- 1General Practice Ward/International Medical Center Ward, General Practice Medical Center, West China Hospital, Sichuan University/West China School of Nursing, Sichuan University, Chengdu, China
- 2Emergency Department of West China Hospital, Sichuan University/West China School of Nursing, Sichuan University, Chengdu, China
Epithelial–mesenchymal transition (EMT) is a complex physiological process that transforms polarized epithelial cells into moving mesenchymal cells. Dysfunction of EMT promotes the invasion and metastasis of cancer. The architectural transcription factor high mobility group AT-hook 2 (HMGA2) is highly overexpressed in various types of cancer (e.g., colorectal cancer, liver cancer, breast cancer, uterine leiomyomas) and significantly correlated with poor survival rates. Evidence indicated that HMGA2 overexpression markedly decreased the expression of epithelial marker E-cadherin (CDH1) and increased that of vimentin (VIM), Snail, N-cadherin (CDH2), and zinc finger E-box binding homeobox 1 (ZEB1) by targeting the transforming growth factor beta/SMAD (TGFβ/SMAD), mitogen-activated protein kinase (MAPK), and WNT/beta-catenin (WNT/β-catenin) signaling pathways. Furthermore, a new class of non-coding RNAs (miRNAs, circular RNAs, and long non-coding RNAs) plays an essential role in the process of HMGA2-induced metastasis and invasion of cancer by accelerating the EMT process. In this review, we discuss alterations in the expression of HMGA2 in various types of cancer. Furthermore, we highlight the role of HMGA2-induced EMT in promoting tumor growth, migration, and invasion. More importantly, we discuss extensively the mechanism through which HMGA2 regulates the EMT process and invasion in most cancers, including signaling pathways and the interacting RNA signaling axis. Thus, the elucidation of molecular mechanisms that underlie the effects of HMGA2 on cancer invasion and patient survival by mediating EMT may offer new therapeutic methods for preventing cancer progression.
1 Introduction
The extracellular matrix (ECM) is a non-cellular support structure that exists in all tissues, and each organ has a unique ECM composition (1). Apart from a support structure for tissue architecture, the ECM is actually a dynamic compartment that modulates and regulates cell functions, such as adhesion, migration, proliferation, and differentiation (2, 3). It is composed of numerous matrix macromolecules, including collagen, laminin, elastin, fibronectin, and hyaluronic acid (4, 5). Epithelial–mesenchymal transition (EMT) is an important physiological process through which polarized epithelial cells are transformed into moving mesenchymal cells (6). The main phenotypic changes in EMT are the downregulation of E-cadherin (CDH1) and the upregulation of vimentin (VIM) (7). EMT is active during embryonic development, invasion, metastasis, and therapeutic resistance in cancer (6).
The progression of EMT is influenced by the expression of multiple transcription factors, such as high mobility group AT-hook 2 (HMGA2) (8, 9). The HMGA proteins act as non-histone components of chromatin (10) and participate in regulating chromatin structure and DNA recombination (11, 12). The HMGA family consists of two members, HMGA1 and HMGA2, which play important roles in several processes (e.g., gene regulation, cell cycle changes) (13, 14). HMGA2 is relatively abundant in the early embryo and most types of cancer; however, it exhibits lower expression in adult tissues (15, 16). Research has shown overexpression of HMGA2 in a variety of cancers [e.g., colorectal cancer (CRC), liver cancer, breast cancer (BC), and uterine leiomyomas], suggesting an essential role in tumor development and invasion (17–20). Furthermore, clinical studies have revealed a significant correlation between the expression of HMGA2 in tissue samples and the grading and metastasis of cancer, as well as the survival rate of patients with cancer (21, 22). Wu et al. demonstrated that high levels of HMGA2 were significantly correlated with poor survival of patients with BC, particularly those with stage II–III disease. In addition, gene set enrichment analysis indicated that HMGA2 expression was positively correlated with gene expression of the mesenchymal phenotype (23). HMGA2 expression is obviously increased in BC, and interference with HMGA2 can inhibit the metastasis and invasion of tumors (24). Moreover, the expression levels of EMT-related proteins were decreased after interfering with HMGA2 expression (24). Similarly, another study has shown that HMGA2 induced metastasis of human epithelial cancers by activating the expression of transforming growth factor beta type II receptor (TGFβRII) (8). Activation of the EMT process was regarded as a major driver of aggravation from tumorigenesis to metastasis (25). Moreover, an in-vitro study strongly suggested a key role of HMGA2 in EMT (26, 27). Most previous studies demonstrated that HMGA2 plays a key role in cancer growth, invasion, and the EMT phenotype, which involved the interactions of multiple signaling proteins and non-coding RNAs (28, 29).
Therefore, targeting HMGA2 for the regulation of EMT may be an important strategy for combating tumor metastasis, recurrence, and drug resistance. In this literature review, we highlight the EMT-induced role of HMGA2 in tumor development and invasion and discuss signaling pathways that may be affected by HMGA2. According to available evidence, HMGA2 suppression may be a promising target for cancer therapy.
2 Physiological function of HMGA2
The HMGA family includes four subtypes, namely, HMGA1a, HMGA1b, HMGA1c, and HMGA2 (30, 31). Among them, HMGA1a–c are different splicing products of the HMGA1 gene, while HMGA2 is a gene product of HMGA2 (32). As a member of the HMGA family, HMGA2 comprises five exons. The first three exons contain DNA-binding domains (termed AT-hook motifs) (15, 33). The “AT-hook” DNA-binding motif contains a unique palindrome sequence PGRGP, surrounded by one or two positively charged amino acids (i.e., lysine or arginine) on each side (34, 35). This special structure facilitates the binding of HMGA2 to the AT-rich regions and an acidic C-terminal tail in the small grooves of DNA. This leads to ordered changes in DNA structure and further affects several processes (e.g., changes in chromatin structure, DNA damage/repair, DNA replication and transcription) (36–38). It is also possible to activate the transcription of target genes by competing with the junction histone H1 to open dense chromatin (39). HMGA2 is highly expressed in the early developmental stage and participates in the differentiation of mesenchymal stem cells during fetal development (40). However, it remains silent in normal mature tissues, except for lung tissue, kidney tissue, and synovial tissue (41, 42). The loss of epithelial markers and the acquisition of mesenchymal markers are typical characteristics of EMT, which play a key role in embryonic development (43). It was reported that high expression of HMGA2 in cancer changes the cell phenotype from epithelial to mesenchymal (44). In non-small cell lung cancer (NSCLC), the protein phosphatase 4 regulatory subunit 1 (PPP4R1) interacts with HMGA2 to promote cell migration and metastasis via activating EMT (45). Furthermore, Kou et al. suggested that HMGA2 facilitated metastasis and the EMT process in renal cell carcinoma cells by the TGFβ/SMAD2 pathway (46). In prostate cancer, AMPK plays a critical role in the promotive effect of HMGA2 on EMT (47). Similarly, another study revealed that HMGA2 was a direct regulatory target for various EMT-related non-coding RNAs (43). Therefore, HMGA2 may be essential in regulating the EMT process in cancer (Figure 1).
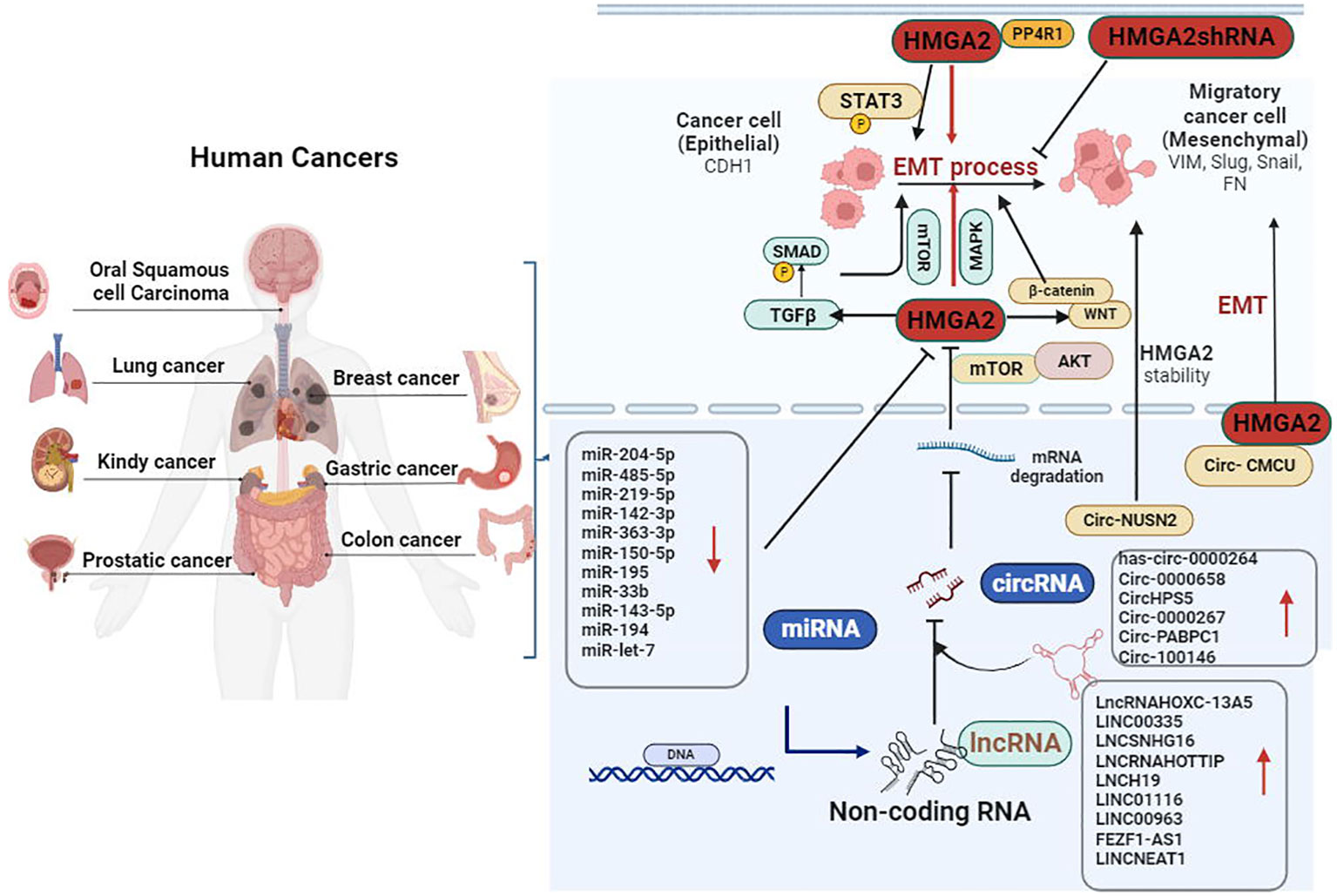
Figure 1 Dysfunction of epithelial–mesenchymal transition (EMT) is an important factor promoting the invasion and metastasis of cancer. HMGA2 overexpression decreased the expression of epithelial marker CDH1, whereas it increased the levels of VIM, Snail, Slug, and FN by targeting the TGFβ/SMAD, MAPK, and WNT/β-catenin signaling pathways. Furthermore, new classes of non-coding RNAs (miRNAs, circRNAs, and lncRNAs) play an essential role in the process of HMGA2-induced metastasis and invasion of cancer by accelerating the EMT process. CDH1, cadherin 1; circRNAs, circular RNAs; β-catenin, beta-catenin; FN, fibronectin; HMGA2, high mobility group AT-hook 2; lncRNAs, long non-coding RNAs; MAPK, mitogen-activated protein kinase; TGFβ, transforming growth factor beta; VIM, vimentin.
