
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 21 February 2024
Sec. Gastrointestinal Cancers: Hepato Pancreatic Biliary Cancers
Volume 14 - 2024 | https://doi.org/10.3389/fonc.2024.1265762
Hepatocellular carcinoma (HCC) represents a leading and fatal malignancy within the gastrointestinal tract. Recent advancements highlight the pivotal role of long non-coding RNAs (lncRNAs) in diverse biological pathways and pathologies, particularly in tumorigenesis. LINC01134, a particular lncRNA, has attracted considerable attention due to its oncogenic potential in hepatoma. Current research underscores LINC01134’s potential in augmenting the onset and progression of HCC, with notable implications in drug resistance. This review comprehensively explores the molecular functions and regulatory mechanisms of LINC01134 in HCC, offering a fresh perspective for therapeutic interventions. By delving into LINC01134’s multifaceted roles, we aim to foster novel strategies in HCC management.
Liver cancer, predominantly hepatocellular carcinoma (HCC), is a gastrointestinal malignancy characterized by its insidious onset, rapid progression, restricted therapeutic efficacy, and dire prognosis. In 2020, liver cancer ranked as the sixth most prevalent cancer globally, with an incidence surpassing 900,000 new diagnoses (1). It also stood as the third leading cause of cancer-related mortality, accounting for over 830,000 deaths annually (1). Remarkably, China bore nearly half of the global liver cancer mortality burden (2, 3). Although the past decade has seen progress in liver cancer therapeutics (4), the long-term prognosis remains bleak, as evidenced by the low 5-year survival rate of 12.1% in China (5), and similarly unsatisfactory rates in affluent nations such as the United States of America (17.4%) (6). Therefore, unraveling the molecular intricacies of HCC and identifying new molecular markers and therapeutic avenues are of paramount importance.
Long noncoding RNAs (lncRNAs) are transcripts defined by their length—typically over 200 nucleotides—and limited protein-coding potential (7, 8). The biogenesis of lncRNAs is complex; they are transcribed by RNA Polymerase II, sharing certain characteristics with mRNAs, yet they also exhibit unique features such as less evolutionary conservation, fewer exons, and lower expression levels (9–11). Some lncRNAs are inefficiently spliced and terminate transcription independently of polyadenylation signals, resulting in nuclear retention and degradation mediated by the RNA exosome (12). Factors including transcriptional regulation, processing, and specific sequence motifs that recruit nuclear factors influence their nuclear localization. U1 small nuclear ribonucleoprotein (snRNP) binding sites and repeat elements play roles in lncRNA localization (13). A significant fraction of lncRNAs is exported to the cytosol via the Nuclear RNA export factor 1 (NXF1) pathway (14), where they undergo sorting processes, potentially associating with ribosomes for translation or localizing to mitochondria or exosomes, broadening their functional diversity. In essence, lncRNA biogenesis involves transcription, splicing, and sequence motifs, governing their nuclear retention and cytoplasmic export (15).
LncRNAs are classified based on their genomic location (intergenic, intronic, sense, and antisense) and size (small, medium, large) (16). Pioneering research has unveiled their crucial roles in various physiological processes, modulating gene expression at the epigenetic (17, 18), transcriptional (19–21), and post-transcriptional levels (20–22). LncRNAs interact with DNA, RNA, and proteins, influencing chromatin structure, gene transcription, RNA splicing, stability, and translation (15). They can serve as scaffolds for protein interactions, guide chromatin modifiers to specific genomic loci, or compete with microRNAs to regulate mRNA expression. Exploring the complexities of lncRNAs is poised to uncover new dimensions of gene regulation and cellular function.
Emerging data highlight the significant role of lncRNAs in human pathologies (23–25), especially in malignant tumors (26–29). Dysregulation of lncRNAs has been closely linked to tumor growth, metastasis, and resistance to chemotherapy (30–34), radiotherapy (35–38), and immune responses (39–41), underscoring their crucial involvement in cancer development.
Homo sapiens long intergenic non-protein-coding RNA 1134 (LINC01134), also known as TLNC1, is a lncRNA gene located on Chromosome 1p36.32 (Figure 1A). This gene, consisting of four exons and spanning a sequence length of 15,044 nucleotides, plays a pivotal role in various biological processes (42–44). To provide context to the significance of LINC01134, it’s essential to consider its neighboring genes, which include C1orf174, LINC01346, LINC02780, and DFFB, as illustrated in Figure 1B. Understanding the genomic context of LINC01134 sheds light on its potential functional importance and its role in gene regulation. Recently, LINC01134 has gained attention for its role in the progression of HCC (42, 45). and it has been identified as a prognostic indicator for HCC patients (42–44). In addition, LINC01134 has been linked to an unfavorable prognosis in ovarian cancer (46), and signatures related to LINC01134 have shown superior predictive value for prognosis in low-grade glioma (47) and gastric cancer (48). Altered expression of LINC01134 has been observed in HCC tissues and cell lines, implicating it in the progression of HCC (42–45, 49–51).
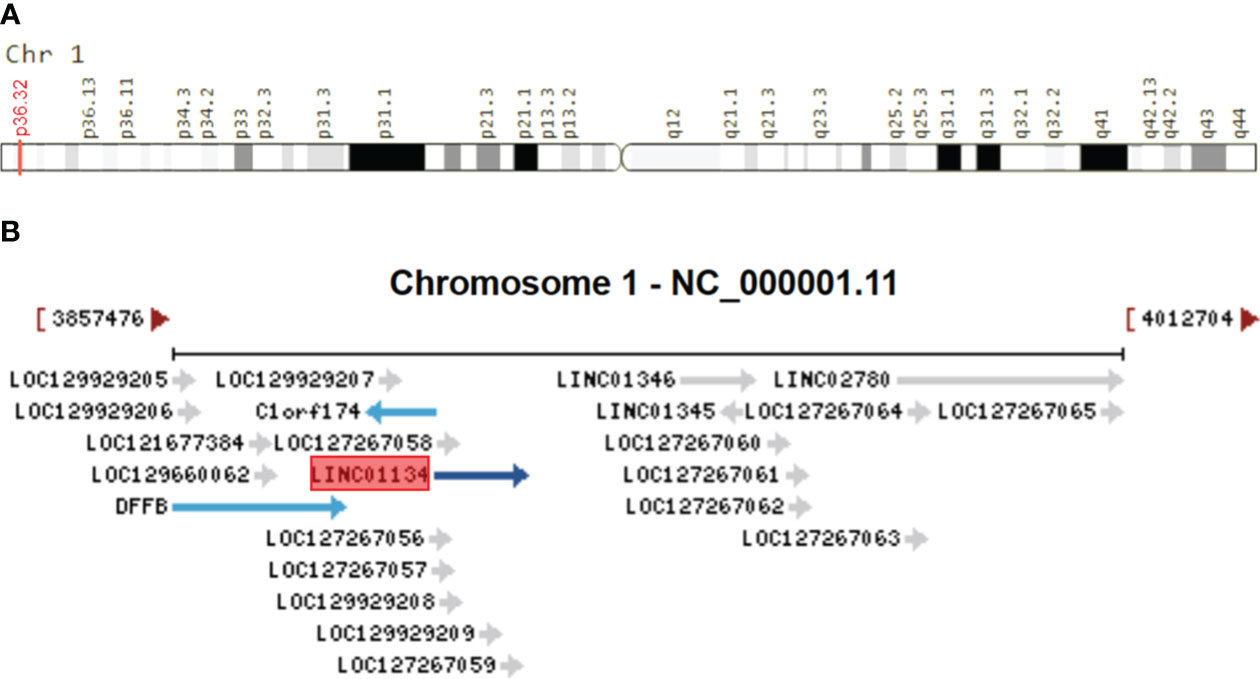
Figure 1 Genomic Overview of LINC01134. (A) Genomic location of LINC01134 sourced from the GeneCaRNA database (https://www.genecards.org/cgi-bin/carddisp.pl?gene=LINC01134), and (B) the genomic context depicting neighboring genes of LINC01134, derived from the NCBI database (https://www.ncbi.nlm.nih.gov/gene/100133612).
To compile pertinent literature for a comprehensive analysis of LINC01134’s involvement in cancer biology and its prospective clinical relevance, we conducted a thorough systematic search across multiple databases. These databases included PubMed, Web of Science, ScienceDirect, Embase, SpringerLink, and Google Scholar, as well as Chinese databases such as China National Knowledge Infrastructure (CNKI), Weipu Information (VIP), and Wanfang Database. Our search utilized the keywords ‘LINC01134’ and ‘TLNC1’. We considered peer-reviewed articles published in English or Chinese up to October 1, 2023. The inclusion criteria were predefined, focusing on original studies that explored LINC01134’s expression, clinicopathological associations, and biological functions in human tumors.
In this review, we provide a comprehensive analysis, addressing aberrant expression patterns, molecular mechanisms, and the clinical significance of LINC01134 in HCC. This review aims to establish a theoretical foundation for potential future clinical applications, predict the functions and regulatory mechanisms of LINC01134, and delineate both the current research gaps and future directions.
Research underscores the pivotal roles of LINC01134 in hepatocarcinogenesis and progression. LINC01134 functions through diverse mechanisms, including interactions with DNA promoters, decoy proteins, mRNA binding, and acting as a miRNA sponge, regulating malignant cellular behaviors, and contributing to tumor therapy resistance (Figure 2). It has also been identified as a promising prognostic and diagnostic tumor marker, along with its potential as a therapeutic target in hepatoma (Figure 2). In the following sections, we reviewed the multifaceted role of LINC01134 in HCC progression, metastasis, and its impact on chemo- and radiotherapy resistance. In addition, we examine the clinical applications of LINC01134 in HCC, focusing on prognosis, diagnosis, and therapy.
