- State Key Laboratory of Reproductive Regulation and Breeding of Grassland Livestock, School of Life Sciences, Inner Mongolia University, Hohhot, Inner Mongolia, China
Background: Bladder cancer stands as the predominant malignant tumor in the urological system, presenting a significant challenge to public health and garnering extensive attention. Recently, with the deepening research into tumor molecular mechanisms, non-coding RNAs (ncRNAs) have emerged as potential biomarkers offering guidance for the diagnosis and prognosis of bladder cancer. However, the definitive role of ncRNAs in bladder cancer remains unclear. Hence, this study aims to elucidate the relevance and significance of ncRNAs through a Meta-analysis.
Methods: A systematic meta-analysis was executed, including studies evaluating the diagnostic performance of ncRNAs and their associations with overall survival (OS) and disease-free survival (DFS). Key metrics such as hazard ratios, sensitivity, specificity, and diagnostic odds ratios were extracted and pooled from these studies. Potential publication bias was assessed using Deeks’ funnel plot, and the robustness of the results was ascertained through a sensitivity analysis.
Results: Elevated ncRNA expression showed a positive correlation with improved OS, evidenced by a hazard ratio (HR) of 0.82 (95% CI: 0.66-0.96, P<0.001). Similarly, a significant association was observed between heightened ncRNA expression and DFS, with an HR of 0.86 (95% CI: 0.73-0.99, P<0.001). Diagnostic performance analysis across 17 articles yielded a pooled sensitivity of 0.76 and a specificity of 0.83. The diagnostic odds ratio was recorded at 2.71, with the area under the ROC curve (AUC) standing at 0.85.
Conclusion: Exosome ncRNAs appear to possess potential significance in the diagnostic and prognostic discussions of bladder cancer. Their relationship with survival outcomes and diagnostic measures suggests a possible clinical utility. Comprehensive investigations are needed to fully determine their role in the ever-evolving landscape of bladder cancer management, especially within the framework of personalized medicine.
1 Introduction
Bladder cancer (BCa), a leading urological malignancy, is characterized by its escalating global incidence and mortality (1–3). Despite advancements in diagnostic and therapeutic interventions, the 5-year overall survival rate for BCa remains suboptimal, primarily due to its predilection for swift recurrence and aggressive metastasis (4, 5). In this evolving landscape, the emergence of molecular biomarkers, especially exosome non-coding RNAs (ncRNAs), offers a promising frontier. Such markers are poised to revolutionize patient management by facilitating treatments tailored to individual molecular signatures, thereby enhancing therapeutic efficacy and survival outcomes (6–8). As we delve deeper into the molecular intricacies of BCa, the imperative to harness these biomarkers becomes paramount, heralding an era dominated by personalized medicine with promising prognostic implications.
Recent molecular biology breakthroughs have underscored the crucial involvement of ncRNAs in diverse cellular pathways, accentuating their role in tumorigenesis and the intricate cascade of cancer progression (9, 10). The vast realm of ncRNAs, which includes microRNAs (miRNAs), long non-coding RNAs (Lnc RNAs), and circular RNAs (circ RNAs), has become a nexus of intense research scrutiny (11–13). However, it’s their association with exosomes, nanoscale extracellular vesicles vital for intercellular communication, that is at the forefront of ongoing research (8, 14, 15). Unraveling the complexities of ncRNAs encapsulated in exosomes can potentially provide unprecedented insights into cancer biology, heralding novel therapeutic and diagnostic paradigms.
A growing corpus of literature has emphatically underscored ncRNAs as paramount molecular sentinels in the BCa diagnostic and prognostic arena (6, 16, 17). he differential expression patterns of ncRNAs in oncogenic pathways earmark them as potent biomarkers (18). Yet, there’s a palpable discord concerning the conclusive prognostic efficacy of exosome ncRNAs (6–8, 14–17, 19–21). Such variances stem from disparate study designs, diverse methodologies, sample sizes, and demographic intricacies, thereby muddying the interpretative waters.
This study aims to systematically investigate the potential prognostic value of exosome ncRNA in bladder cancer patients by conducting a comprehensive meta-analysis that integrates the results of multiple studies. Meta-analysis, through the aggregation of a substantial volume of data, serves to identify and strengthen the consistent associations between exosome ncRNA and bladder cancer prognosis, which may not have been fully revealed by individual studies. Furthermore, by addressing the heterogeneity and methodological differences present in existing research, this study seeks to provide more comprehensive and reliable evidence to support the potential application of exosome ncRNA in the assessment of bladder cancer prognosis.
2 Materials and methods
2.1 Search strategies
A computerized search was conducted in the databases of PubMed, Web of Science, EMBASE, and Cochrane Library for literature related to ncRNA and bladder cancer. The search strategy combined both Mesh terms and free-text words. The principal keywords employed were “Bladder Cancer”, “Non-coding RNA”, “Prognosis”, and “Survival Rate”. To refine the search results, only English language articles were considered, and the search date was updated to September 1, 2023. This search strategy has been registered with the PROSPERO (https://www.crd.york.ac.uk/prospero/) under the registration number CRD42023454417.
2.2 Inclusion and exclusion criteria
Inclusion criteria consisted of: (1) Studies focusing on populations diagnosed with bladder cancer; (2) Research encompassing exosome non-coding RNA; (3) Studies presenting outcomes on diagnostic accuracy and prognostic indices, such as Overall Survival (OS) or Disease-Free Survival (DFS).
Exclusion criteria were: (1) Commentaries, symposium abstracts, case narrations, and expert reviews; (2) Duplicate studies; (3) Studies missing a control group (non-BCa); (4) Studies with significant data absence or unextractable data.
2.3 Literature screening and data extraction
Upon completing the literature search, two researchers carried out systematic literature reviews and data collation to ensure accuracy and consistency. An initial screening was conducted based on article titles and abstracts to exclude manuscripts that were clearly inconsistent with the inclusion criteria or met the exclusion criteria. Subsequently, a thorough reading and assessment of the preliminarily selected literature were performed. Key data were then extracted from the finalized studies, which included research type, sample size, diagnostic precision, and specificity of ncRNAs, as well as prognostic indicators such as OS and DFS Hazard Ratios (HR) along with their respective 95% confidence intervals (CI). In cases of discrepancies or disputes during the literature screening and data extraction processes, resolutions were sought through consultations with a third author.
2.4 Quality assessment
In an evaluation of the incorporated literature, we leveraged the Newcastle-Ottawa Scale (NOS) as our metric of discernment (22). The NOS systematically scrutinizes studies across three pivotal axes: selection, comparability and outcomes. Pp
Studies can accrue a zenithal score of 9 points, with elevated scores emblematic of paramount research caliber. To obviate subjective predilections and bolster the assessment’s rigor, this critical examination was helmed by two discrete authors.
2.5 Statistical analysis
Statistical analyses within this investigation were executed using the Stata 13 software. Initially, pertinent metrics, including sample sizes, counts of negatives, and positives, were extracted from each encompassed study, facilitating the computation of true positives (TP), false positives (FP), true negatives (TN) and false negatives (FP). Armed with this data, we deduced the sensitivity, specificity, positive predictive value, negative predictive value, and the diagnostic odds ratio (DOR). To gauge the cumulative effect magnitude of extracellular non-coding RNAs on bladder cancer prognosis, we amalgamated the HRs and their 95% CIs for OS and DFS from the assimilated studies. The I² statistic was harnessed to probe heterogeneity across the studies, with an I² exceeding 50% signaling pronounced heterogeneity. In the presence of marked heterogeneity, a random-effects model was adopted for meta-analysis; contrarily, a fixed-effects model was favored. Potential publication bias was discerned via funnel plot inspection. All inferential tests were bifurcated, with P<0.05 demarcating statistical significance.
