
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 10 May 2024
Sec. Cancer Genetics
Volume 14 - 2024 | https://doi.org/10.3389/fonc.2024.1374087
Background: Comprehensive molecular profiling of tissue samples that can help guide therapy management is not widely available across the globe.
Methods: Comprehensive molecular profiling through Caris Molecular Intelligence involves the analysis of DNA through next-generation sequencing, chromogenic or fluorescent in situ hybridization, pyrosequencing, and copy number alterations; RNA through whole-transcriptome sequencing and multiplex PCR of RNA; and protein through immunohistochemistry.
Results: Here we describe the experience of molecular profiling of tumor tissue samples from patients diagnosed with advanced solid tumors and treated in two countries, the United Arab Emirates and Thailand. Tumor cancer cases submitted to Caris Life Sciences (Phoenix, Arizona, USA) for molecular profiling from the UAE and Thailand were retrospectively analyzed (data accessed between 2019 and 2020) for their molecular alterations and clinical biomarkers, without regard to ethnicity. A total of 451 samples from 35 distinct types of advanced cancers were examined for mutations, amplifications, overexpression, exon copy number alterations, microsatellite instability, deficient mismatch repair, tumor mutational burden, and fusions. Interrogating each step of the biological pathway, from DNA to RNA to distinct protein, identified an alteration with an associated therapy for 75% of these tumor samples. The most common alterations identified included elevated PDL-1 that can be targeted with an immune checkpoint inhibitors and amplification of HER2 for which a variety of anti HER2 therapies are available.
Conclusion: Comprehensive molecular profiling in patients with advanced malignancies can help optimize therapeutic management allowing for improved prognostic outcome.
Comprehensive molecular profiling has been empowered by rapid advances in molecular technologies examining DNA, RNA, and proteins. Through comprehensive molecular profiling, disease biomarkers can be identified and used to expand patients’ treatment options by revealing effective targeted therapies in what became known as precision medicine. When explored, molecular markers have been shown to carry clinical benefit in several therapeutic areas, such as infectious diseases, cardiovascular diseases, neurodegenerative disorders and most notably, malignancies (1–4).
In oncology, trends in molecular profiling have been increasingly positive as the list of approved biomarker matching targeted drugs continues to grow and new biomarkers continue to be identified. Several molecular markers have been detected in various cancer types that can be targeted with a molecular-matched therapy such as NTRK inhibitors, KRAS G12C inhibitors, and immune checkpoint inhibitors, to name a few. Other gene or protein alterations guiding treatment decisions are ALK, BCL-2, BRAF, BRCA, HER2, EGFR, and RAS mutations, among others (5). Molecular-guided treatment approaches thus allow treatment personalization and targeted cancer management, which was reflected in remarkable improvements in non-small cell lung cancer (NSCLC) outcomes (6, 7). It has become evident that the use of comprehensive molecular profiling to identify various actionable markers carries clinical benefit for cancer patients and improves overall survival (8, 9). Tissue-agnostic clinical trials have thus risen to prominence, constituting a noteworthy paradigm shift in both precision medicine and cancer treatment away from site-driven management (10). As of April 2024, seven therapies have been approved by the US FDA for tissue agnostic indications and include pembrolizumab [microsatellite instability and tumor mutational burden (TMB) ≥10 mutations/megabase (mut/Mb)] and larotrectinib/entrectinib (NTRK fusions) (11). This list is expected to grow as other agnostic biomarkers such as RET are currently being explored in clinical trials. Previous work has examined the relationships between biomarkers and cancer therapeutics, such as immune checkpoint inhibitors (12). However, comprehensive analyses remain necessary to elucidate the interplay between various biomarkers and different cancer types across molecular function levels, including DNA, RNA and proteins.
The use of molecular profiling is rapidly increasing in developed countries, but its systematic adoption remains quite limited around the globe. Molecular profiling relies heavily on next-generation sequencing (NGS), which is not readily available in several less industrialized regions. This study included samples from patients with 35 different types of advanced cancer from the United Arab Emirates (UAE) in the Middle East and Thailand in Asia. Here, we tested DNA, RNA, and protein to comprehensively seek targetable biomarkers. We aim to summarize the experience of these two underreported countries using a CLIA-certified diagnostic service.
Tumor cancer cases submitted to Caris Life Sciences (Phoenix, Arizona, USA) for molecular profiling were retrospectively analyzed (data accessed between 2019 and 2020) for their molecular alterations and clinical biomarkers. In total, 451 individual patient tumor samples were received and analyzed by Caris Life Science. The samples were simply sourced from two separate geographical regions (the UAE and Thailand); sub-analysis by ethnicity was not targeted. Authors had no access to information that could identify individual participants during or after data collection. Caris Molecular Intelligence includes NGS, whole-transcriptome sequencing (WTS), and relevant immunohistochemistry (IHC).
