
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Oncol. , 28 September 2023
Sec. Molecular and Cellular Oncology
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1289756
This article is part of the Research Topic Preclinical Models and Emerging Technologies to Study the Effects of the Tumor Microenvironment on Cancer Heterogeneity and Drug Resistance View all 11 articles
Editorial on the Research Topic
Preclinical models and emerging technologies to study the effects of the tumor microenvironment on cancer heterogeneity and drug resistance
Cancer research has witnessed remarkable advancements in recent years, with molecularly targeted treatments and immunotherapy revolutionizing patient care. However, some cancer types, such as pancreatic ductal adenocarcinoma, continue to defy treatment, and the emergence of drug resistance remains a significant challenge, hampering efforts to achieve lasting remission, especially in metastatic disease. In the past, cancer development was solely attributed to the appearance of genomic alterations in cancer cells. In the last decades, a broad consensus arose about the necessity for cancer cells to cooperate with host cells, such as fibroblasts, vascular cells, lymphatic cells, and immune cells during carcinogenesis and cancer progression. Therefore, the focus shifted toward understanding the tumor microenvironment (TME) heterogeneity and its impact on cancer evolution and drug resistance. In this dynamic landscape, innovative preclinical models and cutting-edge technologies have emerged as indispensable tools in TME characterization and, in general, in the efforts against cancer (Figure 1).
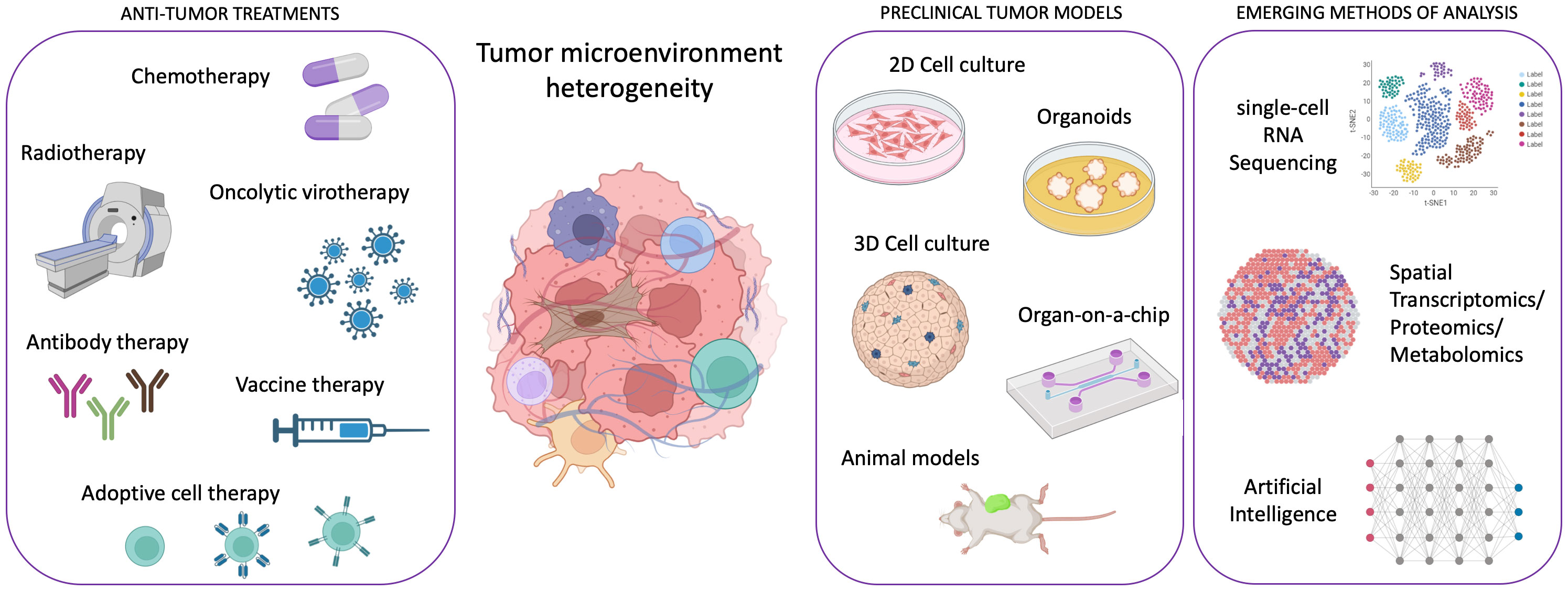
Figure 1 Scheme illustrating the framework of emergent technologies and preclinical tumor models for addressing the complexity of tumor microenvironment heterogeneity in order to enhance the efficacy of anti-tumor therapies. Created with Biorender.com.
Understanding the heterogeneous nature of tumors has presented significant challenges, but recent insights into the TME role have lightened up new paths for therapeutic exploration. This Research Topic examine various facets of the TME influence on cancer heterogeneity and drug resistance, encompassing diverse cancer types, including breast, colorectal, pancreatic, liver, and head and neck cancers. While each article offers a unique perspective, they collectively emphasize the role of the TME in shaping tumor behavior and response to treatment.
