
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 22 December 2023
Sec. Cancer Molecular Targets and Therapeutics
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1327478
This article is part of the Research TopicMolecular Mechanisms and Therapeutic Targets of Cancer Metastasis and Therapy ResistanceView all 5 articles
 Kay K. Myo Min1†
Kay K. Myo Min1† Charlie B. Ffrench1†
Charlie B. Ffrench1† Barbara J. McClure1,2
Barbara J. McClure1,2 Michael Ortiz1
Michael Ortiz1 Emma L. Dorward1
Emma L. Dorward1 Michael S. Samuel1,2,3
Michael S. Samuel1,2,3 Lisa M. Ebert1,4
Lisa M. Ebert1,4 Mỹ G. Mahoney5
Mỹ G. Mahoney5 Claudine S. Bonder1,2*
Claudine S. Bonder1,2*Desmoglein-2 (DSG2) is a calcium-binding single pass transmembrane glycoprotein and a member of the large cadherin family. Until recently, DSG2 was thought to only function as a cell adhesion protein embedded within desmosome junctions designed to enable cells to better tolerate mechanical stress. However, additional roles for DSG2 outside of desmosomes are continuing to emerge, particularly in cancer. Herein, we review the current literature on DSG2 in cancer and detail its impact on biological functions such as cell adhesion, proliferation, migration, invasion, intracellular signaling, extracellular vesicle release and vasculogenic mimicry. An increased understanding of the diverse repertoire of the biological functions of DSG2 holds promise to exploit this cell surface protein as a potential prognostic biomarker and/or target for better patient outcomes. This review explores the canonical and non-canonical functions of DSG2, as well as the context-dependent impacts of DSG2 in the realm of cancer.
Adhesion between individual cells is crucial for structural integrity of tissues, and can regulate the formation and maintenance of a tumor (1). Desmosomes are specialized junctional complexes that can provide cell-to-cell adhesion to maintain tumor mass integrity. A loss of adhesion between cancer cells within a tumor contributes to cancer invasion, dissemination, and metastasis (2). Desmogleins (DSG) and desmocollins (DSC) are the two members of the cadherin superfamily that contribute to desmosomal junctions. Within desmosomes, there are four known isoforms of desmogleins (DSG1-4) and three known isoforms of desmocollins (DSC1-3). Desmoglein-2 (DSG2) is a calcium-binding single pass transmembrane glycoprotein of the cadherin protein family. DSG2 molecules on neighboring cells can dimerize via their extracellular domains (3–5). Within the adhesive unit of desmosomes, preferential heterophilic interactions occur between desmogleins and desmocollins (e.g. DSG2 and DSC2), but homophilic DSG2 interactions have also been documented (6).
DSG2 expression has been observed in several tissue types such as the epidermis, the intestinal mucosa and myocardial cells of the heart (4). Mutations in the DSG2 gene results in a loss of desmosomal adhesion, which has been associated with several autoimmune, infectious and hereditary disease states (7, 8).
There is increasing evidence from us and others that DSG2 regulates many cancer cell functions, including cell adhesion (9, 10), proliferation (11), migration, invasion (12), vasculogenic mimicry (13) and tumor growth (14–16). However, the precise role/s of DSG2 in cancer progression varies across different cancer types. In this article, we review the reported roles of DSG2 in cancer and detail the mechanisms by which DSG2 influences neoplastic behavior.
Research into the adhesive properties of cells began almost half a century ago when Takeichi documented a calcium(Ca2+)-dependent process that facilitated the formation of cell-cell contacts (17). Soon thereafter, the adhesive protein ‘cadherin’ was identified at the joining edge of adjacent epithelial cells, which served to link these junctions to the actin cytoskeleton (18, 19). The discovery of over 100 cadherin-like proteins revealed a superfamily of adhesion molecules (Figure 1) that collectively contribute to cell-cell recognition, intercellular adhesion and signaling pathways that are important for morphogenesis and tissue behavior.
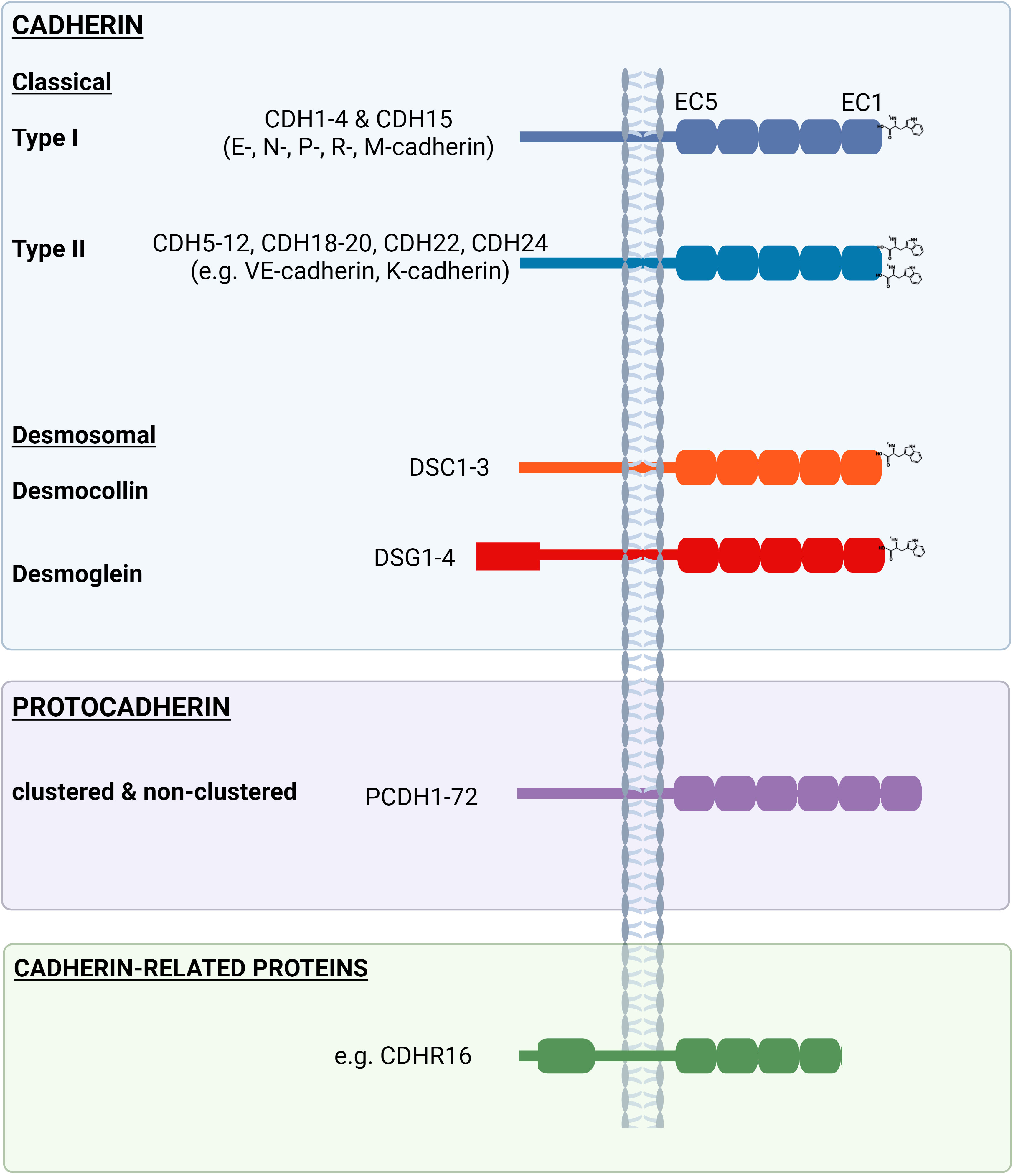
Figure 1 Examples of the ‘cadherin’ family comprising the ‘classical’ (type I and type II) and ‘desmosomal’ (desmogleins and desmocollins) (blue box), as well as ‘protocadherins’ (purple box’) and other ‘cadherin-related proteins’ (green box). Extracellular (EC) domains differ between and across cadherin family members, as do the length of intracellular domains.
Cadherins, such as DSG2, are cell surface proteins with an ectodomain comprised of cadherin repeats, followed by a single pass transmembrane domain and then a relatively short cytoplasmic domain. More specifically, cadherins contain at least two extracellular cadherin (EC) repeats, with each repeat comprised of ~110 amino acids and encoding an immunoglobulin-like fold of seven beta strands that form two beta sheets. The individual EC domains are largely conserved across the cadherin family and are interconnected via highly conserved Ca2+ binding regions. This strand of repeat units are held within the plasma membrane via a single pass transmembrane domain. Notably, the EC domain most proximal to the transmembrane region (i.e., EC5) and the intracellular region are poorly conserved across family members, with exception for the internal catenin-binding motifs. The cadherin superfamily is divided into three major groups; ‘cadherin’, ‘protocadherin’ and ‘cadherin-related proteins’ based on variations in amino acid sequence, structural properties and protein function (reviewed in (20)).
The cadherin family is further subdivided into two main groups, ‘classical’ (type I and type II) and ‘desmosomal’ (DSG and DSCs) (Figure 1). The classical type I (e.g., E-cadherin) cadherins, desmogleins and desmocollins all contain five consecutive EC repeats in their ectodomain and share a highly conserved tryptophan (Trp, W) at position 2 (Trp2) in EC1, which is used during adhesion by strand swapping for either cis- or trans- interactions (6). In contrast, type II cadherins (e.g., VE-cadherin) contain an additional Trp at position 4 (Trp4) in EC1 that orchestrates a homophilic interaction between partner proteins (21).
Compared to the other cadherins, the DSG cytoplasmic domain is considerably longer with an additional C-terminal DSG-specific cytoplasmic region containing an intracellular proline rich linker domain, a variable repeat unit domain (5 repeats in DSG1, 6 in DSG2, 2 in DSG3 and 3 in DSG4) and a DSG terminal domain (with the exception of DSG3, which lacks a terminal domain) (22).
The interdomains between the EC regions are stabilized by conserved Ca2+ binding sites. Three Ca2+ ions within each interface help to maintain the EC domain in an extended rigid configuration to enable adhesive binding (23). Adhesion between cadherin proteins involves strand exchange between the EC1 domains of opposed molecules, mediated by insertion of the hydrophobic side chain of conserved tryptophan residues (Trp2) into hydrophobic pockets on the opposing molecules (22) (Figure 2). Intracellular domains of desmosomal proteins engage with intracellular plakoglobin, plakophilin and desmoplakin, which associates with intermediate filaments, linking the complex to the cytoskeleton, thus forming a ‘hyper adhesive’ desmosome plaque that supports cellular mechanical stress (Figure 2).
