- 1Guangdong Province Key Laboratory of Biotechnology Drug Candidates, School of Life Sciences and Biopharmaceuticals, Guangdong Pharmaceutical University, Guangzhou, Guangdong, China
- 2School of Health Sciences, Guangzhou Xinhua University, Guangzhou, Guangdong, China
Autophagy, a crucial cellular mechanism responsible for degradation and recycling of intracellular components, is modulated by an intricate network of molecular signals. Its paradoxical involvement in oncogenesis, acting as both a tumor suppressor and promoter, has been underscored in recent studies. Central to this regulatory network are the epigenetic modifications of DNA and RNA methylation, notably the presence of N6-methyldeoxyadenosine (6mA) in genomic DNA and N6-methyladenosine (m6A) in eukaryotic mRNA. The 6mA modification in genomic DNA adds an extra dimension of epigenetic regulation, potentially impacting the transcriptional dynamics of genes linked to autophagy and, especially, cancer. Conversely, m6A modification, governed by methyltransferases and demethylases, influences mRNA stability, processing, and translation, affecting genes central to autophagic pathways. As we delve deeper into the complexities of autophagy regulation, the importance of these methylation modifications grows more evident. The interplay of 6mA, m6A, and autophagy points to a layered regulatory mechanism, illuminating cellular reactions to a range of conditions. This review delves into the nexus between DNA 6mA and RNA m6A methylation and their influence on autophagy in cancer contexts. By closely examining these epigenetic markers, we underscore their promise as therapeutic avenues, suggesting novel approaches for cancer intervention through autophagy modulation.
1 Introduction
6mA DNA, which signifies adenine methylation at the sixth position, has been recently spotlighted in research as a key epigenetic marker orchestrating cellular metabolism across a wide spectrum of organisms, from bacteria to mammals (1, 2). Concurrently, there is a growing intrigue surrounding the interplay between m6A RNA methylation and autophagy. A landmark study emphasizes m6A’s pivotal regulatory role in autophagy expression (3). Moreover, researchers discerned a link between type-specific HPV infections and hTERT DNA methylation in patients suffering from invasive cervical cancer, offering an initial insight into how DNA methylation might regulate autophagy (4). On a mechanistic level, 6mA DNA modulates the expression of autophagy-related genes by influencing the accessibility of transcription factors to their target gene promoters (5).
Meanwhile, m6A methylation, characterized by its dynamic reversibility, has also attracted significant attention. Orchestrated by a sophisticated interplay of “writers” or methyltransferases, “erasers” or demethylases, and “readers” or m6A-binding proteins, m6A RNA modification stands out as a key player (6–8). The relationship between autophagy and cancer is intricate and continues to be actively investigated. It plays dual roles, both promoting and inhibiting cancer. This duality stems from the involvement of diverse ATG proteins and core complexes, such as the ULK1/2 kinase core complex, the autophagy-specific class III PI3K complex, and the ATG9A trafficking system, among others (9, 10). These components intricately coordinate a spectrum of tasks within the autophagy pathway, from initiation to degradation. Reflecting on this, it becomes clear that grasping the subtle roles of autophagy in cancer is crucial for propelling therapeutic breakthroughs. Both DNA and RNA methylation are gaining heightened acknowledgment, shaping cellular reactions to assorted stressors and ailments (11). An expanding corpus of evidence endorses the indispensable role of RNA m6A in regulating autophagy, particularly in stabilizing and enhancing the translation of autophagy-related mRNAs (12). Preliminary investigations have begun to shed light on the potential connections between 6mA methylation and its precise regulation of autophagy, notably highlighting its significant relevance in the context of cancer (13).
Autophagy, an intricately orchestrated cellular process, plays a pivotal role in maintaining cellular equilibrium by selectively degrading and recycling intracellular constituents, including damaged organelles and misfolded proteins (14, 15). The dysregulation of autophagy has been implicated in a myriad of human afflictions, among them being cancer, neurodegenerative disorders, and metabolic anomalies (16–18). Significant investigations have unraveled a direct connection between 6mA methylation and the regulation of autophagy (13). Similar to DNA methylation, m6A mRNA methylation also assumes a crucial role in regulating autophagy and adipogenesis, particularly by targeting Atg5 and Atg7. Remarkably, these targets, Atg5 and Atg7, are recognized by the YTHDF2 protein, a known N6-methyladenosine RNA binding entity. Furthermore, the research demonstrates that a deficiency in FTO results in a decrease in white fat mass and impedes autophagy reliant on both ATG5 and ATG7 in vivo (19). These investigations underscore the intricate interplay between mRNA methylation, autophagy, and cancer.
Most current research distinctively explores the roles of 6mA methylation and m6A methylation in modulating gene and protein expression, particularly in the context of autophagy. Yet, a significant gap remains in understanding the synergistic effects of 6mA and m6A methylation on autophagy regulation. Such an understanding is crucial, especially when considering the complex environment of tumor cells. As the expression levels of 6mA and m6A methylation within cells are perpetually changing, it’s pivotal to discern if one form of methylation can impact the other and, in turn, influence broader expression patterns. This holds promising implications for future research.
In this review, our aim is to elucidate the essential roles of 6mA DNA and m6A RNA methylation in autophagy regulation within cancer contexts. We offer a concise overview of autophagy’s role in cancer. Additionally, we delve into the complexities of 6mA methylation and its impact on autophagy, elucidating the underlying mechanisms of 6mA methylation and the dynamic interplay between m6A and autophagy in oncological scenarios. In conclusion, we present an in-depth analysis of m6A methylation’s pivotal role in autophagy regulation, emphasizing the promise of 6mA and m6A-guided autophagy as potential therapeutic strategies in cancer management, potentially heralding novel anticancer treatments.
2 Overview of autophagy in cancer
Macroautophagy, henceforth referred to simply as autophagy, represents an evolutionary conserved cellular process, it not only serves as a pivotal mechanism for preserving intracellular balance by methodically dismantling and subsequently recycling cellular components, but also plays a dual role in cancer (20, 21). Recent studies have expanded our comprehension by illuminating the intricate relationship between autophagy and epigenetic modifications (22, 23). The core of autophagy is the formation of double-membraned structures called autophagosomes, proficiently encapsulating cytoplasmic components, including organelles and proteins, marked for degradation (24). Subsequently, these autophagosomes merge with lysosomes, culminating in the formation of autolysosomes, wherein the engulfed material succumbs to the catabolic prowess of lysosomal hydrolases (25, 26). The ensuing breakdown products are then carefully recycled back into the cytoplasm, providing crucial sustenance for energy production and macromolecular synthesis, thereby furnishing steadfast support to cellular function during periods of exigency (27) (Figure 1A). Autophagy plays a fundamental physiological role, particularly in the elimination of damaged proteins and organelles during periods of stress and aging (28). This process is instrumental in orchestrating organismal development, working in synergy with the adaptive immune system, ensuring energy balance, and maintaining rigorous protein and organelle quality control (29, 30). Recent findings also suggest that autophagy might intensify cancer malignancy by fostering metastatic behaviors (31). Moreover, autophagy is central to enhancing cellular resilience against diverse challenges, from nutrient scarcity and oxidative stress to pathogenic threats, reinforcing cellular strength and integrity (32–34). Recent investigations have elucidated the complex interplay in autophagy regulation, highlighting the nuanced contributions of DNA methylation variants, particularly 6mA, 7mG, and 5mC, in concert with RNA methylation types, including m6A, m1A, and m7G, and synergistic protein modifications (35–37) (Figure 1B). Collectively, these mechanisms ensure the fundamental principles of autophagy regulation, and more importantly, they shed light on the intricate role of autophagy in cancer.
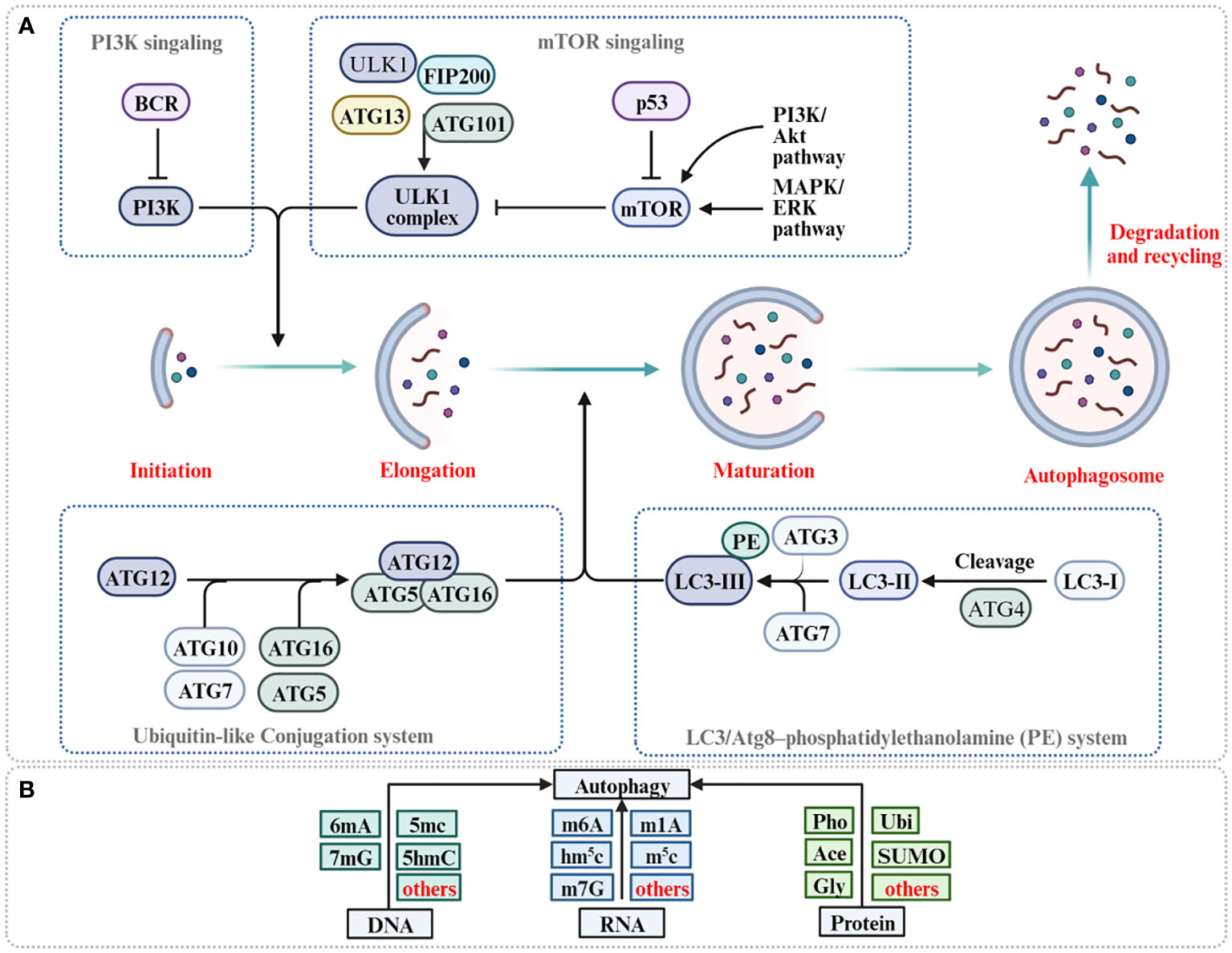
Figure 1 The process of autophagy and its regulation for 6mA methylation, m6A methylation and protein modification. (A) Autophagy is initiated with the formation of ULK1 complex (ULK1-ATG13-FIP200-ATG201) and PI3K complex facilitate the formation of phagophore. ATG5-ATG12-ATG16 complex and PE-conjugated-LC3II promote phagophore elongation and autophagosome formation. Autophagosome fusion with the lysosome results in the degradation of target molecules. (B) Methylation of DNA, methylation of RNA, and protein modification of protein are reported to participate in autophagy regulation (Pho, phosphorylation; Ace, acetylation; Ubi, ubiquitination; Gly, glycosylation).
In the early phases of tumorigenesis, autophagy functions as a protective barrier, eliminating damaged proteins and organelles. This crucial process prevents the accumulation of genetic abnormalities that could potentially trigger malignant transformations (38, 39). However, as tumors progress and mature, autophagy undergoes adaptations to support their growth in nutrient-deprived environments, assuming a dual role in cancer regulation (40). Research indicates that tumors, by harnessing autophagy’s ability to recycle nutrients, can sustain their growth, develop resistance to treatments, and even facilitate metastasis (41). This adaptability empowers tumors to thrive in challenging conditions, rendering them resilient targets for treatments (42). Moreover, the dual role of autophagy, both inhibiting and promoting tumors, adds complexity to therapeutic targeting (43). As cancer progresses, tumors exploit autophagy’s recycling abilities, evolving with heightened aggression, resisting treatments, and potentially spreading (44). Enhancing autophagy in mature tumors can bolster their resilience, suggesting that inhibiting autophagy could be a promising avenue for treatment. Although the precise regulatory mechanisms of m6A in eukaryotic mRNA and 6mA methylation in genomic DNA are not fully understood, the m6A modification, governed by the interplay of methyltransferases and demethylases, likely exerts an influence on mRNA behavior, potentially impacting crucial autophagic genes. Simultaneously, the presence of 6mA in genomic DNA introduces a distinctive epigenetic layer, influencing the transcription of autophagy-related genes (23).