3 Results
3.1 Literature search and characteristics
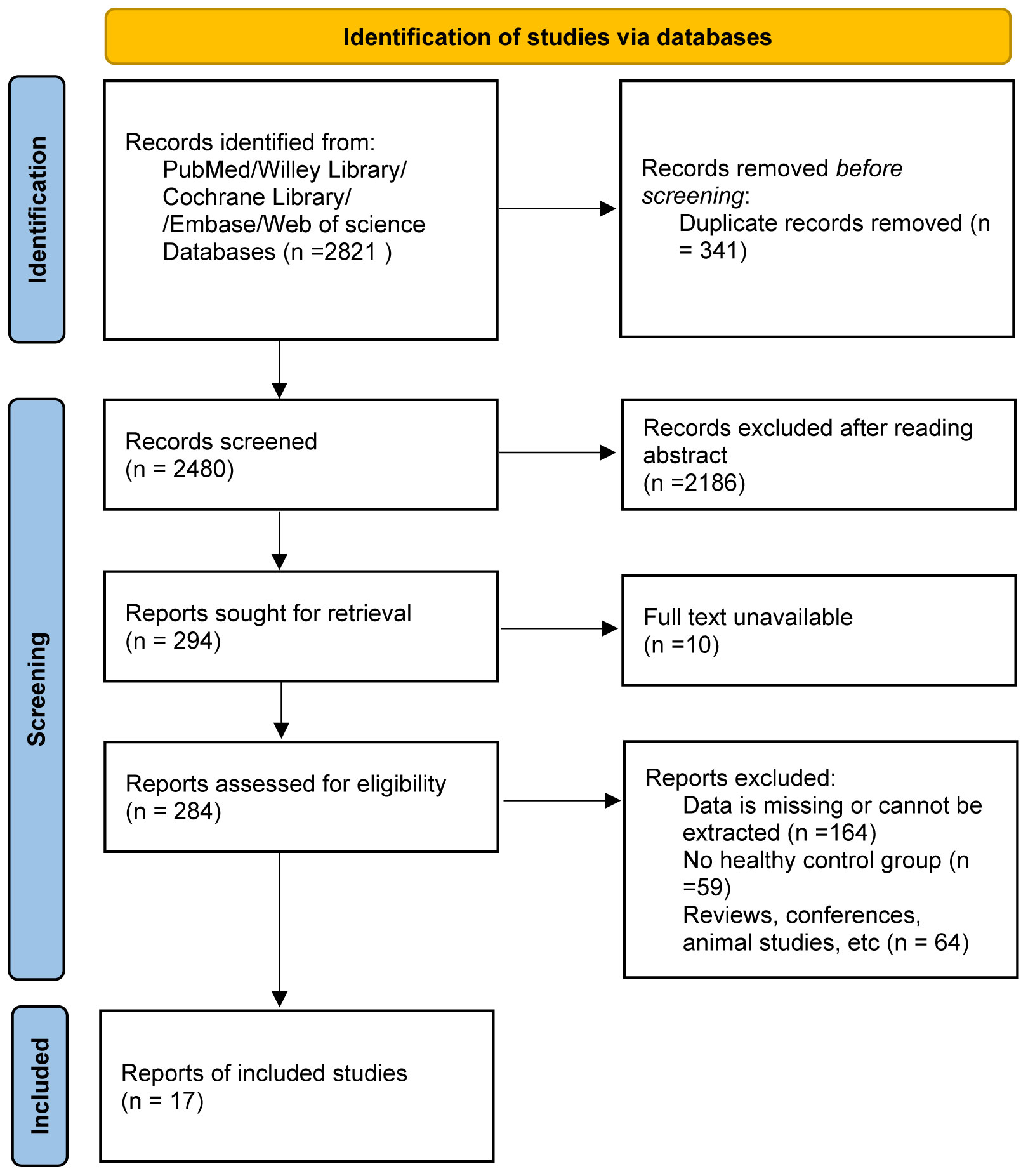
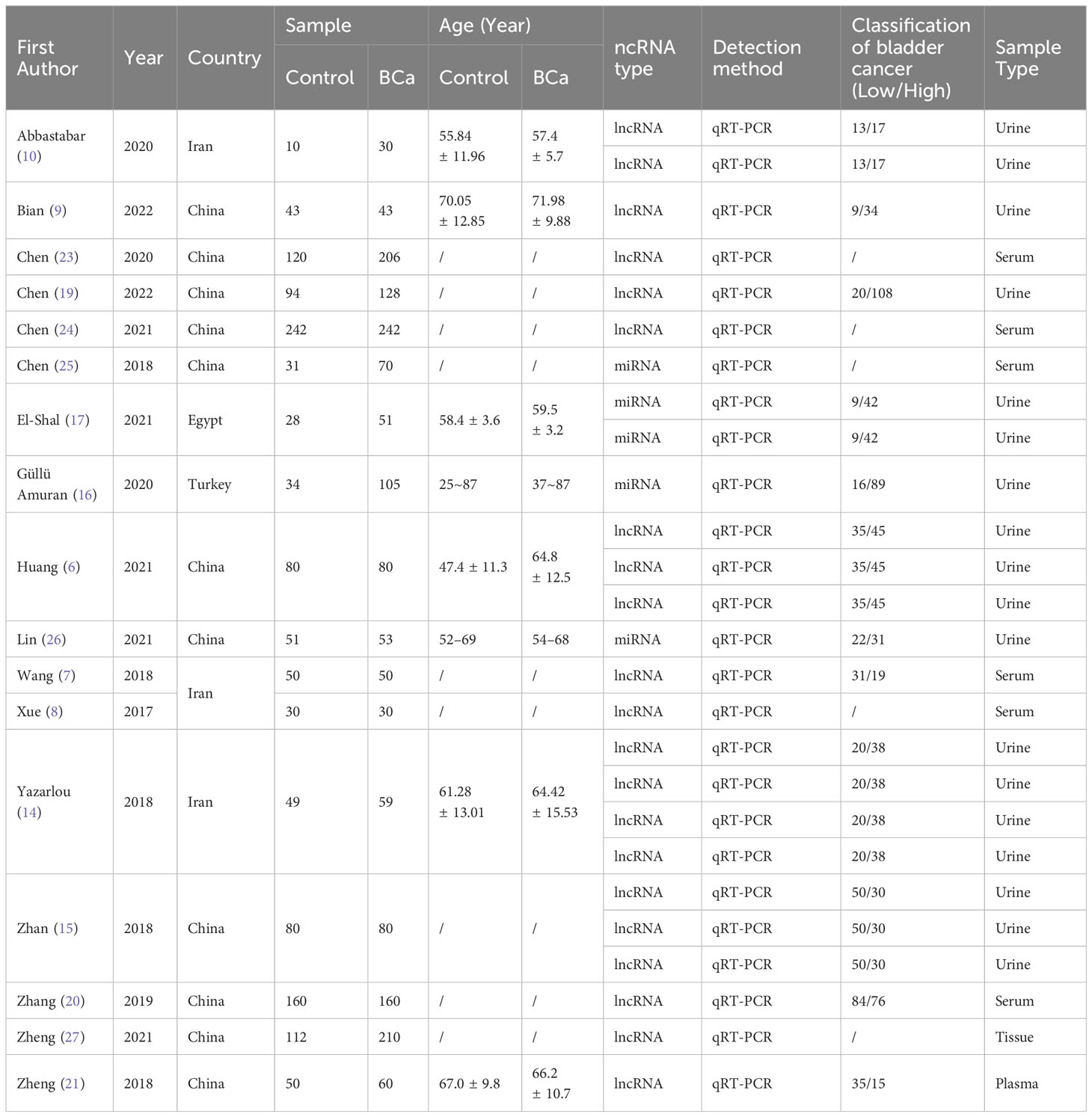
In our search process, we identified 2,821 pertinent publications. Figure 1 provides a visual representation detailing the selection procedure and the rationale for exclusions. Of these, 284 studies were deemed suitable for an in-depth full-text review. Among them, 17 studies rigorously adhered to all inclusion criteria and were consequently selected for further analysis. The characteristics and specific attributes of these incorporated articles are comprehensively delineated in Table 1 and further expounded upon in Supplementary Tables S1, S2. The quality assessment of all the included literatures met the standards set for this research, as demonstrated in Supplementary Table S3.
