- 1Department of Pharmacy, Women’s Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Department of Immunology, School of Basic Medical Sciences, Xinjiang Medical University, Urumqi, China
- 3Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Xinjiang Medical University, Urumqi, Xinjiang, China
- 4Cancer Center, Zhejiang University School of Medicine, Hangzhou, China
- 5Women’s Hospital, Institute of Genetics, and Department of Environmental Medicine, Zhejiang University School of Medicine, Hangzhou, China
- 6State Key Laboratory of Pathogenesis, Prevention and Treatment of High Incidence Diseases in Central Asia, Xinjiang Medical University, Urumqi, China
Breast cancer (BC) is the most common malignancy among women and a leading cause of cancer-related deaths of females worldwide. It is a complex and molecularly heterogeneous disease, with various subtypes that require different treatment strategies. Despite advances in high-resolution single-cell and multinomial technologies, distant metastasis and therapeutic resistance remain major challenges for BC treatment. Long non-coding RNAs (lncRNAs) are non-coding RNAs with more than 200 nucleotides in length. They act as competing endogenous RNAs (ceRNAs) to regulate post-transcriptional gene stability and modulate protein-protein, protein-DNA, and protein-RNA interactions to regulate various biological processes. Emerging evidence suggests that lncRNAs play essential roles in human cancers, including BC. In this review, we focus on the roles and mechanisms of lncRNAs in BC progression, metastasis, and treatment resistance, and discuss their potential value as therapeutic targets. Specifically, we summarize how lncRNAs are involved in the initiation and progression of BC, as well as their roles in metastasis and the development of therapeutic resistance. We also recapitulate the potential of lncRNAs as diagnostic biomarkers and discuss their potential use in personalized medicine. Finally, we provide lncRNA-based strategies to promote the prognosis of breast cancer patients in clinical settings, including the development of novel lncRNA-targeted therapies.
1 Introduction
Breast cancer (BC) may develop due to a variety of factors, including genetic mutations, lifestyle choices, and environmental exposures, with its incidence further influenced by various demographic and socioeconomic elements (1). Despite significant progress in cancer research frontier, BC remains a serious public health issue across the globe. According to latest statistics of GLOBOCAN, an estimated 2.3 million cases of BC were newly diagnosed from 185 countries, accounting for 11.7% of total cancer cases worldwide (1). BC is now recognized as the leading cause of cancer-related mortality in women, with 684,996 deaths reported in 2020. Predominantly attributed to rapid advancements in diagnostic technologies and an increase in the use of mammographic screening, BC has now surpassed lung cancer to become the most commonly diagnosed cancer in women globally. Importantly, it also ranks as the second most common malignancy worldwide, following closely behind lung cancer (1). While less common, BC does occur in men, making up approximately 1% of all BC instances globally (2). Hence, incessant research efforts are crucial to alleviate the significant public health, societal and economic burden posed by BC.
The incidence of BC is governed by a confluence of factors - genetic, epigenetic, and environmental, among others (3). In light of this, BC emerges as a multifaceted disease marked by vast heterogeneity in pathology, genomic alterations, gene expression profiles, and the tumor microenvironment (TME). BC is classified into different biological subtypes, with each subtype displaying unique pathological characteristics and diverse clinical outcomes (4). Indeed, while there have been substantial advancements in diagnostic techniques and treatment strategies over the past decade, the prognosis for BC patients remains unsatisfactory. Survival rates for BC are highly dependent on the stage at which the disease is identified, with early detection correlating to a higher survival advantage (5). Now, it is evident that metastasis accounts for the greatest proportion of BC-related mortality (6). Even though it is detected at an early stage, a notable percentage of women may see their disease evolve into a more aggressive subtype after undergoing initial therapy. This change is likely due to the molecular heterogeneity of BC. Molecular heterogeneity pertains to the genetic variation found within tumor cells, which can result in worse disease progression and resistance to treatment. This is why there is an ongoing focus on creating personalized and targeted therapies.
According to the St. Gallen guidelines, breast cancer (BC) is categorized into four subtypes (7). This categorization is based on the expression status of specific molecular biomarkers including the estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and Ki-67 labeling index, which is a marker of cell proliferation. The four subtypes of BC are Luminal A, Luminal B, HER2-enriched, and Basal-like subtypes (Figure 1). The Basal-like subtype is also known as triple-negative breast cancer (TNBC) due to the absence of ER, PR, and HER2 receptors, which makes this subtype particularly challenging to treat. With high metastatic properties and a lower rate of early detection, TNBC poses a significant challenge to treatment (8). Hence, improving survival rates in breast cancer patients, particularly those with the aggressive triple-negative subtype, necessitates the identification of novel molecular biomarkers. These biomarkers are key in assessing the risk of metastasis and treatment response, and additionally, there is a critical need to develop innovative therapies tailored towards tackling this formidable disease.
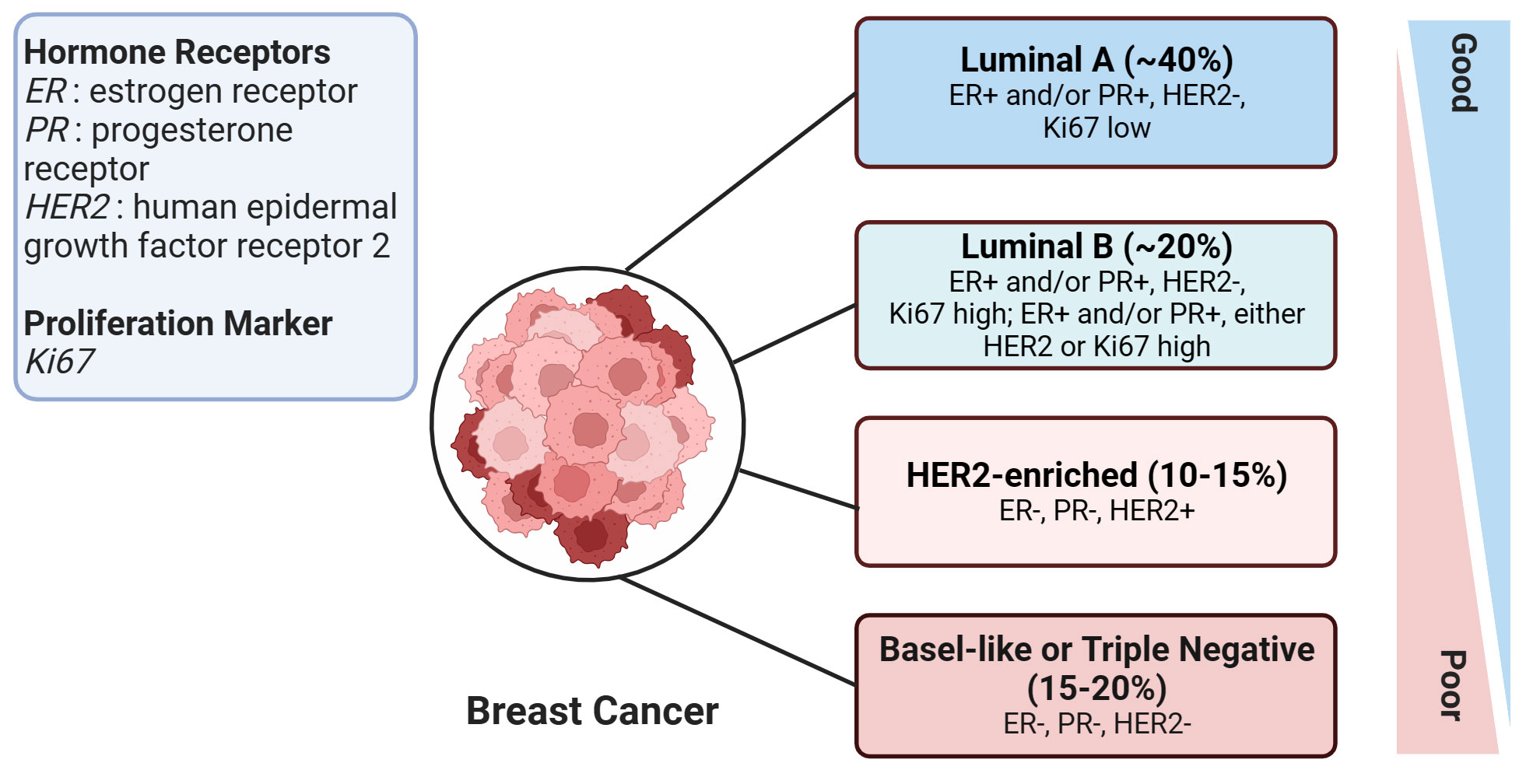
Figure 1 An overview of human breast cancer subtypes. This figure illustrates the various subtypes of breast cancer, with the approximate proportion (%) of each subtype among all breast cancer cases provided in brackets. Prognosis severity increases from top to bottom, signifying that the subtypes at the bottom are associated with worse prognoses. The figure was created in BioRender.com.
Rapidly evolving imaging techniques have prominently emerged as the primary tool for early diagnosis of breast cancer, supported by considerable clinical evidence suggesting their effectiveness in reducing breast cancer mortality (9). Further advancements in machine learning-based digital imaging techniques have notably enhanced diagnostic accuracy (10). However, their widespread application is curtailed by high costs and lack of specificity, rendering them unsuitable for early phase breast cancer detection (11). Similarly, conventional tumor diagnostic markers such as carcinoembryonic antigen (CEA), CA 15-3, and CA 125 prove impractical in detecting early phase breast cancer due to their low sensitivity.
Over the past few decades, there has been a continuous pursuit of diagnostic, prognostic, and predictive biomarkers to enhance breast cancer (BC) management. Numerous large molecules, such as DNA (APC, RARb2) (12), proteins (HER2,P53) (13), autoantibodies(MUC1) (14), and ncRNAs (non-coding RNAs), have been utilized as biomarkers for diagnosis. These biomarkers can be detected from serum samples, other body fluids, or tissues of tumor patients. Liquid biopsy, a more accessible, cost-effective, and repeatable sampling process, serves as a valuable source for such biomarkers, avoiding tumor heterogeneity issues (15). Long non-coding RNAs (lncRNAs) are non-protein-coding transcripts exceeding 200 nucleotides in length (16). Researchers have discovered abnormal lncRNA expression in both BC cell lines and tissues (17), revealing their potential role in breast cancer initiation and development. In this review, we synthesize the existing literature on lncRNAs’ roles in various aspects of BC and possible underlying mechanisms. This analysis offers novel insights into the diagnosis, prognosis, and prediction of BC, paving the way for improved management and treatment strategies.
2 Overview of ncRNAs
The advent of sequencing technologies has drastically revolutionized our understanding of the human genome. We now know that approximately 75% of the human genome is transcribed into RNA, yet only a small fraction— about 3%— translates into protein-coding mRNAs (18). The largest and most critical family of RNAs, non-coding RNAs (ncRNAs), do not code for any proteins. Despite their lack of protein-coding potential, ncRNAs fulfill vital roles in numerous pathophysiological processes, particularly in cancer. NcRNAs can be categorized into two main groups based on their size: long non-coding RNAs (lncRNAs) and small non-coding RNAs (sncRNAs). The latter is a collection of ncRNAs shorter than 200 nucleotides and includes microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs), small interfering RNAs (siRNAs), small nucleolar RNAs (snoRNAs), and small nuclear RNAs (snRNAs) (19). In contrast, lncRNAs, as the name suggests, are longer than 200 nucleotides. Circular RNAs (circRNAs) constitute the third subgroup of ncRNAs. These distinguished by their covalent, close-loop (circular) single-stranded structures (20). Among these, lncRNAs represent the most intricate and challenging class of ncRNAs. These RNAs offer novel routes for research, potentially leading to new diagnoses, treatments, and understandings of diseases like cancer.
2.1 Classification of lncRNAs
LncRNAs were initially discovered as mRNA-like transcripts that possess both RNA- and protein-like functions. Generally, they lack significant open reading frames (ORFs) and are not translated into proteins, except for a few micropeptide-encoding lncRNAs (21). To date, over 27,000 lncRNAs have been annotated and the number continues to grow, while the functions of a large number of lncRNAs remain unexplored (22). Similar to mRNAs, most lncRNAs are transcribed by RNA polymerase II and are subjected to a series of processes including 5’-capping, splicing, and poly-adenylation at the 3’ end (23). LncRNA is the most complex type of ncRNA, with no uniform standard for classification. They can be categorized by length, genomic location, and mechanism of action (24). Based on their genomic localization, they can be classified as intronic, intergenic, sense, antisense, and enhancer lncRNAs. According to their mechanisms of action, they are categorized into four types: guide, scaffold, decoy, and signaling lncRNAs (25). Guide lncRNAs can bind to transcription factors and direct them to specific targets. Scaffold lncRNAs act as a “central platform” that binds different effector molecules simultaneously, like a “scaffold,” to integrate various signal pathways. Silencing guide and scaffold lncRNAs can result in altered localization or even loss of function of effector molecules. Decoy lncRNAs interact with target transcriptional regulators and block downstream signals. Signaling lncRNAs regulate downstream gene transcription without protein translation (26).
LncRNAs play a crucial role in the regulation of the genome and have a high level of tissue specificity, suggesting their integral role in maintaining cellular functions (21). They are involved in numerous biological processes, such as transcription, splicing, and translation. Notably, they can participate in chromatin remodeling and epigenetic regulation (16). Additionally, lncRNAs have been found to be dysregulated in various types of cancers, indicating that their abnormal expression or function could contribute to cancer development or progression (17). Thus, they hold promise as biomarkers for early detection, prognosis, and potential therapeutic targets in cancer treatment.
2.2 Subcellular localization and biological functions of lncRNAs
Though originally characterized as “transcriptional noise” without biological functionality, the biological activity and influence of lncRNAs on various pathophysiological processes have garnered increasing attention. The function of lncRNAs largely depends on their subcellular localization (23). These molecules are known to localize in both the nucleus and the cytoplasm (27). The majority of lncRNAs are found in the nucleus, the site of their biogenesis and processing, where they perform their functions. Nuclear lncRNAs are involved in gene regulation at both the epigenetic and transcriptional levels. They can bind directly to DNA or transcription factors and assist in the regulation of chromatin structure (28). Other lncRNAs require export to the cytoplasm where they target mRNAs, miRNAs, and proteins to regulate gene expression post-transcriptionally and translationally (29). For instance, lncRNAs can operate as “miRNA sponges” or as competitive endogenous RNA (ceRNA). These lncRNAs competitively bind with miRNAs, which allows them to indirectly control the expression of target genes at the post-transcriptional level. Moreover, recent research has begun to explore the subcellular localization of lncRNAs, shedding light on their presence in specific organelles such as mitochondria and the endoplasmic reticulum (ER) (30).
