
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Oncol. , 25 March 2024
Sec. Breast Cancer
Volume 14 - 2024 | https://doi.org/10.3389/fonc.2024.1374674
This article is part of the Research Topic Reviews in Breast Cancer: 2023 View all 28 articles
Backgrounds: The value of circulating microRNA (miR)-155 for breast cancer (BC) diagnosis may differ in different studies. Therefore, we conducted this systematic review and meta-analysis to evaluate the potential application of circulating miR-155 in the diagnosis of BC.
Methods: Articles published before December 2023 and in English were searched in these databases: PubMed, Web of Science, Medline, EMBASE and Google Scholar. A summary of sensitivity, specificity, positive likelihood ratios (PLR), negative likelihood ratios (NLR), and diagnostic odds ratio (DOR) were calculated from the true positive (TP), true negative (TN), false positive (FP) and false negative (FN) of each study. Additionally, the summary receive-operating characteristics (SROC) curve was constructed to summarize the TP and FP rates.
Results: The pooled parameters calculated were as follows: sensitivity, 0.93 (95% CI: 0.83-0.97); specificity, 0.85 (95% CI: 0.74-0.92); PLR, 6.4 (95% CI: 3.4-11.9); NLR, 0.09 (95% CI: 0.04-0.20); and DOR, 74 (95% CI: 22-247). The analysis showed a significant heterogeneity (sensitivity, I2 = 95.19%, p < 0.001; specificity, I2 = 95.29%, p < 0.001; DOR, I2 = 92.9%, p < 0.001). The SROC curve was with an area under curve (AUC) of 0.95 (95% CI: 0.93-0.97).
Conclusion: Circulating miR-155 has a potential in the diagnosis of BC.
Breast cancer (BC) is the most common cancer in women globally, accounting for 31% of all cancers (1). The risk of developing BC over a woman’s lifetime is approximately 1 in 8 (1). It is estimated that there will be 297,790 new cases of invasive BC and 43,170 women will die from BC in the United States in 2023 (2). Almost 20% of global BC patients occurred in China, with an estimated age-standardized incidence rate of approximately 60 cases per 100,000 women in 2040 (3, 4). The incidence of BC in China has increased rapidly in recent decades. Early detection is important for a better prognosis because there are few signs and symptoms in the early stage.
Although mammography is widely considered the gold standard for breast cancer detection, it is not without its drawbacks. It is associated with pain, anxiety, and radiation exposure (5), which can deter some individuals from undergoing regular screenings. Additionally, the effectiveness of mammography is limited in women with dense breasts (6), further reducing its reliability as a screening tool. Furthermore, mammography has been found to be particularly inaccurate in patients below the age of 40, leading to underdiagnosis (7). This is concerning because the incidence of triple-negative tumors, which have worse prognostics, is higher in this age group. Early detection is crucial for improving overall cancer survival rates, as it allows for timely intervention before the cancer has a chance to metastasize (8). To overcome the limitations of imaging techniques, the analysis of biomarkers has emerged as a promising approach for early breast cancer diagnosis. Biomarkers such as the human epidermal growth factor receptor 2 (HER2), the KI-67 protein, and estrogen receptors (ERs) are commonly used for prognosis and to guide systemic treatment decisions. In recent years, miRNAs have also gained momentum as potential biomarkers for breast cancer.
MicroRNAs (miRNAs) are a class of small endogenous RNAs that are 19-25 nucleotides in length (9). MiRNAs contribute to the post-transcription regulation of target messenger RNA (mRNA) via mRNA degradation or translation repression (10). Studies have proven that miRNAs play an important role in a variety of biological processes, including inflammation, cell-cycle regulation, cell differentiation, apoptosis, and migration (11). Besides, various studies have demonstrated that miRNA dysregulation is relevant to cancer progression, such as miR-34a in myeloma and miR-145 in solid tumors (12). And miRNA has the merits such as stable quality, easy acquisition of samples and numerous sources of samples (13). Currently, miRNA biomarkers are not utilized in clinical practice due to the significant challenge of translating them from the laboratory into validated diagnostic tests. Among the miRNAs that have been found to be deregulated in BC, microRNA-21 (miR-21) and miR-155 have been identified as the most commonly associated with BC. However, it is important to note that these miRNAs are not highly specific for diagnostic purposes (14).
MiR-155 is an important oncogenic miRNAs in human cancers including in BC (15). Abnormal expression of miR-155 has been found in multiple cancers, such as lung and cervical cancer (16, 17). The expressions of miR-155 and SOCS3 were opposite in lymphoma and pancreatic cancer cells (18, 19). MiR-155 could affect the proliferation and apoptosis of bladder cancer cells via the GSK-3β/β-catenin pathway (20). These may indicate the important role of miR-155 in cancers. Recent studies have shown that the expression level of circulating miR-155 in BC tissues was significantly higher than in normal tissues, and the level of plasma miR-155 in BC patients was also significantly higher than in healthy controls (21, 22). However, the value of circulating miR-155 for BC diagnosis may differ in different studies. Therefore, we conducted this systematic review and meta-analysis to evaluate the potential application of circulating miR-155 in the diagnosis of BC.
Articles published before December 2023 and in English were searched in these databases: PubMed, Web of Science, Medline, EMBASE and Google Scholar by two researchers (Wang F and Wang J). Search terms used were: (“miR-155” OR “microRNA-155”) AND (“breast cancer”). In addition, duplicates were removed. A total of 384 articles were screened in the study.
The study included all articles based on these inclusion criteria as follows: (i) Studies that involved human patients with BC, (ii) studies in which expression of miR-155 was measured in plasma or serum. Additionally, when true positive (TP), true negative (TN), false positive (FP), and false negative (FN) regarding the diagnostic value of circulating miR-155 for BC could not be acquired or calculated from a study, the study would be excluded. Moreover, we dropped secondary processing of literature such as reviews and meta-analysis articles. Additionally, we excluded case studies without group-level statistics. Two researchers (Wang F and Wang J) independently read the abstracts and full texts.
