
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Oncol., 11 December 2023
Sec. Hematologic Malignancies
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1303677
This article is part of the Research TopicReviews in Hematologic Malignancies: 2023View all 15 articles
Extensive genome-wide sequencing efforts have unveiled the intricate regulatory potential of long non-protein coding RNAs (lncRNAs) within the domain of haematological malignancies. Notably, lncRNAs have been found to directly modulate chromatin architecture, thereby impacting gene expression and disease progression by interacting with DNA, RNA, and proteins in a tissue- or condition-specific manner. Furthermore, recent studies have highlighted the intricate epigenetic control of lncRNAs in cancer. Consequently, this provides a rationale to explore the possibility of therapeutically targeting lncRNAs themselves or the epigenetic mechanisms that govern their activity. Within the scope of this review, we will assess the current state of knowledge regarding the epigenetic regulation of lncRNAs and how, in turn, lncRNAs contribute to chromatin remodelling in the context of multiple myeloma.
Multiple myeloma (MM) is a heterogeneous haematological malignancy characterized by the clonal expansion of malignant plasma cells within the bone marrow (1). It represents the second most prevalent haematological malignancy and it is marked by complex genetic aberrations, including chromosomal translocations, copy number alterations, and somatic mutations, affecting pathways critical to cell cycle regulation, DNA repair, and epigenetic modulation (2–4). Treatment strategies include high-dose chemotherapy regimens, autologous stem cell transplantation, as well as targeted therapies such as proteasome inhibitors and immunomodulatory agents. Despite these therapeutic innovations, disease relapse and drug resistance remain as substantial challenges (5). Thus, treatment of MM is clinically challenging and new therapeutic interventions are required. Prior studies, by us and others, have suggested that the epigenetic machinery plays a crucial role in MM pathogenesis, including aberrant DNA methylation and abnormal histone modification patterns (6–14). Furthermore, more recently, dysregulation of long non-protein coding RNAs (lncRNAs) has been suggested to contribute to MM pathogenesis, patient outcome and drug resistance (15–17). Additionally, dysregulation of lncRNAs has been shown to contribute to disease progression by influencing critical pathways involved in proliferation, apoptosis, immune response, and drug resistance (18, 19). Unravelling the complex network of lncRNA-mediated molecular mechanisms could therefore unveil novel therapeutic targets and diagnostic markers in MM.
lncRNAs represent the largest group of non-protein coding RNAs, however, to date their functions remain largely unexplored. lncRNAs are transcripts exceeding 200 nucleotides in length, and their transcriptional regulation mirrors that of protein-coding genes, including processes such as histone modifications, chromatin compaction, and chromatin remodelling. The biogenesis of lncRNAs encompasses a spectrum of events, including 5’ capping, splicing, variation in exon and intron dimensions, and the addition of polyadenylation (poly(A)+) tails. Notably, features like poly(A)+ tails and 5’ capping play fundamental roles in determining the transcript stability of lncRNAs. In contrast to messenger RNAs (mRNAs), lncRNAs transcripts exhibit a diminished steady-state, as they are commonly less evolutionary conserved (20). lncRNAs can be transcribed from multiple genomic locations, including promoters, enhancers, intergenic regions, as well as in bidirectional and antisense directions. Typically residing within the nucleus, lncRNAs tend to manifest pronounced cell and tissue specificity (21). In addition, a substantial fraction i.e., 81% of lncRNAs, exhibit a limited degree of evolutionary conservation, while 3% of lncRNAs manifest ultra-conservation (22).
Functionally, lncRNAs perform a diverse array of functions both within the nucleus and the cytoplasm. These molecules regulate gene expression by engaging in intricate interactions with RNA, DNA and proteins, including chromatin-modifying enzymes. Within the nuclear domain, lncRNAs have been categorized into four fundamental archetypes: signal, decoy, guide, and scaffold lncRNAs (Figure 1). Signal lncRNAs respond to specific stimuli, promoting integration of signals for the transcription of targeted genes (23). Decoy lncRNAs, on the other hand, can bind proteins, such as transcription factors and chromatin modifiers, resulting in transcriptional control by impeding the binding capacity to their targets (24). Guide lncRNAs have the ability to reposition ribonucleoprotein complexes to designated loci, both in cis and in trans, thereby altering the gene expression patterns. Finally, scaffold-associated lncRNAs engage in temporally and spatially regulated interactions with DNA, different types of RNAs and proteins, thereby bolstering the stability of complexes involved in either transcriptional activation or suppression. Additionally, lncRNAs operate as microRNA (miRNA) sponges, sequestering miRNAs to avert mRNA degradation (25). To date, various lncRNAs have been described to localize with chromatin, where they interact with different chromatin-associated proteins to promote or repress their binding potential to specific DNA locations. These chromatin-associated lncRNAs have been implicated in MM pathogenesis and disease outcome. In addition, lncRNAs do not only act as regulators of the epigenetic landscape but can also themselves be epigenetically regulated by DNA and chromatin modifications as well as by RNA modifications, referred to as the epitranscriptomics. Among these, RNA modifications such as, N6-methyladenosine (m6A), N1-methyladenosine (m1A), 5-methylcytosine (m5C), and 7-methylguanosine (m7G), play a crucial role in regulating various aspects of lncRNA function, structure, stability, localization, and lncRNA-mediated interactions (26–34) (Figure 2).
