- 1Paholpolpayuhasena Hospital, Department of Medical Organization, Kanchanaburi, Thailand
- 2Division of Hematology, Department of Medicine, National Cancer Institute Thailand, Bangkok, Thailand
- 3Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand
- 4Division of Hematology, Department of Medicine, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand
- 5Center of Excellence of Siriraj Adult Acute Myeloid/Lymphoblastic Leukemia, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand
- 6Division of Clinical Pharmacy, Faculty of Pharmaceutical Sciences, Burapha University, Chonburi, Thailand
- 7Department of Medicine, Chao Phraya Yommaraj Hospital, Suphanburi, Thailand
Introduction: Variations in mutation rates among acute myeloid leukemia (AML) patients with myeloid sarcoma (MS) underscore the need for a thorough examination. This meta-analysis was conducted to fill the information gap concerning mutation frequencies in AML patients presenting with MS.
Materials and methods: This study included retrospective and prospective cohorts. It examined genetic alterations in AML patients with and without MS across all age groups. The search strategy employed terms such as “acute myeloid leukemia,” “extramedullary,” “granulocytic sarcoma,” “myeloid sarcoma,” and “leukemic cutis” in the EMBASE, MEDLINE, and Scopus databases. Excluded from the study were reviews, case reports, and case series with fewer than 10 cases. Statistical analyses were performed with Review Manager 5.4 software.
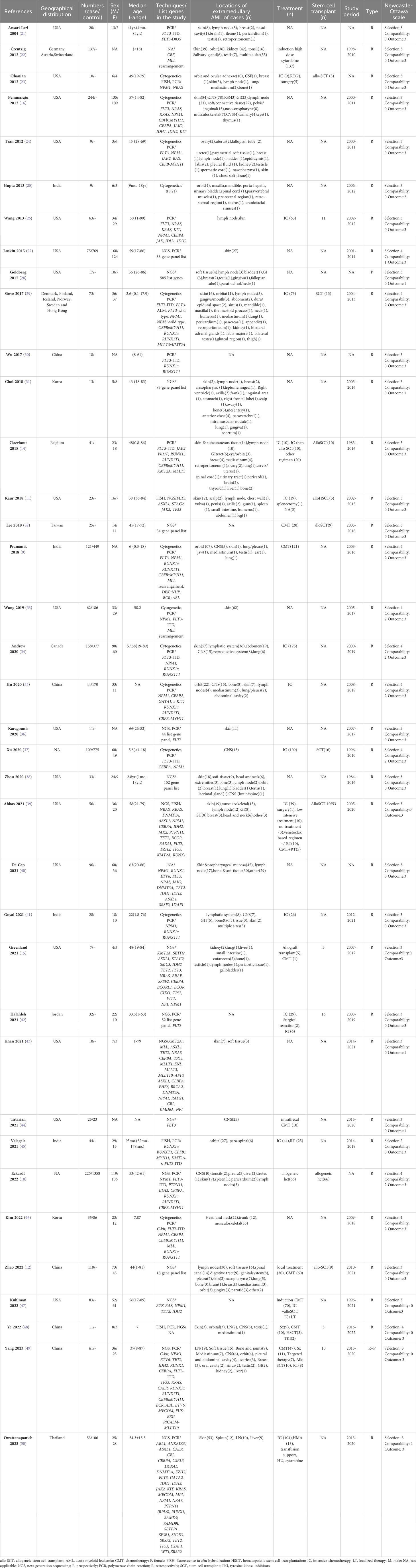
Results: The primary analysis incorporated data from 37 cohorts involving 5646 diagnosed AML patients and revealed a 17.42% incidence of MS. The most prevalent mutation among AML patients with MS was FLT3-ITD, with a pooled prevalence of 17.50% (95% CI 12.60% to 22.50%; I2 82.48%). The dominant fusion gene was RUNX1::RUNX1T1, displaying a pooled prevalence of 28.10% (95% CI 15.10% to 41.20%; I2 96.39%). In comparison, no significant intergroup differences were observed for NPM1, FLT3-ITD, KIT, and IDH2 mutations. Interestingly, the CEBPA mutation exhibited protective effects for MS patients, with an odds ratio of 0.51 (95% CI 0.32 to 0.81; I2 0%). Conversely, the NRAS mutation was associated with an increased risk of MS development, with an odds ratio of 5.07 (95% CI 1.87 to 13.73; I2 0%).
Conclusion: This meta-analysis sheds light on the prevalence of genetic mutations in AML patients with MS, providing insights into the unique characteristics of the mutations and their frequencies. These discoveries are crucial in informing therapeutic and prognostic decisions for individuals with myeloid sarcoma.
Highlights
● Data from 37 cohorts, consisting of 6475 acute myeloid leukemia (AML) patients, were analyzed to determine the genetic profile of AML patients with myeloid sarcoma (MS).
● FLT3-ITD is the most prevalent mutation, and RUNX1::RUNX1T1 is the most common fusion gene in AML patients with MS.
● The CEBPA mutation offers protective effects to MS patients, while the NRAS mutation heightens the risk of MS development.
Introduction
Acute myeloid leukemia (AML) is characterized by the uncontrolled proliferation of myeloid stem cells and impaired differentiation (1). In 2019, the United States observed an estimated total of more than 20 000 AML cases, with certain studies suggesting an age-adjusted incidence rate of 3.43 cases per 100 000 individuals annually (2). Extensive research into AML pathogenesis has identified numerous mutations and cytogenetic abnormalities as pivotal contributors to disease onset (3, 4). In 2016, the World Health Organization classified myeloid sarcoma (MS) as an AML subtype. This classification was retained in the World Health Organization’s updated 2022 classification and the 2022 International Consensus Classification, wherein MS remains a recognized entity (5–7).
MS is a tumor mass formed of myeloblasts outside the bone marrow (3). Predominantly, MS affects patients diagnosed with AML or chronic myeloid leukemia, constituting approximately 9% of these cases (8). Notably, the prevalence of MS is greater in males than females, and the condition predominantly affects individuals aged 46 to 59 years (3, 8, 9). The pathophysiology of MS, especially the migration of cells to extramedullary sites, remains elusive. Prevailing hypotheses suggest that the development of MS may be linked to leukemic cells expressing CD56 (neural cell adhesion molecule) (4). These cells possibly bind to tissues commonly associated with MS manifestations (3).
MS commonly manifests in extramedullary sites such as the skin, bones, soft tissues, and gall bladder (4). However, some studies also document its occurrence in rarer locations, including the pleura, penis, and vulva (10–12). The prognosis for MS patients tends to be unfavorable and can vary based on the location of the lesion and its molecular attributes (1, 8, 13). In modern diagnostic methodologies, next-generation sequencing (NGS) has emerged as a crucial tool for identifying mutations in AML patients, including those with MS (3). The NPM1 mutation is the most common mutation found in MS; other common mutations and fusion genes include KRAS, NRAS, KIT, CEBPA, IDH1, IDH2, RUNX1::RUNX1T1, and CBFB::MYH11 (3, 10, 14–16). However, variations persist in the reported incidence of each mutation in MS among studies (12, 14, 16). Moreover, a previous report indicated variations in the prevalence of chromosomal abnormalities and/or molecular mutations among different countries (17). However, there is currently no available data regarding these variations specifically within the subgroup of AML with MS.