Given autophagy’s dual influence on cancer, it presents both therapeutic challenges and opportunities. Viewing from a genomic lens, autophagy’s diverse roles in cancer unveil complex therapeutic avenues. The deep connection between genomic alterations and autophagy is not only crucial but warrants thorough exploration, as it might redefine our perspective and strategies in cancer therapy.
3 6mA methylation in autophagy regulation
6mA methylation, characterized by the methylation of adenine at its sixth position, has recently gained recognition as a significant epigenetic marker prevalent across the genomes of diverse organisms (45). DNA methylation, a crucial epigenetic mechanism, influences gene expression and various cellular processes without altering the DNA sequence itself (37). One prominent form of methylation is 5-methylcytosine (5mC), which is prevalent in mammals and recognized for its ability to suppress transcription factor-DNA interactions. Apart from 5mC, genomic DNA across different species showcases other methylated bases, such as 6mA and N4-methylcytosine (4mC) (46, 47). Contrary to 5mC, 6mA and 4mC are primarily recognized for their functionality in lower organisms like bacteria and protists (48). Most importantly, 6mA methylation assumes a pivotal role in DNA replication, repair, gene expression, and interactions with pathogens (49). Changes in 6mA methylation might contribute to the development of resistance to cancer therapies. Understanding these mechanisms could lead to new approaches to prevent or overcome resistance, such as combination therapies that include epigenetic drugs. In order to comprehend this indispensable role, we offer a general perspective on the mechanism of 6mA methylation, particularly, we focus on the interplay between 6mA and autophagy to further indicate the relationship between 6mA methylation, autophagy, and cancer.
3.1 Mechanism of 6mA methylation
6mA, a form of DNA methylation characterized the addition of a methyl group to the adenine base, is a well-documented epigenetic feature observed in bacteria and prokaryotes (50). While the presence of 6mA DNA methylation in plants and insects has been acknowledged for some time, its direct involvement in autophagy continues to be elucidated (51, 52). Intriguingly, roles for m6A in stem cell fate determination have also been discovered. For example, research has outlined a mechanism wherein m6A potentially modulates the stability of WUS and STM mRNAs, thereby influencing shoot stem cell fate determination (53). Moreover, scholarly attention has been focused on the 6mA demethylase that operates on dsDNA in eukaryotic organisms. This line of inquiry underscores the unique role of the CcTet D337F mutant protein as a crucial chemical biology tool for in vivo manipulation of 6mA methylation (54). Such discoveries highlight the distinct role of 6mA methylation and its prospective significance in orchestrating subsequent cellular processes. However, its existence and operational function within eukaryotes remain a contentious area within the ambit of scientific exploration. Abnormal patterns of 6mA methylation in cancer cells could serve as biomarkers for early detection, prognosis, or as indicators of therapeutic response. Identifying unique methylation patterns specific to cancer types or stages could lead to more precise diagnostic and prognostic tools. Contemporary research posits that 6mA might exert influence gene expression, transposon silencing, and environmental stress response.
Enzymes such as N6AMT1, METTL4, DDM-1, DAMT, DAMT-1, MTA1c, METTL3, and METTL14 are postulated to be “writers” of this modification. In contrast, ALKBH1, DMAD, NMAD-1, and ALKBH4 function as “erasers”. N6AMT1, identified as the methyltransferase responsible for nuclear 6mA methylation, might potentially shape 6mA DNA patterns through an indirect mechanism (Figure 2) (55). Additionally, Jumu and SSBP1 are categorized as 6mA-DNA-binding factors and are considered “readers” (56, 57). Studies have shown that a decrease in N6AMT1 is correlated with reduced DNA 6mA levels, increased tumor progression, and an unfavorable prognosis for breast cancer (BC) patients. Specifically, silencing N6AMT1 using sh-RNA diminishes DNA 6mA levels and heightened proliferation and migration of BC cells. In contrast, overexpressing N6AMT1 produces the opposite effect (58). Conversely, METTL4, recognized as a member of the adenine methyltransferase clan, has showcased its proficiency in adding 6mA imprints onto DNA (59, 60). Furthermore, researches on METTL3 and METTL14, primarily acknowledged for their RNA m6A transcriptional finesse, has revealed their capability in imprinting 6mA onto DNA (61). In current research focusing on DNA repair, it has been observed that the involvement of METTL3 in regulating homologous recombination repair subsequently impacts the chemotherapeutic response in MCF-7 and MDA-MB-231 cells (62). Notably, METTL3 knockdown heightened the sensitivity of these BC cells to Adriamycin treatment, leading to a pronounced accumulation of DNA damage (63). According to research findings, the METTL3-METTL14 complex exhibits the potential to modify DNA, especially in contexts such as single-stranded DNA regions or areas with DNA damage (64). However, the specific role of METTL3 and METTL14 in 6mA DNA methylation remains unclear. It is imperative to conduct more in-depth research to conclusively determine the involvement of METTL3 and METTL14 in DNA methylation and to elucidate the functional implications of such modifications. ALKBH1, a distinguished member of the renowned AlkB protein family, serves as a pivotal DNA repair enzyme in eukaryotes and acts as a DNA demethylase by excising methyl groups from 6mA in DNA and m6A in RNA (65). Consistent with this, ALKBH4, shown to demethylate DNA 6mA, has the capability to inhibit 6mA methylation. Interestingly, research has provided evidence that the inactivation of BcMETTL4 leads to the downregulation of 13 genes with methylation within their promoter regions. Notably, the autophagy protein Apg6 in B. cinerea was distinctly identified as well (66). Concluding this investigation, the independent disruption of two genes, BcFDH and BcMFS2, while not directly studied within a mammalian context, could nonetheless illuminate potential parallels and offer novel insights into similar genetic mechanisms in mammals. Study have demonstrated that alterations in DNA methylation also play a pivotal role in tumorigenesis and tumor suppression, suggesting the complicated connection between 6mA methylation and autophagy regulation in cancer (67). The intricate interplay between methylation and demethylation processes, and their implications in cellular functions, especially in cancer, underscores the necessity for a deeper understanding of these mechanisms.
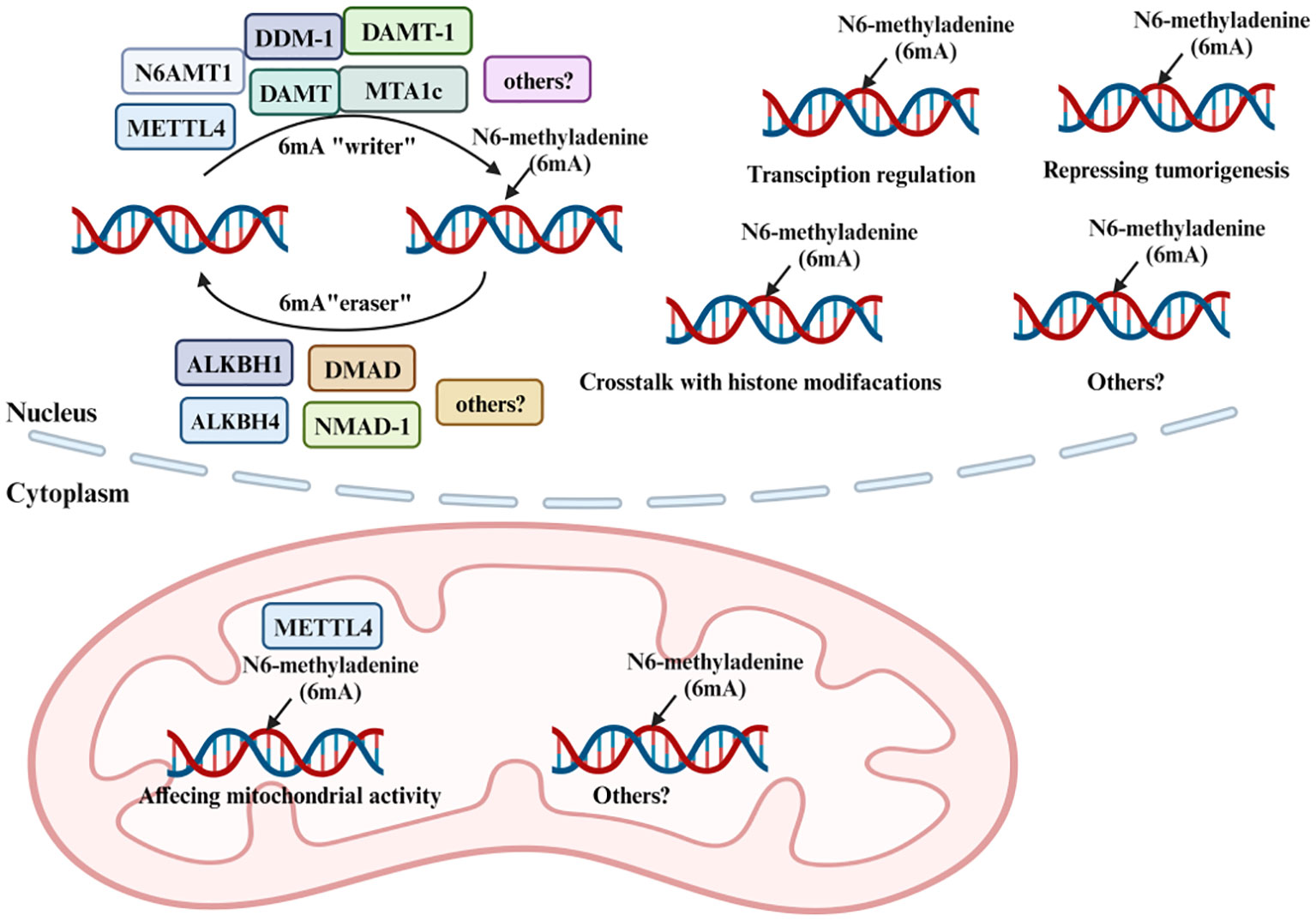
Figure 2 Methylation and demethylation of 6mA. 6mA methylation is a dynamic and reversible process coordinated by a series of methyltransferases (METTL4, N6AMT1, DDM-1, DAMT, DAMT-1, and MTA1c termed as “6mA writers”), demethylases (ALKBH1, ALKBH4, DMAD, and NMAD-1 termed as “6mA erasers”).
3.2 6mA methylation in autophagy and cancer
The intricate relationship between DNA 6mA and autophagy regulation in cancer has garnered significant attention in the field of cellular biology. As previously noted, the 6mA modification on DNA is prevalent across the genomes of both prokaryotes and eukaryotes. In prokaryotes, it plays a regulatory role in various functions, including DNA replication, repair, transcription, and bacterial resistance (68). In humans, 6mA methylation has been linked to several diseases. For instance, a notable example is the role of the ALKBH1-demethylated DNA N6-methyladenine modification, which triggers vascular calcification through osteogenic reprogramming in cases of chronic kidney disease (69). The role of autophagy in cancer is primarily influenced by various molecular regulators, including enzymes associated with 6mA regulation. While articles address the broader aspects of autophagy, one specific area of interest is the connection between enzymes regulating autophagy and 6mA modifications in the context of cancer. A comprehensive approach to understanding 6mA’s role in regulating autophagy would involve examining both its transcriptional regulation and its influence on autophagy-related proteins.
The role of 6mA in autophagy is a rapidly growing area of research, and understanding its precise function requires a comprehensive mapping of 6mA modifications across the genome. METTL4 is part of a subclade of MT-A70 adenine methyltransferases (70). Modifying the epigenetic landscape could potentially alter the immune response to tumors or change the behavior of surrounding stromal cells, which could be leveraged therapeutically. Recently, the human METTL4 protein was reported to demonstrate 6mA methyltransferase activity on mitochondrial DNA (mtDNA) (59). Due to its distinct methylation sites, akin to those of 5mC, the role of 6mA methylation in growth, development, and tumor progression becomes discernible. However, in contrast to 5mC methylation, 6mA methylation appears to demonstrate a reduced correlation with transcription, as observed in mitochondrial transcription in vitro (59). Instead, it exerts a more pronounced influence on cell survival, particularly due to its significant role in genes such as TBC1D3H, CSMD1, and ROBO2, which are characterized by unstable methylation sites (71). The presence of 6mA on the promoter regions of autophagy-related genes can influence their transcription. It has been reported that in glioblastoma, levels of both 6mA and the 6mA demethylase enzyme ALKBH1 are elevated (72). This raises the possibility that other yet-to-be-discovered DNA methyltransferases or demethylase might also contribute to the elevated levels of 6mA observed in glioblastoma. Notably, as recently reported, ALKBH4 impacts protein substrates, although its extensive role in DNA modification has yet to be revealed (73).