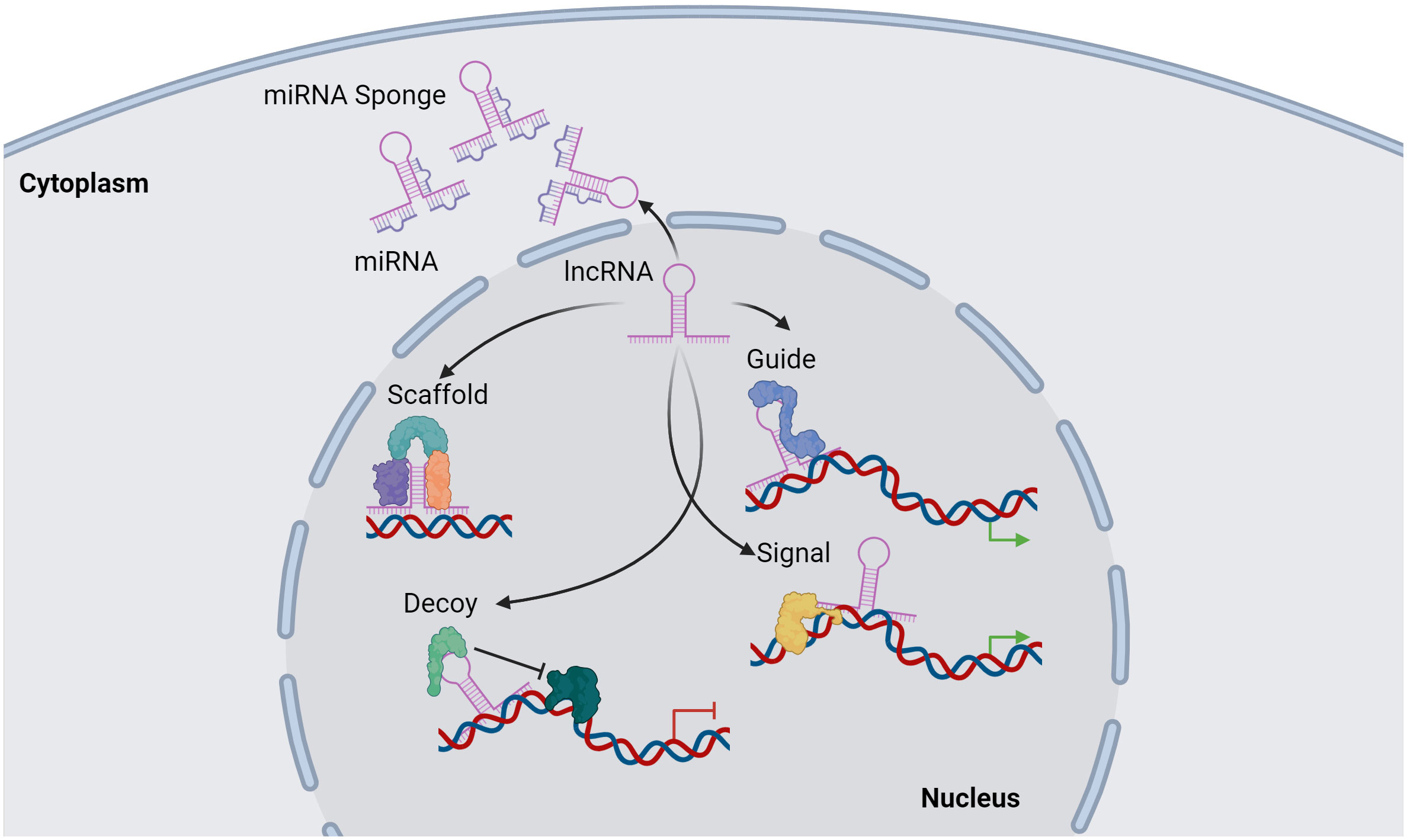
Figure 1 Overview of lncRNA functions. lncRNAs can regulate transcription by acting as a scaffold by binding proteins together in a complex structure. A secondary function of a lncRNA is as a guide of proteins or other molecules to target genomic location. lncRNAs can directly bind to genomic regions within the genome to transduce signal activation of DNA-bound molecules. Furthermore, a lncRNA can act as a decoy, preventing different molecules such as proteins to bind to targeted genomic regions. In addition, lncRNAs can regulate miRNAs function by acting as a miRNA sponge, preventing miRNA-mRNA binding, thus inhibiting mRNAs degradation. Image was created with biorender.com.
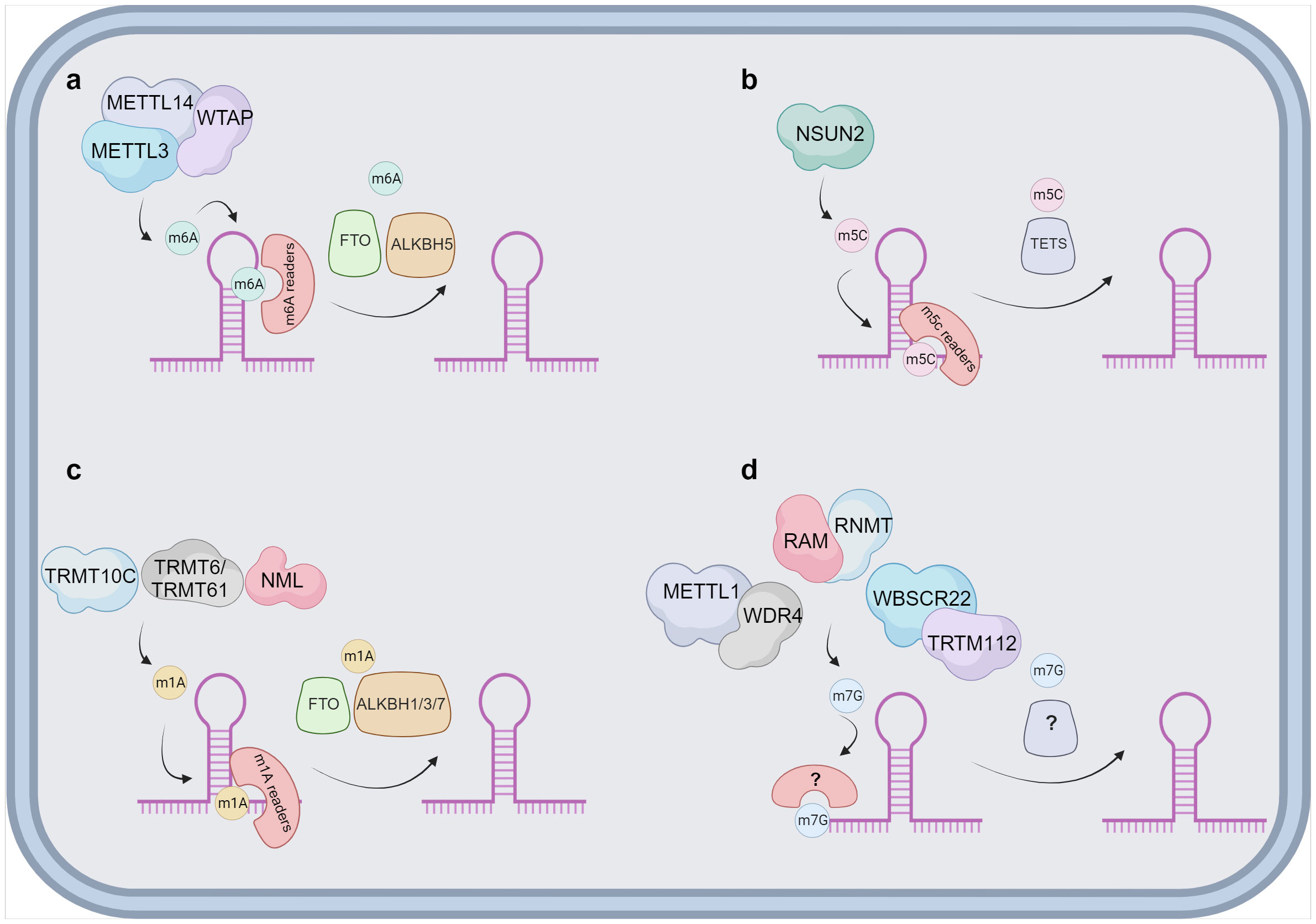
Figure 2 Proposed overview of RNA modifications on lncRNA. Schematic overviews are provided for the different lncRNAs modifications. (A) The m6A modification is deposited by a protein complex constituted mainly by METTL3, METTL14 and the cofactor WTAP. There are multiple m6A readers identified, namely YTHDF1/2/3, IGF2BP1/2/3, HNRNPs and ZC3H13. The demethylation of m6A is catalysed by FTO and ALKBH5. (B) NSUN2 has been reported as the sole writer of the m5C modification in lncRNAs. The m5C can be identified by ALYREF and YBX1 and it is speculated that the removal is catalysed by TETs. (C) m1A is suggested to be deposited by TRMT10C, TRMT6/61 and NML on lncRNAs. Multiple readers have been identified in other types of RNAs, as YTDHF1-3 and YTHDC1 and the removal is mainly associated to FTO and ALKBH1/3/7. (D) The exact manner of how m7G modification is deposited on lncRNAs is currently unknown. Three different complexes can take the role of a writer, METTL1 and WDR4, RAM and RNMT or WBSCR22 and TRTM112. Currently both readers and erasers on m7G are unknown and represented with ? in the figure. Image was created with biorender.com.