Consequently, this systematic review and meta-analysis compiled and analyzed data on the incidence of each mutation from all pertinent sources. Our objective was to better understand the specific characteristics and precise prevalence of genetic mutations in AML patients presenting with MS.
Materials and methods
Data sources and searches
Six researchers (S.U., K.K., S.P., T.K., W.O., and T.R.) independently searched for articles published within the EMBASE, MEDLINE, and Scopus databases from their inception up to August 1, 2023. The search terms included “acute myeloid leukemia,” “extramedullary,” “granulocytic sarcoma,” “myeloid sarcoma,” and “leukemic cutis.” A comprehensive description of the search strategy is provided in Supplementary Data 1. Our systematic review and meta-analysis strictly followed the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines, as elaborated in Supplementary Data 2.
Selection criteria and data extraction
Studies included in the meta-analysis were retrospective or prospective cohort studies on AML with MS and with a primary outcome aligned with our research objective. We excluded reviews, case reports, and case series with fewer than 10 cases. The primary objective of this analysis was to determine the incidence of each mutation in AML patients with MS, while the secondary aim was to compare the mutational statuses of AML patients with and without MS. To ascertain study eligibility, four researchers (S.U., K.K., S.P., and T.K.) independently assessed the titles and abstracts of the retrieved studies. They also reviewed the references in the selected studies to identify any additional pertinent research. In instances of disagreement about the inclusion of specific studies, consensus was reached through mediation with two other investigators (W.O. and T.R.).
Quality assessment
The quality assessment of the included studies was independently conducted by two researchers (S.U. and K.K.) using the Newcastle–Ottawa quality assessment scale (18).
Statistical analysis
We analyzed the data with Review Manager 5.4 software provided by the Cochrane Collaboration (London, United Kingdom). The inverse variance method was employed to compute pooled odds ratios (ORs) and their corresponding 95% confidence intervals (CIs) for each gene across the studies (19). The prevalence of the genetic alterations was meta-analyzed and pooled using the binary random-effects model using the DerSimonian–Laird method (Open Meta–Analyst for Windows 8) (20). Given the anticipated variability among the incorporated studies, a random-effects model was favored over a fixed-effects model for our meta-analysis. We evaluated statistical heterogeneity with Cochran’s Q test and quantified its extent using the I2 statistic. Depending on the I2 values, heterogeneity was classified as either insignificant (0%–25%), low (25%–50%), moderate (50%–75%), or high (75%–100%) (19). For transparency and procedural clarity, we registered our study protocol with the International Platform of Registered Systematic Review and Meta-Analysis Protocols (INPLASY) network (registration number INPLASY202380091).
Results
Search results
A total of 11 145 articles were identified in the search process, with 1934 articles from MEDLINE, 2514 from EMBASE, 6696 from Scopus, and 1 from other sources. Initially, 4876 duplicated articles were removed, and another 6269 were excluded after reviewing the titles and abstracts. The remaining 153 articles underwent a thorough full-text reading. This resulted in a further 116 articles being excluded because they did not meet the inclusion criteria. The remaining 37 articles that met the inclusion criteria were included in our analysis. The data gathering and screening process is depicted in Figure 1.
Baseline patient characteristics
The analysis included 37 cohorts that collectively comprised 6475 diagnosed AML patients. Among these patients, 907 out of 5206 (17.42%) were identified as having AML with MS, resulting in 2199 MS cases. Table 1 details the characteristics and quality assessments of the analyzed cohorts. There was a slight male predominance, accounting for approximately 58.26%, with females comprising 41.74%. When analyzing age demographics across cohorts, 71.87% of the patient population was over 18 years old, while the remaining 28.13% was under 18. Further analysis indicated that the most common sites of MS manifestation were the skin, orbit, central nervous system, and lymph nodes.
Pool prevalence of DNA mutations in AML patients with MS
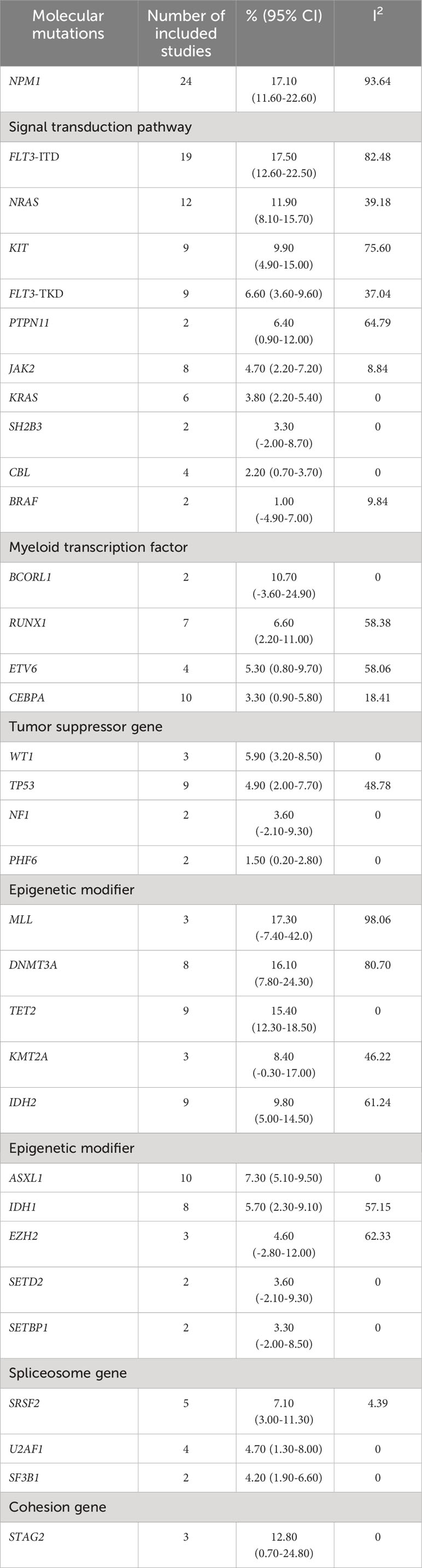
Our detailed analysis of the included articles provided comprehensive insights into the pooled prevalence of molecular mutations among patients diagnosed with MS. Figure 2 presents the significant molecular mutations extracted from this extensive dataset. Among them, the FLT3-ITD mutation was the most prevalent, with a pooled prevalence of 17.50% (95% CI 12.60% to 22.50%; I² 82.48%; Figure 2A) (12, 16, 21, 26, 27, 29–34, 36–38, 42, 44–46, 48, 50). Similarly, the MLL and NPM1 mutations stood out with a prevalence of 17.30% (95% CI -7.40% to 42.0%; I² 98.06%; Figure 2C) (9, 22, 46) and 17.10% (95% CI 11.60% to 22.60%; I² 93.64%; Figure 2B) (9, 12, 15, 16, 23, 24, 26, 27, 29, 31–37, 39–42, 46), respectively. Additionally, the DNMT3A mutation was observed at a prevalence of 16.10% (95% CI 7.80% to 24.30%; I² 80.7%; Figure 2D) (12, 27, 28, 32, 36, 40, 42, 50), while the TET2 mutation had a prevalence of 15.40% (95% CI 12.30% to 18.50%; I² 0%; Figure 2E) (12, 15, 27, 36, 40, 43, 47, 49, 50). Furthermore, the STAG2 and NRAS mutations exhibited a prevalence of 12.80% (95% CI 0.70% to 24.80%; I² 0%; Figure 2F) (11, 15, 36) and 11.9% (95% CI 8.10% to 15.70%; I² 39.18%), respectively. For a more detailed exploration of the pooled prevalence of DNA mutations in MS, please refer to Table 2.