In a seminal investigation, researchers found that the nicotinamide nucleotide transhydrogenase (NNT) gene is silenced through DNA hypermethylation in the cisplatin-resistant A549 phenotype (A549/DDP). This discovery implies that elevating NNT expression in A549/DDP cells might counteract their natural resistance to cisplatin (74). Interestingly, the increased vulnerability of A549/DDP cells to cisplatin, facilitated by NNT, wasn’t mainly due to its traditional role in managing NADPH and ROS balances. Instead, it was largely attributed to NNT’s ability to inhibit protective autophagy in these cells. These detailed insights, derived from CRISPR-based DNA methylation editing, highlight a deeper, perhaps previously underestimated, link between DNA methylation and autophagy. Another insightful study revealed that, upon autophagy activation, the de novo DNA methyltransferase DNMT3A governs DNA methylation of MAP1LC3 loci, leading to a consistent reduction in MAP1LC3 isoforms transcriptionally. Fascinatingly, this reduced MAP1LC3 expression is evident in vivo, as seen in both zebrafish larvae and mice exposed to a brief autophagy stimulus (5). While these observations were particularly significant in zebrafish, a wealth of research has concurrently unveiled a profound association between DNMT3A and oncogenesis (75, 76). The expression profile of the DNMT3A protein in human tissue specimens and its potential susceptibility to DNMT inhibitors remains somewhat unclear, it’s noteworthy to mention that DNA hypomethylation-induced instability predominantly manifests a chromosomal character (77). In a study meticulously delineating the intricate structure and mechanism of DNMT3A protein’s, it was illuminated that the DNMT3A-DNA interplay—encompassing a target recognition domain, a catalytic loop, and a DNMT3A homodimeric interface—revealed that somatic mutations associated with hematological cancers within the substrate-binding residues not only attenuate DNMT3A activity but also precipitate CpG hypomethylation, thereby catalyzing the transformation of hematopoietic cells (78). As a result, understanding DNMT3A-driven methylation changes in autophagy, especially concerning cancer cell evolution, provides crucial insights for discussions on 6mA methylation and its ongoing research.
ALKBH1 belongs to the AlkB family of Fe(II)/α-ketoglutarate-dependent dioxygenases. It is adept at repairing methylation-related DNA damage through a distinctive oxidative demethylation process (79, 80). When the enzyme binds to a methylated base, its active site aligns with a Fe(II) ion and an α-ketoglutarate molecule. Subsequently, molecular oxygen is introduced and activated by the Fe(II) ion, which then targets the methyl group, transforming it into a hydroxymethyl group (81–83). This unstable group swiftly rearranges to form a transient hemiaminal intermediate, which decomposes to release formaldehyde, leaving behind the unmodified base. Simultaneously, the α-ketoglutarate undergoes decarboxylation, yielding succinate and carbon dioxide (84). This process readies the enzyme for future reactions. Through this intricate mechanism, enzymes like ALKBH1 play a crucial role in directly counteracting specific alkylation damages (85). This not only safeguards genomic integrity but also exert influence on gene expression. Elevated intracellular levels of αKG have been demonstrated to inhibit starvation-induced autophagy, suggesting a potential feedback mechanism during nutrient-rich conditions (86). Moreover, αKG derivatives, such as dimethyl α-ketoglutarate, have been shown to influence autophagic responses in specific pathological scenarios. Beyond its primary metabolic roles, αKG serves as a cofactor for dioxygenase enzymes involved in epigenetic modifications. This indirectly affects gene expression patterns associated with autophagy. Research has shown that DMKG, TFMKG, and O-KG elevate intracellular levels of α-ketoglutarate. This elevation, in turn, boosts baseline autophagy in cells cultured in complete medium (87). Such findings indicate that intracellular α-ketoglutarate can inhibit starvation-induced autophagy. Therefore, αKG plays an intricate role within the multifaceted autophagy network, functioning both as a metabolic regulator and an epigenetic influencer. This emphasizes its critical importance in maintaining cellular balance and highlights its potential therapeutic applications. The enzymatic activity of ALKBH1 heavily relies on the presence of αKG, which aids in the hydroxylation of the methyl group on alkylated bases, resulting in base recovery. The interconnected roles of αKG and ALKBH1 highlight their importance in upholding genomic integrity. This connection has potential ramifications in epigenetic alterations and gene regulation. Any disruption in αKG concentrations or ALKBH1 functionality can influence gene expression patterns, suggesting their wider significance in diverse cellular activities and pathological states. Furthermore, research has explicitly shown that the N6-methyladenine DNA demethylase ALKBH1 fosters gastric carcinogenesis by impeding NRF1’s binding capability. Notably, the 6mA sites are abundant in NRF1 binding sequences and are targeted by ALKBH1 for demethylation. The demethylation of 6mA by ALKBH1 hinders NRF1-mediated transcription of subsequent targets, including several genes in the AMP-activated protein kinase (AMPK) signaling pathway (88). Collectively, the intricate interplay between αKG, its derivatives, and ALKBH1 underscores the profound impact of these molecules on cellular processes, ranging from autophagy regulation to epigenetic modifications. The multifaceted roles of αKG, serving as both a metabolic pivot and an epigenetic mediator, along with ALKBH1’s crucial function in DNA repair and gene regulation, highlight the delicate balance that cells must maintain for optimal function. As we delve deeper into comprehending these molecular interactions, it becomes evident that they hold significant promise for therapeutic interventions, especially in the realm of genomic integrity and disease prevention.
After examining the existing frameworks, we suggest that high-resolution mapping could provide more profound insights into the distribution of 6mA modifications. This may illuminate specific genomic regions or genes affected by these modifications, which in turn play pivotal roles in autophagy regulation (89). Targeting 6mA methylation pathways could exploit synthetic lethality, where the combination of a genetic mutation in the tumor and the inhibition of a methylation pathway leads to cancer cell death but spares normal cells. As previously highlighted, the CRISPR/Cas9 genome-editing system emerges as a valuable tool for exploring the functional implications of 6mA modifications (90). Through CRISPR technology, genes linked to 6mA modifications can be either knocked out or overexpressed, providing researchers with a more comprehensive platform to observe the subsequent effects on autophagy. Additionally, the CRISPR system can be harnessed to either introduce or eliminate 6mA modifications at designated genomic locations, providing a straightforward approach to assess their functional significance (91). The identification and in-depth analysis of these enzymes are pivotal for understanding the molecular intricacies of 6mA regulation. Exploring these enzymes can also shed light on the temporal patterns of 6mA modifications, unveiling details about the timing and mechanisms through which these modifications impact autophagy.
4 m6A methylation in autophagy regulation
Methylation, including N1-methylation, 7-methylguanosine, 5-methylcytosine and N6-methyladenosine, involving the addition of a methyl group to a molecule and plays a crucial role in a myriad of biological processes, particularly in regulating the multifaceted functions of RNA (92–96). Similar to the mechanisms governing 6mA methylation, the orchestration of m6A modifications on RNA involves a tripartite regulatory system. Methyltransferases are responsible for the adding the methyl group, demethylases facilitate its removal, and specific binding proteins are responsible for its recognition. This coordinated modulation plays a crucial role in the precise regulation of autophagy processes (97). The m6A “writers”, which encompass METTL3, METTL4, METTL16, WTAP, RBM15/15B, VIRMA, and ZC3H13, are responsible for introducing the m6A modification onto RNA (98–100). These enzymes collaborate to identify specific RNA substrates and catalyze the transfer of a methyl group to the adenosine base. On the other hand, the m6A “readers” are proteins that recognize and bind to the m6A-modified RNA, thereby influencing RNA fate and function. The YTH domain family, which includes YTHDC1, YTHDC2, and YTHDF, constitutes a prominent group of m6A “readers” (101). Other notable m6A readers include Insulin-like Growth Factor 2 mRNA-Binding Proteins (IGF2BPs), the Heterogeneous Nuclear Ribonucleoproteins (HNRNP) family, and Eukaryotic Translation Initiation Factor 3 (eIF3), each of which play distinct roles in RNA metabolism, ranging from splicing to translation (102). Lastly, the m6A “erasers” encompass enzymes like FTO and the ALKBH family members ALKBH3 and ALKBH5, which remove the m6A modification, restoring the RNA to its unmodified state (103). Small molecule inhibitors that affect the activity of these proteins could modulate m6A levels and thereby impact cancer cell behavior. Together, these molecules form a sophisticated regulatory network that fine-tunes RNA function in response to cellular needs.
4.1 Mechanism of m6A methylation
Similar to 6mA methylation mechanism, m6A modification is also governed by a set of enzymes and binding proteins, including methyltransferases (“writers”), demethylases (“erasers”), and m6A-binding proteins (“readers”) (99, 104) (Figure 3). On one hand, the methyltransferases form the m6A methyltransferase complex, catalyzing the methylation reaction on adenine residues in RNA (105), on the other hand, the demethylases remove the methyl group from the N6 position of adenine in RNA, thereby reversing the methylation mark and restoring the unmodified state of RNA (106). Finally, the m6A marks recognized by m6A-binding proteins known as “readers”, play a crucial role in influencing the fate of the methylated RNA, such as regulating mRNA splicing, export, translation efficiency, and stability (107). In an analysis of EC samples, 17 m6A regulators, including YTHDC1, IGF2BP2, FTO, METTL14, and others, exhibited increased expression (108). Alterations in m6A modification have been implicated in the development of resistance to cancer therapies. By understanding these mechanisms, new strategies could be developed to prevent or overcome drug resistance. As research progresses in unraveling the mysteries surrounding these molecules, it paves the way for novel therapeutic strategies and a deeper understanding of the molecular underpinnings of cancer and autophagy.
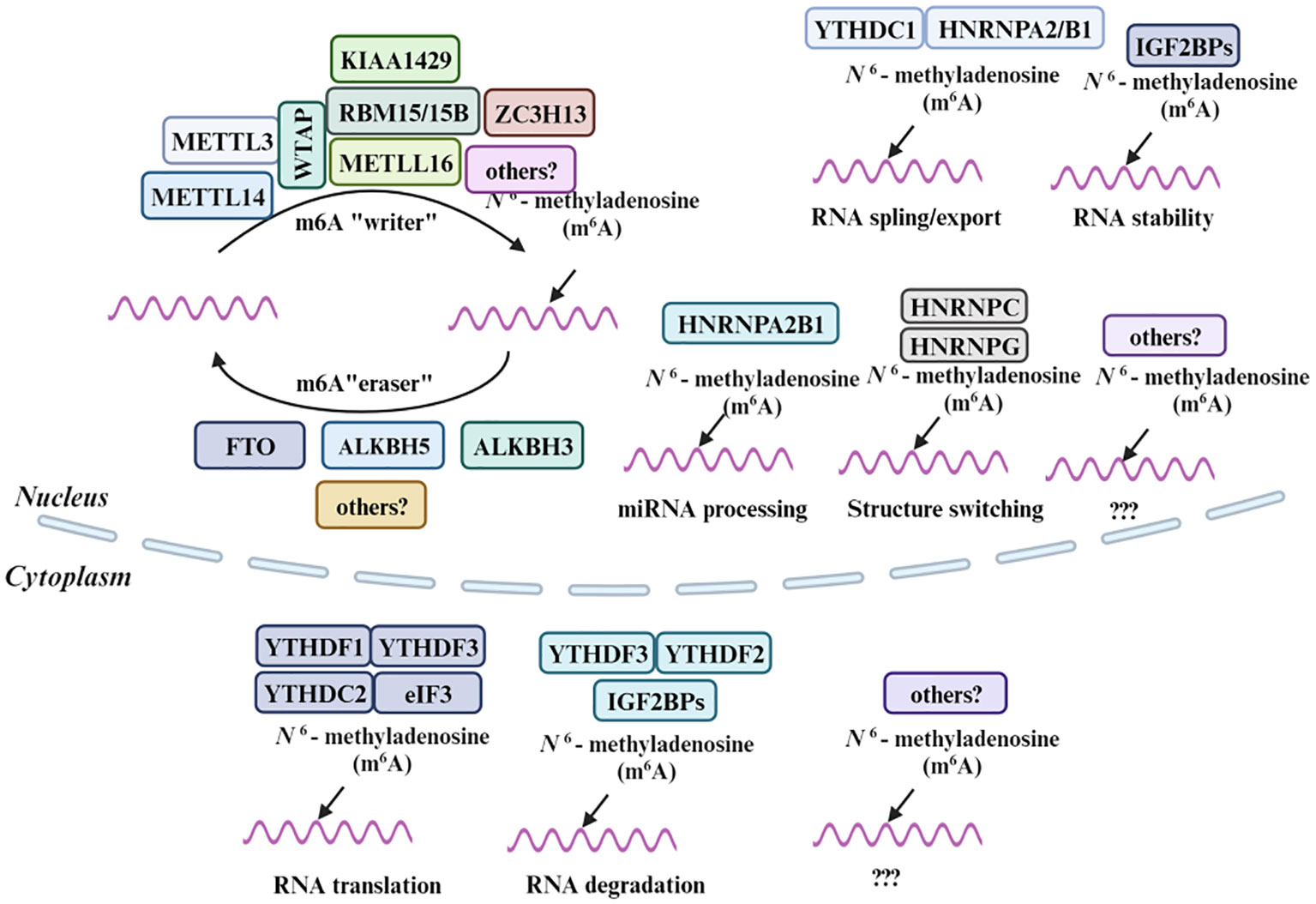
Figure 3 Molecular composition of m6A RNA methylation. M6A methylation is a dynamic and reversible process coordinated by a series of methyltransferases (METTL3/14, WTAP, RBM15/15B, METTL16, KIAA1429, ZC3H13 and termed as “m6A writers”), demethylases (FTO, ALKBH5 and ALKBH3 “m6A erasers”) and identifiers (YTHDF1/2/3, YTHDC1, HNRNPA2B1, HNRNPC, HNRNPG, eIF3, and IGF2BPs, “m6A Readers”).
