- 1Department of Clinical Laboratory, Tianjin Children’s Hospital/Children’s Hospital, Tianjin University, Tianjin, China
- 2Department of Hematology, Tianjin Children’s Hospital/Children’s Hospital, Tianjin University, Tianjin, China
The lysine(K)-specific methyltransferase 2A gene (KMT2A), previously known as mixed lineage leukemia (MLL), frequently rearranged in acute leukemia, belongs to one of the most promiscuous genes and has been found fused to more than 80 different partners. KMT2A::SEPTIN6 fusion is a relatively uncommon rearrangement observed in pediatric acute myeloid leukemia (AML) patients, some of which may harbor other mutations. We herein report a case of AML-M4-infant with KMT2A::SEPTIN6 fusion and DIS3 variant. The 8-month-old girl presented with leukocytosis, anemia and thrombocytopenia. A bone marrow smear disclosed that 64% of the total nucleated cells were blasts. Karyotype analysis showed 46,X,t(X;11)(q24;q23)[10]/46,XX[10]. Fluorescence in situ hybridization analysis suggested a possible break in the KMT2A gene. After whole transcriptome sequencing, Exon 9 of KMT2A was fused in-frame with Exon 2 of SEPTIN6. This is a typical type of chromosomal rearrangement leading to the KMT2A::SEPTIN6 fusion. Meanwhile, DIS3 variant [c.2065C>T, p.R689X, variant allele frequency (VAF): 39.8%] was identified. KMT2A::SEPTIN6 fusion has been associated with the pathogenesis of AML, whereas DIS3 variants are relatively rare genetic events in pediatric AML. Regrettably, the relatives disagreed with the combination chemotherapy, and the patient eventually died of progressive disease. In conclusion, our findings provide a foundation for a better understanding of the genotypic profile of KMT2A::SEPTIN6 associated AML, and the co-existence of KMT2A::SEPTIN6 and DIS3 variant might contribute to the disease progression and transformation of AML.
Introduction
The rearrangements involving the lysine (K)-specific methyltransferase 2A gene (KMT2A), previously known as mixed lineage leukemia (MLL), have been found in more than 70% of the cases of infant leukemia, both acute myeloid leukemia (AML) or acute lymphoblastic leukemia (ALL) (1). The KMT2A gene located at 11q23, belongs to one of the most promiscuous genes and is fused to a variety of partner genes in acute leukemias. Up to now, a total of 107 in-frame KMT2A gene fusions have been identified, including KMT2A::SEPTIN6 and KMT2A::SEPTIN9 (2). SEPTIN6, located at Xq24, is highly conserved in eukaryotes and regulates various biological functions, including filament dynamics, cytokinesis and cell migration (3). Pediatric AML with KMT2A::SEPTIN6 is very rare. To date, KMT2A::SEPTIN6 has so far only been described in 18 pediatric AML in literature (4). The information on the clinical features and treatment strategies of such patients is limited.
DIS3 homolog, exosome endoribonuclease and 3’-5’ exoribonuclease (DIS3) is a highly conserved 3’ to 5’ exoribonuclease, which has a diverse range of functions within RNA metabolism including mRNA quality control, regulation of gene expression and small RNA processing (5). Whole genome sequencing in relapsed AML revealed that DIS3, as a recurrently mutated gene, was associated with AML relapse (6). Desterke et al. reported that DIS3 missense mutations found in the cohort of AML patients affected VacB RNB protein domain, which was implicated in ribonuclease activity and RNA binding of the molecule (7). However, due to the rarity of DIS3 variants in AML, the gene effect on leukemogenesis is still unknown. Here, we report the clinical and molecular characteristics of a KMT2A::SEPTIN6 positive pediatric AML bearing DIS3 variant, and reviewed the relevant literature and cases of AML to expand the current understanding of the genotypic spectrum of this rare form.