Two investigators (Wang F and Zhang H.J) read titles and abstracts of articles. We collected data as follows: Author, publication years, study location, sample type, sample size, sensitivity, specificity, cut-off value, detection method and endogenous control. In each selected article, we collected TP, TN, FP and FN directly or calculated them according to the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV).
All the statistical analysis was conducted using STATA 12.0 software and Meta-Disc Version 1.4. A summary of sensitivity, specificity, positive likelihood ratios (PLR), negative likelihood ratios (NLR), and diagnostic odds ratio (DOR) were calculated from the TP, FP, FN, and TN of each study. Additionally, the summary receive-operating characteristics (SROC) curve was constructed to summarize the TP and FP rates (23). Q test was used to estimate heterogeneity between studies, and computed I2 was used to assess the amount of variation derived from heterogeneity. With invariably high heterogeneity (Q test, p ≤ 0.05), random effects models were used to generate a summary effect size of these studies; Inversely, fixed effects models were performed to summarize effect size in the absence of between-study heterogeneity (Q test, p > 0.05).
Figure 1 showed the initial search results and selection process. Table 1 showed the characteristics of the finally included 16 studies. Data were collected from 16 studies (22, 24–38) for the diagnostic studies with miR-155 for BC (BC group: n = 1,377, control group: n = 716).
Circulating miR-155 showed a diagnostic value for BC. As shown in forest plot (Figures 2, 3), the pooled parameters calculated were as follows: sensitivity, 0.93 (95% CI: 0.83-0.97); specificity, 0.85 (95% CI: 0.74-0.92); PLR, 6.4 (95% CI: 3.4-11.9); NLR, 0.09 (95% CI: 0.04-0.20); and DOR, 74 (95% CI: 22-247). The analysis showed a high heterogeneity (sensitivity, I2 = 95.19%, p < 0.001; specificity, I2 = 95.29%, p < 0.001; DOR, I2 = 92.9%, p < 0.001). Figure 4 shows the SROC curve, with an area under the curve (AUC) of 0.95 (95% CI: 0.93-0.97). The symmetrical Deek’s funnel plot showed a publication bias (p < 0.01, Supplementary Figure 1).
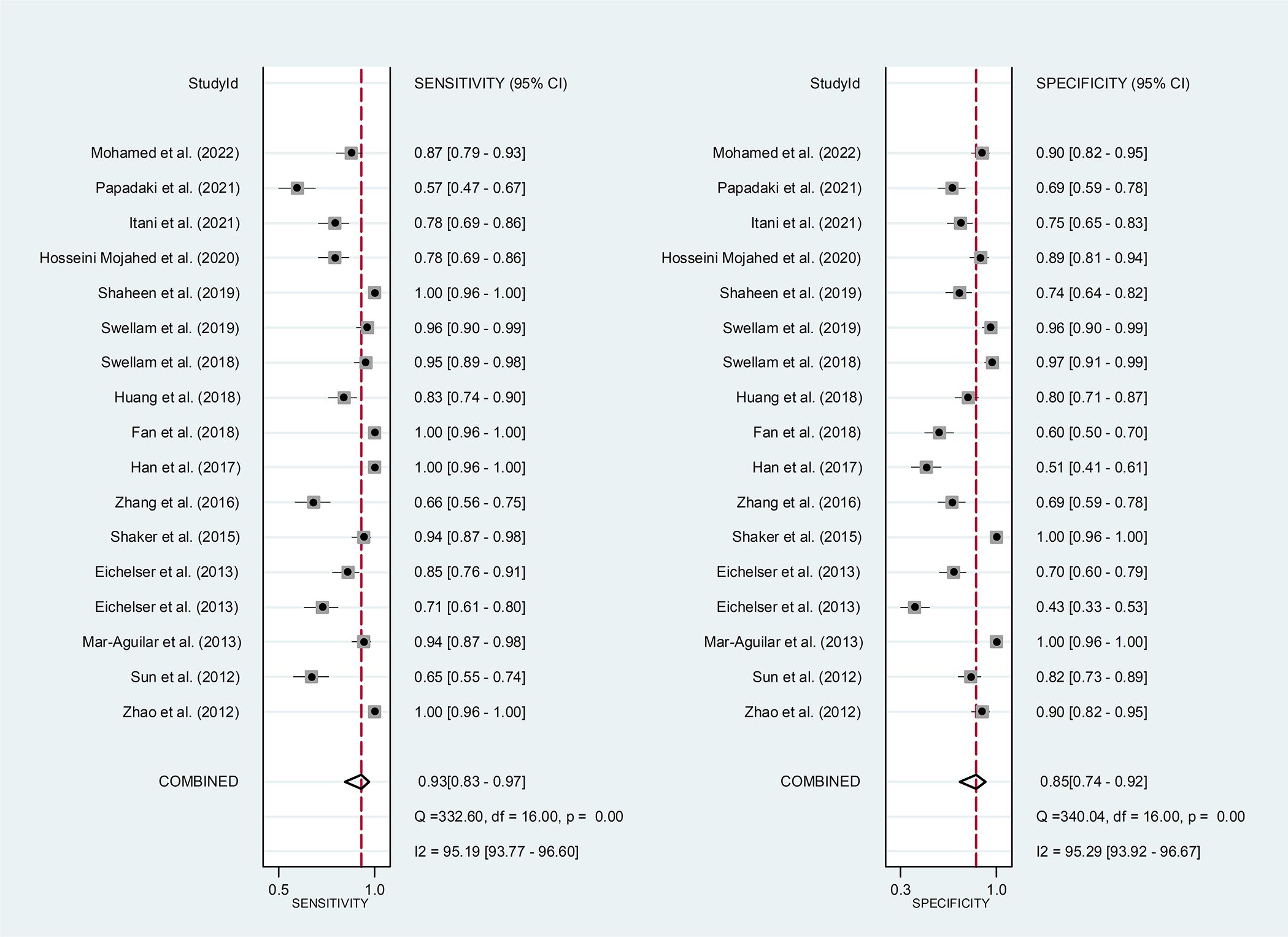
Figure 2 The sensitivity and specificity of circulating miR-155 in the diagnosis of BC. BC, breast cancer; miR-155, microRNA-155.