3.2 Association between ncRNAs expression and prognosis
3.2.1 ncRNAs expression and OS
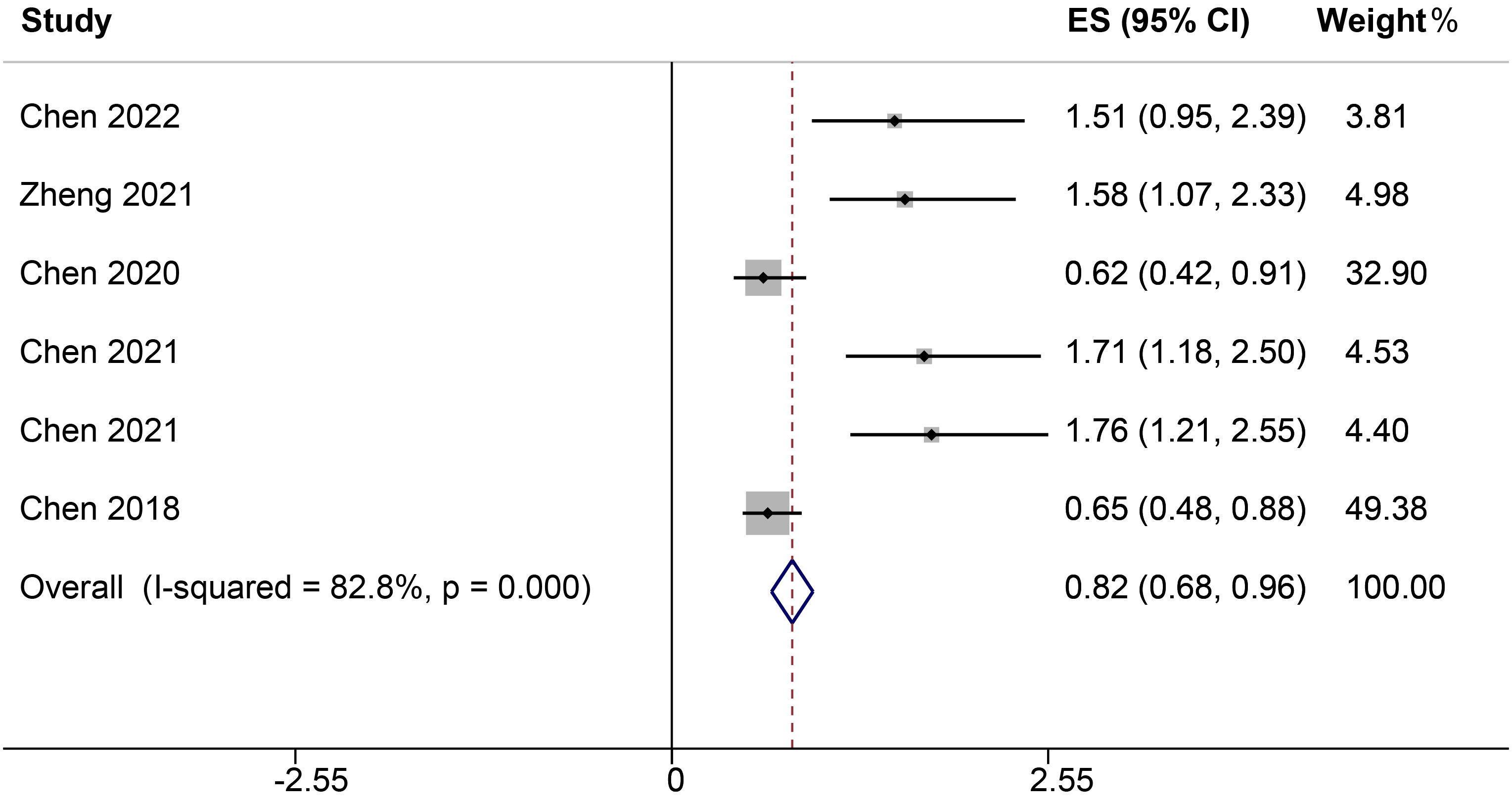
Four studies, encompassing six different ncRNAs, reported on the relationship between low and high ncRNA expression levels and OS. Heterogeneity analysis revealed significant variability among the included publications, with an I² value of 82.8%. Due to this pronounced heterogeneity, a random-effects model was utilized for the analysis. The results indicated a significant correlation between elevated ncRNA expression and OS, with a HR of 0.82 (95% CI: 0.66-0.96, P<0.001, Figure 2).
3.3.2 ncRNAs expression and RFS
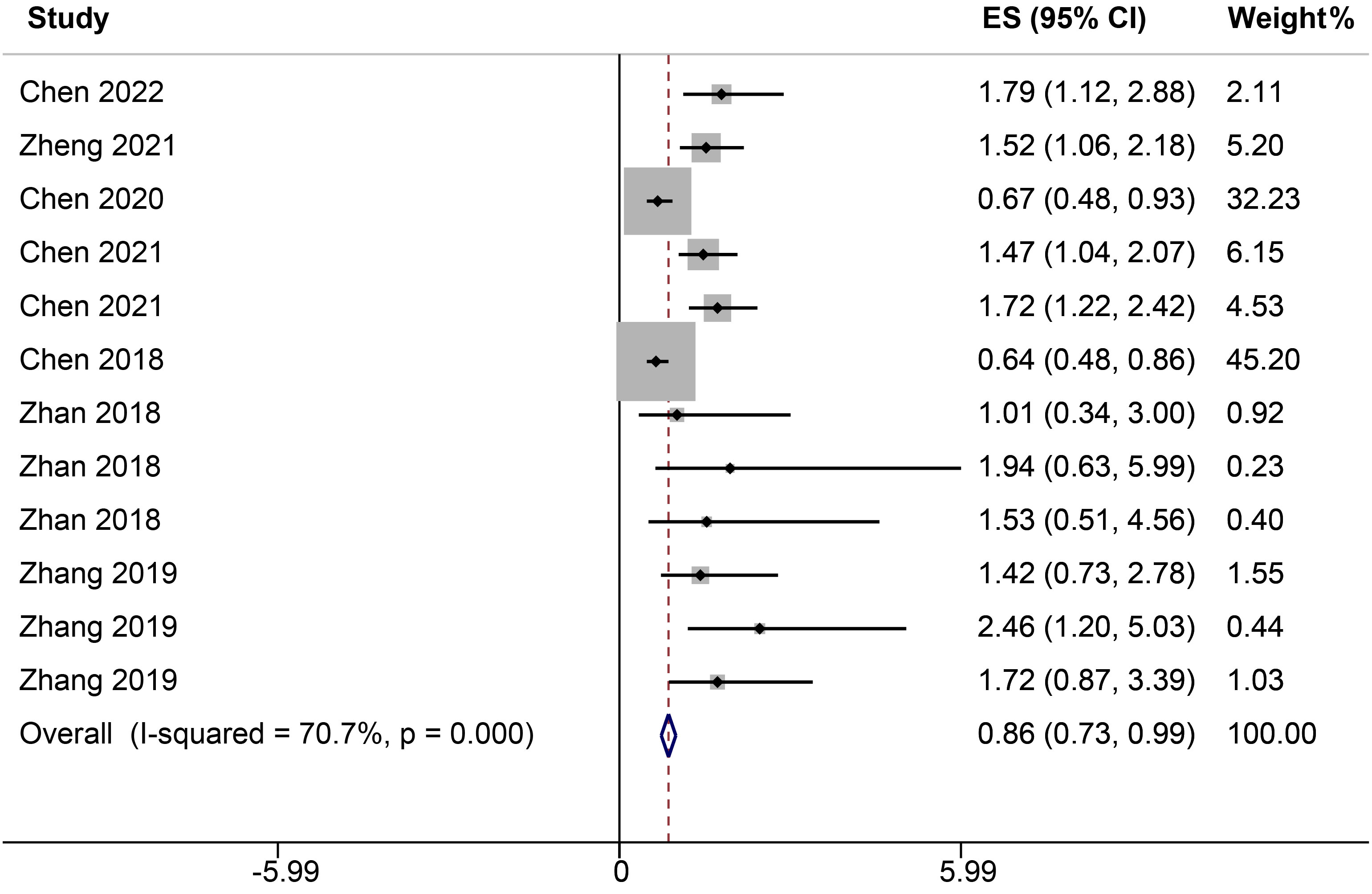
Seven studies, encompassing twelve distinct ncRNAs, delineated the association between varying ncRNA expression levels (low versus high expression) and DFS. A heterogeneity analysis highlighted substantial variability among the selected studies, evidenced by an I² value of 70.7%. Given this pronounced heterogeneity, the analysis was conducted using a random-effects model. The results reveal a significant correlation between elevated ncRNA expression and DFS, with a HR of 0.86 (95% CI: 0.73-0.99, P<0.001, Figure 3).