Materials and methods
Case presentation
The patient, an 8-month-old girl, visited the hospital due to a high-grade fever in June 2021, and a complete blood count showed that WBC (white blood cells): 26.34×109/L (reference range: 5.00-14.20), PLT (platelets): 77×109/L (reference range: 172-601), Hb (hemoglobin level): 99g/L (reference range: 103-138). Physical examination on admission revealed absence of hepatomegaly/splenomegaly or cutaneous lesions. A chest X-Ray was performed showing the inflammatory consolidation of lungs. Owing to the suspicion of acute leukemia, the patient was admitted to the hospital. After admission, laboratory tests confirmed the presence of anemia (Hb: 85g/L), leukocytosis (WBC: 28.2×109/L) with 45% blasts on peripheral blood smear, and thrombocytopenia (PLT: 71×109/L), as well as elevated serum inflammatory markers, including procalcitonin (0.09ng/ml, reference range: 0-0.05) and interleukin-6 (53.57pg/ml, reference range: 0-7.00). There was no evidence of coagulopathy or hyperuricemia. However, lactate dehydrogenase was elevated to 555 U/L (reference range: 120-300). The examinations of urine, stool and cerebrospinal fluid showed no significant abnormalities, and all blood culture tests were negative. A bone marrow (BM) smear disclosed a large number of immature cells and blast cells accounting for 64% of 200 nucleated cells. Cytochemistry analysis reported that most of the blast cells showed a positive reaction for peroxidase, and parts of them were weakly positive for sodium fluoride-sensitive alpha-naphthyl butyrate esterase. Immunophenotypic analysis revealed BM cells expressing HLA-DR, CD4, CD15, CD33, CD64 and MPO, mildly expressing CD11b, CD13, CD38 and CD117. A diagnosis of AML-M4 without neurological involvement was confirmed.