The m6A “writers”, METTL3 initiating the m6A methylation process, while METTL14, another crucial component of the complex whose main function is to ensure the stability of the METTL3-METTL14 complex and facilitate substrate recognition, plays a central role in the transfer of the methyl group to the adenine base in RNA (109). It is noteworthy that the dysregulation of METTL3 and METTL14, and consequently m6A methylation, can have a significant impact on cancer progression. For instance, studies have identified that the Karyopherin Subunit Alpha 2, which is related to METTL3 and METTL14, plays a pivotal role in lung squamous cell carcinoma (110). In contrast to METTL3 and METTL14, METTL16 is responsible for methylating U6 spliceosomal RNA along with a subset of long non-coding RNAs and mRNAs (111). Furthermore, it was observed that METTL16 significantly amplify SOGA1 expression and mRNA stability, accomplished through its interaction with the “reader” protein, insulin-like growth factor 2 mRNA binding protein 1, which will be mentioned later (112). In addition, WTAP, a critical component of the m6A methyltransferase complex, plays an essential role in guiding the localization of the METTL3-METTL14 complex to specific nuclear sites, facilitating m6A methylation (113). Research has also identified WTAP’s association with a spectrum of immune cells in the tumor microenvironment, including cancer-related fibroblasts, myeloid dendritic cells, and various T cells (108). Notably, WTAP and m6A regulators like HNRNPC, YTHDC2, VIRMA, IGF2BP3, and HNRNPA2B1 showed up-regulation in both Esophageal squamous cell carcinoma and esophageal adenocarcinoma compared to normal samples. Furthermore, WTAP has been implicated in the regulation of autophagy in colon cancer cells, acting through the inhibition of FLNA via N6-methyladenosine (114). Specially, mRNA methylation also plays a pivotal role in autophagy of hepatoblastoma cell by enhancing LKB1, subsequently leading to increased phosphorylation of AMPK (115). Moreover, RBM15/15B and VIRMA are proteins that guide the m6A methyltransferase complex to specific sites in the m6A methylation process, ensuring precise and targeted m6A modifications (100). Additionally, ZC3H13, an integral part of the nuclear m6A methyltransferase complex, is essential for RNA methylation and also plays a role in facilitating the complex’s nuclear localization (116, 117).
The m6A “readers”, YTH domain family, which includes YTHDC1, YTHDC2, and YTHDF proteins, specifically recognize and bind to m6A marks on RNA molecules, allowing them to exert various regulatory functions, while the YTHDF proteins, including YTHDF1, YTHDF2, and YTHDF3, are involved in mRNA stability and translation (118–121). YTHDF1 has been distinctly recognized as a pivotal m6A reader protein responsible for BECN1 mRNA stability (122). The suppression of YTHDF1 can counteract the effects of BECN1 plasmid-induced HSC ferroptosis (123). This is further underscored by the role of YTHDF1 in enhancing BECN1 mRNA stability and activating autophagy, accomplished through its recognition of m6A binding site within the BECN1 coding regions (124). Similar to YTH domain family, eIF3 recognizes m6A marks and promotes the translation of m6A-modified mRNAs, thus influencing the efficiency of protein synthesis (125). Moreover, the IGF2BPs, including IGF2BP1, IGF2BP2, and IGF2BP3, constitute another family of m6A readers that recognize and bind to m6A marks and are involved in stabilizing their target mRNAs, protecting them from degradation (126). Members of the HNRNP family, such as HNRNPC and HNRNPG, recognize m6A marks and influence mRNA processing, also help interact with m6A-modified mRNAs, thereby affecting alternative splicing and transcript stability (127, 128). Research suggests that in prostate cancer, the elevation of circCSPP1, potentially catalyzed by HnRNP-L, triggers cellular autophagy via the circCSPP1-miR-520h-EGR1 axis, therefore, the HnRNP-L-regulated circCSPP1/miR-520h/EGR1 axis plays a pivotal role in modulating autophagy and advancing prostate cancer (129).
The “erasers” are FTO and ALKBH5, responsible for removing the m6A mark from RNA molecules (130–132). FTO, belongs to AlkB family, is capable of oxidatively demethylating m6A marks in RNA, converting m6A to adenosine. Consequently, it plays diverse roles in cellular processes, including energy homeostasis, regulating mRNA splicing, and influencing the timing of mitosis and meiosis while ALKBH3 and ALKBH5 exhibit a preference for demethylating m6A in single-stranded RNA regions, affecting mRNA export and metabolism, and also contribute to DNA repair processes (133, 134). Although the exact molecular mechanisms remain to be fully elucidated, emerging evidence suggests that ALKBH5 can influence the expression or activity of PHF20, DDIT4-AS1, and circCPSF6, thereby having an impact on autophagy (135–137). The intricate interplay between molecules like ALKBH5, PHF20, DDIT4-AS1, and circCPSF6 underscores the complexity of cellular processes and their implications in diseases such as cancer.
Absolutely, m6A methylation’s involvement in various aspects of RNA metabolism and its impact on different regions of RNA have profound implications for disease pathogenesis. The interplay between these writers, erasers, and readers ensures a finely-tuned and reversible m6A methylation process. Ongoing research in this field continues to illuminate the precise mechanisms and functional implications of m6A methylation in various biological processes and disease contexts.
4.2 m6A methylation in autophagy and cancer
As mentioned above, m6A plays a critical role in RNA metabolism, encompassing mRNA stability, translation efficiency, and splicing. It stands out as the most prevalent internal modification in eukaryotic messenger RNA and long non-coding RNAs. Moreover, it exerts widespread influence on diverse biological processes, including cell differentiation, tissue development, and stress responses. Particularly, aberrant m6A methylation patterns have been observed in various cancers (106).
METTL4, comparable in its functions to METTL3, is another established enzyme in RNA methylation, specifically involved in m7G methylation, which modulates RNA stability or structure. Consequently, METTL4 exerts an influence on the abundance of proteins essential for autophagy, which is also implicated in both the promotion and suppression of oncogenic pathways (123, 138). Furthermore, WTAP, a crucial component, interacts with METTL3 within the m6A methyltransferase complex, functioning in ensuring the proper localization and consequently implicates it in the regulation of m6A methylation (114). The Aberrant expression of WTAP has been observed in various cancers, including HCC (139). WTAP ensures the nuclear speckle localization of the METTL3-METTL14 heterodimer, thereby enhancing its catalytic activity (32). The significance of WTAP has underscroed by numerous studies, highlighting its pivotal role in a range of cancers, including those affecting liver, esophageal, breast, bladder, lung, and lymphoma system (140). In addition, insulin-like growth factor 2 mRNA-binding proteins are renowned for their role in binding and stabilizing target mRNAs, consequently promoting their translation (141). While their direct involvement in autophagy remains unclear, they theoretically could influence autophagy through their impact on mRNA stability and the translation of autophagy-related mRNAs (142). On the other hand, YTHDC1 is recognized for its capacity to bind m6A-modified RNA, exerting influence on various RNA processes such as splicing, export, and degradation (143). Recent studies have illuminated the multifaceted role of m6A modifications in determining mRNA fate, extending beyond traditional regulatory mechanisms. Specifically, m6A modifications have been demonstrated to facilitate the phase separation of their associated readers (144). A striking example of this is the modulation of cytosolic mRNA fate by m6A through its scaffolding function (145). While a direct association with autophagy has yet to be established, YTHDC1 could potentially impact autophagy by regulating the processing and stability of m6A-modified RNAs that encode proteins involved in autophagy (146).
Demethylases ALKBH3 and ALKBH5, both belonging to the AlkB family of Fe(II) and α-ketoglutarate-dependent dioxygenases, play pivotal roles as “erasers” in the context of RNA m6A modification. They have the potentially to influence the expression of autophagy-related genes through their impact on the RNA methylation (131, 147). Notably, ALKBH5 is not merely a passive marker; it actively modulates oncogenic processes, playing a crucial role in both the proliferation and invasion of cancer cells (148). In the context of gastric cancer, the expression of demethylase genes, FTO and ALKBH1, holds prognostic significance, possibly indicating their involvement in autophagy regulation (149). While an analysis of nine m6A-related genes revealed that elevated mRNA expression of FTO and ALKBH1 correlates with unfavorable overall survival in both the KM and TCGA cohorts, the TMA-IHC data contrasts this finding by showing a pronounced downregulation of FTO and ALKBH1 protein expression in gastric cancer tissues (150). A study revealed that the eIF3, central to protein synthesis initiation, can influence autophagy by controlling the translation of proteins in this process (151). Specifically, the eukaryotic translation initiation factor 3 subunit G (EIF3G) is implicated in the progression of human colorectal cancer (152). Functional assays revealed that EIF3G overexpression enhances HCT-116 cell proliferation, migration, and xenograft tumor growth, whereas xenograft tumors derived from EIF3G-silenced HCT116 cells exhibited reduced weights and volumes compared to those from control cells (151). However, the specifics of these potential interactions necessitate further elucidation. Similar to other complex members, VIRMA’s function in RNA methylation could potentially impact the stability or translational efficiency of autophagy-associated RNAs (100). Given that autophagy requires the intricate coordination of diverse RNAs and proteins, METTL16’s involvement in RNA methylation raises the possibility of it influence on autophagy, potentially through the methylation of RNAs implicated with autophagy-regulating proteins (112). In another study, it was reported that FTO can demethylate the mRNA of ULK1, a significant initiator of autophagy, leading to a decrease in ULK1 protein expression and hence suppressing autophagy (153). Specifically, FTO-driven autophagy, influenced by impaired m6A mRNA demethylation due to low-level arsenic exposure, promotes tumorigenesis and advances renal carcinoma via modulation of SIK2 mRNA stability (154). Concurrently, FTO enhances ovarian cancer cell growth by increasing proliferation, decreasing apoptosis, and activating autophagy (155). Additionally, it has been reported that on a mechanistic level, FTO functions as a tumor suppressor by regulating metastasis-associated protein 1 expression through an m6A-dependent pathway, which promotes metastasis in colorectal cancer under the hypoxia-mediated FTO downregulation (156). RNA-binding proteins RBM15 and its paralog RBM15B, integral components of the m6A methyltransferase complex, play a crucial role in RNA methylation. Targeting m6A regulatory proteins could be used in combination with other therapies to exploit synthetic lethality or to enhance the efficacy of existing treatments. While a direct correlation to autophagy is yet to be unequivocally established, their involvement in RNA methylation implies a potential role in modulating autophagy-associated RNAs, thereby influencing the proteins they encode (157).
The pivotal role of m6A methylation in cancer metabolism is evident in its influence on glucose, amino acid, and fatty acid metabolic processes. This influence extends further to impact metabolism-associated pathways and transcriptional regulators. The research delved into the phosphorylation and localization of YAP/TAZ, uncovering that m6A regulation of LATS1 affects the activity of the Hippo pathway in breast cancer cells (158). This underscores the significance of m6A regulation in modulating cell proliferation and glycolytic metabolism in breast cancer via the Hippo pathway component. 18 genes, including IGF2BP2, IGF2BP1, IGF2BP3, VIRMA, YTHDF1, YTHDF2, YTHDF3, ZC3H13, METTL14, ALKBH5, METTL3, RBMX, WTAP, YTHDC1, FTO, HNRNPC, HNRNPA2B1, and RBM15, were found to be verexpressed in HNSCC, indicating the close connection between m6A methylation and autophagy in cancer (159). Furthermore, another research revealed that METTL3-mediated m6A methylation of the mRNA encoding Transcription Factor EB, a principal orchestrator of lysosomal biogenesis and autophagy, enhances its translation (6).
In conclusion, akin to METTL3, METTL4, METTL16, WTAP, RBM15/15B, VIRMA, and ZC3H13, the potential influence of YTHDC1, YTHDC2, YTHDF, IGF2BPs, the HNRNP family, eIF3, FTO, and ALKBH3/5 on autophagy is intertwined with their roles in RNA metabolism, collectively regulating autophagy in cancer (Table 1). A deeper understanding of these relationships could provide valuable insights for innovative therapeutic strategies targeting conditions involving autophagy dysregulation.
