inflammatory disease, sarcopenia, ophthalmic disease, neurodegenerative disease, cardiovascular disease, and ischemia/reperfusion injury. In some critical diseases (e.g., cancer), Keap1 has also been found to be somatically mutated, resulting in deregulation of its function in mediating ubiquitination, leading to cancer initiation and malignant progression. For example, somatic mutation of Keap1 in lung cancer, causing an increase in Nrf2 protein expression levels, leads to lung cancer initiation and progression. In addition, somatically acquired mutations have also been found in a variety of human cancer tissues, including head and neck (42%), ovarian (37%), gallbladder (30.7%), gastric (11.1%), colorectal (7.8%), clear renal cell (4.7%), liver (2.8%), prostate (1.3%), and glioma (1.7%), leading to the development of cancer (27–30). Keap1 mutations in the somatic fraction are shown in Table 1.
Several reports have indicated that the regulation of Keap1 mainly focuses on the transcription level, protein translation level (e.g., the regulation of miRNAs), and posttranslational modification clipping processing (e.g., oxidative modification, glycosylation modification, and alkylation modification), as shown in Figure 1B.
Keap1, at the transcriptional level, is directly regulated by methylated promoter regions (CpG islands). For example, in prostate cancers (39), non-small cell lung cancers (42), breast cancer (43), and colon cancers (44), where CpG islands act as Keap1 promoter regions, there is high methyl florescence and low expression.
Some microRNAs (also known as microRNAs, miRNAs) have been reported to be regulated at the level of Keap1 protein translation. For example, transfection of miR-223 in HepG2 cells decreased the level of Keap1 protein expression, and transfection of its inhibitor significantly increased the level of Keap1 protein expression; the results showed that miR-223 was able to negatively regulate the protein expression of Keap1 in the cells (45). Another mic RNA, mir-200a, with low expression under fructose induction, activated the expression of Keap1, reduced the antioxidant capacity of the Keap1-Nrf2 pathway, enhanced cellular ROS, and activated the expression of lactamase (NLRP3), resulting in oxidative stress and lipid accumulation in cells, while the use of polydatin, which could effectively enhance the expression level of mir-200a, activated the antioxidant activity of the Keap1-Nrf2 pathway and could serve as one target site for the treatment of fructose-induced related disorders (e.g., liver injury and lipid deposition-like disorders) (46, 47). In addition, mir-432-3p has also been found to inhibit Keap1 expression in ESCC (esophageal squamous cell carcinoma), which in turn regulates the antioxidant activity of the Keap1-Nrf2 pathway (48–50).
Under unstimulated conditions, redox reactions are in stable equilibrium in living cells; however, multiple stress responses are elicited in cells after redox stabilization is disrupted, and this condition is thought to be an important contributor to the development of numerous diseases. In regulating the balance between oxidation and reduction, EPS (Epalrestat), an electrophile, is used to activate relevant defenses against oxidative stress. For example, in the Keap1-Nrf2 are transcriptional pathway, which can be activated by CA (carnosic acid) and CS (carnosol) found in rosemary to rapidly synthesize endogenous antioxidant phase 2 enzymes. Notably, CA and CS are electrophilic only after oxidation and themselves belong to non-electrophilic species (51). In COPD (chronic obstructive pulmonary disease) patients, it was observed that Keap1 changed its conformation due to a long-term stimulation by free radicals or other chemicals in tobacco, leading to abnormal expression of the Keap1-Nrf2 pathway.
O-GlcNAc (O-linked N-acetylglucosamine) is a dynamic posttranslational modification (PTM) that reversibly modifies serine and threonine residues of thousands of nuclear, cytoplasmic, and mitochondrial proteins. It has been documented that the glycosylation (o-GlcNAcylation) modification of Keap1 at S104 is able to regulate the ubiquitination of Nrf2 and proteasomal destruction, and glycosylation at this site is not required for Keap1 to form a dimer. Meanwhile, o-GlcNAcylation at this site can also further optimize the conformation of Keap1 and promote ubiquitination of Keap1 substrates in a manner that enhances keap1-cul3 binding ability (50, 52).
Alkylation of one or more of the 27 cysteine sulfhydryls of human Keap1 has been reported to result in ubiquitination or proteasome-mediated reduction of its substrates. However, alkylation of Keap1 can also occur in the presence of some electrophilic compounds (e.g., quinone methides, carbenium ions, epoxides, quinones, and quinoneimines, among others) (53). Notably, xanthohumol, as a natural compound, is also able to alkylate Keap1 (54, 55). Another endogenous metabolite is itaconate, which has anti-inflammatory metabolic functions and alkylates some of the Cys residues (e.g., C151, c257, c273, c288, and c297, of which C151 has the strongest electrophilic activity) on the Keap1 protein, enhancing the expression of its antioxidant and anti-inflammatory related downstream genes (56–59). The in-depth study of substances such as xanthohumol and itaconic acid may provide a new approach to the pathogenesis of related diseases caused by Keap1.
The Nrf2 transcription factor was first identified in 1994 in a human chronic myelogenous leukemia cell line by MOI et al. and was later determined to be located on human chromosome 2q31.2 (60). Nrf2, also named nfe2l2 (nf-e2-like 2), encodes a total of 605 amino acids and is composed of seven different domains, followed from the N-terminus to the C-terminus by Neh2, Neh4, Neh5, Neh7, Neh6, Neh1, and Neh3 (20, 61–63) (Figure 3A). The functions of each domain are specific and indispensable (64–66).