Cytogenetic analysis
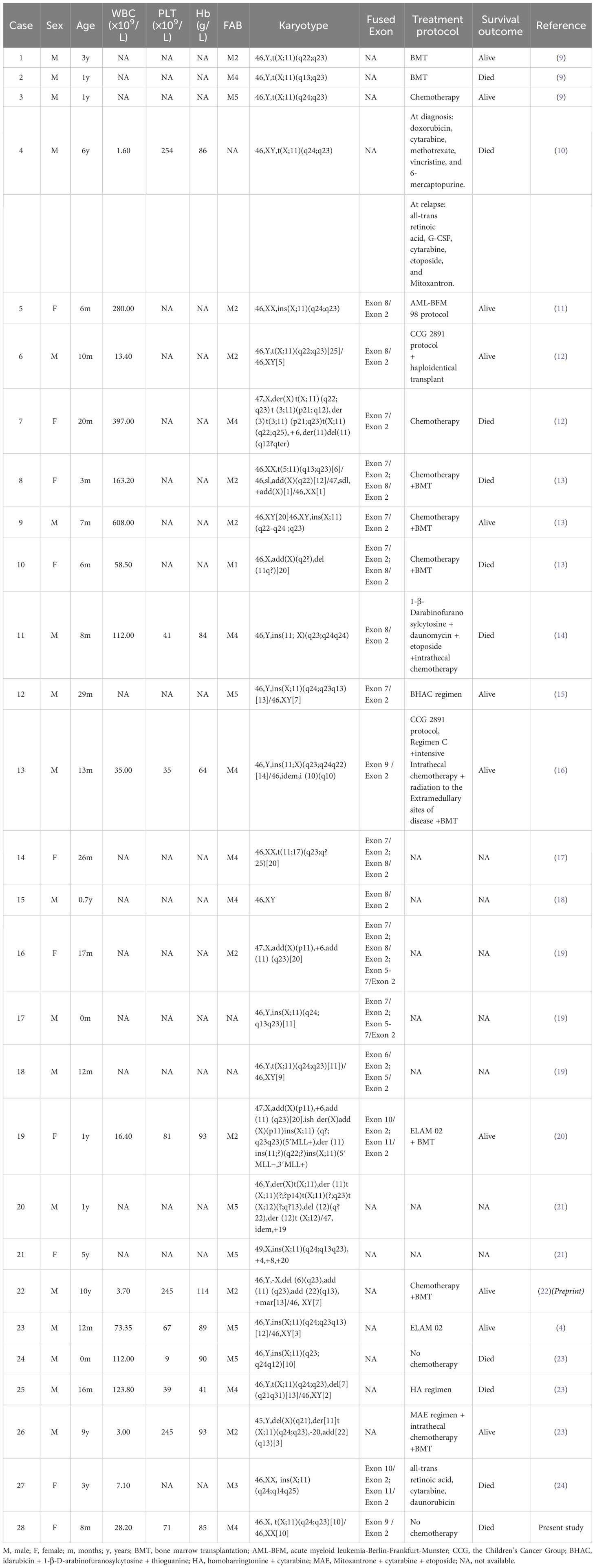
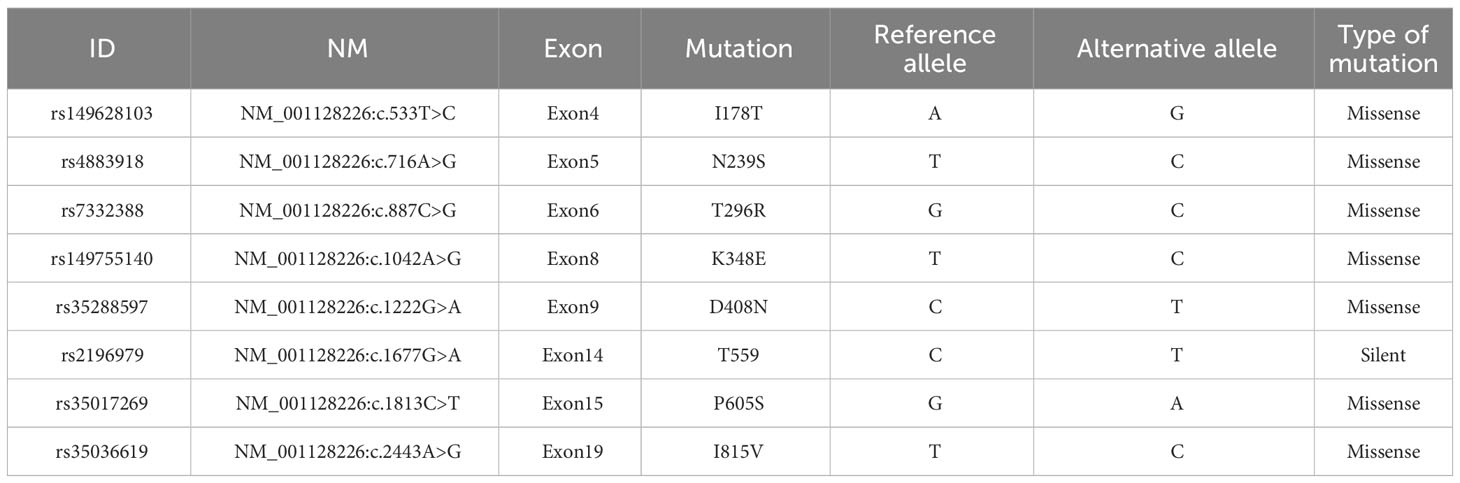
The karyotype was found to be 46,X,t(X;11)(q24;q23)[10]/46,XX[10] (Figure 1A). Fluorescence in situ hybridization (FISH) was used for further understanding a variety of chromosomal abnormalities. FISH with break-apart 11q23 probe (Vysis, Abbott Molecular Inc.) confirmed KMT2A rearrangement showing the typical split signal (Figure 1B).
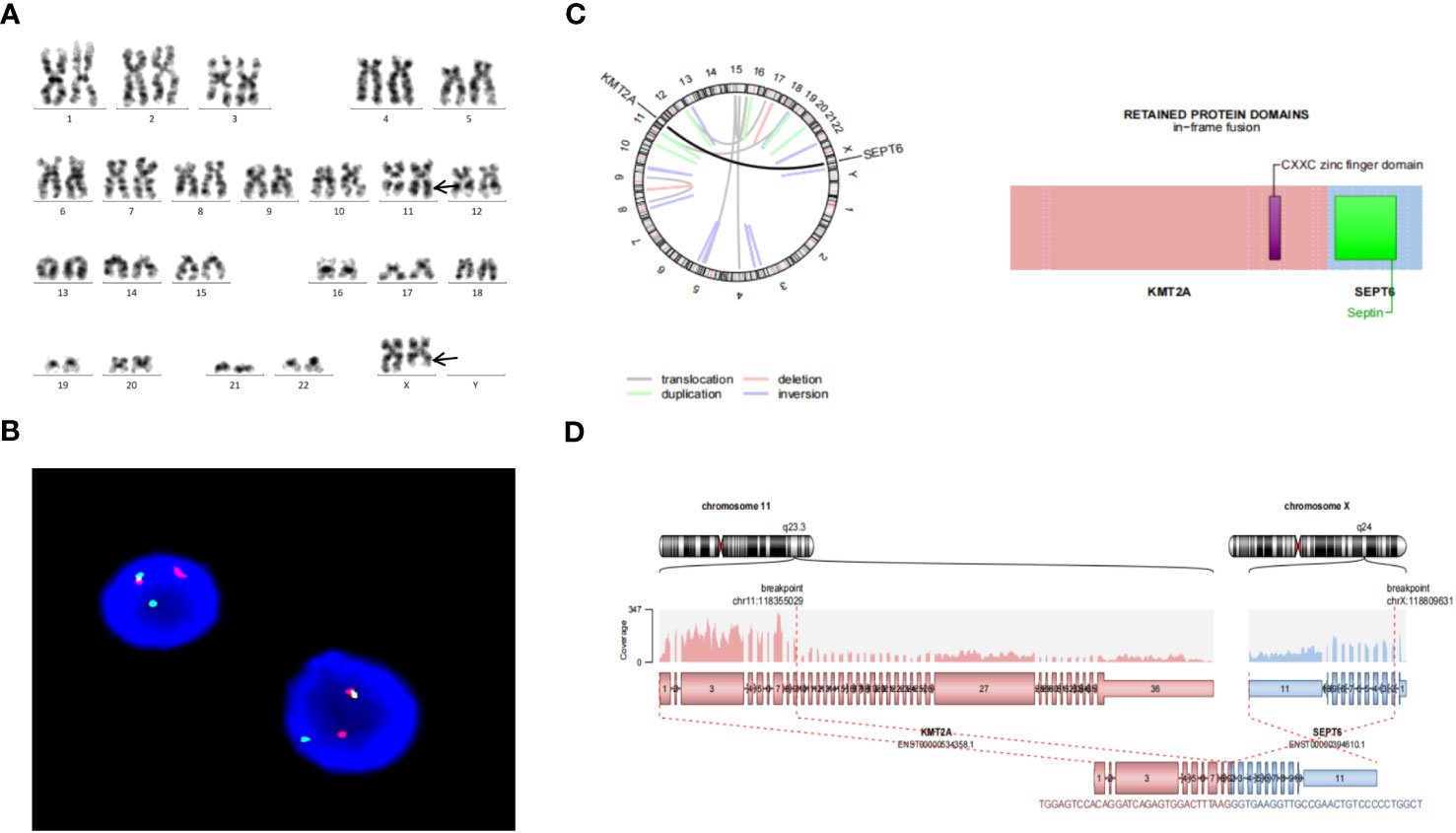
Figure 1 (A) Representative karyotype of the bone marrow cells was 46,X,t(X;11)(q24;q23)[10]/46,XX[10]. (B) FISH analysis using 11q23 break-apart probe. Separation of a green and a red signal revealed KMT2A rearrangement. (C) Circos plot indicating gene fusion between KMT2A and SEPTIN6. (D) KMT2A and SEPTIN6 fusion transcripts. KMT2A Exon 9 was fused in-frame with SEPTIN6 Exon 2.
Molecular analysis
According to the results of clinical diagnosis and cytogenetics, we carried out the whole transcriptome sequencing (WTS), which provided a comprehensive genomic profile. Peripheral blood in EDTA was collected for WTS detection. Ribo-zero based on probe hybridisation was used for depletion of rRNA. The RNA sequencing library was sequenced on the Illumina HiSeq X system, creating 19.55Gb total reads. The RNA-sequencing reads were mapped to the EnsemblGRCh37/hg19 by STAR aligner with the default parameters. The results revealed 5 fusion genes (KMT2A::SEPTIN6, FUS::SETD1A, RBM4::SF1, ADRBK1::RBM4, EDF1::PBX3), and 9 variants in following genes: DIS3, ARID1A, SFPQ, THRAP3, EML4, AFF1, KMT2B, ASXL1 and FLNA (Supplementary Tables S1, S2).
The pathogenicity of the variants were assessed by the Association for Molecular Pathology (AMP), American Society of Clinical Oncology (ASCO), and College of American Pathologists (CAP) proposed standards and guidelines for the interpretation and reporting of sequence variants in cancer (8). Sequence variants were categorized into four categories based on their level of clinical significance: tier I, variants with strong clinical significance (level A and B evidence); tier II, variants with potential clinical significance (level C or D evidence); tier III, variants with unknown clinical significance; and tier IV, variants that are benign or likely benign. In this report, genetic analysis revealed that Exon 9 of KMT2A was fused in-frame with Exon 2 of SEPTIN6 (Figures 1C, D). KMT2A::SEPTIN6 fusion was categorized into tier I, which was pathogenic in AML. Furthermore, DIS3 variant (c.2065C>T, p.R689X) was identified as a nonsense, leading to premature termination of protein coding. DIS3 variant met the criteria for variants with Level C diagnostic/prognostic significance. Variants in this category include those that are diagnostic/prognostic for a group of related cancers or variants that are supportive of a diagnosis along with other genomic variants. DIS3 variant was categorized into tier II, which was likely pathogenic in AML. However, other variants were categorized into tier III, which may have relevant pathological significance in further research.