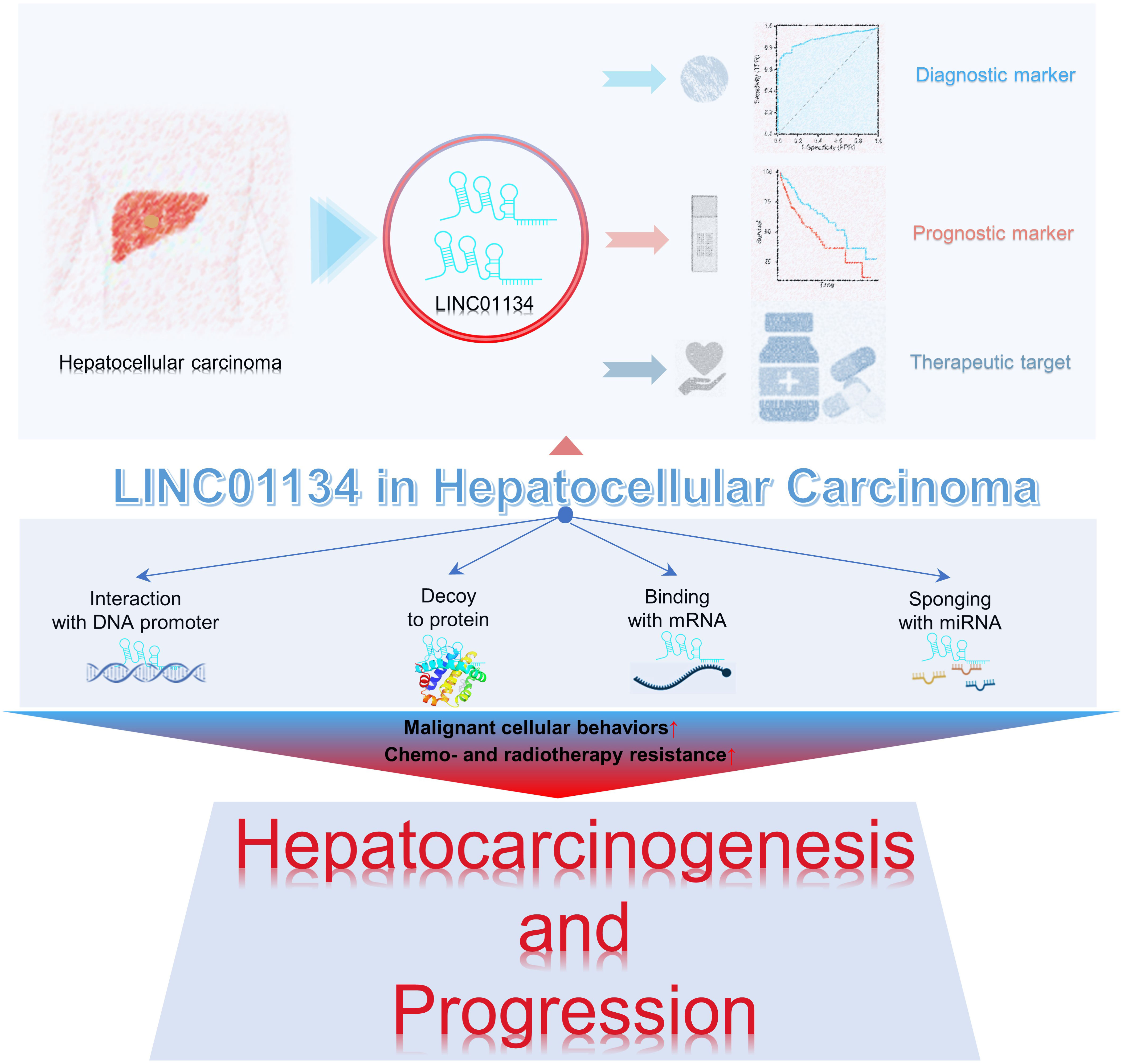
Figure 2 LINC01134 plays a role in the progression of HCC and demonstrates potential clinical significance as both a tumor marker and therapeutic target.
Understanding the expression patterns and functional role of LINC01134 in HCC provides insights into its potential as a valuable biomarker and therapeutic target. Recent experimental studies (Table 1) have provided compelling evidence regarding the significant upregulation of LINC01134 in HCC tissues and HCC cancer cell lines. Consistent associations have been observed between increased expression of LINC01134 and unfavorable clinical outcomes in HCC patients. These findings highlight the potential significance of LINC01134 in HCC, suggesting its viability as a biomarker or therapeutic target for this malignancy.
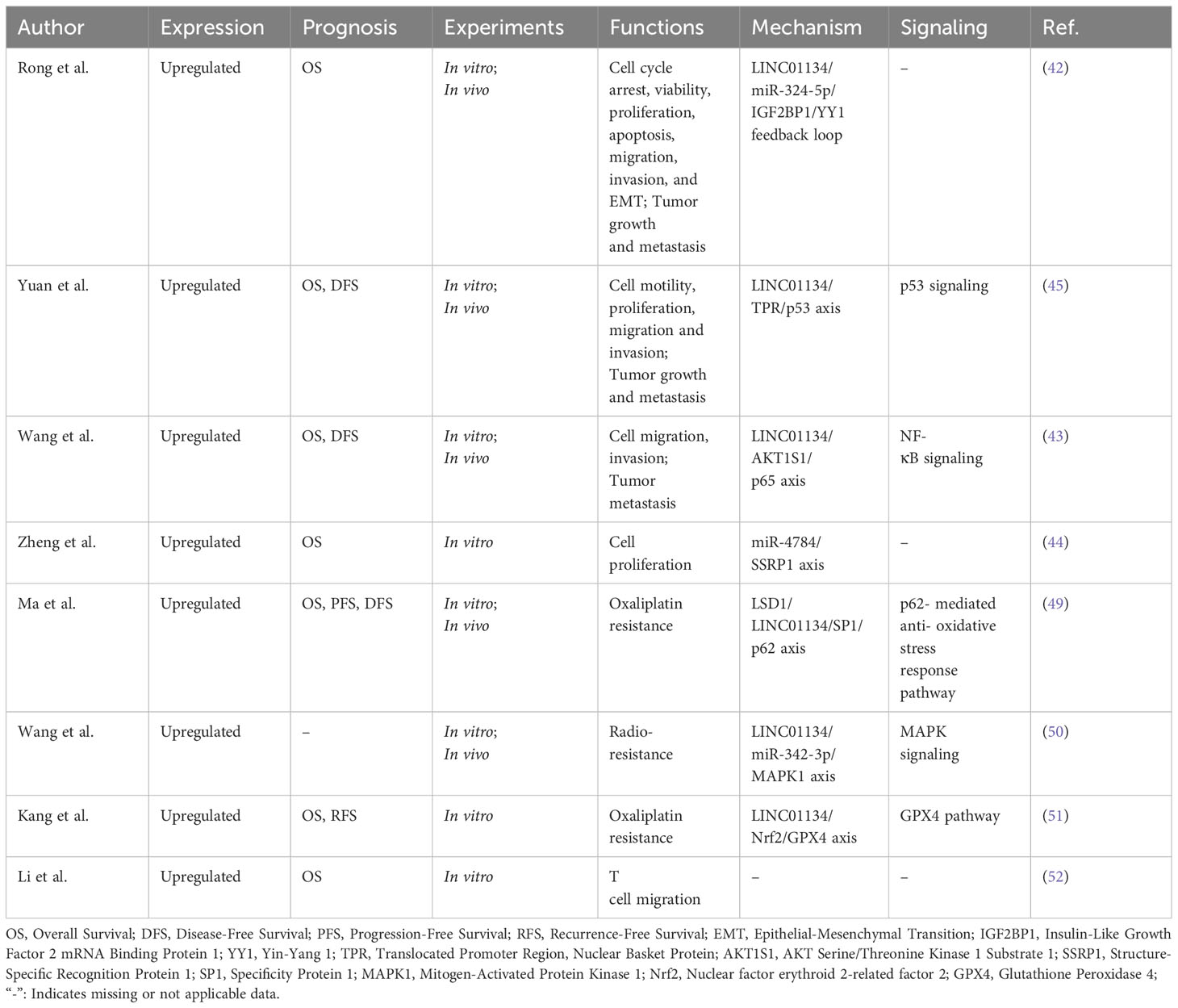
Table 1 LINC01134 expression, prognostic value, and related regulatory roles in hepatocellular carcinoma.
Moreover, elevated LINC01134 expression has been associated with various facets of HCC progression, including enhanced cell proliferation, migration, invasion, and tumor growth and metastasis (Table 1). It has also been implicated in processes such as epithelial-mesenchymal transition (EMT) (42), immune cell infiltration (specifically T cell migration) (52), and resistance to chemotherapy and radiotherapy (49–51) in HCC (Table 1). Furthermore, LINC01134 plays a regulatory role in HCC development by influencing key signaling pathways, including the Nuclear Factor Kappa B (NF-κB) inflammatory pathway (43), the MAPK signaling cascade (50), and the tumor suppressor p53 pathway (45), as well as genes involved in tumorigenesis and disease progression (Table 1). These insights are instrumental in comprehending the essential role of LINC01134 in the functional landscape of HCC.
LINC01134 has risen to prominence as a critical regulator of numerous tumor-associated cellular processes in HCC. The roles played by LINC01134 in modulating malignant biological behaviors, both in vitro and in vivo, are summarized in Figure 3. A series of studies (42–45) have demonstrated that the knockdown of LINC01134 results in decreased HCC cell viability, proliferation, migration, and invasion, and it inhibits the EMT process. Moreover, it induces cell cycle arrest and promotes cell apoptosis in vitro. LINC01134 silencing also inhibits tumor growth and metastasis in vivo. In contrast, ectopic expression of LINC01134 stimulates HCC cell viability, proliferation, migration, and invasion in HCC cell lines. Furthermore, overexpression of LINC01134 facilitates tumor growth and metastasis in mouse models.