3.4 Meta-analysis of diagnostic ncRNA
3.4.1 Diagnostic performance analysis of ncRNA
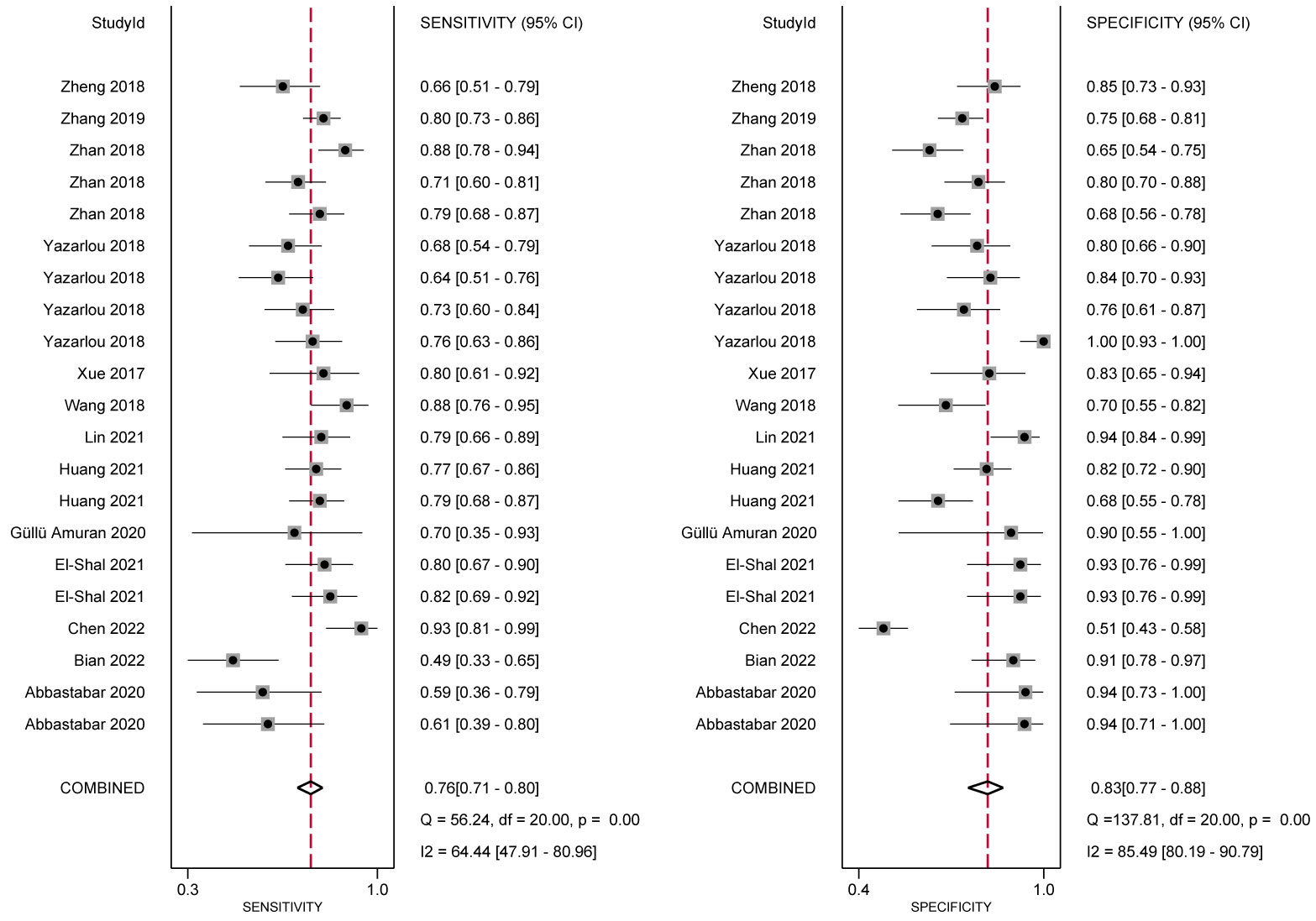
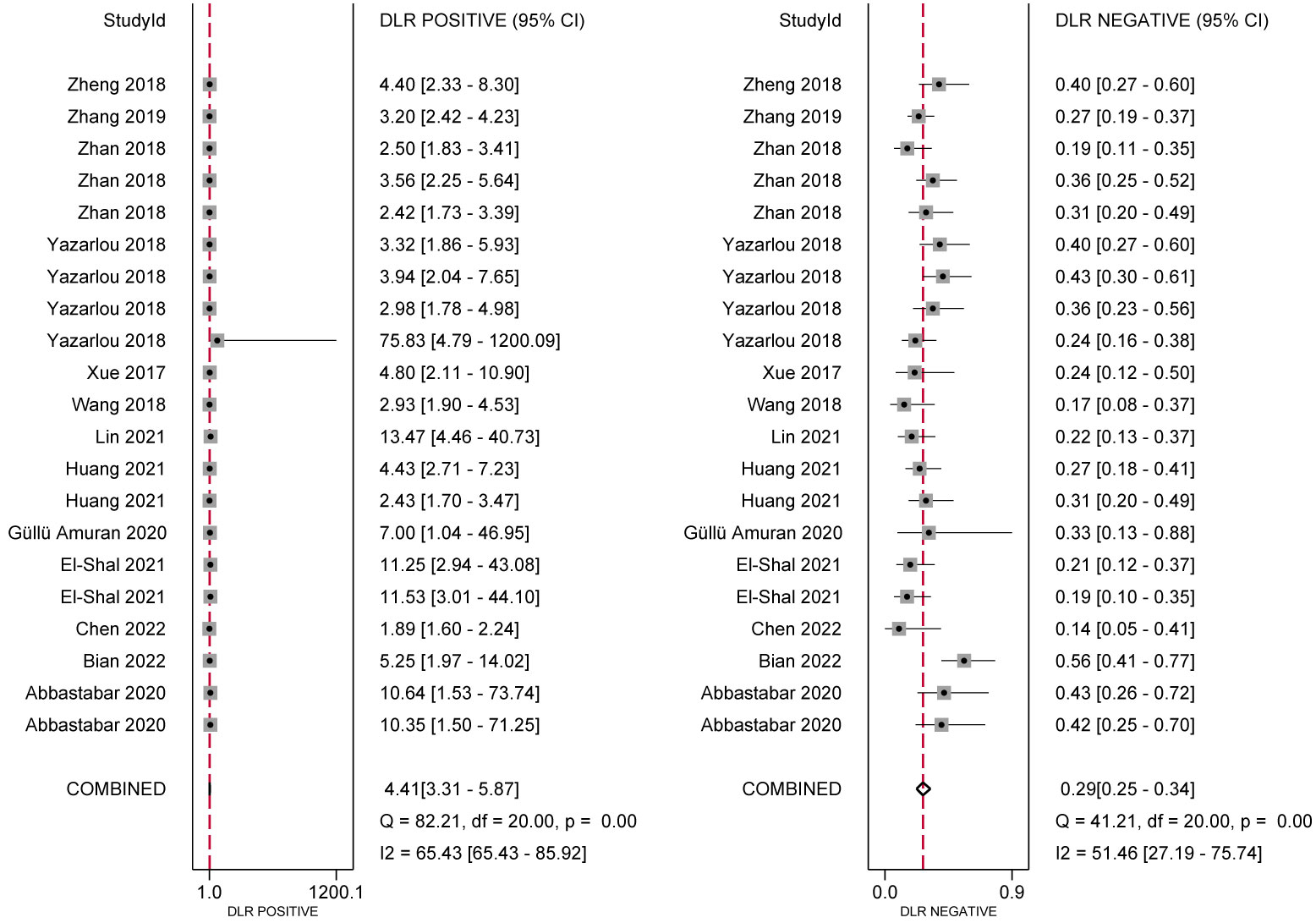
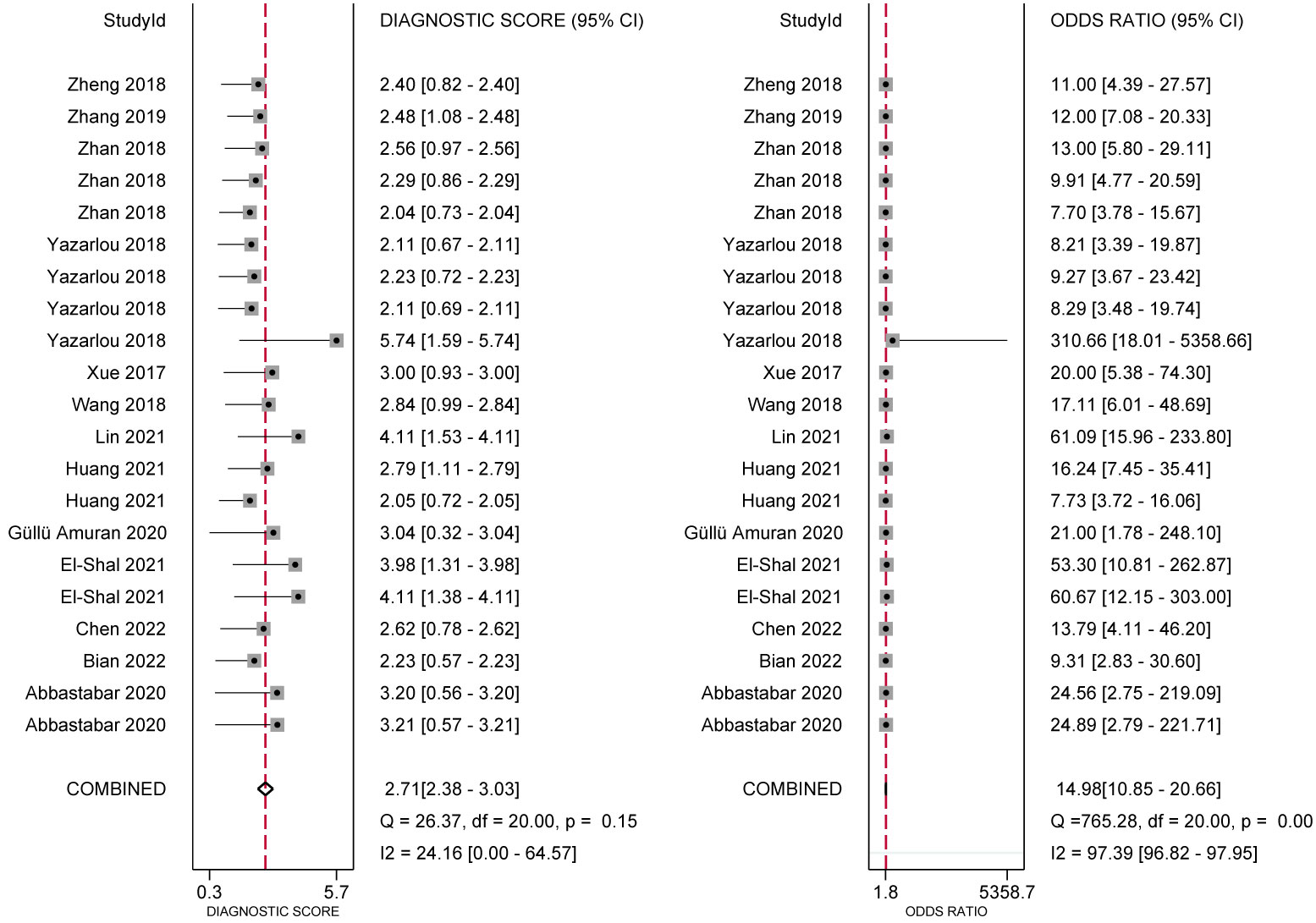
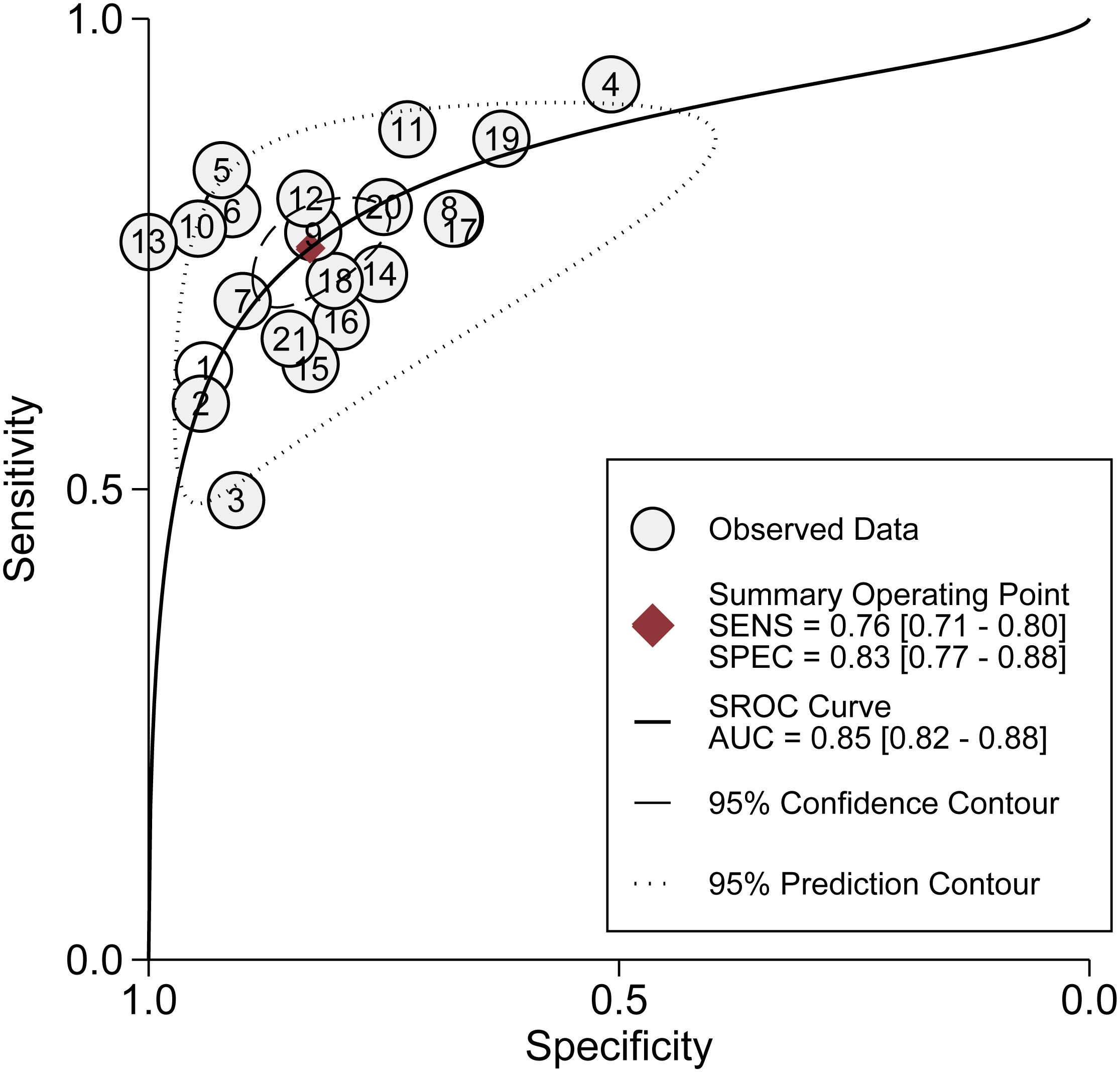
In the analysis of ncRNA diagnostic performance, a total of 17 articles involving 22 different ncRNAs were included. The meta-analysis yielded a pooled sensitivity of 0.76 (95% CI: 0.71–0.80; I²=64.4%) and specificity of 0.83 (95% CI: 0.77–0.88; I² =85.49%). The positive likelihood ratio (PLR) was 4.41 (95% CI: 3.31–5.87; I² =65.43%), while the negative likelihood ratio (NLR) was 0.29 (95% CI: 0.25–0.34; I²=54.46%). The diagnostic odds ratio was 2.71 (95% CI: 2.38–3.03; I²=24.16%), and the diagnostic score was 14.89 (95% CI: 10.85–20.66; I²=97.39%). Further confirmation of the high accuracy of ncRNA in bladder cancer diagnosis was obtained by drawing receiver operator characteristic (ROC) curves and calculating the area under the curve (AUC). The AUC was 0.85 (95% CI: 0.82–0.88), as shown in Figures 4–7.