Treatment and outcome
Due to pneumonia, the patient received intravenous latamoxef sodium (40 mg/kg/day, every 12 h) for anti-infection therapies. The patient was treated with cytoreductive therapy of hydroxyurea to prevent leukocytosis. After 5 days of hospitalization, WBC level dropped to the normal reference value with no metabolic manifestation observed (no evidence for tumor lysis syndrome) and 12% blasts detected in peripheral blood. Because the relatives disagreed with the combination chemotherapy, the patient did not receive medication, and the patient eventually died of progressive disease.
Literature review
PubMed database was searched with keywords including acute myeloid leukemia, KMT2A::SEPT6, MLL::SEPT6, KMT2A::SEPTIN6, MLL::SEPTIN6 and DIS3 to gather related case reports. To date, KMT2A::SEPTIN6 has only been reported in 28 pediatric cases with AML, including one case in present study and twenty-seven cases from the literature (4, 9–24) (Table 1). Clinical information and genetic features of evaluable KMT2A::SEPTIN6-positive pediatric cases with AML are summarized in Table 1. The age of the patients ranged from 0 to 10 years, with a male-female ratio of 1.8:1 (18 males vs. 10 females). All patients were diagnosed with AML according to the former FAB classification: one child with M1, nine children with M2, one child with M3, eight children with M4, six children with M5, and 3 children unknown. Most of them had available cytogenetic information, and complex chromosomal karyotypes with diverse chromosomal abnormalities, including chromosomal translocations (8 patients), chromosomal insertions (8 patients) and chromosomal complex abnormalities (11 patients). As for the treatment, most of them received chemotherapy or bone marrow transplantation (BMT). Ten patients died. AML with DIS3 variants is rare. Among the 108 patients of M1 and M3 AML, 3 cases had the DIS3 variants (25). In 193 patients with AML, 2 patients with AML-M1 presenting a co-association of RUNX1 and DIS3 mutations were found, and one patient with AML-M3 presented an isolated DIS3 variant without co-association with a RUNX1 mutation (8). Furthermore, it has been reported that 8 different types of DIS3 variants were described (26) (Table 2).
Discussion
AML with KMT2A abnormalities represent a subset of pediatric leukemia. Five different SEPTIN genes have been identified as KMT2A fusion partners, giving rise to chimeric fusion proteins. Among them, KMT2A::SEPTIN6 is often complex and sometimes cryptic as a result of the opposite orientation of KMT2A and SEPTIN6 on the respective chromosome arms. KMT2A::SEPTIN6 fusion gene was, to our knowledge, described in some children and very few adult cases diagnosed with AML (5, 23). In this study, we described a pediatric AML with KMT2A::SEPTIN6 fusion. In order to improve the understanding of this rare form of pediatric AML, we reviewed the relevant literature and summarized the characteristics of KMT2A::SEPTIN6-positive cases.
Table 1 lists the detailed clinical features, treatment strategies and prognosis of evaluable patients. Among these cases, nearly 60% were infant patients (≤ 1 year old). It is worth noting that 5 (50%) of the 10 deceased patients were infants. These findings suggest that age is an important factor affecting survival time. Most patients had typical characteristics of leukemia, such as leukocytosis, anemia and low platelet counts. Chen et al. reported that the overall survival of patients with WBC levels ≥ 20.0 ×109/L was much shorter than that of patients with WBC levels < 20.0 ×109/L in KMT2A::SEPTIN6 cases (23). An interesting finding was that, out of the 25 evaluable cases diagnosed with AML according to the former FAB classification, 14 (56%) were classified as FAB M4 or M5 subtypes (AML-M4/M5). Consistent with previous studies, AML-M4/M5 was frequently associated with KMT2A gene rearrangement and its incidence was relatively high among infants (27). In addition to the translocations and insertions, the complex karyotypes were observed in pediatric AML with KMT2A::SEPTIN6 fusion, and there were additional chromosomal abnormalities, such as add, del, der, idem, +4, +6, +8, +10, +19 and +20. Sequencing analysis demonstrated that fusions between SEPTIN6 Exon 2 and KMT2A Exon 7, 8 or both of them to be the most frequent. The fusion between KMT2A Exon 9 and SEPTIN6 Exon 2, as in our study, was a rare occurrence. These acquired genetic abnormalities can play an essential role in the pathogenesis.