Mechanistically (Figure 4), LINC01134 plays a multifaceted role, functioning as an endogenous miRNA sponge, aiding transcriptional activation through interactions with DNA promoters, and modulating protein transport between the nucleus and cytoplasm. These mechanisms contribute to the diverse functional roles of LINC01134 in cellular processes. For instance, Rong et al. (42) demonstrated that LINC01134 acts as a sponge for miR-324-5p and interacts with Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1), thereby increasing the stability of Yin-Yang 1 (YY1) mRNA expression. YY1, a well-established transcription factor, plays a pivotal role in transcriptional regulation (53–55).. Upregulated YY1, in turn, stimulates the expression of LINC01134 by enhancing its promoter activity, establishing a positive feedback loop that drives HCC progression. Similarly, Wang et al. (43) found that LINC01134 directly binds to the promoter of AKT1 Substrate 1 (AKT1S1), further activating the NF-κB pathway and promoting HCC metastasis. Moreover, Zheng et al. (44) reported that LINC01134 may contribute to HCC carcinogenesis, at least partially, through the miR-4784/SSRP1 axis. Additionally, Yuan et al. (45) revealed that LINC01134 promotes HCC growth and metastasis by interacting with TPR and inducing the TPR-mediated transportation of p53 from the nucleus to the cytoplasm. The inactivation of p53 signaling ultimately contributes to the progression of liver cancer. These findings collectively highlight the crucial role of LINC01134 in modulating a spectrum of biological behaviors, offering valuable insights for the development of potential therapeutic strategies against HCC.
Resistance to chemotherapy and radiotherapy is a major obstacle in cancer treatment (56, 57). Recent investigations (49–51) have highlighted the role of LINC01134 in contributing to chemo- and radiotherapy resistance in HCC (Figure 5). LINC01134 has been found to modulate oxaliplatin resistance by influencing cell viability, apoptosis, and mitochondrial homeostasis through the SP1/p62 axis (49), where p62 refers to SQSTM1 or Sequestosome-1, a crucial element in this pathway. Additionally, LINC01134 functions as a negative regulator of ferroptosis and enhances resistance to oxaliplatin in hepatoma cells by activating the Glutathione Peroxidase 4 (GPX4) pathway (51). The GPX4 pathway is a critical defense mechanism against oxidative stress and lipid peroxidation in cells (58–60). Activation of this pathway is achieved through the facilitation of Nuclear Factor Erythroid 2-Related Factor 2 (Nrf2) recruitment to the GPX4 promoter (51). Furthermore, LINC01134 plays a role in the regulation of Mitogen-Activated Protein Kinase 1 (MAPK1) expression by sequestering miR-342-3p and interacting with Insulin-Like Growth Factor 2 mRNA Binding Protein 2 (IGF2BP2). This interaction subsequently activates the MAPK signaling pathway, contributing to radio-resistance in HCC cells (50). Consequently, targeting LINC01134 for downregulation using mimics could potentially enhance the sensitivity of HCC tumor cells to chemoradiotherapy, addressing the issue of treatment resistance.
Early detection is crucial for the successful treatment of HCC. Numerous studies have consistently reported a marked increase in LINC01134 expression in cancerous tissues of HCC patients (42–45, 50, 51), highlighting its potential as a diagnostic biomarker for timely intervention and improved patient outcomes. Notably, a significant elevation in LINC01134 expression has been observed in HCC patients who develop portal vein tumor thrombus (PVTT) (43). PVTT is a prevalent and complex complication in HCC management, linked to high recurrence rates and a poor prognosis (61, 62). These findings emphasize the vital role of LINC01134 in early prediction and prompt treatment, particularly for individuals at risk of developing PVTT.
The expression level of LINC01134 has been significantly correlated with HCC prognosis. Wang et al. (43) disclosed a substantial positive association between elevated LINC01134 levels and both microvascular and macrovascular invasion, critical factors in HCC staging and prognostication. Furthermore, numerous studies (42–45, 49, 51, 52, 63, 64) have reported that HCC patients with high LINC01134 expression in cancerous samples exhibit significantly poorer clinical outcomes, including reduced overall survival, diminished disease-free survival, and lower recurrence-free survival rates. In addition, several lncRNA-based prognostic models containing LINC01134, such as an 11-lncRNAs penal (52), and a 50-lncRNA-pairs (65), have been constructed and showed noticeable potential prognostic value in HCC cases. Collectively, these findings position LINC01134 as a potential prognostic biomarker for HCC.
The potential of lncRNAs as therapeutic targets in cancer treatment is increasingly recognized (24, 28), and LINC01134, a newly identified lncRNA, emerges as a promising candidate for HCC therapy. Experimental evidence from multiple studies (42–45, 49–52) has elucidated the regulatory role of LINC01134 in HCC development, highlighting its influence on key molecules and genes pivotal for tumorigenesis and disease progression (Figure 6).
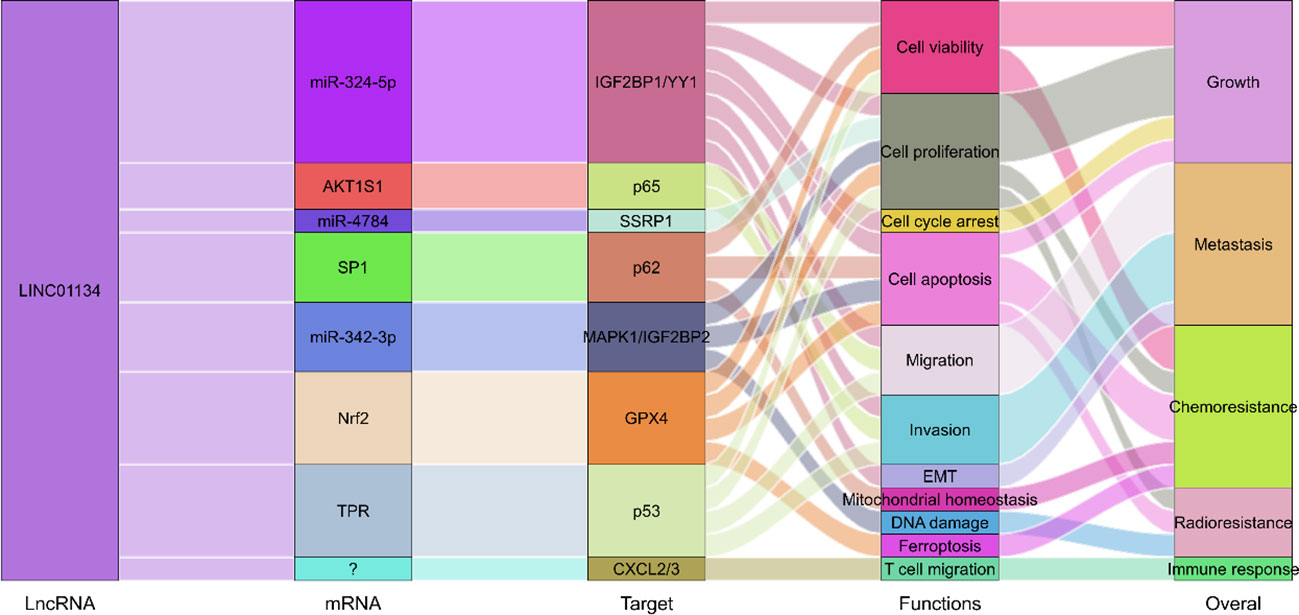
Figure 6 Molecular regulatory mechanisms of LINC01134 in HCC development through involvement in the regulation of multiple biological processes.
Crucially, the downregulation of LINC01134 has been shown to enhance the responsiveness of cancer cells to chemotherapy agents such as oxaliplatin (49, 51) and to mitigate radio-resistance in HCC cells (50). Moreover, a study conducted by Li et al. (52) revealed that a decrease in LINC01134 expression correlates with an upregulation of Chemokine (C-X-C motif) ligand 2 and 3, which are vital regulators in immune responses (66). They also discovered that the medium from HCC cells with reduced LINC01134 expression significantly boosts the migratory ability of Jurkat T cells, known for producing interleukin 2 (67, 68). This cytokine is closely associated with increased sensitivity to cancer drugs (69, 70) and enhanced radiation responses (71, 72).
These findings underscore the potential therapeutic implications of targeting LINC01134 in HCC treatment. Suppressing LINC01134 expression not only inhibits the malignant behaviors of HCC cells but also improves treatment response, modulates immune responses, and enhances overall therapeutic efficacy.
In recent times, bioinformatics has significantly advanced the discovery and annotation of lncRNAs. Mainstream technologies for sequencing and lncRNA profiling include RNA-seq and microarray, which prominently feature in repositories like The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO). This advancement in bioinformatics is crucial for deeper explorations into lncRNAs. Currently, in silico methods, backed by bioinformatic analyses, stand out as powerful tools. Numerous databases are available that can be instrumental in annotating and predicting potential lncRNA functions, such as LncBook 2.0 (73) and RNAInter (74). To enrich this review, we tapped into several online lncRNA databases and high-throughput data, aiming to predict the clinical significance, functional dynamics, and pathway involvement of LINC01134 in tumorigenesis and HCC progression. These insights may provide invaluable guidance for future studies.