Formalin-fixed paraffin-embedded (FFPE) samples were sent for analysis from treating physicians from Thailand and UAE. The tissue diagnoses were submitted based on pathologic assessment of physicians who requested the assays and were further verified by a board-certified oncological pathologist at the Caris laboratory. Disease classification was then determined based on assessment of the submitted tissue along with associated clinical documentation. Age, gender, and specimen source information were available for analysis but demographic and patient clinical information were unavailable. Formalin-fixed paraffin-embedded (FFPE) tissue specimens were analyzed using NGS, IHC, chromogenic or fluorescent in situ hybridization (CISH/FISH), and/or pyrosequencing.
NGS was performed on genomic DNA isolated from FFPE tumor samples using the NextSeq platform (Illumina, Inc., San Diego, CA). Prior to molecular testing, tumor enrichment was achieved by harvesting targeted tissue using manual microdissection techniques. Matched normal tissue was not sequenced. A custom-designed SureSelect XT assay was used to enrich 592 whole-gene targets (Agilent Technologies, Santa Clara, CA). All variants were detected with > 99% confidence based on allele frequency and amplicon coverage, with an average sequencing depth of coverage of > 500-fold and an analytic sensitivity of 5%. Genetic variants identified were interpreted by board-certified molecular geneticists and categorized as ‘pathogenic,’ ‘presumed pathogenic,’ ‘variant of unknown significance,’ ‘presumed benign,’ or ‘benign,’ according to the American College of Medical Genetics and Genomics (ACMG) standards. When assessing mutation frequencies of individual genes, ‘pathogenic,’ and ‘presumed pathogenic’ were counted as mutations while ‘benign’, ‘presumed benign’ variants and ‘variants of unknown significance’ were excluded.
The copy number alteration (CNA) of each exon was determined in DNA by calculating the average depth of the sample along with the sequencing depth of each exon and comparing this calculated result to a pre-calibrated value.
Microsatellite instability (MSI) was examined using over 7,000 target microsatellite loci and comparing those with the reference genome hg19 from the University of California, Santa Cruz (UCSC) Genome Browser database (13). The status was defined as MSI-high (MSI-H) or MSI-low/microsatellite-stable (MSS). The number of microsatellite loci that were altered by somatic insertion or deletion were counted for each sample. Only insertions or deletions that increased or decreased the number of repeats were considered. Genomic variants in the microsatellite loci were detected using the same depth and frequency criteria as used for mutation detection. MSI-NGS results were compared with results from over 2,000 matching clinical cases analyzed with traditional PCR-based methods (14). In order to generate a sensitivity of >95% and specificity of >90%, the threshold to determine MSI by NGS was determined to be 46 or more loci with insertions or deletions. MSI and mismatch repair (MMR) status were determined with a combination of test platforms that included fragment analysis (FA; Promega, Madison, WI), IHC (MLH1: M1 antibody; MSH2: G2191129 antibody; MSH6: 44 antibody; and PMS2: EPR3947 antibody [Ventana Medical Systems, Inc., Tucson, AZ, USA]), and NGS. NGS used the NextSeq platform, which has 7,000 target microsatellite loci that were examined and compared to the reference genome hg19 from the University of California. The status of the tumor regarding MSI and MMR was determined first with IHC, then with FA, and finally with NGS if necessary.
TMB was measured by counting all non-synonymous missense, nonsense, in-frame insertions, in-frame deletions, and frameshift mutations found per tumor that had not been previously described as germline alterations in dbSNP151, Genome Aggregation Database (gnomAD) databases (15), or benign variants identified by Caris geneticists. A cutoff point of ≥ 10 mut/Mb was used based on the KEYNOTE-158 pembrolizumab trial (16), which showed that patients with a TMB of ≥ 10 mut/Mb across several tumor types had higher response rates than patients with a TMB of < 10 mut/Mb. Caris Life Sciences is a participant in the Friends of Cancer Research TMB Harmonization Project (17).
Gene fusions were detected by ArcherDx fusion assay (Archer FusionPlex Solid Tumor panel) on Illumina MiSeq platform or WTS (Agilent SureSelect Human All Exon V7 bait panel) on Illumina NovaSeq platform (Illumina, Inc., San Diego, CA). The formalin-fixed paraffin-embedded tumor samples underwent pathology review and were micro-dissected to enrich the tumor nuclei prior to mRNA extraction. For Archer assay, unidirectional gene-specific primers were used to enrich for target regions, followed by NGS (Illumina MiSeq platform). Targets included 52 genes, and the full list can be found at http://archerdx.com/fusionplex-assays/solid-tumor. For WTS, biotinylated RNA baits were hybridized to the synthesized and purified cDNA targets and the bait-target complexes were amplified in a post capture PCR reaction. The resultant libraries were quantified, normalized and the pooled libraries are denatured, diluted, and sequenced; the reference genome used was GRCh37/hg19.