We and others have shown the importance of lncRNAs in chromatin remodelling and the impact they have on MM patient outcome (6, 15–17). In fact, aberrant lncRNA expression has been demonstrated to have an oncogenic role in MM pathogenesis and progression (35, 36). In this review, we present an overview of the role of lncRNAs in the context of MM through their epigenetic regulation and functional effects on chromatin remodelling.
lncRNAs have been suggested to affect multiple layers of cellular function, encompassing processes such as cellular biogenesis of macromolecules, differentiation, gene expression and chromatin remodelling. The recent establishment of a comprehensive genome-wide lncRNA-chromatin interactome has provided insight into the intricate orchestration of chromatin compaction by lncRNAs, subsequently impacting gene expression patterns (37–39). Notably, the functional implications of lncRNA-mediated chromatin remodelling in the context of cancer have gained considerable interest, however a summary within the domain of MM is currently lacking.
The expression of lncRNAs can be regulated by different epigenetic machineries, such as DNA methylation (40). DNA methylation plays an important role in regulating cell-type specific gene expression. The DNA methylation process consists of the deposition of methyl groups to the 5-carbon position of cytosine in a CpG dinucleotide, resulting in gene suppression when located along the promoter or transcription start site and gene transcription when found in the gene body. This process is catalysed by the DNA methyltransferases, DNMT1 and DNMT3A/B and can be reversed by the DNA demethylase enzymes TET1-3 (41). Disrupted DNA methylation has been shown to promote carcinogenesis and disease progression in multiple cancers (3, 42, 43). In fact, it has previously been suggested that promoter DNA hypermethylation is accountable for decreased expression of 35 lncRNAs in hepatocellular carcinoma (40, 44). Furthermore, patients with lower expression of these lncRNAs had increased expression of the DNA methyltransferase genes DNMT1, DNMT3A, and DNMT3B. In contrast, patients with higher expression of this panel of lncRNAs, exhibited lower expression of the DNA methyltransferases (40). Li et al. reported that the lncRNA BM742401, defined as a tumour suppressor in gastric cancer and chronic lymphocytic leukaemia, undergoes silencing in MM cell lines due to promoter hypermethylation. Notably, decreased BM742401 levels enhanced MM cell migration, while in newly diagnosed MM patients, silencing by elevated DNA methylation levels in the promoter of the lncRNA BM742401 correlated with poor overall survival. This underscores the significant impact that epigenetic regulation of lncRNAs can exert on disease progression (45). Similarly, DNA methylation-mediated silencing of the lncRNA KIAA0495 has been reported in MM cell lines, although it was not found to be relevant for the progression of the disease (46) (Table 1).
An additional level of transcriptional regulation is through chromatin compaction. DNA is packed into chromatin fibres wrapped around a histone octamer, ultimately forming a nucleosome. The nucleosome consists of the four histone proteins H2A, H2B, H3 and H4. Each histone protein has in its N-terminal domain a histone tail that can be reversibly subjected to methylation, acetylation, phosphorylation, ubiquitination, sumoylation and histone tail clipping which control chromatin compaction, thus either promoting or inhibiting transcription factor binding, DNA repair, replication and genomic recombination. The majority of studies have concentrated on examining histone modifications related to protein coding genes and non-protein coding genes such as miRNAs (59–62). Consequently, additional research is warranted to elucidate the influence of histone modifications on lncRNAs’ regulation and the potential implications in various diseases, including MM.
Although, data is largely lacking how regulation of lncRNAs by the deposition of histone modifications may directly influence their expression, there is now emerging data indicating that lncRNAs may act as recruiters, guides and scaffolds for protein complexes including chromatin modifiers, thus epigenetically influencing the expression of other genes. Prior studies have shown that PRC2-mediated gene silencing is important for MM pathogenesis and disease progression, both in vivo and in vitro (7, 8, 10, 43). Furthermore, several lncRNAs have been suggested to regulate the enzymatic activity of PRC2 by binding to the catalytic subunit EZH2. Moreover, lncRNAs can modulate PRC2 activity by acting as a complex recruiter to target genomic locations. For instance, the lncRNA PVT1 was recently described to be overexpressed in primary MM patient samples and associated with poor prognosis, a seemingly independent feature from patients’ cytogenetic background (6). Moreover, PVT1 was shown to interact directly with EZH2, facilitating recruitment of PRC2 to target genomic loci and transcriptional repression of genes associated with pro-apoptotic and tumour suppressor functions (6) (Table 1). Similarly to the function of PVT1, the lncRNA ANRIL, was described to exert a guiding function for PRC1 and PRC2 DNA binding in MM and was demonstrated to promote resistance to conventional therapies such as bortezomib by guiding PRC2 to promote gene silencing of the tumour suppressor gene PTEN. High expression of ANRIL has been associated with poor overall survival in MM (47) (Table 1). Furthermore, upregulation of the lncRNA H19 correlates with worse prognosis and promotes the imbalance of osteogenesis and osteolysis in MM by acting as a miRNA sponge, resulting in upregulation of E2F7, which is a transcriptional activator of EZH2 and thus affecting the suppression of PTEN (48). In addition, increased H19 activity has been shown to activate the chromatin reader protein BRD4 in MM (63). BRD4 is a well-known epigenetic reader of acetylated lysine and assists in the transmission of epigenetic memory during cell division (64, 65). BRD4 has been identified as a therapeutic vulnerability and potential target in MM (66) (Table 1).
The lncRNA CRNDE epigenetically regulates the transcription of DUSP5 and CDKN1A in solid tumours by facilitating PRC2 recruitment (67). Overexpression of CRNDE has been described to be associated with poor prognosis by regulating proliferative capacity through IL6 signalling in MM, however, no direct interaction between CRNDE and PRC2 has been proven (49). Recruitment of the histone H3 lysine 4 methyltransferase MLL has been suggested to occur through the binding to the lncRNA MIAT, which can then guide MLL to the promoter region of the collagen degradation enzyme MMP9. Inhibition of MIAT resulted in the loss of transcriptional activity of MMP9, which is suggested to reduce proliferative capacity and cell migration in non-small cell lung cancer (68). In MM, MIAT is overexpressed and has been associated with sensitivity to bortezomib treatment (50) (Table 1).