In our investigation of LINC01134’s diagnostic potential in HCC samples, we analyzed data from the Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) dataset. Our analysis revealed that LINC01134 exhibits good accuracy in distinguishing between normal and HCC tissues, as evidenced by an area under the curve (AUC) value of 0.833 (Figure 7). Furthermore, LINC01134 demonstrates diagnostic value in distinguishing between N0 and N1 HCC samples (AUC = 0.716) and between M0 and M1 HCC patients (AUC = 0.604) (Figure 7). Turning our attention to the potential of LINC01134 as a minimally invasive diagnostic tool, we initiated an analysis of extracellular vesicle lncRNA expression in blood samples. Notably, exosomes represent a subpopulation of extracellular vesicles with an average diameter of 100 nm (75). Exosomal lncRNAs are currently recognized as promising tumor diagnostic tools (76–79). Utilizing exoRBase (http://www.exorbase.org/), a repository of extracellular vesicle long RNAs derived from RNA-seq data analyses (80), we observed a significant upregulation of LINC01134 expression in the blood of individuals with various cancers, including HCC, when compared to healthy donors (Figure 8). These findings underscore the potential of LINC01134 as a highly promising diagnostic marker for HCC. The prospect of utilizing LINC01134 as a novel circulating oncological biomarker opens up an exciting avenue for future research.
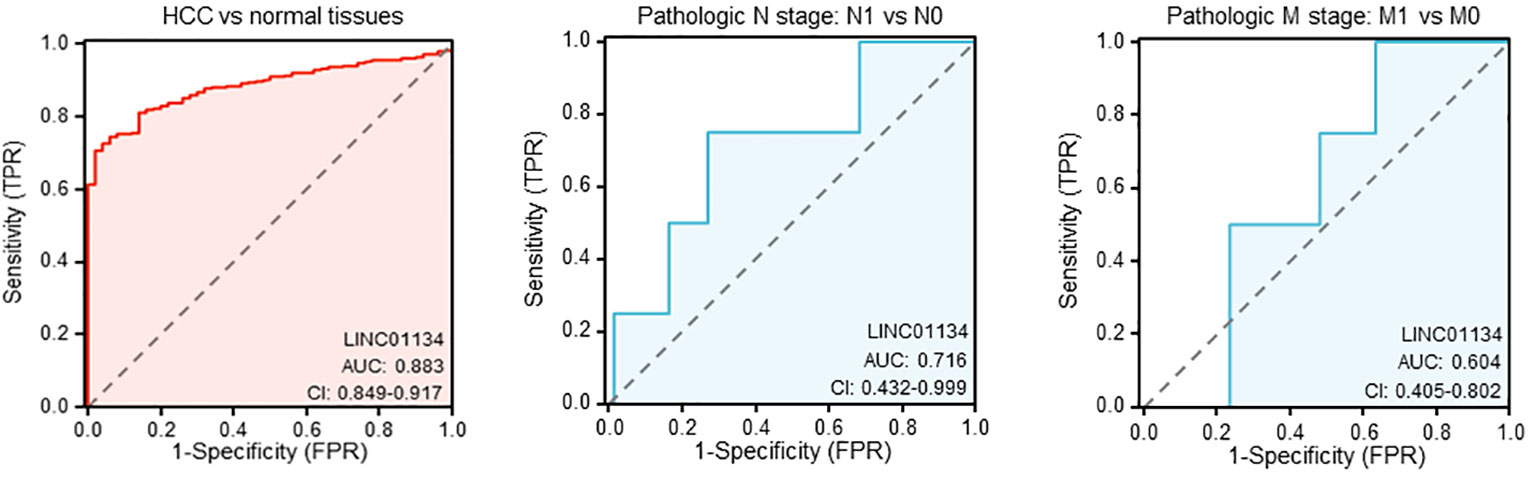
Figure 7 Evaluating the diagnostic utility of LINC01134 in hepatocellular carcinoma samples and their metastatic progressions.
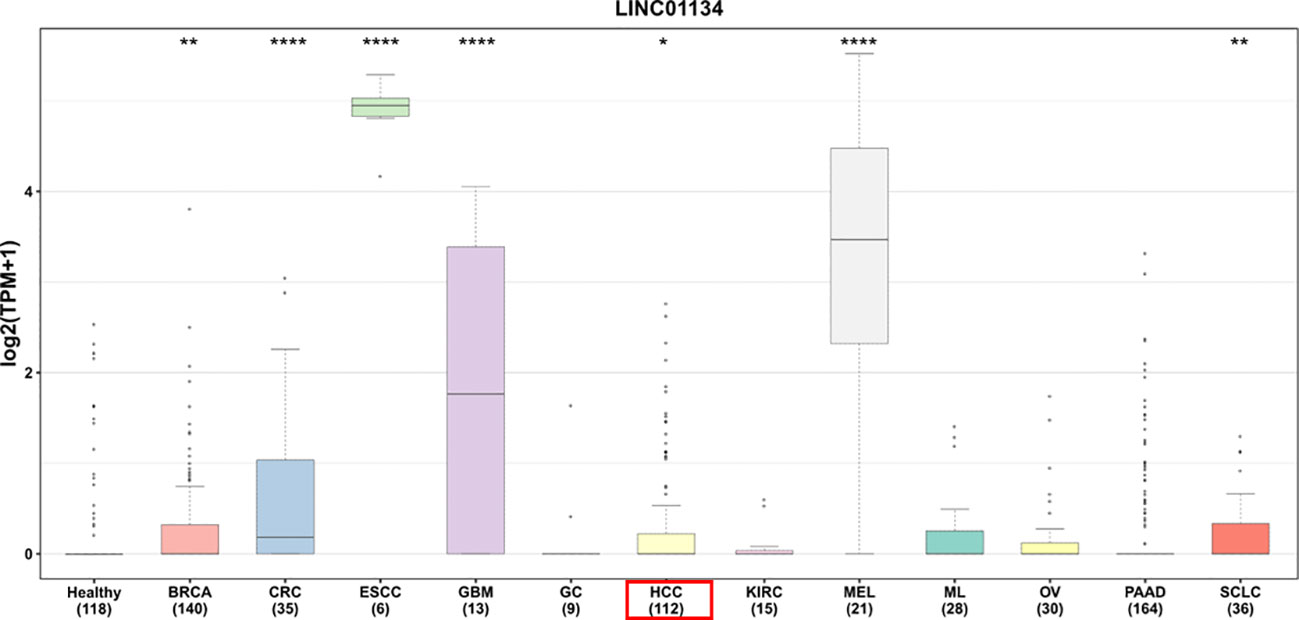
Figure 8 Comparative expression of LINC01134 in extracellular vesicles derived from blood across various cancer types versus healthy donors. This includes: BRCA, breast cancer; CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; GBM, glioblastoma multiforme; GC, gastric cancer; HCC, hepatocellular carcinoma; KIRC, kidney cancer; MEL, melanoma; ML, malignant lymphoma; OV, ovarian cancer; PAAD, pancreatic adenocarcinoma; SCLC, small cell lung cancer. Significance levels are denoted by asterisks above the boxes, with * representing p < 0.05, ** for p < 0.01, and **** for p < 0.0001. Data were sourced from exoRBase (http://www.exorbase.org/).
LncRNAs play crucial roles in modulating chromatin architecture and function, regulating transcription, and influencing RNA processing and stability (81, 82). They achieve these functions through targeting DNA, binding to RNAs, and recruiting proteins (81, 82). Here we found the top significant 25 RNA-DNA interactions, RNA-RNA interactions, and RNA-protein interactions related to LINC01134 (Figure 9), using RNAInter (http://www.rnainter.org/), an RNA interactome database. Accumulating evidence suggests that lncRNAs can behave like competing endogenous RNAs (ceRNAs), and compete with microRNAs to communicate with mRNAs (83–86). LncRNA-associated ceRNA regulation has been implicated in cell-fate determination and tumorigenesis (87–90). Here, we used the LnCeCell database (91) to predict lncRNA-associated ceRNA networks at single cancer cell resolution. We displayed the LINC01134-associated ceRNA-network (Figure 10A) and also identified LINC01134-associated ceRNA-related cancer hallmarks, such as self-sufficiency in growth signals, evading apoptosis, enduring angiogenesis, and tissue infiltration and metastasis (Figure 10B). Furthermore, we conducted an analysis to identify the top 10 enriched functions of LINC01134-associated ceRNA (Figure 10C). The findings indicate that ceRNAs associated with LINC01134 play a role in crucial tumor-related biological processes, such as the “G2-M transition of the mitotic cell cycle”, “maintenance of cellular protein localization” and “MAP kinase activity”. Furthermore, these ceRNAs are linked to several cancer-associated pathways, including the “Aurora B pathway”, “phosphoinositide 3-kinase pathway” and “Wnt non-canonical pathway”. It’s worth noting that these pathways are closely related to the onset and progression of various tumors (92–94), including HCC (95–98).