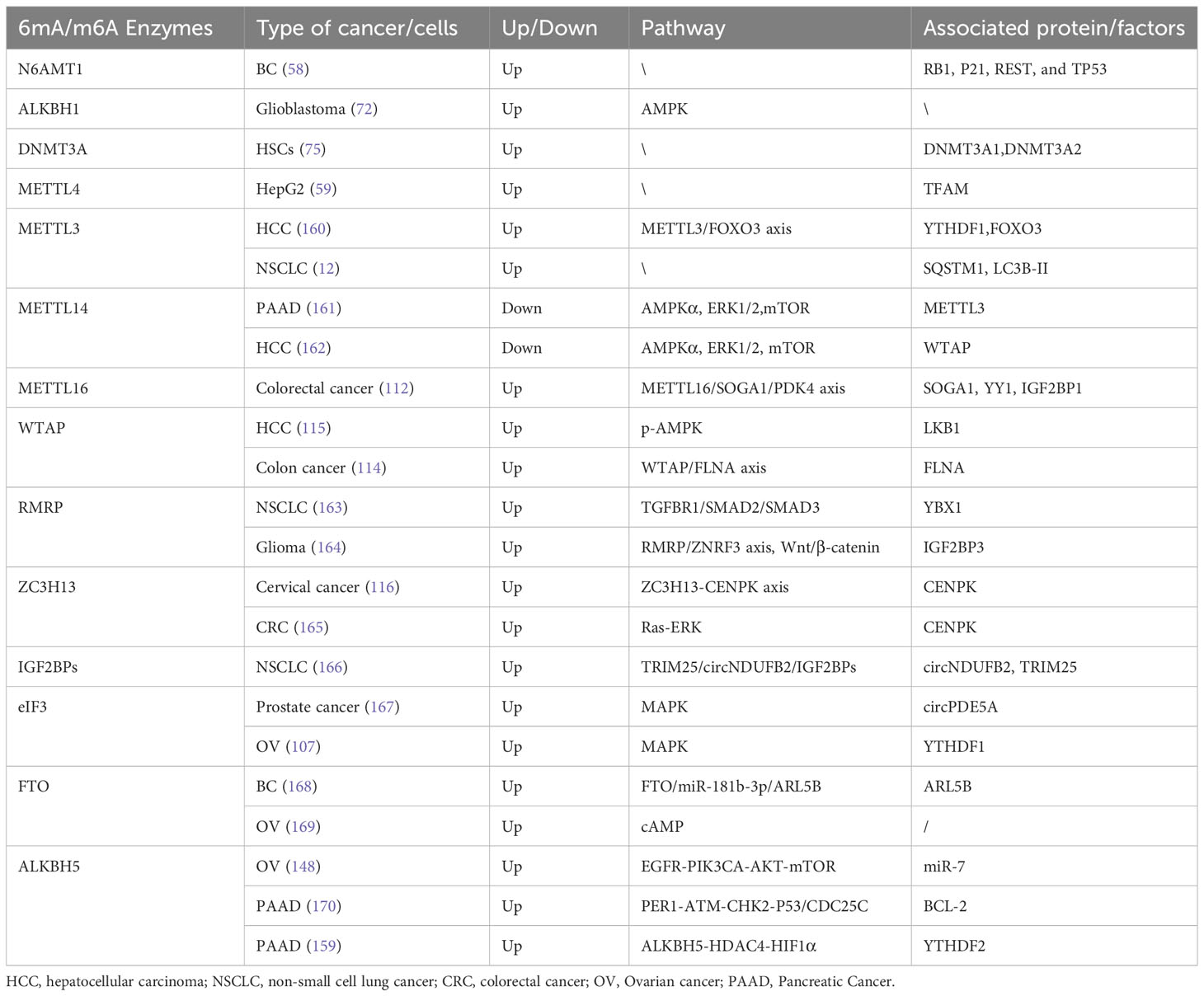
Table 1 6mA, m6A and autophagy associated factors involved in autophagy interaction ad their potential pathway in cancers.
5 Discussion
While the role of m6A in RNA metabolism has been extensively studied, the functions of 6mA, especially in relation to autophagy, remain largely unexplored. Current methodological approaches, despite their advancements, may not be refined to accurately delineate these epigenetic modifications. However, there is optimism in the scientific community, driven by the emergence of cutting-edge mapping techniques, sophisticated functional assays, and the revolutionary capabilities of tools such as CRISPR/Cas9 (91). How to find more methyltransferase and demethylases for 6mA need to worthy of in-depth study. The confluence of integrative omics paradigms, the granularity of single-cell analyses, and the predictive prowess of AI-driven methodologies heralds a renaissance in our comprehension (171). As researchers delve deeper into this field, several pertinent questions arise: How do external stimuli influence these methylation patterns and their subsequent impact on autophagy? Are there specific physiological contexts where these modifications exert a more dominant influence? Furthermore, can we strategically modulate these epigenetic markers for therapeutic purposes in diseases where autophagy is dysregulated? Most importantly, how do these modifications integrate into broader cellular processes such as homeostasis, growth, differentiation, and environmental responses? Are there specific cellular environments where 6mA plays an amplified role in regulating autophagy? how can the CRISPR technology be fine-tuned to elucidate the subtle roles of 6mA in autophagy and other cellular functions? What therapeutic potential lies in altering 6mA levels in diseases marked by autophagy anomalies? Does an intersection between DNA 6mA and RNA m6A in regulating autophagy for cancers warrant further study? In the intricate environment of tumor cells, the levels of 6mA and m6A methylation are in a state of constant flux. Gaining insights into whether one type of methylation can modulate another, and subsequently influence broader gene expression, holds significant potential. Are there innovative strategies we can employ to dynamically decipher these evolving mechanisms? Although the answers to these questions remain uncertain, they hold the potential to significantly reshape our understanding of cellular behavior and pave the way for new treatment options for numerous diseases.
Simultaneously, this intricate interplay between 6mA, m6A, and autophagy, while academically intriguing, could also pave the way for transformative therapeutic interventions (172), offering a unique perspective on autophagy in cancer. The potential of 6mA in DNA, particularly in plants like rice, has been investigated, shedding light on its role in various biological functions (173). On the other hand, the role of m6A in RNA metabolism and its potential impact on autophagy presents an intriguing prospect for therapeutic interventions, especially in conditions marked by autophagy dysregulation (174). Emerging research has also highlighted the potential of targeting the autophagy pathway for therapeutic interventions in diseases like COVID-19 (175). Furthermore, the response of the epigenetic machinery during viral infections, including the involvement of various transcription factors and epifactors, implies that targeting these pathways could provide alternative therapeutic strategies (176). These discoveries may contribute to a deeper understanding of the molecular mechanisms employed by 6mA and m6A-autophagy interaction in inducing human disorders.
In summary, the intricate dynamics of DNA and RNA methylation in relation to autophagy present a burgeoning frontier in scientific research. The potential of harnessing these epigenetic modifications for therapeutic purposes holds immense promise, though comprehensive inferences are constrained by limited findings. As our understanding of these molecular mechanisms, we stand on the cusp of a transformative era in medicine, potentially offering novel solutions for diseases that have previously defied effective treatment.
Author contributions
LL: Formal analysis, Writing – original draft. YZ: Writing – original draft, Data curation. QZ: Writing – original draft. JZ: Writing – original draft. HL: Writing – review & editing. WW: Writing – review & editing, Funding acquisition.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Science and Technology Projects in Guangzhou (Grant # 2023A04J0862), funding by high-level talent introduction into Guangdong Pharmaceutical University (Grant # 51304043013) and Guangdong Province College Students Innovation and Entrepreneurship Training Program (Grant # 202210573044).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wion D, Casadesús J. N6-methyl-adenine: an epigenetic signal for dna-protein interactions. Nat Rev Microbiol (2006) 4:183–92. doi: 10.1038/nrmicro1350
2. Dunn DB, Smith JD. The occurrence of 6-methylaminopurine in deoxyribonucleic acids. Biochem J (1958) 68:627–36. doi: 10.1042/bj0680627
3. Wang X, Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, et al. M(6)a mrna methylation controls autophagy and adipogenesis by targeting atg5 and atg7. Autophagy (2020) 16:1221–35. doi: 10.1080/15548627.2019.1659617
4. Molano M, Moreno-Acosta P, Morales N, Burgos M, Buitrago L, Gamboa O, et al. Association between type-specific hpv infections and htert dna methylation in patients with invasive cervical cancer. Cancer Genomics Proteomics. (2016) 13:483–91. doi: 10.21873/cgp.20011
5. González-Rodríguez P, Cheray M, Füllgrabe J, Salli M, Engskog-Vlachos P, Keane L, et al. The dna methyltransferase dnmt3a contributes to autophagy long-term memory. Autophagy (2021) 17:1259–77. doi: 10.1080/15548627.2020.1816664
6. Meyer KD, Jaffrey SR. The dynamic epitranscriptome: n6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol (2014) 15:313–26. doi: 10.1038/nrm3785
7. Wu R, Jiang D, Wang Y, Wang XN. (6)-methyladenosine (m(6)a) methylation in mrna with a dynamic and reversible epigenetic modification. Mol Biotechnol (2016) 58:450–9. doi: 10.1007/s12033-016-9947-9
8. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A mettl3-mettl14 complex mediates mammalian nuclear rna n6-adenosine methylation. Nat Chem Biol (2014) 10:93–5. doi: 10.1038/nchembio.1432
9. Sain A, Kandasamy T, Naskar D. Targeting unc-51-like kinase 1 and 2 by lignans to modulate autophagy: possible implications in metastatic colorectal cancer. Mol Divers (2023) 27:27–43. doi: 10.1007/s11030-022-10399-4
10. Deng G, Li C, Chen L, Xing C, Fu C, Qian C, et al. Becn2 (beclin 2) negatively regulates inflammasome sensors through atg9a-dependent but atg16l1- and lc3-independent non-canonical autophagy. Autophagy (2022) 18:340–56. doi: 10.1080/15548627.2021.1934270
11. Kleene KC. Connecting cis-elements and trans-factors with mechanisms of developmental regulation of mrna translation in meiotic and haploid mammalian spermatogenic cells. Reproduction (2013) 146:R1–19. doi: 10.1530/REP-12-0362
12. Liu S, Li Q, Li G, Zhang Q, Zhuo L, Han X, et al. The mechanism of m(6)a methyltransferase mettl3-mediated autophagy in reversing gefitinib resistance in nsclc cells by beta-elemene. Cell Death Dis (2020) 11:969. doi: 10.1038/s41419-020-03148-8
13. Chen Y, Wang J, Xu D, Xiang Z, Ding J, Yang X, et al. M(6)a mrna methylation regulates testosterone synthesis through modulating autophagy in leydig cells. Autophagy (2021) 17:457–75. doi: 10.1080/15548627.2020.1720431
14. Wu W, Luo M, Li K, Dai Y, Yi H, Zhong Y, et al. Cholesterol derivatives induce dephosphorylation of the histone deacetylases rpd3/hdac1 to upregulate autophagy. Autophagy (2021) 17:512–28. doi: 10.1080/15548627.2020.1725376
15. Wu W, Li K, Guo S, Xu J, Ma Q, Li S, et al. P300/hdac1 regulates the acetylation/deacetylation and autophagic activities of lc3/atg8-pe ubiquitin-like system. Cell Death Discovery (2021) 7:128. doi: 10.1038/s41420-021-00513-0
16. Kershaw MJ, Talbot NJ. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc Natl Acad Sci U S a. (2009) 106:15967–72. doi: 10.1073/pnas.0901477106
17. Farré JC, Manjithaya R, Mathewson RD, Subramani S. Ppatg30 tags peroxisomes for turnover by selective autophagy. Dev Cell (2008) 14:365–76. doi: 10.1016/j.devcel.2007.12.011
18. Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell (2001) 7:1131–41. doi: 10.1016/s1097-2765(01)00263-5
19. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature (2008) 451:1069–75. doi: 10.1038/nature06639
20. Sui X, Zhu J, Zhou J, Wang X, Li D, Han W, et al. Epigenetic modifications as regulatory elements of autophagy in cancer. Cancer Lett (2015) 360:106–13. doi: 10.1016/j.canlet.2015.02.009
21. Amaravadi RK, Kimmelman AC, Debnath J. Targeting autophagy in cancer: recent advances and future directions. Cancer Discovery (2019) 9:1167–81. doi: 10.1158/2159-8290.CD-19-0292
22. Liu J, Liu Y, Wang Y, Li C, Xie Y, Klionsky DJ, et al. Tmem164 is a new determinant of autophagy-dependent ferroptosis. Autophagy (2023) 19:945–56. doi: 10.1080/15548627.2022.2111635
23. Cui YH, Wilkinson E, Peterson J, He YY. Alkbh4 stabilization is required for arsenic-induced 6ma dna methylation inhibition, keratinocyte Malignant transformation, and tumorigenicity. Water (Basel). (2022) 14:3595. doi: 10.3390/w14223595
24. Cao W, Li J, Yang K, Cao D. An overview of autophagy: mechanism, regulation and research progress. Bull Cancer. (2021) 108:304–22. doi: 10.1016/j.bulcan.2020.11.004
26. Mahapatra KK, Mishra SR, Behera BP, Patil S, Gewirtz DA, Bhutia SK. The lysosome as an imperative regulator of autophagy and cell death. Cell Mol Life Sci (2021) 78:7435–49. doi: 10.1007/s00018-021-03988-3
27. Okamoto K. Organellophagy: eliminating cellular building blocks via selective autophagy. J Cell Biol (2014) 205:435–45. doi: 10.1083/jcb.201402054
28. Xiang J, Gong W, Liu J, Zhang H, Li M, Wang R, et al. Identification of dll3-related genes affecting the prognosis of patients with colon adenocarcinoma. Front Genet (2023) 14:1098190. doi: 10.3389/fgene.2023.1098190
29. Ko SH, Choi JH, Kim JM. Bacteroides fragilis enterotoxin induces autophagy through an ampk and foxo3-pathway, leading to the inhibition of apoptosis in intestinal epithelial cells. Toxins (Basel). (2023) 15:544. doi: 10.3390/toxins15090544
30. Ahn CH, Jeong EG, Lee JW, Kim MS, Kim SH, Kim SS, et al. Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers. Apmis (2007) 115:1344–9. doi: 10.1111/j.1600-0463.2007.00858.x