3 Regulatory role of the HMGA2 axis in the EMT process in cancer
3.1 Gastric cancer
EMT is a key factor in the invasion and metastasis of gastric cancer (GC) (48). It was demonstrated that the transcription factors zinc finger E-box-binding homeobox 1 (ZEB1) and Snail induced EMT by suppressing the expression of CDH1 (49–51). Analysis of surgical specimens of GC and clinical pathological data from patients with cancer showed that HMGA2 overexpression was compared with normal epithelium (52). However, HMGA2 knockdown obviously increased the expression of CDH1 and decreased that of N-cadherin (CDH2), ZEB1, and Snail (52). Moreover, similar research found that HMGA2 decreased the expression levels of Snail and β-catenin in GC cells, indicating that HMGA2 may promote the migratory capacity of GC cells by regulating EMT (53). The key role of the WNT/β-catenin pathway in regulating cell adhesion and migration is closely related to EMT (54, 55). Zha et al. demonstrated that HMGA2 induced the EMT phenotype by targeting the WNT/β-catenin pathway in MKN45 cells (56) (Figure 1). Dysfunction of non-coding RNAs has been associated with the development of malignant tumors (57, 58). Accumulating evidence indicates that HMGA2 primarily acts as a downstream factor of long non-coding RNAs (lncRNAs) for regulating tumor progression (59, 60). Overexpression of circ_0000267 in GC tissues and cell lines is related to cancer progression through regulation of the miR-503-5p/HMGA2 axis (61). In GC stem cells, it was demonstrated that knockdown of FEZF1 antisense RNA 1 (FEZF1-AS1) suppressed GC stem cell progression. Notably, FEZF1-AS1 promoted EMT, invasion, and migration of GC stem cells via the miR-363-3p/HMGA2 pathway (62). Collectively, these results indicated that HMGA2 may be a key target for inhibiting EMT in GC progression.
3.2 Lung cancer
The functions of HMGA2 in lung cancer have been studied extensively (63, 64). For instance, HMGA2 promoted proliferation, apoptosis, and EMT in lung cancer cells and was a biomarker for lung adenocarcinoma (64). Li et al. presented evidence indicating that the loss of three transcription factors (i.e., Foxa2, Cdx2, and Nkx2-1) is sufficient to induce the upregulation of Tks5long, HMGA2, and the EMT mediator Snail (65). In addition, PPP4R1 cooperated with HMGA2 to promote EMT by activating the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signaling pathway, thereby accelerating migration and invasion in NSCLC (66). The interaction between miRNAs and HMGA2 to promote EMT in lung cancer has been demonstrated. It was shown that miR-195 could target and downregulate HMGA2 to induce EMT and proliferation in lung cancer cells (43). As a negative regulator of tumors, fragile histidine triad diadenosine triphosphatase (FHIT) can inhibit metastasis in lung cancer (67). Overexpression of FHIT leads to the downregulation of the EMT-related genes VIM and fibronectin. This effect is exerted by activating miR-30c/HMGA2 to decrease HMGA2 (67). In addition, miR-150-5p expression was significantly suppressed in cancer stem cells, and this observation may be related to disease progression and poor survival in patients with lung cancer. Overexpression of miR-150-5p significantly suppressed the metastasis of NSCLC cells by directly targeting HMGA2 signaling (68). Furthermore, circular RNAs (circRNAs) also play an important regulatory role in the miRNA/HMGA2 pathway. For example, interference of circ_100565 is in favor of NSCLC cell progression by targeting the miR-506-3p/HMGA2 axis (69).
3.3 Colorectal cancer
Most previous studies in CRC demonstrated that HMGA2 contributed to disease progression and was associated with poor patient survival (20, 28, 70). EMT is a key biological process in the progression and invasion of CRC cells (71–73). Research demonstrated that HMGA2 was regulated by directly binding to the fibronectin 1 (FN1) and interleukin 11 (IL11) promoters. This process accelerates EMT in CRC cells via the phosphorylated signal transducer and activator of transcription 3-dependent (STAT3-dependent) pathway (28, 74). In addition, HMGA2 short-hairpin RNA (shRNA) attenuated the proliferation and invasion, while exogenous HMGA2 expression induced an increase in epithelial markers and a reduction in mesenchymal markers. These effects were achieved by binding to the regulatory area of Slug (75). Notably, HMGA2 plays an essential role in the regulation of the EMT phenotype in CRC, which is associated with miRNA regulation (75, 76). In CRC, highly expressed long intergenic non-protein coding RNA 963 (LINC00963) was related to poor prognosis. LINC00963 promoted the expression of EMT-related genes and invasion of CRC through the miR-532-3p/HMGA2 pathway (77). Acting as an inhibitory regulator of upstream genes, overexpression of miR-194 suppressed the expression of HMGA2, thereby improving the cell survival, EMT process, and drug resistance in CRC (78). Similarly, increased miR-330 levels also reduced the phosphorylation of AKT and STAT3 and downregulated the expression of SMAD3, Snail, and vascular endothelial growth factor A (VEGFA) by suppressing HMGA2 (44, 79). Moreover, it was demonstrated that tumor protein p53-induced (TP53-induced) miR-1249 suppressed the disease progression and angiogenesis by regulating the VEGFA-mediated AKT/mechanistic target of rapamycin kinase (AKT/mTOR) pathway. This observation further supported that TP53-induced miR-1249 inhibited the EMT in CRC by targeting VEGFA and HMGA2 (80). Numerous studies reported that dysregulated expression of circRNAs participates in the pathological processes of CRC (26, 45, 81). NOP2/Sun RNA methyltransferase 2 (NSUN2) is an N6-methyladenosine-modified circRNA highly expressed in CRC cells and patients. It has been shown to enhance the stability of HMGA2, increase CDH1 expression, and decrease VIM expression. These findings suggested that circRNA NSUN2 accelerates the EMT process in CRC cells by targeting the HMGA2 pathway (82). Additionally, circRNA 100146 was highly expressed in CRC patients and cells. Knockdown of circRNA 100146 in CRC cells disrupted the proliferation and EMT by sponging the miR-149/HMGA2 pathway (33257506). The circRNA in exosomes participated in intercellular communication and may be closely related to tumor metastasis (83). It was found that exosomal circRNA poly(A) binding protein cytoplasmic 1 (PABPC1) promoted the EMT-mediated CRC liver metastasis by increasing the expression of HMGA2 and bone morphogenetic protein 4/ADAM metallopeptidase domain (19 BMP4/ADAM19) (84). To some extent, these research studies revealed the role of HMGA2 and the potentially involved mechanism in CRC.