Interestingly, additional lncRNAs have been suggested to play important roles in chromatin regulation. The lncRNA HOTAIR has been demonstrated to bind to the PRC2 complex and can further interact with the TF-silencing complex formed by LSD1/CoREST/repressor element 1, promoting gene repression (69). In addition, HOTAIR may function as a stabilizing component of PRC2, as well as a scaffold for complex-complex interactions (69). In MM, HOTAIR has been described to be upregulated in primary patient samples and to contribute to the oncogenic activation of the JAK2/STAT3 signalling pathway (51) (Table 1). Similarly, the MIR17HG-derived lncRNA, RROL, has been demonstrated to act as a chromatin scaffold for protein interactions and to promote MM cell growth (52). lncRNAs such as AIR and HOXB-AS1 have been described to have a guiding function through which they recruit the histone methyltransferases G9a and SET1/MLL to target locations to induce gene repression or activation, respectively (53, 70). Interestingly, HOXB-AS1 has been described to be upregulated in MM, acting as a stabilizer for mRNA (54) (Table 1). In another aspect of epigenetic regulation, DARS-AS1 promotes the recruitment of the histone methyltransferases METTL3 and METTL14 to DARS mRNA to induce m6A modification and enhance translation in cervical cancer (71). In MM, DARS-AS1 has been described to regulate HIF-1α in promoting the mTOR pathway (55) (Table 1).
Increased expression of the lncRNAs GAS5, MALAT1 and NEAT1 in MM patients, is associated with poor outcome and worse overall survival (25) (Table 1). GAS5 has the ability to act as decoy for different molecules by functioning as a DNA mimic, thus preventing DNA motif binding (72). One of the most abundant and most studied lncRNAs is MALAT1 which has been implicated in various functions during MM pathogenesis by acting as a scaffold for proteins involved in DNA repair (56) and as a miRNA sponge (57). Interestingly it has also been described to promote gene silencing by PRC2 recruitment in various cancers (73–75). Recent studies in colorectal cancer have suggested that the lncRNA NEAT1 promotes histone H3 lysine 27 acetylation in genes associated with stemness (76). In addition, NEAT1 has further been implicated in lung cancer by recruiting DNMT1 to the promoter regions of genes regulating cytotoxic T-cell infiltration. In fact, inhibition of NEAT1 leads to loss of DNMT1 binding to these promoter regions and thus activating gene expression (77). In MM, overexpression of NEAT1 has been associated with poor patient outcome. In addition, and further supporting a clinical relevance, inhibition of NEAT1 promoted increased sensitivity to chemotherapeutic treatment (58) (Table 1).
RNA modifications on lncRNAs may influence their stability, subcellular localization, and interactions with DNA, proteins and other RNA molecules. These modifications can also affect lncRNA regulation and contribute to their reported functional diversity (33). Dysregulation of RNA modifications on lncRNAs has been associated with various diseases, including MM (33, 78–80).
The deposition of the N6-methyladenosine (m6A) mark may give rise to structural changes in lncRNAs, thus modifying lncRNA-protein interactions. Additionally, the m6A modification can modulate gene transcription, influence the subcellular localization of lncRNAs and regulate lncRNAs’ stability (81–84). There is an interdependent connection between the m6A modification and lncRNAs. Notably, lncRNAs have the ability to influence the stability and degradation of enzymes involved in m6A, as well as facilitate their integration into protein complexes (85–87). One example of this function is the lncRNA FEZF1-AS1, the knockdown of which led to an increased apoptosis by regulating the signalling of IGF2BP1, an m6A reader protein, in MM (88). Furthermore, dysregulation of m6A-related enzymes has been associated with disease progression, enhancing tumour growth and cell proliferation in MM (89–95). Significantly, m6A studies in MM showed a correlation between exosome-induced drug resistance and high levels of m6A on the lncRNAs LOC606724 and SNHG. Wang et al. identified METTL7A as an additional component of the m6A methyltransferase complex and described how its regulation is mediated by EZH2. Depletion of EZH2 simultaneously reduced METLL7A protein methylation levels, thus altering the m6A levels on the lncRNAs LOC606724 and SNHG (96). Studies in prostate cancer show that high levels of m6A on NEAT1 have been associated with bone metastasis (79, 97). Although no studies of m6A on NEAT1 have been performed in MM, high expression of NEAT1 in patients have been correlated with poor prognosis (98). In addition, NEAT1 can enhance the preservation of DNA integrity, thus promoting survival of MM cells (99). Moreover, knockdown of NEAT1 improved dexamethasone drug response in MM cell lines (100).
5-methylcytosine (m5C) has previously been described to exert important functions on DNA and has also been found to occur on RNA (78). The biological impact of RNA m5C primarily affects RNA localization, stability and transcription efficiency (101). Interestingly, NSUN2 has been reported as the sole writer of the m5C mark on lncRNAs (79). In MM, dysregulated deposition of RNA m5C has been correlated with disease progression and immune microenvironment regulation (102). Furthermore, recent studies have elucidated the importance of this modification in various other cancer types, including lung adenocarcinoma, pancreatic cancer, and colon cancer (79, 103–105).
Modifications of lncRNAs also include the deposition of N1-methyladenosine (m1A), which alters RNA secondary and tertiary structure, subsequently affecting its capacity to interact with RNA binding proteins. However, the function of m1A in lncRNAs is not fully elucidated, and the m1A modification has so far only been reported in the lncRNA MALAT1 (80, 106). Despite the absence of studies focusing on the m1A modification in MM, as previously mentioned, MALAT1 overexpression is correlated with worse prognosis, and the oncogenic role of MALAT1 in promoting MM tumorigenesis has been widely studied (35, 56, 107). MALAT1 dysregulation in MM has been associated with a wide range of processes including cell proliferation, DNA repair mechanisms, metastasis, drug resistance, and angiogenesis pathways (57, 107–109). Nonetheless, if these functions are mediated by chromatin remodelling and regulated via RNA modifications remains to be further investigated.
The N7-methylguanosine (m7G) modification is predominantly found at the 5´cap of mRNA, ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs). However, the impact of m7G on lncRNAs remains uncertain, probably attributed to the absence of 5´cap on less conserved lncRNAs (110, 111). Nevertheless, Yang et al. constructed the first model based on eight m7G-related lncRNAs to predict patient prognosis in colon cancer (112). Similarly, RNA m7G MeRIP-seq uncovered the significance of m7G-enriched lncRNAs in acute myeloid leukemia cells and unravelled a potential role of this modification in modulating gene expression, thereby enhancing drug resistance (111). However, the role of m7G modification in MM remains at present unknown.
The pathogenic impact of lncRNAs in MM and other haematological malignancies is unravelling. Recently, there have been large sequencing efforts in various cancers including MM that have suggested a clinical importance of lncRNAs. In MM, lncRNAs have been implicated in clinically relevant elements such as disease development, progression, drug resistance and patient outcome (25). Studies on the epitranscriptomics of lncRNAs through the addition of methyl groups to the lncRNA transcripts have gained increased attention and have furthered added an additional level of complexity to how lncRNAs contribute to cellular processes, such as RNA stability, translational efficiency of mRNAs and protein complex formation. However, the exact nature of these modifications needs to be further investigated in the context of MM. Moreover, not only can the expression of lncRNAs be epigenetically regulated but can in turn regulate chromatin modifying enzymes. Although lncRNA-chromatin interactions are clearly more dynamically investigated in some areas, such as in the recently shown context of PRC2 recruitment, deep functional evaluation of lncRNAs in MM is still lacking. It is apparent that this field is underdeveloped and a complete picture of how lncRNAs impact the pathophysiological processes in MM remains uncertain. While their functions continue to unfold, targeting lncRNAs arises as compelling innovative treatment option in cancer, including MM.