LncRNAs play crucial roles in various biological processes, including embryonic development, organogenesis, immune function, stem cell differentiation, and pluripotency, by regulating gene expression and protein translation (31). Notably, numerous lncRNAs display dysregulation in a variety of malignancies, with the up- or down-regulation of these lncRNAs either promoting or inhibiting tumor progression. Broadly speaking, lncRNAs can be divided into two categories: oncogenic and tumor-suppressive (32). Oncogenic lncRNAs, which typically exhibit overexpression in tumor tissues compared to healthy samples, play a role in promoting tumorigenesis. As a result, inhibiting these lncRNAs presents a promising potential anticancer strategy. Conversely, lncRNAs with tumor-suppressive properties are generally downregulated in cancerous tissues. Increasing the levels of these lncRNAs could serve as an approach for combating the disease. Moreover, lncRNAs have been found to contribute to tumor metastasis and therapeutic resistance (33). This makes them invaluable subjects of study in cancer research, as many could be developed into novel biomarkers for the diagnosis, progression, invasion, metastasis, and prognosis of cancer, or even as targets for therapeutic intervention.
2.3 Exosomal lncRNAs
Exosomes are tiny, nano-sized extracellular vesicles (EVs) that can be found in various human body fluids like blood, urine, and saliva (34). These exosomes can be secreted by all types of cells, and their formation begins when the plasma membrane invaginates to create intraluminal vesicles (ILVs), which then mature into multivesicular bodies (MVBs). These MVBs either fuse with the plasma membrane to release exosomes into the extracellular space or they get degraded in lysosomes (35). The contents of these exosomes can vary greatly, depending on the specific tissues and organs. They may contain nucleic acids (like mRNAs and ncRNAs), proteins, or even synthetic drugs, demonstrating that exosomes play a critical part in mediating communication between cells. The application of this knowledge extends further into cancer research, where it has been found that exosomes secreted by tumor cells contain tumor-specific long non-coding RNAs (lncRNAs), which reveal the original cellular pathophysiological state (36). These exosomal lncRNAs play a crucial role as regulators in the development of cancer. They are involved in various processes including the growth, proliferation, metastasis of cancer cells, promotion of angiogenesis, drug resistance, and immunomodulation, among other functions.
Indeed, changes in exosomal lncRNAs have been observed in various tumor types, suggesting that they can potentially serve as biomarkers for cancer diagnosis and prognosis (37). One of the advantages of exosomal lncRNAs is their high stability, which can be attributed to the protection provided by their lipid bilayers. This makes them more suitable candidates for developing biomarkers compared to other types of molecules. Moreover, unlike traditional in-situ biopsies, exosomes can be isolated from easily accessible body fluids like blood and urine, without the need for more invasive procedures. This makes the process of monitoring exosomes in body fluids much more convenient, feasible, and less invasive for patients (38). As a result, the utilization of exosomal lncRNAs as biomarkers holds great promise in advancing cancer detection and management.
2.4 LncRNAs-miRNAs-mRNAs network
Micro RNAs (miRNAs) are small non-coding RNAs with lengths ranging from 17 to 25 nucleotides (39). Generally, miRNAs bind to the target mRNAs at their 3’untranslated regions (3’UTRs), subsequently inhibiting translation (40). A single miRNA can target multiple mRNA molecules, and a specific mRNA can be targeted by multiple miRNAs simultaneously. Like lncRNAs, aberrantly expressed miRNAs have been found in various cancer types, including BC (41). The mechanism of lncRNAs-mediated bioprocess regulation often involves miRNAs. In fact, lncRNAs interact with miRNAs through multiple mechanisms to regulate gene expression. Together, they form the lncRNAs-miRNAs-mRNAs axis. In other words, lncRNAs can act as “miRNA sponges” by competitively binding with miRNAs, indirectly regulating the expression of mRNAs (42, 43). These competing endogenous RNAs (ceRNAs) networks are extensively present in all aspects of breast cancer and are not listed separately in this article (Figure 2).
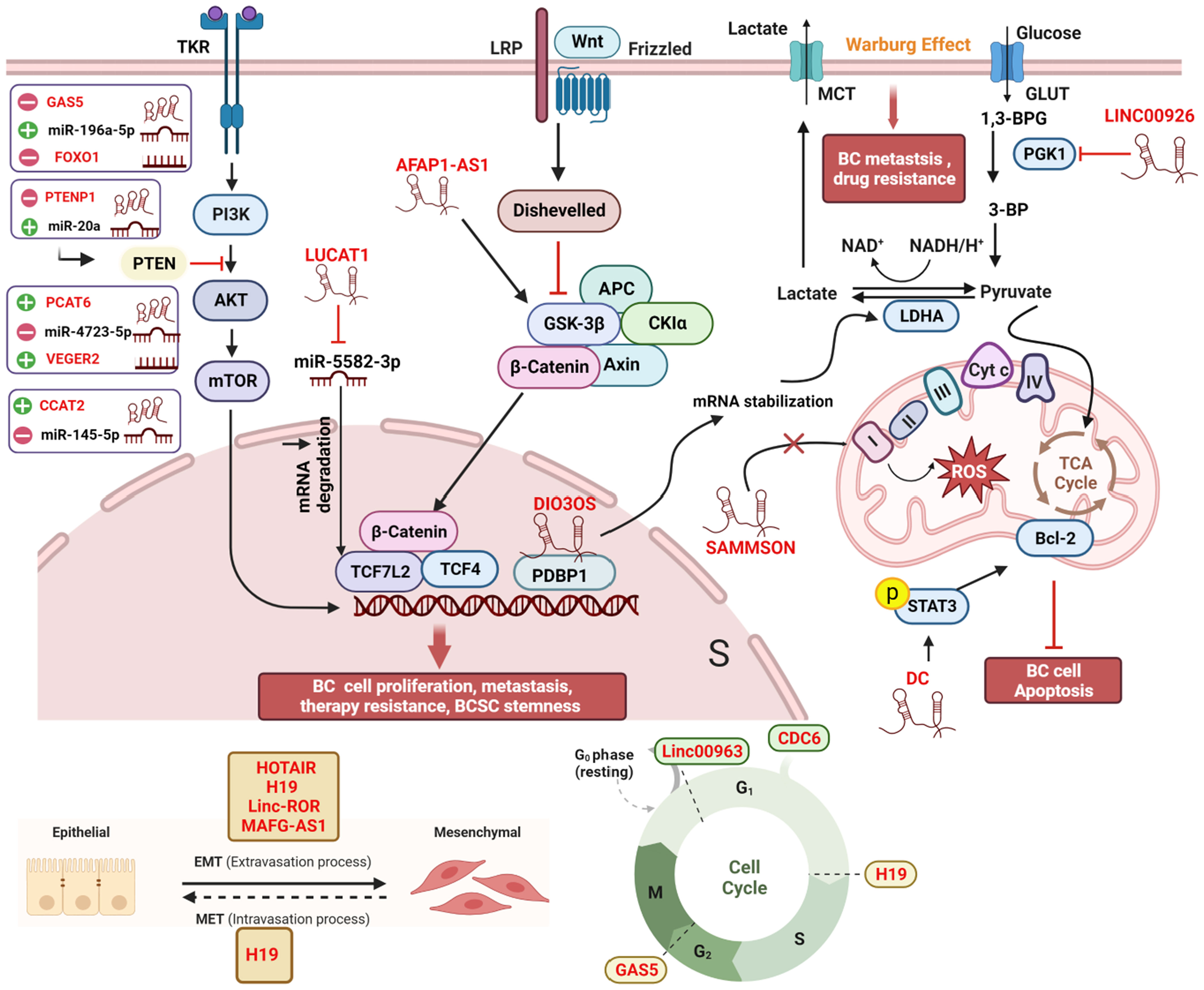
Figure 2 Comprehensive regulatory network of lncRNAs in breast cancer cells. Within the cellular environment, a complex regulatory network involving lncRNAs, their target genes, miRNAs, and interacting proteins is established. This network encompasses diverse signaling pathways, including Wnt/β-catenin, PI3K/AKT, and MAPK/ERK, coordinating various biological processes such as EMT, autophagy, the Warburg effect, oxidative phosphorylation, cell cycle arrest, angiogenesis, and treatment response in breast cancer patients. The lncRNA-miRNA-mRNA axis and ceRNA networks play pivotal roles in modulating gene expression during cancer development and progression, offering potential targets for therapeutic interventions and biomarker discovery. All lncRNAs are colored in red. “-” represents oncogene; “+” represents tumor suppressor; “┴“ represents inhibition; “↑” represents promotion. The figure was created in BioRender.com.
3 LncRNAs in breast cancer
Advancements in targeted therapy, immunotherapy, and innovative combination treatments have significantly enhanced the survival rate of patients with BC (44, 45). Nonetheless, BC still remains the leading cause of cancer-related mortality among women due to its high rates of recurrence, metastasis, and therapeutic resistance. One important reason for this is the complex and molecularly heterogeneous nature of BC. Until now, accumulating studies have demonstrated that lncRNAs play important roles in the progression, metastasis, and even treatment response of BC. Dysregulation of lncRNAs can disrupt normal transcription, leading to abnormal gene expression and ultimately tumor progression through various mechanisms (46). In both cancer and other diseases, a multitude of physiological and pathological processes are intricately linked to lncRNAs. Current studies have shown that the expression of lncRNAs in BC cells and tissues differs significantly from that in normal cells and tissues (47). A large number of lncRNAs have been shown to be involved in tumor progression, metastasis, as well as treatment resistance (Figure 2). Based on their functions and expression patterns in BC, lncRNAs can be classified as tumor suppressor genes or oncogenes, which will be discussed in detail below.
3.1 LncRNAs and BC progression
A large number of studies have provided evidence that lncRNAs are involved in BC progression, making them potential targets for biomarker design and discovery of novel anticancer drugs. In the following sections, we summarize several particular lncRNAs which play important roles in BC.
3.1.1 Hotair
HOTAIR (HOX Transcript Antisense Intergenic RNA) is a long non-coding RNA (lncRNA) composed of 2158 nucleotides. It has the distinction of being the first lncRNA identified in correlation with a poor prognosis in breast cancer (BC). HOTAIR is considered an oncogenic lncRNA, and its overexpression has been reported in nearly all solid tumors (48). Notably, it is observed in both the cytoplasm and the nucleus. In terms of its mode of operation, HOTAIR acts as a “scaffold” that binds to and recruits the Polycomb Repressive Complex 2 (PRC2) to its target genes. Here, the histone methylase activity of PRC2 represses gene transcription (49). HOTAIR’s influence on BC is wide-ranging, contributing to the progression of BC, metastasis, and therapeutic resistance. Its overexpression has been identified in BC tissues and cells (50). In one study, Shi et al. found that HOTAIR enhances the proliferation, invasion, and migration of triple-negative BC cells through the HOTAIR/miR-203/CAV1 axis (51). Another study indicated that a complex of HBXIP, HOTAIR, and LSD1—where HOTAIR serves as a scaffold—can activate the pro-oncogenic transcription factor c-Myc, amplifying the growth of BC cells both in vitro and in vivo (52). Additionally, HOTAIR has been found to regulate the proliferation, migration, apoptosis, and invasion of MCF-7 cells by modulating the p53/Akt/JNK signaling pathway (53).
3.1.2 H19
H19, the first identified long non-coding RNA (lncRNA) with riboregulatory function, is one of the most extensively studied lncRNAs in cancer (54). It is overexpressed in numerous solid tumors, including breast cancer (BC). H19 has been reported to bind with E2F1, a critical factor in the G1/S transition of the cell cycle. This association is linked to increased proliferation of BC cells as well as the progression to a more aggressive phenotype (55). As a miRNA sponge, H19 encourages BC cell proliferation, along with promoting invasiveness and migration. Conversely, the silencing of H19 can induce cell cycle arrest and apoptosis by regulating miR-138 and SOX4 (56).
3.1.3 Other oncogenic lncRNAs
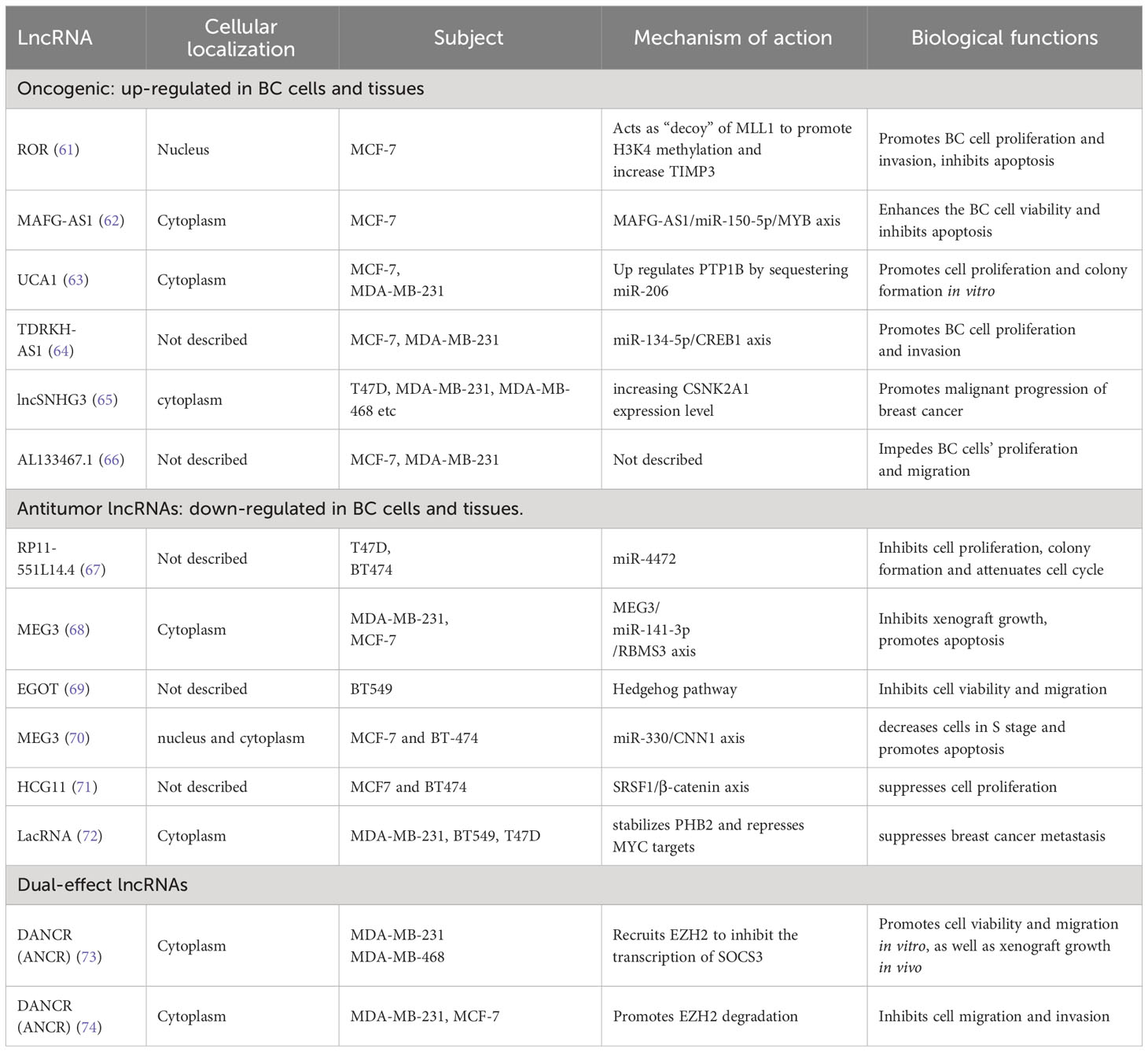
HOTAIR and H19 are classic and widely studied oncogenic lncRNAs in BC. In addition to these, numerous other lncRNAs have been identified to play oncogenic roles in BC progression. For instance, lncRNA CDC6 (cell division cycle 6) has been positively correlated with BC stages. Overexpression of CDC6 deregulates the G1 phase of the cell cycle, promoting BC cell migration. Concurrently, CDC6 acts as a molecular sponge for miR-215, further enhancing BC cell proliferation (57). NEAT1 (nuclear paraspeckle assembly transcript 1) is a structural component of nuclear paraspeckles (58). Although predominantly found in the nucleus, NEAT1 can be translocated to the cytoplasm, a process mediated by Pinin. In the cytoplasm, NEAT1 serves as a “scaffold” for the PGK1/PGAM1/ENO1 complex, promoting the penultimate step of glycolysis via substrate channeling (59). Accelerated glycolysis, also known as the Warburg effect, is a key metabolic change in cancer. Consequently, NEAT1 promotes tumor initiation, growth, and metastasis. GATA3 (GATA Binding Protein 3) is a transcription factor that regulates cell differentiation and acts as a tumor suppressor in BC progression. GATA3-AS1, the antisense RNA1 of GATA3, has been found to promote TNBC cell proliferation and migration by facilitating GATA3 degradation through ubiquitination (60). Additional lncRNAs related to BC initiation and progression are listed in Table 1.