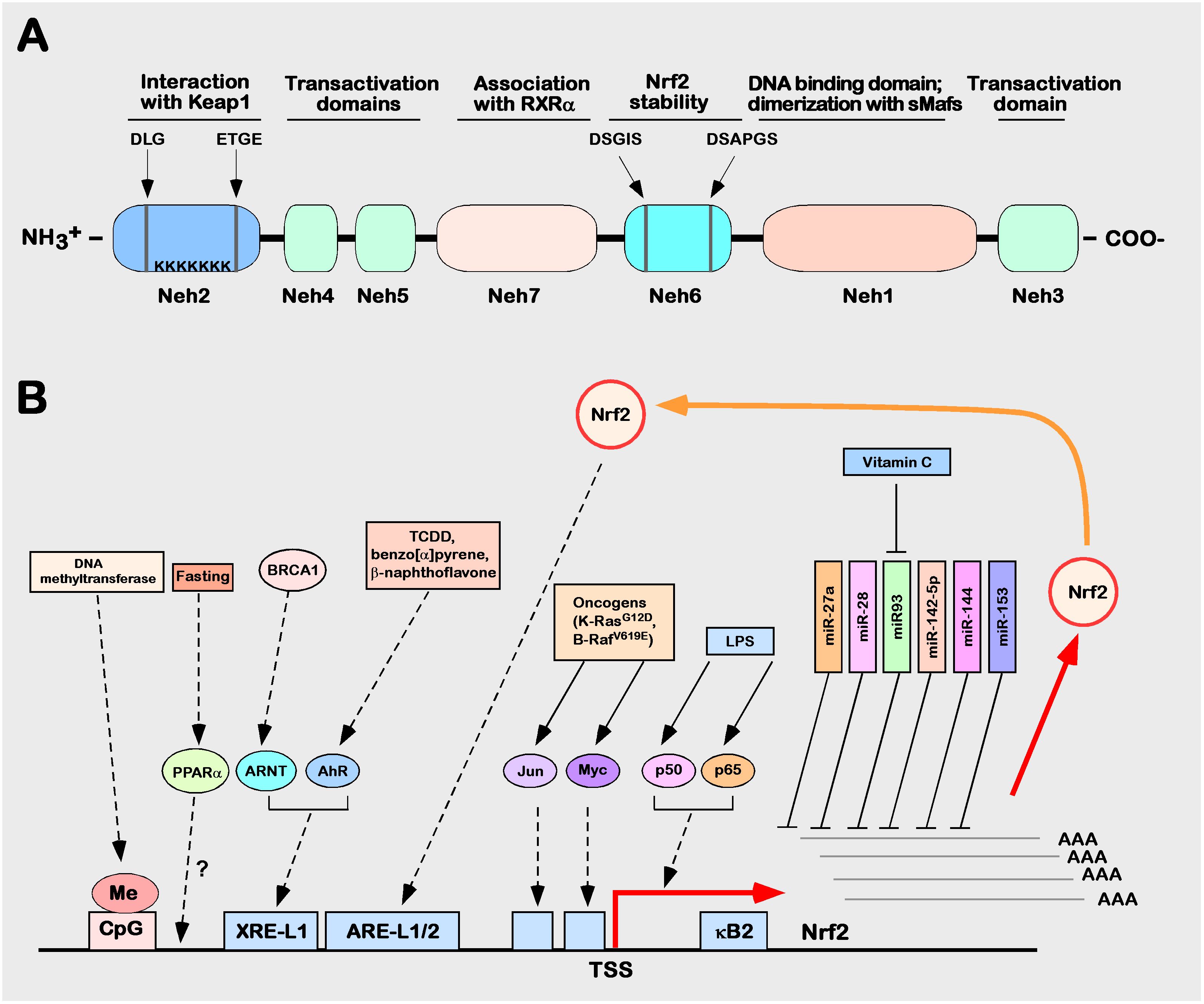
Figure 3 Structure and function of Nrf2. (A) Schematic diagram of the Nrf2 protein structure. (B) Schematic diagram of the regulation of human Nrf2 gene expression. The control mechanism of nuclear factor erythroid 2p45 related factor 2 (Nrf2) gene expression. The Nrf2 gene is depicted as the bottom of a solid black horizontal line graph, and the red right angle arrow represents the transcription start site (TSS). Breast cancer protein (BRCA) 1 increases the expression of Nrf2, which is mediated by ARNT. Lipopolysaccharide (LPS), as a factor promoting inflammation, can induce Nrf2 to recruit TSS (kB2) from the p50–p65 heterodimer through the nuclear factor (NF)-kB binding site. In the process of tumorigenesis and development, Nrf2 can be activated by many factors (such as Jun or Myc). Fasting increases the mRNA expression level of Nrf2, which may be mediated by peroxisome proliferator activation (PPAR). Many miRNAs, such as miR-27a, miR-28, miR-93, miR-142-5p, miR-144, and miR-153, inhibit the expression of Nrf2.
The Neh1 domain, which consists of the conserved CNC and bZIP domains, is critical for Nrf2 binding SMAF proteins in the nucleus to form dimers that recognize DNA sequences of target genes (67, 68). The Neh2 domain is located in the N-terminus of Nrf2 and contains two stretches of highly conserved amino acid sequences (29DLG31 sequence and 79ETGE82 sequence), which are able to bind with Nrf2’s inhibitor protein Keap1 to mediate Nrf2 degradation through the ubiquitination proteasome system (8, 9, 69). The C-terminal neh3 domain, neh4 domain, and neh5 domain are important domains for Nrf2 to exert transcriptional regulation of target gene activity (70–72). The neh6 domain is a serine-rich domain that contains two conserved amino acid motifs (343dsgis347 sequence and 382dsapgs387 sequence) that can be recognized by GSK-3 α/β-Trcp recognition, and deletion or mutation of either of these two sequences reduces β-Trcp-mediated ubiquitination, i.e., this domain is a critical negative regulatory domain that mediates proteasomal degradation of Nrf2 ubiquitination (73, 74). The neh7 domain was only formally defined in 2013 as a domain with a negative regulatory function through the retinal X receptor α (retinoic X receptor α, RXR α) interacting with this region and thereby repressing Nrf2 transcriptional activity (75).
Analysis of Nrf2-related data (transcriptome and proteome data) in the HPA (Human Protein Atlas) database revealed its distribution in most human tissues and organs. Nrf2 expression has been detected in various organs, such as the brain, lung, kidney, liver, male and female reproductive organs, lymphoid tissue gallbladder, and muscle tissue, and in relevant cells, both in the cytoplasm and nucleus. It is worth noting that the expression levels of Nrf2 vary in different tissues or cells due to their functional and structural differences, while it is possible that there are some differences in the expression of protein and RNA levels in the same tissue or corresponding cells. In the brain, bone marrow, gastrointestinal, reproductive, and lymphoid tissues, the protein expression level of Nrf2 is relatively higher than that in other tissues, whereas the RNA expression level of Nrf2 is higher in esophageal tissues than in other tissues. The above data come from the National Center for Biotechnology Information (17).
The related regulation of Nrf2 expression and activity can occur at the transcriptional level, mechanistically through miRNA-mediated regulation and through translational and posttranslational modifications. Because Nrf2 is a soluble protein, its regulation mainly occurs at the protein level, including protein−protein and posttranslational modification level regulation (76).
Nrf2 can be activated or repressed at the transcriptional level through its own or other transcription factors; binding to ARE and XRE sites, such as ppar γ (peroxisome proliferator activated receptor γ) (77), MEF2D (myocyte enhancer factor 2D) (78), and AHR (aryl hydrocarbon receptor) (79), can directly activate Nrf2 expression, while p53 (79), p97 (80), and RXR α (81), among others, can negatively regulate Nrf2. Several miRNAs have been reported to regulate Nrf2 expression at the posttranscriptional level, such as mir27a, mir28, mir-93, mir-142-5p, mir-144, and mir-153. They are able to bind at the three-terminal noncoding region in the mRNA of Nrf2 to inhibit the expression of Nrf2 (76). Five CpG sequences are included in the promoter region of Nrf2, which, after hypermethylation modification occurs, can significantly inhibit Nrf2 expression (82). The mechanisms of the regulation of human Nrf2 gene expression are illustrated (Figure 3B).
The activity regulation of Nrf2 protein can be regulated by several pathways; three pathways are cytoplasmic pathway regulation, endoplasmic reticulum pathway regulation, and nuclear pathway regulation. First, under normal circumstances, Keap1, in a homodimeric manner, recognizes DLG and ETGE sequences in the Neh2 domain of Nrf2 and anchors it in the cytoplasm; meanwhile, the N-terminus of the Keap1 protein recognizes and binds to Cul3, leading to rapid degradation by the proteasome after ubiquitination of Nrf2 (82, 83). There are also studies indicating that Nrf2 in the nucleus undergoes acetylation, leading to its binding to the alkaline region leucine zipper protein to antioxidant response elements, thereby triggering gene transcription (84). Second, in the regulation of ER stress, Nrf2 expression is suppressed through the transcriptional activation of the XBP1-HRD1 arm and the action of E3 ubiquitination-linked enzymes (85–87). Micro-RNAs (miRNA) can serve as a very powerful epigenetic regulator of Nrf2 (88). Third, Nrf2 is a major transcription factor that directly or indirectly significantly regulates over 2,000 genes. Although many of these genes are involved in maintaining redox balance, others are involved in maintaining balance between metabolic pathways that seem unrelated to oxidative stress (89). Nuclear regulation occurs through regulation by β-TrCP. It has been reported that in GSK3 β, following the sequential phosphorylation of dsgis (in the neh6 domain) of Nrf2, β-Trcp is activated; increasing the association of Nrf2 with the interaction force between β-TrCP accelerates the degradation of Nrf2 (73, 90, 91).