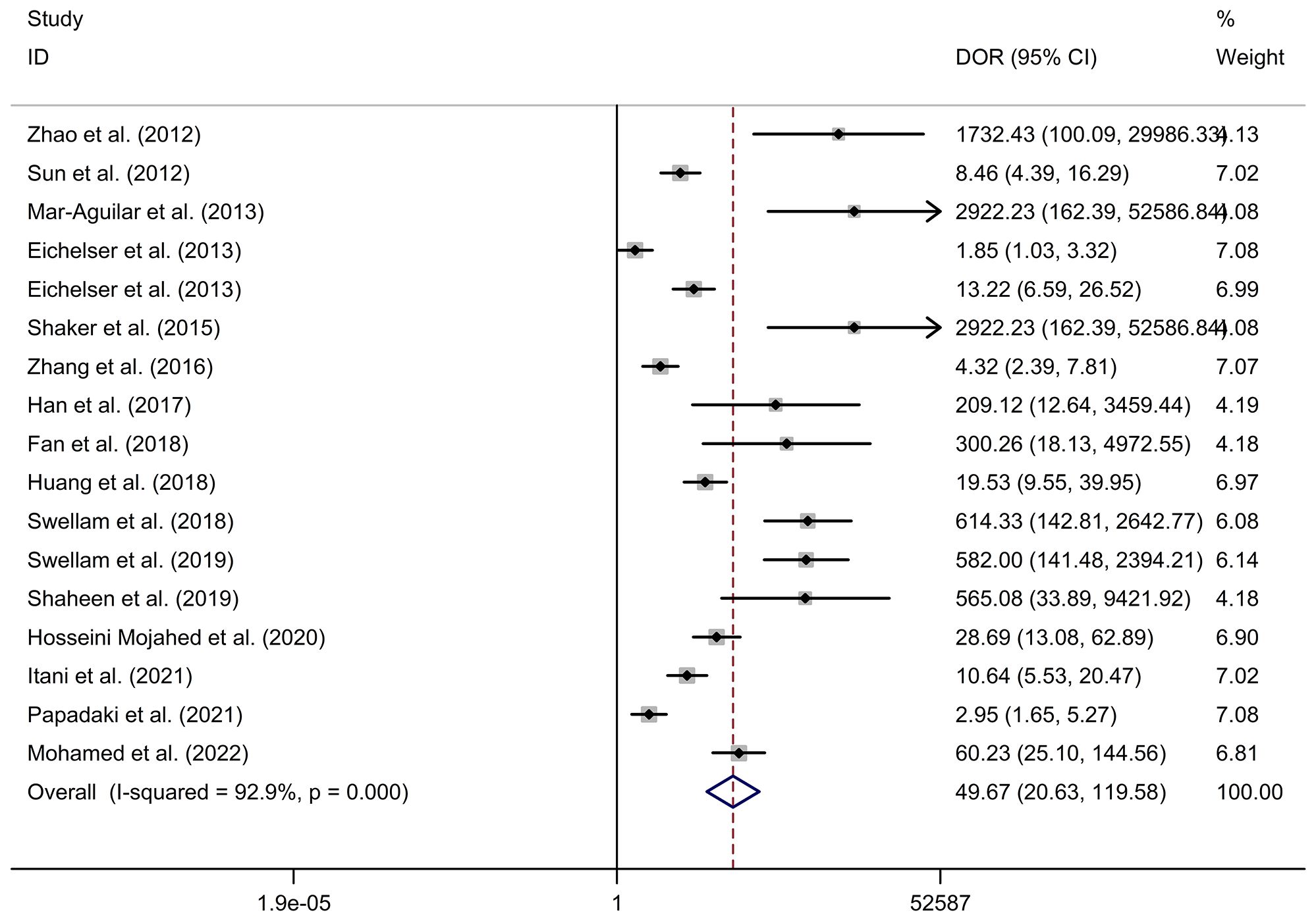
Figure 3 The DOR of circulating miR-155 in the diagnosis of BC. BC, breast cancer; DOR, diagnostic odds ratio; miR-155, microRNA-155.