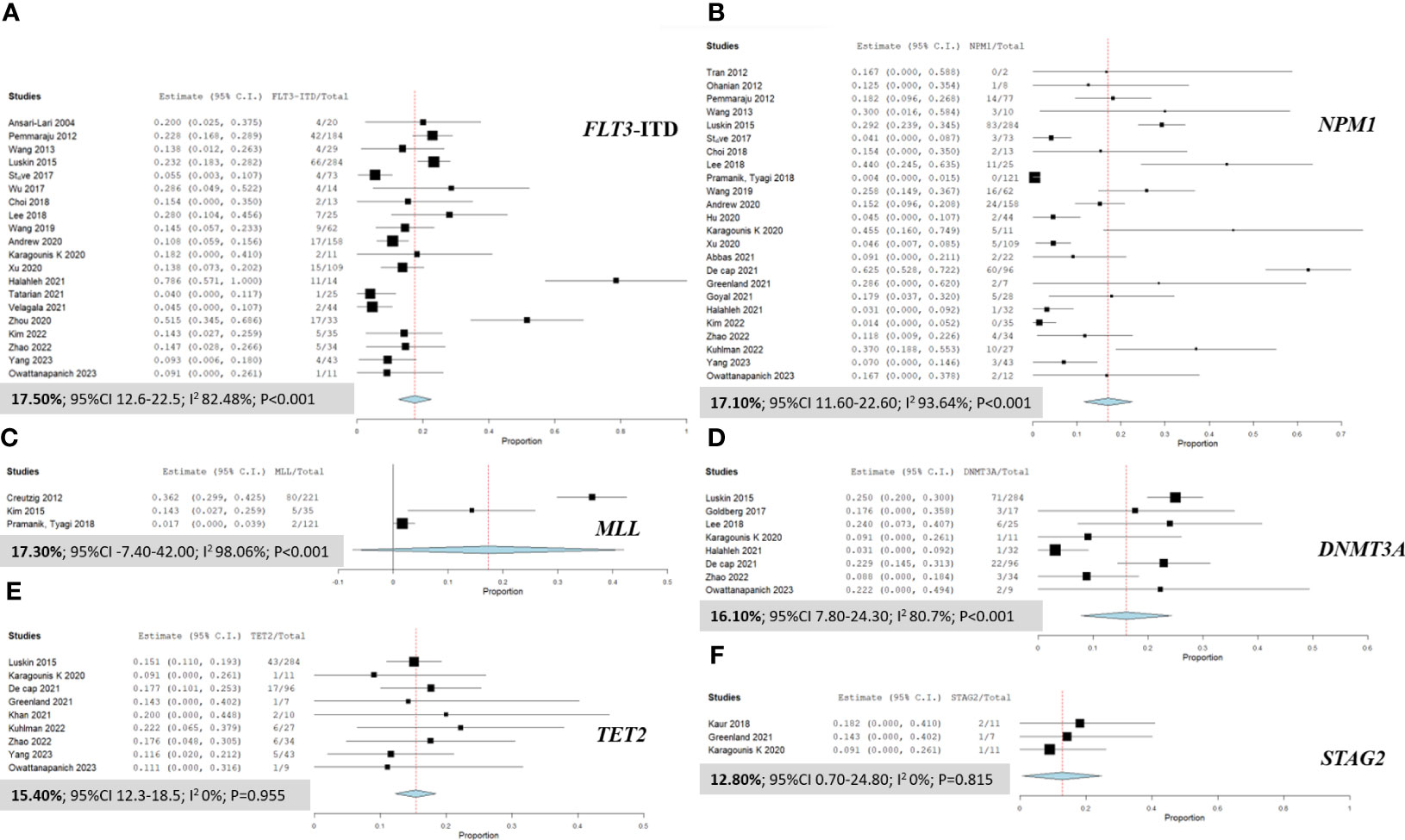
Figure 2 Forest plots illustrating the prevalence of gene mutations in AML patients with myeloid sarcoma. (A) FLT3-ITD; (B) NPM1; (C) MLL; (D) DNMT3A; (E) TET2; and (F) STAG2.
Pool prevalence of fusion genes in AML patients with MS
Figure 3 presents a detailed analysis of the pooled prevalence of fusion genes in patients with MS, highlighting the frequency of various fusion genes within this group. The most predominant fusion gene observed was RUNX1::RUNX1T1, with a remarkable pooled prevalence of 28.10% (95% CI 15.10% to 41.20%; I² 96.39%; Figure 3A) (9, 14, 25, 28, 34, 35, 38, 41, 42, 45, 48–50). The KMT2A::MLLT3 fusion gene was also identified at a pooled prevalence of 19.20% (95% CI -14.60% to 53.00%; I² 79.63%; Figure 3B) (14, 43). Furthermore, the CBFB::MYH11 fusion gene was observed at a pooled prevalence of 10.30% (95% CI 5.40% to 15.10%; I² 84.93%; Figure 3C) (9, 14, 16, 24, 28, 35, 38, 39, 42, 45, 46, 49, 50).