Oxidative stress plays a key role in the pathogenesis of various human cancers (92, 93). Therefore, in some clinical studies, oxidative stress-related reactions have also been used to determine markers of human cancer (94). The Keap1-Nrf2 signaling pathway can prevent organ and cell damage caused by oxidative stress and protects against the occurrence and development of cancer (95). Because oxidative damage is common in carcinogenesis, the Keap1-Nrf2 signaling pathway is widely considered a potential therapeutic target for chemoprevention (93). The inducers of Nrf2 can play the role of chemopreventive agents in the following two ways: first, by preventing carcinogens from reaching their target sites, and second, by preventing carcinogens from interacting with important biological molecules (such as DNA and RNA) and proteins to play the role of chemopreventive agents (93). Although Nrf2 has chemopreventive potential in normal and precancerous tissues, it has also been shown to play a role in tumor cell growth and survival in malignant cells (7, 96). High levels of Nrf2 have been found in several types of human cancer cells. Mutations in Keap1 or Nrf2 lead to the constitutive expression of upregulated genes (97–99). The increased expression of Nrf2 can play a protective role in both normal and cancer cells. The increase in Nrf2 expression levels can lead to an increase in the expression of detoxification enzymes, cytoprotective proteins, and transporters. This allows cancer cells to gain advantages by enhancing cell proliferation and can cause drug resistance to chemotherapy (7, 65, 66, 96–100). Previous studies have shown that inhibiting Nrf2 in malignant cells can inhibit tumor growth and improve the efficacy of chemotherapy (100–102). After interfering with the PI3K/AKT and ERK pathways through natural flavonoids, Nrf2 was reduced at the mRNA and protein levels, making hepatoma cells sensitive to chemotherapy (103, 104). Interestingly, dimethyl fumarate, approved by the FDA as an Nrf2 activator, shows some anticancer activity. In fact, it seems to be an Nrf2 inhibitor at high concentrations (105). The p62 interacting with KEAP1 shows a good effect in HCC by downregulating Nrf2 activation (103).
The Keap1-Nrf2 signaling pathway is involved in both benign and malignant tumor diseases and may be used as a prognostic marker or therapeutic target. Nrf2 has also been shown to have an impact on the drug resistance of cancer (such as lung cancer, liver cancer, and thyroid cancer) to chemotherapy and radiotherapy (106–109). Therefore, in clinical trials, the expected effects of prevention and treatment can be achieved by targeting other components of the Keap1-Nrf2 pathway and its downstream signaling pathway. Clinical studies have shown that the mutation frequency of Keap1 and Nrf2 is approximately 25% in lung cancer patients. The prognosis of lung cancer patients with Keap1 or Nrf2 mutations is worse than that of lung cancer patients without this mutation (99). In addition, some studies have shown that the decrease in the expression level of Keap1 and the increase in the expression level of Nrf2 may also be related to poor prognosis. In general, Nrf2 is believed to contribute to both intrinsic and acquired resistance (110, 111). Nrf2-targeted genes involved in foreign biological metabolism can accelerate the metabolic inactivation of antitumor drugs; genes involved in drug transport can effectively reduce the intracellular drug concentration, and genes involved in thiosulfur synthesis can increase the drug tolerance of tumor cells. These multiple mechanisms together lead to chemotherapy resistance, which is one of the most important carcinogenic functions of Nrf2. In the process of thyroid cancer treatment, there are few alternative drugs (7). Proteasome inhibitors are a substitute for targeted anticancer drugs used in clinical thyroid cancer. Proteasome inhibition usually also leads to Nrf2 activation (112). The mechanism by which Nrf2 promotes thyroid cancer proteasome inhibitor resistance is not limited to the interaction with apoptosis regulators (ATF4, ORP150, etc.) but also includes direct regulation of cell redox status. Nrf2 not only promotes resistance to proteasome inhibitors but also promotes resistance to other experimental therapies (109, 113, 114). Nrf2 is upregulated in head and neck squamous cell carcinoma (HNSCC). Nrf2 reprograms a wide range of cancer metabolic pathways, and the most notable is the pentose phosphate pathway (PPP) (115). In cervical cancer, Nrf2 can activate EMT-related behaviors and promote cancer metastasis (116). There are also studies indicating that Nrf2 acts as a phenotypic stability factor in restricting complete EMT and plays an important role in coordinating collective cancer migration (117). In summary, a better understanding of the relationship between the activation of the Keap1-Nrf2 signaling pathway in cancer and the overall therapeutic effect, and the mode of interaction and the therapeutic relevance of this interaction, will help to further develop therapeutic drugs.
In the Keap1-Nrf2 signaling pathway, newly synthesized Nrf2 protein bypasses Keap1, translocates into the nucleus, and drives the expression of Nrf2 target genes, such as HO-1, NQO1, GCLC, GCLM, and GSTs (7). In Figure 4, five common molecular mechanisms of Nrf2 signaling activation in various cancers have been described (39, 118–123): (1) they are lost after Keap1 has been mutated in cells or Nrf2 has disrupted its binding domain with Keap1; (2) epigenetic silencing of Keap1 expression leads to defective repression of Nrf2; (3) accumulation of interfering proteins (such as p62) leads to dissociation of the Keap1-Nrf2 complex; (4) transcriptional induction of Nrf2 by oncogenic factors (e.g., K-Ras, B-Raf and c-myc); and (5) in familial papillary renal cancer, posttranslational modification of Keap1 cysteines by succinylation due to loss of fumarate hydratase activity deregulates the Keap1-Nrf2 signaling pathway.