Chromogenic in situ hybridization (CISH) was used for Her2/neu (INFORM HER2 Dual ISH DNA Probe Cocktail). Her2 test results were considered amplified if HER2/CEP17 ratio ≥ 2.0 with an average HER2 copy number ≥ 4.0 signals per cell; or if HER2/CEP17 ratio ≥ 2.0 with an average HER2 copy number < 4.0 signals/cell; or HER2/CEP17 ratio < 2.0 with an average HER2 copy number ≥ 6.0 signals/cell.
MGMT promoter methylation was evaluated by pyrosequencing. DNA was extracted from FFPE tumor samples. The five CpG sites (CpGs 74-78) were analyzed by pyrosequencing. All DNA samples underwent a bisulfite treatment and were PCR amplified with primers specific for exon 1 of MGMT (GRCh37/hgl9 – chr10: 131,265,448- 131,265,560). The methylation status of PCR-amplified products was determined using the PyroMark system. Samples with < 7% methylation were considered negative, those with > 9% methylation were considered positive, and those with ≥ 7% and < 9% methylation were considered to equivocal results.
Associations between drugs and biomarkers were determined based on a combination of FDA guidelines, professional society guidelines and review and summary of clinical literatures.
This study was conducted using retrospective, de-identified clinical data submitted to Caris Life Sciences (Phoenix, Arizona, USA), and patient consent was not required for the processed specimens in Thailand. Ethical approval was obtained from the research ethics committee as per local regulations in the UAE.
Of the 451 submitted for molecular testing, 446 (99%) were processed and 405 (91%) were sequenced successfully with all tests of the Caris Molecular Intelligence assays that were deemed relevant for the specific cancer type (Figure 1). Partial, rather than comprehensive molecular testing, was be performed on 41 samples (9%) due to tissue with limited quantities or poor quality. A total of 6 samples (1%) were not tested due to the samples having an insufficient quantity for analysis.
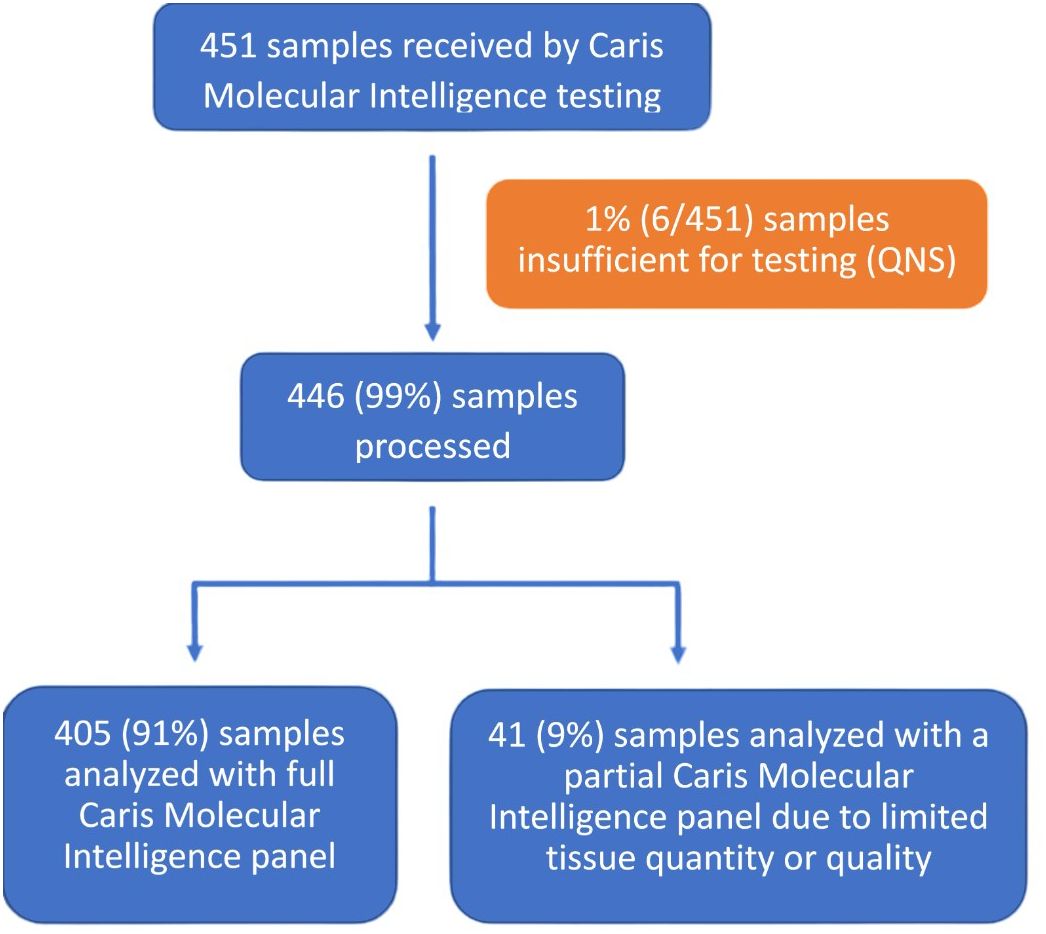
Figure 1 Specimen processing and overall clinical utility. CMI, Caris Molecular Intelligence; QNS, quantity not sufficient.