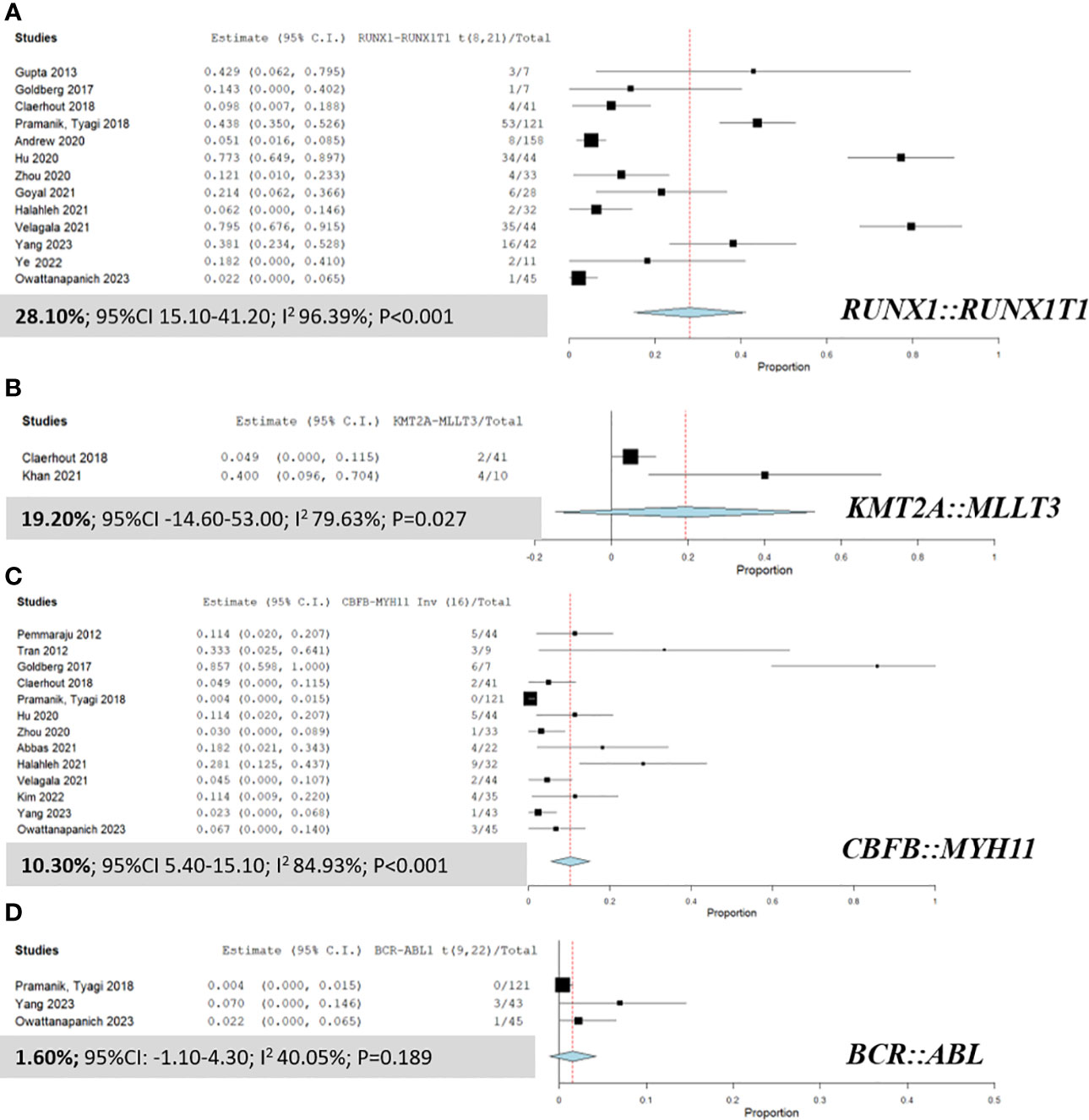
Figure 3 Forest plots displaying the prevalence of fusion genes in AML patients with myeloid sarcoma. (A) RUNX1::RUNX1T1; (B) KMT2A::MLLT3; (C) CBFB::MYH11; and (D) BCR::ABL.
Subgroup analysis
When categorizing patients into two age groups (<40 years and ≥40 years), we observed distinct patterns of mutations. Among patients under 40 years of age, KRAS mutation emerged as the most prevalent, occurring in 50.00% of cases (95% CI 10.00% to 90.00%; I² 0%). Conversely, in individuals aged 40 years and above, SRSF2 mutation was the most commonly observed, with a pooled prevalence of 21.90% (95% CI -0.30% to 44.20%; I² 0%). Supplementary Data 3 provides a detailed breakdown of the prevalence of molecular mutations and fusion genes within these age groups.
Furthermore, we calculated the overall pooled prevalence of AML patients with MS harboring recurrent genetic abnormalities, as per the 2022 World Health Organization classification, to be 36.80% (95% CI: 26.00% to 47.60%; I² 96.85%; Supplementary Data 4) (9, 12, 14–16, 23–29, 31–43, 45–50).
The prevalence of genetic mutations, stratified by geographical distribution, was investigated. Among Western patients, the third most frequently observed mutations were NPM1 (27.50%; 95% CI: 17.80 to 37.30; I² 87.18%), FLT3-ITD (20.50%; 95% CI: 13.90 to 27.10; I² 77.52%), and KMT2A (19.90%; 95% CI: -15.00 to 54.90; I² 72.89%). Conversely, in Eastern patients, the most common mutations were KRAS (20.10%; 95% CI: 8.90 to 31.30; I² 7.93%), FLT3-ITD (18.10%; 95% CI: 5.90 to 27.20; and I² 82.73%), and KIT (15.20%; 95% CI: 9.40 to 21.00; I² 0%) mutations. CBFB::MYH11 emerged as the predominant fusion gene in the Western population, while RUNX1::RUNX1T1 predominated in the Eastern population, with rates of 20.40% (95% CI: 7.20 to 33.50; I² 88.31%) and 21.50% (95% CI: 10.10 to 32.90; I² 95.69%), respectively. The genetic profiling of AML patients with MS in both Western and Eastern countries is presented in Supplementary Data 5, 6.
Comparison of mutational profiles between the AML with and without MS groups
Several noteworthy findings emerged after analyzing gene mutations in patients with MS and non-MS (Figure 4). Specifically, the prevalence of the CEBPA mutation was significantly higher in non-MS patients than in those with MS, with an OR of 0.51 (95% CI 0.32 to 0.81; I² 0%; Figure 4C) (10, 35, 37, 46, 50). Conversely, the NRAS mutation was notably more prevalent in the MS group, with an OR of 5.07 (95% CI 1.87 to 13.73; I² 0%; Figure 4G) (31, 50). However, no significant differences were observed in the prevalence of the NPM1, FLT3-ITD, KIT, and IDH2 mutations between MS and non-MS patients.