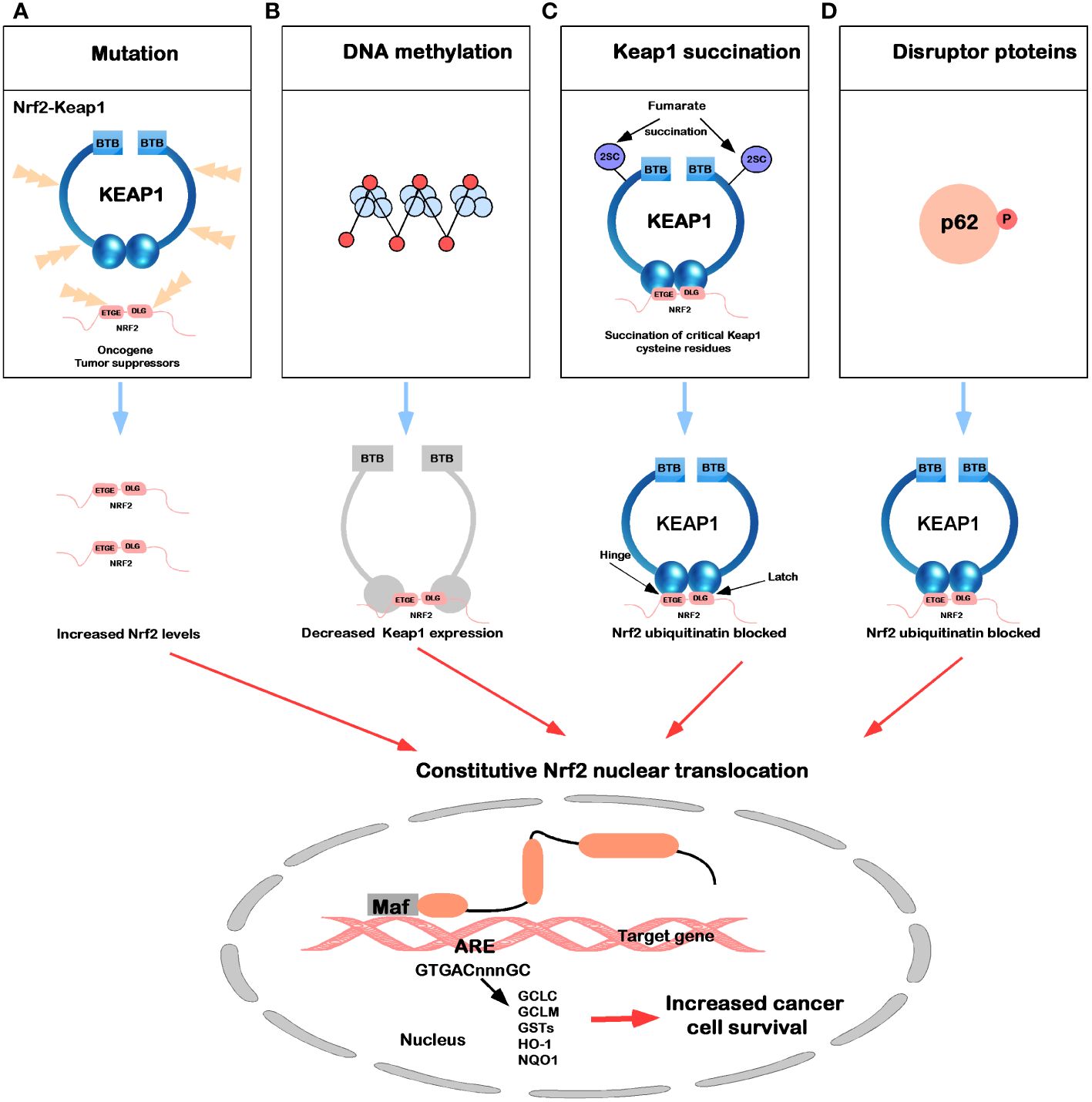
Figure 4 Mechanisms for constitutive nuclear accumulation of Nrf2 in cancer. (A) Somatic mutations in Nrf2 or Keap1 disrupt the interaction of these two proteins. (B) Hypermethylation of the Keap1 promoter in lung cancer and prostate cancer leads to decreased expression of Keap1 mRNA, thereby increasing nuclear accumulation of Nrf2. (C) In familial papillary renal carcinoma, the loss of fumarate hydratase activity leads to the accumulation of fumarate, which in turn leads to the succination of the Keap1 cysteine residue (2SC). (D) The accumulation of interfering proteins such as p62 and p21 can interfere with the binding of Nrf2 to Keap1, leading to an increase in nuclear Nrf2.
The transcription factor Nrf2 has an important function in mediating cellular homeostasis (20, 76, 124, 125), playing a crucial role during tumor development (20, 125–129). It has been shown that tumor proliferation and Nrf1 are distinct and that malignant proliferation is inhibited after interfering with Nrf2 expression or knockdown (130–133). Upon inactivation of the tumor suppressor PTEN, the activity of the PI3K-Akt pathway increases, resulting in the elevated expression of Nrf2, which promotes cell proliferation (134); N-cadherin expression, a marker protein of EMT, was suppressed in cancer cells in which Nrf2 was inhibited or knocked down (135, 136), whereas E-cadherin expression was decreased in cancer cells in which Nrf2 was overexpressed (137). Currently, in most reports, Keap1 is a negative regulator of Nrf2, which is regulated through the ubiquitin proteasome system.
Accumulating evidence indicates that the Nrf2 signaling pathway is deregulated in many cancers, leading to aberrant expression of a Nrf2-dependent gene battery. Therefore, the development of therapies with anti-inflammatory activity mediated by Nrf2 is likely to have a major clinical impact. The ongoing Nrf2 signaling pathway is leading worldwide efforts to develop highly targeted therapeutic agents to control inflammatory symptoms and prevent and treat major diseases such as cancer and neurodegenerative diseases.
FC: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. MX: Writing – review & editing, Data curation. SH: Data curation, Writing – review & editing. MW: Writing – review & editing, Data curation.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by the China Postdoctoral Science Foundation(Certificate Number:2023M740381), Natural Science Foundation of Sichuan (24NSFSC4190), Key Laboratory of Sports Medicine of Sichuan Province (2024-A001), Expert Centre Foundation of Sichuan Province (A2020-SCZJJCC 04) and Chengdu sport university (2023cdtyky15 and 23YJPY16).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Crisman E, Duarte P, Dauden E, Cuadrado A, Rodriguez-Franco MI, Lopez MG, et al. KEAP1-NRF2 protein-protein interaction inhibitors: Design, pharmacological properties and therapeutic potential. Med Res Rev. (2023) 43:237–87. doi: 10.1002/med.21925
2. Kansanen E, Kivela AM, Levonen AL. Regulation of Nrf2-dependent gene expression by 15-deoxy-Delta12,14-prostaglandin J2. Free Radic Biol Med. (2009) 47:1310–7. doi: 10.1016/j.freeradbiomed.2009.06.030
3. Keleku-Lukwete N, Suzuki M, Yamamoto M. An overview of the advantages of KEAP1-NRF2 system activation during inflammatory disease treatment. Antioxid Redox Signal. (2018) 29:1746–55. doi: 10.1089/ars.2017.7358
4. Suzuki T, Yamamoto M. Stress-sensing mechanisms and the physiological roles of the Keap1-Nrf2 system during cellular stress. J Biol Chem. (2017) 292:16817–24. doi: 10.1074/jbc.R117.800169
5. Yamamoto M, Kensler TW, Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol Rev. (2018) 98:1169–203. doi: 10.1152/physrev.00023.2017
6. Kansanen E, Jyrkkanen HK, Levonen AL. Activation of stress signaling pathways by electrophilic oxidized and nitrated lipids. Free Radic Biol Med. (2012) 52:973–82. doi: 10.1016/j.freeradbiomed.2011.11.038
7. Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. (2013) 1:45–9. doi: 10.1016/j.redox.2012.10.001
8. McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. (2003) 278:21592–600. doi: 10.1074/jbc.M300931200
9. Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. (1999) 13:76–86. doi: 10.1101/gad.13.1.76
10. Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. (2004) 24:7130–9. doi: 10.1128/MCB.24.16.7130-7139.2004
11. Dinkova-Kostova AT, Kostov RV, Canning P. Keap1, the cysteine-based mammalian intracellular sensor for electrophiles and oxidants. Arch Biochem Biophys. (2017) 617:84–93. doi: 10.1016/j.abb.2016.08.005
12. Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. (2004) 24:8477–86. doi: 10.1128/MCB.24.19.8477-8486.2004
13. Zipper LM, Mulcahy RT. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J Biol Chem. (2002) 277:36544–52. doi: 10.1074/jbc.M206530200