3.4 Breast cancer
BC remains the main cause of cancer-related disease burden in women (85). EMT leads to the development of drug resistance in BC cells, and inhibition of the EMT process can improve drug sensitivity (86, 87). Accumulating evidence has shown that HMGA2 is highly expressed in patients with BC. This high expression was positively related to advanced tumor grade. Moreover, HMGA2 enhanced the migratory and invasive abilities of BC cells by inducing EMT (23, 88, 89). Multiple signal proteins play essential roles in the process of HMGA2, participating in the metastasis and invasion of BC by accelerating EMT. Kolliopoulos et al. investigated HMGA2-depleted cells, which were stimulated with TGFβ. Genes known to be upregulated during EMT, such as Snail, serpin family E member 1 (SERPINE1), and FN1, were suppressed. This evidence reinforced the key role of HMGA2 in TGFβ-induced EMT (38). In addition, the WNT10B network β-catenin/HMGA2/enhancer of zeste 2 polycomb repressive complex 2 subunit (β-catenin/HMGA2/EZH2) signaling was related to survival and metastasis in triple-negative BC (90). Similarly, in triple-negative BC, HMGA2 suppressed the ubiquitination of Yes-associated protein (YAP) and modulated YAP stability, thereby regulating the EMT in tumors (24). Furthermore, the knockdown of combined HMGA2 and BTB domain and CNC homolog 1 (BACH1) obviously decreased cell migration and EMT. These results suggested that combined targeting of HMGA2 and BACH1 is an effective therapeutic strategy for treating BC (91). It has been indicated that cancer stem cells may arise from non-stem cancer cells upon microenvironment signals (92). A study demonstrated that the Lin-28B/let-7/HMGA2 axis was activated by STAT3/nuclear factor kappa B (STAT3/NFKB) to regulate the EMT/cancer stem cell formation; of note, HMGA2 plays a major role in this axis (93). Recently, the differential expression of clinical pathological factor-related miRNAs has been associated with HMGA2-induced EMT. For instance, miR-33b was lowly expressed in BC tissues and suppressed the EMT progress and invasion of BC despite targeting HMGA2, spalt-like transcription factor 4 (SALL4), and twist family bHLH transcription factor 1 (TWIST1) (94). Furthermore, acting as a negative regulatory factor, miR-143-5p could decrease VIM and CDH2 protein expression and increase CDH1 protein expression by directly targeting HMGA2 (95). Research that focused on the effects of modifications on miRNA has demonstrated that N6-methyladenosine modification of RNAs is crucial for cancer progression (96). Zhao et al. reported that methyltransferase 3, N6-adenosine-methyltransferase complex catalytic subunit (METTL3) regulated EMT in BC by targeting the metastasis-associated lung adenocarcinoma transcript 1/miR-26b/HMGA2 (MALAT1/miR-26b/HMGA2) pathway. This finding may provide an array of new therapeutic targets, including HMGA2, for the treatment of BC (97). In addition, new classes of non-coding RNAs (circRNAs and lncRNAs) play an essential role in the process of HMGA2-induced metastasis and invasion of BC by accelerating the EMT process. In BC, the upregulated expression of circHMCU favored disease progression; circHMCU promotes cell proliferation and metastasis by binding to HMGA2 (98). As an example of regulation of the lncRNA/HMGA2 pathway, the lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) that targeted the miR-211/HMGA2 axis can contribute to the EMT phenotype, thus promoting BC metastasis and chemoresistance (99).
3.5 Esophageal cancer
In esophageal cancer, the high expression of HMGA2 plays an essential role in regulating EMT (100). Esophageal squamous cell carcinoma (ESCC) is currently the most common type of esophageal tumors. Currently, research is mainly focused on non-coding RNAs as the molecular mechanisms related to HMGA2 during EMT in esophageal cancer (100). Mechanistic analysis revealed that miR-490-3p is bound to the 3′-untranslated region of HMGA2 to downregulate its expression. This effect suppressed the invasion, migration, and EMT of ESCC cells (101). It was elucidated that HMGA2 functions as an oncogene; silencing HMGA2 decreased the expression levels of CDH2, VIM, and Snail in ESCC cells by negatively regulating miR-204-5p (72). However, miR-125b-5p acts as an upstream target; its overexpression suppresses cell invasion by decreasing HMGA2 in ESCC (102). In addition, upregulation of hsa_circ_0006948 can promote the proliferation, migration, and invasion of ESCC cells. These effects are exerted by sponging miRNA-490-3p to increase HMGA2 expression (103).
3.6 Other types of cancer
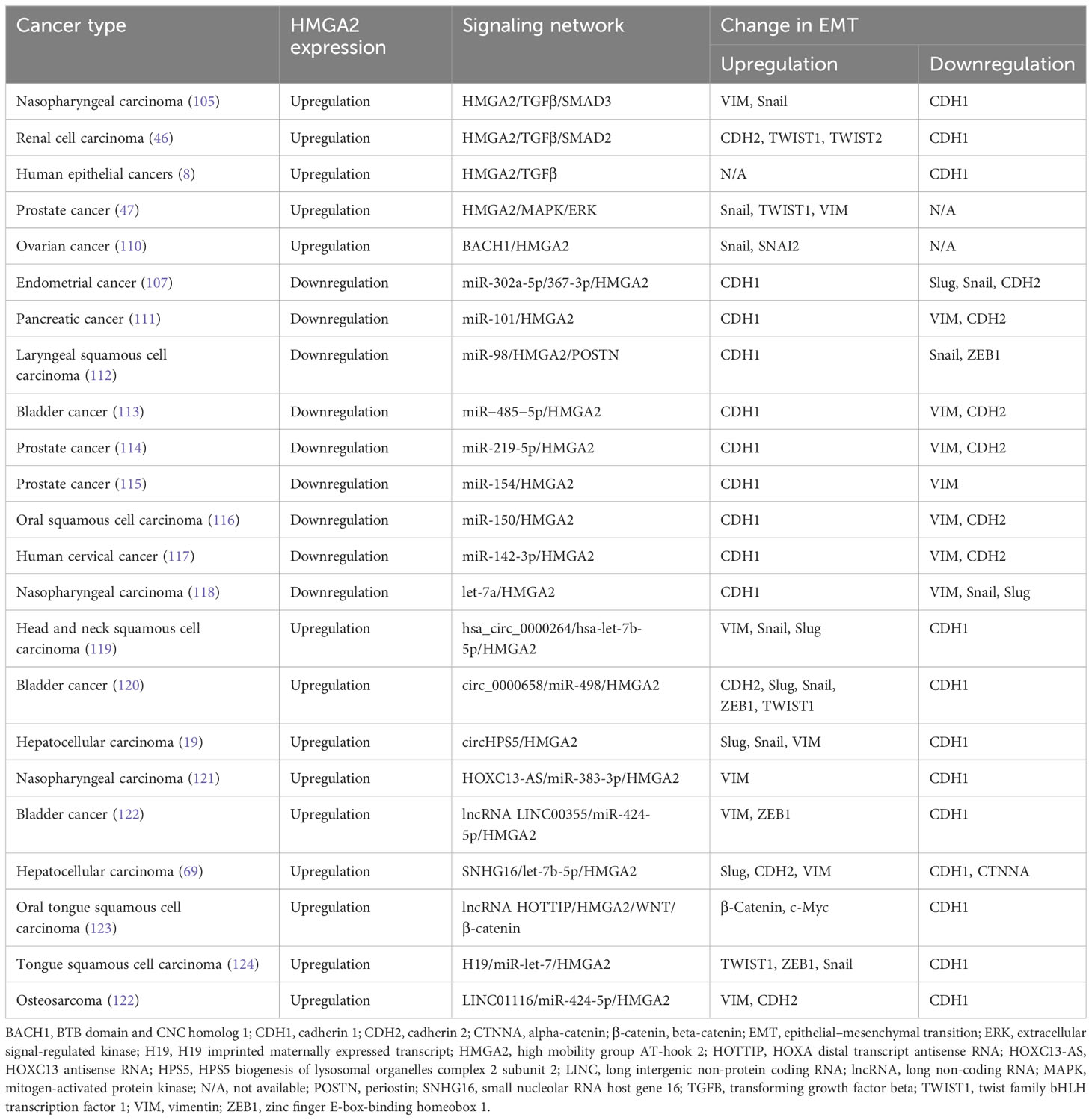
An increasing body of evidence has shown that HMGA2 is overexpressed in most tumor tissues or cancers, except for GC, NSCLC, CRC, BC, and esophageal cancer, also including thyroid cancer, bladder cancer, endometrial cancer, cervical cancer, tongue cancer, and kidney cancer (104–109) (Table 1). High expression of HMGA2 was associated with EMT and metastasis and predicted poor prognosis in patients with cancer (104–106, 125, 126). Recent research showed that exosomal HMGA2 from the Epstein–Barr virus promoted tumor metastasis and EMT (109). Data demonstrated that HMGA2 can regulate the TGFβ/SMAD and MAPK signaling pathways to induce tumor cell invasion and migration. For example, knockdown of HMGA2 significantly inhibited EMT in nasopharyngeal carcinoma cell lines by targeting the TGFβ/SMAD3 signaling pathway (105). In addition, in renal cell carcinoma cells in vitro, overexpression of HMGA2 facilitated the EMT process through the TGFβ/SMAD2 signaling pathway (46). Similarly, activation of the TGFβ signaling pathway may be a key step in EMT, induced by HMGA2 in human epithelial cancers (8). Furthermore, the upregulation of HMGA2 induced EMT phenotypes through regulation of the MAPK pathway (47). Transcription regulatory factors also promoted the transfer and expression of HMGA2, thereby promoting the growth and metastasis of ovarian cancer (110). Massive miRNAs have been clarified to affect EMT and invasion of cancers through regulating the expression of HMGA2. In endometrial cancer, cell proliferation and the EMT were promoted after HMGA2 overexpression which resulted from miR-302a-5p/367-3p downregulation (107). Furthermore, miR-302a-5p/367-3p and miR-142-3p act as tumor-suppressive miRNAs, playing a key role in the regulation of EMT by targeting HMGA2 in human cervical cancer (117). In addition, miR-219-5p and miR-154 suppressed the growth and EMT of prostate cancer cells by directly sponging the expression of HMGA2 (114, 115). In laryngeal squamous cell carcinoma, HMGA2 expression was negatively related to the levels of miR-98, and the miR-98/HMGA2/periostin (miR-98/HMGA2/POSTN) axis played an important role in reversing EMT (112). Furthermore, it has been reported that miR-101, miR-204-5p, miR-485-5p, and miR-150 reverse metastasis and EMT by targeting the HMGA2 (29, 111, 113, 116, 118). Another research study demonstrated that miR-33b reversed the EF24-mediated suppression of EMT by suppressing HMGA2 expression in melanoma (127). It was found that lncRNAs significantly affect the EMT of cancers by modulating the miRNA/HMGA2 axis. In tongue squamous cell carcinoma cells, HOXA distal transcript antisense RNA (HOTTIP) knockdown suppressed the cell migration and EMT by the miR-124-3p/HMGA2 axis, and the H19/let-7a/HMGA2/EMT pathway was also involved in the regulation of EMT (123, 124). In addition, in nasopharyngeal carcinoma, HOXC13 antisense RNA (HOXC13-AS) promoted EMT-induced invasion via regulating the miR-383-3p/HMGA2 pathway (121). Similarly, modulation of the miR-424-5p/HMGA2 pathway by LINC01116 indicated a potential pathway for overcoming the resistance of osteosarcoma to chemotherapy (122). In addition, the small nucleolar RNA host gene 16/let-7b-5p/HMGA2 (SNHG16/let-7b-5p/HMGA2) axis and the lncRNA LINC00355/miR-424-5p/HMGA2 axis play an important role in the EMT of cancers. Suppression of these signaling axes can prevent tumor metastasis (69, 122, 128, 129). Apart from lncRNAs, circRNAs (as upstream regulatory targets) affect EMT in cancers by modulating the miRNA/HMGA2 axis. A study indicated that hsa_circ_0000264 may serve as a target for the treatment of head and neck squamous cell carcinoma-EMT by regulating the hsa-let-7b-5p/HMGA2 pathway (119). Studies showed that circ_0000658 was highly upregulated in bladder cancer; nevertheless, circ_0000658 knockdown reduced the EMT phenotypes by regulating the miR-498/HMGA2 pathway (120).