PN: Conceptualization, Project administration, Visualization, Writing – original draft, Writing – review & editing. BG-Z: Conceptualization, Project administration, Visualization, Writing – original draft, Writing – review & editing. AK: Supervision, Writing – review & editing. HW: Funding acquisition, Supervision, Writing – review & editing, Conceptualization, Visualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The project was supported by grants from the Swedish Research Council (K2019-64X-20102-13-3/KDB 1335/17) and the Swedish Cancer Society (CAN 2016/458, 200727 PjVSF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Fairfield H, Falank C, Avery L, Reagan MR. Multiple myeloma in the marrow: pathogenesis and treatments. Ann N Y Acad Sci (2016) 1364(1):32–51. doi: 10.1111/nyas.13038
2. Kazandjian D. Multiple myeloma epidemiology and survival: A unique Malignancy. Semin Oncol (2016) 43(6):676–81. doi: 10.1053/j.seminoncol.2016.11.004
3. Alzrigat M, Parraga AA, Jernberg-Wiklund H. Epigenetics in multiple myeloma: From mechanisms to therapy. Semin Cancer Biol (2018) 51:101–15. doi: 10.1016/j.semcancer.2017.09.007
4. Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer (2012) 12(5):335–48. doi: 10.1038/nrc3257
5. Pawlyn C, Davies FE. Toward personalized treatment in multiple myeloma based on molecular characteristics. Blood. (2019) 133(7):660–75. doi: 10.1182/blood-2018-09-825331
6. Nylund P, Garrido-Zabala B, Parraga AA, Vasquez L, Pyl PT, Harinck GM, et al. PVT1 interacts with polycomb repressive complex 2 to suppress genomic regions with pro-apoptotic and tumour suppressor functions in multiple myeloma. Haematologica (2023). doi: 10.3324/haematol.2023.282965
7. Kalushkova A, Fryknas M, Lemaire M, Fristedt C, Agarwal P, Eriksson M, et al. Polycomb target genes are silenced in multiple myeloma. PloS One (2010) 5(7):e11483. doi: 10.1371/journal.pone.0011483
8. Agarwal P, Alzrigat M, Parraga AA, Enroth S, Singh U, Ungerstedt J, et al. Genome-wide profiling of histone H3 lysine 27 and lysine 4 trimethylation in multiple myeloma reveals the importance of Polycomb gene targeting and highlights EZH2 as a potential therapeutic target. Oncotarget. (2016) 7(6):6809–23. doi: 10.18632/oncotarget.6843
9. Alzrigat M, Parraga AA, Agarwal P, Zureigat H, Osterborg A, Nahi H, et al. EZH2 inhibition in multiple myeloma downregulates myeloma associated oncogenes and upregulates microRNAs with potential tumor suppressor functions. Oncotarget. (2017) 8(6):10213–24. doi: 10.18632/oncotarget.14378
10. Nylund P, Atienza Parraga A, Haglof J, De Bruyne E, Menu E, Garrido-Zabala B, et al. A distinct metabolic response characterizes sensitivity to EZH2 inhibition in multiple myeloma. Cell Death Dis (2021) 12(2):167. doi: 10.1038/s41419-021-03447-8
11. Walker BA, Wardell CP, Chiecchio L, Smith EM, Boyd KD, Neri A, et al. Aberrant global methylation patterns affect the molecular pathogenesis and prognosis of multiple myeloma. Blood. (2011) 117(2):553–62. doi: 10.1182/blood-2010-04-279539
12. Agirre X, Castellano G, Pascual M, Heath S, Kulis M, Segura V, et al. Whole-epigenome analysis in multiple myeloma reveals DNA hypermethylation of B cell-specific enhancers. Genome Res (2015) 25(4):478–87. doi: 10.1101/gr.180240.114
13. Alaterre E, Ovejero S, Herviou L, de Boussac H, Papadopoulos G, Kulis M, et al. Comprehensive characterization of the epigenetic landscape in multiple Myeloma. Theranostics (2022) 12(4):1715–29. doi: 10.7150/thno.54453
14. Ordoñez R, Kulis M, Russiñol N, Chapaprieta V, Carrasco-Leon A, García-Torre B, et al. Chromatin activation as a unifying principle underlying pathogenic mechanisms in multiple myeloma. Genome Res (2020) 30(9):1217–27. doi: 10.1101/gr.265520.120
15. Zang X, Wang J, Xia Y, Li J, Chen L, Gu Y, et al. LncRNA MEG3 promotes the sensitivity of bortezomib by inhibiting autophagy in multiple myeloma. Leuk Res (2022) 123:106967. doi: 10.1016/j.leukres.2022.106967
16. Pan Y, Zhang Y, Liu W, Huang Y, Shen X, Jing R, et al. LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL-1 via miR-29b-3p. Cell Death Dis (2019) 10(2):106. doi: 10.1038/s41419-018-1219-0
17. Chen T, Sun Z, Cui Y, Ji J, Li Y, Qu X. Identification of long noncoding RNA NEAT1 as a key gene involved in the extramedullary disease of multiple myeloma by bioinformatics analysis. Hematology. (2023) 28(1):2164449. doi: 10.1080/16078454.2022.2164449
18. Meng H, Han L, Hong C, Ding J, Huang Q. Aberrant lncRNA expression in multiple myeloma. Oncol Res (2018) 26(5):809–16. doi: 10.3727/096504017X15123872205507
19. Carrasco-Leon A, Ezponda T, Meydan C, Valcarcel LV, Ordonez R, Kulis M, et al. Characterization of complete lncRNAs transcriptome reveals the functional and clinical impact of lncRNAs in multiple myeloma. Leukemia. (2021) 35(5):1438–50. doi: 10.1038/s41375-021-01147-y
20. Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res (2012) 22(5):885–98. doi: 10.1101/gr.131037.111
21. Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol (2015) 16(1):20. doi: 10.1186/s13059-015-0586-4
22. Butova R, Vychytilova-Faltejskova P, Souckova A, Sevcikova S, Hajek R. Long non-coding RNAs in multiple myeloma. Noncoding RNA (2019) 5(1). doi: 10.3390/ncrna5010013
23. Shi W, Wang Q, Bian Y, Fan Y, Zhou Y, Feng T, et al. Long noncoding RNA PANDA promotes esophageal squamous carcinoma cell progress by dissociating from NF-YA but interact with SAFA. Pathol Res Pract (2019) 215(10):152604. doi: 10.1016/j.prp.2019.152604
24. Chen CL, Tseng YW, Wu JC, Chen GY, Lin KC, Hwang SM, et al. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials. (2015) 44:71–81. doi: 10.1016/j.biomaterials.2014.12.023
25. Carrasco-Leon A, Amundarain A, Gomez-Echarte N, Prosper F, Agirre X. The role of lncRNAs in the pathobiology and clinical behavior of multiple myeloma. Cancers (Basel) (2021) 13(8):1976. doi: 10.3390/cancers13081976