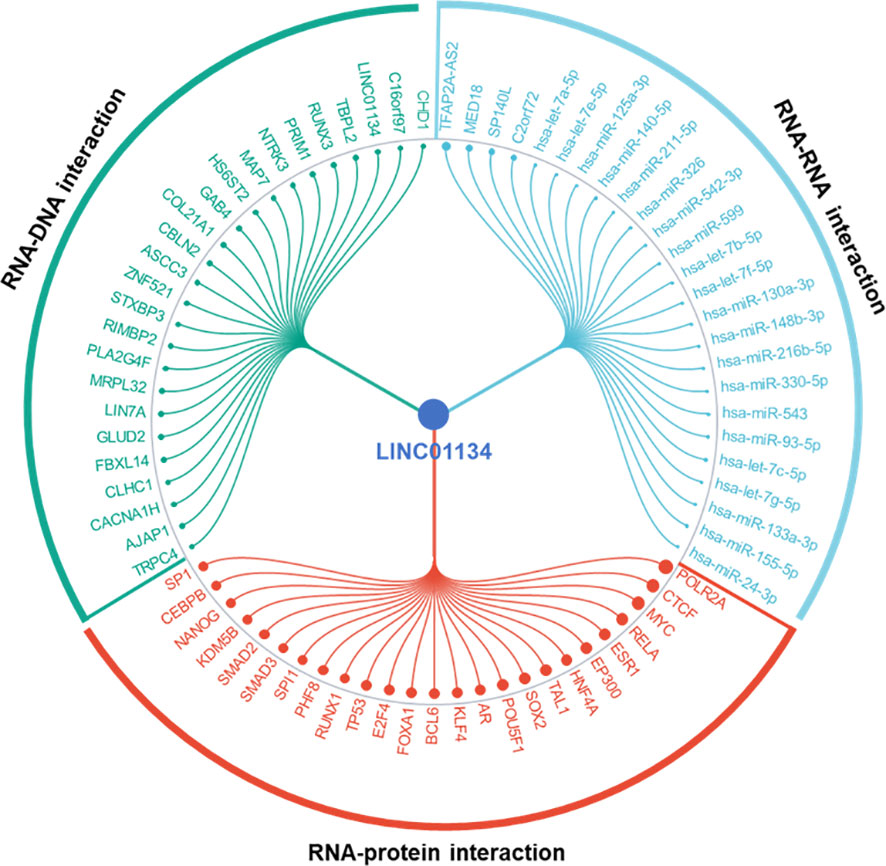
Figure 9 Top 25 RNA-DNA, RNA-RNA, and RNA-protein interactions associated with LINC01134. Data sourced from RNAInter (http://www.rnainter.org/).
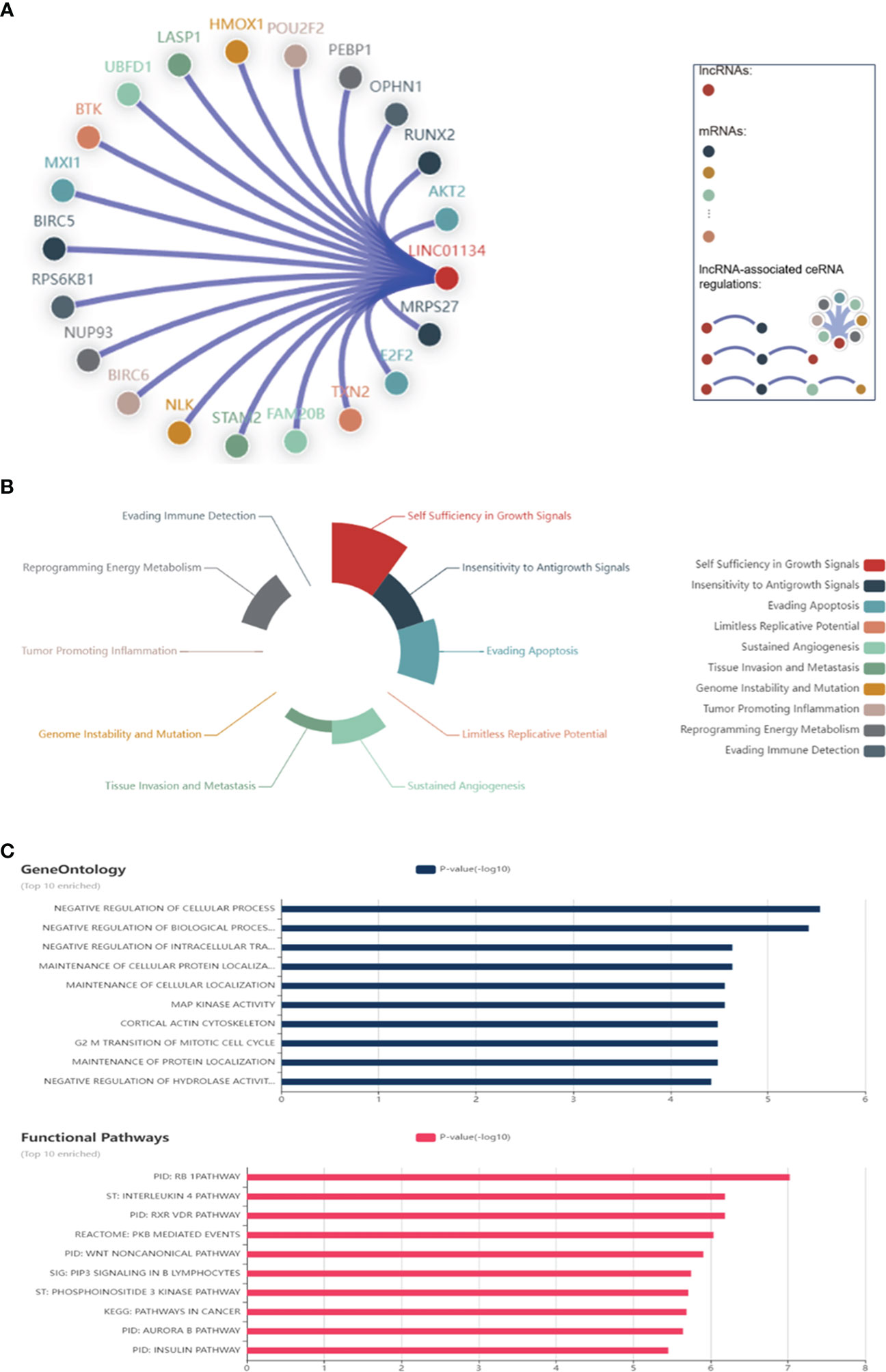
Figure 10 Networks Associated with LINC01134 ceRNA. (A) Interactions between LINC01134 and its associated ceRNA network, including corresponding mRNAs. (B) Principal cancer hallmarks associated with LINC01134 ceRNA, including self-sufficiency in growth signals, evasion of apoptosis, sustained angiogenesis, and tissue infiltration & metastasis. (C) The ten most significant functions of LINC01134-associated ceRNA, derived from gene ontology annotations and functional pathways. Data sourced from LnCeCell (http://bio-bigdata.hrbmu.edu.cn/LnCeCell/).
Moreover, we investigated the role of LINC01134 in immunotherapy. We obtained liver cancer datasets from UCSC (https://xenabrowser.net/) for analysis. Our findings revealed a significant negative correlation between LINC01134 gene expression and immune infiltration scores (including stromal, immune, and ESTIMATE scores) (Figure 11A). This suggests a potential inverse association between LINC01134 and immune infiltration, hinting at its involvement in the immune evasion mechanisms in HCC. However, more experiments and mechanistic studies are required. Additionally, we analyzed the correlation between LINC01134 and 60 key genes related to inhibitory and stimulatory immune checkpoint pathways (Figure 11B). The identification of LINC01134-related regulatory networks enhances our understanding of its role in tumor pathology and aids precision medicine. These predictions offer insights into potential tumor-related mechanisms, requiring further validation.
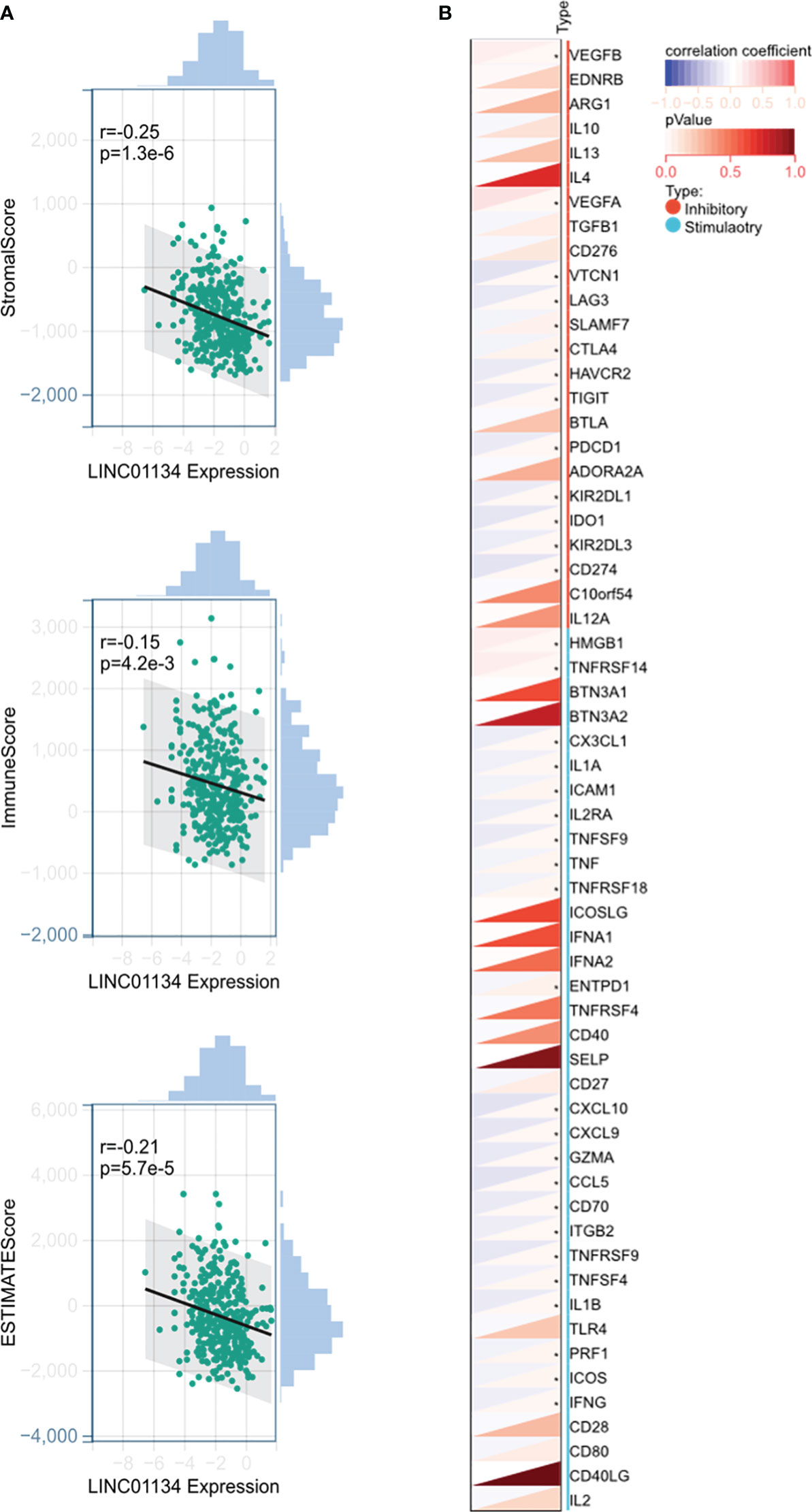
Figure 11 Potential Involvement of LINC01134 in HCC Immunotherapy. (A) Analysis shows a significant negative correlation between LINC01134 expression and immune infiltration scores, (B) Examination of LINC01134’s correlation with 60 genes from inhibitory and stimulatory immune checkpoint pathways. Significance levels are indicated by asterisks, with * denoting p < 0.05.