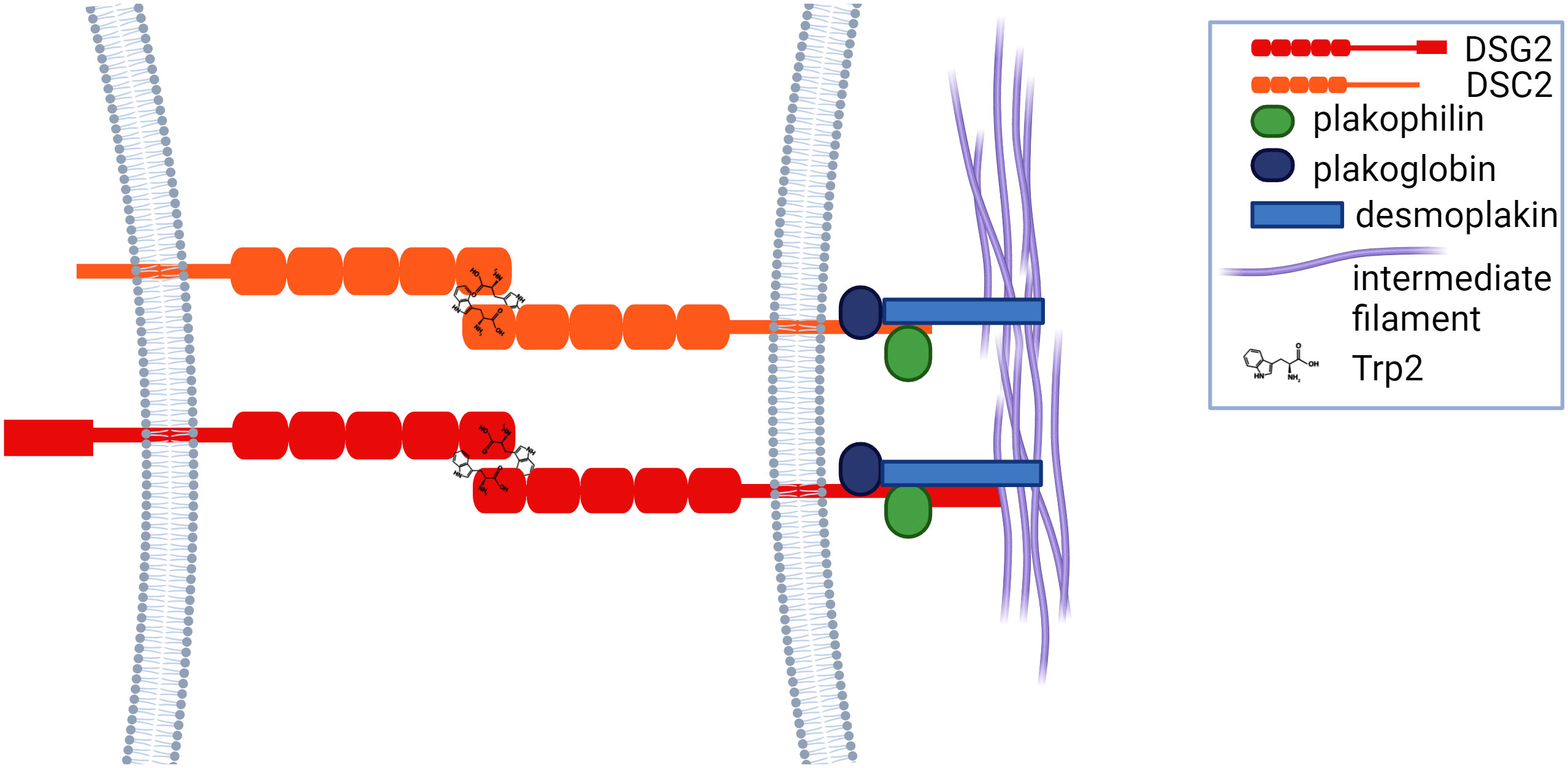
Figure 2 Cadherins adhere via tryptophan residues (Trp2) and strand exchange between the EC1 domains of opposed proteins, after which they engage with plakoglobin, plakophilin and desmoplakin, which is linked to intermediate filaments.
Desmosomal proteins, like other cadherins, can exhibit both homophilic and heterophilic adhesion. Live imaging of fluorescently tagged DSG2 demonstrated its localization to vesicles that, once mobilized to the membrane (24), are stabilized at the membrane via dimerization with plakoglobin proteins (and others) in the cytoplasmic domain (25).
Of particular focus in this review, DSG2 has demonstrated capability far beyond simply binding cells together through desmosomes. Phenotypic features identified in Dsg2 knockout mice, including alterations in cellular differentiation and proliferation (26), are not readily attributable to DSG2’s role in cell-cell adhesion alone. Most notable is the demonstration by Eshkind et al., that complete genetic ablation of Dsg2 in mice is embryonically lethal in the 129/Sv strain background, but not C57BL/6 mice, suggesting a strain dependent lethality (26). Outside of its canonical role in desmosomes and cell-to-cell adhesion, Dsg2 has also been detected in embryonic stem cells lacking desmosomes, where it supports embryonic stem cell proliferation and embryo survival (26). Consistent with this, a related study from our laboratory also demonstrated the viability of a hypomorphic whole body Dsg2 knock out in C57BL/6 mice (wherein low/residual levels of Dsg2 are detectable) (9). This hypomorphic allele is not embryonic lethal but does manifest a pathological response, particularly an enlarged and fibrotic heart (9). DSG2 has also been reported to regulate actin assembly through interaction with integrin-β8 to promote angiogenesis in systemic sclerosis microvascular endothelial cells (27), and to serve a pro-survival role in intestinal epithelial tissue (28).
Further evidence to support DSG2 as a unique desmosomal protein with a biological role outside of desmosomes, is the selective expression of DSG2 by non-desmosome forming cells e.g., endothelial cells (ECs) (9, 29). Indeed, DSG2 is documented to assist in the survival, proliferation, migration, and angiogenesis of endothelial progenitor cells and some ECs (9). Interestingly, a study of microvascular ECs has shown that DSG2 interaction with integrin-β8 delivers pro-angiogenic signals via the Rho family of GTPases, focal adhesion kinase (FAK), Mothers against decapentaplegic homolog1/5(SMAD1/5), and extracellular signal-regulated kinase (ERK) 1/2 (27). More recently, we also documented that DSG2 is expressed by insulin-producing β-cells, with in silico analysis revealing that DSG2 is in the top 10% of abundance of genes expressed by human islets (30).
DSG2 has also been identified on non-desmosome forming haemopoietic progenitor cells. Surface expression of DSG2 has been identified on circulating CD34+CD45dim hematopoietic progenitors (peripheral blood and cord blood), while other desmosomal components were noticeably absent (9). Interestingly, while DSG2 has been documented on progenitor and immature cells, its gene (Figure 3) and cell surface expression diminishes over the course of cellular differentiation (9, 31). The solitary expression of DSG2, and absence of other supporting desmosomal components, is uncharacteristic and suggests an alternative role for DSG2 within the hematological compartment. It is tempting to speculate that important similarities may emerge between DSG2-expressing stem cells and DSG2-expressing cancer cells. For example, the expression of DSG2 on pluripotent stem cells (PSC), is suggested to contribute to PSC self-renewal and pluripotency via regulation of β-catenin localization (32).
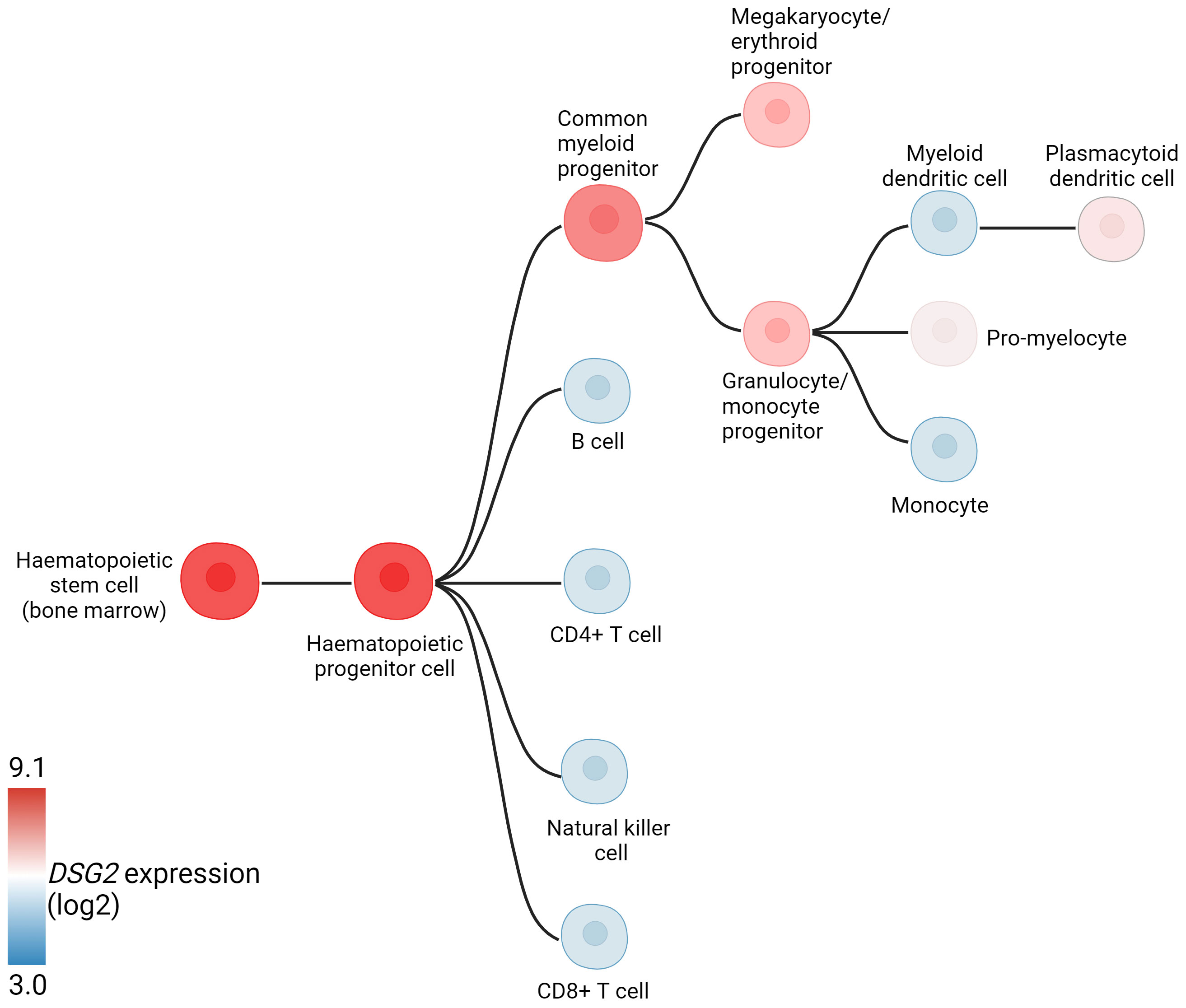
Figure 3 Hematopoietic hierarchical differentiation tree indicating DSG2 expression (log2) at different stages of differentiation. Adapted from BloodSpot (31).
Accumulating evidence suggests that DSG2 is dysregulated in human cancers. This observation creates an opportunity for DSG2 to be a viable biomarker across a myriad of cancers, particularly as a predictive biomarker for disease progression. Figure 4 shows analysis of publicly available in silico RNA seq datasets (Gene Expression Profiling Interactive Analysis, GEPIA2) demonstrating that DSG2 gene expression is significantly upregulated in cancers such as lung squamous cell carcinoma, head and neck adenocarcinoma, pancreatic adenocarcinoma, prostate adenocarcinoma and stomach adenocarcinoma relative to normal healthy tissue (33). Overall survival analyses in GEPIA2 also indicate that high DSG2 expression correlates with poor survival outcomes for patients with cancers including cervix, lung and pancreas (Figure 5) (33). In prognostic studies, high levels of DSG2 resulted in worse clinical outcomes for patients with melanoma, multiple myeloma, basal cell carcinoma (BCC), squamous cell carcinoma (SCC), squamous cell lung carcinoma, ovarian cancer, head and neck SCC, and hepatocellular carcinoma (Table 1). In curious contrast, colon, gastric, prostate, and high-grade serous ovarian carcinoma patients presented with poor outcomes when DSG2 levels were low (Table 1).