26. Helm M, Motorin Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat Rev Genet (2017) 18(5):275–91. doi: 10.1038/nrg.2016.169
27. Wang X, He C. Dynamic RNA modifications in posttranscriptional regulation. Mol Cell (2014) 56(1):5–12. doi: 10.1016/j.molcel.2014.09.001
28. Yang X, Liu M, Li M, Zhang S, Hiju H, Sun J, et al. Epigenetic modulations of noncoding RNA: a novel dimension of Cancer biology. Mol Cancer (2020) 19(1):64. doi: 10.1186/s12943-020-01159-9
29. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. (2012) 485(7397):201–6. doi: 10.1038/nature11112
30. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. (2012) 149(7):1635–46. doi: 10.1016/j.cell.2012.05.003
31. Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res (2017) 27(5):606–25. doi: 10.1038/cr.2017.55
32. Enroth C, Poulsen LD, Iversen S, Kirpekar F, Albrechtsen A, Vinther J. Detection of internal N7-methylguanosine (m7G) RNA modifications by mutational profiling sequencing. Nucleic Acids Res (2019) 47(20):e126. doi: 10.1093/nar/gkz736
33. Haruehanroengra P, Zheng YY, Zhou Y, Huang Y, Sheng J. RNA modifications and cancer. RNA Biol (2020) 17(11):1560–75. doi: 10.1080/15476286.2020.1722449
34. Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. (2016) 530(7591):441–6. doi: 10.1038/nature16998
35. Cho SF, Chang YC, Chang CS, Lin SF, Liu YC, Hsiao HH, et al. MALAT1 long non-coding RNA is overexpressed in multiple myeloma and may serve as a marker to predict disease progression. BMC Cancer (2014) 14:809. doi: 10.1186/1471-2407-14-809
36. Meng YB, He X, Huang YF, Wu QN, Zhou YC, Hao DJ. Long noncoding RNA CRNDE promotes multiple myeloma cell growth by suppressing miR-451. Oncol Res (2017) 25(7):1207–14. doi: 10.3727/096504017X14886679715637
37. Bonetti A, Agostini F, Suzuki AM, Hashimoto K, Pascarella G, Gimenez J, et al. RADICL-seq identifies general and cell type-specific principles of genome-wide RNA-chromatin interactions. Nat Commun (2020) 11(1):1018. doi: 10.1038/s41467-020-14337-6
38. Bell JC, Jukam D, Teran NA, Risca VI, Smith OK, Johnson WL, et al. Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. Elife (2018) 7. doi: 10.7554/eLife.27024
39. Mumbach MR, Granja JM, Flynn RA, Roake CM, Satpathy AT, Rubin AJ, et al. HiChIRP reveals RNA-associated chromosome conformation. Nat Methods (2019) 16(6):489–92. doi: 10.1038/s41592-019-0407-x
40. Recalde M, Garate-Rascon M, Herranz JM, Elizalde M, Azkona M, Unfried JP, et al. DNA methylation regulates a set of long non-coding RNAs compromising hepatic identity during hepatocarcinogenesis. Cancers (Basel) (2022) 14(9):2048. doi: 10.3390/cancers14092048
41. Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet (2013) 14(3):204–20. doi: 10.1038/nrg3354
42. Kulis M, Esteller M. DNA methylation and cancer. Adv Genet (2010) 70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2
43. Kalushkova A, Nylund P, Parraga AA, Lennartsson A, Jernberg-Wiklund H. One omics approach does not rule them all: the metabolome and the epigenome join forces in haematological Malignancies. Epigenomes. (2021) 5(4):22. doi: 10.3390/epigenomes5040022
44. Unfried JP, Serrano G, Suarez B, Sangro P, Ferretti V, Prior C, et al. Identification of coding and long noncoding RNAs differentially expressed in tumors and preferentially expressed in healthy tissues. Cancer Res (2019) 79(20):5167–80. doi: 10.1158/0008-5472.CAN-19-0400
45. Li Z, Kumar S, Jin DY, Calin GA, Chng WJ, Siu KL, et al. Epigenetic silencing of long non-coding RNA BM742401 in multiple myeloma: impact on prognosis and myeloma dissemination. Cancer Cell Int (2020) 20:403. doi: 10.1186/s12935-020-01504-4
46. Wong KY, Li Z, Zhang X, Leung GK, Chan GC, Chim CS. Epigenetic silencing of a long non-coding RNA KIAA0495 in multiple myeloma. Mol Cancer (2015) 14:175. doi: 10.1186/s12943-015-0444-8
47. Yang LH, Du P, Liu W, An LK, Li J, Zhu WY, et al. LncRNA ANRIL promotes multiple myeloma progression and bortezomib resistance by EZH2-mediated epigenetically silencing of PTEN. Neoplasma. (2021) 68(4):788–97. doi: 10.4149/neo_2021_210205N184
48. Guo N, Song Y, Zi F, Zheng J, Cheng J. Abnormal expression pattern of lncRNA H19 participates in multiple myeloma bone disease by unbalancing osteogenesis and osteolysis. Int Immunopharmacol (2023) 119:110058. doi: 10.1016/j.intimp.2023.110058
49. David A, Zocchi S, Talbot A, Choisy C, Ohnona A, Lion J, et al. The long non-coding RNA CRNDE regulates growth of multiple myeloma cells via an effect on IL6 signalling. Leukemia. (2021) 35(6):1710–21. doi: 10.1038/s41375-020-01034-y
50. Fu Y, Liu X, Zhang F, Jiang S, Liu J, Luo Y. Bortezomib-inducible long non-coding RNA myocardial infarction associated transcript is an oncogene in multiple myeloma that suppresses miR-29b. Cell Death Dis (2019) 10(4):319. doi: 10.1038/s41419-019-1551-z
51. Guan R, Wang W, Fu B, Pang Y, Lou Y, Li H. Increased lncRNA HOTAIR expression promotes the chemoresistance of multiple myeloma to dexamethasone by regulating cell viability and apoptosis by mediating the JAK2/STAT3 signaling pathway. Mol Med Rep (2019) 20(4):3917–23. doi: 10.3892/mmr.2019.10603
52. Morelli E, Fulciniti M, Samur MK, Ribeiro CF, Wert-Lamas L, Henninger JE, et al. A MIR17HG-derived long noncoding RNA provides an essential chromatin scaffold for protein interaction and myeloma growth. Blood. (2023) 141(4):391–405. doi: 10.1182/blood.2022016892
53. Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. (2008) 322(5908):1717–20. doi: 10.1126/science.1163802
54. Chen R, Zhang X, Wang C. LncRNA HOXB-AS1 promotes cell growth in multiple myeloma via FUT4 mRNA stability by ELAVL1. J Cell Biochem (2020) 121(10):4043–51. doi: 10.1002/jcb.29573
55. Tong J, Xu X, Zhang Z, Ma C, Xiang R, Liu J, et al. Hypoxia-induced long non-coding RNA DARS-AS1 regulates RBM39 stability to promote myeloma Malignancy. Haematologica. (2020) 105(6):1630–40. doi: 10.3324/haematol.2019.218289