4 Gene therapy methods
4.1 Short-interfering RNA
RNA interference (RNAi) is among the most commonly used and important gene therapy methods, involving the use of short-interfering RNAs (siRNAs) (130). The term siRNA refers to a sequence specifically designed to silence the expression of a target gene. Such sequences are currently used in cancer research (in-vitro cells or in-vivo animal models), providing promising options for the targeted treatment of cancer and other diseases (131, 132). Research demonstrated that the transfection of cells with HMGA2 siRNA markedly suppressed HMGA2 expression, reduced the levels of EMT-related genes, and alleviated the migratory capacity of A549 cells (131). In ACHN cells, the expression of CDH1 was upregulated, whereas that of CDH2 and Snail was downregulated in the tumors treated with HMGA2 shRNA. These results implied that HMGA2 shRNA may be a treatment strategy for renal cell carcinoma (108). In recent years, nanoparticles have attracted attention due to their application in RNAi. The selection of nanoparticles for the molecular delivery of RNAi is mainly attributed to their unique advantages over other carriers (130). Eivazy et al. delivered HMGA2 siRNA by trimethyl chitosan nanoparticles. The delivery of HMGA2 siRNA significantly reduced the expression of HMGA2 and VIM, whereas it increased that of CDH1 (133).
4.2 Clustered regular interval short palindromic repeat sequences/CRISPR-associated protein 9 RNA
Clustered regular interval short palindromic repeat sequences/CRISPR-associated protein 9 (CRISPR/Cas9) RNA nucleases are a powerful reverse-genetic tool that can easily achieve targeted editing of multiple genes, thereby inducing complete gene knockout (134). The use of the CRISPR/Cas9 system for targeted gene therapy against tumors has been widely reported. In doxorubicin-resistant BC cells, flow cytometric analysis showed that targeting MDR1 using the CRISPR/Cas9 system increased drug accumulation within the cell compared with untreated cells (135). Song et al. found an indirect increase in the expression levels of HMGA2 and SRY-box transcription factor 9 (SOX9) after CRISPR/Cas9-based knockout of neurofibromin 1 (NF1; a tumor suppressor mutated in neurofibromatosis). These data suggested that NF1 plays a key role as a liver tumor suppressor by negatively regulating HMGA2 and that NF1 and HMGA2 may be useful prognostic or therapeutic indicators (136). In addition, in papillary thyroid carcinoma cells, the CRISPR/Cas9-mediated knockout of HMGA2 inhibited cell proliferation and invasion. It was suggested that HMGA2 knockout blocked the cell cycle in the G2/M phase and promoted cell necrosis (135). The results mentioned above showed that targeting HMGA2 using CRISPR/Cas9 technology can reduce drug resistance in cancers. Using the CRISPR/Cas9 technology and targeting HMGA2 could inhibit the progression of cancer. However, further investigation should be conducted in various types of cancer. Furthermore, the CRISPR/Cas9 technique has limitations, including safe and efficient cell delivery, off-target mutagenesis, and potential immunogenicity. Hence, effective solutions are required to overcome the limitations of this technique (127, 137, 138).
4.3 Proteolysis-targeting chimeras
At present, proteolysis-targeting chimeras (PROTACs) have been developed as a useful technology for targeted protein degradation (139). Designed hydrophobic tagging (HyT) probes are synthesized by covalently connecting the hydrophobic portion to the ligand of target nuclear proteins [protein of interest (POI)] (140, 141). The binary POI–HyT complex can simulate the partial denaturation state of protein degradation, and the most commonly used hydrophobic parts include adamantane and tert-butyl carbamate (BOC3) arginine (141). PROTACs can induce the dynamic degradation of intracellular proteins or POIs. Thus, they play an important role in addressing drug resistance by degrading the pathogenic protein without compensatory increase or mutation (142). Unlike nucleic acid-based techniques for protein regulation, such as RNAi and CRISPR/Cas9, these low immunogenicity chimeras cause reversible and rapid target depletion (143). In addition, PROTACs can be recovered after POI ubiquitination and degradation, allowing these molecules to recatalyze the elimination of additional POIs (143). Thus far, the treatment strategy involving the use of PROTACs has been successfully applied to conditionally degrade approximately 50 proteins in vitro and in vivo, including bromodomain containing 4-targeting (BRD4-targeting) PROTACs, cereblon-based (CRBN-based) PROTACs, MCL1 apoptosis regulator, BCL2 family member-based (MCL1-based) PROTACs, and STAT3-based PROTACs (144). For example, Wang et al. developed the efficient STAT3 inhibitor SI-109 and used it to develop PROTAC SD-36 targeting STAT3. The results showed that, at low nanomolar concentrations, SD-36 effectively reduces STAT3 in numerous types of leukemia and lymphoma cells (145). Furthermore, Crews et al. synthesized the first PROTAC DAS-2-2-6-CRBN targeting BCR-ABL. This PROTAC resulted in efficient BCR-ABL degradation and growth inhibition in chronic myeloid leukemia K562 cells (146). Research has revealed the presence of >600 E3 ubiquitin ligases in humans; many of those can be used to design PROTACs (147). Thus, further research studies are warranted to identify alternative therapies based on PROTAC-mediated degradation of HMGA2.
5 Conclusions and perspectives
HMGA2 is overexpressed in multiple types of cancer and has been associated with the EMT process and tumor invasion. Thus, targeting HMGA2 may provide multiple benefits in terms of tumor growth, the EMT phenotype, metastasis, and invasion. These effects indicate that HMGA2 is a promising target for enhancing cancer therapy and improving the patient survival rate. Most recent studies have demonstrated that HMGA2 is highly expressed in cancer and linked to the EMT, invasion, and poor prognosis. HMGA2 acts as a key factor in the complex networks of the TGFβ, MAPK, and WNT/β-catenin signaling pathways involved in the EMT process and invasion of tumor cells. Furthermore, most non-coding RNAs (miRNAs, lncRNAs, and circRNAs) participate in the regulation of HMGA2 expression in cancer to affect EMT. In addition, evidence has indicated that HMGA2 siRNA and CRISPR/Cas9-mediated knockout of HMGA2 serve as potential therapeutic approaches by suppressing HMGA2 for the treatment of cancer. However, currently, there are limited treatment options targeting the inhibition of HMGA2 expression to mitigate EMT and invasion of cancer. For example, small molecule inhibitors targeting HMGA2 have not yet been studied or identified. Thus far, there is a lack of drugs targeting HMGA2 to delay EMT and invasion of cancer. Gene modification strategies (e.g., acetylation, methylation, and ubiquitination) targeting HMGA2 are urgently required for the treatment of cancer. This approach may suppress EMT and increase the survival rate of patients with cancer. Furthermore, it is necessary to elucidate the specific molecular mechanism through which HMGA2 mediates the EMT process in cancer. Such knowledge will contribute to the discovery of more effective treatment strategies for inhibiting tumor metastasis and controlling resistance to chemotherapy.
In conclusion, targeting HMGA2 through direct and indirect regulation offers a promising direction for antitumor therapy.
Author contributions
QM: Formal Analysis, Writing – original draft. SY: Writing – original draft, Formal Analysis. HL: Conceptualization, Investigation, Writing – review & editing. YZ: Conceptualization, Writing – review & editing, Supervision. YM: Conceptualization, Writing – review & editing, Investigation. WZ: Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study is supported by the Department of Science and Technology of Sichuan Province (2023YFS0240 to WZ). and Cadre Health Research Project of Sichuan Province (Chuangan Yan2023-114 to QM).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
BC, breast cancer; CRC, colorectal cancer; ECM, extracellular matrix; EMT, epithelial–mesenchymal transition; ESCC, esophageal squamous cell carcinoma; FHIT, fragile histidine triad diadenosine triphosphatase; GC, gastric cancer; HMGA2, high mobility group AT-hook 2; NSCLC, non-small cell lung cancer; PPP4R1, protein phosphatase 4 regulatory subunit 1; ZEB1, zinc finger E-box-binding homeobox 1.