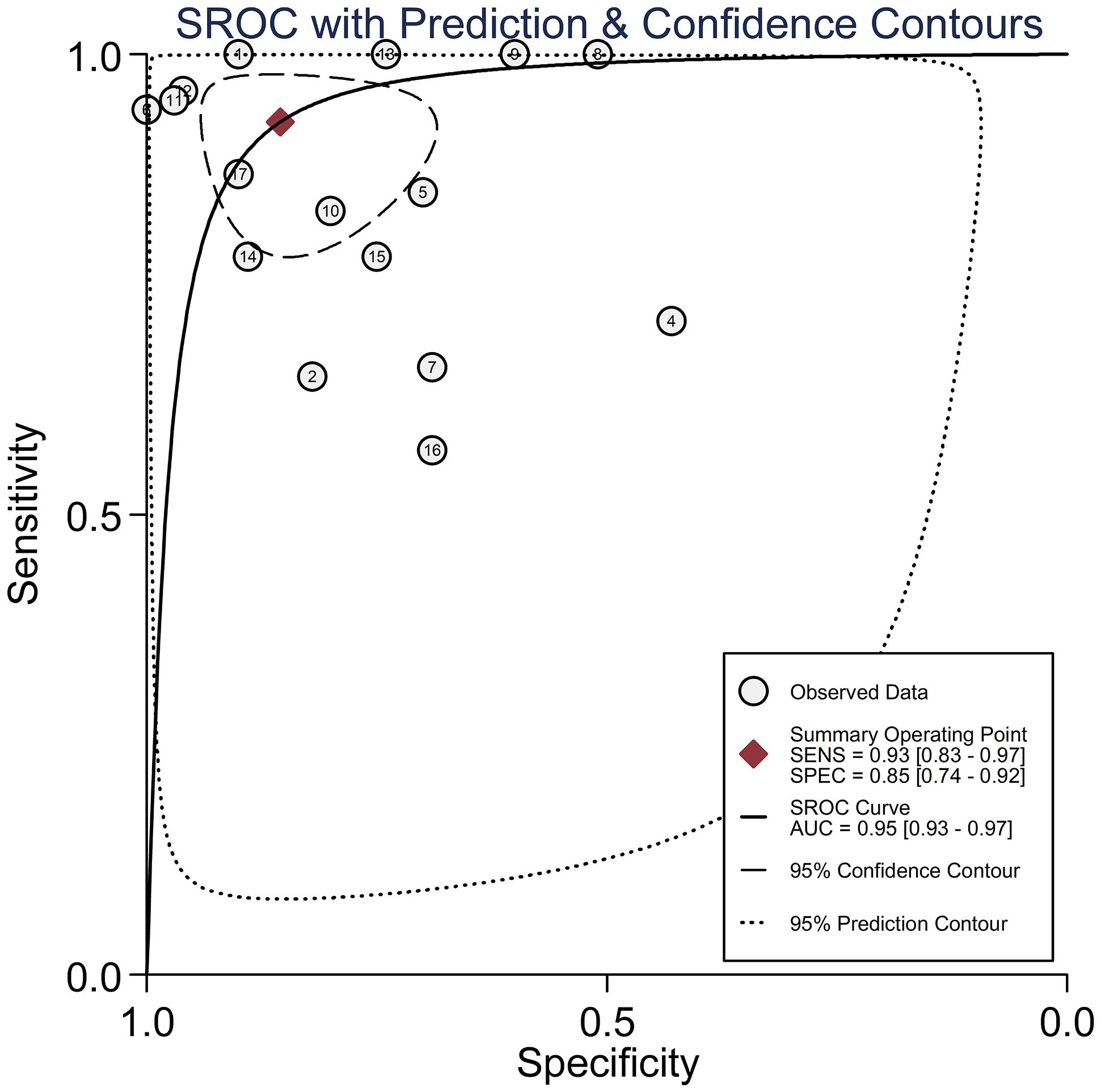
Figure 4 The SROC curve with AUC of circulating miR-155 in the diagnosis of BC. AUC, area under the curve; BC, breast cancer; DOR, diagnostic odds ratio; miR-155, microRNA-155; SROC, summary receiver operator characteristic.
Serum miR-155 showed a diagnostic value for BC. As shown in forest plot (Figures 5A, 6A), the pooled parameters calculated were as follows: sensitivity, 0.94 (95% CI: 0.85-0.97); specificity, 0.89 (95% CI: 0.76-0.96); PLR, 8.6 (95% CI: 3.6-20.4); NLR, 0.07 (95% CI: 0.03-0.17); and DOR, 123 (95% CI: 31-478). The analysis showed a high heterogeneity (sensitivity, I2 = 94.56%, p < 0.001; specificity, I2 = 96.62%, p < 0.001; DOR, I2 = 92.9%, p < 0.001). Supplementary Figure 2 showed the SROC curve, with an AUC of 0.97 (95% CI: 0.95-0.98). The symmetrical Deek’s funnel plot showed a publication bias (p < 0.01, Supplementary Figure 3).
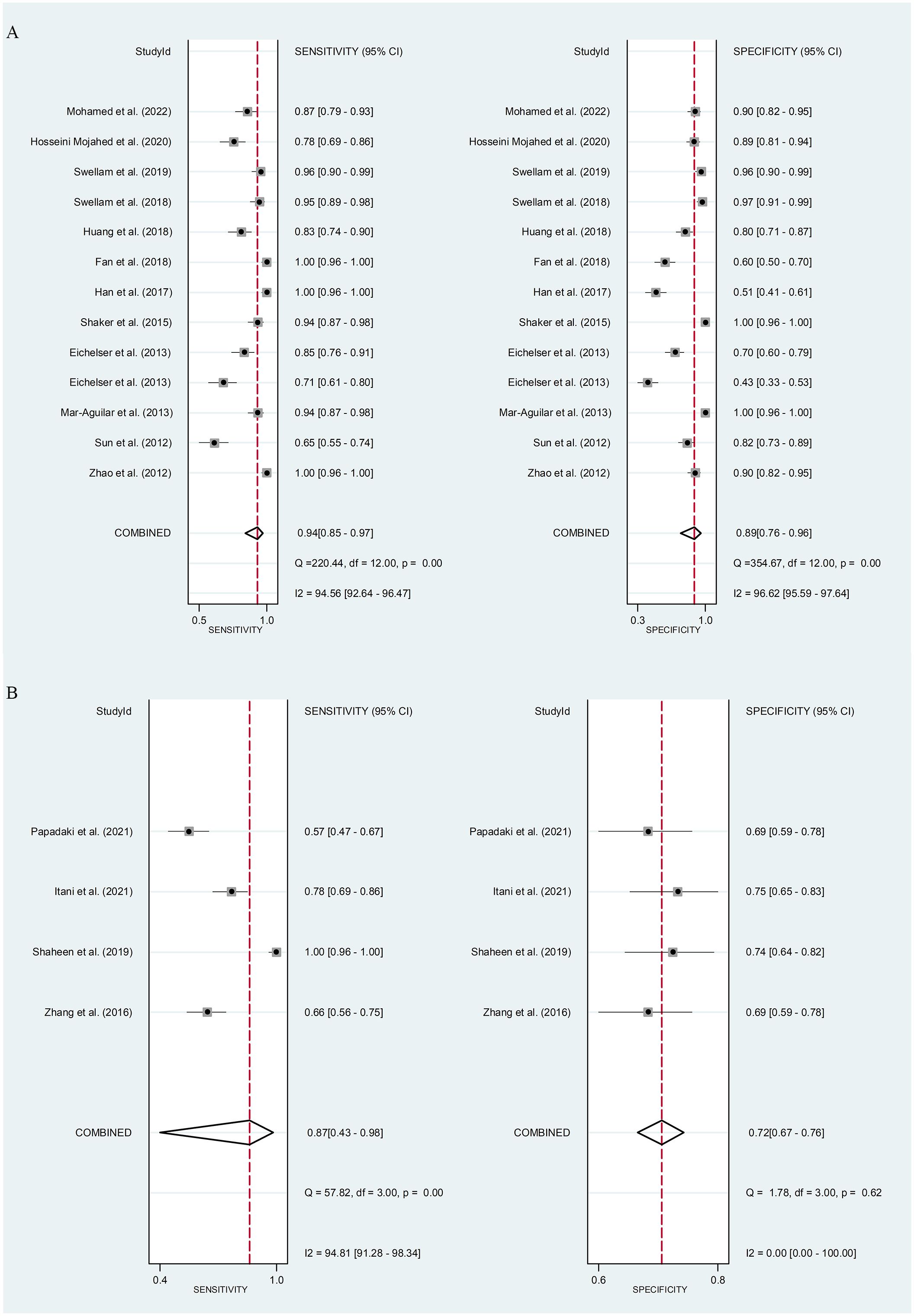
Figure 5 The sensitivity and specificity of serum miR-155 (A) or plasma miR-155 (B) in the diagnosis of BC. BC, breast cancer; miR-155, microRNA-155.