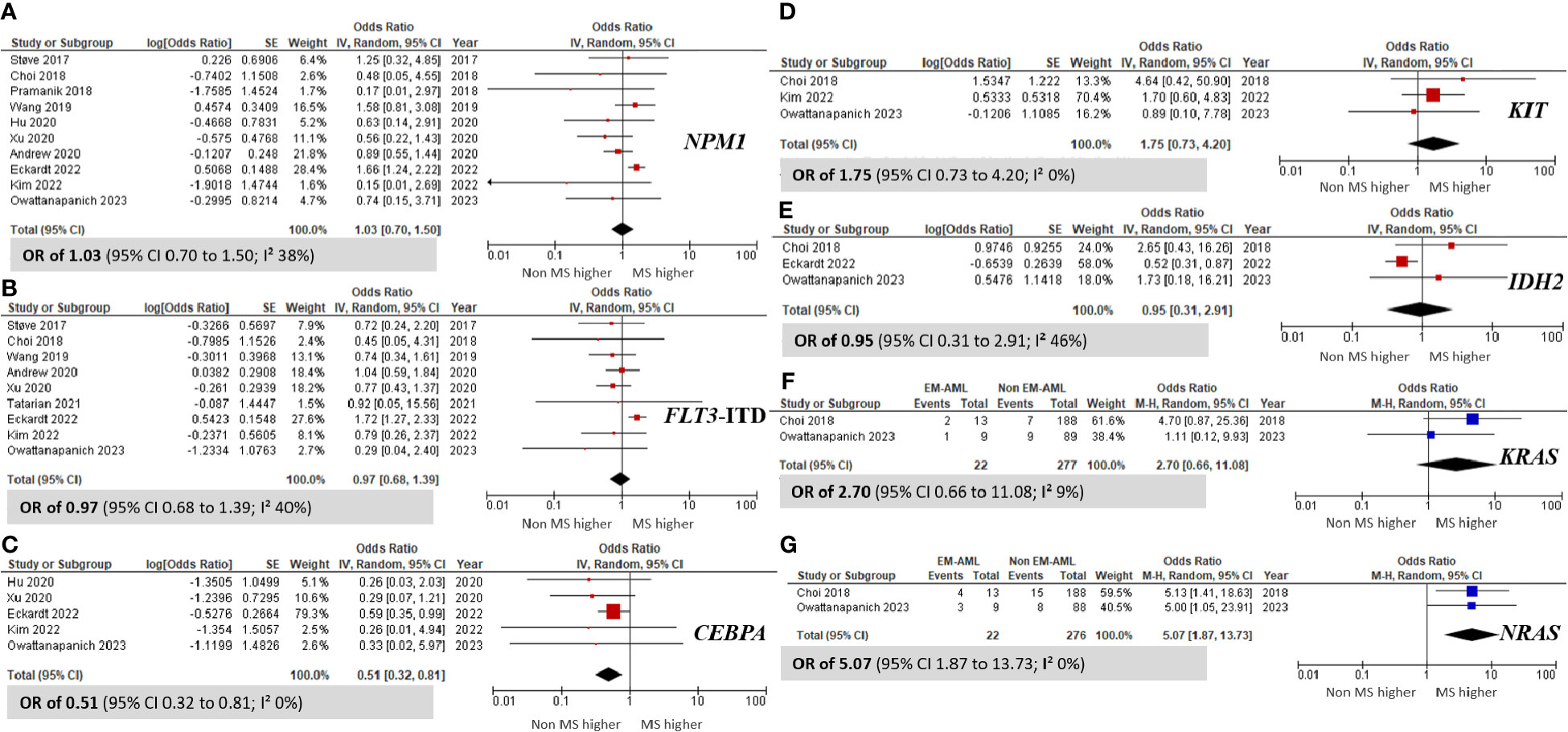
Figure 4 Forest plots indicating the prevalence of gene mutations in AML patients with myeloid sarcoma in comparison to those without myeloid sarcoma. (A) NPM1; (B) FLT3-ITD; (C) CEBPA; (D) KIT; (E) IDH2; (F) KRAS; and (G) NRAS.
Additionally, we assessed the incidence of the RUNX1::RUNX1T1 and CBFB::MYH11 fusion genes in four included studies. The meta-analysis revealed no significant differences in the incidence of these fusion genes between patients with MS and those without MS, with pooled ORs of 1.21 (95% CI 0.53 to 2.75; I² 78%; Figure 5A) (9, 10, 35, 46, 50) for RUNX1::RUNX1T1 and 1.26 (95% CI 0.54 to 2.95; I² 52%; Figure 5B) (9, 10, 35, 46, 50) for CBFB::MYH11. Furthermore, there was no significant correlation between AML patients harboring MS and the presence of recurrent genetic abnormalities (pooled OR 0.74; 95% CI 0.42 to 1.29; I² 82%; Supplementary Data 4) (9, 29, 31, 33, 34, 37, 46, 50).
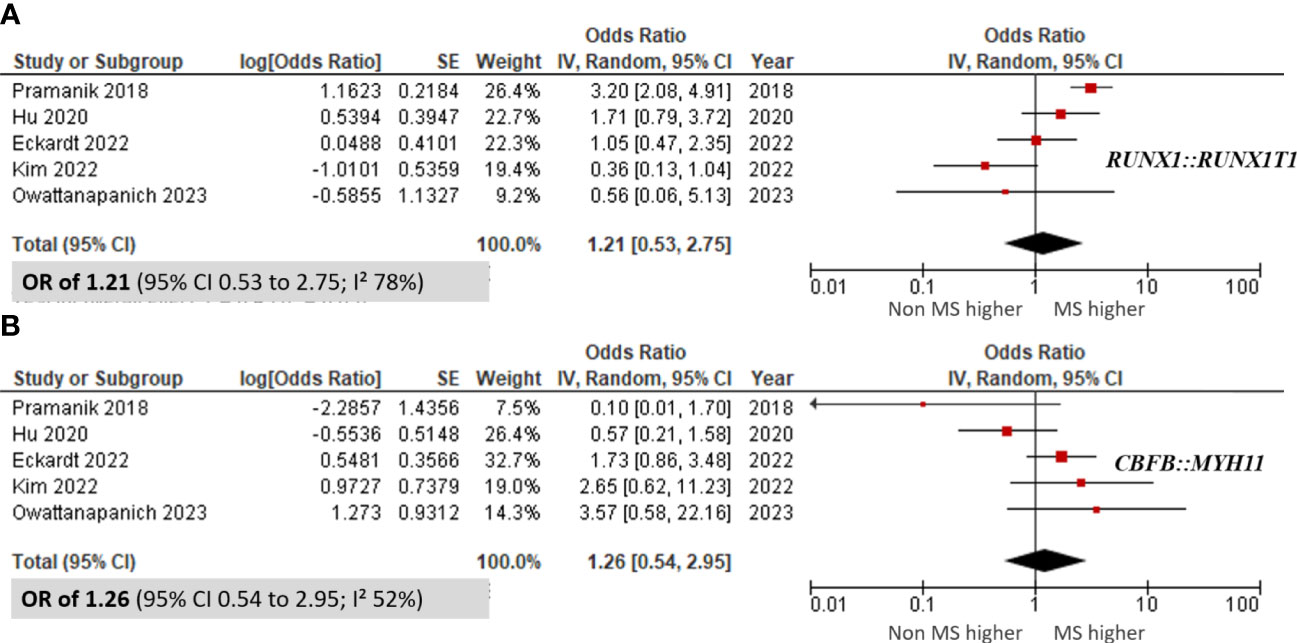
Figure 5 Forest plots showing the prevalence of fusion genes in AML patients with myeloid sarcoma in comparison to those without myeloid sarcoma. (A) RUNX1::RUNX1T1; and (B) CBFB::MYH11.
Discussion
MS, commonly known as extramedullary AML, presents a wide range of clinical manifestations and often poses therapeutic challenges. A prior multicenter cohort study documented an MS incidence of 14.21% among newly diagnosed AML cases. Typically, pivotal therapeutic choices are guided by the genetic alteration profile. This current study marks the inaugural meta-analysis of MS prevalence and its associated genetic abnormalities.
Our study found a cumulative MS incidence of 17.42%, with a slight male predominance. This rate exceeds that reported in earlier research on newly diagnosed cases (10). Our finding aligns with the observations made by Fianchi et al. (51), who reported a decline in MS incidence from 11% to 7% when assessed at the time of AML diagnosis. In terms of molecular genetics, our meta-analysis identified FLT3-ITD mutations as the most frequently linked to MS, with a pooled prevalence of 17.5%. These figures align closely with prior research: Ansari-Lari et al. (21) found these mutations in 15% of MS cases. Pemmaraju et al. (16) and Shallis et al. (3) also reported the FLT3 mutation as the predominant mutation. The second most common mutations were MLL and NPM1, with a pooled prevalence of approximately 17%, as highlighted by the studies of Chang et al. (52) and Eckardt et al. (10). According to Chang et al. (1), the MLL gene mutation, especially the classic 11q23 abnormality but excluding t(9;11), has been associated with extramedullary involvement and remains a poor prognostic factor. Additionally, Falini et al. (3) recorded NPM1 mutations in 14% of 181 MS samples. In a larger cohort of 89 AML patients, Ovcharenko et al. (53) observed mutated NPM1 in 13 out of 15 MS patients. Another key finding from our study is that DNMT3A and TET2 mutations emerged as the third and fourth most common genetic aberrations, respectively.