There is a lack of systematic evaluation on the prognosis of KMT2A::SEPTIN6-associated AML. It was reported that a pediatric AML with the KMT2A::SEPTIN6 fusion initially responded well to multiagent chemotherapy, but a hematologic relapse occurred 2 months later (14). Similarly, the previously published AML-M4 case with KMT2A::SEPTIN6 fusion, received chemotherapy and had complete remission at 8 weeks of treatment; however, one month following the end of therapy, the patient developed a testicular relapse and presented recurrence of the cytogenetic abnormality (16). These results implicate that KMT2A::SEPTIN6-associated AML is easy to relapse and has a poor prognosis. Due to the rarity of this form, the pathogenesis of KMT2A::SEPTIN6 associated AML is not clear. Santos et al. found that a statistically significant down-regulation was observed for the RNA expression of both KMT2A and SEPTIN6 in KMT2A::SEPTIN6 leukemia (28). SEPTIN6 was the component of a core septin hexamer complex (SEPTIN2-SEPTIN6-SEPTIN7 complex), whose formation was thought to be essential to proper cytokinesis. And the correct expression of SEPTIN2, SEPTIN6, and SEPTIN7 seemed to be also relevant for the correct functioning of the cell DNA damage checkpoint (29). These results suggest that a link between aberrant septin expression and deregulation of the cell-cycle machinery is a crucial physiologic cellular mechanism of leukemogenesis. In addition, the fusion partners appeared to convert the rearranged KMT2A protein to a potent transcriptional activator. KMT2A rearrangements resulted in deregulation of KMT2A protein activity causing abnormal patterns of class I homeobox gene expression in hematopoietic stem cells or progenitors, thus promoting leukemogenesis. However, in vitro and in vivo models of the KMT2A::SEPTIN6 fusion have indicated that KMT2A::SEPTIN6 by itself was able to induce lethal myeloproliferative disease but not to induce acute leukemia in mice, implying that secondary genotoxic events related to DNA repair or cell cycle regulation could be required to develop leukemia (30).
The DIS3 gene located at chromosome 13q22.1, encods for a highly conserved ribonuclease indispensable for survival in vertebrates. DIS3 variants have been associated with multiple myeloma. Todoerti et al. reported that the frequency of DIS3 variants in newly diagnosed multiple myeloma was approximately 10% (31). But DIS3 variants have rarely been described in AML. 8 variants in DIS3 gene were previously described in AML (Table 2). The types of DIS3 variants included nonsense, missense and silent. These disruptive events lead to the loss of full-length DIS3 protein and to the lack or low expression of a truncated form of the protein, suggesting an oncogenic potential for DIS3 variants. Furthermore, DIS3 variants were associated with an important down-regulation of genes involved in the cohesin complex and a down-regulation of molecules implicated in DNA double strand repair in M1 patients with double DIS3-RUNX1 mutations, which were thought to indicate an important loss of control for the entry in mitosis (7). These findings indicates that DIS3 variants may play an important role in transformation and progression of AML.
Conclusion
In conclusion, we have described a rare pediatric case with KMT2A::SEPTIN6 associated AML, in particular, bearing DIS3 variant. Since reports of pediatric case are limited, it is difficult to deduce any conclusions involving the prognostic significance of this form. The potential effects require further screening of a large cohort of AML patients with both aberrations. This case extends the mutational spectrum, providing an opportunity to broaden the understanding available of this rare form of AML. Further functional studies are required to better elucidate the pathogenesis of this disease.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by the Medical Ethics Committee of Tianjin Children’s Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author contributions