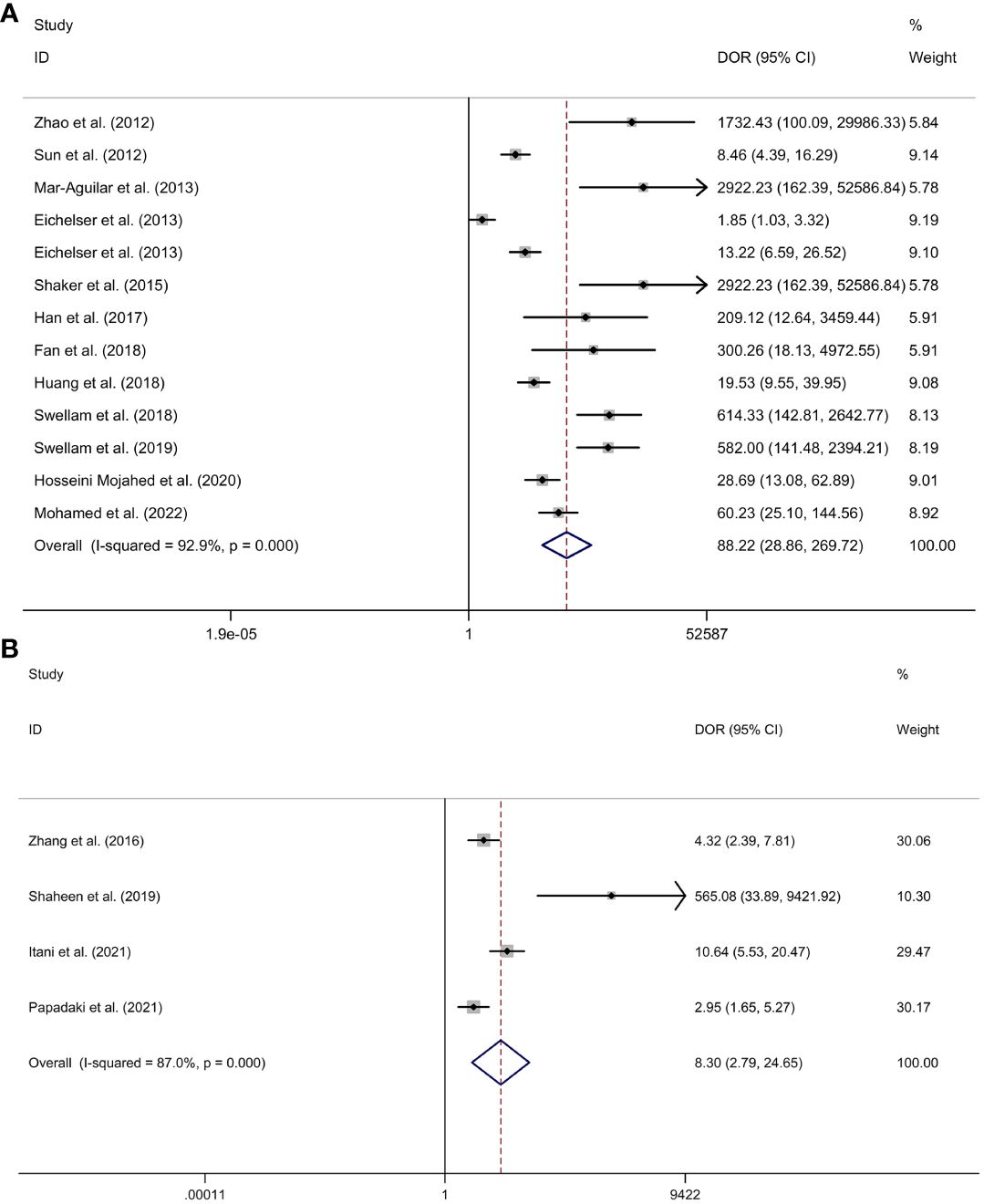
Figure 6 The DOR of serum miR-155 (A) or plasma miR-155 (B) in the diagnosis of BC. Abbreviations: BC, breast cancer; DOR, diagnostic odds ratio; miR-155, microRNA-155.
Plasma miR-155 showed a diagnostic value for BC. As shown in forest plot (Figures 5B, 6B), the pooled parameters calculated were as follows: sensitivity, 0.87 (95% CI: 0.43-0.98); specificity, 0.72 (95% CI: 0.67-0.76); PLR, 3.1 (95% CI: 2.1-4.5); NLR, 0.18 (95% CI: 0.03-1.25); and DOR, 17 (95% CI: 2-163). The analysis showed a high heterogeneity for sensitivity and DOR (sensitivity, I2 = 94.81%, p < 0.001; DOR, I2 = 92.9%, p < 0.001). The analysis showed a low heterogeneity for specificity (I2 = 0.0%, p = 0.062). Supplementary Figure 4 showed the SROC curve, with an AUC of 0.73 (95% CI: 0.95-0.98). The symmetrical Deek’s funnel plot showed a publication bias (p < 0.03, Supplementary Figure 5).