References
1. Hynes RO. The extracellular matrix: not just pretty fibrils. Science (2009) 326:1216–9. doi: 10.1126/science.1176009
2. Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol (2014) 15:786–801. doi: 10.1038/nrm3904
3. Paolillo M, Schinelli S. Extracellular matrix alterations in metastatic processes. Int J Mol Sci (2019) 20:4947. doi: 10.3390/ijms20194947
4. Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, Hynes RO. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol (2016) 49:10–24. doi: 10.1016/j.matbio.2015.06.003
5. Jurj A, Ionescu C, Berindan-Neagoe I, Braicu C. The extracellular matrix alteration, implication in modulation of drug resistance mechanism: friends or foes? J Exp Clin Cancer Res (2022) 41:276. doi: 10.1186/s13046-022-02484-1
6. Ren Y, Mao X, Xu H, Dang Q, Weng S, Zhang Y, et al. Ferroptosis and EMT: key targets for combating cancer progression and therapy resistance. Cell Mol Life Sci (2023) 80:263. doi: 10.1007/s00018-023-04907-4
7. Pastushenko I, Mauri F, Song Y, de Cock F, Meeusen B, Swedlund B, et al. Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature (2021) 589:448–55. doi: 10.1038/s41586-020-03046-1
8. Morishita A, Zaidi MR, Mitoro A, Sankarasharma D, Szabolcs M, Okada Y, et al. HMGA2 is a driver of tumor metastasis. Cancer Res (2013) 73:4289–99. doi: 10.1158/0008-5472.CAN-12-3848
9. Huang Y, Hong W, Wei X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J Hematol Oncol (2022) 15:129. doi: 10.1186/s13045-022-01347-8
10. Goodwin GH, Sanders C, Johns EW. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem (1973) 38:14–9. doi: 10.1111/j.1432-1033.1973.tb03026.x
11. Bai J, Yokomizo-Nakano T, Kubota S, Sun Y, Kanai A, Iimori M, et al. Overexpression of Hmga2 activates Igf2bp2 and remodels transcriptional program of Tet2-deficient stem cells in myeloid transformation. Oncogene (2021) 40:1531–41. doi: 10.1038/s41388-020-01629-w
12. Hashemi M, Rashidi M, Hushmandi K, Ten Hagen TLM, Salimimoghadam S, Taheriazam A, et al. HMGA2 regulation by miRNAs in cancer: Affecting cancer hallmarks and therapy response. Pharmacol Res (2023) 190:106732. doi: 10.1016/j.phrs.2023.106732
13. Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene (2001) 277:63–81. doi: 10.1016/S0378-1119(01)00689-8
14. Young AR, Narita M. Oncogenic HMGA2: short or small? Genes Dev (2007) 21:1005–9. doi: 10.1101/gad.1554707
15. Zhou X, Chada K. HMGI family proteins: architectural transcription factors in mammalian development and cancer. Keio J Med (1998) 47:73–7. doi: 10.2302/kjm.47.73
16. Li M, Zhao H, Zhao SG, Wei DM, Zhao YR, Huang T, et al. The HMGA2-IMP2 pathway promotes granulosa cell proliferation in polycystic ovary syndrome. J Clin Endocrinol Metab (2019) 104:1049–59. doi: 10.1210/jc.2018-00544
17. Zhao W, Geng D, Li S, Chen Z, Sun M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med (2018) 7:842–55. doi: 10.1002/cam4.1353
18. Liu B, Chen G, He Q, Liu M, Gao K, Cai B, et al. An HMGA2-p62-ERα axis regulates uterine leiomyomas proliferation. FASEB J (2020) 34:10966–83. doi: 10.1096/fj.202000520R
19. Rong D, Wu F, Lu C, Sun G, Shi X, Chen X, et al. m6A modification of circHPS5 and hepatocellular carcinoma progression through HMGA2 expression. Mol Ther Nucleic Acids (2021) 26:637–48. doi: 10.1016/j.omtn.2021.09.001
20. Wang X, Wang J, Zhao J, Wang H, Chen J, Wu J. HMGA2 facilitates colorectal cancer progression via STAT3-mediated tumor-associated macrophage recruitment. Theranostics (2022) 12:963–75. doi: 10.7150/thno.65411
21. Meyer B, Loeschke S, Schultze A, Weigel T, Sandkamp M, Goldmann T, et al. HMGA2 overexpression in non-small cell lung cancer. Mol Carcinog (2007) 46:503–11. doi: 10.1002/mc.20235
22. Yang F, Zhao L, Mei D, Jiang L, Geng C, Li Q, et al. HMGA2 plays an important role in Cr (VI)-induced autophagy. Int J Cancer (2017) 141:986–97. doi: 10.1002/ijc.30789
23. Wu J, Wang Y, Xu X, Cao H, Sahengbieke S, Sheng H, et al. Transcriptional activation of FN1 and IL11 by HMGA2 promotes the Malignant behavior of colorectal cancer. Carcinogenesis (2016) 37:511–21. doi: 10.1093/carcin/bgw029
24. Xu J, Fang X, Long L, Wang S, Qian S, Lyu J. HMGA2 promotes breast cancer metastasis by modulating Hippo-YAP signaling pathway. Cancer Biol Ther (2021) 22:5–11. doi: 10.1080/15384047.2020.1832429
25. Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol (2017) 19:518–29. doi: 10.1038/ncb3513
26. Wang J, Zhang Y, Song H, Yin H, Jiang T, Xu Y, et al. The circular RNA circSPARC enhances the migration and proliferation of colorectal cancer by regulating the JAK/STAT pathway. Mol Cancer (2021) 20:81. doi: 10.1186/s12943-021-01375-x
27. Wang X, Wang J, Wu J. Emerging roles for HMGA2 in colorectal cancer. Transl Oncol (2021) 14:100894. doi: 10.1016/j.tranon.2020.100894
28. Wang YC, Liu JS, Tang HK, Nie J, Zhu JX, Wen LL, et al. miR−221 targets HMGA2 to inhibit bleomycin−induced pulmonary fibrosis by regulating TGF−β1/Smad3-induced EMT. Int J Mol Med (2016) 38:1208–16. doi: 10.3892/ijmm.2016.2705
29. Van Branteghem C, Augenlicht A, Demetter P, Craciun L, Maenhaut C. Unraveling the roles of miR-204-5p and HMGA2 in papillary thyroid cancer tumorigenesis. Int J Mol Sci Jun (2023) 28:24. doi: 10.3390/ijms241310764
30. Bianchi ME, Beltrame M. Upwardly mobile proteins. Workshop: the role of HMG proteins in chromatin structure, gene expression and neoplasia. EMBO Rep (2000) 1:109–14. doi: 10.1093/embo-reports/kvd030
31. Dement GA, Treff NR, Magnuson NS, Franceschi V, Reeves R. Dynamic mitochondrial localization of nuclear transcription factor HMGA1. Exp Cell Res (2005) 307:388–401. doi: 10.1016/j.yexcr.2005.04.004
32. Friedmann M, Holth LT, Zoghbi HY, Reeves R. Organization, inducible-expression and chromosome localization of the human HMG-I(Y) nonhistone protein gene. Nucleic Acids Res (1993) 21:4259–67. doi: 10.1093/nar/21.18.4259
33. Unachukwu U, Chada K, D’Armiento J. High mobility group AT-hook 2 (HMGA2) oncogenicity in mesenchymal and epithelial neoplasia. Int J Mol Sci (2020) 21:3151. doi: 10.3390/ijms21093151
34. Reeves R, Nissen MS. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem (1990) 265:8573–82. doi: 10.1016/S0021-9258(19)38926-4
35. Su L, Bryan N, Battista S, Freitas J, Garabedian A, D’Alessio F, et al. Identification of HMGA2 inhibitors by AlphaScreen-based ultra-high-throughput screening assays. Sci Rep (2020) 10:18850. doi: 10.1038/s41598-020-75890-0
36. Cleynen I, Brants JR, Peeters K, Deckers R, Debiec-Rychter M, Sciot R, et al. HMGA2 regulates transcription of the Imp2 gene via an intronic regulatory element in cooperation with nuclear factor-kappaB. Mol Cancer Res (2007) 5:363–72. doi: 10.1158/1541-7786.MCR-06-0331
37. Mansoori B, Mohammadi A, Shirjang S, Baradaran B. HMGI-C suppressing induces P53/caspase9 axis to regulate apoptosis in breast adenocarcinoma cells. Cell Cycle (2016) 15:2585–92. doi: 10.1080/15384101.2016.1190892
38. Divisato G, Chiariello AM, Esposito A, Zoppoli P, Zambelli F, Elia MA, et al. Hmga2 protein loss alters nuclear envelope and 3D chromatin structure. BMC Biol (2022) 20:171. doi: 10.1186/s12915-022-01375-3
39. Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer (2007) 7:899–910. doi: 10.1038/nrc2271
40. Li O, Li J, Dröge P. DNA architectural factor and proto-oncogene HMGA2 regulates key developmental genes in pluripotent human embryonic stem cells. FEBS Lett (2007) 581:3533–7. doi: 10.1016/j.febslet.2007.06.072
41. Broberg K, Tallini G, Höglund M, Lindstrand A, Toksvig-Larsen S, Mertens F. The tumor-associated gene HMGIC is expressed in normal and osteoarthritis-affected synovia. Mod Pathol (2001) 14:311–7. doi: 10.1038/modpathol.3880308
42. Nagaishi M, Nakae R, Matsumoto Y, Fujii Y, Sugiura Y, Takigawa T, et al. High HMGA2 expression without gene rearrangement in meningiomas. Neuropathology (2020) 40:540–5. doi: 10.1111/neup.12670
43. Gao X, Dai M, Li Q, Wang Z, Lu Y, Song Z. HMGA2 regulates lung cancer proliferation and metastasis. Thorac Cancer (2017) 8:501–10. doi: 10.1111/1759-7714.12476
44. Mansoori B, Duijf PHG, Mohammadi A, Najafi S, Roshani E, Shanehbandi D, et al. Overexpression of HMGA2 in breast cancer promotes cell proliferation, migration, invasion and stemness. Expert Opin Ther Targets Mar (2020) 14:1–11. doi: 10.1080/14728222.2020.1736559
45. Wang B, Pan LY, Kang N, Shen XY. PP4R1 interacts with HMGA2 to promote non-small-cell lung cancer migration and metastasis via activating MAPK/ERK-induced epithelial-mesenchymal transition. Mol Carcinog (2020) 59:467–77. doi: 10.1002/mc.23168
46. Kou B, Liu W, Tang X, Kou Q. HMGA2 facilitates epithelial-mesenchymal transition in renal cell carcinoma by regulating the TGF-β/Smad2 signaling pathway. Oncol Rep (2018) 39:101–8. doi: 10.3892/or.2017.6091
47. Hawsawi O, Henderson V, Burton LJ, Dougan J, Nagappan P, Odero-Marah V. High mobility group A2 (HMGA2) promotes EMT via MAPK pathway in prostate cancer. Biochem Biophys Res Commun (2018) 504:196–202. doi: 10.1016/j.bbrc.2018.08.155
48. Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell (2008) 14:818–29. doi: 10.1016/j.devcel.2008.05.009
49. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell (2009) 139:871–90. doi: 10.1016/j.cell.2009.11.007
50. Jing Y, Han Z, Zhang S, Liu Y, Wei L. Epithelial-Mesenchymal Transition in tumor microenvironment. Cell Biosci (2011) 1:29. doi: 10.1186/2045-3701-1-29
51. Macrì S, Simula L, Pellarin I, Pegoraro S, Onorati M, Sgarra R, et al. Hmga2 is required for neural crest cell specification in Xenopus laevis. Dev Biol (2016) 411:25–37. doi: 10.1016/j.ydbio.2016.01.014
52. Lee J, Ha S, Jung CK, Lee HH. High-mobility-group A2 overexpression provokes a poor prognosis of gastric cancer through the epithelial-mesenchymal transition. Int J Oncol (2015) 46:2431–8. doi: 10.3892/ijo.2015.2947
53. Sun J, Sun B, Zhu D, Zhao X, Zhang Y, Dong X, et al. HMGA2 regulates CD44 expression to promote gastric cancer cell motility and sphere formation. Am J Cancer Res (2017) 7:260–74.