14. Kopacz A, Kloska D, Forman HJ, Jozkowicz A, Grochot-Przeczek A. Beyond repression of Nrf2: An update on Keap1. Free Radic Biol Med. (2020) 157:63–74. doi: 10.1016/j.freeradbiomed.2020.03.023
15. Canning P, Cooper CDO, Krojer T, Murray JW, Pike ACW, Chaikuad A, et al. Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J Biol Chem. (2013) 288:7803–14. doi: 10.1074/jbc.M112.437996
16. Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. (2006) 21:689–700. doi: 10.1016/j.molcel.2006.01.013
17. Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. (2014) 13:397–406. doi: 10.1074/mcp.M113.035600
18. Lee DF, Kuo HP, Liu M, Chou CK, Xia W, Du Y, et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell. (2009) 36:131–40. doi: 10.1016/j.molcel.2009.07.025
19. Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. (2015) 22:377–88. doi: 10.1038/cdd.2014.150
20. Rojo de la Vega M, Chapman E, Zhang DD. NRF2 and the hallmarks of cancer. Cancer Cell. (2018) 34:21–43. doi: 10.1016/j.ccell.2018.03.022
21. Karapetian RN, Evstafieva AG, Abaeva IS, Chichkova NV, Filonov GS, Rubtsov YP, et al. Nuclear oncoprotein prothymosin alpha is a partner of Keap1: implications for expression of oxidative stress-protecting genes. Mol Cell Biol. (2005) 25:1089–99. doi: 10.1128/MCB.25.3.1089-1099.2005
22. Camp ND, James RG, Dawson DW, Yan F, Davison JM, Houck SA, et al. Wilms tumor gene on X chromosome (WTX) inhibits degradation of NRF2 protein through competitive binding to KEAP1 protein. J Biol Chem. (2012) 287:6539–50. doi: 10.1074/jbc.M111.316471
23. Sun Z, Zhang S, Chan JY, Zhang DD. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol Cell Biol. (2007) 27:6334–49. doi: 10.1128/MCB.00630-07
24. Slocum SL, Kensler TW. Nrf2: control of sensitivity to carcinogens. Arch Toxicol. (2011) 85:273–84. doi: 10.1007/s00204-011-0675-4
25. Li W, Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog. (2009) 48:91–104. doi: 10.1002/mc.20465
26. Lee Y, Chou TF, Pittman SK, Keith AL, Razani B, Weihl CC. Keap1/Cullin3 Modulates p62/SQSTM1 Activity via UBA Domain Ubiquitination. Cell Rep. (2017) 19:188–202. doi: 10.1016/j.celrep.2017.03.030
27. Wasik U, Milkiewicz M, Kempinska-Podhorodecka A, Milkiewicz P. Protection against oxidative stress mediated by the Nrf2/Keap1 axis is impaired in Primary Biliary Cholangitis. Sci Rep. (2017) 7:44769. doi: 10.1038/srep44769
28. Camina N, Penning TM. Genetic and epigenetic regulation of the NRF2-KEAP1 pathway in human lung cancer. Br J Cancer. (2022) 126:1244–52. doi: 10.1038/s41416-021-01642-0
29. Fabrizio FP, Costantini M, Copetti M, la Torre A, Sparaneo A, Fontana A, et al. Keap1/Nrf2 pathway in kidney cancer: frequent methylation of KEAP1 gene promoter in clear renal cell carcinoma. Oncotarget. (2017) 8:11187–98. doi: 10.18632/oncotarget.14492
30. Fabrizio FP, Sparaneo A, Trombetta D, Muscarella LA. Epigenetic versus genetic deregulation of the KEAP1/NRF2 axis in solid tumors: focus on methylation and noncoding RNAs. Oxid Med Cell Longev. (2018) 2018:2492063. doi: 10.1155/2018/2492063
31. Berger AH, Brooks AN, Wu X, Shrestha Y, Chouinard C, Piccioni F, et al. High-throughput phenotyping of lung cancer somatic mutations. Cancer Cell. (2016) 30:214–28. doi: 10.1016/j.ccell.2016.06.022
32. Cancer Genome Atlas Research, N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. (2012) 489:519–25. doi: 10.1038/nature11404
33. Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. (2008) 135:1358–1368, 1368.e1351-1354. doi: 10.1053/j.gastro.2008.06.082
34. Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. (2017) 45:D777–83. doi: 10.1093/nar/gkw1121
35. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. (2019) 47:D941–7. doi: 10.1093/nar/gky1015
36. Yoo NJ, Kim HR, Kim YR, An CH, Lee SH. Somatic mutations of the KEAP1 gene in common solid cancers. Histopathology. (2012) 60:943–52. doi: 10.1111/j.1365-2559.2012.04178.x
37. Konstantinopoulos PA, Spentzos D, Fountzilas E, Francoeur N, Sanisetty S, Grammatikos AP, et al. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res. (2011) 71:5081–9. doi: 10.1158/0008-5472.CAN-10-4668
38. Shi W, Ling L, Li C, Wu R, Zhang M, Shao F, et al. LncRNA UCA1 promoted cisplatin resistance in lung adenocarcinoma with HO1 targets NRF2/HO1 pathway. J Cancer Res Clin Oncol. (2022) 149(3):1295–311. doi: 10.1007/s00432-022-04152-5
39. Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, et al. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther. (2010) 9:336–46. doi: 10.1158/1535-7163.MCT-09-0589
40. Hintsala HR, Jokinen E, Haapasaari KM, Moza M, Ristimaki A, Soini Y, et al. Nrf2/Keap1 pathway and expression of oxidative stress lesions 8-hydroxy-2'-deoxyguanosine and nitrotyrosine in melanoma. Anticancer Res. (2016) 36:1497–506.