3.1.4 Antitumor lncRNAs
There are several lncRNAs with diverse roles in the progression of BC. Among them, GAS5 (Growth Arrest Specific 5), a tumor suppressor gene, was first isolated from mouse NIH3 cells. It has been observed that the presence of GAS5 is diminished in TNBC tissues, linked to an aggressive disease phenotype. Conversely, the overexpression of GAS5 within TNBC cells considerably promotes apoptosis (programmed cell death) in cancer cells while also inhibiting cell division (75). From a mechanistic standpoint, GAS5 functions as a ceRNA, countering miRNA-196a-5p, thus negating the protumor effects of the miRNA-196a-5p/FOXO1/PI3K/AKT pathway. GAS5’s anticancer effects involve multiple interactions with various miRNAs and proteins to encourage the apoptosis of BC cells via several pathway (76). PTCSC3 (Papillary Thyroid Carcinoma Susceptibility Candidate 3), found to be a tumor suppressor in numerous cancers, may serve as an upstream inhibitor of H19, regulating cell proliferation in TNBC cells. While it’s common for lncRNAs to modulate miRNAs, mRNAs, and chromatin, the regulation of one lncRNA by another is a seldom-witnessed phenomenon (77). PDCD4 (Programmed Cell Death 4) is a well-documented tumor suppressor gene. Its NATs (Natural Antisense Transcripts), known as PDAD4-AS1, can enhance the expression of PDCD4 through stabilizing PDCD4 RNA. Both PDCD4 and PDAD-AS1 negatively regulate BC cell proliferation by inhibiting cell cycle progression (78).
3.1.5 Dual-effects lncRNAs
The above-mentioned lncRNAs have been shown to have either a promoting or inhibitory effect on the progression of BC. However, in certain circumstances, specific lncRNAs exhibit dual effects on BC progression. A single lncRNA can have opposite effects on the same subtype of BC or conflicting effects on different subtypes. For instance, MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) has been identified as an oncogenic lncRNA that promotes the progression and metastasis of BC (79). In contrast, Kim et al. discovered that deficiency of MALAT1 induces BC metastasis, which can be reversed by the exogenous supplementation of MALAT1. This was observed in genetically engineered mouse models and xenograft models (80). Interestingly, another study found no differences in serum MALAT1 levels between BC patients and healthy controls (81). PTENP1 (phosphatase and tensin homolog pseudogene 1) is a pseudogene of the tumor suppressor PTEN, with a highly homologous region upstream of PTEN’s 3’-UTR. PTENP1 represses cell proliferation and promotes apoptosis. Gao et al. discovered that both PTENP1 and PTEN are downregulated in ER-positive cell lines MCF-7 and T47D. Overexpression of PTENP1 suppresses BC progression, while knockdown of PTENP1 enhances malignant behavior in these BC cells (82). Mechanistically, PTENP1 acts as an antitumor lncRNA by sponging miR-20a and regulating BC progression through the PTEN/PI3K/AKT pathway. Similar results were obtained in a previous study (83). However, another article mentions that upregulation of PTENP1 decreased PTEN gene expression in ER-positive MCF-7 and T47D cells and accelerated MCF-7 tumor growth in vivo. On the other hand, PTENP1 upregulation increased PTEN transcript levels and inhibited the growth rate of ER-negative MDA-MB-231 cells, suggesting that PTENP1 influences BC growth depending on the ER status (84).
XIST (X inactive-specific transcript), a key initiator of X chromosome inactivation in female mammals, has been recognized to play important roles in tumor progression regulation. Several studies have shown that XIST is downregulated and acts as an anti-cancer factor in BC. Knockdown of XIST promotes proliferation of MCF-7 cells and ovarian cancer cells. Mechanistically, XIST competes with miR-101 to upregulate C/EBP and KLF6 expression, which inhibits macrophage polarization toward the M2 phenotype, thereby suppressing BC cell proliferation and migration (85). Conversely, Zhao et al. found that XIST expression is upregulated in BC tissues and cell lines, and XIST knockdown significantly represses cell proliferation, migration, invasion, and anti-apoptotic activities in BC cells (86). Mechanistically, XIST acts as a sponge for miR-125b-5p, thereby upregulating the expression of the BC promoter NLRC5.
3.2 LncRNAs and BC metastasis
Metastasis, the most devastating stage of cancer progression, is responsible for the majority of cancer-related deaths. Many cancers, including BC, tend to metastasize preferentially to specific organs, a phenomenon known as organotropism (87). BC tends to metastasize to the brain, bones, lungs, and liver (87). Despite its significance, the process of metastasis is not fully understood, which has hindered the development of early predictive methods and effective treatment options for metastatic BC patients. To gain the ability to survive and metastasize, cancer cells often undergo a series of changes, including genetic and epigenetic alterations, as well as metabolic reprogramming (88). Research has indicated that cancer stem cells (CSCs), epithelial-mesenchymal transition (EMT), and autophagy are the three main mechanisms driving tumor metastasis (89). Within BC, there exists a small subpopulation of cells known as tumor-initiating cells (TICs) or breast cancer stem cells (BCSCs), which have the ability to generate daughter BCSCs (90). These daughter BCSCs possess the capability for unlimited proliferation through self-renewal and differentiation into BC cells. Only BCSCs have the potential to form recurrent or metastatic tumors (91). EMT is a dynamic process in which epithelial cells lose their polarity and intercellular cohesion, transforming into migratory mesenchymal cells. Although EMT is reversible, it provides cancer cells with increased motility and migration capabilities by breaking down intercellular bonds in the epithelial cells (92). EMT plays a crucial role in cancer progression and metastasis. Furthermore, EMT is closely intertwined with CSCs; for example, BCSCs can derive from human mammary epithelial cells through induction of EMT (93). Autophagy is a self-degradative process that can have a dual role in tumorigenesis. In the early stages of tumorigenesis, it exhibits anticancer effects, while in later stages, it contributes to tumor cell proliferation and survival, playing a fundamental role in tumor maintenance (42).
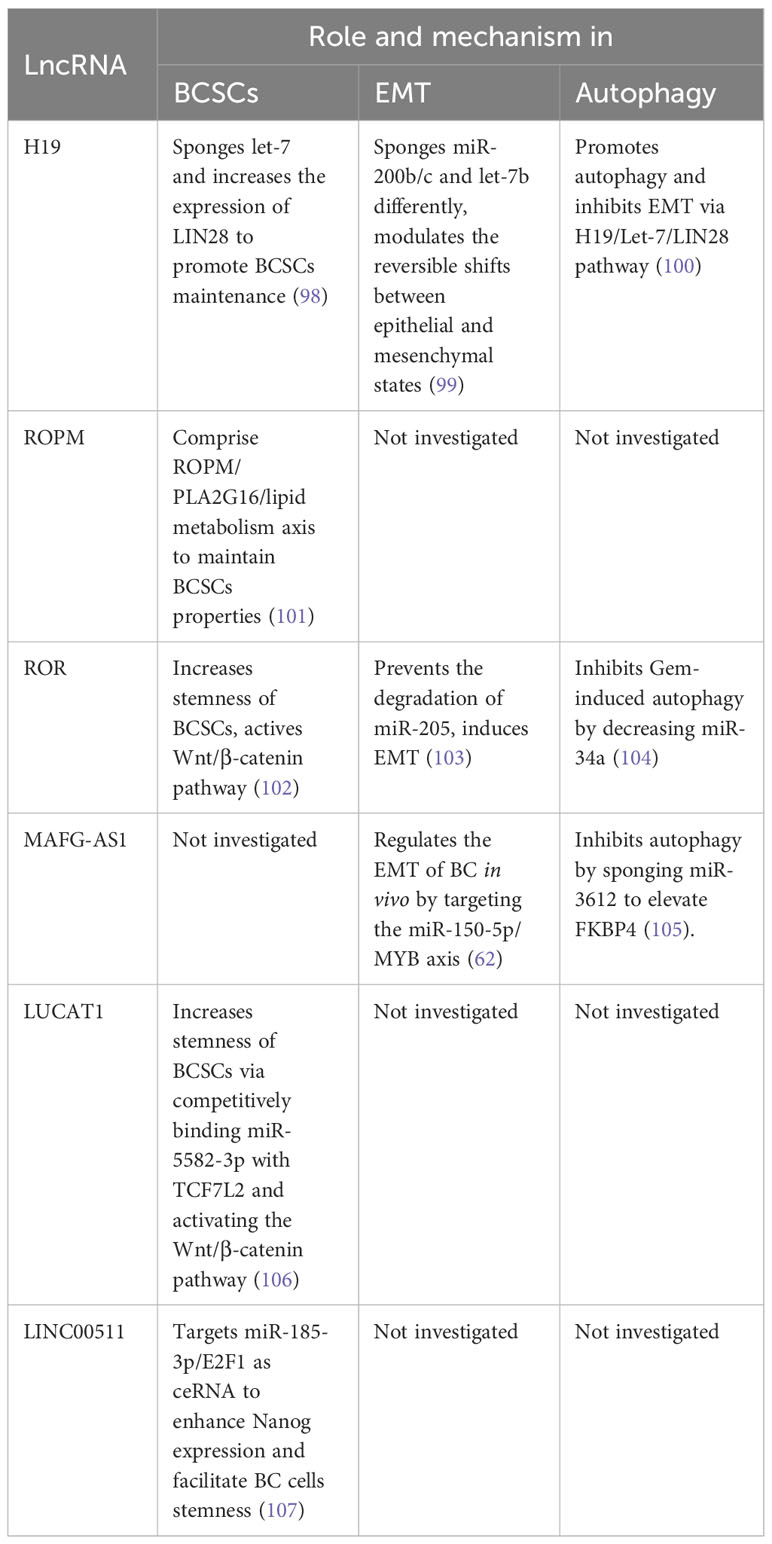
Interestingly, numerous lncRNAs have been identified to be involved in the aforementioned three mechanisms. For instance, HOTAIR is known to play a key role in the invasion, proliferation, colony formation, and self-renewal capacity of BCSCs by regulating SOX2 and NF-kB (94, 95). MiR-7, a metastasis-suppressing miRNA, inhibits both SETDB1 and the cellular EMT process in BCSCs. In MDA-MB231 cells and BC patients, HOTAIR functions by inhibiting miR-7 (96). Furthermore, HOTAIR can regulate autophagy, which is critical for BC cell survival, through its interactions with matrix metalloproteins (97). The functions and mechanisms of lncRNAs involved in these three mechanisms of tumor metastasis are summarized in Table 2.
In addition to the three mechanisms mentioned above, there are other ways in which lncRNAs contribute to metastasis. For example, a novel lncRNA called LINC02273 forms a complex with hnRNPL and activates the oncogene AGR2, thus promoting BC metastasis (108). Cancer cells often undergo metabolic changes, such as increased glucose uptake and glycolysis, to satisfy the energy requirements for their malignant behavior (109). LINC00926 retards BC metastasis by inhibiting the PGK1-mediated Warburg effect, thereby reducing glucose uptake and lactate production (110). Hypoxia is also a hallmark of the tumor microenvironment (TME). In the hypoxic TME, the activation of the HIF (hypoxia-inducible factor) pathway promotes tumor progression and metastasis (111). For instance, HIF-2-induced lncRNA RAB11B-AS1 enhances angiogenic factors VEGFA and ANGPTL4 in hypoxic BC cells, leading to angiogenesis and metastasis (112). Additionally, lncRNA PCAT6 facilitates TNBC metastasis by sponging miR-4723-5p and binding to USP14, resulting in enhanced stability of VEGFR2 protein and activation of the Akt/mTOR pathway (113).
3.3 LncRNAs and therapeutic resistance of BC
In recent years, significant progress has been made in the development of various therapies for the management of BC. These therapies include (i) surgery, (ii) chemotherapy (especially for TNBC patients), (iii) trastuzumab (a HER2-specific monoclonal antibody for HER2-positive patients), (iv) endocrine therapy (e.g., tamoxifen) for ER-positive patients, (v) radiation therapy, and (vi) immunotherapy (114). Unfortunately, the emergence of treatment resistance often leads to metastasis and recurrence of BC, rendering it an incredibly challenging disease to treat. Extensive research has been conducted to understand the mechanisms underlying treatment resistance. The prevailing view is that BC is a stem cell disease, with BC stem cells (BCSCs) being the critical cells responsible for chemo-resistance and radio-resistance during BC therapy (115). Furthermore, the emerging field of lncRNA research has shown that lncRNAs play an essential role in treatment resistance through various molecular pathways, including increasing drug efflux, suppressing apoptosis, promoting BCSCs stemness, and acting as ceRNAs. As a result, lncRNAs have the potential to serve as biomarkers and promising targets to overcome drug/radiation resistance in BC patients.