54. Tang Q, Chen J, Di Z, Yuan W, Zhou Z, Liu Z, et al. TM4SF1 promotes EMT and cancer stemness via the Wnt/β-catenin/SOX2 pathway in colorectal cancer. J Exp Clin Cancer Res (2020) 39:232. doi: 10.1186/s13046-020-01690-z
55. Hu L, Ding M, He W. Emerging therapeutic strategies for attenuating tubular EMT and kidney fibrosis by targeting wnt/β-catenin signaling. Front Pharmacol (2021) 12:830340. doi: 10.3389/fphar.2021.830340
56. Zha L, Zhang J, Tang W, Zhang N, He M, Guo Y, et al. HMGA2 elicits EMT by activating the Wnt/β-catenin pathway in gastric cancer. Dig Dis Sci (2013) 58:724–33. doi: 10.1007/s10620-012-2399-6
57. De Andrade Costa A, Chatterjee J, Cobb O, Sanapala S, Scheaffer S, Guo X, et al. RNA sequence analysis reveals ITGAL/CD11A as a stromal regulator of murine low-grade glioma growth. Neuro Oncol (2022) 24:14–26. doi: 10.1093/neuonc/noab130
58. Tan K, Stupack DG, Wilkinson MF. Nonsense-mediated RNA decay: an emerging modulator of Malignancy. Nat Rev Cancer (2022) 22:437–51. doi: 10.1038/s41568-022-00481-2
59. Mo J, Li B, Zhou Y, Xu Y, Jiang H, Cheng X, et al. LINC00473 promotes hepatocellular carcinoma progression via acting as a ceRNA for microRNA-195 and increasing HMGA2 expression. BioMed Pharmacother (2019) 120:109403. doi: 10.1016/j.biopha.2019.109403
60. Tong H, Zhuang X, Cai J, Ding Y, Si Y, Zhang H, et al. Long noncoding RNA ZFAS1 promotes progression of papillary thyroid carcinoma by sponging miR-590-3p and upregulating HMGA2 expression. Onco Targets Ther (2019) 12:7501–12. doi: 10.2147/OTT.S209138
61. Cai X, Nie J, Chen L, Yu F. Circ_0000267 promotes gastric cancer progression via sponging MiR-503-5p and regulating HMGA2 expression. Mol Genet Genomic Med (2020) 8:e1093. doi: 10.1002/mgg3.1093
62. Hui Y, Yang Y, Li D, Wang J, Di M, Zhang S, et al. LncRNA FEZF1-AS1 modulates cancer stem cell properties of human gastric cancer through miR-363-3p/HMGA2. Cell Transplant (2020) 29:963689720925059. doi: 10.1177/0963689720925059
63. Xu L, Liao WL, Lu QJ, Li CG, Yuan Y, Xu ZY, et al. ANG promotes proliferation and invasion of the cell of lung squamous carcinoma by directly up-regulating HMGA2. J Cancer (2016) 7:862–71. doi: 10.7150/jca.14440
64. Xu L, Ma Y, Zhang H, Lu QJ, Yang L, Jiang GN, et al. HMGA2 regulates circular RNA ASPH to promote tumor growth in lung adenocarcinoma. Cell Death Dis (2020) 11:593. doi: 10.1038/s41419-020-2726-3
65. Li CM, Gocheva V, Oudin MJ, Bhutkar A, Wang SY, Date SR, et al. Foxa2 and Cdx2 cooperate with Nkx2-1 to inhibit lung adenocarcinoma metastasis. Genes Dev (2015) 29:1850–62. doi: 10.1101/gad.267393.115
66. Siddiqui MA, Gollavilli PN, Ramesh V, Parma B, Schwab A, Vazakidou ME, et al. Thymidylate synthase drives the phenotypes of epithelial-to-mesenchymal transition in non-small cell lung cancer. Br J Cancer (2021) 124:281–9. doi: 10.1038/s41416-020-01095-x
67. Suh SS, Yoo JY, Cui R, Kaur B, Huebner K, Lee TK, et al. FHIT suppresses epithelial-mesenchymal transition (EMT) and metastasis in lung cancer through modulation of microRNAs. PloS Genet (2014) 10:e1004652. doi: 10.1371/journal.pgen.1004652
68. Dai FQ, Li CR, Fan XQ, Tan L, Wang RT, Jin H. miR-150-5p inhibits non-small-cell lung cancer metastasis and recurrence by targeting HMGA2 and β-catenin signaling. Mol Ther Nucleic Acids (2019) 16:675–85. doi: 10.1016/j.omtn.2019.04.017
69. Li L, Wei H, Zhang H, Xu F, Che G. Circ_100565 promotes proliferation, migration and invasion in non-small cell lung cancer through upregulating HMGA2 via sponging miR-506-3p. Cancer Cell Int (2020) 20:160. doi: 10.1186/s12935-020-01241-8
70. Chen CH, Hsieh YC, Yang PM, Liu YR, Cho EC. Dicoumarol suppresses HMGA2-mediated oncogenic capacities and inhibits cell proliferation by inducing apoptosis in colon cancer. Biochem Biophys Res Commun (2020) 524:1003–9. doi: 10.1016/j.bbrc.2020.01.147
71. Yang Y, Feng M, Bai L, Liao W, Zhou K, Zhang M, et al. Comprehensive analysis of EMT-related genes and lncRNAs in the prognosis, immunity, and drug treatment of colorectal cancer. J Transl Med (2021) 19:391. doi: 10.1186/s12967-021-03065-0
72. Zhang H, Wu X, Sui Z, Ma Z, Gong L, Meng B, et al. High-mobility group AT-hook 2 promotes growth and metastasis and is regulated by miR-204-5p in oesophageal squamous cell carcinoma. Eur J Clin Invest (2021) 51:e13563. doi: 10.1111/eci.13563
73. Zhang N, Ng AS, Cai S, Li Q, Yang L, Kerr D. Novel therapeutic strategies: targeting epithelial-mesenchymal transition in colorectal cancer. Lancet Oncol (2021) 22:e358–68. doi: 10.1016/S1470-2045(21)00343-0
74. Wang Y, Hu L, Wang J, Li X, Sahengbieke S, Wu J, et al. HMGA2 promotes intestinal tumorigenesis by facilitating MDM2-mediated ubiquitination and degradation of p53. J Pathol (2018) 246:508–18. doi: 10.1002/path.5164
75. Li Y, Zhao Z, Xu C, Zhou Z, Zhu Z, You T. HMGA2 induces transcription factor Slug expression to promote epithelial-to-mesenchymal transition and contributes to colon cancer progression. Cancer Lett (2014) 355:130–40. doi: 10.1016/j.canlet.2014.09.007
76. Cai K, Yang Y, Guo ZJ, Cai RL, Hashida H, Li HX. Amentoflavone inhibits colorectal cancer epithelial-mesenchymal transition via the miR-16-5p/HMGA2/β-catenin pathway. Ann Transl Med (2022) 10:1009. doi: 10.21037/atm-22-3035
77. Ye J, Liu J, Tang T, Xin L, Bao X, Yan Y. LINC00963 affects the development of colorectal cancer via MiR-532-3p/HMGA2 axis. Cancer Cell Int (2021) 21:87. doi: 10.1186/s12935-020-01706-w
78. Chang HY, Ye SP, Pan SL, Kuo TT, Liu BC, Chen YL, et al. Overexpression of miR-194 reverses HMGA2-driven signatures in colorectal cancer. Theranostics (2017) 7:3889–900. doi: 10.7150/thno.20041
79. Mansoori B, Mohammadi A, Naghizadeh S, Gjerstorff M, Shanehbandi D, Shirjang S, et al. miR-330 suppresses EMT and induces apoptosis by downregulating HMGA2 in human colorectal cancer. J Cell Physiol (2020) 235:920–31. doi: 10.1002/jcp.29007
80. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun (2019) 10:4695. doi: 10.1038/s41467-019-12651-2
81. Chen LY, Wang L, Ren YX, Pang Z, Liu Y, Sun XD, et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation. Mol Cancer (2020) 19:164. doi: 10.1186/s12943-020-01272-9
82. Chen X, Zeng K, Xu M, Liu X, Hu X, Xu T, et al. P53-induced miR-1249 inhibits tumor growth, metastasis, and angiogenesis by targeting VEGFA and HMGA2. Cell Death Dis (2019) 10:131. doi: 10.1038/s41419-018-1188-3
83. Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer (2019) 18:116. doi: 10.1186/s12943-019-1041-z
84. Li Y, Hu J, Wang M, Yuan Y, Zhou F, Zhao H, et al. Exosomal circPABPC1 promotes colorectal cancer liver metastases by regulating HMGA2 in the nucleus and BMP4/ADAM19 in the cytoplasm. Cell Death Discovery (2022) 8:335. doi: 10.1038/s41420-022-01124-z
85. Britt KL, Cuzick J, Phillips KA. Key steps for effective breast cancer prevention. Nat Rev Cancer (2020) 20:417–36. doi: 10.1038/s41568-020-0266-x