41. Miura S, Shibazaki M, Kasai S, Yasuhira S, Watanabe A, Inoue T, et al. A somatic mutation of the KEAP1 gene in Malignant melanoma is involved in aberrant NRF2 activation and an increase in intrinsic drug resistance. J Invest Dermatol. (2014) 134:553–6. doi: 10.1038/jid.2013.343
42. Fabrizio FP, Mazza T, Castellana S, Sparaneo A, Muscarella LA. Epigenetic scanning of KEAP1 CpG sites uncovers new molecular-driven patterns in lung adeno and squamous cell carcinomas. Antioxidants (Basel). (2020) 9:904–23. doi: 10.3390/antiox9090904
43. Wang T, Wang P, Wang S, Ma Y, Zhao Z, Long F. Wogonin Diminishes Radioresistance of Breast Cancer via Inhibition of the Nrf2/HIF-1[Formula: see text] Pathway. Am J Chin Med. (2023) 51:2243–62. doi: 10.1142/S0192415X23500969
44. Wang L, Shannar AAF, Wu R, Chou P, Sarwar MS, Kuo HC, et al. Butyrate drives metabolic rewiring and epigenetic reprogramming in human colon cancer cells. Mol Nutr Food Res. (2022) 66:e2200028. doi: 10.1002/mnfr.202200028
45. Ding X, Jian T, Wu Y, Zuo Y, Li J, Lv H, et al. Ellagic acid ameliorates oxidative stress and insulin resistance in high glucose-treated HepG2 cells via miR-223/keap1-Nrf2 pathway. BioMed Pharmacother. (2019) 110:85–94. doi: 10.1016/j.biopha.2018.11.018
46. Zhao XJ, Yu HW, Yang YZ, Wu WY, Chen TY, Jia KK, et al. Polydatin prevents fructose-induced liver inflammation and lipid deposition through increasing miR-200a to regulate Keap1/Nrf2 pathway. Redox Biol. (2018) 18:124–37. doi: 10.1016/j.redox.2018.07.002
47. Zhao XJ, Yu HW, Yang YZ, Wu WY, Chen TY, Jia KK, et al. Corrigendum to "Polydatin prevents fructose-induced liver inflammation and lipid deposition through increasing miR-200a to regulate Keap1/Nrf2 pathway" [Redox Biol. 18 (2018) 124-137]. Redox Biol. (2019) 22:101101. doi: 10.1016/j.redox.2019.101101
48. Ma S, Paiboonrungruan C, Yan T, Williams KP, Major MB, Chen XL. Targeted therapy of esophageal squamous cell carcinoma: the NRF2 signaling pathway as target. Ann N Y Acad Sci. (2018) 1434:164–72. doi: 10.1111/nyas.13681
49. Qiao G, Zhang W, Dong K. Regulation of ferroptosis by noncoding RNAs: a novel promise treatment in esophageal squamous cell carcinoma. Mol Cell Biochem. (2022) 477:2193–202. doi: 10.1007/s11010-022-04441-0
50. Akdemir B, Nakajima Y, Inazawa J, Inoue J. miR-432 induces NRF2 stabilization by directly targeting KEAP1. Mol Cancer Res. (2017) 15:1570–8. doi: 10.1158/1541-7786.MCR-17-0232
51. Satoh T, McKercher SR, Lipton SA. Nrf2/ARE-mediated antioxidant actions of pro-electrophilic drugs. Free Radic Biol Med. (2013) 65:645–57. doi: 10.1016/j.freeradbiomed.2013.07.022
52. Aghagolzadeh P, Radpour R, Bachtler M, van Goor H, Smith ER, Lister A, et al. Hydrogen sulfide attenuates calcification of vascular smooth muscle cells via KEAP1/NRF2/NQO1 activation. Atherosclerosis. (2017) 265:78–86. doi: 10.1016/j.atherosclerosis.2017.08.012
53. Luo Y, Eggler AL, Liu D, Liu G, Mesecar AD, van Breemen RB. Sites of alkylation of human Keap1 by natural chemoprevention agents. J Am Soc Mass Spectrom. (2007) 18:2226–32. doi: 10.1016/j.jasms.2007.09.015
54. Liu G, Eggler AL, Dietz BM, Mesecar AD, Bolton JL, Pezzuto JM, et al. Screening method for the discovery of potential cancer chemoprevention agents based on mass spectrometric detection of alkylated Keap1. Anal Chem. (2005) 77:6407–14. doi: 10.1021/ac050892r
55. Ohnuma T, Nakayama S, Anan E, Nishiyama T, Ogura K, Hiratsuka A. Activation of the Nrf2/ARE pathway via S-alkylation of cysteine 151 in the chemopreventive agent-sensor Keap1 protein by falcarindiol, a conjugated diacetylene compound. Toxicol Appl Pharmacol. (2010) 244:27–36. doi: 10.1016/j.taap.2009.12.012
56. Sun KA, Li Y, Meliton AY, Woods PS, Kimmig LM, Cetin-Atalay R, et al. Endogenous itaconate is not required for particulate matter-induced NRF2 expression or inflammatory response. Elife. (2020) 9:9-31. doi: 10.7554/eLife.54877
57. Zheng Y, Chen Z, She C, Lin Y, Hong Y, Shi L, et al. Four-octyl itaconate activates Nrf2 cascade to protect osteoblasts from hydrogen peroxide-induced oxidative injury. Cell Death Dis. (2020) 11:772. doi: 10.1038/s41419-020-02987-9
58. Otto G. Immunometabolism: Itaconate helps KEAP1's cool. Nat Rev Immunol. (2018) 18:294–5. doi: 10.1038/nri.2018.27
59. Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. (2018) 556:113–7. doi: 10.1038/nature25986
60. Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U.S.A. (1994) 91:9926–30. doi: 10.1073/pnas.91.21.9926
61. Schaue D, Micewicz ED, Ratikan JA, Iwamoto KS, Vlashi E, McDonald JT, et al. NRF2 mediates cellular resistance to transformation, radiation, and inflammation in mice. Antioxidants (Basel). (2022) 11(9):1649–63. doi: 10.3390/antiox11091649
62. Hammad A, Namani A, Elshaer M, Wang XJ, Tang X. "NRF2 addiction" in lung cancer cells and its impact on cancer therapy. Cancer Lett. (2019) 467:40–9. doi: 10.1016/j.canlet.2019.09.016
63. Wang YY, Chen J, Liu XM, Zhao R, Zhe H. Nrf2-mediated metabolic reprogramming in cancer. Oxid Med Cell Longev. (2018) 2018:9304091. doi: 10.1155/2018/9304091
64. Awuah WA, Toufik AR, Yarlagadda R, Mikhailova T, Mehta A, Huang H, et al. Exploring the role of Nrf2 signaling in glioblastoma multiforme. Discovery Oncol. (2022) 13:94. doi: 10.1007/s12672-022-00556-4
65. He F, Ru X, Wen T. NRF2, a transcription factor for stress response and beyond. Int J Mol Sci. (2020) 21(13):4777–99. doi: 10.3390/ijms21134777
66. Tonelli C, Chio IIC, Tuveson DA. Transcriptional regulation by Nrf2. Antioxid Redox Signal. (2018) 29:1727–45. doi: 10.1089/ars.2017.7342
67. Motohashi H, Shavit JA, Igarashi K, Yamamoto M, Engel JD. The world according to Maf. Nucleic Acids Res. (1997) 25:2953–9. doi: 10.1093/nar/25.15.2953
68. Motohashi H, Katsuoka F, Engel JD, Yamamoto M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc Natl Acad Sci U.S.A. (2004) 101:6379–84. doi: 10.1073/pnas.0305902101
69. Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. (2006) 26:2887–900. doi: 10.1128/MCB.26.8.2887-2900.2006
70. Nioi P, Nguyen T, Sherratt PJ, Pickett CB. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol Cell Biol. (2005) 25:10895–906. doi: 10.1128/MCB.25.24.10895-10906.2005
71. Shen G, Hebbar V, Nair S, Xu C, Li W, Lin W, et al. Regulation of Nrf2 transactivation domain activity. The differential effects of mitogen-activated protein kinase cascades and synergistic stimulatory effect of Raf and CREB-binding protein. J Biol Chem. (2004) 279:23052–60. doi: 10.1074/jbc.M401368200
72. Zhang J, Hosoya T, Maruyama A, Nishikawa K, Maher JM, Ohta T, et al. Nrf2 Neh5 domain is differentially utilized in the transactivation of cytoprotective genes. Biochem J. (2007) 404:459–66. doi: 10.1042/BJ20061611
73. Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. (2013) 32:3765–81. doi: 10.1038/onc.2012.388
74. Cuadrado A. Structural and functional characterization of Nrf2 degradation by glycogen synthase kinase 3/beta-TrCP. Free Radic Biol Med. (2015) 88:147–57. doi: 10.1016/j.freeradbiomed.2015.04.029
75. Wang H, Liu K, Geng M, Gao P, Wu X, Hai Y, et al. RXRalpha inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res. (2013) 73:3097–108. doi: 10.1158/0008-5472.CAN-12-3386
76. Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. (2014) 39:199–218. doi: 10.1016/j.tibs.2014.02.002
77. Cho HY, Gladwell W, Wang X, Chorley B, Bell D, Reddy SP, et al. Nrf2-regulated PPAR{gamma} expression is critical to protection against acute lung injury in mice. Am J Respir Crit Care Med. (2010) 182:170–82. doi: 10.1164/rccm.200907-1047OC
78. Miao W, Hu L, Scrivens PJ, Batist G. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem. (2005) 280:20340–8. doi: 10.1074/jbc.M412081200
79. Tung MC, Lin PL, Wang YC, He TY, Lee MC, Yeh SD, et al. Mutant p53 confers chemoresistance in non-small cell lung cancer by upregulating Nrf2. Oncotarget. (2015) 6:41692–705. doi: 10.18632/oncotarget.6150
80. Tao S, Liu P, Luo G, Rojo de la Vega M, Chen H, Wu T, et al. p97 negatively regulates NRF2 by extracting ubiquitylated NRF2 from the KEAP1-CUL3 E3 complex. Mol Cell Biol. (2017) 37(8):e00660–16. doi: 10.1128/MCB.00660-16
81. Wang XJ, Hayes JD, Henderson CJ, Wolf CR. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proc Natl Acad Sci U.S.A. (2007) 104:19589–94. doi: 10.1073/pnas.0709483104
82. Yu S, Khor TO, Cheung KL, Li W, Wu TY, Huang Y, et al. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PloS One. (2010) 5:e8579. doi: 10.1371/journal.pone.0008579
83. Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. (2001) 15:1593–612. doi: 10.1101/gad.891101
84. Kawai Y, Garduño L, Theodore M, Yang J, Arinze IJ. Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J Biol Chem. (2011) 286:7629–40. doi: 10.1074/jbc.M110.208173
85. Egbujor MC, Petrosino M, Zuhra K, Saso L. The role of organosulfur compounds as Nrf2 activators and their antioxidant effects. Antioxidants (Basel). (2022) 11(7):1255–93. doi: 10.3390/antiox11071255
86. Muchtaridi M, Amirah SR, Harmonis JA, Ikram EHK. Role of nuclear factor Erythroid 2 (Nrf2) in the recovery of long COVID-19 using natural antioxidants: A systematic review. Antioxidants (Basel). (2022) 11(8):1551–75. doi: 10.3390/antiox11081551
87. Saito R, Suzuki T, Hiramoto K, Asami S, Naganuma E, Suda H, et al. Characterizations of three major cysteine sensors of Keap1 in stress response. Mol Cell Biol. (2016) 36:271–84. doi: 10.1128/MCB.00868-15
88. Gall Trošelj K, Tomljanović M, Jaganjac M, Matijević Glavan T, Čipak Gašparović A, Milković L, et al. Oxidative stress and cancer heterogeneity orchestrate NRF2 roles relevant for therapy response. Molecules. (2022) 27(5):1468–97. doi: 10.3390/molecules27051468
89. McCord JM, Gao B, Hybertson BM. The complex genetic and epigenetic regulation of the Nrf2 pathways: A review. Antioxidants (Basel). (2023) 12(2):366–83. doi: 10.3390/antiox12020366
90. Jung BJ, Yoo HS, Shin S, Park YJ, Jeon SM. Dysregulation of NRF2 in cancer: from molecular mechanisms to therapeutic opportunities. Biomol Ther (Seoul). (2018) 26:57–68. doi: 10.4062/biomolther.2017.195
91. Dinkova-Kostova AT, Kostov RV, Kazantsev AG. The role of Nrf2 signaling in counteracting neurodegenerative diseases. FEBS J. (2018) 285:3576–90. doi: 10.1111/febs.14379
92. Lee S, Abed DA, Beamer LJ, Hu L. Development of a homogeneous time-resolved fluorescence resonance energy transfer (TR-FRET) assay for the inhibition of Keap1-Nrf2 protein-protein interaction. SLAS Discovery. (2021) 26:100–12. doi: 10.1177/2472555220935816
93. Abed DA, Goldstein M, Albanyan H, Jin H, Hu L. Discovery of direct inhibitors of Keap1-Nrf2 protein-protein interaction as potential therapeutic and preventive agents. Acta Pharm Sin B. (2015) 5:285–99. doi: 10.1016/j.apsb.2015.05.008
94. Arulselvan P, Fard MT, Tan WS, Gothai S, Fakurazi S, Norhaizan ME, et al. Role of antioxidants and natural products in inflammation. Oxid Med Cell Longev. (2016) 2016:5276130. doi: 10.1155/2016/5276130
95. Lewis KN, Mele J, Hayes JD, Buffenstein R. Nrf2, a guardian of healthspan and gatekeeper of species longevity. Integr Comp Biol. (2010) 50:829–43. doi: 10.1093/icb/icq034
96. Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. (2013) 27:2179–91. doi: 10.1101/gad.225680.113
97. Xiang M, Namani A, Wu S, Wang X. Nrf2: bane or blessing in cancer? J Cancer Res Clin Oncol. (2014) 140:1251–9. doi: 10.1007/s00432-014-1627-1
98. Kensler TW, Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. (2010) 31:90–9. doi: 10.1093/carcin/bgp231
99. Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. (2009) 34:176–88. doi: 10.1016/j.tibs.2008.12.008
100. Mata A, Cadenas S. The antioxidant transcription factor Nrf2 in cardiac ischemia-reperfusion injury. Int J Mol Sci. (2021) 22(21):11939–62. doi: 10.3390/ijms222111939
101. Xiang Y, Ye W, Huang C, Yu D, Chen H, Deng T, et al. Brusatol enhances the chemotherapy efficacy of gemcitabine in pancreatic cancer via the Nrf2 signalling pathway. Oxid Med Cell Longev. (2018) 2018:2360427. doi: 10.1155/2018/2360427
102. Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, Toppin HA, et al. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc Natl Acad Sci U.S.A. (2011) 108:1433–8. doi: 10.1073/pnas.1014275108
103. Wang J, Wang H, Sun K, Wang X, Pan H, Zhu J, et al. Chrysin suppresses proliferation, migration, and invasion in glioblastoma cell lines via mediating the ERK/Nrf2 signaling pathway. Drug Des Devel Ther. (2018) 12:721–33. doi: 10.2147/DDDT.S160020
104. Chian S, Thapa R, Chi Z, Wang XJ, Tang X. Luteolin inhibits the Nrf2 signaling pathway and tumor growth in vivo. Biochem Biophys Res Commun. (2014) 447:602–8. doi: 10.1016/j.bbrc.2014.04.039
105. Liu P, Ge M, Hu J, Li X, Che L, Sun K, et al. A functional mammalian target of rapamycin complex 1 signaling is indispensable for c-Myc-driven hepatocarcinogenesis. Hepatology. (2017) 66:167–81. doi: 10.1002/hep.29183
106. Hirose W, Oshikiri H, Taguchi K, Yamamoto M. The KEAP1-NRF2 system and esophageal cancer. Cancers (Basel). (2022) 14(8):e00660–16. doi: 10.3390/cancers14194702
107. Menegon S, Columbano A, Giordano S. The dual roles of NRF2 in cancer. Trends Mol Med. (2016) 22:578–93. doi: 10.1016/j.molmed.2016.05.002
108. Ziros PG, Renaud CO, Chartoumpekis DV, Bongiovanni M, Habeos IG, Liao XH, et al. Mice hypomorphic for Keap1, a negative regulator of the Nrf2 antioxidant response, show age-dependent diffuse goiter with elevated thyrotropin levels. Thyroid. (2021) 31:23–35. doi: 10.1089/thy.2020.0044