Caris Life Science received 451 individual patient tumor samples between 2019 and 2020 (Figure 2), which included 303 from Thailand and 148 from UAE. The median age of the patients was 59 years, with a range from 19 to over age 89 years, and the patients were 51% male and 49% female. A total of 35 distinct tumor types were included, with the most common cancers being colorectal adenocarcinoma (CRC; 13.5%), NSCLC (11.8%), and breast carcinoma (10.6%) in the overall cohort. Following CRC and NSCLC, pancreatic adenocarcinoma and bladder cancer, urothelial were the third most common in Thailand. Breast carcinoma was the most common cancer type in the UAE, followed by CRC and NSCLC.
Of the 451 samples profiled, 436 (97%) had at least one alteration detected (Table 1), and 340 (75%) had at least one alteration with an associated therapy (Figure 3). The frequency of pathogenic and likely-pathogenic genomic alterations varied widely across tumor types. DNA mutations and copy number amplifications were the most commonly observed events. Immunotherapy biomarkers, including elevated PD-L1 and TMB-high, were most common in NSCLC (PD-L1 ≥1% in 63%, ≥10% in 43%, and TMB-high in 17% among 53 samples), head and neck cancer (PD-L1 ≥1% in 100%, ≥10% in 77% among 14 samples), and in urothelial bladder cancer (PD-L1 ≥1% in 88% and TMB-high in 33% among 33 samples).
Biomarkers with potential therapy associations were identified on a transcriptomic and protein level, as appropriate (Table 2); Biomarker alterations associated with a therapy were identified 596 times by NGS, 389 by IHC, and 12 by WTS. Additionally, specific alterations were identified 12 times by CISH for HER2, 5 by pyrosequencing for MGMT-methylation, 2 by CNA for MET alterations, and 10 by the combination of IHC, FA, and, if necessary, NGS for dMMR/MSI-H.
Observed protein alterations included positive expression of ER by IHC (55%, n = 39 of 71 samples), PR by IHC (39%, n = 25 of 65 samples), and AR (48%, n = 29 of 60 samples). NGS identified mutations as commonly occurring in TP53 (60% of 385 samples tested), KRAS (22% of 404 samples tested), APC (13% of 398 samples tested), PIK3CA (13% of 403 samples tested), and KMT2D (9% of 366 samples tested) across the study cohort. CISH identified HER2 amplification in 12 of the 55 samples tested, plus IHC identified HER2 amplification in 10 of 141 samples tested, while CNA identified alterations of HER2 copy numbers in 15 of 400 samples tested, and NGS identified DNA alterations of HER2 in 10 of 141 samples tested, which included 8 samples with HER2 mutations. Actionable BRCA1/2 mutations detected by NGS were not limited to breast and ovarian cancer, but were observed in several tumor types (e.g. colorectal cancer, glioblastoma, head and neck cancer, pancreatic adenocarcinoma, prostate cancer). IHC was also performed in a subset of patients in a reflex situation to detect EGFR in the absence of DNA.
A total of 24 aberrations in RNA were identified by RNA sequencing or by WTS (Table 3). Among the six fusions identified in breast carcinoma samples of the 38 samples examined, three involved NOTCH2, and one each involved FGFR3, NTRK3, and BRAF. Among 53 NSCLC samples examined for fusions, three had fusions involving ALK, two involved RET, and one involved ROS1. Among the 6 fusions identified in NSCLC samples, three involved ALK and two involved RET. Among the nine bone cancer samples examined, three fusions involving EWSR1 were identified. Among 20 cancers of unknown primary examined, two had the CLDN18:ARHGAP26 fusion, which has been reported in gastric carcinomas and may suggest the origin of the unknown primary tumor (18, 19).