In our study, we found that the pooled sensitivity and specificity of circulating miR-155 were 0.93 (95% CI: 0.83-0.97) and 0.85 (95% CI: 0.74-0.92), respectively. The DOR and AUC of miR-155 were 74 (95% CI: 22-247) and 0.95 (95% CI: 0.93-0.97). A recent study showed that the sensitivity and specificity were 0.49 and 0.895 for carcinoembryonic antigen (CEA), 0.521 and 0.837 for carbohydrate antigen (CA)15-3, and the AUC of CEA and CA15-3 was 0.669 (95%CI: 0.595-0737) and 0.839 (95%CI: 0.777-0.889), respectively (35). It is widely recognized that the AUC should be in the region of 0.97 or above to demonstrate excellent accuracy (39). Compared to CEA and CA15-3, miR-155 has better sensitivity and specificity and may probably be suitable for the screening of BC. Therefore, in this meta-analysis, our result showed that miR-155 may be an excellent potential biomarker in the diagnosis of BC. The present study reported that serum miR-155 showed a high diagnostic value for BC (sensitivity, 0.94 (95% CI: 0.85-0.97); specificity, 0.89 (95% CI: 0.76-0.96); AUC, 0.97 (95% CI: 0.95-0.98)), whereas that plasma miR-155 showed a medium diagnostic value for BC (sensitivity, 0.87 (95% CI: 0.43-0.98); specificity, 0.72 (95% CI: 0.67-0.76); AUC, 0.73 (95% CI: 0.95-0.98)). The subgroup analyses based on specimen types revealed that serum had a higher diagnostic value compared to plasma, suggesting that serum may be a more suitable source of clinical specimens for BC detection. Specifically, miR-155 in serum demonstrated a more precise diagnostic value than in plasma. This discrepancy may be attributed to the coagulation process, which can influence the extracellular miRNA spectrum in the blood and consequently lead to varying miRNA expression levels in different specimens (40). Furthermore, differences in detection or normalizing methods between the two specimen types could also influence the diagnostic value (40). However, it is important to note that only four studies were included in the assessment of plasma miR-155 for BC diagnosis, which may have an impact on the overall clinical conclusion. Thus, more studies were essential to explore the diagnostic value of circulating miR-155 for BC.
The association between microRNAs and cancer has been a research focus in recent years, especially some most frequently studied microRNAs, such as miR-155. Recently published basic research results attempted to explain the association between miR-155 and the development of BC. Kim et al. reported that miR-155 was a key regulator of glucose metabolism in breast cancer via phosphoinositide-3-kinase regulatory subunit alpha (PIK3R1)-FOXO3a-cMYC axis and down-regulation of miR-155 could inhibit the growth of tumor in vivo (41). Wang et al. showed that miR-155 played a vital role in regulating the function of dendritic cells in BC, and miR-155 deficiency promoted BC growth in mice (42). MiR-155 was proved to play an important role in the proliferation and migration of BC cells via the down-regulation of suppressors of cytokine signaling (SOCS)1 and up-regulation of matrix metalloproteinase (MMP)16 (43). MiR-155 has also been studied as the potential prediction biomarker of early BC recurrence and therapy resistance (44, 45).
Our meta-analysis result was consistent with previous meta-analysis results published in 2014 (46). However, the previous meta-analysis only included three studies and the sample size of included studies was small. In our meta-analysis, we updated published articles regarding the association between circulating miR-155 and BC to obtain more accurate results. Some limitations still should be noticed. First, the lack of some important information from original published articles limited our research such as TNM-stage, lymph node metastasis, the level of estrogen receptor alpha (ER), progesterone receptor (PR) and human epidermal growth factor receptor (HER)-2. However, Zeng et al. reported that the expression of miR-155 was related to lymph node metastasis, and the status of ER, PR and HER-2 (47). Second, previous studies have demonstrated that circulating miR-155 was also associated with not only cancer, but also some other diseases, such as pre-eclampsia pregnancies (48), coronary artery disease (49), and multiple sclerosis (50). MiR-155-related diseases may influence the expression of miR-155 among the included samples and affect the accuracy of relevant results. So it should be noted that if we apply miR-155 as the screening BC, we should avoid the impact of miR-155-related diseases.
Up until now, we can demonstrate that circulating miR-155 has a potential in the diagnosis of BC. Before circulating miR-155 can be applied to clinical diagnosis, more large-scale clinical studies should be conducted in the future.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
FW: Writing – original draft, Software, Methodology, Investigation, Formal analysis, Data curation. JW: Writing – original draft, Methodology, Investigation, Data curation. HZ: Writing – review & editing, Investigation, Formal analysis, Data curation. BF: Writing – original draft, Formal analysis, Data curation. YZ: Writing – review & editing, Methodology. QJ: Writing – original draft, Methodology, Investigation. YW: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1374674/full#supplementary-material
1. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. (2023) 73:17–48. doi: 10.3322/caac.21763
2. Stephens JA, Fisher JL, Wesolowski R, Paskett ED. Missing components of receptor status among women with invasive breast cancer. JAMA Network Open. (2023) 6:e2330791–e. doi: 10.1001/jamanetworkopen.2023.30791
3. Wu B, Li Y, Shi B, Zhang X, Lai Y, Cui F, et al. Temporal trends of breast cancer burden in the Western Pacific Region from 1990 to 2044: Implications from the Global Burden of Disease Study 2019. J Adv Res. (2023) 6:S2090-1232(23)00182-0. doi: 10.1016/j.jare.2023.07.003
4. Li J, Chen C, Nie J, Wang L, Zhang Z, Li Y. Changes in the disease burden of breast cancer along with attributable risk factors in China from 1990 to 2019 and its projections: An analysis of the global burden of disease study 2019. Cancer Med. (2023) 12:1888–902. doi: 10.1002/cam4.5006
5. Becker S. A historic and scientific review of breast cancer: The next global healthcare challenge. Int J Gynaecol Obstet. (2015) 131 Suppl 1:S36–9. doi: 10.1016/j.ijgo.2015.03.015
6. Wadhwa A, Sullivan JR, Gonyo MB. Missed breast cancer: what can we learn? Curr Probl Diagn Radiol. (2016) 45:402–19. doi: 10.1067/j.cpradiol.2016.03.001
7. Norsuddin NM, Maslan FA. (2020). Association between breast density, lesions characteristic and diagnosis error in mammography, in: 15th International Workshop on Breast Imaging (IWBI2020). SPIE. 2020, 11513: 594–600.
8. DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, et al. Breast cancer statistics, 2019. CA: Cancer J Clin. (2019) 69:438–51. doi: 10.3322/caac.21583
9. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. (2018) 141:1202–7. doi: 10.1016/j.jaci.2017.08.034
10. Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. (2016) 17:1712. doi: 10.3390/ijms17101712
11. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. (2014) 9:287–314. doi: 10.1146/annurev-pathol-012513-104715
12. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. (2017) 16:203–22. doi: 10.1038/nrd.2016.246
13. Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A Compr Review EMBO Mol Med. (2012) 4:143–59. doi: 10.1002/emmm.201100209
14. Grimaldi AM, Incoronato M. Clinical translatability of "Identified" Circulating miRNAs for diagnosing breast cancer: overview and update. Cancers (Basel). (2019) 11:901. doi: 10.3390/cancers11070901
15. Khalighfard S, Alizadeh AM, Irani S, Omranipour R. Plasma miR-21, miR-155, miR-10b, and Let-7a as the potential biomarkers for the monitoring of breast cancer patients. Sci Rep. (2018) 8:17981. doi: 10.1038/s41598-018-36321-3
16. Shao C, Yang F, Qin Z, Jing X, Shu Y, Shen H. The value of miR-155 as a biomarker for the diagnosis and prognosis of lung cancer: a systematic review with meta-analysis. BMC Cancer. (2019) 19:1103. doi: 10.1186/s12885-019-6297-6
17. Park S, Eom K, Kim J, Bang H, Wang HY, Ahn S, et al. MiR-9, miR-21, and miR-155 as potential biomarkers for HPV positive and negative cervical cancer. BMC Cancer. (2017) 17:658. doi: 10.1186/s12885-017-3642-5
18. Wang J, Guo J, Fan H. MiR-155 regulates the proliferation and apoptosis of pancreatic cancer cells through targeting SOCS3. Eur Rev Med Pharmacol Sci. (2019) 23:5168–75. doi: 10.26355/eurrev_201906_18181
19. Li XD, Li XM, Gu JW, Sun XC. MiR-155 regulates lymphoma cell proliferation and apoptosis through targeting SOCS3/JAK-STAT3 signaling pathway. Eur Rev Med Pharmacol Sci. (2017) 21:5153–9. doi: 10.26355/eurrev_201711_13832
20. Dong ZC, Zhang D, Zhang XX, Yao ZQ, Wu H, Chen CH, et al. MiR-155 affects proliferation and apoptosis of bladder cancer cells by regulating GSK-3β/β-catenin pathway. Eur Rev Med Pharmacol Sci. (2019) 23:5682–90. doi: 10.26355/eurrev_201907_18305
21. Soleimanpour E, Babaei E, Hosseinpour-Feizi MA, Montazeri V. Circulating miR-21 and miR-155 as potential noninvasive biomarkers in Iranian Azeri patients with breast carcinoma. J Cancer Res Ther. (2019) 15:1092–7. doi: 10.4103/jcrt.JCRT_1227_16
22. Hosseini Mojahed F, Aalami AH, Pouresmaeil V, Amirabadi A, Qasemi Rad M, Sahebkar A. Clinical evaluation of the diagnostic role of microRNA-155 in breast cancer. Int J Genomics. (2020) 2020:9514831. doi: 10.1155/2020/9514831
23. Moses LE, Shapiro D, Littenberg B. Combining independent studies of a diagnostic test into a summary ROC curve: data-analytic approaches and some additional considerations. Stat Med. (1993) 12:1293–316. doi: 10.1002/sim.4780121403
24. Zhao S, Wu Q, Gao F, Zhang C, Yang X. Serum microRNA-155 as a potential biomarker for breast cancer screening. Chin Sci Bull. (2012) 57:3466–8. doi: 10.1007/s11434-012-5362-1
25. Sun Y, Wang M, Lin G, Sun S, Li X, Qi J, et al. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PloS One. (2012) 7:e47003. doi: 10.1371/journal.pone.0047003
26. Mar-Aguilar F, Mendoza-Ramírez JA, Malagón-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, et al. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers. (2013) 34:163–9. doi: 10.1155/2013/259454
27. Eichelser C, Flesch-Janys D, Chang-Claude J, Pantel K, Schwarzenbach H. Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin Chem. (2013) 59:1489–96. doi: 10.1373/clinchem.2013.205161
28. Shaker O, Maher M, Nassar Y, Morcos G, Gad Z. Role of microRNAs -29b-2, -155, -197 and -205 as diagnostic biomarkers in serum of breast cancer females. Gene. (2015) 560:77–82. doi: 10.1016/j.gene.2015.01.062
29. Zhang J, Jiang C, Shi X, Yu H, Lin H, Peng Y. Diagnostic value of circulating miR-155, miR-21, and miR-10b as promising biomarkers in human breast cancer. Int J Clin Exp Med. (2016) 9:10258–65.