3.3.1 LncRNAs and chemotherapy resistance
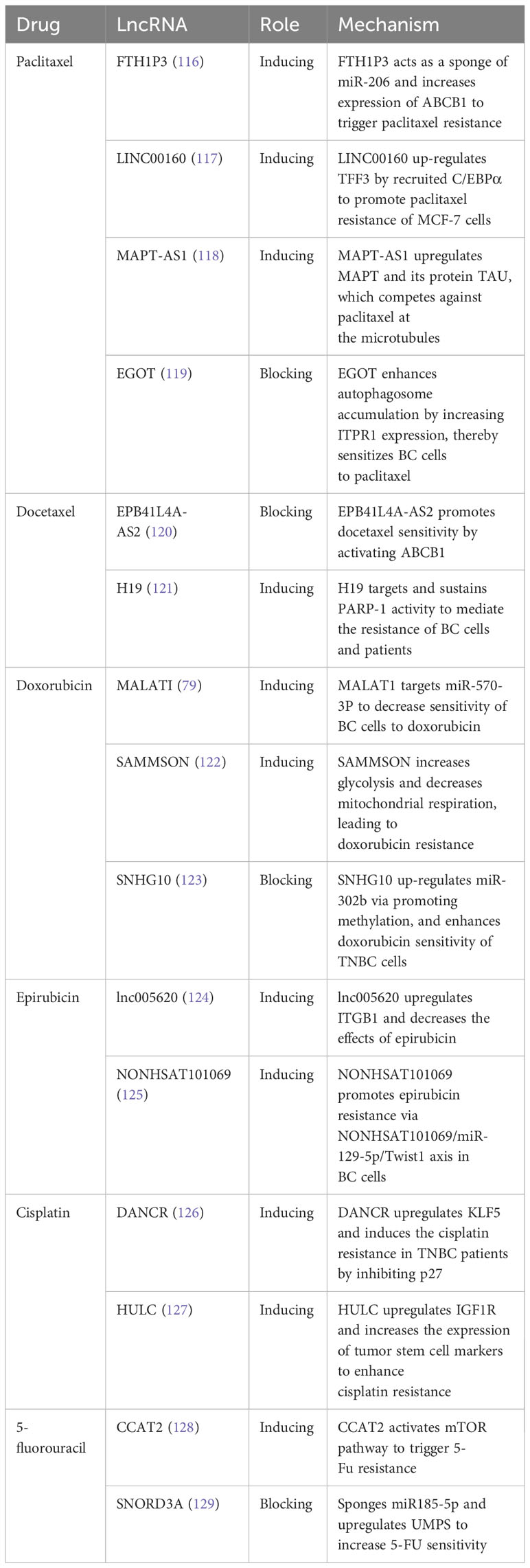
Among the different treatment options for BC, chemotherapy is the most widely used in clinical settings as it can improve patients’ survival rates. Chemotherapy drugs commonly used for BC treatment include taxanes (paclitaxel and docetaxel), anthracyclines (doxorubicin and epirubicin), platinum drugs, and 5-Fluorouracil (5-FU). However, chemotherapy resistance remains a significant challenge in breast cancer treatment. Cancer cells become resistant to chemotherapy drugs, causing cancer growth and spread. Several factors contribute to chemotherapy resistance in breast cancer, such as tumor heterogeneity, genetic mutations, the tumor microenvironment, upregulated drug efflux pumps, metabolic reprogramming, and epigenetic changes, among others. For more details on the lncRNAs involved in chemotherapy resistance, please refer to Table 3.
3.3.2 LncRNAs and tamoxifen resistance
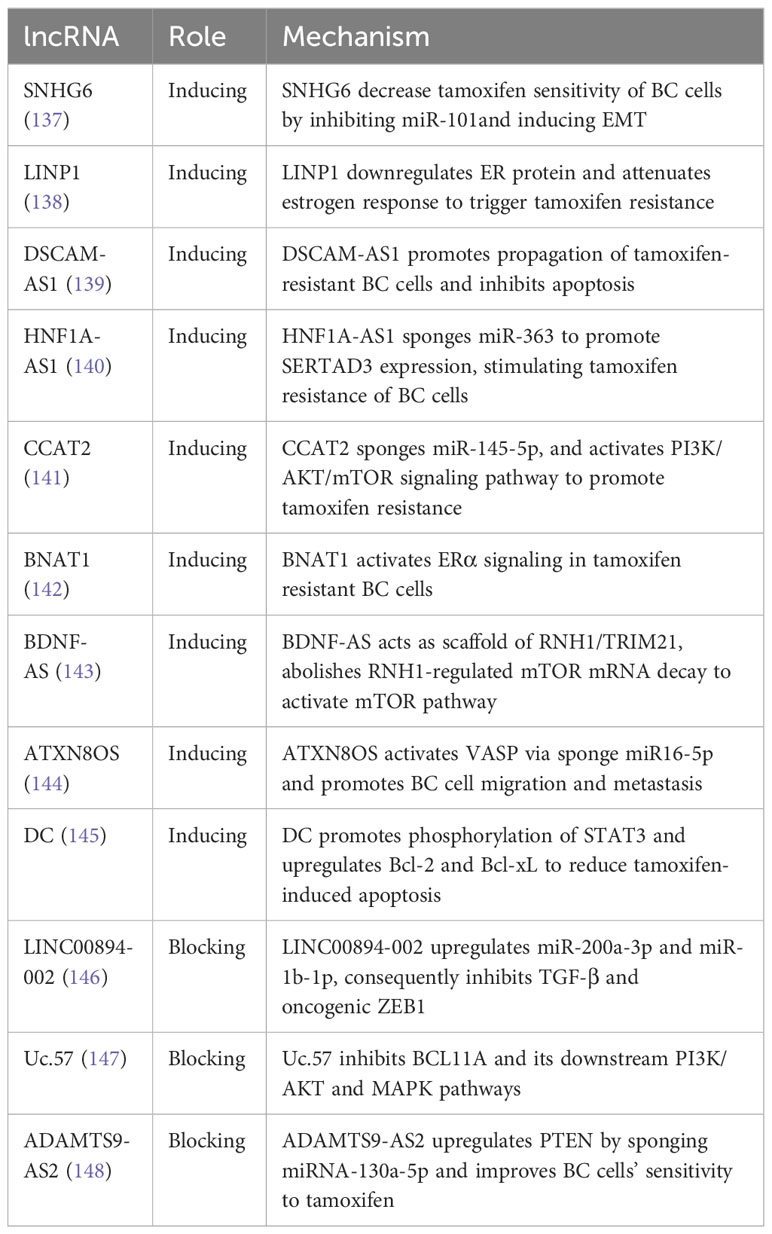
Estrogen receptor-positive (ER-positive) BC accounts for 75% of all BC cases, and ER therapy is crucial to inhibit estrogen-dependent tumor growth (130). ER therapy is the first-line adjuvant therapy for ER-positive BC patients and has been shown to reduce the recurrence and mortality whether chemotherapy is given concurrently (131). Aromatase, a rate-limiting enzyme that converts androgen to estrogen, is a vital target for aromatase inhibitors (AIs) such as tamoxifen, letrozole, and anastrozole. However, AI drug resistance persists in clinical practice, leading to tumor recurrence and metastasis (132). Numerous lncRNAs have been identified to be involved in AI drug resistance. Of particular importance is tamoxifen, one of the most commonly used AI drugs. For example, lncRNA DIO3OS interacts with PTBP1 to upregulate LDHA mRNA stability, activating glycolytic metabolism in tamoxifen-resistant BC cells and promoting ER-independent cell proliferation both in vitro and in vivo (133). Additionally, DILA1 upregulates the oncoprotein Cyclin D1 by inhibiting its phosphorylation and subsequent degradation. The upregulation of Cyclin D1 promotes BC cell proliferation and leads to tamoxifen resistance in both in vivo and in vitro settings (134). Furthermore, HOTAIR is highly expressed in tamoxifen-resistant breast cancer patients compared to newly diagnosed patients before tamoxifen treatment. The upregulation of HOTAIR activates the ER transcriptional program, resulting in increased BC cell proliferation and tamoxifen resistance (135). Overexpression of H19 in BC cells and tamoxifen-resistant BC cells activates autophagy through the H19/SAHH/DNMT3B axis, which contributes to tamoxifen resistance in BC cells (136). For more lncRNAs involved in tamoxifen resistance, please refer to Table 4.
3.3.3 LncRNAs and trastuzumab resistance
In the past, HER2-positive breast cancer patients often had a poor prognosis. However, the discovery of trastuzumab, a recombinant humanized monoclonal antibody that targets the extracellular domain of HER2, has significantly improved the outcomes for these patients (149). Although other anti-HER2 agents like pertuzumab and lapatinib have been developed, trastuzumab remains the gold standard treatment. Unfortunately, the effectiveness of trastuzumab is limited by the emergence of drug resistance (150). Recent research has identified several long non-coding RNAs (lncRNAs) that are closely associated with trastuzumab resistance, including SNHG14, ATB, and AGAP2-AS1 (Table 5).
3.3.4 LncRNAs and radiotherapy resistance
Radiotherapy (RT) is commonly used as an adjuvant treatment after surgery for various types of breast cancer, including TNBC, metastatic BC, and advanced BC, as it has shown great benefits in reducing recurrence (157). The success of radiotherapy depends on the radiosensitivity of the tumor, which is influenced by factors such as cancer stem cells (CSCs), the tumor microenvironment, DNA repair, and gene expression (158). Unfortunately, some breast tumors develop resistance to radiation, leading to treatment failure and recurrence. Understanding the mechanisms of radiation resistance is crucial for improving the efficacy of radiotherapy. Numerous studies have found associations between specific lncRNAs and radiation resistance in breast cancer. For example, LINC00963 has been found to be upregulated in breast cancer tissues and correlated with aggressive tumor characteristics. Knockdown of LINC00963 has been shown to enhance DNA damage and oxidative stress, making breast cancer cells more sensitive to radiation (159). LINC00963 achieves this through its interactions with miR-324-3P, which normally inhibits the expression of ACK1, a driver of tumor progression. Further information on lncRNAs implicated in radiation resistance is summarized in Table 6. These findings highlight the importance of lncRNAs in mediating resistance to trastuzumab and radiotherapy in breast cancer. Further research is necessary to elucidate the underlying mechanisms and identify potential therapeutic targets to overcome drug and radiation resistance in breast cancer patients.
3.4 Circulating lncRNAs as biomarkers of BC
Accurate tumor biomarkers play a crucial role in diagnosing and predicting the prognosis for patients. Continuing efforts are being made to identify new biomarkers with high sensitivity and specificity. These biomarkers are valuable for evaluating tumor stage, metastasis risk, treatment response, and the development of new therapies (166). Circulating nucleic acids, including circulating RNAs such as lncRNAs, miRNAs, and piRNAs, have emerged as a promising class of potential biomarkers for improving tumor diagnosis. Compared to circulating DNAs, circulating RNAs offer higher specificity and sensitivity, garnering significant research attention (167). This review predominantly focuses on circulating lncRNAs as biomarkers for breast cancer, given their stability and abundance in the bloodstream, making them reliable cancer biomarkers (168). As indicated by previous findings on changes in breast cancer cells and tissues, there is a growing focus on circulating lncRNAs. Encouragingly, certain circulating lncRNAs can reflect cancer status, and changes in these lncRNAs are correlated with the degree of tumor progression and clinical features. Furthermore, some of these lncRNAs can identify specific subtypes of breast cancer, while a few others may serve as potential therapeutic agents or treatment targets (168).
For instance, HOTAIR has been found to be significantly elevated in the serum of breast cancer patients compared to healthy individuals, suggesting its potential as a diagnostic biomarker (169). One study indicated that HOTAIR exhibits a stronger diagnostic capability for breast cancer than CEA and CA 15-3, given its association with ER, Her-2, and lymph node metastasis (170). Notably, these researchers have observed a significant decrease in HOTAIR expression levels post-surgery. However, some studies have suggested the existence of potential technical errors in previous experimental results, warranting further investigation to determine whether HOTAIR could be adopted as a prognostic marker (171). Therefore, further studies are needed to assess whether HOTAIR could be adopted as a potential prognostic marker.
Another example is HISLA (HIF-1α-stabilizing long noncoding RNA), which can be transmitted by extracellular vesicles from tumor-associated macrophages to breast cancer cells (172). HISLA is overexpressed in both breast cancer tissues and patient serum and is significantly associated with advanced grade, histological grade, distant metastasis, and poor survival. Moreover, serum levels of HISLA were observed to decrease significantly after surgery, suggesting its potential as a biomarker for diagnosing and prognosticating breast cancer (173). Similarly, H19 is released into the plasma from tumor cells upon breast cancer initiation, leading to an upregulation of plasma H19 levels. Consistently, plasma H19 levels were found to decrease significantly after surgery, and they have been significantly correlated with ER, PR, Her-2, and lymph node metastasis, indicating the potential use of H19 as a diagnostic and monitoring biomarker for breast cancer (174). In addition, a group of researchers detected the serum levels of lncRNAs PVT1, HOTAIR, NEAT1, and MALAT1 from Egyptian breast cancer and fibroadenoma patients, as well as healthy donors, and found that serum PVT1, HOTAIR, and NEAT1 could serve as potential biomarkers for breast cancer. Specifically, HOTAIR and NEAT1 demonstrated feasibility in differentiating between breast cancer and fibroadenoma (81). Moreover, El-Ashmawy et al. identified the upregulation of lncRNAs FAM83H-AS1 and ATB in the serum of breast cancer patients, with ATB exhibiting superior diagnostic accuracy compared to CA 15-3, a well-established serum protein marker (175). Conversely, FAM83H-AS1 demonstrated prognostic rather than diagnostic value, showing a significant association with tumor lymph node metastasis and tumor size (175). A brief overview of circulating lncRNAs with potential applications as biomarkers for breast cancer is presented in Figure 3 (176–181). Some reported lncRNAs may exhibit relatively low specificity or sensitivity when used individually, making it feasible to combine several lncRNAs or use them in conjunction with traditional markers.