31. Peng YF, Shi YH, Ding ZB, Ke AW, Gu CY, Hui B, et al. Autophagy inhibition suppresses pulmonary metastasis of hcc in mice via impairing anoikis resistance and colonization of hcc cells. Autophagy (2013) 9:2056–68. doi: 10.4161/auto.26398
32. Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol (2005) 6:439–48. doi: 10.1038/nrm1660
33. Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. (2007) 7:961–7. doi: 10.1038/nrc2254
34. Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, et al. Autophagic and tumour suppressor activity of a novel beclin1-binding protein uvrag. Nat Cell Biol (2006) 8:688–99. doi: 10.1038/ncb1426
35. Shu F, Xiao H, Li QN, Ren XS, Liu ZG, Hu BW, et al. Epigenetic and post-translational modifications in autophagy: biological functions and therapeutic targets. Signal Transduct Target Ther (2023) 8:32. doi: 10.1038/s41392-022-01300-8
36. Dai X, Ren T, Zhang Y, Nan N. Methylation multiplicity and its clinical values in cancer. Expert Rev Mol Med (2021) 23:e2. doi: 10.1017/erm.2021.4
37. Moore LD, Le T, Fan G. Dna methylation and its basic function. Neuropsychopharmacology (2013) 38:23–38. doi: 10.1038/npp.2012.112
38. Liu Y, Shoji-Kawata S, Sumpter RJ, Wei Y, Ginet V, Zhang L, et al. Autosis is a na+,k+-atpase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci U S a. (2013) 110:20364–71. doi: 10.1073/pnas.1319661110
39. Li X, Li F, Zhang X, Zhang H, Zhao Q, Li M, et al. Caspase-8 auto-cleavage regulates programmed cell death and collaborates with ripk3/mlkl to prevent lymphopenia. Cell Death Differ (2022) 29:1500–12. doi: 10.1038/s41418-022-00938-9
40. Gao W, Wang X, Zhou Y, Wang X, Yu Y. Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor immunotherapy. Signal Transduct Target Ther (2022) 7:196. doi: 10.1038/s41392-022-01046-3
41. Poillet-Perez L, White E. Role of tumor and host autophagy in cancer metabolism. Genes Dev (2019) 33:610–9. doi: 10.1101/gad.325514.119
42. Xia H, Green DR, Zou W. Autophagy in tumour immunity and therapy. Nat Rev Cancer. (2021) 21:281–97. doi: 10.1038/s41568-021-00344-2
43. Han EJ, Choi EY, Jeon SJ, Lee SW, Moon JM, Jung SH, et al. Piperine induces apoptosis and autophagy in hsc-3 human oral cancer cells by regulating pi3k signaling pathway. Int J Mol Sci (2023) 24:13949. doi: 10.3390/ijms241813949
44. Nisticò P, Ciliberto G. Biological mechanisms linked to inflammation in cancer: discovery of tumor microenvironment-related biomarkers and their clinical application in solid tumors. Int J Biol Markers. (2020) 35:8–11. doi: 10.1177/1724600820906155
45. Puertollano R, Ferguson SM, Brugarolas J, Ballabio A. The complex relationship between tfeb transcription factor phosphorylation and subcellular localization. EMBO J (2018) 37:e98804. doi: 10.15252/embj.201798804
46. Lv H, Dao FY, Zhang D, Yang H, Lin H. Advances in mapping the epigenetic modifications of 5-methylcytosine (5mc), n6-methyladenine (6ma), and n4-methylcytosine (4mc). Biotechnol Bioeng. (2021) 118:4204–16. doi: 10.1002/bit.27911
47. Hu J, Othmane B, Yu A, Li H, Cai Z, Chen X, et al. 5mc regulator-mediated molecular subtypes depict the hallmarks of the tumor microenvironment and guide precision medicine in bladder cancer. BMC Med (2021) 19:289. doi: 10.1186/s12916-021-02163-6
48. Liu ZY, Xing JF, Chen W, Luan MW, Xie R, Huang J, et al. Mdr: an integrative dna n6-methyladenine and n4-methylcytosine modification database for rosaceae. Hortic Res (2019) 6:78. doi: 10.1038/s41438-019-0160-4
49. Kong Y, Cao L, Deikus G, Fan Y, Mead EA, Lai W, et al. Critical assessment of dna adenine methylation in eukaryotes using quantitative deconvolution. Science (2022) 375:515–22. doi: 10.1126/science.abe7489
50. Bochtler M, Fernandes H. Dna adenine methylation in eukaryotes: enzymatic mark or a form of dna damage? Bioessays (2021) 43:e2000243. doi: 10.1002/bies.202000243
51. Nolan LL, Kidder GW. Inhibition of growth and purine-metabolizing enzymes of trypanosomid flagellates by n6-methyladenine. Antimicrob Agents Chemother (1980) 17:567–71. doi: 10.1128/AAC.17.4.567
52. Achwal CW, Iyer CA, Chandra HS. Immunochemical evidence for the presence of 5mc, 6ma and 7mg in human, drosophila and mealybug dna. FEBS Lett (1983) 158:353–8. doi: 10.1016/0014-5793(83)80612-7
53. Shen L, Liang Z, Gu X, Chen Y, Teo ZW, Hou X, et al. N(6)-methyladenosine rna modification regulates shoot stem cell fate in arabidopsis. Dev Cell (2016) 38:186–200. doi: 10.1016/j.devcel.2016.06.008
54. Mu Y, Zhang L, Hu J, Zhou J, Lin HW, He C, et al. A fungal dioxygenase cctet serves as a eukaryotic 6ma demethylase on duplex dna. Nat Chem Biol (2022) 18:733–41. doi: 10.1038/s41589-022-01041-3
55. Li X, Zhao Q, Wei W, Lin Q, Magnan C, Emami MR, et al. The dna modification n6-methyl-2’-deoxyadenosine (m6da) drives activity-induced gene expression and is required for fear extinction. Nat Neurosci (2019) 22:534–44. doi: 10.1038/s41593-019-0339-x
56. Guo C, Liu Z, Zhang H. Dna 6ma demethylase alkbh1 regulates ddx18 expression to promote proliferation of human head and neck squamous cell carcinoma. Cell Oncol (Dordr). (2023) 46:1097–111. doi: 10.1007/s13402-023-00800-1
57. Xiao CL, Zhu S, He M, Chen D, Zhang Q, Chen Y, et al. N(6)-methyladenine dna modification in the human genome. Mol Cell (2018) 71:306–18. doi: 10.1016/j.molcel.2018.06.015
58. Chen J, Zhuang Y, Wang P, Ning J, Liu W, Huang Y, et al. Reducing n6amt1-mediated 6ma dna modification promotes breast tumor progression via transcriptional repressing cell cycle inhibitors. Cell Death Dis (2022) 13:216. doi: 10.1038/s41419-022-04661-8
59. Hao Z, Wu T, Cui X, Zhu P, Tan C, Dou X, et al. N(6)-deoxyadenosine methylation in mammalian mitochondrial dna. Mol Cell (2020) 78:382–95. doi: 10.1016/j.molcel.2020.02.018
60. Hsu KW, Lai JC, Chang JS, Peng PH, Huang CH, Lee DY, et al. Mettl4-mediated nuclear n6-deoxyadenosine methylation promotes metastasis through activating multiple metastasis-inducing targets. Genome Biol (2022) 23:249. doi: 10.1186/s13059-022-02819-3
61. Qi S, Mota J, Chan SH, Villarreal J, Dai N, Arya S, et al. Rna binding to human mettl3-mettl14 restricts n(6)-deoxyadenosine methylation of dna in vitro. Elife. (2022) 11:e67150. doi: 10.7554/eLife.67150
62. Li E, Xia M, Du Y, Long K, Ji F, Pan F, et al. Mettl3 promotes homologous recombination repair and modulates chemotherapeutic response in breast cancer by regulating the egf/rad51 axis. Elife (2022) 11:e75231. doi: 10.7554/eLife.75231
63. Lai X, Wei J, Gu XZ, Yao XM, Zhang DS, Li F, et al. Dysregulation of linc00470 and mettl3 promotes chemoresistance and suppresses autophagy of chronic myelocytic leukaemia cells. J Cell Mol Med (2021) 25:4248–59. doi: 10.1111/jcmm.16478
64. Yu D, Horton JR, Yang J, Hajian T, Vedadi M, Sagum CA, et al. Human mettl3-mettl14 rna adenine methyltransferase complex is active on double-stranded dna containing lesions. Nucleic Acids Res (2021) 49:11629–42. doi: 10.1093/nar/gkab460
65. Cai GP, Liu YL, Luo LP, Xiao Y, Jiang TJ, Yuan J, et al. Alkbh1-mediated dna n6-methyladenine modification regulates bone marrow mesenchymal stem cell fate during skeletal aging. Cell Prolif. (2022) 55:e13178. doi: 10.1111/cpr.13178
66. Miao Z, Wang G, Shen H, Wang X, Gabriel DW, Liang W. Bcmettl4-mediated dna adenine n(6)-methylation is critical for virulence of botrytis cinerea. Front Microbiol (2022) 13:925868. doi: 10.3389/fmicb.2022.925868
67. Deng G, Yang J, Zhang Q, Xiao ZX, Cai H. Methcna: a database for integrating genomic and epigenomic data in human cancer. BMC Genomics (2018) 19:138. doi: 10.1186/s12864-018-4525-0
68. Ma C, Niu R, Huang T, Shao LW, Peng Y, Ding W, et al. N6-methyldeoxyadenine is a transgenerational epigenetic signal for mitochondrial stress adaptation. Nat Cell Biol (2019) 21:319–27. doi: 10.1038/s41556-018-0238-5
69. Ouyang L, Su X, Li W, Tang L, Zhang M, Zhu Y, et al. Alkbh1-demethylated dna n6-methyladenine modification triggers vascular calcification via osteogenic reprogramming in chronic kidney disease. J Clin Invest. (2021) 131:e146985. doi: 10.1172/JCI146985
70. Luo Q, Mo J, Chen H, Hu Z, Wang B, Wu J, et al. Structural insights into molecular mechanism for n(6)-adenosine methylation by mt-a70 family methyltransferase mettl4. Nat Commun (2022) 13:5636. doi: 10.1038/s41467-022-33277-x
71. Itoh T, Satoh M, Kanno E, Fukuda M. Screening for target rabs of tbc (tre-2/bub2/cdc16) domain-containing proteins based on their rab-binding activity. Genes Cells (2006) 11:1023–37. doi: 10.1111/j.1365-2443.2006.00997.x
72. Xie Q, Wu TP, Gimple RC, Li Z, Prager BC, Wu Q, et al. N(6)-methyladenine dna modification in glioblastoma. Cell (2018) 175:1228–43. doi: 10.1016/j.cell.2018.10.006
73. Kweon S, Chen Y, Moon E, Kvederaviciutė K, Klimasauskas S, Feldman DE. An adversarial dna n6-methyladenine-sensor network preserves polycomb silencing. Mol Cell (2019) 74:1138–47. doi: 10.1016/j.molcel.2019.03.018
74. Xu C, Jiang S, Ma X, Jiang Z, Pan Y, Li X, et al. Crispr-based dna methylation editing of nnt rescues the cisplatin resistance of lung cancer cells by reducing autophagy. Arch Toxicol (2023) 97:441–56. doi: 10.1007/s00204-022-03404-0
75. Chen BF, Chan WY. The de novo dna methyltransferase dnmt3a in development and cancer. Epigenetics (2014) 9:669–77. doi: 10.4161/epi.28324
76. Na F, Pan X, Chen J, Chen X, Wang M, Chi P, et al. Kmt2c deficiency promotes small cell lung cancer metastasis through dnmt3a-mediated epigenetic reprogramming. Nat Cancer. (2022) 3:753–67. doi: 10.1038/s43018-022-00361-6
77. Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, et al. Chromosomal instability correlates with genome-wide dna demethylation in human primary colorectal cancers. Cancer Res (2006) 66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293
78. Zhang ZM, Lu R, Wang P, Yu Y, Chen D, Gao L, et al. Structural basis for dnmt3a-mediated de novo dna methylation. Nature (2018) 554:387–91. doi: 10.1038/nature25477
79. Falnes PØ, Johansen RF, Seeberg E. Alkb-mediated oxidative demethylation reverses dna damage in escherichia coli. Nature (2002) 419:178–82. doi: 10.1038/nature01048
80. Kuznetsov NA, Kanazhevskaya LY, Fedorova OS. Dna demethylation in the processes of repair and epigenetic regulation performed by 2-ketoglutarate-dependent dna dioxygenases. Int J Mol Sci (2021) 22:10540. doi: 10.3390/ijms221910540
81. Tian LF, Liu YP, Chen L, Tang Q, Wu W, Sun W, et al. Structural basis of nucleic acid recognition and 6ma demethylation by human alkbh1. Cell Res (2020) 30:272–5. doi: 10.1038/s41422-019-0233-9