109. Renaud CO, Ziros PG, Chartoumpekis DV, Bongiovanni M, Sykiotis GP. Keap1/Nrf2 signaling: A new player in thyroid pathophysiology and thyroid cancer. Front Endocrinol (Lausanne). (2019) 10:510. doi: 10.3389/fendo.2019.00510
110. You S, Qian J, Sun C, Zhang H, Ye S, Chen T, et al. An Aza resveratrol-chalcone derivative 6b protects mice against diabetic cardiomyopathy by alleviating inflammation and oxidative stress. J Cell Mol Med. (2018) 22:1931–43. doi: 10.1111/jcmm.13477
111. Lee JA, Son HJ, Choi JW, Kim J, Han SH, Shin N, et al. Activation of the Nrf2 signaling pathway and neuroprotection of nigral dopaminergic neurons by a novel synthetic compound KMS99220. Neurochem Int. (2018) 112:96–107. doi: 10.1016/j.neuint.2017.11.006
112. Putzer D, Gabriel M, Kroiss A, Madleitner R, Eisterer W, Kendler D, et al. First experience with proteasome inhibitor treatment of radioiodine nonavid thyroid cancer using bortezomib. Clin Nucl Med. (2012) 37:539–44. doi: 10.1097/RLU.0b013e31824c5f24
113. Li S, Zhang Y, Zhang J, Yu B, Wang W, Jia B, et al. Neferine exerts ferroptosis-inducing effect and antitumor effect on thyroid cancer through Nrf2/HO-1/NQO1 inhibition. J Oncol. (2022) 2022:7933775. doi: 10.1155/2022/7933775
114. Thanas C, Ziros PG, Chartoumpekis DV, Renaud CO, Sykiotis GP. The Keap1/Nrf2 signaling pathway in the thyroid-2020 update. Antioxidants (Basel). (2020) 9(11):1082–95. doi: 10.3390/antiox9111082
115. Tang YC, Hsiao JR, Jiang SS, Chang JY, Chu PY, Liu KJ, et al. c-MYC-directed NRF2 drives Malignant progression of head and neck cancer via glucose-6-phosphate dehydrogenase and transketolase activation. Theranostics. (2021) 11:5232–47. doi: 10.7150/thno.53417
116. Zhang M, Hong X, Ma N, Wei Z, Ci X, Zhang S. The promoting effect and mechanism of Nrf2 on cell metastasis in cervical cancer. J Transl Med. (2023) 21:433. doi: 10.1186/s12967-023-04287-0
117. Vilchez Mercedes SA, Bocci F, Ahmed M, Eder I, Zhu N, Levine H, et al. Nrf2 modulates the hybrid epithelial/mesenchymal phenotype and notch signaling during collective cancer migration. Front Mol Biosci. (2022) 9:807324. doi: 10.3389/fmolb.2022.807324
118. Ma Q, He X. Molecular basis of electrophilic and oxidative defense: promises and perils of Nrf2. Pharmacol Rev. (2012) 64:1055–81. doi: 10.1124/pr.110.004333
119. Ooi A, Wong JC, Petillo D, Roossien D, Perrier-Trudova V, Whitten D, et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell. (2011) 20:511–23. doi: 10.1016/j.ccr.2011.08.024
120. Adam J, Hatipoglu E, O'Flaherty L, Ternette N, Sahgal N, Lockstone H, et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. (2011) 20:524–37. doi: 10.1016/j.ccr.2011.09.006
121. Hanada N, Takahata T, Zhou Q, Ye X, Sun R, Itoh J, et al. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer. (2012) 12:66. doi: 10.1186/1471-2407-12-66
122. DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. (2011) 475:106–9. doi: 10.1038/nature10189
123. Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer. (2012) 12:564–71. doi: 10.1038/nrc3278
124. Kim HM, Han JW, Chan JY. Nuclear Factor Erythroid-2 Like 1 (NFE2L1): Structure, function and regulation. Gene. (2016) 584:17–25. doi: 10.1016/j.gene.2016.03.002
125. Zhang Y, Xiang Y. Molecular and cellular basis for the unique functioning of Nrf1, an indispensable transcription factor for maintaining cell homoeostasis and organ integrity. Biochem J. (2016) 473:961–1000. doi: 10.1042/BJ20151182
126. Pillai R, Hayashi M, Zavitsanou AM, Papagiannakopoulos T. NRF2: KEAPing tumors protected. Cancer Discovery. (2022) 12:625–43. doi: 10.1158/2159-8290.CD-21-0922
127. Wufuer R, Fan Z, Liu K, Zhang Y. Differential yet integral contributions of Nrf1 and Nrf2 in the human HepG2 cells on antioxidant cytoprotective response against Tert-Butylhydroquinone as a pro-oxidative stressor. Antioxidants (Basel). (2021) 10(10):1610–34. doi: 10.3390/antiox10101610
128. Sekine H, Motohashi H. Roles of CNC transcription factors NRF1 and NRF2 in cancer. Cancers (Basel). (2021) 13(3):541–54. doi: 10.3390/cancers13030541
129. Wu S, Lu H, Bai Y. Nrf2 in cancers: A double-edged sword. Cancer Med. (2019) 8:2252–67. doi: 10.1002/cam4.2101
130. Ren Y, Qiu L, Lu F, Ru X, Li S, Xiang Y, et al. TALENs-directed knockout of the full-length transcription factor Nrf1alpha that represses Malignant behaviour of human hepatocellular carcinoma (HepG2) cells. Sci Rep. (2016) 6:23775. doi: 10.1038/srep23775
131. Chen J, Wang M, Xiang Y, Ru X, Ren Y, Liu X, et al. Nrf1 is endowed with a dominant tumor-repressing effect onto the Wnt/beta-Catenin-Dependent and Wnt/beta-Catenin-Independent signaling networks in the human liver cancer. Oxid Med Cell Longev. (2020) 2020:5138539. doi: 10.1155/2020/5138539
132. Qiu L, Wang M, Hu S, Ru X, Ren Y, Zhang Z, et al. Oncogenic activation of Nrf2, though as a master antioxidant transcription factor, liberated by specific knockout of the full-length nrf1alpha that acts as a dominant tumor repressor. Cancers (Basel). (2018) 10(12):520–61. doi: 10.3390/cancers10120520
133. Wang M, Ren Y, Hu S, Liu K, Qiu L, Zhang Y. TCF11 has a potent tumor-repressing effect than its prototypic Nrf1alpha by definition of both similar yet different regulatory profiles, with a striking disparity from Nrf2. Front Oncol. (2021) 11:707032. doi: 10.3389/fonc.2021.707032
134. Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. (2016) 48:500–9. doi: 10.1038/ng.3547
135. Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, et al. Ensembl 2019. Nucleic Acids Res. (2019) 47:D745–51. doi: 10.1093/nar/gky1113
136. Tasaki M, Kuroiwa Y, Inoue T, Hibi D, Matsushita K, Kijima A, et al. Lack of nrf2 results in progression of proliferative lesions to neoplasms induced by long-term exposure to non-genotoxic hepatocarcinogens involving oxidative stress. Exp Toxicol Pathol. (2014) 66:19–26. doi: 10.1016/j.etp.2013.07.003
Keywords: Keap1, Nrf2, cancer, structure, function, transcriptional regulation, prevention, clinical
Citation: Chen F, Xiao M, Hu S and Wang M (2024) Keap1-Nrf2 pathway: a key mechanism in the occurrence and development of cancer. Front. Oncol. 14:1381467. doi: 10.3389/fonc.2024.1381467
Received: 03 February 2024; Accepted: 19 March 2024;
Published: 03 April 2024.
Edited by:
Tinghe Yu, Chongqing Medical University, ChinaReviewed by:
Debora Decote-Ricardo, Federal Rural University of Rio de Janeiro, BrazilCopyright © 2024 Chen, Xiao, Hu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feilong Chen, Y2hlbmZlaWxvbmdAY2RzdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.