3.4.2 Prior probability and posterior probability
Using Fagan’s nomogram, we evaluated the influence of test outcomes on the posttest probability of diagnosis, based on a predefined pretest probability. The data underscores that ncRNA detection holds considerable diagnostic significance, substantially elevating the posttest probability for bladder cancer recognition, even starting from a relatively low pretest probability (Supplementary Figure S1). The scatter plot delineates a positive correlation between the primary variables (Supplementary Figure S2).
3.5 Publication bias
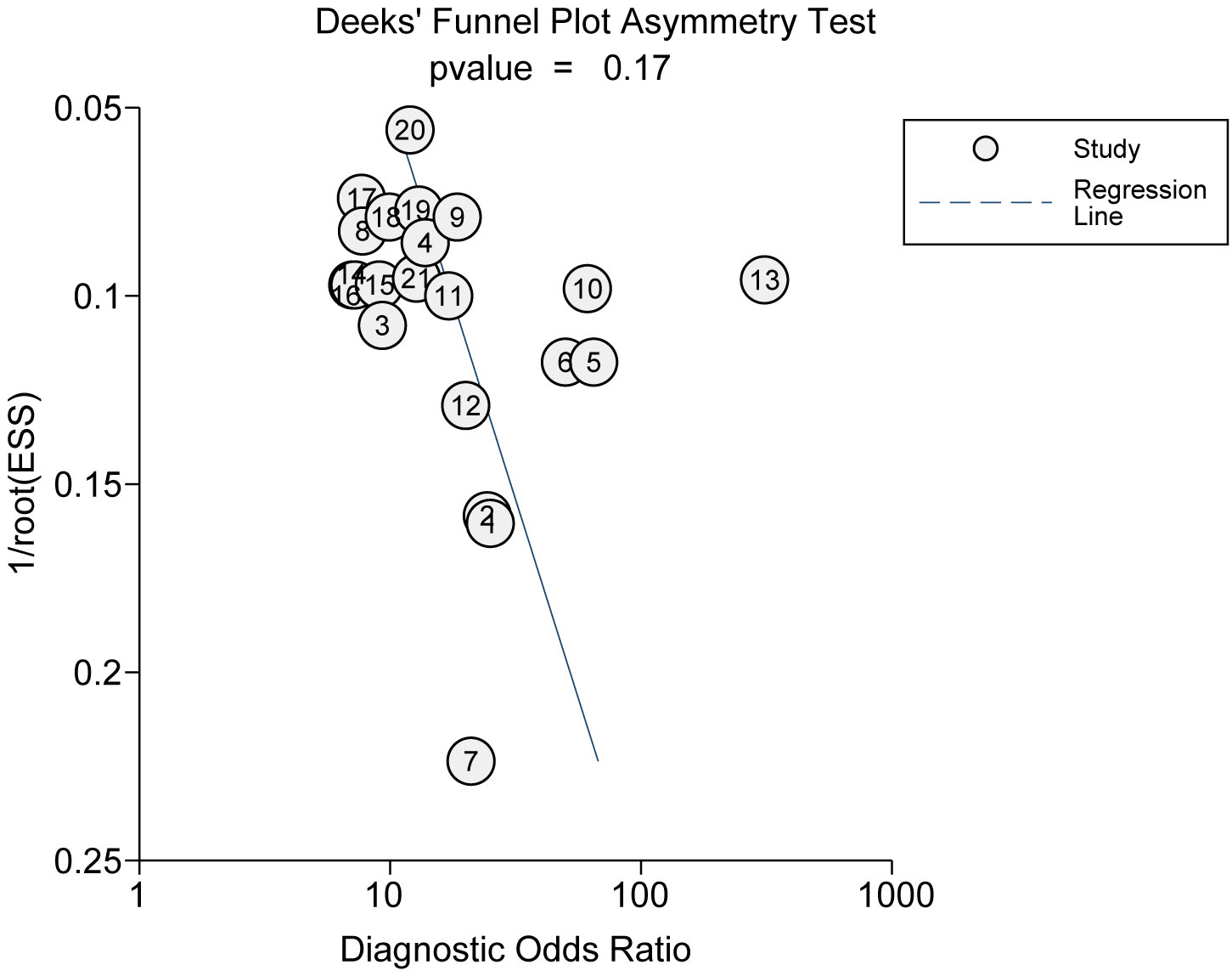
Employing Deeks’ funnel plot, we undertook an assessment of potential publication bias among the studies incorporated. The nearly horizontal slope of the funnel plot, coupled with an associated p-value of 0.33, robustly suggests an absence of notable publication bias (Figure 8).
3.6 Sensitivity analysis
To ensure the robustness of our meta-analysis results, we employed a sensitivity analysis using the one-by-one omission method. The results demonstrated good stability after sequential removal (Supplementary Tables S2, S3).
4 Discussion
The molecular intricacies inherent to oncogenesis consistently highlight the profound influence of non-coding elements in cellular signaling and function (28, 29). Among these, exosome ncRNAs have surfaced as central orchestrators, coordinating myriad processes linked with tumor initiation, progression, and metastasis (24, 30). Among these, exosome ncRNAs have surfaced as central orchestrators, coordinating myriad processes linked with tumor initiation, progression, and metastasis (31, 32). Against this intricate backdrop, the present meta-analysis seeks to provide an exhaustive evaluation of the prognostic significance of exosome ncRNAs within bladder oncology. With meticulous amalgamation and interpretation, we delineate potential clinical trajectories these molecular agents might trace, suggesting a paradigm shift in diagnostic accuracy and therapeutic interventions for bladder cancer care.
4.1 Decoding the prognostic implications of ncRNA expression
The synthesis of various studies accentuates a pronounced correlation between ncRNA expression and survival rates in bladder cancer. This association, mirrored by consistency in DFS outcomes, underscores the cardinal role ncRNAs can play in the clinical evolution of bladder cancer (19, 23–25). This association, reiterated by the congruence in DFS outcomes, emphasizes the pivotal role ncRNAs might play in bladder cancer’s clinical course (15, 20, 25). However, the manifest heterogeneity across the included studies mirrors the multifaceted nature of bladder malignancies. Such heterogeneity may be attributed to a range of factors, including the genetic makeup of patient cohorts, tumor attributes, and methodological differences in ncRNA quantification. Addressing these disparities in subsequent research is paramount for a more integrated comprehension.