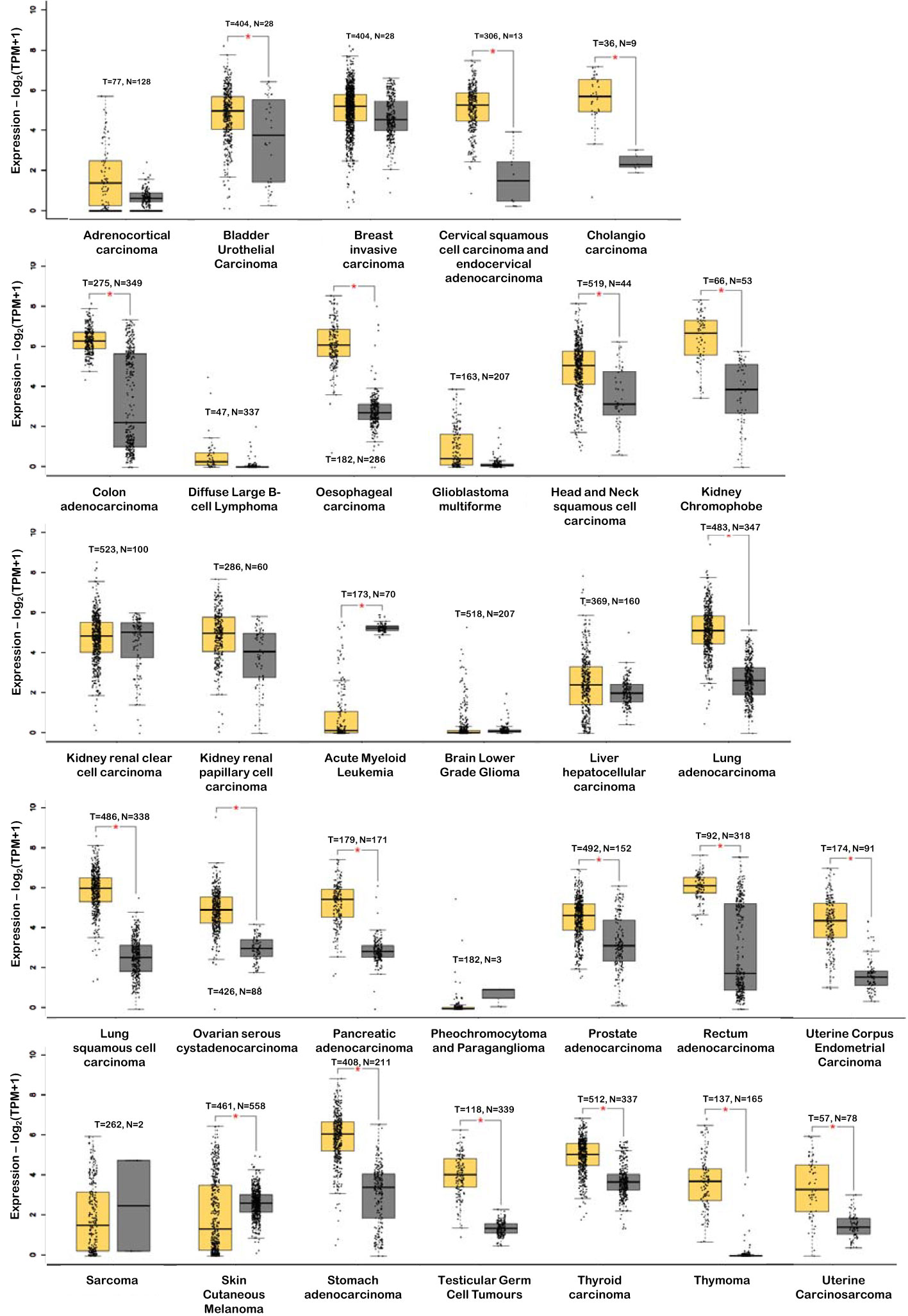
Figure 4 DSG2 gene expression in tumor (T – in yellow) vs normal (N – in grey) tissue. *p<0.05. Data generated from GEPIA2 (33).
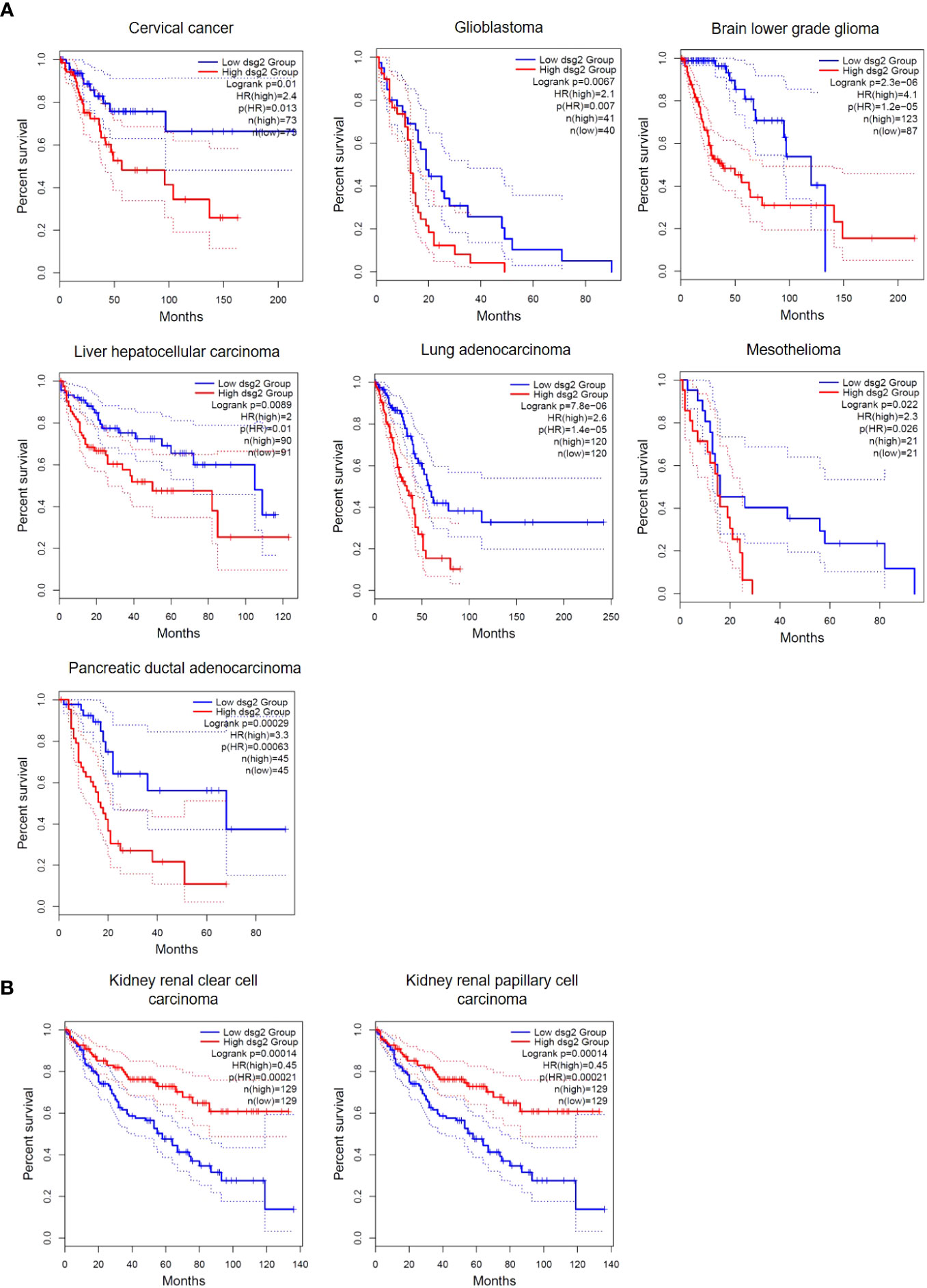
Figure 5 Overall Survival differences in DSG2-low vs DSG2-high expressing patients. (A) cervical cancer, glioblastoma, brain lower grade glioma, liver hepatocellular carcinoma, lung adenocarcinoma, mesothelioma and pancreatic ductal adenocarcinoma patients with significantly reduced Log rank OS in DSG2-high population, (B) kidney renal clear and papillary cell carcinoma patients with significantly increased Log rank OS in DSG2-high populations. Data generated from GEPIA2 (33).
It is well established that the lateral mobility of cell surface proteins like DSG2 is critical for maintaining membrane dynamics (80–82). This highly complex and fluid membrane activity is vital for intracellular signaling through receptor activation as well as intercellular communication through vesicles, and recycling of membrane components. It has been reported by us and others that DSG2 promotes cancer cell migration, invasion, and proliferation in cancers such as SCC, breast, cervical, colon, and lung cancer (Table 2). More recently, we have also demonstrated that DSG2 facilitates cancer cell adhesion in multiple myeloma (9). Here we discuss the different signaling pathways and cellular functions that are regulated by DSG2 and the impact this has on cancer progression.
DSG2 has been previously reported to facilitate EGFR activity; DSG2/EGFR interaction has demonstrated importance in the development of SCC (15), colon cancer (92), and lung adenocarcinoma (16). In response to EGF ligand stimulation, DSG2 is known to be involved in the activation of EGFR, c-Src and Signal transducer and activator of transcription (STAT3), which supports cancer cell growth and migration (15). In SCC, DSG2 co-localizes with the EGFR and promotes the activation of c-Src mediated signaling by displacing EGFR from the inhibitory lipid raft microenvironment, thereby making it available for ligand binding and signaling (104, 111). Overexpression of DSG2 in human SCC cells is also linked to enhanced cancer cell proliferation and migration (also via activation of c-Src) (15). The EGF/EGFR axis directly initiates the phosphorylation of membrane resident catenins that go on to facilitate links between junctional proteins in desmosomes and the actin cytoskeleton or intermediate filaments (112). This impacts cell migration with DSG2/EGF/EGFR-dependent tyrosine phosphorylation of plakoglobin being able to shift the cell from an ‘adhesive’ to a ‘migratory’ phenotype for increased epithelial cell motility (112). The EGFR is also reported to stabilize DSG2 at cellular junctions in an A disintegrin and metalloprotease (ADAM)-dependent manner (113). A study on colon cancer also showed that phosphorylation of the EGFR and downstream signal activation is enhanced by DSG2, where downregulation of DSG2 decreases EGF-induced cell proliferation and suppresses in vivo xenograft tumor growth (92). Additionally, in lung adenocarcinoma, loss of DSG2 (via gene silencing) leads to the nuclear translocation of EGFR, as well as suppression of EGFR signaling via the Src-Rac1-PAK1 pathway (16). Thus, through its role as a regulator of a key protein such as EGFR, DSG2 has far-reaching signaling effects that can potentiate oncogenic outcomes.
Additional pathways that have been linked to DSG2 include the mitogen-activated protein kinase (MAPK) and the sonic hedgehog (SHH) in cervical cancer and BCC respectively. In cervical cancer, DSG2 was observed to promote cell proliferation, migration, and invasion by modulating phospho Mitogen-activated protein kinase kinase (pMEK) and phospho Extracellular signal-regulated kinase (ERK) within the MAPK pathway (12). Whilst in BCC, DSG2 was shown to enhance downstream SHH signaling through the secretion of cytokines, upregulation of receptors (e.g., urokinase plasminogen activator surface receptor (uPAR) and interleukin (IL)-6R) and exosome secretion, leading to STAT3 phosphorylation and activation, and potentiating GLI1 expression (83). DSG2 also has reported roles in the progression of non-small cell lung cancer tumors in vitro and in vivo, with reduced expression of DSG2 causing cell cycle arrest at the G1 phase (11). DSG2 has also been reported to be regulated by Hypoxia-inducible factor 1 (HIF1)-α to support metastasis in breast cancer (85).
While the aforementioned pathways converge to promote tumor growth, there are also reports illustrating that reduced DSG2 levels promote tumorigenesis. For example, in thyroid cancer, DSG2 depletion led to increased cell migration and metastasis via activation of the hepatocyte growth factor receptor (HGFR, c-Met)/Src/Rac1 signaling pathway (110). In gallbladder carcinoma, loss of DSG2 was associated with cancer progression and resistance to EGFR-targeted therapy through activation of Src kinase (95). Furthermore, a study in pancreatic adenocarcinoma suggested that loss of DSG2 promoted cell migration and invasion by regulating the desmosomal linker protein plakoglobin (PG) in an ERK-dependent manner (100). Finally, in gastric cancer, DSG2 was reportedly down-regulated by cell-surface glycoprotein TROP2 to promote invasion and migration through DSG2/PG/β-catenin pathways (96).