LW: Writing – original draft. FQ: Writing – original draft. YS: Writing – original draft. SC: Writing – review & editing. PS: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We thank the patient and the family members for their participation in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1308786/full#supplementary-material
References
1. Daser A, Rabbitts TH. The versatile mixed lineage leukaemia gene MLL and its many associations in leukaemogenesis. Semin Cancer Biol (2005) 15(3):175–88. doi: 10.1016/j.semcancer.2005.01.007
2. Meyer C, Larghero P, Almeida Lopes B, Burmeister T, Gröger D, Sutton R, et al. The KMT2A recombinome of acute leukemias in 2023. Leukemia (2023) 37(5):988–1005. doi: 10.1038/s41375-023-01877-1
3. Fan Y, Du Z, Ding Q, Zhang J, Op Den Winkel M, Gerbes AL, et al. SEPT6 drives hepatocellular carcinoma cell proliferation, migration and invasion via the Hippo/YAP signaling pathway. Int J Oncol (2021) 58(6):25. doi: 10.3892/ijo.2021.5205
4. Chebly A, Djambas Khayat C, Yammine T, Korban R, Semaan W, Bou Zeid J, et al. Pediatric M5 acute myeloid leukemia with MLL-SEPT6 fusion and a favorable outcome. Leuk Res Rep (2021) 16:100277. doi: 10.1016/j.lrr.2021.100277
5. Robinson SR, Oliver AW, Chevassut TJ, Newbury SF. The 3’ to 5’ Exoribonuclease DIS3: from structure and mechanisms to biological functions and role in human disease. Biomolecules (2015) 5(3):1515–39. doi: 10.3390/biom5031515
6. Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature (2012) 481(7382):506–10. doi: 10.1038/nature10738
7. Desterke C, Bennaceur-Griscelli A, Turhan AG. DIS3 mutation in RUNX1-mutated AML1 confers a highly dismal prognosis in AML by repressing sister chromatid cohesion. Blood (2019) 134(Supplement_1):1454. doi: 10.1182/blood-2019-123352
8. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the association for molecular pathology, american society of clinical oncology, and college of american pathologists. J Mol Diagn (2017) 19(1):4–23. doi: 10.1016/j.jmoldx.2016.10.002
9. Harrison CJ, Cuneo A, Clark R, Johansson B, Lafage-Pochitaloff M, Mugneret F, et al. Ten novel 11q23 chromosomal partner sites. European 11q23 workshop participants. Leukemia (1998) 12:811–22. doi: 10.1038/sj.leu.2401017
10. Nakata Y, Mori T, Yamazaki T, Suzuki T, Okazaki T, Kurosawa Y, et al. Acute myeloid leukemia with hypergranular cytoplasm accompanied by t(X;11)(q24;q23) and rearrangement of the MLL gene. Leuk Res (1999) 23:85–8. doi: 10.1016/S0145-2126(98)00131-3
11. Borkhardt A, Teigler-Schlegel A, Fuchs U, Keller C, König M, Harbott J, et al. An ins(X;11)(q24;q23) fuses the MLL and the Septin 6/KIAA0128 gene in an infant with AML-M2. Genes Chromosomes Cancer (2001) 32:82–8. doi: 10.1002/gcc.1169
12. Slater DJ, Hilgenfeld E, Rappaport EF, Shah N, Meek RG, Williams WR, et al. MLL-SEPTIN6 fusion recurs in novel translocation of chromosomes 3, X, and 11 in infant acute myelomonocytic leukemia and in t(X;11) in infant acute myeloid leukemia, and MLL genomic breakpoint in complex MLL-SEPTIN6 rearrangement is a DNA topoisomerase II cleavage site. Oncogene (2002) 21:4706–14. doi: 10.1038/sj.onc.1205572
13. Ono R, Taki T, Taketani T, Kawaguchi H, Taniwaki M, Okamura T, et al. SEPTIN6, a human homolog to mouse Septin6, is fused to MLL in infant acute myeloid leukemia with complex chromosomal abnormalities involving 11q23 and Xq24. Cancer Res (2002) 62:333–7.
14. Fu JF, Liang DC, Yang CP, Hsu JJ, Shih LY. Molecular analysis of t(X;11)(q24;q23) in an infant with AML-M4. Genes Chromosomes Cancer (2003) 38:253–9. doi: 10.1002/gcc.10272
15. Kim HJ, Ki CS, Park Q, Koo HH, Yoo KH, Kim EJ, et al. MLL/SEPTIN6 chimeric transcript from inv ins(X;11)(q24;q23q13) in acute monocytic leukemia:report of a case and review of the literature. Genes Chromosomes Cancer (2003) 38:8–12. doi: 10.1002/gcc.10235
16. Kadkol SS, Bruno A, Oh S, Schmidt ML, Lindgren V. MLL-SEPT6 fusion transcript with a novel sequence in an infant with acute myeloid leukemia. Cancer Genet Cytogenet (2006) 68:162–7. doi: 10.1016/j.cancergencyto.2006.02.020
17. Strehl S, König M, Meyer C, Schneider B, Harbott J, Jäger U, et al. Molecular dissection of t(11;17) in acute myeloid leukemia reveals a variety of gene fusions with heterogeneous fusion transcripts and multiple splice variants. Genes Chromosomes Cancer (2006) 45(11):1041–9. doi: 10.1002/gcc.20372