Challenging clinical cases, such as advanced tumors, relapsed/refractory tumors and cancers of unknown origin, push the boundaries of traditional cancer treatment. Tumor-agnostic therapy bypasses the inherent limitation of treating malignancies based on pathologic classification and tissue of origin through the means of a targeted approach. Molecular screening has been shown to be both clinically feasible and effective in revealing actionable alterations and/or resistance-conferring mutations (20). Tissue-agnostic therapy allows early treatment personalization based on genetic mutations and biomarkers and improves clinical outcomes (e.g. treatment response, overall and progression-free survival) (8, 9). This therapeutic advantage was illustrated in the present study; biomarkers with therapy associations were identified among 75% of samples subjected to comprehensive exome and transcriptome analysis. Notably, 85% of samples from cancer of unknown primary had at least one alteration associated with a therapy identified. In the absence of a clear standard treatment paradigm for cancer of unknown primary, comprehensive molecular profiling clearly expands treatment options beyond systemic therapy or palliative care.
Elevated levels of PD-L1 and amplification of HER2 were frequently observed among the targetable alterations identified in this study. Once high levels of PD-L1 expression are detected, targeted therapy through immune checkpoint inhibitors can be used. In a similar vein, HER2-targeting drugs can be commenced in case of identified HER2 amplification. Targetable HER2 alterations include ERBB2 mutations, amplifications, and changes in HER2 protein expression. The oncogenic potential of HER2 alterations is variable and dependent on primary tumor histology (21). Targeting HER2 has expanded from treating the overexpression of HER2 in breast cancer along to other malignancies. Based on this, HER2 inhibitors could be considered for patients in the study sample with actionable HER2 amplification in their tumors, which included colorectal cancer, stomach cancer and esophageal cancer. Similarly, the PD-L1-directed therapy pembrolizumab provides clinical benefit in patients with advanced cancers of different origins that are heavily pre-treated, have metastatic breast cancer, and high TMB (22). PD-L1 expression ensures candidacy for PD-L1 inhibitors across a wide range of tumor types, such as advanced/metastatic NSCLC, head and neck squamous cell carcinoma, advanced/metastatic urothelial carcinoma, and gastric or gastroesophageal junction adenocarcinoma (23). Both PD-L1 expression and TMB were found to be a promising biomarker of survival and response to precision immunotherapy in NSCLC (24). The predictive efficacy of TMB has recently risen to prominence, with demonstrated prognostic role in malignancies such as NSCLC and CRC (25–28). TMB reflects the total number of somatic mutations per coding area of a tumor genome and shows variability according to tumor type as well as individual patients (29–31). Despite that, TMB seems promising in predicting pathological response to targeted therapy such as immune checkpoint inhibitors, as well as resistance to treatment (25–28). That being said, more prospective trials are needed to fully confirm these associations with treatment efficacy and clinical outcomes.
It is therefore clear, both on a theoretical and clinical level, that an evidence-based approach to identify biomarker-drug associations has the potential to transform patient outcomes. Even though individual markers have low prevalence, comprehensive molecular testing makes use of genomic, transcriptomic, and proteomic insights from the tumor tissue to maximize the chance for target identification. Some examples from the present study show the opportunities provided by molecular profiling in guiding and personalizing cancer therapy even for less common genetic alterations. One of the breast carcinoma samples tested exhibited a fusion in the neurotrophic receptor tyrosine kinase 3 (NTRK3) gene. Fusions of the NTRK gene are clinically actionable, with histology-agnostic responses to larotrectinib and entrectinib occurring in both adult and pediatric patients with cancers exhibiting NTRK fusions (32). Patients whose locally advanced or metastatic urothelial carcinoma carries FGFR3-TACC3 fusions or certain genetic alterations in FGFR3 or FGFR2 can be treated with erdafitinib, with an overall response rate of almost 50% (33). Additional agents targeting multiple FGFRs or selectively targeting FGFR4 are in development (34). Efforts are underway to expand the use of FGFR-targeting molecules to earlier disease stages and other solid tumors. As more evidence emerges, this targeted therapy could prove valuable for patients carrying genetic mutations or fusions in FGFR, such as those observed in our study with high-grade glioma, ovarian cancer and pancreatic cancer.
This study is limited by its lack of follow-up data on the number of patients who received the suggested therapies and on their respective clinical outcomes. Regardless, it is expected that clinical benefit follows the use of therapies identified by Caris Molecular Intelligence. This agrees with recent publications that indicate clinical benefit occurred in 37% of patients who received the suggested therapy (8) and that overall survival was significantly improved for patients who received matched therapies compared with those who received unmatched therapies (9).