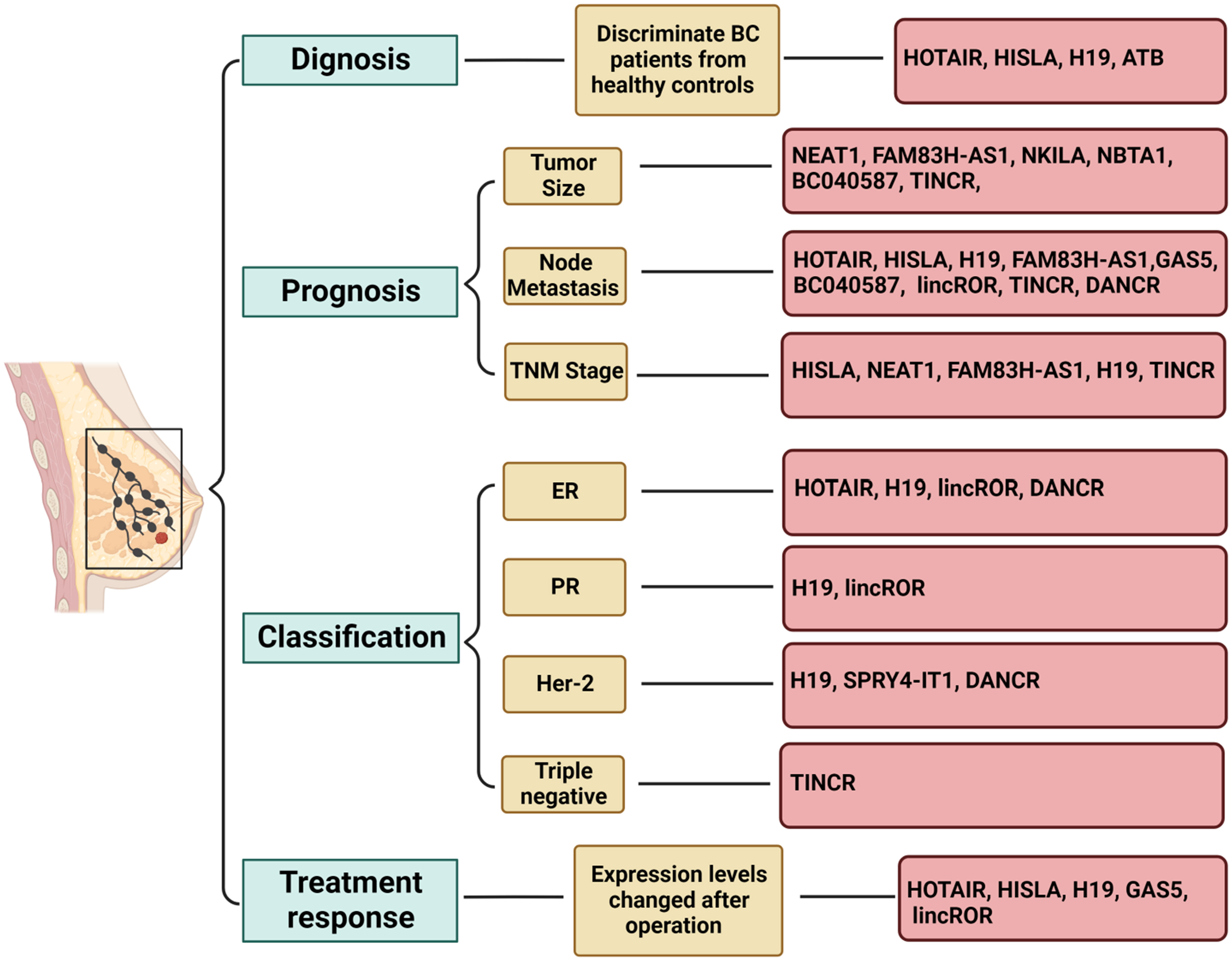
Figure 3 Role of circulating lncRNAs as potential biomarkers in breast cancer. Long non-coding RNAs (lncRNAs) have demonstrated potential as biomarkers in various aspects of breast cancer, including diagnosis, prognosis, subtype classification, and treatment response. Utilizing a combination of multiple lncRNAs may offer a viable strategy for leveraging lncRNA-based biomarkers in clinical applications. The figure was generated using BioRender.com.
4 Therapeutic strategies against lncRNAs in breast cancer
In the ongoing battle against breast cancer, the significance of lncRNAs has come to the forefront of cancer research. Given their crucial role in the etiology and progression of breast cancer, as well as the development of resistance to current therapies, lncRNAs present an enticing target for novel treatments (182). Recent advances have led to the development of various strategies targeting lncRNAs, utilizing antisense oligonucleotides (ASOs), small molecules, and natural compounds.
ASOs represent one of the spearheads in the quest for lncRNA-targeted interventions. These synthetic molecules are engineered to be complementary to specific RNA sequences, allowing them to bind and neutralize lncRNAs through varied mechanisms. Among the mechanisms is the induction of RNase H-dependent degradation of the lncRNA, reducing its oncogenic influence in tumor cells (183). The potential of ASOs is illustrated by their effectiveness against HOTAIR and MALAT1, lncRNAs implicated in breast cancer metastasis and oncogenesis (184, 185). By subduing these lncRNAs, ASOs have demonstrated a capacity to inhibit cancer stem cell properties and reduce metastasis in preclinical models.
Small molecules like quercetin have also emerged as a viable approach to modulate lncRNAs. Quercetin, a widely available dietary flavonoid, has been found to diminish the expression of lncRNA MALAT1 in breast cancer cell lines (186). Through such downregulation, it exerts antitumor effects, likely by disrupting the complex interplay between key cellular pathways, p53, and miRNAs that revolve around MALAT1. This provides a compelling case for the therapeutic repurposing of naturally occurring compounds with regulatory effects on lncRNAs.
Additionally, natural products such as curcumin, recognized for its myriad health benefits, have been shown to regulate the expression of certain lncRNAs. A noteworthy example includes its ability to downregulate lncRNA H19, an action that can suppress the resistance of breast cancer cells to tamoxifen (187). Such findings underscore the potential therapeutic role of natural products in influencing lncRNA expression and combating drug resistance, critical hurdles in current treatment paradigms.
Collectively, the innovative application of ASOs, small molecules, peptides, and natural products holds immense promise for the future of breast cancer therapy, specifically through targeting lncRNAs. These emerging strategies may provide a breakthrough in addressing the challenges BC presents, including drug resistance, tumor recurrence, and metastasis, thereby improving outcomes for patients worldwide. As research progresses, the integration of these treatments into the clinical setting could transform the landscape of breast cancer management, offering hope for more effective and personalized therapeutic options.
5 Discussion and conclusion
Breast cancer (BC) remains a leading cause of cancer-related deaths in women, with different subtypes exhibiting distinct molecular characteristics and treatment responses. While significant progress has been made in improving the prognosis of BC patients through established treatments such as radical surgery and adjuvant therapy, a considerable number of patients still succumb to metastasis or resistance to these treatments. Therefore, there is a continuous need to explore novel therapeutic targets and molecular mechanisms.
The initiation and progression of breast cancer are influenced by a combination of genetic, epigenetic, and non-genetic factors. Previous research has primarily focused on genetic abnormalities and classic epigenetic factors such as histone and DNA modifications (188–191). Recently, ncRNAs, particularly lncRNAs, have emerged as crucial epigenetic regulators in cancer research (28). LncRNAs, which are longer than 200 nucleotides and have limited protein coding potential, play various biological roles in BC development through multiple mechanisms. Dysregulated lncRNAs have been closely associated with BC cell growth, apoptosis, invasion, EMT, autophagy, and therapeutic resistance, contributing to BC progression. Therefore, these lncRNAs hold promise as predictive biomarkers or therapeutic targets for BC patients.
In addition to downstream regulation, the upstream regulatory mechanism of lncRNAs has garnered significant research attention. One such mechanism involves N6-methyladenosine (m6A), the most frequent posttranscriptional modification found on mRNA, which has also been discovered on lncRNAs (192, 193). The interaction between m6A readers and lncRNAs, as well as the reciprocal regulation of m6A modifiers by lncRNAs, plays a role in BC development. For instance, the oncogenic lncRNA MIR210HG is induced by the m6A reader IGF2BP1 in an m6A-dependent manner to promote BC progression and metastasis (194). Another oncogenic lncRNA LNC942 bound to METTL14 (an m6A writer) directly and promoted m6A methylation and stabilization of CXCR4 and CYP1B1, promoting BC cells proliferation and inhibiting apoptosis (195). Collectively, m6A/lncRNAs axis enriches BC regulatory network and provides novel targets for BC prevention and management.
Interestingly, some lncRNAs have been found to encode short peptides that possess functional roles in cancer. For example, the lncRNA HOXB-AS3 encodes a conserved 53-amino acid peptide that suppresses colon cancer growth (196). Similarly, some peptides encoded by lncRNAs are involved in BC initiation and progression. For instance, the lncRNA LINC00908 contains a small open reading frame (ORF) encoding a 60-amino acid polypeptide named ASRPS, which acts as a small regulatory peptide of STAT3 (197). Downregulation of ASRPS was associated with poor clinical outcome, indicating that ASRPS is a potential antitumor peptide. Mechanistically, ASRPS directly binds to STAT3, inhibiting its phosphorylation and downstream VEGF expression, thereby suppressing TNBC angiogenesis (197). Additionally, the lncRNA CTD-2256P15.2 encodes a micropeptide called PACMP that regulates BC progression, multiple drug resistance, and ionizing radiation resistance by maintaining CtIP abundance and promoting PARP-1-dependent poly (ADP-ribosyl)ation (PARylation) through direct binding to DNA damage-induced poly (ADP-ribose) chains (198).
In summary, a substantial number of lncRNAs have been identified as oncogenic in BC, with their overexpression associated with aggressive disease and poor patient survival. Conversely, certain lncRNAs exhibit tumor-suppressive effects and are frequently downregulated in BC. Hence, targeting oncogenic lncRNAs while activating antitumor lncRNAs can provide unique opportunities in the battle against BC. Advances in multiomics sequencing technologies and bioinformatics tools are anticipated to uncover an increasing number of lncRNAs involved in BC progression, expanding the repertoire of diagnostic and prognostic biomarkers. With deeper understanding of the role of lncRNAs in BC initiation and progression, oncogenic lncRNAs may be developed as potential therapeutic targets for BC treatment. However, it is important to note that most studies investigating lncRNAs and BC are still at the preclinical stage, and further research is required to elucidate the underlying mechanisms. Future studies should focus on identifying specific and sensitive lncRNAs for early diagnosis, risk stratification, prognosis monitoring, and personalized treatment of BC.
Author contributions
YW: Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft. BN: Data curation, Investigation, Methodology, Software, Validation, Writing – original draft. XL: Conceptualization, Funding acquisition, Methodology, Resources, Writing – review & editing. QS: Investigation, Methodology, Software, Validation, Writing – original draft. B-FM: Investigation, Software, Visualization, Writing – original draft. WH: Investigation, Validation, Visualization, Writing – review & editing. JY: Investigation, Methodology, Visualization, Writing – original draft. LW: Conceptualization, Methodology, Visualization, Writing – review & editing. XZ: Methodology, Software, Visualization, Writing – review & editing. YM: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the State Key Laboratory of Pathogenesis, Prevention and Treatment of High Incidence Diseases in Central Asia Fund (SKL-HIDCA-2023-JY6), the National Natural Science Foundation of China (82000155), the Natural Science Foundation of Zhejiang Province (LY23H160017), and the China Postdoctoral Science Foundation (No. 2021M702877).
Acknowledgments
We would like to express our sincere gratitude to our colleagues and researchers for their invaluable insights, thought-provoking discussions, and constructive feedback. We would also like to apologize to fellow researchers whose remarkable work could not be referenced due to space constraints.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
BC, breast cancer; ncRNA, non-coding RNA; lncRNA, long non-coding RNA; ceRNA, competitive endogenous RNA; TME, tumor microenvironment; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; CEA, carcinoembryonic antigen; EVs, extracellular vesicles; ILVs, intraluminal vesicles; MVBs, multivesicular bodies; PRC2, Polycomb Repressive Complex 2; CSCs, cancer stem cells; EMT, epithelial-mesenchymal transition; TICs, tumor-initiating cells; BCSCs, breast cancer stem cells; RT, radiotherapy.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
2. Gucalp A, Traina TA, Eisner JR, Parker JS, Selitsky SR, Park BH, et al. Male breast cancer: a disease distinct from female breast cancer. Breast Cancer Res Treat (2019) 173:37–48. doi: 10.1007/s10549-018-4921-9
3. Nolan E, Lindeman GJ, Visvader JE. Deciphering breast cancer: from biology to the clinic. Cell (2023) 186:1708–28. doi: 10.1016/j.cell.2023.01.040
4. Libson S, Lippman M. A review of clinical aspects of breast cancer. Int Rev Psychiatry (2014) 26:4–15. doi: 10.3109/09540261.2013.852971
5. Joyce DP, Kerin MJ, Dwyer RM. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int J Cancer (2016) 139:1443–8. doi: 10.1002/ijc.30179
6. Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther (2020) 5:28. doi: 10.1038/s41392-020-0134-x
7. Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ. Strategies for subtypes–dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol (2011) 22:1736–47. doi: 10.1093/annonc/mdr304
8. Sukumar J, Gast K, Quiroga D, Lustberg M, Williams N. Triple-negative breast cancer: promising prognostic biomarkers currently in development. Expert Rev Anticancer Ther (2021) 21:135–48. doi: 10.1080/14737140.2021.1840984
9. Fowler AM, Strigel RM. Clinical advances in PET-MRI for breast cancer. Lancet Oncol (2022) 23:e32–43. doi: 10.1016/S1470-2045(21)00577-5
10. Lotter W, Diab AR, Haslam B, Kim JG, Grisot G, Wu E, et al. Robust breast cancer detection in mammography and digital breast tomosynthesis using an annotation-efficient deep learning approach. Nat Med (2021) 27:244–9. doi: 10.1038/s41591-020-01174-9
11. Fiorica JV. Breast cancer screening, mammography, and other modalities. Clin Obstet Gynecol (2016) 59:688–709. doi: 10.1097/GRF.0000000000000246
12. Swellam M, Abdelmaksoud MD, Sayed Mahmoud M, Ramadan A, Abdel-Moneem W, Hefny MM. Aberrant methylation of APC and RARβ2 genes in breast cancer patients. IUBMB Life (2015) 67:61–8. doi: 10.1002/iub.1346
13. Yamashita H, Nishio M, Toyama T, Sugiura H, Zhang Z, Kobayashi S, et al. Coexistence of HER2 over-expression and p53 protein accumulation is a strong prognostic molecular marker in breast cancer. Breast Cancer Res (2004) 6:R24–30. doi: 10.1186/bcr738
14. Zaenker P, Gray ES, Ziman MR. Autoantibody production in cancer–the humoral immune response toward autologous antigens in cancer patients. Autoimmun Rev (2016) 15:477–83. doi: 10.1016/j.autrev.2016.01.017
15. Szilágyi M, Pös O, Márton É, Buglyó G, Soltész B, Keserű J, et al. Circulating cell-free nucleic acids: main characteristics and clinical application. Int J Mol Sci (2020) 21(18):6827. doi: 10.3390/ijms21186827
16. Maimaitiyiming Y, Ye L, Yang T, Yu W, Naranmandura H. Linear and circular long non-coding RNAs in acute lymphoblastic leukemia: from pathogenesis to classification and treatment. Int J Mol Sci (2022) 23(8):4442. doi: 10.3390/ijms23084442