Intercellular communication, which is essential for all biological processes, is accomplished by various means, including cell-cell and cell-microenvironment signaling, the release of modulating soluble molecules such as cytokines, chemokines, and growth factors, and the secretion of extracellular vesicles (EVs, cargo-carrying messengers) (114). Subversion of these normal signaling events disrupts normal tissue homeostasis and morphogenesis and underlies pathogenic signaling during cancer progression. EVs are lipid bilayer-bounded organelles with unique membrane-associated proteins and lipids carrying a cargo of proteins, lipids, and/or genetic material (115). The three major subpopulations of EVs include apoptotic bodies, microvesicles, and exosomes, distinguished by their mechanism of biogenesis, size, content, and biological functions. Most importantly, EVs can serve as important biomarkers for disease diagnosis and prognosis in the clinical setting (116, 117).
Exosomes are generated by membrane invagination into the early endosomal compartment and, upon a second round of membrane invagination, mature into multivesicular bodies (MVB), or late endosomes. MVB are degraded following fusion with lysosomes or trafficked back to the plasma membrane for release as encapsulated exosomes. The presence of DSG2 in exosomes was first reported in 2017 (106). A few years later, DSG2 was documented to modulate exosome biogenesis in a palmitoylation-dependent manner, suggesting lipid modification, membrane localization, and protein-protein interaction are critical determinants (14). Furthermore, DSG2 has been shown to regulate proteins, such as flotillin and caveolin, involved in the early steps of endosomal processing (14, 106).
It has been reported that DSG2 plays a role in modulating membrane dynamics through lipid rafts and the endosomal pathway leading to exosome generation and release. Caveolin-1, a known interactor of DSG2, initiates membrane invagination into vesicles of approximately 60-80 nm in diameter and, similar to exosomes, caveolae and lipid rafts are enriched in cholesterols and sphingolipids (118). Additionally, DSG2 undergoes ectodomain shedding mediated by ADAM10 and/or ADAM17, resulting in a 95-kDa ectodomain cleaved product and a 65-kDa membrane-spanning fragment (106, 119). This 65-kDa C-terminal fragment is detected in lipid rafts (104) as well as exosomes (106). Interestingly, DSG2 co-localizes with Sec3, a protein component of the exocyst, an octameric protein complex that facilitates the transport of secretory organelles from the Golgi to the plasma membrane (120). At the plasma membrane, these exocyst organelles dock at desmosomes prior to vesicle fusion. Exocysts do not fuse with the endosomes (121) and are distinct from multivesicular bodies, thus, inhibitors of the endocytic pathways do not affect exocyst biogenesis. These studies highlight many novel critical roles for DSG2 in intracellular signaling and vesicle biogenesis from initiating endosome formation to vesicle docking.
The mechanisms by which exosomes serve as signaling mediators is cargo dependent (122). The list of components detected in exosomes is ever expanding to include lipids, proteins, DNA, and various species of RNA (mRNA, miRNA, lncRNA, and cRNA) (123). DSG2 alters the miRNA levels and cytokine/chemokine profiles not only in cells but also in exosomes (14, 124). The direct impact of these factors modulated by DSG2 requires further study, but early studies show that exosomes derived from DSG2 overexpressing cancer cells enhance dermal fibroblast cell growth, demonstrating that DSG2 expression can modulate the tumor microenvironment. Most importantly, we can demonstrate that intercellular communication through cell-cell adhesion, cytokine release, and secretion of modified EVs are well-coordinated by DSG2. For instance, studies in SCC have also shown that DSG2 is involved in the release of EVs. Briefly, head and neck SCC EVs are enriched for DSG2 and overexpression of DSG2 is shown to increase the release of EVs which can enhance fibroblast cell growth (107). In line with this, EVs released from SCCs overexpressing DSG2 promote SCC tumor growth in vivo and contain cargo that is pro-tumorigenic, e.g. IL-8 and IL-6 (14).
The vasculature plays a pivotal role in tumor growth and metastasis. Tumors can develop a blood supply not only by promoting angiogenesis, where blood vessels are formed by ECs, but also by vessel-like structures directly formed by cancer cells, in a process known as vasculogenic mimicry (VM) (125–127). Abundant VM has been linked to increased invasion, high tumor grade, metastasis, and overall poor survival for several cancer types (128). Of relevance here, DSG2 is an important regulator of both angiogenesis and VM; for example, in a mouse model of melanoma, loss of Dsg2 expression in the host was associated with fewer EC-lined tumor vessels (9). Furthermore, elevated DSG2 on melanoma cells promoted the formation of VM structures in vitro; in support of a pro-vascular phenotype, melanoma cancer cells expressing high levels of DSG2 also overexpress VM-associated genes (13). While further studies are required to fully elucidate the role of DSG2 in VM formation, data to-date suggest that targeting DSG2 in melanoma may inhibit VM formation and suppress tumor growth and metastasis.
Current literature suggests that DSG2 plays a broad and significant role in regulating various cancer cell functions, including cell proliferation, survival, migration, invasion, adhesion, cell cycle progression, vesicle secretion, and tumor development (Table 2; Figure 6). Notably, conflicting roles for DSG2 in cancer progression have emerged and at present appear to coincide with the different cancer types. To the best of our knowledge, Table 2 summarizes the current landscape for DSG2 in cancer and suggests that for the majority of cancers, elevated expression levels of DSG2 promotes signaling pathways that support cancer progression. Briefly, studies related to SCC, BCC, breast cancer, cervical cancer, colon cancer, lung cancer, multiple myeloma, and ovarian cancer are predominantly associated with a pro-tumorigenic function for DSG2. In contrast, reports implicating an anti-tumorigenic function for DSG2 exist for cancers of the gallbladder, thyroid, and the stomach. One potential explanation for these observed differences may involve the interplay between DSG2 and other cell surface expressed proteins associated with cell adhesion. For example, inflammation-induced reduction of DSG2 is linked to alterations in the expression of tight junction proteins, such as Claudin2 (129). Disruption of Claudins facilitates the infiltration of growth factors into the mucosa which are known to promote neoplastic transformation (130). This is supported by Wang et al, who documented that loss of DSG2 on gallbladder cancer cells promoted EMT with reduced expression of E-cadherin and increased expression of Snail (47).
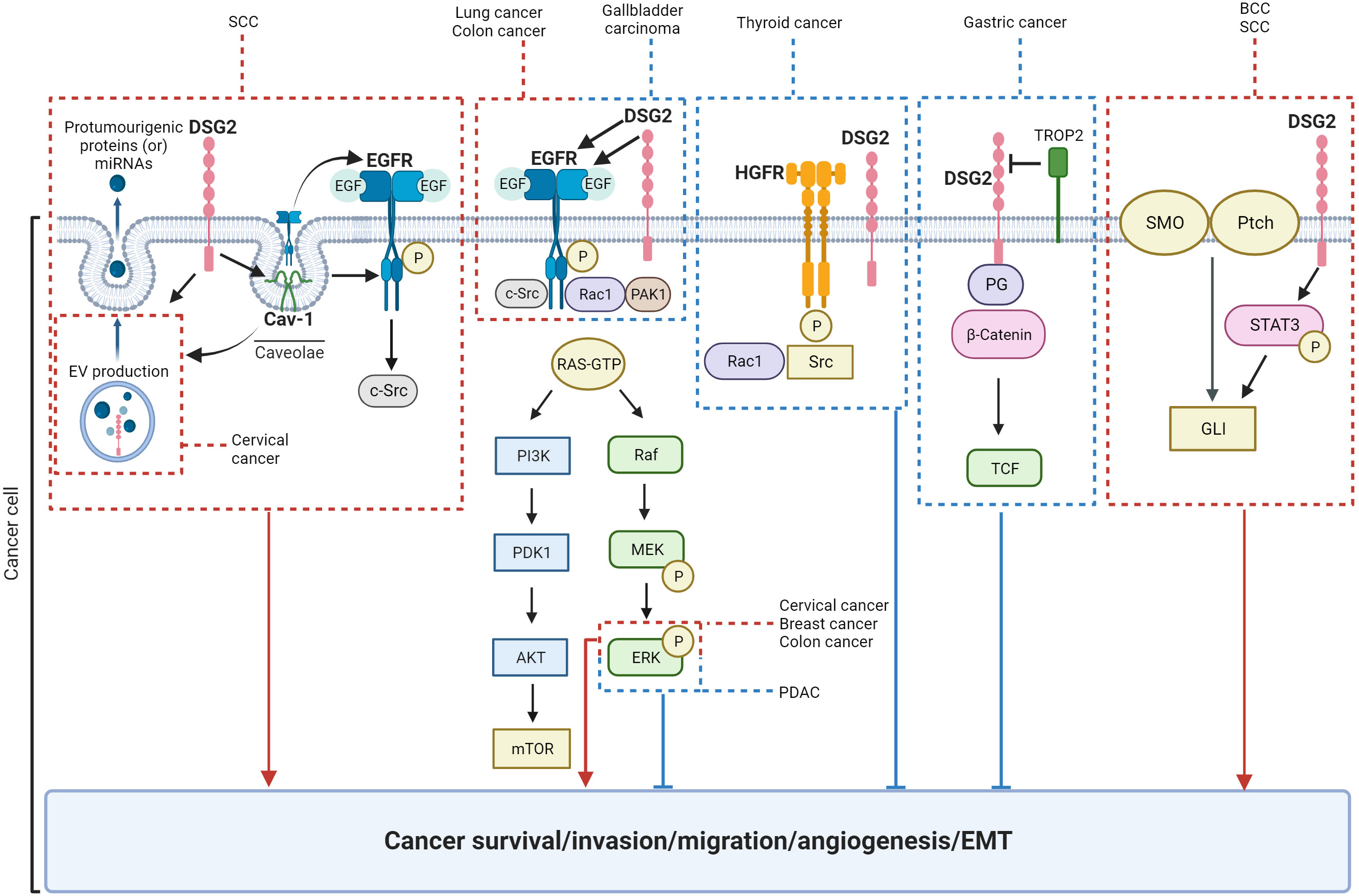
Figure 6 Schematic of signaling pathways known to be regulated by DSG2. Key: red = pro-tumorigenic, blue = anti-tumorigenic.
We speculate that DSG2 may exist as both a vital part of desmosomes facilitating cell-to-cell adhesion as part of its canonical function, as well as an independent signal transducer. This leads to the inference that the mechanisms through which DSG2 influences cancer progression are influenced by the tumor microenvironment and the origin of the cancer itself. DSG2 has been shown to activate a variety of signaling pathways and proteins including MAPK, STAT3, growth factor receptors. To this end, DSG2-mediated EGFR signaling consistently promotes cancer progression whereas DSG2-mediated HGFR signaling suggests an inhibition of cancer growth (Figure 6). Taking the heterogeneity within individual cancers into consideration, it is evident that the role DSG2 plays in tumorigenesis is complex and context dependent. Therefore, the potential for DSG2 to act as a prognostic biomarker is context dependent and influenced by factors such as organ/tissue type, cancer cell location within a tissue, genetic mutations, as well as the broader tumor microenvironment. To date, there is no distinguishing feature that identifies the ‘tumor promoting’ from the ‘tumor suppressing’ role of DSG2 in cancer. To gain clarification, studies that stratify tumor stage, subtype, and genetic drivers based on DSG2 levels across multiple cancers would be informative.
Like other transmembrane junctional proteins, DSG2 can be cleaved by various proteases. DSG2 is predicted to have several cleavage sites; it has been reported that full length DSG2 (150 kDa) can be cleaved into two or three cleaved fragments, i.e. a 110 kDa ectodomain (i.e., extracellular portion) fragment, a 95 kDa ectodomain fragment and a 65 kDa membrane-spanning fragment (104, 119, 131). The ectodomain of DSG2 can be cleaved by proteases including matrix metalloproteinase (MMP) 9 (103, 119), ADAM10 and ADAM17 (103, 113, 119, 132, 133), kallikrein 7 (69), matriptase (134), and γ-secretase (135). Interestingly, the intracellular portion of DSG2 can also be cleaved by caspase 3 and 8 (28, 136, 137) as well as calpain (28). Several studies have also shown that inflammatory mediators such as IL-1β, tissue necrosis factor (TNF)α, interferon (IFN)-γ as well as EGF can induce/increase both the extracellular and intracellular cleavage of DSG2 via activation of the aforementioned proteases (28, 103, 119, 132, 133, 136).