82. Xiong J, Ye TT, Ma CJ, Cheng QY, Yuan BF, Feng YQ. N 6-hydroxymethyladenine: a hydroxylation derivative of n6-methyladenine in genomic dna of mammals. Nucleic Acids Res (2019) 47:1268–77. doi: 10.1093/nar/gky1218
83. Legendre F, MacLean A, Appanna VP, Appanna VD. Biochemical pathways to α-ketoglutarate, a multi-faceted metabolite. World J Microbiol Biotechnol (2020) 36:123. doi: 10.1007/s11274-020-02900-8
84. Joo SH, Pemble CT, Yang EG, Raetz C, Chung HS. Biochemical and structural insights into an fe(ii)/α-ketoglutarate/o(2)-dependent dioxygenase, kdo 3-hydroxylase (kdoo). J Mol Biol (2018) 430:4036–48. doi: 10.1016/j.jmb.2018.07.029
85. Liu PS, Wang H, Li X, Chao T, Teav T, Christen S, et al. A-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol (2017) 18:985–94. doi: 10.1038/ni.3796
86. Liu S, Yang J, Wu Z. The regulatory role of α-ketoglutarate metabolism in macrophages. Mediators Inflamm (2021) 2021:5577577. doi: 10.1155/2021/5577577
87. Baracco EE, Castoldi F, Durand S, Enot DP, Tadic J, Kainz K, et al. A-ketoglutarate inhibits autophagy. Aging (Albany NY). (2019) 11:3418–31. doi: 10.18632/aging.102001
88. Wang X, Wong CC, Chen H, Fu K, Shi L, Su H, et al. The n(6)-methyladenine dna demethylase alkbh1 promotes gastric carcinogenesis by disrupting nrf1 binding capacity. Cell Rep (2023) 42:112279. doi: 10.1016/j.celrep.2023.112279
89. Goh W. Single-nucleotide-resolution sequencing of n(6)-methyldeoxyadenosine. Methods Mol Biol (2021) 2198:369–77. doi: 10.1007/978-1-0716-0876-0_28
90. Hryhorowicz M, Lipiński D, Zeyland J, Słomski R. Crispr/cas9 immune system as a tool for genome engineering. Arch Immunol Ther Exp (Warsz). (2017) 65:233–40. doi: 10.1007/s00005-016-0427-5
91. Guo H, Chen F, Zhou M, Lan W, Zhang W, Shen G, et al. Crispr-cas9-mediated mutation of methyltransferase mettl4 results in embryonic defects in silkworm bombyx mori. Int J Mol Sci (2023) 24:3468. doi: 10.3390/ijms24043468
92. Zhang W, Tao KT, Lin J, Liu P, Guan Z, Deng J, et al. The role of m6a in osteoporosis and the differentiation of mesenchymal stem cells into osteoblasts and adipocytes. Curr Stem Cell Res Ther (2023) 18:339–46. doi: 10.2174/1574888X17666220621155341
93. Shafik AM, Zhou H, Lim J, Dickinson B, Jin P. Dysregulated mitochondrial and cytosolic trna m1a methylation in alzheimer’s disease. Hum Mol Genet (2022) 31:1673–80. doi: 10.1093/hmg/ddab357
94. Huang M, Long J, Yao Z, Zhao Y, Zhao Y, Liao J, et al. Mettl1-mediated m7g trna modification promotes lenvatinib resistance in hepatocellular carcinoma. Cancer Res (2023) 83:89–102. doi: 10.1158/0008-5472.CAN-22-0963
95. Pan J, Huang Z, Xu Y. M5c rna methylation regulators predict prognosis and regulate the immune microenvironment in lung squamous cell carcinoma. Front Oncol (2021) 11:657466. doi: 10.3389/fonc.2021.657466
96. Liu H, Xu Y, Yao B, Sui T, Lai L, Li Z. A novel n6-methyladenosine (m6a)-dependent fate decision for the lncrna thor. Cell Death Dis (2020) 11:613. doi: 10.1038/s41419-020-02833-y
97. Sendinc E, Shi Y. Rna m6a methylation across the transcriptome. Mol Cell (2023) 83:428–41. doi: 10.1016/j.molcel.2023.01.006
98. Li G, Ma L, He S, Luo R, Wang B, Zhang W, et al. Wtap-mediated m(6)a modification of lncrna norad promotes intervertebral disc degeneration. Nat Commun (2022) 13:1469. doi: 10.1038/s41467-022-28990-6
99. Fang Z, Mei W, Qu C, Lu J, Shang L, Cao F, et al. Role of m6a writers, erasers and readers in cancer. Exp Hematol Oncol (2022) 11:45. doi: 10.1186/s40164-022-00298-7
100. Zhu W, Wang JZ, Wei JF, Lu C. Role of m6a methyltransferase component virma in multiple human cancers (review). Cancer Cell Int (2021) 21:172. doi: 10.1186/s12935-021-01868-1
101. Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, et al. The rna m6a reader ythdf2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discovery (2021) 11:480–99. doi: 10.1158/2159-8290.CD-20-0331
102. Zhang N, Shen Y, Li H, Chen Y, Zhang P, Lou S, et al. The m6a reader igf2bp3 promotes acute myeloid leukemia progression by enhancing rcc2 stability. Exp Mol Med (2022) 54:194–205. doi: 10.1038/s12276-022-00735-x
103. Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The role of m6a modification in the biological functions and diseases. Signal Transduct Target Ther (2021) 6:74. doi: 10.1038/s41392-020-00450-x
104. Shi H, Wei J, He C. Where, when, and how: context-dependent functions of rna methylation writers, readers, and erasers. Mol Cell (2019) 74:640–50. doi: 10.1016/j.molcel.2019.04.025
105. Wu Y, Wang Z, Shen J, Yan W, Xiang S, Liu H, et al. The role of m6a methylation in osteosarcoma biological processes and its potential clinical value. Hum Genomics (2022) 16:12. doi: 10.1186/s40246-022-00384-1
106. Li Y, Zhang D, Gao Y, Wang P, Wang Z, Zhang B, et al. Mettl3 exacerbates insulin resistance in hepatocytes by regulating m(6)a modification of cytochrome p450 2b6. Nutr Metab (Lond). (2023) 20:40. doi: 10.1186/s12986-023-00762-z
107. Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The m6a reader ythdf1 promotes ovarian cancer progression via augmenting eif3c translation. Nucleic Acids Res (2020) 48:3816–31. doi: 10.1093/nar/gkaa048
108. Zhao H, Xu Y, Xie Y, Zhang L, Gao M, Li S, et al. M6a regulators is differently expressed and correlated with immune response of esophageal cancer. Front Cell Dev Biol (2021) 9:650023. doi: 10.3389/fcell.2021.650023
109. Wei X, Huo Y, Pi J, Gao Y, Rao S, He M, et al. Mettl3 preferentially enhances non-m(6)a translation of epigenetic factors and promotes tumourigenesis. Nat Cell Biol (2022) 24:1278–90. doi: 10.1038/s41556-022-00968-y
110. Chen T, Liu R, Niu Y, Mo H, Wang H, Lu Y, et al. Hif-1α-activated long non-coding rna kdm4a-as1 promotes hepatocellular carcinoma progression via the mir-411-5p/kpna2/akt pathway. Cell Death Dis (2021) 12:1152. doi: 10.1038/s41419-021-04449-2
111. Su R, Dong L, Li Y, Gao M, He PC, Liu W, et al. Mettl16 exerts an m(6)a-independent function to facilitate translation and tumorigenesis. Nat Cell Biol (2022) 24:205–16. doi: 10.1038/s41556-021-00835-2
112. Wei W, Zhang ZY, Shi B, Cai Y, Zhang HS, Sun CL, et al. Mettl16 promotes glycolytic metabolism reprogramming and colorectal cancer progression. J Exp Clin Cancer Res (2023) 42:151. doi: 10.1186/s13046-023-02732-y
113. Sun HL, Zhu AC, Gao Y, Terajima H, Fei Q, Liu S, et al. Stabilization of erk-phosphorylated mettl3 by usp5 increases m(6)a methylation. Mol Cell (2020) 80:633–47. doi: 10.1016/j.molcel.2020.10.026
114. Huang L, Shao J, Xu X, Hong W, Yu W, Zheng S, et al. Wtap regulates autophagy in colon cancer cells by inhibiting flna through n6-methyladenosine. Cell Adh Migr. (2023) 17:1–13. doi: 10.1080/19336918.2023.2180196
115. Li G, Deng L, Huang N, Cui Z, Wu Q, Ma J, et al. M(6)a mrna methylation regulates lkb1 to promote autophagy of hepatoblastoma cells through upregulated phosphorylation of ampk. Genes (Basel). (2021) 12:1747. doi: 10.3390/genes12111747
116. Lin X, Wang F, Chen J, Liu J, Lin YB, Li L, et al. N(6)-methyladenosine modification of cenpk mrna by zc3h13 promotes cervical cancer stemness and chemoresistance. Mil Med Res (2022) 9:19. doi: 10.1186/s40779-022-00378-z
117. Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 regulates nuclear rna m(6)a methylation and mouse embryonic stem cell self-renewal. Mol Cell (2018) 69:1028–38. doi: 10.1016/j.molcel.2018.02.015
118. Chen L, Gao Y, Xu S, Yuan J, Wang M, Li T, et al. N6-methyladenosine reader ythdf family in biological processes: structures, roles, and mechanisms. Front Immunol (2023) 14:1162607. doi: 10.3389/fimmu.2023.1162607
119. Petri BJ, Klinge CM. M6a readers, writers, erasers, and the m6a epitranscriptome in breast cancer. J Mol Endocrinol (2023) 70:e220110. doi: 10.1530/JME-22-0110
120. Xu Y, Zhang W, Shen F, Yang X, Liu H, Dai S, et al. Yth domain proteins: a family of m(6)a readers in cancer progression. Front Oncol (2021) 11:629560. doi: 10.3389/fonc.2021.629560
121. Widagdo J, Anggono V, Wong JJ. The multifaceted effects of ythdc1-mediated nuclear m(6)a recognition. Trends Genet (2022) 38:325–32. doi: 10.1016/j.tig.2021.11.005
122. Chen Z, Zhong X, Xia M, Zhong J. The roles and mechanisms of the m6a reader protein ythdf1 in tumor biology and human diseases. Mol Ther Nucleic Acids (2021) 26:1270–9. doi: 10.1016/j.omtn.2021.10.023
123. Shen M, Li Y, Wang Y, Shao J, Zhang F, Yin G, et al. N(6)-methyladenosine modification regulates ferroptosis through autophagy signaling pathway in hepatic stellate cells. Redox Biol (2021) 47:102151. doi: 10.1016/j.redox.2021.102151
124. Ma H, Ye D, Liu Y, Wu P, Yu L, Guo L, et al. Propofol suppresses ogd/r-induced ferroptosis in neurons by inhibiting the hif-1α/ythdf1/becn1 axis. Brain Inj (2023) 37, 1285–93. doi: 10.1080/02699052.2023.2237881
125. Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. Mrna circularization by mettl3-eif3h enhances translation and promotes oncogenesis. Nature (2018) 561:556–60. doi: 10.1038/s41586-018-0538-8
126. Liu X, He H, Zhang F, Hu X, Bi F, Li K, et al. M6a methylated epha2 and vegfa through igf2bp2/3 regulation promotes vasculogenic mimicry in colorectal cancer via pi3k/akt and erk1/2 signaling. Cell Death Dis (2022) 13:483. doi: 10.1038/s41419-022-04950-2
127. Bi Z, Liu Y, Zhao Y, Yao Y, Wu R, Liu Q, et al. A dynamic reversible rna n(6) -methyladenosine modification: current status and perspectives. J Cell Physiol (2019) 234:7948–56. doi: 10.1002/jcp.28014
128. Huang XT, Li JH, Zhu XX, Huang CS, Gao ZX, Xu QC, et al. Hnrnpc impedes m(6)a-dependent anti-metastatic alternative splicing events in pancreatic ductal adenocarcinoma. Cancer Lett (2021) 518:196–206. doi: 10.1016/j.canlet.2021.07.016
129. Lu J, Zhong C, Luo J, Shu F, Lv D, Liu Z, et al. Hnrnp-l-regulated circcspp1/mir-520h/egr1 axis modulates autophagy and promotes progression in prostate cancer. Mol Ther Nucleic Acids (2021) 26:927–44. doi: 10.1016/j.omtn.2021.10.006
130. Biswas S, Rao CM. Epigenetic tools (the writers, the readers and the erasers) and their implications in cancer therapy. Eur J Pharmacol (2018) 837:8–24. doi: 10.1016/j.ejphar.2018.08.021
131. Jiang ZX, Wang YN, Li ZY, Dai ZH, He Y, Chu K, et al. The m6a mrna demethylase fto in granulosa cells retards fos-dependent ovarian aging. Cell Death Dis (2021) 12:744. doi: 10.1038/s41419-021-04016-9
132. Hu Y, Gong C, Li Z, Liu J, Chen Y, Huang Y, et al. Demethylase alkbh5 suppresses invasion of gastric cancer via pkmyt1 m6a modification. Mol Cancer. (2022) 21:34. doi: 10.1186/s12943-022-01522-y
133. Gan X, Dai Z, Ge C, Yin H, Wang Y, Tan J, et al. Fto promotes liver inflammation by suppressing m6a mrna methylation of il-17ra. Front Oncol (2022) 12:989353. doi: 10.3389/fonc.2022.989353