17. Dvorská D, Braný D, Ňachajová M, Halašová E, Danková Z. Breast cancer and the other non-coding RNAs. Int J Mol Sci (2021) 22(6):3280. doi: 10.3390/ijms22063280
18. Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem (2021) 65:625–39. doi: 10.1042/EBC20200032
19. Xiong Q, Zhang Y, Li J, Zhu Q. Small non-coding RNAs in human cancer. Genes (Basel) (2022) 13(11):2072. doi: 10.3390/genes13112072
20. Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J (2019) 38:e100836. doi: 10.15252/embj.2018100836
21. Sana J, Faltejskova P, Svoboda M, Slaby O. Novel classes of non-coding RNAs and cancer. J Transl Med (2012) 10:103. doi: 10.1186/1479-5876-10-103
22. Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, et al. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature (2017) 543:199–204. doi: 10.1038/nature21374
23. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol (2021) 22:96–118. doi: 10.1038/s41580-020-00315-9
24. Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol (2013) 10:925–33. doi: 10.4161/rna.24604
25. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell (2018) 172:393–407. doi: 10.1016/j.cell.2018.01.011
26. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell (2011) 43:904–14. doi: 10.1016/j.molcel.2011.08.018
27. Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci (2016) 41:761–72. doi: 10.1016/j.tibs.2016.07.003
28. Tachiwana H, Saitoh N. Nuclear long non-coding RNAs as epigenetic regulators in cancer. Curr Med Chem (2021) 28:5098–109. doi: 10.2174/0929867328666210215114506
29. Rashid F, Shah A, Shan G. Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinf (2016) 14:73–80. doi: 10.1016/j.gpb.2016.03.005
30. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol (2021) 220(2):e202009045. doi: 10.1083/jcb.202009045
31. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci (2016) 73:2491–509. doi: 10.1007/s00018-016-2174-5
32. Jin KT, Yao JY, Fang XL, Di H, Ma YY. Roles of lncRNAs in cancer: Focusing on angiogenesis. Life Sci (2020) 252:117647. doi: 10.1016/j.lfs.2020.117647
33. Liu SJ, Dang HX, Lim DA, Feng FY, Maher CA. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer (2021) 21:446–60. doi: 10.1038/s41568-021-00353-1
34. Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci (2019) 9:19. doi: 10.1186/s13578-019-0282-2
35. Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci (2018) 75:193–208. doi: 10.1007/s00018-017-2595-9
36. Zhang W, Wang Q, Yang Y, Zhou S, Zhang P, Feng T. The role of exosomal lncRNAs in cancer biology and clinical management. Exp Mol Med (2021) 53:1669–73. doi: 10.1038/s12276-021-00699-4
37. Huang X, Huang L, Xie Q, Zhang L, Huang S, Hong M, et al. LncRNAs serve as novel biomarkers for diagnosis and prognosis of childhood ALL. biomark Res (2021) 9:45. doi: 10.1186/s40364-021-00303-x
38. Zheng M, Han R, Yuan W, Chi H, Zhang Y, Sun K, et al. Circulating exosomal lncRNAs in patients with chronic coronary syndromes. Arch Med Sci (2023) 19:46–56. doi: 10.5114/aoms/128014
39. Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol (2013) 14:475–88. doi: 10.1038/nrm3611
40. Zhou P, Xu W, Peng X, Luo Z, Xing Q, Chen X, et al. Large-scale screens of miRNA-mRNA interactions unveiled that the 3’UTR of a gene is targeted by multiple miRNAs. PloS One (2013) 8:e68204. doi: 10.1371/journal.pone.0068204
41. Petri BJ, Klinge CM. Regulation of breast cancer metastasis signaling by miRNAs. Cancer Metastasis Rev (2020) 39:837–86. doi: 10.1007/s10555-020-09905-7
42. Luo W, Wang J, Xu W, Ma C, Wan F, Huang Y, et al. LncRNA RP11-89 facilitates tumorigenesis and ferroptosis resistance through PROM2-activated iron export by sponging miR-129-5p in bladder cancer. Cell Death Dis (2021) 12:1043. doi: 10.1038/s41419-021-04296-1
43. Karagkouni D, Karavangeli A, Paraskevopoulou MD, Hatzigeorgiou AG. Characterizing miRNA-lncRNA interplay. Methods Mol Biol (2021) 2372:243–62. doi: 10.1007/978-1-0716-1697-0_21
44. Lau KH, Tan AM, Shi Y. New and emerging targeted therapies for advanced breast cancer. Int J Mol Sci (2022) 23(4):2288. doi: 10.3390/ijms23042288
45. Esteva FJ, Hubbard-Lucey VM, Tang J, Pusztai L. Immunotherapy and targeted therapy combinations in metastatic breast cancer. Lancet Oncol (2019) 20:e175–e86. doi: 10.1016/S1470-2045(19)30026-9
46. Ahadi A. Functional roles of lncRNAs in the pathogenesis and progression of cancer. Genes Dis (2021) 8:424–37. doi: 10.1016/j.gendis.2020.04.009
47. Sideris N, Dama P, Bayraktar S, Stiff T, Castellano L. LncRNAs in breast cancer: a link to future approaches. Cancer Gene Ther (2022) 29:1866–77. doi: 10.1038/s41417-022-00487-w
48. Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med (2015) 12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006
49. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science (2010) 329:689–93. doi: 10.1126/science.1192002
50. Mozdarani H, Ezzatizadeh V, Rahbar Parvaneh R. The emerging role of the long non-coding RNA HOTAIR in breast cancer development and treatment. J Transl Med (2020) 18:152. doi: 10.1186/s12967-020-02320-0
51. Shi F, Chen X, Wang Y, Xie Y, Zhong J, Su K, et al. HOTAIR/miR-203/CAV1 crosstalk influences proliferation, migration, and invasion in the breast cancer cell. Int J Mol Sci (2022) 23(19):11755. doi: 10.3390/ijms231911755
52. Li Y, Wang Z, Shi H, Li H, Li L, Fang R, et al. HBXIP and LSD1 Scaffolded by lncRNA Hotair Mediate Transcriptional Activation by c-Myc. Cancer Res (2016) 76:293–304. doi: 10.1158/0008-5472.CAN-14-3607
53. Yu Y, Lv F, Liang D, Yang Q, Zhang B, Lin H, et al. HOTAIR may regulate proliferation, apoptosis, migration and invasion of MCF-7 cells through regulating the P53/Akt/JNK signaling pathway. BioMed Pharmacother (2017) 90:555–61. doi: 10.1016/j.biopha.2017.03.054
54. Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol (1990) 10:28–36. doi: 10.1128/mcb.10.1.28-36.1990
55. Berteaux N, Lottin S, Monté D, Pinte S, Quatannens B, Coll J, et al. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem (2005) 280:29625–36. doi: 10.1074/jbc.M504033200
56. Si H, Chen P, Li H, Wang X. Long non-coding RNA H19 regulates cell growth and metastasis via miR-138 in breast cancer. Am J Transl Res (2019) 11:3213–25.
57. Kong X, Duan Y, Sang Y, Li Y, Zhang H, Liang Y, et al. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol (2019) 234:9105–17. doi: 10.1002/jcp.27587
58. Taiana E, Ronchetti D, Todoerti K, Nobili L, Tassone P, Amodio N, et al. LncRNA NEAT1 in paraspeckles: A structural scaffold for cellular DNA damage response systems? Noncoding RNA (2020) 6(3):26. doi: 10.3390/ncrna6030026
59. Park MK, Zhang L, Min KW, Cho JH, Yeh CC, Moon H, et al. NEAT1 is essential for metabolic changes that promote breast cancer growth and metastasis. Cell Metab (2021) 33:2380–97.e9. doi: 10.1016/j.cmet.2021.11.011
60. Zhang M, Wang N, Song P, Fu Y, Ren Y, Li Z, et al. LncRNA GATA3-AS1 facilitates tumour progression and immune escape in triple-negative breast cancer through destabilization of GATA3 but stabilization of PD-L1. Cell Prolif (2020) 53:e12855. doi: 10.1111/cpr.12855
61. Hu A, Hong F, Li D, Jin Y, Kon L, Xu Z, et al. Long non-coding RNA ROR recruits histone transmethylase MLL1 to up-regulate TIMP3 expression and promote breast cancer progression. J Transl Med (2021) 19:95. doi: 10.1186/s12967-020-02682-5
62. Jia H, Wu D, Zhang Z, Li S. Regulatory effect of the MAFG−AS1/miR−150−5p/MYB axis on the proliferation and migration of breast cancer cells. Int J Oncol (2021) 58:33–44. doi: 10.3892/ijo.2020.5150
63. Li Y, Zeng Q, Qiu J, Pang T, Xian J, Zhang X. Long non-coding RNA UCA1 promotes breast cancer by upregulating PTP1B expression via inhibiting miR-206. Cancer Cell Int (2019) 19:275. doi: 10.1186/s12935-019-0958-z
64. Ding Y, Huang Y, Zhang F, Gong L, Liang C, Ding K, et al. LncRNA TDRKH-AS1 promotes breast cancer progression via the miR-134-5p/CREB1 axis. J Transl Med (2023) 21:854. doi: 10.1186/s12967-023-04640-3
65. Nie Z, Xu M, Zhou L, Pan B, Xu T, He B, et al. lncSNHG3 drives breast cancer progression by epigenetically increasing CSNK2A1 expression level. Aging (Albany NY) (2023) 15:5734–50. doi: 10.18632/aging.204824
66. Dong W, Li J, Zhuang Z. Deciphering the prognostic significance of anoikis-related lncRNAs in invasive breast cancer: from comprehensive bioinformatics analysis to functional experimental validation. Aging (Albany NY) (2024) 16:402–30. doi: 10.18632/aging.205376
67. Wang B, Chen H, Yang R, Xing L, Chen C, Chen J. LncRNA RP11-551L14.4 suppresses breast cancer development by inhibiting the expression of miR-4472. PeerJ (2022) 10:e14482. doi: 10.7717/peerj.14482
68. Dong S, Ma M, Li M, Guo Y, Zuo X, Gu X, et al. LncRNA MEG3 regulates breast cancer proliferation and apoptosis through miR-141-3p/RBMS3 axis. Genomics (2021) 113:1689–704. doi: 10.1016/j.ygeno.2021.04.015
69. Qiu S, Chen G, Peng J, Liu J, Chen J, Wang J, et al. LncRNA EGOT decreases breast cancer cell viability and migration via inactivation of the Hedgehog pathway. FEBS Open Bio (2020) 10:817–26. doi: 10.1002/2211-5463.12833
70. Yi D, Wang Z, Yang H, Wang R, Shi X, Liu Z, et al. Long non-coding RNA MEG3 acts as a suppressor in breast cancer by regulating miR-330/CNN1. Aging (Albany NY) (2024) 16:1318–35. doi: 10.18632/aging.205419
71. Xie D, Li S, Wang X, Fang L. lncRNA HCG11 suppresses cell proliferation in hormone receptor-positive breast cancer via SRSF1/β-catenin. Aging (Albany NY) (2023) 15:179–92. doi: 10.18632/aging.204468
72. Guo R, Su Y, Zhang Q, Xiu B, Huang S, Chi W, et al. LINC00478-derived novel cytoplasmic lncRNA LacRNA stabilizes PHB2 and suppresses breast cancer metastasis via repressing MYC targets. J Transl Med (2023) 21:120. doi: 10.1186/s12967-023-03967-1
73. Zhang KJ, Tan XL, Guo L. The long non-coding RNA DANCR regulates the inflammatory phenotype of breast cancer cells and promotes breast cancer progression via EZH2-dependent suppression of SOCS3 transcription. Mol Oncol (2020) 14:309–28. doi: 10.1002/1878-0261.12622
74. Li Z, Hou P, Fan D, Dong M, Ma M, Li H, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ (2017) 24:59–71. doi: 10.1038/cdd.2016.95
75. Li S, Zhou J, Wang Z, Wang P, Gao X, Wang Y. Long noncoding RNA GAS5 suppresses triple negative breast cancer progression through inhibition of proliferation and invasion by competitively binding miR-196a-5p. BioMed Pharmacother (2018) 104:451–7. doi: 10.1016/j.biopha.2018.05.056
76. Filippova EA, Fridman MV, Burdennyy AM, Loginov VI, Pronina IV, Lukina SS, et al. Long noncoding RNA GAS5 in breast cancer: epigenetic mechanisms and biological functions. Int J Mol Sci (2021) 22(13):6810. doi: 10.3390/ijms22136810
77. Wang N, Hou M, Zhan Y, Sheng X. LncRNA PTCSC3 inhibits triple-negative breast cancer cell proliferation by downregulating lncRNA H19. J Cell Biochem (2019) 120:15083–8. doi: 10.1002/jcb.28769
78. Jadaliha M, Gholamalamdari O, Tang W, Zhang Y, Petracovici A, Hao Q, et al. A natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability. PloS Genet (2018) 14:e1007802. doi: 10.1371/journal.pgen.1007802
79. Yue X, Wu WY, Dong M, Guo M. LncRNA MALAT1 promotes breast cancer progression and doxorubicin resistance via regulating miR-570-3p. BioMed J (2021) 44:S296–s304. doi: 10.1016/j.bj.2020.11.002
80. Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet (2018) 50:1705–15. doi: 10.1038/s41588-018-0252-3
81. El-Fattah AAA, Sadik NAH, Shaker OG, Mohamed Kamal A, Shahin NN. Serum long non-coding RNAs PVT1, HOTAIR, and NEAT1 as potential biomarkers in Egyptian women with breast cancer. Biomolecules (2021) 11(2):301. doi: 10.3390/biom11020301
82. Gao X, Qin T, Mao J, Zhang J, Fan S, Lu Y, et al. PTENP1/miR-20a/PTEN axis contributes to breast cancer progression by regulating PTEN via PI3K/AKT pathway. J Exp Clin Cancer Res (2019) 38:256. doi: 10.1186/s13046-019-1260-6
83. Shi X, Tang X, Su L. Overexpression of Long Noncoding RNA PTENP1 Inhibits Cell Proliferation and Migration via Suppression of miR-19b in Breast Cancer Cells. Oncol Res (2018) 26:869–78. doi: 10.3727/096504017X15123838050075
84. Yndestad S, Austreid E, Skaftnesmo KO, Lønning PE, Eikesdal HP. Divergent activity of the pseudogene PTENP1 in ER-positive and negative breast cancer. Mol Cancer Res (2018) 16:78–89. doi: 10.1158/1541-7786.MCR-17-0207
85. Zhao Y, Yu Z, Ma R, Zhang Y, Zhao L, Yan Y, et al. lncRNA-Xist/miR-101-3p/KLF6/C/EBPα axis promotes TAM polarization to regulate cancer cell proliferation and migration. Mol Ther Nucleic Acids (2021) 23:536–51. doi: 10.1016/j.omtn.2020.12.005
86. Zong Y, Zhang Y, Hou D, Xu J, Cui F, Qin Y, et al. The lncRNA XIST promotes the progression of breast cancer by sponging miR-125b-5p to modulate NLRC5. Am J Transl Res (2020) 12:3501–11.