86. Raza U, Saatci Ö, Uhlmann S, Ansari SA, Eyüpoğlu E, Yurdusev E, et al. The miR-644a/CTBP1/p53 axis suppresses drug resistance by simultaneous inhibition of cell survival and epithelial-mesenchymal transition in breast cancer. Oncotarget (2016) 7:49859–77. doi: 10.18632/oncotarget.10489
87. Hashemi M, Arani HZ, Orouei S, Fallah S, Ghorbani A, Khaledabadi M, et al. EMT mechanism in breast cancer metastasis and drug resistance: Revisiting molecular interactions and biological functions. BioMed Pharmacother (2022) 155:113774. doi: 10.1016/j.biopha.2022.113774
88. Mansoori B, Terp MG, Mohammadi A, Pedersen CB, Ditzel HJ, Baradaran B, et al. HMGA2 supports cancer hallmarks in triple-negative breast cancer. Cancers (Basel) (2021) 16:13. doi: 10.3390/cancers13205197
89. Wu J, Zhang S, Shan J, Hu Z, Liu X, Chen L, et al. Elevated HMGA2 expression is associated with cancer aggressiveness and predicts poor outcome in breast cancer. Cancer Lett (2016) 376:284–92. doi: 10.1016/j.canlet.2016.04.005
90. El Ayachi I, Fatima I, Wend P, Alva-Ornelas JA, Runke S, Kuenzinger WL, et al. The WNT10B network is associated with survival and metastases in chemoresistant triple-negative breast cancer. Cancer Res (2019) 79:982–93. doi: 10.1158/0008-5472.CAN-18-1069
91. Mansoori B, Mohammadi A, Asadzadeh Z, Shirjang S, Minouei M, Abedi Gaballu F, et al. HMGA2 and Bach-1 cooperate to promote breast cancer cell Malignancy. J Cell Physiol (2019) 234:17714–26. doi: 10.1002/jcp.28397
92. Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell (2011) 146:633–44. doi: 10.1016/j.cell.2011.07.026
93. Guo L, Cheng X, Chen H, Chen C, Xie S, Zhao M, et al. Induction of breast cancer stem cells by M1 macrophages through Lin-28B-let-7-HMGA2 axis. Cancer Lett (2019) 452:213–25. doi: 10.1016/j.canlet.2019.03.032
94. Lin Y, Liu AY, Fan C, Zheng H, Li Y, Zhang C, et al. MicroRNA-33b inhibits breast cancer metastasis by targeting HMGA2, SALL4 and twist1. Sci Rep (2015) 5:9995. doi: 10.1038/srep09995
95. Mansoori B, Kiani S, Mezajin AA, Zandi P, Banaie H, Rostamzadeh D, et al. MicroRNA-143-5p suppresses ER-positive breast cancer development by targeting oncogenic HMGA2. Clin Breast Cancer (2023) 23:e480–e490. doi: 10.1016/j.clbc.2023.07.011
96. Coker H, Wei G, Brockdorff N. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim Biophys Acta Gene Regul Mech (2019) 1862:310–8. doi: 10.1016/j.bbagrm.2018.12.002
97. Zhao C, Ling X, Xia Y, Yan B, Guan Q. The m6A methyltransferase METTL3 controls epithelial-mesenchymal transition, migration and invasion of breast cancer through the MALAT1/miR-26b/HMGA2 axis. Cancer Cell Int (2021) 21:441. doi: 10.1186/s12935-021-02113-5
98. Song X, Liang Y, Sang Y, Li Y, Zhang H, Chen B, et al. circHMCU promotes proliferation and metastasis of breast cancer by sponging the let-7 family. Mol Ther Nucleic Acids (2020) 20:518–33. doi: 10.1016/j.omtn.2020.03.014
99. Li X, Wang S, Li Z, Long X, Guo Z, Zhang G, et al. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int J Biol Macromol (2017) 105:346–53. doi: 10.1016/j.ijbiomac.2017.07.053
100. Palumbo A Jr., Da Costa NM, Esposito F, De Martino M, D’Angelo D, de Sousa VP, et al. HMGA2 overexpression plays a critical role in the progression of esophageal squamous carcinoma. Oncotarget (2016) 7:25872–84. doi: 10.18632/oncotarget.8288
101. Kang NN, Ge SL, Zhang RQ, Huang YL, Liu SD, Wu KM. MiR-490-3p inhibited the proliferation and metastasis of esophageal squamous cell carcinoma by targeting HMGA2. Eur Rev Med Pharmacol Sci (2018) 22:8298–305. doi: 10.26355/eurrev_201812_16527
102. Mei LL, Wang WJ, Qiu YT, Xie XF, Bai J, Shi ZZ. miR-125b-5p functions as a tumor suppressor gene partially by regulating HMGA2 in esophageal squamous cell carcinoma. PloS One (2017) 12:e0185636. doi: 10.1371/journal.pone.0185636
103. Pan Z, Lin J, Wu D, He X, Wang W, Hu X, et al. Hsa_circ_0006948 enhances cancer progression and epithelial-mesenchymal transition through the miR-490-3p/HMGA2 axis in esophageal squamous cell carcinoma. Aging (Albany NY) (2019) 11:11937–54. doi: 10.18632/aging.102519
104. Liu Y, Fu QZ, Pu L, Meng QG, Liu XF, Dong SF, et al. HMGA2 expression in renal carcinoma and its clinical significance. J Med Biochem (2015) 34:338–43. doi: 10.2478/jomb-2014-0036
105. Xia YY, Yin L, Jiang N, Guo WJ, Tian H, Jiang XS, et al. Downregulating HMGA2 attenuates epithelial-mesenchymal transition-induced invasion and migration in nasopharyngeal cancer cells. Biochem Biophys Res Commun (2015), 463:357–63. doi: 10.1016/j.bbrc.2015.05.068
106. Zhao XP, Zhang H, Jiao JY, Tang DX, Wu YL, Pan CB. Overexpression of HMGA2 promotes tongue cancer metastasis through EMT pathway. J Transl Med (2016) 14:26. doi: 10.1186/s12967-016-0777-0
107. Ma J, Li D, Kong FF, Yang D, Yang H, Ma XX. miR-302a-5p/367-3p-HMGA2 axis regulates Malignant processes during endometrial cancer development. J Exp Clin Cancer Res (2018) 37:19. doi: 10.1186/s13046-018-0686-6
108. Liu Y, Lv G, Bai J, Song L, Ding E, Liu L, et al. Effects of HMGA2 on the epithelial-mesenchymal transition-related genes in ACHN renal cell carcinoma cells-derived xenografts in nude mice. BMC Cancer (2022) 22:421. doi: 10.1186/s12885-022-09537-w
109. Yan J, Dai P, Qin X, He Y, Zhang Y. HMGA2 promotes the migration and invasion of gallbladder cancer cells and HMGA2 knockdown inhibits angiogenesis via targeting VEGFA. Mol Med Rep (2022) 25:54. doi: 10.3892/mmr.2021.12570
110. Han W, Zhang Y, Niu C, Guo J, Li J, Wei X, et al. BTB and CNC homology 1 (Bach1) promotes human ovarian cancer cell metastasis by HMGA2-mediated epithelial-mesenchymal transition. Cancer Lett (2019) 445:45–56. doi: 10.1016/j.canlet.2019.01.003
111. Jiang W, Gu W, Qiu R, He S, Shen C, Wu Y, et al. miRNA-101 suppresses epithelial-to-mesenchymal transition by targeting HMGA2 in pancreatic cancer cells. Anticancer Agents Med Chem (2016) 16:432–9. doi: 10.2174/1871520615666150507122142
112. Zhu M, Zhang C, Chen D, Chen S, Zheng H. MicroRNA-98-HMGA2-POSTN signal pathway reverses epithelial-to-mesenchymal transition in laryngeal squamous cell carcinoma. BioMed Pharmacother (2019) 117:108998. doi: 10.1016/j.biopha.2019.108998
113. Chen Z, Li Q, Wang S, Zhang J. miR−485−5p inhibits bladder cancer metastasis by targeting HMGA2. Int J Mol Med (2015) 36:1136–42. doi: 10.3892/ijmm.2015.2302
114. Huang WT, Zhang H, Jin Z, Li K, Hu C, Li ML, et al. MiR-219-5p inhibits prostate cancer cell growth and metastasis by targeting HMGA2. Eur Rev Med Pharmacol Sci (2020) 24:4710–8. doi: 10.26355/eurrev_202005_21159
115. Zhu C, Li J, Cheng G, Zhou H, Tao L, Cai H, et al. miR-154 inhibits EMT by targeting HMGA2 in prostate cancer cells. Mol Cell Biochem (2013) 379:69–75. doi: 10.1007/s11010-013-1628-4
116. Liu DK, Yu S, Li JP, Song WW, Li JH. MiR-150 suppressed cell viability, invasion and EMT via HMGA2 in oral squamous cell carcinoma. Eur Rev Med Pharmacol Sci (2021) 25:3981–9. doi: 10.26355/eurrev_202106_26038
117. Sharma P, Yadav P, Jain RP, Bera AK, Karunagaran D. miR-142-3p simultaneously targets HMGA1, HMGA2, HMGB1, and HMGB3 and inhibits tumorigenic properties and in-vivo metastatic potential of human cervical cancer cells. Life Sci (2022) 291:120268. doi: 10.1016/j.lfs.2021.120268