30. Han JG, Jiang YD, Zhang CH, Yang YM, Pang D, Song YN, et al. A novel panel of serum miR-21/miR-155/miR-365 as a potential diagnostic biomarker for breast cancer. Ann Surg Treat Res. (2017) 92:55–66. doi: 10.4174/astr.2017.92.2.55
31. Fan T, Mao Y, Sun Q, Liu F, Lin JS, Liu Y, et al. Branched rolling circle amplification method for measuring serum circulating microRNA levels for early breast cancer detection. Cancer Sci. (2018) 109:2897–906. doi: 10.1111/cas.13725
32. Huang SK, Luo Q, Peng H, Li J, Zhao M, Wang J, et al. A panel of serum noncoding RNAs for the diagnosis and monitoring of response to therapy in patients with breast cancer. Med Sci Monit. (2018) 24:2476–88. doi: 10.12659/MSM.909453
33. Swellam M, Zahran RFK, Abo El-Sadat Taha H, El-Khazragy N, Abdel-Malak C. Role of some circulating MiRNAs on breast cancer diagnosis. Arch Physiol Biochem. (2019) 125:456–64. doi: 10.1080/13813455.2018.1482355
34. Shaheen J, Shahid S, Shahzadi S, Akhtar MW, Sadaf S. Identification of circulating miRNAs as non-invasive biomarkers of triple negative breast cancer in the population of Pakistan. Pakistan J Zool. (2019) 51:1113. doi: 10.17582/journal.pjz/2019.51.3.1113.1121
35. Swellam M, Ramadan A, El-Hussieny EA, Bakr NM, Hassan NM, Sobeih ME, et al. Clinical significance of blood-based miRNAs as diagnostic and prognostic nucleic acid markers in breast cancer: Comparative to conventional tumor markers. J Cell Biochem. (2019) 120:12321–30. doi: 10.1002/jcb.28496
36. Itani MM, Nassar FJ, Tfayli AH, Talhouk RS, Chamandi GK, Itani ARS, et al. A signature of four circulating microRNAs as potential biomarkers for diagnosing early-stage breast cancer. Int J Mol Sci. (2021) 22:6121. doi: 10.3390/ijms22116121
37. Papadaki C, Thomopoulou K, Monastirioti A, Koronakis G, Papadaki MA, Rounis K, et al. MicroRNAs regulating tumor and immune cell interactions in the prediction of relapse in early stage breast cancer. Biomedicines. (2021) 9:421. doi: 10.3390/biomedicines9040421
38. Mohamed AA, Allam AE, Aref AM, Mahmoud MO, Eldesoky NA, Fawazy N, et al. Evaluation of expressed microRNAs as prospective biomarkers for detection of breast cancer. Diagnostics (Basel). (2022) 12:789. doi: 10.3390/diagnostics12040789
39. Jones CM, Athanasiou T. Summary receiver operating characteristic curve analysis techniques in the evaluation of diagnostic tests. Ann Thorac Surgery. (2005) 79:16–20. doi: 10.1016/j.athoracsur.2004.09.040
40. Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. Comparing the MicroRNA spectrum between serum and plasma. PloS One. (2012) 7:e41561. doi: 10.1371/journal.pone.0041561
41. Kim S, Lee E, Jung J, Lee JW, Kim HJ, Kim J, et al. microRNA-155 positively regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast cancer. Oncogene. (2018) 37:2982–91. doi: 10.1038/s41388-018-0124-4
42. Wang J, Iwanowycz S, Yu F, Jia X, Leng S, Wang Y, et al. microRNA-155 deficiency impairs dendritic cell function in breast cancer. Oncoimmunology. (2016) 5:e1232223. doi: 10.1080/2162402X.2016.1232223
43. Zhang W, Chen CJ, Guo GL. MiR-155 promotes the proliferation and migration of breast cancer cells via targeting SOCS1 and MMP16. Eur Rev Med Pharmacol Sci. (2018) 22:7323–32. doi: 10.26355/eurrev_201811_16269
44. Bašová P, Pešta M, Sochor M, Stopka T. Prediction Potential of Serum miR-155 and miR-24 for Relapsing Early Breast Cancer. Int J Mol Sci. (2017) 18:2116. doi: 10.3390/ijms18102116
45. Zhang Z, Zhang L, Yu G, Sun Z, Wang T, Tian X, et al. Exosomal miR-1246 and miR-155 as predictive and prognostic biomarkers for trastuzumab-based therapy resistance in HER2-positive breast cancer. Cancer Chemother Pharmacol. (2020) 86: 761–72. doi: 10.1007/s00280-020-04168-z
46. Wang F, Hou J, Jin W, Li J, Yue Y, Jin H, et al. Increased circulating microRNA-155 as a potential biomarker for breast cancer screening: a meta-analysis. Molecules (Basel Switzerland). (2014) 19:6282–93. doi: 10.3390/molecules19056282
47. Zeng H, Fang C, Nam S, Cai Q, Long X. The clinicopathological significance of microRNA-155 in breast cancer: a meta-analysis. BioMed Res Int. (2014) 2014:724209. doi: 10.1155/2014/724209
48. Gan L, Liu Z, Wei M, Chen Y, Yang X, Chen L, et al. MiR-210 and miR-155 as potential diagnostic markers for pre-eclampsia pregnancies. Medicine. (2017) 96:e7515. doi: 10.1097/MD.0000000000007515
49. Faccini J, Ruidavets JB, Cordelier P, Martins F, Maoret JJ, Bongard V, et al. Circulating miR-155, miR-145 and let-7c as diagnostic biomarkers of the coronary artery disease. Sci Rep. (2017) 7:42916. doi: 10.1038/srep42916
Keywords: breast cancer, miR-155, meta-analysis, microRNA, systematic review
Citation: Wang F, Wang J, Zhang H, Fu B, Zhang Y, Jia Q and Wang Y (2024) Diagnostic value of circulating miR-155 for breast cancer: a meta-analysis. Front. Oncol. 14:1374674. doi: 10.3389/fonc.2024.1374674
Received: 22 January 2024; Accepted: 07 March 2024;
Published: 25 March 2024.
Edited by:
Maria Rosaria De Miglio, University of Sassari, ItalyReviewed by:
Amal Ramadan, National Research Centre, EgyptCopyright © 2024 Wang, Wang, Zhang, Fu, Zhang, Jia and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Wang, WW9uZ1dhbmczM18xQG91dGxvb2suY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.