18. Shih LY, Liang DC, Fu JF, Wu JH, Wang PN, Lin TL, et al. Characterization of fusion partner genes in 114 patients with de novo acute myeloid leukemia and MLL rearrangement. Leukemia (2006) 20(2):218–23. doi: 10.1038/sj.leu.2404024
19. Cerveira N, Micci F, Santos J, Pinheiro M, Correia C, Lisboa S, et al. Molecular characterization of the MLL-SEPT6 fusion gene in acute myeloid leukemia: identification of novel fusion transcripts and cloning of genomic breakpoint junctions. Haematologica (2008) 93:1076–80. doi: 10.3324/haematol.12594
20. Cerveira N, Lisboa S, Correia C, Bizarro S, Santos J, Torres L, et al. Genetic and clinical characterization of 45 acute leukemia patients with MLL gene rearrangements from a single institution. Mol Oncol (2012) 6:553–64. doi: 10.1016/j.molonc.2012.06.004
21. Bager N, Juul-Dam KL, Sandahl JD, Abrahamsson J, Beverloo B, de Bont ESJM, et al. Complex and monosomal karyotype are distinct cytogenetic entities with an adverse prognostic impact in paediatric acute myeloid leukaemia. A NOPHO-DBH-AML study. Br J Haematol (2018) 183(4):618–28. doi: 10.1111/bjh.15587
22. Chen Y, Pan Y, Zhang L, Chen X, Yang W, Guo Y, et al. Acute myelogenous leukemia associated with MLL-SEPT6 rearrangement and TRAF3, FGFR3 Mutation: A Pediatric Case Report and Review of the Literature. Preprints (2021). doi: 10.20944/preprints202103.0601.v1
23. Chen F, Yang Y, Fu S. Clinical profile in KMT2A-SEPT6-positive acute myeloid leukemia: Does it often co-occur with NRAS mutations? Front Med (Lausanne) (2022) 9:890959. doi: 10.3389/fmed.2022.890959
24. Borkovskaia A, Bogacheva S, Konyukhova T, Dadakhanova E, Gaskova M, Soldatkina O, et al. Molecular heterogeneity of pediatric AML with atypical promyelocytes accumulation in children-A single center experience. Genes (Basel) (2023) 14(3):675. doi: 10.3390/genes14030675
25. Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell (2012) 150(2):264–78. doi: 10.1016/j.cell.2012.06.023
26. Hirsch P, Zhang Y, Tang R, Joulin V, Boutroux H, Pronier E, et al. Genetic hierarchy and temporal variegation in the clonal history of acute myeloid leukaemia. Nat Commun (2016) 7:12475. doi: 10.1038/ncomms12475
27. Jo A, Tsukimoto I, Ishii E, Asou N, Mitani S, Shimada A, et al. Age-associated difference in gene expression of paediatric acute myelomonocytic lineage leukaemia (FAB M4 and M5 subtypes) and its correlation with prognosis. Br J Haematol (2009) 144(6):917–29. doi: 10.1111/j.1365-2141.2008.07531.x
28. Santos J, Cerveira N, Bizarro S, Ribeiro FR, Correia C, Torres L, et al. Expression pattern of the septin gene family in acute myeloid leukemias with and without MLL-SEPT fusion genes. Leuk Res (2010) 34(5):615–21. doi: 10.1016/j.leukres.2009.08.018
29. Kremer BE, Haystead T, Macara IG. Mammalian septins regulate microtubule stability through interaction with the microtubule-binding protein MAP4. Mol Biol Cell (2005) 16(10):4648–59. doi: 10.1091/mbc.e05-03-0267
30. Ono R, Nakajima H, Ozaki K, Kumagai H, Kawashima T, Taki T, et al. Dimerization of MLL fusion proteins and FLT3 activation synergize to induce multiple-lineage leukemogenesis. J Clin Invest (2005) 115(4):919–29. doi: 10.1172/JCI22725
Keywords: acute myeloid leukemia, gene fusion, KMT2A, SEPTIN6, DIS3
Citation: Wang L, Qiu F, Shen Y, Chen S and Si P (2023) Co-existence of KMT2A::SEPTIN6 fusion and DIS3 variant in a pediatric case with acute myeloid leukemia: a case report and literature review. Front. Oncol. 13:1308786. doi: 10.3389/fonc.2023.1308786
Received: 07 October 2023; Accepted: 29 November 2023;
Published: 13 December 2023.
Edited by:
Kathleen Sakamoto, Stanford University, United StatesReviewed by:
Nathalie Douet-Guilbert, Centre Hospitalier Regional Universitaire (CHU) de Brest, FranceIrina Golovleva, Umeå University, Sweden
Copyright © 2023 Wang, Qiu, Shen, Chen and Si. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sen Chen, Y2hlbnNlbmJsb29kQDEyNi5jb20=; Ping Si, a2FuZ3NpcGluZ0BsaXZlLmNu
†These authors have contributed equally to this work
 Liang Wang
Liang Wang Fangzhou Qiu1†
Fangzhou Qiu1†