Within cancer, the ectodomain cleavage of DSG2 has been associated with decreased cell-cell adhesion (134) and decreased cell proliferation (119, 136). Yulis et al. (136) and Nava et al. (28) showed that IFNγ/TNFα-induced cleavage of DSG2 caused cancer cell apoptosis. Reports, including our own study, indicate that the cleaved ectodomain of DSG2 can be detected in cell culture supernatant and serum (human and mouse) by ELISA (54, 61, 62, 65, 88, 104, 138). Liu et al. (60) demonstrated that the levels of serum DSG2 are significantly higher in patients with esophageal squamous cell carcinoma and esophagogastric junction adenocarcinoma. Kim et al. (61). reported that patients with ovarian cancer who had high circulating levels of soluble DSG2 had poorer progression-free survival compared to healthy controls (i.e., a median survival of 16 months vs. 26 months). Notably, while proteases that cleave DSG2 have been identified, little is known about how the circulating DSG2 fragments cause progression of cancer, and addressing this requires further research.
Also of interest is the ability of DSG2 to act as a high-affinity receptor for some species B human adenoviruses (e.g. HAd3, 7, 11 and 14), with the second and third ectodomains (EC2 and EC3) of DSG2 binding to the fiber knob protein of the virus (139). When bound to DSG2, the adenovirus activates the MAPK pathway to upregulate the proteases ADAM17 and MMP9 that then cleave the extracellular domain of DSG2 releasing it from the cell surface (140). The significance of this process is yet to be determined, but it is evident that adenovirus binding to DSG2 leads to the transient opening of intercellular junctions. Recent reports have shown that Ad3 fiber knob-containing recombinant junction opening proteins (JO-1,2,3,4) are able to bind to and trigger DSG2 shedding, causing a junctional opening (141). Relevant to this review, when administered in vivo in combination with chemotherapy, JO1/4 increased the efficacy of chemotherapeutic drugs, resulting in decreased tumor growth and delayed drug resistance (142–144). Capitalizing on this biological function for better cancer outcomes is yet to be fully realized and warrants careful consideration.
Furthermore, evidence supports a correlation between DSG2 and human papillomavirus (HPV) linked cancers. Zhao et al. showed that DSG2 expression levels were related to HPV status, with all HPV-positive cervical cancer patients exhibiting high DSG2 levels (40). This is supported by a recent study wherein DSG2 levels were elevated in HPV-positive head and neck SCCs when compared to HPV-negative tumors (107). Moreover, DSG2 expression was four times higher in HPV-positive patients who failed to respond to immune checkpoint inhibitors (ICI) compared to those who did respond (107). These results suggest that DSG2 may play an underappreciated role in HPV-induced cervical cancer.
From a potential therapeutic standpoint, since there is evidence to suggest that blocking DSG2 could achieve tumor destruction (Table 2), there may be therapeutic benefit in utilizing DSG2 as either as a monotherapy (e.g., monoclonal antibody, small molecule inhibitors etc.) or in combination with chemotherapy, radiation, ICI, or other targeted therapies for select cancers. For instance, given that DSG2 promotes EGFR expression and EGF-mediated signaling in cancer cells, inhibition of DSG2 in combination with EGFR targeted therapy (e.g., Erlotinib) could be of clinical benefit. There is also building evidence suggesting that DSG2 plays a role in modulating ICI efficacy. More specifically, a study in head and neck SCC suggested that DSG2-mediated signaling negatively impacted ICI responses (107). Based on these observations, targeting DSG2, prior to (or in combination) with ICI treatment, may improve clinical outcomes for current ICI non-responders. Alternatively, given that DSG2 is a functional receptor for select adenoviruses, the possibility exists for its use to improve the systemic delivery of oncolytic viruses (145). The use of JO proteins that target DSG2 and cause junctional opening is yet another promising therapeutic option that could be considered in combination with chemotherapeutics (144). Notably, given the current consensus that DSG2 has varied roles in cancer progression (as described in Table 2), careful consideration is warranted.
In summary, this review presents evidence that DSG2 is unlike other desmosomal components in its capacity to act as a solitary protein with a broader repertoire of biological roles. This is of clinical interest as DSG2 is increasingly implicated in the poor prognosis of patients with cancer. To capitalize on DSG2 as a potential biomarker for cancer prognosis and treatment, we continue to acquire knowledge of its association with (and consequences of) key intracellular signaling pathways. The physical positioning of DSG2 within the plasma membrane of cells provides ample opportunity for it to directly engage with various factors in the local microenvironment (e.g. growth factors and integrins) that initiate cell signaling pathways. These non-canonical roles for DSG2 are undoubtedly advantageous for cancerous cells who thrive on improved cell survival, proliferation and migration. Intersecting these cellular advantages via targeting DSG2 in cancer could provide clinical benefit.
KM: Conceptualization, Writing – original draft, Writing – review & editing. CF: Conceptualization, Writing – original draft, Writing – review & editing. BM: Writing – original draft, Writing – review & editing. MO: Writing – original draft. ED: Writing – review & editing. MS: Writing – review & editing. LE: Writing – review & editing. MM: Writing – original draft, Writing – review & editing. CB: Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This review was supported by grants to CB: a Tour de Cure (Pioneering Grant RSP-358-2020), Cancer Council SA (Beat Cancer Project Support GNT2013221) and NHMRC (GNT2021009 & GNT2013460). KM, CF and MO were supported by a RTP/USAPA post-graduate scholarship. CF was also supported by the Australian Rotary Health and Phillip & Glenis Hayes PhD scholarship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Brooke MA, Nitoiu D, Kelsell DP. Cell–cell connectivity: desmosomes and disease. J Pathol (2012) 226(2):158–71. doi: 10.1002/path.3027
2. Martin TA, Ye L, Sanders AJ, Lane J, Jiang WG. Cancer Invasion and Metastasis: Molecular and Cellular Perspective. Austin (TX: Landes Bioscience: Madame Curie Bioscience Database (2000-2013).
3. Kowalczyk AP, Bornslaeger EA, Norvell SM, Palka HL, Green KJ. Desmosomes: intercellular adhesive junctions specialized for attachment of intermediate filaments. In: Jeon KW, editor. International Review of Cytology. Netherlands: Academic Press (1998). p. 237–302.
4. Chitaev NA, Troyanovsky SM. Direct Ca2+-dependent heterophilic interaction between desmosomal cadherins, desmoglein and desmocollin, contributes to cell–cell adhesion. J Cell Biol (1997) 138(1):193–201. doi: 10.1083/jcb.138.1.193
5. Wang CE, Yumul RC, Lin J, Cheng Y, Lieber A, Pun SH. Junction opener protein increases nanoparticle accumulation in solid tumors. J Control Rel (2018) 272:9–16. doi: 10.1016/j.jconrel.2017.12.032
6. Harrison OJ, Brasch J, Lasso G, Katsamba PS, Ahlsen G, Honig B, et al. Structural basis of adhesive binding by desmocollins and desmogleins. Proc Natl Acad Sci (2016) 113(26):7160–5. doi: 10.1073/pnas.1606272113
7. Awad MM, Dalal D, Cho E, Amat-Alarcon N, James C, Tichnell C, et al. DSG2 mutations contribute to arrhythmogenic right ventricular dysplasia/cardiomyopathy. Am J Hum Genet (2006) 79(1):136–42. doi: 10.1086/504393
8. Brennan D, Mahoney MG. Increased expression of Dsg2 in Malignant skin carcinomas. Cell Adhesion Migration (2009) 3(2):148–54. doi: 10.4161/cam.3.2.7539
9. Ebert LM, Tan LY, Johan MZ, Min KK, Cockshell MP, Parham KA, et al. A non-canonical role for desmoglein-2 in endothelial cells: implications for neoangiogenesis. Angiogenesis (2016) 19(4):463–86. doi: 10.1007/s10456-016-9520-y
10. Gupta A, Nitoiu D, Brennan-Crispi D, Addya S, Riobo NA, Kelsell DP, et al. Cell cycle- and cancer-associated gene networks activated by Dsg2: evidence of cystatin A deregulation and a potential role in cell-cell adhesion. PLoS One (2015) 10(3):e0120091. doi: 10.1371/journal.pone.0120091
11. Cai F, Zhu Q, Miao Y, Shen S, Su X, Shi Y. Desmoglein-2 is overexpressed in non-small cell lung cancer tissues and its knockdown suppresses NSCLC growth by regulation of p27 and CDK2. J Cancer Res Clin Oncol (2017) 143(1):59–69. doi: 10.1007/s00432-016-2250-0
12. Zhou BX, Li Y. Significance of desmoglein-2 on cell Malignant behaviors via mediating MAPK signaling in cervical cancer. Kaohsiung J Med Sci (2020) 36(5):336–43. doi: 10.1002/kjm2.12182
13. Tan LY, Mintoff C, Johan MZ, Ebert BW, Fedele C, Zhang YF, et al. Desmoglein 2 promotes vasculogenic mimicry in melanoma and is associated with poor clinical outcome. Oncotarget (2016) 7(29):46492–508. doi: 10.18632/oncotarget.10216
14. Flemming JP, Hill BL, Haque MW, Raad J, Bonder CS, Harshyne LA, et al. miRNA- and cytokine-associated extracellular vesicles mediate squamous cell carcinomas. J Extracell Vesicles (2020) 9(1):1790159. doi: 10.1080/20013078.2020.1790159
15. Overmiller AM, McGuinn KP, Roberts BJ, Cooper F, Brennan-Crispi DM, Deguchi T, et al. c-Src/Cav1-dependent activation of the EGFR by Dsg2. Oncotarget (2016) 7(25):37536–55. doi: 10.18632/oncotarget.7675
16. Jin R, Wang X, Zang R, Liu C, Zheng S, Li H, et al. Desmoglein-2 modulates tumor progression and osimertinib drug resistance through the EGFR/Src/PAK1 pathway in lung adenocarcinoma. Cancer Lett (2020) 483:46–58. doi: 10.1016/j.canlet.2020.04.001
17. Takeichi M. Functional correlation between cell adhesive properties and some cell surface proteins. J Cell Biol (1977) 75(2 Pt 1):464–74. doi: 10.1083/jcb.75.2.464
18. Urushihara H, Takeichi M. Cell-cell adhesion molecule: identification of a glycoprotein relevant to the Ca2+-independent aggregation of Chinese hamster fibroblasts. Cell (1980) 20(2):363–71. doi: 10.1016/0092-8674(80)90622-4
19. Volk T, Geiger B. A 135-kd membrane protein of intercellular adherens junctions. EMBO J (1984) 3(10):2249–60. doi: 10.1002/j.1460-2075.1984.tb02123.x
20. Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol (2009) 41(2):349–69. doi: 10.1016/j.biocel.2008.09.027
21. Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, et al. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell (2006) 124(6):1255–68. doi: 10.1016/j.cell.2005.12.046
22. Garrod D, Chidgey M. Desmosome structure, composition and function. Biochim Et Biophys Acta-Biomembr (2008) 1778(3):572–87. doi: 10.1016/j.bbamem.2007.07.014
23. Pokutta S, Herrenknecht K, Kemler R, Engel J. Conformational changes of the recombinant extracellular domain of E-cadherin upon calcium binding. Eur J Biochem (1994) 223(3):1019–26. doi: 10.1111/j.1432-1033.1994.tb19080.x
24. Nekrasova OE, Amargo EV, Smith WO, Chen J, Kreitzer GE, Green KJ. Desmosomal cadherins utilize distinct kinesins for assembly into desmosomes. J Cell Biol (2011) 195(7):1185–203. doi: 10.1083/jcb.201106057
25. Chen J, Nekrasova OE, Patel DM, Klessner JL, Godsel LM, Koetsier JL, et al. The C-terminal unique region of desmoglein 2 inhibits its internalization via tail-tail interactions. J Cell Biol (2012) 199(4):699–711. doi: 10.1083/jcb.201202105
26. Eshkind L, Tian Q, Schmidt A, Franke WW, Windoffer R, Leube RE. Loss of desmoglein 2 suggests essential functions for early embryonic development and proliferation of embryonal stem cells. Eur J Cell Biol (2002) 81(11):592–8. doi: 10.1078/0171-9335-00278
27. Giusti B, Margheri F, Rossi L, Lapini I, Magi A, Serratì S, et al. Correction: desmoglein-2-integrin Beta-8 interaction regulates actin assembly in endothelial cells: deregulation in systemic sclerosis. PLoS One (2013) 8(7). doi: 10.1371/annotation/b41766f2-c23d-455e-8d6e-e4bce5ae1d80
28. Nava P, Laukoetter MG, Hopkins AM, Laur O, Gerner-Smidt K, Green KJ, et al. Desmoglein-2: a novel regulator of apoptosis in the intestinal epithelium. Mol Biol Cell (2007) 18(11):4565–78. doi: 10.1091/mbc.e07-05-0426
29. Giusti B, Margheri F, Rossi L, Lapini I, Magi A, Serrati S, et al. Desmoglein-2-integrin Beta-8 interaction regulates actin assembly in endothelial cells: deregulation in systemic sclerosis. PLoS One (2013) 8(7):e68117. doi: 10.1371/journal.pone.0068117
30. Myo Min KK, Rojas-Canales D, Penko D, DeNichilo M, Cockshell MP, Ffrench CB, et al. Desmoglein-2 is important for islet function and beta-cell survival. Cell Death Dis (2022) 13(10):911. doi: 10.1038/s41419-022-05326-2
31. Bagger FO, Kinalis S, Rapin N. BloodSpot: a database of healthy and Malignant haematopoiesis updated with purified and single cell mRNA sequencing profiles. Nucleic Acids Res (2018) 47(D1):D881–5. doi: 10.1093/nar/gky1076
32. Park J, Son Y, Lee NG, Lee K, Lee DG, Song J, et al. DSG2 is a functional cell surface marker for identification and isolation of human pluripotent stem cells. Stem Cell Rep (2018) 11(1):115–27. doi: 10.1016/j.stemcr.2018.05.009
33. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res (2019) 47(W1):W556–60. doi: 10.1093/nar/gkz430
34. Brennan D, Mahoney MG. Increased expression of Dsg2 in Malignant skin carcinomas: A tissue-microarray based study. Cell Adhesion Migration (2009) 3(2):148–54. doi: 10.4161/cam.3.2.7539
35. Gornowicz-Porowska J, Bowszyc-Dmochowska M, Seraszek-Jaros A, Kaczmarek E, Dmochowski M. Loss of correlation between intensities of desmoglein 2 and desmoglein 3 expression in basal cell carcinomas. Acta Dermatovenerol Croat (2011) 19(3):150–5.