Recent years, exosomes have emerged as a non-invasive biomarker with immense potential in the early diagnosis and monitoring of bladder cancer. Exosomes are small vesicles released by cells into bodily fluids, capable of carrying a variety of molecules including ncRNAs, thus reflecting the biological state of their source cells (7, 33, 34). The use of exosomes for diagnosing bladder cancer presents distinct advantages, including their presence in various bodily fluids which facilitates easy and non-invasive sample collection, and the high stability of molecules within exosomes, contributing to the accuracy and reliability of detection (15, 20). However, this method also faces challenges, including the need for further optimization of exosome isolation and purification techniques to enhance efficiency and purity. Additionally, the current understanding of the relationship between specific ncRNAs within exosomes and the development of bladder cancer remains limited, necessitating more foundational and clinical research to deepen our knowledge.
4.2 Diagnostic potential: ncRNA’s forefront role
The evolving diagnostic landscape of bladder cancer has increasingly gravitated towards the burgeoning potential of ncRNAs as primary molecular markers (8, 14, 35). Propelled by advancements in molecular biology and oncogenomic, this shift is anchored in the substantial roles ncRNAs assume at the cellular stratum (36). Functioning beyond mere transcriptional regulators, ncRNAs have been identified to modulate diverse signaling pathways, impact gene expression, and partake in DNA repair mechanisms, highlighting their multifarious role in tumorigenesis (37–39).
Our study, through the assessment of ncRNA diagnostic accuracy, highlighted its significance in the early detection of bladder cancer, where a notable area under the curve (AUC) indicates high sensitivity and specificity. Additionally, through metrics such as positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio, we have reinforced the rigor and reliability of these findings. Compared to other diagnostic methods in bladder cancer management, ncRNA-based detection techniques demonstrate significant advantages. Traditional bladder cancer diagnostic methods, such as urine cytology and cystoscopy, despite their widespread clinical use, have certain limitations (40); urine cytology shows lower sensitivity for low-grade tumors, while cystoscopy, an invasive procedure, may cause discomfort to patients. In contrast, ncRNA-based detection methods, due to their non-invasiveness, high sensitivity, and specificity, show greater potential in early diagnosis and disease monitoring. Nonetheless, the practical application of these emerging technologies still faces challenges, including cost-effectiveness, technical standardization, and validation. As we continually unravel the intricate mechanisms via which ncRNAs function, it becomes conceivable to envisage a horizon where these entities transition from research confines to pervasive clinical applications, augmenting diagnostic precision and guiding tailored therapeutic approaches. Within the continuum of bladder cancer research, pivoting towards an understanding and leveraging of ncRNAs could well represent the next vanguard, melding molecular insights with tangible clinical implications.
4.3 Limitations and future directions
While this investigation offers pivotal insights, certain limitations necessitate acknowledgment. Firstly, the relatively limited number of studies included, especially those with significantly low sample sizes, could restrict the broad applicability and external validity of our conclusions. Moreover, inherent to any meta-analysis, is the latent risk of publication bias, although our evaluations suggest its minimal presence. Concerning the sources of heterogeneity, the biological heterogeneity of bladder cancer itself cannot be overlooked. Tumors in different patients may exhibit substantial differences in molecular characteristics, genetic backgrounds, and responses to treatment, which could be reflected in the variability of ncRNA expression patterns. Additionally, the differences in the sensitivity, specificity, and quantification capabilities of ncRNA detection and quantification methods might significantly contribute to the observed heterogeneity in study outcomes. Given the preliminary nature of our findings, future research should incorporate more high-quality studies to explore the role of ncRNAs in various bladder cancer subtypes, their potential value in disease prognosis, and how these molecular markers can be more effectively utilized for disease management through emerging technologies.
5 Conclusion
This meta-analysis accentuates the prognostic potential of exosomal ncRNAs in bladder cancer, elevated ncRNA expressions correlate with enhanced survival outcomes, advocating for their assimilation into diagnostic and therapeutic frameworks. Further research is imperative to fully capitalize on the potential of ncRNAs in the management of bladder cancer.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
YC: Conceptualization, Data curation, Investigation, Methodology, Software, Writing – review & editing. KS: Data curation, Formal analysis, Methodology, Project administration, Supervision, Writing – original draft. XF: Data curation, Formal analysis, Methodology, Supervision, Writing – original draft. HG: Formal analysis, Funding acquisition, Project administration, Resources, Validation, Visualization, Writing – original draft. TG: Data curation, Formal analysis, Methodology, Software, Supervision, Writing – original draft. HY: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Supervision.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the Key Technology Research Plan Project of Inner Mongolia Autonomous Region (2021GG0153), and the National Natural Science Foundation of China (31760333) to HY.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1336375/full#supplementary-material
References
1. Li P, Mi Q, Yan S, Xie Y, Cui Z, Zhang S, et al. Characterization of circSCL38A1 as a novel oncogene in bladder cancer via targeting ILF3/TGF-β2 signaling axis. Cell Death Dis. (2023) 14:59. doi: 10.1038/s41419-023-05598-2
2. Teoh JY, Huang J, Ko WY, Lok V, Choi P, Ng CF, et al. Global trends of bladder cancer incidence and mortality, and their associations with tobacco use and gross domestic product per capita. Eur Urol. (2020) 78:893–906. doi: 10.1016/j.eururo.2020.09.006
3. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
4. Mondal I, Kulshreshtha R. Potential of microRNA based diagnostics and therapeutics in glioma: a patent review. Expert Opin Ther Pat. (2021) 31:91–106. doi: 10.1080/13543776.2021.1837775
5. Yang C, Wu S, Mou Z, Zhou Q, Dai X, Ou Y, et al. Exosome-derived circTRPS1 promotes Malignant phenotype and CD8+ T cell exhaustion in bladder cancer microenvironments. Mol Ther. (2022) 30:1054–70. doi: 10.1016/j.ymthe.2022.01.022
6. Huang H, Du J, Jin B, Pang L, Duan N, Huang C, et al. Combination of Urine Exosomal mRNAs and lncRNAs as Novel Diagnostic Biomarkers for Bladder Cancer. Front Oncol. (2021) 11:667212. doi: 10.3389/fonc.2021.667212
7. Wang J, Yang K, Yuan W, Gao Z. Determination of serum exosomal H19 as a noninvasive biomarker for bladder cancer diagnosis and prognosis. Med Sci Monit. (2018) 24:9307–16. doi: 10.12659/msm.912018
8. Xue M, Chen W, Xiang A, Wang R, Chen H, Pan J, et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer. (2017) 16:143. doi: 10.1186/s12943-017-0714-8