It is therefore essential to improve access to molecular profiling across the globe. Broad molecular profiling continues to face significant challenges, such as lack of infrastructure, lack of funding and low awareness; this in turn contributes to the poor accessibility of genomic testing and the limited understanding of its benefit and cost effectiveness among clinical oncologists and stakeholders (35, 36). Sporadic drug availability also hinders the implementation of molecular profile-based precision medicine in clinical practice (37). Policy changes are imperative for comprehensive multidisciplinary cancer management and should aim to promote equitable access to comprehensive molecular profiling, while also ensuring that both targeted therapies and adequate clinical expertise are available (38, 39).
Comprehensive molecular profiling identified biomarkers associated with therapies for 75% of the samples tested in this population of patients from countries where molecular profiling is not widely used. Molecular profiling offers the majority of these patients an option for a therapy that is targeted to their tumor. This is expected to lead to improved clinical outcomes. Large prospective trials remain necessary to confirm these findings and demonstrate the benefit of integrating comprehensive molecular profiling into clinical practice, particularly in developing countries with limited molecular resources.
The datasets presented in this article are not readily available because of the terms of use of Caris Molecular Intelligence. Requests to access the datasets should be directed to the corresponding author.
The studies involving humans were approved by Dubai Scientific Research Ethics Committee (DSREC), Dubai Health Authority Dubai, UAE. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because the study used retrospective, de-identified clinical data submitted to Caris Life Sciences (Phoenix, Arizona, USA). Ethical approval was waived in Thailand.
SD: Conceptualization, Investigation, Supervision, Writing – review & editing. VN: Conceptualization, Funding acquisition, Project administration, Writing – review & editing. PD: Conceptualization, Formal analysis, Investigation, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study received funding from Caris Life Sciences. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication. All authors declare no other competing interests.
Kathy Boltz, PhD, of On Point Scientific, LLC, provided medical writing support, which was funded by Caris Life Sciences, Phoenix, Arizona, USA. Nancy Al Akkary, MSc, BSc and Racha Aaraj, Pharm D, MSc, MPH from Phoenix Clinical Research, Lebanon provided medical writing assistance for the preparation of this manuscript. Pilar Ramos, PhD, Klaus Schuster, MD and Michael Foss, PhD provided support in compiling the data for the study.
Author SD has performed paid consultancy services for the following commercial entities: Newbridge Pharmaceuticals, Caris Life Sciences, Janssen, BMS, Lilly, MSD, AZ, Pfizer, Novartis, Ipsen, AZ and AMGEN. These services were provided independently. Author PD has performed paid consultancy services for the following commercial entities: MSD, Roche, Astra Zeneca, Astellas; Lectures for MSD, Roche, Astra Zeneca, Astellas, Janssen, BMS. These services were provided independently. He has also performed paid consultancy services for the following commercial entities: clinical studies: MSD, Roche, Astra Zeneca; Research University Network of Thailand; These services were provided on behalf of Mahidol University, Thailand. Author VN was employed by Caris Life Sciences.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Wijerathne H, Witek MA, Baird AE, Soper SA. Liquid biopsy markers for stroke diagnosis. Expert Rev Mol Diagn. (2020) 20:771–88. doi: 10.1080/14737159.2020.1777859
2. Saarenheimo J, Andersen H, Eigeliene N, Jekunen A. Gene-guided treatment decision-making in non-small cell lung cancer – A systematic review. Front Oncol. (2021) 11:754427/BIBTEX. doi: 10.3389/fonc.2021.754427
3. Banerjee U, Baloni P, Singh A, Chandra N. Immune subtyping in latent tuberculosis. Front Immunol. (2021) 12:595746. doi: 10.3389/FIMMU.2021.595746