In this review, we elucidated the biological function and regulatory mechanism of LINC01134 in hepatocellular carcinoma and highlighted its potential as a diagnostic and prognosis marker, as well as a therapeutic target for HCC. However, it is imperative to emphasize that investigations into LINC01134 are at a nascent stage, and its involvement in most human tumors remains largely unexplored. Currently, LINC01134 has only been reported in the context of liver cancer, While the existing reports are limited in number, this review’s synthesis of LINC01134 research in liver cancer can serve as a valuable foundation and inspiration for future investigations in other human tumors.
Based on an in-depth analysis of previous studies, there are still some significant questions regarding the role of LINC01134 in HCC that warrant further investigation. This includes a need for further research into the biological impact of LINC01134 on other RNA-binding proteins (RBPs) (42). Furthermore, the oncogenic role of LINC01134 in HCC requires a thorough assessment through genetic knockout mouse models. Moreover, further exploration is warranted to understand the role and functions of LINC01134 in tumor stem cells of HCC (45).
Additionally, to elucidate LINC01134’s role in tumor pathogenesis, particularly in the context of drug and radiotherapy resistance, and immunotherapy impact, in-depth experimental studies of its functional and biological mechanisms are required. Furthermore, it is also necessary to investigate the expression profiles of homologous protein-coding genes associated with LINC01134. Given the significant role of endogenous siRNAs (esiRNAs) in regulating RNA silencing (99–101), future research should also focus on unraveling the interactions between esiRNAs derived from LINC01134 and their target genes. Moreover, there is a need to investigate LINC01134’s expression profiles in both tumor tissues and body fluids, aiming to assess its diagnostic potential using minimally invasive samples such as serum, which could facilitate distinguishing cancer patients from healthy individuals. Extensive research is also warranted to evaluate the prognostic value of LINC01134 across diverse populations and various pathological stages of different tumors. Undertaking these research initiatives will provide invaluable insights, paving the way for personalized cancer management in terms of diagnosis, risk stratification, and therapeutic interventions.
In conclusion, our study emphasizes LINC01134 as a multifaceted oncogenic lncRNA exerting a substantial impact on HCC growth, metastasis, and therapy resistance. We recognize its potential as a promising biomarker and therapeutic target in hepatoma, offering a valuable avenue for targeted interventions. However, it is imperative to recognize that further experimental investigations are necessary to comprehensively unravel the underlying roles and mechanisms of LINC01134 in hepatocarcinogenesis, progression, resistance, and immunotherapy, establishing a robust theoretical foundation. To enhance the clinical relevance of LINC01134, future clinical studies focused on this lncRNA are essential for uncovering innovative ideas and approaches in HCC diagnosis, risk stratification, and treatment.
YY: Visualization, Data curation, Formal Analysis, Writing – original draft. JW: Visualization, Writing – original draft, Data curation, Formal Analysis. QG: Writing – original draft. HL: Visualization, Conceptualization, Supervision, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Zheng R, Zhang S, Zeng H, Wang S, Sun K, Chen R, et al. Cancer incidence and mortality in China, 2016. J Natl Cancer Center (2022) 2(1):1–9. doi: 10.1016/j.jncc.2022.02.002
3. Shan T, Ran X, Li H, Feng G, Zhang S, Zhang X, et al. Disparities in stage at diagnosis for liver cancer in China. J Natl Cancer Center (2023) 3(1):7–13. doi: 10.1016/j.jncc.2022.12.002
4. Chen Z, Xie H, Hu M, Huang T, Hu Y, Sang N, et al. Recent progress in treatment of hepatocellular carcinoma. Am J Cancer Res (2020) 10(9):2993–3036.
5. Zeng H, Chen W, Zheng R, Zhang S, Ji JS, Zou X, et al. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Global Health (2018) 6(5):e555–67. doi: 10.1016/S2214-109X(18)30127-X
6. Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet (Lond Engl) (2018) 391(10125):1023–75. doi: 10.1016/S0140-6736(17)33326-3
7. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet (2009) 10(3):155–9. doi: 10.1038/nrg2521
8. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev (2009) 23(13):1494–504. doi: 10.1101/gad.1800909
9. Guo CJ, Ma XK, Xing YH, Zheng CC, Xu YF, Shan L, et al. Distinct processing of lncRNAs contributes to non-conserved functions in stem cells. Cell (2020) 181(3):621–36.e22. doi: 10.1016/j.cell.2020.03.006
10. Quinn JJ, Zhang QC, Georgiev P, Ilik IA, Akhtar A, Chang HY. Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes Dev (2016) 30(2):191–207. doi: 10.1101/gad.272187.115
11. Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep (2015) 11(7):1110–22. doi: 10.1016/j.celrep.2015.04.023
12. Schlackow M, Nojima T, Gomes T, Dhir A, Carmo-Fonseca M, Proudfoot NJ. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol Cell (2017) 65(1):25–38. doi: 10.1016/j.molcel.2016.11.029
13. Yin Y, Lu JY, Zhang X, Shao W, Xu Y, Li P, et al. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature (2020) 580(7801):147–50. doi: 10.1038/s41586-020-2105-3
14. Zuckerman B, Ron M, Mikl M, Segal E, Ulitsky I. Gene architecture and sequence composition underpin selective dependency of nuclear export of long RNAs on NXF1 and the TREX complex. Mol Cell (2020) 79(2):251–67.e6. doi: 10.1016/j.molcel.2020.05.013
15. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol (2021) 22(2):96–118. doi: 10.1038/s41580-020-00315-9
16. Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol (2013) 10(6):925–33. doi: 10.4161/rna.24604
17. Cusenza VY, Tameni A, Neri A, Frazzi R. The lncRNA epigenetics: The significance of m6A and m5C lncRNA modifications in cancer. Front Oncol (2023) 13:1063636. doi: 10.3389/fonc.2023.1063636
18. Alsayed R, Sheikhan K, Alam MA, Buddenkotte J, Steinhoff M, Uddin S, et al. Epigenetic programing of cancer stemness by transcription factors-non-coding RNAs interactions. Semin Cancer Biol (2023) 92:74–83. doi: 10.1016/j.semcancer.2023.04.005
19. Roberts TC, Morris KV, Weinberg MS. Perspectives on the mechanism of transcriptional regulation by long non-coding RNAs. Epigenetics (2014) 9(1):13–20. doi: 10.4161/epi.26700
20. Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol (2019) 21(5):542–51. doi: 10.1038/s41556-019-0311-8
21. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet (2016) 17(1):47–62. doi: 10.1038/nrg.2015.10
22. Pisignano G, Ladomery M. Post-transcriptional regulation through long non-coding RNAs (lncRNAs). Non-coding RNA (2021) 7(2):29. doi: 10.3390/ncrna7020029
23. Li K, Wang Z. lncRNA NEAT1: Key player in neurodegenerative diseases. Ageing Res Rev (2023) 86:101878. doi: 10.1016/j.arr.2023.101878
24. Srinivas T, Mathias C, Oliveira-Mateos C, Guil S. Roles of lncRNAs in brain development and pathogenesis: Emerging therapeutic opportunities. Mol Ther (2023) 31(6):1550–61. doi: 10.1016/j.ymthe.2023.02.008
25. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci (2016) 73(13):2491–509. doi: 10.1007/s00018-016-2174-5
26. Afra F, Mahboobipour AA, Salehi Farid A, Ala M. Recent progress in the immunotherapy of hepatocellular carcinoma: Non-coding RNA-based immunotherapy may improve the outcome. Biomed Pharmacother Biomed Pharmacother (2023) 165:115104. doi: 10.1016/j.biopha.2023.115104
27. Fonseca-Montaño MA, Vázquez-Santillán KI, Hidalgo-Miranda A. The current advances of lncRNAs in breast cancer immunobiology research. Front Immunol (2023) 14:1194300. doi: 10.3389/fimmu.2023.1194300
28. Tang J, Zhang J, Lu Y, He J, Wang H, Liu B, et al. Novel insights into the multifaceted roles of m(6)A-modified LncRNAs in cancers: biological functions and therapeutic applications. biomark Res (2023) 11(1):42. doi: 10.1186/s40364-023-00484-7