56. Hu Y, Lin J, Fang H, Fang J, Li C, Chen W, et al. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia. (2018) 32(10):2250–62. doi: 10.1038/s41375-018-0104-2
57. Sun Y, Jiang T, Jia Y, Zou J, Wang X, Gu W. LncRNA MALAT1/miR-181a-5p affects the proliferation and adhesion of myeloma cells via regulation of Hippo-YAP signaling pathway. Cell Cycle (2019) 18(19):2509–23. doi: 10.1080/15384101.2019.1652034
58. Taiana E, Favasuli V, Ronchetti D, Todoerti K, Pelizzoni F, Manzoni M, et al. Long non-coding RNA NEAT1 targeting impairs the DNA repair machinery and triggers anti-tumor activity in multiple myeloma. Leukemia. (2020) 34(1):234–44. doi: 10.1038/s41375-019-0542-5
59. Agirre X, Martinez-Climent JA, Odero MD, Prosper F. Epigenetic regulation of miRNA genes in acute leukemia. Leukemia. (2012) 26(3):395–403. doi: 10.1038/leu.2011.344
60. Malumbres M. miRNAs and cancer: an epigenetics view. Mol Aspects Med (2013) 34(4):863–74. doi: 10.1016/j.mam.2012.06.005
61. Bure IV, Nemtsova MV, Kuznetsova EB. Histone modifications and non-coding RNAs: mutual epigenetic regulation and role in pathogenesis. Int J Mol Sci (2022) 23(10):5801. doi: 10.3390/ijms23105801
62. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res (2011) 21(3):381–95. doi: 10.1038/cr.2011.22
63. Zheng JF, Guo NH, Zi FM, Cheng J. Long noncoding RNA H19 promotes tumorigenesis of multiple myeloma by activating BRD4 signaling by targeting microRNA 152-3p. Mol Cell Biol (2020) 40(3). doi: 10.1128/MCB.00382-19
64. Wu SY, Lee AY, Lai HT, Zhang H, Chiang CM. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell (2013) 49(5):843–57. doi: 10.1016/j.molcel.2012.12.006
65. Wang R, Li Q, Helfer CM, Jiao J, You J. Bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J Biol Chem (2012) 287(14):10738–52. doi: 10.1074/jbc.M111.323493
66. Stubbs MC, Burn TC, Sparks R, Maduskuie T, Diamond S, Rupar M, et al. The novel bromodomain and extraterminal domain inhibitor INCB054329 induces vulnerabilities in myeloma cells that inform rational combination strategies. Clin Cancer Res (2019) 25(1):300–11. doi: 10.1158/1078-0432.CCR-18-0098
67. Ding J, Li J, Wang H, Tian Y, Xie M, He X, et al. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis (2017) 8(8):e2997. doi: 10.1038/cddis.2017.328
68. Lai IL, Yang CA, Lin PC, Chan WL, Lee YT, Yen JC, et al. Long noncoding RNA MIAT promotes non-small cell lung cancer proliferation and metastasis through MMP9 activation. Oncotarget. (2017) 8(58):98148–62. doi: 10.18632/oncotarget.21465
69. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. (2010) 329(5992):689–93. doi: 10.1126/science.1192002
70. Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv (2017) 3(9):eaao2110. doi: 10.1126/sciadv.aao2110
71. Shen W, Zhu M, Wang Q, Zhou X, Wang J, Wang T, et al. DARS-AS1 recruits METTL3/METTL14 to bind and enhance DARS mRNA m(6)A modification and translation for cytoprotective autophagy in cervical cancer. RNA Biol (2022) 19(1):751–63. doi: 10.1080/15476286.2022.2079889
72. Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal (2010) 3(107):ra8. doi: 10.1126/scisignal.2000568
73. Huo Y, Li Q, Wang X, Jiao X, Zheng J, Li Z, et al. MALAT1 predicts poor survival in osteosarcoma patients and promotes cell metastasis through associating with EZH2. Oncotarget. (2017) 8(29):46993–7006. doi: 10.18632/oncotarget.16551
74. Jing Z, Liu Q, Xie W, Wei Y, Liu J, Zhang Y, et al. NCAPD3 promotes prostate cancer progression by up-regulating EZH2 and MALAT1 through STAT3 and E2F1. Cell Signal (2022) 92:110265. doi: 10.1016/j.cellsig.2022.110265
75. Wang D, Ding L, Wang L, Zhao Y, Sun Z, Karnes RJ, et al. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget. (2015) 6(38):41045–55. doi: 10.18632/oncotarget.5728
76. Zhu Y, Hu H, Yuan Z, Zhang Q, Xiong H, Hu Z, et al. LncRNA NEAT1 remodels chromatin to promote the 5-Fu resistance by maintaining colorectal cancer stemness. Cell Death Dis (2020) 11(11):962. doi: 10.1038/s41419-020-03164-8
77. Ma F, Lei YY, Ding MG, Luo LH, Xie YC, Liu XL. LncRNA NEAT1 Interacted With DNMT1 to Regulate Malignant Phenotype of Cancer Cell and Cytotoxic T Cell Infiltration via Epigenetic Inhibition of p53, cGAS, and STING in Lung Cancer. Front Genet (2020) 11:250. doi: 10.3389/fgene.2020.00250
78. Dinescu S, Ignat S, Lazar AD, Constantin C, Neagu M, Costache M. Epitranscriptomic signatures in lncRNAs and their possible roles in cancer. Genes (Basel) (2019) 10(1):52. doi: 10.3390/genes10010052
79. Cusenza VY, Tameni A, Neri A, Frazzi R. The lncRNA epigenetics: The significance of m6A and m5C lncRNA modifications in cancer. Front Oncol (2023) 13:1063636. doi: 10.3389/fonc.2023.1063636
80. Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer (2020) 20(6):303–22. doi: 10.1038/s41568-020-0253-2
81. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. (2016) 537(7620):369–73. doi: 10.1038/nature19342
82. Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, et al. m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer (2019) 18(1):87. doi: 10.1186/s12943-019-1014-2
83. Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer (2019) 18(1):143. doi: 10.1186/s12943-019-1079-y
84. He RZ, Jiang J, Luo DX. The functions of N6-methyladenosine modification in lncRNAs. Genes Dis (2020) 7(4):598–605. doi: 10.1016/j.gendis.2020.03.005
85. Wei W, Huo B, Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manag Res (2019) 11:1177–87. doi: 10.2147/CMAR.S181058
86. Du M, Zhang Y, Mao Y, Mou J, Zhao J, Xue Q, et al. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem Biophys Res Commun (2017) 482(4):582–9. doi: 10.1016/j.bbrc.2016.11.077
87. Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S, et al. MicroRNA-145 modulates N(6)-methyladenosine levels by targeting the 3'-untranslated mRNA region of the N(6)-methyladenosine binding YTH domain family 2 protein. J Biol Chem (2017) 292(9):3614–23. doi: 10.1074/jbc.M116.749689