36. Pietkiewicz P, Gornowicz-Porowska J, Bowszyc‐Dmochowska M, Jagielska J, Helak‐Łapaj C, Kaczmarek E, et al. Discordant expression of desmoglein 2 and 3 at the mRNA and protein levels in nodular and superficial basal cell carcinoma revealed by immunohistochemistry and fluorescent in situ hybridization. Clin Exp Dermatol (2014) 39(5):628–35. doi: 10.1111/ced.12355
37. Wild PJ, Catto JW, Abbod MF, Linkens DA, Herr A, Pilarsky C, et al. Artificial intelligence and bladder cancer arrays. Verh Dtsch Ges Pathol (2007) 91:308–19.
38. Qin S, Liao Y, Du Q, Wang W, Huang J, Liu P, et al. DSG2 expression is correlated with poor prognosis and promotes early-stage cervical cancer. Cancer Cell Int (2020) 20:206. doi: 10.1186/s12935-020-01292-x
39. Meng H, Liu J, Qiu J, Nie S, Jiang Y, Wan Y, et al. Identification of key genes in association with progression and prognosis in cervical squamous cell carcinoma. DNA Cell Biol (2020) 39(5):848–63. doi: 10.1089/dna.2019.5202
40. Zhao M, Huang W, Zou S, Shen Q, Zhu X. A five-genes-based prognostic signature for cervical cancer overall survival prediction. Int J Genomics (2020) 2020. doi: 10.1155/2020/8347639
41. An R, Meng S, Qian H. Identification of key pathways and establishment of a seven-gene prognostic signature in cervical cancer. J Oncol (2022) 2022. doi: 10.1155/2022/4748796
42. Wang J, Zheng H, Han Y, Wang G, Li Y. A novel four-gene prognostic signature as a risk biomarker in cervical cancer. Int J Genomics (2020) 2020. doi: 10.1155/2020/4535820
43. Molika P, Bissanum R, Surachat K, Pattanapanyasat K, Hanprasertpong J, Chotigeat W, et al. Exploration of extracellular vesicle long non-coding RNAs in serum of patients with cervical cancer. Oncology (2023). doi: 10.1159/000533145
44. Yang T, Gu X, Jia L, Guo J, Tang Q, Zhu J, et al. DSG2 expression is low in colon cancer and correlates with poor survival. BMC Gastroenterol (2021) 21(1):7–7. doi: 10.1186/s12876-020-01588-2
45. Zhu J, O'Mara TA, Liu D, Setiawan VW, Glubb D, Spurdle AB, et al. Associations between genetically predicted circulating protein concentrations and endometrial cancer risk. Cancers (Basel) (2021) 13(9). doi: 10.3390/cancers13092088
46. Xu S, Huang S, Li D, Zou Q, Yuan Y, Yang Z. Negative expression of DSG1 and DSG2, as prognostic biomarkers, impacts on the overall survival in patients with extrahepatic cholangiocarcinoma. Analytical Cell Pathol (2020) 2020. doi: 10.1155/2020/9831646
47. Wang L, Lv Y, Li J, Nan Y, Piao L, Liang Z. Downregulation of desmoglein 2 promotes EMT progression in gallbladder cancer. Histol Histopathol (2022) p:18535–5. doi: 10.14670/HH-18-535
48. Yashiro M, Nishioka N, Hirakawa K. Decreased expression of the adhesion molecule desmoglein-2 is associated with diffuse-type gastric carcinoma. Eur J Cancer (2006) 42(14):2397–403. doi: 10.1016/j.ejca.2006.03.024
49. Zhang HY, Chang H, Fan XN, Zhang KD, Yu L, Cao Y. Expression of adhesion molecules and mucins in human and rhesus macaque gastrointestinal epithelial cells. Histol Histopathol (2011) 26(11):1405–13. doi: 10.14670/HH-26.1405
50. Biedermann K, Vogelsang H, Becker I, Plaschke S, Siewert JR, Höfler H, et al. Desmoglein 2 is expressed abnormally rather than mutated in familial and sporadic gastric cancer. J Pathol (2005) 207(2):199–206. doi: 10.1002/path.1821
51. Oshima T, Yoshikawa T, Miyagi Y, Morita S, Yamamoto M, Tanabe K, et al. Biomarker analysis to predict the pathological response to neoadjuvant chemotherapy in locally advanced gastric cancer: an exploratory biomarker study of COMPASS, a randomized phase II trial. Oncotarget (2020) 11(30):2906. doi: 10.18632/oncotarget.27658
52. Bates EA, Lovatt C, Plein AR, Davies JA, Siebzehnrubl FA, Parker AL, et al. Engineering adenoviral vectors with improved GBM selectivity. Viruses (2023) 15(5). doi: 10.3390/v15051086
53. Han CP, Yu YH, Wang AG, Tian Y, Zhang HT, Zheng ZM, et al. Desmoglein-2 overexpression predicts poor prognosis in hepatocellular carcinoma patients. Eur Rev Med Pharmacol Sci (2018) 22(17):5481–9. doi: 10.26355/eurrev_201809_15808
54. Cury SS, Lapa RML, de Mello JBH, Marchi FA, Domingues MAC, Pinto CAL, et al. Increased DSG2 plasmatic levels identified by transcriptomic-based secretome analysis is a potential prognostic biomarker in laryngeal carcinoma. Oral Oncol (2020) 103:104592. doi: 10.1016/j.oraloncology.2020.104592
55. Sun R, Ma C, Wang W, Yang S. Upregulation of desmoglein 2 and its clinical value in lung adenocarcinoma: a comprehensive analysis by multiple bioinformatics methods. PeerJ (2020) 8:e8420. doi: 10.7717/peerj.8420
56. Weng C-F, Huang C-J, Wu M-H, Lee HH-C, Ling T-Y. Co-expression of Coxsackievirus/adenovirus receptors and desmoglein 2 in lung adenocarcinoma: A comprehensive analysis of bioinformatics and tissue microarrays. J Clin Med (2020) 9(11):3693. doi: 10.3390/jcm9113693
57. Su C, Liu W-X, Wu L-S, Dong T-J, Liu J-F. Screening of hub gene targets for lung cancer via microarray data. Combinatorial Chem High Throughput Screening (2021) 24(2):269–85. doi: 10.2174/1386207323666200808172631
58. Ebert LM, Vandyke K, Johan MZ, DeNichilo M, Tan LY, Myo Min KK, et al. Desmoglein-2 expression is an independent predictor of poor prognosis patients with multiple myeloma. Mol Oncol (2022) 16(6):1221–40. doi: 10.1002/1878-0261.13055
59. Roy Choudhury S, Byrum SD, Alkam D, Ashby C, Zhan F, Tackett AJ, et al. Expression of integrin β-7 is epigenetically enhanced in multiple myeloma subgroups with high-risk cytogenetics. Clin Epigenet (2023) 15(1):18. doi: 10.1186/s13148-023-01433-9
60. Liu Y-Q, Chu L-Y, Yang T, Zhang B, Zheng Z-T, Xie J-J, et al. Serum DSG2 as a potential biomarker for diagnosis of esophageal squamous cell carcinoma and esophagogastric junction adenocarcinoma. Biosci Rep (2022) 42(5):BSR20212612. doi: 10.1042/BSR20212612
61. Kim J, Beidler P, Wang H, Li C, Quassab A, Coles C, et al. Desmoglein-2 as a prognostic and biomarker in ovarian cancer. Cancer Biol Ther (2020) 21(12):1154–62. doi: 10.1080/15384047.2020.1843323
62. Hüttenhain R, Choi M, de la Fuente LM, Oehl K, Chang C-Y, Zimmermann A-K, et al. A targeted mass spectrometry strategy for developing proteomic biomarkers: A case study of epithelial ovarian cancer*[S]. Mol Cell Proteomics (2019) 18(9):1836–50. doi: 10.1074/mcp.RA118.001221
63. Xie H, Xu H, Hou Y, Cai Y, Rong Z, Song W, et al. Integrative prognostic subtype discovery in high-grade serous ovarian cancer. J Cell Biochem (2019) 120(11):18659–66. doi: 10.1002/jcb.29049
64. Chen L, Liu X, Zhang J, Liu Y, Gao A, Xu Y, et al. Characterization of desmoglein 2 expression in ovarian serous tumors and its prognostic significance in high-grade serous carcinoma. Int J Clin Exp Pathol (2018) 11(10):4977–86.