AML with t(8;21)(q22;q22.1); RUNX1::RUNX1T1 represents a distinct subtype of AML. Classified as a core-binding factor leukemia, this form of AML is characterized by frequent genetic recurrence and generally has a favorable prognosis (54). Saia et al. delved into the RUNX1::RUNX1T1 rearrangement in mouse models, shedding light on its frequent association with extramedullary disease (55). Consistent with our data, the predominant fusion gene detected in MS was RUNX1::RUNX1T1, demonstrating a cumulative prevalence of 28.10%. This finding accords with the work of Hu et al. and Velagala et al., and it emphasizes the significance of the RUNX1::RUNX1T1 fusion gene (5, 7). In contrast, certain studies have underscored the sporadic nature of the RUNX1::RUNX1T1 fusion, noting its presence in just 2% to 3% of MS cases (56). It is noteworthy that significant statistical heterogeneity was observed throughout all fusion gene analyses, likely amplified by varying baseline characteristics among the considered studies.
Geographical variations also influence the genetic profiling in MS. The NPM1 mutation was prominently observed among patients in Western regions, whereas KRAS predominated in those from Eastern countries. Additionally, FLT3-ITD was identified as a commonly occurring mutation in both populations. Core-binding fusion genes were frequently observed in AML with MS across continents, albeit with differences in specific fusion genes (CBFB::MYH11 in the Western population and RUNX1::RUNX1T1 in the Eastern population).
When comparing genetic abnormalities between non-MS and MS cases, our analysis revealed no significant differences in the prevalence of the RUNX1::RUNX1T1 and CBFB::MYH11 fusion genes. However, we found that the NRAS mutation was significantly associated with the MS group. Our findings suggest that the CEBPA mutation might confer a protective effect against MS, supported by an OR of 0.51 (95% CI 0.32 to 0.81).
This study examined the prevalence of mutations and fusion genes in AML with multiple MS. The findings revealed variations in the incidence rates of certain mutations between the MS group and AML patients without MS. Furthermore, age and geographical disparities emerged as significant factors influencing the genetic profiling in MS cases. Consequently, a mutational workup should be conducted in all newly diagnosed AML patients with MS, as the results offer valuable insights for risk stratification, guiding treatment decisions, and potentially introducing novel therapeutic options targeting specific mutations.
However, this study has several limitations. First, it drew upon published data, potentially introducing publication bias since studies with positive or novel outcomes are more likely to be published than those with negative or neutral findings. Second, numerous analyses indicated elevated I2 values, which signify substantial heterogeneity among studies. This heterogeneity might have undermined the reliability of our combined results. Third, some included studies lacked details of patients’ baseline characteristics, and there was evident statistical inconsistency in the genetic testing methods used. Additionally, the source of genetic data, whether derived from bone marrow or blood samples, was not always clearly specified, potentially leading to inaccurate representation of genetic variations. Fourth, the limited number of studies comparing gene mutations between MS and non-MS may have resulted in insufficient statistical power to establish significant differences. Fifth, the included studies utilized a range of techniques, such as conventional cytogenetics, Fluorescence In Situ Hybridization, polymerase chain reaction, and NGS, each with varying sensitivities in detecting mutations. This diversity in methods may have influenced the accurate assessment of mutation prevalence. Lastly, the potential relationship between MS and gene mutations is an intriguing area of study; however, we were unable to perform such an analysis in this study due to insufficient data.
Conclusion
This study underscores the importance of three gene mutations—FLT3-ITD, MLL, and NPM1—which were commonly observed in cases of MS. The fusion gene RUNX1::RUNX1T1 emerged as the principal genetic fusion associated with MS. Intriguingly, although the CEBPA mutation appeared to confer some protection against MS, the presence of the NRAS mutation was associated with an elevated risk of developing MS. In essence, this meta-analysis substantially augments our comprehension of the genetic mutation characteristics of MS.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
SU: Conceptualization, Data curation, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft. ST: Conceptualization, Data curation, Investigation, Methodology, Resources, Visualization, Writing – original draft, Writing – review & editing. KK: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft. TR: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft. NL: Conceptualization, Formal analysis, Investigation, Writing – review & editing. SP: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft. TK: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft. WO: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1325431/full#supplementary-material
References
1. De Kouchkovsky I, Abdul-Hay M. Acute myeloid leukemia: a comprehensive review and 2016 update. Blood Cancer J. (2016) 6:e441. doi: 10.1038/bcj.2016.50
2. Shallis RM, Wang R, Davidoff A, Ma X, Zeidan AM. Epidemiology of acute myeloid leukemia: Recent progress and enduring challenges. Blood Rev. (2019) 36:70–87. doi: 10.1016/j.blre.2019.04.005
3. Shallis RM, Gale RP, Lazarus HM, Roberts KB, Xu ML, Seropian SE, et al. Myeloid sarcoma, chloroma, or extramedullary acute myeloid leukemia tumor: a tale of misnomers, controversy and the unresolved. Blood Rev. (2021) 47:100773. doi: 10.1016/j.blre.2020.100773
4. Solh M, Solomon S, Morris L, Holland K, Bashey A. Extramedullary acute myelogenous leukemia. Blood Rev. (2016) 30:333–9. doi: 10.1016/j.blre.2016.04.001
5. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. (2016) 127:2391–405. doi: 10.1182/blood-2016-03-643544
6. Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data. Blood. (2022) 140:1200–28. doi: 10.1182/blood.2022015850
7. Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. (2022) 36:1703–19. doi: 10.1038/s41375-022-01613-1
8. Almond LM, Charalampakis M, Ford SJ, Gourevitch D, Desai A. Myeloid sarcoma: presentation, diagnosis, and treatment. Clin Lymphoma Myeloma Leuk. (2017) 17:263–7. doi: 10.1016/j.clml.2017.02.027
9. Pramanik R, Tyagi A, Chopra A, Kumar A, Vishnubhatla S, Bakhshi S. Myeloid sarcoma predicts superior outcome in pediatric AML; can cytogenetics solve the puzzle? Clin Lymphoma Myeloma Leuk. (2018) 18:e249–54. doi: 10.1016/j.clml.2018.03.013
10. Eckardt JN, Stölzel F, Kunadt D, Röllig C, Stasik S, Wagenführ L, et al. Molecular profiling and clinical implications of patients with acute myeloid leukemia and extramedullary manifestations. J Hematol Oncol. (2022) 15:60. doi: 10.1186/s13045-022-01267-7
11. Kaur V, Swami A, Alapat D, Abdallah AO, Motwani P, Hutchins LF, et al. Clinical characteristics, molecular profile and outcomes of myeloid sarcoma: a single institution experience over 13 years. Hematology. (2018) 23:17–24. doi: 10.1080/10245332.2017.1333275
12. Zhao H, Dong Z, Wan D, Cao W, Xing H, Liu Z, et al. Clinical characteristics, treatment, and prognosis of 118 cases of myeloid sarcoma. Sci Rep. (2022) 12:6752. doi: 10.1038/s41598-022-10831-7
13. Wilson CS, Medeiros LJ. Extramedullary manifestations of myeloid neoplasms. Am J Clin Pathol. (2015) 144:219–39. doi: 10.1309/AJCPO58YWIBUBESX
14. Claerhout H, Van Aelst S, Melis C, Tousseyn T, Gheysens O, Vandenberghe P, et al. Clinicopathological characteristics of de novo and secondary myeloid sarcoma: A monocentric retrospective study. Eur J Haematol. (2018) 100:603–12. doi: 10.1111/ejh.13056
15. Greenland NY, Van Ziffle JA, Liu YC, Qi Z, Prakash S, Wang L. Genomic analysis in myeloid sarcoma and comparison with paired acute myeloid leukemia. Hum Pathol. (2021) 108:76–83. doi: 10.1016/j.humpath.2020.11.005
16. Pemmaraju N, Kantarjian HM, Tran KM, Kazmi SM, Kadia TM, Borthakur G, et al. Cytogenetic and molecular characterization of extramedullary disease (EMD) in patients (pts) with acute myeloid leukemia (AML). Blood. (2012) 120:2592. doi: 10.1182/blood.V120.21.2592.2592
17. De Braekeleer M, De Braekeleer E, Douet-Guilbert N. Geographic/ethnic variability of chromosomal and molecular abnormalities in leukemia. Expert Rev Anticancer Ther. (2015) 15:1093–102. doi: 10.1586/14737140.2015.1068123
18. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. (2010) 25:603–5. doi: 10.1007/s10654-010-9491-z
19. Borenstein M, Hedges LV, Higgins JT, Rothstein HR. “Meta-analysis methods based on direction and p-values,” in Introduction to Meta-Analysis. Chichester: Wiley. (2009), 325–30. doi: 10.1002/9780470743386.ch36
20. Wallace B, Schmid C, Lau J, Trikalinos T, Wallace BC, Schmid CH, et al. Analyst: software for meta-analysis of binary, continuous and diagnostic data. BMC Med Res Methodol. (2009) 9:80. doi: 10.1186/1471-2288-9-80
21. Ansari-Lari MA, Yang CF, Tinawi-Aljundi R, Cooper L, Long P, Allan RH, et al. FLT3 mutations in myeloid sarcoma. Br J Haematol. (2004) 126:785–91. doi: 10.1111/j.1365-2141.2004.05124.x
22. Creutzig U, Zimmermann M, Dworzak M, Bourquin J-P, von Neuhoff C, Sander A, et al. Additional prognostic impact of specific sites of extramedullary leukemia in pediatric AML with MLL-rearrangements but not in core-binding factor (CBF) AML. Blood. (2012) 120:2621. doi: 10.1182/blood.V120.21.2621.2621
23. Ohanian M, Alattar ML, Estrov Z, Quintas-Cardama A, Manning JT, Ravandi F, et al. Myeloid sarcoma of the orbit and ocular adnexae. Blood. (2012) 120:1463. doi: 10.1182/blood.V120.21.1463.1463
24. Tran KM, Kantarjian HM, Kazmi SM, Quintás-Cardama A, Cortes JE, Estrov Z, et al. Cytogenetic and molecular characterization of genitourinary extramedullary disease in acute myeloid leukemia. Blood. (2012) 120:4326. doi: 10.1182/blood.V120.21.4326.4326
25. Gupta AK, Radhakrishan N, Kumar KA, Chinnabhandar V, Diand V, Sachdeva A, et al. Myeloid sarcomas: A single centre experience from India. Indian J Hematol Blood Transfus. (2013) 29:286. doi: 10.1007/s12288-013-0302-z
26. Wang XI, Patel K, Lin P, Zuo Z, Bueso-Ramos CE, Raja L, et al. Clinicopathologic and molecular genetic features of de novo myeloid sarcoma. Lab Invest. (2013) 93:365.
27. Luskin MR, Huen AO, Brooks SA, Stewart C, Watt CD, Morrissette JJD, et al. NPM1 mutation is associated with leukemia cutis in acute myeloid leukemia with monocytic features. Haematologica. (2015) 100:e412–4. doi: 10.3324/haematol.2015.129346
28. Goldberg AD, Farnoud N, Goldberger C, Patel M, Famulare C, Medina J, et al. Molecular profiling in extramedullary acute myeloid leukemia reveals distinct subgroups defined by mutations in KIT and RUNX1. Blood. (2017) 130:2663. doi: 10.1182/blood.V130.Suppl_1.2663.2663
29. Stove HK, Sandahl JD, Abrahamsson J, Asdahl PH, Forestier E, Ha SY, et al. Extramedullary leukemia in children with acute myeloid leukemia: A population-based cohort study from the Nordic Society of Pediatric Hematology and Oncology (NOPHO). Pediatr Blood Cancer. (2017) 64:1–9. doi: 10.1002/pbc.26520
30. Wu Y, Yang Y, Linu T. Next-generation RNA sequencing in myeloid sarcoma tissue sample: benefit on risk stratification of myeloid sarcoma. Blood. (2017) 130:4968. doi: 10.1182/blood.V130.Suppl_1.4968.4968
31. Choi M, Jeon YK, Sun CH, Yun HS, Hong J, Shin DY, et al. RTK-RAS pathway mutation is enriched in myeloid sarcoma. Blood Cancer J. (2018) 8:43. doi: 10.1038/s41408-018-0083-6
32. Lee SH, Huang T-C, Chang-Tsu Y, Kuo Y-Y, Chiou R-J, Chuang Y-K, et al. Discrepant mutational composition between myeloid sarcoma and bone marrow leukemia revealed through targeted next generation sequencing. Blood. (2018) 132:1395. doi: 10.1182/blood-2018-99-116637
33. Wang CX, Pusic I, Anadkat MJ. Association of leukemia cutis with survival in acute myeloid leukemia. JAMA Dermatol. (2019) 155:826–32. doi: 10.1001/jamadermatol.2019.0052
34. Andrews C, Daher-Reyes G, Young T, Atenafu E, Murphy T, McNamara C, et al. Long term data demonstrating inferior survival outcomes in patients with myeloid sarcoma. HemaSphere. (2020) 4:252–3. doi: 10.1097/HS9.0000000000000404