118. Wu A, Wu K, Li J, Mo Y, Lin Y, Wang Y, et al. Let-7a inhibits migration, invasion and epithelial-mesenchymal transition by targeting HMGA2 in nasopharyngeal carcinoma. J Transl Med (2015) 13:105. doi: 10.1186/s12967-015-0462-8
119. Xue F, Feng H, Wang T, Feng G, Ni N, Wang R, et al. hsa_circ_0000264 promotes tumor progression via the hsa-let-7b-5p/HMGA2 axis in head and neck squamous cell carcinoma. Oral Dis (2023) 29:2677–88. doi: 10.1111/odi.14399
120. Qiu F, Liu Q, Xia Y, Jin H, Lin Y, Zhao X. Circ_0000658 knockdown inhibits epithelial-mesenchymal transition in bladder cancer via miR-498-induced HMGA2 downregulation. J Exp Clin Cancer Res (2022) 41:22. doi: 10.1186/s13046-021-02175-3
121. Gao C, Lu W, Lou W, Wang L, Xu Q. Long noncoding RNA HOXC13-AS positively affects cell proliferation and invasion in nasopharyngeal carcinoma via modulating miR-383-3p/HMGA2 axis. J Cell Physiol (2019) 234:12809–20. doi: 10.1002/jcp.27915
122. Li R, Ruan Q, Zheng J, Zhang B, Yang H. LINC01116 promotes doxorubicin resistance in osteosarcoma by epigenetically silencing miR-424-5p and inducing epithelial-mesenchymal transition. Front Pharmacol (2021) 12:632206. doi: 10.3389/fphar.2021.632206
123. Xiong L, Tang Y, Tang J, Liu Z, Wang X. Downregulation of lncRNA HOTTIP suppresses the proliferation, migration, and invasion of oral tongue squamous cell carcinoma by regulation of HMGA2-mediated wnt/β-catenin pathway. Cancer Biother Radiopharm (2020) 35:720–30. doi: 10.1089/cbr.2019.3017
124. Kou N, Liu S, Li X, Li W, Zhong W, Gui L, et al. H19 Facilitates Tongue Squamous Cell Carcinoma Migration and Invasion via Sponging miR-let-7. Oncol Res (2019) 27:173–82. doi: 10.3727/096504018X15202945197589
125. Sakata J, Hirosue A, Yoshida R, Kawahara K, Matsuoka Y, Yamamoto T, et al. HMGA2 contributes to distant metastasis and poor prognosis by promoting angiogenesis in oral squamous cell carcinoma. Int J Mol Sci May (2019) 19:20. doi: 10.3390/ijms20102473
126. Xia YY, Yin L, Tian H, Guo WJ, Jiang N, Jiang XS, et al. HMGA2 is associated with epithelial-mesenchymal transition and can predict poor prognosis in nasopharyngeal carcinoma. Onco Targets Ther (2015) 8:169–76. doi: 10.2147/OTT.S74397
127. Zhang P, Bai H, Liu G, Wang H, Chen F, Zhang B, et al. MicroRNA-33b, upregulated by EF24, a curcumin analog, suppresses the epithelial-to-mesenchymal transition (EMT) and migratory potential of melanoma cells by targeting HMGA2. Toxicol Lett (2015) 234:151–61. doi: 10.1016/j.toxlet.2015.02.018
128. Li S, Peng F, Ning Y, Jiang P, Peng J, Ding X, et al. SNHG16 as the miRNA let-7b-5p sponge facilitates the G2/M and epithelial-mesenchymal transition by regulating CDC25B and HMGA2 expression in hepatocellular carcinoma. J Cell Biochem (2020) 121:2543–58. doi: 10.1002/jcb.29477
129. Li WJ, Li G, Liu ZW, Chen ZY, Pu R. LncRNA LINC00355 promotes EMT and metastasis of bladder cancer cells through the miR-424-5p/HMGA2 axis. Neoplasma (2021) 68:1225–35. doi: 10.4149/neo_2021_210427N574
130. Xin Y, Huang M, Guo WW, Huang Q, Zhang LZ, Jiang G. Nano-based delivery of RNAi in cancer therapy. Mol Cancer (2017) 16:134. doi: 10.1186/s12943-017-0683-y
131. Naghizadeh S, Mansoori B, Mohammadi A, Kafil HS, Mousavi Z, Sakhinia E, et al. Effects of HMGA2 gene downregulation by siRNA on lung carcinoma cell migration in A549 cell lines. J Cell Biochem (2019) 120:5024–32. doi: 10.1002/jcb.27778
132. Setten RL, Rossi JJ, Han SP. The current state and future directions of RNAi-based therapeutics. Nat Rev Drug Discovery (2019) 18:421–46. doi: 10.1038/s41573-019-0017-4
133. Eivazy P, Atyabi F, Jadidi-Niaragh F, Aghebati Maleki L, Miahipour A, Abdolalizadeh J, et al. The impact of the codelivery of drug-siRNA by trimethyl chitosan nanoparticles on the efficacy of chemotherapy for metastatic breast cancer cell line (MDA-MB-231). Artif Cells Nanomed Biotechnol (2017) 45:889–96. doi: 10.1080/21691401.2016.1185727
134. Naldini L. Gene therapy returns to centre stage. Nature (2015) 526:351–60. doi: 10.1038/nature15818
135. Jin S, Yong H, Liu Y, Bao W. CRISPR/Cas9-mediated high-mobility group A2 knockout inhibits cell proliferation and invasion in papillary thyroid carcinoma cells. Adv Med Sci (2023) 68:409–16. doi: 10.1016/j.advms.2023.10.001
136. Song CQ, Li Y, Mou H, Moore J, Park A, Pomyen Y, et al. Genome-wide CRISPR screen identifies regulators of mitogen-activated protein kinase as suppressors of liver tumors in mice. Gastroenterology (2017) 152:1161–1173.e1161. doi: 10.1053/j.gastro.2016.12.002
137. Yin H, Kauffman KJ, Anderson DG. Delivery technologies for genome editing. Nat Rev Drug Discovery (2017) 16:387–99. doi: 10.1038/nrd.2016.280
138. Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/cas9-mediated genome engineering. Mol Ther Nucleic Acids (2015) 4:e264. doi: 10.1038/mtna.2015.37
139. Békés M, Langley DR, Crews CM. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discovery (2022) 21:181–200. doi: 10.1038/s41573-021-00371-6
140. Kubota H. Quality control against misfolded proteins in the cytosol: a network for cell survival. J Biochem (2009) 146:609–16. doi: 10.1093/jb/mvp139
141. Long MJ, Gollapalli DR, Hedstrom L. Inhibitor mediated protein degradation. Chem Biol (2012) 19:629–37. doi: 10.1016/j.chembiol.2012.04.008
142. Salami J, Crews CM. Waste disposal-An attractive strategy for cancer therapy. Science (2017) 355:1163–7. doi: 10.1126/science.aam7340
143. Chen Y, Tandon I, Heelan W, Wang Y, Tang W, Hu Q. Proteolysis-targeting chimera (PROTAC) delivery system: advancing protein degraders towards clinical translation. Chem Soc Rev (2022) 51:5330–50. doi: 10.1039/D1CS00762A
144. Li X, Song Y. Proteolysis-targeting chimera (PROTAC) for targeted protein degradation and cancer therapy. J Hematol Oncol (2020) 13:50. doi: 10.1186/s13045-020-00885-3
145. Bai L, Zhou H, Xu R, Zhao Y, Chinnaswamy K, McEachern D, et al. A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell (2019) 36:498–511.e417. doi: 10.1016/j.ccell.2019.10.002
146. Lai AC, Toure M, Hellerschmied D, Salami J, Jaime-Figueroa S, Ko E, et al. Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew Chem Int Ed Engl (2016) 55:807–10. doi: 10.1002/anie.201507634
Keywords: HMGA2, epithelial-mesenchymal transition, extracellular matrix, cancer, gene therapy
Citation: Ma Q, Ye S, Liu H, Zhao Y, Mao Y and Zhang W (2024) HMGA2 promotes cancer metastasis by regulating epithelial–mesenchymal transition. Front. Oncol. 14:1320887. doi: 10.3389/fonc.2024.1320887
Received: 13 October 2023; Accepted: 09 January 2024;
Published: 01 February 2024.
Edited by:
Jixin Dong, University of Nebraska Medical Center, United StatesReviewed by:
Jing Wang, The Ohio State University, United StatesXiaoxiao Ge, Tongji University, China
Copyright © 2024 Ma, Ye, Liu, Zhao, Mao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Zhang, emhhbmd5aWppYTg0ODY1NUAxNjMuY29t
†These authors share first authorship
 Qing Ma1†
Qing Ma1† Wei Zhang
Wei Zhang