65. Kosanam H, Prassas I, Chrystoja CC, Soleas I, Chan A, Dimitromanolakis A, et al. Laminin, gamma 2 (LAMC2): a promising new putative pancreatic cancer biomarker identified by proteomic analysis of pancreatic adenocarcinoma tissues. Mol Cell Proteomics (2013) 12(10):2820–32. doi: 10.1074/mcp.M112.023507
66. Ormanns S, Altendorf-Hofmann A, Jackstadt R, Horst D, Assmann G, Zhao Y, et al. Desmogleins as prognostic biomarkers in resected pancreatic ductal adenocarcinoma. Br J Cancer (2015) 113(10):1460–6. doi: 10.1038/bjc.2015.362
67. Tang P, Qu W, Wu D, Chen S, Liu M, Chen W, et al. Identifying and validating an acidosis-related signature associated with prognosis and tumor immune infiltration characteristics in pancreatic carcinoma. J Immunol Res (2021) 2021. doi: 10.1155/2021/3821055
68. Chen D, Wirth KM, Kizy S, Muretta JM, Markowski TW, Yong P, et al. Desmoglein 2 functions as a receptor for fatty acid binding protein 4 in breast cancer epithelial cells. Mol Cancer Res (2023) 21(8):836–48. doi: 10.1158/1541-7786.MCR-22-0763
69. Ramani VC, Hennings L, Haun RS. Desmoglein 2 is a substrate of kallikrein 7 in pancreatic cancer. BMC Cancer (2008) 8(1):1–10. doi: 10.1186/1471-2407-8-373
70. Reichert ZR, Kasputis T, Nallandhighal S, Abusamra SM, Kasputis A, Haruray S, et al. Multigene profiling of circulating tumor cells (CTCs) for prognostic assessment in treatment-naïve metastatic hormone-sensitive prostate cancer (mHSPC). Int J Mol Sci (2021) 23(1):4. doi: 10.3390/ijms23010004
71. Barber AG, Castillo-Martin M, Bonal DM, Rybicki BA, Christiano AM, Cordon-Cardo C. Characterization of desmoglein expression in the normal prostatic gland. Desmoglein 2 is an independent prognostic factor for aggressive prostate cancer. PloS One (2014) 9(6):e98786. doi: 10.1371/journal.pone.0098786
72. Barber AG, Castillo-Martin M, Bonal DM, Jia AJ, Rybicki BA, Christiano AM, et al. PI3K/AKT pathway regulates E-cadherin and Desmoglein 2 in aggressive prostate cancer. Cancer Med (2015) 4(8):1258–71. doi: 10.1002/cam4.463
73. Trojan L, Schaaf A, Steidler A, Haak M, Thalmann G, Knoll T, et al. Identification of metastasis-associated genes in prostate cancer by genetic profiling of human prostate cancer cell lines. Anticancer Res (2005) 25(1a):183–91. doi: 10.1016/S1569-9056(05)80090-9
74. Kurzen H, Münzing I, Hartschuh W. Expression of desmosomal proteins in squamous cell carcinomas of the skin. J Cutaneous Pathol (2003) 30(10):621–30. doi: 10.1034/j.1600-0560.2003.00122.x
75. Xin Z, Yamaguchi A, Sakamoto K. Aberrant expression and altered cellular localization of desmosomal and hemidesmosomal proteins are associated with aggressive clinicopathological features of oral squamous cell carcinoma. Virchows Archiv (2014) 465(1):35–47. doi: 10.1007/s00428-014-1594-6
76. Saaber F, Chen Y, Cui T, Yang L, Mireskandari M, Petersen I, et al. Expression of desmogleins 1-3 and their clinical impacts on human lung cancer. Pathol Res Pract (2015) 211(3):208–13. doi: 10.1016/j.prp.2014.10.008
77. Thangaraj SV, Shyamsundar V, Krishnamurthy A, Ramshankar V. Deregulation of extracellular matrix modeling with molecular prognostic markers revealed by transcriptome sequencing and validations in Oral Tongue squamous cell carcinoma. Sci Rep (2021) 11(1):1–15. doi: 10.1038/s41598-020-78624-4
78. Yang W, Zhou W, Zhao X, Wang X, Duan L, Li Y, et al. Prognostic biomarkers and therapeutic targets in oral squamous cell carcinoma: a study based on cross-database analysis. Hereditas (2021) 158(1):1–17. doi: 10.1186/s41065-021-00181-1
79. Fang WK, Gu W, Liao LD, Chen B, Wu ZY, Wu JY, et al. Prognostic significance of desmoglein 2 and desmoglein 3 in esophageal squamous cell carcinoma. Asian Pacific J Cancer Prev (2014) 15(2):871–6. doi: 10.7314/APJCP.2014.15.2.871
80. Fielding CJ, Fielding PE. Molecular physiology of reverse cholesterol transport. J Lipid Res (1995) 36(2):211–28. doi: 10.1016/S0022-2275(20)39898-9
81. Alenghat FJ, Golan DE. Membrane protein dynamics and functional implications in mammalian cells. Curr Top Membr (2013) 72:89–120. doi: 10.1016/B978-0-12-417027-8.00003-9
82. Paul D, Stern O, Vallis Y, Dhillon J, Buchanan A, McMahon H. Cell surface protein aggregation triggers endocytosis to maintain plasma membrane proteostasis. Nat Commun (2023) 14(1):947. doi: 10.1038/s41467-023-36496-y
83. Brennan-Crispi DM, Overmiller AM, Tamayo-Orrego L, Marous MR, Sahu J, McGuinn KP, et al. Overexpression of desmoglein 2 in a mouse model of Gorlin syndrome enhances spontaneous basal cell carcinoma formation through STAT3-mediated Gli1 expression. J Invest Dermatol (2019) 139(2):300–7. doi: 10.1016/j.jid.2018.09.009
84. Rieger-Christ KM, Ng L, Hanley RS, Durrani O, Ma H, Yee AS, et al. Restoration of plakoglobin expression in bladder carcinoma cell lines suppresses cell migration and tumorigenic potential. Br J Cancer (2005) 92(12):2153–9. doi: 10.1038/sj.bjc.6602651
85. Davies E, Cochrane R, Hiscox S, Jiang W, Sweetland H, Mansel R. The role of desmoglein 2 and E-cadherin in the invasion and motility of human breast cancer cells. Int J Oncol (1997) 11(2):415–9. doi: 10.3892/ijo.11.2.415
86. Chang P-H, Chen M-C, Tsai Y-P, Tan GY, Hsu P-H, Jeng Y-M, et al. Interplay between desmoglein2 and hypoxia controls metastasis in breast cancer. Proc Natl Acad Sci (2021) 118(3):e2014408118. doi: 10.1073/pnas.2014408118
87. Al-Awadhi FH, Salvador LA, Law BK, Paul VJ, Luesch H, et al. Kempopeptin C, a novel marine-derived serine protease inhibitor targeting invasive breast cancer. Mar Drugs (2017) 15(9):290. doi: 10.3390/md15090290
88. Santiago-Josefat B, Esselens C, Bech-Serra JJ, Arribas J. Post-transcriptional up-regulation of ADAM17 upon epidermal growth factor receptor activation and in breast tumors*. J Biol Chem (2007) 282(11):8325–31. doi: 10.1074/jbc.M608826200
89. Chen L, Liu Y, Xu Y, Afify SM, Gao A, Du J, et al. Up-regulation of Dsg2 confered stem cells with Malignancy through wnt/β-catenin signaling pathway. Exp Cell Res (2023) 422(1):113416. doi: 10.1016/j.yexcr.2022.113416
90. Koyama-Nasu R, Takahashi R, Yanagida S, Nasu-Nishimura Y, Oyama M, Kozuka-Hata H, et al. The cancer stem cell marker CD133 interacts with plakoglobin and controls desmoglein-2 protein levels. PLoS One (2013) 8(1):e53710. doi: 10.1371/journal.pone.0053710
91. Chen Y, Stagg C, Schlessinger D, Nagaraja R. PLAC1 affects cell to cell communication by interacting with the desmosome complex. Placenta (2021) 110:39–45. doi: 10.1016/j.placenta.2021.06.001
92. Kamekura R, Kolegraff KN, Nava P, Hilgarth RS, Feng M, Parkos CA, et al. Loss of the desmosomal cadherin desmoglein-2 suppresses colon cancer cell proliferation through EGFR signaling. Oncogene (2013) 33:4531. doi: 10.1038/onc.2013.442
93. Wei Z, Ma W, Qi X, Zhu X, Wang Y, Xu Z, et al. Pinin facilitated proliferation and metastasis of colorectal cancer through activating EGFR/ERK signaling pathway. Oncotarget (2016) 7(20):29429. doi: 10.18632/oncotarget.8738
94. Saias L, Gomes A, Cazales M, Ducommun B, Lobjois V. Cell–cell adhesion and cytoskeleton tension oppose each other in regulating tumor cell aggregation. Cancer Res (2015) 75(12):2426–33. doi: 10.1158/0008-5472.CAN-14-3534
95. Lee S-H, Kim J-M, Lee DG, Lee J, Park J-G, Han T-S, et al. Loss of desmoglein-2 promotes gallbladder carcinoma progression and resistance to EGFR-targeted therapy through Src kinase activation. Cell Death Differ (2021) 28(3):968–84. doi: 10.1038/s41418-020-00628-4
96. Yang T, Jia L, Bian S, Chang X, Zhang Q, Tang Q, et al. TROP2 down-regulated DSG2 to promote gastric cancer cell invasion and migration by EGFR/AKT and DSG2/PG/β-catenin pathways. Curr Cancer Drug Targets (2022) 22(8):691–702. doi: 10.2174/1568009622666220407111013
97. Peitsch WK, Doerflinger Y, Fischer-Colbrie R, Huck V, Bauer AT, Utikal J, et al. Desmoglein 2 depletion leads to increased migration and upregulation of the chemoattractant secretoneurin in melanoma cells. PLoS One (2014) 9(2):e89491. doi: 10.1371/journal.pone.0089491
98. Brito JL, Walker B, Jenner M, Dickens NJ, Brown NJ, Ross FM, et al. MMSET deregulation affects cell cycle progression and adhesion regulons in t(4;14) myeloma plasma cells. Haematologica (2009) 94(1):78–86. doi: 10.3324/haematol.13426
99. Kurrey NK, A. K, Bapat SA. Snail and Slug are major determinants of ovarian cancer invasiveness at the transcription level. Gynecologic Oncol (2005) 97(1):155–65. doi: 10.1016/j.ygyno.2004.12.043
100. Hütz K, Zeiler J, Sachs L, Ormanns S, Spindler V, et al. Loss of desmoglein 2 promotes tumorigenic behavior in pancreatic cancer cells. Mol Carcinogen (2017) 56(8):1884–95. doi: 10.1002/mc.22644
101. Lin Z, Du Y, Duan Y, Zhang W, Huang Z, Yin X, et al. Clinicopathological Features, Immune Infiltration Landscape and Involved Signaling Pathways of the desmogleins family in Pancreatic Adenocarcinoma. United States: Research Square (2023).