35. Hu GH, Lu AD, Jia YP, Zuo YX, Wu J, Zhang LP. Prognostic impact of extramedullary infiltration in pediatric low-risk acute myeloid leukemia: A retrospective single-center study over 10 years. Clin Lymphoma Myeloma Leuk. (2020) 20:e813–20. doi: 10.1016/j.clml.2020.06.009
36. Karagounis TK, Rotemberg V, Geskin LJ, Jurcic JG, Mdauthors GN. NPM1 and FLT3-TKD mutations are enriched in patients with leukemia cutis. Dermatol Online J. (2020) 26:13030/qt5gc4c36x. doi: 10.5070/D3267049552
37. Xu LH, Wang Y, Chen ZY, Fang JP. Myeloid sarcoma is associated with poor clinical outcome in pediatric patients with acute myeloid leukemia. J Cancer Res Clin Oncol. (2020) 146:1011–20. doi: 10.1007/s00432-020-03128-7
38. Zhou T, Bloomquist MS, Ferguson LS, Reuther J, Marcogliese AN, Elghetany MT, et al. Pediatric myeloid sarcoma: a single institution clinicopathologic and molecular analysis. Pediatr Hematol Oncol. (2020) 37:76–89. doi: 10.1080/08880018.2019.1683107
39. Abbas HA, Reville PK, Geppner A, Rausch CR, Pemmaraju N, Ohanian M, et al. Clinical and molecular characterization of myeloid sarcoma without medullary leukemia. Leuk Lymphoma. (2021) 62:3402–10. doi: 10.1080/10428194.2021.1961235
40. De Cap MR, Wu L, Pihan G, Patel S, Tam W, Bueso-Ramos C, et al. Clinical and genomic findings of myeloid sarcoma with an emphasis on NPM1 mutations: A study from the bone marrow pathology group. Lab Invest. (2021) 101:884–5. doi: 10.1038/s41379-021-00763-4
41. Goyal K, Bhattacharyya S, Jha K, Bose D, Saha S, Singha N, et al. Extramedullary involvement in acute myeloid leukemia; A tertiary care centre experience. Indian J Hematol Blood Transfus. (2021) 37:34–5. doi: 10.1007/s12288-021-01510-0
42. Halahleh K, Alhalaseh Y, Al-Rimawi D, Da’na W, Alrabi K, Kamal N, et al. Extramedullary acute myeloid leukemia (eAML): Retrospective single center cohort study on clinico-pathological, molecular analysis and survival outcomes. Ann Med Surg (Lond). (2021) 72:102894. doi: 10.1016/j.amsu.2021.102894
43. Khan J, Nassiri M. Molecular findings in bone marrow samples of myeloid sarcoma. Am J Clin Pathol. (2021) 156:S92–3. doi: 10.1093/ajcp/aqab191.197
44. Tatarian J, Male HJ, Byrd KP, Lin TL. Incidence and characteristics of central nervous system involvement in adult acute myeloid leukemia patients. Blood. (2021) 138:2296. doi: 10.1016/j.leukres.2022.106882
45. Velagala S, Dhamne C, Srinivasan S, Gollamudi VRM, Moulik NR, Khanna N, et al. Clinical profile and outcomes of childhood acute myeloid leukemia with myeloid sarcoma. Pediatr Blood Cancer. (2021) 68:195.
46. Kim EH, Im SA, Lee JW, Kim S, Cho B. Extramedullary infiltration in pediatric acute myeloid leukemia on surveillance magnetic resonance imaging and its relationship with established risk factors. J Pediatr Hematol Oncol. (2022) 44:e713–8. doi: 10.1097/MPH.0000000000002353
47. Kuhlman JJ, McCormick BJ, Badar T, Foran JM, Rahman ZHA, Murthy HS. Clinicopathologic characteristics and treatment outcomes of de novo myeloid sarcoma. J Clin Oncol. (2022) 40. doi: 10.1200/JCO.2022.40.16_suppl.e19002
48. Ye F, Zhang H, Zhang W, Dong J, Deng W, Yang L. Clinical characteristics, pathology features and outcomes of pediatric myeloid sarcoma: A retrospective case series. Front Pediatr. (2022) 10:927894. doi: 10.3389/fped.2022.927894
49. Yang Y, Shu Y, Tang Y, Zhao S, Jia Y, Ji J, et al. RNA sequencing of myeloid sarcoma, shed light on myeloid sarcoma stratification. Cancer Med. (2023) 12:9156–66. doi: 10.1002/cam4.5654
50. Kanitthamniyom C, Wannaphut C, Pattanaprichakul P, Kungwankiattichi S, Owattanapanich W. Clinical characteristics and clinical outcome in acute myeloid leukemia patients with leukemic cutis. Thai Clin Trials Registry. (2023) 19:1–15. doi: 10.1371/journal.pone.0297805
51. Fianchi L, Quattrone M, Criscuolo M, Bellesi S, Dragonetti G, Maraglino AME, et al. Extramedullary involvement in acute myeloid leukemia. A single center ten years’ experience. Mediterr J Hematol Infect Dis. (2021) 13:e2021030. doi: 10.4084/MJHID.2021.030
52. Chang H, Brandwein J, yi Q-L, Chun K, Patterson B, Brien B. Extramedullary infiltrates of AML are associated with CD56 expression, 11q23 abnormalities and inferior clinical outcome. Leuk Res. (2004) 28:1007–11. doi: 10.1016/j.leukres.2004.01.006
53. Ovcharenko D, Stölzel F, Poitz D, Fierro F, Schaich M, Neubauer A, et al. miR-10a overexpression is associated with NPM1 mutations and MDM4 downregulation in intermediate-risk acute myeloid leukemia. Exp Hematol. (2011) 39:1030–42.e7. doi: 10.1016/j.exphem.2011.07.008
54. Al-Harbi S, Aljurf M, Mohty M, Almohareb F, Ahmed SOA. An update on the molecular pathogenesis and potential therapeutic targeting of AML with t (8,21)(q22;q22.1);RUNX1-RUNX1T1. Blood Adv. (2020) 4:229–38. doi: 10.1182/bloodadvances.2019000168
55. Saia M, Termanini A, Rizzi N, Mazza M, Barbieri E, Valli D, et al. AML1/ETO accelerates cell migration and impairs cell-to-cell adhesion and homing of hematopoietic stem/progenitor cells. Sci Rep. (2016) 6:34957. doi: 10.1038/srep34957
Keywords: acute myeloid leukemia, extramedullary blast, genetic, mutation, myeloid sarcoma
Citation: Untaaveesup S, Trithiphen S, Kulchutisin K, Rungjirajittranon T, Leelakanok N, Panyoy S, Kaokunakorn T and Owattanapanich W (2024) Genetic alterations in myeloid sarcoma among acute myeloid leukemia patients: insights from 37 cohort studies and a meta-analysis. Front. Oncol. 14:1325431. doi: 10.3389/fonc.2024.1325431
Received: 27 October 2023; Accepted: 13 February 2024;
Published: 01 March 2024.
Edited by:
Carlo Finelli, Sant’Orsola-Malpighi Polyclinic, ItalyReviewed by:
Ana Xavier, University of Alabama at Birmingham, United StatesSandeep Singh, Central University of Punjab, India
Copyright © 2024 Untaaveesup, Trithiphen, Kulchutisin, Rungjirajittranon, Leelakanok, Panyoy, Kaokunakorn and Owattanapanich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weerapat Owattanapanich, d2VlcmFwYXRvMzY3MzNAZ21haWwuY29t
†These authors share first authorship
 Suvijak Untaaveesup
Suvijak Untaaveesup Sasinipa Trithiphen
Sasinipa Trithiphen Kamolchanok Kulchutisin
Kamolchanok Kulchutisin Tarinee Rungjirajittranon
Tarinee Rungjirajittranon Nattawut Leelakanok
Nattawut Leelakanok Sujitra Panyoy
Sujitra Panyoy Thanapon Kaokunakorn4
Thanapon Kaokunakorn4 Weerapat Owattanapanich
Weerapat Owattanapanich