88. Long X, Wen F, Li J, Huang X. LncRNA FEZF1-AS1 accelerates multiple myeloma progression by regulating IGF2BP1/BZW2 signaling. Hematol Oncol (2023). doi: 10.1002/hon.3157
89. Che F, Ye X, Wang Y, Wang X, Ma S, Tan Y, et al. METTL3 facilitates multiple myeloma tumorigenesis by enhancing YY1 stability and pri-microRNA-27 maturation in m(6)A-dependent manner. Cell Biol Toxicol (2022) 39:2033–50. doi: 10.1007/s10565-021-09690-1
90. Huang X, Yang Z, Li Y, Long X. m6A methyltransferase METTL3 facilitates multiple myeloma cell growth through the m6A modification of BZW2. Ann Hematol (2023) 102(7):1801–10. doi: 10.1007/s00277-023-05283-6
91. Xu A, Zhang J, Zuo L, Yan H, Chen L, Zhao F, et al. FTO promotes multiple myeloma progression by posttranscriptional activation of HSF1 in an m(6)A-YTHDF2-dependent manner. Mol Ther (2022) 30(3):1104–18. doi: 10.1016/j.ymthe.2021.12.012
92. Wang C, Li L, Li M, Wang W, Jiang Z. FTO promotes Bortezomib resistance via m6A-dependent destabilization of SOD2 expression in multiple myeloma. Cancer Gene Ther (2023) 30(4):622–8. doi: 10.1038/s41417-022-00429-6
93. Qu J, Hou Y, Chen Q, Chen J, Li Y, Zhang E, et al. RNA demethylase ALKBH5 promotes tumorigenesis in multiple myeloma via TRAF1-mediated activation of NF-kappaB and MAPK signaling pathways. Oncogene. (2022) 41(3):400–13. doi: 10.1038/s41388-021-02095-8
94. Yu T, Yao L, Yin H, Teng Y, Hong M, Wu Q. ALKBH5 Promotes Multiple Myeloma Tumorigenicity through inducing m(6)A-demethylation of SAV1 mRNA and Myeloma Stem Cell Phenotype. Int J Biol Sci (2022) 18(6):2235–48. doi: 10.7150/ijbs.64943
95. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell (2013) 49(1):18–29. doi: 10.1016/j.molcel.2012.10.015
96. Wang Z, He J, Bach DH, Huang YH, Li Z, Liu H, et al. Induction of m(6)A methylation in adipocyte exosomal LncRNAs mediates myeloma drug resistance. J Exp Clin Cancer Res (2022) 41(1):4. doi: 10.1186/s13046-021-02209-w
97. Wen S, Wei Y, Zen C, Xiong W, Niu Y, Zhao Y. Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol Cancer (2020) 19(1):171. doi: 10.1186/s12943-020-01293-4
98. Qin J, Ke B, Liu T, Kong C, Li A, Fu H, et al. Aberrantly expressed long noncoding RNAs as potential prognostic biomarkers in newly diagnosed multiple myeloma: A systemic review and meta-analysis. Cancer Med (2023) 12(3):2199–218. doi: 10.1002/cam4.5135
99. Taiana E, Bandini C, Favasuli VK, Ronchetti D, Silvestris I, Puccio N, et al. Activation of long non-coding RNA NEAT1 leads to survival advantage of multiple myeloma cells by supporting a positive regulatory loop with DNA repair proteins. Haematologica. (2023) 108(1):219–33. doi: 10.3324/haematol.2022.281167
100. Wu Y, Wang H. LncRNA NEAT1 promotes dexamethasone resistance in multiple myeloma by targeting miR-193a/MCL1 pathway. J Biochem Mol Toxicol (2018) 32(1). doi: 10.1002/jbt.22008
101. Chen YS, Yang WL, Zhao YL, Yang YG. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip Rev RNA (2021) 12(4):e1639. doi: 10.1002/wrna.1639
102. Ren H, Liu C, Wu H, Wang Z, Chen S, Zhang X, et al. m(5)C Regulator-mediated methylation modification clusters contribute to the immune microenvironment regulation of multiple myeloma. Front Genet (2022) 13:920164. doi: 10.3389/fgene.2022.920164
103. Liu X, Wang D, Han S, Wang F, Zang J, Xu C, et al. Signature of m5C-Related lncRNA for Prognostic Prediction and Immune Responses in Pancreatic Cancer. J Oncol (2022) 2022:7467797. doi: 10.1155/2022/7467797
104. Pan J, Huang Z, Xu Y. m5C-related lncRNAs predict overall survival of patients and regulate the tumor immune microenvironment in lung adenocarcinoma. Front Cell Dev Biol (2021) 9:671821. doi: 10.3389/fcell.2021.671821
105. Bai M, Sun C. M5C-related lncRNA predicts lung adenocarcinoma and tumor microenvironment remodeling: computational biology and basic science. Front Cell Dev Biol (2022) 10:885568. doi: 10.3389/fcell.2022.885568
106. Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. (2017) 551(7679):251–5. doi: 10.1038/nature24456
107. Goyal B, Yadav SRM, Awasthee N, Gupta S, Kunnumakkara AB, Gupta SC. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim Biophys Acta Rev Cancer (2021) 1875(2):188502. doi: 10.1016/j.bbcan.2021.188502
108. Liu N, Feng S, Li H, Chen X, Bai S, Liu Y. Long non-coding RNA MALAT1 facilitates the tumorigenesis, invasion and glycolysis of multiple myeloma via miR-1271-5p/SOX13 axis. J Cancer Res Clin Oncol (2020) 146(2):367–79. doi: 10.1007/s00432-020-03127-8
109. Gu Y, Xiao X, Yang S. LncRNA MALAT1 acts as an oncogene in multiple myeloma through sponging miR-509-5p to modulate FOXP1 expression. Oncotarget. (2017) 8(60):101984–93. doi: 10.18632/oncotarget.21957
110. Chen Y, Lin H, Miao L, He J. Role of N7-methylguanosine (m(7)G) in cancer. Trends Cell Biol (2022) 32(10):819–24. doi: 10.1016/j.tcb.2022.07.001
111. Han J, Liu Q, Zhou Y, Li D, Wang R. Landscape of internal N7-methylguanosine of long non-coding RNA modifications in resistant acute myeloid leukemia. BMC Genomics (2023) 24(1):425. doi: 10.1186/s12864-023-09526-8
Keywords: lncRNA, chromatin regulation, multiple myeloma, epigenetics, RNA modifications, haematological malignancies
Citation: Nylund P, Garrido-Zabala B, Kalushkova A and Wiklund HJ (2023) The complex nature of lncRNA-mediated chromatin dynamics in multiple myeloma. Front. Oncol. 13:1303677. doi: 10.3389/fonc.2023.1303677
Received: 28 September 2023; Accepted: 27 November 2023;
Published: 11 December 2023.
Edited by:
Felix Seyfried, Ulm University Medical Centre, GermanyReviewed by:
Valentina S. Caputo, London South Bank University, United KingdomCopyright © 2023 Nylund, Garrido-Zabala, Kalushkova and Wiklund. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Helena Jernberg Wiklund, aGVsZW5hLmplcm5iZXJnX3dpa2x1bmRAaWdwLnV1LnNl
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.