87. Gao Y, Bado I, Wang H, Zhang W, Rosen JM, Zhang XH. Metastasis organotropism: redefining the congenial soil. Dev Cell (2019) 49:375–91. doi: 10.1016/j.devcel.2019.04.012
88. Bergers G, Fendt SM. The metabolism of cancer cells during metastasis. Nat Rev Cancer (2021) 21:162–80. doi: 10.1038/s41568-020-00320-2
89. Babaei G, Aziz SG, Jaghi NZZ. EMT, cancer stem cells and autophagy; The three main axes of metastasis. BioMed Pharmacother (2021) 133:110909. doi: 10.1016/j.biopha.2020.110909
90. Fico F, Bousquenaud M, Rüegg C, Santamaria-Martínez A. Breast cancer stem cells with tumor- versus metastasis-initiating capacities are modulated by TGFBR1 inhibition. Stem Cell Rep (2019) 13:1–9. doi: 10.1016/j.stemcr.2019.05.026
91. Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. (2016) 113:E2047–56. doi: 10.1073/pnas.1602883113
92. Park M, Kim D, Ko S, Kim A, Mo K, Yoon H. Breast cancer metastasis: mechanisms and therapeutic implications. Int J Mol Sci (2022) 23(12):6806. doi: 10.3390/ijms23126806
93. Morel AP, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PloS One (2008) 3:e2888. doi: 10.1371/journal.pone.0002888
94. Deng J, Yang M, Jiang R, An N, Wang X, Liu B. Long non-coding RNA HOTAIR regulates the proliferation, self-renewal capacity, tumor formation and migration of the cancer stem-like cell (CSC) subpopulation enriched from breast cancer cells. PloS One (2017) 12:e0170860. doi: 10.1371/journal.pone.0170860
95. Wang J, Liu X, Li P, Wang J, Shu Y, Zhong X, et al. Long noncoding RNA HOTAIR regulates the stemness of breast cancer cells via activation of the NF-κB signaling pathway. J Biol Chem (2022) 298:102630. doi: 10.1016/j.jbc.2022.102630
96. Zhang H, Cai K, Wang J, Wang X, Cheng K, Shi F, et al. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells (2014) 32:2858–68. doi: 10.1002/stem.1795
97. Pawłowska E, Szczepanska J, Blasiak J. The long noncoding RNA HOTAIR in breast cancer: does autophagy play a role? Int J Mol Sci (2017) 18(11):2317. doi: 10.3390/ijms18112317
98. Peng F, Li TT, Wang KL, Xiao GQ, Wang JH, Zhao HD, et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis (2017) 8:e2569. doi: 10.1038/cddis.2016.438
99. Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li LY, et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal (2017) 10(483):eaak9557. doi: 10.1126/scisignal.aak9557
100. Xiong H, Shen J, Chen Z, Yang J, Xie B, Jia Y, et al. H19/let−7/Lin28 ceRNA network mediates autophagy inhibiting epithelial−mesenchymal transition in breast cancer. Int J Oncol (2020) 56:794–806. doi: 10.3892/ijo.2020.4967
101. Liu S, Sun Y, Hou Y, Yang L, Wan X, Qin Y, et al. A novel lncRNA ROPM-mediated lipid metabolism governs breast cancer stem cell properties. J Hematol Oncol (2021) 14:178. doi: 10.1186/s13045-021-01194-z
102. Jiang B, Zhu H, Tang L, Gao T, Zhou Y, Gong F, et al. Apatinib inhibits stem properties and Malignant biological behaviors of breast cancer stem cells by blocking wnt/β-catenin signal pathway through downregulating lncRNA ROR. Anticancer Agents Med Chem (2022) 22:1723–34. doi: 10.2174/1871520621666210412103849
103. Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis (2014) 5:e1287. doi: 10.1038/cddis.2014.249
104. Chen YM, Liu Y, Wei HY, Lv KZ, Fu PF. Large intergenic non-coding RNA-ROR reverses gemcitabine-induced autophagy and apoptosis in breast cancer cells. Oncotarget (2016) 7:59604–17. doi: 10.18632/oncotarget.10730
105. Gao Z, Zheng G, Gong X, Hu H, Shao L, Pang Y, et al. LncRNA MAFG-AS1 deregulated in breast cancer affects autophagy and progression of breast cancer by interacting with miR-3612 and FKBP4 in vitro. Biochem Biophys Res Commun (2022) 616:95–103. doi: 10.1016/j.bbrc.2022.05.020
106. Zheng A, Song X, Zhang L, Zhao L, Mao X, Wei M, et al. Long non-coding RNA LUCAT1/miR-5582-3p/TCF7L2 axis regulates breast cancer stemness via Wnt/β-catenin pathway. J Exp Clin Cancer Res (2019) 38:305. doi: 10.1186/s13046-019-1315-8
107. Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q, et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res (2018) 37:289. doi: 10.1186/s13046-018-0945-6
108. Xiu B, Chi Y, Liu L, Chi W, Zhang Q, Chen J, et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol Cancer (2019) 18:187. doi: 10.1186/s12943-019-1115-y
109. Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer (2021) 21:669–80. doi: 10.1038/s41568-021-00378-6
110. Chu Z, Huo N, Zhu X, Liu H, Cong R, Ma L, et al. FOXO3A-induced LINC00926 suppresses breast tumor growth and metastasis through inhibition of PGK1-mediated Warburg effect. Mol Ther (2021) 29:2737–53. doi: 10.1016/j.ymthe.2021.04.036
111. Cowman SJ, Koh MY. Revisiting the HIF switch in the tumor and its immune microenvironment. Trends Cancer (2022) 8:28–42. doi: 10.1016/j.trecan.2021.10.004
112. Niu Y, Bao L, Chen Y, Wang C, Luo M, Zhang B, et al. HIF2-induced long noncoding RNA RAB11B-AS1 promotes hypoxia-mediated angiogenesis and breast cancer metastasis. Cancer Res (2020) 80:964–75. doi: 10.1158/0008-5472.CAN-19-1532
113. Dong F, Ruan S, Wang J, Xia Y, Le K, Xiao X, et al. M2 macrophage-induced lncRNA PCAT6 facilitates tumorigenesis and angiogenesis of triple-negative breast cancer through modulation of VEGFR2. Cell Death Dis (2020) 11:728. doi: 10.1038/s41419-020-02926-8
114. McDonald ES, Clark AS, Tchou J, Zhang P, Freedman GM. Clinical diagnosis and management of breast cancer. J Nucl Med (2016) 57(Suppl 1):9s–16s. doi: 10.2967/jnumed.115.157834
115. Dittmer J. Breast cancer stem cells: Features, key drivers and treatment options. Semin Cancer Biol (2018) 53:59–74. doi: 10.1016/j.semcancer.2018.07.007
116. Wang R, Zhang T, Yang Z, Jiang C, Seng J. Long non-coding RNA FTH1P3 activates paclitaxel resistance in breast cancer through miR-206/ABCB1. J Cell Mol Med (2018) 22:4068–75. doi: 10.1111/jcmm.13679
117. Wu H, Gu J, Zhou D, Cheng W, Wang Y, Wang Q, et al. LINC00160 mediated paclitaxel-And doxorubicin-resistance in breast cancer cells by regulating TFF3 via transcription factor C/EBPβ. J Cell Mol Med (2020) 24:8589–602. doi: 10.1111/jcmm.15487
118. Pan Y, Pan Y, Cheng Y, Yang F, Yao Z, Wang O. Knockdown of LncRNA MAPT-AS1 inhibites proliferation and migration and sensitizes cancer cells to paclitaxel by regulating MAPT expression in ER-negative breast cancers. Cell Biosci (2018) 8:7. doi: 10.1186/s13578-018-0207-5
119. Xu S, Wang P, Zhang J, Wu H, Sui S, Zhang J, et al. Ai-lncRNA EGOT enhancing autophagy sensitizes paclitaxel cytotoxicity via upregulation of ITPR1 expression by RNA-RNA and RNA-protein interactions in human cancer. Mol Cancer (2019) 18:89. doi: 10.1186/s12943-019-1017-z
120. Huang P, Li F, Li L, You Y, Luo S, Dong Z, et al. lncRNA profile study reveals the mRNAs and lncRNAs associated with docetaxel resistance in breast cancer cells. Sci Rep (2018) 8:17970. doi: 10.1038/s41598-018-36231-4
121. Wang Y, Zhou P, Li P, Yang F, Gao XQ. Long non-coding RNA H19 regulates proliferation and doxorubicin resistance in MCF-7 cells by targeting PARP1. Bioengineered (2020) 11:536–46. doi: 10.1080/21655979.2020.1761512
122. Orre C, Dieu X, Guillon J, Gueguen N, Ahmadpour ST, Dumas JF, et al. The long non-coding RNA SAMMSON is a regulator of chemosensitivity and metabolic orientation in MCF-7 doxorubicin-resistant breast cancer cells. Biol (Basel) (2021) 10(11):1156. doi: 10.3390/biology10111156
123. Aini S, Bolati S, Ding W, Liu S, Su P, Aili S, et al. LncRNA SNHG10 suppresses the development of doxorubicin resistance by downregulating miR-302b in triple-negative breast cancer. Bioengineered (2022) 13:11430–9. doi: 10.1080/21655979.2022.2063592
124. Wang F, Yang S, Lv M, Chen F, Yin H, Gao S, et al. Novel long noncoding RNA 005620 induces epirubicin resistance in triple-negative breast cancer by regulating ITGB1 expression. Front Oncol (2021) 11:592215. doi: 10.3389/fonc.2021.592215
125. Yao N, Fu Y, Chen L, Liu Z, He J, Zhu Y, et al. Long non-coding RNA NONHSAT101069 promotes epirubicin resistance, migration, and invasion of breast cancer cells through NONHSAT101069/miR-129-5p/Twist1 axis. Oncogene (2019) 38:7216–33. doi: 10.1038/s41388-019-0904-5
126. Su A, Yao K, Zhang H, Wang Y, Zhang H, Tang J. DANCR induces cisplatin resistance of triple-negative breast cancer by KLF5/p27 signaling. Am J Pathol (2022) 193(3):248–58. doi: 10.1016/j.ajpath.2022.11.007
127. Zhou L, Li H, Sun T, Wen X, Niu C, Li M, et al. HULC targets the IGF1R-PI3K-AKT axis in trans to promote breast cancer metastasis and cisplatin resistance. Cancer Lett (2022) 548:215861. doi: 10.1016/j.canlet.2022.215861
128. Zhou D, Gu J, Wang Y, Luo B, Feng M, Wang X. Long noncoding RNA CCAT2 reduces chemosensitivity to 5-fluorouracil in breast cancer cells by activating the mTOR axis. J Cell Mol Med (2022) 26:1392–401. doi: 10.1111/jcmm.17041
129. Luo L, Zhang J, Tang H, Zhai D, Huang D, Ling L, et al. LncRNA SNORD3A specifically sensitizes breast cancer cells to 5-FU by sponging miR-185-5p to enhance UMPS expression. Cell Death Dis (2020) 11:329. doi: 10.1038/s41419-020-2557-2
130. Hanker AB, Sudhan DR, Arteaga CL. Overcoming endocrine resistance in breast cancer. Cancer Cell (2020) 37:496–513. doi: 10.1016/j.ccell.2020.03.009
131. Chen YC, Yu J, Metcalfe C, De Bruyn T, Gelzleichter T, Malhi V, et al. Latest generation estrogen receptor degraders for the treatment of hormone receptor-positive breast cancer. Expert Opin Investig Drugs (2022) 31:515–29. doi: 10.1080/13543784.2021.1983542
132. Davies C, Godwin J, Gray R, Clarke M, Cutter D, Darby S, et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet (2011) 378:771–84. doi: 10.1016/S0140-6736(11)60993-8
133. Chen X, Luo R, Zhang Y, Ye S, Zeng X, Liu J, et al. Long noncoding RNA DIO3OS induces glycolytic-dominant metabolic reprogramming to promote aromatase inhibitor resistance in breast cancer. Nat Commun (2022) 13:7160. doi: 10.1038/s41467-022-34702-x
134. Shi Q, Li Y, Li S, Jin L, Lai H, Wu Y, et al. LncRNA DILA1 inhibits Cyclin D1 degradation and contributes to tamoxifen resistance in breast cancer. Nat Commun (2020) 11:5513. doi: 10.1038/s41467-020-19349-w
135. Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene (2016) 35:2746–55. doi: 10.1038/onc.2015.340
136. Wang J, Xie S, Yang J, Xiong H, Jia Y, Zhou Y, et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol (2019) 12:81. doi: 10.1186/s13045-019-0747-0
137. Khan MI, Ahmad A. LncRNA SNHG6 sponges miR-101 and induces tamoxifen resistance in breast cancer cells through induction of EMT. Front Oncol (2022) 12:1015428. doi: 10.3389/fonc.2022.1015428
138. Ma T, Liang Y, Li Y, Song X, Zhang N, Li X, et al. LncRNA LINP1 confers tamoxifen resistance and negatively regulated by ER signaling in breast cancer. Cell Signal (2020) 68:109536. doi: 10.1016/j.cellsig.2020.109536
139. Ma Y, Bu D, Long J, Chai W, Dong J. LncRNA DSCAM-AS1 acts as a sponge of miR-137 to enhance Tamoxifen resistance in breast cancer. J Cell Physiol (2019) 234:2880–94. doi: 10.1002/jcp.27105
140. Li Y, Liu L, Lv Y, Zhang Y, Zhang L, Yu H, et al. Silencing long non-coding RNA HNF1A-AS1 inhibits growth and resistance to TAM of breast cancer cells via the microRNA-363/SERTAD3 axis. J Drug Target (2021) 29:742–53. doi: 10.1080/1061186X.2021.1878362
141. Moradi F, Mohajerani F, Sadeghizadeh M. CCAT2 knockdown inhibits cell growth, and migration and promotes apoptosis through regulating the hsa-mir-145-5p/AKT3/mTOR axis in tamoxifen-resistant MCF7 cells. Life Sci (2022) 311:121183. doi: 10.1016/j.lfs.2022.121183
142. Horie K, Takagi K, Takeiwa T, Mitobe Y, Kawabata H, Suzuki T, et al. Estrogen-inducible lncRNA BNAT1 functions as a modulator for estrogen receptor signaling in endocrine-resistant breast cancer cells. Cells (2022) 11(22):3610. doi: 10.3390/cells11223610
143. Lin X, Dinglin X, Cao S, Zheng S, Wu C, Chen W, et al. Enhancer-Driven lncRNA BDNF-AS Induces Endocrine Resistance and Malignant Progression of Breast Cancer through the RNH1/TRIM21/mTOR Cascade. Cell Rep (2020) 31:107753. doi: 10.1016/j.celrep.2020.107753
144. Zhang H, Zhang J, Dong L, Ma R. LncRNA ATXN8OS enhances tamoxifen resistance in breast cancer. Open Med (Wars) (2021) 16:68–80. doi: 10.1515/med-2021-0012