29. Verma S, Sahu BD, Mugale MN. Role of lncRNAs in hepatocellular carcinoma. Life Sci (2023) 325:121751. doi: 10.1016/j.lfs.2023.121751
30. Jiang Y, Guo H, Tong T, Xie F, Qin X, Wang X, et al. lncRNA lnc-POP1-1 upregulated by VN1R5 promotes cisplatin resistance in head and neck squamous cell carcinoma through interaction with MCM5. Mol Ther (2022) 30(1):448–67. doi: 10.1016/j.ymthe.2021.06.006
31. Hu CY, Su BH, Lee YC, Wang CT, Yang ML, Shen WT, et al. Interruption of the long non-coding RNA HOTAIR signaling axis ameliorates chemotherapy-induced cachexia in bladder cancer. J Biomed Sci (2022) 29(1):104. doi: 10.1186/s12929-022-00887-y
32. Yan H, Luo B, Wu X, Guan F, Yu X, Zhao L, et al. Cisplatin induces pyroptosis via activation of MEG3/NLRP3/caspase-1/GSDMD pathway in triple-negative breast cancer. Int J Biol Sci (2021) 17(10):2606–21. doi: 10.7150/ijbs.60292
33. Sheng X, Dai H, Du Y, Peng J, Sha R, Yang F, et al. LncRNA CARMN overexpression promotes prognosis and chemosensitivity of triple negative breast cancer via acting as miR143-3p host gene and inhibiting DNA replication. J Exp Clin Cancer Res CR (2021) 40(1):205. doi: 10.1186/s13046-021-02015-4
34. Chen Y, Li F, Li D, Liu W, Zhang L. Atezolizumab and blockade of LncRNA PVT1 attenuate cisplatin resistant ovarian cancer cells progression synergistically via JAK2/STAT3/PD-L1 pathway. Clin Immunol (Orlando Fla) (2021) 227:108728. doi: 10.1016/j.clim.2021.108728
35. Kozłowska-Masłoń J, Guglas K, Paszkowska A, Kolenda T, Podralska M, Teresiak A, et al. Radio-lncRNAs: biological function and potential use as biomarkers for personalized oncology. J Personalized Med (2022) 12(10):1605. doi: 10.3390/jpm12101605
36. Yao PA, Wu Y, Zhao K, Li Y, Cao J, Xing C. The feedback loop of ANKHD1/lncRNA MALAT1/YAP1 strengthens the radioresistance of CRC by activating YAP1/AKT signaling. Cell Death Dis (2022) 13(2):103. doi: 10.1038/s41419-022-04554-w
37. Liu S, Zhan N, Gao C, Xu P, Wang H, Wang S, et al. Long noncoding RNA CBR3-AS1 mediates tumorigenesis and radiosensitivity of non-small cell lung cancer through redox and DNA repair by CBR3-AS1 /miR-409-3p/SOD1 axis. Cancer Lett (2022) 526:1–11. doi: 10.1016/j.canlet.2021.11.009
38. Zhang F, Sang Y, Chen D, Wu X, Wang X, Yang W, et al. M2 macrophage-derived exosomal long non-coding RNA AGAP2-AS1 enhances radiotherapy immunity in lung cancer by reducing microRNA-296 and elevating NOTCH2. Cell Death Dis (2021) 12(5):467. doi: 10.1038/s41419-021-03700-0
39. Xu X, Ju Y, Zhao X, Yang P, Zhu F, Fang B. SMG7-AS1 as a prognostic biomarker and predictor of immunotherapy responses for skin cutaneous melanoma. Genomics (2023) 115(3):110614. doi: 10.1016/j.ygeno.2023.110614
40. Lin Q, Liu T, Wang X, Hou G, Xiang Z, Zhang W, et al. Long noncoding RNA HITT coordinates with RGS2 to inhibit PD-L1 translation in T cell immunity. J Clin Invest (2023) 133(11):e162951. doi: 10.1172/JCI162951
41. Bukhari I, Khan MR, Li F, Swiatczak B, Thorne RF, Zheng P, et al. Clinical implications of lncRNA LINC-PINT in cancer. Front Mol Biosci (2023) 10:1097694. doi: 10.3389/fmolb.2023.1097694
42. Rong Z, Wang Z, Wang X, Qin C, Geng W. Molecular interplay between linc01134 and YY1 dictates hepatocellular carcinoma progression. J Exp Clin Cancer Res CR (2020) 39(1):61. doi: 10.1186/s13046-020-01551-9
43. Wang C, Chen Y, Chen K, Zhang L. Long noncoding RNA LINC01134 promotes hepatocellular carcinoma metastasis via activating AKT1S1 and NF-κB signaling. Front Cell Dev Biol (2020) 8:429. doi: 10.3389/fcell.2020.00429
44. Zheng S, Guo Y, Dai L, Liang Z, Yang Q, Yi S. Long intergenic noncoding RNA01134 accelerates hepatocellular carcinoma progression by sponging microRNA-4784 and downregulating structure specific recognition protein 1. Bioengineered (2020) 11(1):1016–26. doi: 10.1080/21655979.2020.1818508
45. Yuan K, Lan J, Xu L, Feng X, Liao H, Xie K, et al. Long noncoding RNA TLNC1 promotes the growth and metastasis of liver cancer via inhibition of p53 signaling. Mol Cancer (2022) 21(1):105. doi: 10.1186/s12943-022-01578-w
46. Reyes-González JM, Quiñones-Díaz BI, Santana Y, Báez-Vega PM, Soto D, Valiyeva F, et al. Downstream effectors of ILK in cisplatin-resistant ovarian cancer. Cancers (2020) 12(4):880. doi: 10.3390/cancers12040880
47. Zhang N, Zhang H, Wu W, Zhou R, Li S, Wang Z, et al. Machine learning-based identification of tumor-infiltrating immune cell-associated lncRNAs for improving outcomes and immunotherapy responses in patients with low-grade glioma. Theranostics (2022) 12(13):5931–48. doi: 10.7150/thno.74281
48. Li G, Huo D, Guo N, Li Y, Ma H, Liu L, et al. Integrating multiple machine learning algorithms for prognostic prediction of gastric cancer based on immune-related lncRNAs. Front Genet (2023) 14:1106724. doi: 10.3389/fgene.2023.1106724
49. Ma L, Xu A, Kang L, Cong R, Fan Z, Zhu X, et al. LSD1-demethylated LINC01134 confers oxaliplatin resistance through SP1-induced p62 transcription in HCC. Hepatol (Baltimore Md) (2021) 74(6):3213–34. doi: 10.1002/hep.32079
50. Wang Z, Wang X, Rong Z, Dai L, Qin C, Wang S, et al. LncRNA LINC01134 contributes to radioresistance in hepatocellular carcinoma by regulating DNA damage response via MAPK signaling pathway. Front Pharmacol (2021) 12:791889. doi: 10.3389/fphar.2021.791889
51. Kang X, Huo Y, Jia S, He F, Li H, Zhou Q, et al. Silenced LINC01134 enhances oxaliplatin sensitivity by facilitating ferroptosis through GPX4 in hepatocarcinoma. Front Oncol (2022) 12:939605. doi: 10.3389/fonc.2022.939605
52. Li X, Zhang Z, Liu M, Fu X, Jun A, Chen G, et al. Establishment of a lncRNA-based prognostic gene signature associated with altered immune responses in HCC. Front Immunol (2022) 13:880288. doi: 10.3389/fimmu.2022.880288
53. Verheul TCJ, van Hijfte L, Perenthaler E, Barakat TS. The why of YY1: mechanisms of transcriptional regulation by yin yang 1. Front Cell Dev Biol (2020) 8:592164. doi: 10.3389/fcell.2020.592164
54. Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, et al. Transcription factor trapping by RNA in gene regulatory elements. Sci (New York NY) (2015) 350(6263):978–81. doi: 10.1126/science.aad3346
55. Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, et al. YY1 is a structural regulator of enhancer-promoter loops. Cell (2017) 171(7):1573–88.e28. doi: 10.1016/j.cell.2017.11.008
56. Alfarouk KO, Stock CM, Taylor S, Walsh M, Muddathir AK, Verduzco D, et al. Resistance to cancer chemotherapy: failure in drug response from ADME to P-gp. Cancer Cell Int (2015) 15:71. doi: 10.1186/s12935-015-0221-1
57. Guo J, Li L, Guo B, Liu D, Shi J, Wu C, et al. Mechanisms of resistance to chemotherapy and radiotherapy in hepatocellular carcinoma. Translational Cancer Research (2018) 7(3):765–81. doi: 10.21037/tcr.2018.05.20
58. Maiorino M, Conrad M, Ursini F. GPx4, lipid peroxidation, and cell death: discoveries, rediscoveries, and open issues. Antioxid Redox Signal (2018) 29(1):61–74. doi: 10.1089/ars.2017.7115
59. Forcina GC, Dixon SJ. GPX4 at the crossroads of lipid homeostasis and ferroptosis. Proteomics (2019) 19(18):e1800311. doi: 10.1002/pmic.201800311
60. Liu Y, Wan Y, Jiang Y, Zhang L, Cheng W. GPX4: The hub of lipid oxidation, ferroptosis, disease and treatment. Biochim Biophys Acta Rev Cancer (2023) 1878(3):188890. doi: 10.1016/j.bbcan.2023.188890
61. Luo F, Li M, Ding J, Zheng S. The progress in the treatment of hepatocellular carcinoma with portal vein tumor thrombus. Front Oncol (2021) 11:635731. doi: 10.3389/fonc.2021.635731
62. Mähringer-Kunz A, Steinle V, Düber C, Weinmann A, Koch S, Schmidtmann I, et al. Extent of portal vein tumour thrombosis in patients with hepatocellular carcinoma: The more, the worse? Liver Int (2019) 39(2):324–31. doi: 10.1111/liv.13988
63. Feng Y, Hu X, Ma K, Zhang B, Sun C. Genome-wide screening identifies prognostic long noncoding RNAs in hepatocellular carcinoma. BioMed Res Int (2021) 2021:6640652. doi: 10.1155/2021/6640652
64. Zhang J, Lou W. A key mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network linked to diagnosis and prognosis of hepatocellular carcinoma. Front Oncol (2020) 10:340. doi: 10.3389/fonc.2020.00340