4. Nabais MF, Laws SM, Lin T, Vallerga CL, Armstrong NJ, Blair IP, et al. Meta-analysis of genome-wide DNA methylation identifies shared associations across neurodegenerative disorders. Genome Biol. (2021) 22(1):90. doi: 10.1186/S13059-021-02275-5
5. Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. Molecular profiling for precision cancer therapies. Genome Med 2020 121. (2020) 12:1–19. doi: 10.1186/S13073-019-0703-1
6. Yuan M, Huang LL, Chen JH, Wu J, Xu Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct Target Ther 2019 41. (2019) 4:1–14. doi: 10.1038/s41392-019-0099-9
7. Xu Y, Wang Q, Xie J, Chen M, Liu H, Zhan P, et al. The predictive value of clinical and molecular characteristics or immunotherapy in non-small cell lung cancer: A meta-analysis of randomized controlled trials. Front Oncol. (2021) 11:732214/BIBTEX. doi: 10.3389/fonc.2021.732214
8. Cobain EF, Wu Y-M, Vats P, Chugh R, Worden F, Smith DC, et al. Assessment of clinical benefit of integrative genomic profiling in advanced solid tumors. JAMA Oncol. (2021) 7(4):525–533. doi: 10.1001/jamaoncol.2020.7987
9. Kopetz S, Mills Shaw KR, Lee JJ, Zhang J, Litzenburger B, Holla V, et al. Use of a targeted exome next-generation sequencing panel offers therapeutic opportunity and clinical benefit in a subset of patients with advanced cancers. JCO Precis Oncol. (2019) 3:1–14. doi: 10.1200/PO.18.00213
10. Seligson ND, Knepper TC, Ragg S, Walko CM. Developing drugs for tissue-agnostic indications: A paradigm shift in leveraging cancer biology for precision medicine. Clin Pharmacol Ther. (2021) 109:334–42. doi: 10.1002/CPT.1946
11. Adashek JJ, Subbiah V, Kurzrock R. From tissue-agnostic to N-of-one therapies: (R)Evolution of the precision paradigm. Trends Cancer. (2021) 7:15–28. doi: 10.1016/J.TRECAN.2020.08.009
12. Vanderwalde A, Spetzler D, Xiao N, Gatalica Z, Marshall J. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med. (2018) 7:746–56. doi: 10.1002/cam4.1372
13. Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. (2002) 12:996–1006. doi: 10.1101/gr.229102
14. Saul M, Poorman K, Tae H, Vanderwalde A, Stafford P, Spetzler D, et al. Population bias in somatic measurement of microsatellite instability status. Cancer Med. (2020) 9:6452–60. doi: 10.1002/CAM4.3294
15. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. (2020) 581:434–43. doi: 10.1038/s41586-020-2308-7
16. Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. (2020) 21:1353–65. doi: 10.1016/S1470-2045(20)30445-9
17. Merino DM, McShane LM, Fabrizio D, Funari V, Chen S-J, White JR, et al. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the Friends of Cancer Research TMB Harmonization Project. J Immunother Cancer. (2020) 8:e000147. doi: 10.1136/jitc-2019-000147
18. Shu Y, Zhang W, Hou Q, Zhao L, Zhang S, Zhou J, et al. Prognostic significance of frequent CLDN18-ARHGAP26/6 fusion in gastric signet-ring cell cancer. Nat Commun. (2018) 9:2447. doi: 10.1038/s41467-018-04907-0
19. Yao F, Kausalya JP, Sia YY, Teo ASM, Lee WH, Ong AGM, et al. Recurrent fusion genes in gastric cancer: CLDN18-ARHGAP26 induces loss of epithelial integrity. Cell Rep. (2015) 12:272–85. doi: 10.1016/j.celrep.2015.06.020
20. Flaherty KT, Gray RJ, Chen AP, Li S, McShane LM, Patton D, et al. Molecular landscape and actionable alterations in a genomically guided cancer clinical trial: national cancer institute molecular analysis for therapy choice (NCI-MATCH). J Clin Oncol. (2020) 38:3883–94. doi: 10.1200/JCO.19.03010
21. Oh D-Y, Bang Y-J. HER2-targeted therapies — a role beyond breast cancer. Nat Rev Clin Oncol. (2020) 17:33–48. doi: 10.1038/s41571-019-0268-3
22. Alva AS, Mangat PK, Garrett-Mayer E, Halabi S, Hansra D, Calfa CJ, et al. Pembrolizumab in patients with metastatic breast cancer with high tumor mutational burden: results from the targeted agent and profiling utilization registry (TAPUR) study. J Clin Oncol. (2021) 39:2443–51. doi: 10.1200/JCO.20.02923
23. Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S, et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol. (2021) 18:345–62. doi: 10.1038/s41571-021-00473-5
24. Yu Y, Zeng D, Ou Q, Liu S, Li A, Chen Y, et al. Association of survival and immune-related biomarkers with immunotherapy in patients with non-small cell lung cancer: A meta-analysis and individual patient-level analysis. JAMA Netw Open. (2019) 2(7):e196879. doi: 10.1001/JAMANETWORKOPEN.2019.6879