102. Lorch JH, Thomas TO, Schmoll H-J. Bortezomib inhibits cell-cell adhesion and cell migration and enhances epidermal growth factor receptor inhibitor–induced cell death in squamous cell cancer. Cancer Res (2007) 67(2):727–34. doi: 10.1158/0008-5472.CAN-06-2162
103. Lorch JH, Klessner J, Park JK, Getsios S, Wu YL, Stack MS, et al. Epidermal growth factor receptor inhibition promotes desmosome assembly and strengthens intercellular adhesion in squamous cell carcinoma cells. J Biol Chem (2004) 279(35):37191–200. doi: 10.1074/jbc.M405123200
104. Brennan D, Peltonen S, Dowling A, Medhat W, Green KJ, Wahl JK 3rd, et al. A role for caveolin-1 in desmoglein binding and desmosome dynamics. Oncogene (2012) 31(13):1636–48. doi: 10.1038/onc.2011.346
105. Brennan-Crispi DM, Hossain C, Sahu J, Brady M, Riobo NA, Mahoney MG. Crosstalk between Desmoglein 2 and Patched 1 accelerates chemical-induced skin tumorigenesis. Oncotarget (2015) 6(11):8593–605. doi: 10.18632/oncotarget.3309
106. Overmiller AM, Pierluissi JA, Wermuth PJ, Sauma S, Martinez‐Outschoorn U, Tuluc M, et al. Desmoglein 2 modulates extracellular vesicle release from squamous cell carcinoma keratinocytes. FASEB J (2017) 31(8):3412–24. doi: 10.1096/fj.201601138RR
107. Hill BL, Calder AN, Flemming JP, Guo Y, Gilmore SL, Trofa MA, et al. IL-8 correlates with nonresponse to neoadjuvant nivolumab in HPV positive HNSCC via a potential extracellular vesicle miR-146a mediated mechanism. Mol Carcinogenesis (2023) 62(9):1428–43. doi: 10.1002/mc.23587
108. Teh M-T, Ken Parkinson E, Thurlow JK, Liu F, Fortune F, Wan H, et al. A molecular study of desmosomes identifies a desmoglein isoform switch in head and neck squamous cell carcinoma. J Oral Pathol Med (2011) 40(1):67–76. doi: 10.1111/j.1600-0714.2010.00951.x
109. Kume K, Haraguchi M, Hijioka H, Ishida T, Miyawaki A, Nakamura N, et al. The transcription factor Snail enhanced the degradation of E-cadherin and desmoglein 2 in oral squamous cell carcinoma cells. Biochem Biophys Res Commun (2013) 430(3):889–94. doi: 10.1016/j.bbrc.2012.12.060
110. Lee K, Lee S-H, Kim W, Lee J, Park J-G, Kim J-S, et al. Dsg2-mediated c-Met activation in anaplastic thyroid cancer motility and invasion. Endocr-Related Cancer (2020) 27(11):601–14. doi: 10.1530/ERC-19-0403
111. Zimmer SE, Kowalczyk AP. The desmosome as a model for lipid raft driven membrane domain organization. Biochim Biophys Acta Biomembr (2020) 1862(9):183329. doi: 10.1016/j.bbamem.2020.183329
112. Gaudry CA, Palka HL, Dusek RL, Huen AC, Khandekar MJ, Hudson LG, et al. Tyrosine-phosphorylated Plakoglobin Is Associated with Desmogleins but Not Desmoplakin after Epidermal Growth Factor Receptor Activation. J Biol Chem (2001) 276(27):24871–80. doi: 10.1074/jbc.M102731200
113. Klessner JL, Desai BV, Amargo EV, Getsios S, Green KJ. EGFR and ADAMs cooperate to regulate shedding and endocytic trafficking of the desmosomal cadherin desmoglein 2. Mol Biol Cell (2009) 20(1):328–37. doi: 10.1091/mbc.e08-04-0356
114. Armingol E, Officer A, Harismendy O, Lewis NE. Deciphering cell-cell interactions and communication from gene expression. Nat Rev Genet (2021) 22(2):71–88. doi: 10.1038/s41576-020-00292-x
115. Doyle LM, Wang MZ. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells (2019) 8(7). doi: 10.3390/cells8070727
116. Xu K, Liu Q, Wu K, Liu L, Zhao M, Yang H, et al. Extracellular vesicles as potential biomarkers and therapeutic approaches in autoimmune diseases. J Transl Med (2020) 18(1):432. doi: 10.1186/s12967-020-02609-0
117. Lee IT, Shen CH, Tsai FC, Chen CB, Ma KS, et al. Cancer-derived extracellular vesicles as biomarkers for cutaneous squamous cell carcinoma: A systematic review. Cancers (Basel) (2022) 14(20). doi: 10.3390/cancers14205098
118. Busija AR, Patel HH, Insel PA. Caveolins and cavins in the trafficking, maturation, and degradation of caveolae: implications for cell physiology. Am J Physiol Cell Physiol (2017) 312(4):C459–c477. doi: 10.1152/ajpcell.00355.2016
119. Kamekura R, Nava P, Feng M, Quiros M, Nishio H, Weber DA, et al. Inflammation-induced desmoglein-2 ectodomain shedding compromises the mucosal barrier. Mol Biol Cell (2015) 26(18):3165–77. doi: 10.1091/mbc.E15-03-0147
120. Andersen NJ, Yeaman C. Sec3-containing exocyst complex is required for desmosome assembly in mammalian epithelial cells. Mol Biol Cell (2010) 21(1):152–64. doi: 10.1091/mbc.e09-06-0459
121. Wang J, Ding Y, Wang J, Hillmer S, Miao Y, Lo SW, et al. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell (2010) 22(12):4009–30. doi: 10.1105/tpc.110.080697
122. Simeone P, Bologna G, Lanuti P, Pierdomenico L, Guagnano MT, Pieragostino D, et al. Extracellular vesicles as signaling mediators and disease biomarkers across biological barriers. Int J Mol Sci (2020) 21(7). doi: 10.3390/ijms21072514
123. Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer (2019) 18(1):75. doi: 10.1186/s12943-019-0991-5
124. Flemming JP, Hill BL, Anderson-Pullinger L, Harshyne LA, Mahoney MG, et al. Cytokine profiling in low- and high-density small extracellular vesicles from epidermoid carcinoma cells. JID Innov (2021) 1(4):100053. doi: 10.1016/j.xjidi.2021.100053
125. Hendrix MJ, Seftor EA, Meltzer PS, Gardner LM, Hess AR, Kirschmann DA, et al. Expression and functional significance of VE-cadherin in aggressive human melanoma cells: role in vasculogenic mimicry. Proc Natl Acad Sci USA (2001) 98(14):8018–23. doi: 10.1073/pnas.131209798
126. Delgado-Bellido D, Serrano-Saenz S, Fernández-Cortés M, Oliver FJ. Vasculogenic mimicry signaling revisited: focus on non-vascular VE-cadherin. Mol Cancer (2017) 16:65. doi: 10.1186/s12943-017-0631-x
127. Chung AS, Lee J, Ferrara N. Targeting the tumour vasculature: insights from physiological angiogenesis. Nat Rev Cancer (2010) 10(7):505–14. doi: 10.1038/nrc2868
128. Yang JP, Liao YD, Mai DM, Xie P, Qiang YY, Zheng LS, et al. Tumor vasculogenic mimicry predicts poor prognosis in cancer patients: a meta-analysis. Angiogenesis (2016) 19(2):191–200. doi: 10.1007/s10456-016-9500-2
129. Burkard N, Meir M, Kannapin F, Otto C, Petzke M, Germer C-T. Desmoglein2 regulates Claudin2 expression by sequestering PI-3-kinase in intestinal epithelial cells. Front Immunol (2021) 12. doi: 10.3389/fimmu.2021.756321
130. Soler AP, Miller RD, Laughlin KV, Carp NZ, Klurfeld DM, Mullin JM. Increased tight junctional permeability is associated with the development of colon cancer. Carcinogenesis (1999) 20(8):1425–31. doi: 10.1093/carcin/20.8.1425
131. Kolegraff K, Nava P, Laur O, Parkos CA, Nusrat A. Characterization of full-length and proteolytic cleavage fragments of desmoglein-2 in native human colon and colonic epithelial cell lines. Cell adhesion migration (2011) 5(4):306–14. doi: 10.4161/cam.5.4.16911
132. Bech-Serra JJ, Santiago-Josefat B, Esselens C, Saftig P, Baselga J, Arribas J, et al. Proteomic identification of desmoglein 2 and activated leukocyte cell adhesion molecule as substrates of ADAM17 and ADAM10 by difference gel electrophoresis. Mol Cell Biol (2006) 26(13):5086–95. doi: 10.1128/MCB.02380-05
133. Esselens C, Bech-Serra JJ, Arribas J. Post-transcriptional up-regulation of ADAM17 upon epidermal growth factor receptor activation and in breast tumors. J Biol Chem (2007) 282(11):8325–31. doi: 10.1074/jbc.M608826200
134. Wadhawan V, Kolhe YA, Sangith N, Gautam AKS, Venkatraman P. From prediction to experimental validation: desmoglein 2 is a functionally relevant substrate of matriptase in epithelial cells and their reciprocal relationship is important for cell adhesion. Biochem J (2012) 447(1):61–70. doi: 10.1042/BJ20111432
135. Hemming ML, Elias JE, Gygi SP, Selkoe DJ. Proteomic profiling of γ-secretase substrates and mapping of substrate requirements. PLoS Biol (2008) 6(10):e257. doi: 10.1371/journal.pbio.0060257
136. Yulis M, Quiros M, Hilgarth R, Parkos CA, Nusrat A, et al. Intracellular Desmoglein-2 cleavage sensitizes epithelial cells to apoptosis in response to pro-inflammatory cytokines. Cell Death Dis (2018) 9(3):1–13. doi: 10.1038/s41419-018-0380-9
137. Cirillo N, Lanza M, De Rosa A, Cammarota M, La Gatta A, Gombos F, et al. The most widespread desmosomal cadherin, desmoglein 2, is a novel target of caspase 3-mediated apoptotic machinery. J Cell Biochem (2008) 103(2):598–606. doi: 10.1002/jcb.21431
138. Greening DW, Kapp EA, Ji H, Speed TP, Simpson RJ. Colon tumour secretopeptidome: insights into endogenous proteolytic cleavage events in the colon tumour microenvironment. Biochim Biophys Acta (BBA)-Proteins Proteomics (2013) 1834(11):2396–407. doi: 10.1016/j.bbapap.2013.05.006
139. Vassal-Stermann E, Effantin G, Zubieta C, Burmeister W, Iseni F, Wang H, et al. CryoEM structure of adenovirus type 3 fibre with desmoglein 2 shows an unusual mode of receptor engagement. Nat Commun (2019) 10(1):1181. doi: 10.1038/s41467-019-09220-y
140. Wang H, Ducournau C, Saydaminova K, Richter M, Yumul R, Ho M, et al. Intracellular signaling and desmoglein 2 shedding triggered by human adenoviruses Ad3, Ad14, and Ad14P1. J Virol (2015) 89(21):10841–59. doi: 10.1128/JVI.01425-15
141. Bahlmann NA, Tsoukas RL, Erkens S, Wang H, Jönsson F, Aydin M, et al. Properties of adenovirus vectors with increased affinity to DSG2 and the potential benefits of oncolytic approaches and gene therapy. Viruses (2022) 14(8). doi: 10.3390/v14081835
142. Beyer I, Cao H, Persson J, Song H, Richter M, Feng Q, et al. Coadministration of epithelial junction opener JO-1 improves the efficacy and safety of chemotherapeutic drugsJunction opener improves chemotherapy. Clin Cancer Res (2012) 18(12):3340–51. doi: 10.1158/1078-0432.CCR-11-3213
143. Richter M, Yumul R, Wang H, Saydaminova K, Ho M, May D, et al. Preclinical safety and efficacy studies with an affinity-enhanced epithelial junction opener and PEGylated liposomal doxorubicin. Mol Ther - Methods Clin Dev (2015) 2:15005. doi: 10.1038/mtm.2015.5
144. Kim J, Li C, Wang H, Kaviraj S, Singh S, Savergave L, et al. Translational development of a tumor junction opening technology. Sci Rep (2022) 12(1):7753. doi: 10.1038/s41598-022-11843-z
Keywords: desmoglein-2 (DSG2), cancer, cadherin, intercellular junctions, prognostic biomarker, vasculogenic mimicry, non-canonical role
Citation: Myo Min KK, Ffrench CB, McClure BJ, Ortiz M, Dorward EL, Samuel MS, Ebert LM, Mahoney MG and Bonder CS (2023) Desmoglein-2 as a cancer modulator: friend or foe? Front. Oncol. 13:1327478. doi: 10.3389/fonc.2023.1327478
Received: 25 October 2023; Accepted: 04 December 2023;
Published: 22 December 2023.
Edited by:
Peng Su, Shandong University, ChinaReviewed by:
Nagaraja Sethuraman Balakathiresan, National Institute on Alcohol Abuse and Alcoholism (NIH), United StatesCopyright © 2023 Myo Min, Ffrench, McClure, Ortiz, Dorward, Samuel, Ebert, Mahoney and Bonder. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claudine S. Bonder, Y2xhdWRpbmUuYm9uZGVyQHVuaXNhLmVkdS5hdQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.