145. Peng WX, Koirala P, Zhou H, Jiang J, Zhang Z, Yang L, et al. Lnc-DC promotes estrogen independent growth and tamoxifen resistance in breast cancer. Cell Death Dis (2021) 12:1000. doi: 10.1038/s41419-021-04288-1
146. Zhang X, Wang M, Sun H, Zhu T, Wang X. Downregulation of LINC00894-002 contributes to tamoxifen resistance by enhancing the TGF-β Signaling pathway. Biochem (Mosc) (2018) 83:603–11. doi: 10.1134/S0006297918050139
147. Zhang CH, Wang J, Zhang LX, Lu YH, Ji TH, Xu L, et al. Shikonin reduces tamoxifen resistance through long non-coding RNA uc. 57. Oncotarget (2017) 8:88658–69. doi: 10.18632/oncotarget.20809
148. Shi YF, Lu H, Wang HB. Downregulated lncRNA ADAMTS9-AS2 in breast cancer enhances tamoxifen resistance by activating microRNA-130a-5p. Eur Rev Med Pharmacol Sci (2019) 23:1563–73. doi: 10.26355/eurrev_201902_17115
149. Burguin A, Furrer D, Ouellette G, Jacob S, Diorio C, Durocher F. Trastuzumab effects depend on HER2 phosphorylation in HER2-negative breast cancer cell lines. PloS One (2020) 15:e0234991. doi: 10.1371/journal.pone.0234991
150. Swain SM, Shastry M, Hamilton E. Targeting HER2-positive breast cancer: advances and future directions. Nat Rev Drug Discov (2023) 22:101–26. doi: 10.1038/s41573-022-00579-0
151. Han J, Qu H, Han M, Ding Y, Xie M, Hu J, et al. MSC-induced lncRNA AGAP2-AS1 promotes stemness and trastuzumab resistance through regulating CPT1 expression and fatty acid oxidation in breast cancer. Oncogene (2021) 40:833–47. doi: 10.1038/s41388-020-01574-8
152. Zhang H, Zhang XY, Kang XN, Jin LJ, Wang ZY. LncRNA-SNHG7 enhances chemotherapy resistance and cell viability of breast cancer cells by regulating miR-186. Cancer Manag Res (2020) 12:10163–72. doi: 10.2147/CMAR.S270328
153. Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget (2015) 6:11652–63. doi: 10.18632/oncotarget.3457
154. Han M, Qian X, Cao H, Wang F, Li X, Han N, et al. lncRNA ZNF649-AS1 induces trastuzumab resistance by promoting ATG5 expression and autophagy. Mol Ther (2020) 28:2488–502. doi: 10.1016/j.ymthe.2020.07.019
155. Dong H, Wang W, Mo S, Liu Q, Chen X, Chen R, et al. Long non-coding RNA SNHG14 induces trastuzumab resistance of breast cancer via regulating PABPC1 expression through H3K27 acetylation. J Cell Mol Med (2018) 22:4935–47. doi: 10.1111/jcmm.13758
156. Ma Y, Yu L, Yan W, Qiu L, Zhang J, Jia X. lncRNA GAS5 sensitizes breast cancer cells to ionizing radiation by inhibiting DNA repair. BioMed Res Int (2022) 2022:1987519. doi: 10.1155/2022/1987519
157. Shah C, Al-Hilli Z, Vicini F. Advances in breast cancer radiotherapy: implications for current and future practice. JCO Oncol Pract (2021) 17:697–706. doi: 10.1200/OP.21.00635
158. Masoudi-Khoram N, Abdolmaleki P. Role of non-coding RNAs in response of breast cancer to radiation therapy. Mol Biol Rep (2022) 49:5199–208. doi: 10.1007/s11033-022-07234-2
159. Zhang N, Zeng X, Sun C, Guo H, Wang T, Wei L, et al. LncRNA LINC00963 promotes tumorigenesis and radioresistance in breast cancer by sponging miR-324-3p and inducing ACK1 expression. Mol Ther Nucleic Acids (2019) 18:871–81. doi: 10.1016/j.omtn.2019.09.033
160. Lei C, Li S, Fan Y, Hua L, Pan Q, Li Y, et al. LncRNA DUXAP8 induces breast cancer radioresistance by modulating the PI3K/AKT/mTOR pathway and the EZH2-E-cadherin/RHOB pathway. Cancer Biol Ther (2022) 23:1–13. doi: 10.1080/15384047.2022.2132008
161. Zhang S, Wang B, Xiao H, Dong J, Li Y, Zhu C, et al. LncRNA HOTAIR enhances breast cancer radioresistance through facilitating HSPA1A expression via sequestering miR-449b-5p. Thorac Cancer (2020) 11:1801–16. doi: 10.1111/1759-7714.13450
162. Zhou Y, Wang C, Liu X, Wu C, Yin H. Long non-coding RNA HOTAIR enhances radioresistance in MDA-MB231 breast cancer cells. Oncol Lett (2017) 13:1143–8. doi: 10.3892/ol.2017.5587
163. Li J, Lei C, Chen B, Zhu Q. LncRNA FGD5-AS1 facilitates the radioresistance of breast cancer cells by enhancing MACC1 expression through competitively sponging miR-497-5p. Front Oncol (2021) 11:671853. doi: 10.3389/fonc.2021.671853
164. Bi Z, Li Q, Dinglin X, Xu Y, You K, Hong H, et al. Nanoparticles (NPs)-meditated lncRNA AFAP1-AS1 silencing to block wnt/β-catenin signaling pathway for synergistic reversal of radioresistance and effective cancer radiotherapy. Adv Sci (Weinh) (2020) 7:2000915. doi: 10.1002/advs.202000915
165. Wang Y, Liu M, Liu X, Guo X. LINC00963-FOSB-mediated transcription activation of UBE3C enhances radioresistance of breast cancer cells by inducing ubiquitination-dependent protein degradation of TP73. J Transl Med (2023) 21:321. doi: 10.1186/s12967-023-04153-z
166. Tarighati E, Keivan H, Mahani H. A review of prognostic and predictive biomarkers in breast cancer. Clin Exp Med (2023) 23:1–16. doi: 10.1007/s10238-021-00781-1
167. Hosseinalizadeh H, Mahmoodpour M, Ebrahimi A. Circulating non-coding RNAs as a diagnostic and management biomarker for breast cancer: current insights. Mol Biol Rep (2022) 49:705–15. doi: 10.1007/s11033-021-06847-3
168. Badowski C, He B, Garmire LX. Blood-derived lncRNAs as biomarkers for cancer diagnosis: the Good, the Bad and the Beauty. NPJ Precis Oncol (2022) 6:40. doi: 10.1038/s41698-022-00283-7
169. Jiao ZY, Tian Q, Li N, Wang HB, Li KZ. Plasma long non-coding RNAs (lncRNAs) serve as potential biomarkers for predicting breast cancer. Eur Rev Med Pharmacol Sci (2018) 22:1994–9. doi: 10.26355/eurrev_201804_14727
170. Zhang Y, Zhang K, Luo Z, Liu L, Wu L, Liu J. Circulating long non-coding HOX transcript antisense intergenic ribonucleic acid in plasma as a potential biomarker for diagnosis of breast cancer. Thorac Cancer (2016) 7:627–32. doi: 10.1111/1759-7714.12373
171. Ezzatizadeh V, Mozdarani H. Does HOTAIR expression level in the peripheral blood have veritably predictive/prognostic impact on breast cancer patients? J Transl Med (2019) 17:404. doi: 10.1186/s12967-019-02158-1
172. Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol (2019) 21:498–510. doi: 10.1038/s41556-019-0299-0
173. Hu H, Hu J, Yang Y, Zhou W, Ye C. Assessment of circulating HISLA as a potential biomarker for breast cancer diagnosis and prognosis. Clin Exp Med (2021) 21:29–34. doi: 10.1007/s10238-020-00670-z
174. Zhang K, Luo Z, Zhang Y, Zhang L, Wu L, Liu L, et al. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer biomark (2016) 17:187–94. doi: 10.3233/CBM-160630
175. El-Ashmawy NE, Hussien FZ, El-Feky OA, Hamouda SM, Al-Ashmawy GM. Serum LncRNA-ATB and FAM83H-AS1 as diagnostic/prognostic non-invasive biomarkers for breast cancer. Life Sci (2020) 259:118193. doi: 10.1016/j.lfs.2020.118193
176. Pourramezan Z, Attar FA, Yusefpour M, Azizi M, Oloomi M. Circulating LncRNAs landscape as potential biomarkers in breast cancer. Cancer Rep (Hoboken) (2023) 6:e1722. doi: 10.1002/cnr2.1722
177. Han L, Ma P, Liu SM, Zhou X. Circulating long noncoding RNA GAS5 as a potential biomarker in breast cancer for assessing the surgical effects. Tumour Biol (2016) 37:6847–54. doi: 10.1007/s13277-015-4568-7
178. Zhong G, Wang K, Li J, Xiao S, Wei W, Liu J. Determination of serum exosomal H19 as a noninvasive biomarker for breast cancer diagnosis. Onco Targets Ther (2020) 13:2563–71. doi: 10.2147/OTT.S243601
179. Zhao T, Wu L, Li X, Dai H, Zhang Z. Large intergenic non-coding RNA-ROR as a potential biomarker for the diagnosis and dynamic monitoring of breast cancer. Cancer biomark (2017) 20:165–73. doi: 10.3233/CBM-170064
180. Wang X, Li S, Xiao H, Deng X. Serum lncRNA TINCR serve as a novel biomarker for predicting the prognosis in triple-negative breast cancer. Technol Cancer Res Treat (2020) 19:1533033820965574. doi: 10.1177/1533033820965574
181. Shi W, Jin X, Wang Y, Zhang Q, Yang L. High serum exosomal long non-coding RNA DANCR expression confers poor prognosis in patients with breast cancer. J Clin Lab Anal (2022) 36:e24186. doi: 10.1002/jcla.24186
182. Maharati A, Moghbeli M. Long non-coding RNAs as the critical regulators of PI3K/AKT, TGF-β, and MAPK signaling pathways during breast tumor progression. J Transl Med (2023) 21:556. doi: 10.1186/s12967-023-04434-7
183. Crooke ST, Baker BF, Crooke RM, Liang XH. Antisense technology: an overview and prospectus. Nat Rev Drug Discov (2021) 20:427–53. doi: 10.1038/s41573-021-00162-z
184. Wang W, Fang F, Ozes A, Nephew KP. Targeting ovarian cancer stem cells by dual inhibition of HOTAIR and DNA methylation. Mol Cancer Ther (2021) 20:1092–101. doi: 10.1158/1535-7163.MCT-20-0826
185. Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev (2016) 30:34–51. doi: 10.1101/gad.270959.115
186. Abdel-Latif M, Riad A, Soliman RA, Elkhouly AM, Nafae H, Gad MZ, et al. MALAT-1/p53/miR-155/miR-146a ceRNA circuit tuned by methoxylated quercitin glycoside alters immunogenic and oncogenic profiles of breast cancer. Mol Cell Biochem (2022) 477:1281–93. doi: 10.1007/s11010-022-04378-4
187. Cai J, Sun H, Zheng B, Xie M, Xu C, Zhang G, et al. Curcumin attenuates lncRNA H19−induced epithelial−mesenchymal transition in tamoxifen−resistant breast cancer cells. Mol Med Rep (2021) 23(1):13. doi: 10.3892/mmr.2020.11651
188. Stecklein SR, Jensen RA, Pal A. Genetic and epigenetic signatures of breast cancer subtypes. Front Biosci (Elite Ed) (2012) 4:934–49. doi: 10.2741/E431
189. Lee A, Moon BI, Kim TH. BRCA1/BRCA2 pathogenic variant breast cancer: treatment and prevention strategies. Ann Lab Med (2020) 40:114–21. doi: 10.3343/alm.2020.40.2.114
190. Wang G, Ren Z, Zhao Y, Li Y. A nine-gene signature as prognostic biomarker in gastric cancer by bioinformatics analysis. Clin Transl Oncol (2023) 25(11):3296–306. doi: 10.1007/s12094-023-03180-y
191. Garcia-Martinez L, Zhang Y, Nakata Y, Chan HL, Morey L. Epigenetic mechanisms in breast cancer therapy and resistance. Nat Commun (2021) 12:1786. doi: 10.1038/s41467-021-22024-3
192. Wang L, Zhan G, Maimaitiyiming Y, Su Y, Lin S, Liu J, et al. m(6)A modification confers thermal vulnerability to HPV E7 oncotranscripts via reverse regulation of its reader protein IGF2BP1 upon heat stress. Cell Rep (2022) 41:111546. doi: 10.1016/j.celrep.2022.111546
193. Lv W, Wang Y, Zhao C, Tan Y, Xiong M, Yi Y, et al. Identification and Validation of m6A-Related lncRNA Signature as Potential Predictive Biomarkers in Breast Cancer. Front Oncol (2021) 11:745719. doi: 10.3389/fonc.2021.745719
194. Shi W, Tang Y, Lu J, Zhuang Y, Wang J. MIR210HG promotes breast cancer progression by IGF2BP1 mediated m6A modification. Cell Biosci (2022) 12:38. doi: 10.1186/s13578-022-00772-z
195. Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W, et al. LNC942 promoting METTL14-mediated m(6)A methylation in breast cancer cell proliferation and progression. Oncogene (2020) 39:5358–72. doi: 10.1038/s41388-020-1338-9
196. Huang JZ, Chen M, Chen D, Gao XC, Zhu S, Huang H, et al. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell (2017) 68:171–84.e6. doi: 10.1016/j.molcel.2017.09.015
197. Wang Y, Wu S, Zhu X, Zhang L, Deng J, Li F, et al. LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J Exp Med (2020) 217(3):jem. doi: 10.1084/jem.20190950
Keywords: breast cancer, metastasis, therapy resistance, long non-coding RNA (LncRNA), competitive endogenous RNA (ceRNA), liquid biopsy
Citation: Wang Y, Bu N, Luan X-f, Song Q-q, Ma B-F, Hao W, Yan J-j, Wang L, Zheng X-l and Maimaitiyiming Y (2024) Harnessing the potential of long non-coding RNAs in breast cancer: from etiology to treatment resistance and clinical applications. Front. Oncol. 14:1337579. doi: 10.3389/fonc.2024.1337579
Received: 13 November 2023; Accepted: 19 February 2024;
Published: 05 March 2024.
Edited by:
Maria Rosaria De Miglio, University of Sassari, ItalyReviewed by:
Mihir Khambete, Yale University, United StatesQihang Yuan, Dalian Medical University, China
Copyright © 2024 Wang, Bu, Luan, Song, Ma, Hao, Yan, Wang, Zheng and Maimaitiyiming. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasen Maimaitiyiming, eWFzaW5qYW5Aemp1LmVkdS5jbg==; eXNqbUB4am11LmVkdS5jbg==
†These authors have contributed equally to this work
 Yun Wang1†
Yun Wang1† Na Bu
Na Bu Jing-jing Yan
Jing-jing Yan Li Wang
Li Wang Xiao-ling Zheng
Xiao-ling Zheng Yasen Maimaitiyiming
Yasen Maimaitiyiming