9. Bian B, Li L, Ke X, Chen H, Liu Y, Zheng N, et al. Urinary exosomal long non-coding RNAs as noninvasive biomarkers for diagnosis of bladder cancer by RNA sequencing. Front Oncol. (2022) 12:976329. doi: 10.3389/fonc.2022.976329
10. Abbastabar M, Sarfi M, Golestani A, Karimi A, Pourmand G, Khalili E. Tumor-derived urinary exosomal long non-coding RNAs as diagnostic biomarkers for bladder cancer. Excli J. (2020) 19:301–10. doi: 10.17179/excli2019-1683
11. Sung WJ, Hong J. Targeting lncRNAs of colorectal cancers with natural products. Front Pharmacol. (2022) 13:1050032. doi: 10.3389/fphar.2022.1050032
12. Stachowiak Z. B narożna and A szczepankiewicz. Non-coding RNAs in pulmonary diseases: comparison of different airway-derived biosamples. Int J Mol Sci. (2023) 24:2006. doi: 10.3390/ijms24032006
13. Li Q, Li Z, Fan Z, Yang Y, Lu C. Involvement of non−coding RNAs in the pathogenesis of myocardial ischemia/reperfusion injury (Review). Int J Mol Med. (2021) 47:42. doi: 10.3892/ijmm.2021.4875
14. Yazarlou F, Modarressi MH, Mowla SJ, Oskooei VK, Motevaseli E, Tooli LF, et al. Urinary exosomal expression of long non-coding RNAs as diagnostic marker in bladder cancer. Cancer Manag Res. (2018) 10:6357–65. doi: 10.2147/cmar.S186108
15. Zhan Y, Du L, Wang L, Jiang X, Zhang S, Li J, et al. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol Cancer. (2018) 17:142. doi: 10.1186/s12943-018-0893-y
16. Güllü Amuran G, Tinay I, Filinte D, Ilgin C, Peker Eyüboğlu I, Akkiprik M. Urinary micro-RNA expressions and protein concentrations may differentiate bladder cancer patients from healthy controls. Int Urol Nephrol. (2020) 52:461–68. doi: 10.1007/s11255-019-02328-6
17. El-Shal AS, Shalaby SM, Abouhashem SE, Elbary EHA, Azazy S, Rashad NM, et al. Urinary exosomal microRNA-96-5p and microRNA-183-5p expression as potential biomarkers of bladder cancer. Mol Biol Rep. (2021) 48:4361–71. doi: 10.1007/s11033-021-06451-5
18. Robles V, Valcarce DG, Riesco MF. Non-coding RNA regulation in reproduction: Their potential use as biomarkers. Noncoding RNA Res. (2019) 4:54–62. doi: 10.1016/j.ncrna.2019.04.001
19. Chen C, Shang A, Sun Z, Gao Y, Huang J, Ping Y, et al. Urinary exosomal long noncoding RNA TERC as a noninvasive diagnostic and prognostic biomarker for bladder urothelial carcinoma. J Immunol Res. (2022) 2022:9038808. doi: 10.1155/2022/9038808
20. Zhang S, Du L, Wang L, Jiang X, Zhan Y, Li J, et al. Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. (2019) 23:1396–405. doi: 10.1111/jcmm.14042
21. Zheng R, Du M, Wang X, Xu W, Liang J, Wang W, et al. Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer. (2018) 17:143. doi: 10.1186/s12943-018-0880-3
22. Lo CK, Mertz D, Loeb M. Newcastle-Ottawa Scale: comparing reviewers' to authors' assessments. BMC Med Res Methodol. (2014) 14:45. doi: 10.1186/1471-2288-14-45
23. Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu H, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest. (2020) 130:404–21. doi: 10.1172/jci130892
24. Chen C, Zheng H, Luo Y, Kong Y, An M, Li Y, et al. SUMOylation promotes extracellular vesicle-mediated transmission of lncRNA ELNAT1 and lymph node metastasis in bladder cancer. J Clin Invest. (2021) 131:e146431. doi: 10.1172/jci146431
25. Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L, et al. PRMT5 Circular RNA Promotes Metastasis of Urothelial Carcinoma of the Bladder through Sponging miR-30c to Induce Epithelial-Mesenchymal Transition. Clin Cancer Res. (2018) 24:6319–30. doi: 10.1158/1078-0432.Ccr-18-1270
26. Lin H, Shi X, Li H, Hui J, Liu R, Chen Z, et al. Urinary Exosomal miRNAs as biomarkers of bladder Cancer and experimental verification of mechanism of miR-93-5p in bladder Cancer. BMC Cancer. (2021) 21:1293. doi: 10.1186/s12885-021-08926-x
27. Zheng H, Chen C, Luo Y, Yu M, He W, An M, et al. Tumor-derived exosomal BCYRN1 activates WNT5A/VEGF-C/VEGFR3 feedforward loop to drive lymphatic metastasis of bladder cancer. Clin Transl Med. (2021) 11:e497. doi: 10.1002/ctm2.497
28. Khan FB, Uddin S, Elderdery AY, Goh KW, Ming LC, Ardianto C, et al. Illuminating the molecular intricacies of exosomes and ncRNAs in cardiovascular diseases: prospective therapeutic and biomarker potential. Cells. (2022) 11:3664. doi: 10.3390/cells11223664
29. Yan L, Li Q, Sun K, Jiang F. MiR-4644 is upregulated in plasma exosomes of bladder cancer patients and promotes bladder cancer progression by targeting UBIAD1. Am J Transl Res. (2020) 12:6277–89.
30. Yin X, Zheng X, Liu M, Wang D, Sun H, Qiu Y, et al. Exosomal miR-663b targets Ets2-repressor factor to promote proliferation and the epithelial-mesenchymal transition of bladder cancer cells. Cell Biol Int. (2020) 44:958–65. doi: 10.1002/cbin.11292
31. Xu Z, Chen Y, Ma L, Chen Y, Liu J, Guo Y, et al. Role of exosomal non-coding RNAs from tumor cells and tumor-associated macrophages in the tumor microenvironment. Mol Ther. (2022) 30:3133–54. doi: 10.1016/j.ymthe.2022.01.046
32. Ramón YCS, Segura MF, Hümmer S. Interplay between ncRNAs and cellular communication: A proposal for understanding cell-specific signaling pathways. Front Genet. (2019) 10:281. doi: 10.3389/fgene.2019.00281
33. Xia B, Liu Y, Wang J, Lu Q, Lv X, Deng K, et al. Emerging role of exosome-shuttled noncoding RNAs in gastrointestinal cancers: From intercellular crosstalk to clinical utility. Pharmacol Res. (2023) 195:106880. doi: 10.1016/j.phrs.2023.106880
34. D'Anca M, Fenoglio C, Serpente M, Arosio B, Cesari M, Scarpini EA, et al. Exosome determinants of physiological aging and age-related neurodegenerative diseases. Front Aging Neurosci. (2019) 11:232. doi: 10.3389/fnagi.2019.00232
35. Baumgart S, Meschkat P, Edelmann P, Heinzelmann J, Pryalukhin A, Bohle R, et al. MicroRNAs in tumor samples and urinary extracellular vesicles as a putative diagnostic tool for muscle-invasive bladder cancer. J Cancer Res Clin Oncol. (2019) 145:2725–36. doi: 10.1007/s00432-019-03035-6
36. Micheel J, Safrastyan A, Wollny D. Advances in non-coding RNA sequencing. Noncoding RNA. (2021) 7:70. doi: 10.3390/ncrna7040070
37. Arjumand W, Asiaf A, Ahmad ST. Noncoding RNAs in DNA damage response: opportunities for cancer therapeutics. Methods Mol Biol. (2018) 1699:3–21. doi: 10.1007/978-1-4939-7435-1_1
38. Khan N, Umar MS, Haq M, Rauf T, Zubair S, Owais M. Exosome-encapsulated ncRNAs: Emerging yin and yang of tumor hallmarks. Front Genet. (2022) 13:1022734. doi: 10.3389/fgene.2022.1022734
39. Ray SK, Mukherjee S. Interaction among noncoding RNAs, DNA damage reactions, and genomic instability in the hypoxic tumor: is it therapeutically exploitable practice? Curr Mol Med. (2023) 23:200–15. doi: 10.2174/1566524022666220120123557
Keywords: bladder cancer, non-coding RNAs, prognosis, diagnosis, meta-analysis
Citation: Chen Y, Shi K, Fu X, Guo H, Gao T and Yu H (2024) Diagnostic and prognostic potential of exosome non-coding RNAs in bladder cancer: a systematic review and meta-analysis. Front. Oncol. 14:1336375. doi: 10.3389/fonc.2024.1336375
Received: 10 November 2023; Accepted: 19 February 2024;
Published: 04 March 2024.
Edited by:
So Hee Kwon, Yonsei University, Republic of KoreaReviewed by:
Kulbhushan Thakur, University of Delhi, IndiaFabio Grizzi, Humanitas Research Hospital, Italy
Copyright © 2024 Chen, Shi, Fu, Guo, Gao and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiquan Yu, aHl1QGltdS5lZHUuY24=
 Yani Chen
Yani Chen Haiquan Yu
Haiquan Yu