25. Deng H, Zhao Y, Cai X, Chen H, Cheng B, Zhong R, et al. PD-L1 expression and Tumor mutation burden as Pathological response biomarkers of Neoadjuvant immunotherapy for Early-stage Non-small cell lung cancer: A systematic review and meta-analysis. Crit Rev Oncol Hematol. (2022) 170:103582. doi: 10.1016/J.CRITREVONC.2022.103582
26. Galvano A, Gristina V, Malapelle U, Pisapia P, Pepe F, Barraco N, et al. The prognostic impact of tumor mutational burden (TMB) in the first-line management of advanced non-oncogene addicted non-small-cell lung cancer (NSCLC): a systematic review and meta-analysis of randomized controlled trials. ESMO Open. (2021) 6(3):100124. doi: 10.1016/J.ESMOOP.2021.100124
27. Randon G, Intini R, Cremolini C, Elez E, Overman MJ, Lee J, et al. Tumour mutational burden predicts resistance to EGFR/BRAF blockade in BRAF-mutated microsatellite stable metastatic colorectal cancer. Eur J Cancer. (2022) 161:90–8. doi: 10.1016/J.EJCA.2021.11.018
28. Li Y, Ma Y, Wu Z, Zeng F, Song B, Zhang Y, et al. Tumor mutational burden predicting the efficacy of immune checkpoint inhibitors in colorectal cancer: A systematic review and meta-analysis. Front Immunol. (2021) 12:751407. doi: 10.3389/FIMMU.2021.751407
29. Yarchoan M, Albacker LA, Hopkins AC, Montesion M, Murugesan K, Vithayathil TT, et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight. (2019) 4(6):e126908. doi: 10.1172/jci.insight.126908
30. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. (2013) 500:415–21. doi: 10.1038/nature12477
31. Shao C, Li G, Huang L, Pruitt S, Castellanos E, Frampton G, et al. Prevalence of high tumor mutational burden and association with survival in patients with less common solid tumors. JAMA Netw Open. (2020) 3:e2025109–e2025109. doi: 10.1001/JAMANETWORKOPEN.2020.25109
32. Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. (2018) 15:731–47. doi: 10.1038/s41571-018-0113-0
33. Zengin ZB, Chehrazi-Raffle A, Salgia NJ, Muddasani R, Ali S, Meza L, et al. Targeted therapies: Expanding the role of FGFR3 inhibition in urothelial carcinoma. Urol Oncol. (2022) 40:25–36. doi: 10.1016/J.UROLONC.2021.10.003
34. De Luca A, Esposito Abate R, Rachiglio AM, Maiello MR, Esposito C, Schettino C, et al. FGFR fusions in cancer: from diagnostic approaches to therapeutic intervention. Int J Mol Sci. (2020) 21:6856. doi: 10.3390/ijms21186856
35. Thomas DM, Hackett JM, Plestina S. Unlocking access to broad molecular profiling: benefits, barriers, and policy solutions. Public Health Genomics. (2021) 25(3-4):70–79. doi: 10.1159/000520000
36. Qoronfleh MW, Chouchane L, Mifsud B, Al Emadi M, Ismail S. THE FUTURE OF MEDICINE, healthcare innovation through precision medicine: policy case study of Qatar. Life Sci Soc Policy. (2020) 16(1):12. doi: 10.1186/S40504-020-00107-1
37. Zhang Q, Fu Q, Bai X, Liang T. Molecular profiling–based precision medicine in cancer: A review of current evidence and challenges. Front Oncol. (2020) 10:532403/BIBTEX. doi: 10.3389/fonc.2020.532403
38. Russo A, Incorvaia L, Capoluongo E, Tagliaferri P, Galvano A, Del Re M, et al. The challenge of the Molecular Tumor Board empowerment in clinical oncology practice: A Position Paper on behalf of the AIOM- SIAPEC/IAP-SIBioC-SIC-SIF-SIGU-SIRM Italian Scientific Societies. Crit Rev Oncol Hematol. (2022) 169:103567. doi: 10.1016/J.CRITREVONC.2021.103567
Keywords: next-generation sequencing, RNA fusions, cancer molecular profiling, cancer biomarkers, tumor mutational burden, immune checkpoint inhibitors, HER2 alteration
Citation: Dawood S, Natarajan V and Danchaivijitr P (2024) Comprehensive molecular profiling identifies actionable biomarkers for patients from Thailand and the United Arab Emirates with advanced malignancies. Front. Oncol. 14:1374087. doi: 10.3389/fonc.2024.1374087
Received: 21 January 2024; Accepted: 03 April 2024;
Published: 10 May 2024.
Edited by:
Gianluca Russo, University of Naples Federico II, ItalyReviewed by:
Pasquale Pisapia, University of Naples Federico II, ItalyCopyright © 2024 Dawood, Natarajan and Danchaivijitr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaheenah Dawood, U2hhaGVlbmFoQHBvc3QuaGFydmFyZC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.