Genta et al., Huang and Tu, and Proietto et al. highlight the advances of preclinical models in deciphering the complexities of the TME. Genetically engineered mouse models, patient-derived organoids and xenografts, and three-dimensional (3D) cell cultures have all emerged as essential tools to study cancer progression, acquired heterogeneity, and predict drug efficacy with an enhanced clinical relevance. Genta et al. encompass both in vitro and in vivo platforms for drug testing, stressing the relevance of patient-derived models to maintain the molecular affinity with the parental tumors and predict patient response. However, the authors also draw attention to the challenge that, despite their use in co-clinical trials and their value in understanding drug resistance, the data generated often arrive too late to modify the therapeutic plans effectively. Huang and Tu dive into the specific challenges posed by tumor heterogeneity and the microenvironment in vascularized tumor models, widely discussing the advantages and limitations of organ-on-a-chip models in replicating the vascularized tumor microenvironment. The addition of different types of supporting cells, namely pericytes, astrocytes, or lymphatic cells, makes those organ-on-a-chip models closely mimic the heterotypic cellular interactions within tumors. Further, the inclusion of organ-specific normal cells allows for assessing general drug toxicity. However, the authors also comment on some limitations, such as the accessibility to quantify cellular forces and cell stiffness. Proietto et al. extensively review the TME preclinical models and broaden the discussion to include the role of emerging single-cell, spatial genomic, and metabolomic technologies in understanding tumor complexity and evolution to design better targeted therapies. The authors highlight the need for multidisciplinary approaches to combat the challenges posed by tumor heterogeneity. Lee et al. remarkably contribute to further highlight the growing significance of cutting-edge spatial omics technologies in dissecting TME heterogeneity, also introducing the value added by artificial intelligence in unraveling the complex interactions within the TME.
The other articles in the Research Topic offer innovative insights on specific cancers, discussing advances in their modeling for preclinical studies and their organotypic TME role in drug resistance. Yau et al. look at the importance of incorporating diverse cell populations within colorectal cancer spheroid models to mimic the intricate signaling pathways and heterogeneity found in vivo. Salemme et al. extensively discuss the characteristics of breast cancer, a highly heterogeneous disease, that leads to drug resistance and encompass the advances in developing in vivo and in vitro models to recapitulate breast cancer complexity. Another work about breast cancer by Lamouline et al. focuses on in vitro models for breast cancer metastasis in bone, elegantly discussing the cellular heterogeneity of this specific microenvironment, shedding light on the critical need to account for the different microenvironments in in vitro models dedicated to the study of metastasis to accurately mimic tissue-specific cell-cell and cell-extracellular matrix (ECM) interactions when assessing drug sensitivity. Indeed, van Tienderen et al. present a novel in vitro model of cholangiocarcinoma metastasis by ingeniously combining patient-derived organoids with decellularized human tissues, confirming the intricate interplay between cancer cells and the ECM in driving dissemination to distant organs such as lung and lymph nodes. Dedicated to studying drug pharmacological effects and tissue physiology is the model proposed by Greier et al. in their paper, in which they demonstrate the promising potential of 3D primary slice cultures to provide preclinical models for studying head and neck cancer and obtain data in a week frame to guide clinicians in the choice of treatments. Giustarini et al. establish human 3D heterocellular tumor spheroids as models mirroring the complexity of the pancreatic ductal adenocarcinoma environment. Notably, these tissue-specific spheroids reveal immunosuppressive tendencies and mimic spatial and cytokine signatures observed in patients, confirming their value for better therapy testing. The more we advance our ability to reproduce complex high-throughput systems in vitro, particularly through the consistent and standardized utilization of patient-derived samples for drug library testing, the sooner we will have a time-saving and cost-effective alternative to in vivo mouse models. While very powerful and extensively used, these animal models present ethical and variance concerns in rigorously reflecting human tumor evolution and heterogeneity, delaying drug development processes.
This Research Topic presents a comprehensive picture of the recent advancements in understanding the TME influence on cancer heterogeneity and drug resistance. As these studies collectively suggest, it is clear that an accurate understanding of the TME role is pivotal for designing effective cancer therapies. This Research Topic highlights how the essential collaboration between clinicians, researchers, and technologists in developing advanced preclinical models and cutting-edge technologies is uncovering the subtle interactions between cancer cells, immune cells, and the surrounding microenvironment to develop effective therapeutics that will tackle drug resistance and improve patient outcomes worldwide.
GA: Writing – original draft. PC: Writing – review & editing. SL: Writing – review & editing.
We extend our sincere gratitude to the remarkable group of authors whose dedication and intellectual contributions have made this research topic a reality. We thank the institutions for providing resources to share advances and scientific knowledge in cancer research. This work was supported by the AIRC-IG grant (grant number 26341) and the Fondazione CRT grant (grant number CRT-2020.0719) to PC, by the 1st Italy-Singapore Science and Technology Cooperation grant (grant number R23I0IR036) to GA and PC, by the AIRC-Start-Up grant (grant number 24750), the European Union’s Horizon 2020 research and innovation programme under Marie Sklodowska-Curie grant agreement (grant number 101029427) to SL.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: tumor microenvironment, tumor heterogeneity, scRNA seq, spatial biology, preclinical tumor models, 3D tumor models, in vivo tumor models
Citation: Adriani G, Cappello P and Lovisa S (2023) Editorial: Preclinical models and emerging technologies to study the effects of the tumor microenvironment on cancer heterogeneity and drug resistance. Front. Oncol. 13:1289756. doi: 10.3389/fonc.2023.1289756
Received: 06 September 2023; Accepted: 07 September 2023;
Published: 28 September 2023.
Edited and Reviewed by:
Tao Liu, University of New South Wales, AustraliaCopyright © 2023 Adriani, Cappello and Lovisa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giulia Adriani, R2l1bGlhX2FkcmlhbmlAaW1tdW5vbC5hLXN0YXIuZWR1LnNn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.