65. Bu X, Ma L, Liu S, Wen D, Kan A, Xu Y, et al. A novel qualitative signature based on lncRNA pairs for prognosis prediction in hepatocellular carcinoma. Cancer Cell Int (2022) 22(1):95. doi: 10.1186/s12935-022-02507-z
66. Ozga AJ, Chow MT, Luster AD. Chemokines and the immune response to cancer. Immunity (2021) 54(5):859–74. doi: 10.1016/j.immuni.2021.01.012
67. Pawelec G, Borowitz A, Krammer PH, Wernet P. Constitutive interleukin 2 production by the JURKAT human leukemic T cell line. Eur J Immunol (1982) 12(5):387–92. doi: 10.1002/eji.1830120506
68. Abraham RT, Weiss A. Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat Rev Immunol (2004) 4(4):301–8. doi: 10.1038/nri1330
69. Allavena P, Grandi M, D'Incalci M, Geri O, Giuliani FC, Mantovani A. Human tumor cell lines with pleiotropic drug resistance are efficiently killed by interleukin-2 activated killer cells and by activated monocytes. Int J Cancer (1987) 40(1):104–7. doi: 10.1002/ijc.2910400119
70. Merchant R, Galligan C, Munegowda MA, Pearce LB, Lloyd P, Smith P, et al. Fine-tuned long-acting interleukin-2 superkine potentiates durable immune responses in mice and non-human primate. J Immunother Cancer (2022) 10(1):e003155. doi: 10.1136/jitc-2021-003155
71. Raeber ME, Sahin D, Karakus U, Boyman O. A systematic review of interleukin-2-based immunotherapies in clinical trials for cancer and autoimmune diseases. EBioMedicine (2023) 90:104539. doi: 10.1016/j.ebiom.2023.104539
72. Lee J, Moran JP, Fenton BM, Koch CJ, Frelinger JG, Keng PC, et al. Alteration of tumour response to radiation by interleukin-2 gene transfer. Br J Cancer (2000) 82(4):937–44. doi: 10.1054/bjoc.1999.1022
73. Li Z, Liu L, Feng C, Qin Y, Xiao J, Zhang Z, et al. LncBook 2.0: integrating human long non-coding RNAs with multi-omics annotations. Nucleic Acids Res (2023) 51(D1):D186–91. doi: 10.1093/nar/gkac999
74. Kang J, Tang Q, He J, Li L, Yang N, Yu S, et al. RNAInter v4.0: RNA interactome repository with redefined confidence scoring system and improved accessibility. Nucleic Acids Res (2022) 50(D1):D326–32. doi: 10.1093/nar/gkab997
75. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Sci (New York NY) (2020) 367(6478):eaau6977. doi: 10.1126/science.aau6977
76. Entezari M, Ghanbarirad M, Taheriazam A, Sadrkhanloo M, Zabolian A, Goharrizi M, et al. Long non-coding RNAs and exosomal lncRNAs: Potential functions in lung cancer progression, drug resistance and tumor microenvironment remodeling. Biomed Pharmacother Biomed Pharmacother (2022) 150:112963. doi: 10.1016/j.biopha.2022.112963
77. Zeng Y, Hu S, Luo Y, He K. Exosome cargos as biomarkers for diagnosis and prognosis of hepatocellular carcinoma. Pharmaceutics (2023) 15(9):2365. doi: 10.3390/pharmaceutics15092365
78. Lin H, Li J, Wang M, Zhang X, Zhu T. Exosomal long noncoding RNAs in NSCLC: dysfunctions and clinical potential. J Cancer (2023) 14(10):1736–50. doi: 10.7150/jca.84506
79. Jiang C, Zhang J, Wang W, Shan Z, Sun F, Tan Y, et al. Extracellular vesicles in gastric cancer: role of exosomal lncRNA and microRNA as diagnostic and therapeutic targets. Front Physiol (2023) 14:1158839. doi: 10.3389/fphys.2023.1158839
80. Lai H, Li Y, Zhang H, Hu J, Liao J, Su Y, et al. exoRBase 2.0: an atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids. Nucleic Acids Res (2022) 50(D1):D118–28. doi: 10.1093/nar/gkab1085
81. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL, et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol (2023) 24(6):430–47. doi: 10.1038/s41580-022-00566-8
82. Gao N, Li Y, Li J, Gao Z, Yang Z, Li Y, et al. Long non-coding RNAs: the regulatory mechanisms, research strategies, and future directions in cancers. Front Oncol (2020) 10:598817. doi: 10.3389/fonc.2020.598817
83. Qi X, Chen X, Zhao Y, Chen J, Niu B, Shen B. Prognostic roles of ceRNA network-based signatures in gastrointestinal cancers. Front Oncol (2022) 12:921194. doi: 10.3389/fonc.2022.921194
84. Xiong G, Pan S, Jin J, Wang X, He R, Peng F, et al. Long noncoding competing endogenous RNA networks in pancreatic cancer. Front Oncol (2021) 11:765216. doi: 10.3389/fonc.2021.765216
85. Huang Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med (2018) 22(12):5768–75. doi: 10.1111/jcmm.13866
86. Zhou T, Li M, Xiao Z, Cai J, Zhao W, Duan J, et al. Chronic stress-induced gene changes in vitro and in vivo: potential biomarkers associated with depression and cancer based on circRNA- and lncRNA-associated ceRNA networks. Front Oncol (2021) 11:744251. doi: 10.3389/fonc.2021.744251
87. Xu J, Xu J, Liu X, Jiang J. The role of lncRNA-mediated ceRNA regulatory networks in pancreatic cancer. Cell Death Discov (2022) 8(1):287. doi: 10.1038/s41420-022-01061-x
88. Niu ZS, Wang WH, Dong XN, Tian LM. Role of long noncoding RNA-mediated competing endogenous RNA regulatory network in hepatocellular carcinoma. World J Gastroenterol (2020) 26(29):4240–60. doi: 10.3748/wjg.v26.i29.4240
89. Li K, Yao T, Wang Z. lncRNA-mediated ceRNA network in bladder cancer. Noncoding RNA Res (2023) 8(2):135–45. doi: 10.1016/j.ncrna.2022.12.002
90. Shi Y, Liu JB, Deng J, Zou DZ, Wu JJ, Cao YH, et al. The role of ceRNA-mediated diagnosis and therapy in hepatocellular carcinoma. Hereditas (2021) 158(1):44. doi: 10.1186/s41065-021-00208-7
91. Wang P, Guo Q, Hao Y, Liu Q, Gao Y, Zhi H, et al. LnCeCell: a comprehensive database of predicted lncRNA-associated ceRNA networks at single-cell resolution. Nucleic Acids Res (2020) 49(D1):D125–33. doi: 10.1093/nar/gkaa1017
92. Chieffi P. Aurora B: A new promising therapeutic target in cancer. Intractable Rare Dis Res (2018) 7(2):141–4. doi: 10.5582/irdr.2018.01018
93. Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase–AKT pathway in human cancer. Nat Rev Cancer (2002) 2(7):489–501. doi: 10.1038/nrc839
94. Chen Y, Chen Z, Tang Y, Xiao Q. The involvement of noncanonical Wnt signaling in cancers. Biomed Pharmacother (2021) 133:110946. doi: 10.1016/j.biopha.2020.110946
95. Zhao H, Wang Y, Yang Z, Wei W, Cong Z, Xie Y. High expression of aurora kinase B predicts poor prognosis in hepatocellular carcinoma after curative surgery and its effects on the tumor microenvironment. Ann Transl Med (2022) 10(21):1168. doi: 10.21037/atm-22-4798
96. Luo Y, Liu L, Zhao J, Jiao Y, Zhang M, Xu G, et al. PI3K/AKT1 signaling pathway mediates sinomenine-induced hepatocellular carcinoma cells apoptosis: an in vitro and in vivo study. Biol Pharm Bull (2022) 45(5):614–24. doi: 10.1248/bpb.b21-01063
97. Selvaggi F, Catalano T, Cotellese R, Aceto GM. Targeting Wnt/β-catenin pathways in primary liver tumours: from microenvironment signaling to therapeutic agents. Cancers (2022) 14(8):1912. doi: 10.3390/cancers14081912
98. Pez F, Lopez A, Kim M, Wands JR, Caron de Fromentel C, Merle P. Wnt signaling and hepatocarcinogenesis: molecular targets for the development of innovative anticancer drugs. J Hepatol (2013) 59(5):1107–17. doi: 10.1016/j.jhep.2013.07.001
99. Nilsen TW. Endo-siRNAs: yet another layer of complexity in RNA silencing. Nat Struct Mol Biol (2008) 15(6):546–8. doi: 10.1038/nsmb0608-546
100. Maida Y, Kyo S, Lassmann T, Hayashizaki Y, Masutomi K. Off-target effect of endogenous siRNA derived from RMRP in human cells. Int J Mol Sci (2013) 14(5):9305–18. doi: 10.3390/ijms14059305
Keywords: hepatocellular carcinoma, LINC01134, tumorigenesis, tumor biomarker, therapeutic target
Citation: Yu Y, Wang J, Guo Q and Luo H (2024) LINC01134: a pivotal oncogene with promising predictive maker and therapeutic target in hepatocellular carcinoma. Front. Oncol. 14:1265762. doi: 10.3389/fonc.2024.1265762
Received: 23 July 2023; Accepted: 29 January 2024;
Published: 21 February 2024.
Edited by:
John Gibbs, Hackensack Meridian Health, United StatesReviewed by:
Patricia Schoenlein, Augusta University, United StatesCopyright © 2024 Yu, Wang, Guo and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongliang Luo, MTMwOTcyODAwMDFAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.