134. Cui Y, Zhang C, Ma S, Li Z, Wang W, Li Y, et al. Rna m6a demethylase fto-mediated epigenetic up-regulation of linc00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res (2021) 40:294. doi: 10.1186/s13046-021-02096-1
135. Zhang Z, Wang L, Zhao L, Wang Q, Yang C, Zhang M, et al. N6-methyladenosine demethylase alkbh5 suppresses colorectal cancer progression potentially by decreasing phf20 mrna methylation. Clin Transl Med (2022) 12:e940. doi: 10.1002/ctm2.940
136. Zhang Y, Liu X, Wang Y, Lai S, Wang Z, Yang Y, et al. The m(6)a demethylase alkbh5-mediated upregulation of ddit4-as1 maintains pancreatic cancer stemness and suppresses chemosensitivity by activating the mtor pathway. Mol Cancer. (2022) 21:174. doi: 10.1186/s12943-022-01647-0
137. Chen Y, Ling Z, Cai X, Xu Y, Lv Z, Man D, et al. Activation of yap1 by n6-methyladenosine-modified circcpsf6 drives Malignancy in hepatocellular carcinoma. Cancer Res (2022) 82:599–614. doi: 10.1158/0008-5472.CAN-21-1628
138. Guan Q, Lin H, Miao L, Guo H, Chen Y, Zhuo Z, et al. Functions, mechanisms, and therapeutic implications of mettl14 in human cancer. J Hematol Oncol (2022) 15:13. doi: 10.1186/s13045-022-01231-5
139. Chen Y, Peng C, Chen J, Chen D, Yang B, He B, et al. Wtap facilitates progression of hepatocellular carcinoma via m6a-hur-dependent epigenetic silencing of ets1. Mol Cancer. (2019) 18:127. doi: 10.1186/s12943-019-1053-8
140. Fan Y, Li X, Sun H, Gao Z, Zhu Z, Yuan K. Role of wtap in cancer: from mechanisms to the therapeutic potential. Biomolecules (2022) 12:1224. doi: 10.3390/biom12091224
141. Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, et al. Insulin-like growth factor 2 mrna-binding proteins (igf2bps): post-transcriptional drivers of cancer progression? Cell Mol Life Sci (2013) 70:2657–75. doi: 10.1007/s00018-012-1186-z
142. He M, Lei H, He X, Liu Y, Wang A, Ren Z, et al. Mettl14 regulates osteogenesis of bone marrow mesenchymal stem cells via inducing autophagy through m6a/igf2bps/beclin-1 signal axis. Stem Cells Transl Med (2022) 11:987–1001. doi: 10.1093/stcltm/szac049
143. Liang D, Lin WJ, Ren M, Qiu J, Yang C, Wang X, et al. M(6)a reader ythdc1 modulates autophagy by targeting sqstm1 in diabetic skin. Autophagy (2022) 18:1318–37. doi: 10.1080/15548627.2021.1974175
144. Gao Y, Pei G, Li D, Li R, Shao Y, Zhang QC, et al. Multivalent m(6)a motifs promote phase separation of ythdf proteins. Cell Res (2019) 29:767–9. doi: 10.1038/s41422-019-0210-3
145. Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, et al. M(6)a enhances the phase separation potential of mrna. Nature (2019) 571:424–8. doi: 10.1038/s41586-019-1374-1
146. Yuan W, Hou Y, Wang Q, Lv T, Ren J, Fan L, et al. Newcastle disease virus activates methylation-related enzymes to reprogram m(6)a methylation in infected cells. Vet Microbiol (2023) 281:109747. doi: 10.1016/j.vetmic.2023.109747
147. Zheng C, Yu G, Su Q, Wu L, Tang J, Lin X, et al. The deficiency of n6-methyladenosine demethylase alkbh5 enhances the neurodegenerative damage induced by cobalt. Sci Total Environ (2023) 881:163429. doi: 10.1016/j.scitotenv.2023.163429
148. Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. Alkbh5 inhibited autophagy of epithelial ovarian cancer through mir-7 and bcl-2. J Exp Clin Cancer Res (2019) 38:163. doi: 10.1186/s13046-019-1159-2
149. Li Y, Zhou D, Liu Q, Zhu W, Ye Z, He C. Gene polymorphisms of m6a erasers fto and alkbh1 associated with susceptibility to gastric cancer. Pharmgenomics Pers Med (2022) 15:547–59. doi: 10.2147/PGPM.S360912
150. Li Y, Zheng D, Wang F, Xu Y, Yu H, Zhang H. Expression of demethylase genes, fto and alkbh1, is associated with prognosis of gastric cancer. Dig Dis Sci (2019) 64:1503–13. doi: 10.1007/s10620-018-5452-2
151. Yang C, Zhang Y, Du W, Cheng H, Li C. Eukaryotic translation initiation factor 3 subunit g promotes human colorectal cancer. Am J Transl Res (2019) 11:612–23.
152. Yang C, Liu X, Li C, Li S, Du W, Yang D. Eukaryotic translation initiation factor 3 subunit g (eif3g) resensitized hct116/5-fu to 5-fluorouracil (5-fu) via inhibition of mrp and mdr1. Onco Targets Ther (2018) 11:5315–24. doi: 10.2147/OTT.S170854
153. Zhang Y, Gao LX, Wang W, Zhang T, Dong FY, Ding WP. M(6) a demethylase fat mass and obesity-associated protein regulates cisplatin resistance of gastric cancer by modulating autophagy activation through ulk1. Cancer Sci (2022) 113:3085–96. doi: 10.1111/cas.15469
154. Xu Y, Zhou J, Li L, Yang W, Zhang Z, Zhang K, et al. Fto-mediated autophagy promotes progression of clear cell renal cell carcinoma via regulating sik2 mrna stability. Int J Biol Sci (2022) 18:5943–62. doi: 10.7150/ijbs.77774
155. Zhao L, Kong X, Zhong W, Wang Y, Li P. Fto accelerates ovarian cancer cell growth by promoting proliferation, inhibiting apoptosis, and activating autophagy. Pathol Res Pract (2020) 216:153042. doi: 10.1016/j.prp.2020.153042
156. Ruan DY, Li T, Wang YN, Meng Q, Li Y, Yu K, et al. Fto downregulation mediated by hypoxia facilitates colorectal cancer metastasis. Oncogene (2021) 40:5168–81. doi: 10.1038/s41388-021-01916-0
157. Nie G, Tang B, Lv M, Li D, Li T, Ou R, et al. Hpv e6 promotes cell proliferation of cervical cancer cell by accelerating accumulation of rbm15 dependently of autophagy inhibition. Cell Biol Int (2023) 47:1327–43. doi: 10.1002/cbin.12020
158. Xu Y, Song M, Hong Z, Chen W, Zhang Q, Zhou J, et al. The n6-methyladenosine mettl3 regulates tumorigenesis and glycolysis by mediating m6a methylation of the tumor suppressor lats1 in breast cancer. J Exp Clin Cancer Res (2023) 42:10. doi: 10.1186/s13046-022-02581-1
159. Liu X, Feng M, Hao X, Gao Z, Wu Z, Wang Y, et al. M6a methylation regulates hypoxia-induced pancreatic cancer glycolytic metabolism through alkbh5-hdac4-hif1α positive feedback loop. Oncogene (2023) 42:2047–60. doi: 10.1038/s41388-023-02704-8
160. Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen X, et al. Rna m(6) a methylation regulates sorafenib resistance in liver cancer through foxo3-mediated autophagy. EMBO J (2020) 39:e103181. doi: 10.15252/embj.2019103181
161. Kong F, Liu X, Zhou Y, Hou X, He J, Li Q, et al. Downregulation of mettl14 increases apoptosis and autophagy induced by cisplatin in pancreatic cancer cells. Int J Biochem Cell Biol (2020) 122:105731. doi: 10.1016/j.biocel.2020.105731
162. Liu X, Qin J, Gao T, Li C, Chen X, Zeng K, et al. Analysis of mettl3 and mettl14 in hepatocellular carcinoma. Aging (Albany NY). (2020) 12:21638–59. doi: 10.18632/aging.103959
163. Yin H, Chen L, Piao S, Wang Y, Li Z, Lin Y, et al. M6a rna methylation-mediated rmrp stability renders proliferation and progression of non-small cell lung cancer through regulating tgfbr1/smad2/smad3 pathway. Cell Death Differ (2023) 30:605–17. doi: 10.1038/s41418-021-00888-8
164. Liu T, Hu J, Han B, Tan S, Jia W, Xin Y. A positive feedback loop of lncrna-rmrp/znrf3 axis and wnt/β-catenin signaling regulates the progression and temozolomide resistance in glioma. Cell Death Dis (2021) 12:952. doi: 10.1038/s41419-021-04245-y
165. Zhu D, Zhou J, Zhao J, Jiang G, Zhang X, Zhang Y, et al. Zc3h13 suppresses colorectal cancer proliferation and invasion via inactivating ras-erk signaling. J Cell Physiol (2019) 234:8899–907. doi: 10.1002/jcp.27551
166. Li B, Zhu L, Lu C, Wang C, Wang H, Jin H, et al. Circndufb2 inhibits non-small cell lung cancer progression via destabilizing igf2bps and activating anti-tumor immunity. Nat Commun (2021) 12:295. doi: 10.1038/s41467-020-20527-z
167. Ding L, Wang R, Zheng Q, Shen D, Wang H, Lu Z, et al. Circpde5a regulates prostate cancer metastasis via controlling wtap-dependent n6-methyladenisine methylation of eif3c mrna. J Exp Clin Cancer Res (2022) 41:187. doi: 10.1186/s13046-022-02391-5
168. Xu Y, Ye S, Zhang N, Zheng S, Liu H, Zhou K, et al. The fto/mir-181b-3p/arl5b signaling pathway regulates cell migration and invasion in breast cancer. Cancer Commun (Lond). (2020) 40:484–500. doi: 10.1002/cac2.12075
169. Huang H, Wang Y, Kandpal M, Zhao G, Cardenas H, Ji Y, et al. Fto-dependent n (6)-methyladenosine modifications inhibit ovarian cancer stem cell self-renewal by blocking camp signaling. Cancer Res (2020) 80:3200–14. doi: 10.1158/0008-5472.CAN-19-4044
170. Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, et al. Rna demethylase alkbh5 prevents pancreatic cancer progression by posttranscriptional activation of per1 in an m6a-ythdf2-dependent manner. Mol Cancer. (2020) 19:91. doi: 10.1186/s12943-020-01158-w
171. Xu H, Cong F, Hwang TH. Machine learning and artificial intelligence-driven spatial analysis of the tumor immune microenvironment in pathology slides. Eur Urol Focus. (2021) 7:706–9. doi: 10.1016/j.euf.2021.07.006
172. Shirin A, Klickstein IS, Feng S, Lin YT, Hlavacek WS, Sorrentino F. Prediction of optimal drug schedules for controlling autophagy. Sci Rep (2019) 9:1428. doi: 10.1038/s41598-019-38763-9
173. Rahman CR, Amin R, Shatabda S, Toaha M. A convolution based computational approach towards dna n6-methyladenine site identification and motif extraction in rice genome. Sci Rep (2021) 11:10357. doi: 10.1038/s41598-021-89850-9
174. Mitra ED, Suderman R, Colvin J, Ionkov A, Hu A, Sauro HM, et al. Pybionetfit and the biological property specification language. Iscience (2019) 19:1012–36. doi: 10.1016/j.isci.2019.08.045
175. Zhu H, Chen CZ, Sakamuru S, Zhao J, Ngan DK, Simeonov A, et al. Mining of high throughput screening database reveals ap-1 and autophagy pathways as potential targets for covid-19 therapeutics. Sci Rep (2021) 11:6725. doi: 10.1038/s41598-021-86110-8
Keywords: 6mA methylation, m6A methylation, autophagy, cancers, therapy
Citation: Lin L, Zhao Y, Zheng Q, Zhang J, Li H and Wu W (2023) Epigenetic targeting of autophagy for cancer: DNA and RNA methylation. Front. Oncol. 13:1290330. doi: 10.3389/fonc.2023.1290330
Received: 07 September 2023; Accepted: 28 November 2023;
Published: 08 December 2023.
Edited by:
Fabio Caradonna, University of Palermo, ItalyReviewed by:
Kamalika Mojumdar, University of Texas MD Anderson Cancer Center, United StatesChang Gu, Tongji University, China
Wuliang Wang, Second Affiliated Hospital of Zhengzhou University, China
Copyright © 2023 Lin, Zhao, Zheng, Zhang, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huaqin Li, aHVhcWlubGkxMTE4QDE2My5jb20=; Wenmei Wu, d3V3ZW5tZWlAZ2RwdS5lZHUuY24=
 Luobin Lin
Luobin Lin Yuntao Zhao
Yuntao Zhao Qinzhou Zheng
Qinzhou Zheng Jiayang Zhang1
Jiayang Zhang1 Huaqin Li
Huaqin Li Wenmei Wu
Wenmei Wu